Abstract
The timing of early mitotic events during preimplantation embryo development is important for subsequent embryogenesis in many mammalian species, including mouse and human, but, to date, no study has closely examined mitotic timing in equine embryos from oocytes obtained by ovum pick-up. Here, cumulus–oocyte complexes were collected by transvaginal follicular aspiration, matured in vitro and fertilised via intracytoplasmic sperm injection. Each fertilised oocyte was cultured up to the blastocyst stage and monitored by time-lapse imaging for the measurement of cell cycle intervals and identification of morphological criteria indicative of developmental potential. Of the 56 fertilised oocytes, 35 initiated mitosis and 11 progressed to the blastocyst stage. Analysis of the first three mitotic divisions in embryos that formed blastocysts determined that typical blastocyst timing (median ± IQR) is 30.0 ± 17.5 min, 8.8 ± 1.7 h and 0.6 ± 1.4 h respectively. Frequent cellular fragmentation, multipolar divisions and blastomere exclusion suggested that equine embryos likely contend with a high incidence of chromosomal missegregation. Indeed, chromosome-containing micronuclei and multinuclei with extensive DNA damage were observed throughout preimplantation embryogenesis. This indicates that time-lapse image analysis may be used as a non-invasive method to assess equine embryo quality in future studies.
Keywords: blastomere, cytokinesis, IVF, micronuclei, mitosis, preimplantation
Introduction
Following IVF, human preimplantation embryos are traditionally scored for quality at the zygote stage and then again at the 4- to 8-cell stage approximately 48–72 h later. Morphological criteria, including cell number, degree of cellular fragmentation and blastomere symmetry, are typically used to evaluate embryo viability at the cleavage stage (Racowsky et al. 2010; Luke et al. 2014). If embryos are allowed to proceed in culture to the blastocyst stage, additional characteristics, such as blastocoel expansion, cohesiveness of the inner cell mass (ICM) and trophectoderm (TE) and hatching ability, are considered in the decision of which to select for transfer (Gardner et al. 2000; Luke et al. 2014). Although these snapshots in time help identify human embryos with poor developmental potential, other informative cellular features may be missed during the several hours between designated embryo assessments. These screening measures also likely require removal of the embryos from the physiological conditions of an incubator, as well as increased exposure to light during visualisation on a stereomicroscope, both of which could negatively affect subsequent embryogenesis.
Based on the limitations of evaluation at static time points and the risks associated with removing embryos from the incubator, non-invasive time-lapse imaging technologies have been established to measure the timing of key cellula revent sand morphological features contributing to blastocyst formation (Daughtry and Chavez 2016). After validating the safety and efficacy of time-lapse monitoring (TLM) in human embryos (Nakahara et al. 2010), several imaging parameters, such as the time to polar body extrusion or first cleavage division, were identified with some success in predicting embryo competency (Payne et al. 1997; Lundin et al. 2001; Hardarson et al. 2002; Lemmen et al. 2008). The first morphokinetic studies, or those that measured the duration or intervals between cell cycles, then arose and the first three mitotic divisions were shown to be highly prognostic of which embryos will likely arrest versus those that form blastocysts (Wong et al. 2010; Meseguer et al. 2011). By combining mitotic timing with assessment of cellular fragmentation dynamics during early cleavage divisions, it was also demonstrated that blastomere behaviour is directly correlated with overall embryo chromosomal composition (Chavez et al. 2012; Daughtry et al. 2019). Additional imaging parameters, including the time to the appearance of the fifth cell, reverse or multipolar cleavage, the initiation of compaction and the completion of blastulation (Meseguer et al. 2011; Azzarello et al. 2012; Cruz et al. 2012; Dal Canto et al. 2012; Hlinka et al. 2012; Rubio et al. 2012; Athayde Wirka et al. 2014; Liu et al. 2014b), have also been used to differentiate between human embryos of low and high quality, as well as ploidy status (Campbell et al. 2013; Basile et al. 2014; Yang et al. 2014; Chawla et al. 2015; Balakier et al. 2016; Minasi et al. 2016; Del Carmen Nogales et al. 2017; Desai et al. 2018).
Although there are several reports of TLM in other mammalian species besides human (Pribenszky et al. 2010; Sugimura et al. 2010, 2012; Weinerman et al. 2016), only recently was TLM used to evaluate equine preimplantation development to the blastocyst stage (Marzano et al. 2019). However, the primary focus of that study was on the effects of a specific toxic chemical present in ovarian follicular fluid and IVM media on embryo morphokinetics. In addition, the authors used oocytes from abattoir obtained ovaries, only those from draft breeds and the time of intracytoplasmic sperm injection (ICSI) for assessment of certain imaging parameters (Marzano et al. 2019). This can have confounding effects on the measurement of other overlapping cell cycle-related characteristics, as shown previously by a report that implemented a biological start point instead of a procedure such as ICSI (Liu et al. 2014a). Thus, a close examination of morphokinetics in non-abattoir-derived oocytes collected by ovum pick-up (OPU) from the more common breeds of mares that serve as oocyte donors and surrogates for embryo transfer is still needed. In the present study, oocytes were obtained by OPU and the first three mitotic divisions evaluated by TLM following fertilisation based on high blastocyst prediction capabilities in other mammals (Pribenszky et al. 2010; Sugimura et al. 2010, 2012; Wong et al. 2010; Chen et al. 2013; Weinerman et al. 2016). A comparison of mitotictiming between arrested embryos and blastocysts was also correlated to identify morphological criteria that should be considered for equine embryo classification and potential ranking.
Materials and methods
Experimental design
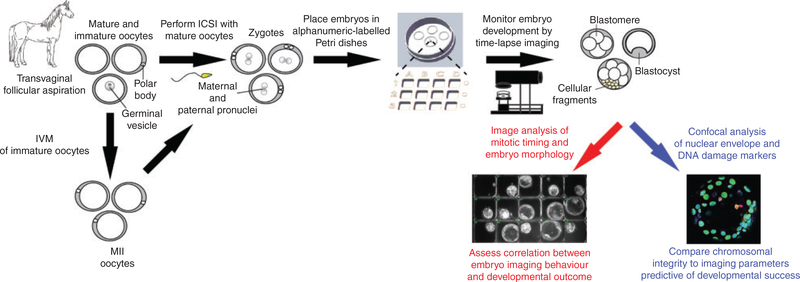
To comprehensively examine early equine embryogenesis under normal culture conditions, we designed an experimental approach using OPU for oocyte collection and TLM to non-invasively assess equine embryos throughout preimplantation development to the blastocyst stage (Fig. 1). Cumulus–oocyte complexes (COCs) obtained by transvaginal aspiration (TVA) from reproductive-age mares were matured in vitro and mature MII oocytes underwent ICSI. Presumed zygotes were analysed by TLM to evaluate mitotic divisions, the absence or presence of cellular fragmentation and other imaging parameters and/or morphological features indicative of embryo competency. After 3–10 days in culture, zygotes that did not divide, arrested cleavage stage embryos and blastocysts were removed and fixed for assessment of nuclear structure and DNA damage by multicolour confocal imaging. The effects of chromosome-containing micronuclei, cellular fragmentation and double-stranded DNA breaks on embryo arrest versus successful progression to the blastocyst stage were evaluated by correlating the imaging parameters to embryo immunostaining and developmental outcome (Fig. 1).
Fig. 1.
Experimental approach to assess equine embryo developmental potential by time-lapse monitoring. Equine oocytes were obtained by transvaginal aspiration from cross-bred mares (n = 9) undergoing ovum pick-up and matured in vitro. Mature MII oocytes were fertilised via intracytoplasmic sperm injection (ICSI) and presumed zygotes (n = 56) were placed in alphanumeric-labelled Petri dishes for embryo tracking and non-invasive time-lapse image analysis of preimplantation development to the blastocyst stage. Early mitotic timing and morphological criteria predictive of blastocyst formation were measured and correlated with embryo developmental outcome. Confocal imaging was used to assess nuclear structure and DNA integrity in arrested cleavage stage embryos versus those that reached the blastocyst stage.
Equine oocyte collection and embryo culture
The oocyte donors were nine non-pregnant mares, one of which was aspirated twice, aged between 4 and 11 years, of multiple breeds (both draft and non-draft) and residing in an embryo transfer recipient herd. An additional five mares of similar age and breed were used to test the prediction model. Between one and three mares were aspirated per day for an average of four oocytes obtained per mare. Equine COCs were collected by ultrasound-guided TVA using a sterile needle guide attached to an 8- to 5-MHz human transvaginal ultrasound probe (Fujifilm Sonsite). The guide was carefully placed intravaginally next to thecervix ipsilateral totheovary to be aspirated and the contents of each follicle were scraped and lavaged with heparinised (5IUmL−1) Complete Flush Solution (Vigro Vetriquinol USA) using a 12-gauge double-lumen collection needle (Mila International). Oocytes were identified in aspirates and placed in equilibrated holding medium (Emcare; ICPbio Reproduction) until overnight shipment in equilibrated medium (all SigmaAldrich), consisting of40% M199 with Hanks’ salts, 40% M199 with Earle’s salts and 20% newborn calf serum (NCS), in a portable incubator (Micro QTechnologies) at 228C as previously described (Foss et al. 2013). COCs recovered from dominant gonadotropin-stimulated follicles were placed in equilibrated transport medium, consisting of 45% M199 with Earle’s salts, 45% Dulbecco’s modified Eagle’s medium (DMEM)/F12, 10% NCS, 0.01% pyruvate, and 25μgmL−1 gentamycin (all Sigma Aldrich) and shipped overnight in another incubator at 38.28C. Immature oocytes underwent incubation at 38.58C in equilibrated maturation medium consisting of 49.5% DMEM/F12 with 15mM HEPES and 25μgmL−1 gentamicin, 32.5% Global Medium (LifeGlobal), 4.5% NCS, 4.5% serum substitute (Life Global),0.1 mM sodium pyruvate, 5 mUmL−1 FSH(Sioux Biochemical), 1mUmL−1 somatotropin (Sioux Biochemical), 10μLmL−1 insulin–transferrin–selenium solution (Sigma Aldrich) and 9% frozen–thawed pooled equine follicular fluid from dominant stimulated preovulatory follicles for 28–32 h, as described by Foss et al. (2013). Mature MII oocytes were denuded following exposure to 80IU hyaluronidase (Sage; Cooper Surgical) and fertilised by ICSI in the afternoon or early evening using thawed spermatozoa from one of two stallions with proven fertility. Immediately following ICSI, each fertilised oocyte was placed in an alphanumeric-labelled microwell Petri dish that holds up to 12 samples (Progyny) to help with embryo tracking. Embryos were incubated at 38.58C with 5% CO2, 5% O2 and 90% N2 for up to 10 days in 100μL culture medium (54% DMEM/F12 with 15 mM HEPES and 25μgmL−1 gentamicin, 36% Global Medium, 5% NCS, 5% serum substitute and 0.1mM sodium pyruvate) under mineral oil, as described previously (Foss et al. 2013).
All animal procedures used in this study adhered to the Principles of Veterinary Medical Ethics of the American Veterinary Medical Association (AVMA).
Time-lapse imaging
Embryos underwent time-lapse imaging until they arrested or reached the blastocyst stage using an Eeva darkfield or bimodal (brightfield-darkfield) microscope system from Progyny, and were collected the morning after arrest or blastocyst formation. Images were taken every 5min and compiled into a movie with well identification labels and time stamps for the measurement of imaging parameters using FIJI software version 2.0.0 (NIH, Bethesda, MD; Schindelin et al. 2012). The timing intervals of the first three mitotic divisions and other imaging parameters predictive of successful development and/or ploidy status, such as the timing, degree and pattern of cellular fragmentation, were measured manually by two independent reviewers (K. E. B. and S. L. C.) based on previous findings of intra- and interobserver variability (Sundvall et al. 2013). For those that formed blastocysts, we also determined whether the embryo exhibited imaging behaviours indicative of potential aneuploidy correction, including, but not limited to, multipolar divisions, resorption or exclusion of cellular fragments and blastomere exclusion, as per previously published results in other mammalian species(Chavez et al. 2012; Daughtry and Chavez 2018; Daughtry et al. 2019).
Immunofluorescence confocal imaging
The zona pellucida (ZP) was removed by a 1- to 2-min incubation in EmbryoMax Acidic Tyrode’s Solution (EMD Millipore) and ZP-free embryos were washed in phosphate-buffered saline (PBS; AlfaAesar)containing 0.1% bovine serum albumin (BSA; Sigma Aldrich) plus 0.1% Tween 20 (Sigma-Aldrich) termed PBST. Each embryo was fixed with 4% para formaldehyde in PBS for 20 min at room temperature. Once fixed, the embryos were washed three times with gentle shaking in PBST for a total of 15min to remove residual fixative. Embryos were permeabilised in 1% Triton-X (Calbiochem) for 1 h at room temperature and washed in PBST as described above. To block non-specific binding, embryos were transferred to a solution of 7% donkey serum (Jackson ImmunoResearch Laboratories) in PBST overnight at 48C. Antibodies against lamin B1 (LMNB1; rabbit; Catalogue no. ab16048; Abcam), a nuclear envelope marker, and the histone variant H2AX phosphorylated at serine 139 (γH2AX; mouse; Clone no. JBW301; EMD Millipore), a marker of double-stranded DNA breaks, were diluted 1:1000 and 1:100 respectively in PBST with 1% donkey serum, and embryos were sequentially stained over two nights at 48C. Primary antibodies were detected using Alexa Fluor 488- or 647-conjugated donkey anti-rabbit LMNB1 or antimouse γH2AX secondary antibodies (Thermo Fisher Scientific) respectively at a dilution of 1 : 250 in 1% donkey serum in PBST at room temperature for 1 h in the dark. Between antibody incubations, embryos were washed in PBST and the DNA was stained with 1 μgmL−1 4′,6′-diamidino-2-phenylindole (DAPI) for 15min. Immunofluorescence was visualised using glass-bottomed Petri dishes (Mattek) and a Leica SP5 AOBS spectral confocal system. Z-stacks, 1–5mM apart, were imaged one fluorophore at a time to avoid spectral overlap between channels. Stacked images and individual channels for each colour were combined into composite images using FIJI software version 2.0.0 (NIH, Bethesda, MD; Schindelin et al. 2012).
Statistical analysis
The time intervals for each imaging parameter are given as the median interquartile range (IQR) calculated by IQR5 Q32Q1, where the first quartile (Q1) is the median of the smallest values and third quartile (Q3) is the median of the largest values. Median values for mitotic timing were compared between arrested embryos and those that formed blastocysts using a Mann–Whitney– Wilcoxon test. Variations in the time intervals between the two embryo groups were evaluated using Levene’s test. Two-sided P,0.05 was considered significant. All statistical analyses were performed using R version s3.5.1 (R Foundation for Statistical Computing).
Pruned decision tree
To predict the probability of an embryo reaching the blastocyst stage using mitotic timing, we built a pruned tree using the ‘tree’ package in R. Briefly, the number of mis classifications was used as the measure of node heterogeneity to guide cost-complexity pruning using 10-fold cross-validation. The optimal cost-complexity parameter determined was then used to build the final classification tree using all data. Prediction accuracy was calculated as a proportion of correctly classified observations.
Results
Comparison of early mitotic timing between equine blastocysts and arrested embryos
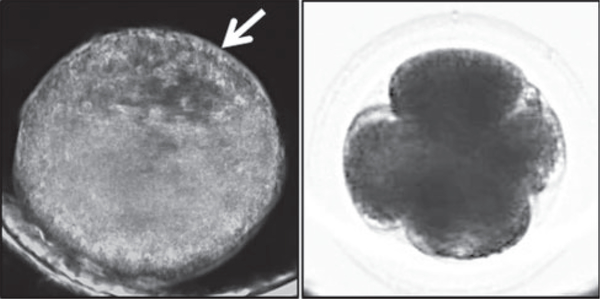
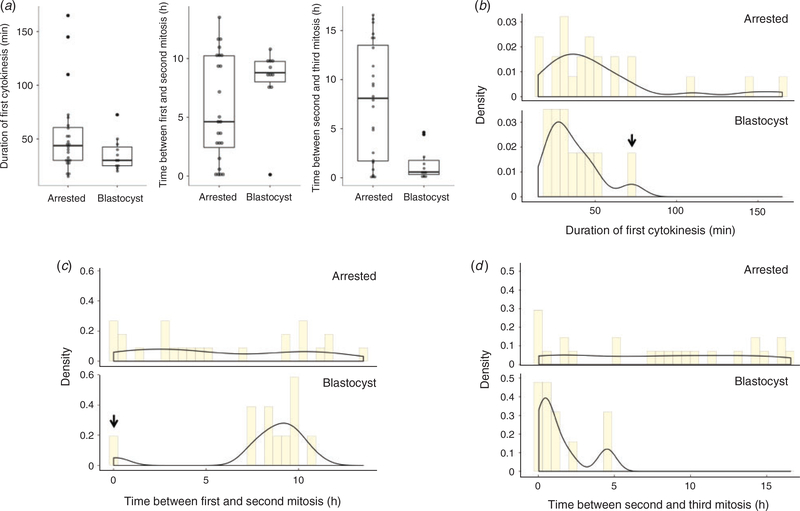
The first objective of this study was to determine whether the time intervals between the first three mitotic divisions are prognostic of blastocyst formation in equine embryos. From the TVA COCs, 56 fertilised oocytes identified by polar body extrusion were obtained, of which 35 embryos underwent mitosis and 11 successfully reached the blastocyst stage (Table 1). Note that blastocyst formation was more easily recognised by visualising the blastocoel(see Fig. 2a) in embryos with dark field time lapse imaging, but bright field imaging was more suitable for assessing early mitotic timing (Fig. 2b). Rather than measuring the time to the first cleavage division, we measured the duration of the first cytokinesis, or the time between the appearance of cleavage furrows and the completion of division as determined by the appearance of two distinct blastomeres with ‘pinched’ membranes as 30.0 ± 17.5min (Q1 = 25.0 min; Q3 = 42.5min), the time between the first and second mitotic division as 8.8 ± 1.7 h (Q1 = 8.02 h; Q3 = 9.75 h) and the time between the second and third mitotic division as 0.6 ± 1.4 h (Q1 = 0.33 h; Q3 = 1.77 h) in those embryos that formed blastocysts (Table 2). Blastocyst formation was typically observed within 8 days of culture and no differences in morphological characteristics or mitotic timing were detected in the two blastocysts that reached this stage by the morning of Day 10. The time intervals between the first three mitotic divisions in equine embryos that arrested before reaching the blastocyst stage were 43.8 ± 30.6min (Q1 = 30.0min; Q3 = 60.6min), 4.67.8 h (Q1 = 2.44 h; Q3 = 10.2 h) and 8.1 ± 11.8 h (Q1 = 1.71 h; Q3 = 13.5 h) respectively. Thus, embryos that reached the blastocyst stage tended to exhibit more similar mitotic timing than the arrested embryos, with the exception of one outlier that was the same embryo for the first two divisions and two different embryo outliers for the third division(Fig. 3a). Althoughthe firstembryo underwent a 1:4 multipolar division at the zygote stage to completely bypass the 2- and 3-cell stages, which explains the reasoning behind its isolation, the two other embryos exhibited normal bipolar divisions but a longer time interval between the second and third mitosis. Statistical analyses showed that the median and variance of the first cytokinesis duration were not significantly different between embryo groups, likely due to the large range in mitotic timing observed in the arrested embryos and/or the primary outlier in the blastocyst group (Table 2; Fig. 3b). However, the variance (P = 0.015, Levene’s test) in time between the first and second mitotic divisions (Fig. 3c) and both the median (P = 0.004, Mann–Whitney–Wilcoxon test)and variance (P,0.001) in time between the second and third mitosis were significantly different between embryo groups (Fig. 3d).
Table 1. Correlation between equine embryo developmental outcome and time-lapse monitoring (TLM).
Data show the number and calculated percentage of equine embryos that initiated mitosis and arrested versus those that reached the blastocyst stage, with the most common imaging behaviours observed. Note that there were five embryos that began fragmenting at the zygote stage but never actually divided
| TLM feature | Total no. embryos | No. embryos exhibiting behaviour (%) |
|---|---|---|
| Initiated cleavage | 56 | 35 (62.5) |
| Blastocyst formation | 56 | 11 (19.6) |
| Cellular fragmentation | 56 | 34 (60.7) |
| Multipolar divisions | 35 | 5 (14.3) |
| Blastomere exclusion | 11 | 2 (18.1) |
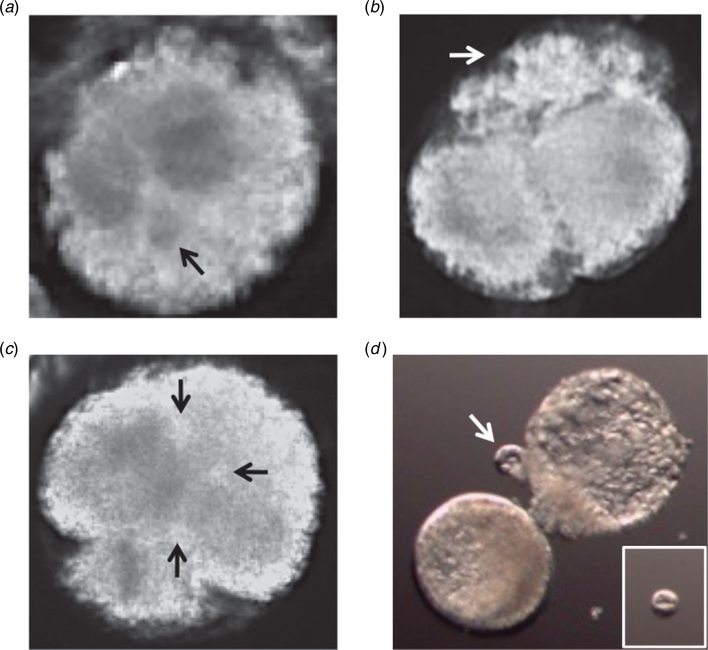
Fig. 2.
Comparison of darkfield and brightfield imaging of equine preimplantation embryogenesis. (a) Image frame of an equine blastocyst monitored by darkfield time-lapse imaging throughout preimplantation development. The white arrow indicates the blastocoel of the blastocyst. (b) A 4-cell equine embryo visualised using the bimodal microscope shows brightfield imaging is more suitable for assessing early mitotic timing.
Table 2.
Comparison of mitotic timing between equine blastocysts and arrested embryos
| Equine blastocysts (n=11) | Arrested equine embryos (n=24) | |
|---|---|---|
| Duration of first cytokinesis (min) | 30.0±17.5 | 43.8±30.6 |
| Time between first and second mitosis (h) | 8.8±1.7* | 4.6±7.8 |
| Time between second and third mitosis (h) | 0.6±1.4†‡ | 8.1±11.8 |
Data show the median±interquartile range of time intervals between the first three mitotic divisions (see Fig. 3) of equine blastocysts and embryos that arrested before reaching this stage. Significant differences in the variance in time between the first and second mitotic division (*P=0.015, Levene’s test) and in both the median (†P=0.004, Mann–Whitney–Wilcoxon test) and variance (‡P<0.001) in time between the second and third mitotic divisions indicated by different symbols
Fig. 3.
Arrested equine embryos exhibit highly variable mitotic timing compared with blastocysts. (a) Box and whisker plots of the time intervals between the first three mitotic divisions in arrested cleavage stage embryos and blastocysts show stricter mitotic timing in embryos that successfully developed to the blastocyst stage. The boxes show the interquartile range, with the median value indicated by the horizontal line; whiskers show the range. Individual symbols show outliers. (b–d) Analysis of the variance in the duration of the first cytokinesis (b) and the time between the first and second mitotic division (c) and the second and third mitosis (d) revealed highly variable early mitotic timing in arrested embryos compared with blastocysts. Columns show the density, with the solid lines indicating trends. Note that the one outlier in the blastocyst group (black arrows in (b) and (c)) was the same embryo for the first two divisions.
Early mitotic timing as a predictor of blastocyst development in equine embryos
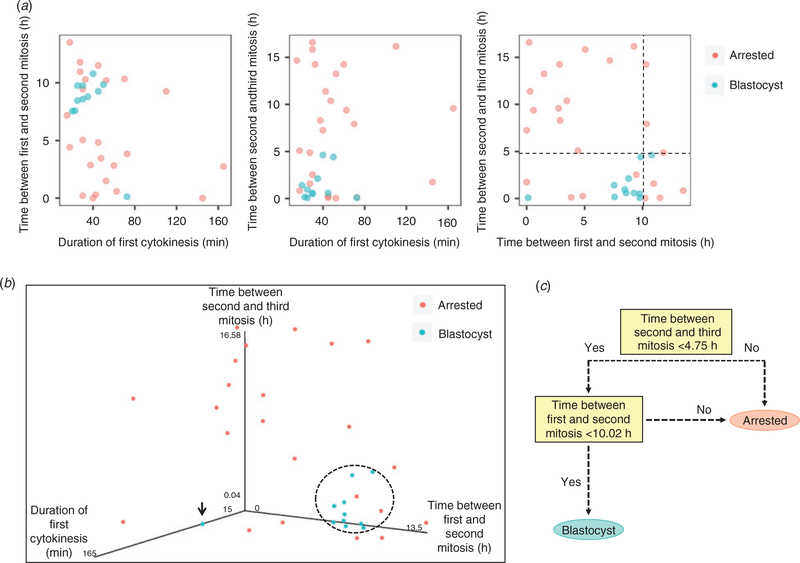
Because the combination of multiple imaging parameters, rather than individual cell cycle timing, is predictive of blastocyst formation in human embryos (Wong et al. 2010; Chen et al. 2013), we next sought to determine whether there was a pairwise relationship between the first three mitotic divisions in equine embryos. As shown in Fig. 4a, there was no clear separation of embryo groups in two-dimensional scatter plots due to some overlap in mitotic timing between blastocysts and arrested embryos. When the time intervals of all three mitotic divisions were graphed in a three-dimensional (3D) parameter plot, with the exception of the one outlier embryo, the blastocysts tended to cluster together in a similar region of mitotic timing (Fig. 4b). In contrast, most (91.7%; n = 22/24) equine embryos that did not reach the blastocyst stage exhibited highly variable mitotic timing and did not cluster with the blastocysts in the 3D plot. Initial observations using a pruned decision tree suggested (with 85.7% accuracy) that if the time between the second and third division was less than 4.75 h and the time between the first and second division was less than 10.02 h, then the embryo had a high probability of progressing to the blastocyst stage (Fig. 4c). To test these preliminary findings, we examined five additional embryosthat formed blastocysts with oocytes obtained from five new mares and determined that four of the five embryos exhibited mitotic timing that was predictive of blastocyst formation (i.e. time between first and second division = 8.12.6 h; time between second and third division = 2.11.2 h). Similar to the outlier embryo described above, the one blastocyst that would have not been predicted to reach this stage also underwent a multipolar first division, but from one to three cells. Although these abnormal divisions are discernible by time-lapse imaging for embryo de-selection, further analyses with an independent dataset using embryos cultured under different conditions are required to validate this prediction model.
Fig. 4.
Combined assessment of mitotic timing is predictive of equine blastocyst development. (a) Two-dimensional scatter plots showing the pairwise correlation between each mitotic division in arrested embryos and blastocysts. The black dotted lines provide decision boundaries for defining the time intervals of the second and third mitosis that predict embryo arrest versus successful development to the blastocyst stage. (b) Three-dimensional scatter plot of the time between the first three mitotic divisions demonstrates that, with the exception of one embryo (black arrow), equine blastocysts tend to cluster together in a similar region (indicated by the dotted circle). (c) Pruned decision tree for determining the likelihood than an embryo will form a blastocyst based on early mitotic timing. If the time between the second and third mitosis is less than 4.75 h and the time between the first and second mitosis is less than 10.02 h, then the embryo has a high probability of progressing to the blastocyst stage (accuracy, 85.7%).
Imaging features indicative of equine embryo aneuploidy generation and/or resolution
Given that mitotic timing in conjunction with assessment of cellular fragmentation dynamics is indicative of chromosomal composition during early cleavage divisions in other mammalian species (Chavez et al. 2012; Daughtry et al. 2019), another aim of the present study was to evaluate the incidence of fragmentation and other cellular events that may be reflective of overall embryo ploidy. As indicated in Table 1, cellular fragmentation was observed in most (60.7%; n = 34) equine embryos at the zygote stage, including several(n = 5)that fragmented but never actually divided. Although we did not directly assess aneuploidy, we suspect that these embryos inherited a meiotic error based on previous observations that most embryos with meiotic chromosomal missegregation, including those that are polyploid, begin fragmenting at the zygote stage (Chavez et al. 2012). This was supported by findings of micronuclei formation around presumably missegregated chromosomes in certain fragmented zygotes by TLM (Fig. 5a). The remaining embryos (n = 29) underwent fragmentation during or after the first cleavage division, which suggests that they had incurred mitotic errors (Fig. 5b). As mentioned above, we also detected a relatively high frequency (14.3%; n = 5) of multipolar divisions identified by more than two cleavage furrows in zygotes (Fig. 5c), four of which arrested following the abnormal division. It is important to note that the one multipolar embryo that did progress to the blastocyst stage was the same outlier described above in Fig. 4b and exhibited a 1:4 cell division without fragmentation, which we have shown is often associated with blastomere exclusion in primate embryos (Daughtry and Chavez 2018; Daughtry et al. 2019). Indeed, the exclusion of a large non-dividing blastomere (Fig. 5d) from the early cleavage stage was observed in this embryo and at least one other upon blastocyst formation and hatching (Table 1).
Fig. 5.
Morphological criteria used to assess equine embryo developmental potential by time-lapse monitoring. Image frames of the most common morphological features observed in equine embryos, including (a) micronuclei formation (black arrow adjacent to pronuclei in this zygote), (b) cellular fragmentation (white arrow) during or immediately after the first cleavage division, (c) multipolar division at the zygote stage (black arrows indicate three cleavage furrows) and (d) blastomere exclusion (white arrow) upon blastocyst formation and hatching. The inset in (d) shows isolation of the large excluded blastomere from the rest of the embryo.
Prevalence of chromosome-containing micronuclei from the zygote to blastocyst stage
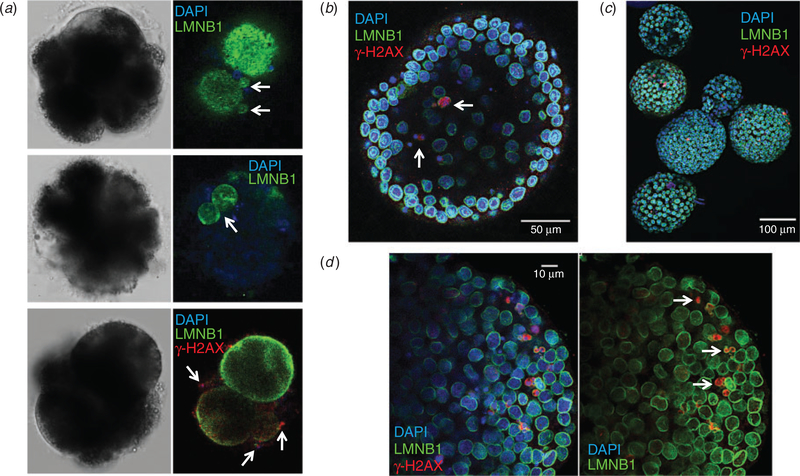
Because aneuploidy is often, but not always, associated with cellular fragmentation and the formation of micronuclei tends to precede the appearance of fragments (Chavez et al. 2012; Daughtry et al. 2019), we examined nuclear structure and DNA integrity in the embryos. Immunolabelling with the nuclear envelope marker LMNB1 revealed micronuclei (Fig. 6a, top panel) and multinuclei (Fig. 6a, middle panel) in arrested cleavage-stage embryos stained with DAPI. Based on several reports of DNA fragility in the micronuclei of both somatic and embryo cells (Crasta et al. 2012; Hatch et al. 2013; Liu et al. 2018; Daughtry et al. 2019), we evaluated whether the micronuclei were susceptible to DNA damage. This was accomplished by immunostaining the embryos for γ-H2AX phosphorylated at Ser139, a marker commonly used to assess double-stranded DNA breaks (Rogakou et al. 1998). Indeed, γ-H2AX-positive foci were associated with micronuclei formation and extensive DNA degradation, which was likely due to embryo arrest. We also observed that blastocysts tended to retain micronuclei (Fig. 6b) but, because it is difficult to distinguish the ICM from the TE of in vitro-produced equine embryos (Hinrichs et al. 2007; Choi et al. 2009), whether there was preferential allocation of micronuclei to one of the two lineages could not be determined. Most blastocysts contained multiple micronuclei with γ-H2AX-positive immunolabels (Fig. 6c), and these structures often lacked or exhibited a defective nuclear envelope (Fig. 6d) that predisposed the DNA to increased and rapid damage, as shown previously (Hatch et al. 2013; Liu et al. 2018).
Fig. 6.
Micronuclei are prevalent and susceptible to DNA damage in equine embryos. (a) Confocal imaging of 4′,6′-diamidino-2-phenylindole (DAPI)-stained (blue) arrested cleavage stage embryos immunolabelled with an antibody against the nuclear envelope marker lamin B1 (LMNB1; green) revealed chromosome-containing micronuclei (top panel) and multinuclei (middle panel). Additional immunostaining with an antibody against the histone variant H2AX phosphorylated at serine 139 (red), a marker of double-stranded DNA breaks, in arrested embryos demonstrated that γ-H2AX-positive foci are associated with micronuclei formation (bottom panel). (b)Similar imaging of DAPI-stained blastocyst stage embryos immunolabelled for LMNB1 and γ-H2AX demonstrated that blastocysts can retain micronuclei. (c) Micronuclei were detected in most blastocysts and exhibited DNA damage, as indicated by the intensity of γ-H2AX immunostaining. (d) The prevalence of DNA breakage in blastocysts was likely due to the lack of, or a defect in, nuclear envelope surrounding micronuclei (white arrows).
Discussion
Time-lapse image analysis of human oocyte maturation, fertilisation and preimplantation embryogenesis was initially reported several years ago (Payne et al. 1997; Hardarson et al. 2002) but only recently used to evaluate embryos from other mammalian species (Pribenszky et al. 2010; Sugimura et al. 2010, 2012; Weinerman et al. 2016). Using abattoir-derived equine ovaries from draft mares, Marzano et al. (2019) showed differences in the time of second polar body extrusion and duration of the second cell cycle between control and di-(2ethylhexyl)phthalate (DEHP)-treated oocytes. To thebest of our knowledge, the present study is the first to closely examine equine preimplantation development to the blastocyst stage via TLM with oocytes obtained by OPU from mares of various breeds (both draft and non-draft) that are commonly used as oocyte donors and surrogates for embryo transfer. Marzano et al. (2019) also determined that the duration of the second cell cycle, or the time between the first and second mitosis, as reported here, was, 17 h in controls, whereas the time between second and third mitotic division was, 6 h, both of which had large standard deviations. In the present study we demonstrated that the time intervals between the first and second, as well as the second and third, divisions in those equine embryos that formed blastocysts was, 7–10 h and, 1–3 h respectively, with significantly less variation and closer to that observed in human embryos (Wong et al. 2010; Chen et al. 2013). This discrepancy is most likely the result of differences in mitotic timing between embryos from diverse breeds of horses, but could also be due to the origins of OPU versus abattoir-obtained oocytes, culture media conditions and/or the stallions used for fertilisation.
In addition to the approximate time between early mitotic divisions, equine embryos appear to undergo cellular fragmentation at a frequency more comparable to humans than ruminants such as cows, which typically exhibit approximately 15–25% fragmentation during preimplantation development (Somfai et al. 2010; Sugimura et al. 2012). We also identified a similar incidence of multipolar divisions in equine embryos as in humans (Kalatova et al. 2015) and, given the association between this cellular event and chromosome sequestration by cellular fragments (Daughtry et al. 2019), it will be important to assess whether the fragments from multipolar equine embryos contain missegregated chromosomes lost from blastomeres. Although aneuploidy was not directly evaluated in the present study, we did examine nuclear structure and DNA integrity and determined that chromosome-containing micronuclei are prevalent throughout early equine embryogenesis, which suggests frequent chromosome missegregation. However, because these oocytes underwent IVM, we suspect that they have increased chromosomal instability over invivo-matured oocytes based on comparisons between these two groups in other mammals (Treff et al. 2016). Current work is focused on determining the incidence of aneuploidy in cleavage stage equine embryos using high-resolution techniques, with plans to apply this technology to the assessment of chromosome missegregation in equine blastocysts at the single-cell level.
Analogous to previous findings with both human and nonhuman primate embryos (Daughtry and Chavez 2018; Daughtry et al. 2019), we did identify at least two equine embryos that exhibited the exclusion of large, non-dividing blastomeres from the early cleavage stage during blastocyst formation. Although the first three mitotic divisions were normal in one of these embryos, the other underwent a 1:4 multipolar division without cellular fragmentation at the zygote stage, as recently described with rhesus monkeys (Daughtry et al. 2019). Therefore, we suspect that blastomere exclusion is conserved across higher mammalian species and likely represents one mechanism to overcome aneuploidy in certain instances, but further work is required to confirm this hypothesis. Nevertheless, the findings of this study suggest that time-lapse image analysis of early mitotic timing in conjunction with morphological assessment may be used as a non-invasive method to effectively evaluate equine embryo developmental competence to the blastocyst stage in future studies. This information can be used to predict embryo developmental success, as well as help determine the best embryo for transfer, especially when multiple blastocysts are obtained from a mare.
Acknowledgements
This study was primarily supported by the Equine IVF Fund in the Oregon Health & Science University (OHSU) Foundation, as well as in part by OHSU/Oregon National Primate Research Center (ONPRC) start-up funds (to Shawn L. Chavez) and the National Institutes of Health (NIH)/National Institute of Child Health and Development (NICHD; R01HD086073-A1). The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors also gratefully acknowledge Richard Beck, Robert Foss and Nancy Cook for COC aspiration and collection.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- Athayde Wirka K, Chen AA, Conaghan J, Ivani K, Gvakharia M, Behr B, Suraj V, Tan L, and Shen S (2014). Atypical embryo phenotypes identified by time-lapse microscopy: high prevalence and association with embryo development.Fertil.Steril 101,1637–1648.e1. doi: 10.1016/J.FERTNSTERT.2014.02.050 [DOI] [PubMed] [Google Scholar]
- Azzarello A, Hoest T, and Mikkelsen AL (2012). The impact of pronuclei morphology and dynamicity on live birth outcome after time-lapse culture. Hum. Reprod 27, 2649–2657. doi: 10.1093/HUMREP/DES210 [DOI] [PubMed] [Google Scholar]
- Balakier H, Sojecki A, Motamedi G, and Librach C (2016). Impact of multinucleated blastomeres on embryo developmental competence, morphokinetics, and aneuploidy. Fertil. Steril 106, 608–614e2. doi: 10.1016/J.FERTNSTERT.2016.04.041 [DOI] [PubMed] [Google Scholar]
- Basile N, Nogales Mdel C, Bronet F, Florensa M, Riqueiros M, Rodrigo L, Garcia-Velasco J, and Meseguer M (2014). Increasing the probability of selecting chromosomally normal embryos by time-lapse morphokinetics analysis. Fertil. Steril 101, 699–704.e1. doi: 10.1016/J.FERTNSTERT.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, and Thornton S (2013). Retrospective analysis of outcomes after IVF using an aneuploidy risk model derived from time-lapse imaging without PGS. Reprod. Biomed. Online 27, 140–146. doi: 10.1016/J.RBMO.2013.04.013 [DOI] [PubMed] [Google Scholar]
- Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, Behr B, and Reijo Pera RA (2012). Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat. Commun 3, 1251. doi: 10.1038/NCOMMS2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla M, Fakih M, Shunnar A, Bayram A, Hellani A, Perumal V, Divakaran J, andBudak E (2015).Morphokinetic analysisof cleavage stage embryos and its relationship to aneuploidy in a retrospective time-lapse imaging study. J. Assist. Reprod. Genet 32, 69–75. doi: 10.1007/S10815-014-0372-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AA, Tan L, Suraj V, Reijo Pera R, and Shen S (2013). Biomarkers identified with time-lapse imaging: discovery, validation, and practical application. Fertil. Steril 99, 1035–1043. doi: 10.1016/J.FERTNSTERT.2013.01.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YH,Harding HD,Hartman DL,Obermiller AD,Kurosaka S, McLaughlin KJ, and Hinrichs K (2009). The uterine environment modulates trophectodermal POU5F1 levels in equine blastocysts. Reproduction 138, 589–599. doi: 10.1530/REP-08-0394 [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, and Pellman D (2012). DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58. doi: 10.1038/NATURE10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz M,Garrido N,Herrero J,Perez-Cano I,Munoz M,andMeseguer M (2012). Timing of cell division in human cleavage-stage embryos is linked with blastocyst formation and quality. Reprod. Biomed. Online 25, 371–381. doi: 10.1016/J.RBMO.2012.06.017 [DOI] [PubMed] [Google Scholar]
- Dal Canto M, Coticchio G, Mignini Renzini M, De Ponti E, Novara PV, Brambillasca F, Comi R, and Fadini R (2012). Cleavage kinetics analysis of human embryos predicts development to blastocyst and implantation. Reprod. Biomed. Online 25, 474–480. doi: 10.1016/J.RBMO.2012.07.016 [DOI] [PubMed] [Google Scholar]
- Daughtry BL, and Chavez SL (2016). Chromosomal instability in mammalian pre-implantation embryos: potential causes, detection methods, and clinical consequences. Cell Tissue Res. 363, 201–225. doi: 10.1007/S00441-015-2305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughtry BL, and Chavez SL (2018). Time-lapse imaging for the detection of chromosomal abnormalities in primate preimplantation embryos. Methods Mol. Biol 1769, 293–317. doi: 10.1007/978-14939-7780-2_19 [DOI] [PubMed] [Google Scholar]
- Daughtry BL, Rosenkrantz JL, Lazar NH, Fei SS, Redmayne N, Torkenczy KA, Adey A, Yan M, Gao L, Park B, Nevonen KA, Carbone L,andChavez SL(2019).Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion. Genome Res. 29, 367–382. doi: 10.1101/GR.239830.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Carmen Nogales M, Bronet F, Basile N, Martinez EM, Linan A, Rodrigo L, and Meseguer M (2017). Type of chromosome abnormality affect sembryo morphology dynamics.Fertil.Steril 107,229–235e2. doi: 10.1016/J.FERTNSTERT.2016.09.019 [DOI] [PubMed] [Google Scholar]
- Desai N, Goldberg JM, Austin C, and Falcone T (2018). Are cleavage anomalies, multinucleation, or specific cell cycle kinetics observed with time-lapse imaging predictive of embryo developmental capacity or ploidy? Fertil. Steril 109, 665–674. doi: 10.1016/J.FERTNSTERT.2017.12.025 [DOI] [PubMed] [Google Scholar]
- Foss R, Ortis H, and Hinrichs K (2013). Effect of potential oocyte transport protocols on blastocyst rates after intracytoplasmic sperm injection in the horse. Equine Vet. J. Suppl 45, 39–43. doi: 10.1111/EVJ.12159 [DOI] [PubMed] [Google Scholar]
- Gardner DK, Lane M, Stevens J, Schlenker T, and Schoolcraft WB (2000). Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer.Fertil.Steril 73,1155–1158.doi: 10.1016/S0015-0282(00)00518-5 [DOI] [PubMed] [Google Scholar]
- Hardarson T, Lofman C, Coull G, Sjogren A, Hamberger L, and Edwards RG (2002). Internalization of cellular fragments in a human embryo: time-lapse recordings. Reprod. Biomed. Online 5, 36–38. doi: 10.1016/S1472-6483(10)61594-5 [DOI] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, and Hetzer MW (2013). Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60. doi: 10.1016/J.CELL.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs K, Choi YH, Walckenaer BE, Varner DD, and Hartman DL(2007).Invitro-produced equine embryos: production of foals after transfer, assessment by differential staining and effect of medium calcium concentrations during culture. The riogenology 68, 521–529. doi: 10.1016/J.THERIOGENOLOGY.2007.04.046 [DOI] [PubMed] [Google Scholar]
- Hlinka D, Kalatova B, Uhrinova I, Dolinska S, Rutarova J, Rezacova J, Lazarovska S, and Dudas M (2012). Time-lapse cleavage rating predicts human embryo viability. Physiol. Res 61, 513–525. [DOI] [PubMed] [Google Scholar]
- Kalatova B, Jesenska R, Hlinka D, and Dudas M (2015). Tripolar mitosis in human cells and embryos: occurrence, pathophysiology and medical implications. Acta Histochem 117, 111–125. doi: 10.1016/J.ACTHIS.2014.11.009 [DOI] [PubMed] [Google Scholar]
- Lemmen JG, Agerholm I, and Ziebe S (2008). Kinetic markers of human embryo quality using time-lapse recordings of IVF/ICSI fertilized oocytes. Reprod. Biomed. Online 17, 385–391. doi: 10.1016/S1472-6483(10)60222-2 [DOI] [PubMed] [Google Scholar]
- Liu Y, Chapple V, Roberts P, Ali J,andMatson P(2014a).Time-lapse videography of human oocytes following intracytoplasmic sperm injection: events up to the first cleavage division. Reprod. Biol 14, 249–256. doi: 10.1016/J.REPBIO.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Liu Y, Chapple V, Roberts P, and Matson P (2014b). Prevalence, consequence, and significance of reverse cleavage by human embryos viewedwith theuse of the Embryo scope time-lapsevideo system.Fertil. Steril 102, 1295–1300e2. doi: 10.1016/J.FERTNSTERT.2014.07.1235 [DOI] [PubMed] [Google Scholar]
- Liu S, Kwon M, Mannino M, Yang N, Renda F, Khodjakov A, and Pellman D (2018). Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 561, 551–555. doi: 10.1038/S41586018-0534-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Brown MB, Stern JE, Jindal SK, Racowsky C, and Ball GD (2014). Using the Society for Assisted Reproductive Technology Clinic Outcome System morphological measures to predict live birth after assisted reproductive technology. Fertil. Steril 102, 1338–1344. doi: 10.1016/J.FERTNSTERT.2014.07.1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin K, Bergh C, and Hardarson T (2001). Early embryo cleavage is a strong indicator of embryo quality in human IVF. Hum. Reprod 16, 2652–2657. doi: 10.1093/HUMREP/16.12.2652 [DOI] [PubMed] [Google Scholar]
- Marzano G, Mastrorocco A, Zianni R, Mangiacotti M, Chiaravalle AE, Lacalandra GM, Minervini F, Cardinali A, Macciocca M, Vicenti R, Fabbri R, Hinrichs K, Dell’Aquila ME, and Martino NA (2019). Altered morphokinetics in equine embryos from oocytes exposed to DEHP during IVM. Mol. Reprod. Dev 86, 1388–1404. doi: 10.1002/MRD.23156 [DOI] [PubMed] [Google Scholar]
- Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, and Remohi J (2011). The use of morphokinetics as a predictor of embryo implantation. Hum. Reprod 26, 2658–2671. doi: 10.1093/HUMREP/DER256 [DOI] [PubMed] [Google Scholar]
- Minasi MG, Colasante A, Riccio T, Ruberti A, Casciani V, Scarselli F, Spinella F, Fiorentino F, Varricchio MT, and Greco E(2016).Correlation between ane uploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: a consecutive case series study. Hum. Reprod 31, 2245–2254. doi: 10.1093/HUMREP/DEW183 [DOI] [PubMed] [Google Scholar]
- Nakahara T, Iwase A, Goto M, Harata T, Suzuki M, Ienaga M, Kobayashi H, Takikawa S, Manabe S, Kikkawa F, and Ando H (2010). Evaluation of the safety of time-lapse observations for human embryos. J. Assist. Reprod. Genet. 27, 93–96. doi: 10.1007/S10815-010-9385-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D, Flaherty SP, Barry MF, and Matthews CD (1997). Preliminary observations on polar body extrusion and pronuclear formation in human oocytes using time-lapse video cinematography. Hum. Reprod 12, 532–541. doi: 10.1093/HUMREP/12.3.532 [DOI] [PubMed] [Google Scholar]
- Pribenszky C, Losonczi E, Molnar M, Lang Z, Matyas S, Rajczy K, Molnar K, Kovacs P, Nagy P, Conceicao J, and Vajta G (2010). Prediction of in-vitro developmental competence of early cleavage-stage mouse embryos with compact time-lapse equipment. Reprod. Biomed. Online 20, 371–379. doi: 10.1016/J.RBMO.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Racowsky C, Vernon M, Mayer J, Ball GD, Behr B, Pomeroy KO, Wininger D, Gibbons W, Conaghan J, and Stern JE (2010). Standardization of grading embryo morphology. Fertil. Steril 94, 1152–1153. doi: 10.1016/J.FERTNSTERT.2010.05.042 [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, and Bonner WM (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem 273, 5858–5868. doi: 10.1074/JBC.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Rubio I, Kuhlmann R, Agerholm I, Kirk J, Herrero J, Escriba MJ, Bellver J, and Meseguer M (2012). Limited implantation success of direct-cleaved human zygotes: a time-lapse study. Fertil. Steril 98, 1458–1463. doi: 10.1016/J.FERTNSTERT.2012.07.1135 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, and Cardona A (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/NMETH.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somfai T, Inaba Y, Aikawa Y, Ohtake M, Kobayashi S, Konishi K, and Imai K (2010). Relationship between the length of cell cycles, cleavage pattern and developmental competence in bovine embryos generated by in vitro fertilization or parthenogenesis. J. Reprod. Dev 56, 200–207. doi: 10.1262/JRD.09-097A [DOI] [PubMed] [Google Scholar]
- Sugimura S, Akai T, Somfai T, Hirayama M, Aikawa Y, Ohtake M, Hattori H, Kobayashi S, Hashiyada Y, Konishi K, and Imai K (2010). Time-lapse cinematography-compatible polystyrene-based microwell culture system: a novel tool for tracking the development of individual bovine embryos. Biol. Reprod 83, 970–978. doi: 10.1095/BIOLREPROD.110.085522 [DOI] [PubMed] [Google Scholar]
- Sugimura S, Akai T, Hashiyada Y, Somfai T, Inaba Y, Hirayama M, Yamanouchi T, Matsuda H, Kobayashi S, Aikawa Y, Ohtake M, Kobayashi E, Konishi K, and Imai K (2012). Promising system for selecting healthy in vitro-fertilized embryos in cattle. PLoS One 7, e36627. doi: 10.1371/JOURNAL.PONE.0036627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvall L, Ingerslev HJ,Breth Knudsen U, andKirkegaard K (2013). Inter- and intra-observer variability of time-lapse annotations. Hum. Reprod 28, 3215–3221. doi: 10.1093/HUMREP/DET366 [DOI] [PubMed] [Google Scholar]
- Treff NR, Krisher RL, Tao X, Garnsey H, Bohrer C, Silva E, Landis J, Taylor D, Scott RT, Woodruff TK, and Duncan FE (2016). Next generation sequencing-based comprehensive chromosome screening in mouse polar bodies, oocytes, and embryos. Biol. Reprod 94, 76. doi: 10.1095/BIOLREPROD.115.135483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinerman R, Feng R, Ord TS, Schultz RM, Bartolomei MS, Coutifaris C, and Mainigi M (2016). Morphokinetic evaluation of embryo development in a mouse model: functional and molecular correlates. Biol. Reprod 94, 84. doi: 10.1095/BIOLREPROD.115.134080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, and Reijo Pera RA (2010). Non-invasive imaging of human embryos before embryonic genome activation predicts development tothe blastocyst stage. Nat. Biotechnol 28, 1115–1121. doi: 10.1038/NBT.1686 [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang J, Salem SA, Liu X, Kuang Y, Salem RD, and Liu J (2014). Selection of competent blastocysts for transfer by combining time-lapse monitoring and array CGH testing for patients undergoing preimplantation genetic screening: a prospective study with sibling oocytes. BMC Med. Genomics 7, 38. doi: 10.1186/1755-8794-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]