The emergence and spread of antibiotic resistance along the food chain can be influenced by the different antimicrobial strategies used from farm to fork. This study evidences that two novel, not yet widely used, nonthermal microbial decontamination techniques, UV light and nonthermal atmospheric plasma, can select variants with increased resistance to various clinically relevant antibiotics, such as ciprofloxacin, streptomycin, tetracycline, and erythromycin. Whole-genome analysis of the resistant variants obtained for Salmonella spp. allowed identification of the genetic changes responsible for the observed phenotypes and suggested that some antimicrobial classes are more susceptible to the cross-resistance phenomena observed. This information is relevant, since these novel decontamination techniques are being proposed as possible alternative green techniques for the decontamination of environments and equipment in food and clinical settings.
KEYWORDS: antimicrobial resistance, ultraviolet light, plasma, resistant variants
ABSTRACT
This study was aimed at assessing whether the repeated exposure of 12 strains of Salmonella spp., Escherichia coli, and Listeria monocytogenes to alternative nonthermal decontamination techniques with UV light (UV-C) and nonthermal atmospheric plasma (NTAP) may cause the emergence of variants showing increased resistance to clinically relevant antibiotics (ampicillin, cefotaxime, ciprofloxacin, gentamicin, streptomycin, tetracycline, erythromycin, vancomycin, and colistin). UV-C and NTAP treatments were applied on the surface of inoculated brain heart infusion (BHI) agar plates. Survivors were recovered and after 24 h of growth in BHI broth were again subjected to the decontamination treatment; this was repeated for 10 consecutive cycles. A total of 174 strain/decontamination technique/antibiotic combinations were tested, and 12 variant strains with increased resistance to one of the antibiotics studied were identified, with the increases in the MICs in Mueller-Hinton broth ranging from 2- to 256-fold. The variant strains of Salmonella spp. isolated were further characterized through phenotypic screenings and whole-genome sequencing (WGS) analyses. Most changes in susceptibility were observed for antibiotics that act at the level of protein synthesis (aminoglycosides, tetracyclines, and glycylcyclines) or DNA replication (fluoroquinolones), as well as for polymyxins. No changes in resistance to β-lactams were detected. WGS analyses showed the occurrence of sequence alterations in some antibiotic cellular targets (e.g., gyrA for ciprofloxacin-resistant variants, rpsL for a streptomycin-resistant variant), accompanied by variations in stress response regulators and membrane transporters likely involved in the nonselective efflux of antibiotics, which altogether resulted in a low- to medium-level increase in microbial resistance to several antibiotics.
IMPORTANCE The emergence and spread of antibiotic resistance along the food chain can be influenced by the different antimicrobial strategies used from farm to fork. This study evidences that two novel, not yet widely used, nonthermal microbial decontamination techniques, UV light and nonthermal atmospheric plasma, can select variants with increased resistance to various clinically relevant antibiotics, such as ciprofloxacin, streptomycin, tetracycline, and erythromycin. Whole-genome analysis of the resistant variants obtained for Salmonella spp. allowed identification of the genetic changes responsible for the observed phenotypes and suggested that some antimicrobial classes are more susceptible to the cross-resistance phenomena observed. This information is relevant, since these novel decontamination techniques are being proposed as possible alternative green techniques for the decontamination of environments and equipment in food and clinical settings.
INTRODUCTION
In Europe alone, antimicrobial resistance (AMR) is responsible for 25,000 deaths annually, accounting for €1.5 billion in health care-associated costs, and it is predicted that by 2050 it could cause even more deaths than cancer (1).
In the last few decades, there has been a growing concern over the possibility of AMR transmission via the food chain (2–4). In this regard, research carried out in the last few years points toward the relevance of food-processing environments as potential hot spots for AMR acquisition and spread and as AMR reservoirs (5). In particular, exposure to some biocides and disinfection agents has been characterized as a selective pressure that may favor the emergence of resistance to certain clinically relevant antibiotics (6, 7).
Several authors have been able to isolate stable mutant strains with increased resistance to one or several antibiotics after serial or prolonged exposure to sublethal concentrations of biocides frequently used for the decontamination and sanitation of surfaces and equipment in health care and food industry-related settings. For instance, Langsrud and coauthors (8) reported that serial cultivation of two Escherichia coli strains in the presence of subinhibitory concentrations of benzalkonium chloride resulted in increased resistance to various antibiotics (ampicillin, penicillin G, norfloxacin, nalidixic acid, kanamycin, gentamicin, chloramphenicol, tetracycline, and erythromycin), with the MICs being 1.5- to 20-fold higher than those observed for control cultures. Similarly, for Salmonella enterica serovar Typhimurium, several authors have described that exposure to widely used disinfectants (e.g., aldehyde-based disinfectants, blends of oxidizing compounds, quaternary ammonium compounds, biocides composed of organic acids and surfactants, halogenated tertiary amine compounds) selected multidrug-resistant mutants with decreased susceptibility to clinically relevant antibiotics, such as ciprofloxacin, chloramphenicol, tetracycline, nalidixic acid, and/or ampicillin (7, 9–11). In addition, it has also been suggested that microbial exposure to other common stresses prevailing in the food industry might also facilitate AMR emergence (12).
These phenomena can be a result of cross-resistance or coresistance events. In cross-resistance events, a single determinant (a single gene or enzyme or a unique substrate modification) confers at the same time resistance to different antibiotics and/or antimicrobial substances, including biocides. For instance, some multidrug efflux pumps, which have more than one substrate that might be chemically unrelated to each other, can simultaneously confer resistance to antibiotics and biocides when they are overexpressed (13). On the other hand, in coresistance events, various different resistance determinants (including both antibiotic resistance and biocide resistance determinants) are associated together on common genetic elements, like plasmids, phages, integrons, or transposons, which can spread to other strains, species, or genera (14). Then, all these determinants closely associated in a single mobile genetic element can be coselected when one of the antimicrobials acts as a selective pressure (15).
Further research efforts are therefore warranted to identify novel ecofriendly decontamination and sanitation strategies which maintain a high level of antimicrobial activity while avoiding the generation of AMR. UV light (UV-C) and nonthermal atmospheric plasma (NTAP) are among those alternative technologies which have shown great potential to be used as surface decontamination techniques. UV-C radiations, which have wavelengths in the range of 220 to 280 nm, inhibit bacterial multiplication by inducing the formation of thymine dimers, thus preventing DNA replication and transcription (16, 17). In addition, they promote the formation of toxic free radicals. NTAP is a recently developed technology which is based on the ionization of a carrier gas through the application of electric discharges at atmospheric pressure and room temperature, with the consequent generation of electrons, ions, UV photons, charged particles, and free radicals (including reactive oxygen and nitrogen species), which cause direct effects that damage microbial cell membranes, DNA, and proteins (18–20). However, although UV light lamps are already implemented in some food-processing facilities, mainly as insect light traps and for some specific decontamination uses, such as in water decontamination, nonthermal atmospheric plasma is still being validated as an effective food and surface decontamination technique (21). In addition, the ability of UV-C and NTAP treatments to induce cross-resistance or coresistance to clinically relevant antibiotics has not been studied yet.
The objectives of this study were (i) to assess whether the exposure of representative strains of the foodborne pathogens Salmonella spp., Escherichia coli, and Listeria monocytogenes to (sub)lethal UV-C and NTAP treatments may cause the emergence of variants showing increased resistance to antibiotics of clinical interest belonging to different antimicrobial classes, i.e., aminoglycosides, β-lactams, glycopeptides, macrolides, quinolones, and tetracyclines, and (ii) to deeply characterize the AMR variants of Salmonella spp. through phenotypic screenings, with the aim of identifying multidrug-resistant phenotypes, and whole-genome sequencing (WGS) analyses, with the aim of identifying the polymorphisms responsible for the observed resistance phenotypes.
RESULTS AND DISCUSSION
Process of adaptation to UV-C and NTAP treatments.
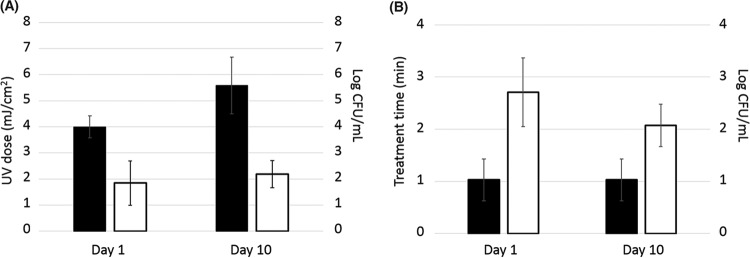
The repeated exposure of E. coli, L. monocytogenes, and Salmonella spp. to UV-C and NTAP gave rise to a gradual decrease in bacterial susceptibility to these decontamination techniques. The aim was to induce a severe stress to microbial cells while the recovery of survivors with potential adaptive mutations was still possible. Thus, for UV-C, it was necessary to gradually increase the treatment intensity, which ranged from 3 to 5 mJ/cm2 at day 1 and from 4 to 8 mJ/cm2 at day 10 of the adaptation process, to achieve similar reduction levels of 1.84 ± 0.85 and 2.19 ± 0.52 log CFU/ml, respectively (Fig. 1A). On the other hand, for NTAP, the treatment time remained unchanged for each of the strains throughout the adaptation process, since the log reductions achieved were always limited to the 1- to 3-log range. Nevertheless, although a decrease in the effectiveness of NTAP treatments as the evolution process progressed was observed, the differences in log reductions found between day 1 (2.71 ± 0.66) and day 10 (2.07 ± 0.41) of the adaptation process were not significant (Fig. 1B).
FIG 1.
(A) Changes in UV dose (black bars) and log reductions (white bars) obtained from day 1 to day 10 of the evolution process. (B) Changes in nonthermal atmospheric plasma (NTAP) treatment time (black bars) and log reductions (white bars) obtained from day 1 to day 10 of the evolution process.
It is worth highlighting that the selected treatment intensities are likely lower than the ones expected to be used in practice in food industries. For instance, the U.S. Environmental Protection Agency (EPA) (22) and Austrian standards (23) recommend a UV-C dose of 40 mJ/cm2 for complete water disinfection. Nevertheless, exposure to lower, sublethal UV-C or NTAP doses may occur in selected niches (e.g., under objects or in cracks and crevices and other harborage sites or due to the presence of organic matter) or for cells within microbial biofilms. Previous research studies which followed a similar direct evolution approach have shown that adaptive mutagenesis can occur upon recurrent microbial exposure to lethal stress conditions, leading to the generation and proliferation of hyperresistant clonal variants in stressed populations (12). Thus, selection of resistant variants after exposure to different food-processing stresses has already been reported for various foodborne pathogens, including E. coli for high hydrostatic pressure (24, 25), Salmonella spp. for acid stress (26), and L. monocytogenes for high hydrostatic pressure (27), heat processing (28), or acid (29) stress. The progressive increase in the UV-C and NTAP resistance of the adapted populations along the evolution process observed in our study suggests that UV-C- and NTAP-resistant variants may also be generated through adaptive mutagenesis following repeated microbial exposure to these decontamination technologies at (sub)lethal intensities.
Isolation of variants with increased AMR.
Assessment of the MICs of the antibiotics ampicillin, cefotaxime, ciprofloxacin, colistin, erythromycin, gentamicin, streptomycin, tetracycline, and vancomycin for the 12 E. coli, L. monocytogenes, and Salmonella reference strains tested on Mueller-Hinton (MH) agar plates showed that all reference strains could be considered sensitive to the different antibiotics according to the EUCAST epidemiological cutoff (ECOFF) values, with the exception of cefotaxime for E. coli CECT 515, for which the MIC was 8 μg/ml, and streptomycin for S. Typhimurium CECT 4595, for which the MIC was 128 μg/ml (see Table S1 in the supplemental material). It is nevertheless important to highlight that the MIC of an antibiotic diluted in agar may vary from its MIC in broth, which is the medium used for the standard method followed by EUCAST (30, 31). Taking all this into account, the selected reference strains were considered good candidates for evaluation of the emergence of new AMR phenotypes following repeated exposure to the UV-C and NTAP decontamination technologies.
In total, 174 strain/decontamination technique/antibiotic combinations were tested in order to identify newly developed AMR variants following the UV-C or NTAP treatments. By following this approach, it was possible to identify 12 variant strains with increased resistance to one of the antibiotics tested (6.9% rate); 5 of these were generated upon UV-C exposure, and 7 were generated through NTAP treatments (Table 1). These included two tetracycline-resistant E. coli variants; four Salmonella variants, two of which showed increased resistance to ciprofloxacin and two of which showed increased resistance to streptomycin; and six L. monocytogenes variants, four of which showed increased resistance to ciprofloxacin, one of which showed increased resistance to streptomycin, and one of which showed increased resistance to erythromycin. The increases in MICs observed in MH broth for these variants ranged from 2-fold (for two L. monocytogenes strains, adapted to UV-C and NTAP, respectively, and resistant to ciprofloxacin) to 256-fold (for an E. coli variant strain selected through NTAP exposure and resistant to streptomycin) (Table 1).
TABLE 1.
Variant strains with reduced susceptibility to antibiotics obtained through repeated exposure to UV light and NTAP
| Strain | Reference strain | CECT no. | Treatment | Antibiotic to which resistance was increased | MIC (μg/ml) |
|
|---|---|---|---|---|---|---|
| Before adaptation | After adaptation | |||||
| 1-EVOL | S. Typhimurium | 4594 | UV light | Streptomycin | 32 | 512 |
| 2-EVOL | S. Enteritidis | 4300 | UV light | Ciprofloxacin | 0.25 | 1 |
| 3-EVOL | S. Typhimurium | 722 | NTAP | Ciprofloxacin | 0.06 | 1 |
| 4-EVOL | S. Typhimurium | 722 | NTAP | Streptomycin | 16 | >2,048 |
| 5-EVOL | E. coli | 99 | NTAP | Tetracycline | 8 | >2,048 |
| 6-EVOL | E. coli | 516 | NTAP | Tetracycline | 8 | 32 |
| 7-EVOL | L. monocytogenes | 911 | NTAP | Ciprofloxacin | 1 | 8 |
| 8-EVOL | L. monocytogenes | 940 | UV light | Ciprofloxacin | 2 | 4 |
| 9-EVOL | L. monocytogenes | 911 | UV light | Erythromycin | 0.25 | 4 |
| 10-EVOL | L. monocytogenes | 911 | NTAP | Streptomycin | 2 | 64 |
| 11-EVOL | L. monocytogenes | 940 | NTAP | Ciprofloxacin | 2 | 4 |
| 12-EVOL | L. monocytogenes | 911 | UV light | Ciprofloxacin | 1 | 4 |
Although the number of AMR variants obtained was relatively limited to carry out a systematic statistical analysis of the factors influencing the acquisition of AMR, some general conclusions could be drawn in this regard. The microbial species which showed a higher number of variants was L. monocytogenes, which had a 12.5% AMR emergence rate (6 out of 48 strain/decontamination technique/antibiotic combinations), while the AMR emergence rates observed for E. coli and Salmonella spp. were 3.5% and 5.7%, respectively. The genomic background of the strains also seems to be important, as there existed intraspecies variability in AMR emergence. Thus, for 5 strains (E. coli CECT 515, E. coli CECT 4267, L. monocytogenes CECT 4031, S. Typhimurium CECT 443, and S. Senftenberg CECT 4565), it was not possible to identify AMR variants, while several variants showing increased resistance to at least one antibiotic could be obtained for other strains, such as L. monocytogenes CECT 911 (Table 1). Likewise, the number of AMR variants obtained was strongly dependent on the type of antibiotic tested. Thus, for ciprofloxacin, a 25% resistance emergence rate (6 out of 24 strain/decontamination technique combinations) was observed, while the resistance emergence rates found for erythromycin, streptomycin, and tetracycline were 16.6%, 12.5%, and 8.3%, respectively, and no AMR variants were obtained for the rest of the antibiotics tested (ampicillin, cefotaxime, colistin, gentamicin, and vancomycin). These findings suggest that the risk of AMR selection through exposure to UV-C or NTAP may depend on the mechanism of action of the antibiotic and, therefore, on the antibiotic family. Thus, it was possible to isolate AMR variants for antibiotics of the fluoroquinolone family, which act by inhibiting DNA replication and cell division by targeting the DNA gyrase and topoisomerases, and for antibiotics of the tetracycline, macrolide, and aminoglycoside families, which all act by inhibiting protein synthesis through binding to different ribosomal subunits. On the other hand, no AMR variants could be isolated from agar plates supplemented with β-lactam or glycopeptide antibiotics, which act by inhibiting cell wall synthesis, or polymyxins, which act by binding to the lipopolysaccharide and phospholipids in the outer cell membrane of Gram-negative bacteria. In relation to the technological treatments, similar numbers of variant strains were isolated for the UV-C and NTAP treatments, which suggests that both treatments have an equal impact on the acquisition of AMR.
The isolation of variants with reduced susceptibility to antibiotics through direct evolution in the presence of other microbial inactivation agents has previously been reported for various biocides commonly used for the decontamination and sanitation of surfaces and equipment in health care and food industry-related settings (7, 12, 32). Our study shows that repeated exposure to alternative decontamination techniques, such as UV-C and NTAP, can also give rise to the emergence of variants with increased resistance to antibiotics of clinical relevance, ciprofloxacin, streptomycin, tetracycline, and erythromycin.
Phenotypic characterization of Salmonella antibiotic-resistant variants.
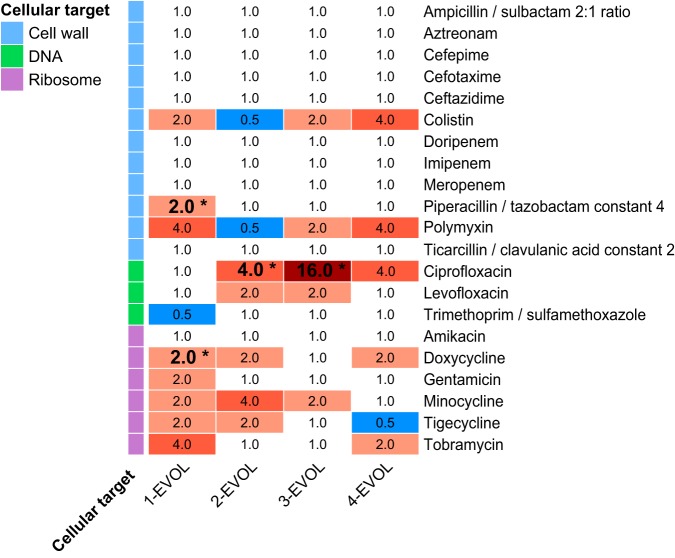
Due to the relevance of Salmonella spp. as a leading foodborne hazard worldwide, a more detailed analysis was carried out for the variant strains of Salmonella spp. with increased AMR, which allowed the identification of changes in the patterns of susceptibility to a wide range of antibiotics (Fig. 2; Table S2). Thus, the variant strain obtained from S. Typhimurium CECT 4594 through UV light exposure (strain 1-EVOL), which showed increased resistance to streptomycin, was also more resistant than its wild-type parental strain to the aminoglycosides gentamicin and tobramycin, the tetracyclines doxycycline (it became resistant according to the ECOFF) and minocycline, the glycylcycline tigecycline, the penicillin piperacillin-tazobactam (it became resistant according to the ECOFF), and the polypeptides colistin and polymyxin, while it was more sensitive to trimethoprim. It is important to highlight that streptomycin was not included in the Sensititre panel.
FIG 2.
Fold change in antimicrobial susceptibility of the Salmonella resistant variants. Increases in resistance are shown in red, while increases in susceptibility are shown in blue. *, a change in the ECOFF, with a transition from a susceptible phenotype in the wild-type strain to a resistant phenotype in the EVOL strain, occurred.
The variant strain obtained from S. Enteritidis CECT 4300 through UV light exposure (strain 2-EVOL), which showed increased resistance to ciprofloxacin, was also more resistant than its parental strain to the tetracyclines doxycycline and minocycline, the glycylcycline tigecycline, and the fluoroquinolone levofloxacin, while it was more sensitive to the polypeptides colistin and polymyxin. Interestingly, the variant strains obtained from S. Typhimurium CECT 722 through exposure to NTAP (strains 3-EVOL and 4-EVOL), which showed increased resistance to ciprofloxacin and streptomycin, respectively, showed different susceptibility patterns, which suggests that a parallel divergent evolution occurred. In fact, the variant strain isolated in MH agar medium supplemented with ciprofloxacin was more resistant than its parental strain to the tetracycline minocycline, the fluoroquinolones ciprofloxacin and levofloxacin, and the polypeptides colistin and polymyxin, while the variant strain isolated in MH agar medium supplemented with streptomycin was more resistant than its parental strain to the aminoglycoside tobramycin, the tetracycline doxycycline, the fluoroquinolone ciprofloxacin, and the polypeptides colistin and polymyxin. It is worth highlighting that although both variant strains showed increased resistance to ciprofloxacin, the MIC was four times higher for the first variant strain (isolated in medium with ciprofloxacin), which also showed a higher level of resistance to levofloxacin, than for the second variant strain (isolated in medium with streptomycin), which was not more resistant to levofloxacin than its parental strain.
The main changes in susceptibility to antimicrobials described above were observed for antibiotics that act at the level of protein synthesis (aminoglycosides, tetracyclines, and glycylcyclines) or DNA replication (fluoroquinolones), as well as for polymyxins, which act by depolarizing the cellular envelopes. However, practically no changes in resistance to β-lactams were detected. It is worth highlighting that resistance to β-lactams is commonly transmissible (mediated through the acquisition of β-lactamase genes), while for other antibiotics, nucleotide polymorphisms causing structural changes in their cellular targets have been shown to frequently result in variants with intrinsic resistance toward them (e.g., single nucleotide polymorphisms [SNPs] in gyrA and ciprofloxacin resistance) (33, 34). In this regard, UV-C light is well-known to be strongly mutagenic through the generation of dimers between adjacent pyrimidine bases on the same DNA strand, which prevent proper replication and which can result in point mutations, deletions, and chromosomal rearrangements (32). On the other hand, although the mutagenic potential of NTAP treatments is not known, plasma contains UV photons and oxidizing free radicals (including reactive oxygen and nitrogen species), which might also cause point mutations and rearrangements in the DNA sequence. Therefore, it seems reasonable to consider that repeated UV-C and NTAP treatments can induce mutagenesis, leading to nucleotide polymorphisms associated with increased resistance to certain antibiotics.
Assays with efflux pump inhibitors (EPIs).
When the susceptibility of the Salmonella variant strains to the antibiotics used at the selection step (streptomycin for 1-EVOL and 4-EVOL and ciprofloxacin for 2-EVOL and 3-EVOL) in comparison to that of their wild-type counterparts was assessed in the presence of the EPIs verapamil, reserpine and phenylalanine-arginine β-naphthylamide (PAβN), no changes in the estimated MICs were observed, with the exception of the MIC for the variant strain 3-EVOL, for which a 4-fold reduction in the MIC of ciprofloxacin was found in the presence of 50 μg/ml of PAβN. However, ciprofloxacin resistance in this variant strain was not affected by supplementation of the culture medium with verapamil or reserpine.
These findings suggest that for 1-EVOL, 2-EVOL, and 4-EVOL, overexpression or the increased activity of multidrug efflux pumps is not responsible for the AMR phenotype observed, while the resistance profile obtained for the variant strain 3-EVOL is at least partly associated with multidrug efflux. The efficacy with which PAβN, a broad-spectrum EPI, increases susceptibility to fluoroquinolones, observed here for 3-EVOL, is well documented in the literature (35, 36). Interestingly, the presence of PAβN in the culture medium was not associated with increased susceptibility to ciprofloxacin for 2-EVOL.
WGS of Salmonella antibiotic-resistant variants.
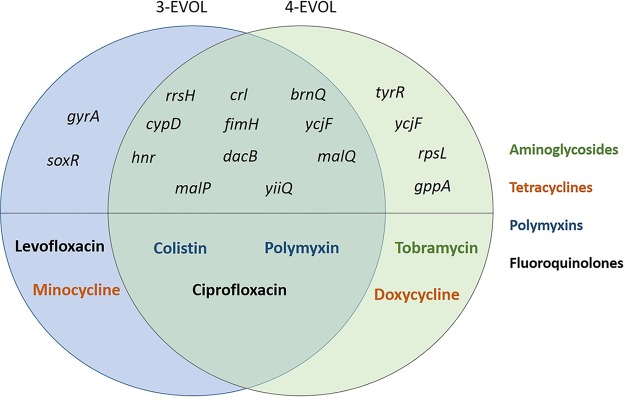
The mapping of the genomes of the four Salmonella AMR variants with respect to the genomes of their reference parental strains allowed the identification of various single nucleotide polymorphisms (SNPs), multiple nucleotide polymorphisms (MNPs), nucleotide insertions. and nucleotide deletions, which are summarized in Table 2 and which are described in detail in Tables S3 to S6. WGS analysis of the two variant strains obtained from S. Typhimurium CECT 722 through exposure to NTAP (strains 3-EVOL and 4-EVOL) confirmed a parallel divergent evolution for both variant strains, which had partially different antibiotic susceptibility patterns, as both isolates had several mutations in common and some differential mutations responsible for their different phenotypic traits (Fig. 3). It is worth highlighting that the variant strain initially isolated in agar medium supplemented with ciprofloxacin (3-EVOL), which showed a higher level of resistance to this antibiotic than the wild-type strain, combined with a higher level of resistance to levofloxacin, showed a missense Pro83Ser SNP in the gyrA gene, which encodes subunit A of DNA gyrase, which is the target of fluoroquinolone antibiotics (37). In addition, it also presented a missense Val20Leu SNP in the transcriptional regulator soxR, which is involved in the microbial response to oxidative stress and which has been shown to affect antibiotic resistance and fitness and to contribute to multidrug resistance in Enterobacteriaceae (38). On the other hand, the variant strain isolated in agar medium with streptomycin (4-EVOL), which also showed increased resistance to another aminoglycoside (tobramycin), had a missense Lys88Glu SNP in rpsL, which encodes the ribosomal protein S12 and which is involved in streptomycin resistance (39), together with other missense mutations in other genes (gppA, which encodes a guanosine pentaphosphatase and which is involved in the stringent response; tyrR, which encodes a tyrosine aminotransferase; and ycjF, which encodes an inner membrane protein) which, to the best of our knowledge, have not been associated before with AMR phenotypes. In addition, both variant strains shared mutations in various stress response regulators (crl, hnr), genes encoding membrane-associated and transport proteins (brnQ, yiiQ, ycjF), virulence factors (fimH, encoding a fimbrial adhesin), genes encoding enzymes involved in protein (cypD) and carbohydrate (malP, malQ) metabolism, and the 16S rRNA gene (rrsH). Both variants also showed an Ala161Val SNP in dacB, encoding a penicillin-binding protein. However, they did not show reduced susceptibility to β-lactam antibiotics. Overall, these findings evidence a common evolution for various generations followed by a parallel divergent evolution which gave rise to relevant phenotype shifts.
TABLE 2.
Mutations identified in genes associated with AMR or the response to stress through WGS analysis of Salmonella antibiotic-resistant variantsa
| Strain | No. of genes with mutations |
AMR-related genes |
Stress response-associated genes |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Complex | Del | Ins | MNP | SNP | Total | Gene | Type | Gene | Type (mutation) | |
| 1-EVOL | 1 | 4 | 4 | 0 | 21 | 29 | crl | Del (Ser35fs) | ||
| rcsC | SNP (Ser394Leu) | |||||||||
| ptsJ | SNP (Pro138Ser) | |||||||||
| cytR | SNP (Asn219Tyr) | |||||||||
| recE-like gene | Complex (SerIleTrp511ArgIleArg) | |||||||||
| recE-like gene | SNP (Ala117Thr), SNP (Val119Ala) | |||||||||
| 2-EVOL | 1 | 1 | 5 | 0 | 46 | 53 | gyrA | SNP (Met86Ile) | uvrB | SNP (Asn624Lys) |
| zntA | SNP (Leu43Pro) | |||||||||
| 3-EVOL | 0 | 2 | 1 | 0 | 18 | 21 | gyrA | SNP (Pro83Ser) | crl | Del (Ser35fs) |
| dacB | SNP (Ala161Val) | hnr | SNP (Leu102Val) | |||||||
| soxR | SNP (Val20Leu) | |||||||||
| 4-EVOL | 1 | 2 | 1 | 0 | 19 | 23 | rpsL | SNP (Lys88Glu) | crl | Del (Ser35fs) |
| dacB | SNP (Ala161Val) | hnr | SNP (Leu102Val) | |||||||
| gppA | Complex (IlePro125MetPro) | |||||||||
Complex, the genome contains two or more of the following mutation types: deletion (Del), insertion (Ins), multiple nucleotide polymorphism (MNP), and single nucleotide polymorphism (SNP).
FIG 3.
Distribution of genes harboring nonsynonymous mutations in the resistant variants obtained for S. Typhimurium CECT 722 (strains 3-EVOL and 4-EVOL) upon repeated exposure to nonthermal atmospheric plasma and the antibiotics to which they showed increased resistance.
The variant strain obtained from S. Typhimurium CECT 4594 through UV light exposure (strain 1-EVOL), which showed increased resistance to streptomycin, showed mutations in several response regulators (crl, rcsC, ptsJ, cytR), genes encoding membrane-associated and transport proteins (nuoL, proV), a gene encoding virulence factors (fimH), and genes encoding proteins involved in the SOS response (RecE and RecE-like proteins) and some hypothetical proteins of still unknown function. No mutations related to genes already known to result in resistance to streptomycin (e.g., rpsL) were detected to explain the decreased streptomycin susceptibility in this variant strain.
The variant strain obtained from S. Enteritidis CECT 4300 through UV light exposure (strain 2-EVOL), which showed increased resistance to ciprofloxacin, had a Met86Ile SNP in the gyrA gene, which was probably responsible for its reduced susceptibility to fluoroquinolones. It also had an Asn624Lys SNP in the uvrB gene, which encodes the excinuclease ABC subunit B, which is involved in nucleotide excision repair and which was previously reported to be required for the repair of UV light-induced DNA photoproducts (40), and a Leu43Pro SNP in the zntA gene, which encodes a zinc/cadmium/mercury/lead-transporting ATPase involved in resistance to heavy metals (41). In addition, it showed mutations in the 23S rRNA gene and genes encoding membrane-associated and transport proteins (ccmH, tbpA, lrgB, and genes encoding an ABC transporter permease and a fimbrial chaperone protein), chemotaxis and flagellar proteins (fliG, motA), enzymes involved in protein metabolism (tuf, astA, a gene encoding a 4-hydroxybutyrate coenzyme A transferase) and nucleotide metabolism (guaA, rnb), proteins involved in cell division (ftsK), and some hypothetical proteins of yet unknown function (Table S4).
Most Salmonella variant strains showed mutations in several regulators involved in the response to stress, such as the oxidative stress response (soxR) or the stringent response (gppA) and the general stress response (crl and hnr, as regulators of the alternative sigma factor RpoS), as well as in the SOS response (RecE and RecE-like proteins). Indeed, interestingly, three S. Typhimurium AMR variants shared a Ser35 frameshift (Ser35fs) mutation in the crl gene. In addition, some variant strains also showed SNPs in genes encoding membrane transporter proteins, which might be involved in the nonselective efflux of antimicrobials. Thus, it is likely that the accumulation of several subtle changes in genes which are not recognized by their role in AMR but which are involved in the response to stress conditions and in the transport of compounds through the cell envelopes may result in a low to medium increase in the level of microbial resistance to several antibiotics. Nevertheless, the assays with efflux pump inhibitors suggest that the AMR profile obtained only for 3-EVOL might be at least partly associated with the overexpression or the increased activity of multidrug efflux pumps.
To sum up, the results obtained in this study evidence that repeated exposure to UV-C and NTAP can lead to the emergence of variant strains with reduced susceptibility to clinically relevant antibiotics. Thus, persistent strains colonizing food-processing environments, which can survive multiple cycles of sanitation, decontamination, and food processing, can develop an AMR phenotype that is not transmissible horizontally but that is transmissible vertically. The development of the phenotype most likely involves the induction of structural/conformational changes in the antibiotic cellular targets (e.g., gyrA for ciprofloxacin, rpsL for streptomycin) accompanied by variations in stress response regulators and membrane transporters likely involved in the nonselective efflux of antibiotics. This type of resistance, not susceptible to horizontal transmission, is, in principle, less worrying than horizontally acquired antimicrobial resistance but can contribute to the spread of AMR along the food chain, contributing to the failure of treatments further ahead in clinical settings.
MATERIALS AND METHODS
Bacterial strains.
The following reference strains from the Spanish Type Culture Collection (CECT) were used: Escherichia coli CECT 99, E. coli CECT 515, E. coli CECT 516, E. coli CECT 4267, Listeria monocytogenes CECT 911, L. monocytogenes CECT 940, L. monocytogenes CECT 4031, Salmonella Typhimurium CECT 443, S. Typhimurium CECT 722, S. Typhimurium CECT 4594, S. Enteritidis CECT 4300, and S. Senftenberg CECT 4565.
Antibiotics.
The following clinically relevant antibiotics were chosen to monitor the generation of AMR through microbial exposure to UV-C and NTAP decontamination processes: ampicillin, cefotaxime, ciprofloxacin, gentamicin, streptomycin, tetracycline, erythromycin (only for L. monocytogenes), vancomycin (only for L. monocytogenes), and colistin (only for E. coli and Salmonella spp.). All antibiotics were purchased from Merck (Darmstadt, Germany).
Decontamination techniques.
Two different decontamination technologies were tested, namely, UV-C light and NTAP. UV-C treatments were applied using a UVPCX-2000 crosslinker (UVP, Upland, CA, USA), which generates UV-C radiation at a wavelength of 254 nm. NTAP treatments were carried out in a specially designed lab-scale plasma jet (CP121 plasma demonstrator; OMVE BV, Netherlands), which consists of a wide-bandwidth copper electrode, configured as a high-impedance voltage probe, whose electrical potential is disturbed when it passes through a gaseous flow in the excited state produced by a high-voltage discharge at 1 kHz.
Preliminary experiments allowed the selection of UV-C and NTAP treatment intensities (in millijoules per square centimeter and seconds or minutes, respectively) which produced similar inactivation rates (∼2-log reductions in the number of CFU per milliliter) for the different bacterial strains tested (data not shown). Treatments at the selected intensities, which ranged from 3 to 5 mJ/cm2 for UV-C treatments and from 10 s to 1.15 min for NTAP treatments, were subsequently used to initiate the process of evolution through exposure to UV-C and NTAP.
Adaptation process.
Bacterial stock cultures were maintained in brain heart infusion (BHI; Merck) agar plates kept under refrigeration. Broth cultures were prepared by inoculating isolated colonies in tubes with 10 ml of BHI broth, which were incubated at 37°C for 24 h. Subsequently, serial dilutions (1:10) were made for each strain in Ringer’s buffer solution (Sigma-Aldrich), and the dilutions were then spread plated (100 μl) onto BHI agar plates to achieve a final load of ∼104 CFU/ml. The agar plates were subsequently subjected to UV-C and NTAP treatments at the predetermined treatment intensities required to obtain a ∼2-log reduction in the number of CFU per milliliter and, finally, were incubated at 37°C for 48 h. In parallel, BHI agar plates were also surface inoculated with 100 μl of a bacterial suspension with approximately 103 CFU/ml, which served as a negative control not subjected to UV-C or NTAP treatments. Following incubation, the survivors were counted, and for each plate, an isolated colony was recovered and used to inoculate a new culture tube with 10 ml of BHI broth, which, after incubation at 37°C for 24 h, was again used to start a new cycle of UV-C or NTAP treatment. In total, 10 treatment cycles were carried out for each strain and decontamination technology. For those cases in which an increase in the number of survivors was observed as the evolution process progressed, the intensity of the treatment was progressively increased (Fig. 1). At the end of the last treatment cycle, for each strain and decontamination technology, an isolated colony was recovered and was grown in BHI broth. The grown culture was preserved with 40% glycerol (Amresco, Solon, OH, USA) at −80°C.
Identification of antibiotic-resistant variants.
In order to evaluate changes in the resistance of the adapted cultures to the different antibiotics, it was first necessary to determine their MICs for the CECT collection of E. coli, Salmonella, and L. monocytogenes strains using the agar dilution method on Mueller-Hinton (MH) agar plates (Merck).
Microbial overnight cultures for each control reference strain and the UV-C-adapted and NTAP-adapted derivative strains were then obtained in BHI broth after incubation at 37°C for 24 h. Subsequently, 100 μl of the bacterial suspensions obtained was spread plated on the surface of MH agar plates supplemented with the different antibiotics at their MIC for the corresponding CECT collection strain. After incubation of the plates at 37°C for 48 h, the presence or absence of growth was visually evaluated in order to identify and isolate possible variant strains with greater resistance to the tested antibiotics than the collection parental strains. For those adapted cultures for which growth was appreciated, individual colonies were isolated and their identity was confirmed by plating on selective or differential medium (agar Listeria according to Ottaviani and Agosti [ALOA agar; Merck] for L. monocytogenes, xylose lysine deoxycholate agar [Merck] and brilliant green agar [Merck] for Salmonella spp., and eosin methylene blue agar [Merck] for E. coli) in order to rule out possible cross-contaminations during the 10-cycle evolution process. Finally, an aliquot of each confirmed variant strain was stored frozen by following the methodology described above.
Phenotypic characterization of Salmonella antibiotic-resistant variants.
A deeper study of the profile of resistance to a wide range of antibiotics for the Salmonella variants isolated after exposure to UV-C or NTAP treatments in comparison to that of the corresponding CECT collection strains was carried out to identify multidrug-resistant variants by using Sensititre plates for Gram-negative bacteria (catalog number YGNX3F; Thermo Scientific, Waltham, MA, USA) following the recommendations of the provider. Briefly, a 0.5 McFarland standard suspension was prepared for each strain in Ringer’s buffer and was serially diluted in MH broth to achieve a microbial load of 1 × 104 to 1 × 105 CFU/ml. Then, each well of the Sensititre test plates was filled with 50 μl of this bacterial suspension. Subsequently, the Sensititre panel plates were incubated at 37°C for 24 h, and the absence/presence of growth in each well was visually evaluated.
Assays with EPIs.
The efflux pump inhibitors (EPIs) verapamil, PaβN, and reserpine (Alfa Aesar, Kandel, Germany) were used in order to determine the contribution of multidrug efflux pumps to the antibiotic resistance phenotypes emerging after exposure to UV-C and NTAP. Microbial overnight cultures for each control reference strain and its UV-C-adapted or NTAP-adapted derivative variant strains were obtained in BHI broth after incubation at 37°C for 24 h. Then, a 0.5 McFarland standard suspension was prepared for each strain in Ringer’s buffer and was serially diluted in MH broth to achieve a microbial load of 1 × 104 to 1 × 105 CFU/ml. Subsequently, 50 μl of the bacterial suspensions was added to 96-well plates containing 50 ml of MH broth supplemented with 2-fold serial dilutions of the corresponding antibiotic and with the EPI reserpine (100 μg/ml), verapamil (100 μg/ml), or PAβN (50 μg/ml). Wells filled in with MH broth supplemented only with 2-fold serial dilutions of the corresponding antibiotic were used as controls. The MICs of the antibiotics were estimated for the wild-type and variant strains after incubation of the plates overnight at 37°C. The assays were carried out on two different days in triplicate.
Whole-genome sequencing of antibiotic-resistant variants.
The Salmonella variant strains were subsequently analyzed through whole-genome sequencing in order to identify the polymorphisms responsible for the increased resistance observed. For this purpose, total genomic DNA was isolated using a DNeasy PowerSoil kit (Qiagen) following the provider’s instructions. The quality of the isolated DNA was checked by capillary electrophoresis using a fragment analyzer (genome DNA 50-kb analysis kit; Advanced Analytical Technologies, Inc.), and the DNA concentration was measured using a Qubit double-stranded DNA high-sensitivity assay kit (Thermo Fisher).
Library preparation was carried out using a TruSeq DNA PCR-free kit (Illumina) following the provider’s instructions, after DNA fragmentation using a Covaris E220 ultrasonicator to obtain ∼550-bp fragments. Then, a quality control step was again completed using the fragment analyzer and the Qubit kit for DNA quantification. Finally, a quantitative PCR was performed to prepare the samples for sequencing using a complete Roche LightCycler 480 kit (Kappa). The Illumina HiSeq 1500 platform was used for sequencing, with 100- to 150-bp paired-end reads being obtained.
The quality of the sequences was analyzed by using the FastQC program (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and adapters were trimmed using the Trim Galore tool (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), followed by a second quality check. Snippy software (https://github.com/tseemann/snippy) was used to detect single nucleotide polymorphisms (SNPs), multiple nucleotide polymorphisms (MNPs), and other sequence alterations, such as deletions or insertions, by comparing the reads obtained for the variant strains with the sequence of the whole genome of their parental reference strains.
Data availability.
The paired-end reads and isolate information were submitted to the NCBI database under BioProject accession number PRJNA546492. The BioSample and SRA accession numbers are as follows: SAMN11938261 and SRR9203580, respectively, for strain 1-EVOL, SAMN11938944 and SRR9203581, respectively, for strain 2-EVOL, SAMN11938747 and SRR9203582, respectively, for strain 3-EVOL, and SAMN11939167 and SRR9203583, respectively, for strain 4-EVOL.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Fundación BBVA and the Ministry of Science, Innovation and Universities of the Spanish Government under grant number AGL2016-78085-P. A.A.-M. is grateful to the Ministry of Science, Innovation, and Universities of the Spanish Government for awarding him a predoctoral grant (FPI BES-2017-079753).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.ECDC. 2009. The bacterial challenge, time to react: a call to narrow the gap between multidrug-resistant bacteria in the EU and the development of new antibacterial agents. ECDC, Stockholm, Sweden. [Google Scholar]
- 2.Capita R, Alonso-Calleja C. 2013. Antibiotic-resistant bacteria: a challenge for the food industry. Crit Rev Food Sci Nutr 53:11–48. doi: 10.1080/10408398.2010.519837. [DOI] [PubMed] [Google Scholar]
- 3.Teale CJ. 2002. Antimicrobial resistance and the food chain. J Appl Microbiol 92:85S–89S. doi: 10.1046/j.1365-2672.92.5s1.20.x. [DOI] [PubMed] [Google Scholar]
- 4.Verraes C, Van Boxstael S, Van Meervenne E, Van Coillie E, Butaye P, Catry B, de Schaetzen M-A, Van Huffel X, Imberechts H, Dierick K, Daube G, Saegerman C, De Block J, Dewulf J, Herman L. 2013. Antimicrobial resistance in the food chain: a review. Int J Environ Res Public Health 10:2643–2669. doi: 10.3390/ijerph10072643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Founou LL, Founou RC, Essack SY. 2016. Antibiotic resistance in the food chain: a developing country-perspective. Front Microbiol 7:1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curiao T, Marchi E, Grandgirard D, León-Sampedro R, Viti C, Leib SL, Baquero F, Oggioni MR, Martinez JL, Coque TM. 2016. Multiple adaptive routes of Salmonella enterica Typhimurium to biocide and antibiotic exposure. BMC Genomics 17:491. doi: 10.1186/s12864-016-2778-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webber MA, Whitehead RN, Mount M, Loman NJ, Pallen MJ, Piddock LV. 2015. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemother 70:2241–2248. doi: 10.1093/jac/dkv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langsrud S, Sundheim G, Holck AL. 2004. Cross-resistance to antibiotics of Escherichia coli adapted to benzalkonium chloride or exposed to stress-inducers. J Appl Microbiol 96:201–208. doi: 10.1046/j.1365-2672.2003.02140.x. [DOI] [PubMed] [Google Scholar]
- 9.Randall LP, Cooles SW, Coldham NG, Penuela EG, Mott AC, Woodward MJ, Piddock LV, Webber MA. 2007. Commonly used farm disinfectants can select for mutant Salmonella enterica serovar Typhimurium with decreased susceptibility to biocides and antibiotics without compromising virulence. J Antimicrob Chemother 60:1273–1280. doi: 10.1093/jac/dkm359. [DOI] [PubMed] [Google Scholar]
- 10.Karatzas KAG, Webber MA, Jorgensen F, Woodward MJ, Piddock LV, Humphrey TJ. 2007. Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J Antimicrob Chemother 60:947–955. doi: 10.1093/jac/dkm314. [DOI] [PubMed] [Google Scholar]
- 11.Whitehead RN, Overton TW, Kemp CL, Webber MA. 2011. Exposure of Salmonella enterica serovar Typhimurium to high level biocide challenge can select multidrug resistant mutants in a single step. PLoS One 6:e22833. doi: 10.1371/journal.pone.0022833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alvarez-Ordóñez A, Broussolle V, Colin P, Nguyen-The C, Prieto M. 2015. The adaptive response of bacterial food-borne pathogens in the environment, host and food: implications for food safety. Int J Food Microbiol 213:99–109. doi: 10.1016/j.ijfoodmicro.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Hernández A, Ruiz FM, Romero A, Martínez JL. 2011. The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia. PLoS Pathog 7:e1002103. doi: 10.1371/journal.ppat.1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjorland J, Steinum T, Kvitle B, Waage S, Sunde M, Heir E. 2005. Widespread distribution of disinfectant resistance genes among staphylococci of bovine and caprine origin in Norway. J Clin Microbiol 43:4363–4368. doi: 10.1128/JCM.43.9.4363-4368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle MP, Loneragan GH, Scott HM, Singer RS. 2013. Antimicrobial resistance: challenges and perspectives. Compr Rev Food Sci Food Saf 12:234–248. doi: 10.1111/1541-4337.12008. [DOI] [Google Scholar]
- 16.Bintsis T, Litopoulou-Tzanetaki E, Robinson RK. 2000. Existing and potential applications of ultraviolet light in the food industry—a critical review. J Sci Food Agric 80:637–645. doi:. [DOI] [PubMed] [Google Scholar]
- 17.Elasri MO, Miller RV. 1999. Study of the response of a biofilm bacterial community to UV radiation. Appl Environ Microbiol 65:2025–2031. doi: 10.1128/AEM.65.5.2025-2031.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patil S, Moiseev T, Misra NN, Cullen PJ, Mosnier JP, Keener KM, Bourke P. 2014. Influence of high voltage atmospheric cold plasma process parameters and role of relative humidity on inactivation of Bacillus atrophaeus spores inside a sealed package. J Hosp Infect 88:162–169. doi: 10.1016/j.jhin.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Fernández A, Shearer N, Wilson DR, Thompson A. 2012. Effect of microbial loading on the efficiency of cold atmospheric gas plasma inactivation of Salmonella enterica serovar Typhimurium. Int J Food Microbiol 152:175–180. doi: 10.1016/j.ijfoodmicro.2011.02.038. [DOI] [PubMed] [Google Scholar]
- 20.Perni S, Shama G, Hobman JL, Lund PA, Kershaw CJ, Hidalgo-Arroyo GA, Penn CW, Deng XT, Walsh JL, Kong MG. 2007. Probing bactericidal mechanisms induced by cold atmospheric plasmas with Escherichia coli mutants. Appl Phys Lett 90:e073902. doi: 10.1063/1.2458162. [DOI] [Google Scholar]
- 21.Sakudo A, Misawa T, Yagyu Y. 2019. Equipment design for cold plasma disinfection of food products, p 289–307. In Bermudez-Aguirre D. (ed), Advances in cold plasma applications for food safety and preservation. Academic Press, Cambridge, MA. [Google Scholar]
- 22.Pirnie M, Linden K, Malley J. 2006. Ultraviolet disinfection guidance manual for the final long term 2 enhanced surface water treatment rule. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 23.Sommer R, Cabaj A, Haider T, Hirschmann G. 2004. UV drinking water disinfection—requirements, testing and surveillance: exemplified by the Austrian national standards M 5873-1 and M 5873-2. IUBA News 6:27–35. [Google Scholar]
- 24.Vanlint D, Mitchell R, Bailey E, Meersman F, McMillan PF, Michiels CW, Aertsen A. 2011. Rapid acquisition of gigapascal-high-pressure resistance by Escherichia coli. mBio 2:e00130-10. doi: 10.1128/mBio.00130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanlint D, Rutten N, Michiels CW, Aertsen A. 2012. Emergence and stability of high-pressure resistance in different food-borne pathogens. Appl Environ Microbiol 78:3234–3241. doi: 10.1128/AEM.00030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karatzas KAG, Hocking PM, Jørgensen F, Mattick K, Leach S, Humphrey TJ. 2008. Effects of repeated cycles of acid challenge and growth on the phenotype and virulence of Salmonella enterica. J Appl Microbiol 105:1640–1648. doi: 10.1111/j.1365-2672.2008.03909.x. [DOI] [PubMed] [Google Scholar]
- 27.Van Boeijen IKH, Chavaroche AAE, Valderrama WB, Moezelaar R, Zwietering MH, Abee T. 2010. Population diversity of Listeria monocytogenes LO28: phenotypic and genotypic characterization of variants resistant to high hydrostatic pressure. Appl Environ Microbiol 76:2225–2233. doi: 10.1128/AEM.02434-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Boeijen IKH, Francke C, Moezelaar R, Abee T, Zwietering MH. 2011. Isolation of highly heat-resistant Listeria monocytogenes variants by use of a kinetic modeling-based sampling scheme. Appl Environ Microbiol 77:2617–2624. doi: 10.1128/AEM.02617-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metselaar KI, den Besten HMW, Abee T, Moezelaar R, Zwietering MH. 2013. Isolation and quantification of highly acid resistant variants of Listeria monocytogenes. Int J Food Microbiol 166:508–514. doi: 10.1016/j.ijfoodmicro.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Tomida J, Oumi A, Okamoto T, Morita Y, Okayama A, Misawa N, Hayashi T, Akaike T, Kawamura Y. 2013. Comparative evaluation of agar dilution and broth microdilution methods for antibiotic susceptibility testing of Helicobacter cinaedi. Microbiol Immunol 57:353–358. doi: 10.1111/1348-0421.12044. [DOI] [PubMed] [Google Scholar]
- 31.Luber P, Bartelt E, Genschow E, Wagner J, Hahn H. 2003. Comparison of broth microdilution, E test, and agar dilution methods for antibiotic susceptibility testing of Campylobacter jejuni and Campylobacter coli. J Clin Microbiol 41:1062–1068. doi: 10.1128/jcm.41.3.1062-1068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oniciuc E-A, Likotrafiti E, Alvarez-Molina A, Prieto M, López M, Alvarez-Ordóñez A. 2019. Food processing as a risk factor for antimicrobial resistance spread along the food chain. Curr Opin Food Sci 30:21–26. doi: 10.1016/j.cofs.2018.09.002. [DOI] [Google Scholar]
- 33.Nosova EY, Bukatina AA, Isaeva YD, Makarova MV, Yu K, Moroz AM. 2013. Analysis of mutations in the gyrA and gyrB genes and their association with the resistance of Mycobacterium tuberculosis to levofloxacin, moxifloxacin and gatifloxacin. J Med Microbiol 62:108–113. doi: 10.1099/jmm.0.046821-0. [DOI] [PubMed] [Google Scholar]
- 34.Farhat MR, Jacobson KR, Franke MF, Kaur D, Sloutsky A, Mitnick CD, Murray M. 2016. Gyrase mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis. J Clin Microbiol 54:727–733. doi: 10.1128/JCM.02775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maleki M-H, Jalilian FA, Khayat H, Mohammadi M, Pourahmad F, Asadollahi K, Pakzad I, Sadeghifard N, Soroush S, Emaneini M, Taherikalani M. 2014. Detection of highly ciprofloxacin resistance Acinetobacter baumannii isolated from patients with burn wound infections in presence and absence of efflux pump inhibitor. Maedica (Buchar) 9:162–167. [PMC free article] [PubMed] [Google Scholar]
- 36.Huguet A, Pensec J, Soumet C. 2013. Resistance in Escherichia coli: variable contribution of efflux pumps with respect to different fluoroquinolones. J Appl Microbiol 114:1294–1299. doi: 10.1111/jam.12156. [DOI] [PubMed] [Google Scholar]
- 37.Gopal M, Elumalai S, Arumugam S, Durairajpandian V, Kannan MA, Selvam E, Seetharaman S. 2016. GyrA ser83 and ParC trp106 mutations in Salmonella enterica serovar Typhi isolated from typhoid fever patients in tertiary care hospital. J Clin Diagn Res 10:DC14–DC18. doi: 10.7860/JCDR/2016/17677.8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Telke AA, Olaitan AO, Morand S, Rolain J-M. 2017. soxRS induces colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrAB-tolC efflux pump. J Antimicrob Chemother 72:2715–2721. doi: 10.1093/jac/dkx215. [DOI] [PubMed] [Google Scholar]
- 39.Smittipat N, Juthayothin T, Billamas P, Jaitrong S, Rukseree K, Dokladda K, Chaiyasirinroje B, Disratthakit A, Chaiprasert A, Mahasirimongkol S, Yanai H, Yamada N, Tokunaga K, Palittapongarnpim P. 2016. Mutations in rrs, rpsL and gidB in streptomycin-resistant Mycobacterium tuberculosis isolates from Thailand. J Glob Antimicrob Resist 4:5–10. doi: 10.1016/j.jgar.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 40.Crowley DJ, Boubriak I, Berquist BR, Clark M, Richard E, Sullivan L, DasSarma S, McCready S. 2006. The uvrA, uvrB and uvrC genes are required for repair of ultraviolet light induced DNA photoproducts in Halobacterium sp. NRC-1. Saline Systems 2:11. doi: 10.1186/1746-1448-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidhyaparkavi A, Osborne J, Babu S. 2017. Analysis of zntA gene in environmental Escherichia coli and additional implications on its role in zinc translocation. 3 Biotech 7:9. doi: 10.1007/s13205-017-0613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The paired-end reads and isolate information were submitted to the NCBI database under BioProject accession number PRJNA546492. The BioSample and SRA accession numbers are as follows: SAMN11938261 and SRR9203580, respectively, for strain 1-EVOL, SAMN11938944 and SRR9203581, respectively, for strain 2-EVOL, SAMN11938747 and SRR9203582, respectively, for strain 3-EVOL, and SAMN11939167 and SRR9203583, respectively, for strain 4-EVOL.