Abstract
The use of charge-reducing reagents to generate lower-charge ions has gained popularity in the field of native mass spectrometry (MS) and ion mobility mass spectrometry (IM-MS). This is because the lower number of charged sites decreases the propensity for Coulombic repulsions and unfolding, helping to preserve the native-like structure. Furthermore, lowering the charge state consequently increases the mass-to-charge values (m/z), effectively increasing spacing between signals originating from small mass differences, such as different proteoforms or protein-drug complexes. IM-MS yields collision cross section (CCS, Ω) values that provide information about the three-dimensional structure of the ion. Travelling wave IM (TWIM) is an established and expanding technique within the native MS field. TWIM measurements require CCS calibration, which is achieved via the use of standard species of known CCS. Current databases for native-like proteins and protein complexes provide CCS values obtained using normal (i.e., non-charge-reducing) conditions. Herein, we explored the validity of using “normal” charge calibrants to calibrate for charge-reduced proteins and show cases where it is not appropriate. Using a custom linear field drift cell that enables the determination of ion mobilities from first principles, we directly determined CCS values for 19 protein calibrant species under 3 solution conditions (yielding a broad range of charge states) and 2 drift gases. This has established a database of CCS and reduced-mobility (K0) values, along with their associated uncertainties, for proteins and protein complexes over a large m/z range. TWIM validation of this database shows improved accuracy over existing methods in calibrating charge-reduced proteins.
Graphical Abstract

INTRODUCTION
Within native mass spectrometry, ion mobility (IM) has developed as a useful gas-phase technique capable of providing structural information about macromolecules and macromolecular complexes. In IM experiments, ions drift through a gas-filled cell, typically under the influence of a weak electric field. Ions with different mobilities (K) will attain different drift velocities, affecting temporal separation at the exit of the cell. An ion’s mobility is a property of both the ion and buffer gas molecules and is representative of the frequency of collisions between them.1 Numerous factors can contribute to differences in mobility such as shape, charge state, and masses of ion and gas molecules. Ions with high charge state or small CCS traverse the ion mobility cell faster (higher K) than those with lower charge state or higher CCS (lower K). Similarly, increasing the size of the buffer gas molecule would result in a lower measured K value. Ions that are “elongated” (e.g., an unfolded protein) will undergo more collisions with the buffer gas and thus traverse the cell slower than a compact protein of comparable size and charge. With the development of commercial IM instrumentation expanding the user base, the use of this technology to answer structural biology questions has dramatically increased.1–5 Interrogation of arrival time distributions (ATDs) can provide additional information regarding the ion population present. For example, a broad ATD or the presence of multiple peaks within an ATD can indicate different shapes/ion structures, protonation isomers (present at coincident m/z), and different charge states (e.g. m/z-coincident oligomers).6–8 Coupling IM separators with quadrupole-time-of-flight mass spectrometry platforms (IM-MS) enables separations orthogonal to mass spectrometry analysis. The ability to convert K values obtained from IM experiments into collision cross sections (CCS) can provide direct information on the conformations adopted and can be compared to theoretical CCS generated from structural models.9–11 Calculating CCS using drift time-derived mobilities provides a value that represents the momentum transfer cross section (Ω), though is often more simply thought of as a rotationally-averaged cross sectional area of the molecule’s 3-dimensional structure. More detail regarding the approximations used in obtaining CCS values from ion mobilities can be found in a comprehensive review from Gabelica et al.12
Several types of IM techniques currently exist: drift tube ion mobility spectrometry (DTIMS)13–15, traveling-wave IMS (TWIMS)15, field asymmetric waveform IMS (FAIMS)16, and trapped ion mobility spectrometry (TIMS)17, to name a few. In TWIMS and TIMS experiments, both of which are available commercially and afford good sensitivity and resolving power18, calibration using previously-established reference compounds is necessary to obtain K and CCS values. While it may theoretically be possible to obtain K values directly from these IM methods without calibrating, the uncertainty and accuracy in this type of approach has yet to be fully understood.19,20 Despite the need to calibrate these measurements, there is not a current consensus for a single primary standard compound.12 Thus, the use of established calibrant databases across a range of analyte classes is needed to obtain useful values from these methods. When selecting reference compounds to be used as calibrants, only robust, stable analytes should be chosen (i.e. those that are stable in storage, produce stable ion current upon ionization, and those that are less susceptible to restructuring upon ion activation).12 Once these calibrant databases have been created, the choice in calibrant for a particular unknown requires matching the calibrants with the analyte in terms of molecular class, mass and charge (i.e. factors influencing mobility).12,21,22 While this requires some advance knowledge of the unknown protein, matching all of these features is necessary in order to minimize the need for unnecessary extrapolations.12,23
Taking advantage of the higher gas-phase basicity of charge-reducing solution additives, it is possible to generate protein ions at lower charge states via positive mode (nano)-electrospray ionization when compared with ammonium acetate, the most common native mass spectrometry (MS) electrolyte. In recent years, charge-reducing reagents have grown in popularity within the native MS community for many reasons.24–26 Charge-reduced ions are thought to better preserve the native-like conformations of the protein ions because they have less Coulombic repulsion and have been shown to be less susceptible to activation.26–30 Recent work has also shown that minimizing conformational disruptions to proteins when they undergo dissociation can be utilized to investigate structures of subcomplexes or subunits in their native-like forms and the use of charge-reducing reagents can help to accomplish this.26 It is important to note, however, that care should always be taken to tune the mass spectrometer to reduce the likelihood of unintended activation when studying proteins and protein complexes in the gas phase, regardless of this proposed characteristic of charge-reduced species. Reduction of charge state when studying proteins via native MS has also been used to increase the separation between peaks; a method especially useful when investigating small molecule binding.24,25,30,31
In addition to work with soluble proteins, the use of tetraethylene glycol monooctyl ether (C8E4) has been shown to demonstrate a charge-reducing effect and has proven useful in studying membrane proteins while minimizing perturbation to the native-like structure.23,32 In addition to charge-reduction, C8E4 is also more easily removed with low activation energies.23 Other detergents such as C12E9 and C12E8 have charge-reducing abilities but are not as easy to remove. Even without the use of this charge-reducing detergent, certain classes of proteins such as membrane protein complexes, can have a lower average charge than soluble proteins. In work from 2016, Allison et al. showed that using soluble proteins to calibrate for CCS measurements of membrane proteins in charge-reducing detergents proved challenging when the calibrants were selected based on mass and CCS alone.23 Because these membrane proteins had lower mobilities than soluble proteins (due to their 30% lower average charge state), the use of larger soluble protein calibrants was required to avoid erroneous extrapolation of the calibration function.23 Additional work has shown that some proteins may also possess higher charge states than their size alone may suggest, as in the case of DNA-bound SgrAI oligomers, which held a total charge greater than that of globular proteins of comparable molecular weight.33 This, along with the growing use of charge-reducing reagents in native MS, highlights the need for calibrants with more comparable charge densities and mobility values.
As illustrated in the literature, it is necessary to choose IM calibrants with comparable K values rather than only mass or CCS.12,23 Thus, we have established a new K and CCS database of proteins and protein complexes with a broad range of K values. To accomplish this, we utilized a custom, linear drift field ion mobility cell installed within a commercial mass spectrometer to obtain ion mobility (K) values from “first principles.” Utilizing the Mason-Schamp equation, we then determined the CCS values for each calibrant. We report here this database of collision cross section and mobility values for a variety of proteins and protein complexes generated from three different solution conditions (one “normal-charge” – from ammonium acetate – and two “charge-reducing” – EDDA or AmAc/TEAA – conditions) and two different drift gases (He and N2), allowing for a comprehensive database that spans a wide range of calibrant masses, charge states, and subsequently mobility values, expanding upon previously-established native-like ion CCS databases. These values are reported according to the guidelines set forth by a comprehensive review from many experts in the field of ion mobility in an effort to provide clear values and uncertainties that can be utilized in experiments across laboratories with confidence.12
EXPERIMENTAL
Sample preparation
Proteins were purchased from suppliers as listed in SI Table S1. Those that arrived as lyophilized powders were reconstituted in ultrapure water (Sartorius Arium® Pro, Gӧttingen, Germany) and stored as aliquots at −20°C until needed for analysis to prevent freeze-thaw cycles that could alter protein structure. GroEL was refolded according to a procedure described previously34 and stored at −80°C until needed for analysis. Prior to mass spectrometry analysis, samples were thawed and immediately buffer exchanged using Micro Bio-Spin P6 spin columns with a 6 kDa cutoff (Bio-Rad, Hercules, CA, USA) into the respective electrolyte solution. Samples prepared under “normal-charge” conditions labeled “AmAc” were buffer exchanged into 200 mM ammonium acetate (99.99%, Millipore Sigma). Samples prepared under “charge-reducing” conditions and labeled “EDDA”, were buffer exchanged into 200 mM ethylenediamine diacetate (98%, Millipore Sigma). Samples prepared under “charge-reducing” conditions and labeled “TEAA” were buffer exchanged into 200 mM ammonium acetate and subsequently adjusted with triethylammonium acetate (TEAA) (1M, Millipore Sigma) and ultrapure water for a final concentration of 160 mM ammonium acetate + 40 mM TEAA. AmAc and TEAA solutions had a measured pH of 6.8 (a word of caution: from experience, different purities of ammonium acetate may result in different solution pH so it is recommended to check the pH prior to use in IM calibration experiments) and EDDA had a measured pH of 6.2. For all experiments, unless indicated otherwise, the pH of the buffer or final solution was not externally adjusted. Following buffer exchange, protein concentrations were measured using a Nanodrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and diluted to a final protein complex concentration of 5–10 µM using the respective final electrolyte solution.
Instrumentation
For all experiments in this work, ions were generated via nanoelectrospray ionization. Borosilicate capillaries were pulled in-house using a micropipette tip puller (Sutter Instruments model P-97, Novato, CA). All linear field drift cell measurements were performed on a Waters Synapt G2 HDMS (Waters Corporation, Wilmslow, UK) instrument. The mass spectrometer was modified to replace the commercial traveling wave-ion mobility (TWIM) cell with an RF-confining linear field drift cell as described in detail previously.35,36 In brief, this linear field drift cell contains a 25.05 cm drift region and was designed in a manner to allow operation utilizing the existing electrical inputs from the TWIM cell, which allows for control of the drift voltage via the instrument software (MassLynx v4.2, Waters Corporation). Buffer gas is introduced utilizing an alternative inlet system which delivers the drift gas (either nitrogen >99.998% or helium >99.999%) to the center of the linear field drift cell, minimizing the net flow of gas throughout the drift region. The pressure of the drift gas was stabilized each day prior to initiating experiments and measured using a calibrated absolute pressure transducer (MKS Baratron model 626C, Wilmington, MA). Pressure readings were recorded every minute using the procedure outlined in the SI. The temperature of the drift gas and linear drift cell region was measured using a Type K vacuum thermocouple (Omega Engineering, Norwalk, CT, USA). Temperature readings were recorded every five minutes as outlined in the SI. The thermocouple was placed immediately next to the linear field drift cell to obtain accurate recordings of the temperature that ions experience within the cell. A diagram of the Baratron and thermocouple locations with respect to the linear field drift cell can be observed in SI Figure S1.
For all experiments obtaining K0 values from first principles, tune settings throughout the mass spectrometer were adjusted to minimize ion activation from the point of ion generation throughout the instrument to mass analysis while still allowing for sufficient signal. For most proteins, the cone voltage was set to 20 V. Ion gating and injection were controlled with a mobility trap height of 5 to 10 V. For all proteins with the exception of glutamine synthetase (GS) and GroEL, the peak-to-peak RF amplitude in the linear drift cell was set to 150 Vpp; for GS and GroEL, the amplitude was adjusted to 250 Vpp. A more detailed list of tune settings are displayed in SI Tables S2, S3, and S4. The drift voltages used in these experiments ranged from 20 to 255 V in 0.8 to 2 torr drift gas for both drift gases; the voltages and pressures were adjusted based on the protein of interest to optimize ion transmission through the drift cell. Corresponding drift cell field energies for these experiments span 0.8 to 13 V cm−1 Torr−1, which falls well within the low-field limit (20–45 V cm−1 Torr−1) as reported for peptide ions37 and consequently falls well within the higher low-field limit for proteins and protein complexes.38,39 Drift voltages and additional variable instrument parameters used in linear field drift cell experiments for each specific protein standard are outlined in SI Tables S5 and S6.
All traveling wave-ion mobility (TWIM) experiments were performed on a Waters Synapt G2 HDMS. All proteins were prepared in the same manner as described above prior to IM-MS analysis. For all TWIM experiments, the cone voltage was set to 20 V, the extraction cone was set to 1 V, trap and transfer CID were both set to 0 V, the peak-to-peak RF amplitude in the TWIM was set to 150 V, the backing pressure was approximately 4 mbar, the IM wave height (W.H.) was set to 17 and wave velocity (W.V.) was set to 350, the transfer W.H. was set to 2 and W.V. was set to 100. Argon was used as the trap/transfer gas at a 2 mL/min flow rate, nitrogen (≥99.998%) was used as the ion mobility drift gas with a 60 mL/min flow rate and helium (≥99.998%) was used in the helium cell at a 120 mL/min flow rate. Temperature was measured in the same manner as the linear field drift cell experiments (SI Figure S1) and averaged 299.4 K each day of the triplicate TWIM experiments.
RESULTS & DISCUSSION
Collision cross sections
Proteins and protein complexes used in this database were selected based on commercial availability, ease of sample preparation, ability to produce stable spray via nanoelectrospray ionization, and thorough coverage of a wide mass and charge state range. Additionally, no pH adjustments were made to the electrolyte solutions within this database to provide a simple and consistent sample preparation approach.
Arrival time distributions were extracted using TWIMExtract40 and ion mobilities were calculated using the procedure outlined in the SI. Collision cross section values (CCS) were calculated using the reduced-mobility values as indicated by the Mason-Schamp equation38,41–43, Equation 1:
| (1) |
in which e is the elementary charge, z is the charge state of the ion, N is the drift gas number density at STP, µ is the reduced mass of the ion and drift gas, kB is the Boltzmann constant, and T is the experimental drift gas temperature.
All CCS values reported in this work are averages from triplicate mobility measurements conducted on different days. The standard deviations associated with the reported CCS values are ≤1% of the reported average. Combined standard uncertainty for each CCS and K0 value was calculated as outlined in the SI and equations S1 and S2, respectively. The combined standard uncertainty considers uncertainties within the Gaussian peak fit, slopes, pressure measurements (Baratron manufacturer-reported accuracy is 0.25% of the measured value), and experimental temperature (thermocouple manufacturer-reported accuracy is ~0.75% of the measured value). Uncertainties are reported in the final database alongside each CCS and K0 value. In all, the combined standard uncertainties for K0 and drift tube primary CCS values (DT,1ryCCS) reported within this database are estimated to be <2% and <1.7%, respectively. The average effective density of all proteins (across both drift gases and all solution conditions) included within this database was calculated to be 0.60 ± 0.08 g·cm−3 which is well below the solvent-excluded regions of proteins (>1.2 g cm−3), confirming that proteins selected for this database retain their native-like structure rather than collapsing in the gas phase.44
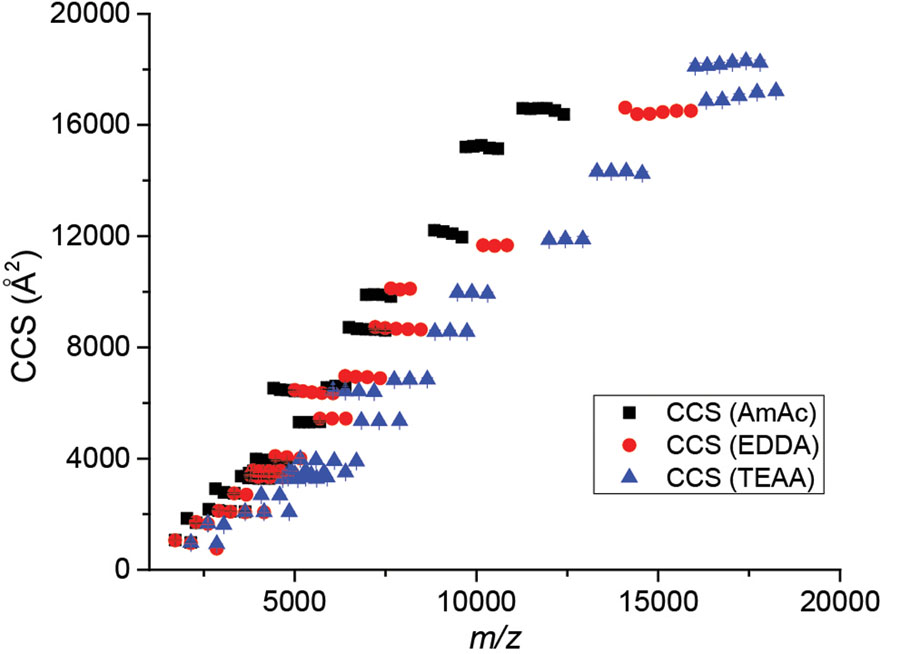
In all, a large database of K0 and DT,1ryCCS values were collected utilizing native-like proteins and protein complexes, providing calibrants for ion mobility experiments that include 19 protein species, three different solution conditions, and two drift gases. A summary of all collected DT,1ryCCSHe and DT,1ryCCSN2 values is shown in Figure 1 below and the corresponding CCS, K0 values, and uncertainties of each data point are listed in SI Tables S7 and S8. We note greater charge reduction for larger molecular weights shown in Figure 1.
Figure 1.
Summary of DT,1ryCCSHe values collected for this database. The values collected here encompass a wide range of protein size and charges. This database provides CCS values ranging from 930–20,540 Å2 corresponding to reduced-mobility values ranging from 0.38–2.7 cm2/V·s across both drift gases. Error bars are included but typically fit within the data point size.
Comparison with prior results
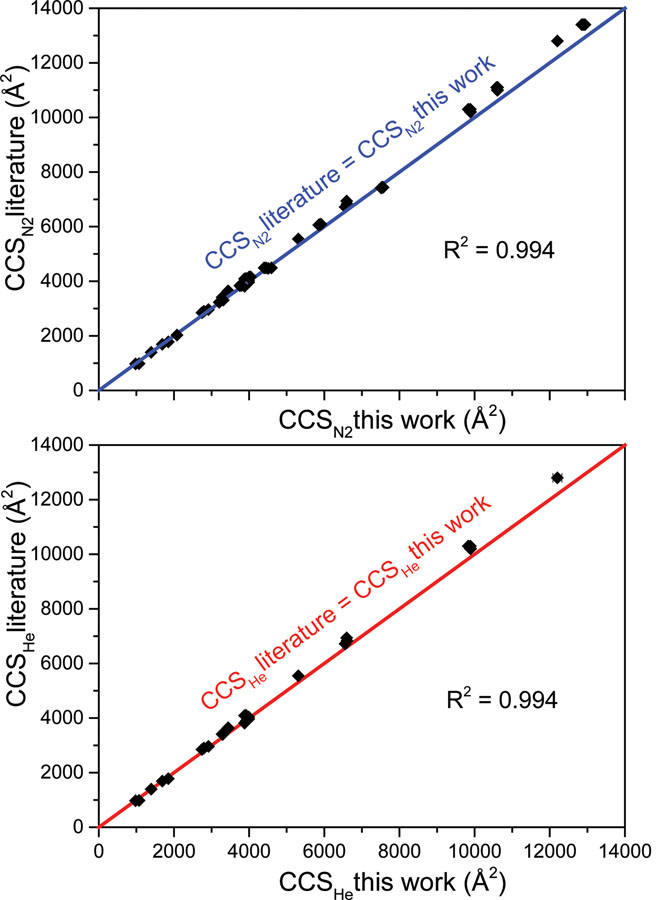
Previously-published collision cross-section datasets for proteins and protein complexes assumed drift gas temperature equal to an average ambient laboratory temperature of 293 K.35 While it was not expected that the assumption of temperature would create a significant error (previous estimates of error caused by temperature assumptions ≈0.5%),35 a difference between labs of 5 K can propagate to cause CCS errors of >1%. This, along with the lack of reported reduced mobility values, provided motivation to recollect most of the CCS values stated in the literature to incorporate experimentally measured temperature using the thermocouple described above and to provide corresponding uncertainty values for K0 and CCS values. Comparison of literature CCS values from multiple publications with those determined from this work in both nitrogen (Figure 2A) and helium (Figure 2B) drift gases in AmAc solution conditions showed good agreement. Temperature is likely the greatest contributing factor to these differences. Such agreement gives confidence in the measurements reported here for alternative solution conditions.
Figure 2.
Comparison of CCS values from “normal charge” ammonium acetate experiments in this work with numerous literature values using comparable solution conditions6,35,36 (from Waters Synapt DTIMS and Agilent 6560 DTIMS platforms; most in He drift gas) in (A) nitrogen drift gas and (B) helium drift gas. The colored linear fit line in each plot represents perfect agreement between the datasets and data points fit to these lines have high R2 values in each case. Error bars on CCS values from this work are shown but typically remain within the data point size.
Solution and gas phase factors affecting protein conformation and CCS
After establishing the database of DT,1ryCCS values for three different solution conditions, it was apparent that for some protein complexes that contained overlapping charge states between different solution conditions, the CCS values did not always provide identical values. While 83% of the charge states that overlapped between AmAc and EDDA or between EDDA and TEAA provided ≤2% difference in CCS from one another (including both helium and nitrogen drift gas values), there was a general trend of CCS(EDDA)>CCS(AmAc) for those that differed by >2%. This discrepancy may be due to a combination of both solution and gas phase effects.
Firstly, differences in ionic strength and pH between different solution conditions may alter the protein conformation, which translates onto the determined CCS. Database CCS(AmAc) values utilized an electrolyte solution with 200 mM total ionic strength and a pH of 6.8. Database CCS(EDDA) values utilized an electrolyte solution with 600 mM total ionic strength and a pH of 6.2. To investigate these possible solution condition effects, we selected a set of four protein complexes that showed larger (>2%) differences in overlapping charge state CCS values when using EDDA and AmAc solution conditions: bovine serum albumin (BSA), avidin (AV), phosphorylase B (PHB), and transthyretin (TTR). We prepared each protein under two different solution conditions. Firstly, to test for CCS(EDDA) differences in ionic strength while keeping pH consistent with the database, we prepared proteins in 66.7 mM EDDA solution (total ionic strength of 200 mM) at a pH of 6.2. Secondly, to test for CCS(EDDA) differences in pH while keeping ionic strength consistent with the database, we prepared proteins in 200 mM EDDA solution with an ethylenediamine-adjusted pH of 6.8 while ensuring a final total ionic strength of 600 mM. This allowed us to test one variable at a time to determine if either ionic strength or pH influenced the increased CCS(EDDA) values when using the database EDDA conditions of 200 mM (600 mM ionic strength) and pH 6.2. The results of these experiments are summarized in Figure 3A and 3B along with SI Figure S2. Overall, the effect of adjusting the ionic strength or pH of EDDA to match that of the AmAc depended on the protein. For example, TTR and PHB DTCCS values more closely aligned with those of AmAc values for the overlapping charge states when either the ionic strength or the pH of the EDDA solution was adjusted to match that of AmAc. For TTR and PHB, adjusting either EDDA ionic strength or pH resulted in DTCCS values <0.65% different from the AmAc DTCCS values which is within the standard deviation of the DT,1ryCCS obtained within this database.
Figure 3.
Left panel (A and B): impact of ionic strength and pH on CCS values for overlapping charge states using AmAc and EDDA solutions. Database CCS(AmAc) are displayed in black (200 mM ionic strength, pH 6.8), database CCS(EDDA) are in red (200 mM EDDA, 600 mM ionic strength, pH 6.2), and CCS(EDDA) adjusted for A) ionic strength (66.7 mM EDDA, 200 mM ionic strength) or B) pH (6.8) are in blue. Neither ionic strength nor pH had strong effects on the CCS of BSA; adjusting ionic strength shifted CCS(EDDA) from an average 4.3% difference with CCS(AmAc) to 3.3% and adjusting pH shifted CCS(EDDA) to an average 2.7% difference with CCS(AmAc). Right panel (C-F): impact of source activation in removal of adducts in the gas phase. C) and E) illustrate the mass spectrum and ATD (14+) of BSA (200 mM EDDA, 600 mM ionic strength, pH 6.2) with “cool” source voltages (sampling cone 20 V, extraction cone 1 V). D) and F) illustrate the mass spectrum and ATD (14+) of the same BSA solution with increased source activation to remove EDDA adducts (sampling cone 120 V, extraction cone 5 V). Increasing source activation removes more adducts, but results in restructuring as evidenced by the increased width in the corresponding ATD. The shoulder observed in F) is likely an additional structure: further evidence of restructuring upon cone activation. A more complete series of source activation voltages is displayed in SI Figure S5.
Secondly, the efficiency of ion desolvation (dependent on solution conditions) may induce changes in the volume and/or mass of detected species, in turn affecting the observed CCS. The latter effect was readily observed when generating ions from EDDA: peaks in the mass spectra were often substantially broader and shifted to higher m/z due to the presence of adducts. The extent of adduction can be readily observed in the experimental masses of each protein under varying solution conditions, displayed in Table S9. For the proteins where the ionic strength or pH did not have as much impact, such as in the case of BSA, the differences in CCS between AmAc and EDDA solution conditions are likely due to these salt adducts that remain bound when analyzing these proteins. To interrogate this further, a Gaussian peak function was fitted to the selected mass spectral peaks. Taking an average of the individual charge states between triplicate measurements of BSA, AV, TTR, and PHB, the full-width half-max (FWHM) of the EDDA peaks were 1.6 ± 0.4 times the FWHM of their AmAc counterparts. Importantly, the use of EDDA (200 mM, 600 mM ionic strength, pH 6.2) resulted in an average of 807 ± 173 Da increase in mass with these proteins, indicating more salt adducts on some proteins when using EDDA solution conditions. Figure S3 illustrates this with the bovine serum albumin 14+ peak. All DT,1ryCCS database calculations incorporated the experimentally observed mass of each protein (Table S9). While the molecular weight contributes only a small portion within the Mason-Schamp equation to determine CCS, the increase in mass from EDDA adducts likely contribute to the overall volume and, consequently, CCS of the protein. For all overlapping charge states, the change in experimentally observed molecular weight was plotted against the change in CCS and shows a positive trend between extent of adduction and observed CCS (Figure S4). Generally, the smaller proteins within the database do not accumulate as many adducts and consequently do not demonstrate these changes in CCS between solution conditions. The experimentally observed masses for each protein often correlate with differences observed in CCS (e.g. more adduction results in higher experimental mass and CCS) of overlapping charge states, though it is likely that adduction, pH, and ionic strength all contribute.
Ion desolvation can be enhanced at the potential cost of structural changes. For example, increasing the cone voltage resulted in reduction of the amount of EDDA bound to BSA (Figure 3C and 3D), but also caused a degree of protein unfolding, as evidenced by the change in arrival time distribution centroids and widths shown in Figure 3E and 3F with more detail illustrated in SI Figure S5. These experiments illustrate the importance of following sample preparation procedures carefully when utilizing CCS calibrant databases as differences in the resulting calibrant structure may arise from differences in sample preparation. Additionally, keeping instrument conditions cool to retain native-like structures is also an important aspect of accurately calibrating for CCS values. The latter may be at a cost of reduced sensitivity and mass spectral quality. In all, if the standard proteins are prepared in the same manner and instrument conditions are tuned to minimize activation of the proteins throughout IM analysis, the structures produced in the gas phase should match those produced within this database for correlation with the listed CCS values. In order for users of this database to best reproduce the CCS of included calibrants, the experimentally observed mass for each charge state is provided in Table S9 and representative mass spectra for each protein are provided in the SI.
Importance in matching K0 when calibrating IM: two case studies
An ion’s mobility is related to its charge and CCS, with CCS and molecular weight broadly correlated with one another.45–47 While it is clear that, at a given molecular weight, a variety of structures may be possible as represented by differing CCS, there is a gross trend between weight and CCS for globular proteins.48 Because of this, it is common practice to select CCS calibrants based on their molecular weight and CCS values because this will typically provide calibrants with mobility values similar to that of the measurand.49 However, differing charges on the same ion can result in a very different mobility through the IM cell. When working with analytes using charge-reducing additives or those that have a lower charge per surface area than normal-charge soluble protein calibrants, selecting these calibrants using molecular weight and CCS criterion alone results in a mobility mismatch. This mismatch results in the need for extrapolation during calibration, which can cause significant errors.49 Previous work has demonstrated that a mismatch of both molecular class and charge state result in greater errors upon CCS calibration than when one or both of these features are appropriately matched to the analyte of interest.50 While work is underway to understand TWIM on a more fundamental level, leading to the opportunity to obtain CCS measurements with fewer calibrants or without calibrating, more work is required to develop and further test these results with high mass, native-like protein ions.51,52 Differences in mobility due to charge and molecular weight are illustrated in Figure S6, which demonstrates the effect of different charge states (generated from different solution conditions) when analyzing the same proteins by IM. The differences in measured mobility between solution conditions are more pronounced at higher molecular weights.
Calibrating for membrane proteins and other charge-reducing reagents:
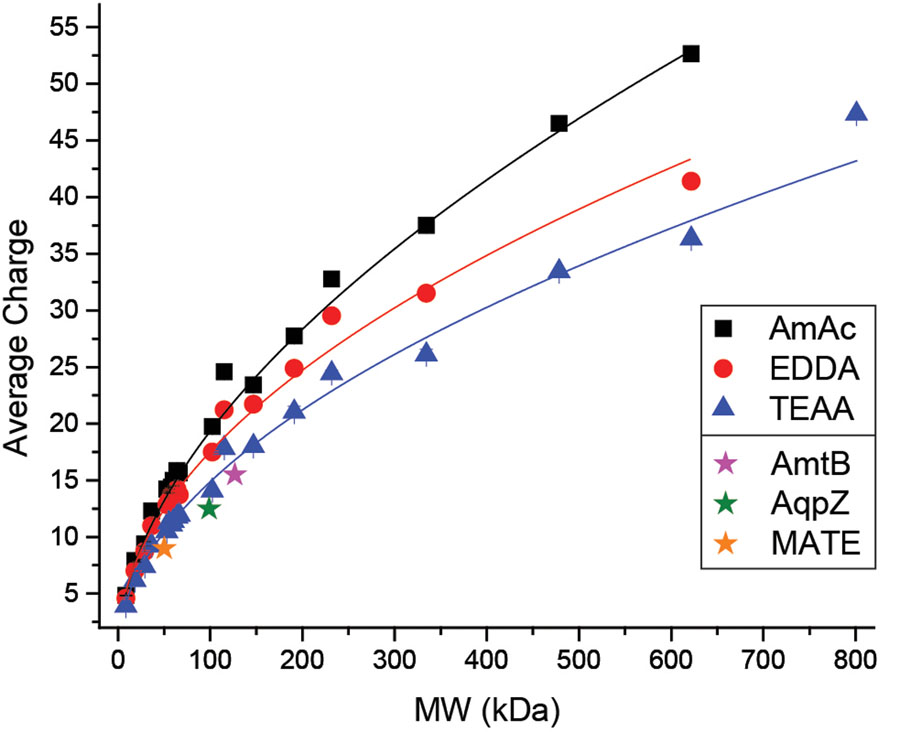
The mobilities of three standard membrane proteins in C8E4 (charge-reducing) from Allison, et al. (2016) are also included in Figure S6 alongside these curves and show mobility values most similar to the TEAA calibrants from this database. In this 2016 study, these membrane proteins were calibrated using soluble protein data and results showed that larger calibrants were required to avoid long-range extrapolations that result from this mismatch in mobility.3 Utilizing larger soluble protein calibrants was effective because they have lower mobilities and therefore bracket the mobility of the membrane proteins better than their lower-mass soluble protein counterparts.3 While this result illustrates an alternative solution using existing normal-charge database values, it is clear from previous work that it is important to select native-like protein calibrants as similar to the analyte as possible (i.e. shape, mass and charge).12,49,50 Thus, IM analysis of membrane proteins with lower mobilities, using both charge-reducing and normal-charge detergents, could benefit from this charge-reduced calibrant database. In addition, numerous other reagents or methods can be utilized to charge-reduce proteins (both membrane and soluble) such as imidazole and its derivatives or acetonitrile vapors.29,29,53 The charge-reduced calibrants generated within this database represent a 14–31% decrease in average charge from their normal-charged AmAc counterparts. If the charge is reduced by a greater percentage, then a similar method to that used by Allison, et al. (2016) may be required for appropriate mobility-matching. In the cases where the properties of a protein are unknown, it may be challenging to select appropriate calibrants. Then, molecular weight and charge, which can be obtained from a mass spectrum, should be utilized to select appropriate calibrants. A summary of the average charge vs. molecular weight for all calibrants generated for this database is shown in Figure 4 (for He drift gas) and Figure S7 (for N2 drift gas).
Figure 4.
Average charge vs. molecular weight for all proteins and protein complexes generated for this CCS database in He drift gas. Star data points illustrate membrane proteins utilized in work from Allison, et al. (2016) as a comparison.23 The mass spectrum of a protein with unknown CCS or mobility can be used to compare with this figure in selecting appropriate mobility-matched calibrants. Trendlines are used to guide the eye.
Calibrating for charge-reduced soluble proteins:
The need to mobility-match calibrants to an unknown can also be observed when calibrating a charge-reduced soluble protein against existing normal-charge soluble protein calibrants. Figure S8 demonstrates a TWIM calibration that emphasizes the need for matching mobility between calibrants and the measurand for smaller “unknown” proteins. Calculations of the CCS values from the TWIM calibrations were performed as described previously.21 Utilizing calibrants under normal-charge AmAc conditions to determine the CCS of either β-lactoglobulin A or transthyretin charge-reduced with TEAA results in significantly greater error than using calibrants under the same charge-reducing conditions (i.e., mobility-matching). This same experiment was conducted with the slightly less charge-reducing EDDA (Figure S9) and yielded similar, but less drastic, results. Exploring this further, additional TWIM experiments were conducted for higher-mass “unknowns” while also investigating the ability to use either a “narrow” (i.e. close window of mobility data points) or a “wide” (i.e. spread out mobility data points) set of calibrants (Figure 5). For these experiments, streptavidin (53 kDa), concanavalin A (103 kDa), and C-reactive protein (115 kDa) were treated as “unknowns”. These results demonstrate the importance in mobility-matching to obtain an accurate CCS calibration. The effect of using a “wide” versus “narrow” set of calibrant ions is relatively minor as long as the mobility values are appropriately matched. Combining the observations from previous work and those from this study shows the general importance in matching charge along with molecular weight when selecting appropriate CCS calibrants.
Figure 5.
TWIM calibration of streptavidin (green), concanavalin A (blue), and C-reactive protein (red) using mobility-matched (closed data points) or mobility-mismatched (open data points) calibrants. Calibrants were also selected to contain either a “narrow” or “wide” window of mass and mobility values as shown in Table S10. Use of this database to calibrate lower-charge proteins is more accurate when compared with traditional methods of calibrating using “normal charge” calibrants.
CONCLUSIONS
The use of charge-reducing solution additives is gaining popularity in the field of native mass spectrometry because of their propensity to minimize perturbation to the native-like protein structure and the ability to resolve small differences in mass through lower charge states. Ion mobility-mass spectrometry offers valuable information regarding the architecture of proteins and protein complexes for use in structural biology. Commercial ion mobility mass spectrometers afford increased sensitivity and resolution but require calibration in order to obtain TWCCS or TIMSCCS values. Using the appropriate calibrants is critical to minimizing error in such experiments. While traditional calibration procedures typically utilize molecular weight and CCS to determine the most appropriate calibrants, our results, along with previously published, work show that charge and mobility are also important factors. Utilizing calibrants that bracket in molecular weight but do not have comparable mobility values results in greater error than using calibrants appropriate in both molecular weight and charge (i.e. mobility). With the growing use of charge-reducing reagents, in addition to increased emphasis on membrane protein mass spectrometry which often utilizes charge-reducing detergents, this new database provides additional protein and protein complex calibrants and charge states compared to already-existing datasets. This database provides DT,1ryCCS values in both He and N2 drift gases across an extensive range of protein masses and mobilities by using “normal-charge” ammonium acetate and “reduced-charge” ethylenediamine diacetate and triethylammonium acetate solution conditions. This comprehensive database focuses on providing calibrants that are widely available and easy to prepare and analyze for a broad range of users. Special care was taken to calculate and provide CCS and K0 uncertainties and details regarding data acquisition and processing as specified by the ion mobility community to contribute to standardized reporting and increased transparency in such database values. Increasing the range of calibrant mobility values with which IM experiments can be calibrated allows for determination of more accurate CCS values if the appropriate care is taken to prepare calibrants in the same manner and retain “cool” instrument conditions that do not alter the native-like structure of these calibrants.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Sophie R. Harvey for advice regarding implementation of the linear field drift cell and helpful discussions regarding appropriate protein complex selection and data analysis, Dr. Jing Yan for advice regarding implementation of the linear field drift cell, and Dr. Florian Busch for helpful discussions regarding appropriate protein complex selection. The authors also gratefully acknowledge funding from the National Institutes of Health (NIH P41GM128577 and NIH R01GM113658 both awarded to V.H.W.).
Footnotes
Supporting Information: Standard protein sources, instrument parameters, IMS measurement details, CCS calculation, final CCS and K0 values along with uncertainties, and additional TWIM experiment results.
COI
The author(s) declare the following competing financial interests: Jakub Ujma and Kevin Giles are employees of Waters Corporation, which manufactures and sells T-wave IM-MS instruments.
References
- (1).Campuzano IDG; Giles K Historical, Current and Future Developments of Traveling Wave Ion Mobility Mass Spectrometry: A Personal Perspective. TrAC Trends Anal. Chem 2019, 115620 10.1016/j.trac.2019.115620. [DOI]
- (2).Seo J; Hoffmann W; Warnke S; Bowers MT; Pagel K; von Helden G Retention of Native Protein Structures in the Absence of Solvent: A Coupled Ion Mobility and Spectroscopic Study. Angew. Chem. Int. Ed 2016, 55 (45), 14173–14176. 10.1002/anie.201606029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Jurneczko E; Barran PE How Useful Is Ion Mobility Mass Spectrometry for Structural Biology? The Relationship between Protein Crystal Structures and Their Collision Cross Sections in the Gas Phase. Analyst 2010, 136 (1), 20–28. 10.1039/C0AN00373E. [DOI] [PubMed] [Google Scholar]
- (4).Robinson CV From Molecular Chaperones to Membrane Motors: Through the Lens of a Mass Spectrometrist. Biochem. Soc. Trans 2017, 45 (1), 251–260. 10.1042/BST20160395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Shoemaker GK; Duijn E. van; Crawford SE; Uetrecht C; Baclayon M; Roos WH; Wuite GJL; Estes MK; Prasad BVV; Heck AJR Norwalk Virus Assembly and Stability Monitored by Mass Spectrometry. Mol. Cell. Proteomics 2010, 9 (8), 1742–1751. 10.1074/mcp.M900620-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Marchand A; Livet S; Rosu F; Gabelica V Drift Tube Ion Mobility: How to Reconstruct Collision Cross Section Distributions from Arrival Time Distributions? Anal. Chem 2017, 89 (23), 12674–12681. 10.1021/acs.analchem.7b01736. [DOI] [PubMed] [Google Scholar]
- (7).Kune C; Far J; De Pauw E Accurate Drift Time Determination by Traveling Wave Ion Mobility Spectrometry: The Concept of the Diffusion Calibration. Anal. Chem 2016, 88 (23), 11639–11646. 10.1021/acs.analchem.6b03215. [DOI] [PubMed] [Google Scholar]
- (8).Sivalingam GN; Yan J; Sahota H; Thalassinos K Amphitrite: A Program for Processing Travelling Wave Ion Mobility Mass Spectrometry Data. Int. J. Mass Spectrom 2013, 345–347, 54–62. 10.1016/j.ijms.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Sahasrabuddhe A; Hsia Y; Busch F; Sheffler W; King NP; Baker D; Wysocki VH Confirmation of Intersubunit Connectivity and Topology of Designed Protein Complexes by Native MS. PNAS 2018, 115 (6), 1268–1273. 10.1073/pnas.1713646115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Quintyn RS; Yan J; Wysocki VH Surface-Induced Dissociation of Homotetramers with D2 Symmetry Yields Their Assembly Pathways and Characterizes the Effect of Ligand Binding. Chem. Biol 2015, 22, 583–592. [DOI] [PubMed] [Google Scholar]
- (11).Politis A; Park AY; Hall Z; Ruotolo BT; Robinson CV Integrative Modelling Coupled with Ion Mobility Mass Spectrometry Reveals Structural Features of the Clamp Loader in Complex with Single-Stranded DNA Binding Protein. J. Mol. Biol 2013, 425 (23), 4790–4801. 10.1016/j.jmb.2013.04.006. [DOI] [PubMed] [Google Scholar]
- (12).Gabelica V; Shvartsburg AA; Afonso C; Barran PE; Benesch JLP; Bleiholder C; Bowers MT; Bilbao A; Bush MF; Campbell JL; et al. Recommendations for Reporting Ion Mobility Mass Spectrometry Measurements. Mass Spectrom. Rev 2019, 9999, 1–30. 10.1002/mas.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Eiceman GA; Karpas Z; Hill HH Jr. Ion Mobility Spectrometry, 3rd Ed.; CRC Press/Taylor & Francis, 2013. [Google Scholar]
- (14).Hill HH; Siems WF; Louis RH St.; McMinn DG. ION MOBILITY SPECTROMETRY. Anal. Chem 1990, 62 (23), 1201A–1209A. 10.1021/ac00222a716. [DOI] [PubMed] [Google Scholar]
- (15).Giles K; Pringle SD; Worthington KR; Little D; Wildgoose JL; Bateman RH Applications of a Travelling Wave-Based Radio-Frequency-Only Stacked Ring Ion Guide. Rapid Commun. Mass Spectrom 2004, 18 (20), 2401–2414. 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- (16).Guevremont R; Purves RW High Field Asymmetric Waveform Ion Mobility Spectrometry-Mass Spectrometry: An Investigation of Leucine Enkephalin Ions Produced by Electrospray Ionization. J. Am. Soc. Mass Spectrom 1999, 10 (6), 492–501. 10.1016/S1044-0305(99)00016-1. [DOI] [PubMed] [Google Scholar]
- (17).Fernandez-Lima FA; Kaplan DA; Park MA Note: Integration of Trapped Ion Mobility Spectrometry with Mass Spectrometry. Rev. Sci. Instrum 2011, 82 (12), 126106 10.1063/1.3665933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Dodds JN; May JC; McLean JA Correlating Resolving Power, Resolution and Collision Cross Section: Unifying Cross Platform Assessment of Separation Efficiency in Ion Mobility Spectrometry. Anal. Chem 2017, 89 (22), 12176–12184. 10.1021/acs.analchem.7b02827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Giles K; Wildgoose JL; Langridge DJ; Campuzano I A Method for Direct Measurement of Ion Mobilities Using a Travelling Wave Ion Guide. Int. J. Mass Spectrom 2010, 298 (1), 10–16. 10.1016/j.ijms.2009.10.008. [DOI] [Google Scholar]
- (20).Mortensen DN; Susa AC; Williams ER Collisional Cross-Sections with T-Wave Ion Mobility Spectrometry without Experimental Calibration. J. Am. Soc. Mass Spectrom 2017, 28 (7), 1282–1292. 10.1007/s13361-017-1669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ruotolo BT; Benesch JLP; Sandercock AM; Hyung S-J; Robinson CV Ion Mobility–Mass Spectrometry Analysis of Large Protein Complexes. Nat. Protoc 2008, 3 (7), 1139–1152. 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- (22).McLean JA The Mass-Mobility Correlation Redux: The Conformational Landscape of Anhydrous Biomolecules. J. Am. Soc. Mass Spectrom 2009, 20 (10), 1775–1781. 10.1016/j.jasms.2009.06.016. [DOI] [PubMed] [Google Scholar]
- (23).Allison TM; Landreh M; Benesch JLP; Robinson CV Low Charge and Reduced Mobility of Membrane Protein Complexes Has Implications for Calibration of Collision Cross Section Measurements | Analytical Chemistry. Anal. Chem 2016, 88 (11), 5879–5884. 10.1021/acs.analchem.6b00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lössl P; Snijder J; Heck AJ Boundaries of Mass Resolution in Native Mass Spectrometry. J. Am. Soc. Mass Spectrom 2014, 25 (6), 906–917. [DOI] [PubMed] [Google Scholar]
- (25).Dyachenko A; Gruber R; Shimon L; Horovitz A; Sharon M Allosteric Mechanisms Can Be Distinguished Using Structural Mass Spectrometry. Proc. Natl. Acad. Sci 2013, 110, 7235–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhou M; Dagan S; H. Wysocki V. Impact of Charge State on Gas-Phase Behaviors of Noncovalent Protein Complexes in Collision Induced Dissociation and Surface Induced Dissociation. Analyst 2013, 138 (5), 1353–1362. 10.1039/C2AN36525A. [DOI] [PubMed] [Google Scholar]
- (27).Hall Z; Politis A; Bush MF; Smith LJ; Robinson CV Charge-State Dependent Compaction and Dissociation of Protein Complexes: Insights from Ion Mobility and Molecular Dynamics. J. Am. Chem. Soc 2012, 134 (7), 3429–3438. 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- (28).Pagel K; Hyung S-J; Ruotolo BT; Robinson CV Alternate Dissociation Pathways Identified in Charge-Reduced Protein Complex Ions. Anal. Chem 2010, 82 (12), 5363–5372. 10.1021/ac101121r. [DOI] [PubMed] [Google Scholar]
- (29).Mehmood S; Marcoux J; Hopper JTS; Allison TM; Liko I; Borysik AJ; Robinson CV Charge Reduction Stabilizes Intact Membrane Protein Complexes for Mass Spectrometry. J. Am. Chem. Soc 2014, 136 (49), 17010–17012. 10.1021/ja510283g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pacholarz KJ; Barran PE Use of a Charge Reducing Agent to Enable Intact Mass Analysis of Cysteine-Linked Antibody-Drug-Conjugates by Native Mass Spectrometry. EuPA Open Proteomics 2016, 11, 23–27. 10.1016/j.euprot.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Patrick JW; Laganowsky A Generation of Charge-Reduced Ions of Membrane Protein Complexes for Native Ion Mobility Mass Spectrometry Studies. J. Am. Soc. Mass Spectrom 2019, 30 (5), 886–892. 10.1007/s13361-019-02187-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Reading E; Liko I; Allison TM; Benesch JLP; Laganowsky A; Robinson CV The Role of the Detergent Micelle in Preserving the Structure of Membrane Proteins in the Gas Phase. Angew. Chem. Int. Ed Engl 2015, 54 (15), 4577–4581. 10.1002/anie.201411622. [DOI] [PubMed] [Google Scholar]
- (33).Ma X; Shah S; Zhou M; Park CK; Wysocki VH; Horton NC Structural Analysis of Activated SgrAI-DNA Oligomers Using Ion Mobility Mass Spectrometry. Biochemistry 2013, 52, 4373–4381. 10.1021/bi3013214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Zhou M; Jones CM; Wysocki VH Dissecting the Large Noncovalent Protein Complex GroEL with Surface-Induced Dissociation and Ion Mobility-Mass Spectrometry. Anal. Chem 2013, 85, 8262–8267. [DOI] [PubMed] [Google Scholar]
- (35).Bush MF; Hall Z; Giles K; Hoyes J; Robinson CV; Ruotolo BT Collision Cross Sections of Proteins and Their Complexes: A Calibration Framework and Database for Gas-Phase Structural Biology. Anal. Chem 2010, 82 (22), 9557–9565. 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- (36).Salbo R; Bush MF; Naver H; Campuzano I; Robinson CV; Pettersson I; Jørgensen TJD; Haselmann KF Traveling-Wave Ion Mobility Mass Spectrometry of Protein Complexes: Accurate Calibrated Collision Cross-Sections of Human Insulin Oligomers. Rapid Commun. Mass Spectrom 2012, 26 (10), 1181–1193. 10.1002/rcm.6211. [DOI] [PubMed] [Google Scholar]
- (37).Valentine SJ; Counterman AE; Clemmer DE A Database of 660 Peptide Ion Cross Sections: Use of Intrinsic Size Parameters for Bona Fide Predictions of Cross Sections. J. Am. Soc. Mass Spectrom 1999, 10 (11), 1188–1211. 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- (38).Mason EA; McDaniel EW Transport Properites of Ions in Gases.
- (39).Verbeck GF; Ruotolo BT; Gillig KJ; Russell DH Resolution Equations for High-Field Ion Mobility. J. Am. Soc. Mass Spectrom 2004, 15 (9), 1320–1324. 10.1016/j.jasms.2004.06.005. [DOI] [PubMed] [Google Scholar]
- (40).Haynes SE; Polasky DA; Dixit SM; Majmudar JD; Neeson K; Ruotolo BT; Martin BR Variable-Velocity Traveling-Wave Ion Mobility Separation Enhancing Peak Capacity for Data-Independent Acquisition Proteomics. Anal. Chem 2017, 89 (11), 5669–5672. 10.1021/acs.analchem.7b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Kanu AB; Dwivedi P; Tam M; Matz L; Hill HH Ion Mobility-Mass Spectrometry. J. Mass Spectrom. JMS 2008, 43 (1), 1–22. 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- (42).Bohrer BC; Merenbloom SI; Koeniger SL; Hilderbrand AE; Clemmer DE Biomolecule Analysis by Ion Mobility Spectrometry. Annu. Rev. Anal. Chem. Palo Alto Calif 2008, 1, 293–327. 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wyttenbach T; Bowers MT Gas-Phase Conformations: The Ion Mobility/Ion Chromatography Method. Mod. Mass Spectrom 2003, No. 225, 207–232.
- (44).Kaddis CS; Lomeli SH; Yin S; Berhane B; Apostol MI; Kickhoefer VA; Rome LH; Loo JA Sizing Large Proteins and Protein Complexes by Electrospray Ionization Mass Spectrometry and Ion Mobility. J. Am. Soc. Mass Spectrom 2007, 18, 1206–1216. 10.1016/j.jasms.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Testa L; Brocca S; Grandori R Charge-Surface Correlation in Electrospray Ionization of Folded and Unfolded Proteins. Anal. Chem 2011, 83 (17), 6459–6463. 10.1021/ac201740z. [DOI] [PubMed] [Google Scholar]
- (46).Fernandez de la Mora J Electrospray Ionization of Large Multiply Charged Species Proceeds via Dole’s Charged Residue Mechanism. Anal. Chim. Acta 2000, 406 (1), 93–104. 10.1016/S0003-2670(99)00601-7. [DOI] [Google Scholar]
- (47).Kaltashov IA; Mohimen A Electrospray Ionization Mass Spectrometry Can Provide Estimates of Protein Surface Areas in Solution. Anal. Chem 2005, 77 (16), 5370–5379. 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Konijnenberg A; Butterer A; Sobott F Native Ion Mobility-Mass Spectrometry and Related Methods in Structural Biology. Biochim. Biophys. Acta BBA - Proteins Proteomics 2013, 1834 (6), 1239–1256. 10.1016/j.bbapap.2012.11.013. [DOI] [PubMed] [Google Scholar]
- (49).Shvartsburg AA; Smith RD Fundamentals of Traveling Wave Ion Mobility Spectrometry. Anal. Chem 2008, 80 (24), 9689–9699. 10.1021/ac8016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Gelb AS; Jarratt RE; Huang Y; Dodds ED A Study of Calibrant Selection in Measurement of Carbohydrate and Peptide Ion-Neutral Collision Cross Sections by Traveling Wave Ion Mobility Spectrometry. Anal. Chem 2014, 86 (22), 11396–11402. 10.1021/ac503379e. [DOI] [PubMed] [Google Scholar]
- (51).Richardson K Fundamentals of Travelling Wave Ion Mobility Revisited: Towards Universal Calibration, 2018.
- (52).Richardson K An Improved Calibration Approach for Travelling Wave Ion Mobility Spectrometry: Robust, High-Precision Collision Cross Sections, 2019. [DOI] [PubMed]
- (53).Hopper JTS; Sokratous K; Oldham NJ Charge State and Adduct Reduction in Electrospray Ionization-Mass Spectrometry Using Solvent Vapor Exposure. Anal. Biochem 2012, 421 (2), 788–790. 10.1016/j.ab.2011.10.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.