As d-amino acids play an important role in cell wall synthesis, we analyzed the effects of 19 different d-amino acids on L. lactis F44, demonstrating that d-Met and d-Phe can participate in peptidoglycan (PG) synthesis and improve the acid resistance and nisin yield of this strain. murF overexpression further increased the levels of d-Met and d-Phe incorporated into PG and contributed to the acid resistance of the strain. These findings suggest that d-Met and d-Phe can be incorporated into PG to improve the acid resistance and nisin yield of L. lactis, and this study provides new ideas for the enhancement of nisin production.
KEYWORDS: d-amino acid, Lactococcus lactis, acid resistance, cell wall, nisin
ABSTRACT
Lactococcus lactis encounters various environmental challenges, especially acid stress, during its growth. The cell wall can maintain the integrity and shape of the cell under environmental stress, and d-amino acids play an important role in cell wall synthesis. Here, by analyzing the effects of 19 different d-amino acids on the physiology of L. lactis F44, we found that exogenously supplied d-methionine and d-phenylalanine increased the nisin yield by 93.22% and 101.29%, respectively, as well as significantly increasing the acid resistance of L. lactis F44. The composition of the cell wall in L. lactis F44 with exogenously supplied d-Met or d-Phe was further investigated via a vancomycin fluorescence experiment and a liquid chromatography-mass spectrometry assay, which demonstrated that d-Met could be incorporated into the fifth position of peptidoglycan (PG) muropeptides and d-Phe could be added to the fourth and fifth positions. Moreover, overexpression of the PG synthesis gene murF further enhanced the levels of d-Met and d-Phe involved in PG and increased the survival rate under acid stress and the nisin yield of the strain. This study reveals that the exogenous supply of d-Met or d-Phe can change the composition of the cell wall and influence acid tolerance as well as nisin yield in L. lactis.
IMPORTANCE As d-amino acids play an important role in cell wall synthesis, we analyzed the effects of 19 different d-amino acids on L. lactis F44, demonstrating that d-Met and d-Phe can participate in peptidoglycan (PG) synthesis and improve the acid resistance and nisin yield of this strain. murF overexpression further increased the levels of d-Met and d-Phe incorporated into PG and contributed to the acid resistance of the strain. These findings suggest that d-Met and d-Phe can be incorporated into PG to improve the acid resistance and nisin yield of L. lactis, and this study provides new ideas for the enhancement of nisin production.
INTRODUCTION
Lactococcus lactis, widely used in the food industry, is a Gram-positive bacterium. Nisin, an antimicrobial peptide produced by L. lactis, is used as a safe food preservative and antimicrobial (1, 2). Many studies have focused on methods to increase nisin yield, such as metabolic regulation, fermentation optimization, and genetic modifications (2–8). It is widely known that L. lactis encounters several environmental stresses, especially lactate, during fermentation, and the acidic medium inhibits its growth and nisin production (9), so we previously utilized a range of strategies to improve the acid resistance of L. lactis, which contributes to increases in nisin yield (10, 11).
The cell wall, a reticular polymer located outside the cell membrane, can maintain the integrity and shape of the cell. In Gram-positive bacteria, the peptide polysaccharide layer is thick and provides anchor points for other components of the cell wall, such as wall phosphoric acid and capsule polysaccharide (12, 13). Peptidoglycan (PG), an essential component of the cell wall, is a polymer of N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc) cross-linked by peptide bridges. The d-Ala–d-Ala dipeptide (catalyzed by the d-alanine ligase Ddl) is incorporated into the monosaccharide tripeptide (generated by MurABCDE) through MurF (UDP–N-acetylmuramoyl–tripeptide–d-alanyl–d-alanine ligase). After the formation of monosaccharide pentapeptide, MraY catalyzes the transfer of phospho-MurNAc-pentapeptide to the lipid carrier undecaprenyl phosphate to form a lipid-linked N-acetylmuramic acid derivative (lipid I), and MurG couples GlcNAc to the C-4 hydroxyl of lipid I to generate the beta-linked disaccharide (lipid II). d-Asp is then attached to the third l-Lys to form a peptide bridge under the action of YxbA ligase (14). The membrane protein MurJ, Amj, or FtsW serves as the lipid II flippase that can transport lipid II across the membrane (15–17). The polymerization of lipid II to generate individual PG strands that are subsequently cross-linked to form PG layers is catalyzed by the penicillin-binding proteins and glycosyltransferases (18). The newly synthesized PG matures through the modification of synthetases and hydrolases (such as l,d-transpeptidase and carboxypeptidase) and is eventually hydrolyzed by PG hydrolases during cell growth and division. Partially hydrolyzed PG fragments are transported to the cytoplasm and reused in lipid II biosynthesis during PG recycling (19, 20).
Our previous studies showed that the cell wall modification could improve the acid resistance of L. lactis. The overexpression of asnH increased the acylation level of the cell wall peptide bridge d-Asp, helped to maintain the rigidity of PG, and promoted the acid tolerance and nisin yield of L. lactis (10). Moreover, the increase in the O-acetylation of MurNAc and the N-acetylation of GlcNAc in the cell wall contributed to the integrity of the cell wall in L. lactis F44 and increased its acid resistance (11).
The d-amino acids involved in the PG synthesis are normally d-alanine and d-glutamate in bacteria. However, Lam and coworkers and Cava and coworkers found that Vibrio cholerae could produce d-methionine and d-leucine, called noncanonical d-amino acids (NCDAAs), which influenced the composition, amount, and flexibility of PG (21, 22). The presence of NCDAAs in PG was confirmed by adding NCDAAs to the medium (23–25). Other studies found that Gly was exchanged with d-Ala at the first, fourth, or fifth position of the peptide chain in bacterial PG (26) and that d-Met, d-Trp, or d-Phe replaced d-Ala at the fourth position of the peptide chain in the PG of Escherichia coli. l-Isomer addition to the external source affected neither the structure of the PG nor the synthesis (27). So far, NCDAAs have been found to participate in PG synthesis in four ways: (i) l,d-carboxypeptidase catalyzes exogenous NCDAAs to exchange with the d-Ala residue of a disaccharide tetrapeptide (27, 28); (ii) l,d-transpeptidase, like LdtA and LdtB, directly replaces d-Ala with NCDAAs at the fourth or fifth position of the cross-linked peptide chain (22); (iii) d,d-transpeptidase has the potential to mediate NCDAAs to replace d-Ala at the fifth position of the stem peptide (24); and (iv) NCDAAs participate in PG synthesis through two steps catalyzed by the enzymes Ddl and MurF (22).
Although NCDAAs which can be incorporated into the PG have been extensively studied in several Gram-negative strains, little information is available about the effects of NCDAAs on the Gram-positive species L. lactis. This study gives insights into the pivotal role of d-amino acids in acid resistance and nisin yield and provides biological evidence that d-Met and d-Phe can participate in cell wall synthesis in L. lactis F44.
RESULTS
Effects of different d-amino acids on acid resistance of L. lactis F44.
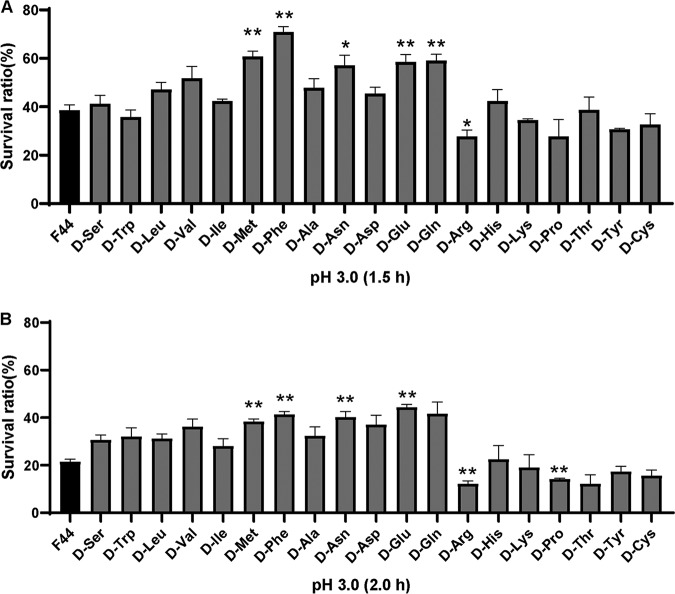
To investigate the effects of various d-amino acids on L. lactis F44, we added the 19 different d-amino acids to the medium separately and detected the acid resistance of F44 in each sample. The experimental groups were L. lactis F44 with the separate addition of various d-amino acids (50 mM) in the medium, and the control group was F44 without any d-amino acid addition. In the mid-log phase, the cells were exposed to pH 3.0 for 1.5 or 2 h. As shown in Fig. 1, the survival rates of F44 added with d-Met, d-Phe, d-Asn, or d-Glu/Gln were significantly higher than that of the control (P < 0.01). Among them, the survival rate of the strain with added d-Phe was the highest—70.82% ± 2.26% and 41.33% ± 1.25% at 1.5 h and 2 h, respectively, which were 1.84 and 1.92 times higher than that of the control (38.47% ± 2.37% at 1.5 h; 21.52% ± 1.00% at 2.0 h). d-Met addition increased the survival rate by 58.33% and 79.53% after 1.5 h and 2 h of acid stress, respectively. Therefore, the separate addition of d-Met, d-Phe, d-Asn, and d-Glu in the medium could improve the survival of F44 under acid stress.
FIG 1.
(A) Survival rate of L. lactis F44 with the addition of various d-amino acids after 1.5 h acid shock. (B) Survival rate of L. lactis F44 with the addition of various d-amino acids after 2.0 h acid shock. Error bars indicate standard deviations (SD) for three independent experiments. *, P < 0.01; **, P < 0.001 (t test).
The influence of d-amino acids on nisin yield is different from that of l-amino acids.
The growth properties, optical density at 600 nm (OD600), pH, and nisin production per cell were detected during fermentation. With the separate addition of 19 different d-amino acids, the final OD600 value of L. lactis F44 was lower, and correspondingly the pH value was higher, than that of the control (see Fig. S1 and S2 in the supplemental material). Notably, d-Cys significantly inhibited strain growth. Figure 2 and Fig. S3 show that the addition of d-Met, d-Phe, d-Asn, d-Trp, or d-Leu to the medium significantly increased nisin production. Among them, d-Met addition enhanced the nisin yield per cell to 5.99 × 10−7 ± 1.08 × 10−8 IU/CFU, which was 93.22% higher than that of the control (3.10 × 10−7 ± 2.02 × 10−8 IU/CFU), and the yield obtained with the d-Phe addition (6.24 × 10−7 ± 2.65 × 10−8 IU/CFU) was 101.29% higher. Therefore, the addition of d-Met or d-Phe could improve the nisin yield of L. lactis F44.
FIG 2.
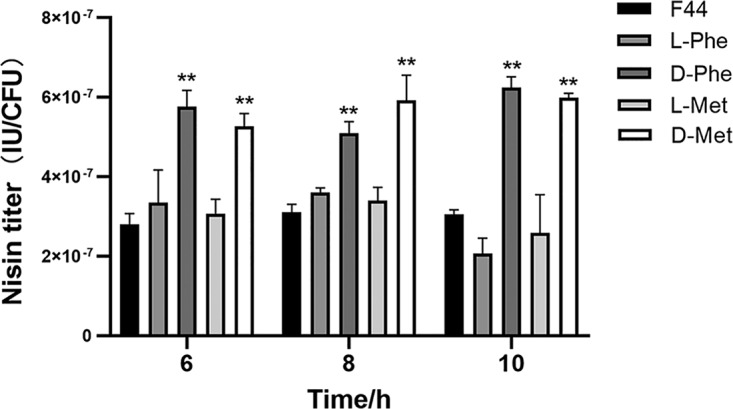
Effects of d-amino acids and l-amino acids on the nisin titer per cell of L. lactis F44. Samples were taken every 2 h from 6 to 10 h. Error bars indicate the SD for three independent experiments. **, P < 0.01 (t test).
l-Amino acids were added to the fermentation medium to investigate the effects of l-Met and l-Phe on nisin yield, which helped to illuminate the question of whether d-Met and d-Phe could be converted to l-Met and l-Phe to influence F44 nisin yield. As shown in Fig. 2, the nisin yield of L. lactis F44 with the 50 mM l-Met or l-Phe addition did not show an obvious change, which was different from the result with the d-amino acid addition.
Preliminary verification of d-Met and d-Phe involved in PG synthesis by fluorescence detection.
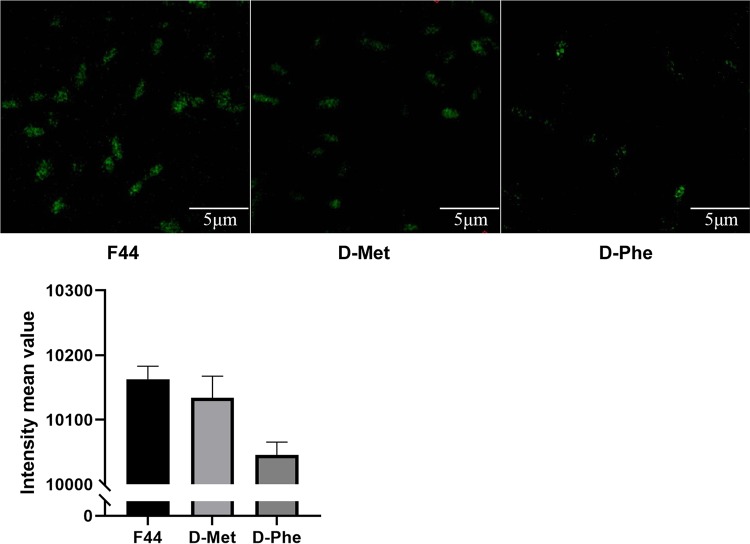
NCDAAs were found to be involved in the formation of PG in the fourth or fifth position of the peptide chain in V. cholerae (22). Here, to investigate whether d-Met and d-Phe could be incorporated into the PG muropeptides to influence the acid resistance and nisin yield of L. lactis, a fluorescent derivative of vancomycin (Van-FL) was used to detect the insertion of NCDAAs (29, 30). According to the previous study, Van-FL could interact with d-Ala–d-Ala at the fourth and fifth positions of the peptide chain through hydrogen bonding to form stable complexes (31), and its binding could be initially observed by the fluorescence of cell walls (21). Given that Van-FL could inhibit the growth of this strain, the tolerance of L. lactis F44 to Van-FL was determined, and 300 ng/ml was found to be a relatively high tolerance concentration (at which no visible inhibition of growth occurred) (see Fig. S4 in the supplemental material).
A fluorescence detection assay was performed with 300 ng/ml Van-FL. As shown in Fig. 3, the fluorescence intensities of the strains with either d-Met or d-Phe in the medium were weaker than that of the control, and the intensity of the sample with the d-Phe addition was the weakest. Therefore, we hypothesized that more d-Phe than d-Met was incorporated into the cell wall muropeptides, thus possibly leading to a more significant increase in acid resistance and nisin yield.
FIG 3.
Verification of NCDAAs involved in PG synthesis through fluorescence detection. Fluorescence intensity of L. lactis F44 was measured after growth in the medium containing d-Met and d-Phe for 6 h. The control group did not have any added d-amino acid. Van-FL (300 ng/ml) was used to detect the insertion of NCDAAs. The fluorescence value is the fluorescence intensity of a single cell, and the fluorescence values of 50 cells in the field were taken to calculate the mean value.
d-Met and d-Phe can be incorporated into the cell wall muropeptides, as verified by LC-MS.
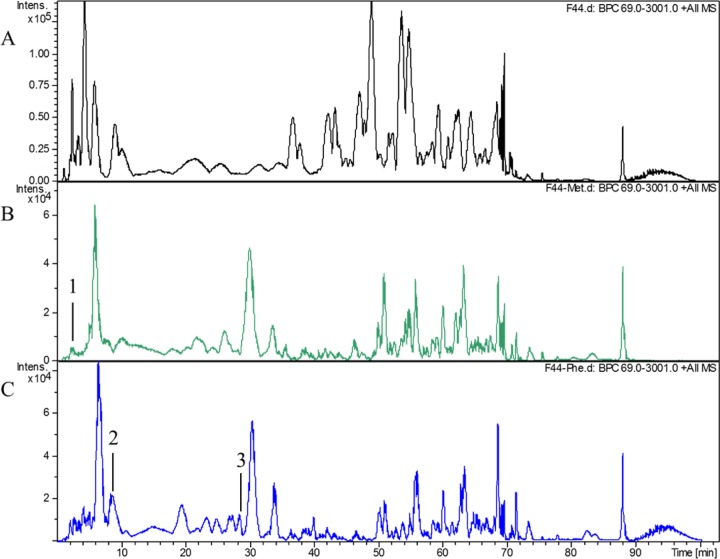
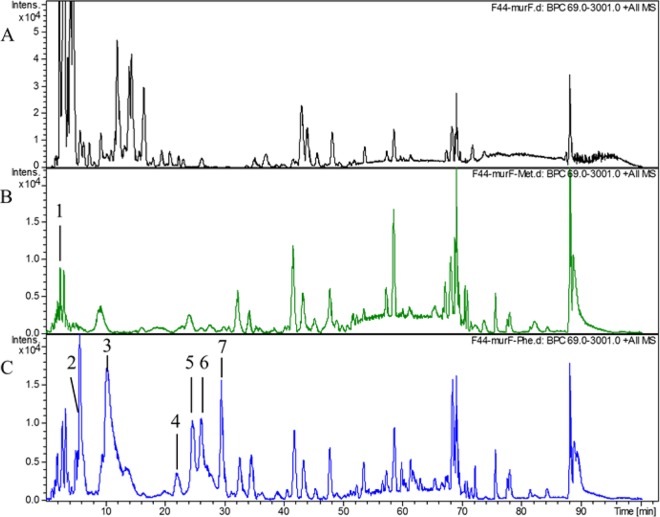
To test the above hypothesis, we extracted and detected the PG after L. lactis F44 had been grown in the fermentation medium containing d-Met or d-Phe by liquid chromatography-mass spectrometry (LC-MS). As shown in Fig. 4 and Table 1, the monomer PentaMet (N-acetylglucosamine–N-acetylmuramyl pentapeptide with the asparagine peptide bridge) was found in the d-Met sample, which indicated that d-Met was incorporated at the fifth position of muropeptides in PG. According to the previous study, d-Met could be incorporated at two locations within PG, namely, the fourth and fifth positions of muropeptides (22). However, in the present study, d-Met was found only at the fifth position of muropeptides, and it accounted for 0.42% of the total PG in L. lactis F44 (Fig. 4).
FIG 4.
Analysis of L. lactis F44 PG by LC-MS. (A) LC-MS analysis of L. lactis F44 without the addition of any d-amino acid. (B) LC-MS analysis of L. lactis F44 PG after growth in medium containing d-Met for 6 h. Peak 1 represents d-Met binding to a disaccharide pentapeptide with the asparagine-peptide bridge, namely, PentaMet (N-acetylglucosamine-N-acetylmuramyl pentapeptide with the asparagine peptide bridge). The structures and parameters are shown in Table 1. (C) LC-MS analysis of L. lactis F44 PG after growth in medium containing d-Phe for 6 h. Peak 2 represents d-Phe binding to PentaPhe, a disaccharide (N-acetylglucosamine-N-acetylmuramyl) pentapeptide with the asparagine peptide bridge. Peak 3 represents d-Phe binding to a TetraPhe, a disaccharide (N-acetylglucosamine-N-acetylmuramyl) tetrapeptide with the aspartate-peptide bridge. The structures and parameters are shown in Table 1.
TABLE 1.
Analysis of L. lactis F44 PG by LC-MS
| Additive | Structurea | Mol wt | Retention time (min) | Area (%) |
|---|---|---|---|---|
| d-Met | PentaMet | 1,140.6 | 2.5 | 0.42 |
| d-Phe | PentaPhe | 1,156.6 | 8.7 | 6.75 |
| d-Phe | TetraPhe | 1,074.5 | 28.3 | 0.85 |
PentaMet, G–M–l-Ala–d-Gln–l-Lys (d-Asn)–d-Ala–d-Met; PentaPhe, G–M–l-Ala–d-Gln–l-Lys (d-Asn)–d-Ala–d-Phe; TetraPhe, G–M–l-Ala–d-Gln–l-Lys (d-Asp)–d-Phe (d-Asn or d-Asp is the peptide bridge bound to l-Lys, G is N-acetylglucosamine [GlcNAc], and M is N-acetylmuramic acid [MurNAc]).
The monomers TetraPhe (N-acetylglucosamine–N-acetylmuramyl tetrapeptide with the aspartate peptide bridge) and PentaPhe (N-acetylglucosamine–N-acetylmuramyl pentapeptide with the asparagine peptide bridge) were found in the d-Phe sample, indicating that d-Phe replaced the d-Ala in the fourth and fifth positions of muropeptides, respectively. In our analysis, TetraPhe and PentaPhe constituted 0.85% and 6.75% of total PG, respectively, demonstrating that the d-Phe sample showed a higher level of incorporation than the d-Met sample, which was consistent with the results of vancomycin fluorescence detection.
Fluorescence assay of recombinant strains L. lactis F44-ddl and F44-murF with the addition of d-amino acids.
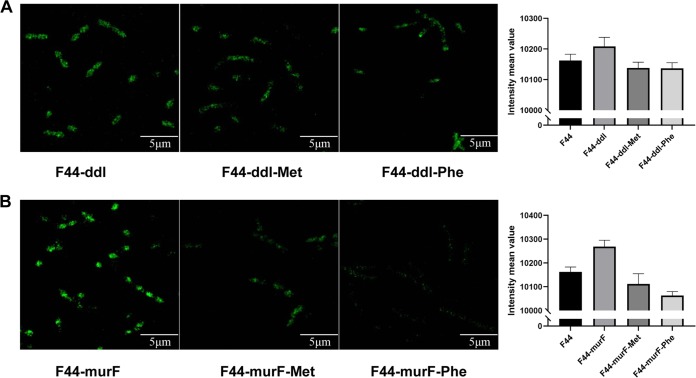
According to a previous study, two cell wall synthesis enzymes, Ddl and MurF, are related to the addition of NCDAAs into the fifth position of muropeptides (22). In the present work, the effects of the two enzymes on the incorporation of d-Phe and d-Met in PG synthesis were further explored. The recombinants L. lactis F44-ddl and F44-murF, which overexpress the ddl and murF genes, respectively, were constructed, and then a Van-FL fluorescence assay was performed.
As shown in Fig. 5, ddl or murF overexpression enhanced the binding of Van-FL to the tripeptides, leading to an increase in fluorescence intensity. Compared with F44, the increase in F44-murF was more significant than that in F44-ddl. Furthermore, the fluorescence intensity decreased when d-Met or d-Phe was added to the medium of F44-murF, and the influence of d-Phe was greater than that of d-Met. Therefore, it was suggested that murF or ddl overexpression could promote the conjugation of d-Ala–d-Ala with monosaccharide tripeptides to form monosaccharide pentapeptides. Notably, murF influenced the PG synthesis more significantly than ddl, and we further explored the changes in the composition of the PG of F44-murF by LC-MS.
FIG 5.
Fluorescence detection of ddl- and murF-overexpressing strain. The fluorescence intensities of F44-ddl and F44-murF after growth in medium containing d-Met and d-Phe for 6 h are shown. The control group was L. lactis F44. Van-FL (300 ng/ml) was used to detect the insertion of NCDAAs. The fluorescence value was the fluorescence intensity of a single cell, and the fluorescence values of 50 cells in the field were taken to calculate the mean value.
MurF catalyzes the formation of the precursors replaced by d-Met or d-Phe, as verified by LC-MS.
By using LC-MS analysis, the monomer PentaMet was identified (Fig. 6), and it accounted for 0.72% of its total PG extracted by F44-murF with the addition of d-Met. As shown in Table 2, with the d-Met addition, F44-murF had 71.42% more PG than F44. Meanwhile, the PG of F44-murF with the d-Phe addition was extracted, and the monomers PentaPhe, PentaPhe(X) (N-acetylglucosamine–N-acetylmuramyl pentapeptide with the aspartate peptide bridge and a molecule of water lost), and TetraPhe accounted for 10.48%, 1.18%, and 9.35% of their total PGs, respectively. Compared with L. lactis F44, PentaPhe and TetraPhe were increased by 55.25% and approximately 10-fold, respectively, revealing that the d-Phe sample had a higher level of binding than the d-Met sample, and the amount of d-Ala–d-Phe participating in the synthesis of pentapeptides was higher than that of d-Ala–d-Met. From these results, we came to the conclusion that murF plays an important role in the incorporation of d-Met or d-Phe into PG.
FIG 6.
Analysis of L. lactis F44 PG by LC-MS. (A) Result for L. lactis F44-murF without addition of any d-amino acids. (B) LC-MS analysis of L. lactis F44-murF PG after growth in medium containing d-Met for 6 h. Peak 1 represents d-Met binding to PentaMet, a disaccharide pentapeptide with the asparagine-peptide bridge. The structures and parameters are shown in Table 2. (C) LC-MS analysis of L. lactis F44-murF PG after growth in medium containing d-Phe for 6 h. Peaks 2 and 3 represent d-Phe binding to PentaPhe, a disaccharide pentapeptide with the asparagine peptide bridge. Peak 4 represents d-Phe binding to PentaPhe(X), a disaccharide (N-acetylglucosamine-N-acetylmuramyl) tetrapeptide with the aspartate peptide bridge and a molecule of water loss. Peaks 5, 6, and 7 represent d-Phe binding to TetraPhe, a disaccharide tetrapeptide with an aspartate peptide bridge. The structures and parameters are shown in Table 2.
TABLE 2.
Analysis of L. lactis F44-murF PG by LC-MS
| Additive | Structurea | Mol wt | Retention time (min) | Area (%) |
|---|---|---|---|---|
| D-Met | PentaMet | 1,140.6 | 2.03 | 0.72 |
| D-Phe | PentaPhe | 1,156.6 | 5.6/10.4 | 10.48 |
| D-Phe | TetraPhe | 1,074.5 | 24.5/24.7/29.4 | 9.35 |
| D-Phe | PentaPhe(X) | 1,139.5 | 21.7 | 1.18 |
PentaMet, G–M–l-Ala–d-Gln–l-Lys (d-Asn)–d-Ala–d-Met; PentaPhe, G–M–l-Ala–d-Gln–l-Lys (d-Asn)–d-Ala–d-Phe; TetraPhe, G–M–l-Ala–d-Gln–l-Lys (d-Asp)–d-Phe; PentaPhe(X), G–M–l-Ala–d-Gln–l-Lys (d-Asp)–d-Ala–d-Phe (d-Asn) or d-Asp is the peptide bridge bound to l-Lys, G is N-acetylglucosamine [GlcNAc], M is N-acetylmuramic acid [MurNAc], and X is the loss of an H2O molecule (18 Da).
Overexpressing murF enhances the effects of d-Met or d-Phe on acid resistance and nisin yield.
The effect of murF on the incorporation of d-Met or d-Phe to PG was further determined by the acid resistance and nisin yield assays, through which we found that, without any d-amino acid addition, the survival rate under acid stress of F44-murF was not obviously different from that of F44 (Fig. 7A). Compared with that of F44 without any d-amino acid addition, the survival rate of F44-murF with the d-Met addition was enhanced 2.10-fold or 2.91-fold after 1.5 h or 2.0 h acid shock, respectively, and that of F44 with the d-Phe addition increased 1.92-fold (1.5 h) and 2.44-fold (2.0 h). As shown in Fig. 7B, the addition of d-Met and d-Phe in the medium further increased the nisin yield of F44-murF 2.02-fold and 2.34-fold, respectively, compared with that of F44 without any d-amino acid addition. These results suggested that overexpressing murF could further enhance the effects of d-Met or d-Phe on the acid resistance and nisin yield.
FIG 7.
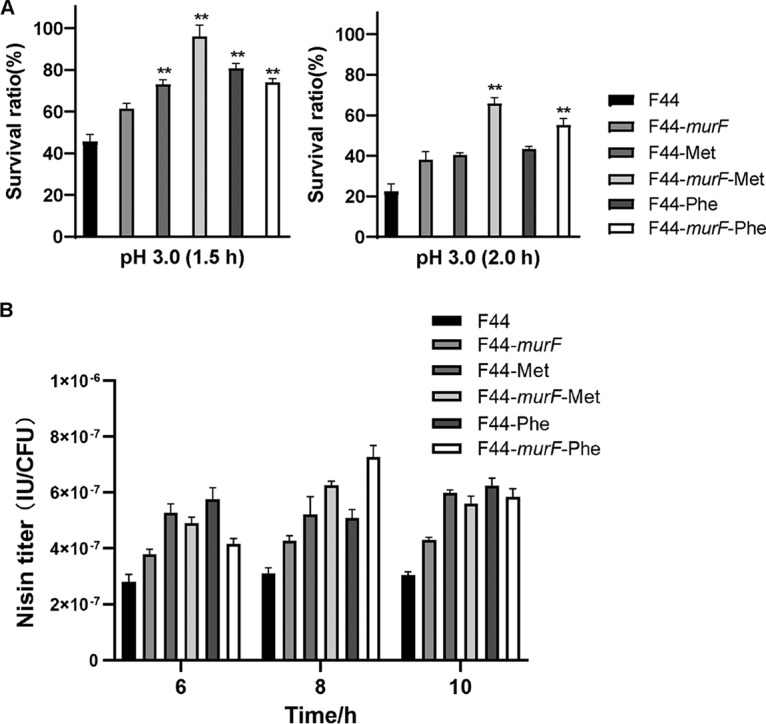
(A) Survival rate of L. lactis F44 and F44-murF with added d-Met or d-Phe after 1.5 h and 2.0 h acid shock. **, P < 0.001 (t test). (B) Nisin titer of L. lactis F44 and F44-murF with added d-Met or d-Phe. Samples were taken every 2 h from 6 to 10 h. Error bars indicate the SD for three independent experiments.
DISCUSSION
Microorganisms communicate with the environment by producing diverse metabolites (32). For instance, many bacteria have been found to produce NCDAAs during growth: V. cholerae produces d-Met, d-Leu, and d-Arg; Bacillus subtilis generates d-Tyr and d-Phe (21, 29, 33); and a variety of lactic acid bacteria species produce d-branched-chain amino acids (34). Bacteria utilize NCDAAs to support their growth, regulate spore germination, and reshape their cell walls to be resistant to several environmental stresses (21, 25, 35, 36).
This study demonstrated that two types of NCDAAs, d-Met and d-Phe, significantly increased the survival rate and nisin yield of L. lactis F44 (Fig. 1 and 2). NCDAAs were identified as being involved in PG composition and made the strain highly resistant to environmental stresses in V. cholerae (21, 22). Our previous studies showed that the changes in cell wall modification could increase acid resistance and indirectly enhance nisin yield (10, 11).
In V. cholerae and B. subtilis, NCDAAs can bind to the fourth or fifth position of the cell wall peptide chain, which affects the function of the cell wall (22). Vancomycin is a clinically important antibiotic that can specifically bind to d-Ala–d-Ala at the fourth and fifth positions of the newly synthesized PG (31). The fluorescence derivatives of vancomycin can provide technical support for the exploration of the topological structure of PG synthesis (30). In this study, the fluorescence derivative Van-FL was used to investigate the incorporation of d-amino acids, and the fluorescence intensity of strains with the d-Met or d-Phe addition was weaker than that of the control (Fig. 3). We speculated that this was because d-Met or d-Phe was incorporated into the fourth or fifth position of the peptide chain, and this consequently hindered the binding of the Van-FL to the peptide chain (31). Moreover, an LC-MS assay showed that d-Met was incorporated at the fifth position of the PG peptide chain and d-Phe at the fourth and fifth positions of (Fig. 4 and Table 1). Our results are consistent with previous studies in V. cholerae and B. subtilis (21, 22), which indicates that the incorporation of NCDAAs in PG is a common phenomenon in bacteria.
In V. cholerae, the incorporation of d-Met into the fourth position in muropeptides is generated by two enzymes involved in the synthesis of PG precursors, Ddl and MurF (22). Ddl connects two d-Ala to form d-Ala–d-Ala, and MurF connects the monosaccharide tripeptide with the dipeptide d-Ala–d-Ala (or d-Ala–d-Phe or d-Ala–d-Met) to form a conventional (or unconventional) monosaccharide pentapeptide (12, 13). In this study, the overexpression of ddl and murF increased the fluorescence intensity (Fig. 5), because the enhanced synthesis of d-Ala–d-Ala provided more substrates binding to the monosaccharide tripeptides. However, the fluorescence intensities of the recombinant strains were weakened when d-Met or d-Phe was added, and that seen with d-Phe was lower, indicating a higher binding level of d-Ala–d-Phe. This result was further confirmed by the LC-MS assay for the PG of F44-murF (Fig. 6 and Table 2). Moreover, the addition of d-Met or d-Phe further increased the acid resistance of F44-murF (Fig. 7). A previous study found that strains producing NCDAAs were more resistant to osmotic stress, because NCDAAs could cause alterations in cell wall structure, such as changing the length of glycan chains and the degree of cross-linking, leading to a stronger PG in the strain (21). Hence, we suggest that overexpressing murF in L. lactis F44 could contribute to the incorporation of d-Met and d-Phe into the PG and, therefore, increase the acid resistance and indirectly enhance the nisin titer of L. lactis F44.
In addition, two Ldts (LdtA and LdtB), inner membrane-anchored periplasmic proteins, contributed to the incorporation of d-Met in the fourth position of the PG peptide chain (22). In L. lactis F44, only an Ldt (ATY88872.1) containing an ErfK (E. coli LdtA) conserved domain was annotated, but it showed a low similarity (25.81% identity), and the function was unknown. E. coli is a non-NCDAA producer, but it can incorporate d-Met into the PG muropeptides by exogenous addition, and similar results were also obtained with some strains of the Gram-positive organisms B. subtilis, Enterococcus faecalis, and S. aureus (22). Whether the Gram-positive strain F44 can produce NCDAAs is still unclear and remains to be further verified. However, in this study, we determined that F44 could also incorporate exogenously supplied d-Met and d-Phe into muropeptides. These results suggest that NCDAAs may serve as signals for microbial communication in microbial communities under the changing environmental threats (22).
In conclusion, this study found that the NCDAAs d-Met and d-Phe participate in the synthesis of PG and affected the survival rate and nisin yield of L. lactis F44. Moreover, overexpressing ddl or murF enhanced the level of d-Met and d-Phe participating in cell wall synthesis, and murF played an important role in the incorporation process. This study gives insight into the pivotal role of NCDAAs in the acid resistance and provides biological evidence that d-Met and d-Phe can participate in L. lactis F44 cell wall synthesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains are listed in Table 3. L. lactis F44 strain (accession number PRJNA419050) was cultured in seed medium or fermentation medium separately supplemented with different d-amino acids when needed. The seed medium was prepared as follows (wt/vol): yeast extract (1.5%), peptone (1.5%), KH2PO4 (2.0%), sucrose (2.0%), NaCl (0.15%), and MgSO4·7H2O (0.015%), pH 7.2. The fermentation medium was similar to the seed medium except for the addition of corn steep liquor (0.3%) and cysteine (0.26%). The constructed vectors were transformed into E. coli for enrichment and then electrotransformed into L. lactis F44 (9). All the L. lactis strains were incubated at 30°C, and E. coli strains were grown in liquid Luria-Bertani (LB) medium at 37°C, with shaking at 180 rpm. Micrococcus flavus, an indicator in the nisin activity assay, was incubated in LB solid medium at 37°C. The media for agar diffusion in the nisin activity assay were as follows (wt/vol): tryptone (0.8%), yeast extract (0.25%), glucose (0.5%), Na2HPO4 (0.2%), NaCl (0.5%), and agar powder (1.5%). All chemicals were purchased from Sangon Biotech, Shanghai, China.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| L. lactis F44 | Evolved from L. lactis YF11 (accession no. PRJNA419050) | 6 |
| F44-ddl | Overexpression of ddl in WT | This study |
| F44-murF | Overexpression of murF in WT | This study |
| F44-pLEB124 | Introduction of expression vectors in WT | Laboratory stock |
| M. flavus | Indicator strains for nisin yield diffusion | Laboratory stock |
| Plasmids | ||
| pLEB124·PP45 | E. coli-L. lactis shuttle and expression vector, Emr; P45 promoter, low copy no. | Laboratory stock |
| pLEB124·PP45ddl | ddl cloned in pLEB124·PP45 | This study |
| pLEB124·PP45murF | murF cloned in pLEB124·PP45 | This study |
Emr, erythromycin resistant.
Construction of overexpressing strains.
Primers used in this study are listed in Table 4. L. lactis F44 was used for the transformation of pLEB124 with the P45 promoter upstream of the multiple cloning sites. The genes ddl and murF were amplified from the L. lactis F44 genome. PCR was performed using TransStart FastPfu DNA polymerase (TransGen, Beijing, China) using the protocol recommended by the manufacturer. The PCR products were digested with HindIII plus BamHI or with BamHI plus NcoI (NEB, Beijing, China) and were ligated to pLEB124 at the same restriction sites. The recombinant plasmids were transformed into E. coli and electrically transformed into L. lactis F44, generating the overexpressing strains F44-ddl and F44-murF. The sequences were confirmed by DNA sequencing (Genewiz, Suzhou, China).
TABLE 4.
Primers used in this study
| Primer | Sequence |
|---|---|
| ddl-F | CCCAAGCTTATGCTGTCAGTAAAAAGAAGTG |
| ddl-R | CGCGGATCCAAATCTGCCAGTTGCGAC |
| murF-F | CGCGGATCCACGATTGCTGACAGATTTTTTT |
| murF-R | CATGCCATGGTCTGCTTTCTCCAAATCGTAA |
| p124-F | AGGGAACCTAGAATAGTGAA |
| p124-R | TTCATTCTGCTAACCAGTAAGGC |
Acid resistance assay.
The acid resistance assay was performed as described in a previous study (8). The F44 strains were incubated in the seed medium (control) and the seed medium with the d-amino acid addition (experimental samples) for three generations before being used in acid tolerance assays. After L. lactis F44 grew to the mid-log phase, cells were centrifuged (2,604 × g, 5 min), washed twice with normal saline, and then exposed to the pH 3.0 seed medium for 1.5 h or 2.0 h. The number of viable cells was detected via plating on agar plates after proper dilution. Plates were incubated at 30°C for 24 h. The number of viable cells before the acid shock was defined as the T0 (0 h) prestress cell number (CFU). The number after the acid shock was defined as the T1.5 (1.5 h) or T2 (2.0 h) poststress cell number (CFU). The survival rate was calculated as follows: (T1.5 or T2 poststress cell number [CFU]/T0 prestress cell number [CFU]) × 100. The relative survival rate is the survival rate of the experimental sample divided by the survival rate of the control.
Fermentation and nisin titer assay.
Samples were taken every 2 h to measure the extracellular pH and the optical density at 600 nm (OD600). Nisin titers were determined using the agar diffusion method (9). The stock solution of nisin (106 IU/ml) was prepared by adding standard nisin (2.5%; balance, sodium chloride; Sigma, St. Louis, MO) to 0.02 M HCl and boiling for 5 min. The stock solution was diluted by adding 0.02 M HCl to standard nisin solutions (50, 100, 200, and 500 IU/ml). Every 2 h, 500 μl of fermentation liquid was added to the same volume of 0.02 M HCl. After boiling for 5 min and centrifugation (8,228 × g for 5 min), the supernatant was serially diluted with 0.02 M HCl. The nisin titer assay was performed as described previously (9). Meanwhile, 100-μl fermentation samples were serially diluted in the normal saline and spread on the seed medium plate. After cultivation at 30°C, the number of viable cells was counted. Each sample was tested in triplicate. The nisin yield per cell was calculated as the nisin titer divided by the number of viable cells.
Fluorescent detection.
A fluorescent vancomycin derivative (Van-FL) was prepared as in a previous study (37). The 500-μl vancomycin solution (Sigma; 10 mg/ml in water) was mixed with 50 μl 5(6)-carboxyfluorescein-N-hydroxy succinimide ester (Fluos [Sigma]; 5 mg/ml in dimethyl sulfoxide) and kept at 4°C overnight. After this solution was mixed with 450 μl 0.1 M Tris (pH 8.0) and reheated for 1 h at 22°C, the resulting solution was stored at –20°C away from light.
The L. lactis culture was diluted to an OD600 of about 0.8 and inoculated into the seed medium. Then Van-FL was added to the seed medium, with the concentrations ranging from 1 ng/ml to 500 μg/ml. The OD600 was measured every 2 h with a microplate reader (Thermo Fisher Scientific, Vantaa, Finland). The highest concentration tolerated (at which no visible inhibition of growth occurred) was determined by the growth curve (see Fig. S4 in the supplemental material).
Van-FL was added to the seed medium at a final concentration of 0.3 μg/ml (30°C for 15 to 20 min) when L. lactis F44 grew to stationary phase. After centrifugation to remove the supernatant (8,228 × g, 5 min), the pellet was washed with precooled phosphate buffer solution (pH 7.0) three times. A 20-μl sample was placed on the microscope slide with 30 μl of anti-fluorescence quencher. Images were taken with a confocal laser scanning microscope (LSM 880; Carl Zeiss AG, Oberkochen, Germany). The digital images were analyzed with Carl Zeiss software version 2.3.
HPLC coupled with quadrupole time of flight MS analysis of PG.
PG was extracted as previously described (10). A 500-ml portion of L. lactis F44 cells at stationary phase were harvested by centrifugation (7,500 × g, 10 min, 4°C). The pellets were resuspended in sterile water and boiled for 10 min. Samples were centrifuged at 40,000 × g for 10 min at 4°C, resuspended in 5% sodium dodecyl sulfate (SDS) solution, and boiled and stirred for 25 min. The previous steps were repeated with 4% SDS solution, and then the pellets were washed with ultrapure water to remove SDS. The pellets were treated with 2 mg/ml pronase for 90 min at 40°C and then with 200 μg/ml trypsin for 16 h at 37°C. After centrifugation, they were resuspended in 40% hydrofluoric acid (16 h, 4°C). Then they were washed with Tris-HCl (0.25 M, pH 7.0) and ultrapure water several times. The final pellets were freeze-dried and stored at –20°C.
After muramidase digestion (18 h) and filtration, the solutions were determined by high-performance LC-MS (HPLC-MS; Agilent 1200 HPLC system; Agilent, Santa Clara, CA) using a C18 column (3.5 μm; 100 × 2.1 mm; Waters, Ireland) at a flow rate of 0.2 ml/min. For the flow phase, buffer A was 0.1% formic acid and buffer B was 100% acetonitrile. Parameters were as follows: 0 to 63 min, 95% to 80% A; 63 to 83 min, 80% to 10% A; 83 to 100 min, 10% to 95% B. Muropeptide peaks were collected by electrospray ionization–tandem mass spectrometry (ESI–MS/MS) by using a mass spectrometer (micrOTOF-QII; Bruker Daltonics, Hamburg, Germany). The parameters were set as previously described (10).
Statistical analysis.
Three independent experiments, each containing three replicates, were performed in the acid resistance experiment. To evaluate the statistical significance of the survival rate under acid stress, a t test was carried out.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Funds for Creative Research Groups of China (21621004), the National Key R&D Program of China (2017YFD0201405), and the National Natural Science Foundation of China (31770076). Jianjun Qiao was supported by the 131 innovative personnel training project of Tianjin (China) and the New Century Outstanding Talent Support Program, Education Ministry of China.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Mierau I, Kleerebezem M. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol 68:705–717. doi: 10.1007/s00253-005-0107-6. [DOI] [PubMed] [Google Scholar]
- 2.Özel B, Şimşek Ö, Akçelik M, Saris PE. 2018. Innovative approaches to nisin production. Appl Microbiol Biotechnol 102:6299–6307. doi: 10.1007/s00253-018-9098-y. [DOI] [PubMed] [Google Scholar]
- 3.Papagianni M, Avramidis N. 2012. Engineering the central pathways in Lactococcus lactis: functional expression of the phosphofructokinase (pfk) and alternative oxidase (aox1) genes from Aspergillus niger in Lactococcus lactis facilitates improved carbon conversion rates under oxidizing conditions. Enzyme Microb Technol 51:125–130. doi: 10.1016/j.enzmictec.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Liu W, Zheng H, Wu Z, Wang Y. 2010. Effects of pH profiles on nisin fermentation coupling with foam separation. Appl Microbiol Biotechnol 85:1401–1407. doi: 10.1007/s00253-009-2217-z. [DOI] [PubMed] [Google Scholar]
- 5.Zhou XX, Pan YJ, Wang YB, Li WF. 2008. Optimization of medium composition for nisin fermentation with response surface methodology. J Food Sci 73:M245–M249. doi: 10.1111/j.1750-3841.2008.00836.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Liu S, Du Y, Feng W, Liu J, Qiao JJ. 2014. Genome shuffling of Lactococcus lactis subspecies lactis YF11 for improving nisin Z production and comparative analysis. J Dairy Sci 97:2528–2541. doi: 10.3168/jds.2013-7238. [DOI] [PubMed] [Google Scholar]
- 7.Ni ZJ, Zhang XY, Liu F, Wang M, Hao RH, Ling PX, Zhu XQ. 2017. Effect of co-over-expression of nisin key genes on nisin production improvement in Lactococcus lactis LS01. Probiotics Antimicrob Proteins 9:204–212. doi: 10.1007/s12602-017-9268-8. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, Liu J, Miao S, Zhao Y, Zhu H, Qiao M, Saris PEJ, Qiao JJ. 2018. Contribution of YthA, a PspC family transcriptional regulator of Lactococcus lactis F44 acid tolerance and nisin yield: a transcriptomic approach. Appl Environ Microbiol 84:e02483-17. doi: 10.1128/AEM.02483-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Caiyin Q, Feng W, Zhao X, Qiao B, Zhao G, Qiao JJ. 2016. Enhance nisin yield via improving acid-tolerant capability of Lactococcus lactis F44. Sci Rep 6:27973. doi: 10.1038/srep27973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao P, Liang D, Cao L, Qiao B, Wu H, Caiyin Q, Zhu H, Qiao JJ. 2017. Promoting acid resistance and nisin yield of Lactococcus lactis F44 by genetically increasing D-Asp amidation level inside cell wall. Appl Microbiol Biotechnol 101:6137–6153. doi: 10.1007/s00253-017-8365-7. [DOI] [PubMed] [Google Scholar]
- 11.Cao L, Liang D, Hao P, Song Q, Xue E, Caiyin Q, Cheng Z, Qiao JJ. 2018. The increase of O-acetylation and N-deacetylation in cell wall promotes acid resistance and nisin production through improving cell wall integrity in Lactococcus lactis. J Ind Microbiol Biotechnol 45:813–825. doi: 10.1007/s10295-018-2052-2. [DOI] [PubMed] [Google Scholar]
- 12.Vollmer W, Blanot D, De Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 13.Chapot-Chartier M-P, Kulakauskas S. 2014. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact 13:S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veiga P, Piquet S, Maisons A, Furlan S, Courtin P, Chapot-Chartier MP, Kulakauskas S. 2006. Identification of an essential gene responsible for d‐Asp incorporation in the Lactococcus lactis peptidoglycan crossbridge. Mol Microbiol 62:1713–1724. doi: 10.1111/j.1365-2958.2006.05474.x. [DOI] [PubMed] [Google Scholar]
- 15.Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. 2015. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A 112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz N. 2015. Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights 8:LPI.S31783. doi: 10.4137/LPI.S31783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammadi T, Van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, Diepeveen-de Bruin M, Nguyen-Distèche M, De Kruijff B, Breukink E. 2011. Identification of FtsW as a transporter of lipid‐linked cell wall precursors across the membrane. EMBO J 30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egan AJF, Biboy J, van't Veer I, Breukink E, Vollmer W. 2015. Activities and regulation of peptidoglycan synthases. Philos Trans R Soc B 370:20150031. doi: 10.1098/rstb.2015.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auer GK, Weibel DB. 2017. Bacterial cell mechanics. Biochemistry 56:3710–3724. doi: 10.1021/acs.biochem.7b00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell‐wall recycling. Ann N Y Acad Sci 1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam H, Oh D-C, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. 2009. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cava F, De Pedro MA, Lam H, Davis BM, Waldor MK. 2011. Distinct pathways for modification of the bacterial cell wall by non‐canonical D‐amino acids. EMBO J 30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fura JM, Kearns D, Pires MM. 2015. d-Amino acid probes for penicillin binding protein-based bacterial surface labeling. J Biol Chem 290:30540–30550. doi: 10.1074/jbc.M115.683342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupoli TJ, Tsukamoto H, Doud EH, Wang TS, Walker S, Kahne D. 2011. Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J Am Chem Soc 133:10748–10751. doi: 10.1021/ja2040656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horcajo P, de Pedro MA, Cava F. 2012. Peptidoglycan plasticity in bacteria: stress-induced peptidoglycan editing by noncanonical D-amino acids. Microb Drug Resist 18:306–313. doi: 10.1089/mdr.2012.0009. [DOI] [PubMed] [Google Scholar]
- 26.Hammes W, Schleifer K, Kandler O. 1973. Mode of action of glycine on the biosynthesis of peptidoglycan. J Bacteriol 116:1029–1053. doi: 10.1128/JB.116.2.1029-1053.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caparros M, Pisabarro A, De Pedro M. 1992. Effect of D-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J Bacteriol 174:5549–5559. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammes WP. 1978. The LD‐carboxypeptidase activity in Gaffkya homari: the target of the action of d‐amino acids or glycine on the formation of wall‐bound peptidoglycan. Eur J Biochem 91:501–507. doi: 10.1111/j.1432-1033.1978.tb12703.x. [DOI] [PubMed] [Google Scholar]
- 29.Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S. 2006. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc Natl Acad Sci U S A 103:11033–11038. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bisicchia P, Bui NK, Aldridge C, Vollmer W, Devine KM. 2011. Acquisition of VanB‐type vancomycin resistance by Bacillus subtilis: the impact on gene expression, cell wall composition and morphology. Mol Microbiol 81:157–178. doi: 10.1111/j.1365-2958.2011.07684.x. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds PE. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis 8:943–950. doi: 10.1007/bf01967563. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto M, Kunisawa A, Hattori T, Kawana S, Kitada Y, Tamada H, Kawano S, Hayakawa Y, Iida J, Fukusaki E. 2018. Free D-amino acids produced by commensal bacteria in the colonic lumen. Sci Rep 8:17915. doi: 10.1038/s41598-018-36244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cava F, Lam H, De Pedro MA, Waldor MK. 2011. Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell Mol Life Sci 68:817–831. doi: 10.1007/s00018-010-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mutaguchi Y, Kasuga K, Kojima I. 2018. Production of D-branched-chain amino acids by lactic acid bacteria carrying homologs to isoleucine 2-epimerase of Lactobacillus buchneri. Front Microbiol 9:1540. doi: 10.3389/fmicb.2018.01540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubota T, Kobayashi T, Nunoura T, Maruyama F, Deguchi S. 2016. Enantioselective utilization of d-amino acids by deep-sea microorganisms. Front Microbiol 7:511. doi: 10.3389/fmicb.2016.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrestha R, Lockless SW, Sorg JA. 2017. A Clostridium difficile alanine racemase affects spore germination and accommodates serine as a substrate. J Biol Chem 292:10735–10742. doi: 10.1074/jbc.M117.791749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776. doi: 10.1016/S0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.