Abstract
SsITL, a secretory protein of the necrotrophic phytopathogen Sclerotinia sclerotiorum, was previously reported to suppress host immunity at the early stages of infection. However, the molecular mechanism that SsITL uses to inhibit plant defence against S. sclerotiorum has not yet been elucidated. Here, we report that SsITL interacted with a chloroplast‐localized calcium‐sensing receptor, CAS, in chloroplasts. We found that CAS is a positive regulator of the salicylic acid signalling pathway in plant immunity to S. sclerotiorum and CAS‐mediated resistance against S. sclerotiorum depends on Ca2+ signalling. Furthermore, we showed that SsITL could interfere with the plant salicylic acid (SA) signalling pathway and SsITL‐expressing transgenic plants were more susceptible to S. sclerotiorum. However, truncated SsITLs (SsITL‐NT1 or SsITL‐CT1) that lost the ability to interact with CAS do not affect plant resistance to S. sclerotiorum. Taken together, our findings reveal that SsITL inhibits SA accumulation during the early stage of infection by interacting with CAS and then facilitating the infection by S. sclerotiorum.
Keywords: calcium‐sensing receptor, effector, salicylic acid signalling pathway, Sclerotinia sclerotiorum, SsITL
The secretory protein SsITL inhibits salicylic acid accumulation during the early stage of infection by interacting with CAS (calcium‐sensing receptor) and then facilitating S. sclerotiorum infection.
1. INTRODUCTION
Sclerotinia sclerotiorum is a necrotrophic fungus that infects more than 400 plant species worldwide. Most of these hosts are dicotyledonous, such as soybean, rapeseed, sunflower, and bean, and a few agriculturally important monocotyledonous plants are also hosts of S. sclerotiorum, such as onion and tulip (Boland and Hall, 1994; Bolton et al., 2006). Sclerotinia disease is responsible for considerable damage to many crops and is difficult to control (Zhou et al., 2014; Hou et al., 2018). Research on the pathogenesis of S. sclerotiorum can provide new insights for the development of sclerotinia disease prevention and control strategies.
As a typical necrotrophic pathogenic fungus, the pathogenesis of S. sclerotiorum is more complicated than we originally thought. Early research focused on the cell wall‐degrading enzymes (CWDEs) and toxic metabolite oxalic acid (OA). This fungus secretes a wide array of CWDEs, which can macerate plant tissues, degrade plant cell wall components, and ultimately promote infection (Riou et al., 1991; Issam et al., 2004; Ellouze et al., 2011). OA plays multiple functions in numerous physiological processes, such as deregulation of guard cells, sequestration of calcium, dampening the plant oxidative burst, induction of apoptotic‐like programmed cell death (PCD), and suppression of autophagy (Marciano et al., 1983; Cessna et al., 2000; Guimaraes and Stotz, 2004; Kim et al., 2008; Williams et al., 2011; Heller and Witt‐Geiges, 2013; Kabbage et al., 2013). Despite the multiple roles of OA in the pathogenesis of S. sclerotiorum, recent studies have shown that oxalate is not required for S. sclerotiorum to cause disease on some host plants; the authors proposed that it is the low pH environment that plays an important role in Sclerotinia pathogenesis (Xu et al., 2015). The genome of S. sclerotiorum encodes more than 600 secreted proteins, 70 putative effectors were predicted and 61 of these have not been reported yet (Amselem et al., 2011; Derbyshire et al., 2017), and only a few of the secreted proteins have been investigated in relation to pathogenesis. For example, secreted protein Ss‐Caf1 is required for appressorium formation and disruption of Ss‐Caf1 completely abolishes the virulence of S. sclerotiorum on host plants, but the mutant produces even more OA than the wild‐type strain (Xiao et al., 2014). Furthermore, some secreted proteins play roles in a subtler way during S. sclerotiorum infection. For instance, the small secreted protein SsSSVP1 interacts with QCR8 and disturbs the subcellular localization of QCR8 in mitochondria, which may disable its biological function and hence interfere with plant energy metabolism to facilitate the infection of S. sclerotiorum (Lyu et al., 2016). A cerato‐platanin protein SsCP1 interacts with plant PR1 and induces cell death in a dose‐dependent manner in the host plant. Interestingly, SsCP1 also triggers plant defence responses through the salicylic acid (SA) signalling pathway (Yang et al., 2018).
The plant hormones SA and jasmonic acid (JA) play important roles in disease resistance. In general, it is known that the JA signalling pathway mainly resists the infection of necrotrophic pathogens, while the SA signalling pathway regulates the resistance response associated with biotrophic pathogens (Glazebrook, 2005; Pieterse et al., 2012). Extensive cross‐talk between the SA and JA signalling pathways was found in plants and this cross‐talk plays a crucial role in plant defence (Glazebrook, 2005; Mur et al., 2006; Spoel et al., 2007; El Oirdi et al., 2011; Robert‐Seilaniantz et al., 2011). Previous research has shown that the SA signalling pathway also plays an important role in plant resistance to the necrotrophic pathogen Botrytis cinerea (Murphy et al., 2000). Furthermore, although S. sclerotiorum is a necrotrophic fungal pathogen, at the early stages of infection S. sclerotiorum grows in the apoplast without crossing the plant cell wall and the host cells are still alive at that stage, suggesting that a short biotrophic interaction between plants and S. sclerotiorum should exist (Kabbage et al., 2015). Many recent studies have shown that the defence against S. sclerotiorum in Arabidopsis and oilseed rape is also associated with SA signalling (Guo and Stotz, 2007; Wang et al., 2012; Nováková et al., 2014; Yang et al., 2018). In short, SA signalling possibly acts as a positive regulator in plant immunity at the early stage of S. sclerotiorum infection.
We previously demonstrated that an integrin‐like protein SsITL, a potential effector of S. sclerotiorum, suppresses host immunity at the early stage of infection (Zhu et al., 2013). However, the molecular mechanism that SsITL uses to suppress plant defence against S. sclerotiorum has not yet been illuminated. Here we report that SsITL interacts with the Arabidopsis calcium‐sensing receptor CAS in chloroplasts. CAS is a chloroplast‐localized protein that acts upstream of SA accumulation and is involved in plant innate immunity (Nomura et al., 2012). Overexpression of CAS in Arabidopsis increased plant resistance to S. sclerotiorum, suggesting that CAS positively regulates plant defence against S. sclerotiorum infection. Ectopic expression of SsITL in Arabidopsis reduced SA concentration after inoculation and enhanced susceptibility to S. sclerotiorum, but the overexpression of truncated SsITLs that cannot interact with CAS do not affect plant resistance to S. sclerotiorum. Our results suggest that SsITL suppresses plant defence through interaction with CAS in chloroplasts and then interferes with SA accumulation during S. sclerotiorum infection.
2. RESULTS
2.1. SsITL interacts with Arabidopsis calcium‐sensing receptor CAS in the chloroplasts
We previously reported that a secretory protein SsITL suppresses host resistance at the early stage of S. sclerotiorum infection (Zhu et al., 2013), while the underlying mechanisms by which SsITL modulates plant immunity have not yet been elucidated. To further clarify the mechanism of SsITL in the virulence of S. sclerotiorum, an immunoprecipitation (IP) combined with liquid chromatography coupled with tandem mass spectrometry (LC‐MS/MS) assay was performed to screen for its plant interactors. IP‐LC‐MS/MS results indicated that Arabidopsis CAS might interact with SsITL (Figure S1 and Table S1). The interaction of SsITL∆SP (lacking the signal peptide) with CAS was investigated using the GAL4‐based yeast two‐hybrid (Y2H) system. The plasmids pGADT7‐CAS, pGADT7‐CAS (Nt, 1–187 amino acids) and pGADT7‐CAS (Ct, 211–387 amino acids) were co‐transformed with pGBKT7‐SsITL ∆SP to yeast strain Y2H Gold. The Y2H results showed that SsITL∆SP interacts with the N terminus of CAS (1–187 amino acids), but cannot interact with the C terminus (211–387 amino acids) (Figure 1a). To further validate the interaction of SsITL with CAS in planta, a co‐immunoprecipitation (Co‐IP) assay was carried out. FLAG‐tagged CAS (pCNF3‐CAS) was co‐expressed with GFP‐tagged SsITL (pCNG‐SsITL) or green fluorescent protein (GFP) (2 × 35S‐MCS‐eGFP, pCNG) in Nicotiana benthamiana. Then the total proteins were extracted from infiltrated leaves and incubated with GFP‐antibody beads. The results show that FLAG‐tagged CAS was significantly enriched in the GFP‐tagged SsITL precipitates, but not in the GFP precipitates, indicating that SsITL interacts with CAS in planta (Figure 1b). Direct physical interaction between SsITL and CAS in vitro was also observed in glutathione‐S‐transferase (GST) pull‐down assays (Figure 1c).
Figure 1.
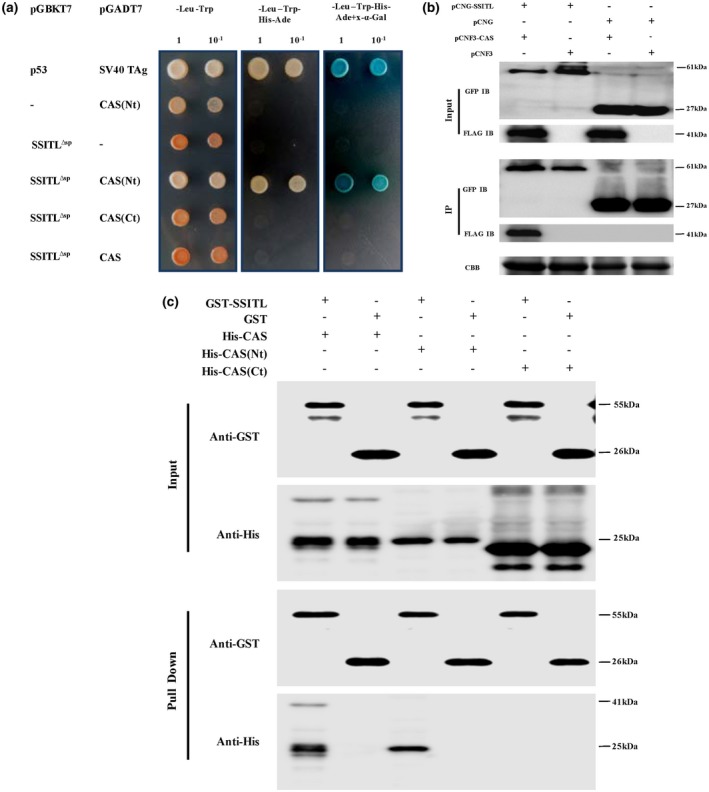
SsITL interacts with Arabidopsis thaliana CAS. (a) Yeast two‐hybrid (Y2H) assay showed that SsITL interacted with CAS(Nt) in yeast. Co‐expression of pGBKT7‐p53 and pGADT7‐SV40 TAg as positive control. –, corresponding empty vectors. The negative controls showed that both SsITL∆sp and CAS(Nt) were not self‐activated. The working concentration of X‐α‐Gal was 40 µg/ml. The plates were photographed 4 days after inoculation and experiments were repeated three times. (b) Co‐immunoprecipitation (Co‐IP) assay confirmed that SsITL interacts with CAS in planta. SsITL‐green fluorescent protein (GFP) was expressed in Nicotiana benthamiana together with CAS‐3 × FLAG and the corresponding empty vectors were set as the negative controls. Input, total proteins of N. benthamiana leaves; IP, protein samples immunoprecipitated with monoclonal GFP antibody; IB, immunoblot. The presence of FLAG proteins after immunoprecipitation was detected by western blot using anti‐FLAG antibody. CBB, total protein stained with Coomassie brilliant blue. (c) Physical interaction of SsITL and CAS in vitro was verified by glutathione‐S‐transferase (GST) pull‐down assay. GST‐SsITL was incubated in binding buffer containing glutathione‐agarose beads with or without CAS‐His, CAS(Nt)‐His or CAS(Ct)‐His, and agarose beads were washed for five times and eluted. Lysis of Escherichia coli (Input) and eluted proteins (Pull Down) from beads was immublotted using anti‐His and anti‐GST antibodies
CAS is known to localize in the chloroplast thylakoid membrane, and the N terminus of CAS appears to be exposed to the stromal side of the thylakoid membrane (Friso et al., 2004; Nomura et al., 2008). To determine the precise intracellular location of SsITL and CAS, SsITL‐GFP and CAS‐YFP (yellow fluorescent protein) fusion proteins were transiently expressed in N. benthamiana leaves using the Agrobacterium infiltration method. Consistent with previous studies, CAS localized in chloroplasts; meanwhile, we found that SsITL was also localized in chloroplasts (Figure 2a). Furthermore, when SsITL‐GFP and CAS‐YFP were co‐expressed in N. benthamiana, a perfect overlap of the YFP and GFP signals indicated that SsITL and CAS co‐localized in chloroplasts (Figure 2b).
Figure 2.
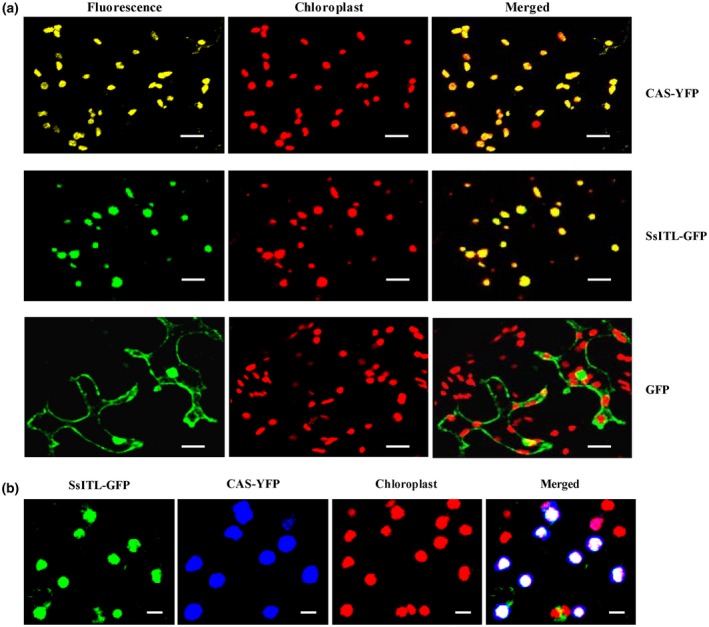
Subcellular localization of SsITL and CAS in Nicotiana benthamiana epidermal cells. (a) Both SsITL‐green fluorescent protein (GFP) and CAS‐yellow fluorescent protein (YFP) localized in the chloroplasts. (b) Co‐localization of SsITL‐GFP and CAS‐YFP to the chloroplasts in N. benthamiana. Agrobacterium tumefaciens GV3101 carrying SsITL‐GFP or CAS‐YFP constructs were agroinfiltrated separately or in combination. Pictures were taken 72 hr post‐agroinfiltration with confocal laser scanning microscopy. Bars = 20 µm
2.2. CAS is a positive regulator of the SA signalling pathway in plant immunity to S. sclerotiorum
A previous study indicated that CAS acts upstream of SA accumulation and is responsible for the pathogen‐associated molecular pattern (PAMP)‐induced innate immune system and effector‐triggered immunity, enhancing the resistance of a plant to a bacterial pathogen (Nomura et al., 2012). To investigate the role of CAS in regulating the defence response of plants to S. sclerotiorum, we generated CAS constitutive overexpression transgenic Arabidopsis lines. The candidate lines 35S:AtCAS‐1 and 35S:AtCAS‐2 were verified by reverse transcription (RT) PCR (Figure S2a). The results show that overexpression of CAS had no impact on plant morphology, growth, and development (Figure S3). However, CAS overexpression significantly enhanced plant resistance to both S. sclerotiorum wild‐type strain Ep‐1PNA367 (Figure 3a, top) and the SsITL‐silenced transformant A10 (Figure 3a, bottom). For example, the lesion areas produced by A10 were 40.62 mm2 on the leaves of Arabidopsis thaliana Col‐0 (wild‐type), but only 20.38 mm2 on the leaves of 35S:AtCAS‐1 at 36 hours post‐inoculation (hpi) (Figure 3a,b), indicating an important role of CAS in the regulation of plant resistance to S. sclerotiorum. Previous research reported that CAS is involved in the regulation of expression of SA biosynthesis‐related genes, such as ICS1, PAD4, PBS3, and EDS5 (Nomura et al., 2012). Therefore, the transcript levels of these genes in CAS overexpression Arabidopsis lines were measured using quantitative reverse transcription PCR (RT‐qPCR). Consistent with expectation, when leaves were challenged with either wild‐type Ep‐1PNA367 or SsITL‐silenced A10, the expressions of PAD4, PBS3, EDS5, and defence marker gene PR1 were up‐regulated in 35S:AtCAS‐1 and 35S:AtCAS‐2 compared to Col‐0, but ICS1 was up‐regulated in overexpressed CAS lines only when inoculated with A10 (Figure 3c). Additionally, although the SA concentration in wild‐type Col‐0 was increased on inoculation with Ep‐1PNA367 and A10, especially when inoculated with A10, the SA concentrations in 35S:AtCAS‐1 and 35S:AtCAS‐2 were much higher than those in Col‐0 when inoculated with Ep‐1PNA367 and A10 (Figure 3d). However, the expression levels of these genes and SA concentrations in uninoculated plants exhibited no significant changes (Figure 3c,d). These data indicate that CAS is involved in the defence response of Arabidopsis to S. sclerotiorum. During S. sclerotiorum infection, CAS positively regulates the accumulation of SA through promoting the expression of SA biosynthesis‐related genes, thereby enhancing the plant resistance to S. sclerotiorum.
Figure 3.
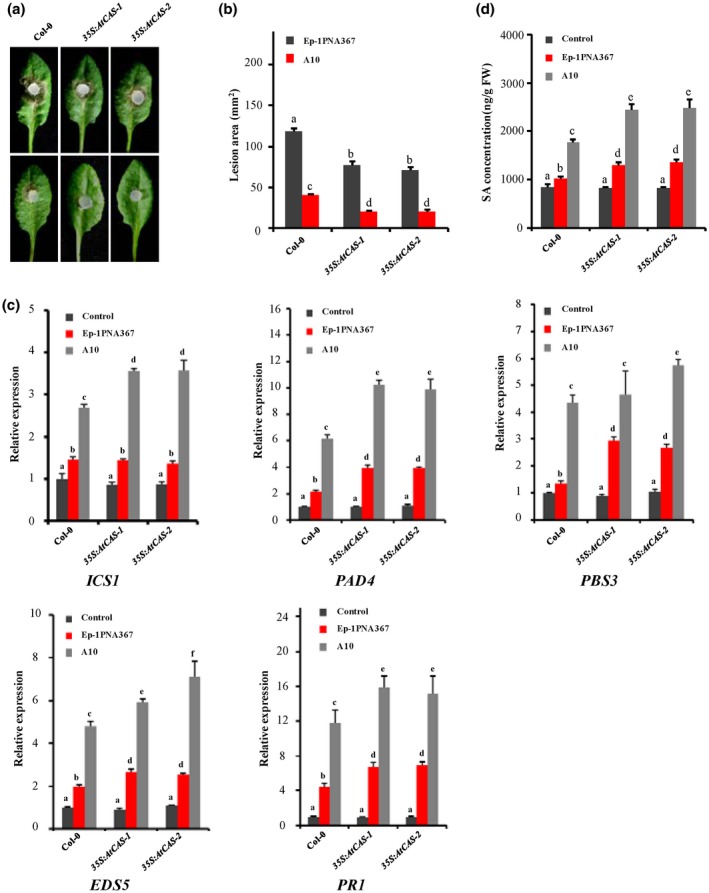
Overexpression of CAS enhances resistance of Arabidopsis to Sclerotinia sclerotiorum. (a) The wild‐type plant Col‐0 and CAS overexpression plants 35S:AtCAS‐1 and 35S:AtCAS‐2 were challenged with S. sclerotiorum strains Ep‐1PNA367 (top) and A10 (bottom). Photographs were taken at 36 hours post‐inoculation (hpi). (b) Statistical analysis of the lesion area induced by Ep‐1PNA367 or A10 on each plant at 36 hpi. Values are means ± SE. (c) The relative expression levels of the salicylic acid (SA) signalling pathway‐related genes (ICS1, PAD4, PBS3, EDS5, and PR1) in each plant were analysed at 12 hpi. The expression levels of GAPDH were used to normalize the expression levels of these genes in the different samples. The expression level in the wild‐type plant Col‐0 without inoculation was set as 1. Values are means ± SD. (d) SA concentrations in each plant were measured at 12 hpi. The wild‐type plant Col‐0 without inoculation was used as control. In all experiments, three independent replicates were performed. Values are means ± SE. Different letters on the same graph indicate statistical significance (one‐way analysis of variance; post hoc = Duncan α [0.05])
cas‐1 knockout plant was employed to further examine the roles of CAS in plant immunity to S. sclerotiorum (Figure S2b). Loss of function of CAS in Arabidopsis had no impact on plant morphology, growth, and development (Figure S3). When inoculated with Ep‐1PNA367 (Figure 4a, top) or A10 (Figure 4a, bottom), the cas‐1 mutant showed greater susceptibility than wild‐type Col‐0 (Figure 4a,b). Compared to the wild‐type Col‐0, the SA concentration and transcription levels of the related genes were also significantly reduced in the cas‐1 mutant when challenged with the SsITL‐silenced strain A10 (Figure 4c,d). The results further confirm that CAS is a positive regulator of the SA signalling pathway in plant immunity to S. sclerotiorum.
Figure 4.
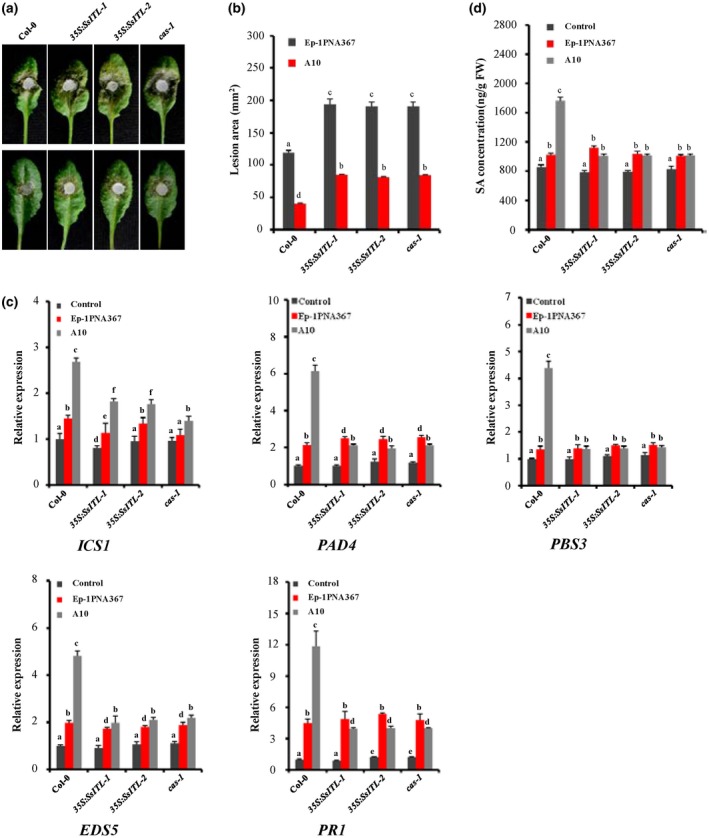
Plant resistance to Sclerotinia sclerotiorum and the salicylic acid (SA) accumulation were impaired in SsITL transgenic and cas‐1 mutant Arabidopsis thaliana plants. (a) The wild‐type plant Col‐0, SsITL transgenic plants 35S:SsITL‐1, 35S:SsITL‐2 and cas‐1 plant were challenged with S. sclerotiorum strains Ep‐1PNA367 (top) or A10 (bottom). Photographs were taken at 36 hours post‐inoculation (hpi). (b) Statistical analysis of leaf lesion area induced by Ep‐1PNA367 or A10 on each plant at 36 hpi. Values are means ± SE. (c) The relative expression levels of the SA signalling pathway‐related genes (ICS1, PAD4, PBS3, EDS5, and PR1) in each plant were analysed at 12 hpi. The expression levels of GAPDH were used to normalize the expression levels of these genes in different samples. The expression level in the wild‐type plant Col‐0 without inoculation was set to 1. Values are means ± SD. (d) SA concentration in each plant was measured at 12 hpi. The wild‐type plant Col‐0 without inoculation was used as control. In all experiments, three independent replicates were performed. Values are means ± SE. Different letters on the same graph indicate statistical significance (one‐way analysis of variance; post hoc = Duncan α [0.05])
2.3. SsITL contributes to plant susceptibility through interaction with CAS
To further investigate the function of SsITL in Sclerotinia–plant interactions, SsITL was constitutively expressed in transgenic plants 35S:SsITL‐1 and 35S:SsITL‐2 (Zhu et al., 2013). Expression of SsITL had no impact on plant morphology, growth, and development (Figure S3). Consistent with our previous results, the lesion areas induced by Ep‐1PNA367 (Figure 4a, top) or A10 (Figure 4a, bottom) were obviously increased in SsITL‐transgenic plants 35S:SsITL‐1 and 35S:SsITL‐2 (Figure 4a,b). Moreover, the size of the lesions produced by S. sclerotiorum in SsITL overexpression plants was almost the same as that of the cas‐1 mutant plant. When inoculated with A10, the SA concentration and transcription levels of related genes were also suppressed in 35S:SsITL‐1 and 35S:SsITL‐2 compared with the wild‐type Col‐0 (Figure 4d). These results suggest that SsITL could interfere with the plant SA signalling pathway, contributing to plant susceptibility.
To further investigate the biological function of the interaction between SsITL and CAS, we performed SsITL deletion screening. Alignment of the amino acid sequence of the SsITL protein revealed that it contains five highly conserved repeat peptides (Zhu et al., 2013). In order to clarify the interaction of these conserved regions with CAS, SsITL∆SP was truncated from the N terminal or the C terminal and then subjected to Y2H assay. The results show that only the full length of SsITL could interact with CAS in yeasts (Figure 5a), all the N‐terminus and C‐terminus truncated SsITLs lost the ability to interact with CAS, although all the truncated proteins were well expressed (Figure S5), suggesting that the complete structure of SsITL might be essential for this interaction.
Figure 5.
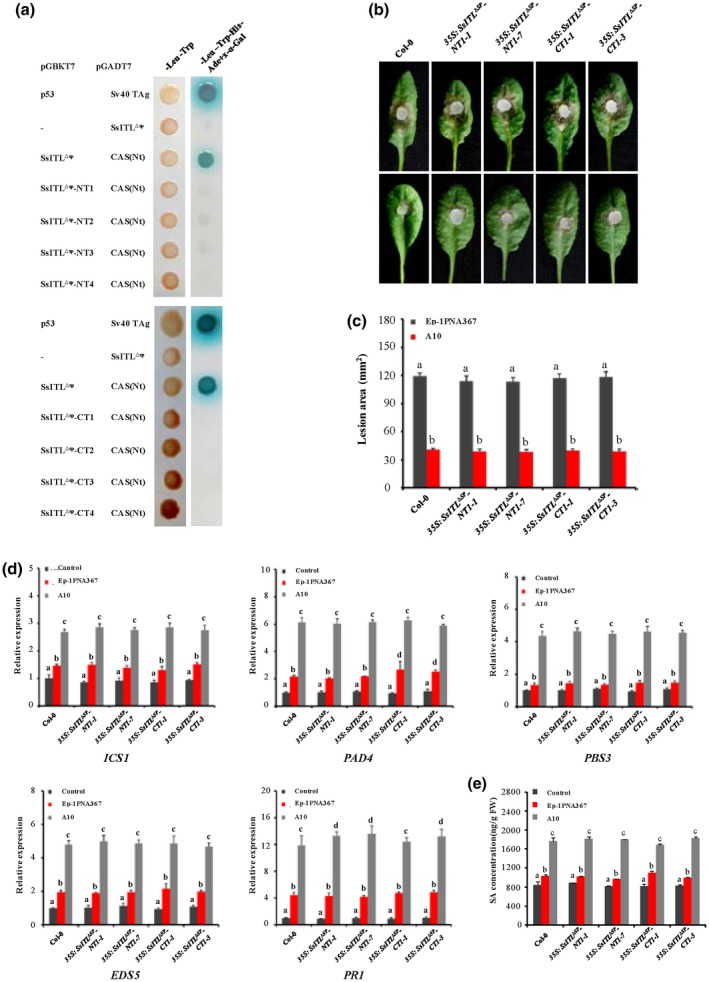
Truncated SsITLs have no effect on plant resistance to Sclerotinia sclerotiorum. (a) Yeast two‐hybrid (Y2H) assay showed that truncated SsITLs (SsITL∆SP‐NT and SsITL∆SP‐CT) cannot interact with CAS. The truncated SsITLs were introduced to the pGBKT7 vector and then were co‐transformed with pGADT7‐CAS(Nt) to the yeast Y2H Gold strain. Co‐expression of pGBKT7‐p53 and pGADT7‐SV40 TAg were used as positive controls. –, corresponding empty vectors. The plates were photographed 4 days after inoculation and experiments were repeated three times. (b) The wild‐type plant Arabidopsis thaliana Col‐0 and truncated SsITL transgenic plants were challenged with S. sclerotiorum strains Ep‐1PNA367 (top) or A10 (bottom). Photographs were taken at 36 hours post‐inoculation (hpi). Values are means ± SE. (c) Statistical analysis of leaf lesion area induced by Ep‐1PNA367 or A10 at 36 hpi. Values are means ± SE. (d) Relative expression levels of the salicylic acid (SA) signalling pathway‐related genes (ICS1, PAD4, PBS3, EDS5, and PR1) in each plant were analysed at 12 hpi. The expression levels of GAPDH were used to normalize the expression levels of these genes in different plants. The expression level in the wild‐type plant Col‐0 without inoculation was set to 1. Values are means ± SD. (e) SA concentration in each plant was measured at 12 hpi. The wild‐type plant Col‐0 without inoculation was used as control. In all experiments, three independent replicates were performed. Values are means ± SE. Different letters on the same graph indicate statistical significance (one‐way analysis of variance; post hoc = Duncan α [0.05])
The biological significance of the interaction between SsITL and CAS was further demonstrated by expressing the truncated SsITL proteins SsITL∆SP‐NT1 (84–302 amino acids) and SsITL∆SP‐CT1 (18–249 amino acids) in Arabidopsis lines, 35S:SsITL ∆SP ‐NT1 and 35S:SsITL ∆SP ‐CT1. The truncated genes were expressed as verified with RT‐PCR (Figure S2c,d), and the transgenic lines exhibited no significant difference in plant morphology and growth compared to the wild‐type Col‐0 (Figure S3). The pathogenicity test showed that expression of SsITL ∆SP ‐NT1 or SsITL ∆SP ‐CT1 had no impact on plant resistance against S. sclerotiorum (Figure 5b,c). In addition, the expression levels of ICS1, PAD4, PBS3, EDS5, and PR1 and concentrations of SA in the transgenic plants 35S:SsITL ∆SP ‐NT1 and 35S SsITL ∆SP ‐CT1 on inoculation with Ep‐1PNA367 or A10 were similar to those in the wild‐type Col‐0 (Figure 5d,e). These data indicated that SsITL truncated proteins were unable to interact with CAS and consequently lost the function to increase plant susceptibility. Together, these results strongly imply that SsITL–CAS interaction is essential for the biological function of SsITL during infection.
2.4. SsITL interferes with chitin‐elicited CAS‐associated SA signalling pathway
As a major component of fungal cell wall, chitin oligomers are a typical PAMP, which plays a critical role in the recognition of potential pathogens and the initiation of basic immune responses in plants and animals (Heath, 2000; Nürnberger et al., 2004). Previous studies have suggested that chitin can elicit a series of defence responses such as the SA signalling and mitogen‐activated protein kinase cascade pathways in plants against invading pathogens (Zhang et al., 2002; Jia et al., 2016). To investigate whether chitin elicits the expression of CAS‐associated genes, leaves of the wild‐type Arabidopsis Col‐0 were infiltrated with 50 µg/ml chitin (Sigma‐Aldrich) and the relative expressions of ISC1, PAD4, PBS3, EDS5, and PR1 were analysed by RT‐qPCR at different time points. Our results showed that all of those genes were significantly up‐regulated at 6 and 12 hr post‐infiltration (Figure S6), indicating that CAS‐associated signal can be activated by chitin. To further confirm that SsITL suppresses the CAS‐mediated SA signal pathway, the transgenic plants 35S:SsITL and 35S:AtCAS and cas‐1 knockout plant were infiltrated with 50 µg/ml chitin. The results of RT‐qPCR reveal that induction of those genes by chitin were significantly impaired in cas‐1 and 35S:SsITL plants at 12 hr post‐infiltration (Figure 6). On the contrary, the expressions of ISC1, PBS3, and EDS5 were obviously enhanced in 35S:AtCAS compared to Col‐0 (Figure 6). These results suggest that CAS‐mediated SA signalling can be activated by recognition of chitin; however, the interaction between SsITL and CAS efficiently suppressed this immune response, which is consistent with the previous study that SsITL suppresses host defence at the early stage of infection (Zhu et al., 2013).
Figure 6.
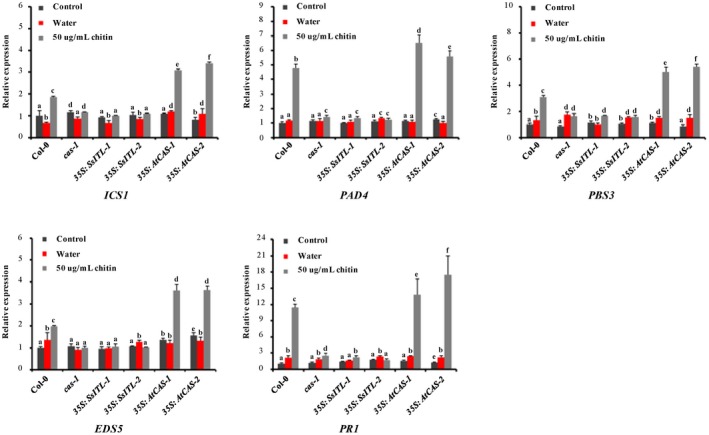
Analysis of the expression levels of five salicylic acid signalling pathway‐related genes after chitin treatment. Relative transcript accumulation of ICS1, PAD4, PBS5, EDS5, and PR1 genes determined by quantitative reverse transcription PCR at 12 hr post‐infiltration with 50 µg/ml chitin. The expression levels of GAPDH were used to normalize the expression levels of these genes in the different plants. The expression level in the wild‐type plant Col‐0 without treatment was set to 1. Three independent replicates were performed. Values are means ± SD. Different letters on the same graph indicate statistical significance (one‐way analysis of variance; post hoc = Duncan α [0.05])
2.5. CAS‐mediated defence response against S. sclerotiorum depends on Ca2+ signalling
CAS is well known as a plant‐specific putative Ca2+‐binding protein that contains low‐affinity/high‐capacity Ca2+ binding sites on the N terminus (Han et al., 2003). Subsequent studies further demonstrated that CAS plays a crucial role in regulating stomatal movement, as well as the generation and fine‐tuning of cytoplasmic Ca2+ (Nomura et al., 2008; Weinl et al., 2008), suggesting that regulation of plant physiological process by CAS is possibly dependent on Ca2+ signals. To illuminate the association between Ca2+ signals and CAS‐mediated resistance against S. sclerotiorum, the virulence test of Ep‐1PNA367 and A10 on 35S:AtCAS transgenic lines and Col‐0 was performed with application of 1 mM LaCl3, which is a putative plasma‐membrane Ca2+ channel blocker and widely used in studying plant Ca2+ signals (Knight et al., 1996; Gao et al., 2013; Choi et al., 2014; Behera et al., 2017). Our results showed that a lesion induced by Ep‐1PNA367 on the wild‐type Col‐0 leaves became larger when 1 mM LaCl3 was applied exogenously. The lesion induced by Ep‐1PNA367 on LaCl3‐pretreated 35S:AtCAS transgenic lines was even larger than the lesion on H2O‐pretreated Col‐0 (Figure 7a). Meanwhile, the lesion induced by A10 on the wild‐type Col‐0 leaves was also larger when plants were pretreated with LaCl3, and CAS‐mediated resistance in 35S:AtCAS transgenic lines to A10 was completely suppressed by exogenous application of LaCl3 (Figure 7b). However, exogenous application of 1 mM LaCl3 had no impact on the growth of S. sclerotiorum (Figure S7). These results suggest that exogenous application of LaCl3 significantly enhances the susceptibility of Arabidopsis to S. sclerotiorum, indicating that Ca2+ signals play a critical role in plant resistance against S. sclerotiorum. On the contrary, CAS‐mediated resistance in 35S:AtCAS transgenic lines to S. sclerotiorum was partially abolished by LaCl3 (Figure 8), suggesting that CAS‐mediated immunity to S. sclerotiorum is mainly dependent on the Ca2+ signalling pathway.
Figure 7.
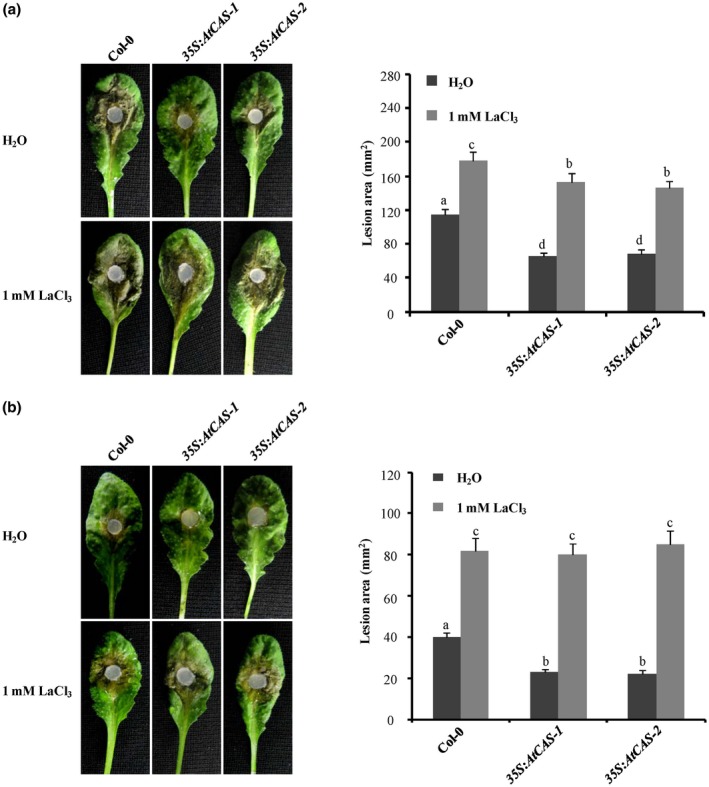
Effect of LaCl3 treatment on plant resistance to Sclerotinia sclerotiorum. (a) Effect of LaCl3 treatment on plant resistance to the wild‐type strain EP‐1PNA367. (b) Effect of LaCl3 treatment on plant resistance to SsITL‐silenced strain A10. Before inoculation, 1 mM LaCl3 solution was uniformly sprayed on the Arabidopsis leaves, and deionized water was used as control. The wild‐type plant Col‐0 and CAS overexpression plants 35S:AtCAS‐1 and 35S:AtCAS‐2 were challenged with S. sclerotiorum strains Ep‐1PNA367 or A10. Photographs were taken at 36 hr post‐inoculation. Statistical analysis of the lesion area induced by Ep‐1PNA367 or A10 on each plant was performed. In all experiments, three independent replicates were performed. Values are means ± SE. Different letters on the same graph indicate statistical significance (one‐way analysis of variance; post hoc = Duncan α [0.05])
Figure 8.
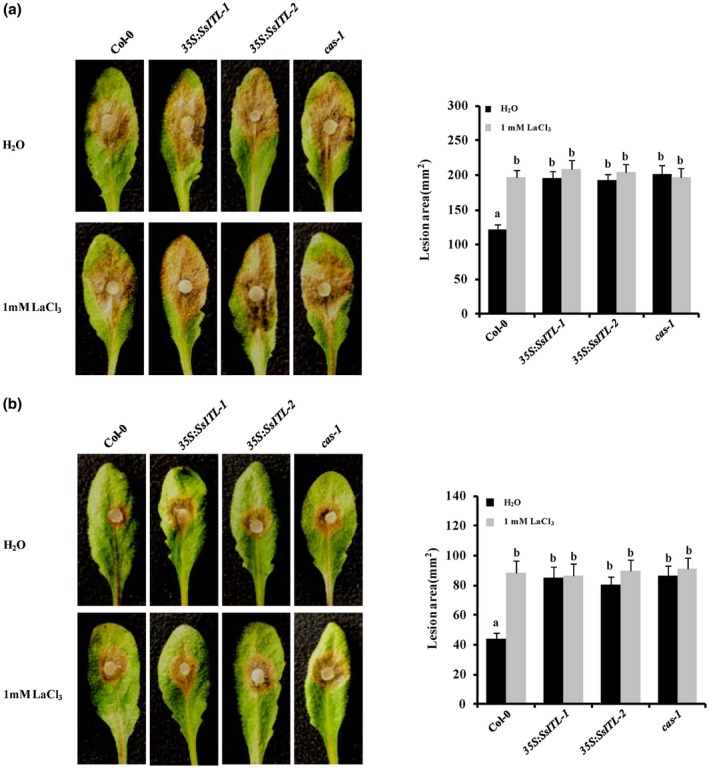
Effect of LaCl3 treatment on 35S:SsITL and cas‐1 mutant plant susceptibility to Sclerotinia sclerotiorum. (a) Effect of LaCl3 treatment on plant susceptibility to the wild‐type strain EP‐1PNA367. (b) Effect of LaCl3 treatment on plant susceptibility to SsITL‐silenced strain A10. Before inoculation, 1 mM LaCl3 solution was uniformly sprayed on the Arabidopsis leaves and deionized water was used as control. The wild‐type plant Col‐0, 35S:SsITL‐1, 35S:SsITL‐2 and cas‐1 mutant plant were challenged with S. sclerotiorum strains Ep‐1PNA367 or A10. Photographs were taken at 36 hr post‐inoculation. Statistical analysis of the lesion area induced by Ep‐1PNA367 or A10 on each plant was performed. In all experiments, three independent replicates were performed. Values are means ± SE. Different letters on the same graph indicate statistical significance (one‐way analysis of variance; post hoc = Duncan α [0.05])
3. DISCUSSION
As an aggressive phytopathogen, S. sclerotiorum possesses abundant powerful weapons, such as CWDEs and OA, which can directly kill host tissues and subsequently establish infection. Recently, the roles of secreted proteins in the pathogenesis of S. sclerotiorum have received increasing attention. Genomic and secretome analysis revealed that S. sclerotiorum encodes approximate 600 secreted proteins, and more than 400 secreted proteins are expressed during infection. Moreover, a large number of effector candidates were predicted by multiple analyses (Amselem et al., 2011; Guyon et al., 2014; Hahn et al., 2014; Derbyshire et al., 2017), suggesting that secreted proteins may play important roles in the pathogenesis of S. sclerotiorum. However, only a few pathogenesis‐related secretory proteins have been identified and functionally characterized so far (Zhu et al., 2013; Xiao et al., 2014; Zhang et al., 2014; Lyu et al., 2016; Yang et al., 2018). We previously reported that the secreted protein SsITL from S. sclerotiorum can inhibit the host immune response in the early stage of infection (Zhu et al., 2013), but the mechanism by which SsITL manipulates plant immunity remains largely unknown. Here, we demonstrated that SsITL interacts with CAS to suppress plant immunity through inhibiting SA signalling pathways, thereby facilitating infection by S. sclerotiorum.
SsITL is an integrin‐like protein and plays very important roles in the virulence of S. sclerotiorum. Our previous study showed that SsITL can enter the host plant cells and inhibit the JA/ethylene (ET)‐mediated signalling pathway at the early stage of infection. The effects of SsITL on the SA signalling pathway were also evaluated by detecting the expression of PR1 and the virulence of wild‐type and SsITL‐silenced strains A10 on Arabidopsis mutant pad4 and NahG, and the results suggest that SsITL might also suppress SA‐mediated resistance (Zhu et al., 2013). CAS is an important functional protein that localizes in the chloroplast thylakoid membrane, and the N terminus of CAS appears to be exposed to the stromal side of the thylakoid membrane (Friso et al., 2004; Nomura et al., 2008). Evidence has emerged that CAS is responsible for both PAMP‐triggered immunity (PTI) and effector‐triggered immunity (ETI) and probably acts upstream of SA accumulation (Nomura et al., 2012). The SA accumulation responds to flg22 (a PAMP from bacterial flagellin) and resistance against Pseudomonas syringae was impaired in the cas‐1 mutant (Nomura et al., 2012). Previous studies have shown that SA signalling is also involved in the plant defence against S. sclerotiorum (Guo and Stotz, 2007; Wang et al., 2012; Novakova et al., 2014; Yang et al., 2018). In this study, we found that SsITL interacts with CAS in the chloroplasts of plant cells. This led us to further investigate the potential roles of CAS in plant defence to the fungal pathogen S. sclerotiorum. We found that overexpression of CAS in wild‐type plant Col‐0 increased the resistance to S. sclerotiorum. At the same time, the SA concentration and the expression level of SA signalling pathway‐related genes (ICS1, PAD4, PBS3, EDS5, and PR1) increased significantly after inoculation with S. sclerotiorum, especially after inoculation with the SsITL‐silenced transformant A10 (Figure 3c). Moreover, the resistance to S. sclerotiorum in cas‐1 mutant was significantly reduced, and the SA accumulation induced by S. sclerotiorum infection was also significantly inhibited in cas‐1 mutant. However, we also noticed that the expression of SA genes maintained a similar level in each plant genotype unless there was pathogen or chitin stimulation, suggesting that CAS is probably involved in the regulation of SA accumulation rather than the biosynthesis itself. Notably, exogenous application of 0.5 mM SA significantly suppressed the infection of S. sclerotiorum while a corresponding concentration of SA had no effect on the growth of S. sclerotiorum (Figure S4), which suggests that it is the SA‐mediated signal as opposed to the compound itself that contributes to plant resistance to S. sclerotiorum. Our results indicate that CAS is a positive regulator of SA biosynthesis and plays important roles in the plant immunity response to S. sclerotiorum.
We found that SsITL co‐localizes with CAS to the chloroplast. The chloroplast has been conventionally viewed as the organelle that conducts photosynthesis, but the chloroplast also plays crucial roles in the plant immune response against multiple invaders (Caplan et al., 2015; de Torres Zabala et al., 2015; Stael et al., 2015; Sugano et al., 2016; Kumar et al., 2018). Corresponding to this, there is growing evidence that pathogen‐delivered effectors can target to chloroplasts and act as virulence factors by manipulating chloroplast functions (Li et al., 2014; de Torres Zabala et al., 2015; Petre et al., 2015). For example, P. syringae virulence effector HopI1, which localizes to chloroplasts, causes chloroplast thylakoid structure remodelling and suppresses SA accumulation (Jelenska et al., 2007). The cysteine protease effector HopN1, which is also secreted by P. syringae, localizes to chloroplasts and suppresses the production of defence‐associated reactive oxygen species by degrading PsbQ (Rodriguez‐Herva et al., 2012). Chloroplasts are the major source for continual production of SA during defence responses (Wildermuth et al., 2001; Asada, 2006; Galvez‐Valdivieso and Mullineaux, 2010). CAS localizes in the chloroplast thylakoid membrane and is a positive regulator of SA accumulation; however, the accumulation of some other plant hormones, such as JA, abscisic acid, indole acetic acid, and cytokinins, was not affected in cas‐1 mutant on flg22 treatment (Nomura et al., 2012). We speculate that SsITL possibly has other targets that are related to the JA/ET signal pathway. Our results show that SsITL interacts with CAS in chloroplasts and inhibits SA accumulation. Combined with previous research (Jelenska et al., 2007; Pecrix et al., 2019; Xu et al., 2019), these findings highlight the chloroplast as a high‐value target potentially attacked by various invaders. Indeed, SsITL transgenic plants are more susceptible to S. sclerotiorum. Correspondingly, the SA accumulation and expression of related genes induced by inoculation in SsITL‐transgenic plants were also significantly lower than those in wild‐type plant Col‐0. Therefore, we speculate that SsITL suppresses plant defence to S. sclerotiorum through interacting with CAS and then affecting the normal biological function of CAS. Consistent with that expectation, the truncated SsITL (SsITL‐NT1 or SsITL‐CT1), which lost the ability to interact with CAS, does not affect plant resistance to S. sclerotiorum. These results further demonstrate a crucial role for the interaction between SsITL and CAS in the pathogenesis of S. sclerotiorum.
CAS was originally considered a primary Ca2+ transducer that was involved in the regulation of extracellular Ca2+‐induced cytosolic Ca2+ oscillation and stomatal closure (Han et al., 2003; Nomura et al., 2008; Wang and Zheng, 2012). Ca2+ signalling is important in the early stages of activation of plant immune responses (Blume et al., 2000; Fromm and Finkler, 2015; Yuan et al., 2017). Previous studies have also shown that the Ca2+ signalling plays an important role in plant immunity to S. sclerotiorum. For example, an endopolygalacturonase (PG) can induce a rapid elevation of cytosolic Ca2+ in plant cells and subsequently programmed cell death (PCD) when S. sclerotiorum infects soybean (Zuppini et al., 2005). The calcium and calmodulin‐dependent protein kinase (SlCCaMK) in Solanum lycopersicum has been demonstrated to function in plant disease resistance against S. sclerotiorum (Wang et al., 2015). We also found that Arabidopsis is more susceptible to S. sclerotiorum after LaCl3 treatment, and CAS‐mediated resistance in 35S:AtCAS transgenic plants was largely blocked by application with LaCl3. These results indicate that the CAS‐mediated resistance response is associated with Ca2+ signalling (Figure 7). A previous study has also shown that CAS‐dependent defence gene expression and SA accumulation are dependent on at least one Ca2+ signalling pathway (Nomura et al., 2012). Furthermore, there is no significant difference between LaCl3‐ and H2O‐treated 35S:SSITL transgenic plants. This result, and the lack of difference between LaCl3‐ and H2O‐treated cas‐1, suggests that the interaction between SsITL and CAS may affect CAS perception and transmission of calcium signalling. This evidence not only supports the idea that calcium signalling plays important roles in plant immunity in resisting S. sclerotiorum infection, but also provides clues for further revealing the molecular mechanism by which SsITL and CAS interact to regulate host resistance. Whether the interaction influences signal transduction by blocking the calcium binding of CAS remains to be investigated.
4. EXPERIMENTAL PROCEDURES
4.1. Fungal strains, plants, and culture conditions
The S. sclerotiorum wild‐type strain Ep‐1PNA367 (Xie et al., 2006) and SsITL‐silenced transformant A10 (Zhu et al., 2013) were used in this study. Fungal strains were cultured on potato dextrose agar (PDA) at 20 °C and stored on PDA slants at 4 °C. S. sclerotiorum transformants were cultured on PDA amended with hygromycin B at 50 µg/ml (Calbiochem). Arabidopsis wild‐type Col‐0 ecotype, cas‐1 knockout plant (SALK_070416 obtained from the Arabidopsis Biological Resource Center), CAS‐overexpressing plants, SsITL transgenic plants (Zhu et al., 2013), SsITL‐NT1 transgenic plants, and SsITL‐CT1 transgenic plants were germinated and grown on one‐half strength Murashige and Skoog (MS) medium containing 0.8% (wt/vol) agar (with or without 50 µg/ml kanamycin) at 22 °C under a 16 hr light (80–100 µmol⋅m−2⋅s−1)/8 hr dark cycles for 10–12 days, then seedlings were transferred to soil and grown in chambers or greenhouse at 20–22 °C under a 14 hr light (140–160 µmol⋅m−2⋅s−1)/10 hr dark cycles with 60%–80% relative humidity. The wild‐type N. benthamiana plants were germinated and grown in the greenhouse at 20 °C under a 14 hr light (140–160 μmol⋅m−2⋅s−1)/10 hr dark cycle with 60%–80% relative humidity.
4.2. Generation of plant expression plasmids
The plasmids pCNF3 and pCNG (Yang et al., 2018) were used for construction of a series of plant expression vectors, and all oligonucleotides and PCR primers used in this study are listed in Table S2. For the IP experiment, the full‐length coding sequence of SsITL (without terminator codon) was amplified from cDNA of S. sclerotiorum with primers SsITL‐SmaI‐F/SsITL∆T‐SmaI‐R, and ligated into the SmaI site of pCNF3 to generate pCNF3‐SsITL construct. To study the subcellular localization of SsITL and CAS in plant cells, the full‐length coding sequence of SsITL (without terminator codon) was amplified from cDNA of S. sclerotiorum with primer pair SsITL‐SmaI‐F/SsITL∆T‐SmaI‐R, and then ligated into SmaI‐digested pCNG to generate pCNG‐SsITL construct. The full‐length coding sequence of CAS (without terminator codon) was amplified from cDNA of Arabidopsis with primer pair CAS‐BamHI‐F/CAS∆T‐SmaI‐R, and then ligated into BamHI/SmaI‐digested pCNF3 to generate pCNF3‐CAS construct. Then, full‐length YFP was amplified with primer pair YFP‐SmaI‐F/YFP‐SmaI‐R, and cloned into SmaI‐digested pCNF3‐CAS to generate pCNY‐CAS construct. Truncated SsITL N‐terminus (84–302 amino acids) and SsITL C‐terminus (1–249 amino acids) were amplified from plasmid pCNF3‐SsITL with primer pair SsITL‐NT1‐BamHI‐F/SsITL‐SmaI‐R and SsITL‐BamHI‐F/SsITL‐CT1‐SmaI‐R, respectively, then ligated into BamHI/SmaI‐digested pCNF3 to generate pCNF3‐SsITL‐NT1 and pCNF3‐SsITL‐CT1 constructs. All the plasmids were confirmed by sequencing analysis.
4.3. Protein extraction, western blot, IP, and LC‐MS/MS assays
For protein extraction, plant tissue was ground in liquid nitrogen and mixed with an equal volume of radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime) with 1 mM phenylmethanesulfonyl fluoride and 1% proteinase inhibitor cocktail (Sigma), then incubated on ice for 30 min and centrifuged at 13,000 × g for 15 min at 4 °C. The supernatant was transferred to a new tube and boiled in sodium dodecyl sulphate (SDS) loading buffer for 5 min. Proteins were separated by SDS‐polyacrylamide gel electrophoresis (PAGE) (12%) followed by electroblotting onto a 0.22 μm polyvinylidene fluoride membrane (Millipore) with a Trans‐Blot SD Semi‐Dry Electrophoretic Transfer Cell (Bio‐Rad). Several monoclonal antibodies, including anti‐FLAG M2 mAb (Sigma‐Aldrich), anti‐HA mAb (Sigma‐Aldrich), anti‐Myc mAb (Cell Signalling Technology), and anti‐GFP mAb (Abmart), were used as primary antibodies, and horseradish peroxidase‐conjugated goat antimouse IgG (H + L) was used as the secondary antibody. The signals on blots were visualized by chemiluminescence using Pierce ECL western blotting substrate (Thermo Scientific) with ChemiDoc XRS + system (Bio‐Rad).
An IP assay was performed for screening of the targets of SsITL in plants. Plasmid pCNF3‐SsITL was transferred into A. tumefaciens GV3101 with electroporation, then the bacteria were cultured, pelleted, and resuspended in infiltration buffer (10 mM 2‐(N‐morpholino) ethanesulphonic acid, 10 mM MgCl2, and 200 mM acetosyringone) for 3–5 hr and the OD600 adjusted to 0.8 before infiltration into N. benthamiana leaves. Total proteins were extracted from N. benthamiana leaves with RIPA lysis buffer (0.5 g leaves/ml) 3 days post‐infiltration. The mixture was vortexed vigorously for 30 s and then incubated on ice for 30 min and centrifuged at 13,000 × g for 15 min at 4 °C. The supernatants were collected and filtered through a 0.22 μm filter. To immunoprecipitate FLAG‐tagged SsITL, 1 ml of supernatant was incubated overnight with 10 μg of anti‐FLAG M2 antibody and 40 μl of protein G plus Agarose (Santa Cruz Biotechnology, Inc.) at 4 °C with gentle shaking. The beads were collected with centrifugation at 1,000 × g for 5 min and then washed five times with RIPA lysis buffer. The co‐immunoprecipitated proteins were eluted from beads by boiling in protein sample buffer for 5 min and separated with SDS‐PAGE, then stained with Coomassie brilliant blue and analysed by western blot with an anti‐FLAG antibody. The gels were then cut into pieces and digested with trypsin to prepare peptides for liquid chromatography‐electrosprayionization tandem mass spectrometry (LC‐ESI‐MS/MS) in a Q Exactive (Thermo Scientific). Identification of proteins was performed by using MASCOT v. 2.3.02.
4.4. Y2H, Co‐IP assay, GST pull‐down, and subcellular localization
The GAL4‐based Matchmaker Gold Yeast Two‐Hybrid System (Clontech,) was applied to screen and verify SsITL candidate targets from IP‐LC‐MS/MS. The coding sequence of SsITL (without signal peptide) was PCR amplified and cloned into the pGBKT7 to generate the bait vector, while the full‐length coding sequence, the N‐terminus (1–187 amino acids) and the C‐terminus (211–387 amino acids) of CAS, were introduced into the pGADT7 to generate the bait vectors (Table S2). The bait and prey plasmids were co‐transformed into yeast strain Y2H Gold according to the manufacturer's instructions. Yeast transformation was performed according to the manufacturer's instructions. Transformed cells were assayed for growth on synthetic dropout (SD)/−Trp −Leu plates for 3–4 days, and single‐colony cells were transferred to 2 ml liquid SD/−Trp −Leu medium for 24 hr. Cells were collected by centrifugation and the concentration was adjusted to 106 cells/ml with sterile water, then 2 μl of yeast suspension was assayed for growth on SD/−Trp −Leu −His −Ade plates containing 5‐bromo‐4‐chloro‐3‐indolyl α‐D‐galactopyranoside (X‐α‐gal). Truncated SsITL mutants (SsITL‐NT2 140–302 amino acids, SsITL‐NT3 196–302 amino acids, SsITL‐NT4 250–302 amino acids, SsITL‐CT2 18–195 amino acids, SsITL‐CT3 18–139 amino acids, SsITL‐CT4 18–83 amino acids) were introduced into pGBKT7 (Table S2) and Y2H assays were performed as described above.
For Co‐IP assay, A. tumefaciens GV3101 harbouring the correct constructs pCNF3‐SsITL‐GFP and pCNF3‐CAS were transiently co‐expressed in N. benthamiana by agroinfiltration. Samples were collected and proteins extracted with RIPA buffer 3 days post‐infiltration. GFP‐tagged SsITL fusions were immunoprecipitated with anti‐GFP antibody, and the eluted proteins were separated by SDS‐PAGE and subjected to immunoblot analysis with anti‐FLAG monoclonal antibody. Approximately 15 μl of RIPA buffer containing the total proteins was loaded as input control.
For GST pull‐down, plasmids pGEX‐6p‐1, pGEX‐6p‐1‐SsITL, and pET28a‐CAS were introduced (separately) into Escherichia coli BL21 (DE3), and the expression of each protein was induced with 0.6 mM isopropyl β‐D‐1‐thiogalactopyranoside at 28 °C for 10 hr. Equal amounts of GST‐SsITL and His‐CAS sonicated lysates were mixed with high‐affinity GST resin (GenScript) and incubated at 4 °C overnight with rotation. The bound proteins were then eluted with fresh 10 mM glutathione elution buffer. Next, the proteins were separated by SDS‐PAGE and immunoblotted with anti‐GST or anti‐His antibody (Proteintech).
To observe the subcellular localization of SsITL and CAS in plant cells, pCNF3‐SsITL‐GFP and pCNF3‐CAS‐YFP constructs were transferred into A. tumefaciens GV3101 by electroporation. Fluorescence in N. benthamiana leaves was observed 2–3 days post‐infiltration using a confocal laser scanning microscope (Olympus FluoView FV1000), with GFP (excitation wavelength of 488 nm, emission wavelength of 495–510 nm) and YFP (excitation wavelength of 514 nm, emission wavelength of 530–560 nm), and chloroplast autofluorescence was detected at 650–707 nm.
4.5. Quantification of endogenous levels of SA
Levels of endogenous SA were analysed by ultra‐fast LC‐electrospray ionization tandem MS with a modification of the method as reported previously (Liu et al., 2012). Briefly, leaf samples were ground two or three times to powder with liquid nitrogen and transferred to 1.5‐ml tubes (three replicates per sample, approximately 0.1 g per replicate). Extraction buffer I (750 μl) (methanol:water:acetic acid, 80:18:2, vol/vol/vol) with internal standard (3 ng/μl naphthaleneacetic acid) was added to the tubes. The samples were shaken at 4 °C, 200 rpm for 16 hr in the dark, and then centrifuged at 4 °C, 13,000 × g for 15 min. The supernatants were transferred to new 2‐ml tubes, and 400 μl of extraction buffer II (methanol:water:acetic acid, 80:19:1, vol/vol/vol) was added to the pellet, samples were shaken at 4 °C, 200 rpm for 4 hr and centrifuged at 13,000 × g for 15 min to collect supernatants. The supernatants were then mixed and filtered with 0.22 μm nylon filters. The filtrates were dried at room temperature with a nitrogen blower, 500 μl of methanol was added to dissolve the precipitate, and the dissolved matter was centrifuged at 13,000 × g for 15 min at 4 °C. The supernatant was diluted 500‐fold with methanol for quantification.
4.6. RNA extraction, cDNA synthesis, and RT‐qPCR
Plant and fungal samples were ground to a powder in liquid nitrogen, and total RNA was extracted using the RNAiso Plus regent (Takara) with a RNase‐free recombinant DNase I (Takara) digestion treatment. The concentration of total RNA was determined with spectrophotometric analysis and 500 ng to 2 µg of total RNA was used to synthesize the first‐strand cDNA using Easy Script One‐Step gDNA Removal and cDNA Synthesis SuperMix (Transgen Biotech). RT‐qPCR assays were performed using the CFX96 Real‐Time PCR Detection System (Bio‐Rad) with iTaq universal SYBR Green supermix (Bio‐Rad). Each sample had three independent replicates, and the RT‐qPCR assay for each gene was performed at least twice. Statistical analyses were performed using IBM SPSS Statistics 19 software and one‐way ANOVA were run with the post hoc style Duncan (α = 0.05).
CONFLICT OF INTEREST
The authors declare that no competing interests exist.
Supporting information
ACKNOWLEDGMENTS
This research was supported by the National Nature Science Foundation of China (31571954), the National Key R & D Program of China (2017YFD0200600), Fundamental Research Funds for the Central Universities (2662017PY010), Open Funds of the State Key Laboratory of Agricultural Microbiology (AMLKF201707), and the earmarked fund of China Agriculture Research System (CARS‐13).
Tang L, Yang G, Ma M, et al. An effector of a necrotrophic fungal pathogen targets the calcium sensing receptor in chloroplasts to inhibit host resistance. Molecular Plant Pathology. 2020;21:686–701. 10.1111/mpp.12922
Funding information
This research was supported by the National Nature Science Foundation of China (31571954), the National Key R & D Program of China (2017YFD0200600), Fundamental Research Funds for the Central Universities (2662017PY010), Open Funds of the State Key Laboratory of Agricultural Microbiology (AMLKF201707), and the earmarked fund of China Agriculture Research System (CARS‐13)
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Amselem, J. , Cuomo, C.A. , van Kan, J.A.L. , Viaud, M. , Benito, E.P. , Couloux, A. et al (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genetics, 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada, K. (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiology, 141, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera, S. , Long, Y. , Schmitz‐Thom, I. , Wang, X.P. , Zhang, C. , Li, H. et al (2017) Two spatially and temporally distinct Ca(2+) signals convey Arabidopsis thaliana responses to K(+) deficiency. New Phytologist, 213, 739–750. [DOI] [PubMed] [Google Scholar]
- Blume, B. , Nürnberger, T. , Nass, N. and Scheel, D. (2000) Receptor‐mediated increase in cytoplasmic free calcium required for activation of pathogen defence in parsley. The Plant Cell, 12, 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland, G.J. and Hall, R. (1994) Index of plant hosts of Sclerotinia sclerotiorum . Canadian Journal of Plant Pathology, 16, 93–108. [Google Scholar]
- Bolton, M.D. , Thomma, B.P. and Nelson, B.D. (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Molecular Plant Pathology, 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Caplan, J.L. , Kumar, A.S. , Park, E. , Padmanabhan, M.S. , Hoban, K. , Modla, S. et al (2015) Chloroplast stromules function during innate immunity. Developmental Cell, 34, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cessna, S.G. , Sears, V.E. , Dickman, M.B. and Low, P.S. (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. The Plant Cell, 12, 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W.G. , Toyota, M. , Kim, S.H. , Hilleary, R. and Gilroy, S. (2014) Salt stress‐induced Ca2+ waves are associated with rapid, long‐distance root‐to‐shoot signaling in plants. Proceedings of the National Academy of Sciences of the United States of America, 111, 6497–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire, M. , Denton‐Giles, M. , Hegedus, D. , Seifbarghy, S. , Rollins, J. , van Kan, J. et al (2017) The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biology and Evolution, 9(3), 593–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oirdi, M. , El Rahman, T.A. , Rigano, L. , El Hadrami, A. , Rodriguez, M.C. , Daayf, F. et al (2011) Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. The Plant Cell, 23, 2405–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellouze, O.E. , Loukil, S. and Marzouki, M.N. (2011) Cloning and molecular characterization of a new fungal xylanase gene from Sclerotinia sclerotiorum S2. BMB Reports, 44, 653–658. [DOI] [PubMed] [Google Scholar]
- Friso, G. , Giacomelli, L. , Ytterberg, A.J. , Peltier, J.B. , Rudella, A. , Sun, Q. et al (2004) In‐depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. The Plant Cell, 16, 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm, H. and Finkler, A. (2015) Repression and de‐repression of gene expression in the plant immune response: the complexity of modulation by Ca2+ and calmodulin. Molecular Plant Pathology, 8, 671–673. [DOI] [PubMed] [Google Scholar]
- Galvez‐Valdivieso, G. and Mullineaux, P.M. (2010) The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiologia Plantarum, 138, 430–439. [DOI] [PubMed] [Google Scholar]
- Gao, X. , Chen, X. , Lin, W. , Chen, S. , Lu, D. , Niu, Y. et al (2013) Bifurcation of Arabidopsis NLR immune signaling via Ca(2)(+)‐dependent protein kinases. PLoS Pathogens, 9, e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defence against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology, 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Guimaraes, R.L. and Stotz, H.U. (2004) Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiology, 136, 3703–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X. and Stotz, H.U. (2007) Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Molecular Plant‐Microbe Interactions, 20, 1384. [DOI] [PubMed] [Google Scholar]
- Guyon, K. , Balagué, C. , Roby, D. and Raffaele, S. (2014) Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum . BMC Genomics, 15, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. , Viaud, M. and van Kan, J. (2014) The Genome of Botrytis cinerea, a ubiquitous broad host range necrotroph In: Dean R.A., Lichens‐Park A. and Kole C. (Eds.) Genomics of Plant‐Associated Fungi and Oomycetes: Dicot Pathogens. Berlin, Germany: Springer, pp. 19–44. [Google Scholar]
- Han, S. , Tang, R. , Anderson, L.K. , Woerner, T.E. and Pei, Z.‐M. (2003) A cell surface receptor mediates extracellular Ca2+ sensing in guard cells. Nature, 425, 196. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000) Nonhost resistance and nonspecific plant defenses. Current Opinion in Plant Biology, 3, 315–319. [DOI] [PubMed] [Google Scholar]
- Heller, A. and Witt‐Geiges, T. (2013) Oxalic acid has an additional, detoxifying function in Sclerotinia sclerotiorum pathogenesis. PLoS ONE, 8, e72292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y.P. , Mao, X.W. , Qu, X.P. , Wang, J.X. , Chen, C.J. and Zhou, M.G. (2018) Molecular and biological characterization of Sclerotinia sclerotiorum resistant to the anilinopyrimidine fungicide cyprodinil. Pesticide Biochemistry and Physiology, 146, 80–89. [DOI] [PubMed] [Google Scholar]
- Issam, S.M. , Mohamed, G. , Dominique, L.M. , Thierry, M. , Farid, L. and Nejib, M. (2004) A β‐glucosidase from Sclerotinia sclerotiorum: biochemical characterization and use in oligosaccharide synthesis. Applied Biochemistry and Biotechnology, 112, 63–78. [DOI] [PubMed] [Google Scholar]
- Jelenska, J. , Yao, N. , Vinatzer, B.A. , Wright, C.M. , Brodsky, J.L. and Greenberg, J.T. (2007) A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Current Biology, 17, 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, X. , Meng, Q. , Zeng, H. , Wang, W. and Yin, H. (2016) Chitosan oligosaccharide induces resistance to tobacco mosaic virus in Arabidopsis via the salicylic acid‐mediated signalling pathway. Scientific Reports, 6, 26144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage, M. , Williams, B. and Dickman, M.B. (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum . PLoS Pathogens, 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage, M. , Yarden, O. and Dickman, M.B. (2015) Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Science, 233, 53–60. [DOI] [PubMed] [Google Scholar]
- Kim, K.S. , Min, J.‐Y. and Dickman, M.B. (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Molecular Plant‐Microbe Interactions, 21, 605–612. [DOI] [PubMed] [Google Scholar]
- Knight, H. , Trewavas, A.J. and Knight, M.R. (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. The Plant Cell, 8, 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A.S. , Park, E. , Nedo, A. , Alqarni, A. , Ren, L. , Hoban, K. et al (2018) Stromule extension along microtubules coordinated with actin‐mediated anchoring guides perinuclear chloroplast movement during innate immunity. eLife, 7, e23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Froehlich, J.E. , Elowsky, C. , Msanne, J. , Ostosh, A.C. , Zhang, C. et al (2014) Distinct Pseudomonas type‐III effectors use a cleavable transit peptide to target chloroplasts. The Plant Journal, 77, 310–321. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Li, X. , Xiao, J. and Wang, S. (2012) A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice–bacterium interaction. Plant Methods, 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu, X. , Shen, C. , Fu, Y. , Xie, J. , Jiang, D. , Li, G. et al (2016) A small secreted virulence‐related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathogens, 12, e1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano, P. , Di Lenna, P. and Magro, P. (1983) Oxalic acid, cell wall‐degrading enzymes and pH in pathogenesis and their significance in the virulence of two Sclerotinia sclerotiorum isolates on sunflower. Physiological Plant Pathology, 22, 339–345. [Google Scholar]
- Mur, L.A. , Kenton, P. , Atzorn, R. , Miersch, O. and Wasternack, C. (2006) The outcomes of concentration‐specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiology, 140, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A.M. , Holcombe, L.J. and Carr, J.P. (2000) Characteristics of salicylic acid‐induced delay in disease caused by a necrotrophic fungal pathogen in tobacco. Physiological and Molecular Plant Pathology, 57, 47–54. [Google Scholar]
- Nürnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunological Reviews, 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Nomura, H. , Komori, T. , Kobori, M. , Nakahira, Y. and Shiina, T. (2008) Evidence for chloroplast control of external Ca2+‐induced cytosolic Ca2+ transients and stomatal closure. The Plant Journal, 53, 988–998. [DOI] [PubMed] [Google Scholar]
- Nomura, H. , Komori, T. , Uemura, S. , Kanda, Y. , Shimotani, K. , Nakai, K. et al (2012) Chloroplast‐mediated activation of plant immune signalling in Arabidopsis . Nature Communications, 3, 926. [DOI] [PubMed] [Google Scholar]
- Nováková, M. , Sasek, V. , Dobrev, P.I. , Valentova, O. and Burketova, L. (2014) Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum‐reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiology and Biochemistry, 80, 308–317. [DOI] [PubMed] [Google Scholar]
- Pecrix, Y. , Buendia, L. , Penouilh‐Suzette, C. , Marechaux, M. , Legrand, L. , Bouchez, O. et al (2019) Sunflower resistance to multiple downy mildew pathotypes revealed by recognition of conserved effectors of the oomycete Plasmopara halstedii . The Plant Journal, 97, 730–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre, B. , Saunders, D.G. , Sklenar, J. , Lorrain, C. , Win, J. , Duplessis, S. and et al (2015) Candidate effector proteins of the rust pathogen Melampsora larici‐populina target diverse plant cell compartments. Molecular Plant‐Microbe Interactions, 28, 689–700. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Does, D.V.D. , Zamioudis, C. , Leonreyes, A. and Wees, S.C.M.V. (2012) Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology, 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Riou, C. , Freyssinet, G. and Fevre, M. (1991) Production of cell wall‐degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum . Applied and Environmental Microbiology, 57, 1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annual Review of Phytopathology, 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Herva, J.J. , González‐Melendi, P. , Cuartas‐Lanza, R. , Antúnez‐Lamas, M. , Río‐Alvarez, I. , Li, Z. et al (2012) A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cellular Microbiology, 14, 669–681. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. , Johnson, J.S. and Dong, X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proceedings of the National Academy of Sciences of the United States of America, 104, 18842–18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stael, S. , Kmiecik, P. , Willems, P. , Van Der Kelen, K. , Coll, N.S. , Teige, M. et al (2015) Plant innate immunity—sunny side up? Trends in Plant Science, 20, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano, S. , Hayashi, N. , Kawagoe, Y. , Mochizuki, S. , Inoue, H. , Mori, M. et al (2016) Rice OsVAMP714, a membrane‐trafficking protein localized to the chloroplast and vacuolar membrane, is involved in resistance to rice blast disease. Plant Molecular Biology, 91, 81–95. [DOI] [PubMed] [Google Scholar]
- de Torres Zabala, M. , Littlejohn, G. , Jayaraman, S. , Studholme, D. , Bailey, T. , Lawson, T. et al (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nature Plants, 1, 15074. [DOI] [PubMed] [Google Scholar]
- Wang, J.P. , Munyampundu, J.P. , Xu, Y.P. and Cai, X.Z. (2015) Phylogeny of plant calcium and calmodulin‐dependent protein kinases (CCaMKs) and functional analyses of tomato CCaMK in disease resistance. Frontiers in Plant Science, 6, 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.H. and Zheng, H.L. (2012) Mechanisms for calcium sensing receptor‐regulated stomatal closure in response to the extracellular calcium signal. Plant Signaling & Behavior, 7, 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Tan, X. , Zhang, Z. , Gu, S. , Li, G. and Shi, H. (2012) Defense to Sclerotinia sclerotiorum in oilseed rape is associated with the sequential activations of salicylic acid signaling and jasmonic acid signaling. Plant Science, 184, 75–82. [DOI] [PubMed] [Google Scholar]
- Weinl, S. , Held, K. , Schlucking, K. , Steinhorst, L. , Kuhlgert, S. , Hippler, M. et al (2008) A plastid protein crucial for Ca2+‐regulated stomatal responses. New Phytologist, 179, 675–686. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562. [DOI] [PubMed] [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.J. , Britt, R. and Dickman, M.B. (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathogens, 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X. , Xie, J. , Cheng, J. , Li, G. , Yi, X. , Jiang, D. et al (2014) Novel secretory protein Ss‐Caf1 of the plant‐pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Molecular Plant‐Microbe Interactions, 27, 40–55. [DOI] [PubMed] [Google Scholar]
- Xie, J. , Wei, D. , Jiang, D. , Fu, Y. , Li, G. , Ghabrial, S. et al (2006) Characterization of debilitation‐associated mycovirus infecting the plant‐pathogenic fungus Sclerotinia sclerotiorum . Journal of General Virology, 87, 241–249. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Xiang, M. , White, D. and Chen, W. (2015) pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate‐minus mutants of Sclerotinia sclerotiorum . Environmental Microbiology, 17, 2896–2909. [DOI] [PubMed] [Google Scholar]
- Xu, Q. , Tang, C. , Wang, X. , Sun, S. , Zhao, J. , Kang, Z. et al (2019) An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nature Communications, 10, 5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, G. , Tang, L. , Gong, Y. , Xie, J. , Fu, Y. , Jiang, D. et al (2018) A cerato‐platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum . New Phytologist, 217, 739–755. [DOI] [PubMed] [Google Scholar]
- Yuan, P. , Jauregui, E. , Du, L. , Tanaka, K. and Poovaiah, B.W. (2017) Calcium signatures and signaling events orchestrate plant–microbe interactions. Current Opinion in Plant Biology, 38, 173–183. [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Ramonell, K. , Somerville, S. and Stacey, G. (2002) Characterization of early, chitin‐induced gene expression in Arabidopsis . Molecular Plant‐Microbe Interactions, 15, 963–970. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Wu, Q. , Cao, S. , Zhao, T. , Chen, L. , Zhuang, P. et al (2014) A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Molecular Biology, 86, 495–511. [DOI] [PubMed] [Google Scholar]
- Zhou, F. , Zhang, X.‐L. , Li, J.‐L. and Zhu, F.‐X. (2014) Dimethachlon resistance in Sclerotinia sclerotiorum in China. Plant Disease, 98, 1221–1226. [DOI] [PubMed] [Google Scholar]
- Zhu, W. , Wei, W. , Fu, Y. , Cheng, J. , Xie, J. , Li, G. et al (2013) A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS ONE, 8, e53901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuppini, A. , Navazio, L. , Sella, L. , Castiglioni, C. , Favaron, F. and Mariani, P. (2005) An endopolygalacturonase from Sclerotinia sclerotiorum induces calcium‐mediated signaling and programmed cell death in soybean cells. Molecular Plant‐Microbe Interactions, 18, 849–855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


