Abstract
Telomeres are specific nucleoprotein structures that are located at the ends of linear eukaryotic chromosomes and play crucial roles in genomic stability. Telomere DNA consists of simple repeats of a short G-rich sequence: TTAGGG in mammals and TTTAGGG in most plants. In recent years, the mammalian telomeric G-rich repeats have been shown to form G-quadruplex (G4) structures, which are crucial for modulating telomere functions. Surprisingly, even though plant telomeres are essential for plant growth, development, and environmental adaptions, only few reports exist on plant telomeric G4 DNA (pTG4). Here, using bulk and single-molecule assays, including CD spectroscopy, and single-molecule FRET approaches, we comprehensively characterized the structure and dynamics of a typical plant telomeric sequence, d[GGG(TTTAGGG)3]. We found that this sequence can fold into mixed G4s in potassium, including parallel and antiparallel structures. We also directly detected intermediate dynamic transitions, including G-hairpin, parallel G-triplex, and antiparallel G-triplex structures. Moreover, we observed that pTG4 is unfolded by the AtRecQ2 helicase but not by AtRecQ3. The results of our work shed light on our understanding about the existence, topological structures, stability, intermediates, unwinding, and functions of pTG4.
Keywords: single-molecule biophysics, G-quadruplex, DNA helicase, DNA-protein interaction, DNA topology, DNA intermediate, genome maintenance, genome stability, nucleoprotein, plant telomere
Introduction
In eukaryotes, special nucleoprotein structures called telomeres are located at the end of linear chromosomes. They perform two essential functions in genomic maintenance (1, 2). First, they protect chromosomes from end-to-end fusions and the chromosome ends from being recognized as double-strand breaks. Second, they prevent chromosomes from shortening caused by lagging-strand incomplete replication and replication fork collapse. Telomeres, referred to as the “telomere clock,” have close links to aging and cancer in humans and growth, development, and adaptions to environmental stresses in plants (3).
There are two conserved mechanisms that deal with chromosome shortening for maintaining telomere length in mammals, plants, and yeasts (4). One depends on telomerase, and the other is the alternative lengthening of telomeres (ALT)3 based on homologous recombination (5). Canonical telomeric DNA is composed of long tandem repeats of a short G-rich sequence: TTAGGG in vertebrates and TTTAGGG in most plants (4). The sequence d[GGG(TTAGGG)3] is able to fold into human telomeric G-quadruplex (hTG4) structures, which are four-stranded non–B-form secondary nucleic acid structures held together by Hoogsteen hydrogen bonds and further stabilized by monovalent cations (typically K+ or Na+) (6). The formation of hTG4 during telomeric replication can result in replication fork arrest and breakage (7). In addition, it will not only inhibit telomerase activity during extension (8) but also could present a barrier to ALT (9). Therefore, this structure must be modulated by telomere-associated proteins (2, 4), especially by helicases (10).
In recent decades, because of its essential biological functions, hTG4 has been widely researched using theoretical and experimental methods. Thus, many details about hTG4 have been revealed, including its topological structures (11–13), intermediates (14), dynamics (15–18), and unfolding (19–22). Surprisingly, there is little pTG4 information reported in the literature. Understanding of pTG4's biochemical and biophysical properties is crucial to determining its cellular functions. In this research, we set out to address three main questions: 1) Can the plant telomeric repeat motif d[GGG(TTTAGGG)3] fold into G4 structures? If so, what are the topological structures? 2) How do monovalent ions (such as K+) interact with pTG4, and what are the intermediates during pTG4 folding/unfolding? 3) If there is a pTG4 structure, can it be unfolded by helicases?
In this report, we used CD spectroscopy to measure the existence and structures of pTG4 and found that pTG4 formed antiparallel G4 in sodium and a mixture of parallel and antiparallel G4 in potassium. The existence was further confirmed by using a G-quadruplex–specific antibody, BG4 (23). Afterward, the basic binding parameters during pTG4-K+ interactions were measured and calculated using microscale thermophoresis (MST) (24). In addition, the single-molecule florescence resonance energy transfer (smFRET) method was used to detect the intermediates and dynamics of pTG4. We directly captured six folding dynamic states: ssDNA, G-hairpin, parallel G-triplex, antiparallel G-triplex, parallel G4, and antiparallel G4. Interestingly, the magnesium ion was found to play an important role in the stability of pTG4. Further, we showed that AtRecQ2, but not AtRecQ3, can unwind pTG4. Our results may be helpful for understanding the functions of pTG4 and focusing attention on G4 in the field of plant studies.
Results
pTG4 can fold into G4 structures
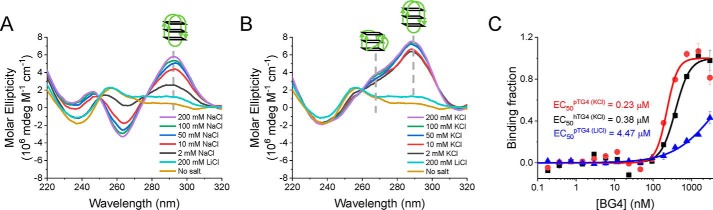
CD spectroscopy was widely used to identify the existence and topological structures of G4. Because K+ and Na+ are the physiologically relevant monovalent ions in plants and have great influences on the stability and topology of G4s (25), we measured the CD spectra of pTG4 (sequence listed in Table S1) in buffers containing 2–200 mm NaCl, 2–200 mm KCl, 200 mm LiCl, and no salt (Fig. 1, A and B). The CD spectrum in NaCl showed an ∼290-nm peak and an ∼260-nm valley, characterizing the typical CD spectrum of antiparallel G4 (13). However, the spectrum in KCl exhibited a peak at ∼290 nm with a small shoulder peak at ∼260 nm, indicating that there were mixed G4 structures containing both parallel and antiparallel (17) or hybrid G4 (26). We were more in favor of the first assumption (please see the section 'pTG4 shows six states in KCl'). The peaks in the CD spectrum of both NaCl and KCl became increasingly significant with the increasing salt concentration, confirming that the typical peaks came from the interaction of pTG4 with monovalent ions. In addition, just as expected, under the LiCl and no-salt conditions, the CD spectra represented no structured oligonucleotides (Fig. 1, A and B). Surprisingly, the CD patterns of pTG4 in NaCl and KCl were similar to those of hTG4, which also believed to have an antiparallel structure in NaCl and the mixed G4 structures (which included parallel and antiparallel structures (Fig. S1) (17)). The structural similarity suggests they may play a similar role in biological progresses, because structure determines function.
Figure 1.
pTG4 sequence is well-folded into G4 structures. A and B, CD spectra of pTG4 in different concentrations of the Na+ buffer (A) and K+ buffer (B). C, detection of G4 structures using BG4 antibody by the MST method. BG4 bound pTG4 (red line) and hTG4 (black line) structures with EC50 values of 0.23 ± 0.02 μm for pTG4 and 0.38 ± 0.09 μm for hTG4 in 100 mm KCl and 10 mm MgCl2, implying their comparable binding affinity. BG4 bound pTG4 (blue line) with EC50 values of 4.47 ± 0.09 μm in 100 mm LiCl, which did not support the formation of G4 structures, implying its good specificity.
To further confirm the folding of pTG4, a highly G4-specific monoclonal antibody, BG4 (23), was used. The hTG4 sequence has been reported to fold into stable G4 structures in 100 mm KCl (17–19). Using it as a control, the half-maximal effective concentration (EC50) values of 0.23 ± 0.02 μm for pTG4 and 0.38 ± 0.09 μm for hTG4 were measured by MST (Fig. S2B and Fig. 1C). The comparable binding affinity further indicated the formation of pTG4. In addition, the binding of BG4 with pTG4 sequence was checked in 100 mm LiCl (Fig. 1C), which did not support the formation of G4 structures (Fig. 1A). The EC50 value of 4.47 ± 0.09 μm in LiCl was ∼20 times higher than that in KCl, reflecting its good specificity. Thus, the reason why the binding affinities in this study are lower than their initial reports (23) may come from different experimental conditions.
The basic binding parameters during G4–K+ interactions
G4–cation interactions are the key processes of G4 folding/unfolding, during which ions bind to the center of the quartet arrangement channel to screen the charges and then stabilize the structure (25); the binding of an ion depends on its size. The smaller Na+ ion can bind to the quartet plane, whereas the larger K+ sits in the area between two quartets (27). Thus, three-layer G4 can maintain three Na+ ions but only two K+ ions. Under physiological conditions, plants maintain a relatively high K+ concentration (100–200 mm) and low Na+ concentration (1–10 mm) (28, 29); thus, the K+ structure should be biologically more relevant.
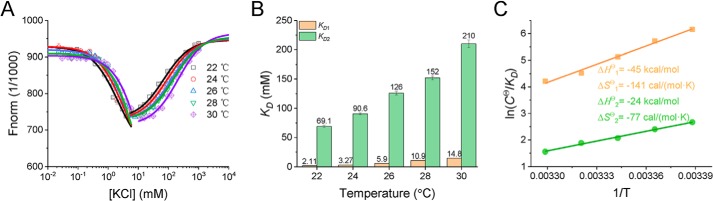
Thermodynamic parameters are essential to understand the interaction process. Recently, we developed an MST-based method to simultaneously and accurately monitor the K+ binding to and structural changes of hTG4. Thus, the basic binding parameters, including the equilibrium dissociation constants (KD), standard enthalpy change (ΔHθ), standard entropy change (ΔSθ), and standard Gibbs free energy change (ΔGθ), can be measured or calculated (24). Here, we used the same method and condition to detect the pTG4-K+ interaction. Fluorescently labeled pTG4 (FAM-pTG4) was mixed with increasing concentrations of KCl (0.03–1000 mm) at different temperatures (22–30 °C) and then the thermophoretic changes were plotted (Fig. 2A). Thereafter, the biphasic thermophoretic changes corresponding to the first and second K+ binding were fitted, allowing us to find their KD values (Fig. 2B). It should be noted that the KD value of pTG4 at the same temperature is larger than that of hTG4 (24), meaning that pTG4 is less stable than hTG4, because the smaller KD value represents a greater binding affinity and more energetically stability. This fact is consistent with the reports that an increase in the loop length decreases the thermodynamic stability (30, 31). Furthermore, the functions of ln (cθ/KD) versus 1/T were plotted and fitted by the van't Hoff equation (Fig. 2C). Therefore, the ΔHθ and ΔSθ of −45 ± 3 kcal mol−1 and −141 ± 8 cal mol−1 K−1 for the first K+ binding (Fig. 2C, orange) and −24 ± 1 kcal mol−1 and −77 ± 3 cal mol−1 K−1 for the second K+ binding (Fig. 2C, green) were achieved. In addition, from ΔGθ = ΔHθ − TΔSθ, the values of ΔGθ were calculated as −3.6 kcal mol−1 and −1.6 kcal mol−1 at 22 °C for the first and second K+ binding, respectively. Under the same experimental conditions, the ΔGθ values of hTG4 were reported to be −4.3 and −2.0 kcal mol−1 (24), which are more negative than these of pTG4, further supporting the idea that pTG4 is less stable than hTG4.
Figure 2.
Determination of the thermodynamic parameters of the pTG4-K+ interactions during pTG4 folding by MST. A, binding curves of K+ with pTG4 represented by normalized fluorescence (1/1000) were fitted using the law of mass action over the temperature range of 22–30 °C. B, KD values of K+ binding to pTG4 at different temperatures. KD1 and KD2 correspond to the first and second K+ binding, respectively. C, fitting of pTG4-K+ interaction parameters using the van't Hoff equation.
pTG4 shows six states in KCl
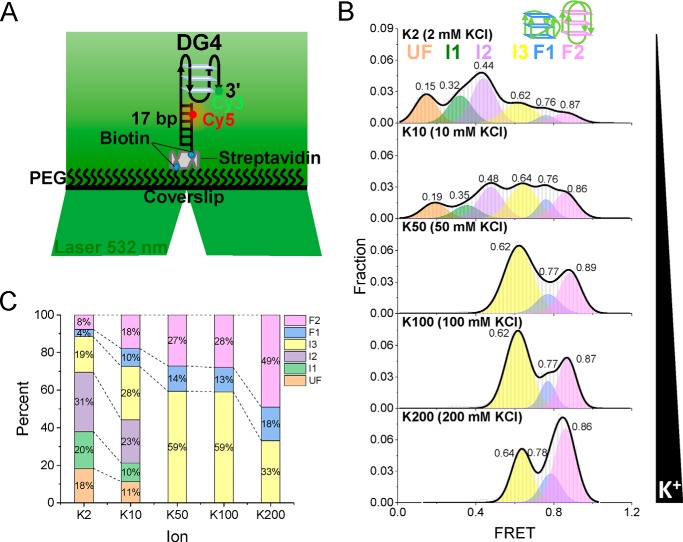
After detecting the binding parameters during the gradual folding of pTG4, the smFRET method was applied to monitor the structural change in real time at the single-molecule level to directly dissect the intermediates and dynamics of pTG4. A fluorescently labeled substrate (referred to as DG4) was designed (Fig. 3A). It was constructed with a single-strand oligonucleotide containing a Cy3 (donor) attached to the 3′ end of the pTG4 motif, and its 5′ tail was hybridized with a complementary stem strand modified by biotin at the 3′ end for immobilization and labeled by a Cy5 (acceptor) at the fifth nucleotide from the 5′ end. The fluorophores were spaced so that the FRET signal could sensitively reflect the conformational change of pTG4. With the increase of the K+ concentration, the G4 structures were gradually induced by the monovalent cations (Figs. 1B and 2A) (24, 27), resulting in an increase of the FRET efficiency (14). This fact gave us an opportunity to detect the intermediates and dynamics during G4 folding.
Figure 3.
Six states of pTG4 in K+ are distinguished from FRET. A, schematic view of the smFRET assay. The pTG4 containing overhang DNA was Cy3-labeled (green) and Cy5-labeled (red). A 532-nm laser was used to light up Cy3. Biotin was used for immobilization. B, FRET histograms for different concentrations of K+. Multipeak Gaussian distributions were used to fit those histograms. C, the fractions of the different folding structures at increasing concentrations of K+.
The pTG4 structures under various K+ concentrations were examined. The FRET efficiency histograms were shown in Fig. 3B, and the corresponding proportions were shown in Fig. 3C. As the K+ concentration increased, the population of low FRET decreased, and the proportion of high FRET increased. Surprisingly, at lower K+ concentrations (2 and 10 mm KCl), the FRET histograms could be well-fitted by a multipeak Gaussian distribution with six peaks. Representative traces were also recognized as the six states using the step-finding algorithm based on hidden Markov modeling (32) (Fig. S3, A and B). Furthermore, we built the transition density plots for 2 mm KCl from 47 single-molecule traces (14, 32). From transition density plots, the information of direct transitions between these states was captured (Fig. S4). Above 50 mm KCl, there were only three states left, and the corresponding representative traces were shown in Fig. S3 (C–E). This fact was consistent with the gradually increasing levels of G4 folding that were achieved with the increasing K+ concentration in the CD and MST experiments (Figs. 1B and 2A, respectively). Therefore, we assigned the lowest state (FRET ∼ 0.15–0.19) to the completely unfolded G4 and the highest two folding peaks of FRET ∼ 0.76 (F1) and ∼ 0.87 (F2) to the parallel and antiparallel G4 structures, respectively. Thus, the other three states should be intermediate states, referred to as I1, I2, and I3. These were very likely the G-hairpins and G-triplexes hypothesized in a number of recent studies (18, 19, 33–41). We assigned FRET of ∼0.76 (F1) to the parallel structures and FRET of ∼0.87 (F2) to the antiparallel structures for two reasons: 1) the fraction of parallel G4 was less than that of the antiparallel G4 in CD assay (Fig. 1B); and 2) the Cy3-Cy5 spatial interval of parallel G4 is farther than that of antiparallel G4 (17, 42), thus leading to a smaller FRET (43). Why was the folded G4 not a hybrid structure? If pTG4 folded into hybrid G4 under the experimental conditions, 1) there would be no FRET more than ∼ 0.75 (14, 44), and 2) there would at most four states captured, corresponding to the unfolded, G-hairpin, G-triplex, and G4 states. Considering the structural similarity of pTG4 with hTG4 in K+ (Fig. 1B and Fig. S1) and the state assignment of hTG4 (18, 19, 33–41), this outcome was reasonable.
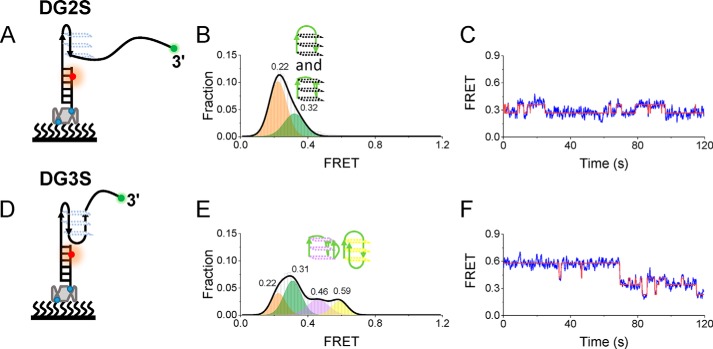
To further confirm the assignment states, putative G-hairpin and G-triplex structures were constructed by replacing the GGG column of DG4 with TTT (Fig. 4, A and D, where the structures were referred to as DG2S and DG3S). Under 100 mm KCl, the FRET distributions consisted of two Gaussian peaks at 0.22 and 0.32 for DG2S (Fig. 4B) and four Gaussian peaks at 0.22, 0.31, 0.46, and 0.59 for DG3S (Fig. 4E), consistent with the dynamic transitions shown in the representative traces (Fig. 4, C and F). DG2S can only fold into the G-hairpin second structure, so the lowest peak ∼ 0.22 should be a completely unfolded single-stranded DNA, and the peak 0.32 should be a G-hairpin. Therefore, considering that DG3S can fold into a G-hairpin and G-triplex, the peaks ∼0.46 and ∼0.59 should be parallel and antiparallel G-triplexes, respectively, which was proved recently by smFRET (36). It should be noted that the FRET values of DG2S and DG3S were close to the four lower FRET values of DG4 (Fig. 3B), supporting that intermediate states should be G-hairpins and G-triplexes in Fig. 3B, and the missing of FRET of ∼0.76 (F1) and ∼0.87 (F2) further supported the fact that F1 and F2 should be well-folded G4s.
Figure 4.
Formation of the G-hairpin and G-triplex. A and D, schematic diagram of DG2S (A) and DG3S (D). B and E, FRET distribution of DG2S (B) and DG3S (E) in 100 mm K+. Multiple-peak Gaussian distributions were used to fit their histograms. C and F, representative FRET traces of DG2S and DG3S in 100 mm K+. Two (C) and four (F) states were determined by hidden Markov modeling (red).
One may ask about why, in early smFRET research about G4s, there are no parallel and antiparallel G-triplexes identified at the same time (14, 19). We think this discrepancy comes from the difference in the substrates. In this study, the parallel (FRET = ∼0.46) and antiparallel (FRET = ∼0.59) G-triplexes were distinguished clearly because at FRET levels of ∼0.5, the change of the FRET is the most sensitive (43) (Fig. S5). In addition, because FRET = ∼0.32 is less sensitive, we cannot distinguish the potential parallel and antiparallel G-hairpins in our experiment. Therefore, the G-hairpin– and G-triplex–involved folding pathway of pTG4 was proposed (Fig. 5). During folding, both parallel and antiparallel pTG4 can go through the corresponding G-hairpin and G-triplex folds. It should be emphasized that in addition to the step-by-step transitions shown in Fig. 5, there were also some other direct transitions between these states (Fig. S4). These transitions may not require step-by-step transitions. Alternatively, intermediate states in these transitions may not be captured under the time resolution of our experiments (100 ms). It should also be noted that we had confirmed the intermediate states of pTG4; however, we could not rule out the existence of other second structures.
Figure 5.
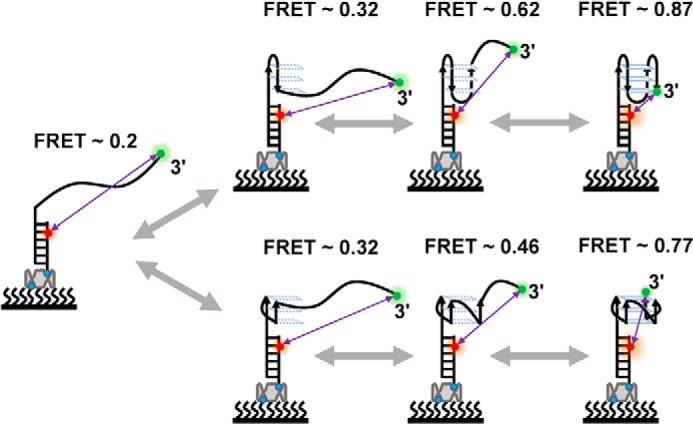
Proposed folding pathways of pTG4. The upper pathway is for the antiparallel pTG4 structure, and the lower one is for the parallel pTG4 structure. Purple lines indicate the distance between Cy3 (green dot) and Cy5 (red dot). The folding processes of both parallel and antiparallel pTG4 include ssDNA, G-hairpin, G-triplex, and G4, corresponding to the FRET values of ∼0.2, 0.32, 0.62, and 0.87 (antiparallel pTG4) and 0.2, 0.32, 0.46, and 0.77 (parallel pTG4), respectively.
Mg2+ can stabilize the pTG4 structure
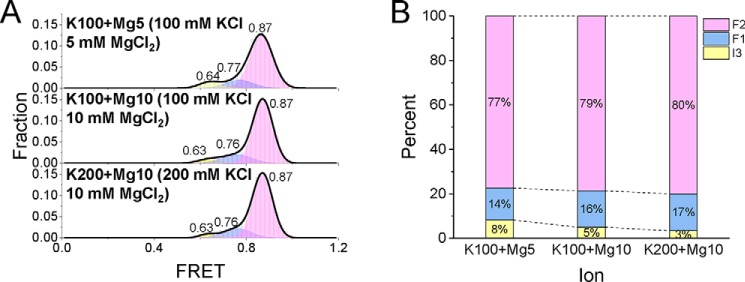
hTG4 can almost fold into stable G4 structures under a physiological K+ concentration (17, 42). However, the above results in this research showed that under a typical plant K+ concentration up to 200 mm, there was still ∼33% in the incompletely folded states (Fig. 3C). This fact is also consistent with the fact that pTG4 is less stable than hTG4, as shown in the MST result (Fig. 2). So why does one-third of the pTG4 not fold into a stable G4 structure? From the literature, we found that the most abundant divalent Mg2+ is one of most studied cations that assists with G4 stabilization (19, 34), and the concentration of Mg2+ is also equivalent to the millimolar level in the plant cell (45). Thus, a relatively low concentration of MgCl2 was added to the same smFRET system, yielding the FRET distributions (Fig. 6A) and corresponding fractions (Fig. 6B) in the simultaneous presence of Mg2+ and K+. The G4 folding fractions were obviously higher than the corresponding fractions in the absence of Mg2+ (compare Fig. 6B with Fig. 3C), indicating that Mg2+ can stabilize the pTG4 structure and may play a modulating role in plants.
Figure 6.
Mg2+ can promote the folding of pTG4. A, FRET distributions of DG4 in different concentrations of K+ and Mg2+. Multiple-peak Gaussian distributions were used to fit their histograms. B, the fractions of different folding states at increasing concentrations of K+ and Mg2+.
pTG4 can be unwound by AtRecQ2 but not AtRecQ3
The formation of stable G4s at the telomere will block the extension of the telomere through telomerase and ALT pathways (8, 9, 46), leading to the shortening of the telomere. Therefore, various specialized helicases are needed for unwinding them. Those helicases include, but are not limited to, RecQ family helicases, Pif1 family helicases, and iron–sulfur helicases (10). Even G4 is thought to play an important regulatory role in plants (47). However, to the best of our knowledge, no plant G4 unwinding helicases have been identified. Humans have five RecQ family members (HsRecQ1–5). The deficiencies of HsBLM (HsRecQ2), HsWRN (HsRecQ3), and HsRTS (HsRecQ4) cause the Bloom, Werner, and Rothmund–Thomson syndromes, respectively (48). Interestingly, these three helicases can robustly unwind G4 structures and join in telomere maintenance (48).
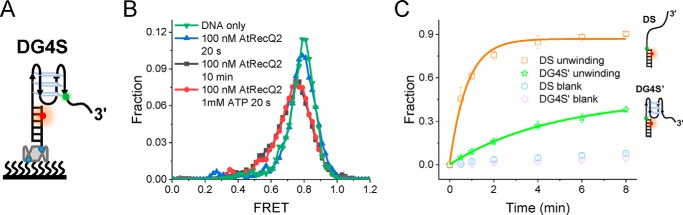
In Arabidopsis, there are seven RecQ-like helicases (49), but only AtRecQ2 and AtRecQ3 have been comprehensively studied in vitro experiments, showing they can unwind different recombination intermediates (50, 51). However, whether they can disrupt G4 is still unknown. We first expressed and purified these two helicase to homologues (Fig. S6A) and prepared a substrate by adding 10T to the 3′ of DG4, which was referred to as DG4S (Fig. 7A). This 3′ ssDNA has been shown to be indispensable for AtRecQ2 and AtRecQ3 unwinding (50, 51). In the presence of 10 mm Mg2+ and 100 mm K+, DG4S almost folded into G4 structures (Fig. 7B), which was consistent with Fig. 6. A previous smFRET study suggested HsBLM and HsWRN can disrupt hTG4 in the absence of ATP, just from binding (52). To check this possibility, we incubated 100 nm AtRecQ2 with DG4S and then recorded the FRET traces directly after 20 s and 10 min. After 20 s of incubation, only a negligible FRET change was observed. However, after 10 min of incubation, there was a significant FRET change captured (Fig. 7B). Interestingly, in the presence of 100 nm AtRecQ2 and 1 mm ATP, it only took 20 s to achieve the 10-min effect (Fig. 7B). The corresponding representative traces were shown in Fig. S6B. These results suggested that AtRecQ2 can indeed unfold pTG4 in the absence of ATP, but it is less efficient than in the presence of ATP, implying its potential role in telomere metabolism. However, no FRET change was detected in AtRecQ3 unwinding under the same experimental conditions of AtRecQ2 (Fig. S7A), indicating AtRecQ3 might have no (or a very weak) G4 unwinding ability. This difference may come from the lack of the helicase and RNaseD C-terminal (HRDC) domain in AtRecQ3 (49), which has been shown to be important for G4 disruption (53). Interestingly, human HRDC-lacking RecQ helicase HsRecQ5 has a partial G4 unwinding activity (54), and HsRecQ1 cannot even unwind G4 (55). These results indicate the importance of the HRDC domain during G4 unwinding.
Figure 7.
AtRecQ2 can unwind pTG4, and pTG4 hinders AtRecQ2 translocation in turn. A, schematic diagram of DG4S. B, FRET histograms for DG4S alone, in the presence of 100 nm AtRecQ2 and of 100 nm AtRecQ2 with 1 mm ATP at different times. C, unwinding fractions of DS and DG4S′ at different times after addition of 100 nm AtRecQ2 and 1 mm ATP in the presence of 100 mm KCl and 10 mm MgCl2.
In addition, we prepared an overhang substrate, replacing pTG4 with the same length of polyT (named DS) to measure the effect of pTG4 on AtRecQ2 (Fig. 7C). The full unwinding of the dsDNA of DS will lead to the release of Cy3-labeled ssDNA, showing the disappearance of both Cy3 and Cy5 fluorescent spots (20, 56). Thus, the unwinding fractions at different time were obtained from counting the number of Cy5 spots (Fig. 7C) in the presence of 100 nm AtRecQ2 and 1 mm ATP. Fitting the data to a single-exponential decay yielded the corresponding unwinding rate constant value of 1.1 ± 0.2 s−1. However, when pTG4 was restored, the unwinding fraction of DG4S′ at each time was less than that of DS, resulting in a rate constant value of 0.10 ± 0.01 s−1. The rate constant of DG4S′ was ∼10 times less than that of DS under the same experimental conditions, indicating that pTG4 hindered the translocation of AtRecQ2, because pTG4 must be first unfolded into ssDNA (53, 57). Without a 3′-ssDNA tail next to the duplex DNA, neither AtRecQ2 nor AtRecQ3 can unwind it (50, 51). For AtRecQ3, its translocation was also blocked by pTG4, just as expected (Fig. S7B). From another perspective, the blocking phenomenon further confirmed the formation of pTG4. The above results showed that the pTG4 structure was able to be efficiently unwound by AtRecQ2 but not AtRecQ3, and pTG4 acted as a translocation block for AtRecQ2 and AtRecQ3.
Discussion
We comprehensively studied the existence, stability, intermediates, and unwinding of pTG4, mainly using the single-molecule method (58); some unprecedented details were revealed. These results should be helpful for understanding the cellular functions of pTG4.
Recently, G4s were thought to play a key role in plants by bioinformatics (47, 59), and their existence was verified from G4-seq (60). However, this insight did not get as much attention as the same kind of insight about G4s in mammals. Telomeric G4 DNA is one of most studied G4 structures, because of the importance of telomeres for cells. Here, we used CD and BG4 to confirm the existence of pTG4 in vitro (Fig. 1). Surprisingly, the folded structures of pTG4 are very similar to those of mammals under the same conditions, strongly suggesting that they may share analogous molecular mechanisms during telomeric metabolism. Further, by using the MST method (24), the basic thermodynamic parameters of pTG4 sequence with K+ were revealed (Fig. 2) and compared with these of hTG4.
The dynamic folding/unfolding processes of pTG4 are essential for understanding its functions. Using G4-forming fluorescently labeled plant telomere sequence and mutations, six states were captured for the first time and assigned to different unfolding states, including ssDNA, G-hairpins, parallel G-triplex, antiparallel G-triplex, parallel G4, and antiparallel G4 at the single-molecule level (Figs. 3 and 4). These results allowed us to propose the folding/unfolding pathways of pTG4 (Fig. 5). It is worth mentioning that this is the first time that parallel and antiparallel G4s' folding/unfolding pathway was well-defined in one system, profiting from the sensitivity of Cy3-Cy5 FRET of ∼0.5 (Fig. S5). Before this study, there were only up to four states captured (14), and the parallel and antiparallel G-triplex states were monitored using different substrates (36). In addition, Mg2+ was proved to promote the stability of pTG4 and may play an essential role in pTG4 maintenance (Fig. 6).
During plant telomere extension, pTG4 must be unfolded, mainly by helicases (10). However, no G4 unfolding helicases have been identified before this study. In this report, we provided direct evidence that AtRecQ2 can unfold intramolecular pTG4, but AtRecQ3 cannot (Fig. 7 and Fig. S7). On the other hand, pTG4 acted as a translocation block for AtRecQ2 and AtRecQ3. AtRecQ2 and AtRecQ3 have been considered to be the homologues of the HsWRN and HsRecQ5β helicases, respectively (50, 51), which is consistent with the fact that HsWRN has strong G4 unwinding abilities, but HsRecQ5β has only partial G4 unwinding abilities (48, 54). There are incompatible reports that HsBLM can unwind human telomeric G4 dependent (19, 61) or independent (52) of ATP. We demonstrated that AtRecQ2 can unwind pTG4 in the absence of ATP, but it is less efficient than in the presence of ATP (Fig. 7). Studying the function of the AtRECQ2–pTG4 interaction in cells will be our next step.
Based on the above analysis, it can be optimistically expected that pTG4 may play a role in regulating reactions in telomeres. This research serves as a reference to study other types of G4s in plants, and it would be interesting to further study the function of G4 DNA in plant cells.
Materials and methods
Buffers
The reaction buffer contained 20 mm Tris-HCl (pH 8.0) and different concentrations of KCl and MgCl2, as specified in the text. For single-molecule imaging, the oxygen scavenging system (0.8% d-glucose, 1 mg/ml glucose oxidase (266600 units/g, Sigma), 0.4 mg/ml catalase (2000–5000 units/mg, Sigma)), and 4 mm Trolox were added.
DNA preparations
All the oligonucleotides were purchased from Sangon Biotech (Shanghai, China) and listed in Table S1. In the smFRET experiments, DNA constructs were composed of a secondary structure (or an ssDNA) containing a strand and a complementary stem strand. DNA substrates were annealed in a mixture with a 3:2 ratio for the smFRET experiments. The strand without a biotin label was used in excess to reduce the possibility of a nonannealed strand anchoring at the coverslip surface. The concentration of the stem strands was 2 μm. In the CD and MST measurements, the substrate was only a single-strand G4, and the annealing concentrations were 10 and 1 μm, respectively. All annealing was carried out by incubation at 95 °C for 5 min and then cooled to room temperature for ∼3 h in the corresponding reaction buffer.
Protein purification
The plasmid encoding BG4 with the expression and purification methods was from Professor Shankar Balasubramanian's lab (23, 62). AtRecQ2 and AtRecQ3 were expressed and purified as previously described (50, 51). The purified BG4, AtRecQ2, and AtRecQ3 were >90% pure, according to the analysis by SDS/PAGE (Figs. S2A and S6A).
CD spectropolarimetry
CD experiments were performed on an AVIV model 420SF optical system (Aviv Biomedical Inc.) equipped with a temperature-controlled cell holder using a quartz cell with a 1-mm path length. A 10 μm solution of G4 was prepared in 20 mm Tris-HCl, pH 8.0, containing a corresponding concentration of salt. The CD spectra were recorded in the UV (200–320 nm) region in 1-nm increments at 25 °C.
MST
For the measurement of the basic binding parameters of pTG4–K+ interactions, the reaction buffer contained 20 mm Tris-HCl (pH 8.0) and different concentrations of KCl (0.03–1000 mm). For the equilibrium measurement of DNA binding with BG4, the reaction buffer was 20 mm Tris-HCl (pH 8.0), 100 mm KCl, and 10 mm MgCl2. The concentration of the DNA substrates was 20 nm. The MST assays were performed according to the manufacturer's protocol using a NanoTemper monolith NT.115 (NanoTemper Technologies). A particular DNA substrate was incubated with varying concentrations of KCl (at 22–30 °C) or BG4 (at 22 °C) for 30 min. Afterward, the experiments were carried out using 60% LED power and 40% MST, Laser-On time 30 s and Laser-Off time 5 s, at specific temperatures. The original fluorescence data were collected from the thermophoresis signal via NTAnalysis from three independent experiments.
smFRET data acquisition and analysis
A homebuilt objective-type total-internal-reflection microscope was used. The Cy3 (donor) was excited by a 532-nm laser (Coherent). An oil immersion objective (100×, N.A. 1.49) was used to generate an evanescent field of illumination. Fluorescence signals from Cy3 and Cy5 were split by a dichroic mirror and collected by an EMCCD (Photometrics). Fluorescence signals were controlled and recorded by a NIS-Elements AR (Nikon). The coverslips (Fisher Scientific) and slides were cleaned thoroughly with a mixture of sulfuric acid, hydrogen peroxide, acetone, and sodium ethoxide; then the surfaces of the coverslips were coated with a mixture of 99% mPEG (m-PEG-5000, Laysan Bio, Inc.) and 1% of biotin-PEG (biotin-PEG-5000, Laysan Bio, Inc.). Streptavidin (10 μg/ml) in the reaction buffer was added to the PEG-coated chamber and incubated for 10 min. After washing, 50 pm DNA was added to the chamber and allowed to be immobilized for 10 min. Then the free DNA was removed by washing with the reaction buffer. Subsequently, the reaction chamber was equilibrated with the reaction buffer containing the oxygen scavenging system. We used an exposure time of 100 ms for all measurements at a constant temperature of 22 °C. To obtain the fraction of DNA unwinding with time, a series of movies were recorded with 2-s durations at different times, and the Cy5 (acceptor) spots were counted to represent the number of remaining DNA molecules from three independent experiments. The FRET efficiency was calculated using IA/(ID + IA), where ID and IA represent the intensity of the donor and acceptor, respectively. Hidden Markov modeling was employed to characterize the states (32). Single-molecule FRET histograms were generated by picking 100 frames of each trace from ∼150 molecules. Basic data analysis was carried out by scripts written in R, and all data fitting was generated by OriginPro 2017.
Data availability
All data presented are contained within the manuscript.
Author contributions
W.-Q. W., M.-L. Z., and C.-P. S. conceptualization; W.-Q. W., M.-L. Z., and C.-P. S. data curation; W.-Q. W. and M.-L. Z. funding acquisition; W.-Q. W., M.-L. Z., and C.-P. S. investigation; W.-Q. W., M.-L. Z., and C.-P. S. writing-original draft; W.-Q. W. and C.-P. S. project administration; W.-Q. W. and C.-P. S. writing-review and editing.
Supplementary Material
Acknowledgment
We thank Dr. Xi-Miao Hou for critical reading of the manuscript.
This work was supported by the Key Scientific Research Projects in Colleges and Universities in Henan Province through Grant 19A180014 and the National Natural Science Foundation of China Grant 31800644. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S7.
- ALT
- alternative lengthening of telomeres
- G4
- G-quadruplex
- pTG4
- plant telomeric G4 DNA
- hTG4
- human telomeric G-quadruplex
- MST
- microscale thermophoresis
- smFRET
- single-molecule florescence resonance energy transfer
- ssDNA
- single-stranded DNA
- HRDC
- the helicase and RNaseD C-terminal.
References
- 1. Webb C. J., Wu Y., and Zakian V. A. (2013) DNA repair at telomeres: keeping the ends intact. Cold Spring Harb. Perspect. Biol. 5, a012666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gilson E., and Géli V. (2007) How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8, 825–838 10.1038/nrm2259 [DOI] [PubMed] [Google Scholar]
- 3. Prochazkova Schrumpfova P., Fojtova M., and Fajkus J. (2019) Telomeres in plants and humans: not so different, not so similar. Cells 8, E58 10.3390/cells8010058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prochazková Schrumpfová P., Schorová S., and Fajkus J. (2016) Telomere- and telomerase-associated proteins and their functions in the plant cell. Front. Plant Sci. 7, 851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cesare A. J., and Reddel R. R. (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330 10.1038/nrg2763 [DOI] [PubMed] [Google Scholar]
- 6. Bochman M. L., Paeschke K., and Zakian V. A. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 10.1038/nrg3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vannier J. B., Pavicic-Kaltenbrunner V., Petalcorin M. I., Ding H., and Boulton S. J. (2012) RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149, 795–806 10.1016/j.cell.2012.03.030 [DOI] [PubMed] [Google Scholar]
- 8. Zahler A. M., Williamson J. R., Cech T. R., and Prescott D. M. (1991) Inhibition of telomerase by G-quartet DNA structures. Nature 350, 718–720 10.1038/350718a0 [DOI] [PubMed] [Google Scholar]
- 9. Clynes D., Jelinska C., Xella B., Ayyub H., Scott C., Mitson M., Taylor S., Higgs D. R., and Gibbons R. J. (2015) Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 6, 7538 10.1038/ncomms8538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mendoza O., Bourdoncle A., Boulé J. B., Brosh R. M. Jr., and Mergny J. L. (2016) G-quadruplexes and helicases. Nucleic Acids Res. 44, 1989–2006 10.1093/nar/gkw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parkinson G. N., Lee M. P., and Neidle S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417, 876–880 10.1038/nature755 [DOI] [PubMed] [Google Scholar]
- 12. Dai J., Carver M., Punchihewa C., Jones R. A., and Yang D. (2007) Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 35, 4927–4940 10.1093/nar/gkm522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y., and Patel D. J. (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1, 263–282 10.1016/0969-2126(93)90015-9 [DOI] [PubMed] [Google Scholar]
- 14. Hou X. M., Fu Y. B., Wu W. Q., Wang L., Teng F. Y., Xie P., Wang P. Y., and Xi X. G. (2017) Involvement of G-triplex and G-hairpin in the multi-pathway folding of human telomeric G-quadruplex. Nucleic Acids Res. 45, 11401–11412 10.1093/nar/gkx766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mashimo T., Yagi H., Sannohe Y., Rajendran A., and Sugiyama H. (2010) Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J. Am. Chem. Soc. 132, 14910–14918 10.1021/ja105806u [DOI] [PubMed] [Google Scholar]
- 16. Kogut M., Kleist C., and Czub J. (2016) Molecular dynamics simulations reveal the balance of forces governing the formation of a guanine tetrad: a common structural unit of G-quadruplex DNA. Nucleic Acids Res. 44, 3020–3030 10.1093/nar/gkw160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tippana R., Xiao W., and Myong S. (2014) G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 42, 8106–8114 10.1093/nar/gku464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang A. Y., and Balasubramanian S. (2012) The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J. Am. Chem. Soc. 134, 19297–19308 10.1021/ja309851t [DOI] [PubMed] [Google Scholar]
- 19. Wu W.-Q., Hou X.-M., Li M., Dou S.-X., and Xi X.-G. (2015) BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 43, 4614–4626 10.1093/nar/gkv361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu W. Q., Hou X. M., Zhang B., Fossé P., René B., Mauffret O., Li M., Dou S. X., and Xi X. G. (2017) Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci. Rep. 7, 43954 10.1038/srep43954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hou X.-M., Wu W.-Q., Duan X.-L., Liu N.-N., Li H.-H., Fu J., Dou S.-X., Li M., and Xi X.-G. (2015) Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 466, 189–199 10.1042/BJ20140997 [DOI] [PubMed] [Google Scholar]
- 22. Wu C. G., and Spies M. (2016) G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 44, 8742–8753 10.1093/nar/gkw574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Biffi G., Tannahill D., McCafferty J., and Balasubramanian S. (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186 10.1038/nchem.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang M. L., Xu Y. P., Kumar A., Zhang Y., and Wu W. Q. (2019) Studying the potassium-induced G-quadruplex DNA folding process using microscale thermophoresis. Biochemistry 58, 3955–3959 10.1021/acs.biochem.9b00447 [DOI] [PubMed] [Google Scholar]
- 25. Bhattacharyya D., Mirihana Arachchilage G., and Basu S. (2016) Metal cations in G-quadruplex folding and stability. Front. Chem. 4, 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ambrus A., Chen D., Dai J., Bialis T., Jones R. A., and Yang D. (2006) Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 34, 2723–2735 10.1093/nar/gkl348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lane A. N., Chaires J. B., Gray R. D., and Trent J. O. (2008) Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 36, 5482–5515 10.1093/nar/gkn517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Higinbotham N. (1973) Electropotentials of plant cells. Annu. Rev. Plant Physiol. 24, 25–46 10.1146/annurev.pp.24.060173.000325 [DOI] [Google Scholar]
- 29. Binzel M. L., Hess F. D., Bressan R. A., and Hasegawa P. M. (1988) Intracellular compartmentation of ions in salt adapted tobacco cells. Plant Physiol. 86, 607–614 10.1104/pp.86.2.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bugaut A., and Balasubramanian S. (2008) A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry 47, 689–697 10.1021/bi701873c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guédin A., Gros J., Alberti P., and Mergny J.-L. (2010) How long is too long?: Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 38, 7858–7868 10.1093/nar/gkq639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McKinney S. A., Joo C., and Ha T. (2006) Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 10.1529/biophysj.106.082487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Limongelli V., De Tito S., Cerofolini L., Fragai M., Pagano B., Trotta R., Cosconati S., Marinelli L., Novellino E., Bertini I., Randazzo A, Luchinat C., and Parrinello M. (2013) The G-triplex DNA. Angew. Chem. Int. Ed. Engl. 52, 2269–2273 10.1002/anie.201206522 [DOI] [PubMed] [Google Scholar]
- 34. Rajendran A., Endo M., Hidaka K., and Sugiyama H. (2014) Direct and single-molecule visualization of the solution-state structures of G-hairpin and G-triplex intermediates. Angew. Chem. Int. Ed. Engl. 53, 4107–4112 10.1002/anie.201308903 [DOI] [PubMed] [Google Scholar]
- 35. Rajendran A., Endo M., Hidaka K., Teulade-Fichou M. P., Mergny J. L., and Sugiyama H. (2015) Small molecule binding to a G-hairpin and a G-triplex: a new insight into anticancer drug design targeting G-rich regions. Chem. Commun. (Camb.) 51, 9181–9184 10.1039/C5CC01678A [DOI] [PubMed] [Google Scholar]
- 36. Lu X. M., Li H., You J., Li W., Wang P. Y., Li M., Dou S. X., and Xi X. G. (2018) Folding dynamics of parallel and antiparallel G-triplexes under the influence of proximal DNA. J. Phys. Chem. B 122, 9499–9506 10.1021/acs.jpcb.8b08110 [DOI] [PubMed] [Google Scholar]
- 37. Gray R. D., Trent J. O., and Chaires J. B. (2014) Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 426, 1629–1650 10.1016/j.jmb.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koirala D., Mashimo T., Sannohe Y., Yu Z., Mao H., and Sugiyama H. (2012) Intramolecular folding in three tandem guanine repeats of human telomeric DNA. Chem. Commun. 48, 2006–2008 10.1039/c2cc16752b [DOI] [PubMed] [Google Scholar]
- 39. Li W., Hou X.-M., Wang P.-Y., Xi X.-G., and Li M. (2013) Direct measurement of sequential folding pathway and energy landscape of human telomeric G-quadruplex structures. J. Am. Chem. Soc. 135, 6423–6426 10.1021/ja4019176 [DOI] [PubMed] [Google Scholar]
- 40. Zhou R., Zhang J., Bochman M. L., Zakian V. A., and Ha T. (2014) Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife 3, e02190 10.7554/eLife.02190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bessi I., Jonker H. R., Richter C., and Schwalbe H. (2015) Involvement of long-lived intermediate states in the complex folding pathway of the human telomeric G-quadruplex. Angew. Chem. Int. Ed. Engl. 54, 8444–8448 10.1002/anie.201502286 [DOI] [PubMed] [Google Scholar]
- 42. Lee J. Y., Okumus B., Kim D. S., and Ha T. (2005) Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. U.S.A. 102, 18938–18943 10.1073/pnas.0506144102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Joo C., and Ha T. (2012) Single-molecule FRET with total internal reflection microscopy. Cold Spring Harbor Protoc. 12, pii: pdb.top072058 10.1101/pdb.top072058 [DOI] [PubMed] [Google Scholar]
- 44. Aznauryan M., Søndergaard S., Noer S. L., Schiøtt B., and Birkedal V. (2016) A direct view of the complex multi-pathway folding of telomeric G-quadruplexes. Nucleic Acids Res. 44, 11024–11032 10.1093/nar/gkw1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hermans C., Chen J., and Verbruggen N. (2013) Magnesium in plants. In Encyclopedia of Metalloproteins (Kretsinger R. H., Uversky V. N., and Permyakov E. A., eds) pp. 1269–1276, Springer New York, New York [Google Scholar]
- 46. Wang Q., Liu J.-Q., Chen Z., Zheng K.-W., Chen C.-Y., Hao Y.-H., and Tan Z. (2011) G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 39, 6229–6237 10.1093/nar/gkr164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yadav V., Hemansi, Kim N., Tuteja N., and Yadav P. (2017) G quadruplex in plants: a ubiquitous regulatory element and its biological relevance. Front. Plant Sci. 8, 1163 10.3389/fpls.2017.01163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Croteau D. L., Popuri V., Opresko P. L., and Bohr V. A. (2014) Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83, 519–552 10.1146/annurev-biochem-060713-035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hartung F., and Puchta H. (2006) The RecQ gene family in plants. J. Plant Physiol. 163, 287–296 10.1016/j.jplph.2005.10.013 [DOI] [PubMed] [Google Scholar]
- 50. Kobbe D., Blanck S., Demand K., Focke M., and Puchta H. (2008) AtRECQ2, a RecQ helicase homologue from Arabidopsis thaliana, is able to disrupt various recombinogenic DNA structures in vitro. Plant J. 55, 397–405 10.1111/j.1365-313X.2008.03511.x [DOI] [PubMed] [Google Scholar]
- 51. Kobbe D., Blanck S., Focke M., and Puchta H. (2009) Biochemical characterization of AtRECQ3 reveals significant differences relative to other RecQ helicases. Plant Physiol. 151, 1658–1666 10.1104/pp.109.144709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Budhathoki J. B., Ray S., Urban V., Janscak P., Yodh J. G., and Balci H. (2014) RecQ-core of BLM unfolds telomeric G-quadruplex in the absence of ATP. Nucleic Acids Res. 42, 11528–11545 10.1093/nar/gku856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chatterjee S., Zagelbaum J., Savitsky P., Sturzenegger A., Huttner D., Janscak P., Hickson I. D., Gileadi O., and Rothenberg E. (2014) Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat. Commun. 5, 5556 10.1038/ncomms6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Budhathoki J. B., Maleki P., Roy W. A., Janscak P., Yodh J. G., and Balci H. (2016) A comparative study of G-quadruplex unfolding and DNA reeling activities of human RECQ5 helicase. Biophys. J. 110, 2585–2596 10.1016/j.bpj.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Popuri V., Bachrati C. Z., Muzzolini L., Mosedale G., Costantini S., Giacomini E., Hickson I. D., and Vindigni A. (2008) The human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 283, 17766–17776 10.1074/jbc.M709749200 [DOI] [PubMed] [Google Scholar]
- 56. Zhang B., Wu W.-Q., Liu N.-N., Duan X.-L., Li M., Dou S.-X., Hou X.-M., and Xi X.-G. (2016) G-quadruplex and G-rich sequence stimulate Pif1p-catalyzed downstream duplex DNA unwinding through reducing waiting time at ss/dsDNA junction. Nucleic Acids Res. 44, 8385–8394 10.1093/nar/gkw669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mendoza O., Gueddouda N. M., Boulé J.-B., Bourdoncle A., and Mergny J.-L. (2015) A fluorescence-based helicase assay: application to the screening of G-quadruplex ligands. Nucleic Acids Res. 43, e71–e71 10.1093/nar/gkv193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu W. Q., Zhu X., and Song C. P. (2019) Single-molecule technique: a revolutionary approach to exploring fundamental questions in plant science. New Phytol. 223, 508–510 10.1111/nph.15928 [DOI] [PubMed] [Google Scholar]
- 59. Griffin B. D., and Bass H. W. (2018) Plant G-quadruplex (G4) motifs in DNA and RNA; abundant, intriguing sequences of unknown function. Plant Sci. 269, 143–147 10.1016/j.plantsci.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 60. Marsico G., Chambers V. S., Sahakyan A. B., McCauley P., Boutell J. M., Antonio M. D., and Balasubramanian S. (2019) Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 47, 3862–3874 10.1093/nar/gkz179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sun H., Karow J. K., Hickson I. D., and Maizels N. (1998) The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273, 27587–27592 10.1074/jbc.273.42.27587 [DOI] [PubMed] [Google Scholar]
- 62. Hänsel-Hertsch R., Spiegel J., Marsico G., Tannahill D., and Balasubramanian S. (2018) Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 13, 551–564 10.1038/nprot.2017.150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented are contained within the manuscript.