Increasing concerns of antibiotic-resistant Campylobacter spp. with regard to public health emphasize the importance of efficient and fast detection. This study described the timely identification and antimicrobial susceptibility testing of Campylobacter spp. by using a microfluidic device. Our developed method not only reduced the total analysis time, but it also simplified food sample preparation and chip operation for end users. Due to the miniaturized size of the lab-on-a-chip platform, the detection was achieved by using up to 1,000 times less of the reagents than with standard reference methods, making it a competitive approach for rapid screening and surveillance study in food industries. In addition, multiple clinically important Campylobacter species (C. jejuni, C. coli, and C. lari) could be tested by our device. This device has potential for wide application in food safety management and clinical diagnostics, especially in resource-limited regions.
KEYWORDS: Campylobacter, antimicrobial susceptibility testing, colorimetric, detection, food safety, lab-on-a-chip, multidrug resistance
ABSTRACT
Campylobacter spp. have been recognized as major foodborne pathogens worldwide. An increasing frequency of antibiotic-resistant pathogens, including Campylobacter spp., have been identified to transmit from food products to humans and cause severe threats to public health. To better mitigate the antibiotic resistance crisis, rapid detection methods are required to provide timely antimicrobial resistance surveillance data for agri-food systems. Herein, we developed a polymer-based microfluidic device for the identification and antimicrobial susceptibility testing (AST) of Campylobacter spp. An array of bacterial incubation chambers were created in the microfluidic device, where chromogenic medium and antibiotics were loaded. The growth of Campylobacter spp. was visualized by color change due to chromogenic reactions. This platform achieved 100% specificity for Campylobacter identification. Sensitive detection of multiple Campylobacter species (C. jejuni, C. coli, and C. lari) was obtained in artificially contaminated milk and poultry meat, with detection limits down to 1 × 102 CFU/ml and 1 × 104 CFU/25 g, respectively. On-chip AST determined Campylobacter antibiotic susceptibilities by the lowest concentration of antibiotics that can inhibit bacterial growth (i.e., no color change observed). High coincidences (91% to 100%) of on-chip AST and the conventional agar dilution method were achieved against several clinically important antibiotics. For a presumptive colony, on-chip identification and AST were completed in parallel within 24 h, whereas standard methods, including biochemical assays and traditional culture-based AST, take several days for multiple sequential steps. In conclusion, this lab-on-a-chip device can achieve rapid and reliable detection of antibiotic-resistant Campylobacter spp.
IMPORTANCE Increasing concerns of antibiotic-resistant Campylobacter spp. with regard to public health emphasize the importance of efficient and fast detection. This study described the timely identification and antimicrobial susceptibility testing of Campylobacter spp. by using a microfluidic device. Our developed method not only reduced the total analysis time, but it also simplified food sample preparation and chip operation for end users. Due to the miniaturized size of the lab-on-a-chip platform, the detection was achieved by using up to 1,000 times less of the reagents than with standard reference methods, making it a competitive approach for rapid screening and surveillance study in food industries. In addition, multiple clinically important Campylobacter species (C. jejuni, C. coli, and C. lari) could be tested by our device. This device has potential for wide application in food safety management and clinical diagnostics, especially in resource-limited regions.
INTRODUCTION
Campylobacter spp. are the leading cause of bacterial gastroenteritis in humans, with the incidence of infections being about 2 to 65 times higher than that with other foodborne pathogens, such as Salmonella spp., Shiga toxin-producing Escherichia coli, and Listeria monocytogenes (1, 2). Globally, 400 million to 500 million cases of diarrhea are estimated to be caused by Campylobacter spp. per year, resulting in severe burdens to public health and economic growth (3). The transmission of Campylobacter spp. to humans occurs mainly through the consumption of contaminated food products, especially poultry meat and unpasteurized milk (4). Most foodborne campylobacteriosis cases are either self-limiting or treated with fluid replenishment, but antibiotic therapy is still warranted if severe or prolonged infections occur, particularly for young, elderly, and immunocompromised patients (3). Campylobacter spp. obtain increasing resistance to clinically important antibiotics such as fluoroquinolones, tetracyclines, and ampicillin (5). The rise of antibiotic resistance contributes to the failure of clinical treatments, extended recovery spans, and doubling mortality rates. Several international and national authorities have emphasized an urgent need to mitigate antibiotic-resistant Campylobacter spp. For example, the Centers for Disease Control and Prevention (CDC) lists antibiotic-resistant Campylobacter spp. as serious hazard-level threats to public health (6). The World Health Organization suggests that multidrug-resistant Campylobacter spp. should receive a high priority for investigation because of their high prevalence of resistance and frequent transmissibility from foods to humans (7).
To reduce antibiotic-resistant threats, one of the major approaches is to monitor the prevalence of resistant Campylobacter spp. in agri-food systems. A better understanding of the transmission between foods and humans can thus be achieved, followed by the development of proper control measures and antibiotic stewardship. In surveillance studies, a gold standard protocol is generally applied based on the growth of bacterial culture, including sequential steps of bacterial isolation, identification, and antimicrobial susceptibility testing (AST) (8). This protocol has several limitations, as follows: (i) the entire analysis takes at least 7 to 9 days (8); (ii) the protocol involves multiple steps of labor-intensive operations, such as preparation of the growth media and biochemical assays; (iii) although conventional culture-based methods readily detect the major gastroenteritis-related Campylobacter species, they may be biased toward C. jejuni and C. coli and thus cause the underreporting of other species such as C. lari (9); and (iv) samples must be transported to centralized microbiology laboratories that are not accessible in resource-limited settings. As a result, current detection methods are not optimal for a wide application of surveillance systems in different regions. A rapid, easy-to-operate, and portable detection approach needs to be developed for multiple clinically important Campylobacter species.
The microfluidic “lab-on-a-chip” technique is a promising tool to detect antibiotic-resistant bacteria in various fields, especially clinical diagnostics and environmental monitoring (10, 11). This technique offers many advantages over macroscale methods in terms of its rapid analysis, precise controlling of fluids, low cost, lower sample volume, portability, and high throughput. Microfluidic-based detection can be categorized into two types, namely, genotypic and phenotypic assays. Genotypic on-chip assays (e.g., PCR and isothermal amplification) target on genetic markers (e.g., 16S rRNA genes and antibiotic resistance genes), which circumvent bacterial proliferation progress and can be finished within several hours (12, 13). These methods provide direct information on bacterial identification but cannot determine bacterial antibiotic susceptibility profiles in a broad-spectrum manner if antibiotic resistance mechanisms are unknown. Moreover, antibiotic susceptibility is not always correlated with the presence or absence of genetic markers, resulting in false-positive and false-negative results (14). In contrast, phenotypic on-chip assays monitor bacterial growth in the presence of antibiotics, resulting in accurate AST results. Generally, bacterial cells were confined in a small volume (e.g., channels, chambers, or droplets) (15, 16), captured by antibodies on membranes or magnetic beads (17, 18), or encapsulated in agarose chambers (19). With the presence of antibiotics, changes in cell numbers, sizes, and morphologies could be monitored using either optical or spectroscopic techniques. The confining of bacteria reduced the antibiotic diffusion distances and allowed the tracking of single-cell replication, significantly improving the detection sensitivity and shortening the analysis time. However, these microfluidic platforms involved expensive and bulky microscopies (e.g., phase-contrast microscopy and fluorescence microscopy) or vibrational spectroscopies (e.g., infrared spectroscopy and Raman spectroscopy) (16, 17, 19, 20). To be free of external equipment, micromechanical or electrochemical-based microfluidic platforms have been proposed to study the mass or electrical transduction changes of bacterial cells under antibiotic stress (20, 21). However, antibodies were required to immobilize bacteria in the detection zones, which were not cost-effective and had limited shelf stability.
To eliminate the use of external detectors and expensive bacterium-capturing elements, colorimetric-based microfluidic platforms have been established to achieve the visualization of results in several recent studies. For example, Cira and coauthors utilized a pH indicator to determine the antibiotic susceptibilities of human pathogens (e.g., E. coli and Enterococcus faecalis) (22). Unlike sensitive strains, resistant isolates survived under antibiotic stress and hydrolyzed glucose into acids, leading to a pH decrease and color change. Elavarasan and others visualized bacterial survival in antibiotics based on viability-dependent resazurin dye reduction (color change from blue to pink and colorless) (23). However, these studies only determined antibiotic susceptibility profiles, whereas bacterial identification had to be performed separately. To solve this problem, chromogenic medium can be applied to selectively support the growth of targeted bacterial genera. With the addition of antibiotics, both identification and AST can be completed simultaneously in microfluidic devices. For example, Xu and colleagues developed a one-step identification and AST for urinary tract infection pathogens using chromogenic agar medium (24). To the best of our knowledge, no microfluidic platform is available for the identification and AST of Campylobacter spp., although this pathogen has significant agri-food and clinical importance.
This research study is a novel example of developing a microfluidic device for the identification and AST of Campylobacter spp. We used chromogenic agar as selective cultivation medium for Campylobacter spp. exclusively. We employed an advanced design of air vents and zigzag channels to prevent the cross-contamination of antibiotics in different testing chambers, ensuring accurate AST results. Rapid detection of three primary Campylobacter species (C. jejuni, C. coli, and C. lari) was completed in various food models. The MICs and susceptibility profiles of Campylobacter isolates were tested by on-chip AST. This new microfluidic device represents a rapid, portable, and cost-effective approach to detect antibiotic-resistant Campylobacter spp. in agri-food systems.
RESULTS AND DISCUSSION
Design and characterization of the microfluidic device.
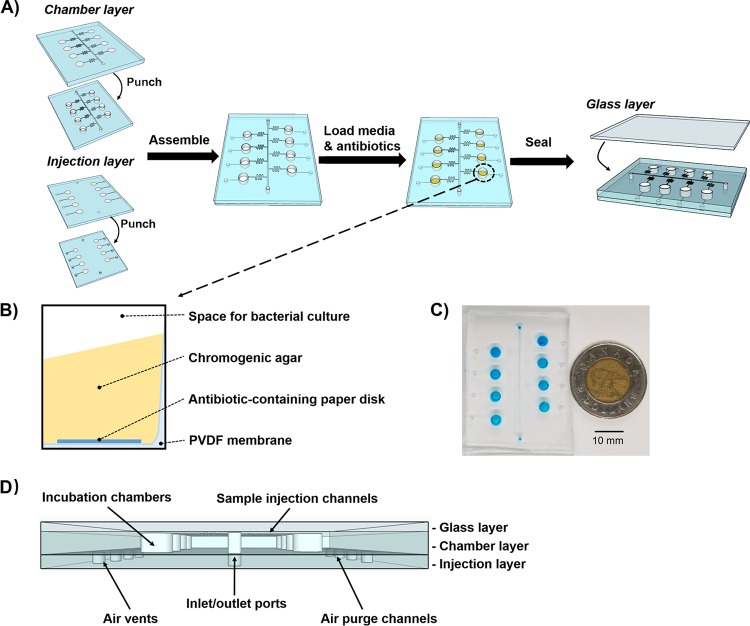
In this study, a colorimetric-based microfluidic chip was prototyped for Campylobacter identification and AST. This device was made of hybrid materials, including one layer of glass slide and two layers of polydimethylsiloxane (PDMS) slabs (Fig. 1). The assembly of multiple chip layers is described in Fig. 1A. In the PDMS-based chamber layer, a main channel was connected to 8 separately positioned incubation chambers via side channels. The side channels were designed in a zigzag shape to eliminate the backflow of sample fluids into the neighboring incubation chambers. The injection layer was also made of PDMS that contained air vents downstream of each incubation chamber. During sample injection, air was purged out of the incubation chambers via air vents to decrease the inner pressure of incubation chambers and facilitate the distribution of bacteria into the incubation chambers. After assembling the chamber injection layer, a polyvinylidene difluoride (PVDF) membrane was deposited into the incubation chambers to support the addition of chromogenic agar and antibiotic-preloaded paper disks (Fig. 1B). Besides, PVDF membranes can function as a white background to improve the visualization of color changes due to bacterial chromogenic reactions. A glass layer was casted onto the chamber injection layer to generate a sealed microfluidic device. This microfluidic device was highly portable, with dimensions of 50 mm in length by 40 mm in width by 5 mm in height (Fig. 1C and D).
FIG 1.
(A) Schematic illustration of device assembly. (B) Side view of the incubation chamber. (C) Representative image of the microfluidic chip. Blue food dye was used for the visualization of chip patterns. The dimensions of microfluidic chip are 50 mm in length, 40 mm in width, and 5 mm in height. (D) Front view of the microfluidic chip.
To achieve efficient sample injection, pump-driven force was applied. As shown in Fig. S1 in the supplemental material, blue food dye was injected into the microfluidic chip at a flow rate of 0.05 ml/min. Once the main channel was filled with food dye (after ∼10 s), we blocked the outlet tubing using a metal clip. Flow resistance in the main channel was increased to exceed the ones in the side channels, leading to favored distribution of samples into the incubation chambers. This sample loading step was completed within 3 min. The absence of bubbles or leakage in the microfluidic chip ensured accurate and equal sample volumes in individual incubation chambers, which was necessary to provide defined antibiotic concentrations for AST study.
We proposed an on-chip AST to simultaneously investigate bacterial susceptibilities against different classes and/or concentrations of antibiotics in a single device. During sample injection, antibiotics on paper disks might diffuse into sample fluids and be further transferred to neighboring incubation chambers (22). To prevent the cross-contamination of antibiotics among incubation chambers, we employed zigzag-shaped side channels and air vents to inhibit the backflow of sample fluids into the main channels. A reagent diffusion test was conducted to verify the chip design (Fig. 2). Instead of antibiotics, cresol red, a pH indicator appearing yellow at pH below 7.2 and red at pH above 8.8, was deposited onto the paper disks and embedded into the incubation chambers. Green food dye (pH 9.5) was introduced into the microfluidic device. With the presence of the alkaline solution, paper disks containing cresol red turned from yellow to red, whereas blank paper disks remained white. After incubation at 22°C for 48 h, no color change was observed in the chambers without cresol red (Fig. 2), indicating that our microfluidic chip effectively prevented cross-contamination between incubation chambers.
FIG 2.
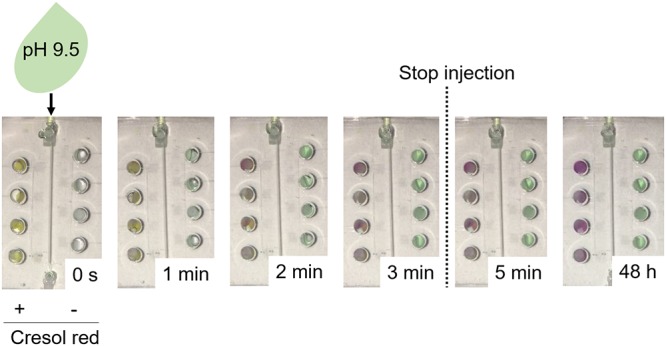
Assessment of cross-contamination among incubation chambers. Paper disks with or without cresol red were deposited into the incubation chambers of a microfluidic chip. A green alkaline solution (pH 9.5) was injected into the microfluidic chip until the incubation chambers were fully filled (∼3 min). The diffusion of cresol red was investigated by keeping the microfluidic chip still at room temperature for 48 h. Cresol red is a pH indicator that appears yellow at pH below 7.2 and red at pH above 8.8. In this case, incubation chambers with a color change from yellow to red indicate an appropriate injection of alkaline solution, whereas the incubation chambers in green suggest that no cresol red diffused from the neighboring chambers.
Optimizing medium concentration and amount for visible chromogenic reaction.
The chromogenic agar medium CHROMagar Campylobacter was used as the selective and differential medium for Campylobacter detection. It is composed of powder base as the nutrient source to support bacterial growth and supplements containing chromogenic substrates and selective agents. With the presence of selective agents (e.g., antibiotics), competing microbiota from complexed food or clinical samples could be inhibited so that a better identification of Campylobacter spp. can be achieved. A chromogenic reaction between Campylobacter cells and chromogenic substrates results in a color change from light yellow to red.
To enhance the visibility and sensitivity of chromogenic reactions, we optimized the concentrations of chromogenic substrates. An aliquot of 20 μl chromogenic agar was prepared in a PCR tube to mimic bacterial cultivation in the miniaturized system (i.e., a microfluidic device). C. jejuni F38011 at an initial concentration of 106 CFU/ml was incubated with different concentrations of supplements at 42°C for 48 h. No color change was recognized by adding 0.21 g/liter of supplements (Fig. S2), which was the recommended concentration according to the manufacturer’s instructions. A higher concentration of supplements generated a higher red signal intensity. When the concentration of supplements reached 0.63 g/liter, we observed a wide spread of chromogenic products in the PCR tube from the bottom view, which was the monitoring view for on-chip chromogenic reactions. Therefore, we selected 0.63 g/liter as the working concentration of supplements for further study.
Since the total amount of chromogenic substrates could influence the visualization of chromogenic reactions, we explored the required amount of chromogenic agar medium in the microfluidic device. In general, a larger amount of agar resulted in a more intense red signal (Fig. 3). Compared to the negative-control group without chromogenic agar, a significant (P < 0.05) color change was obtained when the agar amount fell in a range of 12 to 20 μl. An incubation chamber in the microfluidic chip had a volume of 27 μl. With a larger amount of chromogenic agar in the incubation chamber, a lower volume of samples could be implemented. Thus, we used 20 μl chromogenic agar for on-chip identification and AST to minimize the potential interferences from food matrices during chromogenic reactions. It is worth mentioning that too high of a volume of chromogenic agar (>20 μl) could form a bump in the incubation chamber, which attached to the glass layer after chip assembly and blocked the injection of bacterial culture into the incubation chambers.
FIG 3.
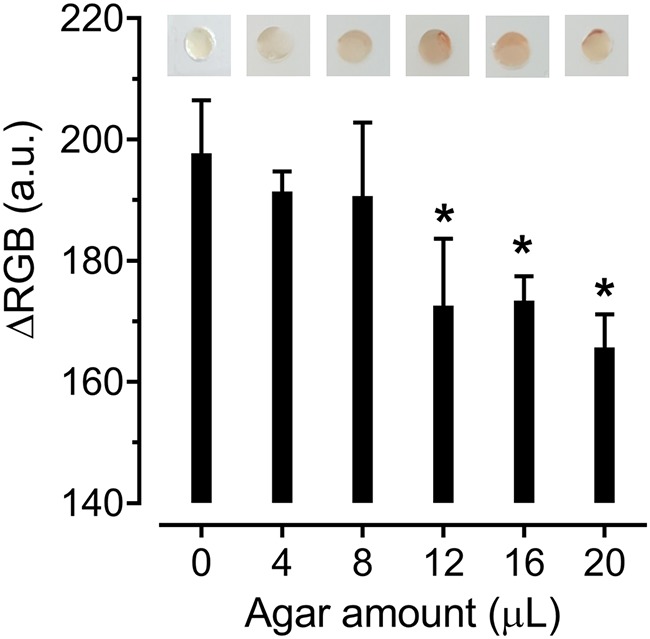
Optimization of the amount of chromogenic agar. Different amounts of chromogenic agar were deposited into the microfluidic chip. C. jejuni F38011 (108 CFU/ml) was incubated in the microfluidic chip at 42°C for 24 h. Photos of incubation chambers were captured using an iPhone 6. We employed ImageJ to analyze the average color intensity of each entire incubation chamber and expressed the intensity in ΔRGB value. The results are presented as the mean standard deviation of four biological replicates. One-way ANOVA followed by post hoc Dunnett’s test was conducted for statistical analysis. Asterisks (*) indicate significant differences (P < 0.05) between the negative-control and test groups.
Feasibility of on-chip bacterial cultivation.
The PDMS-based microfluidic chip is an ideal cultivation platform for biological samples due to its excellent gas permeability and lack of cytotoxicity (25). We evaluated bacterial growth in both microfluidic chips and conventional glass culture tubes by using C. jejuni F38011 as a bacterial model. Bacterial cell counts continuously increased in the microfluidic chip (Fig. S3), indicating that this device provided the appropriate growth conditions for Campylobacter spp. Compared to glass culture tubes, a significantly slower growth (P < 0.05) was observed in the microfluidic chip within 8 h of incubation, indicating that Campylobacter spp. experienced a longer lag phase in the device. The lower cell densities in the microfluidic chips might be due to antibiotic stresses from the chromogenic agar (26). In comparison, nutritious Mueller-Hinton broth (MHB) was used as a growth medium in glass culture tubes. In a previous study, a slightly lower bacterial count of Staphylococcus aureus in PDMS-based microfluidic chips than that in glass flasks was also identified (24). Similar or even significantly higher (P < 0.05) cell counts were identified in our microfluidic chips than in glass culture tubes between 24 and 48 h. Overall, comparable bacterial growth environments were achieved between the microfluidic chip and the conventional platform.
On-chip identification.
A detection device should be both specific and sensitive to the targeted Campylobacter spp., considering that diverse microbiota coexist with this microbe in agri-food products (27) and that it has a low infectious dose (400 to 800 cells) (4). After 48 h of incubation, no color change was observed with the presence of S. aureus, L. monocytogenes, E. coli, Salmonella enterica, Arcobacter butzleri, and Helicobacter pylori (Fig. 4). In contrast, three representative isolates of C. jejuni, C. coli, and C. lari generated red signals in the microfluidic device (Fig. 4). To further assess if on-chip identification is feasible for a range of Campylobacter spp., we incubated a total of 11 Campylobacter isolates (Table 1) in the microfluidic devices. All tested Campylobacter isolates exhibited prominent red signals compared to no signal from the negative control (MHB only) (Fig. S4), indicating that our developed assay could be used for these Campylobacter species. Although the color intensity of each strain might vary, positive readouts could be clearly identified based on the presence of red signals and generated a yes or no answer. Previous studies also discovered the bias on the recovery of Campylobacter isolates by using the conventional detection media (e.g., Preston broth and mExeter broth) due to different tolerance levels of Campylobacter strains to selective antibiotics (28, 29). Taken together, on-chip identification was specific to thermophilic Campylobacter species, including C. jejuni, C. coli, and C. lari.
FIG 4.
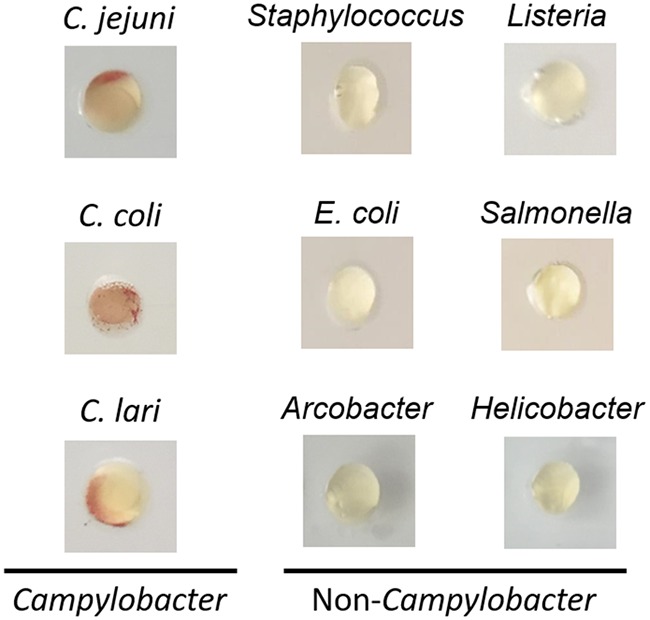
Specificity test. Campylobacter spp. and other foodborne pathogens (∼108 CFU/ml) were incubated in the microfluidic chips at 42°C for 48 h. The tested bacterial strains included C. jejuni F38011, C. coli 314, C. lari RM2818, Staphylococcus aureus S-FF10, Listeria monocytogenes ATCC 7644, Escherichia coli K-12, Salmonella enterica Enteritidis 43353, Arcobacter butzleri CCUG 30485, and Helicobacter pylori ATCC 43504.
TABLE 1.
Campylobacter isolates used in this study
| Strain | Source of isolation | Reference or source |
|---|---|---|
| Campylobacter jejuni F38011 | Clinical | 43 |
| C. jejuni ATCC 33560 | Bovine | 43 |
| C. jejuni NCTC 11168 | Clinical | 43 |
| C. jejuni 1143 | Chicken | This study |
| C. jejuni 1173 | Chicken | This study |
| C. jejuni 1329 | Cat | This study |
| C. coli 171 | Pig | This study |
| C. coli 314 | Pig | This study |
| C. coli 1148 | Pig | This study |
| C. coli 1330 | Cat | This study |
| C. lari RM2818 | Clinical | 48 |
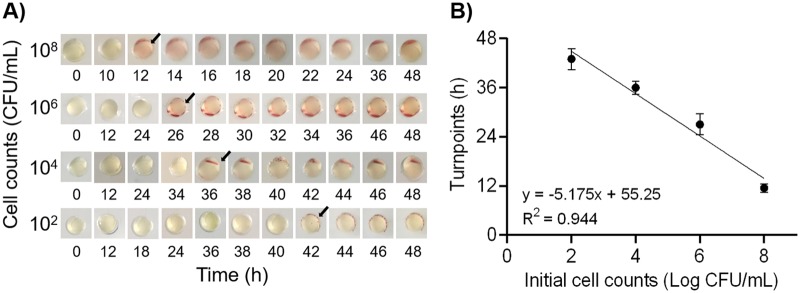
For the sensitivity study, a wide range of concentrations (102 to 108 CFU/ml) of C. jejuni were prepared in MHB for on-chip detection. Red signals in positive samples were easily identified regardless of color intensity (Fig. 5A). Interpretations of color changes were confirmed by 10 nontrained and 2 trained personnel, resulting in the same and correct readouts (data not shown). This demonstrated the objective and easy readouts of bacterial growth in a chromogenic assay-based microfluidic chip that could be accomplished with the naked eye. Typical red signals appeared after 12 h of incubation when the initial bacterial concentration was 1 × 108 CFU/ml (Fig. 5A). The limit of detection was less than 1 × 102 CFU/ml, and its corresponding turnaround analysis time was still shorter than the routine isolation procedure time (48 to 72 h) (8). Besides, a linear correlation (R2 = 0.944) was established between initial bacterial concentrations and color change time points (Fig. 5B). Using this linear regression model (y = −5.175x + 55.25, where y is the turn point [in h] and x is initial cell count [in log CFU/ml]), we could quantify the initial concentration of Campylobacter spp. in the tested samples.
FIG 5.
Sensitivity test. (A) Representative images of C. jejuni F38011 on-chip detection. Arrows indicate the time points when visible color changes were observed. (B) Linear regression model to quantify the initial bacterial counts (n = 3).
Detection of Campylobacter spp. in foods.
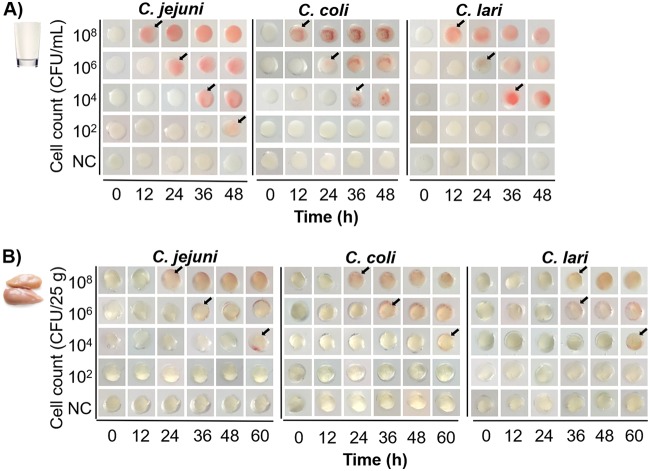
Campylobacter spp. are usually transmitted to humans through the consumption of undercooked chicken and raw milk (4). To demonstrate the practical application of this microfluidic device, we tested the performance of the on-chip assay for identifying Campylobacter spp. in food products. C. jejuni, C. coli, and C. lari were spiked in milk and chicken breast meat to allow the initial bacterial loads to be 102 to 108 CFU/ml and 102 to 108 CFU/25 g, respectively. All tested Campylobacter species produced red signals in the microfluidic device regardless of the food matrix (Fig. 6). No visible color change was obtained in the negative control throughout the analysis, demonstrating the high selectivity of this microfluidic device toward the detection of Campylobacter spp.
FIG 6.
Detection of Campylobacter spp. in whole milk (A) and fresh chicken meat (B) using the microfluidic device. The tested Campylobacter strains were C. jejuni F38011 (clinical isolate), C. coli 314 (pig isolate), and C. lari RM2818 (clinical isolate). Visible color changes are indicated by arrows.
Both the turnaround times and limits of detection varied in the two food models. For whole milk, the detection limit of 1 × 102 CFU/ml C. jejuni could be obtained within 48 h (Fig. 6A). This result was comparable to the detection of C. jejuni in MHB (Fig. 5A), suggesting that the effect of the food matrix on the sensitivity of our device was negligible. For chicken meat, a concentration as low as 1 × 104 CFU of Campylobacter spp. per chicken (25 g) could generate positive results after the analysis time was extended to 60 h (Fig. 6B). Compared to milk, the detection of Campylobacter spp. in chicken meat experienced at least a 12-h delay for the identical bacterial loads. The longer turnaround time and lower sensitivity of the spiked chicken sample were due to the 25× dilution of cells on the chicken surface into the meat rinse water, whereas bacterial cells in milk were directly tested using the microfluidic device. Although the detection limit of Campylobacter spp. in chicken meat was relatively high (400 CFU/g), it still meets the requirement of sampling plan regulation (no. 02005R2073-20190228) in the European Union, in which the maximal detection cutoff value is set as 1,000 CFU/g carcasses of broilers (30). In the United States, the USDA Food Safety and Inspection Service (FSIS) has proposed the chicken-related microbiological safety practices based on the presence/absence of Campylobacter in products (document no. FSIS-2018-0044) (49). In other words, the limit of detection should be <1 CFU/g. Our on-chip detection can be used as a rapid screening method in a high-load sampling plan. Optimizations, such as increasing the sampling size, concentrating the chicken rinse water, and including an enrichment step, can be performed in the future to improve the detection limit.
To further validate the specificity of Campylobacter detection in food samples with a multispecies bacterial background, we conducted an on-chip detection assay for the mixed culture of C. jejuni, Salmonella enterica, and Staphylococcus aureus, as these bacteria are commonly identified in the same food products, including raw chicken meat and milk (31–34). The cocktails of C. jejuni F38011, S. enterica serovar Enteritidis 43353, and S. aureus S-FF10 were prepared in either pasteurized milk or raw chicken breast meat and incubated in the microfluidic chips at 42°C for 24 h. S. Enteritidis in foods, S. aureus in foods, or original food samples were used as the negative controls. As shown in Fig. S5, an obvious red signal was identified in the multispecies bacterial cocktail. No color change was observed in single S. Enteritidis, single S. aureus, or original food samples. Besides the artificially spiked S. Enteritidis and S. aureus, the original food samples also contained natural microflora (Fig. S6). This result demonstrated that our on-chip detection assay has the potential to specifically detect Campylobacter spp. within a high concentration and complex bacterial background.
Antimicrobial susceptibility testing.
Theoretically, on-chip AST shares the principle of the conventional agar dilution method. In the individual incubation chambers, a defined amount of antibiotics was preloaded onto a paper disk, followed by the addition of chromogenic agar. A droplet of bacterial culture (∼7 μl) was then inoculated into each incubation chamber. During bacterial cultivation, antibiotic was expected to diffuse from the paper disk to the chromogenic agar and thus inhibit bacterial growth. The MIC was defined as the lowest antibiotic concentration at which no visible chromogenic reaction was observed.
To verify the accuracy of the on-chip AST, we determined the MIC of C. jejuni F38011 against three types of antibiotics using colorimetric-based microfluidic chips (namely, the on-chip MIC test). Ampicillin, tetracycline, and ciprofloxacin were selected due to their wide usage in both humans and food-producing animals (8) and high frequency of antimicrobial resistance to Campylobacter spp. (5). Two-fold serial dilutions of selected antibiotics were prepared in the microfluidic device. The MICs of all tested antibiotics were clearly distinguished based on the sudden absence of red signals as antibiotic concentrations increased to certain levels (Fig. 7).
FIG 7.
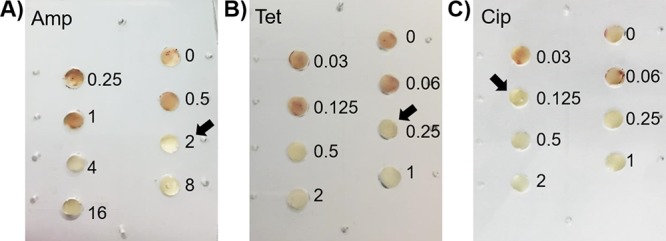
Determination of MICs of a clinical isolate C. jejuni F38011 against ampicillin (Amp) (A), tetracycline (Tet) (B), and ciprofloxacin (Cip) (C) using microfluidic devices. Numbers represent the concentrations (in μg/ml) of selected antibiotics. Arrows indicate the MICs.
Compared to the conventional agar dilution method, the on-chip MIC test had the same or 2-fold difference in MICs for certain antibiotic classes (Table S2). In other words, the on-chip MIC test achieved essential agreement with the reference method, a critical indicator to evaluate the accuracy for commercial AST (35). This result also indirectly validated that no cross-contamination of antibiotics occurred among different incubation chambers in the microfluidic device so that different antibiotic concentrations or types could be customized for the on-chip AST based on the need.
Campylobacter multidrug resistance (MDR) is regarded as a serious threat to public health (3, 6), calling for high-throughput characterization of bacterial multidrug susceptibility in clinical diagnosis and agri-food monitoring. Herein, we implemented on-chip multiplexed AST for ampicillin, tetracycline, and ciprofloxacin, according to susceptibility interpretative criteria established by the Clinical and Laboratory Standards Institute (CLSI) (5, 36). Instead of testing the exact MIC values, our on-chip MDR test only involved 2 levels of antibiotic concentrations, which are defined as susceptibility and resistance breakpoints by CLSI (37, 38). There were 3 possible readings using this colorimetric-based microfluidic device, as follows: (i) “susceptible” strains were determined when negative chromogenic results were generated at both the susceptibility and resistance breakpoints; (ii) “nonsusceptible” strains produced a positive result at the susceptibility breakpoint but a negative result at the resistance breakpoint; and (iii) “resistant” strains were confirmed by positive chromogenic signals at the susceptibility and resistance breakpoints. Different interpretative categories could be determined using an on-chip MDR test (Fig. 8). The appearance of red signals in the control groups without antibiotics suggested that the microfluidic device provided appropriate cultivation conditions for Campylobacter strains (Fig. 8). Taken together, AST and identification could be conducted simultaneously in a single device.
FIG 8.
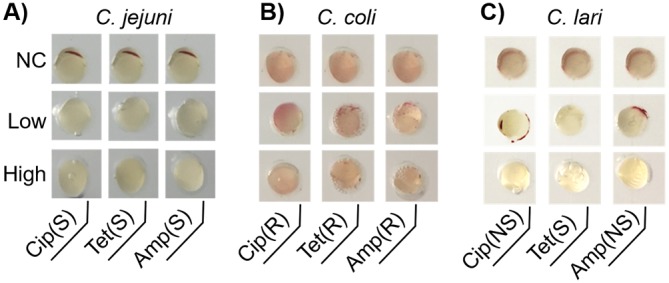
On-chip multidrug resistance (MDR) test to determine MDR profiles of C. jejuni F38011 (A), C. coli 314 (B), and C. lari RM2818 (C). Each column is deposited with selected types of antibiotics, ciprofloxacin (Cip), tetracycline (Tet), and ampicillin (Amp). Each row represents different concentrations of antibiotics, as follows: NC, no antibiotic; low, CLSI breakpoints for susceptible strains; high, CLSI breakpoints for resistant strains. The concentrations for each antibiotic are listed as follows: (i) for ciprofloxacin, the low and high concentrations were 1 and 4 μg/ml, respectively; (ii) for tetracycline, the low and high concentrations were 4 and 16 μg/ml, respectively; and (iii) for ampicillin, the low and high concentrations were 8 and 32 μg/ml, respectively. The AST results are expressed as susceptible (S), resistant (R), or nonsusceptible (NS).
To assess the accuracy of the on-chip MDR test, we calculated the categorical agreement rates between on-chip AST and the conventional agar dilution method. Categorical agreement is defined as obtaining the same categorical interpretation using newly developed approaches and reference methods (35). After testing the 11 animal and clinical isolates of 3 Campylobacter species listed in Table 1, we confirmed the categorical agreement rates to be 100%, 100%, and 90.9% for ciprofloxacin, tetracycline, and ampicillin, respectively (Table 2). These results met the requirement of acceptable categorical agreement (≥90%) by the U.S. Food and Drug Administration (35). Among all bacterium-antibiotic combinations, only one major error case was obtained (Table 2), in which the tested strain was regarded as ampicillin sensitive by the agar dilution method but resistant by the on-chip MDR test. Other than that, no on-chip MDR results were scored as very major errors (if reported as resistant by reference methods but susceptible by new methods) or minor errors (if reported as intermediate by reference methods but resistant or susceptible by new methods, or vice versa) (35). Besides the antibiotics tested in this study, we aim to expand the on-chip AST assay to other clinically important antibiotics in the future, particularly the drugs recommended for human campylobacteriosis (e.g., macrolides) (39).
TABLE 2.
Comparison of on-chip AST results with the conventional agar dilution method for Campylobacter isolates
| Antibiotic | AST results (no. correct/total no. of isolates) |
Categorical agreement rate (%) | No. (%) of errors by type |
||||
|---|---|---|---|---|---|---|---|
| C. jejuni | C. coli | C. lari | Very major | Major | Minor | ||
| Ciprofloxacin | 6/6 | 4/4 | 1/1 | 100 | 0 (0) | 0 (0) | 0 (0) |
| Tetracycline | 6/6 | 4/4 | 1/1 | 100 | 0 (0) | 0 (0) | 0 (0) |
| Ampicillin | 5/6 | 4/4 | 1/1 | 90.9 | 0 (0) | 1 (16.6) | 0 (0) |
In addition, our on-chip ASTs only took half the analysis time of that with the conventional agar dilution method (40). Lee and colleagues also observed this phenomenon when they assessed the vancomycin resistance profiles of Enterococcus spp. using an on-chip pH-based AST (41). There are two possible explanations. First, a high surface-to-volume ratio was provided in the microfluidic incubation chambers that facilitated the interaction between bacterial cells and chromogenic agar to exert chromogenic reactions more quickly. Second, red signals produced by the chromogenic reaction are more easily visualized than are the gray-colored colonies observed with the agar dilution method.
Advantages of on-chip identification and AST.
In this study, we established identification and AST for Campylobacter spp. using a colorimetric-based microfluidic device that provided results as accurate and sensitive as those obtained by gold standard methods. Beyond this, this microfluidic device could offer additional advantages over the conventional methods.
First, simultaneous identification and AST by using the microfluidic device reduced the turnaround time for Campylobacter study. The on-chip study was completed within 24 h once the presumptive Campylobacter isolate was ready. In contrast, standardized Campylobacter identification and AST are conducted separately, taking at least several days to collect the results. Briefly, traditional bacterial identification involves a 1-day shift of performing multiple biochemical assays, including Gram stain, oxidase, and catalase tests, for presumptive colonies collected on selective agar plates (8). The conventional AST approaches (e.g., broth dilution, agar dilution, and disk diffusion) provide definitive results after a 48-h analysis (5, 40). In total, the on-chip approach is expected to take only 30% of the analysis time of that with conventional methods. It is worth mentioning that a negligible operation touch time is required with the microfluidic chip because sample loading (3 min) is the only operation step for the end users.
Second, our device consumed less of the reagents and growth media than did the conventional study platforms, such as petri dishes and 96-well plates. For example, 20 μl of agar is required to test one bacterium-antibiotic combination, while at least 75 μl or 20 ml of growth medium is required for 96-well plates or petri dishes, respectively. Our method saves around 4 to 1,000 times the antibiotic agents and growth media that lead to more cost-effective detection.
Finally, portable and automated detection could be realized if a real-time imaging system (e.g., camera) is developed to monitor the color change of the microfluidic device.
Conclusions.
We established a colorimetric-based microfluidic device for timely identification and AST of Campylobacter spp. This device exhibits high specificity (i.e., shows no cross-reaction with other foodborne pathogens) and sensitivity (detection limit, ∼102 CFU/ml) toward Campylobacter spp. Accurate antibiotic resistance profiles of Campylobacter spp. can be obtained within 24 h using on-chip AST. This miniaturized platform can save 70% turnaround time and up to 1,000 times the reagents compared to those with the standard methods. In addition, on-chip results can be visualized by the naked eye, making the test free from expensive and bulky detectors and thus suitable for in-field studies, especially in resource-limited regions. A personalized AST can also be achieved by loading desired antibiotic-containing paper disks into the microfluidic device. This device has the potential to be used to monitor the prevalence of antibiotic-resistant pathogens in agri-foods, the environment, and clinical settings.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
A diverse panel of Campylobacter strains were tested in this study, including C. jejuni (n = 6), C. coli (n = 4), and C. lari (n = 1). The strain names and isolation sources are listed in Table 1. For routine cultivation, all Campylobacter strains were grown on Mueller-Hinton agar supplemented with 5% defibrinated sheep blood (MHBA) at 37°C under microaerobic conditions (85% N2, 10% CO2, 5% O2) for 48 h. To prepare Campylobacter overnight culture, bacterial colonies from MHBA plates were harvested and suspended in Mueller-Hinton broth (MHB) and then incubated at 37°C under microaerobic conditions for 16 to 18 h in a rotatory manner (at 175 rpm).
Antibiotics.
Ampicillin sodium salt, tetracycline hydrochloride, and ciprofloxacin were purchased from Sigma-Aldrich (Canada). According to solvent solubility, the stock solutions (640 μg/ml) of ampicillin and tetracycline were prepared in distilled water, while ciprofloxacin was dissolved in distilled water supplemented with 0.1 M HCl. The stock solutions were sterilized by filtering through 0.22-μm sterile nylon syringe filters and were stored at –20°C until use. For on-chip AST, an antibiotic working solution was freshly prepared by diluting the stock solution to 8× final concentrations using sterile distilled water. An aliquot of 2.5 μl working solution was added onto a sterile paper disk (Whatman no. 1 filter paper; Ø = 2 mm), air dried in the biosafety cabinet, and added into the microfluidic chips. Then, 20 μl of Campylobacter chromogenic agar medium (i.e., CHROMagar Campylobacter; CHROMagar, USA) was pipetted into each incubation chamber before solidification to achieve a 1× final concentration of antibiotics.
Design and fabrication of a microfluidic device.
The development of a microfluidic device was completed by a procedure consisting of 3 steps, namely, chip pattern design, master fabrication, and construction of a hybrid polydimethylsiloxane (PDMS)/glass microfluidic chip.
We designed the patterns of microfluidic device using the AutoCAD 2016 version (Autodesk, Inc., USA). Our device consists of three layers: an injection layer, a chamber layer, and a glass layer. The glass layer is a plain microscopic slide (75-mm length, 50-mm width, 1-mm thickness; Corning, USA), whereas the other two PDMS layers are featured with certain patterns. The chamber layer was developed with an inlet port (Ø = 1.5 mm), 8 incubation chambers (Ø = 4 mm), and an outlet port (Ø = 1.5 mm). A main channel (500 μm wide and 40 mm long) connected the inlet and outlet ports, and 8 zigzag-shaped side channels (100 μm wide and 7 mm long) were created to direct fluids from the main channel to 8 individual incubation chambers. The injection layer contained inlet and outlet ports (Ø = 1.5 mm), 8 incubation chambers (Ø = 4 mm), and 8 air vents (Ø = 1.5 mm), with a 100-μm-wide channel to connect the incubation chambers and air vents. These two patterns were printed on a transparency film (i.e., photomask) by CAD/Art Services, Inc. (Bandon, OR, USA).
The master was fabricated using standard photolithography (42). In brief, 4 ml of SU-8 2025 photoresist (MicroChem Corp., USA) was poured onto a clean silicon wafer (Ø = 100 mm; University Wafer, USA). To achieve an 80-μm thickness of chip patterns on the master, the SU-8 photoresist reagent was spin coated at 500 rpm for 10 s with an acceleration rate of 100 rpm/s, followed by 1,000 rpm for 30 s with an acceleration rate of 300 rpm/s. The photoresist reagent was soft baked at 65°C for 3 min and then at 95°C for 9 min. After cooling down to room temperature, the photoresist-coated wafer was exposed to UV light (dose, 215 mJ/cm2) through the photomask containing specific chip patterns. A postexposure bake was conducted by heating the photoresist-coated wafer at 65°C for 2 min and then at 95°C for 7 min. Unexposed photoresist was developed in SU-8 developer (MicroChem, USA) and rinsed off from the master.
After obtaining the masters, injection and chamber layers were made by PDMS using soft lithography (16). PDMS prepolymer (Dow Sylgard 184 silicone encapsulant clear kit; Ellsworth Adhesives, Canada) was prepared by mixing silicone elastomer and a curing agent at a ratio of 10:1 (wt/wt), according to the manufacturer’s protocol. A total of 16.5 g and 22 g of PDMS prepolymer was dispensed over the masters for the injection layer and chamber layer, respectively. After degassing, PDMS was cured on a hot plate (80°C) for 8 min, resulting in the defined chip patterns. The cured PDMS replica was carefully peeled off from the masters and punched through the whole thickness at the positions of the inlet/outlet ports (for both the chamber and injection layers), incubation chambers (chamber layer only), and air vents (injection layer only) using a Miltex biopsy punch (Ted Pella, Inc., USA). The assembly of three layers is described in Fig. 1A. Briefly, the PDMS-based injection layer and chamber layer were bonded together after treatment in a plasma cleanser (Harrick Plasma, USA), generating 8 individual reservoirs (i.e., incubation chambers) in the injection chamber slab. A piece of PVDF membrane (Ø = 4 mm) was deposited into each reservoir, serving as the supporting substrate of Campylobacter chromogenic agar. For on-chip AST, paper disks preloaded with specific antibiotics were put on the top of the PVDF membrane, and then 20 μl of chromogenic agar was added. To seal the incubation chambers, a glass slide was bonded to the chamber layer using a plasma-assisted method.
Operation of the microfluidic device.
The microfluidic device system contains a syringe pump, syringe, polyvinyl chloride (PVC) tubing, waste container, outlet blocker, and microfluidic chip. Before assembly, all components of the device were sterilized under UV light for 30 min. The inlet and outlet ports of microfluidic chip were separately connected to the syringe and waste container through capillary PVC tubing. A bacterial suspension was injected into the microfluidic device at a flow rate of 0.05 ml/min. Once the main channel was fully filled, the capillary tubing of outlet port was blocked by an outlet blocker (i.e., a metal clip). The injection was stopped until all incubation chambers were fully occupied by sample fluids. Agarose solution (0.4% [wt/vol]) was then introduced to expel the bacterial suspension from the main channel, insulate each incubation chamber, and prevent cross-contamination. The inlet, outlet, and air vents were sealed with adhesive tape to eliminate liquid evaporation. Microfluidic chips were maintained in a benchtop CO2 incubator (10% CO2, New Brunswick S41i; Eppendorf, USA) at 42°C for up to 48 h.
Bacterial on-chip growth.
The feasibility of on-chip bacterial cultivation was investigated using C. jejuni F38011 as a bacterial model (43). C. jejuni overnight culture in MHB was adjusted to an initial concentration of 106 CFU/ml. A bacterial suspension was inoculated either into the microfluidic device or the conventional glass culture tube, followed by static incubation at 42°C under a microaerobic environment. Bacterial cell counts were monitored at certain time points (0, 2, 8, 24, and 48 h). To collect a bacterial suspension from the microfluidic device, a sterile syringe with a permanently attached needle was used to penetrate the PDMS-based injection layer and withdraw the liquid from the individual incubation chambers. A plating assay was used to determine the bacterial cell counts.
Specificity and sensitivity tests.
The on-chip identification assay was designed to target three major thermophilic Campylobacter species: C. jejuni, C. coli, and C. lari. The presence of Campylobacter spp. was indicated based on the color change from light yellow to red due to a chromogenic reaction. To evaluate the specificity of on-chip identification, we tested common foodborne pathogens, including Staphylococcus aureus (S-FF10, a clinical methicillin-resistant isolate), Listeria monocytogenes (ATCC 7644), Escherichia coli (K-12), and Salmonella enterica serotype Enteritidis (43353) (44, 50). In addition, Arcobacter butzleri (CCUG 30485) and Helicobacter pylori (ATCC 43504) were tested due to their close phylogenic relativeness to Campylobacter spp. (45). Overnight cultures of S. aureus, L. monocytogenes, E. coli, and S. Enteritidis were prepared in tryptic soy broth at 37°C under aerobic conditions, while A. butzleri and H. pylori were cultivated in MHB at 37°C under microaerobic conditions. The overnight culture (16 to 18 h) of each strain was adjusted to 108 CFU/ml, injected into the microfluidic chips, and incubated at 42°C under a microaerobic environment for 48 h. Photos of the on-chip identification assay were collected by using a smartphone (iPhone 6) at the end of the incubation.
For the sensitivity test, C. jejuni F38011 at a wide range of concentrations (102 to 108 CFU/ml) was incubated in the microfluidic chips at 42°C under a microaerobic environment. The color change of the on-chip identification assay was monitored at a time interval of 2 h for up to 48 h. Photos were obtained by using the iPhone 6 at each time point. The limit of detection was defined as the lowest initial concentration of C. jejuni that could generate visible chromogenic reaction results.
Detection of Campylobacter spp. in food samples.
Whole milk and raw chicken meat were selected as food models because the consumption of these food commodities is the major route of human campylobacteriosis.
We purchased pasteurized whole milk (3.25% fat) from local grocery stores in Vancouver, Canada, and kept the milk at 4°C for further study. The absence of Campylobacter spp. in the milk was confirmed by a plating assay using modified charcoal-cefoperazone-deoxycholate agar (mCCDA). To assess the universal feasibility of an on-chip identification assay for major thermophilic Campylobacter spp., we determined the growth of C. jejuni (F38011), C. coli (314), and C. lari (RM2818) in microfluidic chips. An overnight culture of each strain was adjusted to a concentration of 109 CFU/ml (optical density at 600 nm [OD600], ca. 0.3) using MHB, followed by 10-fold serial dilutions in whole milk to achieve the final inoculation concentrations of 102 to 108 CFU/ml. Spiked milk samples were directly injected into the microfluidic chips without any sample pretreatment. Milk without the addition of Campylobacter spp. was regarded as a negative control. A color change of Campylobacter spp. in the artificially spiked milk was monitored at a time interval of 12 h for up to 48 h.
Raw boneless, skinless chicken breast meat was purchased from local grocery stores as well. Chicken breast meat was cut into pieces of 25 g each, the recommended sampling size by the U.S. Food and Drug Administration BAM protocol (https://www.fda.gov/food/laboratory-methods-food/bam-campylobacter). To control the starting concentrations of Campylobacter spp., we disinfected the surface of the chicken meat by using a 1% sodium hypochlorite aqueous solution (VWR International, Canada) and then spiked chicken products with defined concentrations of Campylobacter cells (102 to 108 CFU/25 g). For the recovery of Campylobacter cells, the spiked chicken samples were transferred into sterile stomach bags, followed by adding 25 ml phosphate-buffered saline (PBS) and hand massaging the chicken meat. The rinse solution was filtered through a sterile Whatman grade 1 filtration paper to remove large meat particles. The rinse solution was then injected into the microfluidic chips for further detection. The results were monitored every 12 h for up to 60 h. In the meanwhile, the spiked concentrations of Campylobacter cells in both milk and chicken samples were confirmed by a conventional plating assay. Briefly, serially diluted bacterial samples were streaked onto mCCDA and incubated at 42°C for 48 h for enumeration.
To assess the specificity of on-chip identification of Campylobacter spp. in foods, we spiked Campylobacter spp. and two representative foodborne pathogens (i.e., S. aureus and S. enterica) in food samples that were purchased in grocery stores in Vancouver and used without any further decontamination treatment (i.e., raw chicken breast meat and pasteurized milk). The spiking procedure was the same as that for the sensitivity test described above. Briefly, equal concentrations (1 × 108 CFU/ml) of C. jejuni F38011, S. Enteritidis 43353, and S. aureus S-FF10 were prepared as a bacterial cocktail and inoculated in foods, followed by incubation in the microfluidic chips at 42°C under microaerobic conditions for 24 h. Nonspiked food samples, S. Enteritidis in foods, or S. aureus in foods were used as the negative controls. The initial concentrations of bacterial cocktails were confirmed by a plating assay. The presence of natural microbiota and Campylobacter spp. in nonspiked foods was determined by streaking 1 ml of samples onto tryptic soy agar (TSA) and mCCDA, respectively. TSA plates were incubated at 37°C under aerobic conditions for 24 h, whereas mCCDA plates were incubated at 42°C under microaerobic conditions for 48 h.
Antimicrobial susceptibility testing.
We developed two types of on-chip ASTs to determine either the MIC or multidrug resistance (MDR) profiles of Campylobacter isolates. These were an on-chip MIC assay and an on-chip MDR assay, respectively. Both assays were conducted following the procedures mimicking the CLSI guidelines, with some modifications (37). For the on-chip MIC assay, 8 different concentrations of certain antibiotics were incorporated into the microfluidic device. In brief, we prepared a 2-fold dilution series of antibiotic working solutions (8× final concentration), added 2.5 μl of each working solution onto a paper disk, and deposited these paper disks into the microfluidic chip after air drying. Afterwards, 20 μl of chromogenic agar was added to an individual incubation chamber so that accurate final concentrations of antibiotic in the chip were generated. An inoculation concentration of 108 CFU/ml of C. jejuni F38011 overnight culture was incubated in the microfluidic chips at 42°C under microaerobic conditions for 24 h (37, 46). The MICs were determined based on the lowest antibiotic concentration that inhibited the generation of color change.
For the on-chip MDR assay, susceptible and resistant breakpoints assigned by the CLSI were selected as the tested concentrations for three antibiotic classes (ampicillin, tetracycline, and ciprofloxacin) (39). As no ampicillin breakpoint is available for Campylobacter spp., we used the CLSI breakpoints for Enterobacteriaceae instead (47). The antibiotic breakpoints are listed in Table S2. A total of 11 isolates of Campylobacter (Table 1) were tested using an on-chip MDR assay. A conventional agar dilution method was conducted in parallel to validate on-chip MIC and on-chip MDR results (37, 46).
Images and data analysis.
All of the experiments were performed in at least three biological replicates. Visual interpretation of bacterial growth in the microfluidic chips was obtained by two independent personnel so as to ensure the absence of individual bias on colorimetric-based readouts. Student's t test or one-way analysis of variance (ANOVA), followed by post hoc Tukey’s or Dunnett’s tests, were conducted to determine the significant differences among groups (P < 0.05). The equations for AST categorical agreement are listed in Table S1.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by the Natural Sciences and Engineering Research Council of Canada in the form of Discovery Grant NSERC RGPIN-2019-03960 and Discovery Accelerator Grant PGPAS-2019-00024.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Tack DM, Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Scallan E, Lathrop S, Muse A, Ryan P, Smith K, Tobin‐D'Angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, Geissler AL. 2019. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 2015–2018. Am J Transplant 19:1859–1863. doi: 10.1111/ajt.15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allos BM. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis 32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 3.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol 4:189–200. doi: 10.2217/17460913.4.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. 2015. Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge B, Wang F, Sjölund-Karlsson M, McDermott PF. 2013. Antimicrobial resistance in Campylobacter: susceptibility testing methods and resistance trends. J Microbiol Methods 95:57–67. doi: 10.1016/j.mimet.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. [Google Scholar]
- 7.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 8.Government of Canada. 2018. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2016 annual report. Public Health Agency of Canada, Guelph, Ontario, Canada: http://publications.gc.ca/collections/collection_2018/aspc-phac/HP2-4-2016-eng.pdf. [Google Scholar]
- 9.Moore JE, Corcoran D, Dooley JSG, Fanning S, Lucey B, Matsuda M, McDowell DA, Mégraud F, Millar BC, O'Mahony R, O'Riordan L, O'Rourke M, Rao JR, Rooney PJ, Sails A, Whyte P. 2005. Campylobacter. Vet Res 36:351–382. doi: 10.1051/vetres:2005012. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Yang X, Zhao W. 2017. Emerging microtechnologies and automated systems for rapid bacterial identification and antibiotic susceptibility testing. SLAS Technol 22:585–608. doi: 10.1177/2472630317727519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonard H, Colodner R, Halachmi S, Segal E. 2018. Recent advances in the race to design a rapid diagnostic test for antimicrobial resistance. ACS Sens 3:2202–2217. doi: 10.1021/acssensors.8b00900. [DOI] [PubMed] [Google Scholar]
- 12.Zhang G, Zheng G, Zhang Y, Ma R, Kang X. 2018. Evaluation of a micro/nanofluidic chip platform for the high-throughput detection of bacteria and their antibiotic resistance genes in post-neurosurgical meningitis. Int J Infect Dis 70:115–120. doi: 10.1016/j.ijid.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Wang C-H, Lien K-Y, Wu J-JJ, Lee G-B. 2011. A magnetic bead-based assay for the rapid detection of methicillin-resistant Staphylococcus aureus by using a microfluidic system with integrated loop-mediated isothermal amplification. Lab Chip 11:1521–1531. doi: 10.1039/c0lc00430h. [DOI] [PubMed] [Google Scholar]
- 14.Tang Y, Fang L, Xu C, Zhang Q. 2017. Antibiotic resistance trends and mechanisms in the foodborne pathogen, Campylobacter. Anim Health Res Rev 18:87–98. doi: 10.1017/S1466252317000135. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto Y, Sakakihara S, Grushnikov A, Kikuchi K, Noji H, Yamaguchi A, Iino R, Yagi Y, Nishino K. 2016. A microfluidic channel method for rapid drug-susceptibility testing of Pseudomonas aeruginosa. PLoS One 11:e0148797. doi: 10.1371/journal.pone.0148797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, Samuelson DR, Xu Y, Zhang H, Wang S, Rasco BA, Xu J, Konkel ME. 2013. Detecting and tracking nosocomial methicillin-resistant Staphylococcus aureus using a microfluidic SERS biosensor. Anal Chem 85:2320–2327. doi: 10.1021/ac303279u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong T, Zhao X. 2015. Rapid identification and susceptibility testing of uropathogenic microbes via immunosorbent ATP-bioluminescence assay on a microfluidic simulator for antibiotic therapy. Anal Chem 87:2410–2418. doi: 10.1021/ac504428t. [DOI] [PubMed] [Google Scholar]
- 18.He J, Mu X, Guo Z, Hao H, Zhang C, Zhao Z, Wang Q. 2014. A novel microbead-based microfluidic device for rapid bacterial identification and antibiotic susceptibility testing. Eur J Clin Microbiol Infect Dis 33:2223–2230. doi: 10.1007/s10096-014-2182-z. [DOI] [PubMed] [Google Scholar]
- 19.Pitruzzello G, Thorpe S, Johnson S, Evans A, Gadêlha H, Krauss TF. 2019. Multiparameter antibiotic resistance detection based on hydrodynamic trapping of individual E. coli. Lab Chip 19:1417–1426. doi: 10.1039/c8lc01397g. [DOI] [PubMed] [Google Scholar]
- 20.Etayash H, Khan MF, Kaur K, Thundat T. 2016. Microfluidic cantilever detects bacteria and measures their susceptibility to antibiotics in small confined volumes. Nat Commun 7:12947–12955. doi: 10.1038/ncomms12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kara V, Duan C, Gupta K, Kurosawa S, Stearns-Kurosawa DJ, Ekinci KL. 2018. Microfluidic detection of movements of Escherichia coli for rapid antibiotic susceptibility testing. Lab Chip 18:743–753. doi: 10.1039/c7lc01019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cira NJ, Ho JY, Dueck ME, Weibel DB. 2012. A self-loading microfluidic device for determining the minimum inhibitory concentration of antibiotics. Lab Chip 12:1052–1059. doi: 10.1039/c2lc20887c. [DOI] [PubMed] [Google Scholar]
- 23.Elavarasan T, Chhina SK, Parameswaran (Ash) M, Sankaran K, Parameswaran M, Sankaran K. 2013. Resazurin reduction based colorimetric antibiogram in microfluidic plastic chip. Sens Actuators B Chem 176:174–180. doi: 10.1016/j.snb.2012.10.011. [DOI] [Google Scholar]
- 24.Xu B, Du Y, Lin J, Qi M, Shu B, Wen X, Liang G, Chen B, Liu D. 2016. Simultaneous identification and antimicrobial susceptibility testing of multiple uropathogens on a microfluidic chip with paper-supported cell culture arrays. Anal Chem 88:11593–11600. doi: 10.1021/acs.analchem.6b03052. [DOI] [PubMed] [Google Scholar]
- 25.Sabhachandani P, Sarkar S, Zucchi PC, Whitfield BA, Kirby JE, Hirsch EB, Konry T. 2017. Integrated microfluidic platform for rapid antimicrobial susceptibility testing and bacterial growth analysis using bead-based biosensor via fluorescence imaging. Microchim Acta 184:4619–4628. doi: 10.1007/s00604-017-2492-9. [DOI] [Google Scholar]
- 26.Jacob P, Mdegela RH, Nonga HE. 2011. Comparison of Cape Town and Skirrow’s Campylobacter isolation protocols in humans and broilers in Morogoro, Tanzania. Trop Anim Health Prod 43:1007–1013. doi: 10.1007/s11250-011-9799-z. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Shin H, Park H, Jung H, Kim J, Cho S, Ryu S, Jeon B. 2019. Microbiota analysis for the optimization of Campylobacter isolation from chicken carcasses using selective media. Front Microbiol 10:1381–1389. doi: 10.3389/fmicb.2019.01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams LK, Sait LC, Cogan TA, Jørgensen F, Grogono-Thomas R, Humphrey TJ. 2012. Enrichment culture can bias the isolation of Campylobacter subtypes. Epidemiol Infect 140:1227–1235. doi: 10.1017/S0950268811001877. [DOI] [PubMed] [Google Scholar]
- 29.Goossens H, De Boeck M, Coignau H, Vlaes L, Van den Borre C, Butzler JP. 1986. Modified selective medium for isolation of Campylobacter spp. from feces: comparison with Preston medium, a blood-free medium, and a filtration system. J Clin Microbiol 24:840–843. doi: 10.1128/JCM.24.5.840-843.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.European Parliament. 2005. Commission regulation (EC) no. 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. European Commission, Brussels, Belgium: https://eur-lex.europa.eu/eli/reg/2005/2073/2019-02-28. [Google Scholar]
- 31.Olsen SJ, Ying M, Davis MF, Deasy M, Holland B, Iampietro L, Baysinger CM, Sassano F, Polk LD, Gormley B, Hung MJ, Pilot K, Orsini M, Van Duyne S, Rankin S, Genese C, Bresnitz EA, Smucker J, Moll M, Sobel J. 2004. Multidrug-resistant Salmonella Typhimurium infection from milk contaminated after pasteurization. Emerg Infect Dis 10:932–935. doi: 10.3201/eid1005.030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kümmel J, Stessl B, Gonano M, Walcher G, Bereuter O, Fricker M, Grunert T, Wagner M, Ehling-Schulz M. 2016. Staphylococcus aureus entrance into the dairy chain: tracking S. aureus from dairy cow to cheese. Front Microbiol 7:1603. doi: 10.3389/fmicb.2016.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antunes P, Mourão J, Campos J, Peixe L. 2016. Salmonellosis: the role of poultry meat. Clin Microbiol Infect 22:110–121. doi: 10.1016/j.cmi.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Wu S, Huang J, Wu Q, Zhang J, Zhang F, Yang X, Wu H, Zeng H, Chen M, Ding Y, Wang J, Lei T, Zhang S, Xue L. 2018. Staphylococcus aureus isolated from retail meat and meat products in China: incidence, antibiotic resistance and genetic diversity. Front Microbiol 9:2767–2780. doi: 10.3389/fmicb.2018.02767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, Koeth L, Sei K, CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing. 2018. CLSI Methods Development and Standardization Working Group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol 56:e01934-17. doi: 10.1128/JCM.01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandenberg O, Houf K, Douat N, Vlaes L, Retore P, Butzler JP, Dediste A. 2006. Antimicrobial susceptibility of clinical isolates of non-jejuni/coli campylobacters and arcobacters from Belgium. J Antimicrob Chemother 57:908–913. doi: 10.1093/jac/dkl080. [DOI] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute (CLSI). 2018. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 5th ed CLSI document VET01. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.Clinical and Laboratory Standards Institute (CLSI). 2016. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria, 3rd ed CLSI document M45. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed] [Google Scholar]
- 39.Shen Z, Wang Y, Zhang Q, Shen J. 2017. Antimicrobial resistance in Campylobacter spp. Microbiol Spectr 6:ARBA-0013-2017. doi: 10.1128/microbiolspec.ARBA-0013-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei B, Cha SY, Kang M, Roh JH, Seo HS, Yoon RH, Jang HK. 2014. Antimicrobial susceptibility profiles and molecular typing of Campylobacter jejuni and Campylobacter coli isolates from ducks in South Korea. Appl Environ Microbiol 80:7604–7610. doi: 10.1128/AEM.02469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee W-B, Fu C-Y, Chang W-H, You H-L, Wang C-H, Lee MS, Lee G-B. 2017. A microfluidic device for antimicrobial susceptibility testing based on a broth dilution method. Biosens Bioelectron 87:669–678. doi: 10.1016/j.bios.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Renner LD, Zan J, Hu LI, Martinez M, Resto PJ, Siegel AC, Torres C, Hall SB, Slezak TR, Nguyen TH, Weibel DB. 2017. Detection of ESKAPE bacterial pathogens at the point of care using isothermal DNA-based assays in a portable degas-actuated microfluidic diagnostic assay platform. Appl Environ Microbiol 83:e02449-16. doi: 10.1128/AEM.02449-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng J, Ma L, Nie J, Konkel ME, Lu X. 2017. Environmental stress-induced bacterial lysis and extracellular DNA release contribute to Campylobacter jejuni biofilm formation. Appl Environ Microbiol 84:e02068-17. doi: 10.1128/AEM.02068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Law JWF, Mutalib NSA, Chan KG, Lee LH. 2014. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol 5:770–788. doi: 10.3389/fmicb.2014.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vandamme P, De Ley J. 1991. Proposal for a new family, Campylobacteraceae. Int J Syst Bacteriol 41:451–455. doi: 10.1099/00207713-41-3-451. [DOI] [Google Scholar]
- 46.Ma H, Su Y, Ma L, Ma L, Li P, Du X, Golz G, Wang S, Lu X. 2017. Prevalence and characterization of Campylobacter jejuni isolated from retail chicken in Tianjin, China. J Food Prot 80:1032–1040. doi: 10.4315/0362-028X.JFP-16-561. [DOI] [PubMed] [Google Scholar]
- 47.Clinical and Laboratory Standards Institute (CLSI). 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 48.Miller WG, On SLW, Wang G, Fontanoz S, Lastovica AJ, Mandrell RE. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J Clin Microbiol 43:2315–2329. doi: 10.1128/JCM.43.5.2315-2329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Food Safety and Inspection Service. 2019. Changes to the Campylobacter Verification Testing Program: revised performance standards for Campylobacter in not-ready-to-eat comminuted chicken and turkey and related agency procedures. https://www.govinfo.gov/content/pkg/FR-2019-08-06/pdf/2019-16765.pdf. [Google Scholar]
- 50.Lu X, Samuelson DR, Xu Y, Zhang H, Wang S, Rasco BA, Xu J, Konkel ME. 2013. Detecting and tracking nosocomial methicillin-resistant Staphylococcus aureus using a microfluidic SERS biosensor. Anal Chem 85:2320–2327. doi: 10.1021/ac303279u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.