Summary
Intracellular replication of the deadly pathogen Mycobacterium tuberculosis relies on the production of small organic molecules called siderophores to scavenge iron from host proteins1. M. tuberculosis produces two classes of siderophores, lipid-bound mycobactin and soluble carboxymycobactin2, 3. Functional studies revealed that iron-loaded carboxymycobactin is imported into the cytoplasm by the ABC transporter IrtAB4, which features an additional cytoplasmic siderophore interaction domain (SID)5. However, IrtAB’s predicted ABC exporter fold seemingly contradicts its import function. Here, we show that membrane-reconstituted IrtAB is sufficient to import mycobactins, which are then reduced by the SID to facilitate iron release. Structure determination by X-ray crystallography and cryo-EM confirms IrtAB’s ABC exporter fold, but also reveals structural peculiarities at the transmembrane region of IrtAB resulting in a partially collapsed inward-facing substrate binding cavity. The SID is positioned in close proximity to the inner membrane leaflet, which allows the reduction of membrane-inserted mycobactin. Enzymatic ATPase activity and in vivo growth assays show that IrtAB prefers mycobactin over carboxymycobactin as its substrate. Our study provides insights into an unusual ABC exporter that evolved as highly specialized siderophore import machinery in mycobacteria.
Upon biosynthesis, mycobactins are exported by the inner-membrane RND transporters MmpL4 and MmpL5, requiring as well the periplasmic adaptor proteins MmpS4 and MmpS56. At the level of the outer membrane, the EXS-3 secretion system is needed for mycobactin-mediated iron acquisition7, likely involving secreted effector proteins8. The ATP binding cassette (ABC) transporter IrtAB finally imports iron-bound cMBT across the inner membrane, and its deletion attenuates M. tuberculosis replication in mice4. ABC transporters constitute a large family of membrane transporters, which couple the binding and hydrolysis of ATP at a pair of highly conserved nucleotide binding domains (NBDs) to conformational changes of variable transmembrane domains (TMDs)9. Curiously, IrtAB’s import function stands in contrast to its predicted fold of an ABC exporter. As the name suggests, ABC exporters are typically involved in the export of substrates out of the cell, whereas canonical ABC importers feature TMDs very distinct from ABC exporters and depend on additional substrate binding domains to accomplish substrate uptake9, 10. As an additional unusual feature, IrtAB contains a siderophore interaction domain (SID) fused to the N-terminus of IrtA, which has been suggested, but not shown in vitro, to reduce Fe(III)-cMBT in order to facilitate iron release inside the bacterial cell5.
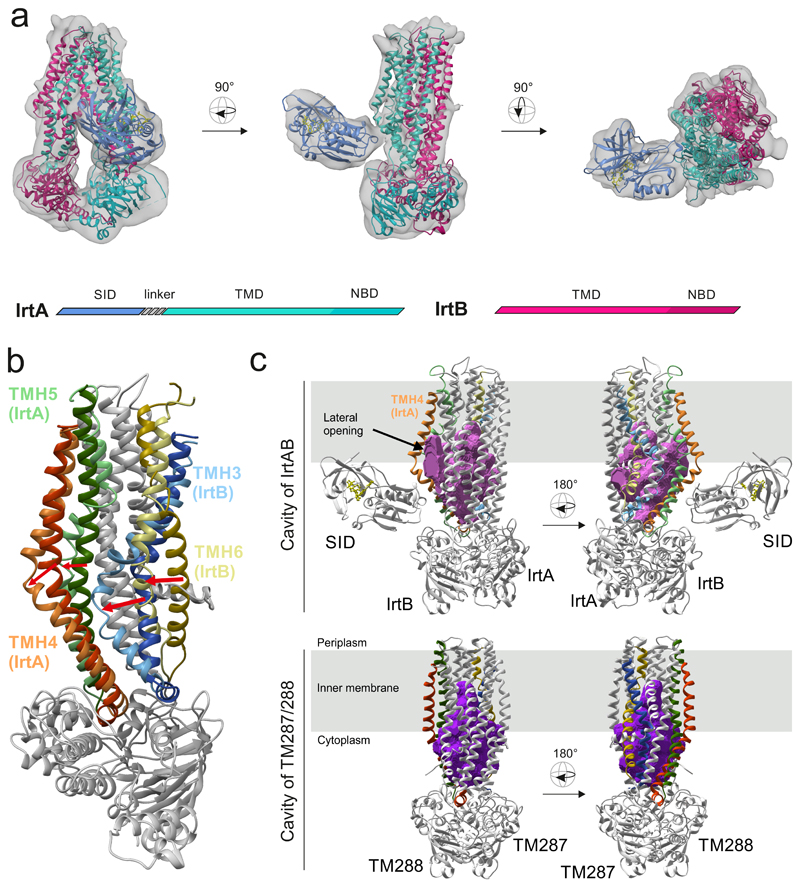
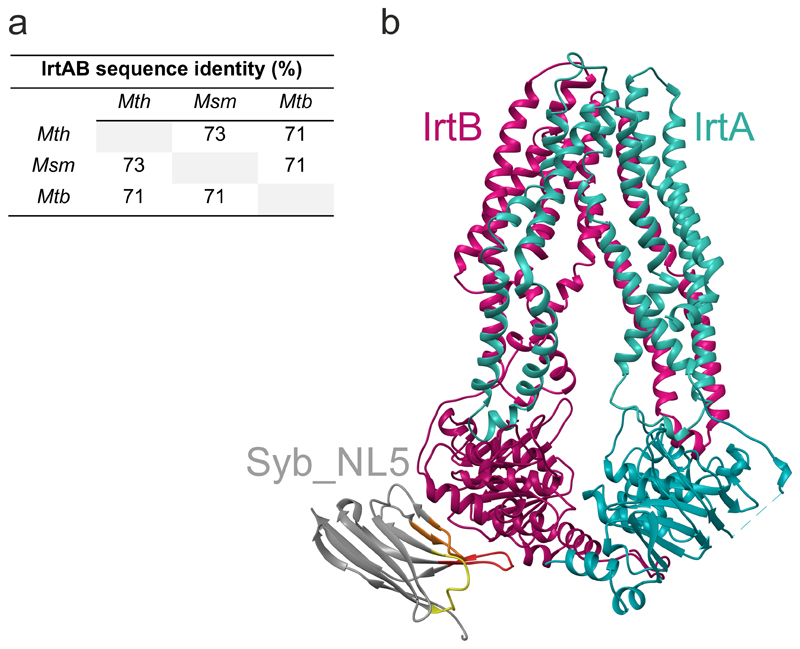
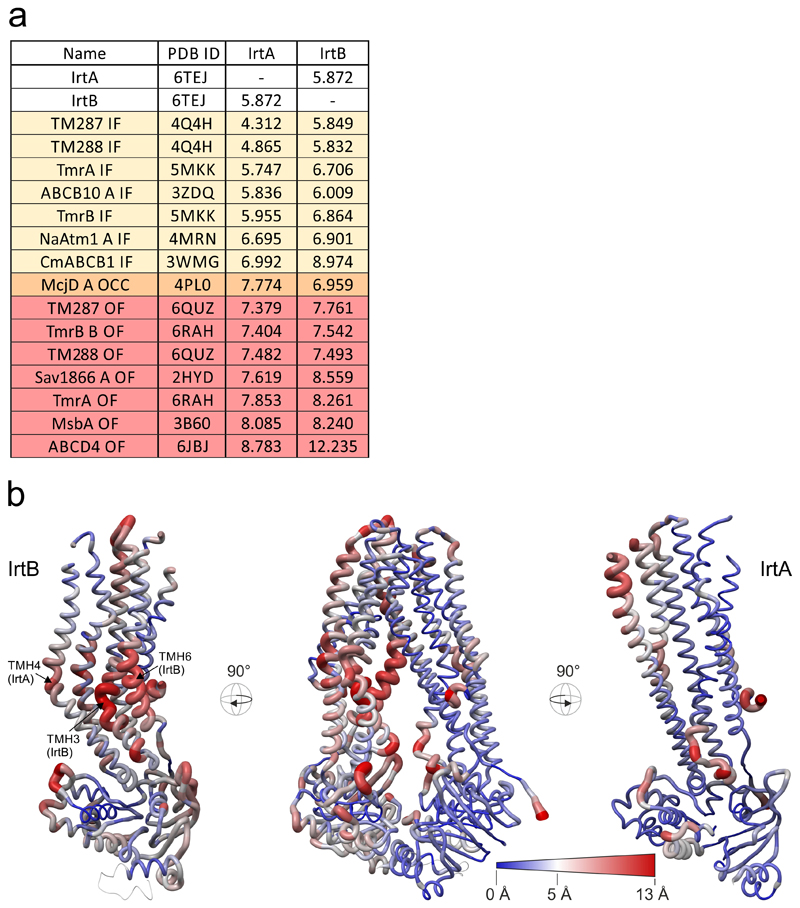
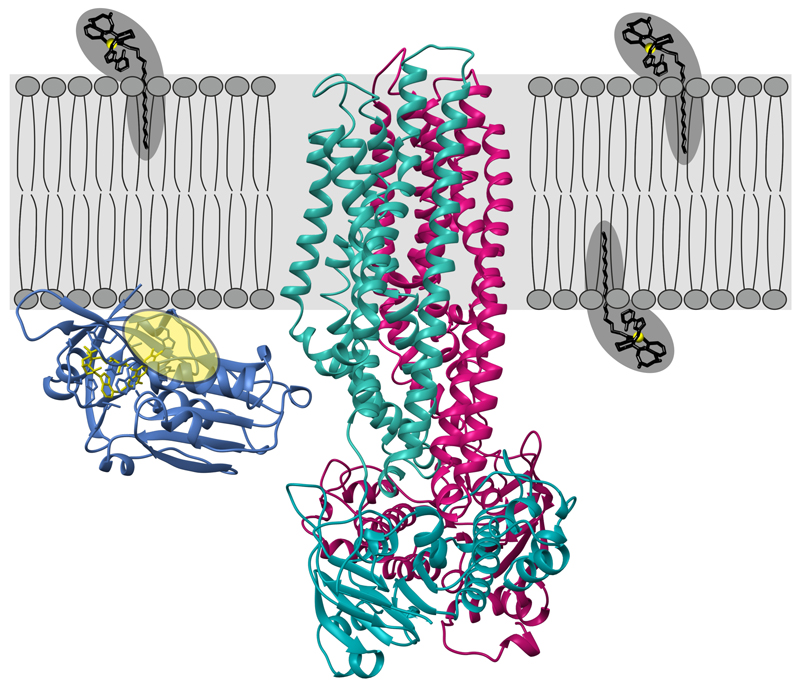
To gain structural insights into this unusual ABC exporter, we combined X-ray crystallography and single particle cryo-EM to determine the structure of the IrtAB protein of Mycobacterium thermoresistibile. IrtAB of this thermophilic mycobacterial species was found to be amenable to structural and biochemical analyses. Importantly, IrtAB of M. thermoresistibile shares a high sequence identity of 71 % with the M. tuberculosis homologue (Extended Data Fig. 1a). A cryo-EM structure of full-length IrtAB was obtained at a global resolution of 6.9 Å and features an unambiguous density for the transporter, as well as for the SID (Fig. 1a, Extended Data Figs. 2 and 3, Extended Data Table 1). With the help of a sybody (synthetic nanobody)11 that was identified by the flycode technology12, the crystal structure of IrtAB devoid of the SID was solved at a resolution of 2.7 Å in its apo state (Extended Data Fig. 1b and Extended Data Table 2). The transporter assumes an inward-facing conformation with the NBDs contacting each other via a C-terminal extension of the NBD of IrtB. A DALI search for similar protein folds clearly marked IrtAB as an ABC exporter 13. With an r.m.s.d. of 5.9 Å over the entire polypeptide chains, the heterodimeric IrtAB complex features major asymmetries14, which allowed to unambiguously assign the identity of the chains within the cryo-EM map (Fig. 1a). Superimpositions with other ABC exporters revealed the highest similarity to the inward-facing structure of TM287/28815 (Extended Data Fig. 4), and further unveiled three conspicuous structural peculiarities of IrtAB: i) transmembrane helix 6 (TMH6) of IrtB protrudes into the central cavity, ii) TMH3 of IrtB threads underneath TMH6 and is strongly kinked and iii) the domain-swap helices (TMH4 and 5) of IrtA are bulged out (Fig. 1b). As a result, the inward-facing cavity of IrtAB is collapsed at the upper region. Further, the bulged-out domain swap helices form a lateral opening from the cavity to the inner leaflet of the lipid bilayer at the side of the transporter, where the SID is located (Fig. 1c). Hence, in contrast to canonical ABC exporters in which the inward-facing conformation represents the high affinity state allowing substrate binding to a deep cavity, the inward-facing IrtAB conformation may represent the low affinity state after mycobactin release towards the SID and the cytoplasm.
Figure 1. Structure of IrtAB.
a, X-ray structures of IrtAB devoid of the SID (2.7 Å, IrtA in turquoise and IrtB in purple) and the isolated SID (1.8 Å, blue with the bound FAD molecule shown in yellow as sticks) were fitted into the cryo-EM map (6.9 Å) of full-length IrtAB. The domain organization of the IrtAB heterodimer is shown at the bottom. b, Superimposition of IrtB plus domain-swap-helices TMH4 and 5 of IrtA with TM288 plus domain-swap-helices TMH4 and 5 of TM287. TMH6 (IrtB/TM288), TMH3 (IrtB/TM288), TMH4 (IrtA/TM287) and TMH5 (IrtA/TM287) are colored as indicated and the rest is shown in grey. Arrows indicate the structural changes from TM287/288 (darker colors) to IrtAB (lighter colors). c, Inward-facing cavities of IrtAB and TM287/288 (PDB: 4Q4A). TMHs with strong differences between IrtAB and TM287/288 are colored as in (b). The inward-facing cavity of IrtAB is collapsed at the upper third as compared to TM287/288 mainly due to the movement of TMH6 of IrtB. The bulged-out domain swap helices TMH4 and TMH5 of IrtA result in a lateral access to the cavity from the inner leaflet of the lipid bilayer (indicated in grey).
The X-ray structure of the SID was determined at 1.8 Å resolution (Fig. 2c, Extended Data Table 2). It exhibits a conserved flavoreductase fold and features an electron-donor pocket, a perpendicularly located substrate pocket and a flavine-adenine dinucleotide (FAD) molecule bound between the pockets16. The SID could be unambiguously placed into the cryo-EM map and is positioned close to the predicted membrane boundary through protein interactions with the TMD of IrtA (Fig. 1a). The 72 amino acid long linker between the SID and the first “elbow“-helix of the TMD of IrtA is not visible in the map, presumably due to its flexibility. Interestingly, the presumed mycobactin binding pocket of the SID is positioned such that it can accommodate Fe-MBT embedded in the cytoplasmic leaflet of the inner membrane, suggesting that the SID plays an important role for iron scavenging via Fe-MBT (Extended Data Fig. 5).
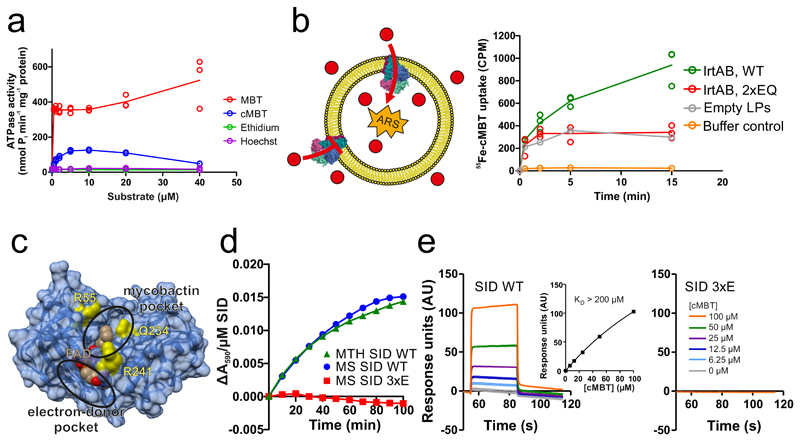
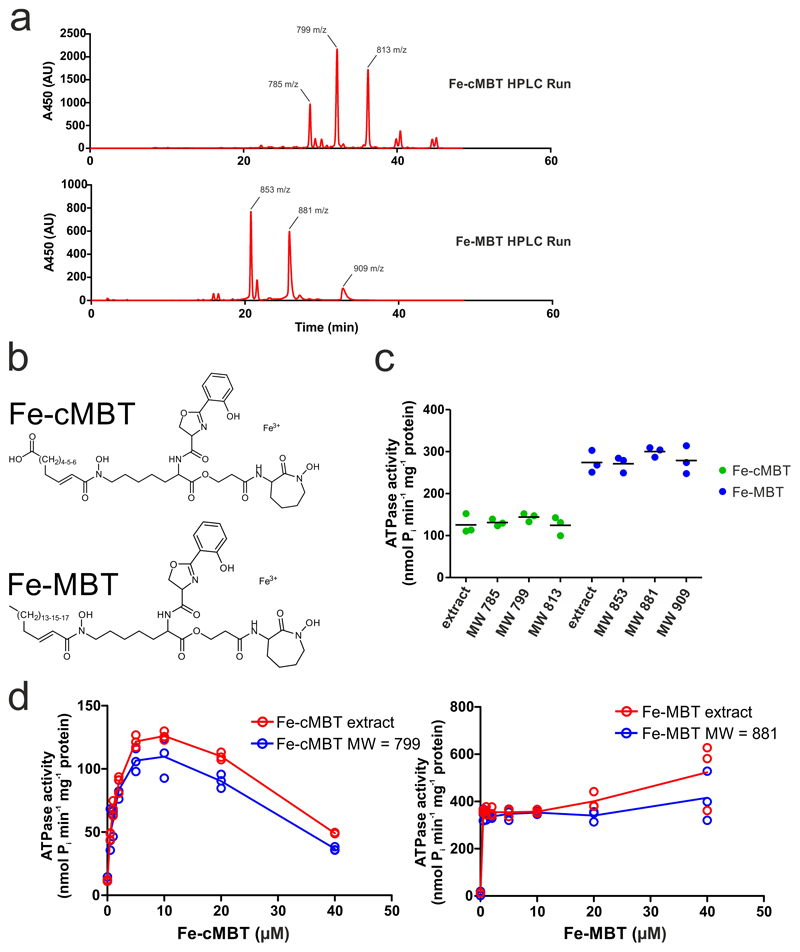
Figure 2. Biochemical characterization of IrtAB.
a, ATPase activity of IrtAB reconstituted into nanodiscs in the presence of increasing concentrations of Fe-MBT, Fe-cMBT, Ethidium and Hoechst. Data points are technical triplicates and curves cross mean values. b, In vitro transport of Fe-cMBT mediated by wildtype (WT) and inactive (2xEQ mutant) IrtAB reconstituted into proteoliposomes. An ATP regenerating system (ARS) was incorporated into the vesicle lumen (left panel). Uptake of radioactively labeled 55Fe-cMBT into the vesicle lumen was measured over time (right panel). As a control, the assay was performed with empty liposomes (LPs) or buffer only. For WT and 2xEQ mutant, data points are technical triplicates and curves cross mean values. c, Structure of the SID with the electron-donor and mycobactin pockets highlighted. FAD is shown as spheres and colored in beige. To generate the 3xE mutant, three conserved residues lining the mycobactin pocket (yellow) were mutated to glutamate. d, Reduction of Fe(III)-cMBT by M. thermoresistibile (MTH) and M. smegmatis (MS) SIDs using NADPH as electron donor and Ferene as a reporter probe of released Fe(II) (Amax = 590 nm). The 3xE mutant served as negative control. Representative data of biological duplicates are shown. e, Binding of Fe-cMBT to the M. smegmatis SID as measured by SPR using Fe-cMBT concentrations as indicated in the right panel. SPR sensorgrams were recorded for the wildtype SID (left panel) or the 3xE mutant (right panel). Fitting of equilibrium binding values indicated a KD > 200 μM (left panel, inset). The SPR experiment was performed once.
In many ABC transporters the ATPase activity is stimulated by the presence of transported substrates17–19. To measure ATPase activities in a lipid environment, the purified M. thermoresistibile IrtAB wildtype (WT) protein was reconstituted into nanodiscs. In analogy to previous studies on the ABC transporter P-glycoprotein20, we mutated the catalytic Walker B glutamates of both nucleotide binding sites to glutamine (E815QIrtA/E493QIrtB, henceforth called 2xEQ mutant), which was used as ATPase-deficient control throughout the study. ATPase activity of reconstituted wildtype IrtAB was stimulated more than 38-fold with Fe-MBT, and more than 10-fold with Fe-cMBT (Fig. 2a). For Fe-cMBT, but not for Fe-MBT, the ATPase activity decreases again at concentrations above 10 μM. This phenomenon of bell-shaped drug stimulation curves is frequently observed for ABC transporters and arises from occupation of a secondary low affinity binding site by the transport substrate, which slows down the ATPase cycle 17, 21. In contrast to mycobactins, ethidium and Hoechst 33342, two benchmark substrates of multidrug ABC exporters17, did not stimulate IrtAB’s ATPase activity. Experiments with HPLC-purified mycobactins revealed that the aliphatic chain length of cMBT or MBT does not have an influence on ATPase stimulation of IrtAB (Extended Data Fig. 6).
Next, we investigated if the ABC exporter IrtAB is sufficient for mycobactin import. To this end, purified M. thermoresistibile IrtAB (wildtype or inactive 2xEQ mutant) was reconstituted into liposomes22. This procedure results in the incorporation of membrane transporters in mixed orientations (i.e. the ATP consuming NBDs are either located in the liposome lumen or point to the bulk buffer) (Fig. 2b). Directionality of transport was imposed by encapsulating an ATP regeneration system (ARS) inside the liposomes. The hydrophobic properties of Fe-MBT prevented us from performing transport assays with liposomes and therefore in vitro transport assays were carried out with Fe-cMBT only. Transport experiments were initiated by adding radioactively labelled 55Fe-cMBT and proteoliposomes were separated from free 55Fe-cMBT by rapid size exclusion chromatography. Liposomes containing wildtype IrtAB showed more uptake of 55Fe-cMBT than the ones containing the inactive 2xEQ mutant. When empty liposomes were included as a control, the trace looked highly similar to the one of liposomes containing the 2xEQ mutant, indicating unspecific interaction of 55Fe-cMBT with the liposome membrane. When the assay was conducted with 55Fe-cMBT in buffer only, the counts were very low, showing minimal leakage during the rapid size exclusion chromatography step. In aggregate, these results show that purified and reconstituted IrtAB is capable of 55Fe-cMBT uptake in an ATPase-dependent fashion.
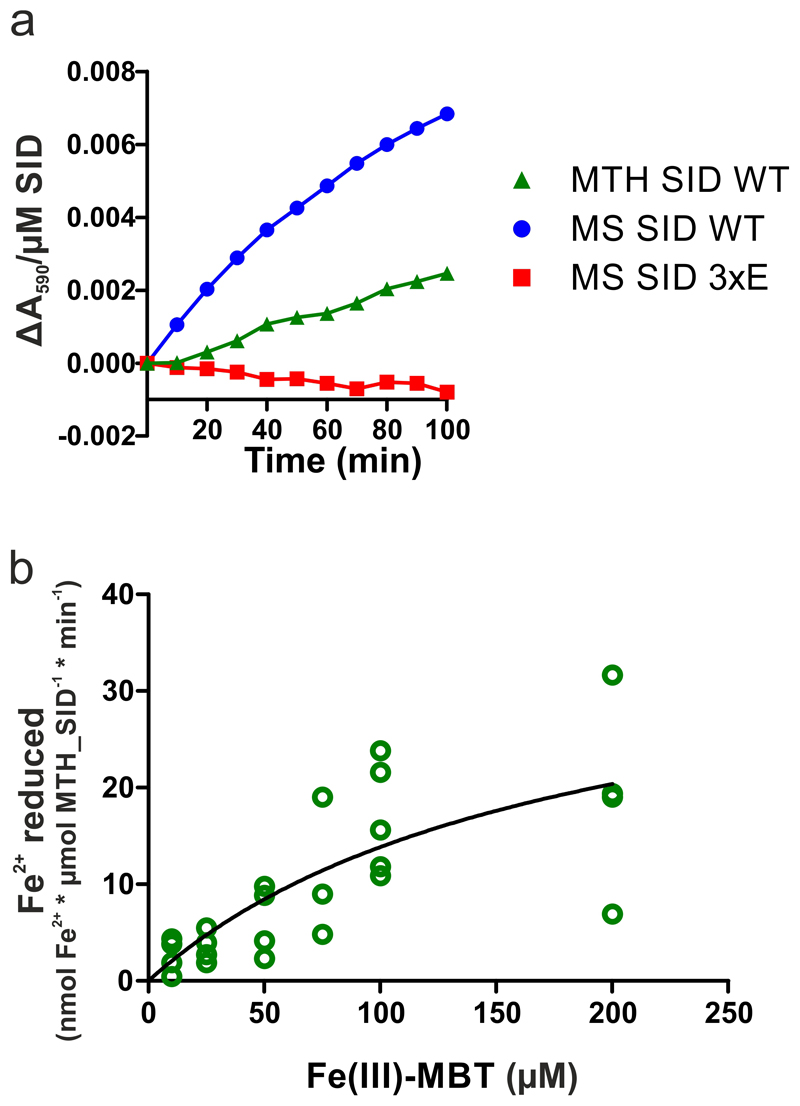
In order to assess if the SID is capable of reducing Fe(III)-cMBT to liberate Fe(II) as suggested by in vivo assays5, we performed redox experiments with the Mycobacterium smegmatis and M. thermoresistibile SIDs under strictly anaerobic conditions. Indeed, Fe(III)-cMBT and Fe(III)-MBT were reduced in the presence of the SIDs using NADPH as electron donor (Fig. 2d, Extended Data Fig. 7a). As a control, three conserved residues lining the presumed mycobactin binding pocket (R55, Q234 and R241) of the M. smegmatis SID were mutated to glutamate (3xE mutant) (Fig. 2c). The purified 3xE mutant eluted as monomeric peak from a size exclusion chromatography column without any indication of misfolding, but was no longer capable of Fe(III)-cMBT and Fe(III)-MBT reduction (Fig. 2d, Extended Data Fig. 7a). We noted that the estimated catalytic rates of the wildtype SID were at least an order of magnitude lower compared to previously published siderophore-interacting proteins16. By measuring reduction rates at different Fe(III)-MBT concentrations, we determined an apparent KM of 185 μM of the M. thermoresistibile SID for mycobactin (Extended Data Fig. 7b). This notion was confirmed in a surface plasmon resonance binding (SPR) experiment, which suggested a KD > 200 μM for Fe-cMBT with M. smegmatis SID (Fig 2e). SPR measurement artifacts could be excluded, because Fe-cMBT binding was not observed when coating the SID 3xE mutant.
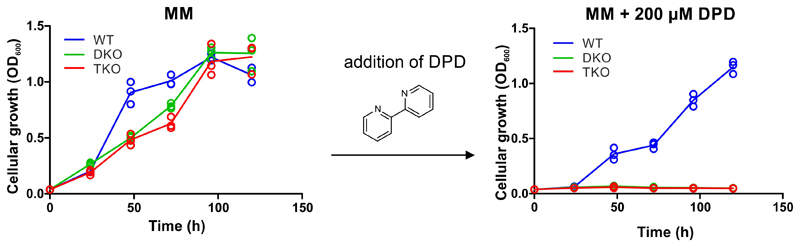
To analyze mycobactin-mediated iron acquisition via IrtAB, we generated the double mutant M. smegmatis ΔfxbAΔmbtD (henceforth called DKO), in which the deletion of the fxbA and mtbD genes abrogate exochelin (an additional siderophore produced by M. smegmatis) and mycobactin biosynthesis, respectively23. Subsequently, we generated the triple mutant M. smegmatis ΔfxbAΔmbtDΔirtAB (henceforth called TKO), which in addition lacks IrtAB. WT, DKO and TKO were grown in minimal medium in the presence of the weak iron chelator 2,2′-dipyridyl (DPD) to achieve conditions in which siderophores become essential for growth7, 23. As expected, WT cells were able to grow under these conditions owing to their capacity to produce siderophores, while no growth was observed for DKO and TKO (Extended Data Fig. 8). When purified Fe-MBT was added to the medium at concentrations ranging from 2- 50 μM as iron source, DKO grew, while TKO lacking the IrtAB transporter did not (Fig. 3a). However, when Fe-cMBT was added at 20 μM or 50 μM, growth of both DKO and TKO were rescued, suggesting the presence of an alternative import protein for Fe-cMBT besides IrtAB (Fig. 3b). The TKO strain only stopped growing at Fe-cMBT concentrations of 2 μM or 5 μM (Fig. 3b). Hence, IrtAB acts as high-affinity importer for Fe-cMBT. In agreement with this notion, IrtAB’s ATPase was maximally stimulated at a concentration of around 5 μM Fe-cMBT (Fig. 2a). Furthermore, for both Fe-cMBT and Fe-MBT, low micromolar concentrations were sufficient to achieve maximal catalytic activity of the transporter in the ATPase stimulation experiments. To ensure the best possible discrimination between DKO and TKO, we added Fe-cMBT and Fe-MBT at 2 μM for subsequent in vivo growth assays.
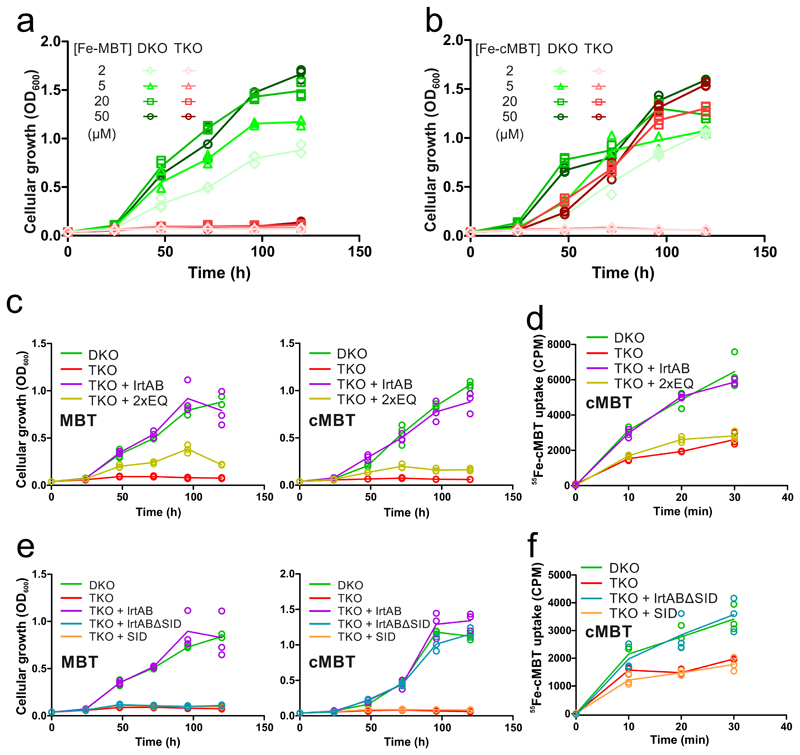
Figure 3. In vivo characterization of IrtAB in M. smegmatis.
a and b, Mycobactin-dependent growth of M. smegmatis ΔfxbAΔmbtD mutant (DKO) and M. smegmatis ΔfxbAΔmbtDΔirtAB mutant (TKO) in the presence of Fe-MBT (a) or Fe-cMBT (b) added at the indicated concentrations. c – f, Complementation of the TKO strain with full-length IrtAB (WT), the IrtAB mutant devoid of ATPase activity (2xEQ), IrtAB devoid of the SID (IrtABΔSID) and the SID alone (SID). c and e, Mycobactin-depended growth determined in the presence of 2 μM Fe-MBT (left) or 2 μM Fe-cMBT (right). d and f, Cellular uptake of radioactive 55Fe-cMBT. Data points are technical triplicates and curves cross mean values.
To further dissect IrtAB at a functional level, the TKO strain was complemented with different IrtAB constructs (M. smegmatis homologue) using the integrative pFLAG vector24. The DKO strain was used as a positive control for growth, while TKO was used as a negative control. To assess whether ATP hydrolysis is required for transport, we complemented TKO with the IrtAB 2xEQ mutant devoid of ATPase activity and wildtype IrtAB. Both for Fe-MBT and Fe-cMBT, complementation with wildtype IrtAB restored growth to the levels of DKO (Fig. 3c). The 2xEQ mutant partially restored growth, suggesting that it either exhibits residual ATPase activity as is the case for other ABC transporters20 or that IrtAB can act as a facilitator for mycobactins (Fig. 3c). To gain insights into shorter time-scales of mycobactin uptake, we measured cellular 55Fe-cMBT accumulation. The TKO strain showed slower 55Fe-cMBT uptake than the DKO strain, and only expression of the wildtype transporter but not the 2xEQ mutant could complement 55Fe-cMBT uptake in the TKO strain (Fig. 3d). Efficient uptake of mycobactins is thus dependent on ATP hydrolysis. To investigate the role of the SID, we complemented the TKO strain with wildtype IrtAB, the isolated SID and the IrtAB transporter without SID (IrtABΔSID). For both Fe-MBT and Fe-cMBT, expression of the SID did not restore growth (Fig. 3e). Interestingly, the IrtABΔSID construct restored growth with Fe-cMBT but not with Fe-MBT, suggesting the requirement of cooperative transport and local reduction for iron utilization via Fe-MBT. Again, we measured cellular 55Fe-cMBT uptake at shorter time scales and confirmed our finding that IrtABΔSID efficiently imports Fe-cMBT (Fig. 3f).
Previous studies suggested that IrtAB is primarily responsible for the import of Fe-cMBT4. Based on our experiments, we show here that Fe-MBT is, over Fe-cMBT, the preferential substrate for IrtAB. Firstly, the ATPase activity of purified and reconstituted IrtAB is stimulated much stronger in the presence of Fe-MBT versus Fe-cMBT and reaches its maximum already at a concentration of 0.5 μM (Fig. 2a). Secondly, IrtAB is essential for the uptake and utilization of Fe-MBT as iron source over a broad range of concentrations, whereas it is dispensable for growth at elevated Fe-cMBT concentrations, most likely due to the presence of an alternative Fe-cMBT import system besides IrtAB (Fig. 3a and b). Thirdly, IrtAB’s SID is required for growth when using Fe-MBT as an iron source, but not when supplying iron via Fe-cMBT (Fig. 3e). The uptake of externally added Fe-cMBT and Fe-MBT appears to function similarly well, because both forms of mycobactin support cellular growth under otherwise iron-deplete conditions in a similar concentration range (Fig. 3a and b). Our findings are in agreement with a previous study showing that both forms of mycobactin are secreted by mycobacteria, namely into the aqueous surroundings in case of cMBT and embedded into secreted membrane vesicles in case of MBT25. Thereby, M. tuberculosis can get access to different iron stores present in the infected cell.
Our experiments unambiguously demonstrate that IrtAB is a mycobactin importer. Yet, IrtAB clearly has the fold of an ABC exporter. To our knowledge, ABC exporters that import substrates have so far only been described for the human N-retinylidene-phosphatidylethanolamine transporter ABCA4 and the cobalamin transporter ABCD426, 27. Our in vitro transport experiments clearly demonstrated that IrtAB does not depend on additional proteins to accomplish ferric mycobactin import. Therefore, the transport mechanism must be distinct from classical ABC importers whose function depends on periplasmic binding proteins.
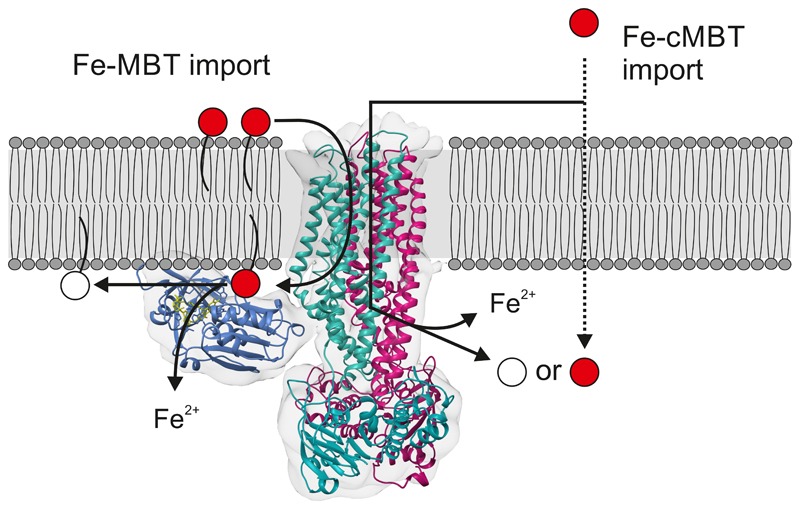
In conclusion, IrtAB imports membrane bound Fe-MBT from the periplasmic to the cytoplasmic leaflet of the inner mycobacterial membrane (Fig. 4). Upon import by IrtAB, the local concentration of Fe-MBT in proximity to the transporter is expected to be higher than the one of Fe-cMBT, as Fe-MBT remains confined to the inner membrane. In this context, the SID is ideally positioned for reduction of Fe-MBT, which explains why the SID is essential for iron uptake via Fe-MBT. This high local concentration likely compensates for the low binding affinities and slow catalytic rates of the SID measured in vitro. In contrast, the SID is dispensable for iron utilization via Fe-cMBT, because this soluble form of mycobactin can freely diffuse into the cytoplasm after import where it may be reduced by other ferric reductases. It is highly likely that IrtAB specifically recognizes the iron-binding core common to Fe-MBT and Fe-cMBT via amino acid side chains within the TMDs that have to be identified in future studies. It is further conceivable that the lipid tail of Fe-MBT remains embedded in the lipid bilayer during the import reaction, in analogy to the “credit card swipe mechanism” described for the glycolipid ABC transporter PglK28. Our findings open avenues to study the transport mechanism of this unusual ABC exporter in more detail while its structural elucidation provides a basis to exploit IrtAB as potential drug target to develop novel antibiotics against M. tuberculosis.
Figure 4. Proposed mechanism of IrtAB-mediated mycobactin uptake.
IrtAB imports both Fe-MBT and Fe-cMBT across the inner membrane. For Fe-cMBT, an alternative import mechanism exists. After import, Fe-MBT remains anchored to the membrane and is thereby ideally positioned for reduction by the SID. The SID is essential for iron utilization via Fe-MBT, but not via Fe-cMBT. Iron-bound mycobactins are shown in red, desferrated mycobactins in white.
Methods
Strains, media and antibiotic concentrations
E. coli XL1-Blue was used for cloning of ORFs, complementation constructs and gene deletions. E. coli MC1061 strain was used for expression of ORFs in E. coli. Luria Broth (LB) was used for liquid cultures, and Luria Broth Agar (LB Agar) for plates. For protein expression, terrific broth (TB) was used for liquid cultures. Antibiotics were used in the following concentrations for E. coli: ampicillin (Amp) 100 μg/ml, kanamycin (Kan) 50 μg/ml, chloramphenicol (Cm) 25 μg/ml, apramycin (Apr) 50 μg/ml and hygromycin (Hyg) 100 μg/ml. The M. smegmatis mc2155 strain was used for all in vivo experiments, for the generation of deletion mutants and generation of complementation strains. When culturing the cells in iron-rich medium, 7H9 with OADC supplement was used for liquid cultures, and 7H10 with OADC supplement was used for plates. Antibiotics were used in the following concentrations for M. smegmatis: kanamycin 25 μg/ml, apramycin 25 μg/ml and hygromycin 50 μg/ml. When culturing the cells under low-iron conditions, minimal medium was prepared according to Ratledge et al.29. 1 liter of minimal medium contained 5 g KH2PO4, 5 g L-asparagine and 60 ml (v/v) glycerol. The pH was set to 7.0 and the medium was autoclaved with 2 % (w/v) aluminium oxide. After autoclaving, the medium was filtered with a 0.22 μM filter (Whatman). The pH was set to 6.8 using HCl and autoclaved again. Before use, the medium was supplemented with 6.88 μM ZnSO4, 1.82 μM MnSO4 and 1.64 mM MgSO4. If not stated otherwise, cells were incubated and grown at 37 °C.
Siderophore production and isolation
For production of MBT, minimal medium in glass flasks was inoculated 1:1’000 with M. smegmatis mc2155 grown to stationary phase in 7H9 medium. The cultures were grown by shaking until they reached stationary phase (typically 5 to 6 days). MBT extraction was performed according to Snow et al.2. In brief, cells were centrifuged at 8’000 g for one hour at 4 °C, and then taken up in 100 % ethanol in 1:10 of the culture volume. MBT extraction was performed overnight by stirring at room temperature. The extract was filtered through a filter paper (Whatman) and one volume of chloroform was added to generate a monophasic solution. 10 % (w/v) FeCl3 in 100 % ethanol was added dropwise to the solution, until no further increase in red color was observed. Water (75 % of solvent volume) was added to form a biphasic solution (chloroform and water/ethanol phases). The chloroform phase containing MBT was washed twice with 1 volume of water, dried with anhydrous MgSO4 and evaporated. The Fe-MBT crude extract was then taken up in 100 % ethanol. For production of cMBT, minimal medium in plastic ware was inoculated 1:1’000 with a dense M. smegmatis ΔfxBC culture (grown in 7H9) and grown to stationary phase. Cells were then inoculated 1:100 into 100 ml minimal medium supplemented with 1.432 μM (NH4)2Fe(II)(SO4)2·6H2O in 1-liter plastic roller bottles (Corning) and grown to stationary phase by shaking. Extraction of cMBT was performed according to Ratledge et al.29. In brief, cells were centrifuged at 8’000 g for one hour at 4 °C, and the supernatant filtered with a 0.22 μM filter (Whatman). The pH of the supernatant was set to 3.5 using HCl, and 10 % (w/v) FeCl3 in 100 % ethanol was added dropwise until formation of precipitate was observed (ferric phosphate). The iron-saturated supernatant was then stirred for one hour at room temperature. The mixture was centrifuged for 15 min at 8’000 g at 4 °C, and the supernatant was extracted with 1 volume of ethyl acetate. The ethyl acetate phase containing Fe-cMBT was washed twice with 1 volume of H2O, dried with MgSO4 and evaporated. The Fe-cMBT crude extract was taken up in 100 % EtOH.
MS analysis and HPLC purification of siderophores
Crude extracts of siderophores were analyzed by ESI-MS/MS with a Q-exactive spectrometer (Thermo Scientific). Single siderophore species were purified by HPLC using a Phenomenex Luna C18(2) reverse phase column. Fe-cMBT was eluted with a 20 to 80 % acetonitrile gradient with a flow rate of 2 ml/min. Fe-MBT was eluted with a 20 to 100 % acetonitrile gradient with a flow rate of 5 ml/min. The masses of the eluted species were confirmed with a Q-exactive spectrometer (Thermo Scientific). The solvent was evaporated and the single species were taken up in 100 % ethanol for further experiments.
Cloning of ORFs
ORFs were amplified from genomic DNA and cloned into the pINIT vector and then into the respective expression vector according to the FX cloning protocol30. Full-length IrtAB of M. thermoresistibile was cloned using primers #1 and #2 (all primers are listed in Supplementary Table 1). The SID of M. thermoresistibile was cloned using primers #1 and #3. The truncated IrtAB construct lacking the SID and linker between SID and TMD was cloned using primers #4 and #2. Full-length IrtAB of M. smegmatis (MSMEG 6553/6554) was cloned using primers #5 and #6. The SID of M. smegmatis was cloned using primers #5 and #7. The truncated IrtAB construct lacking the SID and linker between SID and TMD (IrtABΔSID) was cloned using primers #8 and #6. The 2xEQ mutations (E815QIrtA/E493QIrtB) were introduced into M. thermoresistibile full-length IrtAB in two steps by QuikChange mutagenesis using primer pairs #9/#10 and #11/#12. The corresponding 2xEQ mutations (E770QIrtA/E493QIrtB) were introduced into M. smegmatis full-length IrtAB in two steps by QuikChange mutagenesis using primer pairs #13/#14 and #15/#16. The 3xE mutations (R55E/Q234E/R241E) were introduced into the M. smegmatis SID by QuikChange mutagenesis using primer pairs #17/#18 (R55E) and #19/#20 (Q234E and R241E). All ORFs and mutants cloned into the pINIT vector were completely sequenced. Using FX cloning, the SID constructs were subcloned into the E. coli expression vector pBXNH3 (Addgene #47067). The M. smegmatis SID constructs (wildtype and 3xE mutant) were also subcloned into pBXNH3A (Addgene #47075) for subsequent biotinylation via an Avi-tag. The M. thermoresistibile IrtAB constructs (full-length/ full-length 2xEQ and IrtABΔSID) were subcloned into the E. coli expression vector pBXC3GH (Addgene #47070). For binder selection and SPR experiments the M. thermoresistibile IrtAB full-length construct was subcloned into the E. coli expression vector pBXCA3GH (Addgene #47071). All M. smegmatis ORFs (full-length/ full-length 2xEQ/ IrtABΔSID/ SID) were subcloned into the mycobacterial complementation vector pFLAG (Addgene #110095).
Expression and purification of proteins in E. coli
Proteins were produced in and purified from E. coli MC1061 using TB medium containing 100 μg/ml ampicillin. Cells were grown for 2 hours at 37 °C while shaking. The temperature was lowered to 25 °C and cells were induced after 1 hour with 0.02 % L-arabinose and grown overnight. To purify the SID proteins, cells were harvested by centrifugation at 9’000 g for 15 min at 4 °C. Cells were resuspended in TBS (20 mM Tris/HCl, 150 mM NaCl, pH 7.5) containing a spatula tip of DNAse I. Cells were lysed with a Microfluidizer M-110P (Microfluidics) with 3 passes at 25 KPa. The lysate was first centrifuged for 30 min at 8’000 g at 4°C to remove cell debris. Imidazole was added to a final concentration of 20 mM to the supernatant, which was then loaded onto a Ni2+-NTA gravity flow column (Qiagen). The column was washed with 20 column volumes of wash buffer (50 mM imidazole pH 7.5, 150 mM NaCl, 10 % glycerol), and then eluted with 4 column volumes of elution buffer (200 mM imidazole pH 7.5, 150 mM NaCl, 10 % glycerol). The elution was concentrated to 2.5 ml with a 10 kDa cutoff concentrator (Merck Millipore) and desalted with a PD10 column (GE Healthcare). 3C protease was added and the sample was incubated overnight at 4 °C for cleavage of the His10-tag. Cleaved samples were purified via reverse Ni2+-NTA. Cleaved protein was eluted from the column with 4 volumes of TBS buffer. The samples were concentrated again and loaded onto a Superdex 200 Increase 10/300 GL (GE Healthcare) size exclusion chromatography (SEC) column using TBS as running buffer. To purify the M. thermoresistibile IrtAB membrane protein constructs, cells were broken as described above, centrifuged at 8’000 g, and membrane vesicles were collected by ultracentrifugation at 170’000 g (all purification steps performed at 4 °C). Membrane vesicles were resuspended with TBS, flash frozen and stored at -80 °C until further use. Resuspended membrane vesicles were extracted with 1 % (w/v) n-dodecyl-β-D-maltopyranoside (β-DDM, Glycon Biochemicals) for 2 h, followed by ultracentrifugation at 170’000 g for 30 min. Imidazole was added to a final concentration of 20 mM to the supernatant containing extracted membrane proteins, which was then loaded onto a Ni2+-NTA gravity flow column. The purification was then performed as described for the SID constructs, with the following differences: firstly, 0.03 % β-DDM was included in all buffers; secondly, concentrators with 100 kDa cutoff were used; thirdly, after reverse-Ni2+-NTA purification the sample was diluted 1:10 with ion exchange buffer (15 mM Tris/HCl pH 8.0, 20 mM NaCl, 0.03 % β-DDM) and loaded onto a Resource-Q ion-exchange column (GE Healthcare). The protein was eluted over a NaCl gradient (10 mM – 350 mM, main protein peak eluted at around 150 mM). The main peak was concentrated and separated by SEC using a Superose 6 Increase 10/300 GL (GE Healthcare) column and TBS containing different detergents as running buffer: 0.3 % n-decyl-β-D-maltopyranoside (β-DM) for crystallization of IrtABΔSID; 0.015 % β-DDM for samples used for cryo-EM; 0.03 % β-DDM for subsequent reconstitution of full-length IrtAB into proteoliposomes.
Cryo-EM sample preparation and data acquisition
For structure determination by cryo-EM, 3 μl of detergent-purified M. thermoresistibile IrtAB at a concentration of 2.5 mg/ml were applied to glow-discharged holey carbon grids (Quantifoil, R 2/1 Cu 200 mesh, No. Q32288) and plunge frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) operated at 100 % humidity and 4 °C. Excess liquid was blotted before plunging for 6 s using a blot force of 1. The grids were imaged at 300 kV in a Titan Krios electron microscope (Thermo Fisher Scientific). Data were acquired in energy filtering mode using a postcolumn quantum energy filter (Gatan) with a 20 eV slit and a K2 Summit direct electron detector (Gatan) operating in super-resolution mode. Data were recorded with SerialEM 3.5.8 in low dose mode31. All micrographs were acquired at a nominal magnification of 45’455 with a pixel size of 1.1 Å (0.55 Å in super-resolution) at a defocus between -1.6 and -3.0 μm. Dose-fractionation was used with a frame exposure of 0.2 s at a total exposure time of 10 s (50 frames in total), corresponding to a total electron dose of approximately 85 e/Å2.
Cryo-EM image processing
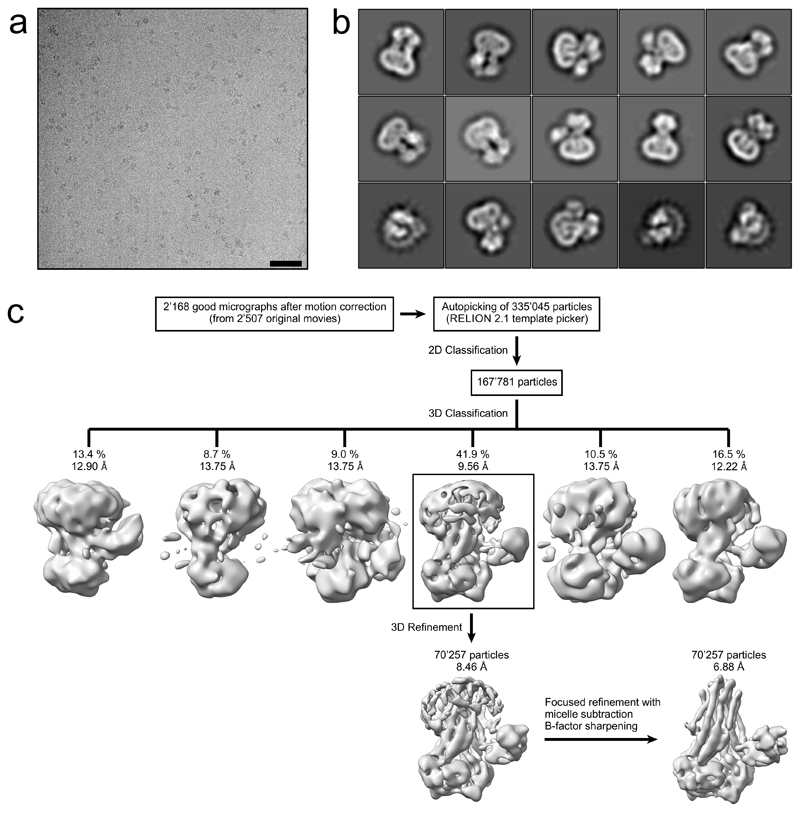
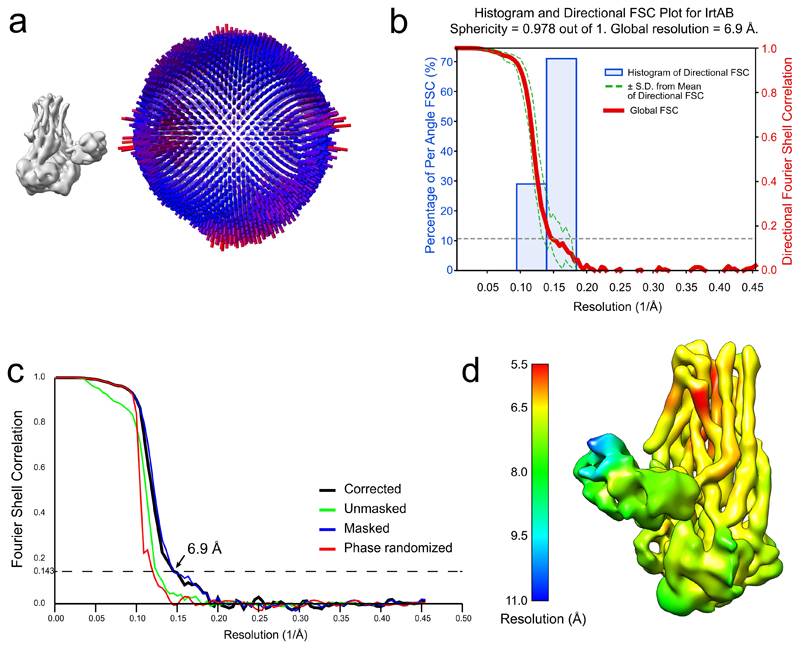
In total, 2’507 micrographs were recorded and processed with RELION 2.1 and RELION 3.032, 33. Frame-based motion correction and dose-weighting were performed with two-times downsampled movies using MotionCor234. The contrast transfer function was estimated using CTFFIND435. Low-quality micrographs showing high defocus, high astigmatism or low resolution were excluded, resulting in 2’168 micrographs used for further processing steps. To generate a template for autopicking, 1’118 IrtAB particles were picked manually, extracted with a box size of 200 pixels and 2D classified into ten classes. These classes served as a reference for automated picking of 335’045 particles. Two rounds of 2D classification using a spherical mask of 160 Å were performed, reducing the number of particles to 167’781. These particles were used for 3D classification into 6 classes, using the mammalian P-glycoprotein (EMDB ID: 600736) at 40 Å as a starting reference. One class with 70’257 particles revealed distinct structural features of IrtAB with its SID and was subjected to auto-refinement. For 3D classification and the first 3D refinement, CTF-amplitude correction was performed only from the first CTF peak onward. The refined structure was used to subtract the micelle from the raw particles, which were subjected to another round of 3D refinement, limiting the rotations to 1.5 degrees. The final unmasked map showed a resolution of 7.86 Å, based on the gold standard Fourier shell correlation (FSC) 0.143 criterion32, 37–39. The structure was sharpened to 6.88 Å using an isotropic B-factor of -319 Å2. The local resolution was estimated using BlocRes from the Bsoft package40, 41. Directional Fourier Shell Correlation curves were calculated with the 3DFSC software 42.
Crystallization, data collection and structure determination of M. thermoresistibile SID
M. thermoresistibile SID crystals were obtained in 400 mM Ammonium-Acetate pH 8.0, 12 % (v/v) PEG2000 at a protein concentration of 30 mg/ml. Crystals were frozen with 30 % (v/v) ethylene glycol as cryo-protecting agent and measured at the Swiss Light Source (SLS) beamline X06SA at λ = 1.0 Å. One dataset diffracting to 1.8 Å was used for structure determination. The diffraction data was processed by the program XDS43 in space group P21. Molecular replacement was performed with a polyA model of the SID from Shewanella putrefaciensis (PDB ID: 2GPJ) using the program Phaser44. The model was then built in Coot45 and refined with the software PHENIX46 (Ramachandran favored/outliers: 98.25 %/0.22 %).
Sybody generation against IrtAB
Sybodies were selected against C-terminally biotinylated M. thermoresistibile IrtAB as described in Zimmermann et al.11. These initial sybodies showed poor affinities, with the best binder displaying an affinity of only 238 nM. Further, a large proportion of the sybodies were specific for the SID. To identify binders with higher affinity, a NestLink experiment was performed according to Egloff et al.12 with the enriched pool of the second round of phage display using the loop and convex sybody libraries. According to the analysis of next generation sequencing (NGS), 578 unique loop sybodies were nested with 47’524 flycodes and 249 unique convex sybodies were nested with 17’628 flycodes. The nested libraries were expressed, purified by Ni2+-NTA and monomeric pool members were isolated by SEC. Monomeric nested sybodies were then mixed with M. thermoresistibile IrtABΔSID and separated again by SEC (running buffer, 20 mM Tris-HCl pH7.4, 150 mM NaCl, 0.3 % β-DM) to isolate sybody binders co-eluting with IrtABΔSID. Flycodes were then isolated, analyzed by LC-MS/MS and ranked as described12. Genes of 11 loop sybodies and 5 convex sybodies were ordered (General Biosystems), expressed in the pBXNPHM3 vector (Addgene #110099) and purified. Affinities of the purified sybodies were measured by surface plasmon resonance (SPR) and were in the range of 74 nM – 2.2 μM. Co-crystallization trails with M. thermoresistibile IrtABΔSID were performed with the two most affine sybodies and sybody Syb_NL5 (KD = 150 nM) of the loop library yielded well-diffracting crystals.
Crystallization, data collection and structure determination of M. thermoresistibile IrtABΔSID
SEC-purified IrtABΔSID was concentrated to 7.9 mg/ml (63 μM). The sybody Syb_NL5 was added to a final concentration of 50 μM, thereby diluting IrtABΔSID to a final concentration of 47 μM. The protein was crystallized at 4 °C by sitting-drop vapor diffusion by mixing 2 μl protein and 2 μl mother liquor containing 28 % (v/v) PEG400, 200 mM NaCl and 100 mM HEPES pH 7.5. Crystals appeared within three days, were cryo-protected using 30 % PEG400 and frozen in liquid nitrogen. Crystals were measured at the Swiss Light Source (SLS) beamline X06DA at λ = 1.0 Å. One dataset diffracting to 2.7 Å was used for structure determination. The diffraction data was processed by the program XDS43 in space group P21. A poly-Ala model of TM287 was manually fitted into the 7 Å cryo-EM density of IrtAB and used as the search model to solve the IrtAB structure by molecular replacement using the program Phaser44. A complete model for IrtABΔSID and Syb_NL5 was then built in Coot45 and refined with BUSTER (www.globalphasing.com) and PHENIX46 (Ramachandran favored/outliers: 94.22 %/0.47 %).
Structural analyses
The X-ray structures of the SID and IrtABΔSID were fitted into the cryo-EM density map using Chimera47 by placing them first manually close to the density and then using the Fit in Map tool. In case of IrtABΔSID, the structures were fitted in both possible orientations regarding the homologous IrtA and IrtB chain and average map values of 0.01218 (correct orientation) and 0.01008 (wrong orientation) were obtained. For the SID, both possible positions were tried and resulted in average map values of 0.01207 (correct orientation) and 0.01039 (wrong orientation). Superimpositions of structures were performed using the MatchMaker tool of Chimera. Nidelmann/Wunsch alignment algorithm and Blossom-62 matrix was used. R.m.s.d. (Å2) values were calculated over the entire polypeptide chains, i.e. without pruning of long atom pairs. Inward-facing cavities of IrtAB and TM287/288 (Fig. 1c) were calculated using the Channel Extract tool of the 3V web server48 using a shell probe radius of 9 Å and a solvent-excluded probe radius of 3 Å. Figures were prepared using Chimera47.
Redox measurements of SIDs
Redox experiments with the purified M. thermoresistibile and M. smegmatis SID proteins (wildtype and 3xE mutant) were performed under anaerobic conditions using a N2-purged continuous-flow anaerobic chamber. All solutions were made oxygen-free on a Schlenk-line by four alternating cycles of vacuum and Argon-flow. Proteins were diluted to final concentration of 5 μM using TBS pH 7.5 and 10 % glycerol. 100 μM Ferene was added to the samples to monitor Fe(III) reduction via the Fe(II)-Ferene complex formation (λmax = 590 nm)16. Fe(III)-cMBT or Fe(III)-MBT were added to a final concentration of 100 μM. Excess NADPH (final concentration 2 mM) was added to initiate ferric reduction. UV-Vis spectra were recorded at 10 min intervals at 37 °C and ΔA590 at time t was obtained by subtracting the initial A590 at time = 0 min. As control, the same reaction was performed with the M. smegmatis SID 3xE mutant. Redox measurements were conducted with two biological replicates and representative data are shown. For Fe-MBT concentration dependence (Extended Data Fig. 7b), the measurements were performed in a 96-well plate format as at least technical triplicates for every data point, sealed with a clear tape and the absorbance at 590 nm were taken using a Tecan Spark 20M at 37°C. The generated Fe(II) was quantified using a Fe(II)-Ferene calibration curve recorded at the same wavelength.
Biotinylation and SPR measurements of M. smegmatis SID
The M. smegmatis SID constructs (wildtype and 3xE mutant) were expressed in the pBXNH3A vector, which allows for N-terminal biotinylation via the Avi-tag49. Expression and purification were performed as described above. Biotinylation was performed simultaneously with 3C protease cleavage overnight. For biotinylation, 5 mM ATP, 10 mM MgOAc, 1.2-fold molar excess of biotin and 16 μg/ml BirA were added. The biotinylated SID constructs were used for SPR measurement using a Biacore T100 instrument (GE Healthcare). Approximately 15’000 response units of the SID constructs were coated onto a Sensor Chip CM5 (GE Healthcare). Afterwards, the coated lanes were saturated by coating with biotin-labelled BSA (Sigma) to avoid unspecific binding of cMBT to the surface of the chip. HPLC purified Fe-cMBT (MW 799 Da) was used as analyte at the concentrations of 0, 6.25, 12.5, 25, 50 and 100 μM. Measurements were conducted as technical duplicates.
Production of 55Fe-cMBT
55Fe-cMBT was prepared according to Ryndak et al.5. 200 μM Fe-cMBT in H2O was mixed with 50 mM EDTA pH 4 in a 1:1 volume ratio. The solution was incubated overnight at room temperature while shaking. The solution was filtered with a 0.22 μM filter (Whatman) and desferrated cMBT was extracted with one volume of chloroform. The chloroform phase was washed twice with one volume of H2O and evaporated. Desferrated cMBT was taken up in 100 % EtOH and 0.4 mCi 55FeCl3 (Perkin-Elmer) was added. Remaining desferrated cMBT was saturated by dropwise addition of cold FeCl3 until no further increase in red color was observed. 55Fe-cMBT was purified again by extraction with one volume of chloroform and washed twice with one volume of H2O to remove free 55Fe3+ and Fe3+.
55Fe-cMBT uptake in M. smegmatis cells
M. smegmatis DKO and TKO cells as well as M. smegmatis TKO cells complemented with different IrtAB constructs using the pFLAG vector24, 50 were grown in 7H9 for 3 nights at 37°C while shaking (140 rpm). 2 ml of these pre-cultures were used to inoculate 100 ml of MM supplemented with 6.88 μM ZnSO4, 1.82 μM MnSO4, 1.64 mM MgSO4 and 0.5% (v/v) Tween 80 (treated with aluminum-oxide) in 1-liter plastic roller bottles (Corning). Cells were grown for approximately 24 hours until they reached an OD600 of 3 – 5. The cultures were adjusted to an OD600 of 3 by diluting with medium. Six 1 ml aliquots of each strain or construct were placed on ice. 2 μl of 55Fe-cMBT stock solution was added to reach final concentrations of 2 μM. Three aliquots (technical triplicates) were then incubated at 37 °C while the remaining three aliquots were kept on ice (for background subtraction). At each time point, 200 μl from each replicate were filtered using a 96-well vacuum manifold equipped with a glass fiber filter (Filtermat A, Perkin Elmer). Filtration was immediately followed with two washing steps using 200 μl ice-cold medium. After all samples had been filtered and washed, the glass fiber filter was removed from the vacuum manifold and fused with a melt-on scintillator sheet (MeltiLex A scintillation gel, Perkin Elmer) at 65°C for 15 min. Radioactivity of incorporated hot iron was measured with MicroBeta Trilux Counter (Perkin Elmer) by counting for 2 min in the “Paralux Low-Background” mode. For each time point, the values obtained of the samples on ice were subtracted from the values obtained of samples incubated at 37 °C 5. Experiments were conducted twice as biological replicates and representative data are shown.
Reconstitution of M. thermoresistibile IrtAB into proteoliposomes and 55Fe-cMBT uptake
Purified M. thermoresistibile IrtAB (wildtype and 2xEQ mutant) was reconstituted into proteoliposomes as described by Geertsma et al.22. In brief, 0.2 ml of purified protein at a concentration of 1 mg/ml was added to detergent-destabilized liposomes in 50 mM KPi pH 7.0 with 20 % (v/v) glycerol. Detergent was then removed by subsequent addition of fresh Bio-Beads (BioRad). Inclusion of an ATP-regenerating system (10 mM ATP, 10 mM MgSO4, 24 mM Na2-creatine phosphate and 2.4 mg/ml creatine kinase) was also performed according to Geertsma et al.22, and the proteoliposomes were kept in 100 mM KPi pH 7.0 after the inclusion. Proteoliposomes at a final lipid concentration of 5 mg/ml were used for the 55Fe-cMBT uptake experiments. 55Fe-cMBT was added at a final concentration of 10 μM (corresponding to approximately 300’000 counts per min (cpm)) to the proteoliposomes, which were pre-incubated for 2 min at 37 °C, and the reaction was continued at 37 °C while shaking. At time intervals of 0.5, 2, 5 and 15 min, 3 aliquots of 40 μl (technical triplicates) were taken from the reaction. The samples were immediately centrifuged with ice-cold Micro Bio-Spin P-30 columns (BioRad, pre-equilibrated twice with 500 μl ice-cold 100 mM KPi pH 7.0 buffer) for 4 min at 1’000 g at 4°C to separate proteoliposomes from free 55Fe-cMBT. The elution containing the proteoliposomes was mixed with 220 μl scintillation liquid. The samples were transferred to a 96-well Isoplate-96 (Perkin-Elmer) and measured with a MicroBeta Trilux Counter (Wallac) by counting for 2 min in the “Paralux Low-Background” mode. The entire assay was performed twice with liposomes charged independently with the ATP regeneration system and representative data are shown.
Reconstitution of M. thermoresistibile IrtAB into nanodiscs and ATPase activity measurements
M. thermoresistibile IrtAB (wildtype and 2xEQ mutant) was concentrated after elution from the Ni2+-NTA gravity flow column and gel-filtrated using Superose 6 Increase 10/300 GL (GE Healthcare) size exclusion column in TBS containing 0.03 % β-DDM. The non-cleaved proteins (still containing GFP fused to the C-terminus of IrtB) were then reconstituted into nanodiscs. To this end, approximately 1 mg protein was mixed with MSP1D1E3 (prepared in house) and E. coli polar lipids in a molar ratio of 240:8:1 of lipids:MSP1D1E3:IrtAB, in a final volume of 200 μl. Lipids were prepared as follows: E. coli polar lipids in chloroform (Avanti) were mixed 3:1 (w/w) with L-α-phosphatidylcholine from egg yolk and evaporated. Dried lipids were dissolved in 20 mM HEPES pH 8.0, 0.5 mM K-EDTA, 100 mM NaCl and 100 mM cholate to a final concentration of 38 mg/ml and filtered using a 0.22 μM filter. The reconstitution mixture was incubated for 20 min at 25°C while shaking. 600 mg (3 mg/μl reconstitution mixture) Bio-Beads (BioRad) were added and the mixture was incubated at 4°C while shaking for 6 hours. Bio-Beads were removed by filtration and the sample containing reconstituted protein diluted 1:20 with TBS. 1.5 ml (3 ml slurry) HiTrap Ni2+-NTA (Qiagen) was added and batch binding was performed for 30 min at 4°C with end-over-end rotation. The resin was washed with 30 volumes of wash buffer (50 mM imidazole pH 7.5, 150 mM NaCl) and then incubated with 3C protease for 2 hours at 4°C with end-over-end rotation. The loaded resin was then transferred into an empty column to elute the cleaved protein reconstituted into nanodiscs. The sample was then gel-filtrated with a Superose 6 Increase 10/300 GL (GE Healthcare) size-exclusion column in TBS. Fractions corresponding to reconstituted IrtAB protein were collected and the concentration determined by A280. ATPase activity was determined by incubation of 10 to 15 nM reconstituted proteins at 37°C for one hour with 2.5 mM ATP and a final concentration of 10 mM MgSO4. A malachite green-molybdate solution was added as described previously to detect the release of inorganic phosphate by measuring the absorbance at 640 nm51. Substrates were added at final concentrations ranging from 0.5 to 40 μM to the protein prior to the incubation with ATP. All measurements were performed as technical triplicates, and the values of the IrtAB 2xEQ mutant were used for background subtraction. The ATPase assays with Fe-cMBT and Fe-MBT added at different concentrations (Fig. 2a) were conducted as three biological replicates and representative data are shown. ATPase assays with HPLC-purified mycobactins (Extended Data Figure 6c and d) were performed as a single biological replicate.
Generation of gene deletions and complementation strains in M. smegmatis
Gene deletions in M. smegmatis were generated as previously described by Arnold et al.24. In brief, upstream and downstream flanking regions of the genes of interest were amplified from genomic DNA. cloned into pINIT and sequenced. The following primers (Supplementary Table 1) were used: #21/#22 and #23/#24 for mbtD (msmeg_4512); #25/#26 and #27/#28 for fxbA (msmeg_0014); #29/#30 and #31/#32 for fxbBC (msmeg_0019); #33/#34 and #35/#36 for irtAB (msmeg_6553/6554). Subsequently, the flanking regions were transferred into the pKO (Apr) vector (Addgene #110088) by FX cloning. Approximately 1 μg of pKO vector containing the flanking regions was transformed into electrocompetent M. smegmatis cells and plated on 7H10 plates with apramycin. Apramycin-resistant clones were tested for the first recombination event by PCR. PCR-positive clones were grown in the absence of apramycin to allow for a second recombination event, and then plated on 7H10 plates containing 20 % sucrose and 0.2 % 2-deoxy-galactose. Clones were screened for deletion of the gene of interest by colony PCR. Colony PCR-positive clones were confirmed by PCR from isolated genomic DNA. For complementation, M. smegmatis IrtAB constructs were cloned in the pFLAG vector24. Approximately 250 to 500 ng of pFLAG vector with the respective construct was co-transformed with pMA_int (Addgene #110096) into electrocompetent M. smegmatisΔmbtDΔfxbAΔirtAB cells and plated on 7H10 plates with apramycin24. Apramycin-resistant clones were confirmed by colony PCR for integration of the pFLAG constructs.
Mycobactin-dependent growth assays in M. smegmatis
M. smegmatis strains with or without integrated pFLAG constructs were grown in 7H9 to stationary phase (3 to 4 nights) by inoculation from glycerol stocks. These precultures were inoculated 1:100 in 1 ml minimal medium with or without 200 μM 2,2′-dipyridyl (DPD, added from a 1’000x stock in ethanol). The cultures were supplemented with 0.5 % (v/v) Tween-80 to avoid cell clumping, and with different concentrations of Fe-cMBT and Fe-MBT crude extracts. 20 % Tween-80 stock solutions were autoclaved with 2 % (w/v) aluminium oxide and then filtered to remove traces of free iron. Antibiotics were not added for strains carrying pFLAG constructs, as pFLAG remains stably integrated into the genome even in the absence of antibiotic selection24. Cultures were grown for 5 days in plastic 96-deep-well plates (Treff Lab), with each strain grown in triplicates. 50 μl aliquots were taken every 24 hours and transferred to a clear 96-well Microplate (Greiner) for OD600 determination. All growth assays shown were performed at least with three biological replicates on different days and representative data are shown.
Extended Data
Extended Data Figure 1. Sequence identity of IrtAB homologues and crystal structure of IrtAB determined with the help of a sybody.
a, Sequence identity matrix of full-length IrtAB from M. thermoresistibile (Mth), M. smegmatis (Msm) and M. tuberculosis (Mtb). b, IrtABΔSID of M. thermoresistibile (lacking the SID and the linker between SID and TMD of IrtA) was crystallized with the help of a synthetic nanobody (sybody). IrtA is colored in turquoise, IrtB in purple and the sybody in grey with CDRs 1, 2 and 3 colored in yellow, orange and red, respectively.
Extended Data Figure 2. Cryo-EM data processing of IrtAB.
a, Typical motion corrected cryo-EM micrograph of the detergent solubilized IrtAB sample. Scale bar: 50 nm. b, Representative 2D class averages used for 3D reconstruction, showing side and bottom views. c, Data processing workflow. 2’168 high-quality micrographs were selected after motion correction and used for autopicking of 335’045 particles. Several rounds of 2D classification were performed to remove false-positives, yielding 167’781 IrtAB particles, which were subjected to 3D classification. One out of six classes (indicated by black box), containing 70’257 particles, revealed distinct structural features of IrtAB with its SID and was subjected to 3D refinement. The resulting structure was used to subtract the detergent micelle from the raw particles. Afterwards, a focused 3D refinement without the detergent micelle was performed. The resulting map was sharpened using a B-factor of -319 Å2. The final structure was resolved to 6.88 Å and reveals details of the TMDs, the NBDs and the SID.
Extended Data Figure 3. Cryo-EM data validation.
a, Angular distribution plot of all IrtAB particles that contributed to the final map. The map and the angular distribution plot have the same orientation. The height and color of the cylinder bars is proportional to the number of particles in those views. b, Plot of the directional Fourier Shell Correlation (FSC) that represents a measure of directional resolution anisotropy. Shown are the global FSC (red line), the spread of directional resolution values defined by ± 1 standard deviation from the mean of the directional resolutions (area encompassed by the green dotted lines) and a histogram of 100 directional resolutions evenly sampled over the 3D FSC (blue bars). A sphericity of 0.978 was determined at an FSC threshold of 0.5, which indicates very isotropic angular distribution (a value of 1 stands for completely isotropic angular distribution). The global resolution was determined to 6.9 Å (0.143 threshold). Directional FSC determination was performed with the 3DFSC software c, Fourier Shell Correlation (FSC) plot of the final density map of IrtAB. The plot shows the unmasked (green), masked (blue), phase randomized (red) and masking-effect corrected (black) FSC curves. The resolution at which the gold-standard FSC curve drops below the 0.143 threshold is indicated. d, Local resolution variations in the cryo-EM map. The resolution ranges from 5.5 to 10.8 Å, as calculated by BlocRes.
Extended Data Figure 4. Superimpositions with ABC exporter structures and structural comparison of IrtAB and TM287/288.
a, Superimposition of ABC exporters using the MatchMaker tool of Chimera. Nidelmann/Wunsch alignment algorithm and Blossom-62 matrix was used. R.m.s.d. (Å2) values were calculated over the entire polypeptide chains, i.e. without pruning of long atom pairs. In case the asymmetric unit contained several polypeptides of identical sequence, chain A was taken for analysis (indicated as “A” in the protein name). IF, inward-facing; OCC, occluded; OF, outward-facing. b, Structural deviations between IrtAB and the ABC exporter TM287/288 (PDB: 4Q4H). Shown is the structure of IrtAB in an “open book representation” with the entire transporter in the middle and the two half-transporters on the left (IrtB with domain-swap-helices TMH4 and 5 of IrtA) and on the right (IrtA with domain-swap-helices of IrtB). The structures are shown as sausage representation in which the thickness and color gradient from blue to red indicate variable degrees of structural deviations between IrtAB and TM287/288. Major structural deviations in the TMDs of IrtB are indicated.
Extended Data Figure 5. Full-length IrtAB structure shown in the membrane context.
MBT molecules with C17 aliphatic chain length are shown in stick representation. The SID is oriented towards the membrane with its mycobactin binding pocket (yellow) facing the inner leaflet.
Extended Data Figure 6. ATPase activity measurements using HPLC-purified mycobactins of defined masses.
a, Reversed-phase chromatography separation by HPLC of Fe-cMBT (top) and Fe-MBT (bottom) isolated from M. smegmatis. Masses of mycobactins giving rise to the main peaks were determined by mass spectrometry. These analyses were performed once. b, Chemical structures of Fe-cMBT and Fe-MBT. c, ATPase activities of nanodisc-reconstituted IrtAB measured in the presence of Fe-cMBT or Fe-MBT of defined mass isolated by HPLC. Crude extracts of mycobactins were included as controls. Mycobactins were added at a concentration of 5 μM. Data points are technical triplicates, which were used to calculate the mean value (black bar). d, ATPase stimulation curves determined over a range of mycobactin concentrations for HPLC-purified samples of defined mass or the respective crude extracts. Data points are technical triplicates and curves cross mean values.
Extended Data Figure 7. Mycobactin reduction by purified SID.
a, Reduction of Fe(III)-MBT (100 μM) by M. thermoresistibile (MTH) and M. smegmatis (MS) SIDs using NADPH as electron donor and Ferene as a reporter probe of released Fe(II) (Amax = 590 nm). The 3xE mutant served as negative control. Representative data of biological duplicates are shown. b, Reduction of Fe(III)-MBT by the M. thermoresistibile SID performed at different Fe(III)-MBT concentrations. The data were fitted using the Michaelis Menten equation. Data points are technical replicates.
Extended Data Figure 8. Siderophore-dependent growth assay.
When grown in minimal medium (MM) under controlled iron concentrations, wildtype M. smegmatis (WT), the M. smegmatis ΔfxbAΔmbtD double mutant (DKO) and the M. smegmatis ΔfxbAΔmbtDΔirtAB triple mutant (TKO) showed no significant differences in growth. Upon the addition of the weak iron chelator DPD, only the WT strain was able to grow owing to its ability to synthesize siderophores that extract iron bound to DPD. Data points are technical triplicates and curves cross mean values.
Extended Data Table 1. Cryo-EM data collection and processing statistics.
| lrtAB full-length (EMD -10319) | |
|---|---|
| Data collection and processing | |
| Magnification | 45455 |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 85 |
| Defocus range (μm) | -1.6 to -3.0 |
| Pixel size (Å) | 0.55 (in super-resolution) 1.10 (for reconstruction) |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 335045 |
| Final particle images (no.) | 70257 |
| Map resolution (Å) | 6.88 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 5.5 to 10.8 |
Extended Data Table 2. X-ray data collection and refinement statistics.
| SID * | lrtAB apo, inward-facing † | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 52.07, 111.7, 52.20 | 103.06, 78.58, 133.57 |
| α, β Y (°) | 90.000, 119.755, 90.000 | 90.000, 98.204, 90.000 |
| Resolution (Å) | 1.8 (1.864 - 1.8) ‡ | 2.7 (2.797- 2.700) |
| Rsym or Rmerge | 11.06 (68.53) | 11.4 (182.5) |
| I / σ/ | 11.44 (2.94) | 15.03 (1.21) |
| Completeness (%) | 95.20 (97.36) | 99.85 (99.69) |
| Redundancy | 6.2 (6.0) | 6.8 (6.9) |
| Refinement | ||
| Resolution (Å) | 45.32 - 1.80 | 47.43-2.70 |
| No. reflections | 45567 (4678) | 58375 (5791) |
| Rwork / Rfree | 0.2229 / 0.2546 | 0.2565 / 0.2907 |
| No. atoms | ||
| Protein | 3674 | 9674 |
| Ligand/ion | 106 | 34 |
| Water | 520 | 54 |
| B-factors | ||
| Protein | 23.39 | 92.27 |
| Ligand/ion | 15.10 | 107.25 |
| Water | 29.60 | 57.20 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.002 | 0.004 |
| Bond angles (°) | 0.53 | 0.99 |
A single crystal was used for the SID structure.
A single crystal was used for the IrtAB apo, inward facing structure.
Values in parenthesis are shown for highest resolution shell.
Supplementary Material
Acknowledgements
We thank all members of the Seeger lab for stimulating discussions. We acknowledge Beat Blattmann, Céline Stutz-Ducommun and Stefanie Eberle of the Protein Crystallization Center UZH for performing the crystallization screening, and the staff of the SLS beamlines X06SA and X06DA for their support during data collection. We thank Jens Sobeck of the Functional Genomics Center Zurich for his support during SPR measurements. Work in the lab of MAS was supported by the European Research Council (ERC) (consolidator grant n° 772190), a SNSF Professorship of the Swiss National Science Foundation (PP00P3_144823), and a grant of the Novartis Foundation for Medical-Biological Research. Work in the lab of OM was supported by the Swiss National Science Foundation (grant 31003A_179418) and the Mäxi Foundation. The work in GM’s laboratory was supported by the Robert A. Welch Foundation (Grant: AT-1935-20170325) and by the National Institute of General Medical Sciences of the National Institutes of Health (R35GM128704). FMA, MSW and IG were supported by three Candoc fellowships of the University of Zurich.
Footnotes
Data Availability
The crystal structures of the SID and IrtAB-Syb_NL5 have been deposited at the protein database (PDB entries 6TEJ and 6TEK). The cryo-EM map has been deposited on the protein database under accession number EMD-10319. Plasmids, strains, the sybody Syb_NL5 and raw data are available from the authors upon reasonable request.
Author Contributions
FMA and MAS conceived the project. FMA cloned all ORFs, established the protein purification protocols, and generated gene deletions in M. smegmatis. FMA established the mycobactin extraction protocols with the help of SA and carried out the great majority of the biochemical and in vivo characterization of IrtAB. FMA, IG and LMH carried out 55Fe-cMBT transport assays with intact cells and proteoliposomes. FMA selected Syb_NL5 with the help of PE and IZ. MSW carried out all cryo-EM work and data analysis under the supervision of OM with samples prepared by FMA. FMA solved the structure of the SID with the support of CAJH. IG crystallized the IrtAB-Syb_NL5 complex with the help of FMA and solved its structure with the help of MAS. IG built the IrtAB model with the support of CAJH and MAS. MJFG carried out the mycobactin reduction experiments under the supervision of GM. EP purified mycobactins by HPLC under the supervision of JP. FMA, MSW, IG, GM, OM and MAS interpreted the data. FMA, MSW, IG and MAS prepared Figures. FMA, MSW and MAS wrote the paper. IG, GM and OM edited the paper.
Competing Interests
The authors declare no competing interests.
References
- 1.De Voss JJ, et al. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci U S A. 2000;97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snow GA, White AJ. Chemical and Biological Properties of Mycobactins Isolated from Various Mycobacteria. Biochem J. 1969;115 doi: 10.1042/bj1151031. 1031-&. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gobin J, et al. Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci U S A. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez GM, Smith I. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J Bacteriol. 2006;188:424–430. doi: 10.1128/JB.188.2.424-430.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryndak MB, Wang SS, Smith I, Rodriguez GM. The Mycobacterium tuberculosis High-Affinity Iron Importer, IrtA, Contains an FAD-Binding Domain. J Bacteriol. 2010;192:861–869. doi: 10.1128/JB.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells RM, et al. Discovery of a Siderophore Export System Essential for Virulence of Mycobacterium tuberculosis. PLoS pathogens. 2013;9 doi: 10.1371/journal.ppat.1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegrist MS, et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tufariello JM, et al. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc Natl Acad Sci U S A. 2016;113:E348–E357. doi: 10.1073/pnas.1523321113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 10.ter Beek J, Guskov A, Slotboom DJ. Structural diversity of ABC transporters. J Gen Physiol. 2014;143:419–435. doi: 10.1085/jgp.201411164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmermann I, et al. Synthetic single domain antibodies for the conformational trapping of membrane proteins. Elife. 2018;7 doi: 10.7554/eLife.34317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egloff P, et al. Engineered peptide barcodes for in-depth analyses of binding protein libraries. Nat Methods. 2019;16:421–428. doi: 10.1038/s41592-019-0389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutter CAJ, et al. The extracellular gate shapes the energy profile of an ABC exporter. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09892-6. 2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohl M, Briand C, Grütter MG, Seeger MA. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat Struct Mol Biol. 2012;19:395–402. doi: 10.1038/nsmb.2267. [DOI] [PubMed] [Google Scholar]
- 16.Miethke M, Hou J, Marahiel MA. The Siderophore-Interacting Protein YqjH Acts as a Ferric Reductase in Different Iron Assimilation Pathways of Escherichia coli. Biochemistry. 2011;50:10951–10964. doi: 10.1021/bi201517h. [DOI] [PubMed] [Google Scholar]
- 17.Hürlimann LM, et al. The Heterodimeric ABC Transporter EfrCD Mediates Multidrug Efflux in Enterococcus faecalis. Antimicrob Agents Chemother. 2016;60:5400–5411. doi: 10.1128/AAC.00661-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambudkar SV, et al. Partial purification and reconstitution of the human multidrug-resistance pump: characterization of the drug-stimulatable ATP hydrolysis. Proc Natl Acad Sci U S A. 1992;89:8472–8476. doi: 10.1073/pnas.89.18.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JY, Yang JG, Zhitnitsky D, Lewinson O, Rees DC. Structural Basis for Heavy Metal Detoxification by an Atm1-Type ABC Exporter. Science. 2014;343:1133–1136. doi: 10.1126/science.1246489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tombline G, Bartholomew LA, Urbatsch IL, Senior AE. Combined mutation of catalytic glutamate residues in the two nucleotide binding domains of P-glycoprotein generates a conformation that binds ATP and ADP tightly. J Biol Chem. 2004;279:31212–31220. doi: 10.1074/jbc.M404689200. [DOI] [PubMed] [Google Scholar]
- 21.Al-Shawi MK, Polar MK, Omote H, Figler RA. Transition state analysis of the coupling of drug transport to ATP hydrolysis by P-glycoprotein. J Biol Chem. 2003;278:52629–52640. doi: 10.1074/jbc.M308175200. [DOI] [PubMed] [Google Scholar]
- 22.Geertsma ER, Nik Mahmood NA, Schuurman-Wolters GK, Poolman B. Membrane reconstitution of ABC transporters and assays of translocator function. Nat Protoc. 2008;3:256–266. doi: 10.1038/nprot.2007.519. [DOI] [PubMed] [Google Scholar]
- 23.Dragset MS, et al. Genome-wide Phenotypic Profiling Identifies and Categorizes Genes Required for Mycobacterial Low Iron Fitness. Sci Rep. 2019;9 doi: 10.1038/s41598-019-47905-y. 11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold FM, et al. A uniform cloning platform for mycobacterial genetics and protein production. Sci Rep. 2018;8 doi: 10.1038/s41598-018-27687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prados-Rosales R, et al. Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol. 2014;196:1250–1256. doi: 10.1128/JB.01090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quazi F, Lenevich S, Molday RS. ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat Commun. 2012;3 doi: 10.1038/ncomms1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu D, et al. Cryo-EM structure of human lysosomal cobalamin exporter ABCD4. Cell Res. 2019 doi: 10.1038/s41422-019-0222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez C, et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 2015;524:433. doi: 10.1038/nature14953. [DOI] [PubMed] [Google Scholar]
- 29.Ratledge C, Ewing M. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology. 1996;142(Pt 8):2207–2212. doi: 10.1099/13500872-142-8-2207. [DOI] [PubMed] [Google Scholar]
- 30.Geertsma ER, Dutzler R. A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry. 2011;50:3272–3278. doi: 10.1021/bi200178z. [DOI] [PubMed] [Google Scholar]
- 31.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moeller A, et al. Distinct Conformational Spectrum of Homologous Multidrug ABC Transporters. Structure. 2015;23:450–460. doi: 10.1016/j.str.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Scheres SHW, Chen SX. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Cardone G, Heymann JB, Steven AC. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan YZ, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat Methods. 2017;14 doi: 10.1038/nmeth.4347. 793-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF, et al. UCSF chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Voss NR, Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38:W555–562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bukowska MA, et al. A Transporter Motor Taken Apart: Flexibility in the Nucleotide Binding Domains of a Heterodimeric ABC Exporter. Biochemistry. 2015;54:3086–3099. doi: 10.1021/acs.biochem.5b00188. [DOI] [PubMed] [Google Scholar]
- 50.Hohl M, et al. Increased drug permeability of a stiffened mycobacterial outer membrane in cells lacking MFS transporter Rv1410 and lipoprotein LprG. Mol Microbiol. 2019 doi: 10.1111/mmi.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hohl M, et al. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc Natl Acad Sci U S A. 2014;111:11025–11030. doi: 10.1073/pnas.1400485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.