Abstract
The linear ubiquitin assembly complex (LUBAC) is an essential component of the innate and adaptive immune system. Modification of cellular substrates with linear polyubiquitin chains is a key regulatory step in signal transduction that impacts cell death and inflammatory signaling downstream of various innate immunity receptors. Loss-of-function mutations in the LUBAC components HOIP and HOIL-1 yield a systemic autoinflammatory disease in humans, whereas their genetic ablation is embryonically lethal in mice. Deficiency of the LUBAC adaptor protein Sharpin results in a multi-organ inflammatory disease in mice characterized by chronic proliferative dermatitis (cpdm), which is propagated by TNFR1-induced and RIPK1-mediated keratinocyte cell death. We have previously shown that caspase-1 and -11 promoted the dermatitis pathology of cpdm mice and mediated cell death in the skin. Here, we describe a reciprocal regulation of caspase-1 and LUBAC activities in keratinocytes. We show that LUBAC interacted with caspase-1 via HOIP and modified its CARD domain with linear polyubiquitin and that depletion of HOIP or Sharpin resulted in heightened caspase-1 activation and cell death in response to inflammasome activation, unlike what is observed in macrophages. Reciprocally, caspase-1, as well as caspase-8, regulated LUBAC activity by proteolytically processing HOIP at Asp-348 and Asp-387 during the execution of cell death. HOIP processing impeded substrate ubiquitination in the NF-κB pathway and resulted in enhanced apoptosis. These results highlight a regulatory mechanism underlying efficient apoptosis in keratinocytes and provide further evidence of a cross-talk between inflammatory and cell death pathways.
Keywords: apoptosis, inflammasome, E3 ubiquitin ligase, keratinocyte, caspase 1 (Casp1), cell death
Introduction
The linear ubiquitin assembly complex (LUBAC)3 is an E3 ligase that has emerged as a critical regulator of both innate (1–4) and adaptive immunity (5–7). LUBAC forms a tripartite oligomeric complex in cells and is composed of the catalytic E3 ligase HOIP, an auxiliary E3 HOIL-1, and the adaptor Sharpin. LUBAC is unique in its ability to assemble linear Met-1–linked polyubiquitin chains (Met1-Ub) that do not utilize any internal lysine residues; instead, the terminal carboxyl group of Gly-76 in the proximal ubiquitin forms a peptide bond with the α-amino group of a distal ubiquitin (8). In the context of innate immunity signaling, Met1-Ub chains act cooperatively with Lys-63–linked polyubiquitin (Lys63-Ub), often synthesized by the cellular inhibitors of apoptosis (cIAP) proteins cIAP1 and cIAP2 E3 ligases, to promote NF-κB activation and curb cell death (9). RIPK1 and the IKK regulatory subunit NEMO are two well-described ubiquitinated substrates in the TNF receptor 1 (TNFR1) pathway that are modified with both Met1-Ub and Lys63-Ub, which act to scaffold downstream signaling factors and facilitate IKKα/β kinase activation (10, 11). Suboptimal Met1-Ub or Lys63-Ub of these substrates promotes caspase-8–driven apoptosis, or RIPK1/RIPK3/MLKL-driven necroptosis when caspases are inhibited (9, 12). Both RIPK1 and caspase-8 have also been reported to be modified with Met1-Ub during necroptosis (13), although the functional consequence of their modification remains poorly understood.
LUBAC is essential for homeostasis and health in both mice and humans. Hypomorphic and/or loss-of-function mutations in HOIP (RNF31) or HOIL (RBCK1) have been identified in patients with a life-threatening systemic disease characterized by recurrent bacterial and viral infections, autoinflammation, amylopectinosis, and lymphangiectasia (14–16). Interestingly, loss-of-function mutations in the Met1-Ub–specific deubiquitinase OTULIN yield an autoinflammatory disease with strikingly similar clinical features (17–19). Of note, the seemingly paradoxical occurrence of autoinflammation, heightened susceptibility to infection, and autoimmunity in these patients has been attributed to the cell type–specific roles for LUBAC and OTULIN (20). In mice, deletion of either HOIP (21) or HOIL-1 (22) results in embryonic lethality at day 10.5 due to TNF-driven endothelial cell death. On the other hand, Sharpin-deficient mice are viable but develop a multi-organ inflammatory disease characterized primarily by chronic proliferative dermatitis (cpdm) (23). Whereas TNF-mediated cell death of keratinocytes is key in disease pathogenesis in the cpdm mice (24–26), we have previously shown that hyperactivation of the Nlrp3-caspase-1 inflammasome is an important initiating signal that promotes downstream apoptotic and necroptotic cell death driving skin pathology (27). However, how LUBAC controlled caspase-1 activity was unclear. Further, how cells regulated LUBAC activity for the proper execution of cell death was incompletely characterized.
Here, we show that caspase-1 is a novel target of Met1-Ub by LUBAC and that LUBAC-deficient keratinocytes display enhanced caspase-1 activation. Upon activation, caspase-1 and -8 cleaved HOIP, which promotes cell death and limits NF-κB–dependent gene expression. Our results describe two novel checkpoints in the regulation of inflammatory cell death pathways in keratinocytes.
Results
LUBAC deficiency in keratinocytes promotes caspase activation
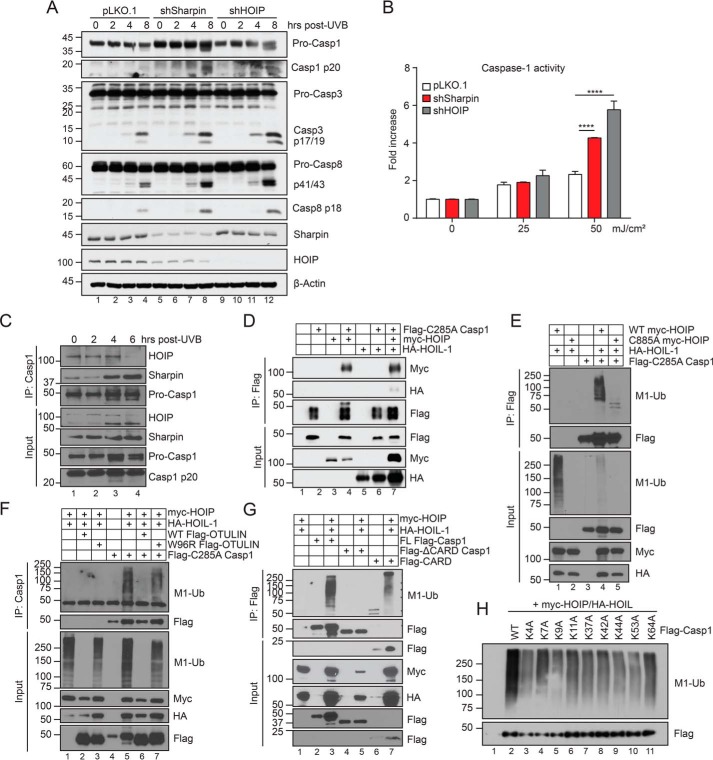
Sharpin deficiency in mice triggers an inflammatory skin disease that is driven by Nlrp3-caspase-1–mediated IL-1β production (27, 28). We and others found that Sharpin-deficient bone marrow–derived macrophages had attenuated caspase-1 activation and IL-1β secretion in response to inflammasome triggers (3, 27). However, it is now well-appreciated that the Met1-Ub machinery possesses distinct functional roles in different cell types (14, 15, 17, 20). Because keratinocytes are thought to be the primary pathogenic cell type involved in cpdm skin disease (24), we sought to investigate the role of LUBAC in controlling caspase-1 activity in keratinocytes. We generated Sharpin-depleted HaCaT keratinocytes using shRNA and investigated their response to UVB irradiation, a known inducer of the Nlrp1-caspase-1 inflammasome in the skin (29, 30). Sharpin-depleted cells displayed enhanced cleavage of caspase-1, -3, and -8 compared with control cells (Fig. 1A). Using a bioluminescent reporter of caspase-1 activity, we confirmed that Sharpin-depleted keratinocytes had hyperactive caspase-1 in response to UVB compared with pLKO.1 control knockdown cells (Fig. 1B). Knockdown of HOIP likewise rendered cells hyperresponsive to UVB-induced caspase activation (Fig. 1, A and B), suggesting that LUBAC is required to limit caspase activity in keratinocytes.
Figure 1.
Sharpin inhibits caspase activation and targets caspase-1 for linear ubiquitination. A, pLKO.1, shSharpin, and shHOIP HaCaT cells were stimulated with 50 mJ/cm2 UVB and lysed at the indicated time points post-irradiation and subjected to immunoblot analysis. Data are representative of three independent experiments. B, caspase-1 activity was measured in the supernatant of pLKO.1, shSharpin, and shHOIP HaCaT cells following UVB irradiation using Caspase-Glo 1 assay. Data shown are representative of two independent experiments and are displayed as mean ± S.E. (error bars). ****, p < 0.0001. C, HaCaT cells were stimulated with 50 mJ/cm2 UVB and harvested at the indicated time points post-irradiation. Lysates were subject to anti-caspase-1 immunoprecipitation, followed by immunoblot analysis. Data are representative of three independent experiments. D, HEK293T cells were transfected with equal amounts of the indicated plasmids. 24 h post-transfection, lysates were harvested and subjected to FLAG M2 immunoprecipitation and immunoblot analysis. E–H, as in D, but cells were lysed in 1% SDS denaturing lysis buffer, followed by dilution to 0.1% SDS for immunoprecipitation with either FLAG M2 affinity gel (E, G, and H) or anti-caspase-1 (F). Data are representative of two independent experiments, except H, which was performed once.
Caspase-1 is a putative linear ubiquitin substrate
To examine the molecular interaction dynamics between LUBAC components and caspase-1, we next performed co-immunoprecipitation experiments in HaCaT keratinocytes at steady state or following UVB irradiation. We observed constitutive binding between HOIP and caspase-1 at steady state, which decreased 6 h post-UVB (Fig. 1C). Conversely, Sharpin recruitment to the complex was enhanced by UVB irradiation (Fig. 1D). HOIP binding to caspase-1 was validated in HEK293T cells (Fig. 1D). Notably, HOIL-1 co-immunoprecipitated with caspase-1 solely in the presence of HOIP, suggesting indirect binding (Fig. 1D). Given this interaction between LUBAC and caspase-1, we wondered whether LUBAC targeted caspase-1 for Met1-Ub. Indeed, using immunoprecipitation under denaturing conditions, we observed that caspase-1 was readily modified with Met1-Ub chains when co-expressed with HOIL-1 and WT HOIP, but not a catalytically inactive (E3-dead) C885A HOIP mutant (Fig. 1E). To confirm the specificity of this modification, we examined the effect of expressing OTULIN, a Met1-Ub–specific deubiquitinase, on the caspase-1 ubiquitination signal. Indeed, expression of WT OTULIN, but not a catalytically suppressed W96R OTULIN mutant (31), led to a complete ablation of the caspase-1 Met1-Ub signal (Fig. 1F, compare lanes 6 and 7). To identify the caspase-1 domain targeted by LUBAC, we generated caspase-1 deletion variants and examined their linear ubiquitination. Such domain mapping revealed that caspase-1 is targeted for Met1-Ub within its CARD (Fig. 1G). To identify the lysine residue modified by Met1-Ub, we systematically mutated all lysine residues within the caspase-1 CARD to alanine. However, whereas individual Lys → Ala mutation reduced the Met1-Ub signal, none led to its complete abrogation (Fig. 1H), suggesting that either multiple sites are targeted or there is compensation following mutation of the bona fide target site. Collectively, these data demonstrate that LUBAC inhibits caspase-1 function in keratinocytes and can target its CARD with Met1-Ub in an overexpression system.
Caspase-1 reciprocally targets HOIP for proteolytic cleavage
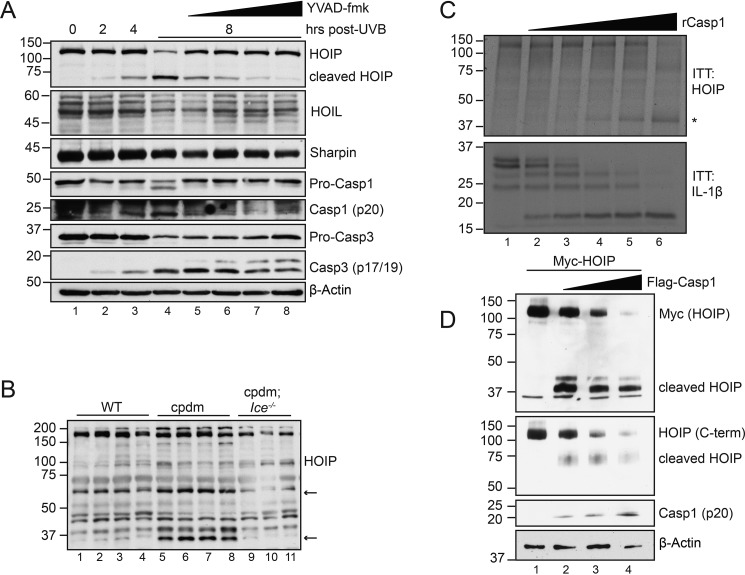
To address whether there may exist a reciprocal regulation of LUBAC function by caspases, we next monitored the expression of LUBAC components following UVB irradiation. Whereas Sharpin and HOIL-1 proteins displayed no changes, as assessed by immunoblot analysis, a lower-molecular-weight HOIP band appeared at 4 h post-UVB irradiation (Fig. 2A). This band dose-dependently disappeared upon treatment with the caspase-1–selective inhibitor YVAD-fmk (Fig. 2A), suggesting a caspase-1–mediated cleavage event. To validate this observation in an in vivo setting, we examined HOIP status in skin homogenates from cpdm mice, which we previously showed to display hyperactivation of caspase-1 (27). We observed similar putative HOIP cleavage fragments (Fig. 2B, arrows) that were not present in either WT mice or cpdm mice deficient in caspase-1/11 (cpdm; Ice−/−) (Fig. 2B). To determine whether HOIP was a direct caspase-1 substrate, we generated in vitro transcribed and translated (ITT) HOIP and incubated the product with increasing doses of recombinant caspase-1, with pro-IL-1β serving as a control for caspase-1 activity. Indeed, recombinant caspase-1 dose-dependently cleaved ITT HOIP (Fig. 2C). Transfected active caspase-1 was likewise capable of cleaving HOIP in HEK293T cells (Fig. 2D). These data identify HOIP as a novel caspase-1 substrate.
Figure 2.
Caspase-1 cleaves HOIP following UVB irradiation. A, HaCaT cells were pretreated with increasing doses of YVAD (2.5, 5, 10, and 20 μm) or DMSO for 45 min before stimulation with 50 mJ/cm2 UVB. Cells were lysed at the indicated time points post-irradiation and subjected to immunoblot analysis. Data are representative of two independent experiments. B, skin homogenates from WT, cpdm, or cpdm;Ice−/− mice were subjected to immunoblot analysis for murine HOIP. Asterisks denote putative cleavage fragments. n = 3–4 mice/genotype. C, in vitro transcribed and translated pro-IL-1β and HOIP were incubated with increasing amounts of recombinant caspase-1 for 1 h at 37 °C and subjected to immunoblot analysis following reaction termination with Laemmli buffer. The asterisk denotes cleaved HOIP. Data are representative of two independent experiments. D, HEK293T cells were transfected with Myc-HOIP and increasing amounts of FLAG-caspase-1. 24 h post-transfection, cells were lysed and subjected to immunoblot analysis. Data are representative of three independent experiments.
HOIP is cleaved at Asp-348 and Asp-387 by caspase-1 and -8 in apoptotic keratinocytes
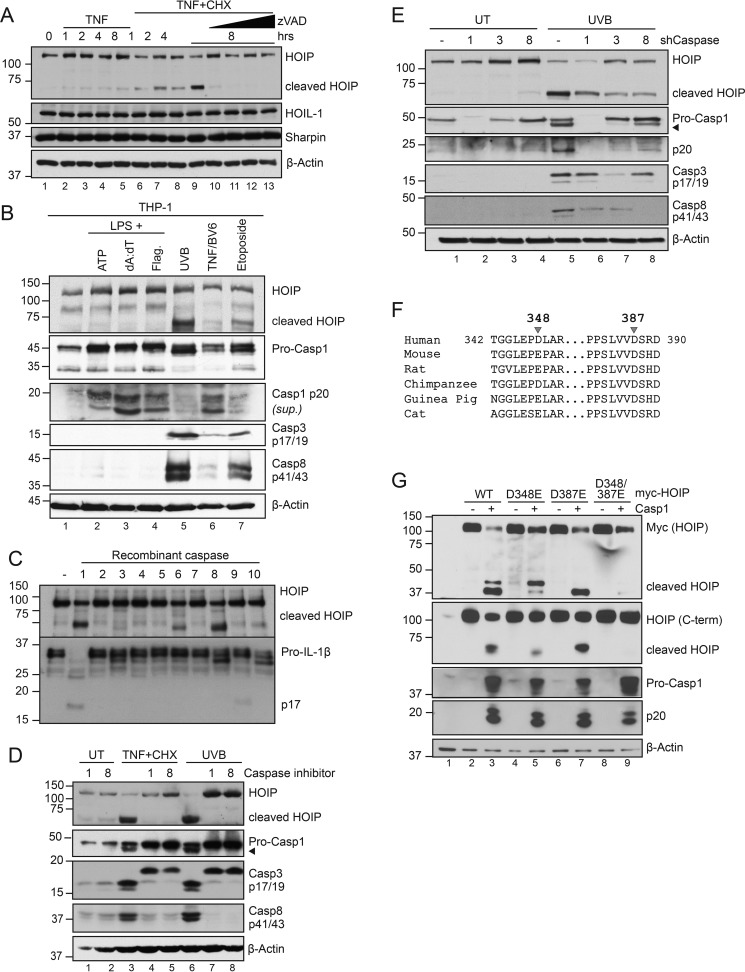
UVB irradiation of keratinocytes activates both the NLRP1-caspase-1 inflammasome and the intrinsic mitochondrial apoptosis pathway (29, 30, 32). We previously noted the concurrent activation of both inflammatory and apoptotic caspases in inflamed cpdm skin (27). To test whether HOIP cleavage is a common feature during cell death, we treated HaCaT keratinocytes with TNF + cycloheximide (CHX) to trigger extrinsic apoptosis. Similar to UVB irradiation, we observed a kinetic processing of HOIP that was blocked with the pan-caspase inhibitor Z-VAD-fmk, whereas both HOIL-1 and Sharpin remained unaffected (Fig. 3A). As caspase-1 is generally not involved in apoptosis in most cell types, we examined HOIP cleavage in THP-1 monocytes in response to both inflammasome and apoptotic triggers. Interestingly, activation of caspase-1 downstream of the NLRP3, AIM2, and NLRC4 inflammasomes via ATP, poly(dA:dT), and flagellin, respectively, in LPS-primed cells did not trigger HOIP cleavage (Fig. 3B). Induction of apoptosis by UVB, TNF + BV6, or etoposide, however, resulted in HOIP cleavage concurrent with caspase-8 activation (Fig. 3B). HOIP processing is thus a common consequence of apoptosis; however, the involvement of caspase-1 seems limited to keratinocytes.
Figure 3.
HOIP is cleaved at Asp-348 and Asp-387 by multiple caspases during apoptosis. A, HaCaT cells were pretreated with CHX (6 μg/ml) with or without Z-VAD (2.5, 5, 10, and 20 μm) for 45 min, followed by TNF (100 ng/ml) for the indicated times. Cells were lysed and subject to immunoblot analysis. Data are representative of two independent experiments. B, PMA-differentiated THP-1 cells were primed with 100 ng/ml LPS for 4 h followed by ATP (5 mm, 1 h), Lipofectamine-transfected poly(dA:dT) (1.5 μg/ml, 6 h), or DOTAP-transfected flagellin (2 μg/ml, 6 h) to engage inflammasomes or with UVB (50 mJ/cm2, 6 h), TNF (100 ng/ml) + BV6 (5 μm) (6 h), or etoposide (200 μm) to engage apoptosis. C, in vitro transcribed and translated HOIP and pro-IL-1β were incubated with a panel of recombinant caspases for 1 h at 37 °C and subjected to immunoblot analysis following reaction termination with Laemmli buffer. 2 units of recombinant caspase was used per reaction (Biovision K233). Data are representative of two independent experiments. D, HaCaT cells were pretreated with YVAD (10 μm; Casp1 inhibitor), IETD (10 μm; Casp8 inhibitor), or CHX (6 μg/ml) for 45 min before treatment with either TNF (100 ng/ml) or UVB (50 mJ/cm2) for 6 h. Cells were lysed and subjected to immunoblot analysis. Data are representative of three independent experiments. E, shCaspase-1, -3, or -8 HaCaT cells were stimulated with 50 mJ/cm2 UVB, lysed 6 h post-irradiation, and subjected to immunoblot analysis. Data are representative of two independent experiments. F, sequence alignment of HOIP (RNF31) from various species. G, HEK293T cells were transfected with equal amounts of the indicated plasmids. 24 h post-transfection, cells were lysed and subjected to immunoblot analysis. Data are representative of two independent experiments.
These data suggest that other caspases are involved in HOIP cleavage. We therefore tested a panel of recombinant caspases for their ability to directly cleave ITT HOIP. Interestingly, caspase-8 and, to a lesser extent, caspase-6 were also capable of cleaving HOIP, generating a fragment that was slightly smaller than that processed by caspase-1 (Fig. 3C). Pretreatment of HaCaT cells with either the caspase-1 inhibitor YVAD-fmk or the caspase-8 inhibitor IETD-fmk was sufficient to block HOIP cleavage in keratinocytes treated with either TNF + CHX or UVB (Fig. 3D). Using shRNA, we stably depleted HaCaT cells of either caspase-1, -3, or -8 and found that loss of any of these caspases attenuated HOIP processing in response to UVB irradiation (Fig. 3E).
Given the size and relative orientation of the cleavage fragments when blotting for either the N-terminal fragment with anti-Myc or the C-terminal fragment with anti-HOIP (Fig. 2D), and with the aid of caspase substrate prediction software (33), we identified Asp-348 and Asp-387 as putative caspase cleavage sites. Interestingly, whereas Asp-387 is conserved across mammalian species, Asp-348 is unique to primates (Fig. 3F). Individual mutation of either of these aspartates to glutamate caused a shift in the size of the HOIP fragments. In contrast, their combination resulted in a HOIP D348E/D387E mutant that was fully resistant to caspase-1 cleavage (Fig. 3G). These data demonstrate that HOIP is targeted by multiple caspases and is cleaved at Asp-348 and Asp-387.
HOIP cleavage limits NF-κB and promotes cell death in keratinocytes
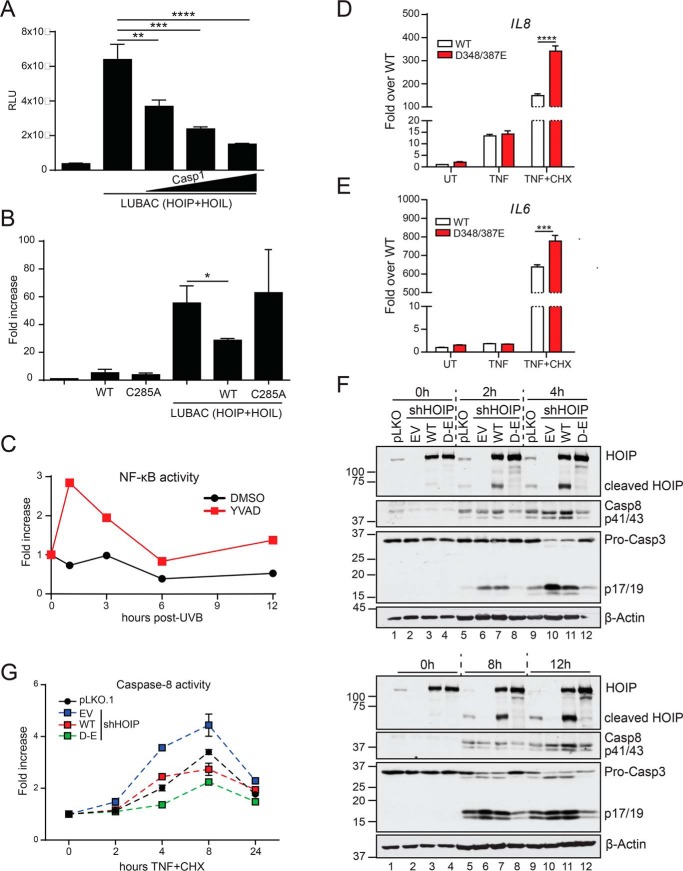
Many caspase substrates identified to date have little biological significance and represent bystander effects of apoptotic cell death (34). To determine whether the cleavage of HOIP had functional consequences, we examined its impact on LUBAC-induced NF-κB activation in HEK293T cells (Fig. 4A). Our results show that expression of WT caspase-1, but not a catalytically inactive C285A caspase-1 mutant, attenuated NF-κB–driven luciferase activity (Fig. 4B). Conversely, inhibition of caspase-1 enzymatic activity enhanced NF-κB activity following UVB irradiation of HaCaT cells (Fig. 4C). To further probe for the functional impact of HOIP cleavage during apoptosis, we depleted endogenous HOIP from HaCaT cells using shRNA and stably reintroduced shRNA-resistant WT HOIP or the noncleavable D348E/D387E HOIP mutant. We found that cells expressing D348E/D387E HOIP had significantly enhanced expression of the NF-κB-target genes IL6 and IL8 in response to TNF + CHX stimulation compared with WT HOIP, whereas no differences were observed in response to TNF alone (Fig. 4, D and E). These data suggest that NF-κB signaling is attenuated during apoptosis in part due to HOIP cleavage. It has been shown that deficiency of HOIP renders a variety of cell types more sensitive to apoptotic cell death (21, 35, 36); thus, we wondered whether cleavage could also impact cell death. We first confirmed that HOIP deficiency in HaCaT cells enhanced TNF + CHX–induced caspase-8 and -3 activation (Fig. 4F), similar to what we observed with UVB irradiation (Fig. 1, A and B). Whereas introduction of WT HOIP attenuated cell death, particularly at early time points, D348E/D387E HOIP potently inhibited caspase activation at all time points studied (Fig. 4, F and G). These data suggest that caspase-mediated cleavage of HOIP impairs LUBAC function in NF-κB activation and in limiting apoptosis.
Figure 4.
HOIP cleavage dampens NF-κB activation and promotes cell death. A and B, HEK293T cells were transfected with β-gal and NF-κB-luciferase reporter plasmids in combination with the indicated plasmids, normalizing for total DNA. Lysates were harvested 24 h later, and luciferase activity was measured and normalized to β-gal activity. Data are representative of three independent experiments. C, HaCaT cells were transfected with 350 ng of β-gal and NF-κB-luciferase reporter plasmids. 24 h post-transfection, cells were pretreated with 20 μm YVAD for 45 min before stimulation with 75 mJ/cm2 UVB. Cells were lysed at the indicated time points post-irradiation, and luciferase activity was measured and normalized to β-gal activity. Data are representative of two independent experiments. D and E, shHOIP HaCaT cells stably expressing either WT or D348E/D387E HOIP were treated for 6 h with the indicated TNF mixtures. RNA was extracted, reverse-transcribed, and subjected to RT-quantitative PCR analysis. Data are representative of two independent experiments and are displayed as mean ± S.E. (error bars). ***, p < 0.001; ****, p < 0.0001. F and G, pLKO.1 or shHOIP HaCaT cells stably expressing either empty vector (EV), WT HOIP, or D348E/D387E HOIP were treated with TNF + CHX for the indicated times, lysed, and subjected to immunoblot analysis (F), or caspase-8 activity was measured (G). Data are representative of two independent experiments and are displayed as mean ± S.E.
Caspase-mediated cleavage of HOIP impedes substrate ubiquitination by LUBAC
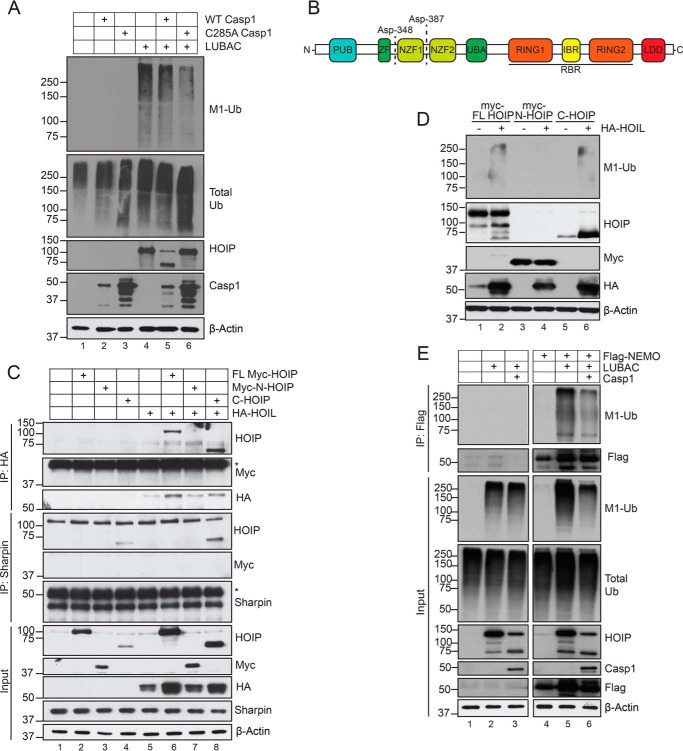
We found that caspase-1–mediated cleavage of HOIP did not affect the levels of total Met1-Ub (Fig. 5, A (compare lanes 4 and 5) and E (compare lanes 2 and 3)). Whereas these data suggest that caspase-mediated cleavage of HOIP does not affect general enzymatic function, it remains possible that targeting of specific LUBAC substrates may be affected. Asp-348 and Asp-387 are located between the Npl4 zinc finger 1 (NZF1) and NZF2 ubiquitin-binding domains in the N terminus of HOIP (Fig. 5B). Caspase targeting of these two residues separates the N-terminal PNGase/UBA or UBX-containing proteins (PUB), zinc finger (ZF), and NZF1 domains from the C-terminal fragment containing the Ub-associated (UBA) domain, the RING-in between-RING (RBR) domain, and the linear ubiquitin chain–determining domain (LDD), which dictate catalytic function. Whereas the PUB domain interacts with deubiquitinases to control LUBAC-dependent signaling (37–39), the UBA domain is necessary for Sharpin and HOIL-1 binding (40). Subcloning of the N- and C-terminal HOIP cleavage fragments revealed that C-HOIP retained the ability to interact with both Sharpin and HOIL-1 (Fig. 5C). Similarly, C-HOIP was able to generate linear ubiquitin chains when co-expressed with HOIL-1, whereas N-HOIP displayed no enzymatic activity (Fig. 5D). To examine whether caspase cleavage of HOIP affected LUBAC-mediated substrate modification, we examined NEMO Met1-Ub as a surrogate readout. Indeed, caspase-1–mediated cleavage of HOIP reduced Met1-Ub of NEMO (Fig. 5E, lanes 5 and 6). Of note, whereas caspase-1 did not affect total Met1-Ub in the absence of NEMO (Fig. 5E, lanes 2 and 3), there was a slight reduction in total Met1-Ub caused by caspase-1 upon overexpression of NEMO (lanes 5 and 6). Collectively, these data demonstrate that in keratinocytes, caspases promote cell death and inhibit NF-κB activation by targeting HOIP for proteolytic cleavage, which may impede substrate ubiquitination by LUBAC.
Figure 5.
Caspase-1 cleavage of HOIP impedes substrate ubiquitination. A, HEK293T cells were transfected with equal amounts of the indicated plasmids. 24 h post-transfection, cells were lysed and subjected to immunoblot analysis. Data are representative of two independent experiments. B, human HOIP domain schematic. C, HEK293T cells were transfected with equal amounts of the indicated plasmids. 24 h post-transfection, lysates were harvested and subjected to either anti-HA (HOIL-1) or anti-Sharpin immunoprecipitation and immunoblot analysis. Data are representative of two independent experiments. Asterisks indicate IgG heavy chain. D, HEK293T cells were transfected with equal amounts of the indicated plasmids. 24 h post-transfection, cells were lysed and subjected to immunoblot analysis. Data are representative of two independent experiments. E, HEK293T cells were transfected with equal amounts of the indicated plasmids. 24 h post-transfection, cells were lysed in 1% SDS lysis buffer, followed by dilution to 0.1% SDS for immunoprecipitation with FLAG M2 affinity gel. Data shown are from one independent experiment.
Discussion
As the first line of defense against invading microorganisms and as a sensor of sterile tissue damage, the skin innate immune system plays a pivotal role in maintaining human health and homeostasis. As such, it has evolved extensive regulatory mechanisms to coordinate a response in duration and magnitude that ensures the elimination of the pathogen, initiation of tissue repair, and/or induction of adaptive immunity while minimizing bystander damage to the host. Pattern recognition receptors and cytokine receptors are key players in innate immunity that, when engaged, rapidly trigger a complex network of intracellular events that can lead to multiple distinct or overlapping signaling outcomes, depending on the cellular context. Here, we describe LUBAC-mediated inhibition of caspase-1 and reciprocal caspase-mediated inhibition of LUBAC as two regulatory checkpoints in keratinocytes that modulate inflammatory signaling and cell death.
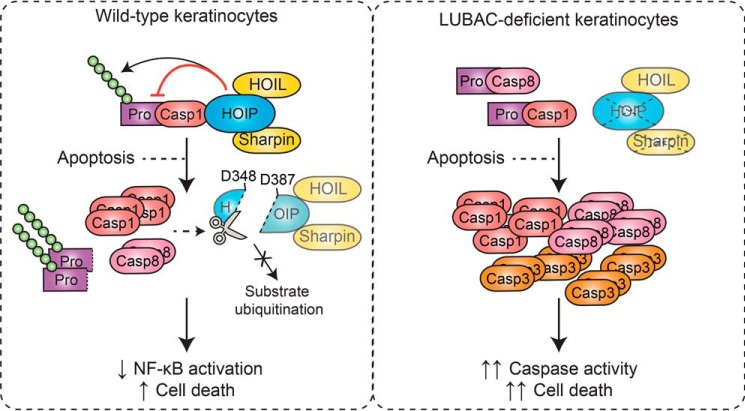
Considering these data and our previous work (27), we propose the following model (Fig. 6). In WT keratinocytes, HOIP forms a constitutive interaction with caspase-1 and mediates the linear ubiquitination of its CARD pro-domain. Upon engagement of apoptosis, caspase-1 and caspase-8 cleave HOIP at Asp-348 and Asp-387, limiting the ability of LUBAC to ubiquitinate substrates. This results in attenuated NF-κB–dependent gene expression and enhanced cell death. Activation of apoptosis in the absence of LUBAC leads to enhanced caspase-1, -3, and -8 activity, promoting pathology in vivo, such as the dermatitis observed in Cpdm mice. It is tempting to speculate that LUBAC directly inhibits caspase-1 activity via linear ubiquitination; however, this remains to be tested empirically. The identification of the lysine residue(s) targeted by LUBAC and subsequent examination of the activity of a nonubiquitinatable caspase-1 mutant will be necessary to test this hypothesis.
Figure 6.
Proposed model. In WT keratinocytes, HOIP forms a constitutive interaction with caspase-1 and mediates the linear ubiquitination of the CARD pro-domain. Upon engagement of apoptosis, caspase-1 and caspase-8 cleave HOIP at Asp-348 and Asp-387, limiting the ability of LUBAC to ubiquitinate substrates. This results in attenuated NF-κB–dependent gene expression and enhanced cell death. Activation of apoptosis in the absence of LUBAC leads to enhanced caspase-1, -3, and -8 activity, which promotes dermatitis in vivo.
Proteolytic regulation of LUBAC activity has been described previously. Upon antigen receptor stimulation in T- and B- cells, LUBAC is recruited to the CARMA1, BCL10, and MALT1 (CBM) complex, where it regulates NF-κB and lymphocyte activation (6). Upon recruitment, the paracaspase MALT1 was shown to specifically cleave HOIL-1 at Arg-165, separating the N-terminal UBL domain (N-HOIL-1), which binds to HOIP, from the C-terminal fragment containing the NZF and RBR domains (C-HOIL-1) (41–43). Although there are discrepancies regarding the functional outcome of HOIL-1 cleavage, it was shown that release of C-HOIL-1 from LUBAC leads to HOIP destabilization and degradation by the proteasome (41). When overexpressed in HEK293T cells, C-HOIL-1 antagonized LUBAC function and NF-κB activity (41, 42). Further, activation of lymphocytes with either phorbol 12-myristate 13-acetate (PMA)/ionomycin or anti-CD3/CD28 leads to a rapid decrease in total Met1-Ub levels in WT cells but not those expressing a catalytically inactive MALT1 mutant (41). One group noted a reduction in NF-κB activation upon overexpression of HOIL-1 in Jurkat T-cells, which was more pronounced when cells expressed the noncleavable mutant (43). Given the variation in experimental design between these reports, it will be important to dissect the functional role of HOIL-1 cleavage by MALT1 by reconstitution of HOIL-1–deficient B/T-cells with the noncleavable mutant. Of note, the deubiquitinase CYLD, which regulates LUBAC function (44, 45), is cleaved by both MALT1 and caspase-8, highlighting a common theme of regulation of the Met1-Ub machinery by caspases.
HOIP has also been described to be cleaved in a variety of cell types during apoptosis (46–48). Joo et al. (46) found that caspase-3 and -6, but not caspase-1 or -8, cleaved HOIP at Asp-348, Asp-387, and Asp-390, and reconstitution of HOIP-deficient Jurkat T-cells with a triple mutant HOIP slightly attenuated TNF-induced apoptosis (46). They observed that although C-HOIP retained enzymatic activity and could target RIPK1 and NEMO with Met1-Ub, it was unable to induce NF-κB activation in parallel overexpression assays. Goto et al. (47) identified Asp-390 as the primary HOIP cleavage site, which likewise enhanced resistance to apoptosis when mutated to glutamate in Jurkat T cells. Unlike Joo et al. (46), they found that C-HOIP is capable of triggering substrate ubiquitination and NF-κB activation as effectively as full-length HOIP. Nevertheless, they found that substrate Met1-Ub decreases during apoptosis, which was attributed to HOIP cleavage. Lafont et al. (48) reported TRAIL-induced HOIP cleavage, predominantly by caspase-8, which can be blocked by mutating Asp-348, Asp-387, and Asp-390. Interestingly, preventing HOIP cleavage had no effect on TRAIL-induced cell death in K562 cells, a myelogenous leukemic cell line. Thus, to date, the functional outcome of HOIP cleavage has only been examined in HEK293T cells and lymphocyte cell lines. The role of LUBAC in cell death and inflammatory signaling varies between cell types (14, 15, 17, 20); for example, unlike keratinocytes (24, 25, 36), LUBAC activity does not limit TNF-induced cell death of primary murine T cells (49). Given the role of LUBAC in mitigating skin inflammation and keratinocyte cell death in vivo (23, 36), we chose to study HaCaT keratinocytes as a relevant model for examining the regulation of LUBAC activity. In these cells, we found that cleavage of HOIP promoted apoptosis and limited NF-κB activity.
It was previously reported that caspase-1 is upstream of apoptotic caspases in UVB-induced cell death of keratinocytes (50). Further, transgenic overexpression of caspase-1 in murine keratinocytes drives massive cutaneous apoptosis and inflammatory skin disease (51). Here, we report that inhibition or silencing of caspase-1 attenuates apoptotic caspase activation downstream of multiple apoptotic triggers in keratinocytes. Thus, whereas the regulation of LUBAC activity and apoptosis by caspase-1 is consistent with the literature, it remains possible that caspase-1 acts indirectly on HOIP through apoptotic caspases. In fact, caspase-1 showed no capacity to process HOIP in previous studies (46–48); this, however, may be in part due to technical differences. Whereas we have used ITT HOIP to examine susceptibility to cleavage by recombinant caspases, Joo et al. (46) and Lafont et al. (48) used immunoprecipitated HOIP from HEK293T cells. These differences may impact caspase sensitivity due to co-precipitation of inhibitory cofactors or endogenous post-translational modifications of HOIP that might prevent its cleavage. Nevertheless, using recombinant proteins, overexpression, pharmacological inhibition, and stable genetic silencing, we found that caspase-1 and caspase-8 cleaved HOIP in response to apoptotic triggers. We observed that inhibition of caspase-1 enhanced NF-κB activity and cytokine production downstream of UVB irradiation, consistent with restored LUBAC function. Interestingly, inhibition of caspase activity revealed the NF-κB–dependent pro-inflammatory and anti-tumorigenic effects of mitochondrial outer membrane permeabilization in vitro and in vivo (52). As mitochondrial outer membrane permeabilization also occurs following UVB irradiation (53), it is tempting to speculate that these reported effects may be in part driven by LUBAC. Indeed, LUBAC has been shown to regulate genotoxic stress–driven NF-κB activation (e.g. following UVB-induced DNA damage) through the linear ubiquitination of NEMO (54).
Using overexpression assays, we identified caspase-1 as a putative LUBAC substrate. In bone marrow–derived macrophages, caspase-1 is modified with polyubiquitin chains of unknown topology following activation of the NLRP1B inflammasome with anthrax lethal toxin (55). As NLRP1 is also the sensor of UVB irradiation in keratinocytes (56), this suggests that LUBAC may play specific roles downstream of NLRP1. Indeed, LUBAC has been shown to target the adaptor ASC with Met1-Ub, which, through an unknown mechanism, promotes inflammasome activation (3). NLRP1, however, possesses a CARD domain, allowing it to bypass ASC and directly bind to and activate caspase-1 (55), supporting the idea that LUBAC may have a distinct inhibitory role in the regulation of the NLRP1 inflammasome. Collectively, these findings expand on the extensive and dynamic regulatory network within innate immunity pathways that control inflammation and cell death signaling.
Experimental procedures
Cell lines
HaCaT cells and THP-1 monocytes were cultured in RPMI + 10% FBS + l-glutamine + penicillin/streptomycin, and HEK293T cells were cultured in Dulbecco's modified Eagle's medium + 10% FBS + l-glutamine. All cell lines were treated with mycoplasma removal agent for 10 days prior to expansion and freezing.
Cell stimulations
Stimulations were performed in serum-free Opti-MEM. All inhibitors were added at least 30 min prior to the addition of TNFα. UVB irradiation was performed using a Transilluminator at 80% intensity. Briefly, medium was removed, and cells were washed with warm PBS. Cells were exposed to direct UVB light for 15 s (equivalent to 50 mJ/cm2) before medium was added back for the indicated times. Caspase-1 and caspase-8 activity was measured using the Caspase Glo-1 (Promega G9951) and Caspase Glo-8 assays (Promega G8201) according to the manufacturer's protocol. Cell viability was measured using the CellTiter-Glo® luminescent cell viability assay (Promega G7570) according to the manufacturer's instructions.
THP-1 monocytes were differentiated overnight using PMA. The following day, medium was replaced with serum-free Opti-MEM, and cells were primed using 100 ng/ml LPS for 4 h before the addition of ATP (5 mm, 1 h), Lipofectamine-transfected poly(dA:dT) (1.5 μg/ml, 6 h), or DOTAP-transfected flagellin (2.0 μg/ml, 6 h). Cells were harvested and lysed in Laemmli buffer, and protein content was precipitated from cell-free supernatants using TCA/acetone, resuspended in 2× loading buffer for immunoblotting.
Membranes were stained with Ponceau S Red before being dried and blocked with either 4% BSA or 5% nonfat dry milk in TBS + 0.1% Tween 20 (TBS-T) for 1 h at room temperature. Membranes were incubated with primary antibody overnight and washed three times for 10 min each with 0.1% TBS-T, and secondary antibody was added for 1 h at room temperature. Following another round of washes, proteins were visualized using enhanced chemiluminescence.
For immunoprecipitation (IP) of FLAG-tagged proteins, cells were washed with ice-cold PBS lysed in ice-cold lysis buffer (1% Triton, 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA) supplemented with 10 mm sodium fluoride, 1 mm sodium orthovanadate, 10 mm β-glycerophosphate, and complete protease inhibitors (Roche Applied Science) and left on ice for 30–60 min, followed by three 15-s bouts of sonication at 65% amplitude. Lysates were centrifuged at 13,000 rpm for 20 min, soluble proteins were collected, and protein concentration was quantified using the Bio-Rad DC Assay. The FLAG M2 affinity gel was washed two times with TBS before a 20-μl packed volume of beads was added to each IP sample. Samples were rotated at 4 °C for 2 h and washed three times with lysis buffer and one time with PBS, and proteins were eluted by boiling in 40 μl of 2× loading buffer. For immunoprecipitation of HA, Sharpin, or caspase-1, lysates were processed as above, and 2 μg of antibody was added to 0.5–1 mg of lysate and rotated overnight at 4 °C. The next day, a 20-μl packed volume of washed Protein G–agarose was added to each IP sample and rotated for an additional 3–4 h. Samples were then washed and eluted as above.
Immunoblotting and immunoprecipitations
Following stimulation, HaCaT cells were scraped into the media and collected on ice. Cells were pelleted by centrifugation, washed with ice-cold PBS, and then lysed in Laemmli buffer and sonicated 3 times for 15 s each at 70% amplitude. Lysates were boiled for 5 min at 95 °C, and an aliquot was used for SDS-PAGE and transferred to nitrocellulose by wet transfer in 10–20% methanol transfer buffer.
When studying linear ubiquitination, cells were resuspended in Ub lysis buffer (1% SDS, 50 mm Tris-HCl, pH 7.5, 5 mm EDTA) supplemented with complete protease inhibitors and 20 mm N-ethylmaleimide and boiled for 15 min at 95 °C. Lysates were diluted 10 times with Buffer G (1% Triton, 50 mm Tris-HCl, pH 7.5) supplemented with N-ethylmaleimide and protease inhibitors and mechanically lysed with a 23-gauge syringe. Lysates were centrifuged at 13,000 rpm for 20 min, soluble proteins were collected, and protein concentration was quantified using the Bio-Rad DC assay. Immunoprecipitations were carried out as above.
The antibodies used in this manuscript were as follows: anti-β-actin (Sigma A1978), anti-caspase-1 (Adipogen AG-20B-0048), anti-caspase-3 (Cell Signaling 9662), anti-caspase-8 (Enzo Alx-804–242; Cell Signaling 9746), anti-FLAG (Sigma E3165), anti-HA (Roche Applied Science ROAHAHA), anti-HOIL-1 (Novus Bio NBP1-88301), anti-HOIP (R&D MA8039), anti-M1-ubiquitin (EMD Millipore MABS451), anti-Myc (Sigma 11 667 149 001), and anti-Sharpin (Proteintech 14626-1-AP).
RT-PCR
RNA was extracted using the RNeasy MINI kit (Qiagen 74104) according to the manufacturer's protocol. 1.0 μg of RNA was reverse-transcribed into cDNA using random hexamers and Moloney murine leukemia virus reverse transcriptase in a total volume of 20 μl, according to the manufacturers' protocol. Quantitative PCR was performed using iTaq SYBR Green supermix, and following normalization to the housekeeping gene RPL32, -fold induction over untreated was calculated using the 2−ΔΔCt method.
Plasmids, cloning, and mutagenesis
All shRNA plasmids were obtained from the MISSION® shRNA library (Sigma-Aldrich) via McGill University. All mutations were performed using the QuikChange Lightning kit (Agilent Technologies 210518) according to the manufacturer's protocol. Subcloning of Myc-HOIP constructs into pLenti-CMV-Blast (Addgene 17486) for reconstitution experiments was performed using the In-Fusion HD Cloning kit (Takara Bio 121416)
Luciferase assay
NF-κB–driven luciferase activity was measured using the Luciferase Reporter Gene Detection Kit (Sigma LUC1) according to the manufacturer's protocol.
In vitro transcription and translation
In vitro transcribed and translated recombinant proteins were synthesized using the Promega TnT Quick Coupled Transcription/Translation System (Promega L1170) using 1 μg of plasmid according to the manufacturer's protocol. 2 μl of reaction product was incubated in CHEGG buffer (50 mm HEPES-KOH, 100 mm NaCl, 2 mm EDTA, 0.10% CHAPS, 10% glycerol, 10 mm DTT) containing 2 units of recombinant caspase (Biovision K233) and incubated at 37 °C for 1 h. The reaction was terminated by the addition of Laemmli buffer, and cleavage events were analyzed by immunoblotting.
Lentiviral transduction
3 × 105 HEK293T cells were seeded on poly-l-lysine–coated 6-well plates in 2 ml of Dulbecco's modified Eagle's medium + 10% FBS. The following day, cells were transfected with 1.5 μg of psPAX2, 0.5 μg of pMD2.G, and 2.0 of μg of shRNA or pLenti constructs for reconstitution using Lipofectamine 2000. 24 h later, medium was collected and stored at 4 °C, and 3 ml of fresh medium was added to cells. The following day, medium was collected, pooled with the 24-h sample, and centrifuged at 2000 rpm for 10-min to pellet cells and debris. 4 ml of virus-containing medium was added to HaCaT cells in 10-cm dishes at 30% confluence and supplemented with penicillin-streptomycin and 8 μg/ml Polybrene. Viral medium was left on for 8 h at 37 °C before cells were washed and replaced with fresh RPMI + 10% FBS + l-glutamine + penicillin/streptomycin for an additional 16 h. Medium was replaced again at 24 h post-transduction, and cells were selected 48 h post-transduction with 2 μg/ml puromycin (for shRNA) or 10 μg/ml blasticidin (for reconstitution) for 10 days.
Statistical analysis
Data are represented as the mean ± S.E. Two-tailed Student's t test was used for evaluating statistical significance between two groups. One-way analysis of variance followed by a Bonferroni post-hoc comparison test was used to evaluate statistical significance among three or more groups. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Data availability
All data are contained within this paper.
Author contributions
T. D. and M. S. conceptualization; T. D. and M. S. data curation; T. D. formal analysis; T. D. investigation; T. D. methodology; T. D. writing-original draft; M. S. resources; M. S. supervision; M. S. funding acquisition; M. S. project administration; M. S. writing-review and editing.
Acknowledgments
We thank Dr. Kazuhiro Iwai (Kyoto University) for the LUBAC and OTULIN plasmids and the Saleh laboratory for reviewing and commenting on this work.
The authors declare that they have no conflicts of interest with the contents of this article.
- LUBAC
- linear ubiquitin assembly complex
- Ub
- ubiquitin
- Met1-Ub and Lys63-Ub
- Met-1–linked and Lys-63–linked polyubiquitin chains, respectively
- TNF
- tumor necrosis factor
- cpdm
- chronic proliferative dermatitis
- fmk
- fluoromethyl ketone
- ITT
- in vitro transcribed and translated
- CHX
- cycloheximide
- Z
- benzyloxycarbonyl
- NZF
- Npl4 zinc finger
- PMA
- phorbol 12-myristate 13-acetate
- LPS
- lipopolysaccharide
- DOTAP
- dioleoyl-3-trimethylammonium propane
- IP
- immunoprecipitation
- HA
- hemagglutinin
- FBS
- fetal bovine serum.
References
- 1. Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., and Iwai K. (2009) Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 11, 123–132 10.1038/ncb1821 [DOI] [PubMed] [Google Scholar]
- 2. Inn K. S., Gack M. U., Tokunaga F., Shi M., Wong L. Y., Iwai K., and Jung J. U. (2011) Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell 41, 354–365 10.1016/j.molcel.2010.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodgers M. A., Bowman J. W., Fujita H., Orazio N., Shi M., Liang Q., Amatya R., Kelly T. J., Iwai K., Ting J., and Jung J. U. (2014) The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. J. Exp. Med. 211, 1333–1347 10.1084/jem.20132486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zinngrebe J., Rieser E., Taraborrelli L., Peltzer N., Hartwig T., Ren H., Kovács I., Endres C., Draber P., Darding M., von Karstedt S., Lemke J., Dome B., Bergmann M., Ferguson B. J., and Walczak H. (2016) LUBAC deficiency perturbs TLR3 signaling to cause immunodeficiency and autoinflammation. J. Exp. Med. 213, 2671–2689 10.1084/jem.20160041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sasaki Y., Sano S., Nakahara M., Murata S., Kometani K., Aiba Y., Sakamoto S., Watanabe Y., Tanaka K., Kurosaki T., and Iwai K. (2013) Defective immune responses in mice lacking LUBAC-mediated linear ubiquitination in B cells. EMBO J. 32, 2463–2476 10.1038/emboj.2013.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dubois S. M., Alexia C., Wu Y., Leclair H. M., Leveau C., Schol E., Fest T., Tarte K., Chen Z. J., Gavard J., and Bidère N. (2014) A catalytic-independent role for the LUBAC in NF-κB activation upon antigen receptor engagement and in lymphoma cells. Blood 123, 2199–2203 10.1182/blood-2013-05-504019 [DOI] [PubMed] [Google Scholar]
- 7. Satpathy S., Wagner S. A., Beli P., Gupta R., Kristiansen T. A., Malinova D., Francavilla C., Tolar P., Bishop G. A., Hostager B. S., and Choudhary C. (2015) Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol. Syst. Biol. 11, 810 10.15252/msb.20145880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirisako T., Kamei K., Murata S., Kato M., Fukumoto H., Kanie M., Sano S., Tokunaga F., Tanaka K., and Iwai K. (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 10.1038/sj.emboj.7601360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haas T. L., Emmerich C. H., Gerlach B., Schmukle A. C., Cordier S. M., Rieser E., Feltham R., Vince J., Warnken U., Wenger T., Koschny R., Komander D., Silke J., and Walczak H. (2009) Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36, 831–844 10.1016/j.molcel.2009.10.013 [DOI] [PubMed] [Google Scholar]
- 10. Peltzer N., Darding M., and Walczak H. (2016) Holding RIPK1 on the Ubiquitin Leash in TNFR1 Signaling. Trends Cell Biol. 26, 445–461 10.1016/j.tcb.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 11. Fujita H., Rahighi S., Akita M., Kato R., Sasaki Y., Wakatsuki S., and Iwai K. (2014) Mechanism underlying IκB kinase activation mediated by the linear ubiquitin chain assembly complex. Mol. Cell. Biol. 34, 1322–1335 10.1128/MCB.01538-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., and Barker P. A. (2008) cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 10.1016/j.molcel.2008.05.014 [DOI] [PubMed] [Google Scholar]
- 13. de Almagro M. C., Goncharov T., Izrael-Tomasevic A., Duttler S., Kist M., Varfolomeev E., Wu X., Lee W. P., Murray J., Webster J. D., Yu K., Kirkpatrick D. S., Newton K., and Vucic D. (2017) Coordinated ubiquitination and phosphorylation of RIP1 regulates necroptotic cell death. Cell Death Differ. 24, 26–37 10.1038/cdd.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boisson B., Laplantine E., Prando C., Giliani S., Israelsson E., Xu Z., Abhyankar A., Israël L., Trevejo-Nunez G., Bogunovic D., Cepika A. M., MacDuff D., Chrabieh M., Hubeau M., Bajolle F., et al. (2012) Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat. Immunol. 13, 1178–1186 10.1038/ni.2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boisson B., Laplantine E., Dobbs K., Cobat A., Tarantino N., Hazen M., Lidov H. G., Hopkins G., Du L., Belkadi A., Chrabieh M., Itan Y., Picard C., Fournet J. C., Eibel H., et al. (2015) Human HOIP and LUBAC deficiency underlies autoinflammation, immunodeficiency, amylopectinosis, and lymphangiectasia. J. Exp. Med. 212, 939–951 10.1084/jem.20141130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oda H., Beck D. B., Kuehn H. S., Sampaio Moura N., Hoffmann P., Ibarra M., Stoddard J., Tsai W. L., Gutierrez-Cruz G., Gadina M., Rosenzweig S. D., Kastner D. L., Notarangelo L. D., and Aksentijevich I. (2019) Second case of HOIP deficiency expands clinical features and defines inflammatory transcriptome regulated by LUBAC. Front. Immunol. 10, 479 10.3389/fimmu.2019.00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Damgaard R. B., Walker J. A., Marco-Casanova P., Morgan N. V., Titheradge H. L., Elliott P. R., McHale D., Maher E. R., McKenzie A. N. J., and Komander D. (2016) The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell 166, 1215–1230.e20 10.1016/j.cell.2016.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou Q., Yu X., Demirkaya E., Deuitch N., Stone D., Tsai W. L., Kuehn H. S., Wang H., Yang D., Park Y. H., Ombrello A. K., Blake M., Romeo T., Remmers E. F., Chae J. J., et al. (2016) Biallelic hypomorphic mutations in a linear deubiquitinase define otulipenia, an early-onset autoinflammatory disease. Proc. Natl. Acad. Sci. U.S.A. 113, 10127–10132 10.1073/pnas.1612594113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nabavi M., Shahrooei M., Rokni-Zadeh H., Vrancken J., Changi-Ashtiani M., Darabi K., Manian M., Seif F., Meyts I., Voet A., Moens L., and Bossuyt X. (2019) Auto-inflammation in a patient with a novel homozygous OTULIN mutation. J. Clin. Immunol. 39, 138–141 10.1007/s10875-019-00599-3 [DOI] [PubMed] [Google Scholar]
- 20. Damgaard R. B., Elliott P. R., Swatek K. N., Maher E. R., Stepensky P., Elpeleg O., Komander D., and Berkun Y. (2019) OTULIN deficiency in ORAS causes cell type-specific LUBAC degradation, dysregulated TNF signalling and cell death. EMBO Mol. Med. 11, e9324 10.15252/emmm.201809324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peltzer N., Rieser E., Taraborrelli L., Draber P., Darding M., Pernaute B., Shimizu Y., Sarr A., Draberova H., Montinaro A., Martinez-Barbera J. P., Silke J., Rodriguez T. A., and Walczak H. (2014) HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 9, 153–165 10.1016/j.celrep.2014.08.066 [DOI] [PubMed] [Google Scholar]
- 22. Peltzer N., Darding M., Montinaro A., Draber P., Draberova H., Kupka S., Rieser E., Fisher A., Hutchinson C., Taraborrelli L., Hartwig T., Lafont E., Haas T. L., Shimizu Y., Böiers C., et al. (2018) LUBAC is essential for embryogenesis by preventing cell death and enabling haematopoiesis. Nature 557, 112–117 10.1038/s41586-018-0064-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seymour R. E., Hasham M. G., Cox G. A., Shultz L. D., Hogenesch H., Roopenian D. C., and Sundberg J. P. (2007) Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 8, 416–421 10.1038/sj.gene.6364403 [DOI] [PubMed] [Google Scholar]
- 24. Kumari S., Redouane Y., Lopez-Mosqueda J., Shiraishi R., Romanowska M., Lutzmayer S., Kuiper J., Martinez C., Dikic I., Pasparakis M., and Ikeda F. (2014) Sharpin prevents skin inflammation by inhibiting TNFR1-induced keratinocyte apoptosis. eLife 3 10.7554/eLife.03422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rickard J. A., Anderton H., Etemadi N., Nachbur U., Darding M., Peltzer N., Lalaoui N., Lawlor K. E., Vanyai H., Hall C., Bankovacki A., Gangoda L., Wong W. W., Corbin J., Huang C., et al. (2014) TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. eLife 3 10.7554/eLife.03464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berger S. B., Kasparcova V., Hoffman S., Swift B., Dare L., Schaeffer M., Capriotti C., Cook M., Finger J., Hughes-Earle A., Harris P. A., Kaiser W. J., Mocarski E. S., Bertin J., and Gough P. J. (2014) Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J. Immunol. 192, 5476–5480 10.4049/jimmunol.1400499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Douglas T., Champagne C., Morizot A., Lapointe J. M., and Saleh M. (2015) The inflammatory caspases-1 and -11 mediate the pathogenesis of dermatitis in Sharpin-deficient mice. J. Immunol. 195, 2365–2373 10.4049/jimmunol.1500542 [DOI] [PubMed] [Google Scholar]
- 28. Gurung P., Sharma B. R., and Kanneganti T. D. (2016) Distinct role of IL-1beta in instigating disease in Sharpin(cpdm) mice. Sci. Rep. 6, 36634 10.1038/srep36634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feldmeyer L., Keller M., Niklaus G., Hohl D., Werner S., and Beer H. D. (2007) The inflammasome mediates UVB-induced activation and secretion of interleukin-1β by keratinocytes. Curr. Biol. 17, 1140–1145 10.1016/j.cub.2007.05.074 [DOI] [PubMed] [Google Scholar]
- 30. Burian M., and Yazdi A. S. (2018) NLRP1 is the key inflammasome in primary human keratinocytes. J. Invest. Dermatol. 138, 2507–2510 10.1016/j.jid.2018.08.004 [DOI] [PubMed] [Google Scholar]
- 31. Rivkin E., Almeida S. M., Ceccarelli D. F., Juang Y. C., MacLean T. A., Srikumar T., Huang H., Dunham W. H., Fukumura R., Xie G., Gondo Y., Raught B., Gingras A. C., Sicheri F., and Cordes S. P. (2013) The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, 318–324 10.1038/nature12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Assefa Z., Garmyn M., Vantieghem A., Declercq W., Vandenabeele P., Vandenheede J. R., and Agostinis P. (2003) Ultraviolet B radiation-induced apoptosis in human keratinocytes: cytosolic activation of procaspase-8 and the role of Bcl-2. FEBS Lett. 540, 125–132 10.1016/S0014-5793(03)00238-2 [DOI] [PubMed] [Google Scholar]
- 33. Wang M., Zhao X. M., Tan H., Akutsu T., Whisstock J. C., and Song J. (2014) Cascleave 2.0, a new approach for predicting caspase and granzyme cleavage targets. Bioinformatics 30, 71–80 10.1093/bioinformatics/btt603 [DOI] [PubMed] [Google Scholar]
- 34. Poreba M., Strózyk A., Salvesen G. S., and Drag M. (2013) Caspase substrates and inhibitors. Cold Spring Harb. Perspect. Biol. 5, a008680 10.1101/cshperspect.a008680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okamura K., Kitamura A., Sasaki Y., Chung D. H., Kagami S., Iwai K., and Yasutomo K. (2016) Survival of mature T cells depends on signaling through HOIP. Sci. Rep. 6, 36135 10.1038/srep36135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taraborrelli L., Peltzer N., Montinaro A., Kupka S., Rieser E., Hartwig T., Sarr A., Darding M., Draber P., Haas T. L., Akarca A., Marafioti T., Pasparakis M., Bertin J., Gough P. J., et al. (2018) LUBAC prevents lethal dermatitis by inhibiting cell death induced by TNF, TRAIL and CD95L. Nat. Commun. 9, 3910 10.1038/s41467-018-06155-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elliott P. R., Leske D., Hrdinka M., Bagola K., Fiil B. K., McLaughlin S. H., Wagstaff J., Volkmar N., Christianson J. C., Kessler B. M., Freund S. M., Komander D., and Gyrd-Hansen M. (2016) SPATA2 links CYLD to LUBAC, activates CYLD, and controls LUBAC signaling. Mol. Cell 63, 990–1005 10.1016/j.molcel.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elliott P. R., Nielsen S. V., Marco-Casanova P., Fiil B. K., Keusekotten K., Mailand N., Freund S. M., Gyrd-Hansen M., and Komander D. (2014) Molecular basis and regulation of OTULIN-LUBAC interaction. Mol. Cell 54, 335–348 10.1016/j.molcel.2014.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schaeffer V., Akutsu M., Olma M. H., Gomes L. C., Kawasaki M., and Dikic I. (2014) Binding of OTULIN to the PUB domain of HOIP controls NF-kappaB signaling. Mol. Cell 54, 349–361 10.1016/j.molcel.2014.03.016 [DOI] [PubMed] [Google Scholar]
- 40. Liu J., Wang Y., Gong Y., Fu T., Hu S., Zhou Z., and Pan L. (2017) Structural insights into SHARPIN-mediated activation of HOIP for the linear ubiquitin chain assembly. Cell Rep. 21, 27–36 10.1016/j.celrep.2017.09.031 [DOI] [PubMed] [Google Scholar]
- 41. Klein T., Fung S. Y., Renner F., Blank M. A., Dufour A., Kang S., Bolger-Munro M., Scurll J. M., Priatel J. J., Schweigler P., Melkko S., Gold M. R., Viner R. I., Régnier C. H., Turvey S. E., and Overall C. M. (2015) The paracaspase MALT1 cleaves HOIL1 reducing linear ubiquitination by LUBAC to dampen lymphocyte NF-κB signalling. Nat. Commun. 6, 8777 10.1038/ncomms9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elton L., Carpentier I., Staal J., Driege Y., Haegman M., and Beyaert R. (2016) MALT1 cleaves the E3 ubiquitin ligase HOIL-1 in activated T cells, generating a dominant negative inhibitor of LUBAC-induced NF-κB signaling. FEBS J. 283, 403–412 10.1111/febs.13597 [DOI] [PubMed] [Google Scholar]
- 43. Douanne T., Gavard J., and Bidère N. (2016) The paracaspase MALT1 cleaves the LUBAC subunit HOIL1 during antigen receptor signaling. J. Cell Sci. 129, 1775–1780 10.1242/jcs.185025 [DOI] [PubMed] [Google Scholar]
- 44. Takiuchi T., Nakagawa T., Tamiya H., Fujita H., Sasaki Y., Saeki Y., Takeda H., Sawasaki T., Buchberger A., Kimura T., and Iwai K. (2014) Suppression of LUBAC-mediated linear ubiquitination by a specific interaction between LUBAC and the deubiquitinases CYLD and OTULIN. Genes Cells 19, 254–272 10.1111/gtc.12128 [DOI] [PubMed] [Google Scholar]
- 45. Draber P., Kupka S., Reichert M., Draberova H., Lafont E., de Miguel D., Spilgies L., Surinova S., Taraborrelli L., Hartwig T., Rieser E., Martino L., Rittinger K., and Walczak H. (2015) LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 13, 2258–2272 10.1016/j.celrep.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Joo D., Tang Y., Blonska M., Jin J., Zhao X., and Lin X. (2016) Regulation of linear ubiquitin chain assembly complex by caspase-mediated cleavage of RNF31. Mol. Cell. Biol. 36, 3010–3018 10.1128/MCB.00474-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goto E., and Tokunaga F. (2017) Decreased linear ubiquitination of NEMO and FADD on apoptosis with caspase-mediated cleavage of HOIP. Biochem. Biophys. Res. Commun. 485, 152–159 10.1016/j.bbrc.2017.02.040 [DOI] [PubMed] [Google Scholar]
- 48. Lafont E., Kantari-Mimoun C., Draber P., De Miguel D., Hartwig T., Reichert M., Kupka S., Shimizu Y., Taraborrelli L., Spit M., Sprick M. R., and Walczak H. (2017) The linear ubiquitin chain assembly complex regulates TRAIL-induced gene activation and cell death. EMBO J. 36, 1147–1166 10.15252/embj.201695699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Teh C. E., Lalaoui N., Jain R., Policheni A. N., Heinlein M., Alvarez-Diaz S., Sheridan J. M., Rieser E., Deuser S., Darding M., Koay H. F., Hu Y., Kupresanin F., O'Reilly L. A., Godfrey D. I., et al. (2016) Linear ubiquitin chain assembly complex coordinates late thymic T-cell differentiation and regulatory T-cell homeostasis. Nat. Commun. 7, 13353 10.1038/ncomms13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sollberger G., Strittmatter G. E., Grossi S., Garstkiewicz M., Auf dem Keller U., French L. E., and Beer H. D. (2015) Caspase-1 activity is required for UVB-induced apoptosis of human keratinocytes. J. Invest. Dermatol. 135, 1395–1404 10.1038/jid.2014.551 [DOI] [PubMed] [Google Scholar]
- 51. Yamanaka K., Tanaka M., Tsutsui H., Kupper T. S., Asahi K., Okamura H., Nakanishi K., Suzuki M., Kayagaki N., Black R. A., Miller D. K., Nakashima K., Shimizu M., and Mizutani H. (2000) Skin-specific caspase-1-transgenic mice show cutaneous apoptosis and pre-endotoxin shock condition with a high serum level of IL-18. J. Immunol. 165, 997–1003 10.4049/jimmunol.165.2.997 [DOI] [PubMed] [Google Scholar]
- 52. Giampazolias E., Zunino B., Dhayade S., Bock F., Cloix C., Cao K., Roca A., Lopez J., Ichim G., Proïcs E., Rubio-Patiño C., Fort L., Yatim N., Woodham E., Orozco S., et al. (2017) Mitochondrial permeabilization engages NF-kappaB-dependent anti-tumour activity under caspase deficiency. Nat. Cell Biol. 19, 1116–1129 10.1038/ncb3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Adrain C., Creagh E. M., and Martin S. J. (2001) Apoptosis-associated release of Smac/DIABLO from mitochondria requires active caspases and is blocked by Bcl-2. EMBO J. 20, 6627–6636 10.1093/emboj/20.23.6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niu J., Shi Y., Iwai K., and Wu Z. H. (2011) LUBAC regulates NF-κB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 30, 3741–3753 10.1038/emboj.2011.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Van Opdenbosch N., Gurung P., Vande Walle L., Fossoul A., Kanneganti T. D., and Lamkanfi M. (2014) Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nat. Commun. 5, 3209 10.1038/ncomms4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fenini G., Grossi S., Contassot E., Biedermann T., Reichmann E., French L. E., and Beer H. D. (2018) Genome editing of human primary keratinocytes by CRISPR/Cas9 reveals an essential role of the NLRP1 inflammasome in UVB sensing. J. Invest. Dermatol. 138, 2644–2652 10.1016/j.jid.2018.07.016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within this paper.