Abstract
Background:
Lead is a ubiquitous toxicant following three compartment kinetics with the longest half-life found in bones. Patella and tibia lead levels—validated measures of cumulative exposure—require specialized X-ray-fluorescence-spectroscopy available only in a few centers worldwide. We developed minimally-invasive biomarkers reflecting individual cumulative lead exposure using blood DNA methylation profiles—obtainable via Illumina450K or IlluminaEPIC bead-chip assays.
Methods:
We developed and tested two methylation-based biomarkers from 348 Normative Aging Study (NAS) elderly men. We selected methylation sites with strong associations with bone lead levels via robust regressions analysis and constructed the biomarkers using elastic nets. Results were validated in a NAS subset, reporting specificity and sensitivity.
Findings:
Participants were 73 years old on average (standard deviation, SD=6), with moderate lead levels of (mean±SD patella:27±18 μg/g; tibia:21±13 μg/g). Methylation-based biomarkers for lead in patella and tibia included 59 and 138 DNA methylation sites, respectively. Estimated lead levels were significantly correlated with actual measured values, (r=0.62 patella, r=0.59 tibia) and had low mean square error (MSE) (MSE=0.68 patella, MSE=0.53 tibia). Means and distributions of the estimated and actual lead levels were not significantly different across patella and tibia bones (p>0.05). Methylation-based biomarkers discriminated participants highly exposed (>median) to lead with a specificity of 74% and 73% for patella and tibia lead levels, respectively, with 70% sensitivity.
Interpretation:
DNA-methylation-based lead biomarkers are novel tools that can be used to reconstruct decades’ worth of individual cumulative lead exposure using only blood DNA methylation profiles and may help identify the consequences of cumulative exposure.
Introduction
Lead exposure is a global public health concern with adverse and chronic effects on the cardiovascular, neuro-cognitive, and renal systems1. Exposure to lead has been also shown to play a critical and detrimental role in both bone maturation and loss.2, 3 Between 2011 and 2012, the United States Occupational Safety and Health Administration (OSHA) estimated that more than 1·6 million employees, mostly men, working in manufacturing and construction have been exposed to lead on an annual basis4, and more than 2,000 U.S. adults had persistently elevated blood lead levels in 20125, 6.
Although blood lead testing repeated over time is widely used as a biomarker for persistent lead exposure, bone (patella and tibia) lead levels are better biomarkers than blood lead levels in capturing cumulative lead exposure and evaluating its long-term health effects in epidemiology studies7-9. Because half-life for lead are over a decade, lead levels in patella and tibia measure cumulative exposure over 8–20 years10 and up to 50 years11, respectively. However, patella and tibia bone lead levels are highly specialized measures and require exposure to radiation. The technology required for scanning bones is available only in a small number of centers worldwide and is often not available for clinical applications. Thus, most patients cannot be easily tested for cumulative lead exposure as this resource is unavailable. Bone lead can be an important adjunct to blood lead data, as blood lead has a half-life of approximately between 30 to 90 days following exposure. Combining blood and bone lead data can help to distinguish whether lead exposure is chronic vs acute.
Lead exposure has been previously associated with alterations in whole blood DNA methylation, one of the most extensively studied epigenetic markers12. DNA methylation is the addition of a methyl-group to a cytosine nucleotide when it is followed by a guanine base (CpG). DNA methylation profiles are relatively stable and inherited during cellular division but are altered by environmental factors, including the presence of lead13, 14. Once established, DNA methylation changes can persist even in the absence of the factors that induced them. Therefore, DNA methylation has unique properties to serve as a biomarker of cumulative exposure15.
Indeed, previous studies have shown that DNA methylation captures cumulative lifetime lifestyle experiences. For instance, studies of tobacco smoking have shown that DNA methylation changes reflect cumulative exposure to tobacco, including years of smoking, pack-years, and years since quitting16. However, no study to date has evaluated whether DNA methylation can serve as a surrogate for other measures of chronic exposures to environmental toxicants. In previous studies, lead levels measured in patella were negatively associated with whole blood DNA methylation in repetitive elements in a cohort of elderly men 12, and individuals exposed to extremely high blood levels of lead (>50 μg/dL) had higher methylation in the promoter region of specific tumor suppressor genes17. These findings suggest that lead exposure may induce specific alterations in blood DNA methylation.
A common methodology for DNA methylation analysis uses microarrays to measure DNA methylation levels at approximately half million CpG sites for each DNA sample. This platform has been used in epigenome-wide association studies in which DNA methylation levels at each CpG site are analyzed individually and has been increasingly employed to uncover biological mechanisms that underlie extrinsic environmental stimuli and adverse health outcomes18-20. However, most previous studies have identified CpGs modified in the presence of environmental exposures or disease states. Because DNA methylation is sensitive to external factors and can show changes at multiple CpG sites in response to environmental toxicants, multiple changes can be combined to build a composite estimator of cumulative environmental exposures.
In the present study, we analyzed whole blood DNA methylation profiles with the Infinium HumanMethylation450 assay in a cohort of men with well characterized lead exposure measured via bone testing. We hypothesized that CpG sites with DNA methylation proportions associated with lead levels could provide information about past environmental lead exposures. Specifically, we assessed whether a machine learning approach, elastic-net, could identify a combination of CpGs to serve as a composite biomarker reflecting bone lead levels.
Materials and Methods
Study sample
The Normative Aging Study (NAS) is an ongoing longitudinal cohort of adult men established in 1963 by the U.S. Department of Veterans Affairs 21. The cohort of 2,280 men was recruited between 1961 and 1970 in eastern Massachusetts. Cohort members were 21 to 80 years old at the time of enrollment; exclusion criteria included chronic medical conditions (e.g., hypertension, heart disease, diabetes, cancer, peptic ulcer, gout, recurrent asthma, bronchitis, or sinusitis). Each NAS participant has returned every 3–5 years for follow-up clinical examinations and, at each visit, provided demographic and lifestyle information 7. During 1999–2012, 579 men provided whole blood draws for DNA methylation measurements. Out of these men, 375 had also undergone bone lead testing. Participants with leukemia or blood cancer (N = 3), participants who were not white (N = 18), and those with missing demographics and lifestyle information were excluded, leaving a total of 354 participants with at least one lead level measurement. All participants in this analysis underwent examination of patella lead levels, while 350 had tibia lead concentration measures. Approval from the Institutional Review Boards at the Veterans Affairs Boston Healthcare System, Brigham and Women’s Hospital, and the Harvard T.H. Chan School of Public Health was obtained prior to study commencement. All subjects provided written informed consent before participating.
Lead exposure assessment
We measured participants’ skeletal lead content via K-shell X-ray fluorescence (KXRF) spectroscopy, a noninvasive technique able to distinguish among very low lead burdens 10, 22. The instrument provided an unbiased estimate of bone lead levels corrected for bone mineral density and expressed as micrograms of lead per gram of bone mineral (μg/g). Bone lead concentrations were measured at the left patella and mid-tibial shaft 7.
DNA methylation measurements
Bisulfite conversion (EZ-96 DNA Methylation Kit, Zymo Research, Orange, CA, USA) was performed on DNA extracted from the buffy coat of whole blood samples, and the Illumina Infinium HumanMethylation450 BeadChip (450k) array was used to measure DNA methylation of ~480,000 CpG sites in each sample. We minimized batch effects with a two-stage age-stratified algorithm. For quality control, we removed samples and probes in which >1% of probes and samples, respectively, had a detection p-value of >0·05. We then preprocessed the remaining samples with background correction, dye-bias, and BMIQ adjustments. We generated DNA methylation beta values at each CpG site as the ratio between the intensity of methylated signal and the intensity of both methylated and un-methylated signals 23. Beta values at each site reflect the methylation percentage for each site in the array. In the analysis, we excluded control probes, non-CpG sites, probes that mapped to allosomal chromosomes, cross-reactive CpGs, probes with an underlying single nucleotide polymorphism (SNP) within 10 bp of the target CpG. Also, to facilitate the lead biomarkers estimation using the Infinium MethylationEPIC BeadChip (EPIC) platform, we included only probes overlapping in both 450k and EPIC platforms24-26. This left a total of 395,005 CpGs to be included in the analysis. We estimated white blood cell counts from DNA methylation using Houseman’s method 27.
Statistical analysis
We ran two separate sets of analyses to examine the association between lead levels in patella and tibia with DNA methylation beta values. We log2-transformed lead concentrations to remove skewness of the exposure distribution in both tissues. In the analysis, we included bone lead measurements taken at the same time or at the closest time visit within an eight-year span of the blood draw, based on the rationale that lead persists in bones for at least eight years 7. Negative estimates of bone lead concentrations occurred for 2% (N = 6) of patella measurements and 1% (N=3) of tibia levels and were discarded from the analysis. We detected lead level outliers using the interquartile range rule and excluded those from the analysis (N = 0 for patella; N = 2 for tibia), thus, leaving a total of 348 men with at least one lead measurement for the analysis.
Epigenome-wide analysis: identification of DNA methylation sites responsive to lead levels independent of predictors and confounders
We performed epigenome-wide robust linear regression to account for potential outliers or heteroskedasticity of DNA methylation beta values 28. We used actual methylation betas to facilitate reproducibility and interpretability of the results as incremental percent change in methylation at each probe for a doubling increase of lead levels and to maintain a linear relationship between DNA methylation levels and white blood cell proportions 29. To select covariates of the epigenome-wide analysis, we performed a principal component analysis of the beta-valued data matrix and list of variables 25. We selected variables showing the major source of variability in the first 25 singular components and adjusted all analyses for age (continuous), education, alcohol consumption (≤2 or >2 drink per day), smoking status (never/ever) and pack-years consumption (continuous), technical variables (plate, position chip on the plate, row), and white cell proportions. Quality checks for the epigenome-wide analysis included lambda genomic-inflation factor and quantile-quantile plots for distribution of p-values. To identify sets of CpGs associated with the exposure variables, we also performed regional analyses via DMRcate 30.
Elastic net approach: creation of lead biomarkers
We selected the most significant CpG sites (p < 0·0001) in agreement with the independent screening approach 31. We then divided the dataset into training and test datasets. We randomly selected 80% of individuals to be in the training dataset and the remaining 20% to be in the test dataset. We performed an elastic-net on the training set with a leave-one-out cross validation for tuning alpha and lambda parameters. We selected parameters that minimized the cross-validated mean squared error and then saved the elastic net regression coefficients. We computed lead biomarkers for bones (patella and tibia) as the linear combination of regression coefficients and DNA methylation beta-values matrix of the test dataset.
Validation of lead biomarkers in subsets of the NAS dataset
We evaluated the quality of lead levels predicted by biosensors by comparing them with actual lead measurements in the testing datasets using Pearson’s correlation coefficients, mean square errors, tests for difference in means, Kolmogorov-Smirnov’s tests, and descriptive plots. We ultimately discriminated high level of exposures using as threshold the median for both patella and tibia lead levels. At those thresholds, we performed receiver operating characteristic (ROC) analyses to assess sensitivity and specificity of the models and computed the area under the curve (AUC) to calculate accuracy.
Assessment of biological pathways associated with methylation sites
We identified biological (KEGG: Kyoto Encyclopedia of Genes and Genomes) pathways of genes targeted by the CpG sites in each epigenetic lead biomarker using the missMethyl package 32. The analysis leveraged on Young et al.’s method,33 which took into account the selection bias of each gene, adjusting for the number of CpGs associated to each gene, and selected enriched pathways based on Wallenius’ noncentral hypergeometric distribution test. For each pathway, we provided the number of unique genes that were selected based on the significant CpG sites, and the p-value indicating statistical significance for over-representation for the pathway. All analyses were performed in R version 3·5·2 and code is available upon reasonable requests.
Results
NAS participants were an average of 73 years old [standard deviation (SD) = 6], and 67% were former or current smokers. Participants had moderate levels of lead in their bones (patella mean ± SD = 27 ± 18 μg/g; tibia mean ± SD = 21 ± 13 μg/g) (Table 1). The top 25 singular principal components of DNA methylation data explained more than 60% of the variance for each dataset (62% with patella lead levels; 61% with tibia lead levels) and showed strong associations with estimated cell type composition, technical variables, and age and moderate associations with phenotype characteristics and the exposure variable (Figures S1,S2).
Table 1.
Descriptive statistics of 348 participants from the Normative Aging Study (NAS)
| Variable | Mean ± SD |
|---|---|
| Age (years) | 72.69 ± 6.18 |
| Education (years) | 15.08 ± 3.09 |
| Pack-years | 20.30 ± 24.82 |
| Smoking status | |
| Never | 115 (0.33) |
| Ever | 233 (0.67) |
| Alcohol consumption | |
| ≤2 drinks per day | 282 (0.81) |
| >2 drinks per day | 66 (0.19) |
| Patella lead levels | 27.36 ± 17.75 |
| Tibia lead levels* | 21.05 ± 12.91 |
Measured among 345 participants.
Epigenome-wide analysis: identification of DNA methylation sites responsive to lead levels independent of predictors and confounders
In the adjusted robust linear regression models and with a Bonferroni significance level of 0.05, one CpG (cg13251292) was differentially methylated relative to patella lead exposure (Table S1) and one CpG (cg20326704) relative to tibia lead exposure (Table S2). We observed a 1% increase in methylation levels of cg13251292 and cg20326704 for every doubling in patella and tibia lead concentrations, respectively. Cg13251292 is annotated to SERPINA6 in chromosome 14 (β = 0·3%; 95% CI: 0·2, 0·5; P = 5·3 × 10−7), while cg20326704 is annotated to TET1 in chromosome 10 (β = 0·3%; 95% CI: 0·2, 0·5; P = 5·3 × 10−7). All epigenome-wide analysis had a genomic inflation factor of ~1 (patella: 1·10; tibia: 1·16), showing no excess of false positive rates (Figure S3). Regional analysis showed null results for both tissues (data not shown).
Elastic net approach: creation of lead biomarkers
For each of the two epigenome-wide analyses (patella lead and tibia lead), we selected CpGs associated with lead concentrations at p < 10−4. We identified 118 CpGs associated with patella lead concentrations (Table S3) and 138 CpGs associated with tibia lead concentrations (Table S4). For each measurement, we randomly created a training dataset, containing 80% of individuals, and a test dataset with the remaining 20%. In total, there were 278 and 274 individuals in the training datasets for patella and tibia, respectively; and 70 and 68 participants in the test datasets for patella and tibia, respectively. In each training dataset, we tuned elastic net parameters via leave-one-out cross-validation and selected parameters that minimized mean square error (Figures S4-S5). The best model for the patella lead biosensor was an elastic net with alpha parameter of 0.1, while for the tibia lead biosensor the best model was a ridge regression with alpha of 0 (Figure S4). We extracted regression coefficients from each model. Biomarkers for patella and tibia included 59 and 138 sites, respectively, with not-null coefficients and a balanced distribution of positive and negative coefficients (% of positive: 50% in patella, 63% in tibia). Among those sites, cg10042319—annotated to NMUR1 in chromosome 2—had positive coefficients in both biomarkers (patella: coefficient = 3·16; tibia: coefficient = 0·84).
To determine biomarkers of lead exposure in the test dataset, we extracted all regression coefficients from elastic nets (Tables S3, S4) and applied those to the corresponding beta DNA methylation matrix.
Validation of lead biomarkers in subsets of the NAS dataset
We represented the relationship between actual and estimated values in each tissue with scatterplots (Figure 1). Pearson’s correlation coefficients between actual and estimated lead levels were moderate for all analyses (r = 0·62 for patella; r = 0·59 for tibia), and mean square errors (MSE) were close to zero for all tissues (MSE = 0·68 for patella; MSE = 0·53 for tibia) (Figure 1). Means of the estimated and actual lead levels were not significantly different across the two types of lead measures (t-tests: p > 0·05) (Figure S6). Kolmogorov-Smirnov tests showed that the estimated and actual lead levels likely came from the same distribution for both patella and tibia (p > 0·05) (Figure S7).
Figure 1.
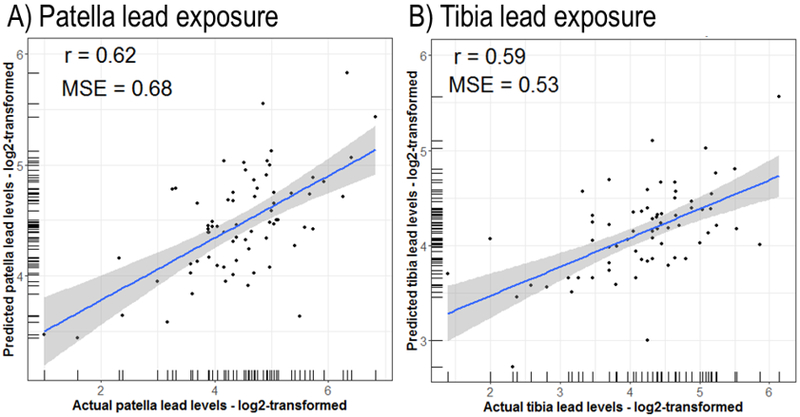
Relationship between actual and estimated (log2-transformed) lead levels in A) patella and B) tibia.
r = Pearson coefficient; MSE = mean square error.
In the test datasets, 32 men were over the median threshold (24 μg/g) for patella lead levels and 35 participants were over the median (19·1 μg/g) for tibia lead levels. With the novel epigenetic lead biomarkers and setting a sensitivity level of 70% for both biomarkers, we discriminated individuals with high lead levels with specificity of 74% and 73% for patella and tibia lead levels, respectively. The accuracy of those models was good for both tissues (79% for patella; 75% for tibia) (Figure 2).
Figure 2.
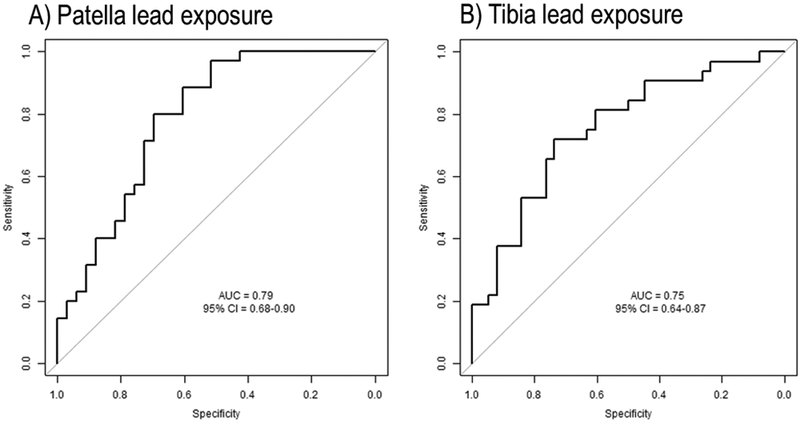
Receiver operating characteristic (ROC) and area under the curve (AUC) with 95% confidence interval (95% CI) for A) patella and B) tibia lead exposure. High levels of exposures were discriminated using the median for both patella (24 μg/g) and tibia (19.1 μg/g) lead levels as a threshold.
Biological pathways associated with methylation sites
To explore the biological significance of genes targeted by CpG sites in each epigenetic lead biomarker, we conducted KEGG pathway enrichment analysis. A suggested enrichment for Alzheimer’s disease was observed among genes mapped to CpGs in the patella epigenetic biomarker (p = 0·055), while the most significantly enriched pathway for genes mapped to CpGs in the tibia epigenetic biomarker was protein digestion and absorption (p = 0·04) (Table 2).
Table 2.
KEGG pathway analysis of methylation sites of patella and tibia lead biomarkers
| Pathway* | Gene count |
Gene Names | p |
|---|---|---|---|
| Patella | |||
| Alzheimer’s disease | 2 | GRIN1; UQCRFS1 | 0.055 |
| Glycosaminoglycan biosynthesis; heparan sulfate/heparin | 1 | HS3ST3A1 | 0.058 |
| Alcoholism | 2 | GRIN1; CAMKK1 | 0.059 |
| Apoptosis | 1 | BIRC2 | 0.066 |
| Huntington’s disease | 2 | GRIN1; UQCRFS1 | 0.068 |
| Tryptophan metabolism | 1 | HAAO | 0.072 |
| Nicotine addiction | 1 | GRIN1 | 0.095 |
| Tibia | |||
| Protein digestion and absorption | 2 | COL6A3; ELN | 0.036 |
All KEGG pathways reported based on p < 0.10 and considered to be enriched pathways based on unadjusted p < 0.05
Discussion
We developed and tested novel and minimally-invasive epigenetic biomarkers of cumulative lead exposure using whole blood DNA methylation profiles measured in men participating in the NAS. Epigenetic biosensors of patella and tibia were linear combinations of 59 and 138 CpGs, respectively, identified by site-by-site analysis and combined via machine learning algorithms. These novel epigenetic biosensors estimated accurately actual lead levels in patella and tibia. Our analysis confirmed that blood DNA methylation can serve as a biomarker of past lead exposure. We also provided detailed information about each CpG site in order to retrospectively estimate past bone lead levels in individuals who had undergone only whole blood methylomic testing via Illumina 450K or Illumina EPIC.
Interestingly, the two sets of CpGs composing epigenetic lead biomarkers were mapped to genes biologically related to diseases or processes previously associated with lead exposure. We identified genes involved in Alzheimer’s disease that were previously associated with both cumulative lead exposure and global changes in whole blood DNA methylation 1. Genes identified by the tibia biomarker were involved in nutritional pathways, including low-protein diets, which have been linked to increased lead absorption 34, 35. These findings support the hypothesis that DNA methylation is an intermediate mechanism between lead exposure and the development of chronic and neuro-cognitive diseases.
While DNA methylomic measures may at first glance seem inefficient to reconstruct exposure to lead, there are research incentives to do so. Methylomic data is commonly assessed as a biological marker of health in many studies. Such data may have within important information on environmental exposures. Such exposure would require the use of a bio-archive to assess, which may or may not exist. Lead is most commonly measured in whole blood, thus cohorts which separated whole blood into plasma and packed red blood cells would not be able to reconstruct blood lead levels. XRF measures of bone lead are highly technical and only a handful of institutions have access to this technology. The ability to reconstruct past exposure to environmental chemicals using extant methylomic data may open the door to a number of research questions that would have prospective data on exposure and health, that otherwise would not exist. Furthermore, these methods may lay the ground work for future research in which other environmental exposure measures from the distant past are reconstructed using epigenomic signatures on extant 450K and EPIC methylomic data.
Our findings showed a good performance of all lead epigenetic biomarkers, with a stronger association between actual and estimated lead levels in patella than in tibia. This may be explained by the heterogeneity of bone tissues—the patella and tibia are trabecular and cortical bones, respectively, and have different bone densities and resorption properties. Trabecular bone is light, porous, filled with marrow and several vascular channels, and resorption takes place along the bone surfaces, while cortical bone forms a dense cylinder down the shaft of the bone surrounding the central marrow cavity, and its resorption tunnels through the bone itself.10 Trabecular bones, such as patella, are more biologically active with higher turnover of lead into the bloodstream than cortical bones like the tibia 10. This is of particular interest for elderly populations, for whom trabecular resorption increases more than in cortical dense bones due to an increase in the surface area exposed to blood flow 10. For this reason, whole blood DNA methylation may be more representative of lead levels in patella than in tibia.
To determine epigenetic lead biomarkers, we employed an ensemble approach, combining site-by-site robust linear regressions and machine learning algorithms. Ensemble approaches are often found to yield better predictive performance than other constituent algorithms 36. In our case, the contribution of this approach was two-fold. First, we reduced complexity with an initial screening of individual sites of interest, providing more insight into the relationship between DNA methylation and lead exposure after removing confounding by cell-proportion shifting, lifestyle, demographic, and technical factors. Thus, the selected sites were associated with lead levels at the net influence of population-specific confounders. Second, we selected the most informative CpGs and uncovered the relationships between correlated CpGs and lead exposures by adopting flexible machine learning approaches. Site-by-site analysis showed no excess of false positive rates and a balance between positive and negative findings, but it was limited by Bonferroni’s correction, which was overly conservative and ignored dependencies among sites 37. We relaxed the significance threshold and elastic nets overcame limitations of the individual analysis, grouping highly correlated sites and estimating the relationships via a regularization regression method. This approach facilitated detection of sites associated with lead exposures that otherwise would be discarded in site-by-site analysis and that contribute jointly with other CpGs to the characterization of past lead exposure.
Other biomarkers for cumulative lead levels have been previously investigated, including biochemical blood parameters such as serum phosphorus, uric acid, total and HDL cholesterols, and socio-demographic data, which are not always retrospectively available 38. Predicted lead measurements from these biomarkers have had correlation coefficients with actual lead levels of <0.5, which was lower than our findings 38. However, none of these other biomarkers included any epigenetic measures.
Based on DNA methylation characteristics of stability and heritability during cellular division and the persistence of alterations after environmental exposures, a few epigenetic biomarkers were constructed based on a combination of CpG sites that reflect age and cumulative smoking. DNA methylation levels of 353 sites predicted age with good correlation between predicted and chronological age (r > 0·8) in several tissues and with correlation similar or worse than that of lead biomarkers in muscles (r = 0·70), fat adipose tissue (r = 0·65), uterine endometrium (r = 0·55), various blood samples (r = 0·46), and cancer tissues (r = 0·16) 39. In addition, cord blood DNA methylation of 28 sites in newborns reflected prenatal exposure to tobacco smoking 16. The smoking biomarker sensitivity showed lower performance than our lead biomarkers when validated on the test dataset 16. Both epigenetic biomarkers of age and tobacco exposure have corresponding demographic (date of birth) and biochemical blood/serum parameters (cotinine) that are inexpensive, easy, and safe to collect even retrospectively from frozen samples. In our study, we identified 59 and 138 CpGs whose whole blood DNA methylation levels reflected two temporal past lead exposures in patella and tibia respectively. Cumulative lead exposure is often under-tested in clinical applications and entails radiation exposure when assessed via X-fluorescence (gold standard). Therefore, our epigenetic lead biomarkers may provide an easier, more accessible, and safer method to assess cumulative exposure to lead, even retrospectively, when only blood testing is available.
Our study population consisted of elderly white men, the majority of whom were retired at the time of the study, limiting the generalizability of our results to populations of other races, sex, and different levels of lead exposure (such as active occupational exposure). However, most NAS participants, having lived in the era of leaded gasoline combustion and being veterans, were exposed to both environmental and occupational lead levels throughout their life-course and thus may reflect exposure trajectories of millions of employees in general industry and construction.40 Other limitations include the use of DNA methylation to impute white blood cell distribution, and residual confounding by cell type distribution could still remain. However, the state of the art method we applied has been shown to accurately reflect leukocyte distribution, especially with an adult reference methylome 27. We included a set of covariates in the analyses, although it is still possible that the observed associations between site-specific DNA methylation levels and lead exposure might be explained by incompletely controlled or uncontrolled confounding factors. However, confounding factors were selected using a principal component analysis, which showed little variability by other factors. We estimated lead biomarkers using methylation profiles from arrays targeting 5-methylcytosine alterations. Other chemical modifications in the cytosines, including 5-hydroxymethylcytosine, could not be detected using those platforms, and we could not rule out the association of lead exposures with those chemical alterations. Further analysis to overcome limitations and improve performance of our biomarkers might include samples from different populations with harmonized covariates, a larger spectrum of tissues, and well-recognized lead predictors.
Conclusions
Whole blood DNA methylation was altered by environmental lead exposure and accurately estimated lead levels in two bone tissues, reflecting two temporal cumulative exposures. These novel epigenetic biomarkers, obtainable from either Illumina450K or IlluminaEPIC platforms, may provide the groundwork for preventive environmental monitoring and diagnostics.
Supplementary Material
Funding and Acknowledgements:
EC and ROW were supported by the National Institute of Environmental Health Sciences (NIEHS) (grant: P30ES023515); ACJ was supported by NIEHS (grant: R00ES023450); MAK was supported by NIEHS (grants: R01ES028805 and P30 ES009089); MW was supported by NIEHS (grant: P30ES000002); JS was supported by NIEHS (grants: R01ES015172, P30ES000002, and R01ES027747); HH was supported by NIH (grants: R01ES021446, and R01ES005257); AAB was supported by NIEHS (grants: P30ES009089, R01ES021733, R01ES025225, and R01ES027747). The VA Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center, Boston, Massachusetts.
Footnotes
Declaration of interests: All authors declare no conflict of interest.
Code and data sharing: Statistical code and de-identified data collected for this study are available from the corresponding author on reasonable request.
References
- 1.Bakulski KM, Rozek LS, Dolinoy DC, Paulson HL, Hu H Alzheimer’s Disease and Environmental Exposure to Lead: The Epidemiologic Evidence and Potential Role of Epigenetics. Current Alzheimer research 2012; 9: 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell JR, Rosier RN, Novotny L, Puzas JE The association between environmental lead exposure and bone density in children. Environmental health perspectives 2004; 112: 1200–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JR, Auinger P The association between blood lead levels and osteoporosis among adults--results from the third national health and nutrition examination survey (NHANES III). Environ Health Perspect 2007; 115: 1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holland MG, Cawthon D, Levels ATFoBL Workplace Lead Exposure. Journal of occupational and environmental medicine 2016; 58: e371–e374. [DOI] [PubMed] [Google Scholar]
- 5.REGISTRY CAFTSAD. CASE STUDIES IN ENVIRONMENTAL MEDICINE, June 12, 2017.
- 6.Alarcon WA Elevated Blood Lead Levels Among Employed Adults - United States, 1994-2013. MMWR Morbidity and mortality weekly report 2016; 63: 59–65. [DOI] [PubMed] [Google Scholar]
- 7.Weisskopf MG, Proctor SP, Wright RO, Schwartz J, Spiro A 3rd, Sparrow D et al. Cumulative lead exposure and cognitive performance among elderly men. Epidemiology 2007; 18: 59–66. [DOI] [PubMed] [Google Scholar]
- 8.Navas-Acien A, Schwartz BS, Rothenberg SJ, Hu H, Silbergeld EK, Guallar E Bone lead levels and blood pressure endpoints: a meta-analysis. Epidemiology 2008; 19: 496–504. [DOI] [PubMed] [Google Scholar]
- 9.Hu H, Shih R, Rothenberg S, Schwartz BS The Epidemiology of Lead Toxicity in Adults: Measuring Dose and Consideration of Other Methodologic Issues. Environmental Health Perspectives 2007; 115: 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu H, Rabinowitz M, Smith D Bone lead as a biological marker in epidemiologic studies of chronic toxicity: conceptual paradigms. Environmental Health Perspectives 1998; 106: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilker E, Korrick S, Nie LH, Sparrow D, Vokonas P, Coull B et al. Longitudinal changes in bone lead levels: the VA Normative Aging Study. Journal of occupational and environmental medicine 2011; 53: 850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect 2010; 118: 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G, Pradhan S Mammalian epigenetic mechanisms. IUBMB life 2014; 66: 240–256. [DOI] [PubMed] [Google Scholar]
- 14.Bird A DNA methylation patterns and epigenetic memory. Genes & development 2002; 16: 6–21. [DOI] [PubMed] [Google Scholar]
- 15.Zhong J, Agha G, Baccarelli AA The Role of DNA Methylation in Cardiovascular Risk and Disease: Methodological Aspects, Study Design, and Data Analysis for Epidemiological Studies. Circulation research 2016; 118: 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reese SE, Zhao S, Wu MC, Joubert BR, Parr CL, Haberg SE et al. DNA Methylation Score as a Biomarker in Newborns for Sustained Maternal Smoking during Pregnancy. Environ Health Perspect 2017; 125: 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovatsi L, Georgiou E, Ioannou A, Haitoglou C, Tzimagiorgis G, Tsoukali H et al. p16 promoter methylation in Pb2+-exposed individuals. Clinical Toxicology 2010; 48: 124–128. [DOI] [PubMed] [Google Scholar]
- 18.Rakyan VK, Down TA, Balding DJ, Beck S Epigenome-Wide Association Studies for common human diseases. Nature reviews Genetics 2011; 12: 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portela A, Esteller M Epigenetic modifications and human disease. Nature biotechnology 2010; 28: 1057–1068. [DOI] [PubMed] [Google Scholar]
- 20.Feinberg AP Epigenomics Reveals a Functional Genome Anatomy and a New Approach to Common Disease. Nature biotechnology 2010; 28: 1049–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell B, Rose CL, Damon A The Veterans Administration longitudinal study of healthy aging. Gerontologist 1966; 6: 179–184. [DOI] [PubMed] [Google Scholar]
- 22.Aro AC, Todd AC, Amarasiriwardena C, Hu H Improvements in the calibration of 109Cd K x-ray fluorescence systems for measuring bone lead in vivo. Physics in medicine and biology 1994; 39: 2263–2271. [DOI] [PubMed] [Google Scholar]
- 23.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics (Oxford, England) 2014; 30: 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y-a, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013; 8: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013; 29: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logue MW, Smith AK, Wolf EJ, Maniates H, Stone A, Schichman SA et al. The correlation of methylation levels measured using Illumina 450K and EPIC BeadChips in blood samples. Epigenomics 2017; 9: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics 2012; 13: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox J, Weisberg S. An R Companion to Applied Regression. SAGE Publications, 2011. [Google Scholar]
- 29.Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC bioinformatics 2010; 11: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R et al. De novo identification of differentially methylated regions in the human genome. Epigenetics & Chromatin 2015; 8: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan J, Lv J Sure independence screening for ultrahigh dimensional feature space. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2008; 70: 849–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phipson B, Maksimovic J, Oshlack A missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics (Oxford, England) 2016; 32: 286–288. [DOI] [PubMed] [Google Scholar]
- 33.Young MD, Wakefield MJ, Smyth GK, Oshlack A Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology 2010; 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barltrop D, Khoo HE The influence of nutritional factors on lead absorption. Postgraduate Medical Journal 1975; 51: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith PJ, Blumenthal JA Dietary Factors and Cognitive Decline. The journal of prevention of Alzheimer’s disease 2016; 3: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.John Lu ZQ The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Journal of the Royal Statistical Society: Series A (Statistics in Society) 2010; 173: 693–694. [Google Scholar]
- 37.Bender R, Lange S Multiple test procedures other than Bonferroni’s deserve wider use. BMJ : British Medical Journal 1999; 318: 600–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SK, Mukherjee B, Xia X, Sparrow D, Weisskopf MG, Nie H et al. Bone lead level prediction models and their application to examine the relationship of lead exposure and hypertension in the Third National Health and Nutrition Examination Survey. Journal of occupational and environmental medicine 2009; 51: 1422–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horvath S DNA methylation age of human tissues and cell types. Genome biology 2013; 14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji JS, Schwartz J, Sparrow D, Hu H, Weisskopf MG Occupational determinants of cumulative lead exposure: analysis of bone lead among men in the VA normative aging study. Journal of occupational and environmental medicine 2014; 56: 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


