The regulation of antibiotic biosynthesis by Streptomyces species is complex, especially for biosynthetic gene clusters with multiple regulatory genes. The biosynthetic gene cluster for the polyene antibiotic candicidin contains four consecutive regulatory genes, which encode regulatory proteins from different families and which form a subcluster within the larger biosynthetic gene cluster in Streptomyces sp. FR-008. Syntenic arrangements of these regulatory genes are widely distributed in polyene gene clusters, such as the amphotericin and nystatin gene clusters, suggesting a conserved regulatory mechanism controlling production of these clinically important medicines. However, the relationships between these multiple regulatory genes are unknown. In this study, we determined that each of these four regulatory genes is critical for candicidin production. Additionally, using transcriptional analyses, bioassays, high-performance liquid chromatography (HPLC) analysis, and genetic cross-complementation, we showed that FscR1 to FscR4 comprise a hierarchical regulatory network that controls candicidin production and is likely representative of how expression of other polyene biosynthetic gene clusters is controlled.
KEYWORDS: Streptomyces, regulation, antibiotic, polyene, regulation of gene expression
ABSTRACT
The four regulatory genes fscR1 to fscR4 in Streptomyces sp. strain FR-008 form a genetic arrangement that is widely distributed in macrolide-producing bacteria. Our previous work has demonstrated that fscR1 and fscR4 are critical for production of the polyene antibiotic candicidin. In this study, we further characterized the roles of the other two regulatory genes, fscR2 and fscR3, focusing on the relationship between these four regulatory genes. Disruption of a single or multiple regulatory genes did not affect bacterial growth, but transcription of genes in the candicidin biosynthetic gene cluster decreased, and candicidin production was abolished, indicating a critical role for each of the four regulatory genes, including fscR2 and fscR3, in candicidin biosynthesis. We found that fscR1 to fscR4, although differentially expressed throughout the growth phase, displayed similar temporal expression patterns, with an abrupt increase in the early exponential phase, coincident with initial detection of antibiotic production in the same phase. Our data suggest that the four regulatory genes fscR1 to fscR4 have various degrees of control over structural genes in the biosynthetic cluster under the conditions examined. Extensive transcriptional analysis indicated that complex regulation exists between these four regulatory genes, forming a regulatory network, with fscR1 and fscR4 functioning at a lower level. Comprehensive cross-complementation analysis indicates that functional complementation is restricted among the four regulators and unidirectional, with fscR1 complementing the loss of fscR3 or -4 and fscR4 complementing loss of fscR2. Our study provides more insights into the roles of, and the regulatory network formed by, these four regulatory genes controlling production of an important pharmaceutical compound.
IMPORTANCE The regulation of antibiotic biosynthesis by Streptomyces species is complex, especially for biosynthetic gene clusters with multiple regulatory genes. The biosynthetic gene cluster for the polyene antibiotic candicidin contains four consecutive regulatory genes, which encode regulatory proteins from different families and which form a subcluster within the larger biosynthetic gene cluster in Streptomyces sp. FR-008. Syntenic arrangements of these regulatory genes are widely distributed in polyene gene clusters, such as the amphotericin and nystatin gene clusters, suggesting a conserved regulatory mechanism controlling production of these clinically important medicines. However, the relationships between these multiple regulatory genes are unknown. In this study, we determined that each of these four regulatory genes is critical for candicidin production. Additionally, using transcriptional analyses, bioassays, high-performance liquid chromatography (HPLC) analysis, and genetic cross-complementation, we showed that FscR1 to FscR4 comprise a hierarchical regulatory network that controls candicidin production and is likely representative of how expression of other polyene biosynthetic gene clusters is controlled.
INTRODUCTION
The actinomycete bacteria, especially the soil-dwelling Streptomyces, are a large family of microbes capable of producing compounds with clinical application, including anti-infectious agents (1). The agents with antifungal activity are mostly polyene antibiotics, which comprise a family of type I polyketide macrolide ring compounds with backbones of 20 to 40 or more carbons containing several conjugated double bonds (2). Polyene antibiotics can bind specifically to ergosterol and alter the cellular functions in which ergosterol is involved (3–6).
In Streptomyces species, genes responsible for the biosynthesis of an antibiotic are usually clustered together, forming a biosynthetic gene cluster (BGC). However, production of antibiotics can be regulated at different levels, including through regulatory genes within the cluster and beyond, forming an apparent regulatory cascade (7). In addition to structural genes that are devoted to the assembly or formation of the main chemical structure, a specific BGC will often contain the regulatory genes that constitute the lower levels of the regulatory cascade controlling production of the metabolite, such as actII-ORF4 for actinorhodin and redD for undecylprodigiosin in Streptomyces coelicolor, as well as other regulators (7–10). However, regulatory factors outside a specific BGC, for example, pleiotropic or global regulatory factors such as signal transduction systems and sigma factors, can also affect the biosynthesis of a specific compound (7, 11–14), and in some circumstances, the pathway-specific regulatory factors mediate the regulatory effects of these pleiotropic or global regulatory factors (15–17).
The best-known polyene antibiotics include amphotericin, candicidin, nystatin, and pimaricin (2, 18–20). Gene clusters for the biosynthesis of these polyene antibiotics have been sequenced (21–28), and multiple regulatory genes were identified within these clusters. The compound FR-008 is a heptaene macrolide antibiotic produced by Streptomyces sp. strain FR-008 and is identical to candicidin in structure (26, 28). The genome of Streptomyces sp. FR-008 has been sequenced (GenBank accession numbers CP009802 to CP009804) (28), and in addition to the linear chromosome, the FR-008 genome harbors two linear plasmids, which altogether comprise a total genome length of 7.2 Mb; this is one of the smallest Streptomyces genomes thus far reported (28, 29). Bioinformatic analysis revealed that the Streptomyces sp. FR-008 genome carries 23 putative secondary metabolic gene clusters, only two of which were annotated as encoding type I polyketide synthases (PKS), including the one for the biosynthesis of candicidin. Streptomyces sp. FR-008 also carries a cluster for the biosynthesis of another antifungal agent, antimycin (26, 28, 30). In the putative BGC for candicidin, which extends more than 100 kb in length and encompasses 21 genes (see Fig. S1 in the supplemental material), four consecutive regulatory genes, fscR1 to fscR4, were found to form a regulatory subcluster (26, 28) encoding transcriptional regulators from two different families in the FR-008 pathway. FscR1 is a relatively small protein of 231 amino acids (aa) containing a PAS domain and a helix-turn-helix (HTH) domain of the LuxR type, for which the archetype is PimM (31, 32); FscR2, FscR3, and FscR4 are relatively large proteins of more than 900 aa and are members of the large ATP-binding family of regulators of the LuxR type (LAL) (33), with each containing an AAA-ATPase domain and an HTH domain of the LuxR type and FscR3 additionally containing a DNA polymerase III domain (see Fig. S2 in the supplemental material). Orthologous genes (see Fig. S3 and S4 in the supplemental material) (34) form syntenic clusters (see Fig. S5 in the supplemental material) in the pathways for multiple polyene antibiotics, such as amphotericin (24, 35), candicidin (25, 27), nystatin (23, 36), and salinomycin (37, 38), implying that the strategy for controlling expression of polyene antibiotics is likely conserved. The roles of the four orthologous regulatory genes in the nystatin biosynthetic cluster have been investigated, and it was found that the four nysR genes are involved in production of nystatin (36, 39, 40). Our previous study showed that at least two of the four regulatory genes in the FR-008 BGC, fscR1 and fscR4, are required for the biosynthesis of candicidin, and interestingly, these two regulatory genes are also cross-regulated (34). However, the relationship among, and the regulatory network formed by, these four regulatory genes had not been characterized. In this study, we further characterized the roles of the remaining two regulatory genes, fscR2 and fscR3, and investigated the relationship among these four regulatory genes in the polyene FR-008 BGC. Our study revealed an intraregulatory network formed by multiple regulatory genes of the same pathway that control polyene production in Streptomyces.
RESULTS
The four regulatory genes fscR1 to fscR4 have similar temporal transcription patterns.
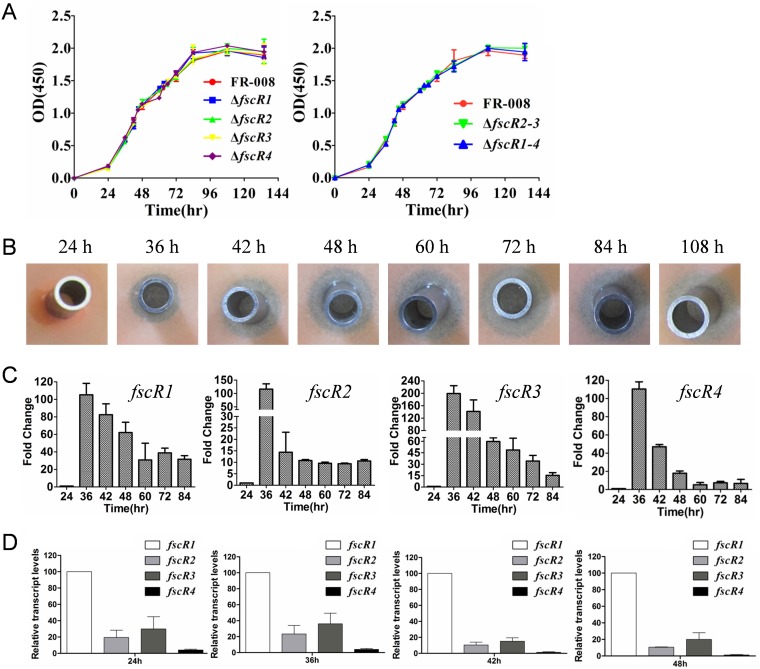
As secondary metabolites, antibiotics are usually produced by Streptomyces species after the organisms enter stationary phase on solid medium, coincident with morphological differentiation (41, 42). To investigate the temporal expression of these regulatory genes and its association with production of candicidin, we first prepared the growth curve of the wild-type strain of Streptomyces sp. FR-008 in liquid YEME, a rich growth medium (43). As shown in Fig. 1A (left panel), the wild-type strain displayed a growth curve typical of Streptomyces (43, 44). Production of candicidin was not yet detectable in the 24-h sample but was detectable for samples taken at 36 h, as reflected by a clear, although small, zone of inhibition of the indicator yeast strain (Fig. 1B). Enhanced levels of candicidin were detected in samples taken at 42 h and thereafter, as judged by the larger zones of inhibition (Fig. 1B), indicating that candicidin is produced during the exponential phase of growth.
FIG 1.
Growth phenotypes of mutants and temporal expression patterns of regulatory genes of the FR-008 pathway. (A) Growth curves of Streptomyces strains in YEME liquid medium. Left panel, wild-type Streptomyces sp. strain FR-008 and single mutant strains with deletion of one regulatory gene; right panel, FR-008 and mutant strains with deletion of more than one regulatory gene. (B) Bioassay of samples of the wild-type strain taken at the indicated time points during growth. (C) Temporal expression pattern of the four regulatory genes fscR1 to fscR4 in Streptomyces sp. FR-008 cultured in YEME liquid medium. RNA samples were isolated at the indicated times, and expression of hrdB, which encodes the major sigma factor, was used as an internal control. The expression level of each gene at 24 h was arbitrarily set to one. The y axis shows the fold change in expression at the indicated time over the level at 24 h. Results are the means (± standard deviations [SD]) from triplicate biological experiments. (D) Comparison of the relative expression of fscR1 to fscR4 in the wild-type strain during growth in YEME liquid medium. In this assay, expression of hrdB, which encodes the major sigma factor, was used as an internal control, and the expression level of fscR1 at each time point was arbitrarily set to 100. The y axis shows the fold change in expression of each gene over the expression of fscR1 at the indicated times. Results are the means (±SD) from triplicate biological experiments.
Streptomyces sp. FR-008 harbors an additional BGC that produces another antifungal agent, antimycin (30), the production of which is also regulated by FscR1 (45). To investigate whether antimycin contributed to the inhibition of the indicator strain (Rhodotorula rubra) in the bioassay, we tested the sensitivity of the indicator strain to antimycin at different concentrations (see Fig. S6 in the supplemental material). Our analysis showed that whereas the wild-type FR-008 strain generated a clear zone of inhibition, antimycin did not produce a detectable zone of inhibition at concentrations up to 10 μg/ml, indicating that under the conditions tested, the indicator strain is not sensitive to antimycin. These results indicate that the bioassay data with Streptomyces sp. FR-008 reflect the level of candicidin.
To examine the temporal expression of the regulatory genes during growth, total RNA was prepared using samples collected at different time points as shown in Fig. 1A, and real-time PCR was performed. To facilitate the analysis, the quantitative PCR (qPCR) value of each gene was first normalized by comparison with hrdB transcription, and then the expression level of each gene at 24 h was arbitrarily set to 1 (Fig. 1C). Similar temporal expression patterns were observed for these four regulatory genes, with maximum expression detected at 36 h and decreasing expression afterwards (Fig. 1C); this pattern was consistent with the detectable production of candicidin at this time point (Fig. 1B), strongly suggesting a role for these regulatory factors in the biosynthesis of candicidin.
The four regulatory genes fscR1 to fscR4 are differentially expressed.
Our above data indicated similar temporal expression patterns for the four regulatory genes, with each exhibiting induction of greater than 100-fold by 36 h. However, as the fold changes shown in Fig. 1C were determined independently for each gene, i.e., relative to the expression level of that gene at 24 h, these results did not allow for direct comparison of the relative expression levels between genes. To compare their relative levels, the expression level of fscR1 at each time point was arbitrarily set to 100 and was used as the control to evaluate the expression of the other three regulatory genes, fscR2 to fscR4, at the same time point (Fig. 1D; see Fig. S7 in the supplemental material). Collectively, our analysis indicated that the four regulatory genes of FR-008 pathway are differentially expressed, with fscR1 expressed at the highest level, followed in order by fscR3, fscR2, and fscR4, suggesting potentially different roles for these regulatory genes in controlling candicidin biosynthesis.
Temporal expression patterns of structural genes in the FR-008 pathway.
In addition to the four regulatory genes fscR1 to fscR4, the FR-008 BGC encompasses another 17 genes involved in the construction, modification, and efflux of the chemical compounds: fscA to fscF (46), encoding the type I PKS subunits; fscMI, fscMII, and fscMIII for biosynthesis and attachment of mycosamine (47); fscP for formation of a carboxyl group (48); fscFE for electron transfer; fscTE for removal of aberrant intermediates (49); pabAB (47), encoding an 4-amino-4-deoxychorismate (ADC) synthetase; fscTI and fscTII, encoding a putative ABC transporter (50); and fscO and pabC, encoding a flavin adenine dinucleotide (FAD)-dependent monooxygenase and an ADC lyase, respectively (26, 28, 51). To investigate the temporal expression pattern of these structural genes, real-time PCR was performed using the same sets of RNAs used for analyzing the regulatory genes, with the level of each gene at 24 h arbitrarily set to one. Our data indicated that all structural genes were obviously induced at 36 h, in a temporal expression pattern similar to that of the regulatory genes and also consistent with the detectable level of candicidin at 36 h. However, based on differences in their induction levels, the structural genes were categorized into three groups. Genes of the first group were only slightly induced at 36 h, as represented by fscO (Fig. 2), the expression of which increased to 5.7 ± 0.4 at 36 h and was then reduced to near-control levels at 42 h and time points thereafter. In the second group, composed of pabC and pabAB (Fig. 2), expression increased to a level between 10- and 20-fold over that of the control by 36 h. The rest of the structural genes fell into the third group (Fig. 2; see Fig. S8 in the supplemental material), which demonstrated a marked level of induction by 36 h, ranging between 60-fold and 90-fold relative to the control sample. In conclusion, the expression of structural genes of the FR-008 pathway was activated by 36 h, although the degree of induction varied dramatically, suggesting that the products of these genes are required at different levels during the biosynthesis of candicidin.
FIG 2.
Temporal expression pattern of structural genes of the FR-008 pathway in the wild-type Streptomyces sp. strain FR-008 cultured in YEME liquid medium. RNA samples were isolated at the indicated times. Expression of hrdB, which encodes the major sigma factor, was used as an internal control. The expression level of each gene at 24 h was arbitrarily set to one. The y axis shows the fold change in expression level at the indicated times over the level at 24 h. Results are the means (±SD) from triplicate biological experiments.
Transcriptional analysis of fscR2 and fscR3.
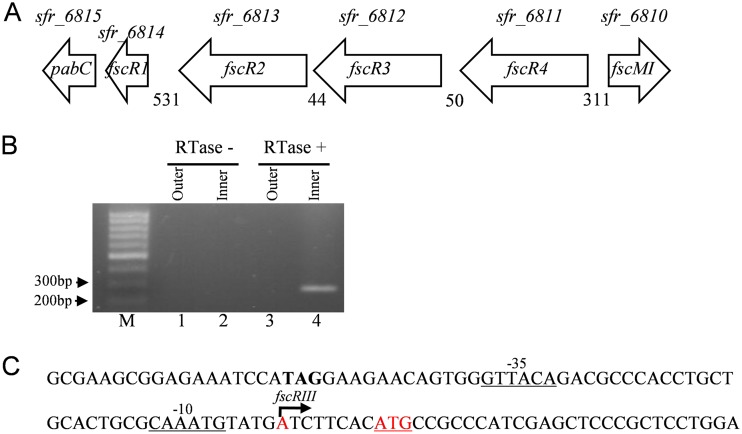
Of the four regulatory genes, the transcriptional start points (TSPs) for fscR1 and fscR4 have been previously mapped (34). To locate the TSPs for fscR2 and fscR3, we performed 5′ rapid amplification of cDNA ends (RACE) analysis. A single TSP was mapped to an adenine at position −8 relative to the fscR3 start codon (Fig. 3). We did not detect a separate TSP for fscR2, possibly due to its lower transcript level, compared to that of fscR3, under the conditions tested or potentially from technical difficulties arising from the specific RNA architecture associated with this region. However, subsequent complementation studies with fscR2 and its upstream intergenic region suggested that fscR2 may have its own promoter in this region.
FIG 3.
Location of the transcriptional start point (TSP) of fscR3. (A) Genetic organization of the four regulatory genes, fscR1 to fscR4, and their flanking genes. Numbers indicate the lengths of the putative intergenic sequences. Gene designations shown above the arrows were obtained from the Streptomyces sp. FR-008 genome sequencing project (28). (B and C) Identification of the TSP of fscR3. (B) TSP localization by 5′-RACE. RNA was incubated without (lanes 1 and 2) or with (lanes 3 and 4) reverse transcriptase (RTase), and the cDNA was ligated to an adaptor. The cDNA ends were amplified using an outer set and then an inner set of primers, and the amplified product was sequenced to identify the TSP. M, 100-bp ladder. (C) Location of the TSP in the fscR3 promoter. The TSP is shown in red type marked by a bent arrow. The predicted fscR3 start codon is shown in underlined red type. The putative −10 and −35 sequences are underlined.
fscR2 and fscR3 are essential for candicidin production.
Of the four regulatory genes, fscR1 and fscR4 have been determined to be essential for normal levels of candicidin biosynthesis (34). To investigate the roles of fscR2 and fscR3 in candicidin biosynthesis, gene replacement mutants were generated. The fscR2 mutant strain (ΔfscR2) was obtained by deleting 2,643 bp of the 2,829-bp coding sequence, corresponding to aa 30 to 910, which contains most of the AAA-ATPase domain and HTH domain. The replacement of this region by an apramycin cassette was verified by PCR analysis (see Fig. S9 in the supplemental material).
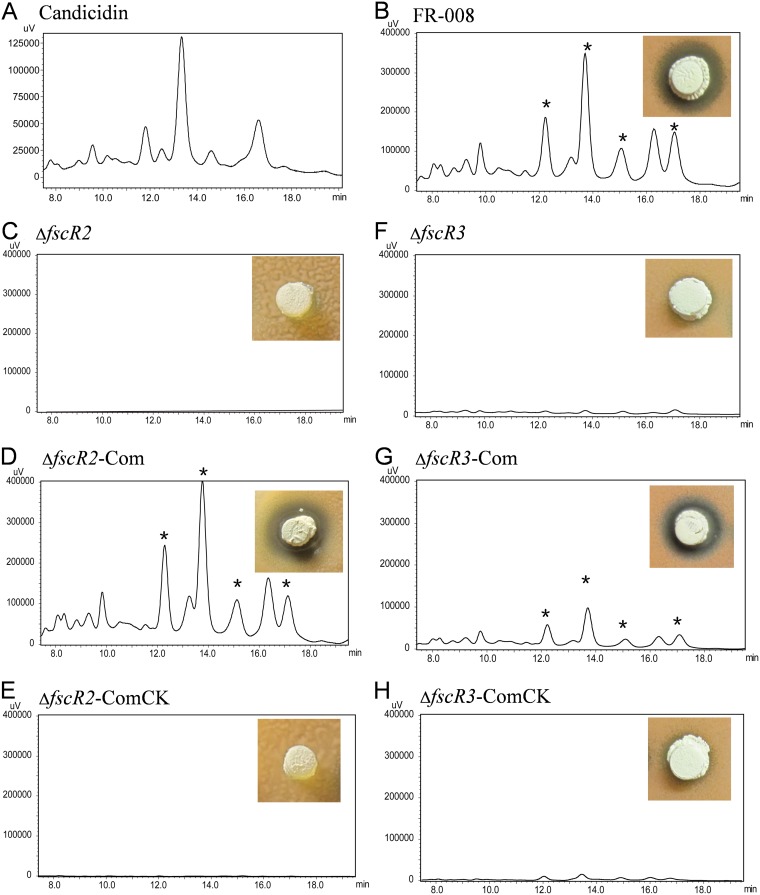
A bioassay was performed to test production of candicidin by the ΔfscR2 strain using Rhodotorula as the indicator strain. While a large clear zone of inhibition was observed for the wild-type FR-008 strain (Fig. 4B), no growth inhibition of the indicator yeast was detected with the ΔfscR2 strain (Fig. 4C), indicating that deletion of fscR2 abrogated candicidin production. We further analyzed the level of candicidin by high-performance liquid chromatography (HPLC) analysis. Five major peaks were detected in the stock solution of candicidin (Fig. 4A), and the same peaks corresponding to the candicidin complex were detected in the extracts of the wild-type strain (Fig. 4B). However, no such peaks were detectable in the ΔfscR2 extracts (Fig. 4C), consistent with the bioassay analysis. These data indicated that fscR2 is critical for candicidin biosynthesis.
FIG 4.
Production of candicidin by mutant and control strains. (A) Candicidin standard; (B) wild-type Streptomyces sp. strain FR-008; (C) ΔfscR2 strain; (D) ΔfscR2-Com (ΔfscR2 complemented with fscR2) strain; (E) ΔfscR2-ComCk (ΔfscR2 complemented with vector) strain; (F) ΔfscR3 strain; (G) ΔfscR3-Com (complemented with fscR3) strain; (H) ΔfscR3-ComCk (ΔfscR3 complemented with vector) strain. The candicidin complex was extracted from strains grown on GS agar medium for 3 days before HPLC analysis. The inset photographs show the growth inhibition of the indicator yeast Rhodotorula rubra by each Streptomyces strain grown on GS agar medium for 3 days. The clear blue zone indicates growth inhibition and thus production of the candicidin complex. *, peaks corresponding to the candicidin complex.
To confirm a direct relation between the deletion of fscR2 and the defect in candicidin production by the ΔfscR2 strain, a DNA segment with the coding sequence of fscR2 and its upstream and downstream intergenic regions was cloned into pMS82, an integrative and conjugative plasmid (52), so that expression of fscR2 would be driven by its native promoter, if present in the upstream region. The resulting plasmid, pCom-fscR2, was introduced into the ΔfscR2 strain to obtain the complemented ΔfscR2-Com strain. The ΔfscR2-ComCk strain, derived by transforming the ΔfscR2 strain with the pMS82 vector alone, was used as a control. The five peaks corresponding to the candicidin complex were detected, and a large clear zone of inhibition was restored in the ΔfscR2-Com strain (Fig. 4D) but not in the ΔfscR2-ComCk strain (Fig. 4E), confirming that FscR2 is directly involved in the regulation of candicidin biosynthesis.
Similarly, a 2,664-bp region of the 3,030-bp coding sequence of fscR3, corresponding to aa 99 to 986 and covering the AAA-ATPase domain, DNA polymerase III domain, and HTH domain, was replaced by an apramycin cassette to generate the ΔfscR3 strain (see Fig. S10 in the supplemental material). Our data from by both the bioassay and HPLC analysis showed that fscR3 is critically required for candicidin production (Fig. 4F). A direct link was established between production of candicidin and the presence of the fscR3 gene by complementation analysis (Fig. 4G and H). These data demonstrated that both FscR2 and FscR3 are essential for candicidin production, as was previously found for FscR1 (34).
We also generated the ΔfscR2 ΔfscR3 mutant strain by deletion of both fscR2 and fscR3 and the ΔfscR1 ΔfscR2 ΔfscR3 ΔfscR4 strain, which has all four regulatory genes deleted, with each strain confirmed by PCR analysis (see Fig. S11 and S12 in the supplemental material). As expected, these two multiple-mutation strains did not produce candicidin (see Fig. 8). We noticed that both the single- and the multiple-mutation strains displayed growth curves similar to that of the wild-type strain (Fig. 1A), and they also exhibited normal morphology (data not shown), suggesting that the four regulatory genes have no role in growth and differentiation.
FIG 8.
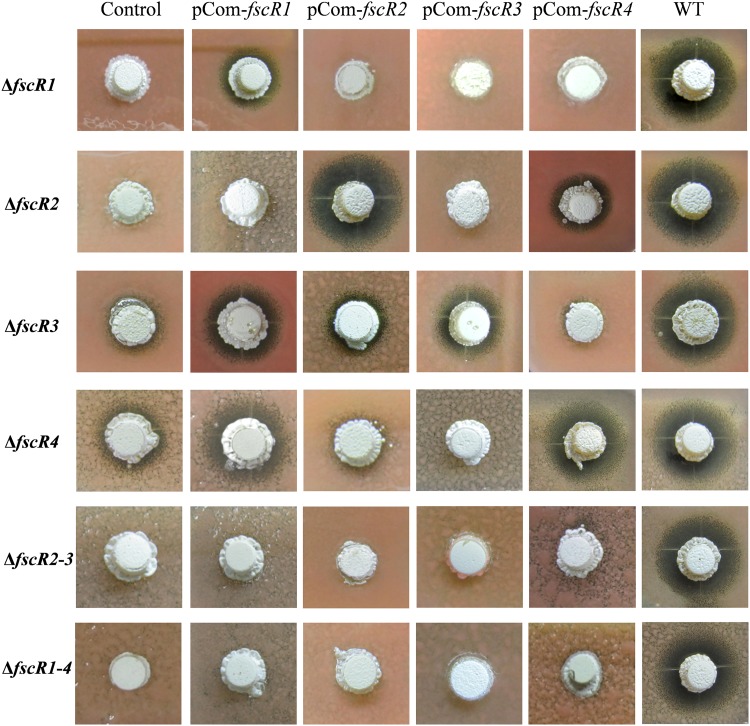
Cross-complementation between regulatory genes. Complementation plasmids (pCom-fscR1 through pCom-fscR4) for each of the four regulatory genes were introduced into each single- and multiple-mutation strain, and the resulting strains were tested for production of candicidin by bioassay after growth for 3 days on MS agar medium. Control column, mutant strains complemented with the vector only; WT, wild-type FR-008 strain.
Reduced expression of structural genes in the ΔfscR2 and ΔfscR3 strains.
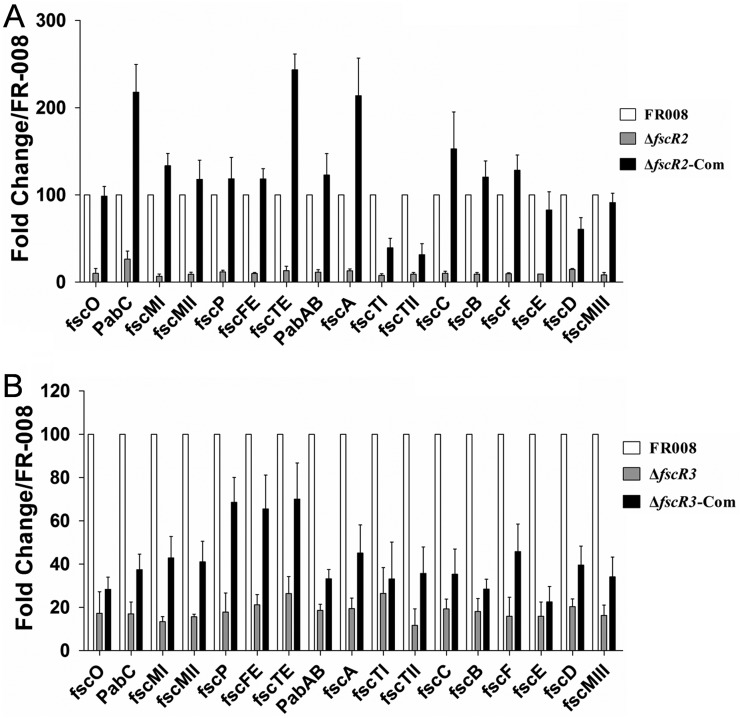
Our previous study indicated that inactivation of fscR1 or fscR4 leads to decreased transcription of structural genes of the FR-008 pathway. To investigate the changes in expression levels caused by deletion of fscR2, we performed real-time PCR analysis using RNA extracted at the 72-h time point from the ΔfscR2 strain, the wild-type strain FR-008, and the complemented ΔfscR2-Com strain, which produces candicidin. The level of expression for each gene in the wild-type strain was arbitrarily set to 100 (Fig. 5A). Eleven structural genes in the ΔfscR2 strain had expression levels of around 10 or less, i.e., less than or equal to 1/10 of control levels: fscO (9.9 ± 5.6), fscMI (6.6 ± 2.6), fscMII (8.9 ± 2.3), fscMIII (8.3 ± 2.6), fscFE (9.9 ± 1.1), fscTI (7.6 ± 1.8), fscTII (9.3 ± 1.8), fscC (10.0 ± 2.4), fscB (9.0 ± 1.9), fscF (9.5 ± 1.1), and fscE (9.2 ± 0.1). Five genes had expression levels of slightly above 10, i.e., over 1/10 of control levels: fscP (11.6 ± 1.9), fscTE (13.1 ± 5.1), pabAB (11.3 ± 2.9), fscA (12.9 ± 2.2), and fscD (14.4 ± 1.2). pabC had the highest expression at 26.4 ± 9.2, about one-fifth of the level in the wild-type strain. These data indicated that transcription of structural genes of the FR-008 pathway is markedly decreased in the ΔfscR2 strain, to about 10 times lower for most of the genes. These low expression levels are consistent with the absence of detectable synthesis of candicidin under the conditions tested.
FIG 5.
Transcriptional analyses of structural genes in the FR-008 pathway. (A) Expression in strain FR-008, the ΔfscR2 strain, and the ΔfscR2 strain complemented with fscR2. (B) Expression in strain FR-008, the ΔfscR3 strain, and the ΔfscR3 strain complemented with fscR3. RNA samples were isolated from strains grown for 3 days in YEME liquid medium. Expression of hrdB, which encodes the major sigma factor, was used as an internal control. The expression level of each gene in the wild-type strain was arbitrarily set to 100. The y axis shows the fold change in expression level relative to the control. Results are the means (±SD) from six biological experiments.
We also determined the expression levels of structural genes in the ΔfscR3 strain, with the expression of each gene in the wild-type strain set to 100. In contrast to the case for the ΔfscR2 strain, levels lower than 10, relative to the control, were not detected for any structural gene in the ΔfscR3 strain. The expression levels of 13 genes were between 10 and 20: fscO (17.2 ± 9.9), pabC (16.9 ± 5.4), fscMI (13.4 ± 2.3), fscMII (15.6 ± 1.2), fscMIII (16.2 ± 4.7), fscP (17.8 ± 8.8), pabAB (18.6 ± 2.8), fscA (19.4 ± 4.8), fscTII (11.6 ± 7.6), fscC (19.3 ± 4.4), fscB (18.1 ± 5.9), fscF (15.9 ± 8.7), and fscE (15.9 ± 6.5). Four genes had expression levels above 20: fscFE (21.1 ± 4.8), fscTE (26.4 ± 7.7), fscTI (26.5 ± 11.9), and fscD (20.4 ± 3.5). The relatively high expression levels of these structural genes, compared to the levels in the ΔfscR2 strain, explain the slightly detectable candicidin activity of the ΔfscR3 strain (Fig. 4F).
We also examined the expression levels of structural genes in the two complemented ΔfscR2-Com and ΔfscR3-Com strains (Fig. 5). In the ΔfscR2-Com strain, expression of three structural genes (fscO, fscE, and fscMIII) was restored to levels comparable to those in the wild-type strain, 11 genes (pabC, fscMI, fscMII, fscP, fscFE, fscTE, pabAB, fscA, fscC, fscB, and fscF) were expressed at levels higher than those in the wild-type control, and only three genes (fscTI, fscTII, and fscD) had expression lower than that in the control, consistent with the near-normal levels of bioactivity for the ΔfscR2-Com strain (Fig. 4C; see Fig. 8). In contrast, in the ΔfscR3-Com strain, only three genes had expression levels restored to between 60 and 70, with all the rest having levels below 50 (Fig. 5B), relative to controls, consistent with the moderately restored bioactivity for this strain (Fig. 4G; see Fig. 8) and further supporting a direct correlation between the relative bioactivity levels among the strains and the comparative expression levels of their structural genes. Altogether, our data indicated that the expression of structural genes in the FR-008 pathway was decreased in both the ΔfscR2 and ΔfscR3 strains, but the levels of reduction varied.
The four regulatory genes exert different levels of control over structural genes.
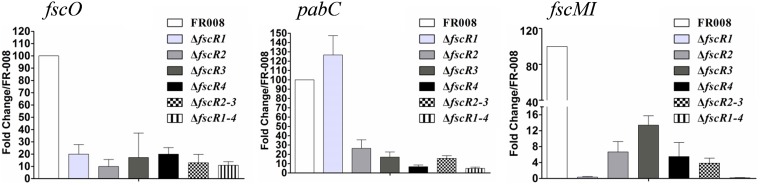
The different transcriptional levels of structural genes in the ΔfscR2 and ΔfscR3 strains reflect different degrees of control over these genes by the regulatory genes. To compare the relative levels of control of the structural genes by each regulatory gene, we performed real-time PCR analysis using our mutant strains for each of the four regulatory genes and arbitrarily set the expression level of each gene in the wild-type FR-008 strain to 100 (Fig. 6; see Fig. S13 in the supplemental material). Based on the data, structural genes could be divided into three groups. fscO is the only gene in group 1, with an expression level ranging between 10 and 20, relative to the control level of 100, in all four single mutant strains (ΔfscR1, ΔfscR2, ΔfscR3, and ΔfscR4), indicating similar levels of control of this gene by the four regulatory genes. Group 2 contains only pabC, whose level in the ΔfscR1 strain (126 ± 26) was even higher than that in the wild-type strain, while its level in the other three single mutants ranged from 6 to 26, indicating negative control of pabC by FscR1 in contrast to positive control by FscR2, FscR3, and FscR4. All the other structural genes are in group 3 (Fig. 6 and S13), in which the expression level of genes in the ΔfscR1 strain was below 1 and expression in the other single mutants varied, but with all levels much lower than in the wild-type strain. Additionally, for group 3, the expression levels in the ΔfscR3 strain were often higher than those in the ΔfscR2 and ΔfscR4 strains, and the expression in the ΔfscR2 strain was generally higher than in the ΔfscR4 strain, indicating overall that FscR1 has the tightest control over these structural genes, followed by FscR2, FscR4, and then FscR3.
FIG 6.
fscR1 to fscR4 exert different levels of control over structural genes in the FR-008 pathway. RNA samples were isolated from Streptomyces sp. FR-008, single-mutation strains, and multiple-mutation strains grown for 3 days in YEME liquid medium. Expression of hrdB, which encodes the major sigma factor, was used as an internal control. The expression level of each gene in the wild-type strain was arbitrarily set to 100. The y axis shows the fold change in expression level relative to the control. Results are the means (±SD) from six biological experiments.
We also evaluated the expression levels of genes in the ΔfscR2 ΔfscR3 and ΔfscR1 ΔfscR2 ΔfscR3 ΔfscR4 multiple-mutation strains. Expression in the ΔfscR2 ΔfscR3 double mutant strain was always lower than that in the ΔfscR2 and ΔfscR3 single mutant strains, and expression in the ΔfscR1 ΔfscR2 ΔfscR3 ΔfscR4 quadruple mutant strain was the lowest and was barely detectable for most genes (Fig. 6 and S13), suggesting a combined effect of the gene deletions.
Cross-regulation between the four regulatory genes.
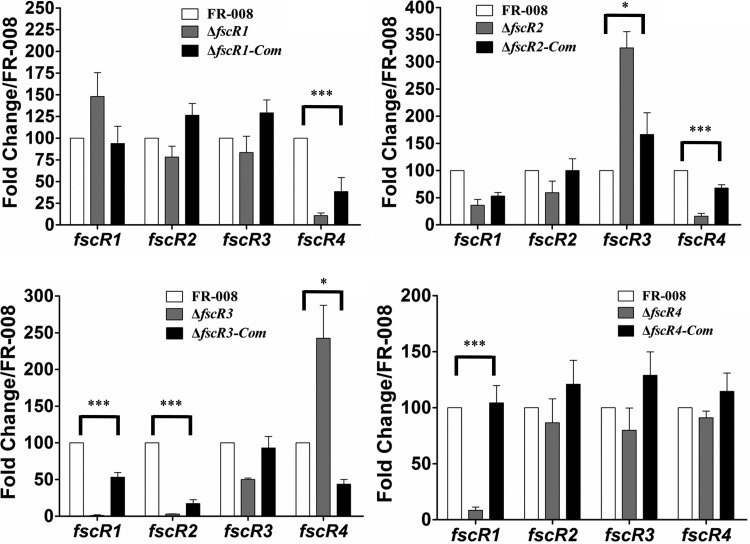
Using the four single mutant strains, we examined the effects of the regulatory genes on each other by real-time PCR analysis using primers specific to the undeleted portion of each regulatory gene (Fig. 7). hrdB was used as an internal control, and the expression level of each gene in the wild-type strain was arbitrarily set to 100 and was used as a control. Transcription of fscR1 was upregulated slightly in the ΔfscR1 strain, indicating a minor autorepressive effect; however, expression of fscR2 and fscR3 in the ΔfscR1 strain was comparable to levels in the wild-type strain, indicating that FscR1 does not impact transcription of fscR2 and fscR3. In contrast, expression of fscR4 was only 10 ± 2, about 1/10 of the wild-type level, indicating a significantly positive regulatory effect on fscR4 by FscR1, in agreement with our previous report (34). Our data showed that transcription of two regulatory genes was significantly affected by mutation of fscR2; expression of fscR3 (325 ± 29) was significantly enhanced, whereas fscR4 expression (15 ± 5) was significantly reduced in the ΔfscR2 strain relative to the control, indicating that FscR2 negatively regulates fscR3 expression but positively regulates fscR4 expression. Basal transcription levels of 1.1 ± 0.9 and 3.0 ± 0.4 were detected for fscR1 and fscR2, respectively, in the ΔfscR3 strain, indicating a profound positive regulatory effect of FscR3 on these two genes. In contrast, markedly enhanced transcription (242 ± 45) was detected for fscR4 in the ΔfscR3 strain (Fig. 7), indicating a significantly negative regulatory effect on this gene by FscR3. In the ΔfscR4 strain, only transcription of fscR1 was significantly impacted, with a decrease to 8.4 ± 2.9, indicating strong regulation of this gene by FscR4, in agreement with our previous report (34). In conclusion, each of the four regulatory factors demonstrated marked regulation over at least one regulatory gene, with both positive and negative effects observed, forming a complex regulatory network.
FIG 7.
Cross-regulation between regulatory genes. The same RNA samples were used as for Fig. 6. The expression level of each gene in the wild-type strain was arbitrarily set to 100. The y axis shows the fold change in expression level relative to the control. Primers for the four regulatory genes were designed to a portion of the coding sequence that was not deleted from the respective mutant. Results are the means (±SD) from six biological experiments. *, P < 0.05; ***, P < 0.01.
Cross-complementation between the four regulatory genes fscR1 to fscR4.
The cross-regulation between the four regulatory genes, fscR1 to fscR4, prompted us to investigate potential functional substitution between these genes. Therefore, we performed a comprehensive cross-complementation analysis by introducing complementation plasmids that contain one copy of each regulatory gene driven by its native promoter into each single mutant and tested candicidin production by each transformant by bioassay (Fig. 8) and by HPLC (see Fig. S14 to S19 in the supplemental material). Our data indicated that fscR1 partially restored candicidin production to the ΔfscR3 and ΔfscR4 strains, indicating functional substitution of FscR3 and FscR4 by FscR1. Similarly, fscR4 restored some candicidin production to the ΔfscR2 strain, indicating that FscR4 can at least partially functionally substitute for FscR2. Notably, the functional substitution (heterocomplementation) appears to be unidirectional. Whereas fscR1 was able to complement the ΔfscR3 and ΔfscR4 strains, neither fscR3 nor fscR4 could complement the ΔfscR1 strain (Fig. 8). Similarly, fscR4 was able to complement the fscR2 deletion, but not vice versa, suggesting a regulatory hierarchy for these genes.
The four complementation plasmids were also transformed into the ΔfscR2 ΔfscR3 double mutant strain and the ΔfscR1 ΔfscR2 ΔfscR3 ΔfscR4 quadruple mutant strain; however, none was able to restore candicidin production to these multiple-mutation strains (Fig. 8). Overall, the findings suggest that optimal production of candicidin requires the presence and coordinated action of all four regulatory genes.
Expression of fscR1 is restored in functionally heterocomplemented strains.
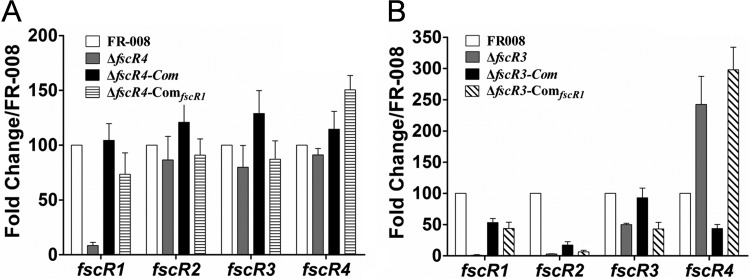
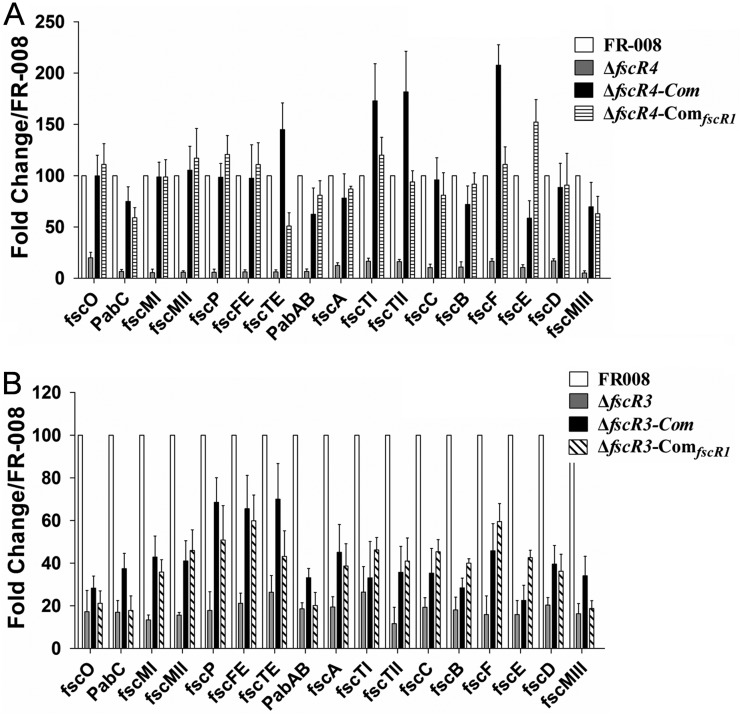
Functional heterocomplementation of the ΔfscR3 and ΔfscR4 strains was detected with fscR1, which restored candicidin production to these two mutant strains (Fig. 8 and S14 to S19). To investigate the mechanisms underlying this restoration in the ΔfscR4 strain, we compared the transcription of genes in the FR-008 pathway using the wild-type FR-008 and the ΔfscR4, ΔfscR4-Com (ΔfscR4 complemented by fscR4 itself), and ΔfscR4-ComfscR1 (ΔfscR4 complemented by fscR1) strains (Fig. 9A). With the expression of fscR1 in the wild-type strain set at 100, the expression of fscR1 in the ΔfscR4 strain was only 8.4 ± 2.9; however, it was restored to 74 ± 19 in the ΔfscR4-ComfscR1 strain, nearly three-quarters of the full expression level. We also noticed that the expression level of fscR4 in the ΔfscR4-ComfscR1 strain was even slightly higher than that in the ΔfscR4-Com strain, supporting a positive effect of FscR1 on fscR4. In contrast, complementation of ΔfscR4 with fscR1 did not have any obvious effects on the expression of fscR2 and fscR3 (Fig. 9A), consistent with our previous data indicating that FscR1 does not impact fscR2 and fscR3 expression (Fig. 7). We also examined the transcription of structural genes of the FR-008 pathway in the ΔfscR4-ComfscR1 strain (Fig. 10A). In general, the markedly lower expression of these genes in the ΔfscR4 strain was restored to levels comparable to those in the wild-type strain in the ΔfscR4-ComfscR1 strain, indicating that fscR1 is responsible for the basal transcription of the structural genes and that the defect in candicidin biosynthesis in the ΔfscR4 strain is largely due to reduced fscR1 transcription.
FIG 9.
Transcription of regulatory genes in heterocomplemented strains. RNA samples were isolated from Streptomyces sp. FR-008, the fscR3 and fscR4 mutants, and complemented strains grown for 3 days in YEME liquid medium. Expression of hrdB, which encodes the major sigma factor, was used as an internal control. The expression level of each gene in the wild-type strain was arbitrarily set to 100. The y axis shows the fold change in expression level relative to the control. Results are the means (±SD) from six biological experiments. Primers for these genes were designed to an undeleted portion of the coding sequence. ΔfscR3-ComfscR1 and ΔfscR4-ComfscR1, ΔfscR3 and ΔfscR4 complemented with fscR1.
FIG 10.
Transcription of structural genes in heterocomplemented strains. RNA samples were isolated from Streptomyces sp. FR-008, the fscR3 and fscR4 mutants, and complemented strains grown 3 days in YEME liquid medium. Expression of hrdB, which encodes the major sigma factor, was used as an internal control. The expression level of each gene in the wild-type strain was arbitrarily set to 100. The y axis shows the fold change in expression level relative to the control. Results are the means (±SD) from six biological experiments. ΔfscR3-ComfscR1 and ΔfscR4-ComfscR1, ΔfscR3 and ΔfscR4 complemented with fscR1.
Similarly, we performed transcription analysis in the ΔfscR3-ComfscR1 strain (ΔfscR3 complemented with fscR1) (Fig. 9B). Expression of fscR1 was barely detectable in the ΔfscR3 strain, but was restored to a relative level of 44 ± 9 in the ΔfscR3-ComfscR1 strain, comparable to the level (53 ± 6) in the ΔfscR3-Com strain (ΔfscR3 complemented with fscR3 itself), although both levels were lower than that in the control strain. The expression of the other three regulatory genes was not altered in the ΔfscR3-ComfscR1 strain, suggesting that FscR1 does not impact these genes under this condition. The expression of most structural genes of the FR-008 pathway in the ΔfscR3-ComfscR1 strain was similar to that in the ΔfscR3-Com strain (Fig. 10B), although both complemented strains had expression levels lower than those in the wild-type control, explaining the partial restoration of candicidin production. In conclusion, the reduced expression level of fscR1 appears to be the main factor in the defect in candicidin biosynthesis in the ΔfscR3 and ΔfscR4 strains.
Functional heterocomplementation was also detected between fscR4 and ΔfscR2 (Fig. 8 and S14). Our transcriptional analysis indicated that introduction of fscR4 into the ΔfscR2 strain restored expression of fscR4 itself and of fscR1, but not of that of fscR2 or fscR3, to levels comparable to those in the ΔfscR2-Com strain (see Fig. S20A in the supplemental material), supporting our results indicating that FscR4 activates fscR1 (Fig. 7). Consistent with the upregulation of fscR1, the expression of structural genes in the ΔfscR2-ComfscR4 strain was partially restored (Fig. S20B), resulting in detectable production of candicidin (Fig. 8 and S14 to S19).
We also investigated the mechanisms by which heterocomplementation failed, in one pairing, by examining the transcription of genes in the ΔfscR1 strain complemented with fscR4, which is downregulated in the ΔfscR1 strain (Fig. 7). Although the ΔfscR1 strain does not contain a functional fscR1 gene, primers were designed to detect transcription of the small remaining portion of the gene. Our transcriptional analysis indicated that introduction of an extra copy of fscR4 into the ΔfscR1 strain did not restore the expression of fscR1 or of fscR4 itself (see Fig. S21A in the supplemental material), indicating that expression of fscR4 needs a functional FscR1. Additionally, expression of the structural genes was not restored in the ΔfscR1-ComfscR4 strain (Fig. S21B), consistent with the absence of functional complementation by fscR4 and our evidence that FscR1 is the key regulatory factor involved in activation of these structural genes.
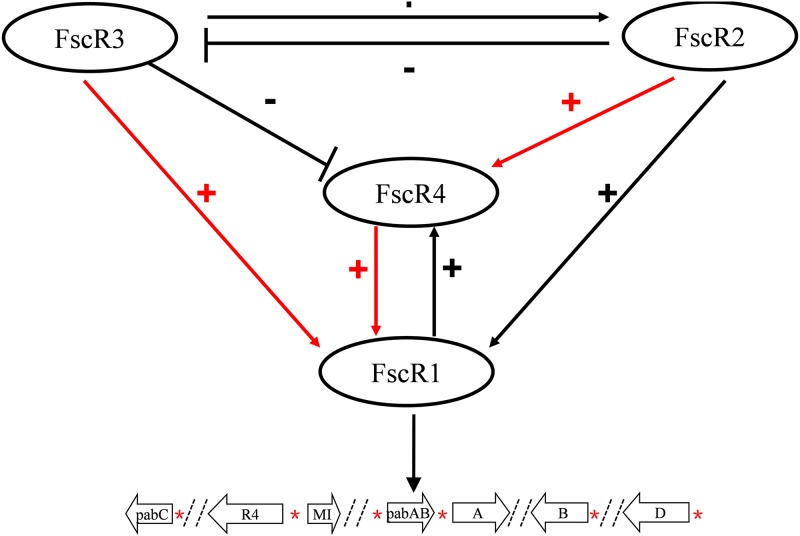
Proposed regulatory network of the four regulatory genes, fscR1 to fscR4, controlling production of the polyene antibiotic candicidin.
Based on our data, we propose a model of the regulatory network comprising fscR1 to fscR4 (Fig. 11). FscR2 and FscR3 are located at the top of the regulatory hierarchy, with FscR3 positively regulating fscR2 but FscR2 negatively regulating fscR3, forming a negative feedback loop. At the next level, FscR1 and FscR4 positively regulate each other, with fscR1 also regulated positively by both FscR2 and FscR3. fscR4 is regulated positively by FscR2 but negatively by FscR3. The functional substitution of fscR1 for fscR3 and fscR4, and of fscR4 for fscR2, is consistent with the model that fscR1 functions downstream of FscR3 and FscR4 and fscR4 functions downstream of FscR2. FscR1 functions at the lowest tier of the hierarchy, where it mediates the regulatory effect of the other three regulators by directly controlling expression of the structural genes for candicidin biosynthesis (34).
FIG 11.
Model of the regulatory network formed by the four regulatory proteins, FscR1 to FscR4, controlling production of the polyene antibiotic candicidin in Streptomyces sp. FR-008. FscR2 and FscR3 are located at the upper level of the network; FscR3 positively regulates fscR2, but FscR2 negatively regulates fscR3, forming a negative feedback loop. The two cross-regulating factors FscR1 and FscR4 are at the lower levels of the regulatory hierarchy, with fscR1 positively regulated by both FscR2 and FscR3 and fscR4 regulated positively by FscR2 but negatively by FscR3. FscR1 mediates the regulatory effect of the other three regulatory factors by directly regulating the structural genes. +, positive regulatory effect; −, negative regulatory effect. A red arrow indicates functional substitution of the upstream gene by the downstream gene, i.e., by the gene at the lower level of the hierarchy. *, locations of FscRI recognition sites in the promoters of the structural genes.
DISCUSSION
One unique feature of the candicidin pathway and other polyene antibiotic pathways is the regulatory subcluster containing four consecutive regulatory genes, which encode transcriptional factors belonging to two different families. The data from this study and a previous study by our group showed that each of these regulatory genes contributes to the optimal production of candicidin by Streptomyces sp. FR-008 (34). The key roles of these regulatory genes in the production of polyene antibiotics are supported by the requirement for their orthologues for nystatin biosynthesis in Streptomyces noursei ATCC 11455 (36).
PAS-LuxR regulators such as FscR1 have been implicated in multiple cellular processes, and even when they are located within a specific gene cluster, they may regulate other pathways (45, 53). FscR1 also regulates production of antimycin (45), although in this study, we focused on the relationship of FscR1 and the other three cluster-associated regulators with production of candicidin. Mutation of the four regulatory genes did not affect growth, morphology, or tested physiological activities other than production of candicidin. Interestingly, these genes displayed similar temporal expression patterns, with all of them activated at an early growth stage during which production of candicidin is initiated. The structural genes of the FR-008 pathway also demonstrated temporal expression patterns similar to those of the four regulatory genes, suggesting coordinated expression of these four regulatory genes in activation of the structural genes and therefore production of the antibiotic. However, surprisingly, the relative expression levels of these four regulatory genes varied to a large extent (Fig. 1D), from the most highly expressed gene, fscR1, to fscR4, which was expressed at a 100-fold-lower level, indicating differential expression for these four genes during growth and suggesting a dominant role for FscR1 in activation of the pathway.
In Streptomyces species, production of an antibiotic is usually coupled with cell differentiation on solid medium at later growth phases (41, 42). However, in our study, both regulatory genes and structural genes of the FR-008 pathway were activated by 36 h, and candicidin production was evident by this relatively early time. These results suggest that Streptomyces FR-008 may be more vulnerable to competition from fungal agents during the early stages of growth, and hence, candicidin is produced earlier than other antibiotics. Additionally, the fast growth of strain FR-008 may also contribute to the early production of antibiotic (28).
Each of the four regulatory genes is required for candicidin biosynthesis, and deletion or mutation of each of them led to reduced expression of the structural genes and, consequently, a defect in candicidin biosynthesis. However, the levels of reduced expression of the structural genes varied among the mutants. The expression of most structural genes decreased by more than 100 times in the ΔfscR1 mutant compared with the wild-type control, explaining the complete abrogation of candicidin production by the ΔfscR1 strain. Although the expression of most structural genes was higher in the ΔfscR2 strain than in the ΔfscR1 strain, the levels were still not high enough for the ΔfscR2 strain to produce enough candicidin compounds for detection under the conditions tested. The highest expression level of structural genes was observed in the ΔfscR3 strain, which demonstrated leaky activity by the bioassay and minor peaks in HPLC analysis, indicating that transcription of the structural genes was just sufficient in this mutant to enable detectable candicidin synthesis. We conclude that the level of candicidin production is closely associated with the expression level of structural genes in the FR-008 pathway.
Although the evolution of the regulatory subcluster with the four regulatory genes is not clear, based on the fact that deletion of each regulatory gene blocks or nearly blocks candicidin biosynthesis, our study indicates that a finely coordinated and simultaneous expression of these four regulatory genes is essential for maintaining a normal level of candicidin biosynthesis, suggesting that the clustered arrangement may be optimal for coordinated regulation.
We revealed that each regulatory gene impacts the expression of at least one other regulatory gene, thus forming a complex regulatory network. Therefore, it should be noted that any changes in expression in a particular mutant strain may result from the combined effect of the specific mutation as well as the altered expression of the regulatory gene(s) that the mutated factor regulates. For example, in the fscR3 mutant, the decrease in transcription of structural genes resulting from the loss of fscR3 and reduced expression of fscR1 and fscR2 may be modulated by the enhanced expression of fscR4 in this mutant (Fig. 7). In a previous study, we characterized the recognition sequence of FscR1 and revealed multiple targets of FscR1, including the first genes of several operons in the FR-008 pathway (34). However, the large size of FscR2, FscR3, and FscR4 made it difficult to express these proteins in Escherichia coli, and thus we did not determine whether the regulation of structural genes by these three regulatory proteins is direct or indirect. However, as fscR1 is located downstream of FscR2, FscR3, and FscR4 in the regulatory network and FscR1 was able to functionally complement fscR3 and fscR4, it is likely that fscR1 mediates the regulatory effect of the other three regulatory genes.
Overall, we obtained substantial insight into understanding the role of, and especially the regulatory network formed by, the four regulatory genes of the FR-008 pathway. We found that the four regulatory genes are differentially expressed throughout growth and that their control over structural genes varies to a large extent. We also revealed cross-regulation between the four regulatory genes, resulting in a complex regulatory network, and we found some functional heterocomplementation among these four regulatory factors, with genes at the lower level of the regulatory hierarchy restoring the loss of activity resulting from mutation of the genes at the upper level. The network formed by the four regulatory genes of the FR-008 pathway can serve as a model for exploring gene regulation in polyene and other complex antibiotic pathways.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The microbial strains and plasmids used in this study are listed in Table 1. The wild-type Streptomyces sp. FR-008 and its derivatives were grown on glycerol soya flour agar for spore preparation, antibiotic production, and conjugation transfer (43) and in YEME liquid medium (43) for growth curve preparation, bioassay, and RNA extraction. Escherichia coli strains were cultivated in Luria-Bertani (LB) liquid or solid agar medium. Antibiotics were added, if necessary, for selection of transformants from E. coli or Streptomyces.
TABLE 1.
Microbial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| Streptomyces | ||
| FR-008 | Wild type | 26 |
| ΔfscR1 strain | Deletion of internal sequence of fscR1, unmarked | 34 |
| ΔfscR2 strain | fscR2::aac(3)IV | This study |
| ΔfscR3 strain | fscR3::aac(3)IV | This study |
| ΔfscR4 strain | Deletion of internal sequence of fscR4, unmarked | 34 |
| ΔfscR2 ΔfscR3 strain | Deletion of sequence from fscR2 to fscR3, unmarked | This study |
| ΔfscR1 ΔfscR2 ΔfscR3 ΔfscR4 strain | Deletion of sequence from fscR1 to fscR4, unmarked | This study |
| ΔfscR1-ComfscR4 strain | ΔfscR1 strain complemented with pCom-fscR4 | This study |
| ΔfscR2-Com strain | ΔfscR2 strain complemented with pCom-fscR2 | This study |
| ΔfscR2-ComfscR4 strain | ΔfscR2 strain complemented with pCom-fscR4 | This study |
| ΔfscR3-Com strain | ΔfscR3 strain complemented with pCom-fscR3 | This study |
| ΔfscR3-ComfscR1 strain | ΔfscR3 strain complemented with pCom-fscR1 | This study |
| ΔfscR4-Com strain | ΔfscR4 strain complemented with pCom-fscR4 | This study |
| ΔfscR4-ComfscR1 strain | ΔfscR4 strain complemented with pCom-fscR1 | This study |
| E. coli | ||
| Novablue | General cloning strain | Novagen |
| ET12567(pUZ8002) | Strain used for conjugation between E. coli and Streptomyces | 43 |
| Rhodotorula rubra | Indicator for candicidin production | |
| Plasmids | ||
| pEASY-Blunt | General cloning vector | Invitrogen |
| pMS82 | Streptomyces integrative vector with hygromycin resistance | 52 |
| pCom-fscR1 | pMS82 with 531-bp upstream sequence, 696-bp coding sequence, and 200-bp downstream sequence of fscR1 | This study |
| pCom-fscR2 | pMS82 with 579-bp upstream sequence, 2,829-bp coding sequence, and 152-bp downstream sequence of fscR2 | This study |
| pCom-fscR3 | pMS82 with 319-bp upstream sequence, 3,030-bp coding sequence, and 95-bp downstream sequence of fscR3 | This study |
| pCom-fscR4 | pMS82 with 404-bp upstream sequence, 2,913-bp coding sequence, and 200-bp downstream sequence of fscR4 | This study |
Mutation of genes in Streptomyces sp. FR-008.
The coding sequence of fscR2 was replaced by an apramycin resistance cassette through homologous recombination using a strategy similar to that described previously (8, 54). Briefly, the DNA fragment serving as the left arm was amplified with RII-LF (with a HindIII site) and RII-LR (with a SpeI site). The amplified DNA fragment was purified and ligated with pEASY-Blunt to generate pRII-HL-Arm. The DNA fragment serving as the right arm was amplified with primers RII-RF (with a SpeI site) and RII-RR (with an XbaI site) and was cloned into pEASY-Blunt to generate pRII-HR-Arm. Primers Apr-Forward (with a SpeI site) and Apr-Reverse (with a SpeI site) were used to amplify a DNA fragment containing the apramycin cassette, using pIJ773 as the template, and the amplified cassette sequence was cloned into pMD-18T to generate pApr-M. pRII-HL-Arm was then digested by SpeI and XbaI (which cut in the vector sequence) and ligated with the right arm, which was released from pRII-HR-Arm by SpeI and HindIII digestion, to generate pRII-HL-R, in which the left and right arms were fused. pRII-HL-R was then cut by SpeI and ligated with the apramycin cassette, which had been removed from pApr-M by SpeI treatment, to generate pRII-L-A-R, thus connecting the left arm, the resistance cassette, and the right arm. The DNA segment with the two arms and apramycin cassette was released from pRII-L-A-R by HindIII and XbaI digestion, purified, and ligated with pJTU1278, pretreated with the same enzymes, to generate pMu-RII. The plasmid pMu-RII was transformed into E. coli ET12567(pUZ8002), and the resulting strain was used as the donor strain in conjugation with Streptomyces sp. FR-008. Exconjugants with resistance to apramycin were selected and verified by PCR using specific primers. One of these confirmed exconjugants was named the ΔfscR2 strain. The same strategy was adapted to obtain the ΔfscR3 strain. All primers used in this study are listed in Table 2.
TABLE 2.
Primers used in this study
| Category and name | Sequence (5′→3′)a |
|---|---|
| fscR2 mutation | |
| RII LF/LR | AAGCTTGGGCACACGGTGTACGGATCACTTG (HindIII)/ACTAGTAGCACCTGACCCACGCCTAC (SpeI) |
| RII RF/RR | ACTAGTTGAGGAGGACGAGGGAGG (SpeI)/TCTAGACCGAACGGCACGAACTGAC (XbaI) |
| Apra F/R | ACTAGTGAGCTCAGCCAATCGACTGG (SpeI)/ACTAGTACAGTGCCGTTGATCGTGCTATGA (SpeI) |
| fscR2 mutation analysis | |
| RII VerF/R | CGACATTCTGGGGGACGCCGGATGT/CAGGTCCTCCCTCGTCCTCCTCACC |
| fscR3 mutation | |
| RIII LF/LR | AAGCTTTGGTGAGGGTGGACTCGG (HindIII)/ACTAGTCGTCTACCGCAAACTCCG (SpeI) |
| RIII RF/RR | ACTAGTTCGTCGGTGAGGAAGAGGC (SpeI)/TCTAGAGGTCTTCGGGTCCTGCG (XbaI) |
| fscR3 mutation analysis | |
| RIII VerF/R | CGAAGGCGAAGTCCTGTTCC/GAACTCCACCTCACCAGCGTC |
| fscR1 to -4 mutation | |
| RI-RIV LF/LR | AAGCTTTGCTCCACCGCTTCCACCTT (HindIII)/ACTAGTGGGCGTCCTCAAGGTCGG (SpeI) |
| RI-RIV RF/RR | ACTAGTCCGACGCGAGGGAGAGCCCTGTG (SpeI)/TCTAGAGTCGAGCCACCTGGTGACCTCGTCG (XbaI) |
| fscR1 to -4 mutation analysis | |
| RI-RIV VerF/R | AAGCCCATGTAGACCACCGACGACT/GGCGGCGATGTTCCCCAGGTTCAGC |
| fscR2 and -3 mutation | |
| RII-RIII LF/LR | AAGCTTCGAGCAGCGTGGTGAAATG (HindIII)/CTCGAGAGCACCTGACCCACGCCTAC (XhoI) |
| RII-RIII RF/RR | CTCGAGCATGTGAAGATCATACATTTGC (XhoI)/TCTAGACACAGCGGTTCAGCTGGGCCTGACC (XbaI) |
| fscR2 and -3 mutation analysis | |
| RII-RIII VerF/R | GCGCTGGAGATCCTCGGCCAGGA/TGGTGACGAACGTTCATCCAGAT |
| fscR1 complementation | |
| RI-MS82-F/R | GAGAACCTAGGATCCAAGCTTTACGCGCCGAGGGAAACG/ACCATGCATAGATCTAAGCTTCGAGCAGCGTGGTGAAATGGCCG |
| fscR2 complementation | |
| RII-MS82-F/R | GAGAACCTAGGATCCAAGCTTCGCCCGGCTGGTCCACGAGGAAGT/ACCATGCATAGATCTAAGCTTGAACGTTCATCCAGATTTCCCGGC |
| fscR3 complementation | |
| RIII-MS82-F/R | GAGAACCTAGGATCCAAGCTTGTCACCGCCTCACCGCTGGACATGCT/ACCATGCATAGATCTAAGCTTCGGTGAGGAGGACGAGGGAGGAC |
| fscR4 complementation | |
| RIV-MS82-F/R | GAGAACCTAGGATCCAAGCTTCCGGGCAAGCTCCCCGGCCAGCA/ACCATGCATAGATCTAAGCTTGCAGCAGATCGTTCTGGCCGAGCCC |
| Rapid amplification of cDNA ends | |
| RIII 5-RACE | TCAGGGGGGTGCCGACGCTCCACT/ACGCCGTACCGCACCTCGTGTTC |
| RII 5-RACE Outer | GAGGATGAGCAGGCGGCT/AAGAGCGGGTTGCCGACGGA |
| RII 5-RACE Inner | GCGTCTCGAAGAGCTGCCGT/CCGAAGGCGAAGTCCTGTT |
| Real-time PCR analysis | |
| sigma RT-F/R | GGGCTACAAGTTCTCCACG/AGGTCCTGGAGCATCTGGC |
| fscA RT-F/R | TGACCGAGATGCTGGAGCG/GTGCGAGTGGAGGCAGATC |
| fscB RT-F/R | CCACCACGGCACCACCTACA/AGGGTGTCGGGCAGGATG |
| fscC RT-F/R | GAGCCCATCGCCGTCATCG/GAAGCCGCCCTCACGGACAT |
| fscD RT-F/R | GACACCGCCTGCTCCTCCT/ACGAAGGTGGTGGGGCTGGC |
| fscE RT-F/R | GGCACCAACGGCCAGGACTA/AGGAGCAGGCGGTGTCGA |
| fscF RT-F/R | ATGCCCGATGACAAGAAGC/TCCAGGAGCCGCCAGTA |
| fscMI RT-F/R | GATGTCGGTGCGGCAGAAG/CGATGCCCAGTTCCTTGCGT |
| fscMII RT-F/R | GACATCGCCTGCTTCTCG/CTTGTGCAGGAAGCTGTGG |
| fscMIII RT-F/R | GCGGTGGAAAGAGGAAAGATG/TGAGGTCGCCGTCCACGAAG |
| pabC RT-F/R | CATTTCACCACGCTGCTCG/AGACGGTGACACGGACGG |
| fscP RT-F/R | GAGCACGACCGTTTCCGC/CGAAGTCGTTCACCAGGTCC |
| fscO RT-F/R | CCCTTCCGGGCCTACAA/TGCTCCACCGCTTCCAC |
| fscFE RT-F/R | TGCGTAGGAGCGGGCCAGTG/CTCCTGGAGGACGACCGA |
| fscTE RT-F/R | GCCAGAACCATCGAAGGGAC/TGGTAGAAGCTGGCGGAGCC |
| pabAB RT-F/R | ACCCGAGGTCATCCGCAACG/GCGCACAGGCCGAAGTCG |
| fscTI RT-F/R | GCGTCAGCAAGGCATACGGC/GTGGTGAGGATGTCGACCAG |
| fscTII RT-F/R | TGGTCATCCTCTTCGTGCTG/GATGCCGTTCTTCTGGTCC |
| fscRI RT-F/R | TGCCTATGTCGCCTGCCTCG/GGGTGGACGAGGTCGCAGAA |
| fscRII RT-F/R | ATCGCATGTTGCTGGAGCGC/GGTGAGGAGGACGAGGGA |
| fscRIII RT-F/R | GAACACGAGGTGCGGTACG/TCGTCGGTGAGGAAGAGGC |
| fscRIV RT-F/R | GCCTGGTGCTCCTGGACGGT/CCACCCTGTACGGCTCCTG |
Underlining indicates restriction sites.
For generation of strains with unmarked mutations, such as the ΔfscR2 ΔfscR3 strain, from which fscR2 and fscR3 were deleted, and the ΔfscR1 ΔfscR2 ΔfscR3 ΔfscR4 strain, from which all four regulatory genes were deleted, the two homologous arms were ligated directly without insertion of resistance genes and cloned into pJTU1278. Selection of exconjugants was performed as described previously (34).
Construction of complemented strains.
To complement the fscR2 mutation, the chromosomal region containing the 579-bp upstream and 152-bp downstream intergenic region and coding sequence of fscR2 was amplified and cloned into the integrating plasmid pMS82 (52) to generate pCom-fscR2 using a homologous recombination strategy; the construct was verified by sequencing and then introduced into the ΔfscR2 strain to generate the complemented ΔfscR2-Com strain. Similarly, plasmid pCom-fscR3, containing the 319-bp upstream and 95-bp downstream intergenic regions and the coding sequence of fscR3, was used to generate the ΔfscR3-Com strain. Other complemented strains were obtained in a similar manner.
RNA isolation, RT, and real-time PCR.
Streptomyces sp. FR-008 and its derivative strains were cultured in liquid YEME medium, and mycelia were collected at the time points indicated. Crude RNA samples were treated with DNase to remove chromosomal DNA. Reverse-transcription (RT) and real-time PCR assays were performed as described previously (8, 11, 55). Gene hrdB, encoding the major sigma factor, was used as an internal control to quantify the relative levels of cDNA.
RNA ligase-mediated RACE.
Streptomyces sp. FR-008 total RNA was extracted and processed using the FirstChoice RLM-RACE kit (Ambion), as described previously (8). The 5´RACE outer primer (Ambion) and FscR3 (FscR2) outer primer were used for outer PCR. For inner PCR, the 5´RACE inner primer (Ambion) was paired with the FscR3 (FscR2) inner primer. PCR products were analyzed on an agarose gel, and specific bands were excised, purified, and cloned into pGEM-T-Easy (Promega). Multiple clones were sequenced to determine the transcriptional start point (TSP).
Bioassay and HPLC.
The bioassay was performed essentially as described previously (34). Briefly, aliquots of supernatant collected during different growth phases in liquid YEME culture or patches of the agar medium were transferred to a thin layer of agar seeded with the indicator yeast strain Rhodotorula rubra, followed by incubation at 30°C for 48 h to test the growth inhibition of the indicator. Extraction and detection of the candicidin antibiotic complex from the wild-type Streptomyces sp. FR-008 were performed as described previously (26). The candicidin standard was purchased from Sigma, and the antimycin standard was purchased from MK Ltd.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Key Research and Development Program of China (2018YFC0310600 to X.P.) and the State Key Laboratory of Microbial Metabolism (MMLKF15-01 to X.P.).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hopwood DA. 2007. Streptomyces in nature and medicine. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 2.Zotchev SB. 2003. Polyene macrolide antibiotics and their applications in human therapy. Curr Med Chem 10:211–223. doi: 10.2174/0929867033368448. [DOI] [PubMed] [Google Scholar]
- 3.Bolard J. 1986. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim Biophys Acta 864:257–304. doi: 10.1016/0304-4157(86)90002-x. [DOI] [PubMed] [Google Scholar]
- 4.Welscher YMT, Ten Napel HH, Balague MM, Souza CM, Riezman H, De Kruijff B, Breukink OE. 2008. Natamycin blocks fungal growth by binding specifically to ergosterol without permeabilizing the membrane. J Biol Chem 283:6393–6401. doi: 10.1074/jbc.M707821200. [DOI] [PubMed] [Google Scholar]
- 5.Aparicio JF, Barreales EG, Payero T, Vicente CM, de Pedro A, Santos-Aberturas J. 2016. Biotechnological production and application of the antibiotic pimaricin: biosynthesis and its regulation. Appl Microbiol Biotechnol 100:61–78. doi: 10.1007/s00253-015-7077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Leeuwen MR, Golovina EA, Dijksterhuis J. 2009. The polyene antimycotics nystatin and filipin disrupt the plasma membrane, whereas natamycin inhibits endocytosis in germinating conidia of Penicillium discolor. J Appl Microbiol 106:1908–1918. doi: 10.1111/j.1365-2672.2009.04165.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P, Wu H, Chen XL, Deng Z, Bai L, Pang X. 2014. Regulation of the biosynthesis of thiopeptide antibiotic cyclothiazomycin by the transcriptional regulator SHJG8833 in Streptomyces hygroscopicus 5008. Microbiology 160:1379–1392. doi: 10.1099/mic.0.076901-0. [DOI] [PubMed] [Google Scholar]
- 9.Arias P, Fernandez-Moreno MA, Malpartida F. 1999. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol 181:6958–6968. doi: 10.1128/JB.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White J, Bibb M. 1997. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J Bacteriol 179:627–633. doi: 10.1128/jb.179.3.627-633.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, Zhang P, Zhu Y, Lu T, Wang Y, Cao G, Shi M, Chen XL, Tao M, Pang X. 2019. Novel two-component system MacRS is a pleiotropic regulator that controls multiple morphogenic membrane protein genes in Streptomyces coelicolor. Appl Environ Microbiol 85. doi: 10.1128/AEM.02178-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He JM, Zhu H, Zheng GS, Liu PP, Wang J, Zhao GP, Zhu GQ, Jiang WH, Lu YH. 2016. Direct involvement of the master nitrogen metabolism regulator GlnR in antibiotic biosynthesis in Streptomyces. J Biol Chem 291:26443–26454. doi: 10.1074/jbc.M116.762476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan H, Lu X, Sun D, Zhuang S, Chen Q, Chen Z, Li J, Wen Y. 2019. BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species. Mol Microbiol doi: 10.1111/mmi.14405. [DOI] [PubMed] [Google Scholar]
- 14.Yu Z, Zhu H, Dang F, Zhang W, Qin Z, Yang S, Tan H, Lu Y, Jiang W. 2012. Differential regulation of antibiotic biosynthesis by DraR-K, a novel two-component system in Streptomyces coelicolor. Mol Microbiol 85:535–556. doi: 10.1111/j.1365-2958.2012.08126.x. [DOI] [PubMed] [Google Scholar]
- 15.Gao C, Hindra, Mulder D, Yin C, Elliot MA. 2012. Crp is a global regulator of antibiotic production in Streptomyces. mBio 3:e00407-12. doi: 10.1128/mBio.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higo A, Horinouchi S, Ohnishi Y. 2011. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol Microbiol 81:1607–1622. doi: 10.1111/j.1365-2958.2011.07795.x. [DOI] [PubMed] [Google Scholar]
- 17.Santos-Beneit F, Rodríguez-García A, Sola-Landa A, Martín JF. 2009. Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol Microbiol 72:53–68. doi: 10.1111/j.1365-2958.2009.06624.x. [DOI] [PubMed] [Google Scholar]
- 18.Martin JF, Aparicio JF. 2009. Enzymology of the polyenes pimaricin and candicidin biosynthesis. Methods Enzymol 459:215–242. doi: 10.1016/S0076-6879(09)04610-2. [DOI] [PubMed] [Google Scholar]
- 19.Kligman AM, Lewis FS. 1953. In vitro and in vivo activity of candicidin on pathogenic fungi. Proc Soc Exp Biol Med 82:399–404. doi: 10.3181/00379727-82-20128. [DOI] [PubMed] [Google Scholar]
- 20.Lechevalier H. 1953. Fungicidal antibiotics, produced by Actinomycetes, candicidin. Presse Med 61:1327–1328. [PubMed] [Google Scholar]
- 21.Aparicio JF, Fouces R, Mendes MV, Olivera N, Martin JF. 2000. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem Biol 7:895–905. doi: 10.1016/S1074-5521(00)00038-7. [DOI] [PubMed] [Google Scholar]
- 22.Aparicio JF, Caffrey P, Gil JA, Zotchev SB. 2003. Polyene antibiotic biosynthesis gene clusters. Appl Microbiol Biotechnol 61:179–188. doi: 10.1007/s00253-002-1183-5. [DOI] [PubMed] [Google Scholar]
- 23.Brautaset T, Sekurova ON, Sletta H, Ellingsen TE, StrŁm AR, Valla S, Zotchev SB. 2000. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem Biol 7:395–403. doi: 10.1016/s1074-5521(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 24.Caffrey P, Lynch S, Flood E, Finnan S, Oliynyk M. 2001. Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem Biol 8:713–723. doi: 10.1016/s1074-5521(01)00046-1. [DOI] [PubMed] [Google Scholar]
- 25.Campelo AB, Gil JA. 2002. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology 148:51–59. doi: 10.1099/00221287-148-1-51. [DOI] [PubMed] [Google Scholar]
- 26.Chen S, Huang X, Zhou X, Bai L, He J, Jeong KJ, Lee SY, Deng Z. 2003. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem Biol 10:1065–1076. doi: 10.1016/j.chembiol.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Gil JA, Campelo-Diez AB. 2003. Candicidin biosynthesis in Streptomyces griseus. Appl Microbiol Biotechnol 60:633–642. doi: 10.1007/s00253-002-1163-9. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Xiao L, Zhou Y, Deng K, Tan G, Han Y, Liu X, Deng Z, Liu T. 2016. Development of Streptomyces sp. FR-008 as an emerging chassis. Synth Syst Biotechnol 1:207–214. doi: 10.1016/j.synbio.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaburannyi N, Rabyk M, Ostash B, Fedorenko V, Luzhetskyy A. 2014. Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genomics 15:97. doi: 10.1186/1471-2164-15-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joynt R, Seipke RF. 2018. A phylogenetic and evolutionary analysis of antimycin biosynthesis. Microbiology 164:28–39. doi: 10.1099/mic.0.000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. doi: 10.1128/MMBR.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antón N, Santos-Aberturas J, Mendes MV, Guerra SM, Martín JF, Aparicio JF. 2007. PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology 153:3174–3183. doi: 10.1099/mic.0.2007/009126-0. [DOI] [PubMed] [Google Scholar]
- 33.Richet E, Raibaud O. 1989. Malt, the regulatory protein of the Escherichia coli maltose system, is an Atp-dependent transcriptional activator. EMBO J 8:981–987. doi: 10.1002/j.1460-2075.1989.tb03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P, Zhao Z, Li H, Chen XL, Deng Z, Bai L, Pang X. 2015. Production of the antibiotic FR-008/candicidin in Streptomyces sp. FR-008 is co-regulated by two regulators, FscRI and FscRIV, from different transcription factor families. Microbiology 161:539–552. doi: 10.1099/mic.0.000033. [DOI] [PubMed] [Google Scholar]
- 35.Carmody M, Byrne B, Murphy B, Breen C, Lynch S, Flood E, Finnan S, Caffrey P. 2004. Analysis and manipulation of amphotericin biosynthetic genes by means of modified phage KC515 transduction techniques. Gene 343:107–115. doi: 10.1016/j.gene.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Sekurova ON, Brautaset T, Sletta H, Borgos SE, Jakobsen MO, Ellingsen TE, Strom AR, Valla S, Zotchev SB. 2004. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J Bacteriol 186:1345–1354. doi: 10.1128/jb.186.5.1345-1354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knirschova R, Novakova R, Feckova L, Timko J, Turna J, Bistakova J, Kormanec J. 2007. Multiple regulatory genes in the salinomycin biosynthetic gene cluster of Streptomyces albus CCM 4719. Folia Microbiol (Praha) 52:359–365. doi: 10.1007/bf02932090. [DOI] [PubMed] [Google Scholar]
- 38.Jiang C, Wang H, Kang Q, Liu J, Bai L. 2012. Cloning and characterization of the polyether salinomycin biosynthesis gene cluster of Streptomyces albus XM211. Appl Environ Microbiol 78:994–1003. doi: 10.1128/AEM.06701-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zotchev S, Haugan K, Sekurova O, Sletta H, Ellingsen TE, Valla S. 2000. Identification of a gene cluster for antibacterial polyketide-derived antibiotic biosynthesis in the nystatin producer Streptomyces noursei ATCC 11455. Microbiology 146:611–619. doi: 10.1099/00221287-146-3-611. [DOI] [PubMed] [Google Scholar]
- 40.Nedal A, Sletta H, Brautaset T, Borgos SE, Sekurova ON, Ellingsen TE, Zotchev SB. 2007. Analysis of the mycosamine biosynthesis and attachment genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455. Appl Environ Microbiol 73:7400–7407. doi: 10.1128/AEM.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliot MA, Buttner MJ, Nodwell JR. 2008. Multicellular development in Streptomyces, p 419–438. In Whitworth DE. (ed), Myxobacteria: multicellularity and differentiation. American Society for Microbiology, Washington, DC. [Google Scholar]
- 42.Chater K. 2011. Differentiation in Streptomyces: the properties and programming of diverse cell-types, p 43–86. In Dyson P. (ed), Streptomyces: molecular biology and biotechnology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 43.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics, 2nd ed John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 44.Pang X, Aigle B, Girardet JM, Mangenot S, Pernodet JL, Decaris B, Leblond P. 2004. Functional angucycline-like antibiotic gene cluster in the terminal inverted repeats of the Streptomyces ambofaciens linear chromosome. Antimicrob Agents Chemother 48:575–588. doi: 10.1128/aac.48.2.575-588.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLean TC, Hoskisson PA, Seipke RF. 2016. Coordinate regulation of antimycin and candicidin biosynthesis. mSphere 1:e00305-16. doi: 10.1128/mSphere.00305-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenke-Kodama H, Börner T, Dittmann E. 2006. Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis. PLoS Comput Biol 2:e132. doi: 10.1371/journal.pcbi.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lei X, Kong L, Zhang C, Liu Q, Yao F, Zhang W, Deng Z, You D. 2013. In vivo investigation of the substrate recognition capability and activity affecting amino acid residues of glycosyltransferase FscMI in the biosynthesis of candicidin. Mol Biosyst 9:422–430. doi: 10.1039/c2mb25464f. [DOI] [PubMed] [Google Scholar]
- 48.Mao X, Wang F, Zhang J, Chen S, Deng Z, Shen Y, Wei D. 2009. The pH shift and precursor feeding strategy in a low-toxicity FR-008/candicidin derivative CS103 fermentation bioprocess by a mutant of Streptomyces sp. FR-008. Appl Biochem Biotechnol 159:673–686. doi: 10.1007/s12010-008-8502-y. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Y, Meng Q, You D, Li J, Chen S, Ding D, Zhou X, Zhou H, Bai L, Deng Z. 2008. Selective removal of aberrant extender units by a type II thioesterase for efficient FR-008/candicidin biosynthesis in Streptomyces sp. strain FR-008. Appl Environ Microbiol 74:7235–7242. doi: 10.1128/AEM.01012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei X, Kong L, Zhang C, You D, Deng Z. 2012. Function of transporter genes fscTI and fscTII in the biosynthetic cluster of candicidin/FR-008. Wei Sheng Wu Xue Bao 52:1458–1466. [PubMed] [Google Scholar]
- 51.Zhou Y, Li J, Zhu J, Chen S, Bai L, Zhou X, Wu H, Deng Z. 2008. Incomplete beta-ketone processing as a mechanism for polyene structural variation in the FR-008/candicidin complex. Chem Biol 15:629–638. doi: 10.1016/j.chembiol.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 52.Gregory MA, Till R, Smith MC. 2003. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J Bacteriol 185:5320–5323. doi: 10.1128/jb.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez-Burgo Y, Santos-Aberturas J, Rodriguez-Garcia A, Barreales EG, Tormo JR, Truman AW, Reyes F, Aparicio JF, Liras P. 2019. Activation of secondary metabolite gene clusters in Streptomyces clavuligerus by the PimM regulator of Streptomyces natalensis. Front Microbiol 10:580. doi: 10.3389/fmicb.2019.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu T, Zhu Y, Zhang P, Sheng D, Cao G, Pang X. 2018. SCO5351 is a pleiotropic factor that impacts secondary metabolism and morphological development in Streptomyces coelicolor. FEMS Microbiol Lett 365:efny150. doi: 10.1093/femsle/fny150. [DOI] [PubMed] [Google Scholar]
- 55.Zhu Y, Zhang P, Zhang J, Xu W, Wang X, Wu L, Sheng D, Ma W, Cao G, Chen XL, Lu Y, Zhang YZ, Pang X. 2019. The developmental regulator MtrA binds GlnR boxes and represses nitrogen metabolism genes in Streptomyces coelicolor. Mol Microbiol 112:29–46. doi: 10.1111/mmi.14252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.