Abstract
Bacterial wilt and tan spot of dry beans (family Fabaceae), caused by Curtobacterium flaccumfaciens pv. flaccumfaciens, is an important emerging disease threatening the edible legume industry around the globe. The management of bacterial wilt has been a major problem since its original description in 1922. This is in part due to the seedborne nature of the pathogen allowing the bacterium to be transmitted long distances via infected seeds, as well as a lack of detailed molecular information concerning the pathogenicity repertoires and virulence determinates of the pathogen. Identification can also be difficult owing to the presence of five different colony colour variants (i.e., yellow, orange, pink, purple, and red) on culture media. In this review, we provide an overview of the aetiology, epidemiology, and management strategies of bacterial wilt disease. First, a comprehensive and comparative symptomology of the disease on different dry bean species is described. Then, the taxonomic history of the causal agent and utility of high‐throughput sequencing‐based approaches in the precise characterization of the pathogen is explained. Furthermore, we provide an updated outline on the global distribution of the pathogen, highlighting expansion of the causal agent into the areas with no history of the disease until the beginning of the current century. Finally, because there are limited options for use of conventional pesticides against the pathogen, we highlight the use of integrated pest management strategies, for example quarantine inspections, resistant cultivars, and crop sanitation, to combat the risk of bacterial wilt disease in the dry bean industry.
Disease symptoms
Interveinal chlorosis on leaflets leading to necrotic areas and systemic wilt. Seed discolouration to yellow, orange, pink, or purple is seen in white‐seeded cultivars.
Host range
Causes bacterial wilt and tan spot disease on edible dry beans in the Fabaceae family, including common bean (Phaseolus vulgaris), cowpea (Vigna unguiculata), mungbean (Vigna radiata), soybean (Glycine max), as well as a number of weed species.
Taxonomic status of the pathogen
Bacteria; phylum Actinobacteria; order Actinomycetales; suborder: Micrococcineae; family Microbacteriaceae; genus Curtobacterium; species Curtobacterium flaccumfaciens.
Synonyms
Corynebacterium flaccumfaciens subsp. flaccumfaciens; Corynebacterium flaccumfaciens pv. flaccumfaciens, Corynebacterium flaccumfaciens, Phytomonas flaccumfaciens, Bacterium flaccumfaciens.
Microbiological properties
Multicoloured (yellow, orange, pink, purple, and red), gram‐positive, aerobic, curved rod, nonspore‐forming, polar flagellated, motile cells.
Distribution
Widespread in America (Brazil, Canada, and the USA), Australia, and Iran. Restricted occurrence in Africa and Europe.
Phytosanitary categorization
EPPO A2 list no. 48, EU Annex II⁄B.
Keywords: actinobacteria, coryneform bacteria, Fabaceae, leguminous, Microbacteriaceae, quarantine pathogen, tan spot
1. DISEASE SYMPTOMS
Bacterial wilt or tan spot of edible dry beans is caused by a gram‐positive bacterium called Curtobacterium flaccumfaciens pv. flaccumfaciens. Field symptoms of the disease include a series of chlorotic (Figure 1a–f) and necrotic (Figure 1g,h) areas on leaves accompanied by overall plant wilt, often permanent, leading to plant mortality (Figure 1i; Harveson et al., 2015). The symptoms first appear as foliar wilting and chlorotic areas leading to necrosis on leaves surrounded by a yellow halo. Subsequent symptoms are leaf wilting during periods of warm and dry weather or periods of moisture stress due to the pathogen's presence within the vascular system, which interferes with normal water movement from roots into the foliage (Huang et al., 2009; Harveson et al., 2015). Hence, the infected plants may recover when the temperature is lower at night or during cloudy or wet weather. Tearing or shredding of the necrotic leaf tissues is common under wind‐driven rain and hailstorms, giving a ragged appearance to leaves with symptoms.
Figure 1.
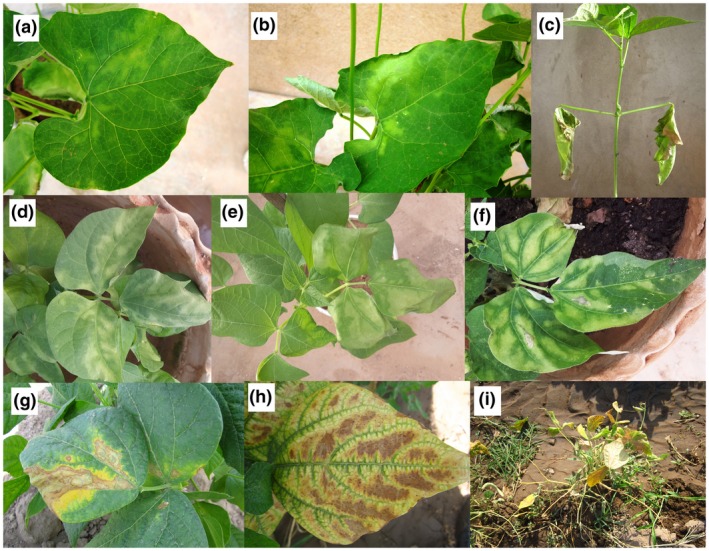
Symptoms of bacterial wilt of dry beans caused by Curtobacterium flaccumfaciens pv. flaccumfaciens on common bean. Symptoms first appear as interveinal chlorotic areas (a,b), leading to leaf wilting (c) and flabby or flaccid areas on the leaves (d,e). Subsequently, tissue necrosis on leaves surrounded by a chlorotic margins are observed (f–h). Severe infections on common bean lead to overall wilting and defoliation as well as plant mortality (i)
Disease severity and plant mortality are often higher on seedlings and young plants, particularly if they originate from infected seeds (Figure 2a–c; Harveson and Schwartz, 2007; Harveson, 2013). Symptoms on seedlings emerged from infected seeds are commonly seen at the cotyledon and/or the second trifoliate leaf stage. In such a situation the seedling rarely survives and will die within a few days post‐emergence (Figure 2d–f; Huang et al., 2009; Harveson et al., 2015). In artificial inoculations of the plants under greenhouse conditions, wilting on cotyledon leaves will be the first symptom observed 4–8 days post‐inoculation. Environmental temperatures above 30 °C enhance growth of the pathogen in vascular tissues, leading to the sudden death of young seedlings (Huang et al., 2009). Young plants affected early usually die, and those surviving remain stunted, showing severe yellowing and wilting symptoms without producing fertile pods.
Figure 2.
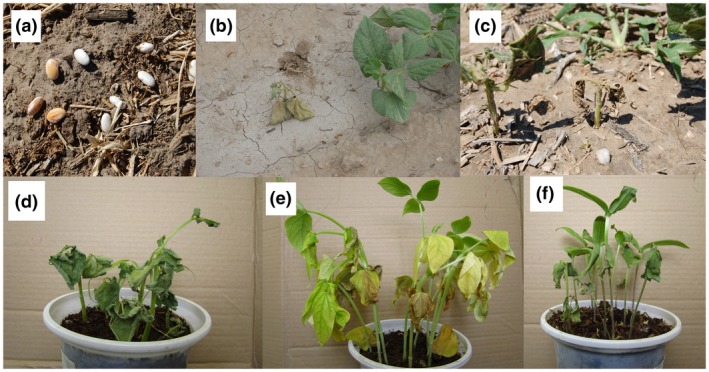
Effect of seed infection with the bacterial wilt pathogen Curtobacterium flaccumfaciens pv. flaccumfaciens on common bean seeds’ appearance (discolouration, a). Water stress will increase the mortality of common bean seedlings emerged from infected seeds (b,c). Common bean (d), cowpea (e), and mungbean (f) seedlings emerging from the infected seeds in greenhouse conditions die within a few days post‐emergence
Seed discolouration is a common symptom in bacterial wilt disease (Figure 2a; Schwartz et al., 2005; Huang et al., 2009). If infected plants survive to produce mature seeds, they are often discoloured as a result of bacterial infection and colonization, particularly in white‐seeded cultivars (Hagedorn and Inglis, 1986; Ishimaru et al., 2005; Schwartz et al., 2005). Because the pathogen is transmitted within the host plant's vascular system, the seed coat may display yellow, orange, pink, or purple discolouration while the exterior sections of pods remain symptomless (Harveson, 2013). In severe infections, flowers are also blighted and under these conditions seed‐set will be severely reduced (Wood and Easdown, 1990).
Economic losses of bacterial wilt disease are attributed not only to substantial decrease in the net crop yield (i.e., the number of pods per plants and the number of mature seeds per pod) but also to a significant decrease in the marketability of the product where visual appearance, shape, and size of the seeds are determinative factors in pricing (Huang et al., 2009). Hence, in visual inspections of dry bean fields it should be noted that seeds may become infected even when pods look healthy (Harveson et al., 2006). When bacterial wilt occurs on older plants, disease and symptom development proceed more slowly and are less severe. In such a case the vascular routes between stems and seeds are blocked, reducing the transmission of the pathogen to the seeds (Harveson et al., 2015). The percentage of common bean seeds possessing bacterial wilt symptoms in a given field (i.e., yellow, orange, pink, or purple discolouration of the seed coat) was reported to vary from 0% to 25% in the seed‐producing fields in Canada and research plots in Nebraska (Harveson et al., 2006; 2015).
Minor differences are seen among the bacterial wilt symptoms on various dry bean species. For instance, on cowpea (Figure 3a–c), chlorotic margins surrounding necrotic areas are more noticeable than on common bean (Figure 1d–f), mungbean (Figure 3d,e), and soybean (Figure 3g–i), resembling golden leaf lesions. High incidence of the disease in cowpea and mungbean fields results in a yellowish appearance of the bulk plants from a distance (Figure 3f), while severely infected common bean plants tend to appear brownish from a distance. On mungbean, overall wilting of the infected plants rarely occurs, while the interveinal necrosis exhibits pale brown to tan colour (Wood and Easdown, 1990). Similar symptoms are observed on soybean, explaining why the disease is called “tan spot” in the latter two crops (Figure 3d–i; Dunleavy, 1983; Wood and Easdown, 1990). In common bean and cowpea, plant defoliation is observed and affected leaves may drop off in windy conditions (Figure 1i), while this is not as common in mungbean and soybean.
Figure 3.
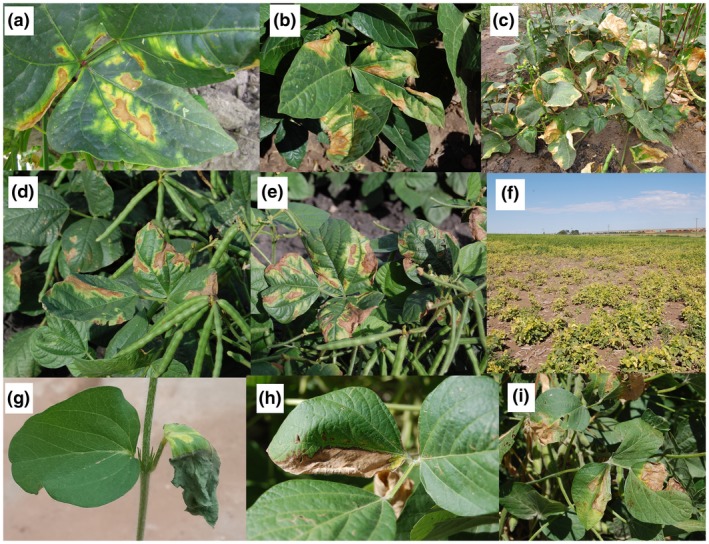
Symptoms of bacterial wilt of dry beans caused by Curtobacterium flaccumfaciens pv. flaccumfaciens on naturally infected cowpea (a–c), mungbean (d–f), and soybean (g–i) plants in field conditions. In all the plant species, interveinal chlorotic areas leading to leaf wilting and tissue necrosis are considered the main symptoms of the disease
Bacterial wilt symptoms may be confused with those caused by the common bacterial blight pathogen Xanthomonas axonopodis pv. phaseoli in the areas where both the pathogens occur in the same field. Bacterial wilt lesions are seen occurring between veins, often accompanied by wilting and death of severely infected plants. Wilting and plant death are less likely to occur in common bacterial blight and halo blight and brown spot caused by Pseudomonas savastanoi pv. phaseolicola and Pseudomonas syringae pv. syringae, respectively (Schwartz et al., 2005; Harveson and Schwartz, 2007).
2. HOST RANGE OF THE PATHOGEN
Common bean is considered the primary host of bacterial wilt pathogen (Huang et al., 2009) and it is the most susceptible host plant to the pathogen among all the agriculturally important dry bean crops within the Fabaceae family (Harveson et al., 2015; Osdaghi et al., 2015a). However, in certain geographic areas the economic impact of the disease on other dry bean species is higher than that observed on common bean. For instance, in Australia, bacterial wilt disease, which is known as tan spot on mungbean, causes substantial crop losses in the Queensland and New South Wales mungbean industry (Wood and Easdown, 1990). The pathogen infects cowpea in northwestern Iran, where the crop is considered the main cultivated dry bean species (Osdaghi et al., 2015a). It has also been identified recently from multiple cowpea production fields in Nebraska (R. M. Harveson, unpublished data). The known host range of the pathogen includes adzuki bean (Vigna angularis), black gram (Vigna mungo), cowpea (Vigna unguiculata), hyacinth bean (Lablab purpureus), lima bean (Phaseolus lunatus), mungbean (Vigna radiata), pea (Pisum sativum), scarlet runner bean (Phaseolus coccineus), soybean (Glycine max), yardlong bean (Vigna unguiculata subsp. sesquipedalis), and Zornia glabra (Dunleavy, 1983; Chavarro et al., 1985; Ishimaru et al., 2005; Harveson and Vidaver, 2007; Osdaghi, 2014; Osdaghi et al., 2015b).
3. TAXONOMIC HISTORY OF THE PATHOGEN
Bacterial wilt of edible dry beans is caused by the actinobacterial pathogen Curtobacterium flaccumfaciens pv. flaccumfaciens (Hedges) Collins & Jones. The pathogen belongs to a group of non‐spore‐forming coryneform bacteria in the family Microbacteriaceae and order Actinomycetales (Evtushenko and Takeuchi, 2006). The term Curtobacterium is derived from a Latin word “curtus” or “shortened” to describe the short and rod‐shaped nature of the bacterial cells (Yamada and Komagata, 1972), while the specific epithet “flaccumfaciens” is a two‐part term (flaccum + faciens) derived from the Latin word “flaccus” meaning “flabby or flaccid” and “faciens” meaning “making”. Hence, the name of the pathogen describes the cell morphology and symptomology of the bacterium as “wilt‐inducing, short, rod‐shaped bacterium” (Hedges, 1922; Harveson et al., 2015).
Bacterial wilt was first reported by Hedges (1922) on common bean plants with symptoms grown in Redfield in South Dakota (USA), and the causal agent was named as Bacterium flaccumfaciens at that time (Hedges, 1922; 1926). Subsequently, Bergey et al. (1939) transferred gram‐positive plant‐pathogenic bacteria into the genus Phytomonas and renamed the wilt pathogen Phytomonas flaccumfaciens. However, because the genus Phytomonas encompassed both gram‐negative, motile, green‐fluorescent (now known as Pseudomonas spp.), and gram‐positive, non‐motile, yellow/orange‐pigmented (now known as Clavibacter spp.), bacteria, the proposed reclassification was not accepted by most bacteriologists at that time. Thus, Dowson (1942) transferred the gram‐positive coryneform plant‐pathogenic bacteria into the genus Corynebacterium (“club” bacterium) (Lehmann and Neumann, 1896) and the bacterial wilt pathogen was named as Corynebacterium flaccumfaciens (Hedges, 1922) Dowson, 1942.
For several decades, the bacterial wilt pathogen was known for possessing only yellow‐pigmented colonies on culture media (Hedges, 1922). However, since that first report, four additional colony variants of the pathogen have been described. In the late 1950s, an orange‐pigmented variant of the pathogen was isolated from discoloured common bean seeds in Nebraska and named at that time as Corynebacterium flaccumfaciens var. aurantiacum (Schuster and Christiansen, 1957). Then, in 1968, another variant of the pathogen was isolated from western Nebraska from a white‐seeded common bean market class (Great Northern) inducing a purple discolouration on seed coats, and the causal agent was described as Corynebacterium flaccumfaciens var. violaceum (Schuster and Sayre, 1967; Schuster et al., 1968).
In the early 1980s, taxonomic studies based on biochemical characteristics, DNA−DNA homology, and cell wall composition were conducted on plant‐pathogenic members of Corynebacterium, resulting in the transfer of several species into the newly erected genus Curtobacterium (Carlson and Vidaver, 1982; Collins and Jones, 1983; Komagata and Suzuki, 1986), including Corynebacterium betae (causing silvering disease of red beet), Corynebacterium flaccumfaciens, Corynebacterium oortii (causing bacterial wilt and spot of tulip), and Corynebacterium poinsettiae (causing bacterial canker of poinsettia). At this time, all the latter species were reclassified as pathovars of Curtobacterium flaccumfaciens (Collins and Jones, 1983; 1984). Hence, the previously mentioned orange and purple colour variants of the bacterial wilt pathogen were classified as C. flaccumfaciens pv. flaccumfaciens. Although Carlson and Vidaver (1982) proposed to change the status of C. flaccumfaciens members from pathovar to subspecies level, it has been determined that pathovars of C. flaccumfaciens are closely related to one another in terms of biochemical and physiological characteristics, while variability in host specificity and bacteriocin production were insufficient to justify their differentiation at the subspecies level (Collins and Jones, 1983). The bacterial wilt pathogen of dry beans is now known as C. flaccumfaciens pv. flaccumfaciens, and will be referred to hereafter as Cff.
In the first two decades of the 21st century, two additional novel colour variants of Cff were identified from two geographically distinct areas. First, a pink‐pigmented variant of the pathogen was isolated from orange‐stained common bean seeds in western Nebraska, USA in 2007 after harvest (Harveson and Vidaver, 2008). In 2014, a red‐pigmented variant of the pathogen was isolated in central Iran (Markazi province) from white‐seeded common bean seed lots possessing deep orange discolouration (Osdaghi et al., 2016). Cff variants differ somewhat in their virulence on various bean species (common bean, cowpea, mungbean, and soybean). Indeed, yellow and orange variants of the pathogen are more aggressive on common bean and cowpea while the red and purple variants showed attenuated level of aggressiveness on all of the evaluated dry bean species (Harveson et al., 2015; Osdaghi et al., 2016).
4. BACTERIOLOGICAL FEATURES
Cff cells are gram‐positive, aerobic, and motile, with short, rod, and/or coryneform‐shaped morphology (typically 0.3–0.5 × 0.6–3.0 µm) that bend with one to three lateral or polar flagella (Evtushenko and Takeuchi, 2006). Colonies on yeast extract‐peptone‐glucose agar (YPGA) medium are circular, smooth, flat or slightly convex, opaque and slightly viscid, with an entire margin. Colony growth on nutrient broth yeast (NBY) extract medium is slow and fluidal. The colony colour varies among yellow, orange, pink, and red pigmentation (Collins and Jones, 1983; Harveson et al., 2015; Osdaghi and Lak, 2015a; Osdaghi et al., 2016). Unlike yellow‐, orange‐, pink‐, and red‐pigmented variants, which are named corresponding to colony colour on culture media, the purple variant does not produce purple‐coloured colonies. Instead, the purple variant produces yellow colonies on culture medium from which water‐soluble purple to deep blue pigment diffuses into the surrounding medium. The same pigment is responsible for common bean seed discolouration to purple or deep blue in naturally infected plants (Harveson and Schwartz, 2007; Harveson et al., 2015). Apart from colony pigmentation, variants of Cff are indistinguishable based on biochemical and physiological characteristics (Osdaghi et al., 2016). However, it has been shown that Cff strains possessing different pigmentation are capable of producing bacteriocins against the other strains on culture media (Figure 4; Osdaghi et al., 2018a). The maximum temperature for growth is 37 °C while the optimum growth occurs at 27–32 °C. Cff cells are positive in the utilization of acetate, citric acid, fumarate, fumaric acid, gluconic acid, lactic acid, and malic acid, and negative for the utilization of glyoxylic acid, propionate, and α‐ketoglutaric acid (Evtushenko and Takeuchi, 2006). Furthermore, the pathogen is positive for H2S production, hydrolysis of aesculin and casein, as well as production of acid from adonitol, l‐rhamnose, and raffinose. The bacterial cell wall in Cff contains galactose, but lacks glucose and fucose. The G + C content of the pathogen was estimated to be 71.1% (Evtushenko and Takeuchi, 2006; Osdaghi et al., 2017), while genome sequencing of an Australian strain gave a G + C content of 69.5% (A. Young, unpublished data).
Figure 4.
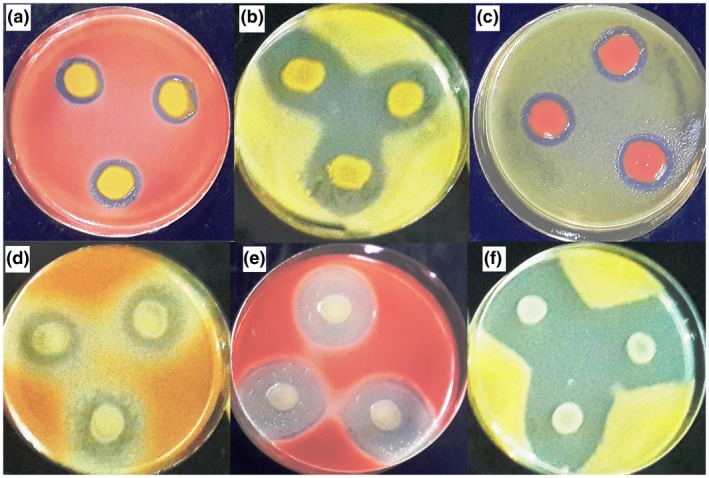
Confrontation of different colony variants of Curtobacterium flaccumfaciens pv. flaccumfaciens on yeast extract‐peptone‐glucose agar (YPGA). Dual culture of orange variant versus red (a) and yellow (b) variants, red variant versus yellow (c) variant, and yellow variant versus orange (d), red (e), and yellow (f) variants of the pathogen show variation in their bacteriocin production scheme. Strains spotted on the corners of an imaginary triangle were potential bacteriocin producers, whereas the strains spread over the surface of the YPGA plates were potential indicators of bacteriocin. Clear zones around the colonies of producer strains indicate inhibition 48 hr post‐incubation. Data obtained from Osdaghi et al. (2018a)
5. POPULATION STRUCTURE AND GENETIC DIVERSITY
Until late in the 20th century, the only reliable method for distinguishing Cff strains from other C. flaccumfaciens pathovars and identification of unknown strains to the pathovar level was to evaluate their pathogenicity and host range on a panel of preselected plant species (Davis, 1986, 2001; Vidaver and Davis, 1988; Davis and Vidaver, 2001). This was due to a lack of bacteriological methods used before that time that could differentiate the diverse pathovars of the species. These tests included biochemical and numerical phenetic analyses (Dye and Kemp, 1977; Harris‐Baldwin and Gudmestad, 1996; Zhao et al., 1997), cell wall analysis and fatty acid/protein profiles (Keddie and Cure, 1977; Carlson and Vidaver, 1982; Henningson and Gudmestad, 1991), temperature‐gradient gel electrophoresis analysis of 16S rRNA (Felske et al., 1999), and immunological characteristics (McDonald and Wong, 2000). These techniques are time‐consuming and required specialized instrumentation and skills. As the genomic era began, the application of molecular methods signalled a significant shift in bacterial taxonomy.
Pathovar differentiation of Cff can now be readily achieved with DNA‐based molecular fingerprinting techniques. Although amplified fragment length polymorphism (AFLP) and repetitive sequence polymerase chain reaction (rep‐PCR) analyses were able to assign some, but not all, Cff strains into pathovar status, all strains were correctly differentiated to the pathovar level using HindIII and XbaI macrorestriction digests in conjunction with pulsed field gel electrophoresis (PFGE) (Guimaraes et al., 2003; Souza et al., 2006). Cluster analysis of a combination of AFLP, PFGE, and rep‐PCR data revealed two distinct clades among Cff strains: clade A included yellow‐, orange‐, and pink‐pigmented strains, while clade B included yellow‐pigmented strains. Strains producing purple extracellular pigment were assigned to both clades (Agarkova et al., 2012).
Unlike other actinobacterial plant pathogens, for example Clavibacter michiganensis (Osdaghi et al., 2018b; Ansari et al., 2019), phylogenetic analysis has revealed that Cff strains are polyphyletic and heterogeneous. The two evolutionary lineages, yellow‐pigmented strains and red‐/orange‐pigmented strains, are distinct. Orange‐ and red‐pigmented strains of Cff were separated from yellow‐pigmented strains using multilocus sequence analysis (MLSA) of five housekeeping genes, atpD, gyrB, ppk, recA, and rpoB (Osdaghi et al., 2018a; 2018c; Gonçalves et al., 2019). Whole‐genome sequencing of Australian Cff isolated from mungbean has shown the presence of multiple lineages (Vaghefi et al., 2019). Interestingly, the phylogenetic distance between yellow‐pigmented Cff strains and red‐/orange‐pigmented strains is higher than that observed among the type strains of C. flaccumfaciens pv. flaccumfaciens, C. flaccumfaciens pv. betae, and C. flaccumfaciens pv. oortii (Osdaghi et al., 2018c). Recently, Gonçalves et al. (2019) analysed a large population of plant‐pathogenic C. flaccumfaciens strains from all over the world and concluded that there is no correlation between the MLSA scheme and the host specificity of the strains. It seems more likely that the evolutionary forces that drive phylogenetic position and bacteriological characteristics, for example colony pigmentation in C. flaccumfaciens, are dominating the plant pathogenicity features and host specification of the strains.
6. GEOGRAPHIC DISTRIBUTION
Bacterial wilt was first observed and described in South Dakota (USA) in 1922 (Hedges, 1922). During the 1920s, the pathogen was isolated from common bean seed lots collected in Maryland, Michigan, Montana, Virginia, and the District of Columbia (Washington, D.C.) (Hedges, 1926). Subsequently, occurrence of the disease was reported in Wyoming (1937), Idaho and Colorado (1947), Nebraska (1948), and Columbia Basin of Washington (1951) (Zaumeyer and Thomas, 1957; Harveson, 2013). Bacterial wilt was reported from Canada and Mexico in 1955 (Patrick, 1954; Yerkes and Crispin, 1956). The disease was restricted to North America until it was observed on mungbean and cowpea plants in Australia in the late 1980s (Wood and Easdown, 1990). The disease has also continued to spread southward into South America, reaching Columbia in 1982 (Torres et al., 1982) and Venezuela in 1990 (EPPO, 2011). Maringoni and Rosa (1997) reported the disease on common bean plants in São Paulo State, Brazil in 1995. Although severe outbreaks of bacterial wilt disease were recorded from the 1930s to the 1960s, the economic importance of the disease was minimal until the end of the 20th century. However, a resurgence of the pathogen in common bean‐growing areas causing economic damage in North America was reported beginning in the mid‐2000s (Harveson et al., 2006; 2015; Huang et al., 2009). Different variants of the pathogens have been chronologically discovered in the USA and Canada. For instance, a purple‐pigmented variant of the pathogen was found in 2006 from infected bean seeds in Alberta and Saskatchewan, Canada (Huang et al., 2006) after first being identified in the USA in 1968. Curiously, the orange, purple, and pink variants were all first identified and reported from Scotts Bluff county, in the panhandle of western Nebraska. In addition to common bean, the disease was first reported on soybean in the USA in Iowa in 1975 and the disease was named bacterial tan spot (Dunleavy, 1983). Subsequently, the disease was reported on soybean in Brazil in 2013 (Soares et al., 2013).
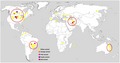
Over the last two decades, the disease has sporadically been reported in some European countries, although economic yield losses on dry beans have not been documented (EPPO, 2011). For instance, the disease was found affecting common bean in southeastern Spain in 2005, while the geographic distribution of the pathogen within Spain was not included in the report (González et al., 2005). Subsequently, Cff was isolated from soybean plants in Germany in 2012 (Sammer and Reiher, 2012). The disease was also reported, but not substantiated, from two distinct areas of Russia (EPPO, 2011). The pathogen was not officially reported from Asia until 2013 when a widespread occurrence of the disease was found in Iran in both common bean and cowpea fields from different provinces of the country (Osdaghi et al., 2015a; 2015b). Subsequently, a red‐pigmented variant of the pathogen was isolated from common bean seeds in central Iran (Osdaghi et al., 2016). Unofficial occurrences of the pathogen have also been recorded in Albania, Belgium, Bulgaria, France, Greece, Hungary, Romania, Switzerland, Tunisia, Turkey, and Yugoslavia (EPPO, 1994; Ishimaru et al., 2005). Although the pathogen has previously been observed infecting common bean in Africa (Kenya, Mauritius, South Africa, and Tunisia; EPPO, 2011), it was not officially documented from Africa until 2019, infecting soybean plants in Zambia (Pawlowski and Hartman, 2019). Currently, economic yield losses due to the bacterial wilt disease have been recorded in Brazil, Canada, eastern Australia, Iran, and the central High Plains of the USA (Figure 5).
Figure 5.
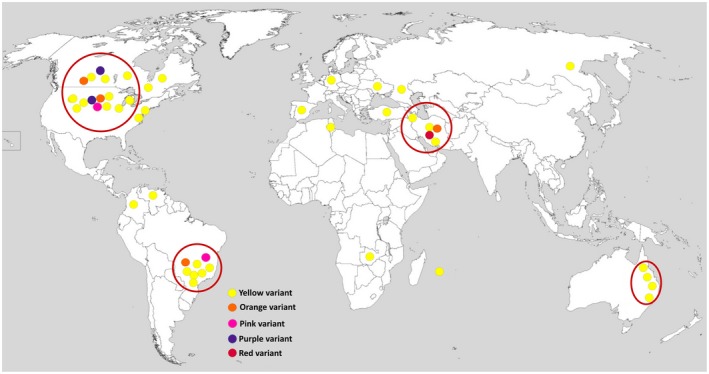
Global distribution of the bacterial wilt pathogen of dry beans Curtobacterium flaccumfaciens pv. flaccumfaciens. Filled small circles indicate the presence of the pathogen in a given area, while the colour of the circles refers to the colony variant of the pathogen: yellow, orange, pink, purple, and red. Large red circles surrounding the filled circles in Brazil, Canada, eastern Australia, Iran, and the central High Plains of the USA indicate the areas where economic yield losses due to the bacterial wilt disease were recorded. The source map is from https://commons.wikimedia.org/wiki/File:A_large_blank_world_map_with_oceans_marked_in_blue.PNG
7. EPIDEMIOLOGY
The epidemiology of bacterial wilt disease is similar to that of other seedborne bacterial diseases in vegetables and annual crops (Figure 6; Gitaitis and Walcott, 2007). Cff is a vascular pathogen that systemically colonizes its host plants. It survives in plant residues and infection may begin from either infected seeds or introduction of the bacterium into roots and leaves via wounds and physical openings (Hedges, 1922; 1926; Zaumeyer, 1932). Mechanical injury of roots by infections from the root‐knot nematode Meloidogyne incognita was shown to provide a mechanism for introducing the pathogen into host plants (Schuster, 1959). Contaminated seeds are considered the main method of introducing Cff inoculum in areas with no history of the disease (Zaumeyer and Thomas, 1957; Saettler and Perry, 1972). The level of seed transmission was reported to range from 6% to 70% in different assays, but transmissibility of the pathogen via seeds depends on the host plant and time of infection (Wood and Easdown, 1990; Camara et al., 2009).
Figure 6.
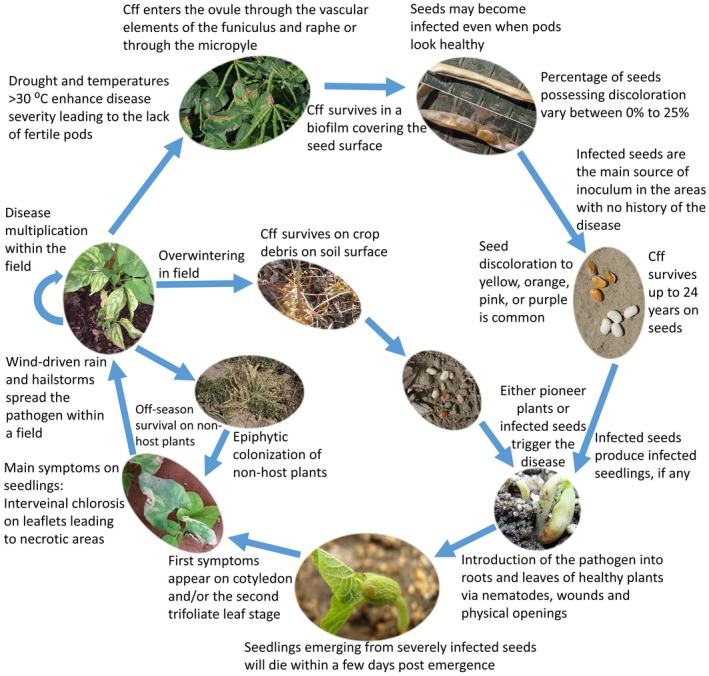
Disease cycle of bacterial wilt of edible dry beans caused by Curtobacterium flaccumfaciens pv. flaccumfaciens
Later infection of plants via aerial parts or foliage when the pods are mature is less likely to result in seed transmission. This is probably due to the fact that seed infection occurs when Cff enters the ovule through the vascular elements of the funiculus and raphe or through the micropyle (Zaumeyer, 1932). All these intraplant transmission routes between the vascular system and pods are closed after the pods mature and become semiripe (Nelson and Dickey, 1970). Furthermore, the rate of transmission in common bean seeds is reported to be higher than in the other hosts of the pathogen. For instance, susceptible and resistant soybean cultivars showed 0%–1% and zero seed transmission, respectively, while up to 70% seed transmission was reported in susceptible common bean cultivars in greenhouse experiments (Camara et al., 2009; Soares et al., 2018). It has been shown that Cff survives up to 24 years saprophytically on common bean seeds stored at room temperature in the laboratory (Burkholder, 1945). Schuster and Sayre (1967) additionally isolated virulent orange‐pigmented Cff strains from 15‐year‐old, and purple‐pigmented strains from 8‐year‐old, common bean seeds kept at 10 °C.
Cff develops a thick bacterial cell layer under the seed coat or superficially on the surface of seeds (Hedges, 1926). Common bean seeds with symptoms evaluated using scanning electron microscopy have also showed extensive biofilms covering the seed surface. The ratio of seed discolouration in dry bean seed lots in the current season is generally correlated with the risk of disease outbreaks in the subsequent cropping season. Hsieh et al. (2006) showed that the degree of seed coat discolouration will affect the seedling emergence, plant height, disease incidence, and severity of bacterial wilt in the resulting plants. Although some of the seedlings derived from seeds with yellow discolouration remain symptomless, Cff can be detected in such seedlings 14 days post‐emergence (Hsieh et al., 2006).
The disease occurs most frequently in areas characterized by hot summers, high precipitation, and humid environmental conditions. High levels of leaf moisture initiated by rain or overhead sprinkler irrigation is considered the leading factor assisting plant‐to‐plant transmission of the pathogen within a field (Ishimaru et al., 2005; Schwartz et al., 2005). Furthermore, the disease spread within a field is enhanced by hailstorms, wind‐driven rain droplets, as well as any form of physical damage caused during field operations, for example scouting, cultivating, and pesticide spraying (Harveson et al., 2015). Environmental stresses, in particular drought and warm temperature during daylight, increase disease progress and severity. The epidemiology of bacterial wilt contrasts with that of halo blight in Australian mungbeans and with common beans in the USA, with hotter and drier conditions most conducive for bacterial wilt disease development, and cooler, moister conditions most likely to result in halo blight epidemics (Harveson and Schwartz, 2007; Noble et al., 2019). Hence, geographic areas with hot summers (>30 °C) are the most favourable environments for bacterial wilt disease infections (Vidaver and Starr, 1981; Vidaver, 1982).
Survival of Cff in intact soil under field conditions varies between 34 and 80 days as a function of temperature and moisture (Gonçalves et al., 2018). Infested common bean debris and straw are important in Cff overwintering. Bacteria in infested dry straw on the soil surface survive far better (up to 22 months) than bacteria in debris buried 20 cm in the soil (Schuster and Coyne, 1974). In Brazil, Cff survived for up to 240 days on bean debris at the soil surface and for 30 days when the residues were buried at 20 cm in the soil (Silva Júnior et al., 2012). Although the pathogen can survive during winter on infected plant residues or weeds, it cannot survive for a long time free in the soil after crop residues decompose (Burkholder, 1948; Burke, 1957; Schuster, 1967; Schuster and Coyne, 1974; 1975). In Nebraska it was determined that the wilt pathogen could survive in soil in the absence of residue for at least 2 years (22–24 months). However, infection percentages of emerging seedlings after planting healthy seeds into these infested soils remained very low (<2%) (Harveson, 2013). Complete genome sequencing of a highly virulent Cff strain (P990) revealed the presence of 1,4‐β‐xylanase. This is the enzyme responsible for breaking down hemicellulose, one of the major components of plant cell walls. Furthermore, these enzymes degrade the linear polysaccharide xylan into xylose, helping the bacterium to survive on plant debris as a saprophyte in the absence of living host (E. Osdaghi, unpublished data). Cff was shown to survive up to 5 years in dried bacterial ooze, hence the viability of the pathogen exceeds that of the bean seeds. Schuster and Coyne (1974) showed a direct correlation between the virulence of a given Cff strain and the length of survival, where the more virulent the strains were, the better adapted they were for survival.
A number of nonleguminous plant species have been shown to harbour the pathogen for overwintering (Schuster, 1970; Saettler, 1991). Gonçalves et al. (2017) showed that under Brazilian field conditions, Cff colonizes barley, black oat, canola, ryegrass, wheat, and white oat where these plant species are cultivated during winter in rotation with common bean. All the Cff strains isolated from these plants were pathogenic to common bean. It has been recommended that common bean cultivation in succession with barley, black oat, canola, ryegrass, wheat, and white oat must be avoided in areas with a history of bacterial wilt occurrence. Cff endophytically colonizes stems of wheat plants inoculated with either wounding or spraying techniques (Silva Júnior et al., 2012). Harveson et al. (2015) have isolated bean‐pathogenic orange and yellow strains of Cff from black chaff‐infected wheat plants. Black chaff is a bacterial disease on small‐grain cereals caused by different pathovars of Xanthomonas translucens (Khojasteh et al., 2019). Dry bean‐pathogenic strains of Cff were also isolated from symptomless eggplant, pepper, and tomato plants in Iran (Osdaghi et al., 2018a). Conversely, highly virulent yellow‐ and pink‐pigmented strains of Cff have been isolated in association with infections of maize plants affected by the Goss’ wilt pathogen Clavibacter nebraskensis (Harveson et al., 2015, R.M. Harveson, unpublished data). The epiphytic population of Cff on nonhost plant species under favourable conditions could be an important source of inoculum for the establishment of the pathogen in the upcoming season. All these examples demonstrate the multifaceted, cosmopolitan nature of the bacterial wilt pathogen, highlighting the potential risk of pathogen survival and distribution through surprisingly diverse methods.
Agronomic practices can significantly affect plant disease dynamics. Harveson and Yonts (2007; Harveson and Urrea, unpublished) have evaluated the effect of different irrigation systems and rotational practices on the incidence and severity of bacterial wilt. They discovered that drip irrigation systems tended to produce higher common bean yields and lower disease severity compared to sprinkler and furrow irrigation systems. Furrow irrigation treatment exhibited an intermediate effect on the disease parameters and crop yields, while plants that were grown under sprinkler irrigation had the most severe disease problems. Crop rotation in plots also played an important role. Dry beans planted following a maize crop resulted in lower disease severity and better yields than those planted following a second crop of dry beans (Harveson and Yonts, 2007; Harveson et al., 2015; Harveson and Urrea, unpublished).
8. DETECTION, ISOLATION, AND IDENTIFICATION OF THE PATHOGEN
Cff can readily be isolated from all the above‐ground parts of dry beans, but leaves, stems, and seeds are the most suitable. Isolation of Cff from either seed lots or plant tissues with symptoms can be performed by cutting the marginal tissues of the visible lesions on leaves and grinding them in sterile distilled water (SDW). Spread a loopful of the resulting suspension on NBY (glucose 5 g, K2HPO4 2 g, KH2PO4 0.5 g, MgSO4 0.25 g, nutrient broth 8 g, yeast extract 2 g, and agar 15 g dissolved in 1 L SDW) or YPGA (glucose 10 g, peptone 5 g, yeast extract 5 g, and agar 15 g dissolved in 1 L SDW) media (EPPO, 1994; 2011). The bacterium also grows on a broad range of media, including Kelman's medium, nutrient agar, and King's medium B (Tegli et al., 2017). Several methods for isolation can be employed. Margins of leaf lesions can be abraded or punctured with a dissecting needle, and streaked onto medium plates (Harveson et al., 2006; 2015). Another technique involves squeezing the sap out of petioles attached to tissues with symptoms and blotting onto medium, followed by streaking with a sterilized inoculating loop (Harveson, 2013). For dried leaf samples leaf tissues can be ground in SDW, shaken for 1–2 hr to release the bacterial cells into the water, and a loopful of the resulting suspension can then be spread onto YPGA or NBY media. Another method of isolation is to place dried tissues in humidity chambers for 12–24 hr. If present, bacteria will ooze out of necrotic tissues and can be streaked onto media (Harveson et al., 2015). Finally, the bacterium can also easily be isolated from infected, discoloured or even symptomless seeds. Discoloured seeds are soaked overnight in SDW and the eluent is streaked onto plates (Harveson and Schwartz, 2007). After streaking on the media surface, fluidal colony growth will be observed on the culture media after incubation at 25–27 °C for 48–72 hr.
Visual inspection of suspected seed lots and isolation of the pathogen were the only available methods for Cff detection prior to the development of semiselective culture media and specific PCR primers (Harveson, 2013; Harveson et al., 2015). Until the late 20th century, the European and Mediterranean Plant Protection Organization (EPPO) recommended a visual examination of seeds, followed by a direct (from bean seeds) or indirect (from the resulting seedlings) isolation of the bacterium, and an immunological test for Cff detection in bean seeds for quarantine purposes (EPPO, 1994). However, these approaches gave false negative results at times. For instance, it was shown that symptomless bean plants could carry both Cff and Xanthomonas axonopodis pv. phaseoli (Thomas and Graham, 1952). Furthermore, both pathogens also possess morphologically similar colonies (yellow, circular, domed, and opaque) on general culture media, leading to misidentification unless the preliminary phenotypic tests (e.g., Gram stain, production of xanthomonadin and hypersensitivity reaction on tobacco leaves) are conducted to differentiate the two pathogens (Thomas and Graham, 1952).
A range of semiselective culture media has been proposed to be used in the isolation of Cff. For instance, Mizuno and Kawai (1993) recommended a semiselective culture medium for isolation of Cff from bean seeds, which included specific carbon sources and antimicrobial reagents. The procedure was accomplished by an immunological test to increase the sensitivity of the method (Mizuno, 1998). Furthermore, Maringoni et al. (2006) developed a semiselective culture medium (MSCFF) that included a basal medium consisting peptone 5 g, meat extract 3 g, sucrose 5 g, and agar 15 g, dissolved in 1 L of SDW. Skim milk powder 5 g, Congo red 50 mg, chlorothalonil 10 mg, thiophanate methyl 10 mg, nalidixic acid 10 mg, nitrofurantoin 10 mg, oxacillin 1 mg, and sodium azide 1 mg were added after autoclaving the basal medium. The MSCFF medium is capable of inhibiting the growth of many saprophytic bacteria, without harming the Cff strains (Maringoni et al., 2006). The MSCFF medium has been successfully used to detect Cff in naturally infected dry bean seeds (Maringoni and Camara, 2006).
Reliable differentiation of pathovars in C. flaccumfaciens species and identification of unknown strains to the pathovar level can be achieved through pathogenicity tests on the natural host plants, including dry beans (Guimaraes et al., 2003; Chen et al., 2007). However, a number of molecular techniques and PCR‐based methods have been developed for accurate and rapid differentiation of the pathovars (Guimaraes et al., 2001; Tegli et al., 2002). The primer pair CF4/CF5 is able to specifically amplify a 198 bp DNA fragment in Cff strains, but not the other pathovars of the species (Guimaraes et al., 2001). However, the ability of CF4/CF5 primer pair to directly detect the pathogen in naturally infected bean seeds has not been evaluated. The primer pair CffFOR2/CffREV4 is sensitive enough to detect the presence of Cff in bean seeds with pathogen concentrations as low as 100 cfu/ml (Tegli et al., 2002). Using these primers in combination with Bio‐PCR and centrifugally concentrated seed extract allows the detection of one artificially Cff‐contaminated bean seed among 999 healthy seeds (Deuner et al., 2012). The primer pair CffFOR2/CffREV4 did not direct the amplification of the expected 306 bp fragment in various nonpathogenic C. flaccumfaciens strains isolated from tomato and eggplant (Osdaghi et al., 2018a). Recently, Tegli et al. (2017) have provided a detail‐oriented step‐by‐step protocol for detection of Cff in dry bean seeds using both general and semiselective culture media as well as conventional PCR.
Although a number of serological techniques have been developed for detection of Cff, they are unable to differentiate nonpathogenic lineages of C. flaccumfaciens from the pathogenic members (Calzolari et al., 1987; Diatloff et al., 1993; McDonald and Wong, 2000). Immunofluorescence staining was successful in the detection of Cff to a concentration of 1.82 × 104 fluorescent cells per millilitre of soaked seed leachate (Calzolari et al., 1987). Immunofluorescence tests using a polyclonal antiserum produced against Cff strain NCPPB 559 lacked adequate sensitivity and did not react with all tested Cff strains (Calzolari et al., 1987; McDonald and Wong, 2000), while a monoclonal antibody developed by Diatloff et al. (1993) was able to detect the pathogen from symptomless mungbean plants. The sensitivity and specificity of serological methods are relatively low when compared to those of pathovar‐specific PCR primers (Guimaraes et al., 2001; Tegli et al., 2002). Thus, due to the morphological diversity within this taxa, more accurate identification of Cff will be achieved when immunofluorescent techniques are integrated with pathovar‐specific PCR primers (Guimaraes et al., 2001; Tegli et al., 2002).
A carbon utilization microplate system developed by Biolog Inc. has proven to be a useful tool for identifying all Curtobacterium spp. strains correctly to the genus level. Harris‐Baldwin and Gudmestad (1996) supplemented the Biolog database for improved detection of plant‐pathogenic coryneform bacteria. Afterwards, the Biolog system was capable of differentiating Cff, C. flaccumfaciens pv. betae and C. flaccumfaciens pv. poinsettiae to the pathovar level with relatively high reliability, while C. flaccumfaciens pv. ootrii was able to be identified only to the species level (Harris‐Baldwin and Gudmestad, 1996). Furthermore, a microarray hybridization Cy3‐labelled probe was developed based on the sequences of the housekeeping gene groEL (heat shock protein) for the identification and specific detection of Cff (Pelludat et al., 2009). However, none of the latter methods were evaluated for their efficiency in detecting the pathogen in naturally infected seed lots.
9. PATHOGENICITY MECHANISMS
Among the economically important gram‐positive bacterial plant pathogens, Cff is the least studied member, with no information from the virulence repertoires and pathogenicity determinates (Osdaghi et al., 2018d; 2020; Thapa et al., 2019). The reason for this is that no genetic manipulation technologies and transformation vectors have been developed yet for the bacterium, although it is possible that platforms for the closely related pathogens C. michiganensis and Leifsonia xyli subsp. xyli could be effective (Brumbley et al., 2006; Thapa et al., 2017). Furthermore, there has not been any complete genome sequence of Cff strains, although genome sequences of a number of nonpathogenic C. flaccumfaciens strains were recently released (Chase et al., 2016). Similar to its actinobacterial relatives, for example C. michiganensis, Clavibacter sepedonicus, Rathayibacter tritici, and L. xyli, Cff colonizes the xylem system of the host plant and moves endophytically (Zaumeyer, 1932; Thapa et al., 2019). There is a lack of type III secretion system (T3SS) in actinobacterial plant pathogens. Instead they use a set of lytic enzymes, toxins, and hormones to disrupt host plant cell walls and acquire nutrients (Thapa et al., 2019). Bacterial wilt pathogens multiply throughout the water‐conducting tissues of the plant and impede water movement, resulting in wilt symptoms. It has been shown that in infected common bean tissues, Cff cell masses are embedded in a slimy matrix that absorbs liquids from surrounding cells, leading to water deficiency in the tissues (Zaumeyer, 1932). Harding et al. (2019) proposed that biofilms play an important role in the pathogenicity and aggressiveness of bacterial wilt pathogen. The biofilm substrates occupy the vessels and occasionally cause complete occlusion, leading to entire plant death (Harding et al., 2019).
Electron microscopy of Cff‐infected plants have shown damage to xylem and decomposition of middle lamella prior to wilting (Vidaver, 1982). Furthermore, high molecular weight glycopeptides were isolated from the pathogen, some of which have been considered to cause plant cell wall disruption (Schuster and Coyne, 1981). Cff harbours lytic enzymes, for example, β‐glucosidase, esterase, peptidases, and lipases, all of which have similarity to C. michiganensis in the API ZYM system of detection of enzymes (De Bruyne et al., 1992). It has been hypothesized that the mechanism of wilting is the restriction of water movement in the xylem by phytotoxic glycopeptides (Gross and Vidaver, 1979; Schuster and Coyne, 1981). Histological studies on Cff‐infected tissues confirmed enzymatic activities (Nelson and Dickey, 1970; Vidaver, 1982). A temperature‐resistant substrate has been derived from the culture filtrate of Cff strains isolated in Australia (Wood and Easdown, 1990). The toxic effects of the substrate were tested with a sterile Cff‐free cell filtrate, which caused leaf wilting and red discolouration of the veins when pricked into the host plant (Wood and Easdown, 1990).
Although the biological structure of the phytotoxic compound produced by Cff remains undetermined, in vitro investigations suggest the putative role of bacteriocins in the pathogenicity of the bacterial wilt pathogen, similar to those reported in other actinobacterial pathogens (Strider, 1969; Durbin, 1972; Gross and Vidaver, 1979). Gross et al. (1979) investigated the biological characteristics of bacteriocins produced by 10 Cff strains and named the resulting substrates flaccumfacin. It has been shown that the ability of Cff strains to produce bacteriocins against the pathogenic and nonpathogenic actinobacterial strains positively correlated with the virulence vigour and aggressiveness of the same strain on common bean. For instance, Osdaghi et al. (2018c) showed that among Iranian Cff strains, the strain P990 had the most vigorous negative effect on the bacterial species evaluated through bacteriocin production, while the same strain had the most aggressiveness on common bean and the most fluidal colonies on culture media in 27 °C. Interestingly, the complete genome sequence of the strain P990 (E. Osdaghi, unpublished data) confirmed the presence of putative bacteriocin‐producing genes in the genome of the pathogen.
Gross et al. (1979) evaluated the plasmid profile of 10 Cff strains and did not find any evidence for the presence of plasmids. Similar results were observed by Osdaghi et al. (2018c) where different colony variants in both the pathogenic and nonpathogenic strains of C. flaccumfaciens were evaluated for the presence of plasmids. Nevertheless, the complete genome sequence of the strain P990 suggested the presence of three circular plasmids in the pathogen (E. Osdaghi, unpublished data). Differences between the in vitro and in silico results might be due to the low number of plasmid copies in Cff cells, which were not detected using agarose gel‐based techniques. Further investigations are warranted to decipher the putative role of plasmids in the pathogenicity of the bacterial wilt pathogen. In addition, it has been shown that a plasmid determines resistance to arsenate, arsenite, and antimony (III) in C. flaccumfaciens pv. oortii (Hendrick et al., 1984). Although no morphological differences were found between bean‐pathogenic and nonpathogenic C. flaccumfaciens strains, the former were resistant to different concentrations of arsenic, while the latter were sensitive to the same concentrations (Osdaghi et al., 2018c). Further investigations are needed to determine whether resistance against arsenic is mediated by the detected plasmids in Cff strains similar to those described for other bacterial species (Silver and Misra, 1988).
10. MANAGEMENT
10.1. Seed sanitation and certification
Due to the seedborne nature of the pathogen, bacterial wilt is a difficult disease to combat. Cff is included in the A2 (high‐risk) list of quarantine pathogens by EPPO, hence it is under strict quarantine control and zero tolerance in the dry bean industry in several countries (EPPO, 2011). Seed inspections during the intra‐ and international transportation of dry beans should be employed to avoid the distribution of the pathogen into new areas with no history of the disease (EPPO, 2011). Farmers must be encouraged to use only certified, disease‐free, and high‐quality seeds for planting purposes. If the disease is already present in an area, a 2‐year rotation with nonhost crops such as small‐grain cereals is recommended, while the possibility of eradication of the pathogen would be improved with strict sanitation of crop debris and volunteer beans to reduce pathogen survival between two bean cropping seasons (Schuster, 1970; Schuster and Coyne, 1975). Viable Cff cells have been found in irrigation systems, hence reuse of drained irrigation water should be avoided (Schuster, 1959). The same strategies can be used for other economically important hosts of the pathogen with some modifications. For instance, Ryley et al. (2010) suggested that the use of clean seed from a certified source and resistant cultivars are equally important in the management of bacterial wilt (tan spot) on mungbean in Australia, while crop rotation played a minor role.
10.2. Resistant cultivars
Development of resistant cultivars is the most practical method for managing bacterial wilt disease in terms of cost effectiveness and durability over time. Common bean cultivars and varieties differ in their reaction to the pathogen, and resistant or tolerant varieties should be planted, if available, in the areas where the disease is established (Hsieh et al., 2005b; Urrea and Harveson, 2014). Furthermore, considering the differences among Cff colony variants in their virulence and aggressiveness, a given cultivar/line should be evaluated for its resistance against different variants of the pathogen (Huang et al., 2007b; Conner et al., 2008; Urrea and Harveson, 2014).
Multiple industries are attempting to develop bacterial wilt resistance. In Canada, a high level of resistance to three variants of the pathogen (yellow, orange, and purple) was observed in the light red kidney bean cultivars AC Litekid, Chinook 2000, and Redkanner as well as dark red kidney bean cultivars Cabernet and Red Hawk, while the white cultivars did not show resistance against the pathogen (Conner et al., 2008). In the central High Plains of the USA, the wilt‐resistant cultivar Emerson is available, but it is a specialized cultivar with restricted target markets in Europe, thus production is limited. Emerson was created via a cross between Great Northern 1140 and PI 165078 from Turkey, and resistance to the pathogen was determined to be inherited quantitatively (Coyne et al., 1965; 1980; Coyne and Schuster, 1971).
Recently, Urrea and Harveson (2014) tested 427 accessions from the North American Phaselous National Plant Germplasm System's bean collection for resistance to the pathogen. They reported that only one accession, PI 325691 (a wild common bean accession collected in Mexico), exhibited disease resistance. However, the seed was too small and not commercially acceptable, thus it still needed to be crossed with others to obtain resistant, high‐yielding cultivars (Urrea and Harveson, 2014). In another study, resistant cultivars or lines were found among black, Great Northern, pink, pinto, small red, and Flor de Mayo bean market classes. The Great Northern line L02E317, the Great Northern cultivar Resolute, and pinto lines L02B662 and 999S‐2A were highly resistant to yellow and orange variants of the pathogen with zero disease severity indices (Hsieh et al., 2005b). Greenhouse experiments provided promising results confirming the source of resistance against different strains of bacterial wilt and common bacterial blight pathogens in Iran (Osdaghi et al., 2009; Osdaghi and Lak, 2015b). In Brazil under greenhouse conditions, resistance to bacterial wilt was observed in dry bean cultivars IAC Diplomata, IAC Alvorada, IAC Imperador, IAC Carioca Tybatã, IAC Carioca Pyatã, IPR Corujinha, and IPR Tangará, and in the lines P5‐4‐4‐1 and C4‐5‐4‐1‐2 (Maringoni et al., 2015).
Different inoculation techniques were proposed to assess the reaction of common bean cultivars/lines against bacterial wilt pathogen. For instance, a seed inoculation method using hilum injury was developed by Hsieh et al. (2003) to screen common bean cultivars and lines for resistance against the pathogen under greenhouse conditions within a short period of time (14 days to be completed). In Nebraska, a highly accurate system was developed for inoculating the wilt pathogen. It consisted of dipping a sterile needle into the 48‐hour‐old cultures and inserting into stems in plants just below the first fully expanded trifoliolate leaf. Symptoms generally appeared 5–7 days post‐inoculation (Harveson and Vidaver, 2007; 2008; Harveson et al., 2015; Urrea and Harveson, 2014). There has been variation within a range of eight mungbean cultivars in reaction to two different inoculation methods (stem prick and leaf infiltration), while Green Diamond cultivar showed tolerance in both tests (Diatloff and Imrie, 2000).
10.3. Biological control
Several studies have investigated the efficiency of biological control in the management of bacterial wilt disease in greenhouse conditions, while none of them has been commercialized on a large scale. Corrêa et al. (2014) applied a combination of Bacillus cereus and Pseudomonas fluorescens strains to reduce the severity of bacterial wilt, fusarium wilt, charcoal rot, and angular leaf spot of dry beans via seed inoculation using an antagonistic bacterial suspension. Significant reduction in the severity of all four diseases was obtained using B. cereus strains DFs093 and DFs769 and P. fluorescens strain DFs831. The bacterial wilt reduction has ranged from 42% to 76% (Martins et al., 2013). Environmental temperature (20 vs. 30 °C) interferes with the colonization of plants with the pathogen and antagonist, while the suppression of bacterial wilt using the B. cereus strain ALB629 was similar at the two temperatures (Martins et al., 2014). Furthermore, soaking common bean seeds in a suspension (3 × 108 cfu/ml) of Pantoea agglomerans resulted in thorough endophytic colonization of the entire bean seedling from root to apical stem 7 days post‐inoculation and resulted in the reduction of bacterial wilt severity in greenhouse conditions (Hsieh et al., 2005a). Application of P. agglomerans as a soil drench 24 hr after planting was effective in suppressing bacterial wilt, but was not as effective as seed treatment (Hsieh et al., 2005a). Furthermore, seed treatment using Rhizobium leguminosarum was reported to reduce the bacterial wilt and common bacterial blight diseases in greenhouse conditions (Huang et al., 2007b; Osdaghi et al., 2011). Bacteriophages were also proposed to be used for the control of bacterial wilt pathogen while no detailed information is available in this regard (Klement, 1957).
10.4. Chemical control
The use of chemical compounds for managing bacterial wilt is more effective when integrated with crop sanitation and cultural practices, but has been erratic in efficacy (Lamichhane et al., 2018). Several studies have proven to be ineffective with the use of copper‐based chemicals (e.g., copper oxychloride, copper hydroxide, and copper sulphate) on common bean (either field spray or seed treatment) against the bacterial wilt pathogen, highlighting the need for alternative, sustainable disease‐control strategies (Martins et al., 2013; Harveson, 2019). Harding et al. (2019) concluded that resistance to chemicals is attributed to the capacity of bacterial wilt pathogen in biofilm production. It has been shown that C. flaccumfaciens strain EF24 associated with Cytisus striatus growing on hexachlorocyclohexane‐contaminated soil is resistant against cadmium (Cd), zinc (Zn), and copper (Cu) compounds (Becerra‐Castro, 2011). The complete genome sequence of the copper‐resistant strain P990 revealed the presence of CopC gene, which is responsible for copper resistance in several plant‐pathogenic bacteria (E. Osdaghi, unpublished data). The Cop proteins are suggested to sequestrate copper compounds outside the cytoplasm to mediate copper resistance (Cha and Cooksey, 1991). Recently, Harveson (2019) reported the results of a 7‐year (2010–2016) field study (13 site years) investigating the performance of newly available antimicrobial chemicals for managing the four major bacterial diseases of dry beans (common blight, halo blight, brown spot, and wilt). These novel products were compared with two conventional copper‐based products, Kocide (copper hydroxide) and MasterCop (copper sulphate), in Nebraska, USA, and several showed great promise for reducing losses due to these diseases. Treatments with SaniDate (hydrogen peroxide 18.5% a.i. and peroxyacetic acid 12% a.i.) and ecoAgra 300 (plant‐based fatty acids, a.i.) consistently produced higher seed yields than the other treatments compared to the controls while not reducing disease incidence. These products do not appear to be curative, but act by slowing or prohibiting disease progress as protectants. Better results in all seasons were achieved from severely affected fields compared to fields with low levels of disease (Harveson, 2019).
Antibiotic seed treatment can reduce surface contamination of seeds. For instance, slurry seed treatments using Streptomycin Agri‐Strep 500 (50,000 ppm) were shown to be effective to combat Cff infection on common bean seeds (Schwartz, 2007). Soaking the seeds in Agrimaicin 500 solution, containing 500 g copper sulphate and 30 g oxytetracyclin per kilogram of the product in concentrations of 10 g/L of water for 30–60 min, eliminated the bacterium from naturally infected seeds; however, it was not effective on the artificially inoculated seeds using 108 cfu/ml suspension of the pathogen (Estefani et al., 2007).
11. CONCLUSION AND FUTURE AVENUES FOR RESEARCH
Since the first description of bacterial wilt published in 1922, many studies have been conducted that have provided a foundation of knowledge, including an understanding of the colony morphology and factors influencing the pathogen spread within the field and in distant areas. Investigations have highlighted the role of quarantine inspections, crop sanitation, and resistant cultivars in the management of bacterial wilt disease. The use of pathogen‐free high‐quality seed lots is considered the cornerstone for disease management in areas where the pathogen has already been established, while strict quarantine rules should prevent distribution of the pathogen into new areas where the disease has not yet been identified. Furthermore, the use of resistant cultivars should be encouraged in areas where the disease occurs. Recent technological advancements in high‐throughput DNA sequencing ensure that in the coming years we will integrate all discoveries into a comprehensive understanding of the genomic, ecological, and virulence repertories of Cff, as well as molecular mechanisms underlying disease development in the host plant. The first complete genome sequence of a highly virulent yellow‐pigmented Cff strain P990 (ICMP 22053) has recently been released (GenBank accession number: CP045287.1; E. Osdaghi, personal communication). Whole‐genome sequencing would allow us to predict and test which genes are involved in the pathogenicity of the wilt pathogen. Furthermore, complete genome sequencing of the pathogen will provide opportunities to develop state‐of‐the‐art genome‐informed detection methods to trace seed infections with less effort and lower cost. In addition, genomics information will allow us to target Cff pathogenicity determinates in the event of developing wilt‐resistant lines and cultivars to aid the dry bean industries of the 21st century around the world.
ACKNOWLEDGEMENTS
This study was supported by Shiraz University, Iran.
Osdaghi E, Young AJ, Harveson RM. Bacterial wilt of dry beans caused by Curtobacterium flaccumfaciens pv. flaccumfaciens: A new threat from an old enemy. Molecular Plant Pathology. 2020;21:605–621. 10.1111/mpp.12926
In this review, we provide an updated overview on the aetiology, epidemiology, and management of bacterial wilt of dry beans and the current taxonomic status of the causal agent Curtobacterium flaccumfaciens pv. flaccumfaciens.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- Agarkova, I.V. , Lambrecht, P.A. , Vidaver, A.K. and Harveson, R.M. (2012) Genetic diversity among Curtobacterium flaccumfaciens pv. flaccumfaciens populations in the American High Plains. Canadian Journal of Microbiology, 58, 788–801. [DOI] [PubMed] [Google Scholar]
- Ansari, M. , Taghavi, S.M. , Hamzehzarghani, H. , Valenzuela, M. , Siri, M.I. and Osdaghi, E. (2019) Multiple introductions of tomato pathogen Clavibacter michiganensis subsp. michiganensis into Iran as revealed by a global‐scale phylogeographic analysis. Applied and Environmental Microbiology, 85, e02098–e2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Castro, C. (2011) Role of plant-associated bacteria in the remediation of contaminated soils. PhD thesis, Instituto De Investigación Agrobiologica De Galicia, Spain. [Google Scholar]
- Bergey, D.H. , Breed, R.S. , Murray, E.G.D. and Hitchens, A.P. (1939) Bergey’s Manual of Determinative Bacteriology, 5th edition London, UK and Williams and Wilkins, Baltimore, USA: Baillière, Tindall and Cox. [Google Scholar]
- Brumbley, S.M. , Petrasovits, L.A. , Hermann, S.R. , Young, A.J. and Croft, B.J. (2006) Recent advances in the molecular biology of Leifsonia xyli subsp. xyli, causal organism for ratoon stunting disease of sugarcane. Australasian Plant Pathology, 35, 681–689. [Google Scholar]
- Burke, D.W. (1957) Bacterial wilt of pinto beans on soils of different types and cropping histories. Plant Disease Reporter, 41, 671–673. [Google Scholar]
- Burkholder, W.H. (1948) Bacteria as plant pathogens. Annual Review of Microbiology, 2, 389–412. [DOI] [PubMed] [Google Scholar]
- Burkholder, W.H. (1945) The longevity of the pathogens causing the wilt of the common bean. Phytopathology, 35, 743–744. [Google Scholar]
- Calzolari, A. , Tomesani, M. and Mazzucchi, U. (1987) Comparison of immunofluorescence staining and indirect isolation for the detection of Corynebacterium flaccumfaciens in bean seeds. EPPO Bulletin, 17, 157–163. [Google Scholar]
- Camara, R.C. , Vigo, S.C. and Maringoni, A.C. (2009) Plant to seed transmission of Curtobacterium flaccumfaciens pv. flaccumfaciens in a dry bean cultivar. Journal of Plant Pathology, 91(3), 549–554. [Google Scholar]
- Carlson, R.R. and Vidaver, A.K. (1982) Taxonomy of Corynebacterium plant pathogens, including a new pathogen of wheat, based on polyacrylamide gel electrophoresis of cellular proteins. International Journal of Systematic Bacteriology, 32, 315–326. [Google Scholar]
- Cha, J.S. and Cooksey, D.A. (1991) Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proceedings of the National Academy of Sciences of the United States of America, 88, 8915–8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, A.B. , Arevalo, P. , Polz, M.F. , Berlemont, R. and Martiny, J.B. (2016) Evidence for ecological flexibility in the cosmopolitan genus Curtobacterium . Frontiers in Microbiology, 7, 1874. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarro, C.A. , Lopez, G.C.A. and Lenne, J.M. (1985) Characteristics and pathogenicity of Corynebacterium flaccumfaciens pv. flaccunfaciens (Hedges) Dows causal agent of bacterial wilt of Zornia spp. and its effect on production of Z. glabra CIAT 7847 and Phaseolus vulgaris . Acta Agronomica, 35, 64–79. [Google Scholar]
- Chen, Y.F. , Yin, Y.N. , Zhang, X.M. and Guo, J.H. (2007) Curtobacterium flaccumfaciens pv. beticola, a new pathovar of pathogens in sugar beet. Plant Disease, 91, 677–684. [DOI] [PubMed] [Google Scholar]
- Collins, M.D. and Jones, D. (1984) Validation of the publication of new names and new combinations previously effectively published outside the IJSB: List No. 14. International Journal of Systematic Bacteriology, 34, 270–271. [Google Scholar]
- Collins, M.D. and Jones, D. (1983) Reclassification of Corynebacterium flaccumfaciens, Corynebacterium betae, Corynebacterium oortii, and Corynebacterium poinsettiae in the genus Curtobacterium as Curtobacterium flaccumfaciens comb. nov. Journal of General Microbiology, 129, 3545–3548. [Google Scholar]
- Conner, R.L. , Balasubramanian, P. , Erickson, R.S. , Huang, H.C. and Mundel, H.H. (2008) Bacterial wilt resistance in kidney beans. Canadian Journal of Plant Science, 88, 1109–1113. [Google Scholar]
- Corrêa, B.O. , Schafer, J.T. and Moura, A.B. (2014) Spectrum of biocontrol bacteria to control leaf, root and vascular diseases of dry bean. Biological Control, 72, 71–75. [Google Scholar]
- Coyne, D.P. and Schuster, M.L. (1971) ‘Emerson’, the new large‐seeded great northern dry bean variety. Bulletin of the Agricultural Experiment Station of Nebraska, 16, 1–11. [Google Scholar]
- Coyne, D.P. , Schuster, M.L. and Young, J.O. (1965) A genetic study of bacterial wilt (Corynebacterium flaccumfaciens var. aurantiacum) tolerance in Phaseolus vulgaris crosses in field beans. Proceedings of the American Society for Horticultural Science, 87, 279–285. [Google Scholar]
- Coyne, D.P. , Schuster, M.L. and Steadman, J.R. (1980) Breeding disease resistant beans in Nebraska. Michigan Dry Bean Digest, 4, 23–24. [Google Scholar]
- Davis, M.J. (1986) Taxonomy of plant‐pathogenic coryneform bacteria. Annual Review of Phytopathology, 24, 115–140. [Google Scholar]
- Davis, M.J. (2001) Coryneform phytobacteria In: Maloy O.C. and Murray T.D. (Eds.) Encyclopdia of Plant Pathology, Vol 1. New York: John Wiley and Sons, Inc; p. 1346. [Google Scholar]
- Davis, M.J. and Vidaver, A.K. (2001) Coryneform plant pathogens In: Schaad N.W., Jones J.B., and Chun W. (Eds.) Laboratory Guide for Identification of Plant Pathogenic Bacteria, 3rd edition St. Paul, MN: APS Press, pp. 218–235. [Google Scholar]
- De Bruyne, E. , Swings, J. and Kersters, K. (1992) Enzymatic relatedness amongst phytopathogenic coryneform bacteria and its potential use for their identification. Systematic and Applied Microbiology, 15, 393–401. [Google Scholar]
- Deuner, C.C. , Magela, R.S. , Zacaroni, A.B. , Figueira, A.R. and Camera, J.N. (2012) Sensitivity of the method of obtaining bacterial cells and PCR for detection of Curtobacterium flaccumfaciens pv. flaccumfaciens in bean seeds. Summa Phytopathologica, 38, 48–53. [Google Scholar]
- Diatloff, A. and Imrie, B.C. (2000) Inoculation techniques for evaluating resistance to Curtobacterium flaccumfaciens pv. flaccumfaciens in mungbean cultivars. Australasian Plant Pathology, 29, 24–28. [Google Scholar]
- Diatloff, A. , Wong, W.C. and Wood, B.A. (1993) Non‐destructive methods of detecting Curtobacterium flaccumfaciens pv. flaccumfaciens in mungbean seeds. Letters in Applied Microbiology, 16, 269–273. [Google Scholar]
- Dowson, W.J. (1942) On the generic name of the Gram‐positive bacterial plant pathogens. Transactions of the British Mycological Society, 25, 311–314. [Google Scholar]
- Dunleavy, J.M. (1983) Bacterial tan spot, a new foliar disease of soybeans. Crop Science, 23, 473–476. [Google Scholar]
- Durbin, R.D. (1972) Bacterial phytotoxins In: Wood R.K.S., Ballio A. and Graniti A. (Eds.) Phytotoxins in Plant Disease. London, New York: Academic Press, pp. 19–23. [Google Scholar]
- Dye, D.W. and Kemp, W.J. (1977) A taxonomic study of plant pathogenic Corynebacterium species. New Zealand Journal of Agricultural Research, 20, 563–582. [Google Scholar]
- EPPO (1994) Curtobacterium flaccumfaciens pv. flaccumfaciens field inspection and seed‐testing methods. EPPO Bulletin, 24, 329–331. [Google Scholar]
- EPPO (2011) Curtobacterium flaccumfaciens pv. flaccumfaciens . EPPO Bulletin, 41, 320–328. [Google Scholar]
- Estefani, R.C.C. , Filho, R.J.M. and Uesugi, C.H. (2007) Tratamentos térmico e químico de sementes de feijoeiro: eficiência na erradicação de Curtobacterium flaccumfaciens pv. flaccumfaciens e efeitos na qualidade fisiológica das sementes. Fitopatologia Brasileira, 32, 434–438. [Google Scholar]
- Evtushenko, L.I. and Takeuchi, M. (2006) The family Microbacteriaceae . The Prokaryotes, 3, 1020–1098. 10.1007/0-387-30743-5_43 [DOI] [Google Scholar]
- Felske, A. , Vancanneyt, M. , Kersters, K. and Akkermans, A.D.L. (1999) Application of temperature‐gradient gel electrophoresis in taxonomy of coryneform bacteria. International Journal of Systematic Bacteriology, 49, 113–121. [DOI] [PubMed] [Google Scholar]
- Gitaitis, R. and Walcott, R. (2007) The epidemiology and management of seed borne bacterial diseases. Annual Review of Phytopathology, 45, 371–397. [DOI] [PubMed] [Google Scholar]
- Gonçalves, R.M. , Balbi‐Peña, M.I. , Soman, J.M. , Maringoni, A.C. , Taghouti, G. , Fischer‐Le, S.M. et al (2019) Genetic diversity of Curtobacterium flaccumfaciens revealed by multilocus sequence analysis. European Journal of Plant Pathology, 154, 189–202. [Google Scholar]
- Gonçalves, R.M. , Schipanski, C.A. , Koguishi, L. , Soman, J.M. , Sakate, R.K. , Silva Júnior, T.A.F. et al (2017) Alternative hosts of Curtobacterium flaccumfaciens pv. flaccumfaciens, causal agent of bean bacterial wilt. European Journal of Plant Pathology, 148, 357–365. [Google Scholar]
- Gonçalves, R.M. , Soman, J.M. , Krause-Sakate, R. , Passos, J.R.S. , Silva Júnior, T.A.F. and Maringoni, A.C. (2018) Survival of Curtobacterium flaccumfaciens pv. flaccumfaciens in the soil under Brazilian conditions. European Journal of Plant Pathology, 152, 213–223. [Google Scholar]
- González, A.J. , Tello, J.C. and Rodicio, M.R. (2005) Bacterial wilt of beans (Phaseolus vulgaris) caused by Curtobacterium flaccumfaciens in southeastern Spain. Plant Disease, 89, 1361. [DOI] [PubMed] [Google Scholar]
- Gross, D.C. and Vidaver, A.K. (1979) Bacteriocins of phytopathogenic Corynebacterium species. Canadian Journal of Microbiology, 25, 367–374. [DOI] [PubMed] [Google Scholar]
- Gross, D.C. , Vidaver, A.K. and Keralis, M.B. (1979) Indigenous plasmids from phytopathogenic Corynebacterium species. Journal of General Microbiology, 115, 479–489. [Google Scholar]
- Guimaraes, P.M. , Palmano, S. , Smith, J.J. , Grossi, M.F.S. , & Saddler, G.S. (2001) Development of a PCR test for the detection of Curtobacterium flaccumfaciens pv. flaccumfaciens . Antonie van Leeuwenhoek Journal of Microbiology, 80, 1–10. [DOI] [PubMed] [Google Scholar]
- Guimaraes, P.M. , Smith, J.J. , Palmano, S. and Saddler, G.S. (2003) Characterization of Curtobacterium flaccumfaciens pathovars by AFLP, rep‐PCR and pulsed‐field gel electrophoresis. European Journal of Plant Pathology, 109, 817–825. [Google Scholar]
- Hagedorn, D.J. and Inglis, D.A. (1986) Handbook of Bean Diseases. Madison, Wisconsin: University of Wisconsin‐Extension, Cooperative Extension Publications. [Google Scholar]
- Harding, M. , Nadworny, P. , Buziak, B. , Omar, A. , Daniels, G. and Feng, J. (2019) Improved methods for treatment of phytopathogenic biofilms: metallic compounds as anti-bacterial coatings and fungicide tank-mix partners. Molecules, 24, 2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris‐Baldwin, A. and Gudmestad, N.C. (1996) Identification of phytopathogenic coryneform bacteria using the Biolog automated microbial identification system. Plant Disease, 80, 874–878. [Google Scholar]
- Harveson, R.M. (2013) The multicolored bacterium. APS Features, 10.1094/APSFeature. [DOI] [Google Scholar]
- Harveson, R.M. (2019) Managing dry bean bacterial diseases in Nebraska with new copper‐alternative chemicals. Plant Health Progress, 20, 14–19. [Google Scholar]
- Harveson, R.M. and Schwartz, H.F. (2007) Bacterial diseases of dry edible beans in the central high plains. Plant Health Progress, 10.1094/PHP-2007-0125-01-DG. [DOI] [PubMed] [Google Scholar]
- Harveson, R.M. and Vidaver, A.K. (2007) First report of the natural occurrence of soybean bacterial wilt isolates pathogenic to dry beans in Nebraska. Online. Plant Health Progress, 10.1094/PHP-2007-0822-01-BR. [DOI] [Google Scholar]
- Harveson, R.M. and Vidaver, A.K. (2008) A new color variant of the dry bean bacterial wilt pathogen (Curtobacterium flaccumfaciens pv. flaccumfaciens) found in western Nebraska. Online. Plant Health Progress, 10.1094/PHP-2008-0815-01-BR. [DOI] [Google Scholar]
- Harveson, R.M. and Yonts, C.D. (2007) Influence of irrigationmethod on incidence and severity of bacterial wilt of dry beans in Nebraska. Phytopathology, 97, S45. [Google Scholar]
- Harveson, R.M. , Schwartz, H.F. , Urrea, C.A. and Yonts, C.D. (2015) Bacterial wilt of dry‐edible beans in the central high plains of the US: Past, present, and future. Plant Disease, 99, 1665–1677. [DOI] [PubMed] [Google Scholar]
- Harveson, R.M. , Schwartz, H.F. , Vidaver, A.K. , Lambrecht, P.A. and Otto, K.L. (2006) New outbreaks of bacterial wilt of dry bean in western Nebraska observed from field infections. Plant Disease, 90, 81. [DOI] [PubMed] [Google Scholar]
- Hedges, F. (1926) Bacterial wilt of beans (Bacterium flaccumfaciens Hedges) including comparison with Bacterium phaseoli . Phytopathology, 16, 1–22. [Google Scholar]
- Hedges, F. (1922) A bacterial wilt of beans caused by Bacterium flaccumfaciens nov. sp. Science, 55, 433–434. [DOI] [PubMed] [Google Scholar]
- Hendrick, C.A. , Haskins, W.P. and Vidaver, A.K. (1984) Conjugative plasmid in Corynebacterium flaccumfaciens subsp. oortii that confers resistance to arsenite, arsenate, and antimony (III). Applied and Environmental Microbiology, 48, 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningson, P.J. and Gudmestad, N.C. (1991) Fatty acid analysis of phytopathogenic coryneform bacteria. Journal of General Microbiology, 137, 427–440. [Google Scholar]
- Hsieh, T.F. , Huang, H.C. , & Erickson, R.S. (2005a) Biological control of bacterial wilt of bean using a bacterial endophyte, Pantoea agglomerans . Journal of Phytopathology, 153, 608–614. [Google Scholar]
- Hsieh, T.F. , Huang, H.C. , Mündel, H.‐H. , Conner, R.L. , Erickson, R.S. and Balasubramanian, P.M. (2005b) Resistance of common bean (Phaseolus vulgaris) to bacterial wilt caused by Curtobacterium flaccumfaciens pv. flaccumfaciens . Journal of Phytopathology, 153, 245–249. [Google Scholar]
- Hsieh, T.F. , Huang, H.C. and Erickson, R.S. (2006) Bacterial wilt of common bean: Effect of seedborne inoculum on disease incidence and seedling vigour. Seed Science and Technology, 34, 57–67. [Google Scholar]
- Hsieh, T.F. , Huang, H.C. , Mündel, H.H. and Erickson, S.R. (2003) A rapid indoor technique for screening common bean (Phaseolus vulgaris L.) for resistance to bacterial wilt [Curtobacterium flaccumfaciens pv. flaccumfaciens (Hedges) Collins and Jones]. Revista Mexicana de Fitopatología, 21, 370–374. [Google Scholar]
- Huang, H.C. , Erickson, R.S. , Yanke, L.J. , Chelle, C.D. and Mündel, H.H. (2006) First report of the purple variant of Curtobacterium flaccumfaciens pv. flaccumfaciens, causal agent of bacterial wilt of bean, in Canada. Plant Disease, 90, 1262. [DOI] [PubMed] [Google Scholar]
- Huang, H.C. , Erickson, R.S. and Hsieh, T.F. (2007a) Control of bacterial wilt of bean (Curtobacterium flaccumfaciens) by seed treatment with Rhizobium leguminosarum . Crop Protection, 26, 1055–1061. [Google Scholar]
- Huang, H.C. , Mundel, H.H. , Erickson, R.S. , Chelle, C.D. , Balasubramanian, P.M. , Kiehn, F. et al (2007b) Resistance of common bean (Phaseolus vulgaris L.) cultivars and germplasm lines to the purple variant of bacterial wilt (Curtobacterium flaccumfaciens pv. flaccumfaciens). Plant Pathology Bulletin, 16, 91–95. [Google Scholar]
- Huang, H.C. , Erickson, R.S. , Balasubramanian, P.M. , Hsieh, T.F. and Conner, R.L. (2009) Resurgence of bacterial wilt of common bean in North America. Canadian Journal of Plant Pathology, 31, 290–300. [Google Scholar]
- Ishimaru, C. , Mohan, S.K. and Franc, G.D. (2005) Bacterial wilt In: Schwartz H.F., Steadman J.R., Hall R. and Forster R.L. (Eds.) Compendium of Bean Diseases, 2nd edition St. Paul, MN: American Phytopathological Society, pp. 50–52. [Google Scholar]
- Keddie, R.M. and Cure, G.L. (1977) The cell wall composition and distribution of free mycolic acids in named strains of coryneform bacteria and in isolates from various natural sources. Journal of Applied Bacteriology, 42, 229–252. [DOI] [PubMed] [Google Scholar]
- Khojasteh, M. , Taghavi, S.M. , Khodaygan, P. , Hamzehzarghani, H. , Chen, G. , Bragard, C. et al (2019) Molecular typing reveals high genetic diversity of Xanthomonas translucens infecting small‐grain cereals in Iran. Applied and Environmental Microbiology, 85, e01518–e1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement, Z. (1957) Two new bacteriophages for bacterial pathogens of the bean. Nature, 180, 41–42. [DOI] [PubMed] [Google Scholar]
- Komagata, K. and Suzuki, K. (1986) Genus Curtobacterium In: Sneath P.H.A., Mair N.A., Sharpe M.E. and Holt J.G. (Eds.) Bergey’s Manual of Systematic Bacteriology. Baltimore, MD: Williams and Wilkins; 2, pp. 1313–1317. [Google Scholar]
- Lamichhane, J.R. , Osdaghi, E. , Behlau, F. , Köhl, J. , Jones, J.B. and Aubertot, J.N. (2018) Thirteen decades of anti‐microbial copper compounds applied in agriculture. A review. Agronomy for Sustainable Development, 38, 28. [Google Scholar]
- Lehmann, K.B. and Neumann, R. (1896) Atlas und Grundriss der Bakteriologie und Lehrbuch der speciellen bakteriologischen Diagnostik. München, Germany: Teil II. J.F. Lehmann. [Google Scholar]
- Maringoni, A.C. and Rosa, E.F. (1997) Occurrence of Curtobacterium flaccumfaciens pv. flaccumfaciens on bean in state of São Paulo, Brazil. Summa Phytopathologica, 23, 160–162. [Google Scholar]
- Maringoni, A.C. , Camara, R.C. and Souza, V.L. (2006) Semi‐selective culture medium for Curtobacterium flaccumfaciens pv. flaccumfaciens isolation from bean seeds. Seed Science and Technology, 34, 117–124. [Google Scholar]
- Maringoni, A.C. and Camara, R.C. (2006) Curtobacterium flaccumfasciens pv. flaccumfasciens detection in bean seeds using a semi‐selective culture medium. Brazilian Journal of Microbiology, 37, 451–455. [Google Scholar]
- Maringoni, A.C. , Ishizuka, M.S. , Silva, A.P. , Soman, J.M. , Moura, M.F. , Silva Júnior, T.A.F. et al (2015) Reaction and colonization of common bean genotypes by Curtobacterium flaccumfasciens pv. flaccumfasciens . Crop Breeding and Applied Biotechnology, 15, 87–93. [Google Scholar]
- Martins, S.J. , Medeiros, F.H.V. , Souza, R.M. and Vilela, L.A.F. (2014) Is curtobacterium wilt biocontrol temperature dependent? Acta Scientiarum. Agronomy Maringá, 36, 409–415. [Google Scholar]
- Martins, S.J. , Medeiros, F.H.V. , Souza, R.M. , Resende, M.L.V. and Junior, P.M.R. (2013) Biological control of bacterial wilt of common bean by plant growth‐promoting rhizobacteria. Biological Control, 66, 65–71. [Google Scholar]
- McDonald, J.G. and Wong, E. (2000) High diversity in Curtobacterium flaccumfaciens pv. flaccumfaciens characterized by serology and rep‐PCR genomic fingerprinting. Canadian Journal of Plant Pathology, 22, 1–6. [Google Scholar]
- Mizuno, A. (1998) Studies on specific detecting methods of two plant pathogenic bacteria. Research Bulletin of the Plant Protection Service Japan, 15, 64–140. [Google Scholar]
- Mizuno, A. and Kawai, A. (1993) Studies on the diagnosis of foreign bacterial diseases of quarantine significance, VI: Curtobacterium flaccumfaciens pv. flaccumfaciens . Research Bulletin of the Plant Protection Service Japan, 29, 27–36. [Google Scholar]
- Nelson, P.E. and Dickey, R.S. (1970) Histopathology of plants infected with vascular bacterial pathogens. Annual Review of Phytopathology, 8, 259–280. [Google Scholar]
- Noble, T.J. , Young, A.J. , Douglas, C.A. , Williams, B. and Mundree, S. (2019) Diagnosis and management of halo blight in Australian mungbeans: A review. Crop & Pasture Science, 70, 195–203. [Google Scholar]
- Osdaghi, E. , Alizadeh, A. , Shams‐Bakhsh, M. and Lak, M.R. (2009) Evaluation of common bean lines for their reaction to the common bacterial blight pathogen. Phytopathologia Mediterranea, 48, 461–468. [Google Scholar]
- Osdaghi, E. , Alizadeh, A. , Shams‐bakhsh, M. , Lak, M.R. and Hatami Maleki, H. (2011) Induction of resistance in common bean by Rhizobium leguminosarum bv. phaseoli and decrease of common bacterial blight. Phytopathologia Mediterranea, 50, 45–54. [Google Scholar]
- Osdaghi, E. (2014) Bacterial wilt of lima bean (Phaseolus lunatus) caused by Curtobacterium flaccumfaciens pv. flaccumfaciens, a new disease in Iran. Journal of Plant Pathology, 96, S4.118. [Google Scholar]
- Osdaghi, E. and Lak, M.R. (2015a) Occurrence of a new orange variant of Curtobacterium flaccumfaciens pv. flaccumfaciens, causing common bean wilt in Iran. Journal of Phytopathology, 163, 867–871. [Google Scholar]
- Osdaghi, E. and Lak, M.R. (2015b) A source of resistance to bacterial wilt in the common bean (Phaseolus vulgaris) in Iran. Crop Protection, 74, 37–41. [Google Scholar]
- Osdaghi, E. , Pakdaman Sardrood, B. , Bavi, M. , Akbari Oghaz, N. , Kimiaei, S. and Hadian, S. (2015a) First report of Curtobacterium flaccumfaciens pv. flaccumfaciens causing cowpea bacterial wilt in Iran. Journal of Phytopathology, 163, 653–656. [Google Scholar]
- Osdaghi, E. , Taghavi, S.M. , Fazliarab, A. , Elahifard, E. and Lamichhane, J.R. (2015b) Characterization, geographic distribution and host range of Curtobacterium flaccumfaciens: An emerging bacterial pathogen in Iran. Crop Protection, 78, 185–192. [Google Scholar]
- Osdaghi, E. , Taghavi, S.M. , Hamzehzarghani, H. , Fazliarab, A. , Harveson, R.M. and Lamichhane, J.R. (2016) Occurrence and characterization of a new red‐pigmented variant of Curtobacterium flaccumfaciens, the causal agent of bacterial wilt of edible dry beans in Iran. European Journal of Plant Pathology, 146, 129–145. [Google Scholar]
- Osdaghi, E. , Forero Serna, N. , Bolot, S. , Fischer‐Le Saux, M. , Jacques, M.‐A. , Portier, P. et al (2017) High‐quality draft genome sequence of Curtobacterium sp. strain Ferrero. Genome Announcements, 5, e01378–e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osdaghi, E. , Taghavi, S.M. , Hamzehzarghani, H. , Fazliarab, A. , Harveson, R.M. , Tegli, S. et al. (2018a) Epiphytic Curtobacterium flaccumfaciens strains isolated from symptomless solanaceous vegetables are pathogenic on leguminous but not on solanaceous plants. Plant Pathology, 67, 388–398. [Google Scholar]
- Osdaghi, E. , Ansari, M. , Taghavi, S.M. , Zarei, S. , Koebnik, R. and Lamichhane, J.R. (2018b) Pathogenicity and phylogenetic analysis of Clavibacter michiganensis strains associated with tomato plants in Iran. Plant Pathology, 674, 957–970. [Google Scholar]
- Osdaghi, E. , Taghavi, S.M. , Calamai, S. , Biancalani, C. , Cerboneschi, M. , Tegli, S. et al (2018c) Phenotypic and molecular‐phylogenetic analysis provide novel insights into the diversity of Curtobacterium flaccumfaciens . Phytopathology, 108, 1154–1164. [DOI] [PubMed] [Google Scholar]
- Osdaghi, E. , Portier, P. , Briand, M. , Taghouti, G. and Jacques, M.‐A. (2018d) Draft genome sequences of the type strains of three Clavibacter subspecies, and atypical peach‐colored strains isolated from tomato. Microbiology Resource Announcements, 7, e01357–e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osdaghi, E. , Rahimi, T. , Taghavi, S.M. , Ansari, M. , Zarei, S. , Portier, P. et al (2020) Comparative genomics and phylogenetic analyses suggest several novel species within the genus Clavibacter, including nonpathogenic tomato‐associated strains. Applied and Environmental Microbiology, 86, e02873–e2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, Z.A. (1954) The antibiotic activity of soil microorganisms as related to bacterial plant pathogens. Canadian Journal of Botany, 32, 705–735. [Google Scholar]
- Pawlowski, M.L. and Hartman, G.L. (2019) First report of Curtobacterium flaccumfaciens pv. flaccumfaciens causing bacterial tan spot on soybean in Africa. Plant Disease, 103, 2665. [DOI] [PubMed] [Google Scholar]
- Pelludat, C. , Duffy, B. and Frey, J.E. (2009) Design and development of a DNA microarray for rapid identification of multiple European quarantine phytopathogenic bacteria. European Journal of Plant Pathology, 2009, 413–423. [Google Scholar]
- Ryley, M. , Douglas, C. , Ryan, M. , Tatnell, J. , Martin, W. , King, K. et al (2010) Integrated management of foliar pathogens of mungbean in Australia In: Proceedings of the 1st Australian Summer Grains Conference, 21–24 June, Gold Coast, Australia: Available at: https://www.yumpu.com/en/document/view/33429631/integrated-management-of-foliar-pathogens-of-mungbean-in-australia [Accessed 21 February 2020]. [Google Scholar]
- Saettler, A.W. and Perry, S.K. (1972) Seed‐transmitted bacterial diseases in Michigan navy (pea) beans, Phaseolus vulgaris . Plant Disease Reporter, 56, 378–381. [Google Scholar]
- Saettler, A.W. (1991) Diseases caused by bacteria In: Hall R. (Ed.) Compendium of Bean Diseases. St. Paul: The American Phytopathological Society, pp. 29–32. [Google Scholar]
- Sammer, U.F. and Reiher, K. (2012) Curtobacterium flaccumfaciens pv. flaccumfaciens on soybean in Germany—A threat for farming. Journal of Phytopathology, 160, 314–316. [Google Scholar]
- Schuster, M.L. and Coyne, D.P. (1974) Survival mechanisms of phytopathogenic bacteria. Annual Review of Phytopathology, 12, 199–221. [Google Scholar]
- Schuster, M.L. and Coyne, D.P. (1981) Biology, epidemiology, genetics and breeding for resistance to bacterial pathogens of Phaseolus vulgaris L. horticultural reviews. The AVI Publishing Company, Inc. Paper 5878, Journal Series, University of Nebraska Agricultural Experiment Station.
- Schuster, M.L. (1959) Relation of root‐knot nematodes and irrigation water to the incidence and dissemination of bacterial wilt of bean. Plant Disease Reporter, 43, 27–32. [Google Scholar]
- Schuster, M.L. (1970) Survival of bacterial pathogens of beans. Bean Improvement Cooperative, 13, 68–70. [Google Scholar]
- Schuster, M.L. and Coyne, D.P. (1975) Survival Factors of Plant Pathogenic Bacteria. Research Bulletin no. 268, USA: University of Nebraska, 53. [Google Scholar]
- Schuster, M.L. and Christiansen, D.W. (1957) An orange‐colored strain of Corynebacterium flaccumfaciens causing bean wilt. Phytopathology, 47, 51–53. [Google Scholar]
- Schuster, M.L. , Vidaver, A.K. and Mandel, M. (1968) A purple pigment‐producing bean wilt bacterium Corynebacterium flaccumfaciens var. violaceum n. var. Canadian Journal of Microbiology, 14, 423–427. [DOI] [PubMed] [Google Scholar]
- Schuster, M.L. (1967) Survival of bean bacterial pathogens in the field and greenhouse under different environmental conditions. Phytopathology, 57, 830. [Google Scholar]
- Schuster, M.L. and Sayre, R.M. (1967) A coryneform bacterium induces purple‐colored seed and leaf hypertrophy of Phaseolus vulgaris and other leguminosae. Phytopathology, 57, 1064–1066. [Google Scholar]
- Schwartz, H.F. (2007). Dry Bean Bacterial Wilt. High Plains IPM Guide, a Cooperative Effort of the University of Wyoming, University of Nebraska, Colorado State University and Montana State University. Nebraska, USA: University of Nebraska. [Google Scholar]
- Schwartz, H.F. , Franc, G.D. , Hanson, L.E. and Harveson, R.M. (2005) Disease management In: Schwartz H.F., Brick M.A., Harveson R.M. and Franc G.D. (Eds.) Dry Bean Production and Pest Management. Bull, pp. 109–143. [Google Scholar]
- Silva Júnior, T.A.F. , Itako, A.T. , Negrão, D.R. and Maringoni, A.C. (2012). Pathogenicity of Curtobacterium flaccumfaciens pv. flaccumfaciens to several plant species. Journal of Plant Pathology, 94, 427–430. [Google Scholar]
- Silver, S. and Misra, T.K. (1988) Plasmid mediated heavy metal resistances. Annual Review of Microbiology, 42, 717–743. [DOI] [PubMed] [Google Scholar]
- Soares, R.M. , Fantinato, G.G.P. , Darben, L.M. , Marcelino‐Guimarães, F.C. , Seixas, C.D.S. and Carneiro, G.E.S. (2013) First report of Curtobacterium flaccumfaciens pv. flaccumfaciens on soybean in Brazil. Tropical Plant Pathology, 38, 452–454. [Google Scholar]
- Soares, R.M. , Fantinato, G.G.P. , Ferreira, E.G.C. and Marcelino‐Guimarães, F.C. (2018) Plant‐to‐seed transmission of Curtobacterium flaccumfaciens pv. flaccumfaciens on soybean. Tropical Plant Pathology, 43, 376. [Google Scholar]
- Souza, V.L. , Maringoni, A.C. and Krause‐Sakate, R. (2006) Genetic variability in Curtobacterium flaccumfaciens isolates. Summa Phytopathologica, 32, 170–176. [Google Scholar]
- Strider, D.L. (1969) Bacterial canker of tomato caused by Corynebacterium michiganense. Technical Bulletin No.193. North Carolina, USA: North Carolina Agricultural Experiment Station. [Google Scholar]
- Tegli, S. , Cerboneschi, M. and Vidaver, A.K. (2017) Detection of Curtobacterium flaccumfaciens pv. flaccumfaciens in bean seeds and in seeds of other Leguminosae crops In: Fatmi M., Walcott R. R. and Schaad N. W. (Eds). Detection of Plant-Pathogenic Bacteria in Seed and Other Planting Material, 2nd edition St Paul, MN: : APS Press, pp. 77–83. [Google Scholar]
- Tegli, S. , Sereni, A. and Surico, G. (2002) PCR‐based assay for the detection of Curtobacterium flaccumfaciens pv. flaccumfaciens in bean seeds. Letters in Applied Microbiology, 35, 331–337. [DOI] [PubMed] [Google Scholar]
- Thapa, S.P. , Davis, E.W. , Lyu, Q. , Weisberg, A.J. , Stevens, D.M. , Clarke, C.R. et al (2019) The evolution, ecology, and mechanisms of infection by gram‐positive, plant‐associated bacteria. Annual Review of Phytopathology, 25, 341–365. [DOI] [PubMed] [Google Scholar]
- Thapa, S.P. , Pattathil, S. , Hahn, M.G. , Jacques, M.A. , Gilbertson, R.L. and Coaker, G. (2017) Genomic analysis of Clavibacter michiganensis reveals insight into virulence strategies and genetic diversity of a gram‐positive bacterial pathogen. Molecular Plant‐Microbe Interactions, 30, 786–802. [DOI] [PubMed] [Google Scholar]
- Thomas, W.D. and Graham, R.W. (1952) Bacteria in apparently healthy pinto beans. Phytopathology, 42, 214–215. [Google Scholar]
- Torres, C. , Lenne, G.M. and Victoria, J. (1982) Bacterial wilt of Zornia spp. caused by Corynebacterium flaccumfaciens In: Lozano G.C. (Ed.). Proceedings of the Fifth International Conference on Plant Pathogenic Bacteria. Cali: Colombia Centra International de Agricuitura Tropical, pp. 74–79. [Google Scholar]
- Urrea, C.A. and Harveson, R.M. (2014) Identification of sources of bacterial wilt resistance in common beans (Phaseolus vulgaris L.). Plant Disease, 98, 973–976. [DOI] [PubMed] [Google Scholar]
- Vaghefi, N. , Adorada, D. , Adorada, E. , Kelly, L. , Young, A. and Sparks, A. (2019) What can we learn from population genomics studies of Curtobacterium flaccumfaciens pv. flaccumfaciens, the cause of tan spot on mungbean? In: Proceedings of the Australasian Plant Pathology Society Biennial Conference, 25–28 November 2019, Melbourne: Abstract available at: http://era.daf.qld.gov.au/id/eprint/7161/ [Accessed 21 February 2020]. [Google Scholar]
- Vidaver, A.K. (1982) The plant pathogenic corynebacteria. Annual Review of Microbiology, 36, 495–517. [DOI] [PubMed] [Google Scholar]
- Vidaver, A.K. and Davis, M.J. (1988) Coryneform plant pathogens In: Schaad N.W. (Ed.) Laboratory Guide for Identification of Plant Pathogenic Bacteria, 2nd edition St. Paul, MN: APS Press, pp. 104–113. [Google Scholar]
- Vidaver, A.K. and Starr, M.P. (1981) Phytopathogenic coryneform and related bacteria In: Starr M.P., Stolp H., Trüper H.G., Balows A. and Schlegel H.G. (Eds). The Prokaryotes, A Handbook on Habitats, Isolation, and Identification of Bacteria. Berlin, Heidelberg, New York: Springer‐Verlag, pp. 1879–1887. [Google Scholar]
- Wood, B.A. and Easdown, W.J. (1990) A new bacterial disease of mungbean and cowpea for Australia. Australasian Plant Pathology, 19, 16–21. [Google Scholar]
- Yamada, K. and Komagata, K. (1972) Taxonomic studies on coryneform bacteria. V: Classification of coryneform bacteria. Journal of General and Applied Microbiology, 18, 417–431. [Google Scholar]
- Yerkes, W.D. and Crispin, A.M. (1956) Bean diseases of importance in Mexico in 1955. Plant Disease Reporter, 40, 222–223. [Google Scholar]
- Zaumeyer, W.J. (1932) Comparative pathological histology of three bacterial diseases of bean. Journal of Agricultural Research, 44, 605–632. [Google Scholar]
- Zaumeyer, W.J. and Thomas, H.R. (1957) A monographic study of bean diseases and methods of their control. U.S. Department Agriculture Technical Bulletin, 868, 84–88. [Google Scholar]
- Zhao, Y.F. , Wei, Y.D. , Gao, C.S. , Zhao, L.Z. , Huang, G.M. and Huang, Q.L. (1997) Using the Biolog identification system for rapid identification of bean wilt bacterium (Curtobacterium flaccumfaciens pv. flaccumfaciens). Acta Phytopathologica Sinica, 27, 139–144. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


