Abstract
Aldehyde oxidases (AOXs) are a small group of enzymes belonging to the larger family of molybdo-flavoenzymes, along with the well-characterized xanthine oxidoreductase. The two major types of reactions that are catalyzed by AOXs are the hydroxylation of heterocycles and the oxidation of aldehydes to their corresponding carboxylic acids. Different animal species have different complements of AOX genes. The two extremes are represented in humans and rodents; whereas the human genome contains a single active gene (AOX1), those of rodents, such as mice, are endowed with four genes (Aox1-4), clustering on the same chromosome, each encoding a functionally distinct AOX enzyme. It still remains enigmatic why some species have numerous AOX enzymes, whereas others harbor only one functional enzyme. At present, little is known about the physiological relevance of AOX enzymes in humans and their additional forms in other mammals. These enzymes are expressed in the liver and play an important role in the metabolisms of drugs and other xenobiotics. In this review, we discuss the expression, tissue-specific roles, and substrate specificities of the different mammalian AOX enzymes and highlight insights into their physiological roles.
Keywords: metalloenzyme, molybdenum, mouse, drug metabolism, flavoprotein, xenobiotic, oxidase, oxygen radicals, iron-sulfur protein, aldehyde oxidase (AOX), enzyme evolution, metal-containing enzyme, molybdenum cofactor (Moco), molybdo-flavoenzyme, 2Fe-2S cluster, flavin adenine dinucleotide (FAD)
Introduction
Mammalian aldehyde oxidases (AOXs)2 are metal-containing enzymes that require the molybdenum cofactor (Moco), two [2Fe-2S] clusters, and FAD for their catalytic activity (1, 2). AOXs belong to the xanthine oxidase family of Moco-containing enzymes along with xanthine oxidoreductase (XOR), bacterial carbon monoxide dehydrogenases, periplasmic aldehyde oxidoreductases, and other molybdoenzymes from prokaryotes. Humans contain a total of four molybdoenzymes, namely aldehyde oxidase 1 (AOX1) (3), xanthine oxidoreductase (4), sulfite oxidase (5), and the mitochondrial amidoxime-reducing component (6). In humans, the only essential molybdoenzyme is sulfite oxidase (7–9).
Eukaryotic AOXs are cytosolic proteins that are expressed in many tissues of various organisms, including insects, rodents, and humans (10, 11). AOXs oxidize purines, pyrimidines, and pteridines and are involved in nicotinic acid metabolism (12). In addition, AOXs bio-transform organic aldehydes into the corresponding carboxylic acids, hydroxylate heteroaromatic rings, and catalyze the reduction of nitro- and sulfo-groups (13–18). Given this broad substrate specificity, human AOX1 is an emerging enzyme in the context of drug metabolism (19, 20). Indeed, the number of drug molecules recognized and metabolized by AOXs is increasing, and this has raised the interest of pharmaceutical companies (21, 22).
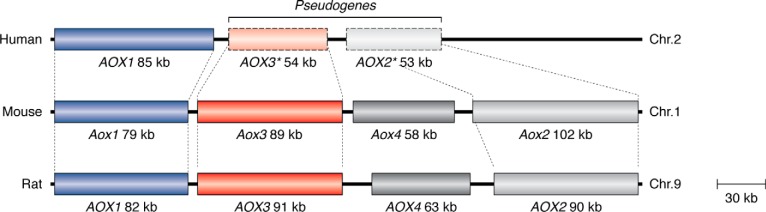
Despite these many characterized activities, the physiological role of mammalian AOXs, is still a matter of debate (23). The genomes of many prokaryotic and eukaryotic organisms contain genes coding for AOX enzymes (11). In mammals, the complement of AOX genes varies according to the animal species considered (10, 11, 24). Mice and rats are characterized by the largest number of active AOX genes (Aox1, Aox2, Aox3, and Aox4),3 each coding for a different AOX enzyme (Fig. 1) (11). In contrast, humans and the majority of primates contain a single active AOX gene, which is the mAox1 orthologue (10). The multiplicity of AOX genes observed in many vertebrates is the result of a series of duplication events from a common ancestor gene. In the mammalian species characterized by multiple AOX genes, the loci cluster on a short region of the same chromosome.
Figure 1.
Organization of the human, mouse, and rat AOX genes. Shown is a schematic representation of the Homo sapiens, Mus musculus, and Rattus norvegicus AOX genes and pseudogenes. Pseudogenes are marked by an asterisk.
The reason why mammals other than humans express multiple AOX enzymes is unknown (25). Nevertheless, the decrease in the number of active AOX isoenzymes from rodents to humans is the result of progressive deletion/inactivation of the corresponding genes (24). It can be speculated that AOX2, AOX3, and AOX4 exert tissue-specific functions in rodents, which are dispensable in humans. The physiological role and demand for several AOX enzymes in certain organisms may find an answer following definition of the physiological function and substrate specificity of the various mammalian AOXs.
In this review, we discuss the evolution, expression, substrate specificities, and structures of the different mammalian AOX enzymes. The major aim is to give an overview on what is known about the potential roles of these different enzyme forms with a focus on the four enzymes present in mice. First insights into the physiological role of one specific enzyme form could be drawn from the physiological studies of mAox4−/− knockout mice. Further, the role of reactive oxygen species (ROS) production by AOX enzymes is discussed, because they were produced in much higher proportions than previously appreciated. For the role of human AOX1 in drug metabolism, the reader is referred to numerous specialized reviews in the literature covering this topic (12, 20, 25–28).
Domain structure and catalytic mechanism of the AOX enzymes
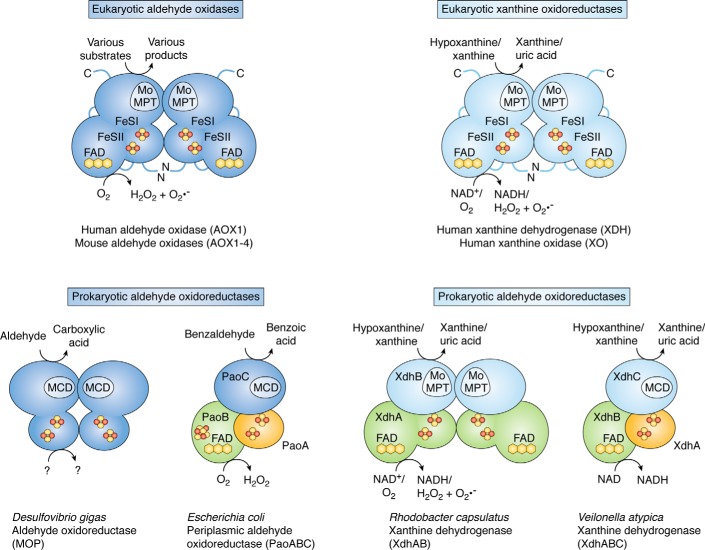
AOX enzymes are present in both eukaryotes and prokaryotes. A common catalytic mechanism has been proposed for all AOXs. Before we focus on the different AOX enzymes present in mammals, we introduce the differences and common principles in the primary structures of AOX enzymes in different organisms. AOXs in general are Moco-containing enzymes that, with some exceptions in prokaryotes, contain two [2Fe-2S] clusters and FAD as additional prosthetic groups. First, it is important to give a brief overview on the structural characteristics of prokaryotic AOX enzymes to highlight the differences from and similarities with the eukaryotic counterparts. In bacteria and archaea, the xanthine oxidase family of molybdoenzymes is complex, as it includes CO dehydrogenases (29), aldehyde oxidoreductases (30–32), and quinoline-metabolizing (31) and nicotinic acid–metabolizing (33) enzymes. In particular, the domain structure, the subunit composition, and the cofactor complement of prokaryotic AOXs is variable (Fig. 2) (30–32). Rhodobacter capsulatus/Veillonella atypica xanthine dehydrogenase (XDH) (34), Desulfovibrio gigas aldehyde oxidoreductase (MOP) (30), and Escherichia coli periplasmic aldehyde oxidoreductase (PaoABC) (35) are the enzymes showing the highest structural similarities with eukaryotic xanthine oxidoreductases and AOXs. The redox-active centers of the bacterial and archaeal members of the xanthine oxidase family are contained in separate subunits, and exceptions to the general cofactor composition have also been identified (Fig. 2). R. capsulatus XDH is organized as an (αβ)2 heterodimer, and it contains the eukaryotic form of Moco (assigned as Mo-molybdopterin (Mo-MPT), Fig. 3). In contrast, V. atypica XDH consists of an αβγ-heterotrimer, and it contains the MPT cytosine dinucleotide (MCD) form of Moco (Fig. 3). In addition, MOP lacks the FAD domain, and it consists of two identical subunits organized as an α2 dimer (30). Each subunit contains the two [2Fe-2S] clusters and the MCD cofactor. Finally, the periplasmic aldehyde oxidoreductase from E. coli is characterized by a structure consisting of an αβγ-heterotrimer, which exceptionally includes an additional [4Fe-4S] cluster proximal to the FAD in the β-subunit (35).
Figure 2.
Overview of the subunit composition and cofactor organization of aldehyde oxidases and xanthine oxidoreductases. In eukaryotes, the catalytically active forms of AOXs and XORs consist of α2 homodimers. Each subunit of the dimer consists of separate domains containing the Moco catalytic site, two distinct [2Fe-2S] redox centers, and the FAD-binding site. In the prokaryotic enzymes, the [2Fe-2S] cluster-binding subunits are shown in orange, the subunits binding the FAD cofactor are colored in green, and the subunits containing the Moco are colored in blue. The FAD domain is absent in the D. gigas aldehyde oxidoreductase. In the R. capsulatus xanthine dehydrogenase, the iron-sulfur and flavin-binding domains of the protein constitute one subunit (XdhA), and the Mo-MPT–binding domain constitutes a second (XdhB) subunit. In the V. atypica xanthine dehydrogenase, the iron-sulfur centers are located in one subunit (XdhA), the flavin in a second (XdhB), and the Moco as MCD in a third (XdhC) subunit. In the PaoABC from E. coli, a [4Fe-4S] cluster is present in proximity to the FAD cofactor on the PaoB subunit. Potential substrates and electron acceptors are indicated.
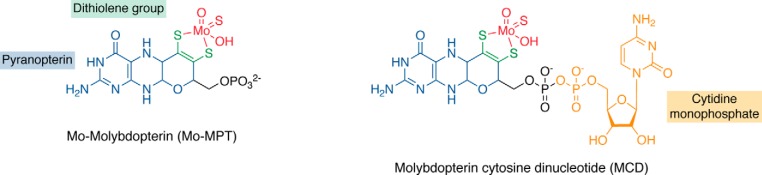
Figure 3.
Forms of the molybdenum cofactor present in enzymes of the xanthine oxidase family. Characteristic to enzymes of the xanthine oxidase family is a sulfido ligand at the equatorial position of the molybdenum atom. In eukaryotic enzymes of this family, the basic form of Moco is a pyranopterin, named Mo-MPT, which coordinates the molybdenum atom (colored in red) by the characteristic dithiolene group (colored in green) at the C1′ and C2′ positions of the pyranopterin ring (colored in blue). In bacteria, the MPT core can be modified by an additional CMP nucleotide (colored in orange) at the phosphate group (colored in black), forming MCD.
In comparison, the xanthine oxidase family of eukaryotic molybdoenzymes consists solely of AOXs (EC 1.2.3.1) and XORs. Two interconvertible forms of the mammalian XOR enzyme are known, xanthine oxidase (XO; EC 1.17.3.2) and XDH (EC 1.17.1.4). The domain structure of eukaryotic AOXs is highly conserved, and the catalytically active enzymes are organized as α2 homodimers (Fig. 2) (36, 37). Each subunit of the catalytically active AOX and XOR homodimer contains three cofactor-binding domains. The N-terminal domain binds two [2Fe-2S] clusters known as FeSI and FeSII. The central domain contains the FAD binding site, whereas the C-terminal domain (4, 38–41) harbors the Moco in the Mo-MPT form (Fig. 3). The three domains are connected by flexible linker regions. The characteristic of the molybdoenzymes belonging to the xanthine oxidase family is the presence of an essential sulfur atom acting as an equatorial sulfido ligand to the molybdenum atom (Mo=S, Fig. 3) (42–45). The presence of this Mo=S ligand is at the basis of the cyanide-dependent inactivation of these molybdoenzymes (46). Cyanide treatment results in the formation of an inactive desulfo form, and these enzymes can be reversibly reactivated by treatment with sulfide and dithionite (42, 43).
The major structural differences among the members of the eukaryotic xanthine oxidase family are observed at the FAD site (3, 38). These differences are particularly evident between the FAD sites of AOXs and XORs, which react with the terminal electron acceptor. The final electron acceptor is molecular oxygen in the case of AOXs and XO, whereas it is NAD+ in the case of XDH (47). The enzymes can be reversibly converted from the XDH form to the XO form, which has been shown to involve the formation of a new disulfide bond, between two cysteine residues at the FAD site (Cys535 and Cys992 in bovine XOR). In addition, the conversion of XDH into XO requires a structural rearrangement of the 11-residue loop (Gln423–Lys433 in bovine XOR) located in close proximity to the FAD cofactor (4). In addition, the movement of this loop leads to a change in the electrostatic environment around the FAD cofactor (48) and blocks the access of NAD+ to its binding site (4). All of these structural elements involved in XDH/XO conversion are absent in mouse and human AOXs. This might explain the preference of AOX toward oxygen as the final electron acceptor (2).
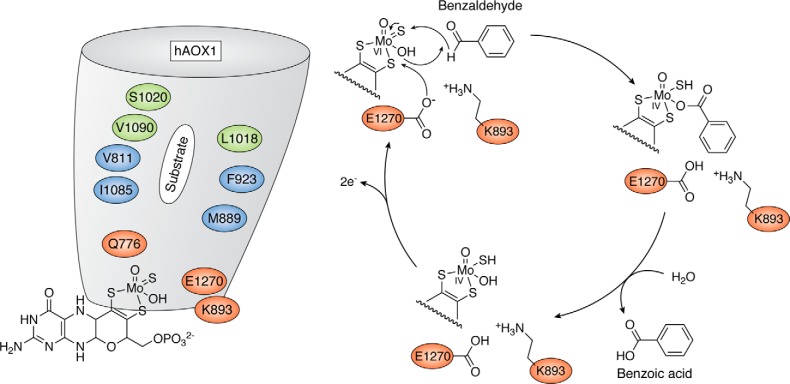
The C-terminal 85-kDa domains of both AOXs and XORs contain the Mo-MPT cofactor, which is the site of substrate conversion. The substrate binds to the active site, and it is converted into its product by an oxygen atom derived from water. There are structural features along the substrate access funnel that might account for the observed differences in substrate specificities between AOX and XOR enzymes. In AOXs, the funnel is wider and more anionic than in XORs. This implies that AOXs are characterized by a better access of larger and bulkier substrates as well as a broader substrate specificity. The role exerted by specific amino acid residues of the active site in the catalytic activity of mammalian AOXs has been elucidated by site-directed mutagenesis studies performed with heterologous expression systems developed in E. coli (49–53). The active site of AOXs contains only a few highly conserved amino residues (Glu1270, Phe923, Lys893, and Gln776; human AOX1 numbering), as many other residues are species-specific and differ in the human and mouse enzymes (Fig. 4) (54). Glu1270 is the essential residue for the catalytic activity of all the enzymes belonging to the xanthine oxidase family (38).
Figure 4.
The catalytic mechanism of human AOX1. Shown is a representation of the proposed reaction mechanism for human AOX1, as exemplified for the substrate benzaldehyde. For details, see “Domain structure and catalytic mechanism of the AOX enzymes.”
The general reaction mechanism proposed for all AOXs is a base-catalyzed mechanism starting with an initial attack of the deprotonated Glu1270 to the -OH ligand of the Mo atom, resulting in the generation of a nucleophilic -O− group (Fig. 4). Subsequently, the activated Mo-O− ligand attacks the substrate, followed by a hydride transfer to the sulfido ligand, leading to the generation of an intermediate species. In human AOX1, this intermediate is stabilized by hydrogen-bonding interactions with the Val811, Met889, and Lys893 residues of the active site. This mechanism is supported by experimental evidence with substituted N-heterocycles and by DFT calculations (55). Whereas the lysine is also conserved in mammalian AOX enzymes, the valine and methionine differ among AOX enzymes, depending on their substrate specificity. The next reaction step involves the release of the product from the reduced Mo site and the binding of a water molecule. The reaction cycle is completed once Mo is reoxidized and the two reducing equivalents are transferred to molecular oxygen via the two [2Fe-2S] clusters and FAD.
The evolution of AOX genes from bacteria to humans
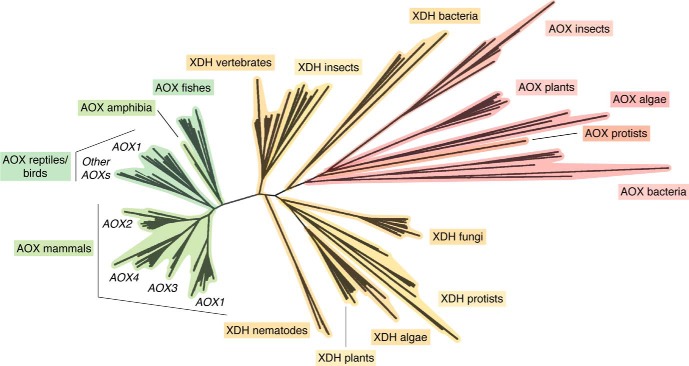
The evolutionary history of AOX and XDH genes has been reconstructed from the DNA-sequencing data available for many organisms from bacteria to humans (11). A phylogenetic analysis of the deduced protein sequences indicates that the extant complement of mammalian AOX genes is likely to originate from two distinct and primordial gene duplication events involving an ancestral XDH gene (24). The first duplication involves a bacterial XDH gene and led to the development of the current set of AOX genes observed in bacteria, protists, algae, and plants (Fig. 5). The second duplication is likely to have originated from a fish XDH gene, and it is at the basis of the present complement of vertebrate AOX genes (Fig. 5).
Figure 5.
Phylogenetic tree of AOX and XDH proteins in prokaryotes and eukaryotes. The unrooted phylogenetic tree was generated from the available prokaryotic as well as eukaryotic AOX and XDH protein sequences. The phylogenetic tree consists of all of the AOX and XDH proteins whose structure could be predicted from the cloning of the corresponding cDNAs or deduced from genome-sequencing data.
Vertebrate AOX and XDH proteins are characterized not only by high levels of similarity in terms of their amino acid sequences, but also by a similar exon-intron organization of the corresponding genes. As already mentioned, the mouse and rat genomes contain four distinct loci, Aox1, Aox2, Aox3, and Aox4, while a single active AOX1 gene is present in humans and higher primates (Fig. 1). All mammalian AOX and XDH genes consist of 35 and 36 exons, respectively. In mammals, the exon-intron junctions of all of the AOX and XDH genes are strictly conserved (10, 24). The Aox1, Aox2, Aox3, and Aox4 genes cluster on a short region of mouse chromosome 1 and rat chromosome 9 in a head-to-tail configuration (Fig. 1). The presence of one or more AOX genes resulted from a process of asynchronous AOX gene duplication events that occurred during the evolution of vertebrates, starting from fishes (24). AOX1 is likely to be the most ancient gene, whereas the order of appearance of AOX3, AOX4, and AOX2 during vertebrate evolution is still unknown. However, a number of considerations led to the proposal that AOX4 is more ancient than AOX3, which, in turn, appeared earlier than AOX2. It is concluded that the presence of only a single AOX enzyme in humans is due to gene deletion and pseudogenization events. In humans, downstream of the AOX1 gene, the AOX4 gene has been completely deleted, whereas the genes for AOX3 and AOX2 were transformed into inactive pseudogenes. Cows seem to have maintained three active aldehyde oxidase genes (AOX1, AOX4 and AOX2) on chromosome 2. The absence of nucleotide sequences with similarity to AOX3 strongly suggests that this gene has been deleted. Deletion of the AOX3 gene seems to be a conserved feature in horses; however, our present view of the aldehyde oxidase cluster in this animal species is still incomplete. Functional inactivation of AOX3 seems to be a common theme. The genome of dogs is characterized by two seemingly active AOX4 and AOX2 loci and two inactive AOX1 and AOX3 pseudogenes clustering on chromosome 37. The vestiges of numerous exons with nucleotide similarity to the rodent Aox1 and Aox3 genes are easily identified on two separate regions slightly upstream of the dog AOX4 and AOX2 loci. It is interesting to note that the dog is currently the only mammalian species that seems to be lacking AOX1, in addition to AOX3. Consequently, this mammal is devoid of aldehyde oxidase activity in the liver.
Tissue-specific expression of different AOX enzymes
The only mammalian species extensively characterized for the profiles of tissue- and cell-specific AOX expression are humans and mice (2). The tissue-specific pattern of human AOX1 mRNA expression, which is based on EST (expressed sequence tag) data, is available in the UNIGENE section of the NCBI site (UniGene Hs.406238). These data indicate that detectable levels of the AOX1 transcript are observed in many tissues. Adrenal glands, adipose tissue, and liver are the richest sources of human AOX1, followed by the trachea, the glandular epithelium of the prostate, bone, kidney, and connective tissue. The presence of human AOX1 in the adrenal gland and glandular epithelium of the prostate is consistent with a potential role of the enzyme in steroid hormone metabolism.
In mice, the expression of the human AOX1 orthologous gene (Aox1, UNIGENE Mm.26787) is more restricted, and it is limited to a selected number of tissues (2). The largest amounts of mAOX1 mRNA are present in the inner ear and the seminal vesicles, although measurable amounts of the transcript are observed also in the liver, the lung, and the central nervous system (56). Expression of the mAox3 gene (UNIGENE Mm.20108) is also relatively restricted, as significant amounts of the corresponding transcript are found only in the liver, lung, oviduct, and testis. Interestingly, mAOX3 and AOX1 are co-expressed in lung and liver. In liver, AOX3 is by far the most abundant enzyme. Significant amounts of the mAOX4 transcript (UNIGENE Mm.244525) are evident in the fertilized ovum, the inner ear, the tongue (2), and the epidermal layer of the skin (57). However, the richest source of mAOX4 is the Harderian gland, where the molybdo-enzyme accounts for ∼2% of all of the cytosolic proteins. The Harderian gland is a large exocrine gland located behind the eye bulb and characterized by the secretion of a lipid-rich fluid, which lubricates the eye surface and the fur (56). The most restricted profile of expression is observed in the case of the mAOX2 enzyme (2), as the corresponding transcript (UNIGENE Mm.414292) is detectable only in the nasal cavity (58). In this location, high levels of AOX2 mRNA are expressed in the Bowman's gland of the submucosal layer, which is responsible for the secretion of the mucous fluid. Nevertheless, AOX2 is also detectable in the sustentacular cells located in the apical layer of the nasal neuroepithelium. Given this localization, we speculate that AOX2 may play a role in the olfactory process.
Aox4 knockout mice give insights into the physiological role of the enzyme
Further insights into the tissue-specific function of the various mammalian AOXs have been and will be provided by the generation of knockout animals. Currently, Aox4−/− mice are the only knockout animals available, whereas the construction of knockouts in the other genes is in progress (59). Aox4−/− mice are viable and fertile and generally do not show any major morphological or behavioral abnormalities. Despite this, Aox4−/− mice are characterized by perturbations in the expression of clock genes, which are accompanied by reduced locomotor activity as well as resistance to diet-induced obesity and hepatic steatosis. All of these effects are observed in both female and male animals. Resistance to obesity is due to diminished fat accumulation resulting from increased energy dissipation. The phenomenon results from the white adipocytes of Aox4−/− mice undergoing trans-differentiation toward thermogenic brown adipocytes. The data obtained in this knockout model indicate that AOX4 contributes to the local synthesis and biodisposition of endogenous retinoids in the Harderian gland and skin, the principal sources of the AOX isoenzyme. The Harderian gland's transcriptome of Aox4−/− mice shows an overall down-regulation of direct retinoid-dependent genes as well as perturbations in the pathways controlling lipid homeostasis and cellular secretion, particularly in sexually immature animals. The skin of knockout mice is characterized by thickening of the epidermis in basal conditions and after UV light exposure. This is confirmed by analysis of the transcriptome, which shows an enrichment and overall up-regulation of genes involved in the process of epidermal thickening.
Metabolomic analyses performed in the Harderian glands of Aox4−/− and WT animals demonstrate the presence of 25 metabolites whose levels are significantly different in the two types of mice. Among the differentially produced metabolites, tryptophan and 5-hydroxyindolacetic acid stand out, because they are part of the serotonin/melatonin biosynthetic pathway, which controls circadian rhythms. Tryptophan is more abundant in Aox4−/− mice than WT mice, whereas 5-hydroxyindolacetic acid is measurable only in Aox4−/− mice. In addition, the levels of serotonin are significantly lower in the Harderian gland of Aox4−/− than WT mice. A similar trend is observed in the serum. Consistent with this, purified Harderian gland AOX4 recognizes tryptophan and 5-hydroxyindolacetic acid, but not serotonin, as substrates. Given the relevance of the tryptophan/serotonin pathway for circadian rhythms and fat deposition, the data support the idea that AOX4-dependent alterations in the levels of tryptophan and 5-hydroxyindolacetic may explain the perturbations in the two processes observed in Aox4−/− mice.
In conclusion, the phenotype of Aox4−/− mouse provides insights into the physiological function of AOX4, demonstrating that the enzyme plays a role in the control of diurnal rhythms, adipogenesis, and locomotor activity, supporting a link between these three processes. Interestingly, the reported effects have no impact on life expectancy, which is the same in Aox4−/− and WT mice. It is possible that human AOX1 plays a similar role as mAOX4 in adipogenesis, as there are data indicating that the enzyme controls fat deposition in adipocyte cultures (60). This point requires further investigations, as human AOX1 may represent a useful molecular target for the development of new anti-obesity agents.
Crystal structures of human AOX1 and mouse AOX3
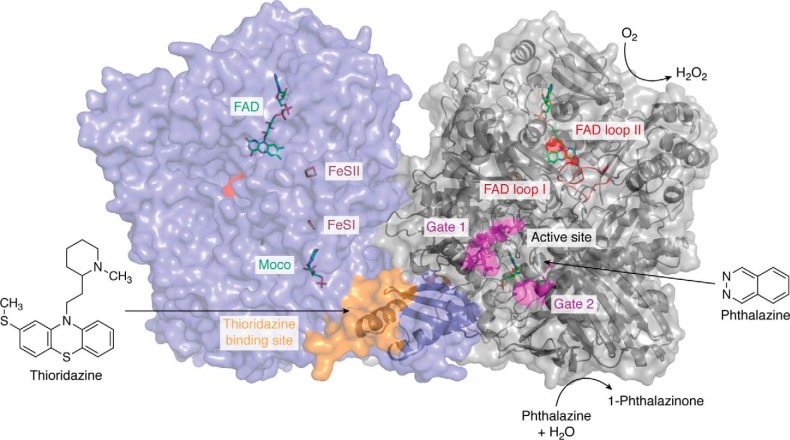
To obtain further insights into differences of the four AOX enzymes, and additionally to reveal substrate- and inhibitor-binding sites, attempts have been made to solve the crystal structures of the four enzymes from mice and AOX1 from humans. So far, only the crystal structures of human AOX1 and mAOX3 are available. However, our knowledge on the molecular details of AOXs has greatly increased based on the 2.5 Å (PDB: 4UHW) structure of human AOX1 in addition to a 2.6 Å structure in complex with the substrate phthalazine and the inhibitor thioridazine (PDB: 4UHX) (Fig. 6) (3). These enzymes were purified after heterologous expression in E. coli. The crystal structure of mAOX3 has been determined at 2.8 Å resolution (38, 50) (PDB: 3ZYV) from an enzyme that was directly purified from mouse livers. Overall, human AOX1 and mAOX3 are characterized by a 65% amino acid sequence identity and are very similar in terms of overall fold.
Figure 6.
The crystal structure of the human AOX1 homodimer in complex with phthalazine (substrate) and thioridazine (inhibitor). The different protein cofactors (FAD, FeSII, FeSI, and Moco) are indicated and are shown in a color-coded stick representation on the left monomer. The phthalazine substrate as well as the substrate-binding site, including the flexible Gates 1 and 2, are marked in pink on the right monomer. The thioridazine-binding site is marked in orange. The flexible loops at the FAD site (loop I and loop II) are marked in red (surface representation on the left monomer, cartoon representation on the right monomer). The figure was generated using PDB entry 1UHW published by Coelho et al. (3).
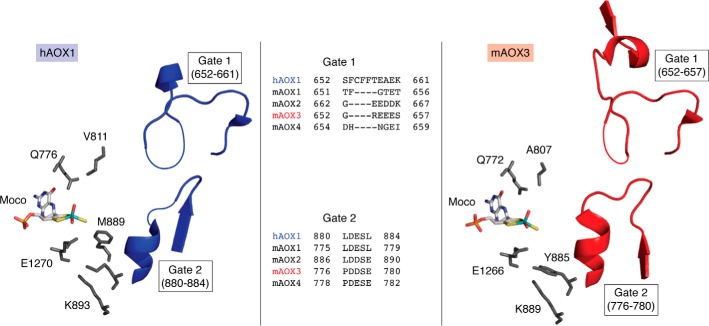
Details on substrate binding at the Mo active site were obtained from the crystal structure of the phthalazine-thioridazine complex cocrystallized with human AOX1 (Fig. 6) (3). The structure is deemed to contain two putative gates controlling access of the substrate and release of the product from the Moco active site. Both gates correspond to mobile, unordered regions, with Gate 1 consisting of the 652SFCFFTEAEK661 and Gate 2 consisting of the 880LDESL884 amino acid sequences (Fig. 7). By comparison, Gate 2 in mAOX3 and also in the more distinctly related bovine XOR is remarkably ordered. In human AOX1, Gate 1 is predominantly composed of bulky hydrophobic residues. With the exception of Glu660 (Asp in mAOX2), Gate 1 amino acid residues are not conserved in other members of the XOR family. In addition, Gate 1 is shorter in mouse AOX2, AOX3, and AOX4. In all AOX enzymes, Asp881 of Gate 2 is strictly conserved, and it is always followed by another acidic residue (Glu or Asp). The side chains of these amino acids point toward the substrate channel and are likely to be involved in binding or orienting polar substrates toward the catalytic center. The active site of human AOX1 contains a few residues (Glu1270, Phe923, Lys893, and Gln776) that are conserved in all mammalian AOXs (Fig. 7). Glu1270 is a key residue present in all members of the xanthine oxidase family, whereas Lys893 is specific to AOXs.
Figure 7.
The active site of hAOX1 and mAOX3. Shown are the residues of hAOX1 (left) and mAOX3 (right) surrounding the Moco and the location of Gate 1 and Gate 2 at the entrance of the substrate funnel. Missing residues in the electron density (and therefore not present in the coordinate files) are indicated by thin lines. An amino acid sequence alignment of Gate 1 and Gate 2 of the human and mouse enzymes is given. The figure was created using PyMOL version 2.1.1.
Substrate specificities of the mammalian AOX enzymes
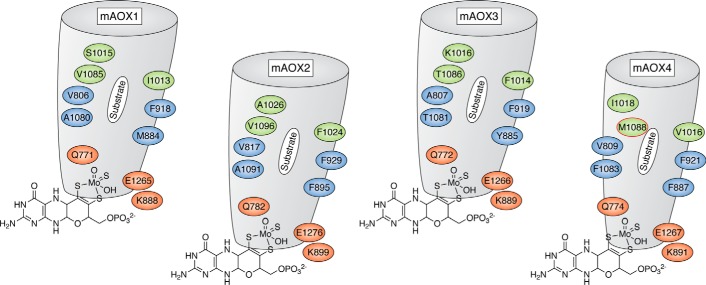
Mouse AOX1, AOX3, AOX4, and AOX2 proteins are characterized by ∼60% sequence similarity and show the same domain and cofactor organization (2). To provide information on differences in the substrate-binding regions and the funnels leading to the active site, homology modeling based on the available mAOX3 crystal structure (PDB: 3ZYV) has been performed on mouse AOX1, AOX2, and AOX4 (54). The results point to major differences among mouse AOX1, AOX2, AOX3, and AOX4 in the substrate-binding region. On the basis of these data, it has been proposed that the substrate-binding site consists of a region that builds the inner active site consisting of the amino acids Gln772, Ala807, Phe919, Phe1014, Lys889, and Glu1266 (mAOX3 numbering) (Fig. 8). The more distal amino acids in the substrate-binding tunnel are predicted to form an enzyme-specific region, which shows remarkable differences in the four mAOX enzymes. From these comparisons, mAOX1 is proposed to have the widest substrate-binding funnel. Conversely, mAOX4 is proposed to have the narrowest funnel, which is shaped by several bulky amino acid side chains. Mouse AOX2 and AOX3 are proposed to contain funnels of similar width, but with the mAOX2 funnel likely to be more hydrophobic (54).
Figure 8.
Active site and substrate binding funnel in mouse AOX enzymes. Representation of the substrate binding sites in mouse AOX1, AOX2, AOX3, and AOX4 in addition to residues in the conserved and nonconserved substrate-binding funnel, which selects the substrate specificity. The representation of mAOX3 is taken from the crystal structure (PDB: 3ZYV) (38), whereas those of mAOX1, mAOX2, and mAOX4 are taken from the modeled structures (54). Residues in red indicate the amino acids whose nature is conserved in all mouse AOX enzymes; residues in blue are hydrophobic residues, partially conserved and involved in substrate orientation; and residues in green are those specific for AOX4, with the AOX4-Met1088 residue highlighted in red. The funnel for mAOX4 is predicted to be smaller compared with the other enzymes.
To test whether the predicted substrate selectivity of the four mouse AOX enzymes can be explained by the shape of the entrance pocket, the kinetic parameters of the four mAOX enzymes have been compared in a study using 30 different structurally related aromatic and aliphatic substrates (61). In this study, the 30 substrates considered were divided into azo-heterocyclic compounds and molecules containing an aldehyde group (benzaldehydes, alkyl aldehydes, and structurally related cinnamaldehydes). As a general outcome, this study showed that the three mouse AOX1, AOX2, and AOX3 enzymes have no significant differences in substrate selectivity (61). Mouse AOX1 and AOX3 are characterized by the highest catalytic activities against the majority of the common substrates considered (a general summary of selected substrates is given in Table 1) (61). Both mouse AOX1 and mouse AOX3 are mainly expressed in the liver and have evolved overlapping substrate specificities with high substrate turnover rates. It has to be highlighted that AOX1 does not use N1-methylnicotinamide as substrate. The mAOX3 enzyme thereby often is more effective than mAOX1 under lower substrate concentrations (related to a lower Km) (61). This role might be beneficial in the liver, where most substrate degradation pathways occur. Thus, mAOX1 and mAOX3 might cover different niches at different substrate concentrations. The mAOX2 enzyme is also active with all substrates tested; however, the enzyme is often more effective under lower substrate concentrations (61). The role of mAOX2 in rodents still needs to be elucidated in more detail in the future. Expression of mAOX2 is highly restricted to the Bowman's gland in the nasal cavity, but no specific substrate for mAOX2 has been identified. Overall, the substrate specificities of mAOX2 are overlapping with those of mAOX1 and mAOX3. Lower substrate concentrations might be present in the Bowman's gland as compared with the liver, explaining the higher catalytic efficiencies of the enzyme, and likely, high turnover rates are also not mandatory in this tissue.
Table 1.
Substrate specificities of the mouse AOX enzymes
ND, not determined; ++++, very good substrate (kcat > 300 min−1); +++, good substrate (kcat = 120–300 min−1); ++, moderate substrate (kcat = 50–120 min−1); +, poor substrate (kcat = 15–50 min−1); −, substrate not converted.
| AOX1 | AOX2 | AOX3 | AOX4 | |
|---|---|---|---|---|
| Former names | AOX1 | AOH3, AOX3L1 | AOH1 | AOH2 |
| Potential functions | ND | ND | ND | Adipogenesis, circadian clock, locomotor activity |
| Predominant expression in tissue | Liver | Bowman's gland | Liver | Harderian gland |
| Rate of O2˙̄ production in moles per mole of substrate | 30% | 40% | 20% | 20% |
| Preferred substrates/products: Aromatic aldehydes | ||||
| Benzaldehyde → benzoic acid | +++ | +++ | +++ | ++ |
| Vanillin → vanillic acid | ++++ | ++ | +++ | + |
| 2-Methoxybenzaldehyde → 2-methoxybenzoic acid | ++++ | + | ++++ | − |
| N-Heterocyclic compounds | ||||
| Phthalazine → phthalazinone | ++ | +++ | +++ | ++++ |
| Phenanthridine → phenantridinone | +++ | +++ | +++ | − |
| Purine → hydroxypurine | ++++ | +++ | ++++ | + |
| N1-Methylnicotinamide → 6-pyridone | − | ++ | + | − |
| Cinnamaldehyde-related compounds | ||||
| Cinnamaldehyde → cinnamic acid | +++ | ++ | +++ | − |
| 4-Dimethylamino cinnamaldehyde → 4-dimethylamino cinnamic acid | +++ | ++ | ++ | + |
| Aliphatic aldehyde | ||||
| Hexanal → hexanoic acid | +++ | + | ++ | + |
Conversely, mAOX4 is generally the least active enzyme with the highest selectivity toward the substrates considered. In particular, mAOX4 is unable to use phenanthridine, N1-methylnicotinamide, ethylvanillin, and methoxybenzaldehyde as substrates and is characterized by a poor reactivity toward purine, whereas these compounds are generally good substrates for most of the other three mouse AOX enzymes (Table 1). Conclusively, mAOX4 is unlikely to play an important role in the oxidation of aromatic heterocyclic compounds. In addition and consistent with the predicted hydrophobicity of the substrate-binding funnel, mAOX4 does not recognize hydrophilic aromatic substrates and some N-heterocyclic compounds (54).
In conclusion, from the overlapping substrate specificities of mouse AOX1, AOX2, and AOX3, similar substrate-binding cavities were predicted according to modeling studies (54) (Fig. 8). The kinetic data show that only mAOX4 has a predicted narrower substrate-binding funnel, suggesting a higher selectivity, lower turnover numbers, and catalytic efficiencies for most substrates tested (Table 1 and Fig. 8). In general, the substrate selectivities predicted by only computationally based analyses were not confirmed by kinetic data (54).
To identify the amino acids that are specific for the substrate selectivity of mAOX4, site-directed mutagenesis was performed. To this purpose, residues Val1016, Ile1018, and Met1088 in AOX4 (Fig. 8) were exchanged into the corresponding amino acids present in the other mAOX enzymes. These residues are not conserved and were identified to be specific for mAOX4 (61). The M1088T exchange had a major effect on the kinetic constants of the AOX4 enzyme. The kcat of this variant was increased with benzaldehyde, salicylaldehyde, phthalazine, and pentanal compared with the corresponding value of WT counterpart (61). Thus, this amino acid largely contributes to substrate binding and turnover. Conclusively, whereas the role of mAOX4 is specific to the Harderian gland, the enzyme apparently has not evolved for high substrate turnover in this tissue, because one selected amino acid exchange in an amino acid specific for mAOX4 led to an increase in activity, whereas the substrate selectivity was unaltered in this variant.
In general, the data discussed above indicate that there is an urgent need to obtain the crystal structures of the three mouse AOX enzymes (AOX1, AOX2, and AOX4), as this is the only way to identify the structural differences at the basis of the experimentally determined profiles of substrate selectivity. In fact, computational modeling data seem to provide pieces of information that are not sufficient to predict the substrate specificity of mouse AOX1, AOX2, AOX3, and AOX4.
The role of mammalian AOXs in the production of reactive oxygen species
AOXs are cytosolic proteins and use molecular oxygen as the sole electron acceptor. During substrate oxidation and molecular oxygen reduction, AOXs generate mainly H2O2 in addition to a proportion of O2˙̄ radicals. So far, XOR in its XO form has been considered to be the only molybdo-flavoenzyme involved in the generation of O2˙̄ (62–64). The fact that also AOXs produce a significant amount of O2˙̄ radicals has been largely overlooked in the past and might be of patho-physiological interest (65). Particularly, it is noticeable that hepatic AOX1 is calculated to generate 24-fold larger amounts of O2˙̄ than XOR. These calculations are based on the relative levels of XO and AOX enzymatic activity in human liver (17, 65). Thus, AOX1 and other mammalian AOXs may represent significant sources of ROS in the cytosol of liver and other tissues and may play a critical role in ROS-mediated tissue injury under specific conditions (66, 67). Given this background, the abilities of the purified mouse AOX1, AOX2, AOX3, and AOX4 enzymes to produce O2˙̄ have been compared (53). The four mAOX enzymes generally show a different rate of O2˙̄ production in relation to the amount of substrate converted. In particular, mAOX2 is the most efficient producer of superoxide anions, with a rate of 40% (Table 1). As the enzyme is located in the nasal mucosa, which is one of the main sites of entrance for infective agents, it might be feasible to speculate that mAOX2 produces ROS from endogenous substrates to protect mice from viral and bacterial infections. A similar role might be played by human and mAOX1 in the liver. Also, mAOX1 was shown to produce a ratio of 30% superoxide radicals, which is higher than the reported value of 15–20% for XO (67). In contrast, mAOX3 and mAOX4 produce a rate of 20% O2˙̄ radicals per mole of substrate converted, a ratio that is significantly lower as compared with mAOX1 and mAOX2. In this and other tissues, it is also possible that AOX-dependent production of O2˙̄ might be involved in some of the toxic effects triggered by drugs and xenobiotics.
Potential physiological functions of mammalian AOXs in endogenous pathways
The functional role of AOXs in the vertebrate organisms is still largely unknown, although the broad substrate specificity supports the idea that these enzymes are involved in numerous metabolic pathways. In addition to the substrates shown in Table 1, it has to be noted that under hypoxic conditions, AOXs can also catalyze the reduction of N-oxides, sulfoxides, nitro-compounds, and heterocycles (12, 68–71). More recently, it was also demonstrated that AOXs hydrolyze amides (12, 72, 73). Despite this, still very little information is available on the endogenous substrates recognized by AOXs and the metabolic pathways these enzymes are involved in.
As mentioned above, AOXs may play a role in tryptophan metabolism, as metabolomic analyses conducted in Aox4 knockout mice demonstrated that tryptophan and 5-hydroxyindolacetic acid levels are higher in the Harderian gland of genetically engineered than in that of control animals (62).
Nicotinamide catabolism is another metabolic pathway that may be partially controlled by AOXs. In fact, AOXs oxidize N1-methylnicotinamide into N1-methyl-2-pyridone-5-carboxamide and N1-methyl-4-pyridone-5-carboxamide. N1-Methylnicotinamide is metabolized by AOX-enriched extracts obtained from human (74), monkey (74), rat (75), rabbit (76), and guinea pig liver (77), suggesting that the compound may be recognized by human and other mammalian AOX1 isoenzymes or mammalian AOX3. In this case, only purified mouse AOX3 and AOX2 proteins have been shown to recognize N1-methyl-nicotinamide as a substrate (44).
AOXs are also suggested to be involved in vitamin A metabolism. In fact, AOXs are implicated in the oxidation of 9-cis- and all-trans-retinal into the corresponding retinoic acids. All-trans-retinoic acid is the active metabolite of vitamin A, and it controls many aspects of vertebrate homeostasis in both the developing and adult organisms. In vertebrate embryos, all-trans-retinoic acid is a well-known morphogen, and it regulates the development of numerous tissues and organs, including the central nervous system. In adult organisms, the molecule influences numerous biological processes, including vision as well as growth/differentiation of skin epithelial and hematopoietic cells. At present, all-trans-retinal is the candidate endogenous substrate of AOXs for which the greatest amount of supporting evidence is available. Purified mouse liver AOX1 (78, 79) and AOX3 (80), Harderian gland AOX4 (57), and nasal AOX2 (81) proteins recognize retinaldehyde as a substrate. The data were confirmed by using purified recombinant enzymes, which demonstrates that mAOX1 is endowed with the highest retinaldehyde-metabolizing activity. In line with this, a marked decrease in the levels of all-trans-retinoic acid is evident in the Harderian gland and skin of Aox4 knockout mice relative to what is observed in WT animals (57).
Roles in drug and xenobiotic metabolism
The presence of AOX1 and AOX1/AOX3 in the hepatic tissue is the basis for the role played by this class of enzymes in the metabolism of exogenous compounds, such as drugs and environmental pollutants humans and other mammals are exposed to (25, 38, 82). AOXs metabolize a variety of anti-tumor, immunosuppressive, anti-malarial, and anti-viral drugs as well as molecules acting in the central nervous system. As for the anti-tumor and immunosuppressive agents, methotrexate and 6-mercaptopurine are two prominent examples of AOX substrates. In the context of antimalarial and anti-viral drugs, liver AOXs have been shown to oxidize cryptolepine into cryptolepine-11-one (83) and quinine into 2-quinone (84).
AOXs metabolize not only drugs, but also molecules of toxicological interest, such as the environmental pollutants, phthalazines, which are classical AOX substrates. Different AOXs oxidize phthalazine into 1-hydroxyphthalazine, which undergoes irreversible isomerization into 1-phthalazinone (3, 16, 85). Phthalazine administration increases rabbit liver AOX activity, demonstrating that the compound is not only a substrate, but it is also an AOX inducer (86). Two other examples of relevant AOX substrates are caffeine (87) and the aromatic aldehyde, vanillin. Vanillin is a sweetener, and it is oxidized into the corresponding carboxylic acid by AOXs. Nitro-compounds are further prototypes of toxic agents with the potential to be metabolized by mammalian AOXs. Indeed, molecules containing a nitro functionality exemplify the ability of AOXs to act not only as oxidases, but also as reductases (13, 71, 88–90). For instance, various AOX isoenzymes have the potential to reduce 2-nitrofluorene, 1-nitropyrene, and 4-nitrobiphenyl, three widespread pollutants, into the corresponding amines. Interestingly, AOX-dependent reduction of these substrates is observed not only in the liver (13), but also in the skin of mice (88, 89). This has important ramifications for the human organism, as our skin contains AOX1 enzymatic activity (91), and human AOX1-dependent reduction of nitro-compounds may represent an important defense reaction against environmental pollutants.
Conclusions
The role of human AOX1 in the metabolism of xenobiotics is of general significance in drug development programs (20, 26, 27). However, the animal models used during preclinical studies represent a major problem, because unlike humans, some of the most popular animal models, such as rodents, are characterized by expression of other AOXs besides AOX1. Further, dogs and cats are not good experimental tools for metabolic studies involving human AOX1 substrates (27) because of inactivation of the AOX1 and AOX3 genes by pseudogenization (24, 92). Thus, it is not surprising that canine studies have failed to detect metabolites of AOX substrates either in vitro or in vivo (93, 94).
The decrease in the number of active AOX isoenzymes from rodents to humans is the result of progressive deletion/inactivation of the corresponding genes. Combining the studies summarized above, it is possible that, at least in part, the functions carried out by rodent AOX1, AOX2, AOX3, and AOX4 are combined in human AOX1, making it a multipurpose enzyme. This may have resulted from evolutionary changes in the substrate-binding funnel of human AOX1, which is wider and consequently has resulted in an increase in the ability of the enzyme to recognize physiological substrates originally specific to AOX2, AOX3, and AOX4. However, a summary of the data present in the literature mainly on the mouse enzymes implies that AOX2 and AOX4 exert specialized functions in organs and tissues that are no longer active or have become dispensable in humans. The presence of two enzymes, mAOX1 and mAOX3, in the liver might be beneficial to mice to cover a larger range of substrate concentrations to be converted by the two enzymes. More detailed future studies in addition to knockout mice in each individual AOX enzyme are expected to provide more information on the patho-physiological and tissue-specific roles of these enzymes. In particular, the role of AOX4 in adipogenesis, as revealed by the Aox4 knockout mouse, provides important information that might be beneficial for the treatment of obesity in humans.
This work was supported by Deutsche Forschungsgemeinschaft Grant Le1171/8-3 (to S. L.) and by FCT-MCTES, research unit UCIBIO (UID/Multi/04378/2019) and project PTDC/BBB-BEP/1185/2014 (to M. J. R.). This work was also supported in part by Fondazione Italo Monzino and the Associazione per la Ricerca sul Cancro (AIRC) (to E. G.). The authors declare that they have no conflicts of interest with the contents of this article.
AOX2 was originally named AOH3 and then renamed as AOX3L1; AOX4 was originally denominated as AOH1; AOX4 was originally denominated as AOH2.
- AOX
- aldehyde oxidase
- Moco
- molybdenum cofactor
- XO
- xanthine oxidase
- XOR
- xanthine oxidoreductase
- XDH
- xanthine dehydrogenase
- Mo-MPT
- molybdopterin with bound Mo
- MCD
- molybdopterin cytosine dinucleotide
- MOP
- D. gigas aldehyde oxidoreductase
- ROS
- reactive oxygen species
- PaoABC
- periplasmic aldehyde oxidoreductase
- PDB
- Protein Data Bank.
References
- 1. Hille R., Hall J., and Basu P. (2014) The mononuclear molybdenum enzymes. Chem. Rev. 114, 3963–4038 10.1021/cr400443z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Terao M., Romão M. J., Leimkühler S., Bolis M., Fratelli M., Coelho C., Santos-Silva T., and Garattini E. (2016) Structure and function of mammalian aldehyde oxidases. Arch. Toxicol. 90, 753–780 10.1007/s00204-016-1683-1 [DOI] [PubMed] [Google Scholar]
- 3. Coelho C., Foti A., Hartmann T., Santos-Silva T., Leimkühler S., and Romão M. J. (2015) Structural insights into xenobiotic and inhibitor binding to human aldehyde oxidase. Nat. Chem. Biol. 11, 779–783 10.1038/nchembio.1895 [DOI] [PubMed] [Google Scholar]
- 4. Enroth C., Eger B. T., Okamoto K., Nishino T., Nishino T., and Pai E. F. (2000) Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion. Proc. Natl. Acad. Sci. U.S.A. 97, 10723–10728 10.1073/pnas.97.20.10723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kisker C., Schindelin H., Pacheco A., Wehbi W. A., Garrett R. M., Rajagopalan K. V., Enemark J. H., and Rees D. C. (1997) Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase. Cell 91, 973–983 10.1016/S0092-8674(00)80488-2 [DOI] [PubMed] [Google Scholar]
- 6. Kubitza C., Bittner F., Ginsel C., Havemeyer A., Clement B., and Scheidig A. J. (2018) Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes. Proc. Natl. Acad. Sci. U.S.A. 115, 11958–11963 10.1073/pnas.1808576115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duran M., Beemer F. A., van de Heiden C., Korteland J., de Bree P. K., Brink M., Wadman S. K., and Lombeck I. (1978) Combined deficiency of xanthine oxidase and sulphite oxidase: a defect of molybdenum metabolism or transport? J. Inher. Metab. Dis. 1, 175–178 10.1007/BF01805591 [DOI] [PubMed] [Google Scholar]
- 8. Shih V. E., Abroms I. F., Johnson J. L., Carney M., Mandell R., Robb R. M., Cloherty J. P., and Rajagopalan K. V. (1977) Sulfite oxidase deficiency. Biochemical and clinical investigations of a hereditary metabolic disorder in sulfur metabolism. N. Engl. J. Med. 297, 1022–1028 10.1056/NEJM197711102971902 [DOI] [PubMed] [Google Scholar]
- 9. Veldman A., Santamaria-Araujo J. A., Sollazzo S., Pitt J., Gianello R., Yaplito-Lee J., Wong F., Ramsden C. A., Reiss J., Cook I., Fairweather J., and Schwarz G. (2010) Successful treatment of molybdenum cofactor deficiency type A with cPMP. Pediatrics 125, e1249–e1254 10.1542/peds.2009-2192 [DOI] [PubMed] [Google Scholar]
- 10. Garattini E., Fratelli M., and Terao M. (2008) Mammalian aldehyde oxidases: genetics, evolution and biochemistry. Cell. Mol. Life Sci. 65, 1019–1048 10.1007/s00018-007-7398-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garattini E., Fratelli M., and Terao M. (2009) The mammalian aldehyde oxidase gene family. Hum. Genomics 4, 119–130 10.1186/1479-7364-4-2-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mota C., Coelho C., Leimkühler S., Garattini E., Terao M., Santos-Silva T., and Romao M. J. (2018) Critical overview on the structure and metabolism of human aldehyde oxidase and its role in pharmacokinetics. Coord. Chem. Rev. 368, 35–59 10.1016/j.ccr.2018.04.006 [DOI] [Google Scholar]
- 13. Tatsumi K., Kitamura S., and Narai N. (1986) Reductive metabolism of aromatic nitro compounds including carcinogens by rabbit liver preparations. Cancer Res. 46, 1089–1093 [PubMed] [Google Scholar]
- 14. Hutzler J. M., Obach R. S., Dalvie D., and Zientek M. A. (2013) Strategies for a comprehensive understanding of metabolism by aldehyde oxidase. Expert. Opin. Drug Metab. Toxicol. 9, 153–168 10.1517/17425255.2013.738668 [DOI] [PubMed] [Google Scholar]
- 15. Beedham C. (1998) Oxidation of carbon via molybdenum hydroxylases. Biomed. Health Res. 25, 39–52 [Google Scholar]
- 16. Beedham C., Bruce S. E., Critchley D. J., and Rance D. J. (1990) 1-Substituted phthalazines as probes of the substrate-binding site of mammalian molybdenum hydroxylases. Biochem. Pharmacol. 39, 1213–1221 10.1016/0006-2952(90)90265-M [DOI] [PubMed] [Google Scholar]
- 17. Krenitsky T. A., Neil S. M., Elion G. B., and Hitchings G. H. (1972) A comparison of the specificities of xanthine oxidase and aldehyde oxidase. Arch. Biochem. Biophys. 150, 585–599 10.1016/0003-9861(72)90078-1 [DOI] [PubMed] [Google Scholar]
- 18. Tatsumi K., Kitamura S., and Yamada H. (1983) Sulfoxide reductase activity of liver aldehyde oxidase. Biochim. Biophys. Acta 747, 86–92 10.1016/0167-4838(83)90125-5 [DOI] [PubMed] [Google Scholar]
- 19. Kitamura S., and Sugihara K. (2014) Current status of prediction of drug disposition and toxicity in humans using chimeric mice with humanized liver. Xenobiotica 44, 123–134 10.3109/00498254.2013.868062 [DOI] [PubMed] [Google Scholar]
- 20. Garattini E., and Terao M. (2012) The role of aldehyde oxidase in drug metabolism. Expert Opin. Drug Metab. Toxicol. 8, 487–503 10.1517/17425255.2012.663352 [DOI] [PubMed] [Google Scholar]
- 21. Obach R. S., Huynh P., Allen M. C., and Beedham C. (2004) Human liver aldehyde oxidase: inhibition by 239 drugs. J. Clin. Pharmacol. 44, 7–19 10.1177/0091270003260336 [DOI] [PubMed] [Google Scholar]
- 22. Barr J. T., and Jones J. P. (2011) Inhibition of human liver aldehyde oxidase: implications for potential drug-drug interactions. Drug Metab. Dispos. 39, 2381–2386 10.1124/dmd.111.041806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garattini E., Mendel R., Romão M. J., Wright R., and Terao M. (2003) Mammalian molybdo-flavoenzymes, an expanding family of proteins: structure, genetics, regulation, function and pathophysiology. Biochem. J. 372, 15–32 10.1042/bj20030121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kurosaki M., Bolis M., Fratelli M., Barzago M. M., Pattini L., Perretta G., Terao M., and Garattini E. (2013) Structure and evolution of vertebrate aldehyde oxidases: from gene duplication to gene suppression. Cell. Mol. Life Sci. 70, 1807–1830 10.1007/s00018-012-1229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanoh S., Tayama Y., Sugihara K., Kitamura S., and Ohta S. (2015) Significance of aldehyde oxidase during drug development: effects on drug metabolism, pharmacokinetics, toxicity, and efficacy. Drug Metab. Pharmacokinet. 30, 52–63 10.1016/j.dmpk.2014.10.009 [DOI] [PubMed] [Google Scholar]
- 26. Garattini E., and Terao M. (2011) Increasing recognition of the importance of aldehyde oxidase in drug development and discovery. Drug Metab. Rev. 43, 374–386 10.3109/03602532.2011.560606 [DOI] [PubMed] [Google Scholar]
- 27. Garattini E., and Terao M. (2013) Aldehyde oxidase and its importance in novel drug discovery: present and future challenges. Expert Opin. Drug Discov. 8, 641–654 10.1517/17460441.2013.788497 [DOI] [PubMed] [Google Scholar]
- 28. Pryde D. C., Dalvie D., Hu Q., Jones P., Obach R. S., and Tran T. D. (2010) Aldehyde oxidase: an enzyme of emerging importance in drug discovery. J. Med. Chem. 53, 8441–8460 10.1021/jm100888d [DOI] [PubMed] [Google Scholar]
- 29. Dobbek H., Gremer L., Meyer O., and Huber R. (1999) Crystal structure and mechanism of CO dehydrogenase, a molybdo iron-sulfur flavoprotein containing S-selanylcysteine. Proc. Natl. Acad. Sci. U.S.A. 96, 8884–8889 10.1073/pnas.96.16.8884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Romão M. J., Archer M., Moura I., Moura J. J. G., LeGall J., Engh R., Schneider M., Hof P., and Huber R. (1995) The crystal structure of xanthine oxidase related aldehyde oxidoreductase. Science 270, 1170–1176 10.1126/science.270.5239.1170 [DOI] [PubMed] [Google Scholar]
- 31. Bonin I., Martins B. M., Purvanov V., Fetzner S., Huber R., and Dobbek H. (2004) Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase. Structure 12, 1425–1435 10.1016/j.str.2004.05.014 [DOI] [PubMed] [Google Scholar]
- 32. Kappl R., Sielker S., Ranguelova K., Wegner J., Parschat K., Hüttermann J., and Fetzner S. (2006) Spectroscopic and biochemical studies on protein variants of quinaldine 4-oxidase: role of E736 in catalysis and effects of serine ligands on the FeSI and FeSII clusters. Biochemistry 45, 14853–14868 10.1021/bi061185a [DOI] [PubMed] [Google Scholar]
- 33. Holcenberg J. S., and Stadtman E. R. (1969) Nicotinic acid metabolism. 3. Purification and properties of a nicotinic acid hydroxylase. J. Biol. Chem. 244, 1194–1203 [PubMed] [Google Scholar]
- 34. Truglio J. J., Theis K., Leimkühler S., Rappa R., Rajagopalan K. V., and Kisker C. (2002) Crystal structures of the active and alloxanthine-inhibited forms of xanthine dehydrogenase from Rhodobacter capsulatus. Structure 10, 115–125 10.1016/S0969-2126(01)00697-9 [DOI] [PubMed] [Google Scholar]
- 35. Correia M. A., Otrelo-Cardoso A. R., Schwuchow V., Sigfridsson Clauss K. G., Haumann M., Romão M. J., Leimkühler S., and Santos-Silva T. (2016) The Escherichia coli periplasmic aldehyde oxidoreductase is an exceptional member of the xanthine oxidase family of molybdoenzymes. ACS Chem. Biol. 11, 2923–2935 10.1021/acschembio.6b00572 [DOI] [PubMed] [Google Scholar]
- 36. Hille R. (2005) Molybdenum-containing hydroxylases. Arch. Biochem. Biophys. 433, 107–116 10.1016/j.abb.2004.08.012 [DOI] [PubMed] [Google Scholar]
- 37. Hille R., Nishino T., and Bittner F. (2011) Molybdenum enzymes in higher organisms. Coord. Chem. Rev. 255, 1179–1205 10.1016/j.ccr.2010.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coelho C., Mahro M., Trincão J., Carvalho A. T., Ramos M. J., Terao M., Garattini E., Leimkühler S., and Romão M. J. (2012) The first mammalian aldehyde oxidase crystal structure: insights into substrate specificity. J. Biol. Chem. 287, 40690–40702 10.1074/jbc.M112.390419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Amaya Y., Yamazaki K., Sato M., Noda K., Nishino T., and Nishino T. (1990) Proteolytic conversion of xanthine dehydrogenase from the NAD-dependent type to the O2-dependent type: amino acid sequence of rat liver xanthine dehydrogenase and identification of the cleavage sites of the enzyme protein during irreversible conversion by trypsin. J. Biol. Chem. 265, 14170–14175 [PubMed] [Google Scholar]
- 40. Ichida K., Amaya Y., Noda K., Minoshima S., Hosoya T., Sakai O., Shimizu N., and Nishino T. (1993) Cloning of the cDNA encoding human xanthine dehydrogenase (oxidase): structural analysis of the protein and chromosomal location of the gene. Gene 133, 279–284 10.1016/0378-1119(93)90652-J [DOI] [PubMed] [Google Scholar]
- 41. Sato A., Nishino T., Noda K., Amaya Y., and Nishino T. (1995) The structure of chicken liver xanthine dehydrogenase: cDNA cloning and the domain structure. J. Biol. Chem. 270, 2818–2826 10.1074/jbc.270.6.2818 [DOI] [PubMed] [Google Scholar]
- 42. Massey V., and Edmondson D. (1970) On the mechanism of inactivation of xanthine oxidase by cyanide. J. Biol. Chem. 245, 6595–6598 [PubMed] [Google Scholar]
- 43. Nishino T., Ito R., and Tsushima K. (1975) Studies on chicken liver xanthine dehydrogenase with reference to the problem of non-equivalence of FAD moieties. Biochim. Biophys. Acta 403, 17–22 10.1016/0005-2744(75)90004-2 [DOI] [PubMed] [Google Scholar]
- 44. Wahl R. C., and Rajagopalan K. V. (1982) Evidence for the inorganic nature of the cyanolyzable sulfur of molybdenum hydroxylases. J. Biol. Chem. 257, 1354–1359 [PubMed] [Google Scholar]
- 45. Okamoto K., Matsumoto K., Hille R., Eger B. T., Pai E. F., and Nishino T. (2004) The crystal structure of xanthine oxidoreductase during catalysis: implications for reaction mechanism and enzyme inhibition. Proc. Natl. Acad. Sci. U.S.A. 101, 7931–7936 10.1073/pnas.0400973101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dixon M., and Keilin D. (1936) The action of cyanide and other respiratory inhibitors on xanthine oxidase. Proceedings of the royal society B 119, 159–190 10.1098/rspb.1936.0004 [DOI] [Google Scholar]
- 47. Nishino T., Okamoto K., Eger B. T., Pai E. F., and Nishino T. (2008) Mammalian xanthine oxidoreductase: mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J. 275, 3278–3289 10.1111/j.1742-4658.2008.06489.x [DOI] [PubMed] [Google Scholar]
- 48. Nishino T., Okamoto K., Kawaguchi Y., Hori H., Matsumura T., Eger B. T., Pai E. F., and Nishino T. (2005) Mechanism of the conversion of xanthine dehydrogenase to xanthine oxidase: identification of the two cysteine disulfide bonds and crystal structure of a non-convertible rat liver xanthine dehydrogenase mutant. J. Biol. Chem. 280, 24888–24894 10.1074/jbc.M501830200 [DOI] [PubMed] [Google Scholar]
- 49. Schumann S., Terao M., Garattini E., Saggu M., Lendzian F., Hildebrandt P., and Leimkühler S. (2009) Site directed mutagenesis of amino acid residues at the active site of mouse aldehyde oxidase AOX1. PLoS One 4, e5348 10.1371/journal.pone.0005348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mahro M., Coelho C., Trincão J., Rodrigues D., Terao M., Garattini E., Saggu M., Lendzian F., Hildebrandt P., Romão M. J., and Leimkühler S. (2011) Characterization and crystallization of mouse aldehyde oxidase 3: from mouse liver to Escherichia coli heterologous protein expression. Drug Metab. Dispos. 39, 1939–1945 10.1124/dmd.111.040873 [DOI] [PubMed] [Google Scholar]
- 51. Hartmann T., Terao M., Garattini E., Teutloff C., Alfaro J. F., Jones J. P., and Leimkühler S. (2012) The impact of single nucleotide polymorphisms on human aldehyde oxidase. Drug Metab. Dispos. 40, 856–864 10.1124/dmd.111.043828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Foti A., Hartmann T., Coelho C., Santos-Silva T., Romão M. J., and Leimkühler S. (2016) Optimization of the expression of human aldehyde oxidase for investigations of single nucleotide polymorphisms. Drug Metab. Dispos. 44, 1277–1285 10.1124/dmd.115.068395 [DOI] [PubMed] [Google Scholar]
- 53. Kücükgöze G., Terao M., Garattini E., and Leimkühler S. (2017) Direct comparison of the enzymatic characteristics and superoxide production of the four aldehyde oxidase enzymes present in mouse. Drug Metab. Dispos. 45, 947–955 10.1124/dmd.117.075937 [DOI] [PubMed] [Google Scholar]
- 54. Cerqueira N. M., Coelho C., Brás N. F., Fernandes P. A., Garattini E., Terao M., Romão M. J., and Ramos M. J. (2015) Insights into the structural determinants of substrate specificity and activity in mouse aldehyde oxidases. J. Biol. Inorg. Chem. 20, 209–217 10.1007/s00775-014-1198-2 [DOI] [PubMed] [Google Scholar]
- 55. Alfaro J. F., and Jones J. P. (2008) Studies on the mechanism of aldehyde oxidase and xanthine oxidase. J. Org. Chem. 73, 9469–9472 10.1021/jo801053u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bendotti C., Prosperini E., Kurosaki M., Garattini E., and Terao M. (1997) Selective localization of mouse aldehyde oxidase mRNA in the choroid plexus and motor neurons. Neuroreport 8, 2343–2349 10.1097/00001756-199707070-00048 [DOI] [PubMed] [Google Scholar]
- 57. Terao M., Kurosaki M., Barzago M. M., Fratelli M., Bagnati R., Bastone A., Giudice C., Scanziani E., Mancuso A., Tiveron C., and Garattini E. (2009) Role of the molybdoflavoenzyme aldehyde oxidase homolog 2 in the biosynthesis of retinoic acid: generation and characterization of a knockout mouse. Mol. Cell. Biol. 29, 357–377 10.1128/MCB.01385-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Terao M., Kurosaki M., Saltini G., Demontis S., Marini M., Salmona M., and Garattini E. (2000) Cloning of the cDNAs coding for two novel molybdo-flavoproteins showing high similarity with aldehyde oxidase and xanthine oxidoreductase. J. Biol. Chem. 275, 30690–30700 10.1074/jbc.M005355200 [DOI] [PubMed] [Google Scholar]
- 59. Terao M., Barzago M. M., Kurosaki M., Fratelli M., Bolis M., Borsotti A., Bigini P., Micotti E., Carli M., Invernizzi R. W., Bagnati R., Passoni A., Pastorelli R., Brunelli L., Toschi I., et al. (2016) Mouse aldehyde-oxidase-4 controls diurnal rhythms, fat deposition and locomotor activity. Sci. Rep. 6, 30343 10.1038/srep30343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weigert J., Neumeier M., Bauer S., Mages W., Schnitzbauer A. A., Obed A., Gröschl B., Hartmann A., Schäffler A., Aslanidis C., Schölmerich J., and Buechler C. (2008) Small-interference RNA-mediated knock-down of aldehyde oxidase 1 in 3T3-L1 cells impairs adipogenesis and adiponectin release. FEBS Lett. 582, 2965–2972 10.1016/j.febslet.2008.07.034 [DOI] [PubMed] [Google Scholar]
- 61. Kücükgöze G., and Leimkühler S. (2018) Direct comparison of the four aldehyde oxidase enzymes present in mouse gives insight into their substrate specificities. PLoS ONE 13, e0191819 10.1371/journal.pone.0191819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Harris C. M., and Massey V. (1997) The reaction of reduced xanthine dehydrogenase with molecular oxygen: reaction kinetics and measurement of superoxide radical. J. Biol. Chem. 272, 8370–8379 10.1074/jbc.272.13.8370 [DOI] [PubMed] [Google Scholar]
- 63. Lee M. C., Velayutham M., Komatsu T., Hille R., and Zweier J. L. (2014) Measurement and characterization of superoxide generation from xanthine dehydrogenase: a redox-regulated pathway of radical generation in ischemic tissues. Biochemistry 53, 6615–6623 10.1021/bi500582r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Terada L. S., Rubinstein J. D., Lesnefsky E. J., Horwitz L. D., Leff J. A., and Repine J. E. (1991) Existence and participation of xanthine oxidase in reperfusion injury of ischemic rabbit myocardium. Am. J. Physiol. 260, H805–H810 10.1152/ajpheart.1991.260.3.H805 [DOI] [PubMed] [Google Scholar]
- 65. Kundu T. K., Hille R., Velayutham M., and Zweier J. L. (2007) Characterization of superoxide production from aldehyde oxidase: an important source of oxidants in biological tissues. Arch. Biochem. Biophys. 460, 113–121 10.1016/j.abb.2006.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kundu T. K., Velayutham M., and Zweier J. L. (2012) Aldehyde oxidase functions as a superoxide generating NADH oxidase: an important redox regulated pathway of cellular oxygen radical formation. Biochemistry 51, 2930–2939 10.1021/bi3000879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hunt J., and Massey V. (1992) Purification and properties of milk xanthine dehydrogenase. J. Biol. Chem. 267, 21479–21485 [PubMed] [Google Scholar]
- 68. Kitamura S., and Tatsumi K. (1984) Reduction of tertiary amine N-oxides by liver preparations: function of aldehyde oxidase as a major N-oxide reductase. Biochem. Biophys. Res. Commun. 121, 749–754 10.1016/0006-291X(84)90742-3 [DOI] [PubMed] [Google Scholar]
- 69. Kitamura S., and Tatsumi K. (1984) Involvement of liver aldehyde oxidase in the reduction of nicotinamide N-oxide. Biochem. Biophys. Res. Commun. 120, 602–606 10.1016/0006-291X(84)91297-X [DOI] [PubMed] [Google Scholar]
- 70. Dick R. A., Kanne D. B., and Casida J. E. (2006) Substrate specificity of rabbit aldehyde oxidase for nitroguanidine and nitromethylene neonicotinoid insecticides. Chem. Res. Toxicol. 19, 38–43 10.1021/tx050230x [DOI] [PubMed] [Google Scholar]
- 71. Ogiso T., Fukami T., Mishiro K., Konishi K., Jones J. P., and Nakajima M. (2018) Substrate selectivity of human aldehyde oxidase 1 in reduction of nitroaromatic drugs. Arch. Biochem. Biophys. 659, 85–92 10.1016/j.abb.2018.10.017 [DOI] [PubMed] [Google Scholar]
- 72. Sodhi J. K., Wong S., Kirkpatrick D. S., Liu L., Khojasteh S. C., Hop C. E., Barr J. T., Jones J. P., and Halladay J. S. (2015) A novel reaction mediated by human aldehyde oxidase: amide hydrolysis of GDC-0834. Drug Metab. Dispos. 43, 908–915 10.1124/dmd.114.061804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lepri S., Ceccarelli M., Milani N., Tortorella S., Cucco A., Valeri A., Goracci L., Brink A., and Cruciani G. (2017) Structure-metabolism relationships in human-AOX: chemical insights from a large database of aza-aromatic and amide compounds. Proc. Natl. Acad. Sci. U.S.A. 114, E3178–E3187 10.1073/pnas.1618881114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sugihara K., Kitamura S., Tatsumi K., Asahara T., and Dohi K. (1997) Differences in aldehyde oxidase activity in cytosolic preparations of human and monkey liver. Biochem. Mol. Biol. Int. 41, 1153–1160 10.1080/15216549700202241 [DOI] [PubMed] [Google Scholar]
- 75. Ohkubo M., Sakiyama S., and Fujimura S. (1983) Increase of nicotinamide methyltransferase and N1-methyl-nicotinamide oxidase activities in the livers of the rats administered alkylating agents. Cancer Lett. 21, 175–181 10.1016/0304-3835(83)90205-7 [DOI] [PubMed] [Google Scholar]
- 76. Stoddart A. M., and Levine W. G. (1992) Azoreductase activity by purified rabbit liver aldehyde oxidase. Biochem. Pharmacol. 43, 2227–2235 10.1016/0006-2952(92)90182-I [DOI] [PubMed] [Google Scholar]
- 77. Yoshihara S., and Tatsumi K. (1985) Guinea pig liver aldehyde oxidase as a sulfoxide reductase: its purification and characterization. Arch. Biochem. Biophys. 242, 213–224 10.1016/0003-9861(85)90495-3 [DOI] [PubMed] [Google Scholar]
- 78. Huang D. Y., Furukawa A., and Ichikawa Y. (1999) Molecular cloning of retinal oxidase/aldehyde oxidase cDNAs from rabbit and mouse livers and functional expression of recombinant mouse retinal oxidase cDNA in Escherichia coli. Arch. Biochem. Biophys. 364, 264–272 10.1006/abbi.1999.1129 [DOI] [PubMed] [Google Scholar]
- 79. Vila R., Kurosaki M., Barzago M. M., Kolek M., Bastone A., Colombo L., Salmona M., Terao M., and Garattini E. (2004) Regulation and biochemistry of mouse molybdo-flavoenzymes: the DBA/2 mouse is selectively deficient in the expression of aldehyde oxidase homologues 1 and 2 and represents a unique source for the purification and characterization of aldehyde oxidase. J. Biol. Chem. 279, 8668–8683 10.1074/jbc.M308137200 [DOI] [PubMed] [Google Scholar]
- 80. Terao M., Kurosaki M., Marini M., Vanoni M. A., Saltini G., Bonetto V., Bastone A., Federico C., Saccone S., Fanelli R., Salmona M., and Garattini E. (2001) Purification of the aldehyde oxidase homolog 1 (AOH1) protein and cloning of the AOH1 and aldehyde oxidase homolog 2 (AOH2) genes: identification of a novel molybdo-flavoprotein gene cluster on mouse chromosome 1. J. Biol. Chem. 276, 46347–46363 10.1074/jbc.M105744200 [DOI] [PubMed] [Google Scholar]
- 81. Kurosaki M., Terao M., Barzago M. M., Bastone A., Bernardinello D., Salmona M., and Garattini E. (2004) The aldehyde oxidase gene cluster in mice and rats: aldehyde oxidase homologue 3, a novel member of the molybdo-flavoenzyme family with selective expression in the olfactory mucosa. J. Biol. Chem. 279, 50482–50498 10.1074/jbc.M408734200 [DOI] [PubMed] [Google Scholar]
- 82. Zientek M. A., and Youdim K. (2015) Reaction phenotyping: advances in the experimental strategies used to characterize the contribution of drug-metabolizing enzymes. Drug Metab. Dispos. 43, 163–181 10.1124/dmd.114.058750 [DOI] [PubMed] [Google Scholar]
- 83. Stell J. G., Wheelhouse R. T., and Wright C. W. (2012) Metabolism of cryptolepine and 2-fluorocryptolepine by aldehyde oxidase. J. Pharm. Pharmacol. 64, 237–243 10.1111/j.2042-7158.2011.01408.x [DOI] [PubMed] [Google Scholar]
- 84. Beedham C., al-Tayib Y., and Smith J. A. (1992) Role of guinea pig and rabbit hepatic aldehyde oxidase in oxidative in vitro metabolism of cinchona antimalarials. Drug Metab. Dispos. 20, 889–895 [PubMed] [Google Scholar]
- 85. Stubley C., Stell J. G., and Mathieson D. W. (1979) The oxidation of azaheterocycles with mammalian liver aldehyde oxidase. Xenobiotica 9, 475–484 10.3109/00498257909087261 [DOI] [PubMed] [Google Scholar]
- 86. Johnson C., Stubley-Beedham C., and Stell J. G. (1984) Elevation of molybdenum hydroxylase levels in rabbit liver after ingestion of phthalazine or its hydroxylated metabolite. Biochem. Pharmacol. 33, 3699–3705 10.1016/0006-2952(84)90159-X [DOI] [PubMed] [Google Scholar]
- 87. Castro G. D., Delgado de Layño A. M., Costantini M. H., and Castro J. A. (2001) Cytosolic xanthine oxidoreductase mediated bioactivation of ethanol to acetaldehyde and free radicals in rat breast tissue: its potential role in alcohol-promoted mammary cancer. Toxicology 160, 11–18 10.1016/S0300-483X(00)00433-9 [DOI] [PubMed] [Google Scholar]
- 88. Ueda O., Kitamura S., Ohashi K., Sugihara K., and Ohta S. (2003) Xanthine oxidase-catalyzed metabolism of 2-nitrofluorene, a carcinogenic air pollutant, in rat skin. Drug Metab. Dispos. 31, 367–372 10.1124/dmd.31.4.367 [DOI] [PubMed] [Google Scholar]
- 89. Ueda O., Sugihara K., Ohta S., and Kitamura S. (2005) Involvement of molybdenum hydroxylases in reductive metabolism of nitro polycyclic aromatic hydrocarbons in mammalian skin. Drug Metab. Dispos. 33, 1312–1318 10.1124/dmd.105.005306 [DOI] [PubMed] [Google Scholar]
- 90. Paragas E. M., Humphreys S. C., Min J., Joswig-Jones C. A., and Jones J. P. (2017) The two faces of aldehyde oxidase: oxidative and reductive transformations of 5-nitroquinoline. Biochem. Pharmacol. 145, 210–217 10.1016/j.bcp.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 91. Manevski N., Balavenkatraman K. K., Bertschi B., Swart P., Walles M., Camenisch G., Schiller H., Kretz O., Ling B., Wettstein R., Schaefer D. J., Pognan F., Wolf A., and Litherland K. (2014) Aldehyde oxidase activity in fresh human skin. Drug Metab. Dispos. 42, 2049–2057 10.1124/dmd.114.060368 [DOI] [PubMed] [Google Scholar]
- 92. Terao M., Kurosaki M., Barzago M. M., Varasano E., Boldetti A., Bastone A., Fratelli M., and Garattini E. (2006) Avian and canine aldehyde oxidases: novel insights into the biology and evolution of molybdo-flavoenzymes. J. Biol. Chem. 281, 19748–19761 10.1074/jbc.M600850200 [DOI] [PubMed] [Google Scholar]
- 93. Kaye B., Offerman J. L., Reid J. L., Elliott H. L., and Hillis W. S. (1984) A species difference in the presystemic metabolism of carbazeran in dog and man. Xenobiotica 14, 935–945 10.3109/00498258409151492 [DOI] [PubMed] [Google Scholar]
- 94. Klecker R. W., Cysyk R. L., and Collins J. M. (2006) Zebularine metabolism by aldehyde oxidase in hepatic cytosol from humans, monkeys, dogs, rats, and mice: influence of sex and inhibitors. Bioorg. Med. Chem. 14, 62–66 10.1016/j.bmc.2005.07.053 [DOI] [PubMed] [Google Scholar]