Abstract
The actin cytoskeleton is extremely dynamic and supports diverse cellular functions in many physiological and pathological processes, including tumorigenesis. However, the mechanisms that regulate the actin-related protein 2/3 (ARP2/3) complex and thereby promote actin polymerization and organization in cancer cells are not well-understood. We previously implicated the proline-rich 11 (PRR11) protein in lung cancer development. In this study, using immunofluorescence staining, actin polymerization assays, and siRNA-mediated gene silencing, we uncovered that cytoplasmic PRR11 is involved in F-actin polymerization and organization. We found that dysregulation of PRR11 expression results in F-actin rearrangement and nuclear instability in non-small cell lung cancer cells. Results from molecular mechanistic experiments indicated that PRR11 associates with and recruits the ARP2/3 complex, facilitates F-actin polymerization, and thereby disrupts the F-actin cytoskeleton, leading to abnormal nuclear lamina assembly and chromatin reorganization. Inhibition of the ARP2/3 complex activity abolished irregular F-actin polymerization, lamina assembly, and chromatin reorganization due to PRR11 overexpression. Notably, experiments with truncated PRR11 variants revealed that PRR11 regulates F-actin through different regions. We found that deletion of either the N or C terminus of PRR11 abrogates its effects on F-actin polymerization and nuclear instability and that deletion of amino acid residues 100–184 or 100–200 strongly induces an F-actin structure called the actin comet tail, not observed with WT PRR11. Our findings indicate that cytoplasmic PRR11 plays an essential role in regulating F-actin assembly and nuclear stability by recruiting the ARP2/3 complex in human non-small cell lung carcinoma cells.
Keywords: actin, nuclear lamina, Arp2/3 complex, cytoskeleton, heterochromatin, lung cancer, actin comet tail, actin-related protein 2/3 (Arp2/3) complex, F-actin polymerization, nuclear lamina, proline-rich 11 (PRR11)
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide. Despite some advances in early detection and recent improvements in treatment, 5-year survival rates for lung cancer remain low (19%) for all stages (1). Thus, unraveling more detailed and special molecular mechanisms underlying lung cancer progression is required. Changes in the pattern of gene expression play an important role in allowing cancer cells to acquire their hallmark characteristics (2, 3). Nuclear lamina interacts with special histones and lamina-associated domains (LADs)3 to help organize chromatin inside the nucleus and associate with gene expression repression (4–6). Alterations in nuclear lamina often result in changes to chromatin organization and genome function (4, 7). The actin cytoskeleton is capable of mediating a huge range of specific cellular processes such as nuclear lamina assembly and chromatin organization (8–11). Recent studies have shown that the cytoskeleton is involved in nuclear morphology and chromatin dynamics (10, 11), but as yet very little is known about cytoplasmic factors involved in modulating nuclear lamina integrity and chromatin organization.
The actin cytoskeleton is outstandingly dynamic, and the polymerization of globular (G)- to filament (F)-actin can generate a variety of architectures, which is tightly regulated and catalyzed by actin-nucleating proteins such as the Arp2/3 complex (actin-related protein 2/3 complex), which nucleates and facilitates branched actin filaments. The Arp2/3 complex drives branched actin filament networks playing critical roles in lamellipodium protrusion, vesicle trafficking, and pathogen motility (12–15). The Arp2/3 complex has an important role in human respiratory syncytial virus–induced filopodia in A549 cells (16). In contrast, formins nucleate and elongate unbranched actin filaments (8, 17). Arp2/3 complex, a seven-subunit protein consisting of Arp2, Arp3, and Arpc1 to Arpc5, is tightly regulated and is activated by actin monomers and Wiskott-Aldrich syndrome protein (WASP) family proteins. WASP family proteins activate the Arp2/3 complex to produce branched actin filaments through the conserved Arp2/3-binding catalytic C-terminal VCA domain (verprolin-homology (WH2), cofilin homology, and acidic domains), which have been well-understood (8). In addition, either surface proteins of the pathogen (ActA or IcsA) recruit N-WASP or WHAMM (WASP homolog-associated protein with actin) is directed to activate the Arp2/3 complex and is able to kinetically stimulate the Arp2/3 complex for actin polymerization resulting in actin comet tails (15, 18, 19). Profilin binds with and inhibits Arp2/3 complex-mediated branched actin assembly. In contrast, it can bind with formins and Ena/VASP, which accelerates unbranched actin filament polymerization (20–23). However, less is known about the other factors stimulating the Arp2/3 complex to promote actin polymerization and arrangement.
Actin cytoskeleton couples with nuclear lamins by the LINC complex, which is composed of KASH domain proteins (Nesprins) in the outer nuclear membrane and SUN domain proteins (SUN1/2) in the inner nuclear membrane (6, 24–26). Nuclear laminae consist of A-type and B-type lamins, which form a meshwork of intermediate filaments underlying the inner nuclear membrane. Nuclear laminae maintain the shape and mechanical stability of the nucleus and also have important roles in chromatin organization, gene expression, and DNA replication (5, 25, 27, 28). Nuclear laminae regulate chromatin organization to provide a transcriptionally-repressive environment via directly interacting with heterochromatin marks, the histone 3 lysine 9 di- and tri-methylation modifications (H3K9me2/3) and histone 3 lysine 27 di- and tri-methylation modifications (H3K27me2/3), as well as the heterochromatin protein 1 (HP1) family, and/or hundreds of LADs (4, 29, 30). In addition, nuclear laminae associate with several transcription factors and are involved in specific regulatory pathways (5, 31). It is clear that perturbations in the nuclear lamina structure can strongly influence chromatin organization and gene expression.
We have previously demonstrated that PRR11 is implicated in non-small cell lung cancer development and cell-cycle progression. PRR11 knockdown caused the dysregulation of multiple critical pathways and various important genes involved in cell cycle, tumorigenesis, and metastasis (32, 33). PRR11 also has critical roles in gastric cancer, breast cancer, and hilar cholangiocarcinoma (34–36). However, the precise molecular mechanism behind PRR11-mediated cellular functions and tumorigenesis remains unclear.
In this report, we demonstrate that cytoplasmic PRR11 promotes Arp2/3 complex-dependent F-actin polymerization, nuclear lamina integrity, and chromatin organization. Mechanistically, PRR11 interacts and recruits the Arp2/3 complex to drive F-actin polymerization. Overexpression of PRR11 in lung cancer cells led to higher F-actin polymerization and F-actin rearrangement, inducing aberrant nuclear lamina integrity and chromatin dynamics, which is tightly associated with altered gene expression and tumorigenesis. Our study reveals that PRR11 serves as a novel cytoplasmic factor regulating F-actin assembly and nuclear stability.
Results
PRR11 associates with and recruits Arp2/3 complex
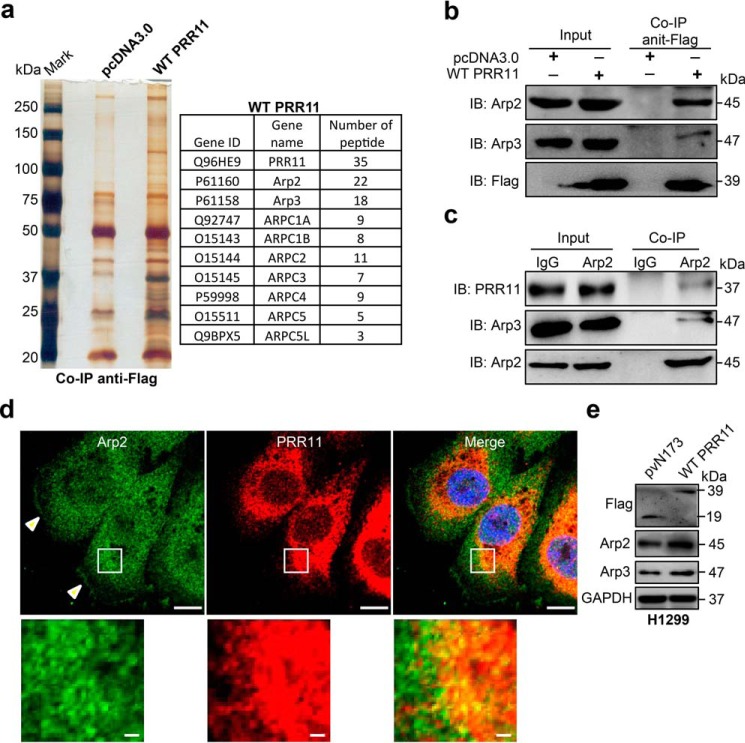
PRR11 is previously identified as a cancer-related gene that is implicated in lung cancer development (32, 37); however, the mechanism behind PRR11-mediated cellular function remains unclear. To explore how PRR11 regulates cellular functions and lung tumorigenesis, we first performed PRR11 protein co-immunoprecipitation to identify its interacting partners. Multiple components of the seven-subunit actin-related protein 2/3 (Arp2/3) complex were identified in the PRR11 immunoprecipitates by mass spectrometry (MS), including Arp2, Arp3, ARPC1A, ARPC1B, ARPC2, ARPC3, ARPC4, ARPC5, and ARPC5L (Fig. 1a). To verify the interaction of PRR11 with the Arp2/Arp3 complex, we carried out co-immunoprecipitation followed by Western blotting assays. Consistent with the result from MS, we detected a significant amount of Arp2 and Arp3 in ectopic Flag-tagged wildtype (WT) PRR11 immunoprecipitates (Fig. 1b). The endogenous PRR11 and Arp2 in H1299 lung cancer cells are also co-immunoprecipitated (Fig. 1c). To further test the notion that PRR11 was associated with the Arp2/3 complex in lung cancer cells, we used immunofluorescence assay to analyze the co-localization of PRR11 and Arp2 in H1299 cells. Notably, WT PRR11-overexpression cells showed PRR11-containing meshwork structures (Fig. S1a). Arp2 was dramatically co-localized with the PRR11 in cytoplasm (Fig. 1d). In addition, endogenous Arp2 was also localized to the PRR11-containing meshwork structures in the PRR11 ectopic expression cells (Fig. S1, b and c). Additionally, immunofluorescence assay also showed that Arp2 expression significantly increased in ectopic PRR11-expressing cells (Fig. S1, b and d), which was consistent with the Western blotting result that overexpression of PRR11 increased Arp2 and Arp3 levels (Fig. 1e). To confirm the specificity of PRR11 immunostaining signals, we have stained the Flag–PRR11-overexpression cells with both PRR11 and Flag antibodies. We showed that PRR11 antibody can detect both endogenous and exogenous Flag–PRR11 (Fig. S1e). Together, these data demonstrate PRR11 is associated with the Arp2/3 complex, and it is required for the recruitment and accumulation of the Arp2/3 complex in lung cancer cells.
Figure 1.
PRR11 associates with and recruits Arp2/3 complex. a, mass spectrometry analysis of PRR11-associated proteins. Lysates from H1299 cells transfected with Flag-WT PRR11 or pcDNA3.0 (empty) were immunoprecipitated with anti-Flag and then analyzed by MS. Subunits of Arp2/3 complex (Arp2, Arp3, ARPC1A, ARPC1B, ARPC2, ARPC3, ARPC4, ARPC5, and ARPC5L) were identified in the PRR11 immunoprecipitates. b, and c, interaction between PRR11 and Arp2 was confirmed by coimmunoprecipitation. b, cells were transfected with plasmids encoding Flag-WT PRR11 or pcDNA3.0. Cell lysates were immunoprecipitated with Flag antibody and blotted with different antibodies as indicated. c, to verify the interaction of endogenous proteins, the lysate of cell was immunoprecipitated with Arp2 antibody. Co-immunoprecipitation (Co-IP) and whole-cell lysates (Input) were subjected to immunoblotting (IB) with different antibodies as indicated. d, endogenous PRR11 co-localizes with Arp2. Cells were fixed and stained for PRR11 and Arp2 antibody. Scale bars, 10 μm. White arrowhead indicates that Arp2 localizes at the lamellipodia, and zoomed images of the boxed region are shown at the bottom-left corner (scale bars, 1 μm). e, Western blot analysis for the indicated proteins in the pvN173- and WT PRR11-transfected cells. Cells were lysed 24 h after transfection and analyzed for the indicated proteins. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
PRR11 facilitates F-actin polymerization via Arp2/3 complex
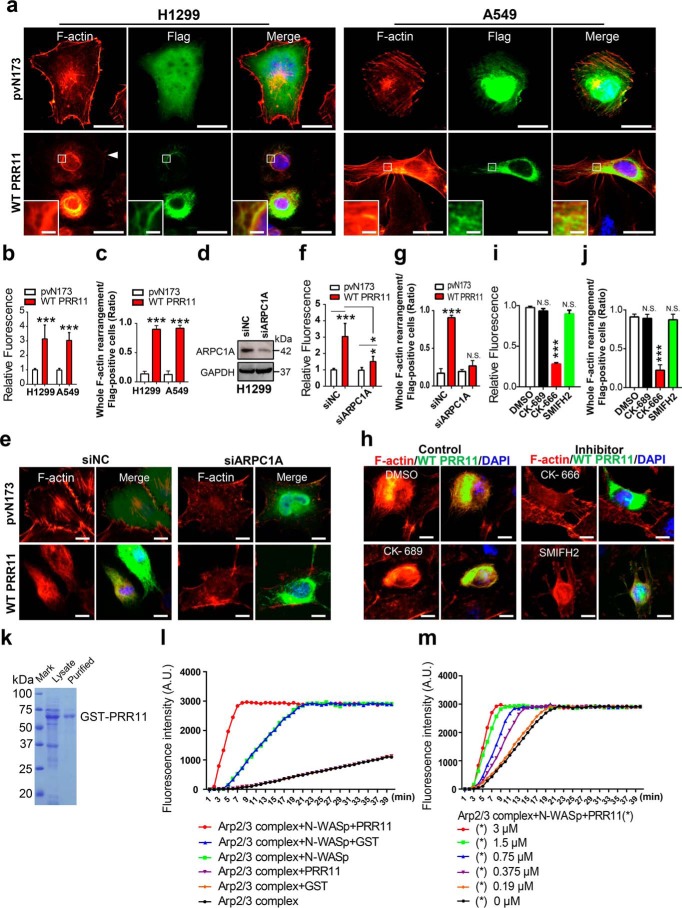
The seven-subunit actin-related protein 2/3 (Arp2/3) complex is the key nucleation factor for the polymerization of branched actin network as opposed to the formins or Ena/VASP, which form bundle actin filaments, and this complex is highly conserved in almost all eukaryotes (17, 20). Recently, a target Arp2-specific siRNA and an Arp2/3 complex inhibitor, CK-666, were employed to inhibit Arp2/3 complex assembling branched actin filaments in A549 cells (16, 38). To test whether PRR11 could promote F-actin polymerization by facilitating the recruitment and accumulation of Arp2/3 complex, H1299 and A549 cells were expressed with WT PRR11, and then phalloidin–TRITC staining was performed to identify F-actin. Interestingly, the phalloidin–TRITC staining not only demonstrated that the fluorescence signal of F-actin was notably increased in WT PRR11-expressing cells compared with pvN173-control cells, but also revealed that the subcellular distribution of F-actin was rearranged into more meshwork actin structures upon PRR11 overexpression, indicating that PRR11 promoted F-actin polymerization and rearrangement (Fig. 2, a–c). To investigate whether PRR11-induced F-actin polymerization and rearrangement depended on the Arp2/3 complex, we inhibited the activity of the Arp2/3 complex by silencing ARPC1A (a subunit of Arp2/3 complex) expression or using CK-666 (the Arp2/3 complex inhibitor), respectively (20, 39). As shown in Fig. 2, d–g, knockdown of ARPC1A restored both the F-actin signal and its rearrangement induced by PRR11 overexpression. In keeping with the siRNA-mediated knockdown experiments, CK-666 also gave rise to an inhibition of PRR11-induced F-actin signal enhancement and rearrangement, whereas DMSO, CK-689 (the Arp2/3 complex negative control reagent), and SMIFH2 (inhibitor of formin homology 2 domains) did not (Fig. 2, h–j). To further confirm that PRR11 promotes the kinetics of Arp2/3 complex-induced F-actin polymerization, we next performed pyrene-labeled actin polymerization assays. GST–PRR11 was produced and purified (Fig. 2k). The recombinant GST–N-WASP was used as the nucleation-promoting factor, which stimulated the activation Arp2/3 complex to induce actin polymerization (Fig. 2l). In the presence of both GST–N-WASP and Arp2/3 complex, GST–PRR11 significantly enhanced actin polymerization compared with GST protein (Fig. 2l). In addition, GST–PRR11 stimulates Arp2/3 complex-mediated actin polymerization in a dose-dependent manner (Fig. 2m). Taken together, these data demonstrate that PRR11 associates with the Arp2/3 complex to promote F-actin polymerization.
Figure 2.
PRR11 promotes F-actin polymerization via Arp2/3 complex. a, PRR11 overexpression stimulates F-actin assembly and reorganization. H1299 and A549 cells were fixed at 24 h after transfection with pvN173 Flag–control or Flag-WT PRR11 and stained with Flag antibody and F-actin–binding phalloidin–TRITC. Scale bars, 20 μm. White arrowhead indicates WT PRR11-expressing cells, and zoomed images of boxed region are shown at the bottom-left corner (scale bars, 2 μm). Representative images are shown. b, relative phalloidin fluorescence (Flag PRR11–positive cells/pvN173–positive cells) was quantitatively analyzed for a. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. c, quantitative analysis of F-actin rearrangement of cells is demonstrated in a. Ratio indicated that the number of cells with whole F-actin rearrangement is relative to the number of Flag-positive cells. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. d, siRNA-mediated silencing of ARPC1A. H1299 cells were transfected with negative control siRNA (siNC) or siRNA against ARPC1A (siARPC). 48 h after transfection, whole-cell lysates were prepared and subjected to immunoblotting. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. e–g, silencing of ARPC1 represses F-actin assembly induced by overexpression of PRR11. e, H1299 cells were transfected with siNC or siARPC1 for 24 h, and then the cells were transfected with Flag–PRR11 or pvN173 Flag–control for another 24 h, respectively. The cells were fixed and stained for F-actin and Flag. Representative images are shown. Scale bars, 10 μm. f, relative fluorescence of F-actin (Flag PRR11-positive cells/pvN173-positive cells) was quantitatively analyzed for e. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. g, quantitative analysis of F-actin rearrangement of cells is demonstrated in e. Ratio indicated that the number of cells with whole F-actin rearrangement is relative to the number of Flag-positive cells. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. h–j, CK-666, the Arp2/3 complex inhibitor, represses F-actin assembly driven by PRR11. h, H1299 cells transfected with Flag–PRR11 or pvN173 Flag–control were treated with CK-666 (84 μm), SMIFH2 (15 μm), or CK-689 (100 μm) for 24 h and then were fixed and stained with Flag antibody and the F-actin–binding phalloidin–TRITC. Representative images are shown. Scale bars, 20 μm. i, relative phalloidin fluorescence (Flag–PRR11-positive cells/pvN173–positive cells) was quantitatively analyzed for h. Results are shown as the mean ± S.E. (n = 3). Greater than 50 cells were counted per condition in every repeat. j, quantitative analysis of F-actin rearrangement of cells demonstrated in h. Ratio indicated that the number of cells with whole F-actin rearrangement is relative to the number of Flag-positive cells. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. k, Coomassie Blue–stained gel shows the purified GST–PRR11. l, effect of PRR11 on Arp2/3-mediated actin polymerization was examined using the pyrene actin assay. N-WASP was added to activate Arp2/3-mediated actin polymerization. The addition of 3 μm purified GST–PRR11 promoted actin assembly. As a control, GST had no effect on the kinetics of actin polymerization mediated by Arp2/3 and N-WASP. m, PRR11 stimulates Arp2/3-mediated actin polymerization in a dose-dependent manner. The graph showed the effects of 0–3 μm PRR11 on Arp2/3-mediated actin polymerization in the presence of N-WASP. A.U., arbitrary units. Cell nuclei were stained with DAPI (blue). *, p < 0.05; ***, p < 0.001; N.S., p > 0.05.
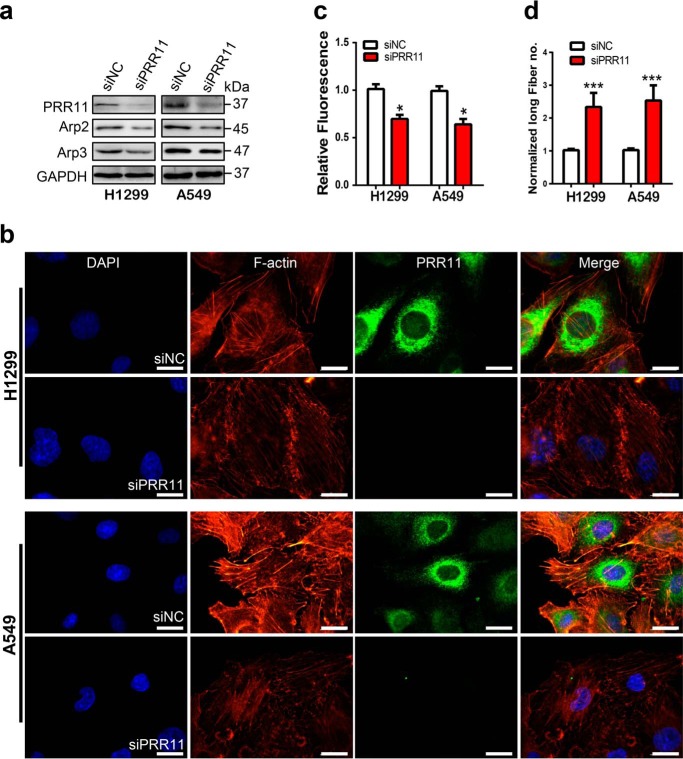
To support the notion that PRR11 is required for F-actin polymerization and arrangement, we further investigated the F-actin polymerization and arrangement in response to siRNA-mediated PRR11 silencing in H1299 and A549 cells. Western blotting showed that silencing of PRR11 reduced the expression levels of Arp2 and Arp3 (Fig. 3a). Immunofluorescence staining also showed a decreased fluorescence signal of F-actin following PRR11 siRNA treatment (Fig. 3, b and c). We also found that the F-actin meshwork structure was inhibited, leading to an increased number of long linear filament fibers in PRR11 siRNA cells (Fig. 3, b and d), suggesting that PRR11 knockdown suppressed F-actin polymerization and induced its rearrangement in lung cancer cells.
Figure 3.
Silencing of PRR11 inhibits F-actin polymerization and arrangement. a, Western blot analysis for the indicated proteins. Cells transfected with siNC or siRNA directed against PRR11 were lysed, and whole-cell lysates were prepared to analyze the expression of the indicated proteins 48 h after transfection. b, PRR11 knockdown suppressed F-actin assembly. Cells treated with siRNA were fixed 48 h after and stained with F-actin–binding phalloidin–TRITC and PRR11 antibody. Representative images are shown. Bars, 20 μm. c, relative phalloidin–TRITC fluorescence in cells demonstrated in b. Results are shown as the mean ± S.E. (n = 3). Greater than 50 cells were counted per condition in every repeat. d, quantification of the number of rearranged long filament actin following the siRNA treatment described as in b. Results are presented as mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. Cell nuclei were stained with DAPI (blue). *, p < 0.05; ***, p < 0.001.
PRR11 promotes F-actin polymerization and rearrangement via both N and C termini
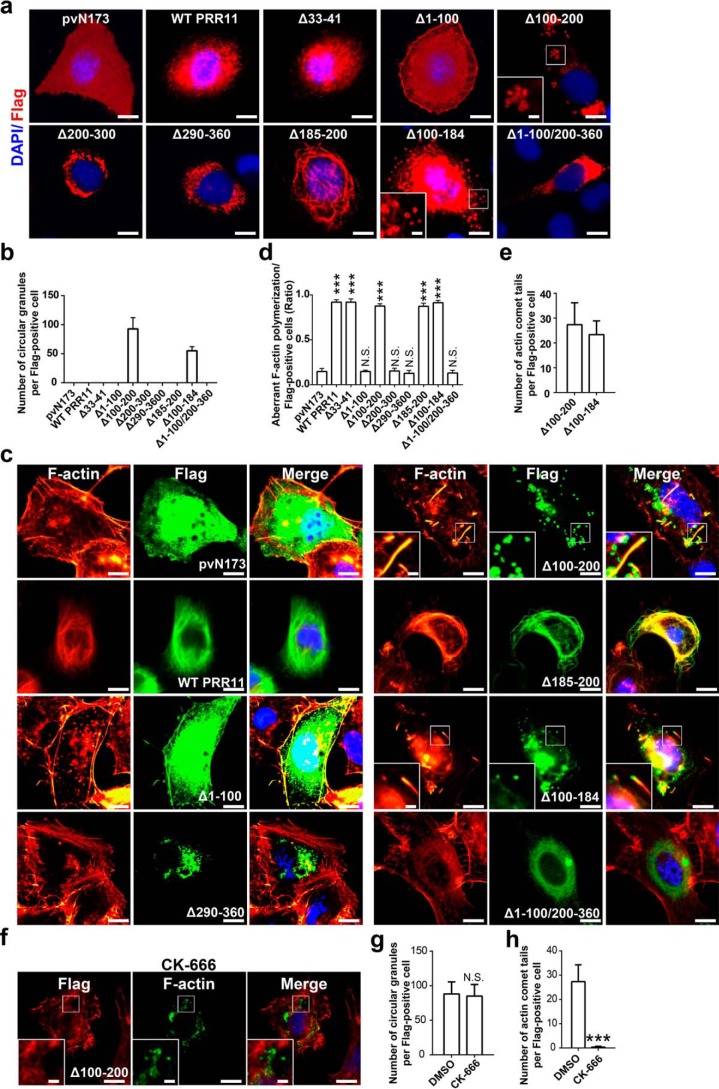
We found that PRR11 overexpression or depletion altered F-actin polymerization and rearrangement. We then sought to determine the regions of PRR11 that are essential for F-actin organization. To this end, we first generated the Δ33–41 mutant by deleting proline-rich region 1 and a series of truncated mutants of PRR11 by deleting the protein regions from the N terminus to the C terminus, including the N-terminal 1–100 amino acids (Δ1–100) and 100–200 amino acids (Δ100–200) and the C-terminal 200–300 amino acids (Δ200–300) and 290–360 amino acids (Δ290–360) (Fig. S2a).
By overexpression of WT PRR11 and the truncated PRR11 mutants, we found that the truncated mutant absence of proline-rich region 1 (Δ33–41) showed similar localization to WT PRR11 (Fig. 4a). The mutant absence of N-terminal Δ1–100 was localized at the pericellular region and dispersed in the cytoplasm. Δ200–300 and Δ290–360 localized mainly in the perinuclear space (Fig. 4a). In contrast, in Δ100–200–expressing cells, a kind of PRR11-containing circular aggregation (circular granules) was identified (Fig. 4a). These data suggested that all of the domains of 1–100, 100–200, 200–300, and 290–360 were essential for PRR11 localization, whereas the proline-rich 1 domain was not.
Figure 4.
PRR11 promotes F-actin polymerization via both N and C termini. a, subcellular distribution of truncated mutations. H1299 cells were overexpressed with Flag-tagged pvN173, WT PRR11, Δ33–41, Δ1–100, Δ100–200, Δ200–300, Δ290–360, Δ185–200, Δ100–184, or Δ1–100/200–360, and then the cells were fixed 24 h after and stained for Flag. Representative images are shown. Bar, 10 μm. Zoomed images of boxed region are shown at the bottom-left corner (scale bars, 5 μm). b, quantitative analysis of circular granules demonstrated in a. The number of circular granules per Flag-positive cells is given. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. c–e, effects of truncated mutations on F-actin polymerization and rearrangement. c, H1299 cells were overexpressed with Flag-tagged pvN173, WT PRR11, Δ1–100, Δ100–200, Δ290–360, Δ185–200, Δ100–184, or Δ1–100/200–360, and then cells were fixed 24 h after and stained for Flag and phalloidin. Representative images are shown. Bar, 10 μm. Zoomed images of boxed region are shown at the bottom-left corner (scale bars, 5 μm). d, quantitative analysis of aberrant F-actin polymerization cells demonstrated in c. Ratio indicated that the number of cells with aberrant F-actin polymerization is relative to the number of Flag-positive cells. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. e, quantitative analysis of actin comet tails demonstrated in c. The number of cells with actin comet tails per Flag-positive cells is shown. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. f, CK-666 inhibited actin comet tails but not circular granules induced by Δ100–200. H1299 cells were overexpressed with Δ100–200 and combined with/without CK-666 (84 μm), and then cells were fixed 24 h after and stained for Flag and phalloidin. Representative images are shown. Bar, 10 μm. Zoomed images of boxed region are shown at the bottom-left corner (scale bars, 5 μm). g, quantitative analysis of circular granules demonstrated in f. The number of circular granules per Flag-positive cells is shown. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. h, quantitative analysis of actin comet tails demonstrated in f. The number of cells with actin comet tails per Flag-positive cells is shown. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. Cell nuclei were stained with DAPI (blue).***, p < 0.001, or N.S., not significant (p > 0.05).
Because of the specific phenotype (circular granules) identified in Δ100–200 overexpression cells, but not in WT PRR11 and other truncated mutant overexpression cells (Fig. 4, a and b), we further generated Δ100–184 and Δ185–200 (Δproline-rich region 2) by deleting shorter domains in the region of 100–200 and a mutant containing 100–200 amino acids (Δ1–110/Δ 200–360) (Fig. S2a). Interestingly, we found that Δ185–200 showed thick filamentous PRR11-containing structures (Fig. 4a), but in Δ100–184–expressing cells, both PRR11-containing meshwork and circular granules were identified (Fig. 4, a and b). However, the 100–200 protein domain (Δ1–110/200–360 construct) was localized to the perinuclear cytoplasm, similar to Δ200–300 and Δ290–360 (Fig. 4a). Taken together, these data demonstrated that domain 100–200 is involved in specific subcellular localization of PRR11, and deletion of residues 100–200 induces PRR11-containing circular aggregation (circular granules).
Next, we examined whether ectopic expression of the truncated mutations can affect F-actin polymerization and rearrangement. The result showed that WT PRR11, Δ33–41, Δ100–184, Δ185–200, and Δ100–200 remarkably facilitate F-actin assembly. The Δ33–41 mutant, which showed the same localization as WT PRR11, caused F-actin polymerization and rearrangement similar to that of WT PRR11 (Fig. S2b). However, the expression of Δ100–184, Δ185–200, and Δ100–200 led to F-actin organization different from that of WT PRR11 (Fig. 4, c–e). In contrast to WT PRR11 that promoted more meshwork F-actin assembly, Δ185–200 mutant stimulated thick actin filaments that highly co-localized with Δ185–200 PRR11 mutant (Fig. 4c). The Δ100–200 mutant, however, led to the formation of actin comet tails associated with PRR11 circular granules (Fig. 4, c and e). Δ100–184 mutant also caused polymerized actin (including actin comet tails) associated with PRR11 (Fig. 4, c and e). These data suggest that the disordered localization of certain PRR11 mutants caused different forms of aberrant F-actin organization.
Actin comet tails are assembled by actin polymerization and mediated by Arp2/3 complex (18). As mentioned above, we have demonstrated that PRR11 recruits Arp2/3 complex to facilitate F-actin polymerization. We therefore examined whether the actin comet tails induced by PRR11 Δ100–200 truncations were also dependent on Arp2/3 activity by treating the cells with CK-666. As expected, the actin comet tails were absolutely inhibited by CK-666 in Δ100–200–expressing cells, although the localization of mutant PRR11 in circular granules was not affected (Fig. 4, f–h). Taken together, these data supported that Arp2/3 complex is required for the formation of actin comet tails in Δ100–200 overexpression cells.
Despite the positive effect of WT PRR11, Δ33–41, Δ100–184, Δ185–200, and Δ100–200 in F-actin polymerization and assembly, the expression of Δ200–300, Δ290–360, and Δ1–100/200–360 mutants, which localized at the perinuclear space, showed little co-localization with F-actin and did not affect F-actin organization (Fig. 4, c–e). Δ1–100 mutant also did not affect the F-actin structure compared with the pvN173 control, although it partially co-localized with F-actin at the pericellular region and cytoplasm (Fig. 4, c–e). These data revealed that N terminus (1–100), C terminus (290–360), and domain 200–300 were required for WT PRR11-mediated F-actin assembly.
Cytoplasmic PRR11 regulates nuclear integrity via Arp2/3 complex
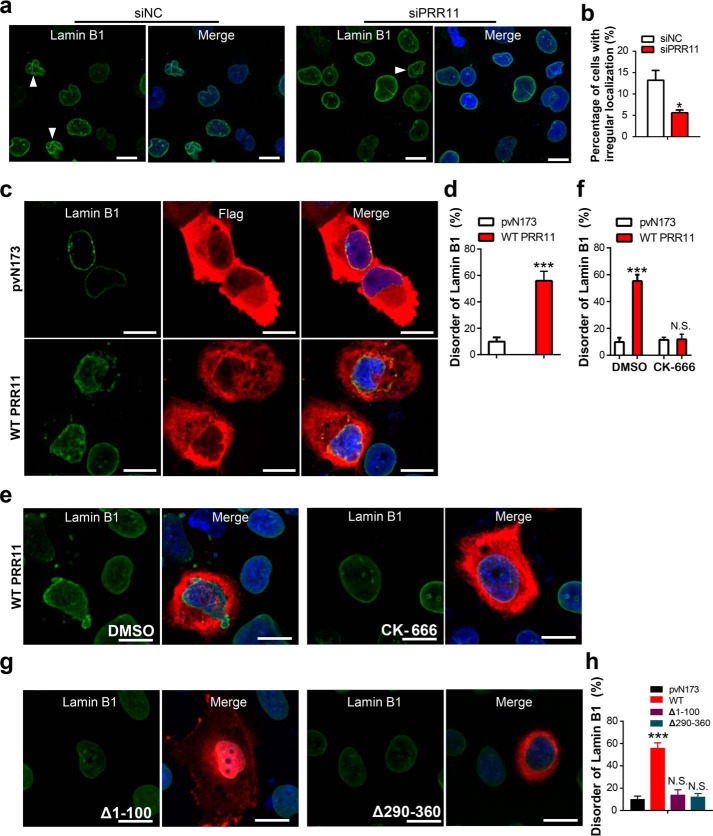
Nuclear lamins form a dense meshwork of filaments termed nuclear lamina at the nuclear periphery that is thought to play a critical role in nuclear stability, chromatin organization, and gene expression (4, 5, 7, 10). The subcellular distribution of nuclear lamins is usually disorderly in cancer cells, and alterations in the nuclear lamina of cells are closely related to tumorigenesis (26). Interestingly, we found that the H1299 lung cancer cells expressing a higher level of PRR11 showed more severe aberrant nuclear laminae as demonstrated by dislocated Lamin A/C compared with its nuclear peripheral localization in PRR11 low cells (Fig. S3a).
The actin cytoskeleton has been shown to physically couple with the nuclear laminae to regulate nuclear morphology, nuclear stability, and chromatin organization (8, 10, 11, 40–42). Also, we have shown above that PRR11 is required for the Arp2/3 complex–dependent F-actin accumulation and cytoskeleton assembly in lung cancer cells. We then examined whether PRR11 could regulate nuclear laminae through the Arp2/3 complex. We first tested whether silencing of PRR11 influences the integrity of nuclear laminae. As shown in Fig. 5, a and b, and Fig. S3, b and c, the irregular distribution of Lamin B1 and lamin A/C in PRR11 knockdown cells (Lamin B1, 4.8 ± 1.3%; Lamin A/C, 4.3 ± 1.9%) was notably reduced compared with that in control knockdown cells (Lamin B1, 13.6 ± 4.2%; Lamin A/C, 12.8 ± 4.7%). In contrast, immunofluorescence staining showed that the number of cells with disordered Lamin B1 and Lamin A/C distributions was remarkably increased in ectopic PRR11 expression cells (Lamin B1, 54.8 ± 9.3%; Lamin A/C, 58.7 ± 10.2%) compared with that in pVN173-control cells (Lamin B1, 14.7 ± 3.3%; Lamin A/C, 15.7 ± 4.2%) (Fig. 5, c and d, and Fig. S3, d and e). Our data show that PRR11 is involved in the novel regulation of lamins' subcellular distribution and nuclear lamina integrity.
Figure 5.
PRR11 controls nuclear lamina assembly via F-actin cytoskeleton organization. a, H1299 cells treated with siNC or siPRR11 were stained for Lamin B1. Representative images are shown. White arrowheads indicate the cells with irregular shape. Bars, 10 μm. b, quantification of cells of irregular shape demonstrated in a. Results are shown as the mean ± S.E. (n = 3). Greater than 50 cells were counted per condition in every repeat. c, immunofluorescence staining for Lamin B1 of cells transfected with Flag-tagged pVN173 or WT PRR11. White arrowheads indicate the WT PRR11-expressing cells with disordered nuclear lamina as shown by Lamin B1. Representative images are shown. Scale bars, 10 μm. d, quantification of cells of disordered Lamin B1 as normalized to total Flag-positive cells demonstrated in c. Results are shown as the mean ± S.E. (n = 3). Greater than 50 cells were counted per condition in every repeat. e and f, CK-666 inhibited nuclear lamina reassembly induced by WT PRR11 overexpression. e, H1299 cells were overexpressed with WT PRR11 and treated with or without CK-666 (84 μm), and then cells were fixed 24 h after and stained for Flag and Lamin B1. Representative images are shown. Bar, 10 μm. f, quantification of cells of disordered Lamin B1 as normalized to total Flag-positive cells demonstrated in e. Results are shown as the mean ± S.E. (n = 3). Greater than 50 cells were counted per condition in every repeat. g and h, effects of PRR11 N- and C-terminal truncated mutations on nuclear lamina assembly. g, H1299 cells were overexpressed with Δ1–100 or Δ290–360, and then cells were fixed 24 h after and stained for Flag and Lamin B1. Representative images are shown. Bar, 20 μm. h, quantitative analysis of aberrant Lamin B1 distribution cells demonstrated in g. Ratio indicated that the number of cells with aberrant Lamin B1 distribution is relative to the number of Flag-positive cells. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. Cell nuclei were stained with DAPI (blue). ***, p < 0.001, or N.S., not significant (p > 0.05).
We have demonstrated that PRR11 is key for the distribution of lamins and nuclear lamina integrity. However, immunofluorescence staining showed PRR11 does not co-localize with the nuclear lamina as demonstrated by Lamin A/C staining (Fig. S3, f and g), and immunoblot analyses identified that PRR11 was expressed in cytoplasm (Fig. S3h). These findings indicate that cytoplasmic PRR11 regulates nuclear lamina assembly independent of its direct interaction with nuclear lamins, supporting the notion that PRR11 might regulate Arp2/3 complex-dependent F-actin polymerization and rearrangement to modulate nuclear lamins. To test this possibility, WT PRR11 was overexpressed in H1299 cells with or without the treatment of the Arp2/3 complex inhibitor CK-666. Remarkably, the number of PRR11-overexpression cells with irregular lamina assembly was completely restored to the level of pvN173 controls upon CK-666 treatment (Fig. 5, e and f), supporting the essential role of Arp2/3 complex activity in the regulation of nuclear laminae by PRR11. We also compared the effect of PRR11 N- and C-terminal truncations Δ1–100 and Δ290–360 in nuclear lamina assembly. Strikingly, the ability of PRR11 to induce disorganization or collapse of nuclear lamina was absolutely abrogated by the Δ1–100 and Δ290–360 mutants, which lack the ability of assembling F-actin cytoskeleton (Fig. 5, g and h). Our data together indicate that PRR11 recruits and activates the Arp2/3 complex to drive F-actin polymerization and rearrangement, thereby regulating the integrity of nuclear laminae. PRR11 serves as a novel cytoplasmic factor involved in the cyto-nucleoskeletal assembly.
PRR11 regulates chromatin organization via Arp2/3 complex
We previously found that overexpression of PRR11 promotes abnormal chromatin remodeling (33). The WT PRR11–expressing cells displayed a highly-abnormal level of chromatin condensation (Fig. S4, a–c). To characterize the reasons for abnormal chromatin remodeling driven by overexpression of PRR11, we first determined whether it depended on the chromatin condensation–related protein. We silenced the chromatin condensation-related gene expression by specific siRNAs and found that abnormal chromatin remodeling caused by PRR11 overexpression was independent of chromatin condensation-related proteins in H1299 and A549 (Fig. S4, d and e). By cleaved caspase 3 staining, we also found the percentage of apoptotic cells was less than the percentage of cells with abnormal chromatin remodeling in the cells that overexpressed PRR11 (Fig. S4, f and g), indicating that PRR11-induced abnormal chromatin remodeling is also independent of apoptosis.
Recent reports have demonstrated that nuclear laminae interact with heterochromatic genomic regions. The dysfunction of nuclear lamina can span large domains of chromatin organization with coordinated chromatin dynamic gene expression, histone modification, and DNA methylation changes (4, 8, 9, 43). As mentioned above, PRR11 is involved in nuclear lamina assembly. We next examined whether PRR11 is also involved in chromatin organization.
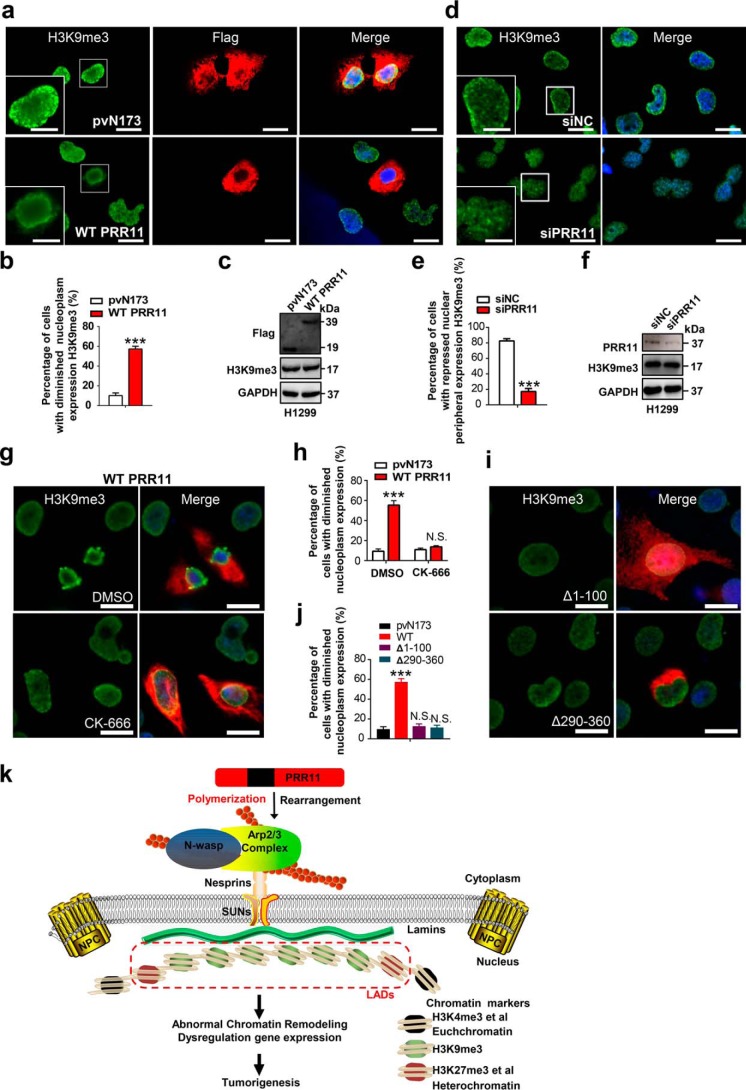
To determine the role of PRR11 in chromatin organization, we examined heterochromatin markers upon PRR11 overexpression and knockdown in lung cancer cells. H3K9me3, a histone modification associated with heterochromatin, contributes to gene regulation by forming large repressive domains on the chromosomes that can be dynamic in mammalian development (4, 44). Immunofluorescence microscopy showed that H3K9me3 was diminished from the nucleoplasm and was enriched at the nuclear periphery in PRR11-positive cells relative to pvN173 controls (Fig. 6, a and b), although the expression of H3K9me3 was not changed (Fig. 6c). However, knockdown of PRR11 also elicited H3K9me3 redistribution to the nucleoplasm but repressed its localization at the nuclear periphery, without affecting H3K9me3 expression level (Fig. 6, d–f). H3K4me3 is often described as an “activating” histone modification and assumed to have an instructive role in the transcription of genes (4, 44). As shown in Fig. S5, a and b, the fluorescence intensity of H3K4me3 significantly decreased in Flag–PRR11-expressing cells. To examine whether PRR11 regulates H3K9me3 localization dependent on nuclear lamins, Lamin B1 was silenced with oligonucleotide siRNA in cells. Western blot analysis verified that the expression of Lamin B1 can be significantly inhibited 48 h post-transfection of Lamin B1 siRNA (Fig. S5c). Similar the PRR11 knockdown, Lamin B1 silencing also showed that H3K9me3 was redistributed, and its nuclear peripheral localization decreased (Fig. S5, d and e). Notably, PRR11 overexpression-induced H3K9me3 enrichment at the nuclear envelope was abrogated in Lamin B1–silenced H1299 cells (Fig. S5, f and g), without changing the expression level of H3K9me3 (Fig. S5h).
Figure 6.
PRR11-mediated cyto-nucleoskeletal assembly regulates heterochromatin organization. a, immunofluorescence staining for H3K9me3 in cells transfected with Flag-tagged pVN173 or WT PRR11. The H3K9me3 localization of WT PRR11–expressing cells was diminished from the nucleoplasm. Representative images are shown. Bar, 20 μm. Zoomed images of boxed region are shown at the bottom-left corner (scale bars, 10 μm). b, quantification of cells with diminished H3K9me3 nucleoplasm distribution as normalized to total Flag-positive cells demonstrated in a. Results are shown as the mean ± S.E. (n = 3). Greater than 50 cells were counted per condition in every repeat. c, Western blot analysis for the indicated proteins. Cells were treated as in a and then were lysed and analyzed for expression of the indicated proteins by Western blotting. d–f, PRR11 silencing altered the localization of H3K9me3. d, immunofluorescence staining for H3K9me3 in cells treated with siNC or siRNA directed against PRR11. Representative images are shown. Bars, 20 μm. Zoomed images of boxed region are shown at the bottom-left corner (scale bars, 10 μm). e, qualification of cells with H3K9me3 nuclear peripheral distribution as normalized to total cells demonstrated in d. Results are shown as the mean ± S.E. (n = 3). Greater than 50 cells were counted per condition in every repeat. f, Western blot analysis for the indicated proteins. Cells were treated as in d and then were lysed and analyzed for expression of the indicated proteins by Western blotting. g and h, CK-666 restored H3K9me3 nucleoplasm distribution in WT PRR11-overexpression cells. g, H1299 cells were transfected with WT PRR11 with or without CK-666 (84 μm) treatment, and then cells were fixed and stained for Flag and H3K9me3. Representative images are shown. Bar, 10 μm. h, quantification of cells with diminished H3K9me3 nucleoplasm distribution as normalized to total Flag-positive cells. Results are shown as the mean ± S.E. (n = 3). Greater than 50 cells were counted per condition in every repeat. i and j, effects of PRR11-truncated mutations on the H3K9me3 redistribution. i, H1299 cells were overexpressed with Flag-tagged Δ1–100 or Δ290–360, and then cells were fixed 24 h after transfection and stained for Flag and H3K9me3. Representative images are shown. Bar, 10 μm. j, quantitative analysis of cells with diminished H3K9me3 nucleoplasm distribution. Ratio indicated that the number of cells with aberrant localization of H3K9me3 is relative to that of total Flag-positive cells. Results are shown as the mean ± S.E. (n = 3). Greater than 20 cells were counted per condition in every repeat. Cell nuclei were stained with DAPI (blue). ***, p < 0.001, or N.S., not significant (p > 0.05). k, proposed working model. PRR11 associates with and recruits Arp2/3 complex to facilitate F-actin assembly and rearrangement via the N and C termini (red regions) and thereby to regulate nuclear lamina integrity, which might be mediated by the LINC complex coupling both nuclear lamina and actin cytoskeleton. Nuclear laminae bind directly to lamina-associated domains and/or heterochromatin-associated proteins, such as H3K9me2/3 and H3K27me3, and this is important for chromatin organization and gene expression repression. Dysfunction of PRR11 disrupts cytoskeleton–nucleoskeleton, chromatin organization, and gene expression to promote lung tumorigenesis.
Next, we asked whether inhibition of Arp2/3 complex–dependent F-actin polymerization and rearrangement can rescue heterochromatin reorganization by treating PRR11-overexpression cells with or without CK-666. The number of PRR11-overexpression cells with dislocated H3K9me3 was restored to the level of pvN173 control cells upon CK-666 treatment, suggesting that PRR11-induced H3K9me3 redistribution depends on Arp2/3 complex activity (Fig. 6, g and h). We further investigated the role of PRR11 truncations Δ1–100 and Δ290–360 in H3K9me3 localization. Notably, the effect on disorganization of H3K9me3 was also abolished in Δ1–100- and Δ290–360–expressing cells (Fig. 6, i and j). Taken together, these data demonstrate that PRR11 regulates chromatin H3K9me3 localization through modulating Arp2/3 complex and nuclear lamins. Overexpression of PRR11 in lung cancer cells led to Arp2/3 recruitment and accumulation, F-actin polymerization, and cytoskeletal rearrangement, thereby disturbing lamina assembly and chromatin organization. Our findings provide a potential mechanism that dysfunction of PRR11 disrupts chromatin organization and gene expression to promote lung tumorigenesis via the Arp2/3 complex/F-actin/nuclear lamins axis (Fig. 6k).
Discussion
PRR11 has been reported to overexpress in lung cancers and demonstrated an important role in cell-cycle progression and tumorigenesis (32, 33). Silencing of PRR11 results in altered expression of various important genes involved in cell cycle, tumorigenesis, and metastasis (32). However, the molecular mechanisms underlying these events remain elusive. In this study, we have found that cytoplasmic PRR11 associates with and recruits Arp2/3 complex, thereby facilitating F-actin polymerization in vivo and in vitro, leading to nuclear lamina instability and chromatin reorganization in lung cancer cells, which is associated with aberrant gene expression and tumorigenesis (Fig. 6k). Our findings have uncovered the molecular mechanisms that cytoplasmic PRR11 regulates Arp2/3 complex–mediated actin polymerization and rearrangement, nuclear integrity, as well as chromatin organization via the cyto-nucleoskeletal assembly.
Actin cytoskeleton controls diverse cellular functions in many physiological and pathological processes, including tumorigenesis (8). Also, actin cytoskeleton is a dynamic structure with continuous polymerization and disassembly. F-actin polymerization pathways are classified as Arp2/3-dependent pathway and -independent pathway, including formins and Ena/VASP, which polymerize F-actin at specific subcellular locations and lead to various cellular responses (17, 20, 21, 45, 46). We know that the dysregulation of the actin cytoskeleton is tightly connected with tumorigenesis, however, the mechanisms by which the Arp2/3 complex is recruited to modulate cytoskeleton assembly in cancer cells remains poorly known. In this paper, we find a novel cytoplasmic protein PRR11 that promotes Arp2 recruitment and accumulation, thereby facilitating cytoskeleton organization. PRR11 overexpression disorders Arp2/3-dependent F-actin polymerization and rearrangement in lung cancer cells. Our data suggest that aberrant expression of PRR11 might participate in tumorigenesis by disturbing the actin cytoskeleton that is involved in diverse cellular processes such as cell division, cellular motility, endocytosis, intracellular nuclear positioning, chromatin dynamics, and gene expression (11, 20). Future investigations into the functions of PRR11–Arp2/3 complex will lead to better understanding of the many biological processes, including tumorigenesis.
Evidence suggests that the association of chromatin with the nuclear periphery could serve as a prime determinant of chromatin modeling. The mechanical integrity of the nuclear periphery is defined by the nuclear laminae where deficiencies in specific components result in nuclear instability, chromatin remodeling, as well as multiple diseases (3, 11). In particular, alterations in the nuclear morphology of tumor cells is the gold standard for cancer diagnosis (4, 5, 24, 47).
Nuclear laminae can directly interact with LADs of the nuclear periphery to regulate chromatin organization and gene expression, as well as DNA replication and repairing (4, 5). LADs are among the most prominent heterochromatin domains in genomes. LADs collectively cover about 30–40% of the genome and are enriched in the histone modifications of H3K9me2, H3K9me3, and H3K27me3. Several thousands of genes are located within LADs, and the majority of these genes are transcriptionally inactive. Loss of nuclear lamins leads to abnormal chromatin organization, which is accompanied by a reduction in the levels or redistribution of heterochromatin marks, because it is believed that LADs may form a repressive chromatin state (5, 48–51). Current studies have demonstrated that alteration of heterochromatin-related proteins, such as H3K9me3, HP1α, or HP1γ, is implicated in lung cancer, breast cancer, and prostate cancer (52–54). In this paper, we find that PRR11 is implicated in the assembly of nuclear lamina, and aberrant PRR11 expression induces nuclear lamina instability and aberrant heterochromatin organization as shown by the disordered H3K9me3 expression via the Arp2/3-mediated cytoskeletal organization in lung cancer cells. We thereby identify that PRR11 serves as a new cytoplasmic factor involved in nuclear laminae and chromatin remodeling via Arp2/3-dependent cyto-nucleoskeletal assembly.
Truncated mutations demonstrate that PRR11 regulates F-actin polymerization and rearrangement dependent on different regions. Both the N and C termini are required for PRR11-mediated F-actin organization. Deletion of the PRR11 N and C termini (Δ1–100 and Δ290–300) completely abolished the effect of PRR11 on F-actin polymerization. In addition, the effects of PRR11 on nuclear lamina assembly and heterochromatin organization are also abrogated by deleting the regions (N and C termini), further demonstrate that the role of PRR11 on nuclear integrity and functions depends on its N and C termini–mediated cytoskeleton assembly.
Interestingly, we show that residues 100–200 are necessary for PRR11 regulating actin cytoskeleton rearrangement, but in a way different from WT PRR11, by forming actin comet tails. It has been shown that actin comet tails are implicated in phagocytosis and endocytosis (18). To date, the mechanism of actin comet tail formation is best understood in Listeria monocytogenes and Shigella flexneri where the surface proteins ActA or IcsA of pathogens recruit N-WASP to stimulate actin nucleation activity of the Arp2/3 complex and form actin comet tails (19). Wiskott-Aldrich syndrome protein family members, the nucleation promoting factors of Arp2/3 complex, direct the Arp2/3 complex to induce actin comet tails (18). However, other proteins inducing actin comet tails are still rarely reported in mammal cells. One of the important findings in our work is not only that PRR11 regulates actin polymerization and rearrangement in lung cancer cells but also deletion of residues 100–200 powerfully promote actin comet tails.
Our data collectively demonstrate for the first time that cytoplasmic PRR11 associates with and recruits Arp2/3 complex to stimulate F-actin polymerization and cytoskeleton rearrangement, consequently influencing nuclear lamina integrity and chromatin organization in lung cancer cells. We thereby uncover a potential novel mechanism driven by the PRR11-Arp2/3 complex-nuclear lamina axis for lung cancer progression, diagnosis, and treatment.
Materials and methods
Cell culture
Human non-small cell lung carcinoma–derived H1299 and A549 cells were obtained from the ATCC. Cells were cultured in RPMI 1640 medium and Dulbecco's modified Eagle's medium (DMEM), respectively, supplemented with 10% heat-inactivated fetal bovine serum (Gibco), and penicillin (100 IU/ml)/streptomycin (100 mg/ml). Cells were maintained at 37 °C in a water-saturated atmosphere of 5% CO2 in air. For the detection of mycoplasma in the cell cultures, we used the Mycoplasma stain kit (Mpbio). CK-689, CK-666, and SMIFH2 were purchased from Sigma-Aldrich.
Construction of expression plasmids and transient transfection
The entire or the truncated coding sequence of human PRR11 was inserted into the mammalian expression plasmid pcDNA3.0 along with an N-terminal Flag-tag. After sequencing to confirm the accuracy of the resulting sequences, these constructs were designated as WT PRR11 (pcDNA–PRR11), Δ33–41 (deletion of residues 33–41), Δ1–100 (deletion of residues 1–100), Δ185–200 (deletion of residues 185–200), Δ100–184 (deletion of residues 100–184), Δ290–360 (deletion of residues 290–360), Δ100–200 (deletion of residues 100–200), Δ200–300 (deletion of residues 200–300), and Δ1–110/200–360 (deletion of residues 1–100/200–360). (The primers used for construction of expression plasmids are listed in Table S1). For fusion protein expression, PRR11 was cloned in pET-GST vector. pvN173 (Flag-tagged N-terminal 173 residues of enhanced GFP) was obtained from Dr. Cheng Lu, State Key Laboratory of Silkworm Genome Biology, Southwest University, China.
For transient transfection, cells were seeded at a density of 0.8 × 105 cells/well in a 24-well tissue culture plate or 2.5 × 105 cells/well in a 6-well tissue culture plate and incubated overnight. Cells were then transiently transfected with the indicated plasmids using Lipofectamine 2000 transfection reagent (Invitrogen) following the manufacturer's protocols.
Immunoprecipitation and MS
For immunoprecipitation assays, cells were lysed with 500 μl of ice-cold lysis buffer (20 mm Tris, pH 7.5; 150 mm NaCl; 1% Triton X-100; protease inhibitors (10 μg/ml aprotinin, 1 × 10−3 mol/liter phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin), and phosphatase inhibitors (50 μmol/liter sodium fluoride, 1 μmol/liter sodium orthovanadate, 10 μmol/liter sodium pyrophosphate)). Then cellular extracts were incubated with the appropriate primary antibodies at 4 °C overnight, followed by addition of protein A/G–agarose beads for 2 h at 4 °C. Beads were then washed, and the immune complexes were subjected to SDS-PAGE followed by immunoblotting with secondary antibodies. Fractions of the bead volume were collected and resolved on SDS-PAGE with subsequent silver staining. To identify proteins associated with Flag–PRR11, LC-MS/MS analysis was performed using a Thermo Finnigan LTQ linear ion trap mass spectrometer in line with a Thermo Finnigan Surveyor MS Pump Plus HPLC system. Consequently, MS data were analyzed using SEQUEST (version 28) against NCBI human protein database (December 14, 2011 download, 33,256 entries). At least three distinct peptides were used to identify each protein.
Indirect immunofluorescence staining
Cells were fixed and incubated with primary antibodies, followed by incubation with Alexa 488/594–conjugated secondary antibodies (Table S2). Cells were then mounted with medium containing 4′,6-diamidino-2-phenylindole (DAPI; Sigma), and the preparations were visualized with a Leica fluorescence microscope and a Zeiss confocal LSM 768 microscope. Pixel quantification was done using ImageJ software. The antibodies used for immunofluorescence assays are listed in Table S2.
Immunoblotting analysis
Cells were lysed in RIPA lysis buffer (50 mm Tris-HCl, pH 7.4; 150 mm NaCl; 0.1% SDS; 1% Nonidet P-40; 0.5% deoxycholate; Santa Cruz Biotechnology) supplemented with protease inhibitor mixture (Roche Applied Science). For nuclear and cytoplasmic separation, the cells were incubated with lysis buffer on ice for 5 min. The samples were spun down at 4 °C for 15 min at full speed, and the supernatants were collected as cytoplasmic fractions. The pellets were washed with lysis buffer twice and sonicated in the lysis buffer to obtain the nuclear fractions. The protein concentration of each lysate was determined by BCA reagent (Applygen Technologies, Inc.). Equal amounts of the lysates (30 μg of protein) were denatured at 100 °C for 5 min, separated by 10% standard SDS-PAGE, and electro-transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline (TBS) containing 0.1% Tween 20 at 4 °C overnight. After blocking, the membranes were then probed with the indicated primary antibodies at room temperature for 1 h, followed by incubation with the corresponding horseradish peroxidase–conjugated secondary antibodies at room temperature for 1 h. The proteins were finally visualized by enhanced chemiluminescence (Amersham Biosciences). The antibodies used in this study are listed in Table S2.
Actin polymerization assay
Protein purification
GST–PRR11 was expressed in Escherichia coli strain BL21-CodonPlus (DE3)-RP and purified using GSH-Sepharose 4B (GE Healthcare). GST–N-WASP (ab132277) was purchased from Abcam.
Pyrene–actin assay
The Actin Polymerization BiochemTM kit and Arp2/3 protein complex were purchased from Cytoskeleton. Pyrene–actin assays were performed as described previously (39). The kinetics of actin polymerization was monitored by pyrene fluorescence with excitation at 365 nm and emission at 407 nm. Polymerization reaction mixtures contained 2 μm G-actin, 20% μm pyrene-labeled actin, 0.2 μm ATP, and various proteins in 80 μl of X buffer (10 mm HEPES, pH 7.0; 100 mm KCl; 1 mm MgCl2; 0.1 mm EDTA; 1 mm DTT) and were preincubated for 5 min. GST–N-WASP and GST–PRR11 were added at final concentrations of 25 and 0–3 μm, respectively. The reaction was initiated by adding a mixture of actin and pyrene-labeled actin to the preincubated protein mixtures.
siRNA-mediated knockdown
The nucleotide sequences of control siRNA and specific against PRR11, Lamin B1, and ARPC1 siRNAs were described previously or were designed by Online tools (Invitrogen) (Table S1) (32, 33, 39). Prior to transfection, cells were seeded at a density of 5 × 104 cells/well in a 24-well tissue culture plate or 2 × 105 cells/well in a 6-well tissue culture plate and allowed to attach overnight. The indicated siRNAs were then transiently transfected into cells using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions.
Quantitative RT-PCR
The expression level of genes was detected as described (55). The primers used for quantitative PCRs are listed in Table S1.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (RRID:SCR_002798). Data were expressed as the means ± S.E. of the values from the independent experiments performed, as indicated in the corresponding figure legends. The numbers of biological replicates, and what they represent, are indicated in each figure legend. Two-tailed Student's t tests were used for single comparison, and two-way analysis of variance was used for multiple comparisons. In all figures, statistical significances between the indicated sample and control or between marked pairs are designated: *, p < 0.05; **, p < 0.01; ***, p < 0.001, or N.S. (p > 0.05, refers to no significant difference).
Author contributions
L. Z. and C. Z. data curation; L. Z., Y. Z., Y. Lei, Z. W., and Y. Li. investigation; L. Z. writing-original draft; Y. W., Y. B., and C. Z. funding acquisition; C. Z. visualization; C. Z. methodology; C. Z. writing-review and editing.
Supplementary Material
Acknowledgment
We are grateful to Dr. Guoxiang Jin for helpful discussions.
This work was supported in part by National Natural Science Foundation of China Grants-in-aid 81501979 (to C. Z.) and 81672301 and 81001097 (to Y. B.), Chongqing Natural Science Foundation Grant cstc2017jcyjAX0464 (to C. Z.), and Scientific and Technological Research Projects of Chongqing Education Commission Grant KJQN201800423 (to C. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Tables S1 and S2.
- LAD
- lamina-associated domain
- NPC
- nuclear pore complex
- DAPI
- 4′,6-diamidino-2-phenylindole
- siNC
- negative control siRNA
- TRITC
- tetramethylrhodamine isothiocyanate
- WASP
- Wiskott-Aldrich syndrome protein.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2018) Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2. Morgan M. A., and Shilatifard A. (2015) Chromatin signatures of cancer. Genes Dev. 29, 238–249 10.1101/gad.255182.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Flavahan W. A., Gaskell E., and Bernstein B. E. (2017) Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 10.1126/science.aal2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Steensel B., and Belmont A. S. (2017) Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dechat T., Adam S. A., Taimen P., Shimi T., and Goldman R. D. (2010) Nuclear lamins. Cold Spring Harb. Perspect. Biol. 2, a000547 10.1101/cshperspect.a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simon D. N., and Wilson K. L. (2011) The nucleoskeleton as a genome-associated dynamic 'network of networks'. Nat. Rev. Mol. Cell Biol. 12, 695–708 10.1038/nrm3207 [DOI] [PubMed] [Google Scholar]
- 7. Burke B. (2019) Chain reaction: LINC complexes and nuclear positioning. F1000Res. 8, F1000 Faculty Rev–136 10.12688/f1000research.16877.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kanellos G., Zhou J., Patel H., Ridgway R. A., Huels D., Gurniak C. B., Sandilands E., Carragher N. O., Sansom O. J., Witke W., Brunton V. G., and Frame M. C. (2015) ADF and cofilin1 control actin stress fibers, nuclear integrity, and cell survival. Cell Rep. 13, 1949–1964 10.1016/j.celrep.2015.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spagnol S. T., and Dahl K. N. (2014) Active cytoskeletal force and chromatin condensation independently modulate intranuclear network fluctuations. Integr. Biol. 6, 523–531 10.1039/C3IB40226F [DOI] [PubMed] [Google Scholar]
- 10. Almonacid M., Terret M. E., and Verlhac M. H. (2019) Nuclear positioning as an integrator of cell fate. Curr. Opin. Cell Biol. 56, 122–129 10.1016/j.ceb.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 11. Ramdas N. M., and Shivashankar G. V. (2015) Cytoskeletal control of nuclear morphology and chromatin organization. J. Mol. Biol. 427, 695–706 10.1016/j.jmb.2014.09.008 [DOI] [PubMed] [Google Scholar]
- 12. Rottner K., and Schaks M. (2019) Assembling actin filaments for protrusion. Curr. Opin. Cell Biol. 56, 53–63 10.1016/j.ceb.2018.09.004 [DOI] [PubMed] [Google Scholar]
- 13. Rottner K., Faix J., Bogdan S., Linder S., and Kerkhoff E. (2017) Actin assembly mechanisms at a glance. J. Cell Sci. 130, 3427–3435 10.1242/jcs.206433 [DOI] [PubMed] [Google Scholar]
- 14. Dimchev G., Steffen A., Kage F., Dimchev V., Pernier J., Carlier M. F., and Rottner K. (2017) Efficiency of lamellipodia protrusion is determined by the extent of cytosolic actin assembly. Mol. Biol. Cell 28, 1311–1325 10.1091/mbc.e16-05-0334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rottner K., and Stradal T. E. (2016) How distinct Arp2/3 complex variants regulate actin filament assembly. Nat. Cell Biol. 18, 1–3 10.1038/ncb3293 [DOI] [PubMed] [Google Scholar]
- 16. Mehedi M., McCarty T., Martin S. E., Le Nouën C., Buehler E., Chen Y. C., Smelkinson M., Ganesan S., Fischer E. R., Brock L. G., Liang B., Munir S., Collins P. L., and Buchholz U. J. (2016) Actin-related protein 2 (ARP2) and virus-induced filopodia facilitate human respiratory syncytial virus spread. PLoS Pathog. 12, e1006062 10.1371/journal.ppat.1006062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blanchoin L., Pollard T. D., and Mullins R. D. (2000) Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10, 1273–1282 10.1016/S0960-9822(00)00749-1 [DOI] [PubMed] [Google Scholar]
- 18. Kast D. J., Zajac A. L., Holzbaur E. L., Ostap E. M., and Dominguez R. (2015) WHAMM directs the Arp2/3 complex to the ER for autophagosome biogenesis through an actin comet tail mechanism. Curr. Biol. 25, 1791–1797 10.1016/j.cub.2015.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fehrenbacher K., Huckaba T., Yang H. C., Boldogh I., and Pon L. (2003) Actin comet tails, endosomes and endosymbionts. J. Exp. Biol. 206, 1977–1984 10.1242/jeb.00240 [DOI] [PubMed] [Google Scholar]
- 20. Davidson A. J., and Wood W. (2016) Unravelling the actin cytoskeleton: a new competitive edge? Trends Cell Biol. 26, 569–576 10.1016/j.tcb.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rotty J. D., Wu C., Haynes E. M., Suarez C., Winkelman J. D., Johnson H. E., Haugh J. M., Kovar D. R., and Bear J. E. (2015) Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev. Cell 32, 54–67 10.1016/j.devcel.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suarez C., Carroll R. T., Burke T. A., Christensen J. R., Bestul A. J., Sees J. A., James M. L., Sirotkin V., and Kovar D. R. (2015) Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev. Cell 32, 43–53 10.1016/j.devcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pernier J., Shekhar S., Jegou A., Guichard B., and Carlier M. F. (2016) Profilin interaction with actin filament barbed end controls dynamic instability, capping, branching, and motility. Dev. Cell 36, 201–214 10.1016/j.devcel.2015.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Isermann P., and Lammerding J. (2013) Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 23, R1113–R1121 10.1016/j.cub.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burke B., and Stewart C. L. (2014) Functional architecture of the cell's nucleus in development, aging, and disease. Curr. Top. Dev. Biol. 109, 1–52 10.1016/B978-0-12-397920-9.00006-8 [DOI] [PubMed] [Google Scholar]
- 26. Dobrzynska A., Gonzalo S., Shanahan C., and Askjaer P. (2016) The nuclear lamina in health and disease. Nucleus 7, 233–248 10.1080/19491034.2016.1183848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimi T., and Goldman R. D. (2014) Nuclear lamins and oxidative stress in cell proliferation and longevity. Adv. Exp. Med. Biol. 773, 415–430 10.1007/978-1-4899-8032-8_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D. K., Solimando L., and Goldman R. D. (2008) Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832–853 10.1101/gad.1652708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Poleshko A., Mansfield K. M., Burlingame C. C., Andrake M. D., Shah N. R., and Katz R. A. (2013) The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 5, 292–301 10.1016/j.celrep.2013.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Solovei I., Wang A. S., Thanisch K., Schmidt C. S., Krebs S., Zwerger M., Cohen T. V., Devys D., Foisner R., Peichl L., Herrmann H., Blum H., Engelkamp D., Stewart C. L., Leonhardt H., and Joffe B. (2013) LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598 10.1016/j.cell.2013.01.009 [DOI] [PubMed] [Google Scholar]
- 31. Andrés V., and González J. M. (2009) Role of A-type lamins in signaling, transcription, and chromatin organization. J. Cell Biol. 187, 945–957 10.1083/jcb.200904124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ji Y., Xie M., Lan H., Zhang Y., Long Y., Weng H., Li D., Cai W., Zhu H., Niu Y., Yang Z., Zhang C., Song F., and Bu Y. (2013) PRR11 is a novel gene implicated in cell cycle progression and lung cancer. Int. J. Biochem. Cell Biol. 45, 645–656 10.1016/j.biocel.2012.12.002 [DOI] [PubMed] [Google Scholar]
- 33. Zhang C., Zhang Y., Li Y., Zhu H., Wang Y., Cai W., Zhu J., Ozaki T., and Bu Y. (2015) PRR11 regulates late-S to G2/M phase progression and induces premature chromatin condensation (PCC). Biochem. Biophys. Res. Commun. 458, 501–508 10.1016/j.bbrc.2015.01.139 [DOI] [PubMed] [Google Scholar]
- 34. Wang Y., Zhang C., Mai L., Niu Y., Wang Y., and Bu Y. (2019) PRR11 and SKA2 gene pair is overexpressed and regulated by p53 in breast cancer. BMB Rep. 52, 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Y., Cha Z., Fang W., Qian B., Yu W., Li W., Yu G., and Gao Y. (2015) The prognostic potential and oncogenic effects of PRR11 expression in hilar cholangiocarcinoma. Oncotarget. 6, 20419–20433 10.18632/oncotarget.3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song Z., Liu W., Xiao Y., Zhang M., Luo Y., Yuan W., Xu Y., Yu G., and Hu Y. (2015) PRR11 is a prognostic marker and potential oncogene in patients with gastric cancer. PLoS ONE 10, e0128943 10.1371/journal.pone.0128943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y., Zhang Y., Zhang C., Weng H., Li Y., Cai W., Xie M., Long Y., Ai Q., Liu Z., Du G., Wang S., Niu Y., Song F., Ozaki T., and Bu Y. (2015) The gene pair PRR11 and SKA2 shares a NF-Y-regulated bidirectional promoter and contributes to lung cancer development. Biochim. Biophys. Acta 1849, 1133–1144 10.1016/j.bbagrm.2015.07.002 [DOI] [PubMed] [Google Scholar]
- 38. Rana M. K., Aloisio F. M., Choi C., and Barber D. L. (2018) Formin-dependent TGF-β signaling for epithelial to mesenchymal transition. Mol. Biol. Cell 29, 1465–1475 10.1091/mbc.E17-05-0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J., Zhao Y., Sun Y., He B., Yang C., Svitkina T., Goldman Y. E., and Guo W. (2012) Exo70 stimulates the Arp2/3 complex for lamellipodia formation and directional cell migration. Curr. Biol. 22, 1510–1515 10.1016/j.cub.2012.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warren D. T., Zhang Q., Weissberg P. L., and Shanahan C. M. (2005) Nesprins: intracellular scaffolds that maintain cell architecture and coordinate cell function? Expert Rev. Mol. Med. 7, 1–15 [DOI] [PubMed] [Google Scholar]
- 41. Roux K. J., Crisp M. L., Liu Q., Kim D., Kozlov S., Stewart C. L., and Burke B. (2009) Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. U.S.A. 106, 2194–2199 10.1073/pnas.0808602106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilhelmsen K., Litjens S. H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., and Sonnenberg A. (2005) Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171, 799–810 10.1083/jcb.200506083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toh K. C., Ramdas N. M., and Shivashankar G. V. (2015) Actin cytoskeleton differentially alters the dynamics of lamin A, HP1alpha and H2B core histone proteins to remodel chromatin condensation state in living cells. Integr. Biol. 7, 1309–1317 10.1039/C5IB00027K [DOI] [PubMed] [Google Scholar]
- 44. Du Q., Bert S. A., Armstrong N. J., Caldon C. E., Song J. Z., Nair S. S., Gould C. M., Luu P. L., Peters T., Khoury A., Qu W., Zotenko E., Stirzaker C., and Clark S. J. (2019) Replication timing and epigenome remodelling are associated with the nature of chromosomal rearrangements in cancer. Nat. Commun. 10, 416 10.1038/s41467-019-08302-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holt M. R., and Koffer A. (2001) Cell motility: proline-rich proteins promote protrusions. Trends Cell Biol. 11, 38–46 10.1016/S0962-8924(00)01876-6 [DOI] [PubMed] [Google Scholar]
- 46. Walders-Harbeck B., Khaitlina S. Y., Hinssen H., Jockusch B. M., and Illenberger S. (2002) The vasodilator-stimulated phosphoprotein promotes actin polymerisation through direct binding to monomeric actin. FEBS Lett. 529, 275–280 10.1016/S0014-5793(02)03356-2 [DOI] [PubMed] [Google Scholar]
- 47. Zink D., Fischer A. H., and Nickerson J. A. (2004) Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677–687 10.1038/nrc1430 [DOI] [PubMed] [Google Scholar]
- 48. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M. B., Talhout W., Eussen B. H., de Klein A., Wessels L., de Laat W., and van Steensel B. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- 49. Goldman R. D., Shumaker D. K., Erdos M. R., Eriksson M., Goldman A. E., Gordon L. B., Gruenbaum Y., Khuon S., Mendez M., Varga R., and Collins F. S. (2004) Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U.S.A. 101, 8963–8968 10.1073/pnas.0402943101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Camps J., Erdos M. R., and Ried T. (2015) The role of Lamin B1 for the maintenance of nuclear structure and function. Nucleus 6, 8–14 10.1080/19491034.2014.1003510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leemans C., van der Zwalm M. C. H., Brueckner L., Comoglio F., van Schaik T., Pagie L., van Arensbergen J., and van Steensel B. (2019) Promoter-intrinsic and local chromatin features determine gene repression in LADs. Cell 177, 852–864.e14 10.1016/j.cell.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ci X., Hao J., Dong X., Choi S. Y., Xue H., Wu R., Qu S., Gout P. W., Zhang F., Haegert A. M., Fazli L., Crea F., Ong C. J., Zoubeidi A., He H. H., et al. (2018) Heterochromatin protein 1α mediates development and aggressiveness of neuroendocrine prostate cancer. Cancer Res. 78, 2691–2704 10.1158/0008-5472.CAN-17-3677 [DOI] [PubMed] [Google Scholar]
- 53. Alam H., Li N., Dhar S. S., Wu S. J., Lv J., Chen K., Flores E. R., Baseler L., and Lee M. G. (2018) HP1γ promotes lung adenocarcinoma by downregulating the transcription-repressive regulators NCOR2 and ZBTB7A. Cancer Res. 78, 3834–3848 10.1158/0008-5472.CAN-17-3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li Q. L., Lei P. J., Zhao Q. Y., Li L., Wei G., and Wu M. (2017) Epigenomic analysis in a cell-based model reveals the roles of H3K9me3 in breast cancer transformation. Epigenomics 9, 1077–1092 10.2217/epi-2016-0183 [DOI] [PubMed] [Google Scholar]
- 55. Bu Y., Suenaga Y., Okoshi R., Sang M., Kubo N., Song F., Nakagawara A., and Ozaki T. (2010) NFBD1/MDC1 participates in the regulation of G2/M transition in mammalian cells. Biochem. Biophys. Res. Commun. 397, 157–162 10.1016/j.bbrc.2010.05.063 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.