Abstract
Glycosylation is one of the most ubiquitous and complex post-translational modifications (PTMs). It plays pivotal roles in various biological processes. Studies at the glycopeptide level are typically considered as a downstream work resulting from enzymatic digested glycoproteins. Less attention has been focused on glycosylated endogenous signaling peptides due to their low abundance, structural heterogeneity and the lack of enabling analytical tools. Here, protocols are presented to isolate and characterize glycosylated neuropeptides utilizing nanoflow liquid chromatography coupled with mass spectrometry (LC-MS). We first demonstrate how to extract neuropeptides from raw tissues and perform further separation/cleanup before MS analysis. Then we describe hybrid MS methods for glycosylated neuropeptides profiling and site-specific analysis. We also include recommendations for data analysis to identify glycosylated neuropeptides in crustaceans where a complete neuropeptide database is still lacking. Other strategies and future directions are discussed to provide readers with alternative approaches and further unravel biological complexity rendered by glycosylation.
Keywords: glycosylation, glycopeptide, LC-MS/MS, MALDI, ESI, enrichment, separation, characterization, derivatization, EThcD
1. Introduction
Proteins and peptides are usually released to the circulating system after cellular enzymatic processes from inactive precursors and multiple post-translational modifications (PTMs) (Hook et al., 2008; Hook, Lietz, Podvin, Cajka, & Fiehn, 2018; McClure, Walls, & Grinnell, 1992). PTMs are of great significance in determining or altering peptide structures and biological behaviors (Duan & Walther, 2015; Seo & Lee, 2004). Glycosylation is one of the most common yet the most challenging PTMs, due to the complexity of attached glycans and the site occupancy on the backbones. It is broadly related to various functional roles in a wide range of biological processes, including signal transduction, ion homeostasis, protein folding stability and kinetics, cell-cell adhesion, protein-protein interaction, and immune defense (Arey, 2012; Arnold, Wormald, Sim, Rudd, & Dwek, 2007; Helenius & Aebi, 2001, 2004; Jayaprakash & Surolia, 2017; Khidekel, Ficarro, Peters, & Hsieh-Wilson, 2004; Ohtsubo & Marth, 2006; Rudd, Elliott, Cresswell, Wilson, & Dwek, 2001). The distribution of glycosylation covers all three domains of life, eukarya, bacteria, and archaea, and especially in eukarya, more than half of proteins are modified by various attached glycans (Apweiler, Hermjakob, & Sharon, 1999; Dell, Galadari, Sastre, & Hitchen, 2010).
The glycans appear as short carbohydrate chains consisting of a single monosaccharide or polysaccharides, exhibiting broad diversities in structure, linkage and branching patterns. The complexity of glycan moieties results in a variety of glycosylated modifications. Among different types of glycosylation, there are two categories that are widely studied, namely N- and O-linked glycosylation. Specifically, an oligosaccharide attached to the side chain amide nitrogen of asparagine (Asn) is recognized as N-linked glycans with the Asn-X-Ser/Thr sequon, where X could be any amino acid except proline. O-linked glycosylation usually occurs on the hydroxyl group of serine and threonine when the carbohydrate is attached to the oxygen. Unlike the N-linked structures, O-linked glycans are found to be associated with no consensus sequence yet. Besides the different residues that glycans are attached to, N- and O-linked glycans also have distinct structures, due to their unique enzymatic pathway during biosynthesis (Lodish et al., 2000). N-glycans start with a conserved chitobiose core, (GlcNAc)2Man3. With the extension of other monosaccharides from the core unit, this group will be further divided into high-mannose, complex and hybrid types. However, O-linked carbohydrates do not share a common core structure, only featured by eight common formations (Alley, Mann, & Novotny, 2013). Challenges of glycosylation studies mostly stem from the sophisticated nature of glycans and their site occupancy throughout the primary amino acid sequences (Kolarich, Jensen, Altmann, & Packer, 2012; Stavenhagen et al., 2013). On a single glycosylated site in a peptide backbone, there is often a set of modifications with multiple glycan isoforms at different length and branch patterns. Throughout the full peptide sequence, there can be multiple glycosylation sites, which are completely, partially or scarcely occupied. Extensive research efforts have been devoted to the development of analytical tools applied in glycomics and glycoproteomics (Z. Chen, Huang, & Li, 2018; Ruhaak, Xu, Li, Goonatilleke, & Lebrilla, 2018; Xiao, Suttapitugsakul, Sun, & Wu, 2018; A. Yu et al., 2018). However, limited studies have focused on the endogenous glycosylated signaling peptides.
Neuropeptides and peptide hormones are main classes of signaling molecules in nervous and endocrine systems that regulate physiological processes and behaviors (Buchberger, Yu, & Li, 2015; Elphick, Mirabeau, & Larhammar, 2018; Heck et al., 2018; Matsubayashi, 2018). They are classified into different families based on the shared sequence motifs (Christie, Stemmler, & Dickinson, 2010; Enman, Sabban, McGonigle, & Van Bockstaele, 2015; Hökfelt et al., 2000). Although isoforms in the same family often share similar sequences, and even with identical masses, they may still have distinct physiological roles (Chung & Zmora, 2008; Nusbaum & Blitz, 2012). Therefore, the similarity in chemical and physical properties does not necessarily imply identical function. Neuropeptides are typically short amino acid chains, however, some peptide families could consist of up to 70 residues (Chung, Zmora, Katayama, & Tsutsui, 2010; Webster, Keller, & Dircksen, 2012). With various PTMs, the structural diversity presents significant challenges in global characterization. Furthermore, the concentration of neuropeptides and peptide hormones are oftentimes at nM-pM level (Adrian et al., 1983; Q. Li, Zubieta, & Kennedy, 2009), which would also cause great difficulties in analysis.
The glycosylated endogenous peptides undertake essential and non-negligible functional roles in many biological processes, although they exist at a further lower concentration level compared to the non-glycosylated counterparts (Halim et al., 2011). For example, glycosylation of opioid peptides (cyclic/linear enkephalin, Met-/Leu-enkephalin, endomorphin, deltorphin) improves the transportation of these pharmacologically active neuropeptides into the brain (Egleton et al., 2000, 2001; Masand et al., 2006; Varamini et al., 2012; Witt & Davis, 2006) by crossing the blood-brain barrier (BBB), a highly selective barrier that prohibits most compounds from penetrating into the brain, the central nervous system (CNS) (Serrano, Ribeiro, & Castanho, 2012; Varamini et al., 2012). Furthermore, the attachments of carbohydrates to peptides would increase the resistance to enzymatic cleavage and degradation (Semenov et al., 2009; T. Yamamoto et al., 2009), lengthen the half-life time in serum (Polt et al., 1994) and enhance the receptor binding (Egleton et al., 2000). The improved factors make the glycosylated analogues a more effective methodology as peptide drugs. For example, studies showed that the additional sugar moieties boost the analgesic activity (Masand et al., 2006; Polt et al., 1994), an antinociceptive effect similar to morphine (Bilsky et al., 2000), both in maximal analgesic level and the analgesia duration (Egleton et al., 2001). On another aspect, radiolabeled glycopeptides affect the biodistribution in the application to cancer cell imaging (Schweinsberg et al., 2008; Watanabe et al., 2012), including less liver and renal accumulation and improvement in tumor uptake, leading to a minimum effective dosage and an increased tumor-to-background ratio in imaging (Haubner et al., 2001; Schottelius, Wester, Reubi, Senekowitsch-Schmidtke, & Schwaiger, 2002). Besides functioning as potential therapeutic drugs, O-glycosylated peptide toxins, Vespulakinins, isolated from yellowjacket venom sacs were the first reported vasoactive glycopeptides and naturally occurring glycosylated derivatives of bradykinin, with an identical C-terminus sequence (Yoshida, Geller, & Pisano, 1976). In the cone snail, two other O-glycopeptides, κA-conotoxin SIVA and Contulakin-G, were found in the venom, as the first evidence of biologically active glycosylated peptides in this model. They were demonstrated to have higher activity compared to nonglycosylated counterparts when administered in vivo (A. G. Craig et al., 1998; A. Grey Craig, Bandyopadhyay, & Olivera, 1999). Notably, Contulakin-G was the first known neurotensin family peptide from a nonvertebrate model. The C-terminus of its glycosylated isoform, due to the high homology to mammalian neuropeptides, was detected to target the mammalian neurotransmitter receptors (A. Grey Craig, Norberg, et al., 1999). Recently, more studies have discovered that the glycosylated endogenous peptides are associated with many diseases, such as diabetes, heart failure and neurodegenerative diseases (Brinkmalm et al., 2012; Halfinger et al., 2017; Halim et al., 2011; Semenov et al., 2009; Q. Yu, Canales, et al., 2017).
To deal with the high complexity and low abundance of the endogenous glycosylated peptides, mass spectrometry (MS)-based approaches have emerged as powerful tools, benefiting from the high sensitivity and high resolution. With the instrumentation advances in ionization sources, mass analyzers and especially, the fragmentation modes, significant advancements in glycopeptide analysis enable revealing peptide backbone structures, site-specific information and glycan types with less instrument time. Meanwhile, the extraction and enrichment of these endogenous peptides from biological samples are also the crucial steps in the entire workflow. Extraction methods may vary depending on the specific properties of the samples and the targeted peptides. Prior to rapid and highly accurate MS detection, glycopeptide enrichment and multidimensional chromatographic separation would facilitate the qualitative and quantitative analysis of glycosylated peptides at a lower detection limit with a larger dynamic range. In this chapter, we summarize the current state of endogenous glycopeptide studies and highlight recent advances in various techniques benefiting glycosylated peptide analysis (Fig. 1). Specifically, we present detailed workflows ranging from tissue collection to MS analysis that enable discovery and identification of glycosylated neuropeptides in crustacean model organisms. Protocols of other strategies are also discussed, including peptide extraction, glycopeptide enrichment and mass spectrometric analysis to provide the readers sufficient details so that effective experiments can be designed and performed according to different sample types, target analytes and available instrument platforms.
Figure 1.
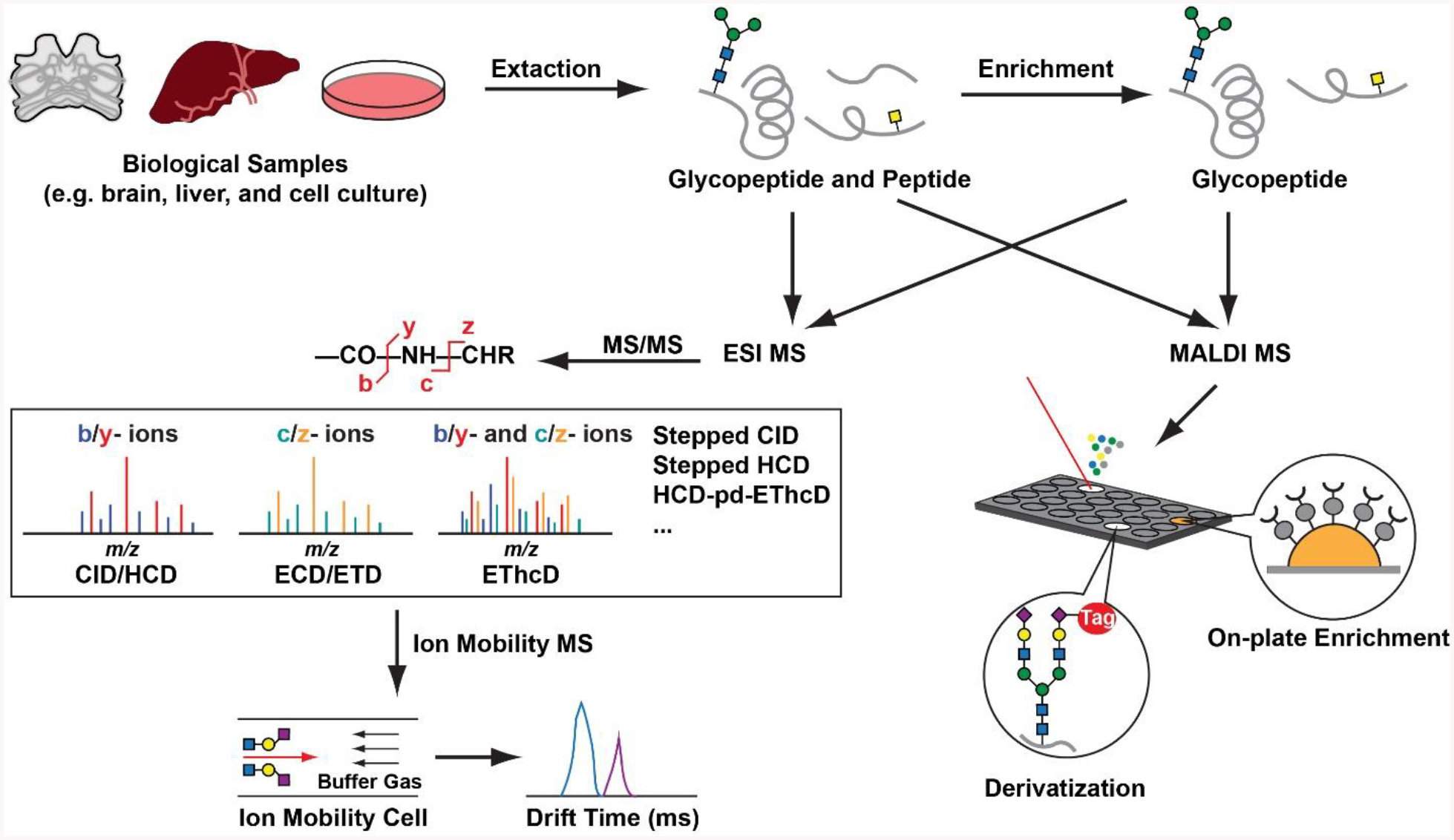
A typical workflow to identify glycopeptides via multifaceted MS approaches in complex biological samples.
2. Neuropeptide Extraction from Crustacean Neuronal Tissues
Besides endogenous peptides, biological samples usually also contain lipids, salts and large proteins (Aebersold & Mann, 2016; Arnaud et al., 2005; Cajka & Fiehn, 2016; Paglia, Kliman, Claude, Geromanos, & Astarita, 2015). They will interfere the ionization efficiency, contaminate the LC platform and suppress the signal intensity of target peptides. Reduction of sample complexity is the primary step in peptidome analysis. Commonly used peptide extraction methods will be introduced here, while the purification approaches specific to glycopeptides will be described. The crustacean nervous system contains rich contents of neuropeptides. Acid extraction is a classical method to easily purify neuropeptides. Here, we first describe how to collect crustacean neuronal tissues and then a detailed workflow to extract neuropeptides. The whole experiment can take 1 day to 2 days, depending on the tissue amount involved.
2.1. Neuronal Tissue Collection
2.1.1. Materials and Buffer Recipes
Physiological saline buffer stored at 4 °C: 440 mM NaCl, 11 mM KCl, 26 mM MgCl2, 13 mM CaCl2, 11 mM Trizma base, 5 mM maleic acid
Ice-cold acidified methanol (methanol/water/glacial acetic acid = 90/9/1, v/v/v)
Dry ice in a sealed box
Dissection tools, including petri dish, spatula, scissors, tweezers, forceps and needles
Microscope
2.1.2. Tissue Harvest via Animal Sacrifice
Anesthetize a crab by burying it in ice for 15 min.
Use forceps to open crab shell, isolate large tissues where neuronal tissues are located and transfer them to a petri dish, add physiological saline buffer and use needles to pin down specific tissues for micro-dissection.
Use scissors and tweezers to collect desired neuronal tissues under a microscope.
Transfer isolated neuronal tissues into centrifuge tubes filled with 100 μL ice-cold acidified methanol and immediately put tubes in dry ice. Tissues can be used for immediate handling or stored in −80 °C freezer until further processing. Repeat dissection and tissue collection until enough samples are collected.
2.2. Acid Extraction
2.2.1. Materials and Buffer Recipes
Ice-cold acidified methanol (methanol/water/glacial acetic acid = 90/9/1, v/v/v)
Probe dismembrator
30 kDa molecular weight cutoff filter
ACN/MeOH/H2O = 20/30/50, v/v/v
Ice
Speed-vacuum sample dry system
2.2.2. Neuropeptide Extraction
Prerinse 30 kDa molecular weight cutoff filter with 80 μL ACN/MeOH/H2O (20/30/50, v/v/v) and spin at 4 °C for 15 min at 14,000g. Repeat once.
Put centrifuge tubes containing neuronal tissues onto ice-water mixture. Use a probe dismemberator in a 4 °C cold room to break large tissues in acidified methanol for efficient extraction.
Centrifuge the tissue content at 4 °C for 5 min at 16,100g.
Transfer supernatant onto the prerinsed 30 kDa molecular weight cutoff filter and centrifuge at 4 °C for 20 min at 14,000g.
Collect the supernatant that has spun through the filter. Repeat step 4 until no more flow-through is observed.
Transfer 50 μL acidified methanol onto the filter and further centrifuge at 4 °C for 20 min at 14,000g. Combine all the supernatants.
Evaporate the solvent under vacuum and store the dried sample at −80 °C for future analysis.
2.3. Neuropeptide Reduction and Alkylation
2.3.1. Materials and Buffer Recipes
50 mM ammonium bicarbonate (ABC)
100 mM DTT
200 mM IAA
Speed-vacuum sample dry system
2.3.2. DTT and IAA Reactions
Resuspend neuropeptide extract in a centrifuge tube with 100 μL 50 mM ABC. Vortex to allow sufficient mixing
Add 5.26 μL DTT into the centrifuge tube and incubate at room temperature for 1 hr.
Add 8.53 μL IAA into the centrifuge tube and incubate in dark at room temperature for 30 min.
Add 5.26 μL DTT into the centrifuge tube and incubate at room temperature for 5 min.
Dry down sample solvent under vacuum and store the dried sample at −80 °C for future analysis.
Our method incorporates both organic solvent precipitation and acid extraction to remove large proteins from neuropeptides in tissue samples. Organic solvent precipitation is one of the most common methods applied to isolate peptides from insoluble proteins (Patel et al., 2018; Simpson & Beynon, 2010; Williams et al., 2010). The addition of organic solvents disrupt the ordered water molecules surrounding the proteins and the hydrate layer around are decreased. Polar interactions between aqueous solvent and the protein are minimized, leading the protein to aggregate and precipitate due to the electrostatic interaction. The reduced solubility can effectively clean out the abundant large proteins, making peptides and small proteins in the solution accessible to analysis in the following steps. Popular choices for precipitants are various alcohols and acetone, among which, acetone is considered superior because of the minimal diluted volume and more dense precipitation (Chertov et al., 2004; Polson, Sarkar, Incledon, Raguvaran, & Grant, 2003).
Adding acid is also a widely adopted approach in precipitating proteins and extracting peptides (Kirkpatrick et al., 2017; S.-I. Park, Kim, & Yoe, 2015; Robert et al., 2015). Proteins, positively charged at pHs below their pI, interact with the acidic reagents and form insoluble salts. Besides inducing precipitant, acids also play another essential role in the preparation of biological samples containing active proteases. The acidic environment causes protein denature, thus curtailing the protease activity (Fink, Calciano, Goto, Kurotsu, & Palleros, 1994) to preserve fast-degrading peptides.
In peptide extraction, the interference of large proteins is not only their high abundance, but also the binding effects with some small peptides. The peptide-protein interaction is fundamental in cell signaling and its prevalence (London, Movshovitz-Attias, & Schueler-Furman, 2010; Schulze & Mann, 2004; M. Yang, Wu, & Fields, 1995) would lead to huge sample loss if peptides are not unbound from the proteins before being precipitated. Organic solvents, together with acidic reagents will dissociate the peptides from binding sites of large proteins, improving the extraction efficiency (Chertov et al., 2004). Another effective method to extract low molecular weight peptides would be differential solubilization (Kawashima et al., 2010). Sample was first mixed with denaturing solutions and then added ice-cold acetone, resulting in precipitation of all proteins and peptides. The precipitate was further washed with 70% ACN containing 12mM HCl, in which peptides were easily dissolved and separated from other proteins. It has been shown that compared to other extraction methods, differential solubilization gave higher yield and better reproducibility, being independent of the composition of the proteins/peptides from the samples.
Our experimental protocol also utilizes ultrafiltration enabled by molecular weight cutoff filter to further separate neuropeptides with soluble proteins. Ultrafiltration is also a frequently used approach in peptide extraction and purification, which retains the high molecular weight solutes while allowing the low molecular weight to pass through the semi-permeable membrane (Greening & Simpson, 2010; Luque-Garcia & Neubert, 2007). Various molecular weight cut-off of the membranes define the boundary between filtrate and retentate, providing more choices to different sample components and separation purposes (Chang, Ismail, Yanagita, Mohd Esa, & Baharuldin, 2015; Kirkpatrick et al., 2017; Zheng, Baker, & Hancock, 2006). The successful separation of centrifugation ultrafiltration relies on the low centrifugation speed and the low sample concentration (Luque-Garcia & Neubert, 2007). Although the centrifugal ultrafiltration method is facile, rapid and cost-effective in peptidome analysis (Finoulst, Pinkse, Van Dongen, & Verhaert, 2011; Greening & Simpson, 2010), it still lacks sharp separation indicated by the molecular weight cut-off membranes (Georgiou, Rice, & Baker, 2001; Picot et al., 2010). The large molecules stuck on the membrane can easily block the pores, affecting the separation efficiency. Electrophoresis is widely known as a high-resolution purification approach. A hybrid method of electrodialysis with insertion of ultrafiltration membrane was developed to enhance the peptide extraction and purification performance (Dlask & Václavíková, 2018; Picot et al., 2010). In electrodialysis ultrafiltration, the electric potential differences act as driving force and the variation in pH can selectively separate acidic, basic and neutral peptides, reducing the accumulation of molecules on membrane surface and clogs of the porous membranes (Dlask & Václavíková, 2018; Poulin, Amiot, & Bazinet, 2006).
To manually operate the protein precipitation would be quite time-consuming when being applied to large sample sets. The development of membrane-based protein precipitation filter plates helps to avoid the tedious workflow. By using a 96-well filter block, the separation of peptides and large proteins become more effective (Biddlecombe & Pleasance, 1999; Kitchen, Wang, Musson, Yang, & Fisher, 2003), with higher throughput and more simplified procedures, due to the elimination of centrifugation and supernatant transfer steps (Watt, Morrison, Locker, & Evans, 2000). In this method, filtrate containing low-molecular-weight proteins and peptides can be easily collected, suffering from less cross contamination, and being compatible to downstream analysis using MS at high accuracy and reproducibility (Walter, Cramer, & Tse, 2001).
2.4. Alternative Strategy - Restricted Access Materials
Restricted access materials (RAMs) are solid sorbents that exclude the access of macromolecules to the surface binding while the low molecular weight particles are selectively retained (Kole, Venkatesh, Kotecha, & Sheshala, 2011). The first silica-based RAM model was proposed in 1986, named as internal surface reverse-phase (ISRP) (Cook & Pinkerton, 1986). In 1991, the modifications on the surface of silica particles were successfully carried out and the concept of RAM was first introduced (Desilets, Rounds, & Regnier, 1991). From then on, various types of RAM have been developed, which can be classified based on the mechanisms how the large proteins are excluded from extraction (de Faria, Abrão, Santos, Barbosa, & Figueiredo, 2017; Souverain, Rudaz, & Veuthey, 2004). These special extraction methods allow automating sample preparation and on-line extraction followed by MS analysis using a RAM column. It enables simplified operation, high throughput and reproducible identification of peptides. Around 400 peptides were identified in 2μl human serum using one-dimensional separation and with the help of a SCX/SEC/RP-MS platform (Fig. 2); identifications of 1286 peptides were achieved by the on-line processing of 20μl of human serum (L. Hu, Boos, Ye, Wu, & Zou, 2009). The same group later exploited a more simplified system, constructed with one switching valve, two gradient pumps and one UV detector, to realize the on-line extraction, desalting and fractionation of peptides in human serum (L. Hu, Boos, Ye, & Zou, 2014).
Figure 2.
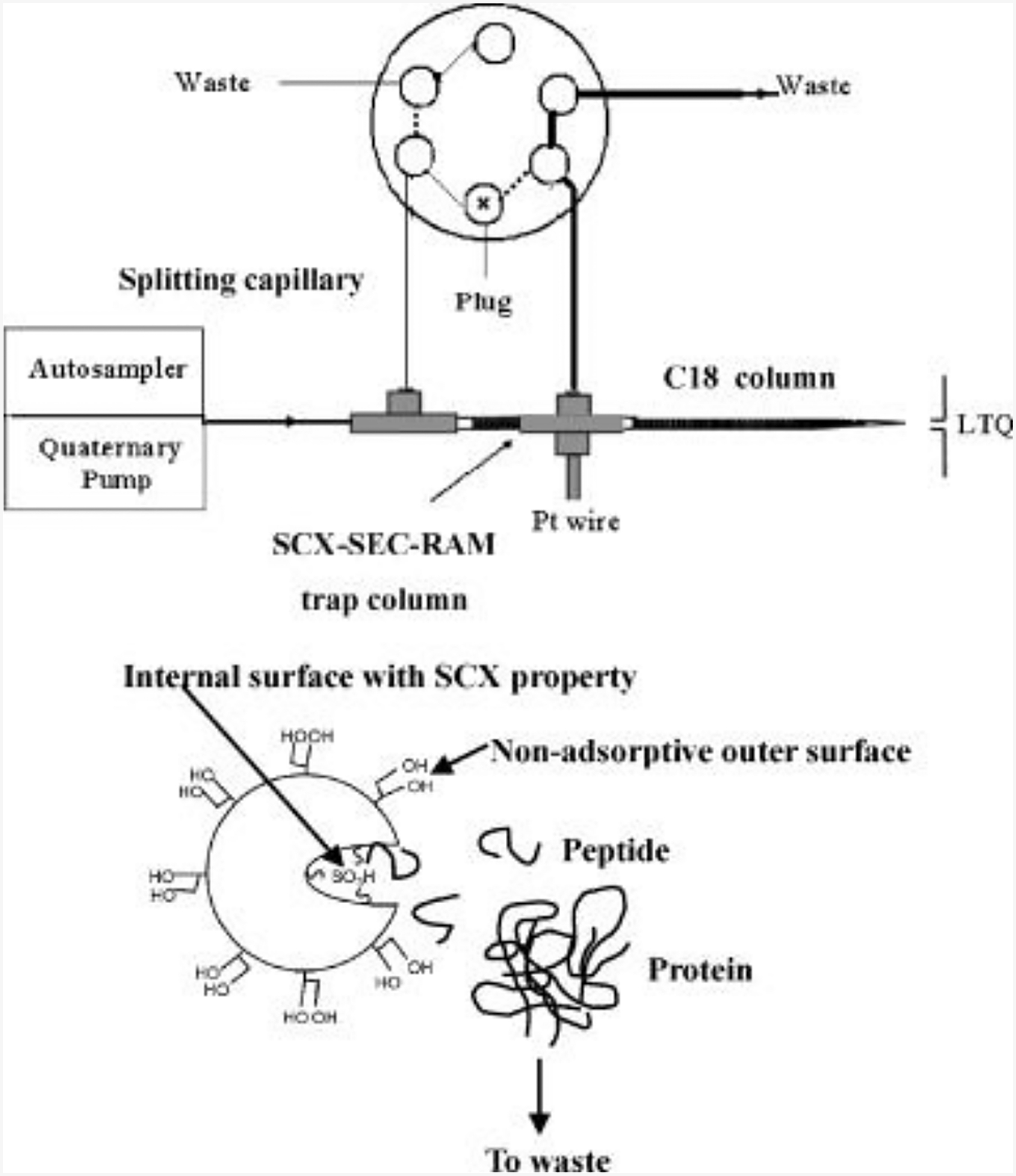
Schematic representation of the set up for the on-line extraction system and chromatographic principles of the bifunctional SCX/SEC-RAM for selective extraction. Solid line represents position for loading sample and dashed line represents position for separation. Adapted from (L. Hu et al., 2009) with permission.
2.5. Alternative Strategy - Magnetic beads
Magnetic bead-based technique is used in peptidome analysis due to its operation simplicity and high selectivity in peptide extraction (Safarik & Safarikova, 2004; Villanueva et al., 2004), especially when conjugated with antibodies (Whiteaker et al., 2007). Magnetic particles usually have affinity ligands or ion-exchange groups which interacts with isolated analytes. They can be directly mixed with crude samples followed by a period of incubation time for targeted compounds to bind with magnetic particles. The complex can be isolated from the sample using magnetic separators and finally the targeted molecules are eluted for further analysis. This effective peptide extraction method was also applied in tissue imaging using mass spectrometry, indicating that more peptides were detected with no compromise in localization information (T. Andrews, B. Skube, & B. Hummon, 2018).
Among many different peptide extraction approaches, each has its pros and cons. In general, non-specific methods, like organic precipitation and ultrafiltration, recovering peptides with no bias, are suitable for comprehensive studies. Methods with selectivity over certain structure or distinct characteristics of target analyte results in more efficient peptide extraction. However, there is no overall gold standard for method selection. Decisions on a single extraction method or a combination of multiple methods mainly depend on the aim of the analysis.
3. Sample Preparation — Glycopeptide Purification and Enrichment
Endogenous glycosylated neuropeptides are often present at low abundance compared to their non-glycosylated counterparts and exhibit a wide dynamic range in biological samples. In our protocol, an enrichment-free method is utilized, which is slightly different from other approaches where glycopeptides are often isolated and enriched via molecular interactions with other materials (Fanayan, Hincapie, & Hancock, 2012; Palmisano, Lendal, & Larsen, 2011; Xu et al., 2009; L. Yu, Li, Guo, Zhang, & Liang, 2009). Those methods can be generally catalogued based on how the glycosylated peptides are recognized and bound, such as interactions based on physicochemical properties of peptides or the specific recognition of the glycans attached to peptide backbones (C.-C. Chen et al., 2014). We first describe high pH fractionation and C18 cleanup workflow for upcoming glycosylated neuropeptide analysis with hybrid MS method and then discuss various efficient enrichment strategies for readers’ reference to select appropriate methods and address their specific experimental needs accordingly.
3.1. Purification with High pH Offline Fractionation
Glycopeptide samples from extraction and enrichment steps usually contain various salts, which requires further cleaning procedures. Common desalting steps performed on pipette tips, cartridges or HPLC column, depending on the loading amount and the consideration of sample loss. When handling relatively large tissue amount (>10 pieces) neuropeptide extracts, it is beneficial to first use offline LC to further separate neuropeptides and reduce sample complexity before MS analysis. As this separation step also helps desalt, the collected fractions are ready for MS characterization.
3.1.1. Materials and Mobile Phase Recipes
Phenomenex Kinetex 5u EVO C18 column (150 mm × 2.1 mm, 100 A, part number 00F-4633-AN)
Mobile phase A: 10 mM ammonium formate in H2O, pH = 10
Mobile phase B: 10 mM ammonium formate in ACN/H2O (90/10, v/v), pH =10
Fraction collector
Speed-vacuum sample dry system
3.1.2. Neuropeptide Separation and Purification
Attach a C18 column to HPLC system. Set up column temperature (30 °C for the column we used). Once temperature is stable, set up the flow rate (0.2 mL/min for the column we used). Allow sufficient equilibration of the column (10 times of column volume with mobile phase A).
Set up the sequence. Run a blank sample before real sample analysis to ensure instrument and column are functioning properly.
Resuspend the neuropeptide sample from Section 2.3.2 in 100 μL mobile phase A and inject onto the LC system.
Run the following gradient: 0–3 min, 1% mobile phase B; 3–50 min, 1–35% mobile phase B; 54–58 min, 60–70% mobile phase B; 59–74 min, 100% mobile phase B; 74.5–90 min, 1% mobile phase B. A typical elution profile is shown in Fig. 3.
Set up the fraction collector for sample collection in 2 min intervals from 6 to 62 min.
Combine the collected fractions into 4~5 centrifuge tubes and dry down under vacuum for solvent removal. Store the dried samples in a −80 °C freezer until MS analysis.
For LC-ESI-MS analysis, resuspend sample into 10 μL 0.1% FA H2O.
Figure 3.
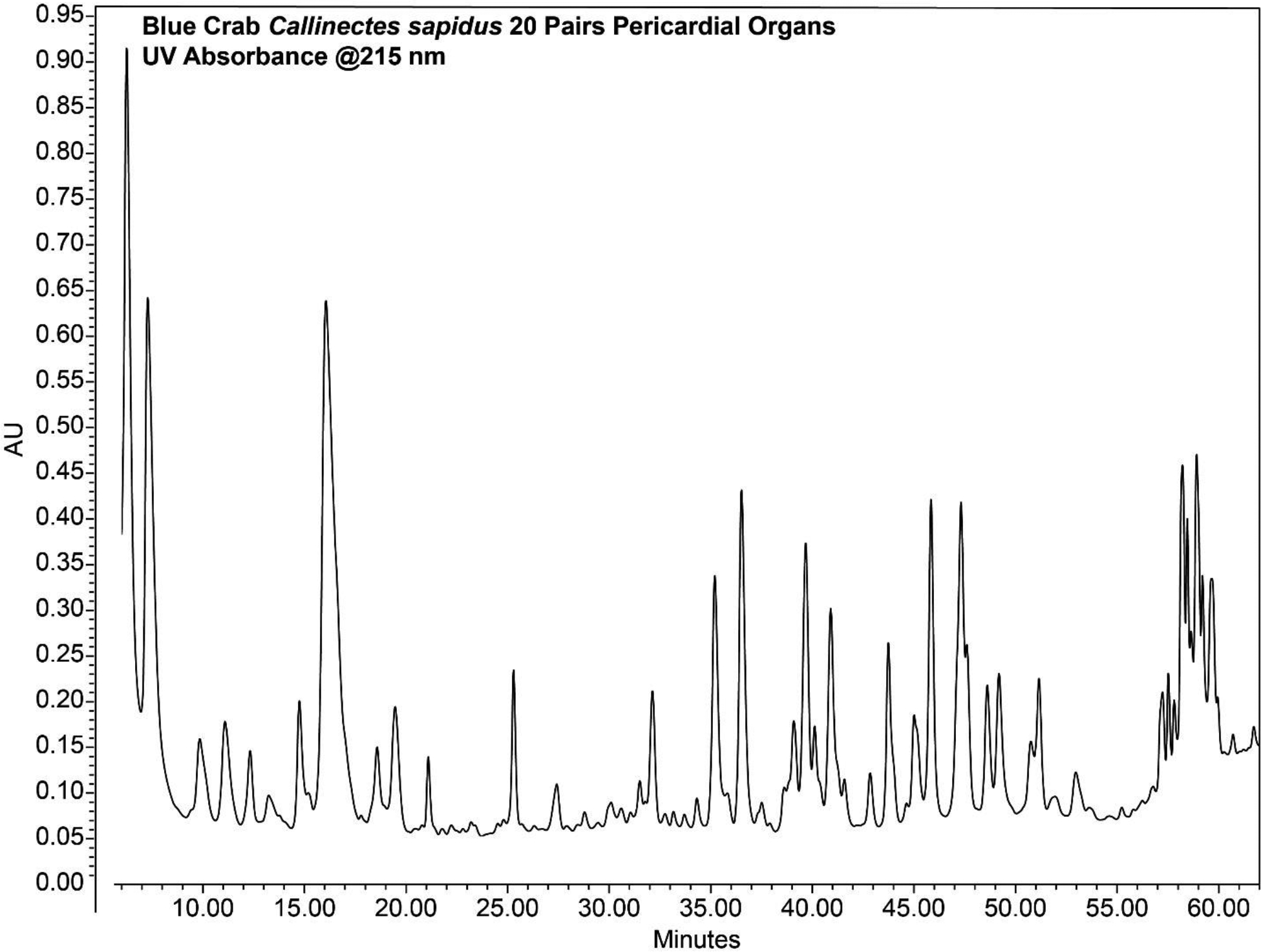
Chromatogram of crustacean neuropeptides separated by offline high pH fractionation.
3.2. Purification with C18 Ziptip Desalting
While high pH fractionation reduces sample complexity, it also causes sample loss during separation. For smaller amount (<10 pieces) of crustacean tissue neuropeptide extract, we prefer using C18 Ziptip to remove salt before MS analysis with no need of further separation, as sample complexity is not the major concern.
3.2.1. Materials and Buffer Recipes
0.1% trifluoroacetic acid (TFA)
Acetonitrile (ACN)
Elution buffer 1: ACN/H2O, 50/50, v/v
Elution buffer 2: ACN/H2O, 75/25, v/v
Agilent C18 Ziptip, 100 μL volume
Speed-vacuum sample dry system
3.2.2. Neuropeptide Desalting
Aspirate the Ziptip with 100 μL ACN and dispense to waste. Repeat 2 times.
Aspirate the Ziptip with 100 μL 0.1% TFA and dispense to waste. Repeat 3 times.
Resuspend neuropeptide sample from Section 2.3.2 in 100 μL 0.1% TFA. Load the sample onto C18 Ziptip binding material by aspirating the resuspended extract for 10 times.
Load 100 μL 0.1% TFA into Ziptip, aspirate and dispense to waste. Repeat 3 times.
Elute neuropeptides with 100 μL ACN/H2O (50/50, v/v) by aspirating for 10 times
Elute neuropeptides with 100 μL ACN/H2O (75/25, v/v) by aspirating for 10 times
Combine the elution and dry down sample under vacuum.
For LC-ESI-MS analysis, resuspend sample into 10 μL 0.1% FA H2O.
3.3. Enrichment with Hydrophilic Interaction Chromatography (HILIC)
The term of hydrophilic interaction chromatography (HILIC), as a variation of normal phase-HPLC, was first proposed and described in 1990 (Alpert, 1990). In HILIC separation, the higher polarity of stationary phase strongly retains hydrophilic analytes and the less polar mobile phase, which usually consists of 50–70% organic solvent, elutes the retained molecules in an order from least hydrophilic to the most hydrophilic (Buszewski & Noga, 2012). Due to the sugar side chains attached, glycopeptides usually exhibit higher hydrophilicity than their counterparts without glycosylation, therefore, they can be fractionated and collected at different retention times (P. G. Wang, He, & He, 2011). The hydrophilicity difference between glycosylated and non-glycosylated peptides can be exaggerated and the overlap in chromatography will diminish with the addition of ion pairing reagents (Ding, Hill, & Kelly, 2007; Furuki & Toyo’oka, 2017; Palmisano et al., 2010). Different types of HILIC methods have been reported using a wide range of solid phase materials, including silica particles (Wan et al., 2011), cellulose (Ohta, Kameda, Matsumoto, & Kawasaki, 2017; Snovida, Bodnar, Viner, Saba, & Perreault, 2010), polysaccharide-based (Xiong et al., 2013), functional group based, such as amide (Palmisano et al., 2012) and amine (Kuo, Wu, Hsiao, & Khoo, 2012), even just simple cotton (Selman, Hemayatkar, Deelder, & Wuhrer, 2011). A recently emerged and largely adopted method is ZIC-HILIC, functioned by sulfobetaine (Dedvisitsakul et al., 2014; Pohlentz, Marx, & Mormann, 2016; Zhao et al., 2017). It has been demonstrated to selectively separate sialylated N-glycan glycopeptides (Takegawa et al., 2006). The composition of mobile phase can largely alter the separation mechanism and selectivity as well (Alagesan, Khilji, & Kolarich, 2017). Electrostatic repulsion hydrophilic interaction chromatography (ERLIC), a combination of HILIC and ion-exchange chromatography, was proposed in 2008 (Alpert, 2008). The high portion organic solvents and the ion-exchange stationary phase enable the retention of both the same and opposite charged analytes as the ion-exchange resins. Glycopeptides with noncharged glycans are mainly subject to hydrophilic interaction, while for those charged glycans with sialic acid, the retention also depends on the charge-based repulsion forces. The sequential HILIC-ERLIC enrichment provided complementary identification of glycopeptides to HILIC-only method (Zacharias et al., 2016). In addition to the enrichment of glycopeptides from their non-glycopeptide counterparts, the HILIC approach is also capable to differentiate the isomeric glycopeptides, with the same backbone but various sugar moieties (Y. Huang, Nie, Boyes, & Orlando, 2016; Takegawa et al., 2006). Improvements in HILIC technique include the combination with other enrichment methods, such as lectin affinity capture (C. Ma et al., 2013) and titanium dioxide (Palmisano et al., 2010) to increase selectivity and manufacture of microcolumns (Boersema, Mohammed, & Heck, 2008; Dedvisitsakul et al., 2014) and microtips (J. Jiang et al., 2014; Selman et al., 2011; Vreeker et al., 2018) to decrease sample loss, especially for low abundance endogenous peptides (Kawahara, Saad, Angeli, & Palmisano, 2016). Most recently, HILIC probes incorporated with magnetic mesoporous materials were successfully applied to capturing endogenous glycopeptides. A zwitterionic hydrophilic mesoporous silica material (FSAu@mSiO2@L-Cys) was synthesized and the pore size was designed to enrich endogenous glycopeptides with large-size proteins excluded (Z. Wang, Wu, Chen, Sun, & Deng, 2018). 40 endogenous glycopeptides were identified from 10 μl human saliva, which was for the first time used as biological sample for endogenous glycopeptidome analysis. Shortly after, the same group developed another hydrophilic probe in mesoporous pore (FE3O4@mSiO2@G6P) with an even smaller pore size. From 10 μl healthy and gastric carcinoma human saliva, a total of 39 and 25 endogenous glycopeptides were detected (Y. Li, Deng, & Sun, 2018). The total number of endogenous glycopeptides found in both studies were comparable (40 and 39 from the same volume of healthy saliva sample), however, there was only one peptide appeared to be overlapped. This could probably be explained by the dynamic change of peptide expression or the different affinity property of each mesoporous material. However, it also reveals that poor reproducibility could present significant barrier in glycosylated neuropeptides studies.
3.3.1. Materials and Buffer Recipes
ZIC-HILIC cartridge.
80% ACN (v/v) 1% TFA (v/v).
0.1% TFA.
Speed-vacuum sample dry system.
3.3.2. Peptide Extraction-ZIC-HILIC Cartridge
Add 1ml 1% TFA in water to balance the cartridge. Repeat for 3 times.
Add 1ml 80% ACN 1%TFA (v/v) to equilibrate the cartridge and repeat for 3 times.
Dissolve sample in 80% ACN 1% TFA (v/v).
Load sample to cartridge and wash with 1ml 80% ACN (v/v) 1% TFA (v/v) for 3 times.
Elute glycopeptide into 0.1% TFA.
Evaporate the solvent using speedvac and store the dried sample at −80 °C.
3.4. Enrichment with Lectin Affinity Chromatography (LAC)
Lectin affinity chromatography (LAC), is another extensively used method in glycopeptide enrichment (F. Zhu, Clemmer, & Trinidad, 2016). Different from HILIC isolating glycopeptide based on their hydrophobicity, lectins are proteins which specifically bind with carbohydrate groups. They recognize and bind to glycans through a combination of hydrogen binding to sugar hydroxyl groups and the van der Waals interaction between sugars and amino acid side chains (Weis & Drickamer, 1996). Therefore, the binding selectivity of lectins to glycopeptides is highly diverse and depends on the structures of both lectin and the glycans. There are at least 160 known species of lectins and more than 60 of them are commercially available. Concanavalin A (ConA), the most widely used lectin, has a high affinity in binding the high mannose core in N-glycans (Y. Liu et al., 2016; Ohyama, Kasai, Nomoto, & Inoue, 1985). Wheat germ agglutinin (WGA) tend to specifically binds to sialic acid and N-acetyl-glucosamine (Bhavanandan & Katlic, 1979; Nagata & Burger, 1974; K. Yamamoto, Tsuji, Matsumoto, & Osawa, 1981). Aleuria aurantia lectin (AAL) is used to recognize α1,6-fucosyl residues (Matsumura et al., 2007; Yamashita, Kochibe, Ohkura, Ueda, & Kobata, 1985). However, some lectins have relative narrow specificity and weak affinity to unique glycans. L-phytohemagglytinin (L-PHA) usually targets the structure of β−1,6-GlcNAc moiety of N-glycans (Abbott et al., 2008; Ahn et al., 2009). Jacalin (JAC) is a seed lectin from jackfruits, which specifically binds with Galβ1–3GalNAc and O-linked glycans (Bourne et al., 2002; Hortin, 1990; Tachibana et al., 2006). These lectins with less robust binding with targeted sugar moieties may result in low recovery and sample loss. A serial lectin affinity chromatography (SLAC) was devised to avoid the coverage limit given by a single lectin (Cummings & Kornfeld, 1982). By applying several lectin enrichment strategies in succession, different glycosylation patterns can be better covered, at the same time, effectively separated (Harada et al., 1987; Lehoux & Ju, 2017). SLAC offers more flexibility in the design of enrichment workflow with some specific goals. For example, in order to identify peptides with O-glycosylation in human serum, N-linked high mannose glycopeptides were removed in the first step by ConA. The “cleaned” sample was then applied to the Jacalin column for exclusive binding to O-glycosylated peptides, which yield an improved selection of broad identification of O-linked glycopeptides compared to that result from Jacalin enrichment alone (Durham & Regnier, 2006). In 2004, a more powerful enrichment method was designed as multi-lectin affinity column (MLAC), which contains several different lectins in the same column (Z. Yang & Hancock, 2004). It has been demonstrated that MLAC can produce reproducible results, higher dynamic ranges and enable more comprehensive capture of glycopeptides than a single-lectin column (Totten et al., 2018; Totten, Kullolli, & Pitteri, 2017; Z. Yang, Harris, Palmer-Toy, & Hancock, 2006). This method was optimized to a high-performance multi-lectin affinity chromatography (HP-MLAC) by incorporating commercial ConA, WGA and JAC to a HPLC column (Kullolli, Hancock, & Hincapie, 2008). The automated HPLC platform integrated with inline MLAC enrichment was shown to generate high recovery, high throughput for clinical studies and disease related biomarker analysis (Kullolli, Hancock, & Hincapie, 2010; Z. Zeng et al., 2011).
3.4.1. Materials and Buffer Recipes
ConA cartridge.
Äkta Purifier (Amersham Biosciences, Piscattaway, NJ) equipped with a UV detector set at 280 nm.
Loading buffer: 10mM TRIS buffer, 500 mM NaCl, 1mM CaCl2, 1mM MnCl2, 0.02% sodium azide adjust pH to 7.4 using HCl.
Elution Buffer: 10mM TRIS buffer, 500 mM NaCl, 1mM CaCl2, 1mM MnCl2, 0.02% sodium azide, 100 mM Methylmannose, adjust pH to 7.4 using HCl.
Speed-vacuum sample dry system.
3.4.2. Peptide Extraction-ConA Microcolumn
Equilibrate the cartridge with loading buffer at 100μL/min with 10 column volumes.
Reduce flow rate to 50 μL/min.
Dissolve 50 μg sample in loading buffer (injection volume depends on the volume held by the sample loop).
Inject sample into the sample loop and load onto the microcolumn.
Wash the microcolumn with 5–10 column volumes or until the A280 returns to baseline value.
Elute peptides using elution buffer at step-gradient 0–100% over 10 column volumes.
Evaporate the solvent using Speedvac and store the dried sample at −80 °C.
3.5. Enrichment with Metal Ion Affinity
3.5.1. Titanium
Enrichment and separation based on the metal ion affinity are extensively adopted in glycopeptide studies, benefiting from the critical interaction between the spare pair electrons of oxygen atoms in carboxylate or hydroxyl groups and the unoccupied orbitals. The most representative example is titanium oxide (TiO2), which is also a tool for phosphopeptide enrichment (Thingholm, Jørgensen, Jensen, & Larsen, 2006; Thingholm & Larsen, 2016; B. Zhang et al., 2011). It is an ideal choice when simultaneous analysis of phosphopeptides and glycopeptides are desired (Melo-Braga, Ibáñez-Vea, Larsen, & Kulej, 2015; Palmisano et al., 2012). For studies focusing on glycosylation, the primary interference would be the competitive binding with phosphopeptides. Therefore, the phosphatase pretreatment is necessary prior to the glycopeptide purification (Larsen, Jensen, Jakobsen, & Heegaard, 2007; Palmisano et al., 2010). TiO2 is usually considered to bind sialic acid-containing glycopeptides preferentially (Larsen et al., 2007; Palmisano et al., 2010); however, it also interacts with peptides containing neutral glycans (Yan et al., 2010) based on the understanding of different enrichment mechanisms of TiO2 towards glycopeptides with different solvent compositions (Sheng et al., 2013).
3.5.1.1. Materials and Buffer Recipes
Alkaline phosphatase.
Denaturing buffer: 6M urea, 2M thiourea and 50 mM NH4HCO3.
TiO2 beads.
TiO2 loading buffer: 1 M glycolic acid in ACN/H2O, 80/20, v/v; 5% TFA, v/v.
TiO2 washing buffer 1: ACN/H2O, 80/20, v/v; 1% TFA, v/v.
TiO2 washing buffer 2: ACN/H2O, 20/80, v/v; 0.1% TFA, v/v.
TiO2 elution buffer: NH4OH/H2O, 25/75, v/v, pH 11.3.
ACN/H2O, 50/50, v/v.
Vortex mixture.
Centrifuge.
Speed-vacuum sample dry system.
3.5.1.2. Peptide extraction-Titanium Dioxide
Dissolve extracted peptides in 100 μL denaturing buffer.
Treat the peptides with 20 U alkaline phosphatase at 30 °C for 2 h to avoid the co-purification of phosphorylated peptides in the following steps.
Prepare TiO2 beads (~10mg) in 100 μL ACN.
Mix 0.6 mg of TiO2 beads with per 100 μg peptides in TiO2 loading buffer. Then incubate 30 min on a mixer.
Centrifuge the mixture for 1 min × 1000g at room temperature and remove supernatant.
Add 500 μL TiO2 loading buffer to the beads and incubate on the mixer for 1 min.
Centrifuge the mixture for 1 min × 1000g at room temperature and remove supernatant.
Add 500 μL TiO2 washing buffer 1 to the beads, vortex 15 s.
Centrifuge for 1 min × 1000g and remove supernatant.
Add 500 μL TiO2 washing buffer 2 to the beads.
Dry the mixture for 5–10 min in SpeedVac.
Incubate the beads with 200 μL TiO2 elution buffer for 20 min on the mixer to elute the sialoglycopeptides.
Remove the beads and wash with 10 μL 50/50 ACN/H2O and repeat twice.
Combine supernatant from steps 12 and 13for future analysis.
3.5.2. Zirconium
Zirconium dioxide (ZrO2) is also commonly used in phosphopeptide enrichment (Dong, Zhou, Wu, Ye, & Zou, 2007; Zhou et al., 2007). Zr belongs to the same group in periodical table as titanium, therefore it has similar electron configuration; thus, similar chemical properties. It is inevitable to explore the advantage of zirconia in glycopeptide enrichment. A method of using zirconia layer coated mesoporous silica microspheres (Fig. 4) was proposed to specifically enrich glycopeptides (Wan et al., 2011). It has been tested on trypsin digested IgG, RNase B and α-casein. Under different solvent compositions, nonglycopeptides and glycopeptides were eluted in different fractions to minimize signal suppression, meanwhile, the phosphopeptides are strongly bound by the Lewis acid-basic action without being eluted to the same fraction as glycopeptides. This method resulted in better site-specific identification and higher selectivity of glycopeptides out of nonglycopeptides, compared to commercial sepharose and silica microspheres. In a more recent study, Zr-magnetic metal-organic framework (MOF) was modified onto the polydopamine (PDA)-coated magnetic microsphere (Fe3O4@PDA@UiO-66-NH2) to build up a “one for two” hydrophilic magnetic amino-functionalized metal-organic framework (Xie & Deng, 2017). Owing to the large surface area of MOF, the strong magnetic responsiveness given by the Fe3O4 core, the hydrophilic interaction of Zr towards glycopeptides and its binding with phosphopeptides, this route exhibited excellent performance in identification of glyco- and phosphopeptides with high sensitivity and selectivity.
Figure 4.
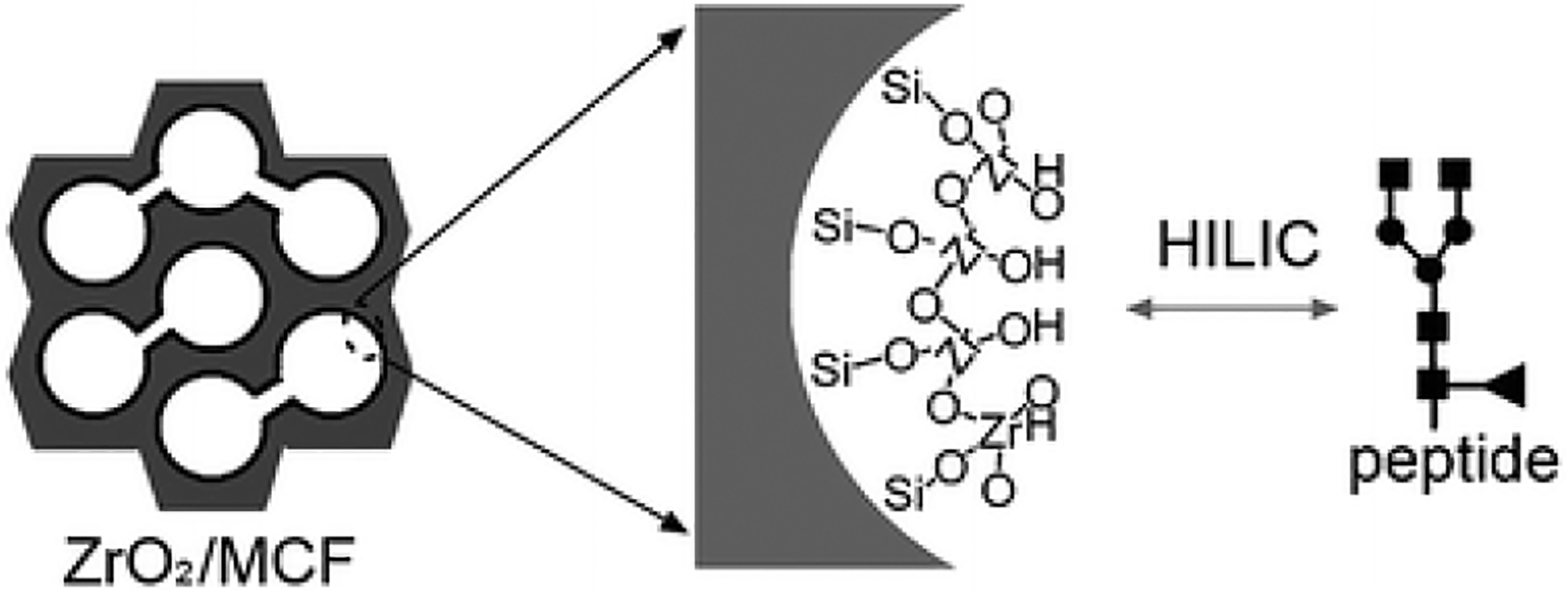
Illustration of glycopeptide enrichment ability with ZrO2/MCF mesoporous microspheres, Adapted from (Wan et al., 2011) with permission.
3.5.3. Silver
Silver nanoparticles (Ag NPs) have been reported to be applied to glycopeptide separation and enrichment (B. Jiang, Wu, Zhang, & Zhang, 2017; W.-F. Ma et al., 2012; Y. Ma, Wang, Zhou, & Zhang, 2019) (Fig. 5). They proposed that the strong affinity between glycopeptides and Ag NPs was the multivalent interactions of hydroxyl groups and the silver surface. The affinity of glycopeptides to Ag NPs is comparable to that of water molecules. Therefore, the binding and elution in glycopeptide enrichment process were realized by adjusting the water content in the buffer. Ag NPs were shown to enable trace level glycopeptide enrichment at high recovery rate.
Figure 5.
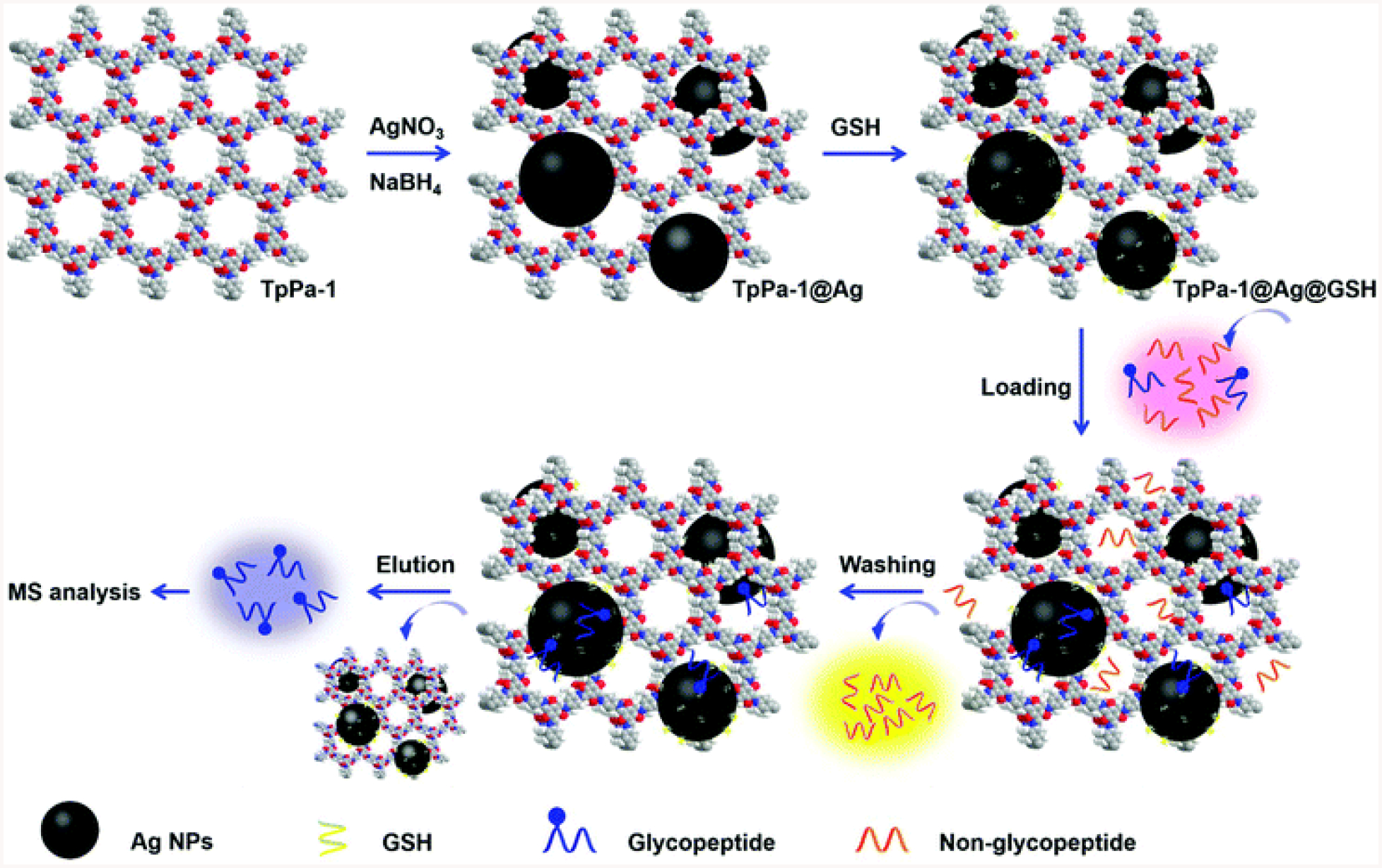
The procedure for the post-synthetic modification of TpPa-1 and the enrichment and detection of the N-linked glycopeptides based on TpPa-1@Ag@GSH. Adapted from (Y. Ma et al., 2019) with permission.
3.6. Covalent Bond Interaction
3.6.1. Boronic acid chemistry
Boronic acid can enrich and isolate a wide range of N- and O-linked glycopeptides with no bias because it reacts with cis-diol-containing saccharides, like mannose, galactose and glucose and forming stable heterocyclic diesters (Sparbier, Koch, Kessler, Wenzel, & Kostrzewa, 2005). The covalent bonding between boronic acid and cis-diol group usually occur under basic aqueous or non-aqueous condition. The covalent linkage is reversible and the cyclic esters dissociate in acidic environment with no alteration to the glycan structures (X. Wang, Xia, & Liu, 2013). In addition to the advantages of high tolerance to glycan structures (branches or linear), saccharide types, as well as monosaccharide modifications, boronic acid can be easily incorporated onto various solid supports. Diboronic acid was grafted onto the surface of ordered mesoporous silica matrix (Xu et al., 2009). The universal covalent bonding of boronic acid and the high surface area of mesoporous silica enabled 2-orders of magnitude improvement in glycopeptide detection limit. Boronic acid-conjugated nanoparticles were designed and have been reported to enrich trace level glycopeptide from complex biological samples rapidly and efficiently (D. Li, Xia, & Wang, 2018; Tang et al., 2009; X. Zhang, Wang, He, Chen, & Zhang, 2015). To enable micro-scale analysis of glycopeptides, boronate affinity monolith columns were developed (M. Chen, Lu, Ma, Guo, & Feng, 2009; Jin, Zhang, Yang, Dai, & Zhou, 2018; H. Li & Liu, 2012). For example, phenylboronic acid (PBA) bound to SiO2 microsphere was synthesized and packed into columns to enrich glycopeptides in HILIC mode (Jianying Chen et al., 2017). This method exhibited high selectivity for both neutral and acidic glycopeptides, with high resistance of interference from 100-fold (in molar) excess of bovine serum albumin digests. MOF with abundant boronic acid sites was designed (S. Li et al., 2018) and has been reported to enable detection of 209 N-glycosylated peptides from human serum digests with enhanced improvement (0.5 fmol/μl) and selectivity (1:100) (Xie, Liu, Li, & Deng, 2018). For endogenous peptidome study, mesoporous silica MCM-41, with pore size as small as 2–3nm, was functionalized with boronic acid groups at inner surface, so that only peptides with molecular weights smaller than 12 kDa were allowed to enter and then glycopeptides were selectively captured by the grafted boronic acid groups (L. Liu et al., 2012). As a result, fifteen unique glycosylation sites mapped to 15 different endogenous glycopeptides were identified from the rat serum.
3.6.1.1. Materials and buffer recipes
Boronic acid conjugated magnetic beads.
Binding buffer: 200mM ammomium acetate buffer (pH=10).
Elution buffer: ACN: H2O: TFA = 50:49:1.
Incubation shaker.
Centrifuge.
3.6.1.2. Peptide extraction-Boronic acid magnetic beads
Store boronic acid (BA) beads in ethanol (6mM).
Wash BA beads with 2ml of binding buffer. Repeat 3 times.
Resuspend BA beads in 500 μL of binding buffer.
Transfer the beads into sample tubes.
Incubate at 37 °C for 1 h with appropriate shaking.
Centrifuge the mixture at 900g for 3min.
Collect the supernatant.
Add 500 μL of binding buffer to the remaining bead.
Repeat step 6–8 to wash the beads for 5 times.
Elute the glycopeptide using 500 μL elution buffer. Repeat 3 times and combine the elute.
3.6.2. Hydrazine capture chemistry
Hydrazine capture of glycopeptides, relying on the covalent bond formation as well, is a selective isolation and enrichment method first introduced in 2003 (H. Zhang, Li, Martin, & Aebersold, 2003). The cis-diol groups in carbohydrates were oxidized to aldehydes by NaIO4 and further coupled with hydrazide groups immobilized on solid support materials via covalent bonds (Fig. 6). Nonglycosylated peptides were removed and the N-linked glycopeptides were released by PNGase F treatment. The choice of solid support can affect the enrichment efficiency and preference (Sajid, Jabeen, Hussain, Ashiq, & Najam-Ul-Haq, 2017). Soluble nanopolymers was modified by hydrazide and followed by filter-aided sample preparation (FASP), resulting in 158 unique glycopeptides mapped to 60 glycoproteins from 5μl human serum (L. Zhang et al., 2014). Magnetic nanoparticles functionalized with multiple hydrazide groups offer a large capacity of binding sites, hence, enabled enrichment and isolation of glycopeptides from non-glycosylated counterparts at even 100-fold greater abundance. The application of this method in analysis of mouse liver concluded in 511 unique glycopeptides from 372 different glycoproteins (Qichen Cao et al., 2013). Hydrazide resin has also been incorporated with pipet tips to achieve rapid, automated, high throughput isolation of N-linked glycopeptides (Jing Chen, Shah, & Zhang, 2013). Although the covalent bonding between hydrazide and carbohydrates are of no discrimination, the recovery of glycopeptides highly depends on the enzymatic release step. For example, the PNGase F detachment efficiency in the original protocol described above is limited by the inherent steric hindrance effect. A hydroxylamine assisted PNGase F deglycosylation (HAPD) method was developed to release glycopeptides through cleavage at hydrazine bonds by transamination followed by the PNGase F treatment (J. Huang et al., 2015). Using the HAPD strategy, the detection of N-glycosylation sites was increased by 27% and the identification of N-terminus glycosylated peptides was improved by 5-fold compared to the conventional methods. Another concern in the application of hydrazide capture in O-linked glycopeptide isolation is limited by the lack of an enzyme to specifically cleave O-linked glycans and release the peptides. Also, according to the original method, glycan structure information is not available since the glycans remained on hydrazide resin. These concerns were partially solved by a method proposed to enrich specifically sialic acid-containing peptides, involving both N- and O-linked glycosylation (Nilsson et al., 2009). Sialic acids were selectively oxidized by mild periodate oxidation, followed by hydrazide capture. After the non-glycosylated peptides being removed, the captured glycopeptides were released by acid hydrolysis rather than PNGase F; because the terminal sialic acid and the penultimate monosaccharide are vulnerable to mild acid hydrolysis and the O-linked glycopeptides can be collected in this way. They optimized this method by applying PNGase F to reduce the interference from N-linked glycopeptides prior to the sialic acid-capture-and-release protocol so that selective characterization of O-glycopeptides was largely improved in the analysis of human cerebrospinal fluid (Halim, Rüetschi, Larson, & Nilsson, 2013). They also detected endogenous neuropeptides containing three different glycopeptide stretches from ProSAAS which were cleaved from the proprotein by convertase. The identical endogenous neuropeptides have also been investigated in previous neuropeptidome study (Gupta et al., 2010) and were found to carry sialylated O-glycans as PTMs in a CSF peptidedome study (Zougman et al., 2008). However, this strategy is still subject to the limitations that information of sialylation and nonsialylation peptides is absent. It was reported that the sialylation information can be preserved by using ice-cold 1M hydrochloride to release the sialic acid-containing peptide from hydrazide beads, leaving the sialic acid intact (Kurogochi et al., 2010).
Figure 6.
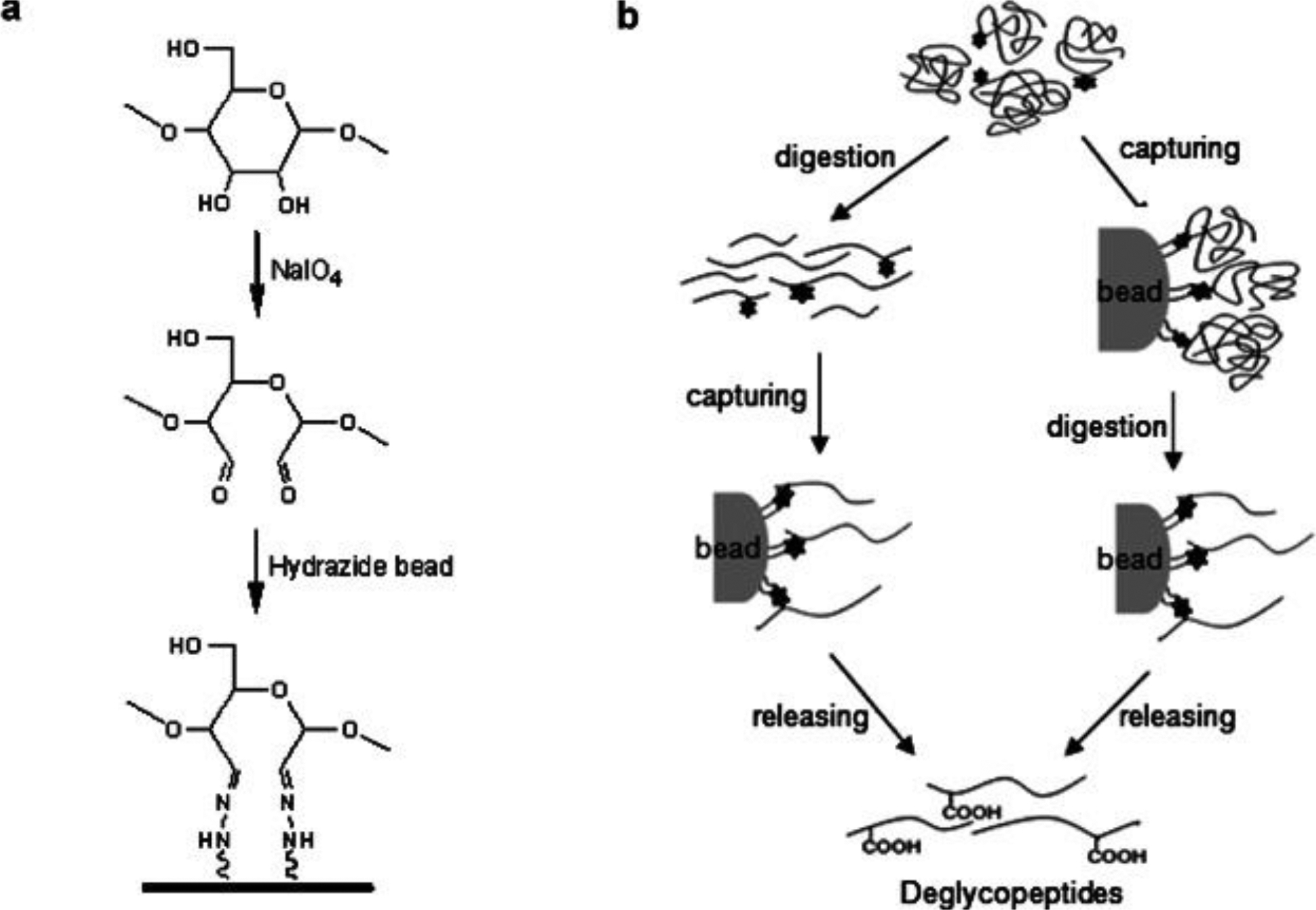
Graphical features in hydrazide-capturing method for glycoproteome enrichment. Adapted from (Ahn, Kim, & Yoo, 2015) with permission.
3.6.2.1. Materials and buffer recipes
Hydrazine resin.
Oxidation buffer (100 mM NaAc, 150 mM NaCl, pH = 5.5).
Methanol.
NaIO4.
Na2SO4.
ACN/H2O, 80/20. v/v.
NaCl, 1.5M.
NH4HCO3, 10mM, 100 mM.
PNGase F.
3.6.2.2. Peptide extraction-Hydrazine capture
Peptides from serum or tissue extract was dissolved in 100 μL of oxidation buffer.
Add NaIO4 to final concentration of 15 mM.
Incubate the mixture at room temperature in dark for 1 hour.
Quench the reaction by adding Na2SO4 to consume excess NaIO4.
Add hydrazine beads into the reaction mixture.
Incubate overnight at room temperature.
Wash glycopeptide-captured hydrazine beads with 80/20 ACN/H2O, 1.5 M NaCl and 100% methanol sequentially for 3 times.
Add NH4HCO3 buffer containing 500U PNGase F enzyme.
Incubate at 37 °C overnight to release glycans from the glycopeptides to produce deglycosylated peptides. This process leaves a 0.9858 Da mass on the glycosylation site which makes it possible for site-specific identification.
Collect supernatant after enzyme digestion.
Wash the beads with 100 μL of 100 mM NH4HCO3. Repeat twice.
Combine supernatant for further analysis.
To select a proper isolation and enrichment method is the crucial step in a glycopeptidome experiment. The decision usually depends on the aim of the study. Method performing nondiscriminatory separation would be an optimal choice in experiments aiming to obtain comprehensive global glycopeptide identifications. For studies concerning a specific or a rare group of peptides, the lectin affinity-based strategies contribute more precise isolation by choosing the corresponding lectin species. It would be an efficient way to combine methods introduced above, taking advantages of multiple interactions in capturing glycopeptides with unique features. Nonetheless, there will be a trade-off between enrichment efficiency and the sample loss when the multi-step experiment design gets more complicated.
4. Glycopeptide Profiling by MS
Mass spectrometry has grown to be a powerful tool to address challenges in the analysis of complex and low-abundance endogenous glycopeptides. Typical MS strategies consist of various ionization methods and fragmentation methods that would contribute to the depth and the breadth of glycopeptide identification/characterization. Matrix-assisted laser desorption/ionization (MALDI) facilitates simple sample preparation, rapid instrument operation and more straightforward spectral interpretation, which offers benefits in high throughput analysis. Electrospray ionization (ESI), which is more compatible with LC separation system, usually produces multiply charged ions that can be further fragmented to generate tandem mass spectra, delivering more information in peptide backbone sequences, glycan structures and glycosylation sites. As the fragmentation methods continue to develop, MS/MS strategies present more choices and solutions to site-specific glycosylation characterization. Here, in addition to the common MS strategies, we also introduced a novel method, using HCD product ion-triggered EThcD fragmentation, specifically applied to tackle glycosylation occurring on crustacean neuropeptides.
4.1. MALDI-MS
4.1.1. Derivatization approaches
Glycosylated peptides are more hydrophilic due to the attached sugar moieties, which cause reduction in ionization efficiency. The signal suppression of high abundance non-glycosylated peptides will further exacerbate glycopeptide detection. The glycosidic bonds, especially oligosaccharides with sialic acid and fucose, are labile to in-source or post-source dissociation. The preferential loss of sugar moieties during mass measurement is a major problem for MALDI analysis (Nishikaze, 2017). In order to enhance the relative signal intensities of glycopeptides, derivatization methods, such as methyl esterification (Powell & Harvey, 1996), permethylation (Z. Wu et al., 2017) and perbenzolylation (P. Chen, Werner-Zwanziger, Wiesler, Pagel, & Novotny, 1999) have been routinely introduced into sample preparation process (Morelle & Michalski, 2007). The derivatization benefits MALDI MS analysis by stabilizing the labile sialic acid moieties, enabling detection of neutral and acidic glycans in the positive mode and improving the ionization efficiency of derivatives (X. Liu, Qiu, Lee, Chen, & Li, 2010). The stabilization can be accomplished by derivatization of the released glycans (Reiding, Blank, Kuijper, Deelder, & Wuhrer, 2014; Sekiya, Wada, & Tanaka, 2005; Thaysen-Andersen, Mysling, & Højrup, 2009). A microplate-based permethylation kit, which could process up to 96 samples in parallel, was developed to be robotics compatible, high-throughput, and capable to provide linkage-specific analysis of glycans (Shubhakar et al., 2016). Another pathway is to derivatize the glycans without detachment from the intact glycopeptides, allowing simultaneous interpretation for both site-specific glycosylation and peptide sequences. In addition to the derivatization of free glycans, the glycopeptide-level reaction requires more attention in the experimental condition control. For example, the carboxylic acids on the peptide backbones may participate in the derivatization, like ethyl or methyl esterification, causing internal or external reaction and producing heterogeneous products. A dimethylamidation protocol which induced mass difference between α2,3- and α2,6-linked sialic acids, was employed in linkage-specific analysis of sialic acids (de Haan et al., 2015). In addition to reveal accurate structures of the glycans, the informative fragmentation spectra of derivatized glycopeptides enable derivation of peptide backbone sequences as well. This method has been proven to be rapid, simple, reproducible and high-throughput compared to other strategies, like LC-MS, CE-MS and the usage of sialidases.
4.1.2. On-plate enrichment
There also have been efforts combining on-plate enrichment with MALDI MS analysis for fast and selective characterization of glycosylated peptides. The MALDI plate was directly fabricated with boronate functional particles or first modified with metal substrates (e.g. gold) then further functionalized by boronic acid-containing molecules to utilize the large surface area and affinity to glycans and to yield rapid and high-throughput analysis of low-abundance glycosylation in complex samples (Tang et al., 2009; Xu, Zhang, Lu, & Yang, 2010; Y.-C. Liu & Chen, 2017; Zhoufang Zeng, Wang, Guo, Wang, & Lu, 2013). These novel and facile strategies are proven to be effective and versatile with ease of use to profile glycopeptides in complex biological mixtures with minimum sample pre-treatment. When metal gold is used as supporting substrate on MALDI plate, it enhances UV absorption thus benefits desorption and ionization of enriched glycopeptides for improved MS sensitivity.
4.2. ESI-MS
As another soft ionization approach, ESI is a prevalent method to produce multiply charged ions before MS analysis. The coupling of LC and ESI-MS has enabled separation and site-specific characterization of glycosylated peptides simultaneously to unravel biological complexity. The advancements on mass spectrometry fragmentation techniques significantly facilitate the understandings toward glycosylation on peptides via systematic elucidation of peptide backbone, glycan composition and glycosylation site. We present various MS/MS fragmentation approaches utilized to identify glycopeptides, with an emphasis on employing hybrid MS/MS techniques to uncover less studied crustacean glycosylated neuropeptides.
4.2.1. Different MS/MS fragmentation techniques
In tandem MS experiments, multiple MS dissociation approaches (Reiding, Bondt, Franc, & Heck, 2018) can be employed to generate structural fragments of precursor ions (Fig. 7). Commonly applied MS/MS fragmentation processes include electron-induced dissociation (ExD), such as electron-transfer dissociation (ETD) and electron-capture dissociation (ECD), and collision-induced dissociation (CID) whose higher energy version is also known as high-energy collisional dissociation (HCD) in Orbitrap instruments. CID and HCD tandem MS spectra often contain peptide b/y-ion series and monosaccharide ions produced from glycosidic bond cleavage. While in ExD events, the major fragments (c/z-ions) are peptide backbone fragments with intact glycan preserved. Although these tandem MS strategies provide meaningful information of the molecular structures, a fast and thorough characterization of glycosylated peptides can be challenging to achieve due to a lack of full understanding of peptide primary structure, site of glycosylation and glycan elucidation (H. Hu, Khatri, Klein, Leymarie, & Zaia, 2016).
Figure 7.
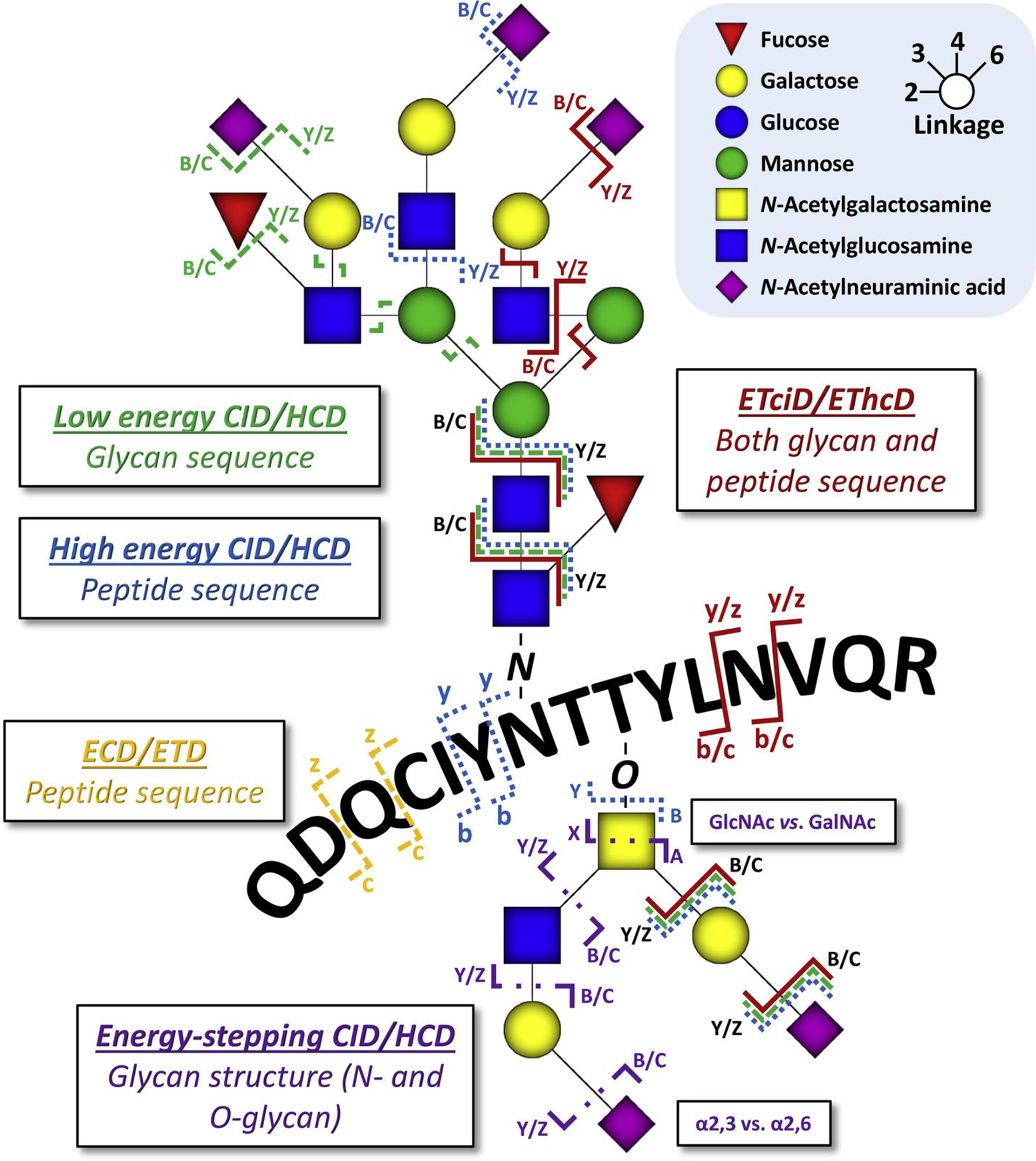
Overview of contemporary glycopeptide fragmentation methods and their preferred sites of action. The displayed peptide sequence matches that of a tryptic N-glycopeptide from alpha-1-acid glycoprotein, whereas the O-glycan has been added for illustrative purposes. Locations of fragmentation are exemplary, and dissociation of glycosidic linkages and peptide bonds can also occur elsewhere on the molecule. The actual observed fragments will depend highly on both the glycan and the peptide in question and the particular energy deposited in the precursor ions. Adapted from (Reiding, Bondt, Franc, & Heck, 2018) with permission.
4.2.2. Integration of MS/MS fragmentation techniques
4.2.2.1. Combination of different MS/MS fragmentation techniques
As various tandem MS dissociation approaches provide different aspects of glycopeptide structure, complementary fragmentation techniques have been utilized to generate comprehensive information and served as effective tools to uncover intact glycopeptides. CID/HCD and ExD have been implemented sequentially into a single online LC-MS experiment (S.-L. Wu, Hühmer, Hao, & Karger, 2007) or in independent LC-MS experiments (Halim, Nilsson, Rüetschi, Hesse, & Larson, 2012; Halim et al., 2012; C. Ma et al., 2016) for the elucidation of glycosylated peptides. CID and HCD have also been integrated to map native glycopeptides through the sequencing of deglycosylated peptides and intact peptides, respectively. (R. Chen, Seebun, Ye, Zou, & Figeys, 2014)
4.2.2.2. Product ion-triggered MS/MS fragmentation techniques
Glycan diagnostic ions produced from HCD scans are beneficial to fast probing the presence of glycosylated peptides and improving instrument duty cycles. In the low m/z region, those diagnostic oxonium ions include HexNAc (m/z 204.09), HexNAcHex (m/z 366.14) and several HexNAc fragmented pieces (m/z 126.06, 138.06, 144.07, 168.07, and 186.08) (Halim et al., 2014). Taking advantage of those signature ions in HCD scans, subsequent MS/MS scans can be selectively triggered to further elucidate glycopeptide structures, with no need of prior knowledge of glycan structures or sample enrichment steps. An HCD-triggered ETD method has been applied to study N-linked glycosylation on ribonuclease B (Lys-C digested) and human immunoglobin G (Trypsin-digested) (Singh, Zampronio, Creese, & Cooper, 2012). This type of method is generally referred as HCD-product dependent-ETD (HCD-pd-ETD) that has been widely applied to elucidate glycopeptide compositions. (Saba, Dutta, Hemenway, & Viner, 2012; S.-W. Wu, Pu, Viner, & Khoo, 2014; Yin et al., 2013)
4.2.2.3. Hybrid MS/MS fragmentation techniques
Recently the advancement of modern MS technologies has enabled several hybrid MS/MS fragmentation approaches to acquire in-depth glycopeptide information in a single tandem MS spectrum. The most appealing strategies include stepping-energy CID, stepped collisional energy HCD, and electron-transfer/higher-energy collision dissociation (EThcD). For instance, stepping-energy CID has been employed to elucidate N-linked and O-linked glycosylation on human C1-inhibitor protein (Stavenhagen et al., 2018). As different HCD collisional energies produce various fragmentation patterns, the utilization of stepped collision energy HCD (normalized collisional energy set to 20%, 30% and 40%) offers a more comprehensive characterization of the N-linked glycopeptides after HILIC enrichment from human serum (H. Yang, Yang, & Sun, 2018). EThcD was first reported by the Heck group for the characterization of phosphosite by incorporating HCD and ETD fragments in one MS/MS spectrum revealing significantly larger amount of fragments. The employment of EThcD in glycosylation studies (Franc, Yang, & Heck, 2017; Khurana et al., 2019; Y. Zhang et al., 2018) has enabled simultaneous acquisition of glycan composition and peptide backbone fragments of the glycosylated peptide precursors. The Li group first reported the optimization of EThcD parameters including tuning the reaction time in ETD scans per precursor charges to improve spectral quality and produce more abundant structural information. The optimized method with improved duty cycle was applied to profile intact N-glycosylated peptides (Fig. 8) and increase our knowledge about in vivo glycoproteomes in various biological systems (Z. Chen, Huang, et al., 2018; Glover et al., 2018; Q. Yu, Wang, et al., 2017; J. Zhu et al., 2018). Subsequently, this method has been further adapted by Yu et al. for an oxonium ion-triggered HCD-pd-EThcD approach to achieve selective and targeted characterization of O-glycosylation on mouse insulin. An EThcD scan was only triggered when prior HCD spectrum detected oxonium ions, in order to save the ETD reaction time and to increase throughput (Q. Yu, Canales, et al., 2017). This approach enabled the discovery of first O-glycosylated insulin peptides in mouse and human pancreatic islets.
Figure 8.
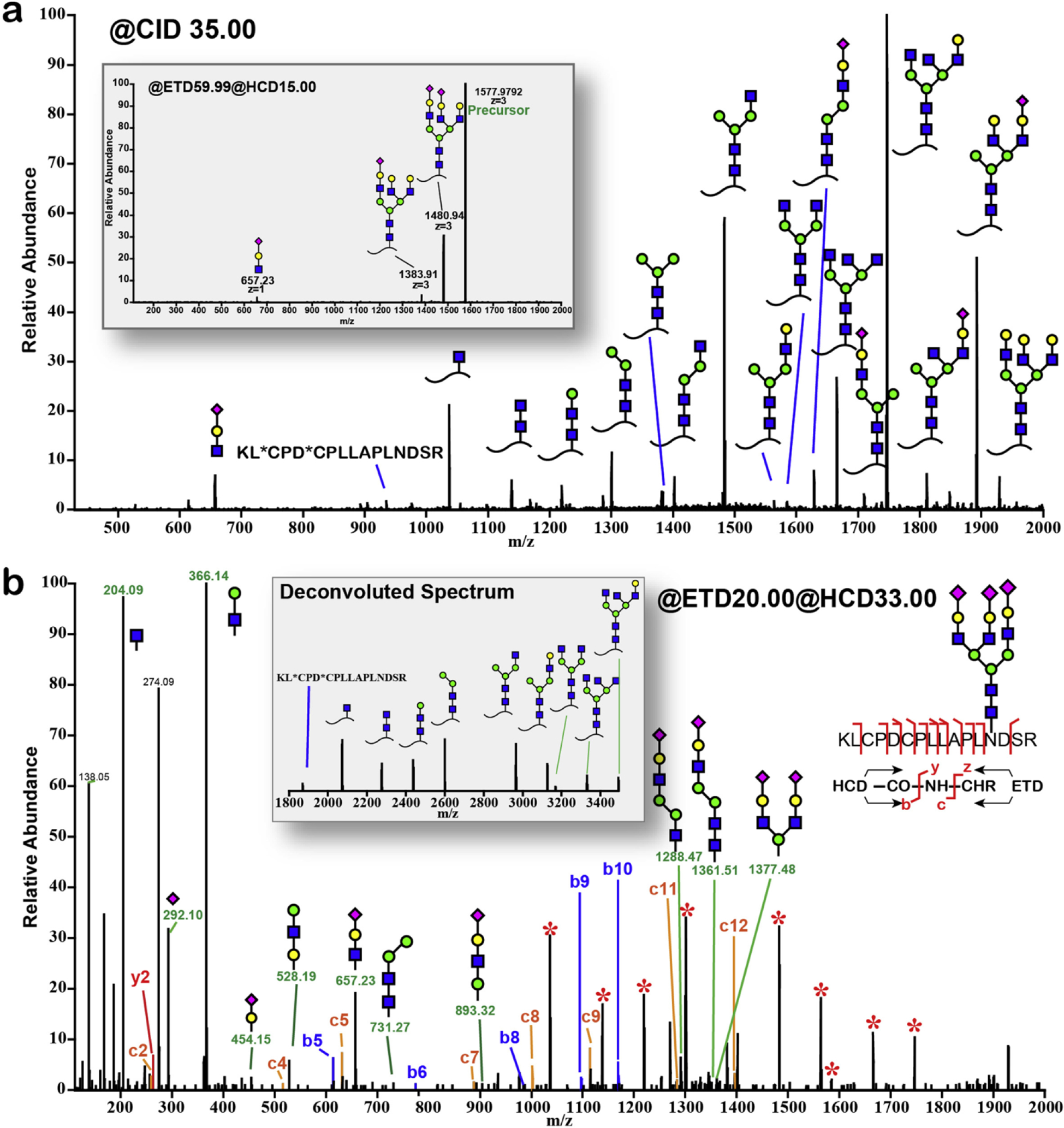
MS/MS of 3+ charge state precursor ion at m/z 1577.9 of bovine fetuin triantennary N-glycopeptide KLCPDCPLLAPLNDSR (AA 126–141). Alternating between CID/ETD/EThcD resulted in different sets of ions. (a) CID and ETD spectra (inset). Asterisk (*) in the peptide sequence indicates carbamidomethylation. (b) EThcD spectrum. Starred peaks (*) in the spectra were deconvoluted and annotated in the inset. Adapted from (Yu, Wang, et al., 2017) with permission.
As prior evidence showed the glycosylation modification on a signaling peptide hormone, insulin, the HCD-pd-EThcD method has been recently applied to study N-linked and O-linked glycosylation of neuropeptides in crustacean nervous system without prior enrichment, to further extend our knowledge of neuromodulation and crustacean peptidome. In addition to all the benefits this HCD-pd-EThcD method offers, this approach is especially beneficial for studies of glycosylation occurring on neuropeptides in crustaceans whose complete neuropeptide database is still missing. Since the HCD scans also produce an overview of peptidome for the sample analyzed, an updated database with novel crustacean neuropeptides can be built and then used in the following data analysis procedures. This approach (Fig. 9) enables the discovery of glycosylation on novel neuropeptides in addition to known neuropeptides with limited amount of neuronal tissue samples (Qinjingwen Cao, Yu, Liu, Chen, & Li, submitted). The detailed MS workflow is shown below:
Figure 9.
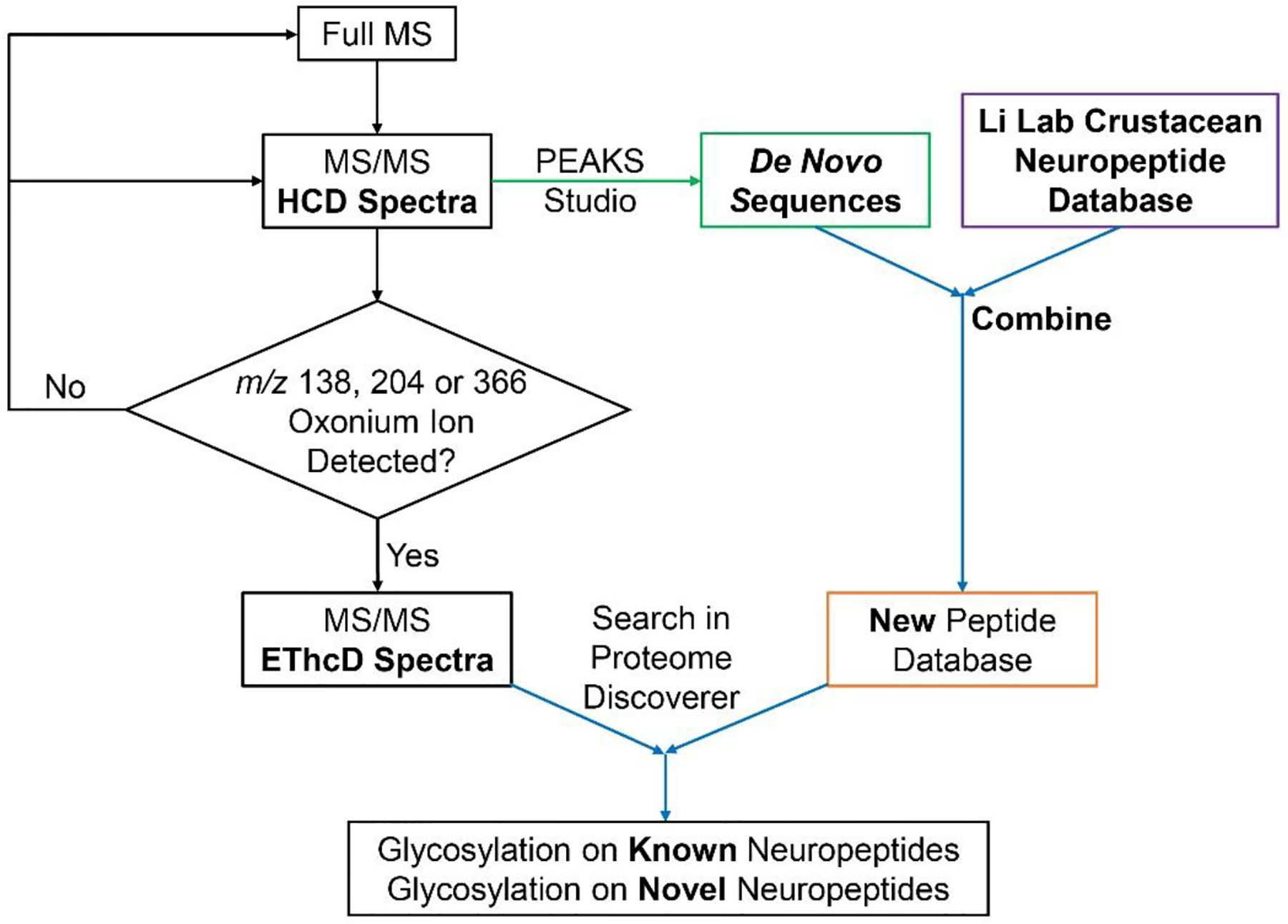
Data analysis workflow for crustacean glyconeuropeptide identification and characterization.
Attach a C18 column (75 μm inner diameter × 15 cm, either home-packed or commercially available) onto the LC system coupled with MS. Mobile phase A is 0.1% FA in water and mobile phase B is 0.1% FA in acetonitrile. Set the flow rate to 0.3 μL/min. Allow sufficient time for column to equilibrate and reach stable pressure.
Inject 2 μL of the crustacean neuropeptide extract from Section 3.1 (starting tissues >10 pieces) or Section 3.2 (starting tissues < 10 pieces).
Set up the following gradient: 0–90 min, 3–30% B; 90.5–110.5 min, 30–75% B; 110–130 min, 75–95% B; and include another 15 min 3% B at the end for column equilibration.
- Set up an HCD-pd-EThcD MS method:
- Acquire full MS scan at Top Speed mode (3s) from m/z 400–1800 at 120 K resolution.
- For data-dependent acquisition (DDA), use mode cycle time (3s) and dynamic exclusion 20 s.
- For HCD scans, use normalized collisional energy 35% with resolution 30 K.
- Include a targeted mass trigger containing m/z 138.0545, m/z 204.0867 and m/z 366.1396 and set up the following criteria: If any of the above three ions is within the top 30 most abundant peaks in the HCD spectra, trigger a subsequent EThcD MS/MS scan.
- For EThcD scans, precursors at different charges are exposed to distinct ETD reaction times followed by same amount of supplemental activation energy (33%) : z = 2, reaction time = 50 ms; z = 3–4, reaction time = 20 ms; z = 5, reaction time = 10 ms; z = 6–15, use instrument charge-dependent ETD parameters. All EThcD spectra are acquired at resolution 60 K.
4.2.3. Ion mobility MS
Ion mobility (IM) MS is a gas phase separation technique that allows differentiation of molecules based on their structural/conformational differences, measured by drift time analytes experienced inside the IM cell filled with buffer gas. IM-MS has drawn increasing attention in glycosylation studies (Z. Chen, Glover, & Li, 2018), because complete structural elucidation of glycopeptides remains to be challenging relying solely on masses obtained from tandem MS events. The challenges imposed mainly are contributed by glycan heterogeneity (Fig. 10), especially sugar structural isomerization, monosaccharide α/β-stereochemistry linkage at multiple potential sites, and anomeric α/β-linkage between amino acid residue and glycan. Often times there are only subtle structural variances among various glycan and glycoconjugate isomers thus it is highly demanding to develop novel techniques to assist differentiation of those molecules. A high field asymmetric wave ion mobility spectrometry (FAIMS) has been developed to separate isomeric O-linked glycopeptides with merely glycosylation site difference (Creese & Cooper, 2012). The two glycoconjugate isomers were indistinguishable in LC but the glycosylation site differences have potentially contributed to gas-phase intramolecular interactions resulting in the structural variance noticeable in drift times. IM MS has also been demonstrated as an efficient strategy resolving N-acetylneuraminic acid α2,3 and α2,6 linkages by measuring signature CID fragmentation ion drift times (Hinneburg et al., 2016). Utilization of IM-MS is anticipated to foster knowledge of glycosciences with the improved technologies and integration of other orthogonal techniques, including capillary electrophoresis (CE), LC and tandem MS.
Figure 10.
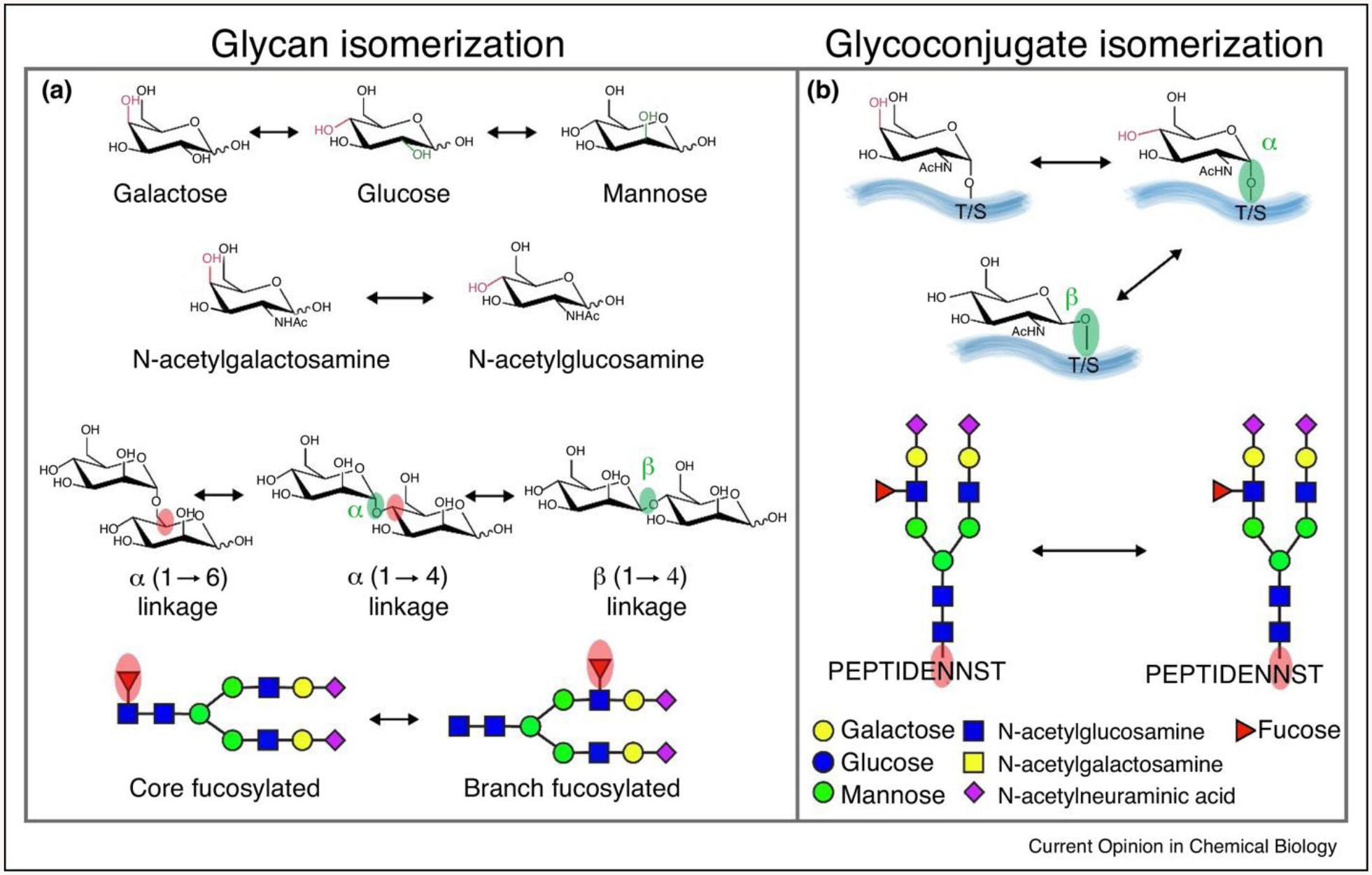
The isomerization of glycan and glycoconjugates. (a) The building blocks (monosaccharides) that compose larger glycans are structural isomers (hexose: galactose, glucose, mannose, N-acetylhexosamine: N-acetylgalactosamine, N-acetylglucosamine); monosaccharides can be connected either α-stereochemistry or β-stereochemistry at multiple potential linkage position; fucose could be either attached to N-glycan core or branches. (b) Epimeric glycoconjugates results from alternative configurations (α- or β-) at the anomeric linkages or the presence of epimeric glycan monomers (galactose or glucose), two isomeric N-glycopeptides differ in the site of N-glycan attachment. Adapted from (Z. Chen, Glover, & Li, 2018) with permission.
4.3. Data Analysis to Uncover Sequence and Structural Information about Intact Glycopeptides
There have been continuous efforts to develop software for MS data interpretation and large-scale glycopeptide identification. A plethora of search engines are available, including Byonic (Cooper, Gasteiger, & Packer, 2001), GlycoMaster DB (He, Xin, Shan, Lajoie, & Ma, 2014), GPQuest (Toghi Eshghi, Shah, Yang, Li, & Zhang, 2015), MAGIC (Lynn et al., 2015), Integrated GlycoProteome Analyzer (I-GPA) (G. W. Park et al., 2016), SweetNET (Nasir et al., 2016), pGlyco (W.-F. Zeng et al., 2016), pGlyco 2.0 (M.-Q. Liu et al., 2017). As glycosylated peptide standards are generally difficult to synthesize, the rich software selections have not only enabled deciphering of complicated glycopeptides but also provided various cross-validation opportunities to consolidate search results generated from different MS platforms. The MS data processing steps typically involve a match between MS/MS spectra and in silico peptide/protein sequences together with a glycan library. For the organisms and species missing a completely characterized genomic and peptide database, such as crustaceans, de novo sequencing can first be applied to the HCD scans in HCD-pd-EThcD spectra to generate a new peptide database to be used in data analysis to achieve better identification rate of intact glycosylated neuropeptides, including novel glycosylated neuropeptides.
A detailed procedure enabling discovery of crustacean glycosylated neuropeptides is provided below. Briefly, PEAKS Studio is first used to analyze the acquired data and generate de novo peptide sequences (typically >5K sequences); then those de novo sequences are combined with existing Li Lab crustacean neuropeptide database (containing ~650 neuropeptides) to generate a new peptide database; following this, the acquired MS data are searched against the new peptide database using Proteome Discoverer embedded with Byonic node. Finally, the identified glycosylated neuropeptide spectra are manually checked to ensure correct assignment.
Use PEAKS Studio to generate de novo sequences. The version we used is PEAKS 7 but there are updated versions available, such as PEAKS X. In data refinement, set full MS precursor tolerance to be 10 ppm and fragment tolerance ion to be 0.02 Da. Select “no enzyme” as no digestion has been involved. In the PTM types, select fixed modification to be Cys carbamidomethylation; for variable modifications, include C-terminal amidation, Met oxidation, pyro-Glu from E, pyro-Glu from Q, Tyr-sulfation, dehydration, methylation and glycan modifications consisting no more than 2 glycans. Import Li Lab crustacean neuropeptide database. After search is finished, go to “de novo only” tab and export all de novo sequences with average local confidence (ALC) > 50%.
Combine de novo sequences with custom-built crustacean neuropeptide database. Use Excel or customized scripts to remove any unnecessary columns and adapt the format of the sequences to be FASTA style. Save the new database into .fasta file. Note: name the Li Lab crustacean neuropeptides and de novo sequences in a distinguishable manner as the de novo sequences are not completely confirmed.
Use Proteome Discoverer with Byonic node to identify glycosylated neuropeptides. The software version used is Proteome Discoverer 2.1. Data are searched against mammalian glycan database and the new database from Step 2. Precursor tolerance is set to be 10 ppm and fragment tolerance ion to be ±0.01 Da. Include modifications used in Step 1.
Manually check the spectra in Byonic Viewer. A glycosylated neuropeptide should have oxonium ions in the low mass region with most of the fragments assigned. Fig. 11 illustrates a tandem MS spectrum annotated and assigned as a novel B-type allatostatin neuropeptide with O-linked HexNAc2Hex2Fuc2 discovered from blue crab pericardial organ.
Figure 11.
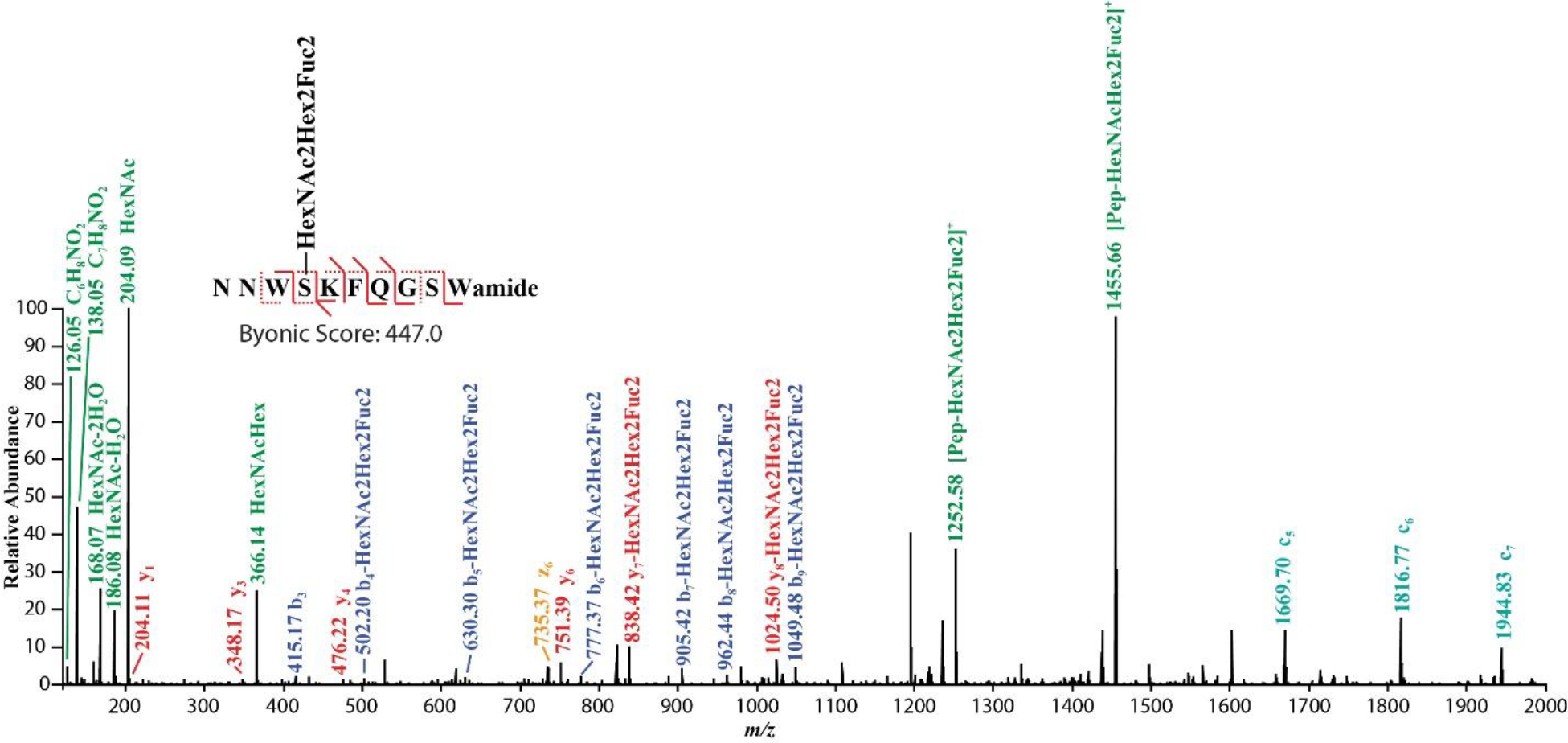
O-glycosylation occurring on a novel B-type allatostatin neuropeptide revealed by EThcD fragmentation.
5. Conclusions and Future Directions
Over the past decades, extensive efforts have been devoted to the large-scale characterization of glycopeptides and glycoproteins due to their important biological functions. The rapid technological innovations, ranging from sample enrichment strategies to MS/MS characterization techniques, have shed light on this complex PTM that is essential in a variety of biological processes, such as cell-cell communication and disease progression. The large-scale profiling of glycopeptidome or even glycoproteome with site-specific elucidation has become more feasible thanks to the development of hybrid MS/MS approaches on highly sensitive instrument platforms. However, to reveal complete glycopeptide information in a complex biological system, more selective and efficient enrichment methods are still in demand to preserve intact glycan structures to the best extent, especially for O-linked glycosylation. Furthermore, current MS/MS fragmentation approaches only enable complete characterization of peptides with relatively simple glycosylation structure. The isomer differentiation and high-order glycan complexity require more diverse tools (e.g. IM-MS and spectroscopy) to differentiate the subtle differences among structural isomers and characterize glycan microheterogeneity, respectively. Another aspect is to understand the roles of glycosylated signaling molecules, such as glycosylated neuropeptides, by incorporating quantitatively comparative studies and electrophysiological studies in the nervous system. The relative abundance changes of glycosylated neuropeptides in response to certain stimuli or perturbation and associated electrical potential alterations are expected to generate new insights into neuromodulation mechanism and signaling transmission. Future advancements should include sample enrichment improvement, streamlined hybrid characterization techniques, improved bioinformatics strategies for glycan sequencing and functional studies to enable more a comprehensive understanding the roles of these glycosylated peptides at the cellular and network levels.
Acknowledgements
This work was supported in part by the National Institutes of Health grants U01CA231081, R01 DK071801, and RF1 AG052324, and National Science Foundation (NSF) grant CHE-1710140. LL acknowledges a Vilas Distinguished Achievement Professorship and a Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Reference
- Abbott KL, Aoki K, Lim J-M, Porterfield M, Johnson R, O’Regan RM, … Pierce M (2008). Targeted glycoproteomic identification of biomarkers for human breast carcinoma. Journal of Proteome Research, 7(4), 1470–1480. 10.1021/pr700792g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian TE, Allen JM, Bloom SR, Ghatei MA, Rossor MN, Roberts GW, … Polak JM (1983). Neuropeptide Y distribution in human brain. Nature, 306(5943), 584–586. [DOI] [PubMed] [Google Scholar]
- Aebersold R, & Mann M (2016). Mass-spectrometric exploration of proteome structure and function. Nature, 537(7620), 347–355. 10.1038/nature19949 [DOI] [PubMed] [Google Scholar]
- Ahn YH, Kim JY, & Yoo JS (2015). QUANTITATIVE MASS SPECTROMETRIC ANALYSIS OF GLYCOPROTEINS COMBINED WITH ENRICHMENT METHODS. Mass Spectrometry Reviews, 34(2), 148–165. 10.1002/mas.21428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn YH, Lee JY, Lee JY, Kim Y-S, Ko JH, & Yoo JS (2009). Quantitative Analysis of an Aberrant Glycoform of TIMP1 from Colon Cancer Serum by L-PHA-Enrichment and SISCAPA with MRM Mass Spectrometry. Journal of Proteome Research, 8(9), 4216–4224. 10.1021/pr900269s [DOI] [PubMed] [Google Scholar]
- Alagesan K, Khilji SK, & Kolarich D (2017). It is all about the solvent: On the importance of the mobile phase for ZIC-HILIC glycopeptide enrichment. Analytical and Bioanalytical Chemistry, 409(2), 529–538. 10.1007/s00216-016-0051-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley WR, Mann BF, & Novotny MV (2013). High-sensitivity Analytical Approaches for the Structural Characterization of Glycoproteins. Chemical Reviews, 113(4), 2668–2732. 10.1021/cr3003714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert AJ (1990). Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. Journal of Chromatography A, 499, 177–196. 10.1016/S0021-9673(00)96972-3 [DOI] [PubMed] [Google Scholar]
- Alpert AJ (2008). Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Analytical Chemistry, 80(1), 62–76. 10.1021/ac070997p [DOI] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, & Sharon N (1999). On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica et Biophysica Acta (BBA) - General Subjects, 1473(1), 4–8. 10.1016/S0304-4165(99)00165-8 [DOI] [PubMed] [Google Scholar]
- Arey BJ (2012). The Role of Glycosylation in Receptor Signaling. Glycosylation. 10.5772/50262 [DOI] [Google Scholar]
- Arnaud IL, Josserand J, Jensen H, Lion N, Roussel C, & Girault HH (2005). Salt removal during Off-Gel electrophoresis of protein samples. Electrophoresis, 26(9), 1650–1658. 10.1002/elps.200410294 [DOI] [PubMed] [Google Scholar]
- Arnold JN, Wormald MR, Sim RB, Rudd PM, & Dwek RA (2007). The Impact of Glycosylation on the Biological Function and Structure of Human Immunoglobulins. Annual Review of Immunology, 25(1), 21–50. 10.1146/annurev.immunol.25.022106.141702 [DOI] [PubMed] [Google Scholar]
- Bhavanandan VP, & Katlic AW (1979). The interaction of wheat germ agglutinin with sialoglycoproteins. The role of sialic acid. The Journal of Biological Chemistry, 254(10), 4000–4008. [PubMed] [Google Scholar]
- Biddlecombe RA, & Pleasance S (1999). Automated protein precipitation by filtration in the 96-well format. Journal of Chromatography. B, Biomedical Sciences and Applications, 734(2), 257–265. [DOI] [PubMed] [Google Scholar]
- Bilsky EJ, Egleton RD, Mitchell SA, Palian MM, Davis P, Huber JD, … Polt R (2000). Enkephalin Glycopeptide Analogues Produce Analgesia with Reduced Dependence Liability. Journal of Medicinal Chemistry, 43(13), 2586–2590. 10.1021/jm000077y [DOI] [PubMed] [Google Scholar]
- Boersema PJ, Mohammed S, & Heck AJR (2008). Hydrophilic interaction liquid chromatography (HILIC) in proteomics. Analytical and Bioanalytical Chemistry, 391(1), 151–159. 10.1007/s00216-008-1865-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y, Astoul CH, Zamboni V, Peumans WJ, Menu-Bouaouiche L, Van Damme EJM, … Rougé P (2002). Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose. The Biochemical Journal, 364(Pt 1), 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmalm G, Portelius E, Öhrfelt A, Mattsson N, Persson R, Gustavsson MK, … Brinkmalm A (2012). An online nano-LC-ESI-FTICR-MS method for comprehensive characterization of endogenous fragments from amyloid β and amyloid precursor protein in human and cat cerebrospinal fluid. Journal of Mass Spectrometry, 47(5), 591–603. 10.1002/jms.2987 [DOI] [PubMed] [Google Scholar]
- Buchberger A, Yu Q, & Li L (2015). Advances in Mass Spectrometric Tools for Probing Neuropeptides. Annual Review of Analytical Chemistry, 8(1), 485–509. 10.1146/annurev-anchem-071114-040210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszewski B, & Noga S (2012). Hydrophilic interaction liquid chromatography (HILIC)—a powerful separation technique. Analytical and Bioanalytical Chemistry, 402(1), 231–247. 10.1007/s00216-011-5308-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajka T, & Fiehn O (2016). Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Analytical Chemistry, 88(1), 524–545. 10.1021/acs.analchem.5b04491 [DOI] [PubMed] [Google Scholar]
- Cao Qichen, Ma C, Bai H, Li X, Yan H, Zhao Y, … Qian X (2013). Multivalent hydrazide-functionalized magnetic nanoparticles for glycopeptide enrichment and identification. Analyst, 139(3), 603–609. 10.1039/C3AN01532G [DOI] [PubMed] [Google Scholar]
- Cao Qinjingwen, Yu Q, Liu Y, Chen Z, & Li L (submitted). Signature-Ion Triggered Mass Spectrometry Approach Enabled Discovery of N- and O-linked Glycosylated Neuropeptides in the Crustacean Nervous System. [DOI] [PMC free article] [PubMed]
- Chang SK, Ismail A, Yanagita T, Mohd Esa N, & Baharuldin MTH (2015). Antioxidant peptides purified and identified from the oil palm (Elaeis guineensis Jacq.) kernel protein hydrolysate. Journal of Functional Foods, 14, 63–75. 10.1016/j.jff.2015.01.011 [DOI] [Google Scholar]
- Chen C-C, Su W-C, Huang B-Y, Chen Y-J, Tai H-C, & Obena RP (2014). Interaction modes and approaches to glycopeptide and glycoprotein enrichment. The Analyst, 139(4), 688–704. 10.1039/c3an01813j [DOI] [PubMed] [Google Scholar]
- Chen Jianying, Li X, Feng M, Luo K, Yang J, & Zhang B (2017). Novel boronate material affords efficient enrichment of glycopeptides by synergized hydrophilic and affinity interactions. Analytical and Bioanalytical Chemistry, 409(2), 519–528. 10.1007/s00216-016-0044-5 [DOI] [PubMed] [Google Scholar]
- Chen Jing, Shah P, & Zhang H (2013). Solid Phase Extraction of N-Linked Glycopeptides Using Hydrazide Tip. Analytical Chemistry, 85(22), 10670–10674. 10.1021/ac401812b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Lu Y, Ma Q, Guo L, & Feng Y-Q (2009). Boronate affinity monolith for highly selective enrichment of glycopeptides and glycoproteins. Analyst, 134(10), 2158–2164. 10.1039/B909581K [DOI] [PubMed] [Google Scholar]
- Chen P, Werner-Zwanziger U, Wiesler D, Pagel M, & Novotny MV (1999). Mass Spectrometric Analysis of Benzoylated Sialooligosaccharides and Differentiation of Terminal α 2→3 and α 2→6 Sialogalactosylated Linkages at Subpicomole Levels. Analytical Chemistry, 71(21), 4969–4973. 10.1021/ac990674w [DOI] [PubMed] [Google Scholar]
- Chen R, Seebun D, Ye M, Zou H, & Figeys D (2014). Site-specific characterization of cell membrane N-glycosylation with integrated hydrophilic interaction chromatography solid phase extraction and LC–MS/MS. Journal of Proteomics, 103, 194–203. 10.1016/j.jprot.2014.03.040 [DOI] [PubMed] [Google Scholar]
- Chen Z, Glover MS, & Li L (2018). Recent advances in ion mobility–mass spectrometry for improved structural characterization of glycans and glycoconjugates. Current Opinion in Chemical Biology, 42, 1–8. 10.1016/j.cbpa.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Huang J, & Li L (2018). Recent advances in mass spectrometry (MS)-based glycoproteomics in complex biological samples. TrAC Trends in Analytical Chemistry. 10.1016/j.trac.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertov O, Biragyn A, Kwak LW, Simpson JT, Boronina T, Hoang VM, … Fisher RJ (2004). Organic solvent extraction of proteins and peptides from serum as an effective sample preparation for detection and identification of biomarkers by mass spectrometry. Proteomics, 4(4), 1195–1203. 10.1002/pmic.200300677 [DOI] [PubMed] [Google Scholar]
- Christie AE, Stemmler EA, & Dickinson PS (2010). Crustacean neuropeptides. Cellular and Molecular Life Sciences: CMLS, 67(24), 4135–4169. 10.1007/s00018-010-0482-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JS, & Zmora N (2008). Functional studies of crustacean hyperglycemic hormones (CHHs) of the blue crab, Callinectes sapidus - the expression and release of CHH in eyestalk and pericardial organ in response to environmental stress. The FEBS Journal, 275(4), 693–704. 10.1111/j.1742-4658.2007.06231.x [DOI] [PubMed] [Google Scholar]
- Chung JS, Zmora N, Katayama H, & Tsutsui N (2010). Crustacean hyperglycemic hormone (CHH) neuropeptidesfamily: Functions, titer, and binding to target tissues. General and Comparative Endocrinology, 166(3), 447–454. 10.1016/j.ygcen.2009.12.011 [DOI] [PubMed] [Google Scholar]
- Cook SE, & Pinkerton TC (1986). Characterization of internal surface reversed-phase silica supports for liquid chromatography. Journal of Chromatography A, 368, 233–248. 10.1016/S0021-9673(00)91066-5 [DOI] [PubMed] [Google Scholar]
- Cooper CA, Gasteiger E, & Packer NH (2001). GlycoMod--a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics, 1(2), 340–349. [DOI] [PubMed] [Google Scholar]
- Craig AG, Zafaralla G, Cruz LJ, Santos AD, Hillyard DR, Dykert J, … Olivera BM (1998). An O-glycosylated neuroexcitatory conus peptide. Biochemistry, 37(46), 16019–16025. 10.1021/bi981690a [DOI] [PubMed] [Google Scholar]
- Grey Craig A, Bandyopadhyay P, & Olivera BM (1999). Post-translationally modified neuropeptides from Conus venoms. European Journal of Biochemistry, 264(2), 271–275. 10.1046/j.1432-1327.1999.00624.x [DOI] [PubMed] [Google Scholar]
- Grey Craig A, Norberg T, Griffin D, Hoeger C, Akhtar M, Schmidt K, … Olivera BM (1999). Contulakin-G, an O-Glycosylated Invertebrate Neurotensin. Journal of Biological Chemistry, 274(20), 13752–13759. 10.1074/jbc.274.20.13752 [DOI] [PubMed] [Google Scholar]
- Creese AJ, & Cooper HJ (2012). Separation and Identification of Isomeric Glycopeptides by High Field Asymmetric Waveform Ion Mobility Spectrometry. Analytical Chemistry, 84(5), 2597–2601. 10.1021/ac203321y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings RD, & Kornfeld S (1982). Fractionation of asparagine-linked oligosaccharides by serial lectin-Agarose affinity chromatography. A rapid, sensitive, and specific technique. The Journal of Biological Chemistry, 257(19), 11235–11240. [PubMed] [Google Scholar]
- de Faria HD, de C. Abrão LC, Santos MG, Barbosa AF, & Figueiredo EC (2017). New advances in restricted access materials for sample preparation: A review. Analytica Chimica Acta, 959, 43–65. 10.1016/j.aca.2016.12.047 [DOI] [PubMed] [Google Scholar]
- de Haan N, Reiding KR, Haberger M, Reusch D, Falck D, & Wuhrer M (2015). Linkage-specific sialic acid derivatization for MALDI-TOF-MS profiling of IgG glycopeptides. Analytical Chemistry, 87(16), 8284–8291. 10.1021/acs.analchem.5b02426 [DOI] [PubMed] [Google Scholar]
- Dedvisitsakul P, Jacobsen S, Svensson B, Bunkenborg J, Finnie C, & Hägglund P (2014). Glycopeptide Enrichment Using a Combination of ZIC-HILIC and Cotton Wool for Exploring the Glycoproteome of Wheat Flour Albumins. Journal of Proteome Research, 13(5), 2696–2703. 10.1021/pr401282r [DOI] [PubMed] [Google Scholar]
- Dell A, Galadari A, Sastre F, & Hitchen P (2010). Similarities and Differences in the Glycosylation Mechanisms in Prokaryotes and Eukaryotes [Research article]. 10.1155/2010/148178 [DOI] [PMC free article] [PubMed]
- Desilets CP, Rounds MA, & Regnier FE (1991). Semipermeable-surface reversed-phase media for high-performance liquid chromatography. Journal of Chromatography, 544(1–2), 25–39. [DOI] [PubMed] [Google Scholar]
- Ding W, Hill JJ, & Kelly J (2007). Selective Enrichment of Glycopeptides from Glycoprotein Digests Using Ion-Pairing Normal-Phase Liquid Chromatography. Analytical Chemistry, 79(23), 8891–8899. 10.1021/ac0707535 [DOI] [PubMed] [Google Scholar]
- Dlask O, & Václavíková N (2018). Electrodialysis with ultrafiltration membranes for peptide separation. Chemical Papers, 72(2), 261–271. 10.1007/s11696-017-0293-6 [DOI] [Google Scholar]
- Dong J, Zhou H, Wu R, Ye M, & Zou H (2007). Specific capture of phosphopeptides by Zr4+− modified monolithic capillary column. Journal of Separation Science, 30(17), 2917–2923. 10.1002/jssc.200700350 [DOI] [PubMed] [Google Scholar]
- Duan G, & Walther D (2015). The Roles of Post-translational Modifications in the Context of Protein Interaction Networks. PLOS Computational Biology, 11(2), e1004049 10.1371/journal.pcbi.1004049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham M, & Regnier FE (2006). Targeted glycoproteomics: Serial lectin affinity chromatography in the selection of O-glycosylation sites on proteins from the human blood proteome. Journal of Chromatography A, 1132(1), 165–173. 10.1016/j.chroma.2006.07.070 [DOI] [PubMed] [Google Scholar]
- Egleton RD, Mitchell SA, Huber JD, Janders J, Stropova D, Polt R, … Davis TP (2000). Improved bioavailability to the brain of glycosylated Met-enkephalin analogs. Brain Research, 881(1), 37–46. [DOI] [PubMed] [Google Scholar]
- Egleton RD, Mitchell SA, Huber JD, Palian MM, Polt R, & Davis TP (2001). Improved blood-brain barrier penetration and enhanced analgesia of an opioid peptide by glycosylation. The Journal of Pharmacology and Experimental Therapeutics, 299(3), 967–972. [PubMed] [Google Scholar]
- Elphick MR, Mirabeau O, & Larhammar D (2018). Evolution of neuropeptide signalling systems. Journal of Experimental Biology, 221(3), jeb151092 10.1242/jeb.151092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enman NM, Sabban EL, McGonigle P, & Van Bockstaele EJ (2015). Targeting the neuropeptide Y system in stress-related psychiatric disorders. Neurobiology of Stress, 1, 33–43. 10.1016/j.ynstr.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanayan S, Hincapie M, & Hancock WS (2012). Using lectins to harvest the plasma/serum glycoproteome. ELECTROPHORESIS, 33(12), 1746–1754. 10.1002/elps.201100567 [DOI] [PubMed] [Google Scholar]
- Fink AL, Calciano LJ, Goto Y, Kurotsu T, & Palleros DR (1994). Classification of Acid Denaturation of Proteins: Intermediates and Unfolded States. Biochemistry, 33(41), 12504–12511. 10.1021/bi00207a018 [DOI] [PubMed] [Google Scholar]
- Finoulst I, Pinkse M, Van Dongen W, & Verhaert P (2011). Sample preparation techniques for the untargeted LC-MS-based discovery of peptides in complex biological matrices. Journal of Biomedicine & Biotechnology, 2011, 245291 10.1155/2011/245291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc V, Yang Y, & Heck AJR (2017). Proteoform Profile Mapping of the Human Serum Complement Component C9 Revealing Unexpected New Features of N-, O-, and C-Glycosylation. Analytical Chemistry, 89(6), 3483–3491. 10.1021/acs.analchem.6b04527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuki K, & Toyo’oka T (2017). Retention of glycopeptides analyzed using hydrophilic interaction chromatography is influenced by charge and carbon chain length of ion-pairing reagent for mobile phase. Biomedical Chromatography: BMC, 31(11). 10.1002/bmc.3988 [DOI] [PubMed] [Google Scholar]
- Georgiou HM, Rice GE, & Baker MS (2001). Proteomic analysis of human plasma: Failure of centrifugal ultrafiltration to remove albumin and other high molecular weight proteins. PROTEOMICS, 1(12), 1503–1506. [DOI] [PubMed] [Google Scholar]
- Glover MS, Yu Q, Chen Z, Shi X, Kent KC, & Li L (2018). Characterization of intact sialylated glycopeptides and phosphorylated glycopeptides from IMAC enriched samples by EThcD fragmentation: Toward combining phosphoproteomics and glycoproteomics. International Journal of Mass Spectrometry, 427, 35–42. 10.1016/j.ijms.2017.09.002 [DOI] [Google Scholar]
- Greening DW, & Simpson RJ (2010). A centrifugal ultrafiltration strategy for isolating the low-molecular weight (<or=25K) component of human plasma proteome. Journal of Proteomics, 73(3), 637–648. 10.1016/j.jprot.2009.09.013 [DOI] [PubMed] [Google Scholar]
- Gupta N, Bark SJ, Lu WD, Taupenot L, O’Connor DT, Pevzner P, & Hook V (2010). Mass spectrometry-based neuropeptidomics of secretory vesicles from human adrenal medullary pheochromocytoma reveals novel peptide products of prohormone processing. Journal of Proteome Research, 9(10), 5065–5075. 10.1021/pr100358b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfinger B, Hammerer-Lercher A, Amplatz B, Sarg B, Kremser L, & Lindner HH (2017). Unraveling the Molecular Complexity of O-Glycosylated Endogenous (N-Terminal) pro–B-Type Natriuretic Peptide Forms in Blood Plasma of Patients with Severe Heart Failure. Clinical Chemistry, 63(1), 359–368. 10.1373/clinchem.2016.265397 [DOI] [PubMed] [Google Scholar]
- Halim A, Brinkmalm G, Rüetschi U, Westman-Brinkmalm A, Portelius E, Zetterberg H, … Nilsson J (2011). Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid β-peptides in human cerebrospinal fluid. Proceedings of the National Academy of Sciences, 108(29), 11848–11853. 10.1073/pnas.1102664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim A, Nilsson J, Rüetschi U, Hesse C, & Larson G (2012). Human Urinary Glycoproteomics; Attachment Site Specific Analysis of N- and O-Linked Glycosylations by CID and ECD. Molecular & Cellular Proteomics : MCP, 11(4). 10.1074/mcp.M111.013649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim A, Rüetschi U, Larson G, & Nilsson J (2013). LC–MS/MS Characterization of O-Glycosylation Sites and Glycan Structures of Human Cerebrospinal Fluid Glycoproteins. Journal of Proteome Research, 12(2), 573–584. 10.1021/pr300963h [DOI] [PubMed] [Google Scholar]
- Halim A, Westerlind U, Pett C, Schorlemer M, Rüetschi U, Brinkmalm G, … Nilsson J (2014). Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. Journal of Proteome Research, 13(12), 6024–6032. 10.1021/pr500898r [DOI] [PubMed] [Google Scholar]
- Harada H, Kamei M, Tokumoto Y, Yui S, Koyama F, Kochibe N, … Kobata A (1987). Systematic fractionation of oligosaccharides of human immunoglobulin G by serial affinity chromatography on immobilized lectin columns. Analytical Biochemistry, 164(2), 374–381. 10.1016/0003-2697(87)90507-0 [DOI] [PubMed] [Google Scholar]
- Haubner R, Wester HJ, Burkhart F, Senekowitsch-Schmidtke R, Weber W, Goodman SL, … Schwaiger M (2001). Glycosylated RGD-containing peptides: Tracer for tumor targeting and angiogenesis imaging with improved biokinetics. Journal of Nuclear Medicine: Official Publication, Society of Nuclear Medicine, 42(2), 326–336. [PubMed] [Google Scholar]
- He L, Xin L, Shan B, Lajoie GA, & Ma B (2014). GlycoMaster DB: Software To Assist the Automated Identification of N-Linked Glycopeptides by Tandem Mass Spectrometry. Journal of Proteome Research, 13(9), 3881–3895. 10.1021/pr401115y [DOI] [PubMed] [Google Scholar]
- Heck AL, Crestani CC, Fernández-Guasti A, Larco DO, Mayerhofer A, & Roselli CE (2018). Neuropeptide and steroid hormone mediators of neuroendocrine regulation. Journal of Neuroendocrinology, 30(10), e12599 10.1111/jne.12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, & Aebi, and M. (2001). Intracellular Functions of N-Linked Glycans. Science, 291(5512), 2364–2369. 10.1126/science.291.5512.2364 [DOI] [PubMed] [Google Scholar]
- Helenius A, & Aebi M (2004). Roles of N-Linked Glycans in the Endoplasmic Reticulum. Annual Review of Biochemistry, 73(1), 1019–1049. 10.1146/annurev.biochem.73.011303.073752 [DOI] [PubMed] [Google Scholar]
- Hinneburg H, Hofmann J, Struwe WB, Thader A, Altmann F, Silva DV, … Kolarich D (2016). Distinguishing N-acetylneuraminic acid linkage isomers on glycopeptides by ion mobility-mass spectrometry. Chemical Communications, 52(23), 4381–4384. 10.1039/C6CC01114D [DOI] [PubMed] [Google Scholar]
- Hökfelt T, Broberger C, Xu Z-QD, Sergeyev V, Ubink R, & Diez M (2000). Neuropeptides — an overview. Neuropharmacology, 39(8), 1337–1356. 10.1016/S0028-3908(00)00010-1 [DOI] [PubMed] [Google Scholar]
- Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, & Hwang S-R (2008). Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annual Review of Pharmacology and Toxicology, 48, 393–423. 10.1146/annurev.pharmtox.48.113006.094812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook V, Lietz CB, Podvin S, Cajka T, & Fiehn O (2018). Diversity of Neuropeptide Cell-Cell Signaling Molecules Generated by Proteolytic Processing Revealed by Neuropeptidomics Mass Spectrometry. Journal of The American Society for Mass Spectrometry, 29(5), 807–816. 10.1007/s13361-018-1914-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortin GL (1990). Isolation of glycopeptides containing O-linked oligosaccharides by lectin affinity chromatography on jacalin-agarose. Analytical Biochemistry, 191(2), 262–267. 10.1016/0003-2697(90)90217-W [DOI] [PubMed] [Google Scholar]
- Hu H, Khatri K, Klein J, Leymarie N, & Zaia J (2016). A review of methods for interpretation of glycopeptide tandem mass spectral data. Glycoconjugate Journal, 33(3), 285–296. 10.1007/s10719-015-9633-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Boos K-S, Ye M, Wu R, & Zou H (2009). Selective on-line serum peptide extraction and multidimensional separation by coupling a restricted-access material-based capillary trap column with nanoliquid chromatography-tandem mass spectrometry. Journal of Chromatography. A, 1216(28), 5377–5384. 10.1016/j.chroma.2009.05.030 [DOI] [PubMed] [Google Scholar]
- Hu L, Boos K-S, Ye M, & Zou H (2014). Analysis of the endogenous human serum peptides by on-line extraction with restricted-access material and HPLC-MS/MS identification. Talanta, 127, 191–195. 10.1016/j.talanta.2014.04.011 [DOI] [PubMed] [Google Scholar]
- Huang J, Wan H, Yao Y, Li J, Cheng K, Mao J, … Zou H (2015). Highly Efficient Release of Glycopeptides from Hydrazide Beads by Hydroxylamine Assisted PNGase F Deglycosylation for N-Glycoproteome Analysis. Analytical Chemistry, 87(20), 10199–10204. 10.1021/acs.analchem.5b02669 [DOI] [PubMed] [Google Scholar]
- Huang Y, Nie Y, Boyes B, & Orlando R (2016). Resolving Isomeric Glycopeptide Glycoforms with Hydrophilic Interaction Chromatography (HILIC). Journal of Biomolecular Techniques: JBT, 27(3), 98–104. 10.7171/jbt.16-2703-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaprakash NG, & Surolia A (2017). Role of glycosylation in nucleating protein folding and stability. Biochemical Journal, 474(14), 2333–2347. 10.1042/BCJ20170111 [DOI] [PubMed] [Google Scholar]
- Jiang B, Wu Q, Zhang L, & Zhang Y (2017). Preparation and application of silver nanoparticle-functionalized magnetic graphene oxide nanocomposites. Nanoscale, 9(4), 1607–1615. 10.1039/C6NR09260H [DOI] [PubMed] [Google Scholar]
- Jiang J, Tian F, Cai Y, Qian X, Costello CE, & Ying W (2014). Site-specific qualitative and quantitative analysis of N- and O-glycoforms in recombinant human erythropoietin. Analytical and Bioanalytical Chemistry, 406(25), 6265–6274. 10.1007/s00216-014-8037-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zhang W, Yang Q, Dai L, & Zhou P (2018). An inorganic boronate affinity in-needle monolithic device for specific capture of cis-diol containing compounds. Talanta, 178, 710–715. 10.1016/j.talanta.2017.10.011 [DOI] [PubMed] [Google Scholar]
- Kawahara R, Saad J, Angeli CB, & Palmisano G (2016). Site-specific characterization of N-linked glycosylation in human urinary glycoproteins and endogenous glycopeptides. Glycoconjugate Journal, 33(6), 937–951. 10.1007/s10719-016-9677-z [DOI] [PubMed] [Google Scholar]
- Kawashima Y, Fukutomi T, Tomonaga T, Takahashi H, Nomura F, Maeda T, & Kodera Y (2010). High-Yield Peptide-Extraction Method for the Discovery of Subnanomolar Biomarkers from Small Serum Samples. Journal of Proteome Research, 9(4), 1694–1705. 10.1021/pr9008018 [DOI] [PubMed] [Google Scholar]
- Khidekel N, Ficarro SB, Peters EC, & Hsieh-Wilson LC (2004). Exploring the O-GlcNAc proteome: Direct identification of O-GlcNAc-modified proteins from the brain. Proceedings of the National Academy of Sciences, 101(36), 13132–13137. 10.1073/pnas.0403471101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Coffey MJ, John A, Uboldi AD, Huynh M-H, Stewart RJ, … Scott NE (2019). Protein O-fucosyltransferase 2-mediated O-glycosylation of the adhesin MIC2 is dispensable for Toxoplasma gondii tachyzoite infection. The Journal of Biological Chemistry, 294(5), 1541–1553. 10.1074/jbc.RA118.005357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick CL, Broberg CA, McCool EN, Lee WJ, Chao A, McConnell EW, … Hicks LM (2017). The “PepSAVI-MS” Pipeline for Natural Product Bioactive Peptide Discovery. Analytical Chemistry, 89(2), 1194–1201. 10.1021/acs.analchem.6b03625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen CJ, Wang AQ, Musson DG, Yang AY, & Fisher AL (2003). A semi-automated 96-well protein precipitation method for the determination of montelukast in human plasma using high performance liquid chromatography/fluorescence detection. Journal of Pharmaceutical and Biomedical Analysis, 31(4), 647–654. 10.1016/S0731-7085(02)00723-9 [DOI] [PubMed] [Google Scholar]
- Kolarich D, Jensen PH, Altmann F, & Packer NH (2012). Determination of site-specific glycan heterogeneity on glycoproteins. Nature Protocols, 7(7), 1285–1298. 10.1038/nprot.2012.062 [DOI] [PubMed] [Google Scholar]
- Kole PL, Venkatesh G, Kotecha J, & Sheshala R (2011). Recent advances in sample preparation techniques for effective bioanalytical methods. Biomedical Chromatography: BMC, 25(1–2), 199–217. 10.1002/bmc.1560 [DOI] [PubMed] [Google Scholar]
- Kullolli M, Hancock WS, & Hincapie M (2008). Preparation of a high-performance multi-lectin affinity chromatography (HP-M-LAC) adsorbent for the analysis of human plasma glycoproteins. Journal of Separation Science, 31(14), 2733–2739. 10.1002/jssc.200800233 [DOI] [PubMed] [Google Scholar]
- Kullolli M, Hancock WS, & Hincapie M (2010). Automated Platform for Fractionation of Human Plasma Glycoproteome in Clinical Proteomics. Analytical Chemistry, 82(1), 115–120. 10.1021/ac9013308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-W, Wu I-L, Hsiao H-H, & Khoo K-H (2012). Rapid glycopeptide enrichment and N-glycosylation site mapping strategies based on amine-functionalized magnetic nanoparticles. Analytical and Bioanalytical Chemistry, 402(9), 2765–2776. 10.1007/s00216-012-5724-1 [DOI] [PubMed] [Google Scholar]
- Kurogochi M, Matsushista T, Amano M, Furukawa J, Shinohara Y, Aoshima M, & Nishimura S-I (2010). Sialic Acid-focused Quantitative Mouse Serum Glycoproteomics by Multiple Reaction Monitoring Assay. Molecular & Cellular Proteomics, 9(11), 2354–2368. 10.1074/mcp.M110.000430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MR, Jensen SS, Jakobsen LA, & Heegaard NHH (2007). Exploring the Sialiome Using Titanium Dioxide Chromatography and Mass Spectrometry. Molecular & Cellular Proteomics, 6(10), 1778–1787. 10.1074/mcp.M700086-MCP200 [DOI] [PubMed] [Google Scholar]
- Lehoux S, & Ju T (2017). Chapter Five - Separation of Two Distinct O-Glycoforms of Human IgA1 by Serial Lectin Chromatography Followed by Mass Spectrometry O-Glycan Analysis. In Shukla AK (Ed.), Methods in Enzymology (pp. 61–75). 10.1016/bs.mie.2016.10.003 [DOI] [PubMed] [Google Scholar]
- Li D, Xia H, & Wang L (2018). Branched polyethyleneimine-assisted boronic acid-functionalized silica nanoparticles for the selective enrichment of trace glycoproteins. Talanta, 184, 235–243. 10.1016/j.talanta.2018.02.021 [DOI] [PubMed] [Google Scholar]
- Li H, & Liu Z (2012). Recent advances in monolithic column-based boronate-affinity chromatography. TrAC Trends in Analytical Chemistry, 37, 148–161. 10.1016/j.trac.2012.03.010 [DOI] [Google Scholar]
- Li Q, Zubieta J-K, & Kennedy RT (2009). Practical Aspects of in Vivo Detection of Neuropeptides by Microdialysis Coupled Off-Line to Capillary LC with Multi-stage MS. Analytical Chemistry, 81(6), 2242–2250. 10.1021/ac802391b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Qin Y, Zhong G, Cai C, Chen X, & Chen C (2018). Highly Efficient Separation of Glycoprotein by Dual-Functional Magnetic Metal–Organic Framework with Hydrophilicity and Boronic Acid Affinity. ACS Applied Materials & Interfaces, 10(33), 27612–27620. 10.1021/acsami.8b07671 [DOI] [PubMed] [Google Scholar]
- Li Y, Deng C, & Sun N (2018). Hydrophilic probe in mesoporous pore for selective enrichment of endogenous glycopeptides in biological samples. Analytica Chimica Acta, 1024, 84–92. 10.1016/j.aca.2018.04.030 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Zhang L, Yan G, Yao J, Yang P, & Lu H (2012). Highly specific revelation of rat serum glycopeptidome by boronic acid-functionalized mesoporous silica. Analytica Chimica Acta, 753, 64–72. 10.1016/j.aca.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Liu M-Q, Zeng W-F, Fang P, Cao W-Q, Liu C, Yan G-Q, … Yang P-Y (2017). pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nature Communications, 8(1), 438 10.1038/s41467-017-00535-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Qiu H, Lee RK, Chen W, & Li J (2010). Methylamidation for Sialoglycomics by MALDI-MS: A Facile Derivatization Strategy for Both α2,3- and α2,6-Linked Sialic Acids. Analytical Chemistry, 82(19), 8300–8306. 10.1021/ac101831t [DOI] [PubMed] [Google Scholar]
- Liu Y, Fu D, Yu L, Xiao Y, Peng X, & Liang X (2016). Oxidized dextran facilitated synthesis of a silica-based concanavalin a material for lectin affinity enrichment of glycoproteins/glycopeptides. Journal of Chromatography. A, 1455, 147–155. 10.1016/j.chroma.2016.05.093 [DOI] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, & Darnell J (2000). Protein Glycosylation in the ER and Golgi Complex. Molecular Cell Biology. 4th Edition Retrieved from https://www.ncbi.nlm.nih.gov/books/NBK21744/ [Google Scholar]
- London N, Movshovitz-Attias D, & Schueler-Furman O (2010). The Structural Basis of Peptide-Protein Binding Strategies. Structure, 18(2), 188–199. 10.1016/j.str.2009.11.012 [DOI] [PubMed] [Google Scholar]
- Luque-Garcia JL, & Neubert TA (2007). Sample preparation for serum/plasma profiling and biomarker identification by mass spectrometry. Journal of Chromatography. A, 1153(1–2), 259–276. 10.1016/j.chroma.2006.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K-S, Chen C-C, Lih TM, Cheng C-W, Su W-C, Chang C-H, … Sung T-Y (2015). MAGIC: An Automated N-Linked Glycoprotein Identification Tool Using a Y1-Ion Pattern Matching Algorithm and in Silico MS2 Approach. Analytical Chemistry, 87(4), 2466–2473. 10.1021/ac5044829 [DOI] [PubMed] [Google Scholar]
- Ma C, Qu J, Li X, Zhao X, Li L, Xiao C, … Wang PG (2016). Improvement of core-fucosylated glycoproteome coverage via alternating HCD and ETD fragmentation. Journal of Proteomics, 146, 90–98. 10.1016/j.jprot.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Zhao X, Han H, Tong W, Zhang Q, Qin P, … Qian X (2013). N-linked glycoproteome profiling of human serum using tandem enrichment and multiple fraction concatenation. Electrophoresis, 34(16), 2440–2450. 10.1002/elps.201200662 [DOI] [PubMed] [Google Scholar]
- Ma W-F, Li L-L, Zhang Y, An Q, You L-J, Li J-M, … Wang C-C (2012). Ligand-free strategy for ultrafast and highly selective enrichment of glycopeptides using Ag-coated magnetic nanoarchitectures. Journal of Materials Chemistry, 22(45), 23981–23988. 10.1039/C2JM35196J [DOI] [Google Scholar]
- Ma Y, Wang L, Zhou Y, & Zhang X (2019). A Facilely Synthesized Glutathione-Functionalized Silver Nanoparticles-Grafted Covalent Organic Framework for Rapid and Highly Efficient Enrichment of N-Linked Glycopeptides. Nanoscale. 10.1039/C9NR00392D [DOI] [PubMed] [Google Scholar]
- Masand G, Hanif K, Sen S, Ahsan A, Maiti S, & Pasha S (2006). Synthesis, conformational and pharmacological studies of glycosylated chimeric peptides of Met-enkephalin and FMRFa. Brain Research Bulletin, 68(5), 329–334. 10.1016/j.brainresbull.2005.09.009 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y (2018). Exploring peptide hormones in plants: Identification of four peptide hormone-receptor pairs and two post-translational modification enzymes. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences, 94(2), 59–74. 10.2183/pjab.94.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K, Higashida K, Ishida H, Hata Y, Yamamoto K, Shigeta M, … Taniguchi N (2007). Carbohydrate Binding Specificity of a Fucose-specific Lectin from Aspergillus oryzae A NOVEL PROBE FOR CORE FUCOSE. Journal of Biological Chemistry, 282(21), 15700–15708. 10.1074/jbc.M701195200 [DOI] [PubMed] [Google Scholar]
- McClure DB, Walls JD, & Grinnell BW (1992). Post-translational processing events in the secretion pathway of human protein C, a complex vitamin K-dependent antithrombotic factor. The Journal of Biological Chemistry, 267(27), 19710–19717. [PubMed] [Google Scholar]
- Melo-Braga MN, Ibáñez-Vea M, Larsen MR, & Kulej K (2015). Comprehensive Protocol to Simultaneously Study Protein Phosphorylation, Acetylation, and N-Linked Sialylated Glycosylation. In Posch A (Ed.), Proteomic Profiling: Methods and Protocols (pp. 275–292). 10.1007/978-1-4939-2550-6_21 [DOI] [PubMed] [Google Scholar]
- Morelle W, & Michalski J-C (2007). Analysis of protein glycosylation by mass spectrometry. Nature Protocols, 2(7), 1585–1602. 10.1038/nprot.2007.227 [DOI] [PubMed] [Google Scholar]
- Nagata Y, & Burger MM (1974). Wheat Germ Agglutinin MOLECULAR CHARACTERISTICS AND SPECIFICITY FOR SUGAR BINDING. Journal of Biological Chemistry, 249(10), 3116–3122. [PubMed] [Google Scholar]
- Nasir W, Toledo AG, Noborn F, Nilsson J, Wang M, Bandeira N, & Larson G (2016). SweetNET: A Bioinformatics Workflow for Glycopeptide MS/MS Spectral Analysis. Journal of Proteome Research, 15(8), 2826–2840. 10.1021/acs.jproteome.6b00417 [DOI] [PubMed] [Google Scholar]
- Nilsson J, Rüetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, & Larson G (2009). Enrichment of glycopeptides for glycan structure and attachment site identification. Nature Methods, 6(11), 809–811. 10.1038/nmeth.1392 [DOI] [PubMed] [Google Scholar]
- Nishikaze T (2017). Sensitive and Structure-Informative N-Glycosylation Analysis by MALDI-MS; Ionization, Fragmentation, and Derivatization. Mass Spectrometry (Tokyo, Japan), 6(1), A0060 10.5702/massspectrometry.A0060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum MP, & Blitz DM (2012). Neuropeptide Modulation of Microcircuits. Current Opinion in Neurobiology, 22(4), 592–601. 10.1016/j.conb.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Kameda K, Matsumoto M, & Kawasaki N (2017). Rapid Glycopeptide Enrichment Using Cellulose Hydrophilic Interaction/Reversed-Phase StageTips. Mass Spectrometry (Tokyo, Japan), 6(1), A0061 10.5702/massspectrometry.A0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K, & Marth JD (2006). Glycosylation in Cellular Mechanisms of Health and Disease. Cell, 126(5), 855–867. 10.1016/j.cell.2006.08.019 [DOI] [PubMed] [Google Scholar]
- Ohyama Y, Kasai K, Nomoto H, & Inoue Y (1985). Frontal affinity chromatography of ovalbumin glycoasparagines on a concanavalin A-sepharose column. A quantitative study of the binding specificity of the lectin. The Journal of Biological Chemistry, 260(11), 6882–6887. [PubMed] [Google Scholar]
- Paglia G, Kliman M, Claude E, Geromanos S, & Astarita G (2015). Applications of ion-mobility mass spectrometry for lipid analysis. Analytical and Bioanalytical Chemistry, 407(17), 4995–5007. 10.1007/s00216-015-8664-8 [DOI] [PubMed] [Google Scholar]
- Palmisano G, Lendal SE, Engholm-Keller K, Leth-Larsen R, Parker BL, & Larsen MR (2010). Selective enrichment of sialic acid-containing glycopeptides using titanium dioxide chromatography with analysis by HILIC and mass spectrometry. Nature Protocols, 5(12), 1974–1982. 10.1038/nprot.2010.167 [DOI] [PubMed] [Google Scholar]
- Palmisano G, Lendal SE, & Larsen MR (2011). Titanium dioxide enrichment of sialic acid-containing glycopeptides. Methods in Molecular Biology (Clifton, N.J.), 753, 309–322. 10.1007/978-1-61779-148-2_21 [DOI] [PubMed] [Google Scholar]
- Palmisano G, Parker BL, Engholm-Keller K, Lendal SE, Kulej K, Schulz M, … Larsen MR (2012). A novel method for the simultaneous enrichment, identification, and quantification of phosphopeptides and sialylated glycopeptides applied to a temporal profile of mouse brain development. Molecular & Cellular Proteomics: MCP, 11(11), 1191–1202. 10.1074/mcp.M112.017509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GW, Kim JY, Hwang H, Lee JY, Ahn YH, Lee HK, … Yoo JS (2016). Integrated GlycoProteome Analyzer (I-GPA) for Automated Identification and Quantitation of Site-Specific N-Glycosylation. Scientific Reports, 6, 21175 10.1038/srep21175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-I, Kim J-W, & Yoe SM (2015). Purification and characterization of a novel antibacterial peptide from black soldier fly (Hermetia illucens) larvae. Developmental & Comparative Immunology, 52(1), 98–106. 10.1016/j.dci.2015.04.018 [DOI] [PubMed] [Google Scholar]
- Patel N, Mohd-Radzman NA, Corcilius L, Crossett B, Connolly A, Cordwell SJ, … Djordjevic MA (2018). Diverse Peptide Hormones Affecting Root Growth Identified in the Medicago truncatula Secreted Peptidome. Molecular & Cellular Proteomics, 17(1), 160–174. 10.1074/mcp.RA117.000168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot L, Ravallec R, Fouchereau-Péron M, Vandanjon L, Jaouen P, Chaplain-Derouiniot M, … Bourseau P (2010). Impact of ultrafiltration and nanofiltration of an industrial fish protein hydrolysate on its bioactive properties. Journal of the Science of Food and Agriculture, 90(11), 1819–1826. 10.1002/jsfa.4020 [DOI] [PubMed] [Google Scholar]
- Pohlentz G, Marx K, & Mormann M (2016). Characterization of Protein N-Glycosylation by Analysis of ZIC-HILIC-Enriched Intact Proteolytic Glycopeptides. In Reinders J (Ed.), Proteomics in Systems Biology: Methods and Protocols (pp. 163–179). 10.1007/978-1-4939-3341-9_12 [DOI] [PubMed] [Google Scholar]
- Polson C, Sarkar P, Incledon B, Raguvaran V, & Grant R (2003). Optimization of protein precipitation based upon effectiveness of protein removal and ionization effect in liquid chromatography-tandem mass spectrometry. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences, 785(2), 263–275. [DOI] [PubMed] [Google Scholar]
- Polt R, Porreca F, Szabò LZ, Bilsky EJ, Davis P, Abbruscato TJ, … Hruby VJ (1994). Glycopeptide enkephalin analogues produce analgesia in mice: Evidence for penetration of the blood-brain barrier. Proceedings of the National Academy of Sciences, 91(15), 7114–7118. 10.1073/pnas.91.15.7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin J-F, Amiot J, & Bazinet L (2006). Simultaneous separation of acid and basic bioactive peptides by electrodialysis with ultrafiltration membrane. Journal of Biotechnology, 123(3), 314–328. 10.1016/j.jbiotec.2005.11.016 [DOI] [PubMed] [Google Scholar]
- Powell AK, & Harvey DJ (1996). Stabilization of Sialic Acids in N-linked Oligosaccharides and Gangliosides for Analysis by Positive Ion Matrix-assisted Laser Desorption/Ionization Mass Spectrometry. Rapid Communications in Mass Spectrometry, 10(9), 1027–1032. [DOI] [PubMed] [Google Scholar]
- Reiding KR, Blank D, Kuijper DM, Deelder AM, & Wuhrer M (2014). High-Throughput Profiling of Protein N-Glycosylation by MALDI-TOF-MS Employing Linkage-Specific Sialic Acid Esterification. Analytical Chemistry, 86(12), 5784–5793. 10.1021/ac500335t [DOI] [PubMed] [Google Scholar]
- Reiding KR, Bondt A, Franc V, & Heck AJR (2018). The benefits of hybrid fragmentation methods for glycoproteomics. TrAC Trends in Analytical Chemistry, 108, 260–268. 10.1016/j.trac.2018.09.007 [DOI] [Google Scholar]
- Robert M, Zatylny-Gaudin C, Fournier V, Corre E, Le Corguillé G, Bernay B, & Henry J (2015). Molecular characterization of peptide fractions of a Tilapia (Oreochromis niloticus) by-product hydrolysate and in vitro evaluation of antibacterial activity. Process Biochemistry, 50(3), 487–492. 10.1016/j.procbio.2014.12.022 [DOI] [Google Scholar]
- Rudd PM, Elliott T, Cresswell P, Wilson IA, & Dwek RA (2001). Glycosylation and the Immune System. Science, 291(5512), 2370–2376. 10.1126/science.291.5512.2370 [DOI] [PubMed] [Google Scholar]
- Ruhaak LR, Xu G, Li Q, Goonatilleke E, & Lebrilla CB (2018). Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses. Chemical Reviews, 118(17), 7886–7930. 10.1021/acs.chemrev.7b00732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba J, Dutta S, Hemenway E, & Viner R (2012). Increasing the Productivity of Glycopeptides Analysis by Using Higher-Energy Collision Dissociation-Accurate Mass-Product-Dependent Electron Transfer Dissociation [Research article]. 10.1155/2012/560391 [DOI] [PMC free article] [PubMed]
- Safarik I, & Safarikova M (2004). Magnetic techniques for the isolation and purification of proteins and peptides. Biomagnetic Research and Technology, 2, 7 10.1186/1477-044X-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid MS, Jabeen F, Hussain D, Ashiq MN, & Najam-Ul-Haq M (2017). Hydrazide-functionalized affinity on conventional support materials for glycopeptide enrichment. Analytical and Bioanalytical Chemistry, 409(12), 3135–3143. 10.1007/s00216-017-0254-5 [DOI] [PubMed] [Google Scholar]
- Schottelius M, Wester H-J, Reubi JC, Senekowitsch-Schmidtke R, & Schwaiger M (2002). Improvement of Pharmacokinetics of Radioiodinated Tyr3-Octreotide by Conjugation with Carbohydrates. Bioconjugate Chemistry, 13(5), 1021–1030. 10.1021/bc0200069 [DOI] [PubMed] [Google Scholar]
- Schulze WX, & Mann M (2004). A Novel Proteomic Screen for Peptide-Protein Interactions. Journal of Biological Chemistry, 279(11), 10756–10764. 10.1074/jbc.M309909200 [DOI] [PubMed] [Google Scholar]
- Schweinsberg C, Maes V, Brans L, Bläuenstein P, Tourwé DA, Schubiger PA, … Garayoa EG (2008). Novel Glycated [99mTc(CO)3]-Labeled Bombesin Analogues for Improved Targeting of Gastrin-Releasing Peptide Receptor-Positive Tumors. Bioconjugate Chemistry, 19(12), 2432–2439. 10.1021/bc800319g [DOI] [PubMed] [Google Scholar]
- Sekiya S, Wada Y, & Tanaka K (2005). Derivatization for stabilizing sialic acids in MALDI-MS. Analytical Chemistry, 77(15), 4962–4968. 10.1021/ac050287o [DOI] [PubMed] [Google Scholar]
- Selman MHJ, Hemayatkar M, Deelder AM, & Wuhrer M (2011). Cotton HILIC SPE Microtips for Microscale Purification and Enrichment of Glycans and Glycopeptides. Analytical Chemistry, 83(7), 2492–2499. 10.1021/ac1027116 [DOI] [PubMed] [Google Scholar]
- Semenov AG, Postnikov AB, Tamm NN, Seferian KR, Karpova NS, Bloshchitsyna MN, … Katrukha AG (2009). Processing of Pro–Brain Natriuretic Peptide Is Suppressed by O-Glycosylation in the Region Close to the Cleavage Site. Clinical Chemistry, 55(3), 489–498. 10.1373/clinchem.2008.113373 [DOI] [PubMed] [Google Scholar]
- Seo J, & Lee K-J (2004). Post-translational modifications and their biological functions: Proteomic analysis and systematic approaches. Journal of Biochemistry and Molecular Biology, 37(1), 35–44. [DOI] [PubMed] [Google Scholar]
- Serrano ID, Ribeiro MMB, & Castanho M a. RB (2012). A focus on glucose-mediated drug delivery to the central nervous system. Mini Reviews in Medicinal Chemistry, 12(4), 301–312. [DOI] [PubMed] [Google Scholar]
- Sheng Q, Li X, Yin W, Yu L, Ke Y, & Liang X (2013). Retention mechanism and enrichment of glycopeptides on titanium dioxide. Analytical Methods, 5(24), 7072–7080. 10.1039/C3AY41294F [DOI] [Google Scholar]
- Shubhakar A, Kozak RP, Reiding KR, Royle L, Spencer DIR, Fernandes DL, & Wuhrer M (2016). Automated High-Throughput Permethylation for Glycosylation Analysis of Biologics Using MALDI-TOF-MS. Analytical Chemistry, 88(17), 8562–8569. 10.1021/acs.analchem.6b01639 [DOI] [PubMed] [Google Scholar]
- Simpson DM, & Beynon RJ (2010). Acetone precipitation of proteins and the modification of peptides. Journal of Proteome Research, 9(1), 444–450. 10.1021/pr900806x [DOI] [PubMed] [Google Scholar]
- Singh C, Zampronio CG, Creese AJ, & Cooper HJ (2012). Higher Energy Collision Dissociation (HCD) Product Ion-Triggered Electron Transfer Dissociation (ETD) Mass Spectrometry for the Analysis of N-Linked Glycoproteins. Journal of Proteome Research, 11(9), 4517–4525. 10.1021/pr300257c [DOI] [PubMed] [Google Scholar]
- Snovida SI, Bodnar ED, Viner R, Saba J, & Perreault H (2010). A simple cellulose column procedure for selective enrichment of glycopeptides and characterization by nano LC coupled with electron-transfer and high-energy collisional-dissociation tandem mass spectrometry. Carbohydrate Research, 345(6), 792–801. 10.1016/j.carres.2010.01.006 [DOI] [PubMed] [Google Scholar]
- Souverain S, Rudaz S, & Veuthey J-L (2004). Restricted access materials and large particle supports for on-line sample preparation: An attractive approach for biological fluids analysis. Journal of Chromatography B, 801(2), 141–156. 10.1016/j.jchromb.2003.11.043 [DOI] [PubMed] [Google Scholar]
- Sparbier K, Koch S, Kessler I, Wenzel T, & Kostrzewa M (2005). Selective isolation of glycoproteins and glycopeptides for MALDI-TOF MS detection supported by magnetic particles. Journal of Biomolecular Techniques: JBT, 16(4), 407–413. [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen K, Hinneburg H, Thaysen-Andersen M, Hartmann L, Varón Silva D, Fuchser J, … Kolarich D (2013). Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: An evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. Journal of Mass Spectrometry: JMS, 48(6), 627–639. 10.1002/jms.3210 [DOI] [PubMed] [Google Scholar]
- Stavenhagen K, Kayili HM, Holst S, Koeleman CAM, Engel R, Wouters D, … Wuhrer M (2018). N- and O-glycosylation Analysis of Human C1-inhibitor Reveals Extensive Mucin-type O-Glycosylation. Molecular & Cellular Proteomics, 17(6), 1225–1238. 10.1074/mcp.RA117.000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana K, Nakamura S, Wang H, Iwasaki H, Tachibana K, Maebara K, … Narimatsu H (2006). Elucidation of binding specificity of Jacalin toward O-glycosylated peptides: Quantitative analysis by frontal affinity chromatography. Glycobiology, 16(1), 46–53. 10.1093/glycob/cwj038 [DOI] [PubMed] [Google Scholar]
- Takegawa Y, Deguchi K, Ito H, Keira T, Nakagawa H, & Nishimura S-I (2006). Simple separation of isomeric sialylated N-glycopeptides by a zwitterionic type of hydrophilic interaction chromatography. Journal of Separation Science, 29(16), 2533–2540. 10.1002/jssc.200600133 [DOI] [PubMed] [Google Scholar]
- Andrews WT, Skube SB, & Hummon AB (2018). Magnetic bead-based peptide extraction methodology for tissue imaging. Analyst, 143(1), 133–140. 10.1039/C7AN00757D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Liu Y, Qi D, Yao G, Deng C, & Zhang X (2009). On-plate-selective enrichment of glycopeptides using boronic acid-modified gold nanoparticles for direct MALDI-QIT-TOF MS analysis. Proteomics, 9(22), 5046–5055. 10.1002/pmic.200900033 [DOI] [PubMed] [Google Scholar]
- Thaysen-Andersen M, Mysling S, & Højrup P (2009). Site-Specific Glycoprofiling of N-Linked Glycopeptides Using MALDI-TOF MS: Strong Correlation between Signal Strength and Glycoform Quantities. Analytical Chemistry, 81(10), 3933–3943. 10.1021/ac900231w [DOI] [PubMed] [Google Scholar]
- Thingholm TE, Jørgensen TJD, Jensen ON, & Larsen MR (2006). Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nature Protocols, 1(4), 1929–1935. 10.1038/nprot.2006.185 [DOI] [PubMed] [Google Scholar]
- Thingholm TE, & Larsen MR (2016). The Use of Titanium Dioxide for Selective Enrichment of Phosphorylated Peptides. Methods in Molecular Biology (Clifton, N.J.), 1355, 135–146. 10.1007/978-1-4939-3049-4_9 [DOI] [PubMed] [Google Scholar]
- Toghi Eshghi S, Shah P, Yang W, Li X, & Zhang H (2015). GPQuest: A Spectral Library Matching Algorithm for Site-Specific Assignment of Tandem Mass Spectra to Intact N-glycopeptides. Analytical Chemistry, 87(10), 5181–5188. 10.1021/acs.analchem.5b00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totten SM, Adusumilli R, Kullolli M, Tanimoto C, Brooks JD, Mallick P, & Pitteri SJ (2018). Multi-lectin Affinity Chromatography and Quantitative Proteomic Analysis Reveal Differential Glycoform Levels between Prostate Cancer and Benign Prostatic Hyperplasia Sera. Scientific Reports, 8(1), 6509 10.1038/s41598-018-24270-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totten SM, Kullolli M, & Pitteri SJ (2017). Multi-Lectin Affinity Chromatography for Separation, Identification, and Quantitation of Intact Protein Glycoforms in Complex Biological Mixtures. In Comai L, Katz JE, & Mallick P (Eds.), Proteomics: Methods and Protocols (pp. 99–113). 10.1007/978-1-4939-6747-6_9 [DOI] [PubMed] [Google Scholar]
- Varamini P, Mansfeld FM, Blanchfield JT, Wyse BD, Smith MT, & Toth I (2012). Synthesis and Biological Evaluation of an Orally Active Glycosylated Endomorphin-1. Journal of Medicinal Chemistry, 55(12), 5859–5867. 10.1021/jm300418d [DOI] [PubMed] [Google Scholar]
- Villanueva J, Philip J, Entenberg D, Chaparro CA, Tanwar MK, Holland EC, & Tempst P (2004). Serum Peptide Profiling by Magnetic Particle-Assisted, Automated Sample Processing and MALDI-TOF Mass Spectrometry. Analytical Chemistry, 76(6), 1560–1570. 10.1021/ac0352171 [DOI] [PubMed] [Google Scholar]
- Vreeker GCM, Nicolardi S, Bladergroen MR, van der Plas CJ, Mesker WE, Tollenaar RAEM, … Wuhrer M (2018). Automated Plasma Glycomics with Linkage-Specific Sialic Acid Esterification and Ultrahigh Resolution MS. Analytical Chemistry, 90(20), 11955–11961. 10.1021/acs.analchem.8b02391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter RE, Cramer JA, & Tse FL (2001). Comparison of manual protein precipitation (PPT) versus a new small volume PPT 96-well filter plate to decrease sample preparation time. Journal of Pharmaceutical and Biomedical Analysis, 25(2), 331–337. [DOI] [PubMed] [Google Scholar]
- Wan H, Yan J, Yu L, Sheng Q, Zhang X, Xue X, … Liang X (2011). Zirconia layer coated mesoporous silica microspheres as HILIC SPE materials for selective glycopeptide enrichment. Analyst, 136(21), 4422–4430. 10.1039/C1AN15554G [DOI] [PubMed] [Google Scholar]
- Wang PG, He W, & He W (2011). Hydrophilic Interaction Liquid Chromatography (HILIC) and Advanced Applications. 10.1201/b10609 [DOI]
- Wang X, Xia N, & Liu L (2013). Boronic Acid-based approach for separation and immobilization of glycoproteins and its application in sensing. International Journal of Molecular Sciences, 14(10), 20890–20912. 10.3390/ijms141020890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wu R, Chen H, Sun N, & Deng C (2018). Synthesis of zwitterionic hydrophilic magnetic mesoporous silica materials for endogenous glycopeptide analysis in human saliva. Nanoscale, 10(11), 5335–5341. 10.1039/C7NR08613J [DOI] [PubMed] [Google Scholar]
- Watanabe A, Nishijima K, Zhao S, Tanaka Y, Itoh T, Takemoto H, … Kuge Y (2012). Effect of glycosylation on biodistribution of radiolabeled glucagon-like peptide 1. Annals of Nuclear Medicine, 26(2), 184–191. 10.1007/s12149-011-0558-z [DOI] [PubMed] [Google Scholar]
- Watt AP, Morrison D, Locker KL, & Evans DC (2000). Higher throughput bioanalysis by automation of a protein precipitation assay using a 96-well format with detection by LC-MS/MS. Analytical Chemistry, 72(5), 979–984. [DOI] [PubMed] [Google Scholar]
- Webster SG, Keller R, & Dircksen H (2012). The CHH-superfamily of multifunctional peptide hormones controlling crustacean metabolism, osmoregulation, moulting, and reproduction. General and Comparative Endocrinology, 175(2), 217–233. 10.1016/j.ygcen.2011.11.035 [DOI] [PubMed] [Google Scholar]
- Weis WI, & Drickamer K (1996). Structural basis of lectin-carbohydrate recognition. Annual Review of Biochemistry, 65, 441–473. 10.1146/annurev.bi.65.070196.002301 [DOI] [PubMed] [Google Scholar]
- Whiteaker JR, Zhao L, Zhang HY, Feng L-C, Piening BD, Anderson L, & Paulovich AG (2007). Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Analytical Biochemistry, 362(1), 44–54. 10.1016/j.ab.2006.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D, Ackloo S, Zhu P, Bowden P, Evans KR, Addison CL, … Marshall JG (2010). Precipitation and selective extraction of human serum endogenous peptides with analysis by quadrupole time-of-flight mass spectrometry reveals posttranslational modifications and low-abundance peptides. Analytical and Bioanalytical Chemistry, 396(3), 1223–1247. 10.1007/s00216-009-3345-0 [DOI] [PubMed] [Google Scholar]
- Witt KA, & Davis TP (2006). CNS drug delivery: Opioid peptides and the blood-brain barrier. The AAPS Journal, 8(1), E76–E88. 10.1208/aapsj080109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-L, Hühmer AFR, Hao Z, & Karger BL (2007). On-Line LC–MS Approach Combining Collision-Induced Dissociation (CID), Electron-Transfer Dissociation (ETD), and CID of an Isolated Charge-Reduced Species for the Trace-Level Characterization of Proteins with Post-Translational Modifications. Journal of Proteome Research, 6(11), 4230–4244. 10.1021/pr070313u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-W, Pu T-H, Viner R, & Khoo K-H (2014). Novel LC-MS2 product dependent parallel data acquisition function and data analysis workflow for sequencing and identification of intact glycopeptides. Analytical Chemistry, 86(11), 5478–5486. 10.1021/ac500945m [DOI] [PubMed] [Google Scholar]
- Wu Z, Li H, Zhang Q, Liu X, Zheng Q, & Li J (2017). Characterization of O-acetylation in sialoglycans by MALDI-MS using a combination of methylamidation and permethylation. Scientific Reports, 7, 46206 10.1038/srep46206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Suttapitugsakul S, Sun F, & Wu R (2018). Mass Spectrometry-Based Chemical and Enzymatic Methods for Global Analysis of Protein Glycosylation. Accounts of Chemical Research, 51(8), 1796–1806. 10.1021/acs.accounts.8b00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, & Deng C (2017). Designed synthesis of a “One for Two” hydrophilic magnetic amino-functionalized metal-organic framework for highly efficient enrichment of glycopeptides and phosphopeptides. Scientific Reports, 7(1), 1162 10.1038/s41598-017-01341-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Liu Q, Li Y, & Deng C (2018). Core-shell structured magnetic metal-organic framework composites for highly selective detection of N-glycopeptides based on boronic acid affinity chromatography. Journal of Chromatography A, 1540, 87–93. 10.1016/j.chroma.2018.02.013 [DOI] [PubMed] [Google Scholar]
- Xiong Z, Qin H, Wan H, Huang G, Zhang Z, Dong J, … Zou H (2013). Layer-by-layer assembly of multilayer polysaccharide coated magnetic nanoparticles for the selective enrichment of glycopeptides. Chemical Communications, 49(81), 9284–9286. 10.1039/C3CC45008B [DOI] [PubMed] [Google Scholar]
- Xu Y, Wu Z, Zhang L, Lu H, Yang P, Webley PA, & Zhao D (2009). Highly Specific Enrichment of Glycopeptides Using Boronic Acid-Functionalized Mesoporous Silica. Analytical Chemistry, 81(1), 503–508. 10.1021/ac801912t [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhang L, Lu H, & Yang P (2010). On-plate enrichment of glycopeptides by using boronic acid functionalized gold-coated Si wafer. PROTEOMICS, 10(5), 1079–1086. 10.1002/pmic.200900097 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Tsuji T, Matsumoto I, & Osawa T (1981). Structural requirements for the binding of oligosaccharides and glycopeptides to immobilized wheat germ agglutinin. Biochemistry, 20(20), 5894–5899. 10.1021/bi00523a037 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nair P, Jacobsen NE, Vagner J, Kulkarni V, Davis P, … Hruby VJ (2009). Improving Metabolic Stability by Glycosylation: Bifunctional Peptide Derivatives That Are Opioid Receptor Agonists and Neurokinin 1 Receptor Antagonists. Journal of Medicinal Chemistry, 52(16), 5164–5175. 10.1021/jm900473p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Kochibe N, Ohkura T, Ueda I, & Kobata A (1985). Fractionation of L-fucose-containing oligosaccharides on immobilized Aleuria aurantia lectin. Journal of Biological Chemistry, 260(8), 4688–4693. [PubMed] [Google Scholar]
- Yan J, Li X, Yu L, Jin Y, Zhang X, Xue X, … Liang X (2010). Selective enrichment of glycopeptides/phosphopeptides using porous titania microspheres. Chemical Communications, 46(30), 5488–5490. 10.1039/C000094A [DOI] [PubMed] [Google Scholar]
- Yang H, Yang C, & Sun T (2018). Characterization of glycopeptides using a stepped higher-energy C-trap dissociation approach on a hybrid quadrupole orbitrap. Rapid Communications in Mass Spectrometry, 32(16), 1353–1362. 10.1002/rcm.8191 [DOI] [PubMed] [Google Scholar]
- Yang M, Wu Z, & Fields S (1995). Protein-peptide interactions analyzed with the yeast two-hybrid system. Nucleic Acids Research, 23(7), 1152–1156. 10.1093/nar/23.7.1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, & Hancock WS (2004). Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. Journal of Chromatography A, 1053(1), 79–88. 10.1016/j.chroma.2004.08.150 [DOI] [PubMed] [Google Scholar]
- Yang Z, Harris LE, Palmer-Toy DE, & Hancock WS (2006). Multilectin Affinity Chromatography for Characterization of Multiple Glycoprotein Biomarker Candidates in Serum from Breast Cancer Patients. Clinical Chemistry, 52(10), 1897–1905. 10.1373/clinchem.2005.065862 [DOI] [PubMed] [Google Scholar]
- Yin X, Bern M, Xing Q, Ho J, Viner R, & Mayr M (2013). Glycoproteomic analysis of the secretome of human endothelial cells. Molecular & Cellular Proteomics: MCP, 12(4), 956–978. 10.1074/mcp.M112.024018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Geller RG, & Pisano JJ (1976). Vespulakinins: New carbohydrate-containing bradykinin derivatives. Biochemistry, 15(1), 61–64. [DOI] [PubMed] [Google Scholar]
- Yu A, Zhao J, Peng W, Banazadeh A, Williamson SD, Goli M, … Mechref Y (2018). Advances in mass spectrometry-based glycoproteomics. ELECTROPHORESIS, 39(24), 3104–3122. 10.1002/elps.201800272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Li X, Guo Z, Zhang X, & Liang X (2009). Hydrophilic Interaction Chromatography Based Enrichment of Glycopeptides by Using Click Maltose: A Matrix with High Selectivity and Glycosylation Heterogeneity Coverage. Chemistry – A European Journal, 15(46), 12618–12626. 10.1002/chem.200902370 [DOI] [PubMed] [Google Scholar]
- Yu Q, Canales A, Glover MS, Das R, Shi X, Liu Y, … Li L (2017). Targeted Mass Spectrometry Approach Enabled Discovery of O-Glycosylated Insulin and Related Signaling Peptides in Mouse and Human Pancreatic Islets. Analytical Chemistry, 89(17), 9184–9191. 10.1021/acs.analchem.7b01926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Wang B, Chen Z, Urabe G, Glover MS, Shi X, … Li L (2017). Electron-Transfer/Higher-Energy Collision Dissociation (EThcD)-Enabled Intact Glycopeptide/Glycoproteome Characterization. Journal of The American Society for Mass Spectrometry, 28(9), 1751–1764. 10.1007/s13361-017-1701-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias LG, Hartmann AK, Song E, Zhao J, Zhu R, Mirzaei P, & Mechref Y (2016). HILIC and ERLIC Enrichment of Glycopeptides Derived from Breast and Brain Cancer Cells. Journal of Proteome Research, 15(10), 3624–3634. 10.1021/acs.jproteome.6b00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W-F, Liu M-Q, Zhang Y, Wu J-Q, Fang P, Peng C, … Yang P (2016). pGlyco: A pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3. Scientific Reports, 6, 25102 10.1038/srep25102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Hincapie M, Pitteri SJ, Hanash S, Schalkwijk J, Hogan JM, … Hancock WS (2011). A Proteomics Platform Combining Depletion, Multi-lectin Affinity Chromatography (M-LAC), and Isoelectric Focusing to Study the Breast Cancer Proteome. Analytical Chemistry, 83(12), 4845–4854. 10.1021/ac2002802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Sheng Q, Li X, Liang Q, Yan J, & Liang X (2011). Selective enrichment of glycopeptides for mass spectrometry analysis using C18 fractionation and titanium dioxide chromatography. Journal of Separation Science, 34(19), 2745–2750. 10.1002/jssc.201100427 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li X, Martin DB, & Aebersold R (2003). Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature Biotechnology, 21(6), 660–666. 10.1038/nbt827 [DOI] [PubMed] [Google Scholar]
- Zhang L, Jiang H, Yao J, Wang Y, Fang C, Yang P, & Lu H (2014). Highly specific enrichment of N-linked glycopeptides based on hydrazide functionalized soluble nanopolymers. Chemical Communications, 50(8), 1027–1029. 10.1039/C3CC47347C [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang J, He X, Chen L, & Zhang Y (2015). Tailor-Made Boronic Acid Functionalized Magnetic Nanoparticles with a Tunable Polymer Shell-Assisted for the Selective Enrichment of Glycoproteins/Glycopeptides. ACS Applied Materials & Interfaces, 7(44), 24576–24584. 10.1021/acsami.5b06445 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xie X, Zhao X, Tian F, Lv J, Ying W, & Qian X (2018). Systems analysis of singly and multiply O-glycosylated peptides in the human serum glycoproteome via EThcD and HCD mass spectrometry. Journal of Proteomics, 170, 14–27. 10.1016/j.jprot.2017.09.014 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chen Y, Xiong Z, Sun X, Zhang Q, Gan Y, … Zhang W (2017). Synthesis of magnetic zwitterionic–hydrophilic material for the selective enrichment of N-linked glycopeptides. Journal of Chromatography A, 1482, 23–31. 10.1016/j.chroma.2016.12.054 [DOI] [PubMed] [Google Scholar]
- Zheng X, Baker H, & Hancock WS (2006). Analysis of the low molecular weight serum peptidome using ultrafiltration and a hybrid ion trap-Fourier transform mass spectrometer. Journal of Chromatography. A, 1120(1–2), 173–184. 10.1016/j.chroma.2006.01.098 [DOI] [PubMed] [Google Scholar]
- Zhou H, Tian R, Ye M, Xu S, Feng S, Pan C, … Zou H (2007). Highly specific enrichment of phosphopeptides by zirconium dioxide nanoparticles for phosphoproteome analysis. ELECTROPHORESIS, 28(13), 2201–2215. 10.1002/elps.200600718 [DOI] [PubMed] [Google Scholar]
- Zhu F, Clemmer DE, & Trinidad JC (2016). Characterization of lectin binding affinities via direct LC-MS profiling: Implications for glycopeptide enrichment and separation strategies. Analyst, 142(1), 65–74. 10.1039/C6AN02043G [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen Z, Zhang J, An M, Wu J, Yu Q, … Lubman DM (2018). Differential Quantitative Determination of Site-Specific Intact N-Glycopeptides in Serum Haptoglobin between Hepatocellular Carcinoma and Cirrhosis Using LC-EThcD-MS/MS. Journal of Proteome Research. 10.1021/acs.jproteome.8b00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zougman A, Pilch B, Podtelejnikov A, Kiehntopf M, Schnabel C, Kumar C, & Mann M (2008). Integrated Analysis of the Cerebrospinal Fluid Peptidome and Proteome. Journal of Proteome Research, 7(1), 386–399. 10.1021/pr070501k [DOI] [PubMed] [Google Scholar]


