Abstract
Lens-epithelium-derived growth-factor (LEDGF/p75)-binding site on HIV-1 integrase (IN), is an attractive target for antiviral chemotherapy. Small-molecule compounds binding to this site are referred as LEDGF-IN inhibitors (LEDGINs). In this study, compound libraries were screened to identify new inhibitors of LEDGF/p75-IN interaction. Ebselen (2-phenyl-1,2-benzisoselenazol-3-one), a reported anti-HIV-1 agent, was identified as a moderate micromolar inhibitor of LEDGF/p75-IN interaction. Ebselen inhibited the interaction by binding to LEDGF/p75 and the ability of ebselen to inhibit the interaction could be reversed by dithiothreitol (DTT). BLI experiment showed that ebselen probably formed selenium-sulphur bonds with reduced thiols in LEDGF/p75. To the best of our knowledge, we showed for the first time that small-molecule compound, ebselen inhibited LEDGF/p75-IN interaction by directly binding to LEDGF/p75. The compound discovered here could be used as probe compounds to design and develop new disrupter of LEDGF/p75-IN interaction.
Keywords: ALLNIs, integrase, LEDGF/p75-integrase interaction, HIV-1
Introduction
HIV-1, the virus that causes acquired immune deficiency syndrome (AIDS), is one of the world’s most serious health and development challenges. There were approximately 36.7 million people worldwide living with HIV/AIDS at the end of 20161. During the last three decades, significant progress has been made in the medical treatment of patients with HIV-1 infection; however, the rapid emergence of drug resistance together with toxicity and patient compliance limit the use of antiviral drugs, there remains a need for discovery of new antiviral agents2,3. HIV-1 integrase (IN), an essential enzyme encoded at the 3′-end of the HIV pol gene, is an attractive target for chemotherapeutic intervention4,5. IN is a multifaceted player in HIV-1 infection. Apart from its catalytic activity composed of 3′ processing and strand transfer, investigation of mutagenesis in IN and the mode of action of allosteric IN inhibitor (ALLINI) revealed that IN also play several other biological roles in HIV-1 life cycle, including virion morphogenesis, virus particle uncoating and PIC nuclear import6.
Currently, there are four approved HIV-1 IN inhibitors, including raltegravir, elvitegravir, dolutegravir and bictegravir, which block the strand-transfer step by binding to the active site in IN and designated as integrase-strand-transfer inhibitor (INSTI)7–11. During the last decades, ALLINI has emerged as a promising and complementary approach to the use of INSTI. ALLINIs, alternatively referred as lens-epithelium-derived growth-factor (LEDGF/p75)-integrase inhibitors (LEDGINs), noncatalytic site integrase inhibitors (NCINIs), IN-LEDGF/p75 allosteric inhibitors (INLAIs) and selective multimeric IN inhibitors (MINIs), are mechanistically distinct from active-site inhibitors INSTIs and therefore provide an important clinical complement to INSTIs in the clinical treatment of HIV-1 infection12–18.
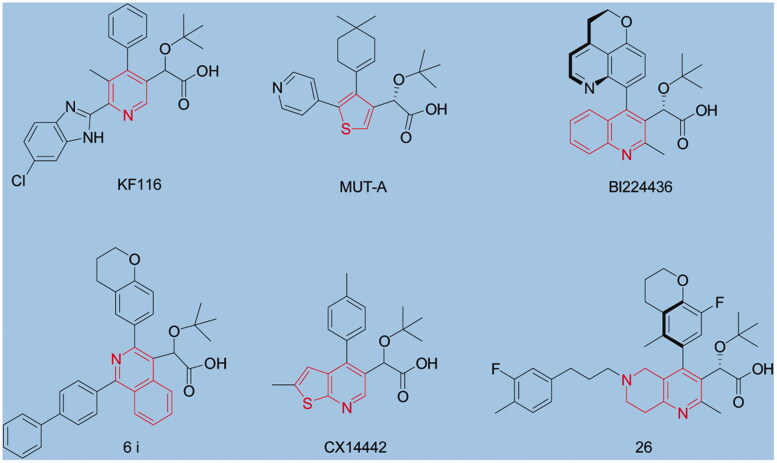
Lens epithelium-derived growth factor (LEDGF/p75) is a cellular cofactor hijacked by HIV-1 virus to tethers the integration complex to chromatin, thereby facilitating viral integration into host-cell DNA19. LEDGF interacts with HIV integrase (IN) through its C-terminal integrase binding domain (IBD, amino-acid residues 347–429)20. Disruption of the LEDGF/p75-integrase protein-protein can efficiently block HIV-1 replication21. During the last decade, several small molecules (representative chemotypes showed in Figure 1) have been identified as disruptors of the LEDGF/p75-HIV-1 IN interaction, which bind to the IN catalytic core domain (CCD) dimer interface in the LEDGF/p75 binding pocket6. Although these agents (ALLINIs) do indeed bind to the LEDGF/p75 interface on IN in vitro, their primary mechanism of action instead is to block viral-particle maturation during the late stages of viral replication16,22–24. And further multiple studies have elucidated the late stage mechanism of ALLINIs: ALLINIs stimulate aberrant or hyper-multimerization of IN thus impairing IN binding to vRNA required for particle morphogenesis and causing the mislocalization of ribonucleoprotein complex (RNPs) outside of the conical core made of capsid proteins25,26.
Figure 1.
Structures of representative ALLINIs. Chemotypes of each ALLNI are highlighted in red colour.
Recently discovered ALLINIs all inhibit HIV-1 replication by occupying the LEDGF/p75 pocket in IN. Although, cyclic peptides targeting LEDF were discovered to inhibit the LEDGF/p75–IN interaction27,28, there are no potent small-molecule inhibitors directly binding to the LEDGF/p75 to date. In this study, we used an HTRF-based assay to identify novel disrupters of LEDGF/p75-IN interaction by screening compound libraries. In a previous study, ebselen was reported as HIV-1 capsid inhibitor, which inhibits the replication of HIV-1 with a 50% effective concentration (EC50) of 1.99 µM and has a half-maximal cytotoxic concentration (CC50) of 25.4 µM. In present study, ebselen was identified as ligand binding to the LEDGF/p75 and inhibitor of LEDGF/p75-IN interaction. This work provides proof-of-concept for direct targeting of LEDGF/p75 by small molecule as novel therapeutic strategy and the compound here serve as leads for future development of new inhibitors of LEDGF/p75-IN interaction in vitro.
Material and methods
Agents and inhibitor libraries
All general biochemical reagents were obtained from AMRESCO (Solon, USA). Ni-NTA resin and GST resin were purchased from Smart-Lifesciences (Changzhou, China). Anti-6His-XL665- and anti-GST-Eu Cryptate antibodies were purchased from Cisbio. 96-well Black microplates were purchased from Greiner Bio-One. White 384-shallow well microplate was purchased from PerkinElmer. Streptavidin (SA) biosensors were purchased from Pall ForteBio. EZ-Link™ Sulfo-NHS-LC-LC-Biotin were purchased from Thermo Fisher Scientific. Ebselen was purchased from MedChem Express (Shanghai, China). Three protein kinase inhibitor libraries (Syn kinase inhibitor library, protein kinase inhibitor library and kinase inhibitor library) and REDOX library were obtained from National Compound Resource Center (Shanghai, China).
In vitro IBD-IN interaction assay
Recombinant IBD with N-terminal glutathione S-transferase (GST-IBD) and IN with N-terminal hexahistidine tag (His6-HIV-1 IN) were expressed and purified as previously described29. An assay based on a homogeneous time-resolved fluorescence resonance energy transfer (HTRF) was used to measure the interaction between HIV-1 IN and IBD according to a previous study30. The assay was performed in white 384-shallow well microplate. Prepare “Protein MIX” by mixing His6-HIV-1 IN (final concentration of 50 nM) and GST-IBD (final concentration of 25) in the assay buffer (25-mM Tris-HCl pH 7.5, 150-mM NaCl, 1 mg/ml BSA, 0.1% NP40, 2-mM MgCl2). 8 µl “Protein Mix” and 2 µl assay buffer were added to the plate and incubated at 25 °C for 30 min, followed by the addition of 10 µl of premixed antibodies (5 µµl anti-His XL665 [4 nM] and 5 µl anti-GST europium cryptate [0.8 nM]) in the assay buffer with 100-mM KF. The plate was incubated in the dark for 1 h at 25 °C. Finally, the plate was read in an Envision 2102 multilabel reader (PerkinElmer Life Sciences). Raw counts (in counts/s) at 665 and 620 nm were collected, and the signal was calculated as the ratio of (cps 665:620 nm) × 10,000.
Screening assay
Add 8-μL protein MIX to each well in a white 384-shallow well microplate. To the 8 μL protein MIX, add and mix thoroughly 2 μL of DMSO or compound (50 µM) dissolved in DMSO. The concentrations of DMSO in the assay should be no more than 4%. Incubate the plate for 30 min at 25 °C. Add 10 μL premixed antibodies and mix. Incubate the assay plate in the dark for 1 h at 25 °C. Perform the HTRF measurement in an Envision 2102 multilabel reader. The Z’ factor were calculated to evaluate the screening results of each plate31. Data were analysed and visualised in GraphPad Prism 5.0.
Dose-response curves
Percentages of inhibition at different concentrations were determined using the same reaction conditions as primary screen. All measurements were performed as 12-point (the range of compound concentration from 0.024 to 50 µM) dose–response curves. BI 224436 was used as a reference inhibitor. Data analysis was performed and visualised using GraphPad Prism 5.0 nonlinear curve fitting.
Biolayer interferometry assay
A protein binding assay was performed by biolayer interferometry (BLI) as described previously32. First, purified recombinant LEDGF/p75 protein was biotinylated using the Thermo EZLink long-chain biotinylation reagent. Then, biolayer interferometry (BLI) assay was performed using an OctetRED96 instrument from PALL/ForteBio. All assays were run at 30 °C with continuous 1000 rpm shaking. PBS with 0.01% Tween-20 was used as the assay buffer. Briefly, biotinylated LEDGF/p75 protein was tethered on Super Streptavidin (SSA) biosensors (ForteBio) by dipping sensors into 200 µl per well 50 µg/ml protein solutions. The measurement processes were all under computer control. Programme procedures were established as follows: For the initial step, biosensors were washed in assay buffer for 300 s to form a baseline; the biosensors labelled with biotin-LEDGF/p75 were exposed to 100 µM compounds for association, and were monitored for 600 s; and then, the biosensors were moved back into assay buffer to disassociate for another 1800s. Data were fit globally and generated automatically by Octet User software (version 9.0.0.10; Fortebio).
Results and discussion
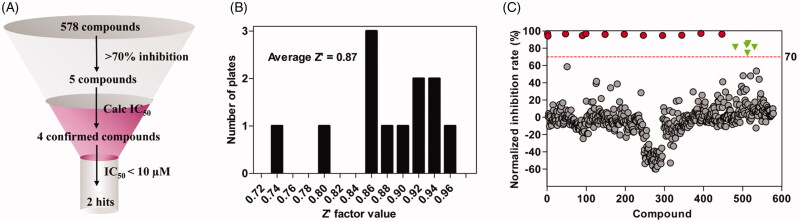
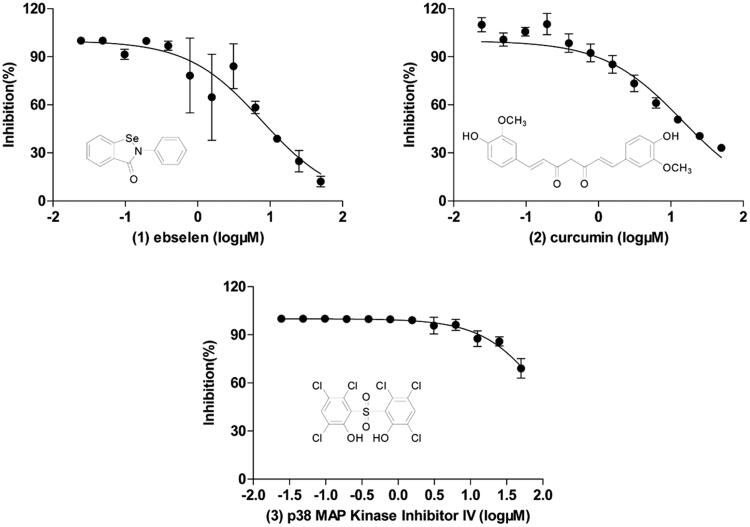
The IBD was previously shown to be necessary and sufficient for the interaction with HIV-1 IN33. In this study, the interaction between IBD (truncated form of LEDGF/p75) and IN was used to screen inhibitors of LEDGF/p75-IN interaction. To discover new chemicals disrupting LEDGF/p75-HIV-1 IN interaction, we screened at 50 µM four libraries (Syn kinase inhibitor library, protein kinase inhibitor library, kinase inhibitor library and REDOX library) of 578 compounds. Overview of the screening process is summarised in Figure 2(A). The average Z’ factor value for assays is 0.61 and no plates failed during screening (Figure 2(B)). Out of these compounds, 5 molecules displayed a percentage of inhibition above threshold (70%) at the screening concentration (Figure 2(C)). Dose–response curves were performed and 3 compounds (1–3) were confirmed (0.17% hit rate), with IC50s ranging from 7.70 µM to 116 µM: curcumin, p38 MAP Kinase Inhibitor IV and ebselen, which display unique structural motifs and activities (Figure 3 and Table 1). This hit rate is comparable to those published with other drug libraries (NIH Clinical Collections libraries, 2P2I-Oriented Chemical Library……) screened on isolated targets (Table 2). Out of these compounds, we selected ebselen (1), for further investigation of mode of action and/or binding.
Figure 2.
Overview and primary screening results. (A) Screening cascade. (B) Z-factor frequency distribution for 12 screening plates. (C) Replicate plot from screening 578 compounds for disruption of LEDGF/p75 IBD-IN interaction at 50 µM. The red dash line indicates our cut-off point of 70% inhibition and 5 compounds inhibited the interaction by more than 70%.
Figure 3.
Structures and dose–response curves of confirmed positives 1–3. Data represent the mean ± SD of three independent experiments.
Table 1.
Reported hit rates for the screening of libraries drugs
| Target | Acronym | Hit rate (%) | Screened library (size) | No. of hits | Reference | |
|---|---|---|---|---|---|---|
| Insulin-degrading enzyme | IDE | 0.45 | APTEEUS-Universite de Lille Library | 1120 | 5 | Leroux et al.42 |
| Human 3-hydroxy-3-methyl-glutaryl-coenzyme A | HMG-CoA reductase | 3.3 | NIH Clinical Collections librarie | 727 | 24 | Bessoff et al.43 |
| Aspergillus fumigatus chitinase A1 | AfChiA1 | 0.08 | Dundee Drug Discovery Unit diversity | 59904 | 48 | Lockhart et al.44 |
| Dengue virus non-structural proteins NS3 and NS interaction | NS3/NS5 | 1.6 | 2P2I-Oriented Chemical Library |
1664 | 26 | Milhas et al.45 |
| HIV virus Nef and Hck kinase interaction | Nef/SH3-Hck | 0.2 | 2P2I-Oriented Chemical Library |
1664 | 2 | Milhas et al.45 |
| HIV virus integrase and LEDGF/p75 IBD domain | IN/IBD | 0.17 | Protein kinase inhibitor library, REDOX library | 578 | 1 | This paper |
Table 2.
Inhibitory potencies of confirmed positives
| Compound | Name | IC50 (µM) |
|---|---|---|
| 1 | Ebselen | 7.70 |
| 2 | Curcumin | 14.52 |
| 3 | p38 MAP kinase inhibitor IV | 116.90 |
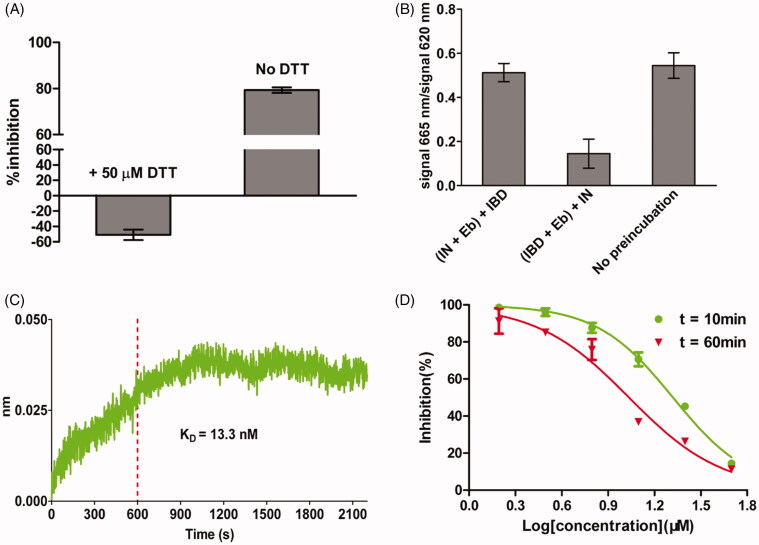
Ebselen is a synthetic organoselenium compound, with anti-inflammatory, anti-oxidant and cytoprotective activity33. It is being investigated as possible treatments for reperfusion injury and stroke, hearing loss and tinnitus, and bipolar disorder34. Ebselen has also been investigated against infectious diseases (Table 3). Importantly, in most cases, ebselen was shown to be a covalent inhibitor of these previously studied proteins. Accordingly, we designed experiments to study the binding mode of ebselen. If ebselen functions by modifying reduced this in proteins, then adding compounds with reduced thiols should abrogate the ability of ebselen to inhibit the interaction. To test the hypothesis, we repeated the HTRF-based LEDGF/p75 IBD-IN interaction assay with ebselen in the presence of 50 µM dithiothreitol (DTT). The inhibitory effect of ebselen was completely abolished in the presence of DTT (Figure 4(A)). In a previous study, DTT was reported to inhibit LEDGF/p75-IN interaction (IC50 = 4.5 mM), but under the concentration (50 µM) used in our experiment, this inhibitory effect of DTT was negligible according to the precious research35. Then, we devised the following experiment to determine which protein (IN or LEDGF/p75) ebselen binds to. When IBD was pre-cubated with ebselen, there was a dramatical decrease in the HFRF signal, whereas no obvious signal change was observed when IN was pre-cubated with ebselen (Figure 4(B)). These results suggested that ebselen probably disrupt the interaction by binding to LEDGF/p75. Using biolayer interferometry (BLI) by Octet Red, we confirmed that ebselen binds to LEDGF/p75 with a KD value of 13.3 nM (Figure 4(C)). If ebselen is a covalent inhibitor, it should show apparent increasing affinity over time36. The results showed that ebselen displayed an increase in its affinity over 1 h, consistent with covalent bond formation. (Figure 4(D)). All these data showed that ebselen probably inhibited the LEDGF/p75-IN interaction by covalently modifying the residues in LEDGF/p75.
Table 3.
Activity of ebselen on targets from pathogen.
| Target | IC50(µM) | Covalent | Reference |
|---|---|---|---|
| Cryptosporidium parvum glucose-6-phosphate isomerase | 8.33 | Yes | Eltahan et al.46 |
| Tumor marker endothelial 8 and protective antigen interaction | 1.7 | Yes | Cryan et al.37 |
| Trypanosoma brucei hexokinase 1 | 0.05 | Yes | Gordhan et al.47 |
| Escherichia coli thioredoxin reductase | 0.52 (Ki) | Yes | Lu et al.48 |
| C-terminal domain of HIV-1 capsid | 0.047 | Yes | Thenin-Houssier et al.39 |
| Bacillus anthracis thioredoxin reductase | 1 | ND | Gustafsson et al.49 |
| Clostridium difficile cysteine protease domain | 0.0069 | Yes | Bender et al.50 |
| New Delhi metallo-β-lactamase | 0.38 | Yes | Chiou et al.51 |
| Mycobacterium tuberculosis antigen 85 | 0.063 | Yes | Favrot et al.52 |
| Hepatitis C Virus NS3 Helicase-Nucleic Acid interaction | 1.1/0.9 | Yes | Mukherjee et al.53 |
| Cryptosporidium parvum Inosine 5′-monophosphate dehydrogenase | 0.71 | Yes | Sarwono et al.54 |
| Pseudomonas aeruginosa Diguanylate Cyclases | 5 | Yes | Lieberman et al.55 |
Figure 4.
Determination of the inhibition mode of ebselen on LEDGF/p75-IN interaction. (A) The inhibition of ebselen on LEDGF/p75-IN interaction was abolished in the presence of 50 µM DTT. (B) Ebselen inhibited the LEDGF/p75-IN interaction by binding to LEDGF/p75. (C) Association/dissociation kinetics of ebselen for LEDGF/p75 determined by Octet. (D) Time-dependent inhibition of the LEDGF/p75-IN interaction with ebselen. The data are representative of results obtained in three independent experiments. Each point is carried out in triplicate; error bars show the mean ± SD.
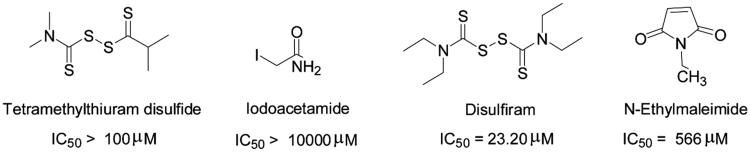
However, ebselen seems to be a promiscuous compound. Data from PubChem reveals that ebselen was found to be active in 193 out of 1023 drug screenings, with 126 of these hits coming from confirmatory screens37. And the promiscuity of ebselen may be partly attributed to its known ability of modifying cysteine residues. If ebselen really bind LEDGF/p75 by modifying reduced thiols, then similar reagents should also bind LEDGF/p75 and inhibit the interaction. To test our hypothesis, we tested the sensitivity of LEDGF/p75-IN interaction to iodoacetamide, N-ethylmaleimide, disulphiram and tetramethylthiuram disulphide, four common reagents used to covalently modify deduced cysteines. Neither was a potent inhibitor of LEDGF/p75-IN interaction except for disulphiram (IC50 = 23.2 µM) (Figure 5). Obviously, ebselen is more potent PPI than other thiol-modifying compounds. Moreover, BLI experiment indicated that ebselen do not show obvious dissociation from LEDGF/p75 (Figure 4(C)). Taken together, as a highly active compound, ebselen probably formed selenium-sulphur bonds with reduced thiols in LEDGF/p75.
Figure 5.
Ability of other thiol-modifying agents to inhibit LEDGF/p75 IBD-IN interaction.
Development of LEDGF/p75-IN interaction inhibitors has been recognised as an attractive target for new antiviral drugs, and multiple studies have identified small-molecule inhibitors, all of which bind to IN. However, ligands binding to the LEDGF/p75 are more desired to avoid resistance resulting from mutation of the viral IN38. To the best of our knowledge, no potent small-molecule inhibitors directly binding to the IBD have been identified to date. In the present study, we showed for the first time that small-molecule compounds, ebselen inhibited LEDGF/p75-IN interaction by directly binding to LEDGF/p75. Although, a previous research indicated that ebselen inhibited replication of HIV-1 by targeting capsid39, the compound discovered here could be used as probe compounds to design and develop new inhibitors of LEDGF/p75-IN interaction. Moreover, LEDGF/p75 plays a critical role as an oncogenic cofactor of MLL fusion proteins40. LEDGF/p75 uses the same site to bind MLL as well as IN41. Therefore, the discovery of two LEDGF/p75-binding compounds will pave the way for development of novel drugs with dual applications for both M HIV and MLL leukaemias.
In summary, this study has identified a compound, ebselen, as novel LEDGF/p75-IN interaction inhibitor with new scaffolds via drug screening of three compound libraries. The research revealed that ebselen disrupted the interaction by covalently binding to LEDGF/p75. This study provides new clues for the development of LEDGF/p75-IN interaction inhibitors with novel scaffolds.
Acknowledgements
The authors thank National Compound Resource Center (Shanghai, China) for supplying the small compounds used in this study.
Funding Statement
This work was supported by the National Natural Science Foundation of China [grant number 31700297], [grant number 81803430], [grant number 81603152], [grant number 81102483]; Innovation and Entrepreneurship Program 2017 of Jiangsu Province; Jiangsu Overseas Visiting Scholar Program for University Prominent Young & Middle-aged Teachers and Presidents; Natural Science Foundation of Jiangsu Province [grant number BE2019650], [grant number BK20170311], [grant number BK20170312]; Qing Lan Project and Six Talent Peaks Project in Jiangsu Province [grant number 2016-XYDXXJS-020].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Zhang D, Guo J, Zhang M, et al. Oxazole-containing diterpenoids from cell cultures of Salvia miltiorrhiza and their anti-HIV-1 activities. J Nat Prod 2017;80:3241–6. [DOI] [PubMed] [Google Scholar]
- 2.Wainberg MA, Zaharatos GJ, Brenner BG.. Development of antiretroviral drug resistance. N Engl J Med 2011;365:637–46. [DOI] [PubMed] [Google Scholar]
- 3.Barton KM, Burch BD, Soriano-Sarabia N, Margolis DM.. Prospects for treatment of latent HIV. Clin Pharmacol Ther 2013;93:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McColl DJ, Chen X.. Strand transfer inhibitors of HIV-1 integrase: bringing IN a new era of antiretroviral therapy. Antiviral Res 2010;85:101–18. [DOI] [PubMed] [Google Scholar]
- 5.Pendri A, Meanwell NA, Peese KM, Walker MA.. New first and second generation inhibitors of human immunodeficiency virus-1 integrase. Expert Opin Ther Pat 2011;21:1173–89. [DOI] [PubMed] [Google Scholar]
- 6.Engelman AN. Multifaceted HIV integrase functionalities and therapeutic strategies for their inhibition. J Biol Chem 2019;294:15137–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Summa V, Petrocchi A, Bonelli F, et al. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIVAIDS infection. J Med Chem 2008;51:5843–55. [DOI] [PubMed] [Google Scholar]
- 8.Rowley M. The discovery of raltegravir, an integrase inhibitor for the treatment of HIV infection. Prog Med Chem 2008;46:1–26. [DOI] [PubMed] [Google Scholar]
- 9.Sato M, Kawakami H, Motomura T, et al. Quinolone carboxylic acids as a novel monoketo acid class of human immunodeficiency virus type 1 integrase inhibitors. J Med Chem 2009;52:4869–82. [DOI] [PubMed] [Google Scholar]
- 10.Johns BA, Kawasuji T, Weatherhead JG, et al. Carbamoyl pyridone HIV-1 integrase inhibitors 3. A diastereomeric approach to chiral nonracemic tricyclic ring systems and the discovery of dolutegravir (S/GSK1349572) and (S/GSK1265744). J Med Chem 2013;56:5901–16. [DOI] [PubMed] [Google Scholar]
- 11.Tsiang M, Jones GS, Goldsmith J, et al. Antiviral activity of bictegravir (GS-9883), a novel potent hiv-1 integrase strand transfer inhibitor with an improved resistance profile. Antimicrob. Agents Chemother 2016;60:7086–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenwick CW, Tremblay S, Wardrop E, et al. Resistance studies with HIV-1 non-catalytic site integrase inhibitors. Antivir Ther 2011;16: (Suppl. 1):A9. [Google Scholar]
- 13.Fader LD, Malenfant E, Parisien M, et al. Discovery of BI224436, a noncatalytic site integrase inhibitor (NCINI) of HIV-1. ACS Med Chem Lett 2014;5:422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christ F, Voet A, Marchand A, et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol 2010;6:442–8. [DOI] [PubMed] [Google Scholar]
- 15.Kessl JJ, Jena N, Koh Y, et al. Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem 2012;287:16801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balakrishnan M, Yant SR, Tsai L, et al. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLoS One 2013;8:e74163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Rouzic E, Bonnard D, Chasset S, et al. Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology 2013;10:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma A, Slaughter A, Jena N, et al. A new class of multimerization selective inhibitors of HIV-1 integrase. PloS Pathog 2014;10:e1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciuffi A, Llano M, Poeschla E, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 2005;11:1287–9. [DOI] [PubMed] [Google Scholar]
- 20.Cherepanov P, Ambrosio AL, Rahman S, et al. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA 2005;102:17308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demeulemeester J, Chaltin P, Marchand A, et al. LEDGINs, non-catalytic site inhibitors of HIV-1 integrase: a patent review (2006–2014). Expert Opin Ther Pat 2014;24:609–32. [DOI] [PubMed] [Google Scholar]
- 22.Jurado KA, Wang H, Slaughter A, et al. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc Natl Acad Sci USA 2013;110:8690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desimmie BA, Schrijvers R, Demeulemeester J, et al. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 2013;10:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontana J, Jurado KA, Cheng N, et al. Distribution and redistribution of HIV-1 nucleocapsid protein in immature, mature, and integrase-inhibited virions: a role for integrase in maturation. J Virol 2015;89:9765–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng L, Sharma A, Slaughter A, et al. The A128T resistance mutation reveals aberrant protein multimerization as the primary mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem 2013;288:15813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kessl JJ, Kutluay SB, Townsend D, et al. HIV-1 integrase binds the viral RNA genome and is essential during virion morphogenesis. Cell 2016;166:1257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desimmie BA, Humbert M, Lescrinier E, et al. Phage display-directed discovery of LEDGF/p75 binding cyclic peptide inhibitors of HIV replication. Mol Ther 2012;20:2064–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao L, Hannon C, Cruz-Mignoni A, et al. Intracellular immunization against HIV infection with an intracellular antibody that mimics HIV integrase binding to the cellular LEDGF protein. Sci Rep 2017;7:16869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang DW, He HQ, Liu MM, Meng ZX, et al. A novel assay for screening inhibitors targeting HIV integrase LEDGF/p75 interaction based on Ni2+ coated magnetic agarose beads. Sci Rep 2016;6:33477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenwick C, Bailey MD, Bethell R, Bös M, et al. Preclinical profile of BI 224436, a novel HIV-1 non-catalytic-site integrase inhibitor. Antimicrob Agents Chemother 2014;58:3233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang JH, Chung TD, Oldenburg KR.. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999;4:67–73. [DOI] [PubMed] [Google Scholar]
- 32.Xu S, Aguilar A, Xu T, et al. Design of the first‐in‐class, highly potent irreversible inhibitor targeting the menin‐MLL protein–protein interaction. Angew Chem Int Ed Engl 2018;57:1601–5. [DOI] [PubMed] [Google Scholar]
- 33.Cherepanov P, Devroe E, Silver PA, Engelman A.. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem 2004;279:48883–92. [DOI] [PubMed] [Google Scholar]
- 34.Azad GK, Tomar RS.. Ebselen, a promising antioxidant drug: mechanisms of action and targets of biological pathways. Mol Biol Rep 2014;41:4865–79. [DOI] [PubMed] [Google Scholar]
- 35.Tsiang M, Jones GS, Hung M, et al. Dithiothreitol causes HIV-1 integrase dimer dissociation while agents interacting with the integrase dimer interface promote dimer formation. Biochemistry 2011;50:1567–81. [DOI] [PubMed] [Google Scholar]
- 36.Singh J, Petter RC, Baillie TA, Whitty A.. The resurgence of covalent drugs. Nat Rev Drug Discov 2011;10:307–17. [DOI] [PubMed] [Google Scholar]
- 37.Cryan LM, Habeshian KA, Caldwell TP, et al. Identification of small molecules that inhibit the interaction of TEM8 with anthrax protective antigen using a FRET assay. J Biomol Screen 2013;18:714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hombrouck A, De Rijck J, Hendrix J, et al. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog 2007;3:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thenin-Houssier S, De Vera IMS, Pedro-Rosa L, et al. Ebselen, a small-molecule capsid inhibitor of HIV-1 replication. Antimicrob Agents Chemother 2016;60:2195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ElAshkar S, Schwaller J, Pieters T, et al. LEDGF/p75 is dispensable for hematopoiesis but essential for MLL-rearranged leukemogenesis. Blood 2018;131:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murai MJ, Pollock J, He S, et al. The same site on the integrase-binding domain of lens epithelium–derived growth factor is a therapeutic target for MLL leukemia and HIV. Blood 2014;124:3730–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leroux F, Bosc D, Beghyn T, et al. Identification of ebselen as a potent inhibitor of insulin degrading enzyme by a drug repurposing screening. Eur J Med Chem 2019;179:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bessoff K, Sateriale A, Lee KK, Huston CD.. Drug repurposing screen reveals FDA-approved inhibitors of human HMG-CoA reductase and isoprenoid synthesis that block Cryptosporidium parvum growth. Antimicrob Agents Chemother 2013;57:1804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lockhart DE, Schuettelkopf A, Blair DE, van Aalten DM.. Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors. FEBS Lett 2014;588:3282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milhas S, Raux B, Betzi S, et al. Protein-protein interaction inhibition (2P2I)-oriented chemical library accelerates hit discovery. ACS Chem Biol 2016;11:2140–8. [DOI] [PubMed] [Google Scholar]
- 46.Eltahan R, Guo F, Zhang H, et al. Discovery of ebselen as an inhibitor of Cryptosporidium parvum glucose-6-phosphate isomerase (CpGPI) by high-throughput screening of existing drugs. Int J Parasitol Drugs Drug Resist 2018;8:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordhan HM, Patrick SL, Swasy MI, et al. Evaluation of substituted ebselen derivatives as potential trypanocidal agents. Bioorg Med Chem Lett 2017;27:537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J, Vlamis-Gardikas A, Kandasamy K, et al. Inhibition of bacterial thioredoxin reductase: an antibiotic mechanism targeting bacteria lacking glutathione. Faseb J 2013;27:1394–403. [DOI] [PubMed] [Google Scholar]
- 49.Gustafsson TN, Osman H, Werngren J, et al. Ebselen and analogs as inhibitors of Bacillus anthracis thioredoxin reductase and bactericidal antibacterials targeting Bacillus species, Staphylococcus aureus and Mycobacterium tuberculosis. Biochim Biophys Acta 2016;1860:1265–71. [DOI] [PubMed] [Google Scholar]
- 50.Bender KO, Garland M, Ferreyra JA, et al. A small-molecule antivirulence agent for treating Clostridium difficile infection. Sci Transl Med 2015;7:306ra148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiou J, Wan S, Chan KF, et al. Ebselen as a potent covalent inhibitor of New Delhi metallo-β-lactamase (NDM-1). Chem Commun (Camb) 2015;51:9543–6. [DOI] [PubMed] [Google Scholar]
- 52.Favrot L, Grzegorzewicz AE, Lajiness DH, et al. Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen. Nat Commun 2013;4:2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee S, Weiner WS, Schroeder CE, et al. Ebselen inhibits hepatitis C virus NS3 helicase binding to nucleic acid and prevents viral replication. ACS Chem Biol 2014;9:2393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarwono AEY, Mitsuhashi S, Kabir MHB, et al. Repurposing existing drugs: identification of irreversible IMPDH inhibitors by high-throughput screening. J Enzyme Inhib Med Chem 2019;34:171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lieberman OJ, Orr MW, Wang Y, Lee VT.. High-throughput screening using the differential radial capillary action of ligand assay identifies ebselen as an inhibitor of diguanylate cyclases. ACS Chem Biol 2014;9:183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]