Abstract
Background: The etiology of pulmonary sarcoidosis is not well established. Although the mechanism triggering pulmonary sarcoidosis remains to be established, inflammatory reactions seem to play an important role in this process. Objectives: The aim of this study was to define the composition of the lower airway microbiota in the bronchoalveolar lavage (BAL) of patients affected by interstitial lung diseases, including sarcoidosis, to determine whether the bacterial signature differs among these diseases. Methods: Ten patients affected by pulmonary sarcoidosis and 9 patients affected by other interstitial lung diseases were enrolled. 16S rRNA next-generation sequencing was used to study BAL microbial composition of these patients, and were also compared with already published microbial content in higher airways of such diseases. Results: Four phyla dominated the lower airway microbiota, Bacteroidetes being the most abundant phylum in both groups (56.9%). Diversity analysis showed no significant differences between the various diseases, particularly between pulmonary sarcoidosis and other interstitial lung diseases affecting lower airways. Conclusions: Our data indicate that the bacterial lower airways microbiota share the same signature and, therefore, cannot be used as a diagnostic tool to discriminate among different interstitial lung diseases, including sarcoidosis, while microbial diversity is present when considering lower or higher respiratory airways. (Sarcoidosis Vasc Diffuse Lung Dis 2018; 35: 354-362)
Keywords: airway microbiota, bronchoalveolar lavage (BAL), pulmonary sarcoidosis, interstitial lung diseases, next generation sequencing
Introduction
Human microbiome estimation in various areas of our body is becoming more and more important for the knowledge of the saprophytes genomes that inhabit on our external surfaces and on the mucosal tissues open to the outside. This has reached even more importance when the study is compared with the pathological aspects of the various diseases that may occur to those different sites. In particular, it is obvious that, inside the body, there are less accessible organs, through lined up by mucosal tissue open to the outside, that are often too much invasive to be reached, but they may be important to be analyzed for pathological aspects among which the microbiome is an important aspect for several pathological situations.
In this context, lower airways are certainly sites that can be reached by an invasive procedure through a bronchoscopy approach and also by collecting in that situation the bronchoalveolar lavage (BAL) fluid. By this approach, the BAL fluid may be analyzed for its content, including the microbiome, and compared with several pathological situations where signatures may be taken both to compare with normal ones and among them, and also to establish signatures at level of microbiome that can be used for indicating or contribute to the clinical diagnosis as, for instance, in pulmonary sarcoidosis.
Sarcoidosis is a chronic granulomatous disorder characterized by multi-organ system impairment that mainly affects the lungs (1, 2). The typical, although not specific, histological finding of sarcoidosis is the presence, within affected tissues, of epitheliod granulomas with inflammatory infiltration (1). Clinical signs are extremely variable depending on the extent of the inflammatory lesions and on the kind of tissues affected (1). In the most severe forms, it can evolve to respiratory insufficiency, severe neurologic impairments, blindness and sudden cardiac death (2).
The etiopathogenesis of sarcoidosis is not well established. The currently accepted hypothesis is that, in genetically predisposed (both HLA and non-HLA) subjects, a persistent host immune response occurs against a specific antigen; viral infections have been implicated in the development of sarcoidosis, although their specific role remains to be established (3-12).
In recent years, next-generation sequencing technologies (NGS) have given an impulse to metagenomics (13-15). These approaches, in addition to the sequence of the entire genome of specific microorganisms (16, 17), by enabling the simultaneous study of all the genomes of a microbial community directly recovered from specific environments, have many advantages over more traditional techniques (14, 15). They have already been successfully used to identify the composition of microbial communities in different ecosystems and to characterize specific human microbial niches, also in relation to different diseases (18-21). Notably, a NGS-based approach has recently been used to investigate the microbial communities of the upper and lower airways of patients affected by different respiratory diseases (22-27).
Here, we describe the use of a multiplexed 16S rRNA amplicon-based approach, coupled with NGS analysis, to characterize the lower airway microbiota of patients with interstitial lung diseases to determine whether bacterial dysbiosis is disease-specific, and the difference with higher airways chronic respiratory diseases.
Materials and methods
Patients’ enrollment and clinical features
Nineteen unrelated subjects were enrolled in this study among patients attending the Respiratory Section of the University Federico II of Naples, Italy. Subjects underwent bronchoscopy for a diagnostic purpose and were diagnosed, with a multidisciplinary approach, as pulmonary sarcoidosis (PS, 10 patients), or interstitial lung disease (ILD, 9 patients), (see table 1). In particular, in such a last group, 6 were nonspecific idiopathic pneumonia (NSIP), and 3 were idiopathic pulmonary fibrosis (IPF): all of them were recently diagnosed (1-2 months), whereby no treatment was administered. In the first group (PS), most of patients were classified as stage 1 (7), where only 3 were assigned to stage 2.
Table 1.
General features (clinical and instrumental data) of the study population
| Groups | ||
| PS | ILD | |
| N. of subjects | 10 | 9 |
| Age, years (mean±SD) | 57±10 | 64±11 |
| Sex F/M | 4F/6M | 3F/6M |
| Clinical diagnosis | Sarcoidosis | Interstitial lung diseases |
| FVC % (range; mean±SD) | 71-98; 94.2±15.6 | 42-88; 63.8±28.8 |
| DLCO % (range; mean±SD) | 25-85; 66.7±17.5 | 34-75; 54.3±15 |
| Recent infections | 0/10 | 0/9 |
| Probiotics assumption | 0/10 | 0/9 |
| Immunosuppressive therapy (N) | 1/10 | 0/9 |
| Corticosteroid therapy (N) | 2/10 | 1/9 |
PS, pulmonary sarcoidosis patients; ILD, interstitial lung diseases; SD, standard deviation; F, female; M, male; FVC, forced vital capacity; DLCO, diffusing capacity of the lung carbon monoxide; N, number
Inclusion criteria were: no use of antibiotics, antivirals or corticosteroids for at least 2 weeks before sampling, no use of probiotics in the 2 months before sampling, no recent bacterial/viral respiratory tract infections 2 weeks before sampling. Notably, we did not enroll a control group of healthy subjects because of the invasiveness of the BAL procedure for which also the Ethics Committee did not allowed. All patients gave their written informed consent to the study; the study was approved by the Ethics Committee of the Ospedale dei Colli, Naples, Italy, to which the mentioned Respiratory Section belongs.
Sample collection, DNA extraction and 16S rRNA sequencing
A BAL sample was collected from each subject during bronchoscopy. An aliquot was used for standard diagnostic evaluations (including traditional microbiological assessment) and an aliquot was immediately cooled in ice and stored at -80°C for metagenomic analysis. DNA was extracted from BAL (600 μl/sample) using the QIAamp DNA mini Kit (Qiagen,Venlo, Netherlands) following manufacturer’s instructions. V4-V6 hypervariable regions of the bacterial 16S rRNA gene were amplified using a specific primer pair (forward primer: 5’-CAGCAGCCGCGGTAATAC-3’; reverse primer: 5’- TGACGACAGCCATGC-3’), as we previously described (28). Sequence data analysis was carried out using the specific metagenomic tool QIIME, v. 1.8.0 (29, 30). A pre-quality filtering step was conducted in QIIME using suitable parameters for 454 sequencing. For a sequence to be retained, the following criteria had to be met: (i) a minimum average quality Phred score of 25; (ii) a minimum and maximum sequence length (200-1000, 454 data set); (iii) no more than six ambiguous bases or homopolymers. A total of 387,357 sequences passed the quality-filtering step (mean: 21,519; Stdev: 13,523), and were clustered to obtain OTU using a subsampled open-reference OTU picking approach. The OTU picking procedure was computed with UCLUST (at 97% identity) using the 16S rRNA GREENGENES database (v. 13_8) as a reference database (31, 32). Sequences that did not match the GREENGENES database were clustered as de novo as to not lose the overall novel diversity. Taxonomic assignment was also computed using UCLUST with a 0.9% similarity against a representative set of 16S rRNA gene sequences from the GREENGENES database. A phylogenetic tree was obtained using PyNAST (33) and used for downstream phylogenetic analysis. The alpha and beta diversity analyses were computed at a rarefaction depth of 8,031 sequences/sample. Alpha diversity was computed for each rarefied OTU table, using the number of observed species metric and the Faith’s Phylogenetic Diversity Index. Beta diversity was performed using weighted and unweighted UniFrac distance matrices and plotted as principal coordinate analysis (PCoA) in Emperor (34). Taxonomic assignment was done, within the bacteria kingdom, according to hierarchical classification from phylum down to genus.
Statistical analysis
Quantitative differences in the relative abundance of each identified taxa within the two groups were evaluated using specific QIIME scripts based on Kruskal Wallis non-parametric ANOVA for the analysis of variance with both Bonferroni and FDR corrections (p≤0.05 after correction indicates significant differences). Alpha diversity significance was assessed using a non-parametric two-sample t-test to compare the alpha diversities for both metrics (i.e. Phylogenetic diversity “PD” and number of observed OTUs) using 999 Monte Carlo permutations. P-values were adjusted using Bonferroni correction to show any difference between the compared groups. Beta diversity was analyzed through the two different statistical tests ADONIS and ANOSIM using UniFrac distance matrices (35, 36).
Results
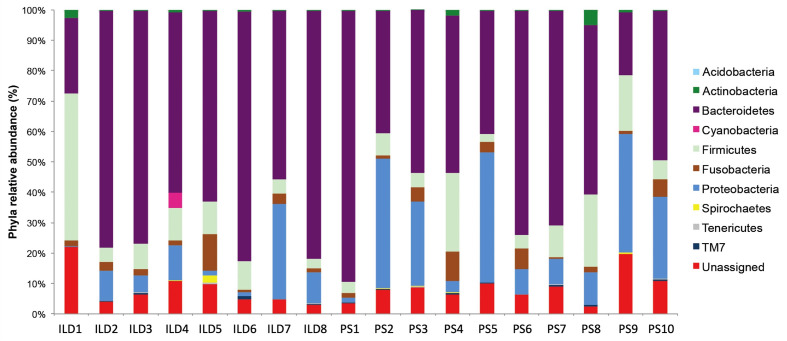
Bronchoalveolar lavage samples from the 19 enrolled patients were analyzed for their microbial qualitative and quantitative richness, as described under Methods. Traditional microbiological evaluation did not reveal any positive cultures in any patient. Next, we carried out a pyrosequencing NGS run of all samples to analyze the entire study population, and obtained more than 400,000 sequences that resulted in an average of 21,878 high quality mapped reads/sample. Taxonomic assignment of reads revealed 18 phyla, of which 10 at an abundance ≥0.1% (fig. 1). Of the phyla present in all patients, the most represented were Bacteroidetes, Firmicutes, Fusobacteria and Proteobacteria, of which the relative abundances were respectively 70%, 10%, 3% and 15.5%. Bacterial composition was similar in all patients except in a patient affected by interstitial lung disease (ILD). The microbiome profile of this patient (ILD9) was dominated by the Proteobacteria phylum; its peculiar profile remains down to genus level where the Moraxella genus represents 98.4% of all the bacterial communities. This profile may have resulted from an acute occasional infection, which rarely invades the lung from the upper airways. Given its extreme peculiarity we did not include it in the downstream comparison between the study groups.
Fig. 1.
Microbial communities identified in the 18 subjects at phylum level using 16S rRNA pyrosequencing and QIIME taxonomic assignment. The specific bacterial distribution of each study subject at phylum level; each bar represents a study subject. Different phyla are assigned to specific colors; Bacteroidetes phylum results the most represented one in the whole set of subjects
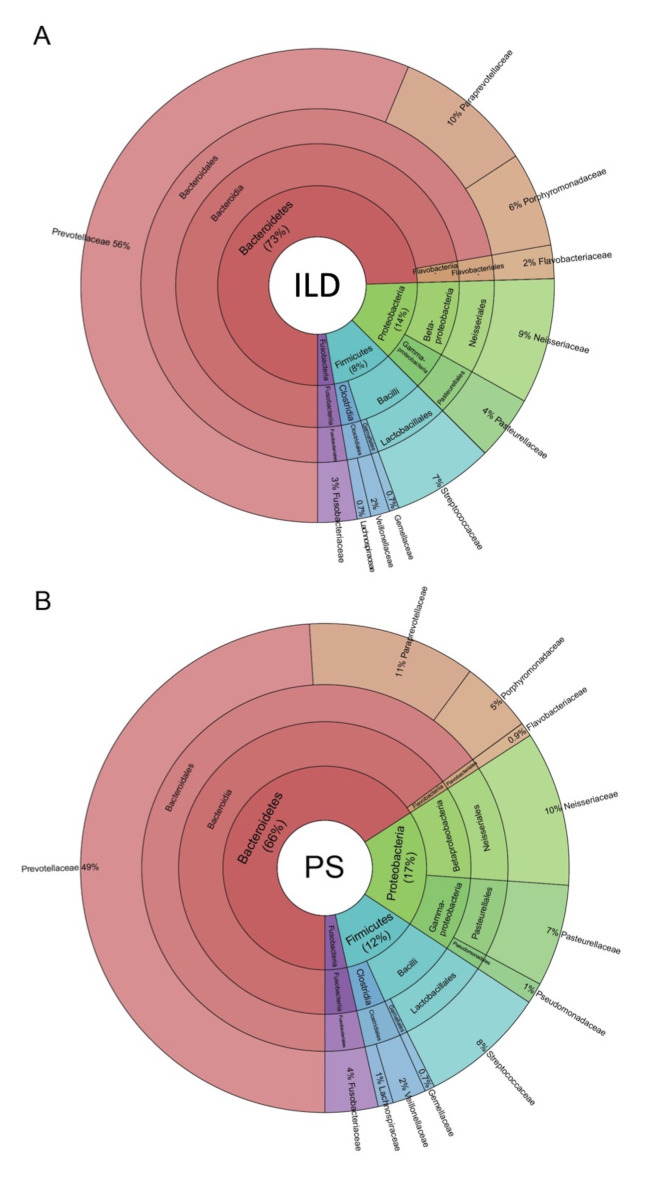
Figure 2 illustrates the composition of the microbial communities in the two groups of patients studied. The taxonomic assignment did not reveal any significant differences between patients affected PS and patients affected by ILD. Four phyla were present at a relative abundance >1%: Bacteroidetes, Proteobacteria, Firmicutes and Fusobacteria. Bacteroidetes was the most abundant in both groups with an average relative abundance of about 56.9% (fig. 2A, B). The relative abundance of different bacteria remained unchanged down to genus level. In detail, within the Bacteroidetes phylum, we found going from class to genus level, a high level of (1) Bacteroidia (class level, 55.5% average relative abundance); (2) Bacteroidales (order level, 55.5% average relative abundance); (3) Prevotellaceae, [Paraprevotellaceae] and Porphyromonadaceae (family level, 42.5%, 8.4% and 4.5% average relative abundance, respectively); and (4) Prevotella, [Prevotella] and Porphyromonas (genus level, 42.5%, 8.4% and 4.4% average relative abundance, respectively) (fig. 2C). Firmicutes and Proteobacteria were also represented in the lower airway microbiota of the two groups.
Fig. 2.
The composition of the lower airway microbiota identified in interstitial lung diseases (ILD) and pulmonary sarcoidosis (PS) patients, from phylum to genus level (from center to peripheral ring). Krona plots show the hierarchical taxonomic distribution in each ring from phylum to family. Taxonomic assignment shows no significant differences between the two studied groups. Bacteroidetes was the most represented phylum with an average relative abundance of 70% in the entire population both in ILD (A) and PS (B) subjects. Percentages are normalized relative abundances of bacteria observed only in the most abundant phyla (>1%)
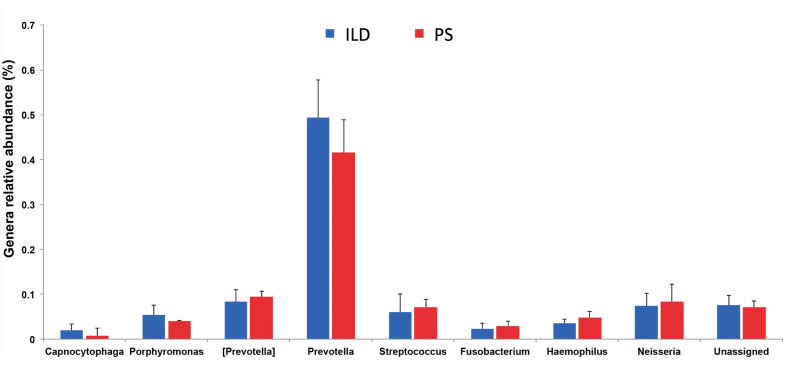
At genus level, the average relative abundance of Streptococcus, (belonging to Firmicutes) was 6.1%, while the average relative abundances of Neisseria and Haemophilus, (belonging to Proteobacteria) were 7.5% and 4%, respectively (fig. 3).
Fig. 3.
Genus-level comparison of BAL microbial communities among the two study groups showed no differences in microbial composition among interstitial lung diseases (ILD) and pulmonary sarcoidosis (PS) patients. At genus level, we found the prevalence of members of the Bacteroidetes phylum being Prevotella (42.5%) and [Prevotella] (8.4%) the most abundant genera. Members of the Firmicutes and Proteobacteria phyla were also represented in both groups; Streptococcus (Firmicutes), Neisseria and Haemophilus (both within Proteobacteria) genera were respectively the most abundant within these two groups of patients, even if at a rather low level
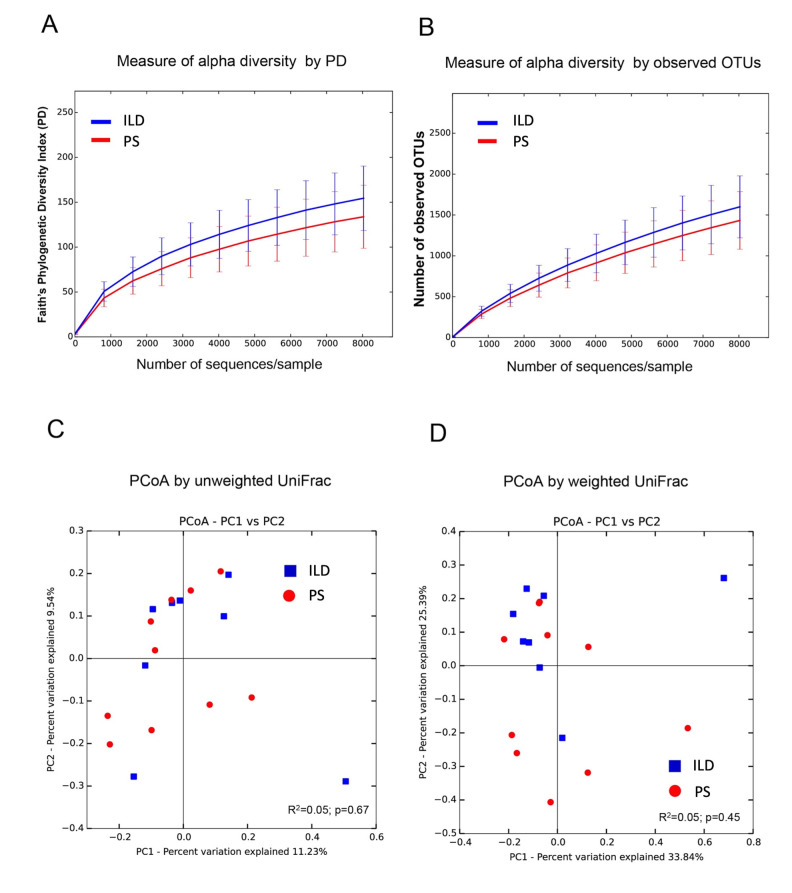
Further, we used alpha diversity, as computed with Faith’s Phylogenetic (fig. 4A) and OTUs (fig. 4B) diversity indices to evaluate bacterial community diversity, and found no differences in bacterial community richness between PS and ILD patients. However, the number of bacterial taxa was lower, albeit not significantly so, in PS patients than in ILD patients (fig. 4A,B). Beta diversity, namely, global community differences, was inferred using both unweighted (quantitative) (fig. 4C) and weighted (qualitative) (fig. 4D) UniFrac distances matrices. The resulting PCoA plots did not reveal any statistical difference between the two groups. In fact, in the PCoA plots, patients were randomly distributed when samples were clustered by disease status.
Fig. 4.
Lower airway microbiota diversity analysis. Alpha diversity of microbial communities was measured using Faith’s phylogenetic diversity index (A) and the number of observed OTUs (B). The error bars represent standard error of the mean. In addition, beta diversity was also computed using the Principal Coordinates Analysis (PCoA) with both unweighted (C) and weighted (D) UniFrac distances. No statistical significance was observed when samples were grouped by disease status, which also showed a very low effect size on the microbiome composition.
Discussion
Various studies have suggested that infection can cause PS and identified different potential disease-causing bacteria (37-42). In detail, mycobacterial DNA has been identified in different PS samples, such as lung biopsies, formalin-fixed paraffin-embedded tissues (37), lymph nodes (38), lung tissue and bronchial lavage fluid (39). All these studies performed polymerase chain reaction (PCR) to detect mycobacterial DNA in clinical samples from patients with sarcoidosis. Similarly, the presence of Propionibacterium acnes was demonstrated in tissue samples of PS patients using bacterial cultures (40), whereas antibodies against P. acnes have been identified in BAL fluid (41). Recently, propionibacterial rRNA was identified through real-time quantitative reverse transcription-polymerase chain reaction in formalin-fixed and paraffin-embedded tissue of lymph node biopsies, and thus proposed as a potential biomarker of sarcoidosis (42).
Thanks to advances in meta-omics technologies, it is now possible to analyze microbial communities in their integrity (13-15). Consequently, the focus has shifted from a single bacterial infection to a more general alteration of microbial communities. It seems that modifications of the mutual relationship among microbial species in a specific environment, as well as the alteration in microbiome composition (i.e., dysbiosis), can affect host physiology. This, in turn, leads to disease development or at least it might trigger the pathogenesis of more complex diseases. Therefore, we used a NGS-based 16S rRNA approach to fully characterize the pulmonary microbiota in the lower airways, and to evaluate whether dysbiosis is specific to a given disease, e.g., PS. The lower airway microbiota of patients affected by interstitial lung disease, including a small number of sarcoidosis patients, have been previously identified and no differences in terms of the composition of microbial communities able to discriminate the different diseases were found (23). Interestingly, in this paper Prevotellaceae and Streptococcaceae were the most represented taxa at family level; similarly, irrespective of disease etiology, Prevotellaceae was the most abundant family also in our datasets, with Prevotella being the most represented bacterial genus in BAL fluid. Conversely, in the same paper, differences between the upper and lower airway microbiota where identified, suggesting that a microbial discontinuity could be present, going from the upper to lower airways. In our study, we focused on PS in order to study a more homogeneous group of patients, and fully characterized their microbiota versus a group of patients affected by a variety of chronic inflammatory diseases. We found that the microbiota of PS patients does not have a specific signature able to discriminate sarcoidosis from other chronic respiratory diseases of the lower airways. Consequently, patients affected by diverse chronic respiratory diseases seem to share the same signature.
We may acknowledge that a limitation of our study is the rather limited number of subjects included in each group. This mainly depends on two factors. First, sarcoidosis, a relatively rare disease, has a high phenotype variability. Thus, to obtain a more homogenous sample, we included only patients with pulmonary signs, in wash-out from corticosteroids, and undergoing a bronchoscopy procedure. The second issue is the difficulty in enrolling healthy subjects as control group due to the invasiveness of sampling procedures. Therefore, we enrolled only patients who required a bronchoscopy for the diagnosis or monitoring of their specific lung disease. Furthermore, we excluded subjects with recent or ongoing infections at the time of examination to avoid bias in the microbiome evaluation. In addition, we analyzed the patients’ microbiome using non-parametric approaches (e.g., adonis, ANOSIM) based on permutation testing (e.g., Monte Carlo), which are frequently used to establish the effect size of a particular condition on the microbiome composition as well as to perform rarefaction analysis to limit sampling variability (43, 44). Given our statistical evaluations, the results are highly reproducible also when applied to higher numbers of samples (45, 46). The use of power analyses and/or parametric models, as applied to metagenomic amplicon studies, is new and currently under investigation (45, 46), therefore we used a more conservative approach based on multiple rarefaction curves.
Globally, our results suggest the absence of a qualitative and/or quantitative microbial signature in the lower airway microbiota able to specifically discriminate PS from other interstitial lung diseases. In addition, our findings are in line with other studies that did not identify distinct microbial signatures in different respiratory diseases (23, 47). Taken together, our results suggest that, unlike other body sites, the microbiome of BAL from lower airways is practically stable and not influenced in its qualitative and quantitative composition by different pathological processes that induce respiratory impairment.
In conclusion, our study shows that there is a similar microbiome signature in the BAL fluid of patients affected by different interstitial lung diseases of the lower airways. However, similar analyses are required to determine the possible effect of viruses and fungi populations in these pulmonary diseases.
Acknowledgments
The authors thank Jean Ann Gilder (Scientific Communication srl., Naples, Italy) for editing the text, and Vittorio Lucignano, CEINGE–Biotecnologie Avanzate for technical assistance.
Financial supports:
This study was supported by grant PON01_02589 (MICROMAP) 2012 from the Ministry of University and Research and grants POR Campania FSE 2007/2013 (CAMPUS-Bioframe and project DIAINTECH) from the Regione Campania, Italy.
References
- 1.Chen ES, Moller DR. Sarcoidosis--scientific progress and clinical challenges. Nat Rev Rheumatol. 2011 Jul 12;7(8):457–67. doi: 10.1038/nrrheum.2011.93. [DOI] [PubMed] [Google Scholar]
- 2.Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007 Nov 22;357(21):2153–65. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- 3.Medica I, Kastrin A, Maver A, Peterlin B. Role of genetic polymorphisms in ACE and TNF-alpha gene in sarcoidosis: a meta-analysis. J Hum Genet. 2007;52(10):836–847. doi: 10.1007/s10038-007-0185-7. [DOI] [PubMed] [Google Scholar]
- 4.Müller-Quernheim J, Schürmann M, Hofmann S, Gaede KI, Fischer A, Prasse A, et al. Genetics of sarcoidosis. Clin Chest Med. 2008 Sep;29(3):391–414. doi: 10.1016/j.ccm.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Sverrild A, Backer V, Kyvik KO, Kaprio J, Milman N, Svendsen CB, et al. Heredity in sarcoidosis: a registry-based twin study. Thorax. 2008 Oct;63(10):894–6. doi: 10.1136/thx.2007.094060. [DOI] [PubMed] [Google Scholar]
- 6.Mortaz E, Masjedi MR, Tabarsi P, Pourabdollah M, Adcock IM. Immunopathology of sarcoidosis. Iran J Allergy Asthma Immunol. 2014 Oct;13(5):300–6. [PubMed] [Google Scholar]
- 7.Newman LS, Rose CS, Bresnitz EA, Rossman MD, Barnard J, Frederick M, et al. A case control etiologic study of sarcoidosis: environmental and occupational risk factors. Am J Respir Crit Care Med. 2004 Dec 15;170(12):1324–30. doi: 10.1164/rccm.200402-249OC. [DOI] [PubMed] [Google Scholar]
- 8.Milman N, Lisby G, Friis S, Kemp L. Prolonged culture for mycobacteria in mediastinal lymph nodes from patients with pulmonary sarcoidosis. A negative study. Sarcoidosis Vasc Diffuse Lung Dis. 2004 Mar;21(1):25–8. [PubMed] [Google Scholar]
- 9.Gupta D, Agarwal R, Aggarwal AN, Jindal SK. Molecular evidence for the role of mycobacteria in sarcoidosis: a meta-analysis. Eur Respir J. 2007 Sep;30(3):508–16. doi: 10.1183/09031936.00002607. [DOI] [PubMed] [Google Scholar]
- 10.Shaykhiev R, Krause A, Salit J, Strulovici-Barel Y, Harvey BG, O’Connor TP, et al. Smoking-dependent reprogramming of alveolar macrophage polarization: implication for pathogenesis of chronic obstructive pulmonary disease. J Immunol. 2009 Aug 15;183(4):2867–83. doi: 10.4049/jimmunol.0900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tchaptchet S, Kirberg J, Freudenberg N, Schamel WW, Galanos C, Freudenberg MA. Innate, antigen-independent role for T cells in the activation of the immune system by Propionibacterium acnes. Eur J Immunol. 2010 Sep;40(9):2506–16. doi: 10.1002/eji.200939860. [DOI] [PubMed] [Google Scholar]
- 12.Broos CE, van Nimwegen M, Hoogsteden HC, Hendriks RW, Kool M, van den Blink B. Granuloma formation in pulmonary sarcoidosis. Front Immunol. 2013 Dec 10;4:437. doi: 10.3389/fimmu.2013.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Precone V, Del Monaco V, Esposito V, De Palma F, Ruocco A, Salvatore F, et al. Cracking the code of human diseases using next-generation sequencing: applications, challenges and perspectives. Biomed Res Int. 2015;2015 doi: 10.1155/2015/161648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Argenio V, Salvatore F. The role of the gut microbiome in the healthy adult status. Clin Chim Acta. 2015 Dec 7;451(Pt A):97–102. doi: 10.1016/j.cca.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 15.D’Argenio V. Human Microbiome Acquisition and Bioinformatic Challenges in Metagenomic Studies. Int J Mol Sci. 2018 Jan 27;19(2):383. doi: 10.3390/ijms19020383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Argenio V, Notomista E, Petrillo M, Cantiello P, Cafaro V, Izzo V, et al. Complete sequencing of Novosphingobium sp. PP1Y reveals a biotechnologically meaningful metabolic pattern. BMC Genomics. 2014 May 19;15:384. doi: 10.1186/1471-2164-15-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Argenio V, Petrillo M, Pasanisi D, Pagliarulo C, Colicchio R, Talà A, et al. The complete 12 Mb genome and transcriptome of Nonomuraea gerenzanensis with new insights into its duplicated “magic” RNA polymerase. Sci Rep. 2016 Dec 21;6(1):18. doi: 10.1038/s41598-016-0025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huebinger RM, Liu MM, Dowd SE, Rivera-Chavez FA, Boynton J, Carey C, et al. Examination with next-generation sequencing technology of the bacterial microbiota in bronchoalveolar lavage samples after traumatic injury. Surg Infect (Larchmt) 2013 Jun;14(3):275–82. doi: 10.1089/sur.2012.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouter KE, van Raalte DH, Groen AK, Nieuwdorp M. Role of the Gut Microbiome in the Pathogenesis of Obesity and Obesity-Related Metabolic Dysfunction. Gastroenterology. 2017 May;152(7):1671–1678. doi: 10.1053/j.gastro.2016.12.048. [DOI] [PubMed] [Google Scholar]
- 20.Bressa C, Bailén-Andrino M, Pérez-Santiago J, González-Soltero R, Pérez M, Montalvo-Lominchar MG, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One. 2017 Feb 10;12(2) doi: 10.1371/journal.pone.0171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Argenio V, Torino M, Precone V, Casaburi G, Esposito MV, Iaffaldano L, et al. The Cause of Death of a Child in the 18th Century Solved by Bone Microbiome Typing Using Laser Microdissection and Next Generation Sequencing. Int J Mol Sci. 2017, Jan 6;18(1) doi: 10.3390/ijms18010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One. 2012;7(10):e47305. doi: 10.1371/journal.pone.0047305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garzoni C, Brugger SD, Qi W, Wasmer S, Cusini A, Dumont P, et al. Microbial communities in the respiratory tract of patients with interstitial lung disease. Thorax. 2013 Dec;68(12):1150–6. doi: 10.1136/thoraxjnl-2012-202917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi H, Yong D, Lee K, Cho YJ, Chun J. Profiling bacterial community in upper respiratory tracts. BMC Infect Dis. 2014 Nov 13;14:583. doi: 10.1186/s12879-014-0583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feigelman R, Kahlert CR, Baty F, Rassouli F, Kleiner RL, Kohler P, et al. Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details. Microbiome. 2017 Feb 10;5(1):20. doi: 10.1186/s40168-017-0234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu W, Yuan X, Xu X, Ding R, Pang L, Liu Y, et al. Reduced airway microbiota diversity is associated with elevated allergic respiratory inflammation. Ann Allergy Asthma Immunol. 2015 Jul;115(1):63–8. doi: 10.1016/j.anai.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 27.Scher JU, Joshua V, Artacho A, Abdollahi-Roodsaz S, Öckinger J, Kullberg S, et al. The lung microbiota in early rheumatoid arthritis and autoimmunity. Microbiome. 2016 Nov 17;4(1):60. doi: 10.1186/s40168-016-0206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Argenio V, Casaburi G, Precone V, Pagliuca C, Colicchio R, Sarnataro D, et al. Metagenomics Reveals Dysbiosis and a Potentially Pathogenic N. flavescens Strain in Duodenum of Adult Celiac Patients. Am J Gastroenterol. 2016 Jun;111(6):879–90. doi: 10.1038/ajg.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010 May;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Argenio V, Casaburi G, Precone V, Salvatore F. Comparative metagenomic analysis of human gut microbiome composition using two different bioinformatic pipelines. Biomed Res Int. 2014;2014 doi: 10.1155/2014/325340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006 Jul;72(7):5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010 Oct 1;26(19):2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 33.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010 Jan 15;26(2):266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013 Nov 26;2(1):16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarke KR. Nonparametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–23. [Google Scholar]
- 36.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 37.Ikonomopoulos JA, Gorgoulis VG, Zacharatos PV, Manolis EN, Kanavaros P, Rassidakis A, et al. Multiplex polymerase chain reaction for the detection of mycobacterial DNA in cases of tuberculosis and sarcoidosis. Mod Pathol. 1999 Sep;12(9):854–62. [PubMed] [Google Scholar]
- 38.Ishige I, Usui Y, Takemura T, Eishi Y. Quantitative PCR of mycobacterial and propionibacterial DNA in lymph nodes of Japanese patients with sarcoidosis. Lancet. 1999 Jul 10;354(9173):120–3. doi: 10.1016/S0140-6736(98)12310-3. [DOI] [PubMed] [Google Scholar]
- 39.Li N, Bajoghli A, Kubba A, Bhawan J. Identification of mycobacterial DNA in cutaneous lesions of sarcoidosis. J Cutan Pathol. 1999 Jul;26(6):271–8. doi: 10.1111/j.1600-0560.1999.tb01844.x. [DOI] [PubMed] [Google Scholar]
- 40.De Brouwer B, Veltkamp M, Wauters CA, Grutters JC, Janssen R. Propionibacterium acnes isolated from lymph nodes of patients with sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis. 2015 Sep 14;32(3):271–4. [PubMed] [Google Scholar]
- 41.Schupp JC, Tchaptchet S, Lützen N, Engelhard P, Müller-Quernheim J, Freudenberg MA, et al. Immune response to Propionibacterium acnes in patients with sarcoidosis--in vivo and in vitro. BMC Pulm Med. 2015 Jul 24;15:75. doi: 10.1186/s12890-015-0070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, Wei YR, Zhang Y, Du SS, Baughman RP, Li HP. Real-time quantitative reverse transcription-polymerase chain reaction to detect propionibacterial ribosomal RNA in the lymph nodes of Chinese patients with sarcoidosis. Clin Exp Immunol. 2015 Sep;181(3):511–7. doi: 10.1111/cei.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White JR, Nagarajan N, Pop M. Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PLoS Comput Biol. 2009 Apr;5(4) doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005, Dec;71(12):8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.La Rosa PS. Diss. Washington University: 2015. Hypothesis testing of metagenomic data. [Google Scholar]
- 46.La Rosa PS, Brooks JP, Deych E, Boone EL, Edwards DJ, Wang Q, et al. Hypothesis testing and power calculations for taxonomic-based human microbiome data. PLoS One. 2012;7(12):e52078. doi: 10.1371/journal.pone.0052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmermann A, Knecht H, Häsler R, Zissel G, Gaede KI, Hofmann S, et al. Atopobium and Fusobacterium as novel candidates for sarcoidosis-associated microbiota. Eur Respir J. 2017 Dec 14;50(6) doi: 10.1183/13993003.00746-2016. [DOI] [PubMed] [Google Scholar]