Abstract
Conventional immunohistochemistry (IHC) is a widely used diagnostic technique in tissue pathology. However, this technique is associated with a number of limitations, including high inter‐observer variability and the capacity to label only one marker per tissue section. This review details various highly multiplexed techniques that have emerged to circumvent these constraints, allowing simultaneous detection of multiple markers on a single tissue section and the comprehensive study of cell composition, cellular functional and cell‐cell interactions. Among these techniques, multiplex Immunohistochemistry/Immunofluorescence (mIHC/IF) has emerged to be particularly promising. mIHC/IF provides high‐throughput multiplex staining and standardized quantitative analysis for highly reproducible, efficient and cost‐effective tissue studies. This technique has immediate potential for translational research and clinical practice, particularly in the era of cancer immunotherapy.
Keywords: immunofluorescence, immunohistochemistry, immunotherapy, multiplex, overview
Abbreviations
- 2D
2‐dimension
- 3D
3‐dimension
- Ce3D
Clearing‐enhanced 3D
- CODEX
CO detection by indEXing
- CRO
Contract Research Organisation
- CV
coefficient of variation
- CyTOF
time‐of‐flight mass cytometry
- DAB
3, 3′‐diaminobenzidine
- DSP
Digital Spatial Profiling
- ER
estrogen receptor
- FDA
Food and Drug Administration
- FFPE
formalin‐fixed paraffin‐embedded
- FOXP3
forkhead box P3
- GEP
gene expression profiling
- HCC
hepatocellular carcinoma
- HDR
High dynamic range
- HER2
human epidermal growth factor receptor 2
- IHC
immunohistochemistry
- IMC
Imaging mass cytometry
- IVD
In Vitro Diagnostic
- IVD
In vitro diagnostic
- MIBI
Multiplexed Ion Beam Imaging
- MICSSS
multiplexed immunohistochemical consecutive staining on single slide
- mIHC/IF
multiplex Immunohistochemistry/Immunofluorescence
- PD‐1
programmed cell death receptor 1
- PD‐L1
programmed cell death ligand 1
- PIPS
Philips IntelliSite Pathology Solution
- PR
progesterone receptor
- QIF
quantitative immunofluorescence
- ROI
region of interest
- TEM
effector memory T cell
- TIL
tumor‐infiltrating lymphocyte
- TMA
tissue microarray
- TMB
tumor mutational burden
- Treg
regulatory T cell
- TRM
tissue‐resident memory T cell
- WSI
whole slide imaging
1. BACKGROUND
Conventional immunohistochemistry (IHC) is commonly used as a diagnostic technique in the field of tissue pathology but suffers from certain limitations. The most critical of these is that this technique only permits the labelling of a single marker per tissue section. This results in missed opportunities to gain important prognostic and diagnostic information from patient samples. Multiplex Immunohistochemistry/Immunofluorescence (mIHC/IF) technologies, which allow the simultaneous detection of multiple markers on a single tissue section, have been introduced and adopted in both research and clinical settings in response to increased demand for improved techniques. A number of highly multiplexed tissue imaging technologies have also emerged, permitting comprehensive studies of cell composition, functional state and cell‐cell interactions which suggest improved diagnostic benefit [1]. These novel imaging techniques are based on cyclic immunofluorescence [2], tyramide‐based mIHC/IF [3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14], epitope‐targeted mass spectrometry [15, 16], or RNA detection [17, 18]. Such techniques provide a comprehensive view of marker distribution and tissue composition, and are poised to solve major questions surrounding the pathogenesis of various complex disorders [19]. The ability to label multiple markers on a single section is of particular significance when studying samples taken from rare donors, where tissues may be of low availability.
The mIHC/IF is also widely recognized to play an important role in the era of cancer immunotherapy for both research and clinical purposes. In a recent publication, Lu et al. [1] demonstrated that mIHC/IF appeared to be associated with improved performance in predicting response to programmed cell death ligand 1 (PD‐L1)/ programmed cell death receptor 1 (PD‐1) treatment in different solid tumor types when compared to PD‐L1 IHC, tumor mutational burden (TMB) or gene expression profiling (GEP) alone. This was shown through a meta‐analysis of studies involving tumor specimen assays of over 10 different solid tumor types in 8135 patients and the results were correlated with anti‐PD‐1/PD‐L1 immunotherapy response.
Several other studies[20, 21, 22] have utilized different types of mIHC/IF to assess their tumor samples and obtained the data required for analysis. However, these methods were not without their limitations. Thus, this review discusses the uses, constraints and accessibility of the more commercialized imaging modalities, with a focus on mIHC/IF, to enable prospective users to make an informed choice when conducting their research or clinical work.
2. THE LIMITATIONS OF IHC
As aforementioned, the inability to label more than one marker per tissue section is the most important limitation of IHC. For example, tumor‐infiltrating CD8+ T cells can be identified through CD8, CD3, forkhead box P3 (FOXP3) and CD20 expression [23]. The expression of certain molecules, such as PD‐L1 [24, 25, 26, 27] and PD‐1 on the surface of these cells [28, 29] predicts responsiveness to treatment with PD‐L1/PD‐1 blockade. These markers may be predictive individually or in combination, and some are also of prognostic value in several cancers [30‐8, 33, 34, 35, 36]. Furthermore, investigating the relative spatial distribution of immune cells and markers, as well as marker expression patterns and interactions between immune cells, cancer cells and stromal cells may lead to improved understanding on cancer progression [37, 38, 39]. Thus, while IHC remains a highly practical and cost‐effective diagnostic and prognostic method, this single‐marker method cannot tell the whole story of complex immune microenvironment.
Another drawback of IHC‐based biomarker assessment is high inter‐observer variability [40, 41, 42, 43, 44, 45, 46, 47, 48]. For instance, Ki‐67 is a widely endorsed marker for a range of cancers [29, 49, 1, 46]. At the recent 2017 St. Gallen International Expert Consensus Conference, an issue of caution was raised concerning the reproducibility of IHC for Ki‐67 and the implications of this variability for clinical decisions [50]. Multiple groups have previously demonstrated that inter‐observer variability can be negated by scoring Ki‐67 in a reproducible and quantitative manner using digital analysis; for instance, using the Definiens Tissue Studio software (Definiens, Munich, Germany) and the Aperio ePathology image analysis system (Leica Biosystems, Wetzlar, Germany) [51, 52, 53, 54]. This mIHC/IF digital analysis can also be performed using the PerkinElmer Vectra platform (PerkinElmer, Inc., Waltham, MA, USA) followed by inForm software (PerkinElmer, Inc., Waltham, MA, USA) [55, 56, 57], which circumvents this limitation as well. However, this does not solve the major issue with conventional IHC, which is the restriction to labelling a single marker per tissue section. Even though Ki‐67 is useful alone, a multiplex setting provides the opportunity to examine panels of several markers simultaneously (for example, Ki‐67 with PD‐L1 and cytokeratin for tumor proportion score, PD‐1, CD3 and CD8). Furthermore, such techniques permit the standardization of staining, application of scoring methods and cut‐offs for all markers.
3. BRIGHTFIELD‐BASED MIHC/IF
3.1. Discovery ultra
DISCOVERY ULTRA (Roche Diagnostics, Basel, Switzerland) is an alternative IHC research platform that allows complex assay development for research purposes [58, 59, 60]. This platform overcomes some of the problems associated with IHC by performing multiplexed analysis of multiple biomarkers, using tyramine chemistry to combine a vast spectrum of new chromogenic dyes as shown in Figure 1A. This is useful for in situ analysis with conventional brightfield microscopes. The dyes, useful both individually and blended to generate novel colors; provide signals similar to the conventional 3, 3′‐diaminobenzidine (DAB) chromogen. They may also enable the analysis of co‐localized biomarkers. These chromogens have broad absorbance spectra which produce dark staining patterns that are supposedly easy to distinguish during light microscopy[61]. More importantly, conventional scanners can acquire images of such stained slides, facilitating biomarker research and the possibility of in vitro diagnostics product development. As shown in Table 1, the system does not come with its own imaging light microscopy nor analytic software. Visualization of co‐localized biomarkers (especially in the same cellular compartment) with other software be challenging or incompatible. In the DISCOVERY ULTRA bright field setting, pathologists can assess the mIHC/IF without any particular software or visualization tool, which is clearly an advantage. However, recognizing more than 2‐3 colors for co‐localization of markers in the same cellular compartment might be beyond what the human eye can achieve (Figure 2). Thus, a dedicated analytic pipeline would be needed for appropriately analyzing and evaluating this platform.
FIGURE 1.
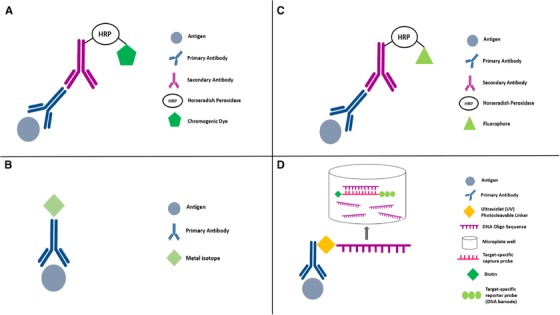
Diagram showing mechanism of each of the mIHC/IF platform. (A) DISCOVERY ULTRA system: after primary antibody incubation, a secondary antibody labelled with HRP is introduced. The HRP is reacted with an appropriate substrate bound to a chromogenic dye, leading to the precipitation of insoluble, coloured precipitates at the site where the antigens are found. (B) Metal‐based IHC techniques such as IMC and MIBI: a primary antibody bound to the target antigen is tagged with a metal isotope of known molecular mass. Analysis is carried out using mass spectrometry in MIBI and laser ablation coupled to mass cytometry in IMC. (C) Vectra: after primary antibody incubation, a secondary antibody labelled with HRP is introduced. A fluorophore‐conjugated tyramide molecule serves as the substrate for HRP, resulting in an antigen‐associated fluorescence signal. (D) Nanostring's DSP: the target antigen will bind the primary antibody which is coupled to a photocleavable oligonucleotide tag. UV light is used to cleave the oligonucleotide tags and is collected using a microcapillary tube and stored in a microplate well. The oligonucleotide tags will bind to the reporter probe via the target‐specific capture probe. Reporter probes are imaged and counted by the nCounter analysis system. Abbreviations: mIHC/IF, multiplex immunohistochemistry/immunofluorescence; HRP, horseradish peroxidase; IHC, immunohistochemistry; IMC, Imaging Mass Cytometry; MIBI, Multiplexed Ion Beam Imaging; DSP, Digital Spatial Profiling
TABLE 1.
Overview and comparison of the different imaging modalities
| Characteristic | Imaging modality | |||||||
|---|---|---|---|---|---|---|---|---|
| Vectra | DISCOVERY ULTRA | CyTOF Imaging | InSituPlex | MIBI | Codex | DSP | Chipcytometry | |
| Vendor | Perkin Elmer/Akoya | Roche | Fluidigm | Ultivue | IonPath | Akoya | NanoString | ZELLKRAFTWERK |
| Plexing | 9+ | 5+ | 37+ | 16+ (claimed) | 40+ | 40+ (claimed) | 30+ | 39 (claimed) |
| Antibody conjugation | Fluorescent based | Fluorescent & chromogenic based | Metal‐based | DNA‐barcoding based | Metal‐based | DNA‐barcoding based | DNA‐barcoding based | Fluorescent based |
| Staining reagents | √√ | √√ | √ | √√ | √ | √√ | ‐ | ‐ |
| Staining machine | √ | √√ | ‐ | ‐ | ‐ | √√ | ‐ | ‐ |
| Imaging microscopy | √√ | ‐ | √√ | ‐ | √√ | ‐ | ‐ | ‐ |
| Analytic software | √√ | ‐ | √√ | ‐ | √√ | √ | √√ | √ |
| RNA detection | √ | √ | √ | ‐ | √ | ‐ | √√ | ‐ |
| FFPE validation | √√ | √√ | √√ | √√ | √√ | √ | √√ | √ |
| Publication | >100 or more | Limited | 36+ | Limited | 5+ | 1 | 2 | limited |
| Commercialized | √√√ | √√ | √√√ | √√ (launched 2018‐2019) | √ (launched 2018‐2019) | √ (launched 2018‐2019) | √ (launched 2018‐2019) | √ (launched 2018‐2019) |
| Cost | $$ | $$ | $$$$ | $ | $$$$$$$$ | $ | $$$ | $ |
| Recommended software | InForm, HALO | ‐ | HistoCat | HALO | QuPath, HALO | “Bundled software” | “Bundled software” | “Bundled software” |
| Scanning speed | +++ | No scanner | + | No scanner | + | ++ | ++ | ++ |
| Resolution | 0.25 μm | No scanner | 0.5 μm | No scanner | 0.38 μm | 260 nm | 10‐20 μm | 0.5μm |
| Scanning capacity | 80 slides | No scanner | 1 slide | No scanner | 1 slide | 1 slide | 1 slide | 1 slide |
| Whole Slide Imaging | Yes | No scanner | Possible but very costly and time consuming | No scanner | Yes | Possible but very costly and time consuming | Possible but very costly and time consuming | No |
| Key advantages | “clinical” relevant; End‐to‐end solution | “clinical” relevant | End‐to‐end solution | DNA barcoding | Promising | DNA barcoding | “clinical” relevant; +700 RNA detection | Microfluidic system |
| Key disadvantages | Not Applicable | Brightfield | Slow throughput | Publication | Cost | Publication | No Image reconstruction | Publication |
Abbreviations: MIBI, Multiplexed Ion Beam Imaging; DSP, Digital Spatial Profiling.
FIGURE 2.
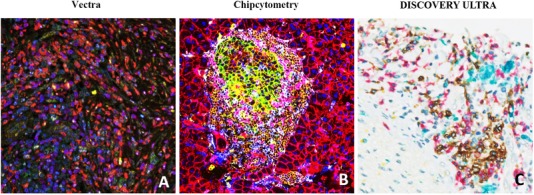
Representative mIHC/IF images captured through the Vectra, Chipcytometry, or DISCOVERY ULTRA imaging system. (A) mIHC/IF of pancreatic adenocarcinoma FFPE sections labelled with DAPI (blue), CD73 (green), CD8 (yellow), CD68 (red), FoxP3 (cyan), CD3 (magenta) and CK (orange) were scanned using the Vectra imaging system. (B) Mouse pancreas FFPE sections labelled with CD45 (brown), CD274 (green), CD3e (purple), CD4 (cyan), CD8a (pink), CD11b (yellow), CD31 (dark brown), CD326/EpCAM (red), B220 (orange), F4/80 (blue), NK1.1 (purple), Pan‐CK (maroon), Hoechst 33342 (dark blue) were scanned using the Chipcytometry imaging system. (C) Cholangiocarcinoma FFPE sections labelled with CD20 (blue), CD8 (red), CD68 (turquoise), CD3 (yellow) were scanned using the DISCOVERY ULTRA imaging system. Abbreviations: mIHC/IF, multiplex immunohistochemistry/immunofluorescence; FFPE, Formalin‐Fixed Paraffin‐Embedded; DAPI, 4′,6‐diamidino‐2‐phenylindole
3.2. Other chromogenic mIHC/IF
There are also other chromogenic mIHC/IF that have been proven to allow easy morphological control, standardized processing of large tissue sample series, whole slide imaging and easy integration into the routine clinical workflow of pathology departments [62]. For instance, Remark et al. [63] mentioned the development of the multiplexed immunohistochemical consecutive staining on a single slide (MICSSS) which utilizes iterative cycles of tagging, image scanning, and destaining of a chromogenic substrate on a single slide. This technology captures the complexity of the immunome and allow for high‐dimensional immunohistochemical analyses of the sample using routine pathology workflow and standards. It is also notable that the chromogenic mIHC/IF has also been seen to be effective and useful for research purposes when combined with color unmixing [62, 63, 64] using new algorithms which allows for a higher accuracy of image analysis. These chromogenic methods are, however, also largely bound by the same limitations as mentioned above. On the other hand, the long processing time [62, 63, 64] is also a limitation of such technique which requires improvement of automation.
3.3. Metal‐based mIHC/IF
3.3.1. Imaging mass cytometry (IMC)
One of the more commercialized and accessible multiplex tissue imaging techniques is imaging mass cytometry (IMC; Fluidigm, South San Francisco, California, USA; Figure 1B). This method combines high‐resolution laser ablation with mass cytometry for the simultaneous evaluation of more than 100 biomarkers (although this is theoretical and based on the availability of additional isotopes which makes the actual value > 40 biomarkers) from tissue sections labelled with metal‐tagged antibodies as shown in Figure 1B, with single‐cell and spatial resolution (Figure 3) [16]. The number of markers that can be examined is the main benefit of IMC but certain constraints remain. For example, the function of the selected antibodies must be thoroughly validated for each IMC run. In addition, the acquisition speed constrains the area that can be imaged by IMC. To overcome this caveat, immunofluorescence is used to select specific regions of a slide for IMC analysis. However, the number of slides that can be imaged also remains limited ‐ which begs the question if the quantity of biomarkers that can be evaluated by IMC is really of true value. Cell segmentation remains the most challenging process in multiplexed image analysis because cell borders are not always clearly defined and individual pixels may contain information from more than one cell [65]. Therefore, tissue imaging data tends to be noisier than the data obtained using suspension analysis methods.
FIGURE 3.
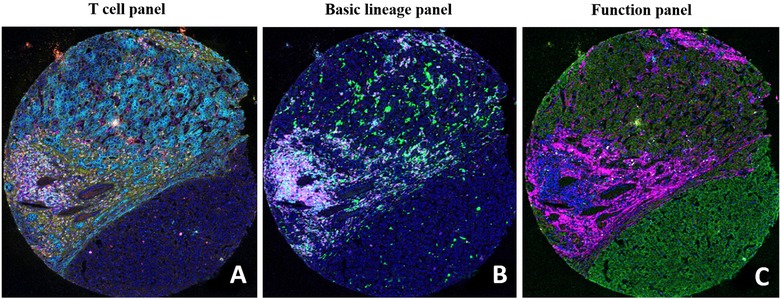
Representative IMC images of human tissue sections. Each image depicts the tumor microenvironment with the following immune cell lineages: T cell panel (A; CD45RO depicted in green, CK in cyan, collagen in yellow, CD8 in red, CD4 in magenta, and Ki67 in white), basic lineage panel (B; CD68 in green, CD20 in cyan, PD‐L1 in yellow, VISTA in red, CD3 in magenta, and CD45 in white), and function panel (C; OX40 in green, CD38 in cyan, Ki67 in yellow, ecadherin in red, collagen in magenta, and granzymeB in white). Abbreviations: IMC, imaging mass cytometry; PD‐L1, programmed cell death ligand 1
As with other platforms, there are also constraints associated with the IMC platform itself. IMC has lower sensitivity than traditional IF as it directly measures antibody abundance and lacks signal amplification or options to increase “exposure time”. This is easily achievable with fluorescence‐based imaging technologies. Furthermore, due to limited precision of the laser spot, the x‐y resolution of IMC is set at 1 μm, which is relatively low compared to other technologies. For example, mIHC/IF, which will be discussed below, offers 0.5 and 0.25 μm resolution [3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14]. This should be sufficient for cell‐level analysis since the average cell size is ∼10 μm in diameter. However, at this resolution, it is difficult to perform subcellular analysis. In addition, the acquisition process is time‐consuming as seen in Table 2. For a 1000 μm2 region of interest (ROI), ablation takes ∼2 h. This slow rate of image acquisition impairs system throughput and may introduce batch effects due to hourly and daily instrument drift. The IMC platform does not provide options to capture colorimetric images, such as hematoxylin and eosin images, and this issue can be only bypassed by examination of consecutive sections before or after IMC. IMC is also disruptive to the tissue as it is ablated and thus, not reusable. However, similar to other commercial multiplex techniques, IMC also offers a quantitative option using two free‐access software packages (Table 1), namely, CellProfiler [66, 67] and histoCAT [68]. These packages can perform cell segmentation and quantitative image analysis. Lastly, just like all the hyperplexed mIHC/IF system, there may be the problem of steric hindrance and non‐specific binding between antibodies and epitopes. Hence, proper staining controls need to be in place to safeguard the quality of the staining and imaging.
TABLE 2.
Some of the commonly adopted analytic platforms for mIHC/IF
| Characteristic | Analytic pipeline | |||
|---|---|---|---|---|
| HALO | Oncotopix | HistoCAT | QuPath | |
| Open source | No | No | Yes | Yes |
| Key advantages |
|
|
|
|
| Key disadvantages | Cost | Cost | Need to couple with other segmentation software such as CellProfiler | Coding/programming based |
| Developer | Indica Labs | Visionpharm | Bodenmiller Lab | P. Bankhead and team |
Abbreviations: mIHC/IF, multiplex immunohistochemistry/immunofluorescence; IMC, Imaging Mass Cytometry.
3.3.2. MIBI
A rather new alternative is Multiplexed Ion Beam Imaging (MIBI, Figure 1B) by IONpath (http://www.ionpath.com) where tissues are stained with up to 40‐100 metal‐labelled antibodies that are ionized by high‐energy beams to generate secondary ions which are then detected by an imaging mass spectrometer over a five‐log dynamic range [15, 69]. Similar to DISCOVERY ULTRA, it is compatible with formalin‐fixed paraffin‐embedded (FFPE) tissues which are the most common type of specimens in clinical settings [70]. However, as shown in Table 1, it can also perform imaging and analyzing of tissue samples with MIBItracker Software (IONpath, Menlo Park, California, USA), something that other platforms do not provide as a bundled analytic tool. Its ability to rescan slides at multiple resolutions helps to make sub‐cellular imaging possible with MIBIscope Software (IONpath, Menlo Park, California, USA). Hence, it not only provides information about the degree of immune cells infiltration, spatial information about cell morphology, and localization at light microscopy resolution but also measures high and low protein levels in individual cells like PD‐1 and FOXP3 which are often missed. This makes MIBI potentially very useful at identifying heterogeneity in the composition of tumors. MIBI can be quite costly as illustrated in Table 1 [71], and as more markers are added, longer acquisition times may be required. Similar to IMC, steric hindrance and non‐specific binding between antibodies and epitopes are still a challenge whereby proper controls are needed for the staining and imaging.
3.3.3. Fluorescence‐based mIHC/IF
Vectra
The recent development of another multiplex technology, TSA‐based mIHC/IF such as Vectra (Akoya Bioscience, Menlo Park, California, USA; Figure 1C), may resolve some of the issues with the use of other mIHC/IF systems. This method allows simultaneous antibody‐based detection and quantification of the expression of up to six protein markers (recently 9 or more markers) [4, 72, 3, 5, 29, 73], plus a nuclear counterstain, on a single tissue section. This provides the opportunity and possibility to acquire gold‐standard diagnostic and prognostic information. mIHC/IF may enable pathologists to refine the diagnostic criteria in lymphoid pathology and to assess the predictive value of biomarkers in specific cell types. This technique has been used to identify quantitative and spatial immune parameters in tumor cells [74], and is reproducible and reliable when used to detect the co‐expression of tumor biomarkers [3, 74].
To achieve this, mIHC/IF requires sequential cycles where individual epitopes are labelled with antibodies, followed by horseradish peroxidase‐catalyzed deposition of fluorophore‐conjugated tyramide molecules around the epitope of interest, as shown in Figure 1C. The deposited fluorophores become covalently bound to tyrosine residues on or immediately around the epitope via the activated tyramide. This allows both the primary and secondary antibodies to be stripped from the section, eliminating the risk of antibody cross‐reactivity when performing the next round of labelling which might be a challenge that many other methods providing “one‐off” staining methodology face [20]. Multiple reports have demonstrated that reproducible mIHC/IF can be achieved with a manual protocol utilizing microwave exposure to strip antibodies between each round of marker labelling [4, 72, 3, 5, 75]. However, these protocols are labor‐intensive, take several days to complete, and introduce the risk of multiple rounds of human error. This may lead to unacceptable levels of staining variability in the final image. For this reason, if mIHC/IF is to be widely adopted as a diagnostic and prognostic tool, staining and imaging protocols need to be standardized, automated and validated. Automated and semi‐automated diagnostic stainers are relatively common equipment in clinical laboratories. Moreover, it is possible to adapt mIHC/IF into existing IHC workflows without extensive re‐optimization. A fully automated staining protocol for mIHC/IF has been reported by Lim et al. [20] using a widely available and accessible clinical diagnostic autostainer, the Leica Bond Max (Leica Biosystems, Wetzlar, Germany). This automated protocol was of great significance to the field, showing that a routine‐use, two‐color diagnostic autostainer can be used to standardize the production of high‐quality, seven‐color mIHC/IF slides. This method provides significant time‐saving and improves staining uniformity compared to manual and semi‐automated staining methods. The Leica Bond Max can process 30 seven‐color mIHC/IF slides within a day while manual staining would take three full days to achieve the same. Furthermore, manual staining is comparatively challenging and may incur the risk of human errors.
This protocol affords laboratories the diagnostic‐use of Leica platform with the opportunity to perform and validate mIHC/IF for translational research and future clinical applications [29, 76, 75]. One caveat is that the run must be restarted every two‐antibody cycle. One mIHC/IF kit manufacturer has since released guidance on automating multiplex staining (PerkinElmer, Inc., Opal 4‐color and 7‐color automation IHC kits, for the Leica Biosystems BOND RX system software version 4.0; user manual 2017)in a single run using the research‐use Leica Bond Rx system (Leica Biosystems, Wetzlar, Germany). mIHC/IF offers a promising tool in this context as it allows the quantitative assessment of antigen co‐expression and spatial relationships with higher precision than other techniques, while conserving scarce tissue samples [3]. When this imaging technology is partnered with digital image analysis software, data interpretation becomes more efficient and this facilitates the study of tumor and microenvironment heterogeneity [74, 77, 23, 78]. For commercially available TSA‐based mIHC/IF, images can be acquired using a Vectra pathology imaging system microscope (Figure 2; Akoya Bioscience, Menlo Park, California, USA.) and analyzed using the inForm software (Akoya Bioscience, Menlo Park, California, USA) [55‐57, 76] and more recently the Halo software (Indica Labs, Albuquerque, New Mexico, USA) [75, 76, 79, 80].
The Vectra system is the most widely adopted mIHC/IF system currently with several notable publications [4, 72, 3, 5, 29, 73, 81, 82] over the past half a decade worldwide. Some institutions and hospitals even utilize this technology as part of their clinical laboratory tests to help clinicians in their clinical decision making and treatment plans.
3.4. Chipcytometry
Chipcytometry is a platform technology designed to control the complete pipeline aiming to optimize data quality from staining‐imaging to analysis. The tissue sections (FFPE or FF) are sectioned on adherence‐enhanced coverslips joined into the microfluidic chip, generating a closed chamber. These can be subjected to a virtually unlimited number of staining processes (iterative staining‐imaging‐bleaching/quenching cycles) with a maximum of 5 colors repetitively to generate a high number of markers. These markers can be analyzed with standard dyes like BUV395, BV421, FITC, PE and PerCP‐Cy5.5, making assay development much easier. Five plex cocktails can be generated to be used in a stepwise manner, allowing the use of different staining protocols in each cycle (buffers, temperature, time) which is very important when handling FFPE samples.
For the subsequent quantitative imaging process, Chipcytometry uses 32 bit/color true high dynamic resolution (HDR) imaging (8 decades‐4.2 billion shades of grey per marker). This is important because immunologists know from flow cytometry that biomarkers are expressed in a wide range, covering several decades of signals. This is on top of autofluorescence which can be significant in certain tissues like the liver or brain to cover an additional 2‐3 decades of fluorescent signals. Standard microscopy imagers only cover 1‐3 decades of fluorescent signals. Thus, having every pixel of the image in a linear/dynamic range in any imaged tissue type without manual interaction is only possible using high dynamic range imaging (Figure 2). Based on HDR imaging, Chipcytometry also enhances imaging and data quality. Images are taken before and after each staining, and since each pixel is in linear range, pixels or biomarker expression data can be subtracted from each other pre/post staining, thus, eliminating autofluorescence and illumination artefacts in the data (Net‐fluorescence, Net‐FL). However, up to date, Chipcytometry is still not commonly reported in literature, especially for the application of FFPE samples as shown in Table 1.
3.5. DNA barcoding‐based mIHC/IF
3.5.1. CO detection by indEXing (CODEX)
CODEX is a multiparametric imaging technology commercialized by Akoya Biosciences, Inc. (http://www.akoyabio.com). Unlike conventional IF and IHC which are limited to measuring few parameters simultaneously and single‐cell technologies like mass cytometry that have limited spatial information, CODEX uses high‐throughput technology to detect more than 50 biomarkers simultaneously in a single tissue sample. In addition, it provides information about the relative abundance and expression of these biomarkers at a spatial level [22].
By using antibodies conjugated to barcodes comprised of unique oligonucleotide sequences, the CODEX assay can target specific barcodes using a dye‐labelled Reporter (fluorophore) for highly specific detection. Cycles of labelling, imaging and removing reporters are performed by a fully automated fluidics system and images are collected and compiled across all the cycles to achieve single‐cell resolution [83]. The platform is compatible with the existing three‐colored fluorescence microscope, enabling the conversion of a simple fluorescence microscope into a tool for multiparametric imaging and cytometry [84].
CODEX antibodies have demonstrated comparable staining patterns when compared to other IF and IHC grade dye‐conjugated antibodies [84, 85]. The compatibility of the system with various tissue types including spleen, tonsil, lymph node and FFPE melanoma has been demonstrated [85]. It has also successfully achieved deep and previously uncharacterized immune profiling of the mouse splenic architecture by comparing normal murine spleens to those with autoimmune disease [86].
Although a study reported minimal steric hindrance when large panels of CODEX antibodies are used for staining, it was found that there are some cases where the signal might be lower [22]. Hence, there is still a need for the users to perform proper controls for specific antibodies and combinations of interest.
3.5.2. Digital Spatial Profiling (DSP)
DSP is a method of high‐plex spatial profiling of proteins and RNA using oligonucleotide detection technologies as shown as Figure 1D. In this method, oligonucleotide barcodes are conjugated to antibodies using a photocleavable UV light‐sensitive linker. UV light is then used to decouple the high‐plex oligo tags from the antibody and retrieved from the surface of the tissue, enabling the sample to be reused. The oligo barcodes undergo quantitative analysis and are mapped back to tissue location to allow spatial profiling at the defined region of interest. It is also a high‐throughput technology that can analyze up to 16‐20 sections per day [21].
DSP has been successfully used to characterize the tumor profile of patients with melanoma on checkpoint blockade therapy and has demonstrated a correlation between baseline immune infiltration and treatment response [87, 88]. A validation study [89] also showed a robust detection of high abundance protein and RNA targets. When compared with IHC, the dynamic range of DSP was found to be significantly greater [89]. DSP has also shown high concordance with quantitative immunofluorescence (QIF) and has been validated based on regression and outcome assessment [90].
However, there are some limitations of the DSP platform as shown in Table 1. Profiling every cell at single‐cell resolution using DSP may be impractical and cost‐prohibitive. Thus, a general advantage of imaging‐based methods compared to DSP is the ability to get multiplexed information on each cell in the tissue slice rather than high‐plex information of regions of interest as in DSP [91]. In other words, the DSP can only be visualized based on its ‘morphology kit’ of up to 3 markers, including cytokeratin and CD45. The multiplexing of that technique (up to 60 markers) does not provide any visualization. Hence, it only provides a 60‐marker snapshot of the selected regions of interest based on the visualization of the ‘morphology kit’, providing limited information on the spatial distribution of cells. Despite this, the protein plus gene expression data harvested from the same regions of interest is attractive to most of the researchers.
3.5.3. Ultivue's InSituPlex
Ultivue's InSituPlex technology is a method that detects a unique DNA barcode conjugated to the primary antibodies. This enables a higher order of multiplexing without damaging the tissue sample. The tissue sample is first incubated with barcoded antibodies. Then, it undergoes a single amplification step that increases the ratio of barcodes per antibody to enable more complementary probe strands to bind. Lastly, complementary fluorescent DNA probes are added which bind to the targets, enabling imaging of the sample.
InSituPlex is particularly promising in the field of immuno‐oncology research. A separate study using 15 different tumor markers on an FFPE tissue sample was able to yield high‐dimensional images and successfully spatially profile the different sub‐populations of immune cells (Table 1) [92]. In another study involving 5 samples from 4 tumor types (breast, lung, colon, melanoma), InSituPlex technology was able to provide analysis on the different phenotypic subtypes of immune cells, proliferative indexes and cell densities [93]. This is all achieved while maintaining the integrity of the tissue samples. Moreover, Insituplex can be adopted easily in laboratories with the usual fluorescent microscope (Figure 4), eliminating the need for any other more costly platforms (Table 1). It is also potentially much more reproducible than other multiplexing techniques available, with a coefficient of variation (CV) of less than 10% as compared to that of other IHC based assays which have a CV of less than 15%, making it suitable for translational applications in the future [94].
FIGURE 4.
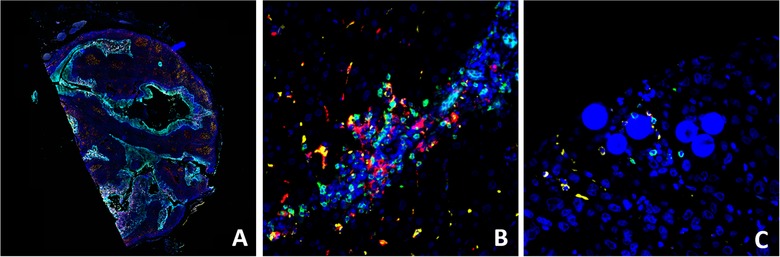
Representative Ultivue's InSituPlex images of human tissue samples labelled with CD8 (green), CD68 (yellow), PD‐L1 (red) and CK/Sox10 (cyan). Whole slide imaging of tonsil section (A), high magnification view of HCC (B), and radioembolization‐treated HCC (C, Y‐90 visible as microspheres). Abbreviations: HCC, hepatocellular carcinoma; PD‐L1, programmed cell death ligand 1
3.6. Analytic solution for multiplex IHC/IF
We have discussed some novel imaging techniques used to label multiple markers on limited samples, hereby we also introduce some of the commonly adopted analytic software that are usually coupled with these imaging platforms to analyze tissue samples. The problem with analyzing large groups of samples in traditional and manual ways by experienced pathologists, is that it is highly laborious, time‐consuming, and prone to human error. To overcome this, algorithms such as digital pathology are created to analyze digitized images of whole slides scanned at high resolution for the purposes of quality improvement, filtering, recording and segmentation of tissues. They are then validated by pathologists and provide researchers with interpretable data which they can correlate clinically, thus identify new predictive or prognostic morphological characteristics or to couple them with other modalities such as genomic analysis to better stratify tumors and potential novel therapeutic targets.
Most multiplex IHC/IF imaging solution come with their own unique bundled analytic platforms such as Inform software for Vectra, Codex Analysis suite for Codex (Table 1) which serves little interest to discuss in this review. For the purpose of this review, we would only discuss two open‐sources as well as two commercialized analytic software that are compatible with most multiplex IHC/IF platforms that we mentioned above (Table 2). Meanwhile, we would also highlight that as of now, the Food and Drug Administration (FDA) only approved the whole slide imaging (WSI) system from Philips IntelliSite Pathology Solution (PIPS) and Aperio AT2 DX System from Leica Biosystems (http://www.leicabiosystems.com), and so, most analytic software we have currently are for research purposes. We reiterate that our goal is not to evaluate the technique and technical details of these software as the key objective of this review is to provide an overview of multiplex IHC/IF technique across the process of staining‐imaging‐interpretation in order to allow readers make informed decisions when they apply multiplex IHC/IF in their translational research.
3.6.1. HALO
HALO (Indica Labs, Albuquerque, New Mexico, USA, ; https://www.indicalab.com/halo/) is an image analysis platform for quantitative tissue analysis in digital pathology, mainly used for analysis in oncology, toxicology, metabolism, neuroscience, and more [90, 91, 92, 93, 94]. It reports the morphology and multiplexed expression data of each and every cell across the entire tissue sections. Its ease of usage comes from maintaining an interactive link between cell data and cell image. Apart from being able to see the analysis outputs for a specific cell, HALO's sorting and filtering tools also allow the user to evaluate millions of other cells in comparison with corresponding cell populations. As such, it can locate cells with the highest intensity of a selected biomarker and can be used in segmenting tumor and stroma among other uses. HALO comes with a myriad of add‐ons such as the Tissue Classifier to automate tissue segmentation, FISH‐IF Quantification to contextualize protein and gene expression profile of every cell, Spatial Analysis to identify relative spatial distribution of cells, Tissue Microarray Add‐on for batch analysis of whole slide TMA images, and Serial Section Analysis to analyze serial section(s) stained for additional IHC markers. These add‐ons are also compatible with a broad spectrum of image and digital slide formats, namely JPG, TIF, ND2, MRXS, QPTIFF, component TIFF, VSI, NDPI, NDPIS, SVS, AFI, CZI, SCN, LIF, and BIF. More importantly, HALO itself is compatible and frequently used with Vectra and Insituplex as the recommended imaging platform [87, 88, 95]. While quantitative biomarker analysis platforms like HALO might accurately inform patient selection for clinicians to better predict response to treatment, the potential clinical translation of the software is shown in Table 2; HALO is limited by its cost and has yet to obtain accreditation for In Vitro Diagnostic (IVD) medical device. Up to date, it is mainly used for research purposes and by Contract Research Organisations (CROs) where it is used to meet their pre‐clinical high‐volume image analysis needs.
3.6.2. Oncotopix
As shown in Table 2 , the other commercially available image analysis software for quantitative digital pathology is Oncotopix (Visiopharm, Hoersholm, Denmark; https://www.visiopharm.com/) which is claimed to have been designed for cancer diagnostics but is also used for research and pharmacological purposes [96, 97]. It uses image analysis, machine learning, and artificial intelligence to analyze, manage and report massive data sets, connect to laboratories and their information management system, and compatible with all image/slide formats. Unlike other software in the market, it has been applied in multiple clinical uses as an in vitro diagnostic (IVD) approved device such as in breast and lung cancer detection [80, 98]. For breast cancer, it is essential to assess for estrogen receptor (ER), progesterone receptor (PR), Ki‐67 and human epidermal growth factor receptor 2 (HER2) invasive cancers and to exclude non‐invasive structures [89, 99‐101]. However, the exclusion of non‐invasive tumor manually can be challenging and labor‐intensive, especially in cancers with a diffuse growth pattern. The Invasive Tumor Detection APP under Oncotopix automatically detects the invasive tumor. The software combines with a full biomarker analysis workflow, to allow automatic hot spot detection, accurate biomarker quantification, and visualization of tumor heterogeneity. This is based on a physical double stain of p63+CK7/19−‐ invasive tumor components are identified and are labelled as regions positive for CK, whereas non‐invasive tumor components (regions positive for both CK and p63) are excluded. Similarly, for lung cancer, the tissue is first categorized into tumor and stroma using cytokeratin staining. It then undergoes tissue segmentation where individual cells are identified by the presence of DNA (DAPI) and membrane proteins. The phenotyping algorithm then scans multiple images, automatically identifies cellular phenotypes, and categorizes them accordingly. Using Phenotypic Matrix, Phenotypic Profile and/or tSNE plots, the phenotypes can be summed up and further visualized for a better understanding of the data. This APP is made to work with amplified fluorescence signals (unmixed with multispectral imaging) where each stain can be spectrally isolated from background autofluorescence and spectrally overlapping fluorophores.
3.6.3. HistoCAT
HistoCAT (Fluidigm, South San Francisco, California, USA) is an interactive free‐access computational platform that makes quantitative analysis of highly multiplexed, single‐cell‐resolved tissues [95, 96]. HistoCAT combines high‐dimensional image visualization, analysis methods for cell phenotype characterization, and novel algorithms for the comprehensive study of cell‐to‐cell interactions and the social networks of cells within complex tissues [95].
HistoCAT operates by first extracting data of individual cells from images, including the quantity of all measured markers for a cell and within the area of interest, morphological features like cell size and shape, and features of the cell's environment such as cell neighbors and crowding. This data is synthesized and represented in a flow cytometry standard format (.fcs) file for further analysis using the HistoCAT or other compatible analytic software. HistoCAT promotes the idea of “Round‐trip” analyses, by comparing the single‐cell phenotypes and their interactions of a specific area of an image to the entire dataset, and back to the visualization of unique cells on images which would help users define and understand important cell populations and their spatial context within tissues. HistoCAT also possess a novel algorithm which can detect proximate cell‐to‐cell interactions that are more frequently detected than expected by chance and identify the more significant interactions and unique cell environments across entire datasets and cohorts. As demonstrated in Table 1 and 2, HistoCAT is also one of the recommended image analysis software for images processed through imaging mass cytometry, as mentioned above [97, 98], where more than 40 unique metal‐isotope‐labeled antibodies can be detected simultaneously at a resolution comparable to fluorescence microscopy.
The software is, however, limited by its ability to perform tissue segmentation, where it is coupled with other cell segmentation software such as the CellProfiler (https://Cellprofiler.org). Besides, a small percentage of cells will overlap in 5‐μm thick tissue sections, leading to only a portion of a cell being present within the analyzed section. Small differences in segmentation between adjacent cells can thus result in a small spillover of signal between neighboring cells. HistoCAT addresses this by allowing for cell boundaries to be defined using a combination of different markers to improve the accuracy of segmentation masks [95]. Overall, HistoCAT is still in the phase of constant upgrade and maintenance with the latest version uploaded in August 2019 by Bodenmiller Labs.
3.6.4. QuPath
QuPath (https://qupath.github.io) is another high‐throughput, free access image analysis software for whole slide image analysis with robust batch‐processing and scripting functionalities [99]. It is capable of handling large whole slide imaging data without needing to crop or down‐sample the image data into a manageable size for subsequent analysis. It also serves as a platform for researchers to develop and share novel algorithms and workflows to analyze multiplexed tissue images as shown in Table 2. A common workflow of analysis in QuPath for a tissue microarray (TMA) sample starts with creating a multi‐slide project with automated TMA dearraying and stain estimation. This is followed by the single‐cell analysis which includes cell detection, feature computation and trainable cell classification. The data is then integrated and further analyzed through batch processing and survival analysis. QuPath also offers supplementary functionalities such as support for whole face tissue sections and fluorescence image analysis, data transfer with existing software (e.g. ImageJ and MATLAB), scriptable data mining, and rapid processing and export of spatial, morphological and intensity‐based data. There is also good interobserver reproducibility of bioimage data using QuPath in the TMA setting [100].
The technology is, however, limited by its ability to perform digital scoring of biomarkers with more complex patterns of staining, such as mismatch repair proteins or immune checkpoint inhibitors like PD‐L1. This arises from the nature of staining multiple cell populations (tumor and immune) and the distinction between tumor and immune cell staining being difficult due to a patchy pattern of staining [100]. Several studies have illustrated the potential of QuPath in TMA biomarker scoring [99, 101, 102], and it is also currently actively utilized by researchers to analyze mIHC/IF samples in the study of cancer immunotherapy [103]. As a whole, QuPath is still a work in progress, has only become publicly available in 2016 and is now being constantly updated with new features by its owner, P. Bankhead.
3.6.5. 3‐dimension (3D) Imaging
An emerging alternative to the 2‐dimension (2D) imaging techniques mentioned previously would be 3D imaging which allows for a more detailed reconstruction of the molecular properties in a 3D tissue sample. Intravital microscopy was the earliest tool used in 3D imaging. It is a technique that enables visualization of the cellular processes in real‐time and has been used extensively to study dynamic processes in live animals [104]. However, the imaging volume is limited, and a significant portion of imaged tissues cannot be analyzed. This was followed by the development of ‘Histo‐cytometry’ which involves staining a sample with up to 14 antibodies at one time and performing quantitative analysis whereby each cell has defined parameters which can be visualized on the tissue sample. ‘Histo‐cytometry’ demonstrated the ability to provide cellular subset discrimination of CD11c+ dendritic cells, CD8, CD4 and B cells that was comparable to the findings from flow cytometric analysis of the dissociated cells[105]. Following this, new tools such as Clearing‐enhanced 3D (Ce3D) imaging have emerged. Ce3D imaging has superior scanning depth and allows for adequate visualization of large tissue volumes, while preserving the capacity for multiplex antibody staining. Furthermore, image analysis can be done using advanced platforms such as volumetric histocytometry to quantitatively characterize the cells with respect to their spatial positioning, phenotypic and functional markers [106]. Ce3D can also be used to examine tissue samples in any plane of interest. This is particularly important as it can reduce sampling errors in samples that have an uneven and asymmetrical distribution of cells, such as in a tumor with immune cell infiltrate [107]. Ce3D has also been used successfully in the visualization of cellular anatomical relationships such as in dense dendritic cell aggregates in the thymic medullary regions [106]. However, Ce3D has some limitations. There may be problems with antibody penetration in thick tissues. For example, in brain tissues that are rich in lipids, longer incubation times of up to 2 weeks are required to penetrate 1 mm of the tissue [107]. While 3D imaging is not as widely accessible to most labs and regions of the world as other 2D imaging techniques, it still remains an exciting area of ongoing development with great potential for application in the era of cancer immunotherapy.
3.7. Is mIHC/IF the more worthwhile technique?
In theory, using conventional IHC, one could perform 6 single IHC staining procedures on 6 consecutive sections to achieve a “multiplex effect” similar to mIHC/IF, averting the need for investment in a multiplex microscopy machine, or the additional costs relating to staining. However, this line of thought is of significant concern. To begin, for a typical cell (10 μm in diameter) [108], the conventional consecutive section method may only produce 3 consecutive sections of the same cell. This would only permit cross‐referencing of a single cell with 3 markers. It is also difficult to achieve this for 6 consecutive sections, with sections primarily standardized to 4‐5 μm.
Furthermore, tissue depletion is a serious concern in clinical practice, especially during clinical trials and when using biopsy samples. Consider a lung cancer patient who is being tested prior to enrolment in immune‐checkpoint molecule therapy. There may be little tissue left after routine diagnostic panels for TTF1, Napsin, p63 and cytokeratin. Additional tissue may be used for molecular testing, such as for EGFR mutations and tumor mutational burden. It may be difficult to obtain even one section of useful tissue to image biomarkers such as PD‐L1, PD‐1, CD68, CD45, CD8 and CD3 [24‐26, 28, 109, 110], or other markers which have been reported or are presently being explored. Cutting 6 consecutive sections may be impossible in this scenario, and this patient would lose the opportunity to benefit from the advances being made in precision medicine. Another consideration is the difficulties that even senior and experienced pathologists may encounter when attempting to “cross‐refer” two or more consecutive slides to observe co‐localization of more than two markers.
Lastly, taking fluorescent‐based mIHC/IF as an example, and based on the listed price provided by the manufacturer (Opal 4‐color and 7‐color automation IHC kits; PerkinElmer, Inc.), one kit, which can be used for 50 multiplex runs, costs ∼2000 USD. This means that a single run costs ∼40 USD in addition to the automated staining protocol. This cost seems affordable compared to conventional IHC on 6 consecutive sections, which needs to factor in the relevant cost of 6 consecutive slides, the cost of 5 extra glass slides and coverslips, and additional manpower. Moreover, the cost of the machine itself represents a one‐off investment. Such a machine has the capacity and throughput to cater to multiple research projects simultaneously be supported by research grants. More importantly, most vendors nowadays seek a business model whereby the machine is provided as a loan or in‐kind contribution if the amount of reagents and consumables purchased reaches certain targets. This requires no upfront investment.
3.8. Prognostic and predictive cancer markers identified by mIHC/IF
Multiple laboratories have utilized mIHC/IF to report prognostic or even predictive values of immune subsets in several types of cancer. Recently, Yeong et al. [81] have demonstrated an optimized multiplex IHC/IF based translational assay to report multiple clones of PD‐L1 for clinical samples for personalized immunotherapy. The group compared the multiplex IHC/IF scoring with manual scoring from 4 pathologists and achieved moderate‐to‐strong correlations which the individual concordance rates ranged from 67%‐100%, with Spearman's rank correlation coefficient values of up to 0.88. The study demonstrated a promising tool in the era of cancer immunotherapy as it could simultaneously detect and quantify PD‐L1 labelling with multiple antibody clones to allow accurate evaluation of tumor and immune cells.
As mentioned above, Lu et al. [1] published a meta‐analysis of different strategies for predicting response to anti PD1/PDL1 treatment which adopted multiple studies of TSA‐based mIHC/IF assays. These studies evaluated parameters such as PD‐1 to PD‐L1 proximity [73, 75], CD8+ cell density within a specific TME compartment (for instance: intratumoral/peritumoral defined by IHC/IF tumor marker [23, 29]), or co‐expressed markers indicating T‐cell activation [49, 111] as summarized in Table 3. For instance, Johnson et al. [112] and Giraldo et al. [113] evaluated PD‐1 to PD‐L1 proximity using TSA‐based mIHC/IF in metastatic melanoma in 166 patients and Merkel cell carcinoma in 42 patients, respectively. Both papers reported that greater PD‐1 to PD‐L1 proximity in patients led to better clinical response to anti‐PD1 therapy, and their overall survival rates were also improved in patients treated with anti‐PD1 therapy for metastatic melanoma. Furthermore, Johnson et al. [112] also mentioned that this can be a factor for reliably selecting patients to undergo immunotherapy in metastatic melanoma (Table 3).
TABLE 3.
List of papers using mIHC/IF in the meta‐analysis of Lu et al. [1]
| Author | Tumor | Year | Marker |
|---|---|---|---|
| Gettinger et al. [125] | Non‐small cell lung cancer | 2018 | CK, CD3, Ki‐67, GZB |
| Johnson et al. [126] | Melanoma | 2018 | PD‐1, PD‐L1, IDO‐1, HLA‐DR, CD11b or S100 |
| Giraldo et al. [127] | Merkel cell carcninoma | 2018 | CD8, CD20, PD‐1, PD‐L1, CD68, FOXP3 and NSE |
| Wong et al. [128] | Melanoma | 2018 | S100 and HMB45, CD3, CD4, CD8, CD20, GZB, Ki67 |
| Tumeh et al. [129] | Melanoma | 2014 | CD8, CD4, Ki‐67, PD‐1, PD‐L1 |
| Mazzaschi et al. [130] | Non‐small cell lung cancer | 2018 | CD3, CD8, CD4, PD‐1, PD‐L1,CD57, FOXP3, CD25, GZB |
Abbreviations: mIHC/IF, multiplex immunohistochemistry/immunofluorescence; GZB, GranzymeB; NSE, neuron‐specific enolase;
PD‐1, Programmed death protein 1; PD‐L1, Programmed death‐ligand 1.
On the other hand, Yeong et al. [34, 114‐118] have previously used mIHC/IF to reveal the prognostic values of multiple tumor‐infiltrating lymphocyte (TIL) subsets in Asian triple negative breast cancer, such as FOXP3+ regulatory T cells (Treg), CD20+ B cells, CD38+ plasma cells, PD‐1+ CD8+ T cells and proliferating immune subsets. The quantitative approach of these works was convincingly validated with manual scoring by pathologists. Savas et al. [119] recently reported that CD8+ tissue‐resident memory T cells (TRM) contributed to breast cancer immunosurveillance and were the key biomarkers of modulation by immune checkpoint inhibitors. In addition, Feng et al. [57] reported that multiparametric immune profiling can be defined using mIHC/IF and may be useful for stratifying patients with oral squamous cell cancer for clinical trials. Most notably, this study provided unprecedented reports on the use of quantitative spatial distribution of immune subsets as a prognostic parameter. The number of FOXP3+ and PD‐L1+ cells present within 30 μm of CD8+ T cells was significantly associated with the presence of a high number of suppressive elements close to CD8+ T cells and reduced overall survival.
Similarly, Garnelo et al. [8] suggested that the distance between tumor‐infiltrating T and B cells represented a functional interaction between them, which was linked to enhanced local immune activation in the microenvironment. This contributed to improved clinical outcomes for patients with hepatocellular carcinoma (HCC) [8]. Subsequently, the same group reported that TRM were enriched in the HCC tumor microenvironment compared to adjacent normal liver tissues, and represented an immune‐exhausted phenotype [120]. Upon combining mIHC/IF with time‐of‐flight mass cytometry (CyTOF), Lim et al. [121] accurately delineated multiple TIL subsets with increasingly suppressive phenotypes into two immunosuppressive subsets: FOXP3+ CD4+ Treg and PD‐1+ CD8+ T cells. Specifically, multiple exhaustion markers were highly expressed in tumor‐infiltrating memory TRM and CD8+ effector memory T cells (TEM), particularly PD‐1, which is known to be associated with cancer immune evasion. In vitro studies revealed that these cells were not only associated with hepatitis B virus‐related HCC [121] but were key subsets that predicted responsiveness to PD‐1 blockade treatment [120]. These findings open up new avenues for the identification of novel biomarkers, especially for immunotherapy in HCC, which was approved by the FDA in 2017 [122, 123, 124].
4. CONCLUSION
In conclusion, emerging multiplex IHC and immunofluorescence technologies are promising in the field of cancer immunotherapy. Unlike conventional IHC which only allows the labelling of one single marker in a tissue sample, multiplex IHC is able to detect multiple markers from a single tissue sample while providing comprehensive information about the cell composition and spatial arrangement.
DISCOVERY ULTRA provides a promising platform to overcome the limitations of conventional IHC by allowing multiplexed analysis of numerous biomarkers. However, it remains limited in the visualization of co‐localized biomarkers. Similarly, metal‐based mIHC/IF such as MIBI and IMC are able to detect up to 100 markers on a single tissue sample but the process is time‐consuming, costly and less sensitive than IF due to the nature of meta‐conjugation. As such, DNA barcoding‐based mIHC/IF such as CODEX, DSP and Insituplex appear to circumvent these problems. In addition to multiplexed analysis, these techniques are able to provide comprehensive cellular spatial information, allowing greater insight into the pathogenesis of cancer and responsiveness to immunotherapy. These techniques also preserve tissue samples, allowing reusability for other further studies. However, cost‐effectiveness and practicality of such detailed spatial imaging remains a concern. Alternatives such as fluorescence‐based mIHC/IF including Vectra and Chipcytometry are also useful especially in the practical setting as these systems provide a complete solution, from staining‐imaging to analysis protocol. On the other hand, 3D imaging for multiplexing remains a promisingly new approach in the foreseeable future. Last but not least, we also discussed that both free and commercialized imaging software have their own advantages and disadvantages in regard to their application in both the clinical and research setting. It is imperative for the user to understand the constraints and goals of their own clinical work or research before they can decide on which software would be most ideal in achieving their goals.
Amongst the numerous mIHC/IF techniques, hopefully, all vendors would work to circumvent some of the aforementioned limitations. In due course, consensus will be reached and there will be a widely adopted, high‐throughput quantitative multiplex staining technique with high reproducibility, reduced turnover time, affordable cost, and a standardized quantitative protocol. We believe that the mIHC/IF technique has great potential in translational research and clinical practice in the immediate future, particularly in this era of cancer immunotherapy.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
Not applicable.
AUTHORS' CONTRIBUTIONS
J.Y. and T.K.H.L. conceived, directed and supervised the review. Y.T.F.W. and J.L.C.T provided scientific inputs from pathology and histology perspectives. T.W.C.C., S.N.N., C.H.Y., H.H.M.N. and D.W. drafted the manuscript with the assistance of JY, with final review from all authors.
All authors read and approved the final manuscript
ACKNOWLEDGEMENTS
We wish to thank Fluidigm Corporation, ZELLKRAFTWERK, Ultivue, Inc., PerkinElmer, Inc. and Dr. Valerie Chew for the collaboration to generate some of the images, as well as Dr. Alice Bridges for critical review of the manuscript.
Tan WCC, Nerurkar SN, Cai HY, et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Communications. 2020;40:135–153. 10.1002/cac2.12023
Contributor Information
Joe Yeong, Email: yeongps@imcb.a-star.edu.sg.
Tony Kiat Hon Lim, Email: lim.kiat.hon@singhealth.com.sg.
REFERENCES
- 1. Lu S, Stein JE, Rimm DL, Wang DW, Bell JM, Johnson DB, et al. Comparison of biomarker modalities for predicting response to PD‐1/PD‐L1 checkpoint blockade: A systematic review and meta‐analysis. JAMA Oncol. 2019. 10.1001/jamaoncol.2019.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zrazhevskiy P, Gao X. Quantum dot imaging platform for single‐cell molecular profiling. Nat Commun. 2013;4:1619 10.1038/ncomms2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70(1):46–58. 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 4. Abel EJ, Bauman TM, Weiker M, Shi F, Downs TM, Jarrard DF, et al. Analysis and validation of tissue biomarkers for renal cell carcinoma using automated high‐throughput evaluation of protein expression. Hum Pathol. 2014;45(5):1092–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lovisa S, LeBleu VS, Tampe B, Sugimoto H, Vadnagara K, Carstens JL, et al. Epithelial‐to‐mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. 2015;21(9):998–1009. 10.1038/nm.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, et al. Interaction between tumour‐infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2015;15(310814):2015–310814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yeong J, Thike AA, Lim JC, Lee B, Li H, Wong SC, et al. Higher densities of Foxp3(+) regulatory T cells are associated with better prognosis in triple‐negative breast cancer. Breast Cancer Res Treat. 2017;163(1):21–35. [DOI] [PubMed] [Google Scholar]
- 8. Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, et al. Interaction between tumour‐infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2017;66(2):342–51. 10.1136/gutjnl-2015-310814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim JCT, Yeong JPS, Lim CJ, Ong CCH, Chew VSP, Ahmed SS, Tan PH, & Iqbal J. An automated staining protocol for 7‐colour immunofluorescence of human tissue sections for diagnostic and prognostic use. Journal of The Royal College of Pathologists of Australasia. In Press. [Google Scholar]
- 10. Esbona K, Inman D, Saha S, Jeffery J, Schedin P, Wilke L, et al. COX‐2 modulates mammary tumor progression in response to collagen density. Breast Cancer Res. 2016;18(1):35 10.1186/s13058-016-0695-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mlecnik B, Bindea G, Kirilovsky A, Angell HK, Obenauf AC, Tosolini M, et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci Transl Med. 2016;8(327). [DOI] [PubMed] [Google Scholar]
- 12. Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD‐1 blockade with pembrolizumab in advanced merkel‐cell carcinoma. New Engl J Med. 2016;374(26):2542–52. 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng Z, Jensen SM, Messenheimer DJ, Farhad M, Neuberger M, Bifulco CB, et al. Multispectral imaging of T and B cells in murine spleen and tumor. J Immunol. 2016;196(9):3943–50. 10.4049/jimmunol.1502635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yeong J, Lim JCT, Lee B, Li H, Chia N, Ong CCH, et al. High densities of tumor‐associated plasma cells predict improved prognosis in triple negative breast cancer. Front Immunol. 2018;9(1209). 10.3389/fimmu.2018.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–42. 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–22. 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 17. Jung E, Lee J, Hong HJ, Park I, Lee Y. RNA recognition by a human antibody against brain cytoplasmic 200 RNA. RNA (New York, NY: ). 2014;20(6):805–14. 10.1261/rna.040899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ke J, Chen RZ, Ban T, Zhou XE, Gu X, Tan MH, et al. Structural basis for RNA recognition by a dimeric PPR‐protein complex. Nat Struct Mol Biol. 2013;20(12):1377–82. 10.1038/nsmb.2710. [DOI] [PubMed] [Google Scholar]
- 19. Bodenmiller B. Multiplexed epitope‐based tissue imaging for discovery and healthcare applications. Cell Syst. 2016;2(4):225–38. 10.1016/j.cels.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 20. Lim JCT, Yeong JPS, Lim CJ, Ong CCH, Wong SC, Chew VSP, et al. An automated staining protocol for seven‐colour immunofluorescence of human tissue sections for diagnostic and prognostic use. Pathology. 2018;50(3):333–41. 10.1016/j.pathol.2017.11.087. [DOI] [PubMed] [Google Scholar]
- 21. Digital Spatial Profiling (DSP) Technology . 2019. https://www.nanostring.com/scientific-content/technology-overview/digital-spatial-profiling-technology.
- 22. Dakshinamoorthy G, Singh J, Kim J, Nikulina N, Bashier R, Mistry S, et al. Abstract 490: Highly multiplexed single‐cell spatial analysis of tissue specimens using CODEX. Cancer Res. 2019;79(13 Supplement):490 10.1158/1538-7445.Am2019-490. [DOI] [Google Scholar]
- 23. Mazzaschi G, Madeddu D, Falco A, Bocchialini G, Goldoni M, Sogni F, et al. Low PD‐1 expression in cytotoxic CD8(+) tumor‐infiltrating lymphocytes confers an immune‐privileged tissue microenvironment in NSCLC with a prognostic and predictive value. Clin Cancer Res. 2018;24(2):407–19. 10.1158/1078-0432.CCR-17-2156. [DOI] [PubMed] [Google Scholar]
- 24. Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti‐PD‐L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–7. 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herbst RS, Baas P, Kim DW, Felip E, Perez‐Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD‐L1‐positive, advanced non‐small‐cell lung cancer (KEYNOTE‐010): a randomised controlled trial. Lancet. 2016;387(10027):1540–50. 10.1016/s0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 26. Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non‐small‐cell lung cancer. N Engl J Med. 2015;372(21):2018–28. 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 27. Yeong J, Lim JCT, Lee B, Li H, Ong CCH, Thike AA, et al. Prognostic value of CD8 + PD‐1+ immune infiltrates and PDCD1 gene expression in triple negative breast cancer. J Immunother Cancer. 2019;7(1):34 10.1186/s40425-019-0499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, et al. A transcriptionally and functionally distinct PD‐1+ CD8+ T cell pool with predictive potential in non‐small‐cell lung cancer treated with PD‐1 blockade. Nat Med. 2018;24(7):994–1004. 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD‐1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–71. 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, et al. Tumor antigen‐specific CD8 T cells infiltrating the tumor express high levels of PD‐1 and are functionally impaired. Blood. 2009;114(8):1537–44. 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Badoual C, Hans S, Merillon N, Van Ryswick C, Ravel P, Benhamouda N, et al. PD‐1‐expressing tumor‐infiltrating T cells are a favorable prognostic biomarker in HPV‐associated head and neck cancer. Cancer Res. 2013;73(1):128–38. 10.1158/0008-5472.can-12-2606. [DOI] [PubMed] [Google Scholar]
- 32. Gehring AJ, Ho ZZ, Tan AT, Aung MO, Lee KH, Tan KC, et al. Profile of tumor antigen‐specific CD8 T cells in patients with hepatitis B virus‐related hepatocellular carcinoma. Gastroenterology. 2009;137(2):682–90. 10.1053/j.gastro.2009.04.045. [DOI] [PubMed] [Google Scholar]
- 33. Muenst S, Hoeller S, Willi N, Dirnhofera S, Tzankov A. Diagnostic and prognostic utility of PD‐1 in B cell lymphomas. Dis Markers. 2010;29(1):47–53. 10.3233/dma-2010-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yeong J, Lim JCT, Lee B, Li H, Chia N, Ong CCH, et al. High densities of tumor‐associated plasma cells predict improved prognosis in triple negative breast cancer. Front Immunol. 2018;9:1209 10.3389/fimmu.2018.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choueiri TK, Figueroa DJ, Fay AP, Signoretti S, Liu Y, Gagnon R, et al. Correlation of PD‐L1 tumor expression and treatment outcomes in patients with renal cell carcinoma receiving sunitinib or pazopanib: results from COMPARZ, a randomized controlled trial. Clin Cancer Res. 2015;21(5):1071–7. 10.1158/1078-0432.ccr-14-1993. [DOI] [PubMed] [Google Scholar]
- 36. Saito T, Nishikawa H, Wada H. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. 2016;22(6):679–84. 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 37. Hainaut P, Plymoth A. Targeting the hallmarks of cancer: towards a rational approach to next‐generation cancer therapy. Curr Opin Oncol. 2013;25(1):50–1. 10.1097/CCO.0b013e32835b651e. [DOI] [PubMed] [Google Scholar]
- 38. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- 39. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 40. Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG, et al. An international Ki67 reproducibility study. J Natl Cancer Inst. 2013;105(24):1897–906. 10.1093/jnci/djt306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Varga Z, Diebold J, Dommann‐Scherrer C, Frick H, Kaup D, Noske A, et al. How reliable is Ki‐67 immunohistochemistry in grade 2 breast carcinomas? A QA study of the Swiss Working Group of Breast‐ and Gynecopathologists. PLoS One. 2012;7(5):e37379 10.1371/journal.pone.0037379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng CL, Thike AA, Tan SYJ, et al. Expression of FGFR1 is an independent prognostic factor in triple‐negative breast cancer. Breast Cancer Res Treat. 2015;151(1). [DOI] [PubMed] [Google Scholar]
- 43. Matsumoto H, Koo SL, Dent R, Tan PH, Iqbal J. Role of inflammatory infiltrates in triple negative breast cancer. J Clin Pathol. 2015;68(7):506–10. 10.1136/jclinpath-2015-202944. [DOI] [PubMed] [Google Scholar]
- 44. Vincent‐Salomon A, Gruel N, Lucchesi C, MacGrogan G, Dendale R, Sigal‐Zafrani B, et al. Identification of typical medullary breast carcinoma as a genomic sub‐group of basal‐like carcinomas, a heterogeneous new molecular entity. Breast Cancer Res. 2007;9(2):R24 10.1186/bcr1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gerdes J, Dallenbach F, Lennert K, Lemke H, Stein H. Growth fractions in malignant non‐Hodgkin's lymphomas (NHL) as determined in situ with the monoclonal antibody Ki‐67. Hematol Oncol. 1984;2(4):365–71. [DOI] [PubMed] [Google Scholar]
- 46. Li LT, Jiang G, Chen Q, Zheng JN. Ki67 is a promising molecular target in the diagnosis of cancer (Review). Molecular Medicine Reports. 2015;11:1566–72. [DOI] [PubMed] [Google Scholar]
- 47. Mohammed ZMA, McMillan DC, Elsberger B, Going JJ, Orange C, Mallon E, et al. Comparison of Visual and automated assessment of Ki‐67 proliferative activity and their impact on outcome in primary operable invasive ductal breast cancer. Br J Cancer. 2012;106:383 10.1038/bjc.2011.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Munzone E, Botteri E, Sciandivasci A, Curigliano G, Nole F, Mastropasqua M, et al. Prognostic value of Ki‐67 labeling index in patients with node‐negative, triple‐negative breast cancer. Breast Cancer Res Treat. 2012;134(1):277–82. 10.1007/s10549-012-2040-6. [DOI] [PubMed] [Google Scholar]
- 49. Gettinger SN, Choi J, Mani N, Sanmamed MF, Datar I, Sowell R, et al. A dormant TIL phenotype defines non‐small cell lung carcinomas sensitive to immune checkpoint blockers. Nat Commun. 2018;9(1):3196 10.1038/s41467-018-05032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Curigliano G, Burstein HJ, Winer EP, Gnant M, Dubsky P, Loibl S, et al. De‐escalating and escalating treatments for early‐stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. 2017;28(8):1700–12. 10.1093/annonc/mdx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tay TKY, Thike AA, Pathmanathan N, Jara‐Lazaro AR, Iqbal J, Sng ASH, et al. Using computer assisted image analysis to determine the optimal Ki67 threshold for predicting outcome of invasive breast cancer. Oncotarget. 2018;9(14):11619–30. 10.18632/oncotarget.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Koopman T, Buikema HJ, Hollema H, de Bock GH, van der Vegt B. Digital image analysis of Ki67 proliferation index in breast cancer using virtual dual staining on whole tissue sections: clinical validation and inter‐platform agreement. Breast Cancer Res Treat. 2018;169(1):33–42. 10.1007/s10549-018-4669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Laurinavicius A, Plancoulaine B, Laurinaviciene A, Herlin P, Meskauskas R, Baltrusaityte I, et al. A methodology to ensure and improve accuracy of Ki67 labelling index estimation by automated digital image analysis in breast cancer tissue. Breast Cancer Research : BCR. 2014;16(2):R35–R. 10.1186/bcr3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhong F, Bi R, Yu B, Yang F, Yang W, Shui R. A comparison of visual assessment and automated digital image analysis of Ki67 labeling index in breast cancer. PLoS One. 2016;11(2):e0150505 10.1371/journal.pone.0150505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fiore C, Bailey D, Conlon N, Wu X, Martin N, Fiorentino M, et al. Utility of multispectral imaging in automated quantitative scoring of immunohistochemistry. J Clin Pathol. 2012;65(6):496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Abel EJ, Bauman TM, Weiker M, Shi F, Downs TM, Jarrard DF, et al. Analysis and validation of tissue biomarkers for renal cell carcinoma using automated high‐throughput evaluation of protein expression. Hum Pathol. 2014;45(5):1092–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Feng Z, Bethmann D, Kappler M, Ballesteros‐Merino C, Eckert A, Bell RB, et al. Multiparametric immune profiling in HPV– oral squamous cell cancer. JCI Insight. 2017;2(14). 10.1172/jci.insight.93652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang W, Hubbard A, Jones T, Racolta A, Bhaumik S, Cummins N, et al. Fully automated 5‐plex fluorescent immunohistochemistry with tyramide signal amplification and same species antibodies. Lab Invest. 2017;97(7):873–85. 10.1038/labinvest.2017.37. [DOI] [PubMed] [Google Scholar]
- 59. Ma Z, Shiao SL, Yoshida EJ, Swartwood S, Huang F, Doche ME, et al. Data integration from pathology slides for quantitative imaging of multiple cell types within the tumor immune cell infiltrate. Diagnostic Pathology. 2017;12(1):69 10.1186/s13000-017-0658-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Day WA, Lefever MR, Ochs RL, Pedata A, Behman LJ, Ashworth‐Sharpe J, et al. Covalently deposited dyes: a new chromogen paradigm that facilitates analysis of multiple biomarkers in situ. Lab Invest. 2017;97(1):104–13. 10.1038/labinvest.2016.115. [DOI] [PubMed] [Google Scholar]
- 61. McNamara G, Difilippantonio MJ, Ried T. Microscopy and image analysis. Curr Protoc Hum Genet. 2005;Chapter 4:Unit‐4. 10.1002/0471142905.hg0404s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ilie M, Beaulande M, Ben Hadj S, Chamorey E, Schiappa R, Long‐Mira E, et al. Chromogenic Multiplex Immunohistochemistry Reveals Modulation of the Immune Microenvironment Associated with Survival in Elderly Patients with Lung Adenocarcinoma. Cancers. 2018;10(9). 10.3390/cancers10090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Remark R, Merghoub T, Grabe N, Litjens G, Damotte D, Wolchok JD, et al. In‐depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Science Immunology. 2016;1(1):aaf6925 10.1126/sciimmunol.aaf6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen T, Srinivas C. Group sparsity model for stain unmixing in brightfield multiplex immunohistochemistry images. Comput Med Imaging Graph. 2015;46:30–9. 10.1016/j.compmedimag.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 65. Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14(9):873–6. 10.1038/nmeth.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, et al. Improved structure, function and compatibility for CellProfiler: modular high‐throughput image analysis software. Bioinformatics. 2011;27(8):1179–80. 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14:873 10.1038/nmeth.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Levenson RM, Borowsky AD, Angelo M. Immunohistochemistry and mass spectrometry for highly multiplexed cellular molecular imaging. Lab Invest. 2015;95(4):397–405. 10.1038/labinvest.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–42. 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Parra ER, Francisco‐Cruz A, Wistuba, II. State‐of‐the‐Art of Profiling Immune Contexture in the Era of Multiplexed Staining and Digital Analysis to Study Paraffin Tumor Tissues. Cancers (Basel). 2019;11(2). 10.3390/cancers11020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Feng Z, Jensen SM, Messenheimer DJ, Farhad M, Neuberger M, Bifulco CB, et al. Multispectral imaging of T and B cells in murine spleen and tumor. J Immunol. 2016;196(9):3943–50. 10.4049/jimmunol.1502635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Johnson DB, Bordeaux J, Kim JY, Vaupel C, Rimm DL, Ho TH, et al. Quantitative spatial profiling of PD‐1/PD‐L1 interaction and HLA‐DR/IDO‐1 predicts improved outcomes of Anti‐PD‐1 therapies in metastatic melanoma. Clin Cancer Res. 2018;24(21):5250–60. 10.1158/1078-0432.CCR-18-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feng Z, Jensen SM. Multispectral imaging of T and B Cells in murine spleen and tumor. J Immunol. 2016;196(9):3943–50. 10.4049/jimmunol.1502635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Giraldo NA, Nguyen P, Engle EL, Kaunitz GJ, Cottrell TR, Berry S, et al. Multidimensional, quantitative assessment of PD‐1/PD‐L1 expression in patients with Merkel cell carcinoma and association with response to pembrolizumab. J Immunother Cancer. 2018;6(1):99 10.1186/s40425-018-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mascaux C, Angelova M, Vasaturo A, Beane J, Hijazi K, Anthoine G, et al. Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature. 2019. 10.1038/s41586-019-1330-0. [DOI] [PubMed] [Google Scholar]
- 77. Cregger M, Berger AJ, Rimm DL. Immunohistochemistry and quantitative analysis of protein expression. Arch Pathol Lab Med. 2006;130(7):1026–30. 10.1043/1543-2165(2006)130[1026:iaqaop]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 78. Simoni Y, Becht E, Fehlings M, Loh CY, Koo SL, Teng KWW, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557(7706):575–9. 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 79. Chen J, Yang H, Teo ASM, Amer LB, Sherbaf FG, Tan CQ, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet. 2020;52(2):177–86. 10.1038/s41588-019-0569-6. [DOI] [PubMed] [Google Scholar]
- 80. Lam JH, Ng HHM, Lim CJ, Sim XN, Malavasi F, Li H, et al. Expression of CD38 on macrophages predicts improved prognosis in hepatocellular carcinoma. Front Immunol. 2019;10(2093). 10.3389/fimmu.2019.02093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yeong J, Tan T, Chow ZL, Cheng Q, Lee B, Seet A, et al. Multiplex immunohistochemistry/immunofluorescence (mIHC/IF) for PD‐L1 testing in triple‐negative breast cancer: a translational assay compared with conventional IHC. J Clin Pathol. 2020. 10.1136/jclinpath-2019-206252. [DOI] [PubMed] [Google Scholar]
- 82. Tan‐Garcia A, Lai F, Yeong JPS, Irac SE, Ng PY, Msallam R, et al. Liver fibrosis and CD206+ macrophage accumulation are suppressed by anti‐GM‐CSF therapy. JHEP Reports. 2020;2(1):100062 10.1016/j.jhepr.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gallina ME, Choksi A, Nikulina N, Singh J, Dakshinamoorthy G, Kim J, et al. Abstract A074: Spatially resolved deep antigen profiling of single cells in FFPE tissue samples through CODEXTM. Cancer Immunol Res. 2019;7(2 Supplement):A074–A. 10.1158/2326-6074.Cricimteatiaacr18-a074. [DOI] [Google Scholar]
- 84. Goltsev Y, Samusik N, Kennedy‐Darling J, Bhate S, Hale M, Vazquez G, et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell. 2018;174(4):968–81 e15. 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gallina ME, Choksi A, Nikulina N, Singh J, Dakshinamoorthy G, Kim J, et al. Abstract A073: CODEXTM: A novel platform for spatially resolved deep antigen profiling of single cells in tissue samples. Cancer Immunol Res. 2019;7(2 Supplement):A073–A. 10.1158/2326-6074.Cricimteatiaacr18-a073. [DOI] [Google Scholar]
- 86. Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of american pathologists clinical practice guideline focused update. J Clin Oncol. 2018;36(20):2105–22. 10.1200/jco.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 87. Amaria RN, Reddy SM, Tawbi HA, Davies MA, Ross MI, Glitza IC, et al. Neoadjuvant immune checkpoint blockade in high‐risk resectable melanoma. Nat Med. 2018;24(11):1649–54. 10.1038/s41591-018-0197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Blank CU, Rozeman EA, Fanchi LF, Sikorska K, van de Wiel B, Kvistborg P, et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat Med. 2018;24(11):1655–61. 10.1038/s41591-018-0198-0. [DOI] [PubMed] [Google Scholar]
- 89. Decalf J, Albert ML, Ziai J. New tools for pathology: a user's review of a highly multiplexed method for in situ analysis of protein and RNA expression in tissue. J Pathol. 2019;247(5):650–61. 10.1002/path.5223. [DOI] [PubMed] [Google Scholar]
- 90. Toki MI, Merritt CR, Wong PF, Smithy JW, Kluger HM, Syrigos KN, et al. High‐plex predictive marker discovery for melanoma immunotherapy treated patients using Digital Spatial Profiling. Clin Cancer Res. 2019:clincanres.0104.2019. 10.1158/1078-0432.ccr-19-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Merritt CR, Ong GT, Church S, Barker K, Geiss G, Hoang M, et al. High multiplex, digital spatial profiling of proteins and RNA in fixed tissue using genomic detection methods. bioRxiv. 2019:559021 10.1101/559021. [DOI] [PubMed] [Google Scholar]
- 92. Mohammed AM, Xia Z, Chatterjee G, Hwang K, Manesse M. Abstract 1183: High‐plex spatial profiling of whole FFPE tissue sections using InSituPlex® technology for discovery applications. Cancer Res. 2019;79(13 Supplement):1183‐ 10.1158/1538-7445.Am2019-1183. [DOI] [Google Scholar]
- 93. Downing S, Patel K, Buell J, Hebert C. Abstract 5657: Use of a novel immunofluorescence multiplexing technology, InSituPlex™, for the simultaneous detection of immune cells in multiple cancer types. Cancer Res. 2018;78(13 Supplement):5657‐ 10.1158/1538-7445.Am2018-5657. [DOI] [Google Scholar]
- 94. Lykkegaard Andersen N, Brügmann A, Lelkaitis G, Nielsen S, Friis Lippert M, Vyberg M. Virtual double staining: A digital approach to immunohistochemical quantification of estrogen receptor protein in breast carcinoma specimens. Appl Immunohistochem Molecul Morphol. 2018;26(9):620–6. 10.1097/pai.0000000000000502. [DOI] [PubMed] [Google Scholar]
- 95. Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14(9):873–6. 10.1038/nmeth.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rashid R, Gaglia G, Chen Y‐A, Lin J‐R, Du Z, Maliga Z, et al. Highly multiplexed immunofluorescence images and single‐cell data of immune markers in tonsil and lung cancer. Scientific Data. 2019;6(1):323 10.1038/s41597-019-0332-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Catena R, Montuenga LM, Bodenmiller B. Ruthenium counterstaining for imaging mass cytometry. J Pathol. 2018;244(4):479–84. 10.1002/path.5049. [DOI] [PubMed] [Google Scholar]
- 98. Hartmann FJ, Bendall SC. Immune monitoring using mass cytometry and related high‐dimensional imaging approaches. Nature Reviews Rheumatology. 2020;16(2):87–99. 10.1038/s41584-019-0338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Loughrey MB, Bankhead P, Coleman HG, Hagan RS, Craig S, McCorry AMB, et al. Validation of the systematic scoring of immunohistochemically stained tumour tissue microarrays using QuPath digital image analysis. Histopathology. 2018;73(2):327–38. 10.1111/his.13516. [DOI] [PubMed] [Google Scholar]
- 101. Gray RT, Cantwell MM, Coleman HG, Loughrey MB, Bankhead P, McQuaid S, et al. Evaluation of PTGS2 Expression, PIK3CA Mutation, Aspirin Use and Colon Cancer Survival in a Population‐Based Cohort Study. Clinical and Translational Gastroenterology. 2017;8(4):e91 10.1038/ctg.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gray RT, Loughrey MB, Bankhead P, Cardwell CR, McQuaid S, O'Neill RF, et al. Statin use, candidate mevalonate pathway biomarkers, and colon cancer survival in a population‐based cohort study. Br J Cancer. 2017;116(12):1652–9. 10.1038/bjc.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Śledzińska A, Vila de Mucha M, Bergerhoff K, Hotblack A, Demane DF, Ghorani E, et al. Regulatory T Cells restrain interleukin‐2‐ and blimp‐1‐dependent acquisition of cytotoxic function by CD4+ T cells. Immunity. 2020;52(1):151–66.e6. 10.1016/j.immuni.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ritsma L, Ponsioen B, van Rheenen J. Intravital imaging of cell signaling in mice. IntraVital. 2012;1(1):2–10. 10.4161/intv.20802. [DOI] [Google Scholar]
- 105. Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo‐cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37(2):364–76. 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Li W, Germain RN, Gerner MY. High‐dimensional cell‐level analysis of tissues with Ce3D multiplex volume imaging. Nat Protoc. 2019;14(6):1708–33. 10.1038/s41596-019-0156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Li W, Germain RN, Gerner MY. Multiplex, quantitative cellular analysis in large tissue volumes with clearing‐enhanced 3D microscopy (Ce3D). PNAS. 2017;114(35):E7321–e30. 10.1073/pnas.1708981114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Shashni B, Ariyasu S, Takeda R, Suzuki T, Shiina S, Akimoto K, et al. Size‐based differentiation of cancer and normal cells by a particle size analyzer assisted by a cell‐recognition PC software. Biol Pharm Bull. 2018;41(4):487–503. 10.1248/bpb.b17-00776. [DOI] [PubMed] [Google Scholar]
- 109. Hegde PS, Karanikas V, Evers S. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res. 2016;22(8):1865–74. 10.1158/1078-0432.ccr-15-1507. [DOI] [PubMed] [Google Scholar]
- 110. Chen DS, Mellman I. Elements of cancer immunity and the cancer–immune set point. Nature. 2017;541:321 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 111. Wong PF, Smithy JW, Blenman KR, Kluger HM, & Rimm DL, editor. Abstract 3638: Quantitative assessment of tumor‐infiltrating lymphocytes and immunotherapy outcome in metastatic melanoma. 2018. [DOI] [PMC free article] [PubMed]
- 112. Johnson DB, Bordeaux J, Kim JY, Vaupel C, Rimm DL, Ho TH. Quantitative spatial profiling of PD‐1/PD‐L1 interaction and HLA‐DR/IDO‐1 predicts improved outcomes of Anti‐PD‐1 therapies in metastatic melanoma. 2018;24(21):5250–60. 10.1158/1078-0432.ccr-18-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Giraldo NA, Nguyen P, Engle EL, Kaunitz GJ, Cottrell TR, Berry S, et al. Multidimensional, quantitative assessment of PD‐1/PD‐L1 expression in patients with Merkel cell carcinoma and association with response to pembrolizumab. 2018;6(1):99 10.1186/s40425-018-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Yeong J, Thike AA, Lim JC, Lee B, Li H, Wong SC, et al. Higher densities of Foxp3(+) regulatory T cells are associated with better prognosis in triple‐negative breast cancer. Breast Cancer Res Treat. 2017;163(1):21–35. 10.1007/s10549-017-4161-4. [DOI] [PubMed] [Google Scholar]
- 115. Yeong J, Lim JCT, Lee B, Li H, Ong CCH, Thike AA, et al. Prognostic value of CD8+PD‐1+ immune infiltrates and PDCD1 gene expression in triple negative breast cancer. J Immunother Cancer. 2019;7(1):34 10.1186/s40425-019-0499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yeong J, Lim JCT, Lee B, Li H, Chia N, Ong CCH, et al. High densities of tumor‐associated plasma cells predict improved prognosis in triple negative breast cancer. Front Immunol. 2018;9:1209‐. 10.3389/fimmu.2018.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Lai CPT, Yeong JPS, Tan AS, Ong CHC, Lee B, Lim JCT, et al. Evaluation of phospho‐histone H3 in Asian triple‐negative breast cancer using multiplex immunofluorescence. 2019. 10.1007/s10549-019-05396-5. [DOI] [PubMed]
- 118. Tan AS, Yeong JPS, Lai CPT, Ong CHC, Lee B, Lim JCT, et al. The role of Ki‐67 in Asian triple negative breast cancers: a novel combinatory panel approach. Virchows Archiv : an International Journal of Pathology. 2019. 10.1007/s00428-019-02635-4. [DOI] [PubMed] [Google Scholar]
- 119. Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single‐cell profiling of breast cancer T cells reveals a tissue‐resident memory subset associated with improved prognosis. Nat Med. 2018;24(7):986–93. 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 120. Chew V, Lai L, Pan L, Lim CJ, Li J, Ong R, et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high‐dimensional proteomic and transcriptomic analyses. Proc Natl Acad Sci. 2017;114(29):E5900–E9. 10.1073/pnas.1706559114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lim CJ, Lee YH, Pan L, Lai L, Chua C, Wasser M, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus‐related hepatocellular carcinoma. Gut. 2018. 10.1136/gutjnl-2018-316510. [DOI] [PubMed] [Google Scholar]
- 122. Stein S, Pishvaian MJ, Lee MS, Lee K‐H, Hernandez S, Kwan A, et al. Safety and clinical activity of 1L atezolizumab + bevacizumab in a phase Ib study in hepatocellular carcinoma (HCC). J Clin Oncol. 2018;36(15_suppl):4074‐ 10.1200/JCO.2018.36.15_suppl.4074. [DOI] [Google Scholar]
- 123. Finn RS, Ducreux M, Qin S, Galle PR, Zhu AX, Ikeda M, et al. IMbrave150: A randomized phase III study of 1L atezolizumab plus bevacizumab vs sorafenib in locally advanced or metastatic hepatocellular carcinoma. J Clin Oncol. 2018;36(15_suppl):TPS4141–TPS. 10.1200/JCO.2018.36.15_suppl.TPS4141. [DOI] [Google Scholar]
- 124. El‐Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open‐label, non‐comparative, phase 1/2 dose escalation and expansion trial. Lancet (London, England). 2017;389(10088):2492–502. 10.1016/s0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Gettinger SN, Choi J, Mani N, Sanmamed MF, Datar I, Sowell R, et al. A dormant TIL phenotype defines non‐small cell lung carcinomas sensitive to immune checkpoint blockers. Nat Commun. 2018;9(1):3196‐ 10.1038/s41467-018-05032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Johnson DB, Bordeaux J, Kim JY, Vaupel C, Rimm DL, Ho TH, et al. Quantitative spatial profiling of PD‐1/PD‐L1 interaction and HLA‐DR/IDO‐1 predicts improved outcomes of Anti–PD‐1 therapies in metastatic melanoma. Clin Cancer Res. 2018;24(21):5250–60. 10.1158/1078-0432.ccr-18-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Giraldo NA, Nguyen P, Engle EL, Kaunitz GJ, Cottrell TR, Berry S, et al. Multidimensional, quantitative assessment of PD‐1/PD‐L1 expression in patients with Merkel cell carcinoma and association with response to pembrolizumab. J Immunother Cancer. 2018;6(1):99 10.1186/s40425-018-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wong PF, Smithy JW, Blenman KR, Kluger HM, Rimm DL. Abstract 3638: Quantitative assessment of tumor‐infiltrating lymphocytes and immunotherapy outcome in metastatic melanoma. Cancer Res. 2018;78(13 Supplement):3638‐ 10.1158/1538-7445.am2018-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, et al. PD‐1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–71. 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mazzaschi G, Madeddu D, Falco A, Bocchialini G, Goldoni M, Sogni F, et al. Low PD‐1 expression in cytotoxic CD8+ Tumor‐Infiltrating lymphocytes confers an immune‐privileged tissue microenvironment in NSCLC with a prognostic and predictive value. Clin Cancer Res. 2018;24(2):407–19. 10.1158/1078-0432.ccr-17-2156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


