Abstract
Background
The 21‐gene recurrence score (RS) assay has been recommended by major guidelines for treatment decision in hormone receptor (HR)‐positive early breast cancer (EBC). However, the genomic assay is not accessible and affordable worldwide. Alternatively, an increasing number of studies have shown that traditional immunohistochemistry (IHC) can partially or even completely replace the role of the 21‐gene genomic assay. Here, we developed and validated a predictive model (IHC3 model) combining the Ki‐67 index, progesterone receptor (PR) expression, histologic grade, and tumor size to predict the recurrence risk of HR‐positive EBC.
Methods
The data from 389 patients (development set) with HR‐positive, human epidermal growth factor receptor 2‐negative, lymph node non‐metastasized invasive breast cancer were used to construct the IHC3 model based on the Surexam® 21‐gene RS and the TAILORx clinical trial criteria. An additional 146 patients with the same characteristics constituted the validation set. The predictive accuracy of the IHC3 model was compared with that of Orucevic et al.’s nomogram. Invasive disease‐free survival (IDFS) was analyzed in the IHC3 predictive low‐recurrence risk (pLR) group and the predictive high‐recurrence risk (pHR) group. The Pearson chi‐square test, Fisher exact test, and log‐rank test were used for analysis.
Results
The pLR and pHR group could be easily stratified using the decision tree model without network dependence. The accuracies of the IHC3 model were 86.1% in the development set and 87.7% in the validation set. The predictive accuracy of the IHC3 model and Orucevic et al.’s nomogram for the whole cohort was 86.5% and 86.9%, respectively. After a 52‐month of median follow‐up, a significant difference was found in IDFS between of the IHC3 pLR and the pHR groups (P = 0.001) but not in the IDFS between the low‐ and high‐recurrence risk groups according to the Surexam® 21‐gene RS and the TAILORx clinical trial criteria (P = 0.556) or 21‐gene binary RS group (P = 0.511).
Conclusions
The proposed IHC3 model could reliably predict low and high recurrence risks in most HR‐positive EBC patients. This easy‐to‐use predictive model may be a reliable replacement for the 21‐gene genomic assay in patients with EBC who have no access to or cannot afford the 21‐gene genomic assay.
Keywords: 21‐gene genomic assay, breast cancer, recurrence score, histologic grade, progesterone receptor, Ki‐67
Abbreviations
- ALND
axillary lymph node dissection
- CART
classification and regression trees
- EBC
early breast cancer
- ER
estrogen receptor
- FISH
fluorescence in situ hybridization
- HER2
human epidermal growth factor receptor 2
- HR
hormone receptor
- IDC
invasive ductal carcinoma
- IDFS
invasive disease‐free survival
- IHC
immunohistochemistry
- ILC
invasive lobular carcinoma
- NPV
negative predictive value
- pHR
predictive high‐recurrence risk
- pLR
predictive low‐recurrence risk
- PPV
positive predictive value
- PR
progesterone receptor
- RS
recurrence score
- SERM
selective estrogen receptor modulator
- SLNB
sentinel lymph node biopsy
1. BACKGROUND
Breast cancer is the most common cancer diagnosed in women worldwide [1]. In 2018, approximately 2,088,849 women were newly diagnosed with breast cancer, representing a cumulative global lifetime risk of 5.03% among those aged 0‐74 years old [2]. Breast cancer‐related mortality rates vary from 11.3 per 100,000 to 30 per 100,000 people among different regions [3]. Importantly, for early breast cancers (EBCs), as recurrences may lead to increased mortality, therefore, the patients will have to receive additional treatments to reduce their risk of recurrence; leading to an increase in their health care expenses.
With increasing awareness of the importance of breast cancer screening, more breast cancers are detected at their early stage, when they are still small and have not yet metastasized to the lymph nodes. More than half of EBCs are hormone receptor (HR)‐positive [4]. The main challenge is selecting patients with non‐metastasized lymph nodes, HR‐positive, and human epidermal growth factor receptor 2 (HER2)‐negative breast cancer who would benefit from adjuvant chemotherapy.
Multigene assays provide prognostic and predictive information beyond histological parameters in EBC. Currently, the Oncotype DX 21‐gene recurrence score (RS) assay (Genomic Health, Redwood City, CA, USA) is the most widely used genomic assay for HR‐positive cancers in the United States and is recommended by the National Comprehensive Cancer Network (NCCN) [5], European Society for Medical Oncology [6], and St. Gallen Consensus [7]. A recent meta‐analysis showed that the prognostic and predictive value of the 21‐gene genomic assay were consistent for patients with non‐metastasized lymph nodes, HR‐positive disease in different countries, despite different local treatment guidelines [8].
According to new results from the prospective TAILORx clinical study published in 2018 [9], patients with a low 21‐gene RS of 0‐25 had no benefit from chemotherapy, except for women aged ≤50 years old with an RS of 16‐25. Subsequently, the RS was categorized as a binary variable (low RS, 0‐25; high RS, 26‐100), and adjuvant chemotherapy was recommended for patients with a high RS.
Nevertheless, the 21‐gene genomic assay is not available and affordable worldwide, particularly in developing countries. In the past 10 years, several multivariable models have been integrated into traditional pathological information including tumor size, tumor histologic grade, the expression of estrogen receptor (ER), progesterone receptor (PR), and HER2, and Ki‐67 index to predict tumor recurrence and replace the RS [10, 11, 12, 13, 14]. Most recurrence predictive models are mathematical equations (e.g., Cuzick et al.’s IHC4 score [10] and Turner et al.’s Magee [11] models) that are available online. Given the increasing incidence of EBC in developing countries, there is a need for a simple and accurate model that could perform similar to and thus replace the expensive 21‐gene genomic assay.
Findings from our previous study [15] showed that tumor histologic grade, PR expression, and Ki‐67 index were significantly different between the three traditional 21‐gene RS risk cohorts (low risk: RS 0‐17, intermediate risk: RS 18‐30, and high risk: RS 31‐100). In this study, we aimed to develop an easy‐to‐use predictive model to classify patients into a high‐ or low‐recurrence risk categories and help the decision making of adjuvant chemotherapy, similar to that by the 21‐gene genomic assay. Towards this goal, we investigated the combination of the above‐mentioned three immunohistochemical (IHC) variables.
2. PATIENTS AND METHODS
2.1. Patient selection and study design
This study included consecutive patients with HR‐positive, HER2‐negative, axillary non‐metastasized lymph nodes invasive breast cancer tested with the domestic Surexam® 21‐gene RS assay (Surexam®, Patent number: CN201010261745.5, Guangzhou, Guangdong, China) and treated in the Breast Surgery Department of Peking Union Medical College Hospital (PUMCH, Beijing, China) between May 2012 and January 2017. The institutional review board of PUMCH approved this study and the need for informed consent was waived by the committee because of the retrospective nature of the study and the use of anonymized data.
The inclusion criteria were 1) primary invasive breast cancer without neoadjuvant chemotherapy and/or endocrine therapy, 2) underwent breast cancer surgery, and 3) no contraindication to receiving systemic chemotherapy and endocrine therapy. Patients were excluded from this study if they had synchronous or metachronous bilateral breast cancer in the past 10 years.
2.2. Data collection
Data regarding the patients’ age, tumor size, histologic grade (Nottingham Combined Histology Grade, based on an assessment of tubule/gland formation, nuclear pleomorphism, and mitotic count), ER status (by IHC staining, ≥1% positive), PR status (by IHC staining, ≥1% positive) [16], HER2/neu status (IHC staining score [0, 1+, 2+, 3+] and the fluorescence in situ hybridization (FISH) ratio when IHC staining score was 2+) [17], Ki‐67 index, Surexam® 21‐gene RS, surgical procedure, and adjuvant treatment (including chemotherapy, radiotherapy, and endocrine therapy) were recorded. The pathological specimens were reviewed by two senior pathologists at the qualified general pathology laboratory of PUMCH. The chemotherapy regimens mainly included the pirarubicin plus cyclophosphamide (AC) regimen (pirarubicin [Main Luck Pharmaceuticals Inc., Shenzhen, China]: intravenously (IV) prescribed at 45 mg/m2 on day 1, cyclophosphamide [SL Pharmaceutical Co., Ltd., Beijing, China]: 600 mg/m2 IV day 1), pirarubicin plus capecitabine (AX) regimen (pirarubicin: 45 mg/m2 IV day 1, capecitabine [Roche Pharmaceuticals Ltd., Shanghai, China]: 1000 mg/ m2 PO days 1‐14) [18], and docetaxel plus cyclophosphamide (TC) regimens (docetaxel [Sanofi‐Aventis Deutschland GmbH, Frankfurt, Germany]: 75 mg/m2 IV day 1, cyclophosphamide: 600 mg/m2 IV day 1). All the regimens were given every 21 days for 4 cycles. Follow‐up information was obtained from the review system of the outpatient clinic or medical charts at 6, 12, 18, and 24 months postoperatively and then annually. Follow‐up was censored on November 30, 2019.
Invasive disease‐free survival (IDFS) was defined as the time between breast cancer surgery and local/regional invasive recurrence, distant recurrence, invasive contralateral breast cancer, second primary invasive cancer (non‐breast), or death; corresponding to the standardized definitions for efficacy endpoints of IDFS [19].
Tumor size and histologic grade were combined into a new variable, named the clinical risk, similar to the TAILORx clinical trial [20]. The clinical risk was referred to as low if the tumor diameter was ≤3 cm with a low histologic grade, ≤2 cm with an intermediate grade, or ≤1 cm with a high grade. If the low‐risk criteria were not met, the clinical risk was referred to as high.
After removing samples with missing data, data from 389 patients (between May 2012 and December 2015; the development set) were used to develop the current predictive IHC3 model, and an additional 146 patients (between January 2016 and January 2017; the validation set) were used to validate the model. The patients were assigned into recurrence risk categories based on the Surexam® 21‐gene RS and the TAILORx clinical trial criteria [20] (modeling reference standard/gold standard for IHC3 model). Recurrence risk was defined as high if patients were >50 years of age with an RS >25, ≤50 years of age with an RS of >20, or ≤50 years old with an RS of 16‐20 and had a high clinical risk. The recurrence risk was defined as low if the high‐recurrence risk criteria were not met. This study adopted the same age cutoff value as the TAILORx trial. Also, considering the average menopause age of Chinese women, 50 years of age was considered as a proper reference to distinguish between premenopausal and postmenopausal patients.
2.3. IHC3 model construction
The IHC3 model was constructed based on the classification and regression trees (CART) (R package “rpart”, Terry Therneau and Beth Atkinson, 2018). It selected the features from candidate IHC predictors, determined the nodes of the decision tree, and classified samples into risk groups. Candidate IHC predictors included 4 categorical variables (i.e., clinical risk, age, tumor size, and HER2 expression) and 2 numeric variables (i.e., Ki‐67 index and PR expression). For the 2 numeric variables, the best cutoff value was determined using CART, which was also used for the split point of a node when performing classification. The predicted variables were the risk categories according to the Surexam® 21‐gene RS and the TAILORx prospective trial criteria [9, 20].
Given that the data were class imbalanced (low: high = 9: 2) and that we prioritized the classification of high‐risk cases correctly, we imposed higher penalizations on high‐to‐low misclassifications. Loss matrix as follows was assigned to CART.
Losses for the high‐to‐low misclassifications were twice as much as those of the low‐to‐high misclassifications. This aimed to overcome the class imbalance problem and increase the accuracy of high recurrence risk classification. The complexity parameter was set to 0.03 while trimming the original decision tree.
Subsequently, to verify that the IHC features selected by CART were stable, that is, the feature selection was not dependent on the samples, we performed 100 random samplings by selecting 75% of samples from the development set with replacement and repeated the decision tree modeling procedure based on these sub‐samples. The times of appearance and importance score of each variable in the 100 decision trees were recorded. The importance score of CART was defined by the R package “rpart,” which used the goodness of split as a measurement. Clinical risk, PR expression, and Ki‐67 index were selected more frequently than other variables, and their importance scores ranked top. We also applied the “varSelRF” package (Diaz‐Uriarte, 2007) in R and calculated the importance score of variables using the mean decrease in accuracy as a measurement. A similar importance rank to “rpart” was obtained, Ki‐67 index (0.0647), PR expression (0.0190), clinical risk (0.0150), age (0.0117), and HER2 expression (0.0029), from highest to lowest.
2.4. IHC3 model validation
The overall accuracy of the IHC3 model was calculated as follows:
where Nlow correct and Nhigh correct represent the number of patients correctly assigned with low‐ and high‐ recurrence risk, respectively, and Ntotal is the total number of patients.
The final IHC3 model required the following three IHC variables: clinical risk, PR expression (express as a percentage), and Ki‐67 index. The cutoff value of each variable was evaluated using CART. Patients were stratified into two risk groups based on the IHC3 model (Figure 1). The predictive low‐recurrence risk (pLR) group included patients (1) with a Ki‐67 index <27.5%, or (2) with a Ki‐67 index ≥27.5%, PR expression ≥55%, and low clinical risk. The predictive high‐recurrence risk (pHR) group included patients (1) with a Ki‐67 index ≥27.5% and PR expression <55%, or (2) with a Ki‐67 index ≥27.5%, PR expression ≥55%, and high clinical risk.
FIGURE 1.
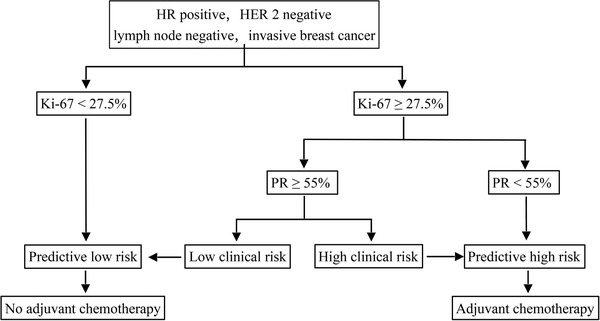
The decision tree for IHC3 model. Clinical risk was defined as low if the tumor diameter was 3 cm or smaller with a low histologic grade, 2 cm or smaller with an intermediate grade, or 1 cm or smaller with a high grade. If the low‐risk criteria were not met, the clinical risk was defined as high.
Abbreviations: IHC immunohistochemistry; HR hormone receptor; HER2 human epidermal growth factor receptor 2; PR progesterone receptor
A recent nomogram published by Orucevic et al. in 2019 [14] was used to predict a high‐risk RS (range, 26‐100) in our study. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the IHC3 model and Orucevic et al.’s nomogram were compared based on the Surexam® 21‐gene RS and the TAILORx clinical trial criteria (gold standard for IHC3 model), and 21‐gene binary RS group (low risk group: RS 0‐25; high risk group: RS 26‐100) (gold standard for Orucevic et al.’s nomogram). The IDFS rates were analyzed and compared between the low‐ and high‐recurrence risk groups according to the IHC3 model, the Surexam® 21‐gene RS and the TAILORx clinical trial criteria (gold standard for IHC3 model), and the 21‐gene binary RS group.
In this study, the patients were stratified into luminal A‐like and luminal B‐like (HER2‐negative) groups based on the 2013 St. Gallen criteria [21]. The luminal A‐like group (deemed to have low‐recurrence risk) included patients with ER‐positive breast cancer, PR expression ≥20%, HER2‐negative breast cancer, and Ki‐67 index <14%. The luminal B‐like group (deemed to have high‐recurrence risk) included patients with ER‐positive breast cancer, HER2‐negative breast cancer, and PR expression <20% or Ki‐67 index ≥14%. The concordance to classify patients into low‐ or high‐recurrence risk categories between the 2013 St. Gallen criteria and the Surexam® 21‐gene RS and the concordance between the 2013 St. Gallen criteria and IHC3 model were both calculated.
2.5. Statistical analysis
We compared the clinical characteristics between the development and validation sets. The sensitivity, specificity, PPV, and NPV of the IHC3 model and Orucevic et al.’s nomogram were calculated separately for the entire dataset. The Pearson chi‐square test and Fisher exact test were used to analyze the categorical variables. IDFS was estimated using the Kaplan‐Meier curves and analyzed using the log‐rank test. A two‐sided P value <0.05 was considered significant. Percentage was used to calculate the concordance. All statistical analyses were performed using the R software version 3.4.0 (R Foundation, Vienna, Austria).
3. RESULTS
3.1. Patient characteristics
This analysis included 535 patients (389 in the development set and 146 in the validation set; Table 1). The median age of the development and the validation set was 48 years (range, 26‐70 years) and 48.5 years (range, 28‐67 years), respectively. These two sets were not exactly balanced with respect to the histologic type and tumor size. The validation set had larger number of invasive ductal cancer (P = 0.047) and smaller tumors (P = 0.003).
TABLE 1.
Baseline characteristics of patients with HR‐positive, HER2‐negative, axillary non‐metastasized lymph node invasive breast cancer in the development and validation sets
| Characteristics | IHC3 development set [cases (%)] | IHC3 validation set [cases (%)] | P value |
|---|---|---|---|
| Total | 389 | 146 | |
| Age category | 0.425 * | ||
| ≤50 years | 224 (57.6) | 78 (53.4) | |
| >50 years | 165 (42.4) | 68 (46.6) | |
| Histologic type | 0.047** | ||
| IDC | 340 (87.4) | 138 (94.5) | |
| ILC | 26 (6.7) | 7 (4.8) | |
| IDC + ILC | 8 (2.1) | 0 (0) | |
| Others | 15 (3.9) | 1 (0.7) | |
| Tumor size | 0.003 * | ||
| ≤2 cm | 348 (89.5) | 143 (97.9) | |
| >2 cm | 41 (10.5) | 3 (2.1) | |
| Histologic grade | 0.568** | ||
| Grade 1 | 94 (24.2) | 33 (22.6) | |
| Grade 2 | 258 (66.3) | 103 (70.5) | |
| Grade 3 | 37 (9.5) | 10 (6.8) | |
| Clinical risk | 0.003 * | ||
| Low | 329 (84.6) | 138 (94.5) | |
| High | 60 (15.4) | 8 (5.5) | |
| Ki‐67 index | 0.981 * | ||
| <27.5% | 282 (72.5) | 105 (71.9) | |
| ≥27.5% | 107 (27.5) | 41 (28.1) | |
| PR expression | 0.201 * | ||
| <55% | 114 (29.3) | 34 (23.3) | |
| ≥55% | 275 (70.7) | 112 (76.7) |
Pearson chi‐square test; ** Fisher exact test; IDC invasive ductal carcinoma; ILC invasive lobular carcinoma; PR progesterone receptor.
3.2. Classification of patients using the IHC3 model
In the IHC3 development set (n = 389) (Table 2), 328 patients (84.3%) were in the pLR group, and 61 (15.7%) in the pHR group. Based on the Surexam® 21‐gene RS and the TAILORx clinical trial criteria, 316 and 73 patients were deemed to have low and high recurrence risks, respectively. The IHC3 model identified 295 (93.4%) low recurrence risk patients correctly from the 316 patients, and 21 patients were upgraded to the pHR group in the IHC3 model. Among the 73 patients with a high recurrence risk, 40 were correctly predicted as having high recurrence risk by the IHC3 model, and 33 were downgraded to the pLR group. The accuracy of the IHC3 model in the development set was 86.1% ([295 + 40] / 389). The IHC3 model had a sensitivity of 54.8%, specificity of 93.4%, PPV of 65.6%, and NPV of 89.9% when predicting high recurrence risk (Table 3).
TABLE 2.
Classification of patients using the IHC3 model and recurrence risk categories based on the Surexam® 21‐gene RS and the TAILORx clinical trial criteria (modeling reference standard / gold standard for IHC3 model)
| Surexam® 21‐gene RS + TAILORx criteria | |||
|---|---|---|---|
| Data set | Low recurrence risk | High recurrence risk | Total cases |
| IHC3 development set | |||
| Total | 316 | 73 | 389 |
| pLR | 295 | 33 | 328 |
| pHR | 21 | 40 | 61 |
| IHC3 validation set | |||
| Total | 120 | 26 | 146 |
| pLR | 115 | 13 | 128 |
| pHR | 5 | 13 | 18 |
IHC immunohistochemistry; pLR predictive low‐recurrence risk; pHR predictive high‐recurrence risk. According to the Surexam® 21‐gene RS and the TAILORx clinical trial criteria, recurrence risk was defined as high if patients were >50 years of age with an RS >25, ≤50 years of age with an RS of >20, or ≤50 years with an RS of 16‐20 and high clinical risk; the recurrence risk was defined as low if the high‐recurrence risk criteria were not met.
TABLE 3.
Comparison of sensitivity, specificity, PPV, and NPV of the IHC3 model and Orucevic et al.’s Nomogram for predicting high recurrence risk patients in development, validation and whole sets based on different reference standard
| Reference standard | Data set | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|
| Surexam® 21‐gene RS + TAILORx criteria | IHC3 model in development set | 54.8 (40/73) | 93.4 (295/316) | 65.6 (40/61) | 89.9 (295/328) |
| IHC3 model in validation set | 50.0 (13/26) | 95.8 (115/120) | 72.2 (13/18) | 89.8 (115/128) | |
| IHC3 model in whole set | 53.5 (53/99) | 94.0 (410/436) | 67.1 (53/79) | 89.9 (410/456) | |
| Orucevic et al.’s Nomogram in whole set | 4.0 (4/99) | 99.5 (434/436) | 66.7 (4/6) | 82.0 (434/529) | |
| 21‐gene binary RS (cutoff: 25) | IHC3 model in development set | 66.7 (36/54) | 92.5 (310/335) | 59.0 (36/61) | 94.5 (310/328) |
| IHC3 model in validation set | 61.1 (11/18) | 94.5 (121/128) | 61.1 (11/18) | 94.5 (121/128) | |
| IHC3 model in whole set | 65.3 (47/72) | 93.1 (431/463) | 59.5 (47/79) | 94.5 (431/456) | |
| Orucevic et al.’s Nomogram in whole set | 5.6 (4/72) | 99.6 (461/463) | 66.7 (4/6) | 87.1 (461/529) |
IHC: Immunohistochemistry; PPV positive predictive value; NPV negative predictive value; RS recurrence score
In the validation set (n = 146) (Table 2), a total of 128 (87.7%) patients were categorized in the pLR group, and 18 (12.3%) in the pHR group. Among the 120 low recurrence risk patients, 115 (95.8%) were correctly stratified as pLR, and 5 (4.2%) were upgraded to the pHR group in the IHC3 model. Among the 26 high recurrence risk patients, 13 (50%) were correctly stratified as pHR, and 13 were downgraded to the pLR group. The accuracy for predicting high and low risk by using the IHC3 model in the validation set was 87.7% ([115 + 13] / 146). The sensitivity was 50.0%, specificity was 95.8%, PPV was 72.2%, and NPV was 89.8% for predicting high recurrence risk (Table 3).
3.3. Comparison of the IHC3 model with Orucevic et al.’s nomogram
Among the 535 patients, only six were predicted to have a high‐risk RS (range, 26‐100) using the Orucevic et al.’s nomogram, when actually there were 72 patients with an RS (Surexam® 21‐gene RS) of 26‐100 in the whole cohort. The whole data set was used to compare the sensitivity, specificity, PPV, and NPV of the IHC3 model and Orucevic et al.’s nomogram for predicting high recurrence risk (Table 3). Based on the Surexam® 21‐gene RS and the TAILORx clinical trial criteria, the estimated sensitivity was 53.5%, specificity was 94.0%, PPV was 67.1%, and NPV was 89.9% for the IHC3 model. For the Orucevic et al.’s nomogram, the sensitivity was 4.0%, specificity was 99.5%, PPV was 66.7%, and NPV was 82.0%. Based on the 21‐gene binary RS group (low‐risk group: RS 0‐25; high‐risk group: RS 26‐100), the sensitivity for predicting high recurrence risk was 65.3% for the IHC3 model and 5.6% for Orucevic et al.’s nomogram. Based on the respective modeling gold standards, the predictive accuracy of the IHC3 model and Orucevic et al.’s nomogram for the whole cohort was 86.5% ([53+410]/535) and 86.9% ([4+461]/535), respectively.
3.4. Survival analysis according to the IHC3 model and 21‐gene genomic assay
The median age of the pLR group (n = 456) and the pHR group (n = 79) was 48 years (range, 26‐69 years) and 53 years (range, 28‐70 years), respectively. Patient characteristics and treatments were mostly different between the pLR and pHR groups when classified using the IHC3 model (Table 4). The chemotherapy rates were 8.3% in the pLR group and 63.3% in the pHR group (P <0.001). All patients receiving breast‐conserving therapy underwent radiotherapy. About 3% of the patients did not receive endocrine therapy in both groups.
TABLE 4.
Baseline characteristics and treatment of the patients in the pLR and pHR groups classified by using the IHC3 model
| Characteristics | IHC3 pLR group (cases [%]) | IHC3 pHR group (cases [%]) | P value |
|---|---|---|---|
| Total | 456 | 79 | |
| Age category | 0.006* | ||
| ≤50 years | 269 (59.0) | 33 (41.8) | |
| >50 years | 187 (41.0) | 46 (58.2) | |
| Histologic type | 0.050* * | ||
| IDC | 401 (87.9) | 77 (97.5) | |
| ILC | 32 (7.0) | 1 (1.3) | |
| IDC + ILC | 7 (1.5) | 1 (1.3) | |
| Others | 16 (3.5) | 0 (0) | |
| Tumor size | 0.002* | ||
| ≤2 cm | 426 (93.4) | 65 (82.3) | |
| >2 cm | 30 (6.6) | 14 (17.7) | |
| Histologic grade | <0.001* * | ||
| Grade 1 | 124 (27.2) | 3 (3.8) | |
| Grade 2 | 315 (69.1) | 46 (58.2) | |
| Grade 3 | 17 (3.7) | 30 (38.0) | |
| Clinical risk | <0.001* | ||
| Low | 425 (93.2) | 42 (53.2) | |
| High | 31 (6.8) | 37 (46.8) | |
| Ki‐67 index | <0.001* | ||
| <27.5% | 387 (84.9) | 0 (0) | |
| ≥27.5% | 69 (15.1) | 79 (100.0) | |
| PR expression | <0.001* | ||
| <55% | 88 (19.3) | 60 (75.9) | |
| ≥55% | 368 (80.7) | 19 (24.1) | |
| Surgery | 0.143* * | ||
| Mastectomy + ALND | 249 (54.6) | 54 (68.4) | |
| Lumpectomy + ALND | 74 (16.2) | 11 (13.9) | |
| Mastectomy + SLNB | 43 (9.4) | 4 (5.1) | |
| Lumpectomy + SLNB | 90 (19.7) | 10 (12.7) | |
| Chemotherapy | <0.001* | ||
| Yes | 38 (8.3) | 50 (63.3) | |
| No | 418 (91.7) | 29 (36.7) | |
| Endocrine therapy | 0.006* | ||
| SERM | 283 (62.1) | 36 (45.6) | |
| Aromatase inhibitor | 159 (34.9) | 41 (51.9) |
Pearson chi‐square test; ** Fisher exact test. pLR predictive low‐recurrence risk; pHR predictive high‐recurrence risk; IDC invasive ductal carcinoma; ILC invasive lobular carcinoma; PR progesterone receptor; ALND axillary lymph node dissection; SLNB sentinel lymph node biopsy; SERM selective estrogen receptor modulator
After a median follow‐up of 52 months (range, 12‐90 months), 41 IDFS events in the whole cohort were reported. The IDFS was significantly different between the pLR and pHR groups classified using the IHC3 model (log‐rank test, P = 0.001) (Figure 2). The 4‐year IDFS rate was 94.7% in the pLR group and 83.5% in the pHR group (chi‐square test, P < 0.001) (Table 5). However, the IDFS difference between the low recurrence risk group and high recurrence group according to Surexam® 21‐gene RS and the TAILORx clinical trial criteria was not significant for the whole cohort (log‐rank test, P = 0.556), age group >50 (P = 0.276) and ≤50 (P = 0.942) (Figure 3). The difference in IDFS between patients with an RS (Surexam® 21‐gene) of 0‐25 (low risk) and those with an RS of 26‐100 (high risk) was not significant either for the whole cohort (log‐rank test, P = 0.511), age group >50 (P = 0.276) and ≤50 (P = 0.954) (Figure 4).
FIGURE 2.
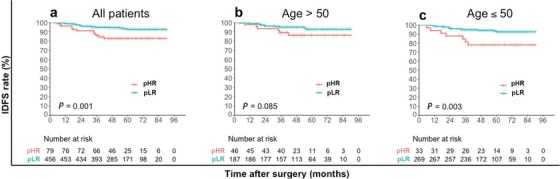
Kaplan–Meier estimates of invasive disease‐free survival curves for patients in IHC3 pLR and pHR groups. a all patients; b patients older than 50 years of age; c patients 50 years of age or younger.
Abbreviations: pLR predictive low‐recurrence risk; pHR predictive high‐recurrence risk
TABLE 5.
The 4‐year IDFS rates in different recurrence risk groups according to different criteria
| Recurrence risk criteria | Low recurrence risk group (%) | High recurrence risk group (%) | P value |
|---|---|---|---|
| IHC3 model | 94.7 (432/456) | 83.5 (66/79) | <0.001 |
| Surexam® 21‐gene RS+ TAILORx criteria | 93.6 (408/436) | 90.9 (90/99) | 0.468 |
| Surexam® 21‐gene binary RS | 93.5 (433/463) | 90.3 (65/72) | 0.448 |
| Orucevic et al.’s Nomogram | 93.2 (493/529) | 83.3 (5/6) | 0.891 |
IDFS invasive disease‐free survival; IHC immunohistochemistry
FIGURE 3.
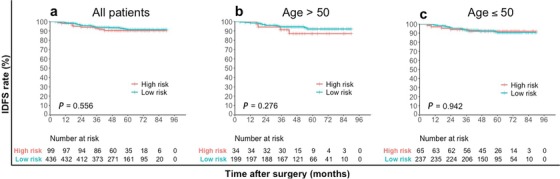
Kaplan–Meier estimates of invasive disease‐free survival for patients in the low recurrence risk group and high recurrence group according to Surexam® 21‐gene RS and TAILORx clinical trial criteria. a all patients; b patients older than 50 years of age; c patients 50 years of age or younger.
Abbreviations: RS recurrence score
FIGURE 4.
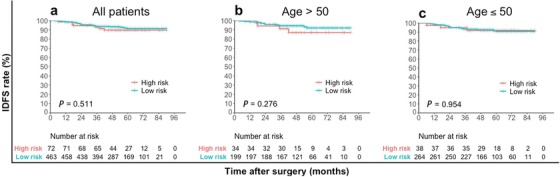
Kaplan–Meier estimates of invasive disease‐free survival curves for patients with an RS (Surexam® 21‐gene) of 0‐25 (low risk) and patients with an RS (Surexam® 21‐gene) of 26‐100 (high risk). a all patients; b patients older than 50 years of age; c patients 50 years of age or younger.
Abbreviations: RS recurrence score
3.5. Predictive risk category of the IHC3 model and benefit of chemotherapy
Of the 456 patients with a pLR according to the IHC3 model (Table 6), only 38 received chemotherapy. There was no significant difference in the IDFS events between the chemotherapy group and the non‐chemotherapy group (5.3% vs. 6.2%, P = 1.000). Among the 79 pHR patients, no significant difference in IDFS events were found between the chemotherapy (9/50, 18.0%) and the non‐chemotherapy groups (4/29, 13.8%) (P = 0.864). The benefit of chemotherapy was not observed in the pLR and pHR groups in our retrospective study.
TABLE 6.
Association between IDFS events and chemotherapy in different IHC3 risk groups
| IHC3 model | Data set | Chemotherapy group (cases [%]) | Non‐chemotherapy group (cases [%]) | P value |
|---|---|---|---|---|
| pLR | Total (n = 456) | 38 (100) | 418 (100) | |
| IDFS event (n = 28) | 2 (5.3) | 26 (6.2) # | 1.000* | |
| No IDFS event (n = 428) | 36 (94.7) | 392 (93.8) | ||
| pHR | Total (n = 79) | 50 (100) | 29 (100) | |
| IDFS event (n = 13) | 9 (18) | 4 (13.8) | 0.864* | |
| No IDFS event (n = 66) | 41 (82) | 25 (86.2) |
pLR predictive low‐recurrence risk; pHR predictive high‐recurrence risk; IDFS invasive disease‐free survival; IHC immunohistochemistry. # Among the 26 events, five were secondary primary cancers. *Pearson chi‐square test.
3.6. Concordance between the 2013 St. Gallen criteria and the Surexam® 21‐gene RS/IHC3 model
Based on the 2013 St. Gallen criteria (Table 7) [18], 205 patients were categorized in the luminal A‐like group, and 330 patients in the luminal B‐like group. When the concordance between the 2013 St. Gallen criteria and Surexam® 21‐gene RS was calculated, 99.0% (203/205) of the patients categorized into the luminal A‐like group were in the RS <26 group, and only 1.0% (2/205) were in the RS ≥26 group. Among the 330 patients categorized in the luminal B‐like group, 21.2% (70/330) were in the RS ≥26 group, and 78.8% (260/330) were in the RS <26 group. When calculating the concordance between the 2013 St. Gallen criteria and IHC3 model, all patients in the luminal A‐like group were in the pLR group. In total, 79 (23.9%) luminal B‐like patients were in the pHR group, while 251 (76.1%) were categorized to the pLR group. In summary, almost all luminal A‐like patients were classified into the low‐recurrence risk group but more than three‐quarters of luminal B‐like patients, which deemed to have high‐recurrence risk, were also classified into the low‐recurrence risk group. From the results of poor concordance, the 2013 St. Gallen criteria was considered not an appropriate tool to distinguish low‐ and high‐ recurrence risk patients.
TABLE 7.
Concordance between the 2013 St Gallen criteria and the Surexam® 21‐gene RS / IHC3 model
| Data set | Luminal A‐like group (cases [%]) | Luminal B‐like group (cases [%]) |
|---|---|---|
| Total | 205 | 330 |
| Surexam® 21‐gene RS <26 group (n = 463) | 203 (99.0) | 260 (78.8) |
| Surexam® 21‐gene RS ≥26 group (n = 72) | 2 (1.0) | 70 (21.2) |
| IHC3 pLR (n = 456) | 205 (100) | 251 (76.1) |
| IHC3 pHR (n = 79) | 0 (0) | 79 (23.9) |
RS recurrence score; IHC immunohistochemistry; pLR predictive low‐recurrence risk; pHR predictive high‐recurrence risk
4. DISCUSSION
The 21‐gene RS provides prognostic information regarding the loco‐regional and distant recurrence risk and predicts chemotherapy benefit in EBC [9, 22, 23]. New results from the prospective TAILORx clinical trial showed that the combination of clinical risk stratification on the basis of tumor size, histologic grade, and 21‐gene RS could provide more accurate prognostic information about recurrence [20]. However, the 21‐gene RS assay is not available in most developing countries and is costly. Our study developed and validated an easy‐to‐use predictive model (IHC3 model) based on the 21‐gene RS assay and clinical risk to stratify patients into a pLR or pHR group by decision tree without network support. The definition of clinical risk used in this present study was consistent with that in the TAILORx clinical trial [20]. Accuracies of the IHC3 model were 86.1% and 87.7% in the development and validation sets, respectively. Although the sensitivity of the IHC3 model was only 50% for predicting the high‐recurrence risk group, the high specificity (95.8%) and high NPV (89.8%) for predicting the high‐recurrence risk group in the validation set of our model indicated the high accuracy of the IHC3 model in predicting low‐recurrence risk patients. These findings may be attributed to the loss matrix we set in the IHC3 model construction. Losses for the high‐to‐low misclassifications were twice as much as those of the low‐to‐high misclassifications in the IHC3 model construction to reduce the probability of patients entering the low risk group and reduce the risk of undertreatment.
The IDFS was significantly different between the pLR and pHR groups in the IHC3 model (P = 0.001). Unfortunately, no IDFS difference was observed when the combination of the Surexam® 21‐gene RS and TAILORx clinical trial criteria (P = 0.556) or the 21 gene binary RS criteria (0‐25 vs. 26‐100) (P = 0.511) was used. Subgroup analyses of IDFS in the IHC3 model showed that the survival difference was significant for younger patients aged ≤50 years (P = 0.003), but not for older patients aged >50 years (P = 0.085) (Figure 2). This difference may be due to the risk setting during model development. We set different standards for different age subgroups. For patients aged 50 years or younger, the risk setting was not determined by the 21‐gene RS alone. Meanwhile, the clinical risk was taken into account for younger patients with an RS of 16‐20 because no chemotherapy benefit was observed in the subgroup with a low clinical risk group and an RS of 16 to 20 in the TAILORx clinical trial [20]. In addition to sensitivity, specificity, PPV, and NPV, the discrepancy in the survival rate of patients with different predictive recurrence risks can also reflect the accuracy of the IHC3 model. From the perspective of IDFS, the predictive accuracy of the IHC3 model was superior to that of Surexam® 21‐gene RS and TAILORx clinical trial criteria or pure 21 gene binary RS criteria.
Recently, Orucevic et al. [14] published an updated nomogram based on the TAILORx clinical trial results. The nomogram was built and validated in a large oncotype DX‐tested cohort. We used this nomogram to predict high‐risk RS (26‐100) in our study. Among the 535 patients investigated, only six patients were predicted to have a high‐risk RS. Our findings showed that the sensitivity of the Orucevic et al.’s nomogram for predicting high‐risk RS (26‐100) was only 5.6% (Table 3). A large percentage (94.4%) of patients with an RS (Surexam® 21‐gene) of 26‐100 were improperly predicted into the RS 0‐25 group by Orucevic et al.’s nomogram. Although the nomogram accuracy validated in our study (86.9%) was similar to the overall accuracy reported by Orucevic et al. (86.8%) [14], the very low sensitivity for predicting high‐risk patients may lead to a large number of undertreatment. However, it was difficult to directly compare the published results using the online tools. The discrepancy in the predictive results between the IHC3 model and Orucevic et al.’s nomogram may have been caused by the following: 1) patient population (Chinese vs. American) with different baseline characteristics, 2) gene testing assay (Surexam® vs. Oncotype DX), 3) statistical methods (CART model [present study] vs. nomogram [Orucevic et al. study]), and 4) clinicopathologic variables used for modeling (with Ki‐67 [present study] vs. without Ki‐67 [Orucevic et al. study]). Considering the high cost of genetic testing, many patients with HR‐positive EBC make chemotherapy treatment decisions based on clinicopathological features observed in clinical practice. These patients did not undergo multigene assay testing and were not enrolled in our study. Therefore, the distribution of the RS in the present study (RS 0‐25, 86.5%; RS 26‐100, 13.5%) was slightly different from that in other studies (Orucevic et al.’s study [14]: RS 0‐25, 84.8%; Lee et al.’s study [13]: RS 0‐25, 84.1%). Because of the unavailability of the Oncotype DX in China, we have adopted the domestic Surexam® 21‐gene assay to measure the RNA in formalin‐fixed paraffin‐embedded tissues with branched‐DNA liquid chip, which is different from the reverse transcription‐polymerase chain reaction (RT‐PCR) used by Oncotype DX. Although the consistency of the testing results has been verified in the Surexam® research laboratory [24], some method differences for the measurement of gene expression may still exist. In Orucevic et al.’s nomogram, tumor grade and PR expression were the most significant predictors for stratification, followed by histologic type, tumor size, and age [14]. In our IHC3 model, the Ki‐67 index, PR expression and clinical risk (including histologic grade and tumor size) were the most important IHC variables. Currently, the quantification of ER, PR [16], and HER2 expressions [17] is increasingly standardized, and reproducibility among laboratories has substantially improved. Nevertheless, the histological grade and Ki‐67 index still have modest inter‐laboratory and inter‐observer concordances [25]. To minimize the impact of this unsatisfactory concordance, the histological grade was combined with tumor size to define the clinical risk in our study, similar to the TAILORx clinical trial. Based on the latest results of TAILORx clinical trial [20], all patients >50 years of age with an RS 0‐25 could get no benefit from chemotherapy with no relationship with the clinical risk, so this variable was used for only risk stratification in younger patients ≤50 years of age. Considering the prognostic and predictive potential of the Ki‐67 index [26] and that the Ki‐67 index is widely used in many published tools [11, 12, 13], the Ki‐67 index was used to develop the IHC3 model. Conversely, the Ki‐67 index was not included in Orucevic et al.’s nomogram, which might be a reason for its relatively low sensitivity for predicting high‐risk (RS 26‐100) patients.
The value of PR expression in predicting the prognosis for HR‐positive, HER2‐negative invasive breast cancer has been previously validated [27, 28, 29]. Prat et al. [30] proposed that the IHC‐based definition of luminal A‐like breast cancer is HR positivity, HER2 negativity, Ki‐67 index <14%, and PR expression >20% which led to the modification of the definition of luminal A‐like breast cancer in the 2013 St. Gallen criteria [21]. When the concordance between the 2013 St. Gallen criteria and the Surexam® 21‐gene RS / IHC3 model were calculated in the present study, we observed that almost all patients with luminal A‐like breast cancer were categorized in the RS <26 and pLR groups. For patients with a luminal B‐like disease, which were supposed to have high‐recurrence risk, only 21.2% (70/330) were in the RS ≥26 group, and 23.9% (79/330) were in the pHR group. The discrepancy between the 2013 St. Gallen criteria and the IHC3 model may be due to the different cutoffs for the Ki‐67 index and PR expression, and the distinct variable combination mode. The lower Ki‐67 index cutoff of the 2013 St. Gallen criteria (14% [St. Gallen criteria] vs 27.5% [IHC3 model]) categorized more patients into luminal B‐like group. Moreover, PR expression was a single determinant in the 2013 St. Gallen criteria, while in the IHC3 model, the PR expression was only a node in the decision tree. Oncologists may more confidently omit chemotherapy for patients with a luminal A‐like breast cancer for its satisfactory PPV when the 21‐gene genomic assay is not available. However, for luminal B‐like breast cancer patients, chemotherapy decision‐making may still depend on the result of the 21‐gene genomic assay. Compared with the St. Gallen criteria, the IHC3 model may provide a better combination of IHC variables and predict more low‐recurrence risk patients accurately. Considering the discordance of IHC variables between different medical institutions, a minor modification of each cutoff value in the predictive model, depending on the target population to be focused (i.e., high or low risk), is recommended to achieve a better predictive accuracy for classifying high‐risk or low‐risk patients.
On the basis of the results from the TAILORx clinical trial [9], the NCCN clinical practice guidelines for breast cancer updated the RS cutoffs in 2019 (RS <18, RS 18‐30, and RS ≥31 [previous version] vs. RS < 26, RS 26‐30, and RS ≥31 [current version]). For patients with an RS of 26‐30, the omission of chemotherapy has not been studied prospectively. To avoid undertreatment, it is reasonable to combine the RS 26‐30 and RS ≥31 into the high‐recurrence risk group, which is also consistent with the definition of high‐recurrence risk group in the TAILORx clinical trial. To further refine the risk stratification, we further divided the patients into two subgroups according to age (≤50 years and >50 years) and included the clinical risk in the model, with reference to the most recent published results from the TAILORx clinical trial [20]. Although many models have been developed for predicting genomic assay results, only three models used the TAILORx cutoffs (0‐25 and 26‐100) as the modeling standard [12‐14, 31]. Irrespective of whether using Kim et al.’s online recurrence estimator tool (http://www.breastrecurrenceestimator.onc.jhmi.edu) [12], Lee et al.’s nomogram [13], or Orucevic et al.’s updated nomogram [14], the prediction process must depend on the network and computer. Compared with the aforementioned models, the major advantage of the IHC3 model is that it is easy to use as it does not require network support. On the basis of ensuring the accuracy of prediction, patients can be simply classified as low‐ or high‐risk by decision tree.
In recent years, the prognostic and predictive values of the 21‐gene genomic assay have been generally recognized. A previous study by our group showed that the Surexam® 21‐gene RS results influenced the chemotherapy decisions of Chinese oncologists and patients more than the standard clinicopathological criteria, such as the St. Gallen Consensus Statement and Adjuvant! algorithm, which is no longer available for clinical use [15]. However, the limited availability and high cost of the 21‐gene genomic assay has always been an obstacle to its widespread use. With the implementation of our IHC3 model, patients can be categorized into a pLR or pHR groups. Chemotherapy could be avoided in pLR patients without the 21‐gene RS. The similar IDFS rate between the chemotherapy and the non‐chemotherapy subgroups might be an evidence that pLR patients might get no benefit from chemotherapy. Among the pHR patients, the IDFS event rate of the subgroup receiving chemotherapy reached 18%. Although the benefit of chemotherapy was not observed in our retrospective study (IDFS event rate: 18% [chemotherapy subgroup] vs. 13.8% [non‐chemotherapy subgroup], P = 0.864), we still recommend chemotherapy for pHR patients because a large number of previous studies [8, 22] have shown that patients with high 21‐gene RS can benefit from chemotherapy. The lack of chemotherapy benefit in the pHR group of our study may be due to the higher mean RS of patients with chemotherapy (RS 33.6 [chemotherapy subgroup] vs. 18.3 [non‐chemotherapy subgroup]) and the small number of overall IDFS events in the pHR group (n = 13).
Our study has some limitations. First, it was a single‐institution study with no external validation of the IHC3 model. The clinical case selection bias was inevitable. If multiple centers were involved and external validation was obtained, the predictive accuracy of the IHC3 model would be further improved. Second, the median follow‐up of 52 months may have still been inadequate, even though a significant difference in IDFS was found between the pLR and pHR groups. For HR‐positive EBC patients, the predictive advantage of the IHC3 model needs to be confirmed by longer follow‐up. Lastly, patients with metastasized lymph nodes, ER‐positive breast cancer were not enrolled in this study because although the 21‐gene RS has been demonstrated to be a predictor of the benefit of chemotherapy for such patients [32], the NCCN guidelines before 2018 recommend chemotherapy for these patients without the need for the 21‐gene assay.
5. CONCLUSIONS
We developed and validated an easy‐to‐use model based on new results of the TAILORx clinical trial. The accuracy of the IHC3 model was 87.7% in the validation set, and there was a significant difference in IDFS between the pLR and pHR groups. Our IHC3 model could be used as a replacement of the 21‐gene genomic assay in patients with breast cancer when the assay is not available or affordable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The institutional review boards of Peking Union Medical College Hospital (PUMCH) approved the study. The need for individual consent was waived by the committee because of the retrospective nature of the study and the anonymous analysis of the data.
CONSENT FOR PUBLICATION
Not applicable.
DATA AVAILABILITY
Not applicable.
COMPETING INTERESTS
All authors declare that they have no competing interests.
FUNDING
This study was funded by the science and technology commission of Beijing: Optimization of breast cancer screening program for 35‐75 years old women in Beijing (Grant No. D161100000816005)
AUTHORS' CONTRIBUTIONS
YNZ conceived and coordinated the study, designed, performed, and analyzed the experiments, wrote the paper. YDZ, FM, RY carried out the data collection, data analysis, and revised the paper. QS designed the experiments, carried out the data analysis, and revised the paper. All authors agreed to be accountable for all aspects of the work, reviewed the results and approved the final version of the manuscript.
ACKNOWLEDGMENTS
Not applicable.
Zhang Y, Zhou Y, Mao F, Yao R, Sun Q. Ki‐67 index, progesterone receptor expression, histologic grade and tumor size in predicting breast cancer recurrence risk: A consecutive cohort study. Cancer Communications. 2020;40:181–193. 10.1002/cac2.12024
REFERENCES
- 1. Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389(10074):1134‐50. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 4. Kamal AH, Loprinzi CL, Reynolds C, Dueck AC, Geiger XJ, Ingle JN, et al. Breast medical oncologists' use of standard prognostic factors to predict a 21‐gene recurrence score. Oncologist. 2011;16(10):1359‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goetz MP, Gradishar WJ, Anderson BO, Abraham J, Aft R, Allison KH, et al. NCCN Guidelines Insights: Breast Cancer, Version 3.2018. J Natl Compr Canc Netw. 2019;17(2):118‐26. [DOI] [PubMed] [Google Scholar]
- 6. Cardoso F, Kyriakides S, Ohno S, Penault‐Llorca F, Poortmans P, Rubio IT, et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol. 2019;30(8):1194‐220. [DOI] [PubMed] [Google Scholar]
- 7. Curigliano G, Burstein HJ, Winer EP, Gnant M, Dubsky P, Loibl S, et al. De‐escalating and escalating treatments for early‐stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. 2017;28(8):1700‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carlson JJ, Roth JA. The impact of the Oncotype Dx breast cancer assay in clinical practice: a systematic review and meta‐analysis. Breast Cancer Res Treat. 2013;141(1):13‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Adjuvant Chemotherapy Guided by a 21‐Gene Expression Assay in Breast Cancer. N Engl J Med. 2018;379(2):111‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cuzick J, Dowsett M, Pineda S, Wale C, Salter J, Quinn E, et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki‐67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol. 2011;29(32):4273‐8. [DOI] [PubMed] [Google Scholar]
- 11. Turner BM, Skinner KA, Tang P, Jackson MC, Soukiazian N, Shayne M, et al. Use of modified Magee equations and histologic criteria to predict the Oncotype DX recurrence score. Mod Pathol. 2015;28(7):921‐31. [DOI] [PubMed] [Google Scholar]
- 12. Kim HS, Umbricht CB, Illei PB, Cimino‐Mathews A, Cho S, Chowdhury N, et al. Optimizing the Use of Gene Expression Profiling in Early‐Stage Breast Cancer. J Clin Oncol. 2016;34(36):4390‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee SB, Kim J, Sohn G, Kim J, Chung IY, Kim HJ, et al. A Nomogram for Predicting the Oncotype DX Recurrence Score in Women with T1‐3N0‐1miM0 Hormone ReceptorPositive, Human Epidermal Growth Factor 2 (HER2)Negative Breast Cancer. Cancer Res Treat. 2019;51(3):1073‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Orucevic A, Bell JL, King M, McNabb AP, Heidel RE. Nomogram update based on TAILORx clinical trial results ‐ Oncotype DX breast cancer recurrence score can be predicted using clinicopathologic data. Breast. 2019;46:116‐25. [DOI] [PubMed] [Google Scholar]
- 15. Zhang YN, Zhou YD, Mao F, Sun Q. Impact of the 21‐Gene Recurrence Score Assay in adjuvant chemotherapy selection for node‐negative, hormone receptor‐positive breast cancer in the Chinese population. Neoplasma. 2015;62(4):658‐65. [DOI] [PubMed] [Google Scholar]
- 16. Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997‐4013. [DOI] [PubMed] [Google Scholar]
- 18. Zhang X, Zhou Y, Mao F, Lin Y, Guan J, Sun Q. Efficacy and safety of pirarubicin plus capecitabine versus pirarubicin plus cyclophosphamide in Chinese node‐negative breast cancer patients: a 4‐year open‐label, randomized, controlled study. Med Oncol. 2015;32(10):240‐6. [DOI] [PubMed] [Google Scholar]
- 19. Hudis CA, Barlow WE, Costantino JP, Gray RJ, Pritchard KI, Chapman JA, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007;25(15):2127‐32. [DOI] [PubMed] [Google Scholar]
- 20. Sparano JA, Gray RJ, Ravdin PM, Makower DF, Pritchard KI, Albain KS, et al. Clinical and Genomic Risk to Guide the Use of Adjuvant Therapy for Breast Cancer. N Engl J Med. 2019;380(25):2395‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garcia Fernandez A, Chabrera C, Garcia Font M, Fraile M, Lain JM, Gonzalez S, et al. Differential patterns of recurrence and specific survival between luminal A and luminal B breast cancer according to recent changes in the 2013 St Gallen immunohistochemical classification. Clin Transl Oncol. 2015;17(3):238‐46. [DOI] [PubMed] [Google Scholar]
- 22. Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node‐negative, estrogen receptor‐positive breast cancer. J Clin Oncol. 2006;24(23):3726‐34. [DOI] [PubMed] [Google Scholar]
- 23. Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, et al. Prognostic and predictive value of the 21‐gene recurrence score assay in postmenopausal women with node‐positive, oestrogen‐receptor‐positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11(1):55‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knudsen BS, Allen AN, McLerran DF, Vessella RL, Karademos J, Davies JE, et al. Evaluation of the branched‐chain DNA assay for measurement of RNA in formalin‐fixed tissues. J Mol Diagn. 2008;10(2):169‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Polley MY, Leung SC, Gao D, Mastropasqua MG, Zabaglo LA, Bartlett JM, et al. An international study to increase concordance in Ki67 scoring. Mod Pathol. 2015;28(6):778‐86. [DOI] [PubMed] [Google Scholar]
- 26. Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11(2):174‐83. [DOI] [PubMed] [Google Scholar]
- 27. Onoda T, Yamauchi H, Yagata H, Tsugawa K, Hayashi N, Yoshida A, et al. The value of progesterone receptor expression in predicting the Recurrence Score for hormone‐receptor positive invasive breast cancer patients. Breast Cancer. 2015;22(4):406‐12. [DOI] [PubMed] [Google Scholar]
- 28. Nishimukai A, Yagi T, Yanai A, Miyagawa Y, Enomoto Y, Murase K, et al. High Ki‐67 Expression and Low Progesterone Receptor Expression Could Independently Lead to a Worse Prognosis for Postmenopausal Patients With Estrogen Receptor‐Positive and HER2‐Negative Breast Cancer. Clin Breast Cancer. 2015;15(3):204‐11. [DOI] [PubMed] [Google Scholar]
- 29. Li AQ, Zhou SL, Li M, Xu Y, Shui RH, Yu BH, et al. Clinicopathologic Characteristics of Oestrogen Receptor‐Positive/Progesterone Receptor‐Negative/Her2‐Negative Breast Cancer According to a Novel Definition of Negative Progesterone Receptor Status: A Large Population‐Based Study from China. PLoS One. 2015;10(5):e0125067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prat A, Cheang MC, Martin M, Parker JS, Carrasco E, Caballero R, et al. Prognostic significance of progesterone receptor‐positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol. 2013;31(2):203‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Orucevic A, Bell JL, McNabb AP, Heidel RE. Oncotype DX breast cancer recurrence score can be predicted with a novel nomogram using clinicopathologic data. Breast Cancer Res Treat. 2017;163(1):51‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jasem J, Fisher CM, Amini A, Shagisultanova E, Rabinovitch R, Borges VF, et al. The 21‐Gene Recurrence Score Assay for Node‐Positive, Early‐Stage Breast Cancer and Impact of RxPONDER Trial on Chemotherapy Decision‐Making: Have Clinicians Already Decided? J Natl Compr Canc Netw. 2017;15(4):494‐503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
COMPETING INTERESTS
All authors declare that they have no competing interests.
FUNDING
This study was funded by the science and technology commission of Beijing: Optimization of breast cancer screening program for 35‐75 years old women in Beijing (Grant No. D161100000816005)
AUTHORS' CONTRIBUTIONS
YNZ conceived and coordinated the study, designed, performed, and analyzed the experiments, wrote the paper. YDZ, FM, RY carried out the data collection, data analysis, and revised the paper. QS designed the experiments, carried out the data analysis, and revised the paper. All authors agreed to be accountable for all aspects of the work, reviewed the results and approved the final version of the manuscript.


