Agroecosystems represent an efficient model for studying fungal adaptation and evolution in anthropogenic environments. In this work, we studied what evolutionary forces shape populations of one of the most important fungal plant pathogens, B. cinerea, in small fruit agroecosystems of the Pacific Northwest. We hypothesized that host, geographic, and anthropogenic factors of agroecosystems structure B. cinerea populations. By combining neutral markers with markers that directly respond to human-induced selection pressures, we show that pathogen populations are highly localized and that selection pressure caused by fungicide use can have a greater effect on population structure than adaptation to host. Our results give a better understanding of population biology and evolution of this important plant pathogen in heterogeneous environments but also provide a practical framework for the development of efficient management strategies by limiting pathogen adaptation to fungicides and other human-induced selection pressures present in agroecosystems of the Pacific Northwest and elsewhere.
KEYWORDS: Botrytis cinerea, population structure, pathogen adaptation, fungal evolution, fungicide resistance, agroecosystems
ABSTRACT
Many fungal pathogens have short generation times, large population sizes, and mixed reproductive systems, providing high potential to adapt to heterogeneous environments of agroecosystems. Such adaptation complicates disease management and threatens food production. A better understanding of pathogen population biology in such environments is important to reveal key aspects of adaptive divergence processes to allow improved disease management. Here, we studied how evolutionary forces shape population structure of Botrytis cinerea, the causal agent of gray mold, in the Pacific Northwest agroecosystems. Populations of B. cinerea from adjacent fields of small fruit hosts were characterized by combining neutral markers (microsatellites) with markers that directly respond to human-induced selection pressures (fungicide resistance). Populations were diverse, without evidence for recombination and association of pathogen genotype with host. Populations were highly localized with limited migration even among adjacent fields within a farm. A fungicide resistance marker revealed strong selection on population structure due to fungicide use. We found no association of resistance allele with genetic background, suggesting de novo development of fungicide resistance and frequent extinction/recolonization events by different genotypes rather than the spread of resistance alleles among fields via migration of a dominant genotype. Overall our results showed that in agroecosystems, B. cinerea populations respond strongly to selection by fungicide use with greater effect on population structure compared to adaptation to host plant species. This knowledge will be used to improve disease management by developing strategies that limit pathogen local adaptation to fungicides and other human-induced selection pressures present in Pacific Northwest agroecosystems and elsewhere.
IMPORTANCE Agroecosystems represent an efficient model for studying fungal adaptation and evolution in anthropogenic environments. In this work, we studied what evolutionary forces shape populations of one of the most important fungal plant pathogens, B. cinerea, in small fruit agroecosystems of the Pacific Northwest. We hypothesized that host, geographic, and anthropogenic factors of agroecosystems structure B. cinerea populations. By combining neutral markers with markers that directly respond to human-induced selection pressures, we show that pathogen populations are highly localized and that selection pressure caused by fungicide use can have a greater effect on population structure than adaptation to host. Our results give a better understanding of population biology and evolution of this important plant pathogen in heterogeneous environments but also provide a practical framework for the development of efficient management strategies by limiting pathogen adaptation to fungicides and other human-induced selection pressures present in agroecosystems of the Pacific Northwest and elsewhere.
INTRODUCTION
Many plant pathogens have high evolutionary potential due to their large populations sizes, short generation times, mixed mating systems, and high potential for gene flow (1). A population genetic structure reflects its evolutionary history and potential to evolve (2). This knowledge is important for the development of efficient and sustainable disease management (3, 4). Over the last few decades, much effort has been put into studying the structure and diversity of populations of different plant pathogens (5). Population diversity is directly related to organismal mating preferences, dispersal strategies, and/or adaptation to local environments (2, 6). Pathogen adaptation in agroecosystems is a major concern for maintaining crop yields and preventing epidemics (6). Adaptation to specific hosts or regions has been identified for many plant pathogens (7–10). In addition to host and geography, the widespread use of pesticides in modern agriculture is a powerful evolutionary force that shapes diversity and the structure of microbial populations in agroecosystems. A number of studies have reported signals for local adaptation through rapid development of resistance to pesticides and drugs in various systems, including insecticides (11–15), herbicides (16, 17), nematicides (18), antibiotics (19, 20), and fungicides (21, 22).
Increasing problems with fungicide resistance in target microorganisms have raised important questions about how fungal pathogens develop, maintain, and spread resistance within and among populations (1, 6). Most modern fungicides affect a specific biochemical target in a fungal cell, usually the product of a single gene (23). Point mutations in these genes are the most common resistance mechanism (24). The extent of adaptation of a new mutation depends on fitness costs and population size (1, 25). In small populations, adaptation results in the increase of frequency of one genotype with a beneficial mutation due to selection. In large populations, by contrast, beneficial mutations can arise independently multiple times that again rise in frequency due to selection (26). Mechanisms of fungicide resistance evolution among different fungal plant pathogens show evidence for its independent development in different pathosystems. Empirical evidence suggests that beneficial mutations repeatedly emerged in a small number of genes that code for fungicide targets. For example, resistance to the quinone inhibitor fungicide was reported to be conferred by the G143A substitution in the mitochondrially encoded target site cytochrome b in several fungal species (27–34) and within populations of the same species (35–37). Succinate dehydrogenase inhibitors (SDHI) represent another class of fungicides being used against many plant pathogens (38). Resistance to SDHIs was found to be associated with multiple point mutations in the B, C, and D subunit genes of the SDH complex in the respiratory chain of fungal cells. Independent development of resistance to SDHIs in various plant pathogen species and geographic regions has been reported (39). Such widespread resistance development in pathogen populations has become a major issue for food production worldwide. Reduced ability to control pathogens in fields due to resistance leads to yield losses and often increases the use of fungicides that have negative effects on the environment and human health. Moreover, limited fungicide chemistries are available for management of plant pathogens in the field. Once pathogen populations evolve resistance to a specific fungicide in the field, farmers are left with fewer tools they can use to protect crops. For all these reasons, studying the forces that shape population structures of plant pathogens in modern agroecosystems is essential for the development of rational strategies for fungicide use and disease management.
Over the last few decades, the ascomycete fungus Botrytis cinerea has become the most extensively studied necrotrophic plant pathogen due to its worldwide importance (40). It causes gray mold disease on >1,400 plant species of vascular plants (41) and can lead to substantial yield losses, especially of soft fruit (42–45). B. cinerea is known to be highly variable and genetically diverse, with signs of sexual recombination in some areas (46–49), but not others (50, 51). Differentiation of B. cinerea populations has been studied in various locations and from various hosts, without a consistent pattern of host specificity observed (47, 48, 50, 52–54). Significant regional differentiation (47, 48, 50, 55) and adaptation to management strategies (56, 57) were also reported. With its ubiquity (58), mixed reproduction system (59), and large population sizes, B. cinerea rapidly develops fungicide resistance in agroecosystems (24). The potential for selection for fungicide resistance in B. cinerea populations was briefly highlighted in some cases (51, 60, 61), suggesting the importance of further investigation of the contribution of selection for fungicide resistance to population subdivision.
In this study, we hypothesized that B. cinerea populations become specialized to small fruit hosts and that geographic and anthropogenic factors of agroecosystems structure B. cinerea populations in the Pacific Northwest (PNW) small fruit cropping systems (Fig. 1). Using both selectively neutral markers (simple sequence repeats [SSRs]) and a fungicide resistance marker under selection, we addressed the following questions. (i) Are B. cinerea populations specialized to small fruit hosts? (ii) What is the structure of B. cinerea populations at regional and local scales? (iii) Finally, does fungicide use affect the genetic variation and structure of B. cinerea populations?
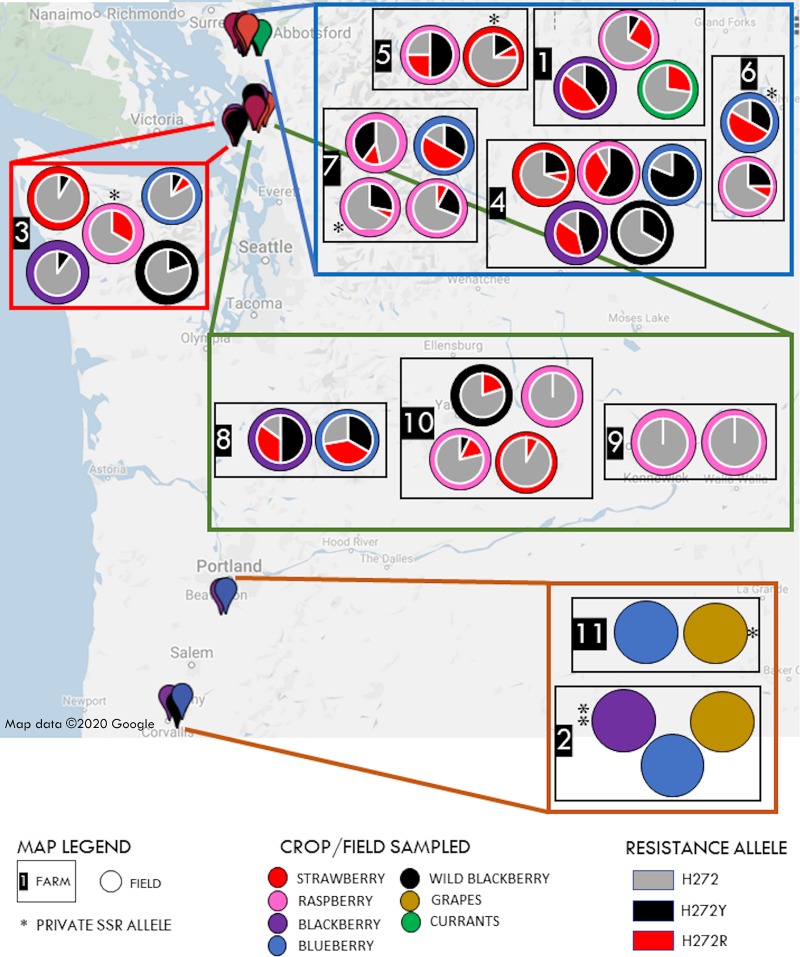
FIG 1.
Sampling map of B. cinerea isolates from 34 fields on 11 farms of small fruit hosts in the Pacific Northwest, USA (PNW), with B. cinerea private SSR alleles (*) and fungicide resistance profiles overlaying fields. Whatcom County, WA, fields are in the blue box. Skagit County, WA, fields are in the green box. Island County, WA, fields are in the red box. Clackamas and Linn Counties, OR, fields are in the orange box.
RESULTS
Population structure of B. cinerea on small fruit.
Sixteen of 679 isolates were identified as B. pseudocinerea using the species-specific PCR and excluded from analysis. The remaining isolates were identified as B. cinerea sensu stricto. SSR repeat motifs from representative alleles were confirmed as homologous by sequencing the microsatellite locus for four isolates carrying each putative allele. Differences in SSR fragment sizes of 1 to 2 bp were considered as changes in flanking regions or technical errors and were binned with alleles of adjacent size. Homoplasy was not detected in any of the sequenced alleles. All SSR loci were polymorphic (Table 1). Locus Bc7 had the highest diversity based on Hexp (0.86) and λ (0.85), while loci Bc4 and Bc6 were the least diverse, with Hexp = 0.57 and λ = 0.55.
TABLE 1.
Allelic diversity of seven simple sequence repeat loci in B. cinerea populations infecting small fruit hosts in the PNW
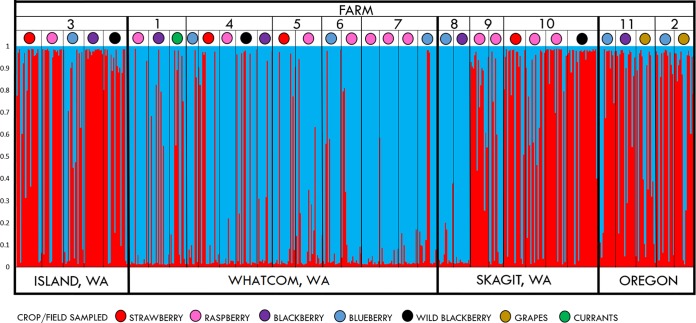
Based on the results of Bayesian clustering analysis, a model with two genetic groups had the highest likelihood (Fig. 2). Analysis of molecular variance (AMOVA) confirmed significant differentiation between these groups, accounting for 21% of the observed variation (P < 0.0001). Many isolates were assigned to more than one genetic group. Totals of 43 and 157 individuals could not be assigned to either group with probabilities >75 and >95%, respectively. Both groups contained isolates of mixed ancestry but populations from seven fields (raspberry fields from farms 5, 6, and 7 and blackberry and blueberry fields from farm 8) were assigned to a single group. Overall, isolates from the same group dominated each farm, except farm 9, where 90% of individuals from one field were assigned to one group, but 67% of individuals from another field were assigned to another group. Group 1 dominated among populations sampled in OR (farms 2 and 11) and Island (farm 3) and Skagit Counties (farms 9 and 10), WA. Populations sampled in Whatcom County, WA, were mostly assigned to group 2. This group also dominated farm 8 in Skagit County, WA.
FIG 2.
Bayesian clustering analysis of B. cinerea populations from 34 fields on 11 small fruit farms in the PNW. Each isolate is represented by a vertical line divided into two color groups (red, group 1; blue, group 2). Each color corresponds to the isolate membership coefficient in each of the two clusters. Each farm is numbered on the top of the graph, with the colored circles representing the specific field/host according to the legend.
Six alleles at three microsatellite loci were highly influential in determining assignment in Structure (Table 2). The alleles differed by more than one repeat unit from each other (Bc6, Bc299 and Bc3 differed by three, six, and two repeats, respectively), suggesting these groups were robust to repeated and/or reverse mutations.
TABLE 2.
Influential loci and alleles in the Structure analysis of B. cinerea populations infecting small fruit hosts in the PNW
| Locus | Allelea | Frequency |
||
|---|---|---|---|---|
| Allele | Group 1 | Group 2 | ||
| Bc6 | 130 | 0.381 | 0.867 | 0.267 |
| 136 | 0.158 | 0.002 | 0.419 | |
| Bc299 | 250 | 0.063 | 0.375 | 0.009 |
| 232 | 0.220 | 0.030 | 0.592 | |
| Bc3 | 222 | 0.126 | 0.431 | 0.054 |
| 218 | 0.232 | 0.044 | 0.519 | |
Only alleles with large representations in one or the other group are shown.
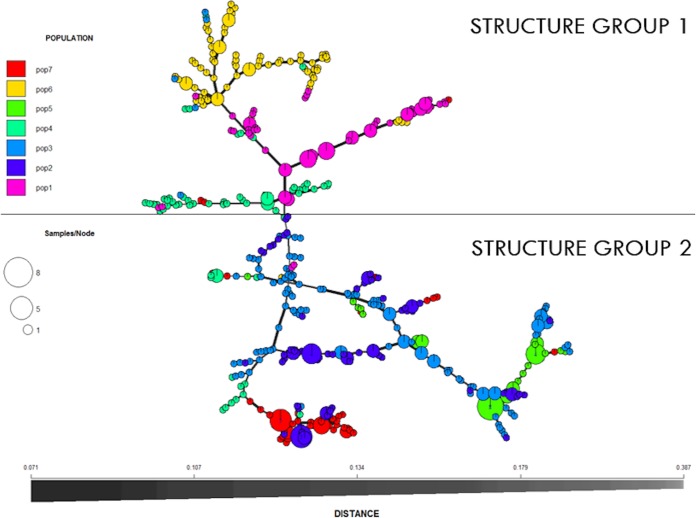
Discriminant analysis of principal components (DAPC) supported 6 to 15 clusters by plotting the mean Bayesian information criterion (BIC) against the number of clusters. The data set with seven clusters assigned 499 individuals of a total of 555 to one of seven clusters which was the largest number of assigned individuals among all DAPC outputs. When k was set to 6, only 432 individuals were assigned to one of six clusters, and with the increase of number of clusters after k = 7 the number of individuals assigned to clusters with a probability of ≥75% decreased (e.g., 354 and 252 individuals of 555 with k = 10 and k = 15, respectively). Therefore, k = 7 was chosen as the most useful summary of the data. AMOVA supported significant differentiation among 7 DAPC clusters (P < 0.001). Each of the 7 DAPC clusters was mostly represented by one group or the other using Structure v.2.3.4 (Fig. 3). One hundred percent of DAPC cluster 1, 92.5% of DAPC cluster 4, and 100% of DAPC cluster 6 were assigned to structure group 1, while 96.4% of DAPC cluster 2, 83.9% of DAPC cluster 3, 100% of DAPC cluster 5, and 96.2% of DAPC cluster 7 were assigned to structure group 2. The minimum spanning network (MSN) supported the division of individuals defined by structure and DAPC (Fig. 3).
FIG 3.
Minimum spanning network (MSN) of B. cinerea isolates from 34 small fruit fields on 11 farms in the PNW based on Bruvo’s distance. Populations for MSN analysis were assigned based on discriminant analysis of principal components output. The size of a node represents the number of individuals of the same multilocus genotype. The thickness of the lines connecting nodes in the MSN represents Bruvo’s distance (thicker line = smaller distance).
We first performed hierarchical AMOVA with the clone-corrected data set with host as the grouping factor (Table 3). The major contribution to genetic variance was due to variation within region (93%). The effect of region within host was significant (P < 0.0001), while host was not statistically significant (P = 0.1773). When location was used as a grouping factor, effects of region (P = 0.0001), host within each region (P < 0.0001), and variation within host (P < 0.0001) were all statistically significant. These results indicated that the effect of geography on B. cinerea populations is greater than the effect of host. The significant effect of host within region indicated potential local population structure. Significant differentiation among 11 farms (P = 0.0002), among populations from fields within farms (P = 0.0013), and within populations (P < 0.0001) was further confirmed with AMOVA using “farm” as a grouping factor (Table 4). When populations from fields within one farm were pulled together, Slatkin’s Rst confirmed significant pairwise differentiation among 11 farms with few exceptions (see Fig. S1a in the supplemental material). When Slatkin’s Rst was estimated within each farm separately, significant pairwise differentiation was also detected among some adjacent fields on a farm scale (Fig. S1b). Seven private SSR alleles were detected among 6 of 34 fields sampled (Fig. 1), suggesting restricted movement of B. cinerea among adjacent fields. Altogether, these results indicate that B. cinerea populations are differentiated on a fine scale with no association with host plant species. Mantel tests were not statistically significant when using the whole data set (R2 = 0.0004, P = 0.141), or when WA and OR fields were analyzed separately (WA: R2 = 0.0071, P = 0.106; OR: R2 = 0.1644, P = 0.061). These results indicated that in addition to geography, other factors shape population differentiation on a fine scale.
TABLE 3.
Hierarchical AMOVA of B. cinerea populations using crop and sampling location as grouping factors
| Source of variation | df | Sum of squares | Variance component | Variation (%) | P |
|---|---|---|---|---|---|
| Among hosts | 5 | 732.77 | 0.43 | 0.82 | 0.1773 |
| Among region within host | 28 | 3,173.14 | 3.97 | 7.55 | <0.0001 |
| Within region | 521 | 25,099.83 | 48.18 | 91.63 | <0.0001 |
| Total | 554 | 29,005.74 | 52.58 | ||
| Region | 3 | 1,094.92 | 2.12 | 3.99 | 0.0001 |
| Host within region | 30 | 2,810.99 | 2.81 | 5.28 | <0.0001 |
| Within host | 521 | 25,099.83 | 48.18 | 90.73 | <0.0001 |
| Total | 554 | 29,005.74 | 53.09 |
TABLE 4.
Clone corrected AMOVA of B. cinerea populations obtained from 34 small fruit fields on 11 farms of adjacent fields in the PNW
| Source of variation | df | Sum of squares | Variance component | Variation (%) | P |
|---|---|---|---|---|---|
| Clone corrected | |||||
| Among farms | 10 | 2,148.19 | 2.79 | 5.30 | 0.0002 |
| Among populations within farms | 23 | 1,757.72 | 1.73 | 3.29 | 0.0013 |
| Within populations | 521 | 25,099.83 | 48.18 | 91.41 | <0.0001 |
| Total | 554 | 29,005.74 | 52.70 |
Tests of recombination in B. cinerea populations from 11 small fruit farms in the PNW.
Due to significant local differentiation among B. cinerea populations, recombination tests were performed at the farm level. Multilocus linkage disequilibrium tests (rd) rejected the null hypothesis that alleles at SSR loci are freely recombining on all 11 farms sampled (rd = 0.090 to 0.160, P = 0.001 to 0.049). In addition, we estimated rd for genetic groups defined by structure, and these results also yielded no evidence for recombination (rd = 0.080 [P = 0.001] and 0.021 [P = 0.001] for group 1 and group 2, respectively). Analysis of mating type distribution showed that both idiomorphs were present on all farms with distributions not significantly different from a 1:1 ratio (χ2 = 0.01 to 2.47, P = 0.18 to 0.99) (data not shown).
Impact of fungicide use on genetic diversity of B. cinerea populations.
In order to determine whether selection by use of fungicide boscalid affects population genetic diversity and structure, we checked for the association between boscalid resistance frequencies per field and several diversity measures of the SSRs. Significant negative correlations were detected between boscalid resistance frequencies and genetic diversity of neutral markers (Hexp, r = −0.465, P = 0.010) and evenness (E5, r = −0.436, P = 0.020). The clonal fraction in each field was positively correlated with boscalid resistance frequencies (r = 0.344, P = 0.068), while boscalid resistance frequencies were negatively correlated with allelic richness (Ar; r = −0.322, P = 0.088), expected multilocus genotypes (eMLGs; r = −0.341, P = 0.069), and the H (r = −0.324, P = 0.087) and G (r = −0.0327, P = 0.084) indices of MLG diversity. Even though the last five Pearson’s r values were not statistically significant, their P values were just above the 0.050 probability threshold.
Since structure genetic group 2 comprised farms with higher frequencies of boscalid resistance compared to group 1 (58 and 20% resistant isolates from total tested in group 2 and group 1, respectively), we estimated genetic and genotypic diversities separately for each group. Reduced Hexp and Ar values were observed in group 2 compared to group 1 (Table 5). H, biased toward rare genotypes, was lower in group 1, whereas G, biased toward common genotypes, was higher in group 1 than in group 2. The 95% confidence interval (CI) for H and G overlapped between the two groups, suggesting a lack of precision in estimations of these two indices. Differences in genotypic diversity can be due to differences in genotypic richness (eMLG) or evenness (E5). In group 2 both measures were significantly lower than in group 1, meaning that group 2 consists of fewer unique MLGs that are less evenly distributed within the sample. Significant differences in evenness between two groups explain higher H, but lower G diversities in group 2 than in group 1. In addition, the genotypic clonal fraction was almost two times higher in group 2 than in group 1, further indicating reduced a genotypic diversity in group 2. Altogether, these results provided evidence that selection pressure due to use of fungicide boscalid is associated with decreased diversity of B. cinerea populations on small fruit in the PNW on both genetic and genotypic levels.
TABLE 5.
Genetic diversity, genotypic richness, diversity and evenness of Botrytis cinerea from small fruit assigned to two genetic groups based on Bayesian clustering analysis implemented in Structure
| Population | Na | MLGb | eMLGc | SEd | Variable (95% CI) |
Clonal fraction | Hexph | Ari | ||
|---|---|---|---|---|---|---|---|---|---|---|
| H′e | Gf | E5g | ||||||||
| Group 1 | 220 | 198 | 198 | 0.00 | 5.25 (5.18–5.32) | 181 (171–189) | 0.95 (0.92–0.98) | 0.10 | 0.72 | 8.90 |
| Group 2 | 292 | 238 | 186 | 3.06 | 5.37 (5.27–5.46) | 177 (159–194) | 0.83 (0.76–0.89) | 0.19 | 0.63 | 6.80 |
| Total | 512 | 436 | 203 | 3.41 | 6.00 (5.94–6.06) | 350 (327–372) | 0.87 (0.82–0.91) | 0.15 | 0.76 | |
Number of isolates.
Number of multilocus genotypes (MLG).
Number of expected multilocus genotypes from the smallest rarefied sample >10 (eMLG).
Standard error based on eMLG.
Shannon-Wiener index of MLG diversity (H) (87). Numbers in parentheses represent 95% confidence intervals (CI) calculated from 1,000 bootstraps.
Stoddart and Taylor’s index of MLG diversity (G) (88).
Nei’s unbiased gene diversity (Hexp) (83).
Allelic richness corrected for sample size with rarefaction (Ar). Groups 1 and 2 refer to the red and blue clusters in Fig. 2, respectively.
Distribution of boscalid resistance alleles in different genetic backgrounds.
Boscalid resistance frequencies in 29 WA fields ranged between 0 and 92% (Fig. 1). Overall, 52% of sampled isolates were resistant to boscalid. Boscalid-resistant isolates were not detected in organically managed fields (farm 9) but were detected in wild blackberries adjacent to commercial farms. Resistance was conferred either by the H272Y (62% of isolates) or the H272R mutations (38% isolates) in the B subunit of the mitochondrial SDH enzyme complex. No additional resistance mutations were identified. Both resistance alleles were present in 18 fields, while a single allele was detected in 9 fields. Allele frequencies differed among fields on the same farm, and there was no association of resistance allele with either crop, geography, or farm. In cases where multiple fields of the same crop were sampled on a farm, resistance frequencies also differed among those fields (e.g., raspberry fields on farm 7 and farm 10), suggesting limited migration of B. cinerea among fields within farms.
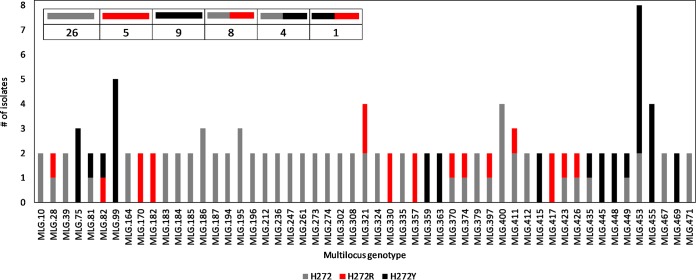
Of 398 MLGs defined using the microsatellite markers and observed among 29 fields in WA, 53 MLGs were observed in more than one field (Fig. 4, n = 125 isolates). Of 53 shared MLGs, 26 were sensitive to boscalid, 12 were both resistant and sensitive to boscalid, and 15 were resistant to boscalid. Although the majority of shared MLGs had the same resistance profiles, suggesting either the presence of migration among fields or the spread of B. cinerea from a common source, 13 shared MLGs carried different SdhB alleles in different fields (Fig. 4). Specifically, eight MLGs were shared among isolates with either H272R or a sensitive allele, four MLGs were shared among isolates with either H272Y or a sensitive allele, and one MLG was shared between isolates carrying either the H272R or H272Y allele. Despite detecting shared MLGs among resistant isolates, we did not find evidence that boscalid resistance alleles are more likely to occur in some genetic backgrounds compared to others. Average pairwise rd among SSR loci (0.026) was significantly higher than pairwise rd among SSRs and boscalid-resistant loci (0.009), suggesting a lack of association of alleles under selection with MLGs. Together, with the lack of evidence for sexual recombination, these results suggest independent development of boscalid resistance mutations in B. cinerea populations with regular recolonization of small fruit fields in the PNW by different fungal genotypes.
FIG 4.
Boscalid resistance profiles of 53 B. cinerea MLGs defined by SSR markers that appeared in more than one of 29 fields on nine small fruit farms sampled in Washington state. The legend indicates the number of MLGs for each unique fungicide resistance profile.
DISCUSSION
Several lines of evidence suggest that genetically diverse B. cinerea populations are structured on a fine scale and adapted to local environments. Using neutral microsatellite markers, no significant association of B. cinerea with host of origin was found, but populations were geographically differentiated at the regional and farm scales, suggesting restricted gene flow, even between adjacent fields on the same farm. In addition, evidence for local adaptation due to selection was detected with boscalid resistance markers and the limited movement of B. cinerea among fields demonstrated with the microsatellite markers was confirmed with the boscalid resistance marker. Different resistance profiles were observed among adjacent fields, and no association among boscalid resistance alleles and B. cinerea genetic backgrounds was detected, suggesting possible de novo mutations to resistance in different genetic backgrounds and frequent extinction/recolonization events by different genotypes carrying resistance alleles rather than the spread of resistance alleles among fields via migration of a dominant genotype. A significant effect of fungicide selection pressure on diversity of B. cinerea populations was detected. Overall, our results show that B. cinerea populations infecting small fruit in the PNW are diverse, but most likely locally structured due to their strong adaptability to anthropogenic environment of agroecosystems, such as fungicide applications.
Diverse populations with lack of evidence for sexual reproduction.
An analysis of SSR polymorphism showed high genotypic variability of B. cinerea populations infecting small fruit crops in the PNW. High B. cinerea genotypic diversity may be a product of cryptic sexual reproduction, even though sexual structures are rarely observed in nature (62). By estimating linkage disequilibrium among neutral markers, some studies have reported signatures of recombination in some B. cinerea populations (46, 48, 61), but not others (51, 52, 57). In this study, B. cinerea populations appear to be asexual but highly diverse. Both mating types were present in populations in 1:1 distributions, but this may not be the result of frequency-dependent selection in mating. It has been suggested that B. cinerea mating genes participate in processes other than mating because they remain transcriptionally active at different developmental stages of the fungus (59). The role of mating genes in fungal development, including hyphal growth, asexual sporulation, and pathogenicity, was previously reported for different filamentous fungi, including plant pathogens (63–67). Hence, even if this organism reproduces asexually, both mating types will still be maintained in a population. Apart from sexual reproduction, another common source of high genetic diversity and plasticity in B. cinerea genome can be the activity of transposable elements. By transposing themselves from one location in the genome to another, they can cause structural rearrangements of DNA sequences and chromosomes, gene duplication, inactivation, and changes in gene expression. In some studies, transposable element composition has been linked to pathogenic behavior of B. cinerea on different hosts (59), suggesting their potential role in fungal adaptation to local environments through constant shuffling of fungal genome.
No evidence for host specificity of B. cinerea populations on small fruits in the PNW.
Structure and DAPC clustering divided populations into two and seven genetic clusters, respectively. MSN supported this differentiation by grouping isolates from the same clusters together. The genetic clustering did not correspond with host of origin or sampling location, despite the fact that small fruit production is well established in the PNW. Inconsistent host association of B. cinerea populations from different host species was reported in France (68). The authors of that study suggested that B. cinerea populations represent collection of specialized strains on certain hosts (tomato and grapevine) and of generalist strains on other hosts (strawberry, bramble, and hydrangea) included in the study. It is possible that lack of association of genetic clusters with host plant species sampled in this study may be due to such partial specialization of B. cinerea mentioned above. We, however, did not find evidence for association of B. cinerea with grapevine host when populations were analyzed on a local scale. It would be beneficial to investigate host association of PNW B. cinerea with other than small fruit hosts in future studies.
AMOVA detected a lack of differentiation by host, but significant differentiation by geography among and within regions and among farms, suggesting the presence of a local population structure. The differentiation among farms belonging to two structure groups was supported with pairwise Rst, except farm 9 (group 1), which was not significantly differentiated from farms 5 and 6 (group 2). There were, however, no shared MLGs among farms 9, 5, and 6. Pairwise Rst within each farm detected inconsistent, but significant differentiation among adjacent fields. Lack of a clear geographic structure at a farm scale may indicate extensive migration of pathogen propagules in space, but it is known that B. cinerea conidia do not move far and usually stay within a field (69). Another explanation of inconsistent geographic structure can be large effective population sizes of B. cinerea making it impossible to accurately assess differentiation using limited sample sizes as is the case in this study. We have not assessed effective population size of B. cinerea in this study, but large effective population sizes were reported before (61). Moreover, the ubiquity and high genetic diversity of B. cinerea were confirmed by analyzing strains from agricultural and nonagricultural habitats in France, suggesting large population sizes that retain their pathogenic potential even without association with hosts (58). Therefore, we conclude that inconsistent population differentiation at a farm scale is most likely caused by local adaptation to anthropogenic factors, such as cropping systems and/or management strategies (4).
Microsatellite and resistance markers reveal limited movement of B. cinerea among adjacent fields.
The fungicide boscalid is widely used to manage gray mold on small fruit in the PNW, and we hypothesized that B. cinerea populations should undergo strong positive selection in response to applications of boscalid and other fungicides. From personal communication with growers, we know that fields in Whatcom County overall receive more fungicide applications compared to Skagit and Island Counties. Even though the information about the boscalid application history among studied fields was not available in this work, different resistance frequencies were detected among small fruit farms in the PNW that reflect high levels of heterogeneity in fungicide programs used on different farms resulting in different levels of selection pressure across the region. In addition to significant pairwise differentiation among populations from adjacent fields that was detected with microsatellites, differences in resistance profiles were also detected among adjacent fields within farms, including fields of the same crop. For example, one of three raspberry fields on farm 7 had higher resistance frequencies than others. It is known that fields on the same farm are managed identically. The field with higher resistance frequencies was 2 years older than the others and experienced selection for longer. These results further suggest that populations remain local on a fine scale. We also detected resistant strains in populations sampled from wild blackberry plants adjacent to commercial farms. One explanation could be limited migration of B. cinerea from cultivated to wild berries. It is, however, not clear how significant the role of migration is in spreading resistance, based on the findings discussed above. Moreover, growers regularly spray wild blackberries adjacent to commercial fields with fungicides as part of their management programs. The latter may select for resistant genotypes and keep them in populations on a wild host. Only two types of mutations conferring resistance to boscalid were revealed within all boscalid-resistant phenotypes from studied populations, H272R and H272Y substitutions in codon 272 of the SdhB gene. Frequencies of the mutations differed among fields within farms, in nine fields resistance was represented by a private allele (either H272R or H272Y), and in one case a resistant private allele was detected on a farm scale (H272Y in farm 10). These substitutions are the most prevalent in B. cinerea sampled from different hosts and regions (70–75). Detection of the same mutations in fungal populations on different hosts and in different regions suggests their parallel development rather than spread via migration. This was consistent with the lack of association of resistant alleles with genetic backgrounds based on rd estimations performed in this study. Moreover, cases of different resistance profiles within shared MLGs from different farms support the hypothesis of parallel development of boscalid resistance in pathogen populations. We, however, realize that the spread of resistant strains from a common source cannot be completely excluded. Fungicide-resistant B. cinerea strains were previously detected in raspberry nurseries in Europe (76) and strawberry nurseries in the United States (77). In this work, we did not investigate B. cinerea in places that supply growers with plants, and fungicide resistance profiles of fungal populations from nursery stock should be studied in future work. As stated above, we did not find evidence for sexual reproduction in studied populations, but we also realize that possible infrequent random mating events cannot be completely excluded. The analysis of linkage disequilibrium and clonal fraction in B. cinerea populations on small fruit hosts in the PNW yielded average values compared to values identified in greenhouse (presumably asexual reproduction) and open-field (presumably sexual reproduction) populations in France (4). It therefore remains unknown how much such possible infrequent random mating events could facilitate in spread of boscalid resistance alleles.
Fungicide use lowers genetic diversity in B. cinerea populations.
Strong selection pressure caused by fungicides leads to allele loss in populations due to a reduction in effective population size (49). A study of B. cinerea adaptation to fungicides in France (61) revealed reduced effective population sizes and genetic diversities in fungicide-treated populations compared to untreated ones. In the current work, we detected significantly reduced genetic and genotypic diversities within a population with almost three times higher boscalid resistance frequencies due to significantly lower allelic and genotypic evenness and richness. These results, together with significant negative correlations between gene, genotypic diversities, and resistance frequencies in each field, support the hypothesis that multiple fungicide treatments, applied during growing season, reduce the pathogen population size and its diversity by selecting for resistant genotypes. The spread and maintenance of resistance mutations are directly related to the fitness of resistant strains. By evaluating fitness penalties for B. cinerea Sdh mutants in laboratory experiments, various degrees of fitness alterations depending on the type of the resistant mutation were reported (78). There was no fitness cost detected in a B. cinerea mutant carrying the H272Y allele, but reduced conidial and sclerotial production was observed in the homologous H272R mutant in vitro (78). We did not assess fitness parameters of resistant B. cinerea field isolates in this work, but we did not detect significant reduction in conidial or sclerotial production by H272R mutants at the culturing stage of the isolates. It is possible that the genetic background of mutants may affect the degree of fitness penalties caused by these mutations. In the future, it would be beneficial to assess maintenance of H272R and H272Y mutations in small fruit B. cinerea populations over multiple generations after selection is relaxed.
Concluding remarks.
Fungal pathogens have short generation times, large population sizes, and mixed reproductive systems, providing high potential to adapt to heterogeneous environments. Due to its ubiquity and its ability to attack various hosts and survive for extended periods without losing its virulence, B. cinerea has a strong capacity for rapid adaptation. Here, we show evidence that in agroecosystems pathogen populations undergo strong local adaptation caused by anthropogenic factors, such as fungicide treatments, that have a significant effect on their structure and diversity at a fine scale. Our results show that selection pressure caused by fungicide (boscalid in particular) use can have a greater effect on B. cinerea population structure than adaptation to host plant species. This adaptation cannot always be detected with neutral markers alone, especially in highly diverse organisms such as B. cinerea. Therefore, combining neutral markers with markers that directly respond to human-induced selection pressures may reveal key aspects of adaptive divergence processes, help in forecasting epidemics, and improve management. In summary, our findings provide a better understanding of the population biology of the most important small fruit pathogen in the PNW and worldwide and will be used to improve gray mold management strategies by developing customized resistance management programs for individual fields based on fungicide resistance profiles in the PNW and elsewhere.
MATERIALS AND METHODS
Collection and identification of B. cinerea from small fruit hosts.
Asymptomatic red raspberry (Rubus idaeus L.), blueberry (Vaccinium corymbosum L.), strawberry (Fragaria ananassa Duchesne ex Rozier), blackberry (Rubus ursinus Cham. and Schltdl.), currant (Ribes nigrum L.), and grape (Vitis vinifera L.) fruit were collected from adjacent commercial fields (n = 200 fruit per field) in Washington (WA) and Oregon (OR) between June and September 2015 (Fig. 1). Three fields of wild blackberry (Rubus armeniacus Focke), adjacent to commercial fields, were sampled in WA. Fruits were surface disinfested in 70% alcohol for 10 s, followed by 1 min in 1% NaOCl, and then washed in deionized water three times for 1 min each time and incubated in humid chambers at 12°C for 14 days until sporulation appeared. Single-spore isolates were transferred to potato dextrose agar (PDA). DNA was extracted using a DNeasy PowerSoil kit (Qiagen, Germantown, MD) according to the manufacturer’s protocol. Samples were verified as B. cinerea and differentiated from its cryptic sister species B. pseudocinerea by using a PCR assay (79).
Identification of new polymorphic SSR markers.
The SSR primers Bc1, Bc2, Bc3, Bc4, Bc5, Bc6, Bc7, and Bc10 previously described by Fournier et al. (80) were tested with 50 isolates, and polymorphisms could not be detected for 20 of 50 isolates (data not shown). Therefore, new primers were designed for this study. Seven loci were selected from markers designed from the B05.10 and T4 genomic sequences (81). The new markers were selected according to the following criteria: repeat motifs of three or more base pairs to augment predominantly 2-bp repeat motifs (80); primer pairs with annealing temperatures between 58 and 60°C for the ability to multiplex; amplification of a single repeat motif; allele sizes of >200 bp; and 11 to 16 repeats to increase the chance of detecting highly polymorphic loci while avoiding hybrid motifs (82) Previously published reverse primers were modified at the 5′ end by the addition of a GTTT PIG tail (Table 6). In addition, Bc2, Bc3, and Bc6 forward primers were adjusted to permit a higher annealing temperature when multiplexing. The M13-tailed microsatellite protocol was used to assess all loci prior to fluorescent labeling of forward primers. To this end, forward primers were modified to include a universal sequence (TGTAAAACGACGGCCAGT) at their 5′ ends. A tailed forward primer was then combined with a reverse primer and a universal M13 6-carboxyfluorescein (6-FAM) fluorescently labeled primer (TGTAAAACGACGGCCAGT). Universal M13-tailed PCRs consisted of 10 μl and included 20 ng of DNA, 1× GenScript Taq buffer, 0.2 mM deoxynucleoside triphosphates, 0.04 μM M13-tailed forward primer, 0.16 μM reverse primer, 0.16 μM universal M13 6-FAM primer, and 0.5 U of GenScript Taq (GenScript, Piscataway, NJ). Thermal cycling conditions were as follows: 94°C for 3 min; followed by 35 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 30 s; with a final extension at 72°C for 10 min (Veriti thermal cycler; Life Technologies, Grand Island, NY). Alleles for each of the seven loci were screened for the expected size on an ABI 3130 genetic analyzer (Applied Biosystems, Foster City, CA). In addition, alleles from isolate 327 were cloned at the seven loci from the genome data and sequenced. Cloning was completed by using a Topo TA cloning kit from Invitrogen supplied with the pCR 4-TOPO plasmid and OneShot TOP10 chemically competent E. coli (Thermo Fisher Scientific, Waltham, MA). After ligation, transformation, and dilution plating, single colonies were picked for analysis, and Luria-Bertani (LB) cultures were started. Topo plasmids containing inserts were extracted from LB cultures using the FastPlasmid minikit (5PRIME, Gaithersburg, MD). Plasmid inserts were sequenced on an ABI 3730 capillary DNA sequencer (Applied Biosystems) in both directions with T7 and T3 primers in the Center for Genome Research and Biocomputing Core Facilities, Oregon State University (Corvallis, OR). Sequences were assembled in Geneious R6 (Biomatters, Ltd., Auckland, New Zealand) and confirmed to be microsatellites. Primer sequences are shown in Table 6.
TABLE 6.
SSR primer sequence final concentrations and multiplex PCR conditions used in microsatellite genotyping of Botrytis cinerea populations infecting small fruit in the Pacific Northwesta
| SSR locus | Dye | Product (bp) | Primer sequenced | Final concn (μM) | Reaction |
|---|---|---|---|---|---|
| Bc5b | VIC | 153–175 | FrwdVIC-CGTTTTCCAGCATTTCAAGT | 0.05 | 2a-plex |
| Rev-GTTTCATCTCATATTCGTTCCTCA | |||||
| Bc2b,e | VIC | 138–172 | FrwdVIC-CATACACGTATTTCTTCCAACTACCAAC | 0.05 | 2b-plex |
| Rev-GTTTACGAGTGTTTTTGTTAGAAT | |||||
| Bc3b,e | NED | 211–229 | FrwdNED-GGATGAATCAGTTGTTTGTGACG | 0.10 | 2a-plex |
| Rev-GTTTCACCTAGGTATTTCCTGGTA | |||||
| Bc4b | NED | 119–131 | FrwdNED-CATCTTCTGGGAACGCACAT | 0.04 | 3-plex |
| Rev-GTTTATCCACCCCCAAACGATTGT | |||||
| Bc6b,e | PET | 118–152 | FrwdPET-ACTAGATTCGAGATTCAGTTATATGAT | 0.16 | 3-plex |
| Rev-GTTTAAGGTGGTATGAGCGGTTTA | |||||
| Bc7b | 6-FAM | 120–134 | Frwd6FAM-CCAGTTTCGAGGAGGTCCAC | 0.11 | 2b-plex |
| Rev-GTTTGCCTTAGCGGATGTGAGGTA | |||||
| Bc299c | 6-FAM | 206–260 | Frwd6FAM-TGATGGAATGTTCTTGGATGA | 0.10 | 3-plex |
| Rev-GTTTCACCAGGACTCCAGTCACCT |
The forward primers for each of the markers were labeled with fluorescent probe using multiplex PCR. Each 5-μl reaction mixture contained 2 μl of genomic DNA (3 to 20 ng), 0.5 μl of 10× primer mix, and 2.5 μl of 2× Qiagen Type-IT PCR mix (Qiagen, Germantown, MD). Thermal cycling conditions included an initial denaturation at 95°C for 5 min, 32 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 90 s, and extension at 72°C for 20 s, with a final extension at 68°C for 10 min. One microliter of 1:100 dilution of PCR products was mixed with a Liz 500 internal size standard-formamide mix (Applied Biosystems). Sizing was performed on an ABI 3730 capillary DNA sequencer (Applied Biosystems).
Data from Fournier et al. (80).
Data from the current study and Amselem et al. (81).
Reverse (Rev) primer includes the PIG tail addition, indicated in italics (101).
The 3′ ends of the original primers were modified (current study), as indicated in italics.
Genetic and genotypic characterization of B. cinerea populations from small fruit.
All isolates were genotyped at seven SSRs—Bc2, Bc3, Bc4, Bc5, Bc6, Bc7 (80), and Bc299—developed in this study (Table 6). Alleles were scored in GeneMarker v.5 (SoftGenetics LLC, State College, PA). Four PCRs were repeatedly genotyped in three separate runs to check for genotyping variation of the sequencer. Repeat motifs were confirmed and potential homoplasy was assessed by sequencing representative alleles of each locus by Eurofins Genomics LLC (Louisville, KY). Primers and PCR conditions used for allele amplification are in Table 7. Sequences were aligned and compared in Geneious v8.1.8 (Biomatters, Ltd., Auckland, New Zealand).
TABLE 7.
Primers used in the studya
| Function and locus | Primer | Sequence (5′–3′) | Annealing temp (°C) | Amplified fragment (bp) |
|---|---|---|---|---|
| PCR amplification and sequencing of B. cinerea alleles among 7 SSRs | ||||
| BC6 | BC6_Fw | CCCTAACCCAAGCGACAAAC | 58 | 807 |
| BC6_Rev | CGCACTCGGTAATACTTTTCACT | |||
| BC2 | BC2_Fw | CATCCCACACCAAATCGCAT | 58 | 801 |
| BC2_Rev | ATTGAGATGGTGCATGGCTG | |||
| BC3 | BC3_Fw | GCTGGCAAGAGAGTGCAATG | 58 | 694 |
| BC3_Rev | GAGGTCGCATGGTGGGTATC | |||
| BC7 | BC7_Fw | GTGAGTGCTAGTAACACAGCTG | 63 | 803 |
| BC7_Rev | TTGCTACCAGTAACTCCGGG | |||
| Bc299 | Bc299_Fw | TGATGGAATGTTCTTGGATGA | 63 | 787 |
| Bc299_Rev | GTTTCACCAGGACTCCAGTCACCT | |||
| BC5 | BC5_Fw | TCGAGCTCACAATATCAGCATT | 63 | 795 |
| BC5_Rev | TGTCCTGGAAATATCTTGAGCTGT | |||
| BC4 | BC4_Fw | ACTCGACGCGACATTGAAGT | 63 | 797 |
| BC4_Rev | ACACACTCCGTACGTTGCTT | |||
| PCR amplification of B. cinerea mating type idiomorphs | ||||
| MAT-1-1 | MAT-1-1_Fw | AAGCTTCGATGACCCTTTGA | 681 | |
| MAT-1-2 | MAT-1-2_Fw | TCGTTGCAGTCTCAGAATTGA | 56 | 486 |
| MAT_1_Rev | CGAACCGATCTCTGGTGGAG | |||
| PCR detection of B. cinerea H272R allele in the SdhB locusd | ||||
| SdhB-H272R | H272R-Fw | GGCAGCTTTGGATAACAGCATGAGTTTGTACAGATGGC | 67 | 600 |
| H272R-Rev | GCCATTTCCTTCTTAATCTCCGC | |||
| Tetra-ARMS PCR detection of H272Y allele in the B. cinerea SdhB locus | ||||
| SdhB-H272Y | Y_Out_Fw | ATCGTAAGAAGCTTGATGGACTTTACGA | 64.5 | 292 |
| Y_Out_Rev | TAGAAAGCCATTTCCTTCTTAATCTCCG | |||
| Y_C | TGGATAACAGCATGAGTTTGTACAGATTTC | 119b | ||
| Y_T | ACATGTCCTCGAGCAGTTGAGAATAGTTTA | 233c |
The final concentrations of each of outer and inner primers were 0.5 and 2 μM, respectively.
Allele H272 (wild allele).
Allele H272Y (resistant allele). Primers were designed with the Primer1 Tetra ARMS-PCR online tool (http://primer1.soton.ac.uk/primer1.html) (102).
Primers were designed with Primer-BLAST software (National Center for Biotechnology Information, Bethesda, MD). All protocols followed the manufacturer’s recommendations (GenScript USA, Inc., Piscataway, NJ) for a 25-μl (except for the SdhB-H272R total volume = 12.5 μl) reaction total volume.
Within-population diversity was assessed for each of two genetic populations defined with Structure analysis (see Results). Gene diversity was estimated using Nei’s unbiased gene diversity (Hexp) (83) and allelic richness (Ar) with rarefaction. Genotypic evenness (E5) (84–86), clonal fraction of multilocus genotypes (MLGs; 1-MLG/N), Shannon-Wiener index (H) (87), and Stoddart and Taylor’s index (G) (88). Indices of MLG diversity were estimated with the number of MLGs observed. The significance of H, G, and E5 was established with bootstrap resampling. Genotypic richness was computed with the number of expected MLGs (eMLGs) using rarefaction. Using the clone corrected data set, an unbiased index of multilocus linkage disequilibrium was assessed with standardized index of association (rd) over all SSR loci. Deviation of rd from 0, corresponding to a null hypothesis of complete panmixia, was estimated using 999 permutations. All analyses were computed with Poppr v.2.8.1 (89) and PopGenReport v.3.0.4 (90) R packages in R Studio v.1.1.463 (RStudio Integrated Development for R; RStudio, Inc., Boston, MA). In addition, the distribution of B. cinerea mating types was determined by multiplex PCR of mating idiomorphs, using primers developed in this study (Table 7). Chi-square (χ2) tests were used to test the hypothesis of idiomorphs 1:1 ratios in sampled populations. Distributions were considered significantly different from 1:1 at P = 0.05.
Population structure of B. cinerea infecting small fruit in the PNW.
To examine the genetic structure of B. cinerea populations infecting small fruit, the Bayesian assignment approach implemented in Structure v.2.3.4 (91) and discriminant analysis of principal components (DAPC) implemented in R package ADEGENET (92) were employed with the clone corrected data set (n = 34 fields). For structure, the admixed model was used with a 50,000 burn-in period and 500,000 Markov chain Monte Carlo iterations. The number of underlying groups (K) varied from 1 to 10 and replicated ten times. The optimal K was estimated using the method of Evanno et al. (93) with STRUCTURE HARVESTER (94). Clustering membership coefficients at the optimal K were averaged across ten replicates using CLUMPP v.1.1.2 (95). For DAPC, the optimal k (92) was chosen by analyzing DAPC outputs for each k from 6 to 15 and calculating the greatest proportion of isolates assigned to each of the clusters based on a membership probability of ≥75%. Genetic differentiation among isolates assigned to k clusters in DAPC and K-groups in Structure was tested with AMOVA. Relationships among B. cinerea MLGs were visualized with minimum spanning network (MSN) using Bruvo’s genetic distance (96) with the Poppr R package (89). In MSN analysis, B. cinerea isolates were assigned to populations using DAPC clusters.
The relative contributions of host and location to the genetic variance were quantified using hierarchical AMOVA in ARLEQUIN 3.5 (97) with 10,000 data permutations and 10,000 pairwise populations permutations. Significance of tests was calculated with 10,100 data permutations. Population structure on a farm scale was assessed using pairwise comparisons of Slatkin’s Rst (98). Pairwise Rst values were also plotted against geographic distances between fields to test for isolation by distance using a Mantel test in GenAlEx v.6.5 (99) with 1,000 permutations among all fields (n = 34) and separately among fields in WA (n = 29) and OR (n = 5).
Distribution of alleles under selection and relationships among population diversities and resistance frequencies.
All B. cinerea isolates from WA fields were screened for sensitivity to the SDHI fungicide boscalid using in vitro assays. Mycelial plugs were cultured on PDA amended with a discriminatory dose of 5 μg/ml formulated boscalid (Endura 70% [wt/wt]; BASF Corporation, Ludwigshafen, Germany) and 100 μg/ml of salicylhydroxamic acid (100). All assays were replicated once, and mycelial measurements were obtained after 48 h of growth at 25°C under a 12-h photoperiod. It was previously determined that under these conditions, B. cinerea isolates that grew <80% on 5 μg/ml discriminatory boscalid concentration were fully inhibited by field rates of the fungicide applied to detached raspberry fruit. Conidial germination and germ tube elongation of these isolates was also completely inhibited on field rates of formulated boscalid applied to agar plates (T. L. Peever, unpublished data). Sequences of the SdhB and SdhD target genes of such isolates revealed the absence of mutations associated with boscalid resistance either in B or D subunits of the SDH enzyme complex, thereby confirming their sensitivity to the fungicide (data not shown). In order to detect mutations conferring resistance to boscalid in studied populations, partial sequencing of the SdhB gene was performed for 20 randomly chosen resistant isolates using previously published primers (75). Among 20 sequences, two types of amino acid substitutions encoded by codon 272 of the SdhB gene were detected: H272R and H272Y. Therefore, all isolates were screened for resistance alleles in the SdhB gene using PCR assays developed in this study (Table 7).
Relationships between boscalid resistance frequencies and population diversity indices in each field estimated with microsatellite markers (Hexp, Ar, H, G, E5, clonal fraction, and eMLGs) were tested with Pearson’s correlation coefficient (Pearson’s r). The significance was assessed with a t distribution.
Test of association between alleles under selection and genetic background.
The hypothesis of boscalid resistance alleles being independent of genetic backgrounds was tested using pairwise linkage disequilibrium (rd) with 999 permutations among SSR alleles alone and among SSR and alleles under selection in Poppr v.2.8.1 (89). The significance of the test was determined by bootstrapping the rd SSR output data with 1,000 replications without replacement. The SSR rd distribution was compared to rd distribution of SSR alleles and alleles under selection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Washington Red Raspberry Commission and The Northwest Center for Small Fruit Research for funding this work.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hawkins NJ, Bass C, Dixon A, Neve P. 2019. The evolutionary origins of pesticide resistance. Biol Rev 94:135–155. doi: 10.1111/brv.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald BA. 1997. The population genetics of fungi: tools and techniques. Phytopathology 87:448–453. doi: 10.1094/PHYTO.1997.87.4.448. [DOI] [PubMed] [Google Scholar]
- 3.Milgroom MG, Fry WE. 1997. Contributions of population genetics to plant disease epidemiology and management. Adv Bot Res 24:1–30. doi: 10.1016/S0065-2296(08)60069-5. [DOI] [Google Scholar]
- 4.Walker A-S, Gladieux P, Decognet V, Fermaud M, Confais J, Roudet J, Bardin M, Bout A, Nicot PC, Poncet C, Fournier E. 2015. Population structure and temporal maintenance of the multihost fungal pathogen Botrytis cinerea: causes and implications for disease management. Environ Microbiol 17:1261–1274. doi: 10.1111/1462-2920.12563. [DOI] [PubMed] [Google Scholar]
- 5.Grunwald NJ, Goss EM. 2011. Evolution and population genetics of exotic and reemerging pathogens: traditional and novel tools and approaches. Annu Rev Phytopathol 49:249–267. doi: 10.1146/annurev-phyto-072910-095246. [DOI] [PubMed] [Google Scholar]
- 6.Croll D, McDonald BA. 2017. The genetic basis of local adaptation for pathogenic fungi in agricultural ecosystems. Mol Ecol 26:2027–2040. doi: 10.1111/mec.13870. [DOI] [PubMed] [Google Scholar]
- 7.Peever TL, Olsen L, Ibanez A, Timmer LW. 2000. Genetic differentiation and host specificity among populations of Alternaria spp. causing brown spot of grapefruit and tangerine × grapefruit hybrids in Florida. Phytopathology 90:407–414. doi: 10.1094/PHYTO.2000.90.4.407. [DOI] [PubMed] [Google Scholar]
- 8.Giraud T, Villaréal LMMA, Austerlitz F, Le Gac M, Lavigne C. 2006. Importance of the life cycle in sympatric host race formation and speciation of pathogens. Phytopathology 96:280–287. doi: 10.1094/PHYTO-96-0280. [DOI] [PubMed] [Google Scholar]
- 9.Goss EM, Larsen M, Chastagner GA, Givens DR, Grunwald NJ. 2009. Population genetic analysis infers migration pathways of Phytophthora ramorum in US nurseries. PLoS Pathog 5:e1000583. doi: 10.1371/journal.ppat.1000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert S, Ravigne V, Zapater MF, Abadie C, Carlier J. 2012. Contrasting introduction scenarios among continents in the worldwide invasion of the banana fungal pathogen Mycosphaerella fijiensis. Mol Ecol 21:1098–1114. doi: 10.1111/j.1365-294X.2011.05432.x. [DOI] [PubMed] [Google Scholar]
- 11.Anthony NM, Brown JK, Markham PG, Ffrench-Constan RH. 1995. Molecular analysis of cyclodiene resistance-associated mutations among populations of the sweet-potato whitefly Bemisia-Tabaci. Pestic Biochem Phys 51:220–228. doi: 10.1006/pest.1995.1022. [DOI] [Google Scholar]
- 12.Andreev D, Kreitman M, Phillips TW, Beeman RW, Ffrench-Constant RH. 1999. Multiple origins of cyclodiene insecticide resistance in Tribolium castaneum (Coleoptera: Tenebrionidae). J Mol Evol 48:615–624. doi: 10.1007/pl00006504. [DOI] [PubMed] [Google Scholar]
- 13.Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, Tijet N, Perry T, Heckel D, Batterham P, Feyereisen R, Wilson TG, Ffrench-Constant RH. 2002. A single P450 allele associated with insecticide resistance in Drosophila. Science 297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- 14.Anstead JA, Williamson MS, Denholm I. 2005. Evidence for multiple origins of identical insecticide resistance mutations in the aphid Myzus persicae. Insect Biochem Mol Biol 35:249–256. doi: 10.1016/j.ibmb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Nardi F, Carapelli A, Vontas JG, Dallai R, Roderick GK, Frati F. 2006. Geographical distribution and evolutionary history of organophosphate-resistant Ace alleles in the olive fly (Bactrocera oleae). Insect Biochem Mol Biol 36:593–602. doi: 10.1016/j.ibmb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Bernasconi P, Woodworth AR, Rosen BA, Subramanian MV, Siehl DL. 1995. A naturally occurring point mutation confers broad range tolerance to herbicides that target acetolactate synthase. J Biol Chem 270:17381–17385. doi: 10.1074/jbc.270.29.17381. [DOI] [PubMed] [Google Scholar]
- 17.Delye C, Straub C, Michel S, Le Corre V. 2004. Nucleotide variability at the acetyl coenzyme A carboxylase gene and the signature of herbicide selection in the grass weed Alopecurus myosuroides (Huds.). Mol Biol Evol 21:884–892. doi: 10.1093/molbev/msh095. [DOI] [PubMed] [Google Scholar]
- 18.Bass C, Denholm I, Williamson MS, Nauen R. 2015. The global status of insect resistance to neonicotinoid insecticides. Pestic Biochem Physiol 121:78–87. doi: 10.1016/j.pestbp.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Barlow M, Hall BG. 2003. Experimental prediction of the natural evolution of antibiotic resistance. Genetics 163:1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall BG, Salipante SJ, Barlow M. 2004. Independent origins of subgroup B1+B2 and subgroup B3 metallo-β-lactamases. J Mol Evol 59:133–141. doi: 10.1007/s00239-003-2572-9. [DOI] [PubMed] [Google Scholar]
- 21.McDonald BA, Mundt CC. 2016. How knowledge of pathogen population biology informs management of Septoria tritici blotch. Phytopathology 106:948–955. doi: 10.1094/PHYTO-03-16-0131-RVW. [DOI] [PubMed] [Google Scholar]
- 22.Delmas CEL, Dussert Y, Delière L, Couture C, Mazet ID, Richart Cervera S, Delmotte F. 2017. Soft selective sweeps in fungicide resistance evolution: recurrent mutations without fitness costs in grapevine downy mildew. Mol Ecol 26:1936–1951. doi: 10.1111/mec.14006. [DOI] [PubMed] [Google Scholar]
- 23.Milgroom MG. 2015. Selection for fungicide resistance, p 112–116. In Population biology of plant pathogens: genetics, ecology, and evolution. American Phytopathological Society, St Paul, MN. [Google Scholar]
- 24.Hahn MJ. 2014. The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J Chem Biol 7:133–141. doi: 10.1007/s12154-014-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charlesworth B. 2009. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet 10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 26.Pennings PS, Hermisson J. 2006. Soft sweeps II: molecular population genetics of adaptation from recurrent mutation or migration. Mol Biol Evol 23:1076–1084. doi: 10.1093/molbev/msj117. [DOI] [PubMed] [Google Scholar]
- 27.Sierotzki H, Wullschleger J, Gisi U. 2000. Point mutation in cytochrome b gene conferring resistance to strobilurin fungicides in Erysiphe graminis f. sp. tritici field isolates. Pestic Biochem Phys 68:107–112. doi: 10.1006/pest.2000.2506. [DOI] [Google Scholar]
- 28.Olaya G, Holm A. 2001. Sensitivity of Didymella bryoniae isolates to azoxystrobin. Phytopathology 91:S67. [Google Scholar]
- 29.Ishii H, Fraaije BA, Sugiyama T, Noguchi K, Nishimura K, Takeda T, Amano T, Hollomon DW. 2001. Occurrence and molecular characterization of strobilurin resistance in cucumber powdery mildew and downy mildew. Phytopathology 91:1166–1171. doi: 10.1094/PHYTO.2001.91.12.1166. [DOI] [PubMed] [Google Scholar]
- 30.Gisi U, Sierotzki H, Cook A, McCaffery A. 2002. Mechanisms influencing the evolution of resistance to Qo inhibitor fungicides. Pest Manag Sci 58:859–867. doi: 10.1002/ps.565. [DOI] [PubMed] [Google Scholar]
- 31.Baumler S, Felsenstein FG, Schwarz G. 2003. CAPS and DHPLC analysis of a single nucleotide polymorphism in the cytochrome b gene conferring resistance to strobilurin in field isolates of Blumeria graminis f. sp. hordei. J Phytopathol 151:149–152. doi: 10.1046/j.1439-0434.2003.00699.x. [DOI] [Google Scholar]
- 32.Kim YS, Dixon EW, Vincelli P, Farman ML. 2003. Field resistance to strobilurin (QoI) fungicides in Pyricularia grisea caused by mutations in the mitochondrial cytochrome b gene. Phytopathology 93:891–900. doi: 10.1094/PHYTO.2003.93.7.891. [DOI] [PubMed] [Google Scholar]
- 33.Ma Z, Felts D, Michailides TJ. 2003. Resistance to azoxystrobin in Alternaria isolates from pistachio in California. Pestic Biochem Phys 77:66–74. doi: 10.1016/j.pestbp.2003.08.002. [DOI] [Google Scholar]
- 34.Ma Z, Michailides TJ. 2005. Advances in understanding molecular mechanisms of fungicide resistance and molecular detection of resistant genotypes in phytopathogenic fungi. Crop Prot 24:853–863. doi: 10.1016/j.cropro.2005.01.011. [DOI] [Google Scholar]
- 35.Chen W-J, Delmotte F, Richard-Cervera S, Douence L, Greif C, Corio-Costet M-F. 2007. At least two origins of fungicide resistance in grapevine downy mildew populations. Appl Environ Microbiol 73:5162–5172. doi: 10.1128/AEM.00507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torriani SFF, Brunner PC, McDonald BA, Sierotzki H. 2009. QoI resistance emerged independently at least 4 times in European populations of Mycosphaerella graminicola. Pest Manag Sci 65:155–162. doi: 10.1002/ps.1662. [DOI] [PubMed] [Google Scholar]
- 37.Estep LK, Torriani SFF, Zala M, Anderson NP, Flowers MD, Mcdonald BA, Mundt CC, Brunner PC. 2015. Emergence and early evolution of fungicide resistance in North American populations of Zymoseptoria tritici. Plant Pathol 64:961–971. doi: 10.1111/ppa.12314. [DOI] [Google Scholar]
- 38.Avenot HF, Michailides TJ. 2010. Progress in understanding molecular mechanisms and evolution of resistance to succinate dehydrogenase inhibiting (SDHI) fungicides in phytopathogenic fungi. Crop Prot 29:643–651. doi: 10.1016/j.cropro.2010.02.019. [DOI] [Google Scholar]
- 39.Sierotzki H, Scalliet G. 2013. A review of current knowledge of resistance aspects for the next-generation succinate dehydrogenase inhibitor fungicides. Phytopathology 103:880–887. doi: 10.1094/PHYTO-01-13-0009-RVW. [DOI] [PubMed] [Google Scholar]
- 40.Williamson B, Tudzynski B, Tudzynski P, van Kan J. 2007. Botrytis cinerea: the cause of grey mould disease. Mol Plant Pathol 8:561–580. doi: 10.1111/j.1364-3703.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 41.Elad Y, Pertot I, Prado AMC, Stewart A. 2016. Plant hosts of Botrytis spp, p415–458. In Fillinger S, Elad Y (ed), Botrytis: the fungus, the pathogen, and its management in agricultural systems. Springer Publishing, New York, NY. [Google Scholar]
- 42.Martinez F, Blancard D, Lecomte P, Levis C, Dubos B, Fermaud M. 2003. Phenotypic differences between vacuma and transposa subpopulations of Botrytis cinerea. Eur J Plant Pathol 109:479–488. doi: 10.1023/A:1024222206991. [DOI] [Google Scholar]
- 43.Droby S, Lichter A. 2007. Post-harvest botrytis infection: etiology, development, and management, p 349–367. In Elad Y, Williamson B, Tudzynski P, Delen N (ed), Botrytis: biology, pathology, and control. Springer, Dordrecht, Netherlands. [Google Scholar]
- 44.Dean R, van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. 2012. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozhar O, Peever TL. 2018. How does Botrytis cinerea infect red raspberry? Phytopathology 108:1287–1298. doi: 10.1094/PHYTO-01-18-0016-R. [DOI] [PubMed] [Google Scholar]
- 46.Giraud T, Fortini D, Levis C, Leroux P, Brygoo Y. 1997. RFLP markers show genetic recombination in Botryotina fuckeliana (Botrytis cinerea) and transposable elements reveal two sympatric species. Mol Biol Evol 14:1177–1185. doi: 10.1093/oxfordjournals.molbev.a025727. [DOI] [PubMed] [Google Scholar]
- 47.Karchani-Balma S, Gautier A, Raies A, Fournier E. 2008. Geography, plants, and growing systems shape the genetic structure of Tunisian Botrytis cinerea populations. Phytopathology 98:1271–1279. doi: 10.1094/PHYTO-98-12-1271. [DOI] [PubMed] [Google Scholar]
- 48.Fournier E, Giraud T. 2008. Sympatric genetic differentiation of a generalist pathogenic fungus, Botrytis cinerea, on two different host plants, grapevine and bramble. J Evol Biol 21:122–132. doi: 10.1111/j.1420-9101.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 49.Walker A-S. 2016. Diversity within and between species of Botrytis, p 91–125. In Fillinger S, Elad Y (ed), Botrytis: the fungus, the pathogen, and its management in agricultural systems. Springer Publishing, New York, NY. [Google Scholar]
- 50.Rajaguru BAP, Shaw MW. 2010. Genetic differentiation between hosts and locations in populations of latent Botrytis cinerea in southern England. Plant Pathol 59:1081–1090. doi: 10.1111/j.1365-3059.2010.02346.x. [DOI] [Google Scholar]
- 51.Wessels BA, Linde CC, Fourie PH, Mostert L. 2016. Genetic population structure and fungicide resistance of Botrytis cinerea in pear orchards in the Western Cape of South Africa. Plant Pathol 65:1473–1483. doi: 10.1111/ppa.12523. [DOI] [Google Scholar]
- 52.Ma Z, Michailides TJ. 2005. Genetic structure of Botrytis cinerea populations from different host plants in California. Plant Dis 89:1083–1089. doi: 10.1094/PD-89-1083. [DOI] [PubMed] [Google Scholar]
- 53.Munoz G, Hinrichsen P, Brygoo Y, Giraud T. 2002. Genetic characterization of Botrytis cinerea populations in Chile. Mycol Res 106:594–601. doi: 10.1017/S0953756202005981. [DOI] [Google Scholar]
- 54.Leyronas C, Bryone F, Duffaud M, Troulet C, Nicot PC. 2015. Assessing host specialization of Botrytis cinerea on lettuce and tomato by genotypic and phenotypic characterization. Plant Pathol 64:119–127. doi: 10.1111/ppa.12234. [DOI] [Google Scholar]
- 55.Isenegger DA, Macleod WJ, Ford R, Taylor P. 2008. Genotypic diversity and migration of clonal lineages of Botrytis cinerea from chickpea fields of Bangladesh inferred by microsatellite markers. Plant Pathol 57:967–973. doi: 10.1111/j.1365-3059.2008.01885.x. [DOI] [Google Scholar]
- 56.Adjebli A, Leyronas C, Aissat K, Nicot PC. 2015. Comparison of Botrytis cinerea populations collected from tomato greenhouses in northern Algeria. J Phytopathol 163:124–132. doi: 10.1111/jph.12289. [DOI] [Google Scholar]
- 57.Campia P, Venturini G, Moreno SP, Casati P, Toffolatti SL. 2017. Genetic structure and fungicide sensitivity of Botrytis cinerea populations isolated from grapevine in northern Italy. Plant Pathol 66:890–899. doi: 10.1111/ppa.12643. [DOI] [Google Scholar]
- 58.Bardin M, Leyronas C, Troulet C, Morris CE. 2018. Striking similarities between Botrytis cinerea from non-agricultural and from agricultural habitats. Front Plant Sci 9:1820. doi: 10.3389/fpls.2018.01820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Miccolis Angelini RM, Pollastro S, Faretra F. 2016. Genetics of Botrytis cinerea, p 127–148. In Fillinger S, Elad Y (ed), Botrytis: the fungus, the pathogen, and its management in agricultural systems. Springer Publishing, New York, NY. [Google Scholar]
- 60.Polat İ, Baysal Ö, Mercati F, Gümrükcü E, Sülü G, Kitapcı A, Araniti F, Carimi F. 2018. Characterization of Botrytis cinerea isolates collected on pepper in southern Turkey by using molecular markers, fungicide resistance genes and virulence assay. Infect Genet Evol 60:151–159. doi: 10.1016/j.meegid.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 61.Walker AS, Ravigne V, Rieux A, Ali S, Carpentier F, Fournier E. 2017. Fungal adaptation to contemporary fungicide applications: the case of Botrytis cinerea populations from Champagne vineyards (France). Mol Ecol 26:1919–1935. doi: 10.1111/mec.14072. [DOI] [PubMed] [Google Scholar]
- 62.Beever RE, Weeds PL. 2004. Taxonomy and genetic variation of Botrytis and Botryotinia, p 29–52. In Elad Y, Williamson B, Tudzynski P, Delan N (ed), Botrytis: biology, pathology, and control. Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
- 63.Zhan J, Torriani SFF, McDonald BA. 2007. Significant difference in pathogenicity between MAT1-1 and MAT1–2 isolates in the wheat pathogen Mycosphaerella graminicola. Fungal Genet Biol 5:339–346. doi: 10.1016/j.fgb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 64.Zheng Q, Hou R, Zhang J, Ma J, Wu Z, Wang G, Wang C, Xu J. 2013. The MAT locus genes play different roles in sexual reproduction and pathogenesis in Fusarium graminearum. PLoS One 8:e66980. doi: 10.1371/journal.pone.0066980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan MX, Zhu GN, Lin SY, Xian XY, Chang CQ, Xi PG, Shen W, Huang W, Cai E, Jiang Z, Deng YZ, Zhang L-H. 2016. The mating-type locus b of the sugarcane smut Sporisorium scitamineum is essential for mating, filamentous growth, and pathogenicity. Fungal Genet Biol 86:1–8. doi: 10.1016/j.fgb.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 66.Wang Q, Wang S, Xiong CL, James TY, Zhang XG. 2017. Mating-type genes of the anamorphic fungus Ulocladium botrytis affect both asexual sporulation and sexual reproduction. Sci Rep 7:7932. doi: 10.1038/s41598-017-08471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Yin Y, Hu P, Yu J, Xia W, Ge Q, Cao Q, Cui H, Yu X, Ye Z. 2019. Mating-type loci of Ustilago esculenta are essential for mating and development. Fungal Genet Biol 125:60–70. doi: 10.1016/j.fgb.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 68.Mercier A, Carpentier F, Duplaix C, Auger A, Pradier J-M, Viaud M, Gladieux P, Walker A-S. 2019. The polyphagous plant pathogenic fungus Botrytis cinerea encompasses host-specialized and generalist populations. Environ Microbiol 21:4808–4821. doi: 10.1111/1462-2920.14829. [DOI] [PubMed] [Google Scholar]
- 69.Carisse O. 2016. Epidemiology and aerobiology of Botrytis spp, p 127–148 In Fillinger S, Elad Y (ed), Botrytis: the fungus, the pathogen, and its management in agricultural systems. Springer Publishing, New York, NY. [Google Scholar]
- 70.Stammler G, Brix H-D, Glaettli A, Semar M, Schoefl U. 2007. Biological properties of the carboxamide boscalid, including recent studies on its mode of action, p 16–21. 16th International Congress of Plant Protection, Session 2A, Glasgow, United Kingdom. [Google Scholar]
- 71.Alberoni G, Ciriani A, Banorri M, Collina M, Brunelli A. 2011. Sensitivity to boscalid of Stemphylium vesicarium and Botrytis cinerea in Italy, p 273–277. In Dehne HW, Deising HB, Gisi U, Kuck KH, Russell PE, Lyr H (ed), Modern fungicides and antifungal compounds VI. DPG, Braunschweig, Germany. [Google Scholar]
- 72.Stammler G, Glättli A, Koch A, Schlehuber S. 2011. Mutations in the target protein conferring resistance to SDHI fungicides, p 195–198. In Dehne HW, Deising HB, Gisi U, Kuck KH, Russell PE, Lyr H (ed), Modern fungicides and antifungal compounds VI. DPG, Braunschweig, Germany. [Google Scholar]
- 73.Veloukas T, Leroch M, Hahn M, Karaoglanidis GS. 2011. Detection and molecular characterization of boscalid-resistant Botrytis cinerea isolates from strawberry. Plant Dis 95:1302–1307. doi: 10.1094/PDIS-04-11-0317. [DOI] [PubMed] [Google Scholar]
- 74.Walker A-S, Gredt M, Leroux P. 2011. Resistance to QoIs and SDHIs in populations of Botrytis cinerea, p 187–194. In Dehne HW, Deising HB, Gisi U, Kuck KH, Russell PE, Lyr H (ed), Modern fungicides and antifungal compounds VI. DPG, Braunschweig, Germany. [Google Scholar]
- 75.Yin YN, Kim YK, Xiao CL. 2011. Molecular characterization of boscalid resistance in field isolates of Botrytis cinerea from apple. Phytopathology 101:986–995. doi: 10.1094/PHYTO-01-11-0016. [DOI] [PubMed] [Google Scholar]
- 76.Weber RWS, Entrop A. 2017. Recovery of Botrytis strains with multiple fungicide resistance from raspberry nursery plants. Eur J Plant Pathol 147:933–936. doi: 10.1007/s10658-016-1046-z. [DOI] [Google Scholar]
- 77.Amiri A, Zuniga IA, Peres NA. 2018. Prevalence of Botrytis cryptic species in strawberry nursery transplants and strawberry and blueberry commercial fields in the eastern United States. Plant Dis 102:398–404. doi: 10.1094/PDIS-07-17-1065-RE. [DOI] [PubMed] [Google Scholar]
- 78.Laleve A, Fillinger S, Walker A-S. 2014. Fitness measurement reveals contrasting costs in homologous recombinant mutants of Botrytis cinerea resistant to succinate dehydrogenase inhibitors. Fungal Genet Biol 67:24–36. doi: 10.1016/j.fgb.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 79.Plesken C, Weber RWS, Rupp S, Leroch M, Hahn M. 2015. Botrytis pseudocinerea is a significant pathogen of several crop plants but susceptible to displacement by fungicide-resistant Botrytis cinerea strains. Appl Environ Microbiol 81:7048–7056. doi: 10.1128/AEM.01719-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fournier E, Giraud T, Loiseau A, Vautrin D, Estoup A, Solignac M, Cornuet JM, Brygoo Y. 2002. Characterization of nine polymorphic microsatellite loci in the fungus Botrytis cinerea (Ascomycota). Mol Ecol Notes 2:253–255. doi: 10.1046/j.1471-8286.2002.00207.x. [DOI] [Google Scholar]
- 81.Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier J-M, Quévillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun M-H, Dickman M. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guichoux E, Lagache L, Wagner S, Chaumeil P, Leger P, Lepais O, Lepoittevin C, Malausa T, Revardel E, Salin F, Petit RJ. 2011. Current trends in microsatellite genotyping. Mol Ecol Resour 11:591–611. doi: 10.1111/j.1755-0998.2011.03014.x. [DOI] [PubMed] [Google Scholar]
- 83.Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pielou EC. 1975. Ecological diversity. Wiley, New York, NY. [Google Scholar]
- 85.Ludwig JA, Reynolds JF. 1988. Statistical ecology: a primer in methods and computing. Wiley, New York, NY. [Google Scholar]
- 86.Grunwald NJ, Goodwin SB, Milgroom MG, Fry WE. 2003. Analysis of genotypic diversity data for populations of microorganisms. Phytopathology 93:738–746. doi: 10.1094/PHYTO.2003.93.6.738. [DOI] [PubMed] [Google Scholar]
- 87.Shannon CE. 2001. A mathematical theory of communication. Sigmobile Mob Comput Commun Rev 5:3–55. doi: 10.1145/584091.584093. [DOI] [Google Scholar]
- 88.Stoddart JA, Taylor JF. 1988. Genotypic diversity: estimation and prediction in samples. Genetics 118:705–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamvar ZN, Tabima JF, Grünwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adamack AT, Gruber B. 2014. PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5:384–387. doi: 10.1111/2041-210X.12158. [DOI] [Google Scholar]
- 91.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 93.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 94.Earl DA, vonHoldt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resour 4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 95.Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 96.Bruvo R, Michiels NK, D’Souza TG, Schulenburg H. 2004. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol Ecol 13:2101–2106. doi: 10.1111/j.1365-294X.2004.02209.x. [DOI] [PubMed] [Google Scholar]
- 97.Excoffier L, Lischer H. 2010. Arlequin suite v3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 98.Slatkin M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leroux P, Gredt M, Leroch M, Walker A-S. 2010. Exploring mechanisms of resistance to respiratory inhibitors in field strains of Botrytis cinerea, the causal agent of gray mold. Appl Environ Microbiol 76:6615–6630. doi: 10.1128/AEM.00931-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brownstein MJ, Carpten JD, Smith JR. 1996. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004–1006. 1008–1010. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- 102.Collins A, Ke X. 2012. Primer1: primer design web service for tetra-primer ARMS-PCR. Open Bioinforma J 6:55–58. doi: 10.2174/1875036201206010055. [DOI] [Google Scholar]
- 103.Simpson EH. 1949. Measurement of diversity. Nature 163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.