Abstract
Evidence suggests that fasting exerts extensive antitumor effects in various cancers, including colorectal cancer (CRC). However, the mechanism behind this response is unclear. We investigate the effect of fasting on glucose metabolism and malignancy in CRC. We find that fasting upregulates the expression of a cholesterogenic gene, Farnesyl-Diphosphate Farnesyltransferase 1 (FDFT1), during the inhibition of CRC cell aerobic glycolysis and proliferation. In addition, the downregulation of FDFT1 is correlated with malignant progression and poor prognosis in CRC. Moreover, FDFT1 acts as a critical tumor suppressor in CRC. Mechanistically, FDFT1 performs its tumor-inhibitory function by negatively regulating AKT/mTOR/HIF1α signaling. Furthermore, mTOR inhibitor can synergize with fasting in inhibiting the proliferation of CRC. These results indicate that FDFT1 is a key downstream target of the fasting response and may be involved in CRC cell glucose metabolism. Our results suggest therapeutic implications in CRC and potential crosstalk between a cholesterogenic gene and glycolysis.
Subject terms: Cancer metabolism, Colorectal cancer
The molecular mechanisms underpinning how fasting inhibits tumourigenesis are not completely elucidated. Here, the authors show that fasting upregulates the cholesterogenic gene FDFT1 which leads to decreased AKT/mTOR/HIF1a signalling and glycolysis reduction in colorectal cancer.
Introduction
Colorectal cancer (CRC) is one of the deadliest diseases globally, ranking third in cancer morbidity and second in cancer mortality worldwide1. The incidence has stabilized or is declining in the US and some other developed countries, but the incidence and mortality are increasing in some developing countries, especially China and Spain2,3. Although great progress has been achieved in surgical techniques and treatment for CRC, the 5-year relative survival rate of CRC patients has not changed significantly in the past decades4,5. Therefore, there is an urgent need to better understand the molecular mechanisms that govern the oncogenesis and progression of CRC.
Fasting, defined as consuming no or minimal amounts of food, usually from 12 h to 3 weeks, is known for extending the lifespan in numerous experimental organisms6–8. Many prospective clinical trials have shown that fasting can reduce risk factors for aging-related diseases, including cardiovascular disease, diabetes, and cancer9–11. Fasting can also increase resistance to various oxidative stresses, such as acute surgical stress12–14. On the molecular level, fasting is not well understood but based on the data from studies on fasting-mediated longevity and stress resistance, it is thought to work at least in part through the inhibition of insulin/IGF-1/mTORC1 signaling15–18. Although fasting exerts extensive antitumor effects in numerous contexts, the impact of fasting on metabolic changes in CRC remains poorly studied.
Aberrant metabolism has been considered a hallmark of cancer cells, and this important research field has recently attracted interest19,20. Unlike normal cells, which derive most of their energy from mitochondrial oxidative phosphorylation, cancer cells rely on aerobic glycolysis as their primary energy resource. This process is recognized as the “Warburg effect”21–23. AKT/mTOR/HIF1α signaling has been suggested to play critical roles in promoting glycolysis and lactate production and thus in the “metabolic reprogramming” of cancer cells24–28. However, fasting could reprogram metabolic derangements to inhibit cancer growth8,29–31. Therefore, an understanding of the effects of fasting on metabolic alterations in CRC could lead to better therapeutic approaches.
Farnesyl-diphosphate farnesyltransferase 1 (FDFT1) encodes a membrane-associated enzyme acting at a branch point in the mevalonate pathway. The encoded protein is the first enzyme in cholesterol biosynthesis and catalyzes the dimerization of two molecules of farnesyl diphosphate via a two-step reaction to form squalene32, which plays an important role in cholesterol biosynthesis33,34. Although increased FDFT1 transcription is associated with increased invasion in prostate cancer, the exact role of FDFT1 in CRC progression has not been investigated35. However, our results indicated that fasting upregulated the expression of FDFT1 during the inhibition of CRC cell glucose metabolism and proliferation. Clinically, high FDFT1 expression in CRC is associated with better prognosis in The Cancer Genome Atlas (TCGA) data sets. This finding prompted us to speculate that FDFT1 may play a negative regulatory role in glucose metabolism, which is a critical aspect in the fasting-mediated suppression of CRC oncogenesis and progression.
In this study, we provide ample evidence that fasting negatively regulates glucose metabolism and proliferation via the FDFT1/AKT-mTOR-HIF1α axis in CRC. Overall, our results indicate that FDFT1 is a key downstream target of the fasting response and involve in CRC cell glucose metabolism. More broadly, our present study also suggests potential therapeutic implications (involving fasting and mTOR) for CRC and implies potential crosstalk between a cholesterogenic gene and glycolysis.
Results
Fasting impairs glycolysis and inhibits proliferation in CRC
To explore the effect of fasting on the proliferation of CRC cells, we examined cell proliferation by using a Cell Counting Kit-8 (CCK8) and EdU assays. Fasting significantly inhibited CRC cell proliferation in the CCK8 assay (Fig. 1a; Supplementary Fig. 2a). In the EdU immunofluorescence staining assay, the fasting group contained a lower relative fold fraction of EdU-positive cells than the control group (Fig. 1b, d; Supplementary Fig. 2b, d). To further investigate the effect of fasting on cell invasion, a Transwell assay was performed. Fasting markedly inhibited CT26 cell invasion (Supplementary Fig. 1a, b). Moreover, to explore the role of fasting in the cell cycle and apoptosis distribution, flow cytometry was performed. Our results demonstrated that fasting-induced cell cycle arrest in the G0/G1 and G2/M phases and induced apoptosis (Supplementary Figs. 1c, d, 2c, e; Supplementary Figs. 28, 29), which probably explains why fasting inhibits the proliferation of CRC cells.
Fig. 1. Fasting impairs glycolysis and proliferation of CT26 cells in vitro and in vivo.
Fasting inhibited CT26 cell proliferation as measured by a CCK8 assay (from left to right: P = 0.0009; P = 0.0006). b Cell proliferation was also evaluated using EdU immunofluorescence staining. Proliferating cells were labeled with EdU. n = 3; scale bar: 100 µm. c Bioinformatics analysis of differentially expressed genes identified via iTRAQ proteomics. We magnified the genes related to the glycolysis pathway particularly. d The graph shows the relative fold fraction of EdU-positive cells. e Fasting reduced glucose uptake in CT26 cells. f Fasting decreased lactate production via glycolysis in CT26 cells. g Fasting downregulated the expression of rate-limiting glycolytic enzymes in glucose metabolism (GLUT1, HK2, LDHA, PGK1, and GPI) by western blot. h ECAR, an indicator of glycolysis, was reduced in CT26 cell cultured in the fasting mimic medium. i OCR, which reflects mitochondrial respiration, was increased in CT26 cell cultured in the fasting mimic medium. j CT26 cells were injected into BALB/c mice. When the tumors were palpable, the mice were randomly assigned to the control group or the fasting mimic diet (FMD) group. Photograph of dissected tumors (upper: FMD group; lower: control group; n = 5). k Fasting downregulated the transcription of rate-limiting glycolytic enzymes in glucose metabolism (GLUT1, HK2, LDHA, PGK1, and GPI) by qRT-PCR. l The tumor volumes were measured every day after the 9th day. The FMD attenuated tumor growth in mice (n = 5). m, o Tumor weights and tumor volumes on the 25th day (n = 5; P = 0.001; P = 0.0292). n Representative 18F-FDG microPET/CT imaging of tumor-bearing mice (n = 3) (upper: control group; lower: FMD group). The tumors are indicated with arrows. p The ratio of the tumor SUVmax in the control group and the FMD group (n = 3; P = 0.0459). Error bars, mean ± SD, the data are from three independent experiments. Two-sided t tests. *P < 0.05, **P < 0.01, ***P < 0.001, compared with the control group.
To further assess the effect of fasting on the proliferation of CRC, an 8-plex iTRAQ proteomic technique and bioinformatics analysis were performed (Fig. 1c; Supplementary Figs. 3a, 4a, b). The heatmap showed 111 differentially upregulated genes and 168 differentially downregulated genes between the control and fasting groups. Gene Ontology analysis of the differentially expressed genes (DEGs) was carried out (Supplementary Fig. 5a–c). Pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) that were enriched in DEGs indicated that the “Glycolysis/gluconeogenesis” pathway was highly downregulated (Supplementary Fig. 6a, b; Fig. 1c). To validate the impact of fasting on glucose metabolism, we examined glucose uptake and lactate production, two primary indicators of the Warburg effect. As expected, fasting reduced glucose uptake and lactate production in cells (Fig. 1e, f; Supplementary Fig. 2g, h). The extracellular acidification rate (ECAR), which is another indicator of glycolysis, was reduced in cells cultured in the fasting mimic medium (Fig. 1h; Supplementary Fig. 2j). The oxygen consumption rate (OCR), which reflects mitochondrial respiration, was increased in in the fasting mimic medium (Fig. 1i; Supplementary Fig. 2k). Aerobic glycolysis was accompanied by the activation of a series of glycolytic genes. Therefore, several key rate-limiting enzymes in glucose metabolism were examined. The transcription and expression of rate-limiting enzymes in glucose metabolism (GLUT1, HK2, LDHA, PGK1, and GPI) were downregulated in CT26 and SW620 cells cultured in the fasting mimic medium (Fig. 1g, k; Supplementary Fig. 2f, i). Our data indicated that fasting plays a vital role in inhibiting glycolysis in CRC cells.
To further verify the effect of fasting on glucose metabolism observed in vitro, we subcutaneously injected CT26 cells into BALB/c mice. When the tumors were palpable, the mice were randomly assigned to a control or the fasting mimic diet (FMD) group. The FMD appreciably attenuated tumor growth in the mice (Fig. 1j, l). The final tumor weights and volumes in the fasting group were markedly lower than those in the control group (Fig. 1m, o). Furthermore, we used an 18F-fluorodeoxyglucose (18F-FDG) microPET/CT imaging system to assess the role of fasting in glucose metabolism. Consistent with the results in vitro, fasting dramatically inhibited 18F-FDG uptake in the in vivo xenograft model (Fig. 1n, p). Weight profiles during the normal diet and FMD cycle are shown in Supplementary Fig. 7. Taken together, these results suggested that fasting impairs glycolysis and inhibits proliferation in CRC.
FDFT1 is upregulated by fasting and correlates with prognosis in CRC
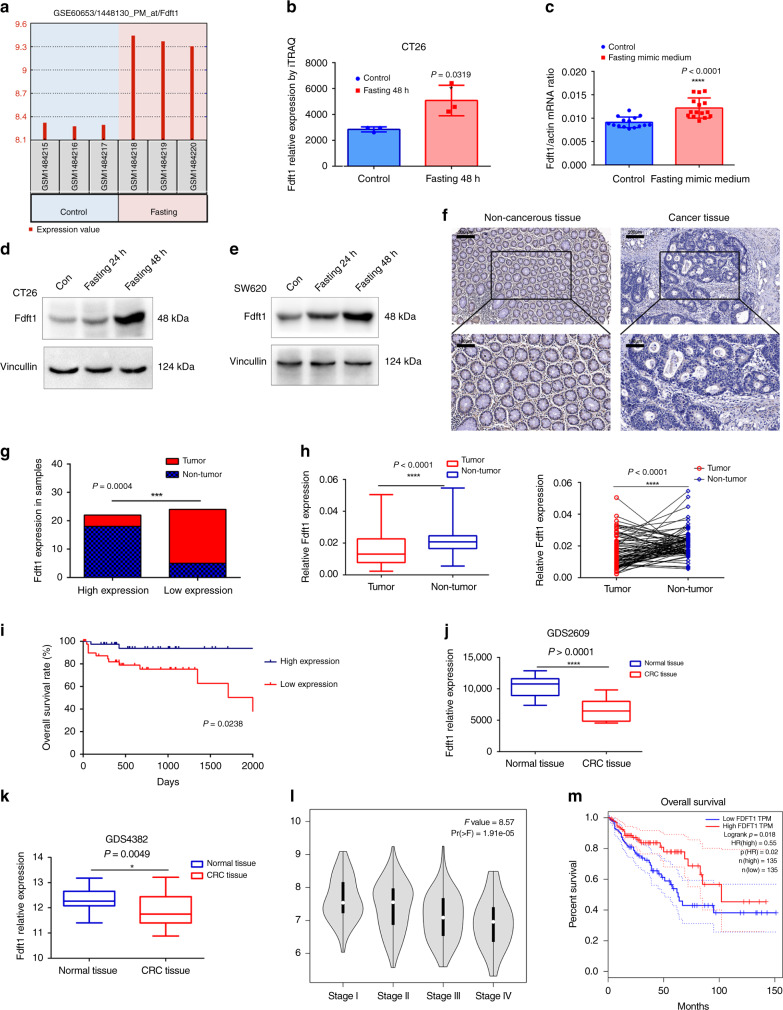
To further explore the effect of fasting on the proliferation of CRC cells, the GSE60653 data set28 (from a study on fasting-induced anti-Warburg effects in CRC) was analyzed to identify DEGs between the control and fasting groups (Supplementary Figs. 8a, b and 9a, b). Gene Ontology and KEGG pathway analyses for the DEGs were performed using FunRich software (http://www.funrich.org/). Surprisingly, the most enriched biological pathway and biological process were the “Cholesterol biosynthesis” pathway and the “Energy pathway” and “Metabolism” processes (Supplementary Fig. 9c–f). Via the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/), the top significantly enriched biological process and KEGG pathway were the “Cholesterol biosynthetic process” and the “Steroid biosynthesis pathway”, respectively (Supplementary Fig. 10a, b). FDFT1 acts at the beginning of the “Steroid biosynthesis” pathway. Therefore, we chose the FDFT1 as our hub gene for further research. First, we validated that fasting can upregulate FDFT1 expression. In the GSE60653 data set, the expression of FDFT1 was increased significantly in the fasting group compared with that in the control group (Fig. 2a). Furthermore, in the iTRAQ proteomics analysis, the relative expression of FDFT1 was greatly elevated in the fasting group compared with that in the control group (Fig. 2b). In addition, the mRNA expression of FDFT1 in dissected tumor samples from the fasting mimic group and the control group was measured by qRT-PCR. The mRNA expression of FDFT1 was markedly increased in the fasting group (Fig. 2c), and western blotting indicated that fasting mimic medium increased the protein level of FDFT1 in cells (Fig. 2d, e). Our results thus showed that fasting upregulates the expression of FDFT1 in CRC.
Fig. 2. Fasting upregulates the level of FDFT1, which is correlated with prognosis in CRC.
a The expression of FDFT1 was increased significantly in the fasting group compared with that in the control group in the GSE60653 data set (n = 3). b The relative expression of FDFT1 was also increased greatly in the fasting group compared with that in the control group by iTRAQ (n = 3; P = 0.0319). c The mRNA expression of FDFT1 in dissected tumor tissue from the fasting mimic group and the control group was measured by qRT-PCR (n = 15; P < 0.0001). d, e Fasting mimic medium also increased the protein level of FDFT1 in CT26 and SW620 cells. f Representative graph of the IHC analysis carried out in human CRC and noncancerous tissues (n = 23; upper: scale bar is 200 µm; lower: scale bar is 100 µm). g The expression of FDFT1 was downregulated in most of the tumor tissues (19/23), but was upregulated in most of the adjacent noncancerous tissues (18/23) (n = 23; P = 0.0004). h The relative expression levels of FDFT1 mRNA in CRC tissues and matched adjacent noncancerous tissues were determined by qRT-PCR (n = 81; both P < 0.0001). i Kaplan–Meier analysis of the overall survival of patients with CRC in the FUSCC cohort according to FDFT1 expression. The median expression level was used as the cutoff. High FDFT1 expression predicted better prognoses for CRC patients in the FUSCC cohort. (high FDFT1 patients = 39, low FDFT1 patients = 42; P = 0.0238, log-rank test) j, k The expression of the FDFT1 gene was significantly lower in CRC tissues than in normal tissues in the GDS2609 and GDS4382 data sets (P < 0.0001; P = 0.0049). l Analysis of the correlation of FDFT1 expression with TNM stage in CRC patients. Lower FDFT1 expression was correlated with higher TNM stage (P = 1.91 × 10−5). m Survival analysis of FDFT1 data from the TCGA database stratified by FDFT1 expression. High FDFT1 expression indicated a better prognosis. (P = 0.018, log-rank test). Error bars, mean ± SD, the data are from three independent experiments. Two-sided t tests. Box denotes 25th to 75th percentile, horizontal bar is median in h, j, and k. Kaplan–Meier analysis and log-rank tests were used in panels i, m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, compared with the control group (or non-tumor/normal tissue).
The expression of FDFT1 was compared in 23 human CRC tissues and matched adjacent noncancerous tissues, by immunohistochemical (IHC) staining and qRT-PCR. The expression of FDFT1 was downregulated and lower in most of the tumor tissues (19/23), but upregulated in most of the adjacent noncancerous tissues (18/23) (Fig. 2f, g). The relative expression levels of FDFT1 mRNA was also assessed in 81 CRC tissues and matched adjacent noncancerous tissues. The levels of FDFT1 in CRC tissues were dramatically lower than those in noncancerous tissues (Fig. 2h). We further examined FDFT1 expression in CRC cell lines (DLD1, HCT116, SW620, SW480, and CT26) and the colorectal mucosal epithelial cell line NCM460 by qRT-PCR. As expected, the expression of FDFT1 was also appreciably decreased in the CRC cell lines compared with NCM460 cells (Supplementary Fig. 11a), consistent with the results in Cancer Cell Line Encyclopedia (CCLE) database (Supplementary Fig. 11b, c). Therefore, FDFT1 is downregulated in CRC tissues and cell lines.
To further determine the role of FDFT1 in CRC, we assessed the association between FDFT1 expression and clinicopathological features in patients with CRC. The median expression level was used as the cutoff. The low expression of FDFT1 was significantly associated with tumor size, histological type, lymph node metastasis, tumor differentiation, invasion, distant metastasis, and clinical stage (Table 1). However, no significant association was found between FDFT1 expression and age, gender, tumor location, or carcinoembryonic antigen (CEA) level. These findings indicated that FDFT1 downregulation is involved in the malignant progression of CRC. To further evaluate the relationship between FDFT1 expression and the survival time of CRC patients, a Kaplan–Meier analysis was performed. Our data showed that high FDFT1 expression predicts better prognoses for patients with CRC in the Fudan University Shanghai Cancer Center (FUSCC) cohort (P = 0.0238, log-rank test) (Fig. 2i).
Table 1.
The relationship between Fdft1 expression and clinicopathological features in 81 CRC patients from FUSCC.
| Fdft1 | |||||
|---|---|---|---|---|---|
| Variable | Number | Low expression | High expression | X2 test | |
| P-value | |||||
| Age (years) | <60 | 51 | 27 | 24 | 0.798 |
| ≥60 | 30 | 15 | 15 | ||
| Gender | Female | 60 | 33 | 27 | 0.338 |
| Male | 21 | 9 | 12 | ||
| Tumor location | Colon | 30 | 12 | 18 | 0.102 |
| Rectum | 51 | 30 | 21 | ||
| Size | <5 cm | 42 | 15 | 27 | 0.003* |
| ≥5 | 39 | 27 | 12 | ||
| Histological type | Adenocarcinoma | 66 | 30 | 36 | 0.016* |
| Mucinous adenocarcinoma | 15 | 12 | 3 | ||
| Lymph node metastasis | No | 42 | 12 | 30 | 0.000*** |
| Yes | 39 | 30 | 9 | ||
| Tumor differentiation | Well, moderate | 60 | 27 | 33 | 0.037* |
| Poor and others | 21 | 15 | 6 | ||
| Invasion | T1–2 | 21 | 6 | 15 | 0.013* |
| T3–4 | 60 | 36 | 24 | ||
| Distant metastasis | No | 75 | 36 | 39 | 0.043* |
| Yes | 6 | 6 | 0 | ||
| Clinical stage | I–II | 30 | 9 | 21 | 0.003** |
| III–IV | 51 | 33 | 18 | ||
| CEA | < 5 ng/ml | 54 | 24 | 30 | 0.059 |
| ≥5 ng/ml | 27 | 18 | 9 | ||
Pearson’s X2 tests were used. The results were considered statistically significant at P < 0.05. *P < 0.05, **P < 0.01, ***P < 0.001.
Moreover, to further validate the role of FDFT1 in CRC, we used GEO and Gene Expression Profiling Interactive Analysis (GEPIA) databases to analyze TCGA data sets. The expression of the FDFT1 gene was significantly lower in CRC tissues than in normal tissues in the GDS2609 and GDS4382 data sets (Fig. 2j, k). Furthermore, the GEPIA database (http://gepia.cancer-pku.cn/) indicated lower FDFT1 correlated with higher TNM stage (P = 1.91 × 10−5) in CRC patients (Fig. 2l). The survival analysis of the data from the TCGA data set indicated that high FDFT1 expression was associated with better prognosis (P = 0.018, log-rank test) (Fig. 2m). These results were consistent with our results in the FUSCC cohort. Altogether, these results indicated that FDFT1 was downregulated in CRC tissues, and was correlated with malignant progression and poor prognosis in patients with CRC.
Subsequently, to gain a deeper understanding of its mechanism, we studied the role of sterol regulatory element-binding protein-2 (SREBP2), a key transcriptional regulator of metabolic genes. The results showed that fasting simulation increased the protein and mRNA levels of SREBP2 in CRC cells (Supplementary Fig. 12a–c). To assess the biological function of SREBP2 in CRC, we used lentiviral-mediated SREBP2 overexpression and knockdown in CT26 cells (Supplementary Fig. 12d, e). We also performed a colony-formation assay when overexpressing or knocking down SREBP2, and the results showed that SREBP2 overexpression decreased, whereas SREBP2 knockdown increased colony-formation capacity (Supplementary Fig. 12f, g). Then, we examined the protein and mRNA levels of FDFT1 when overexpressing or knocking down SREBP2, and found that overexpression of SREBP2 upregulated FDFT1 and vice versa, moreover, the overexpression of SREBP2 was more pronounced during fasting simulation (Supplementary Fig. 12h, i). Subsequently, we used a cloning assay to detect whether SREBP2 reverses cell proliferation caused by knockdown of FDFT1 in CT26 Supplementary Fig. 12j, k). In conclusion, these results indicated that FDFT1 played a role in reducing the proliferation of CRC cells under the action of SREBP2.
FDFT1 negatively regulates the proliferation of CRC cells
To assess the biological function of FDFT1 in CRC, lentivirus-mediated overexpression and knockdown of FDFT1 were performed in CT26 and SW620 cells. Images of GFP expression in CRC cells after lentiviral infection were acquired with a fluorescence microscope (Supplementary Figs. 13a–16a). The efficiency of FDFT1 overexpression (Fig. 3a, b; Supplementary Fig. 15b, c) and knockdown (Fig. 3c, d; Supplementary Fig. 16c, d) were effectively achieved in CT26 and SW620 cells, as indicated by the q-PCR and western blotting results.
Fig. 3. FDFT1 negatively regulates the proliferation of CRC cells.
a, b The efficiency of FDFT1 overexpression in CT26 cells was measured by qRT-PCR and western blotting (P < 0.0001). c, d The efficiency of FDFT1 knockdown in CT26 cells was measured by qRT-PCR and western blotting (both P < 0.0001). e FDFT1 overexpression inhibited CT26 cell proliferation as measured by a CCK8 assay (P = 0.0006). f FDFT1 knockdown increased CT26 cell proliferation as measured by a CCK8 assay (sh1 vs con: P = 0.0008; sh3 vs con: P = 0.0009). g Cell proliferation in control and FDFT1-overexpressing CT26 cells was also evaluated using EdU immunofluorescence staining. Scale bar: 100 µm. h The graph shows the relative fold fraction of EdU-positive cells. i Cell proliferation was also evaluated in control and FDFT1 knockdown CT26 cells using EdU immunofluorescence staining. Scale bar: 100 µm. j The graph shows the relative fold fraction of EdU-positive cells. k FDFT1 overexpression decreased the colony-forming capacity of CT26 cells as measured by a colony-formation assay. l The graph shows the statistical results of the colonies. m FDFT1 knockdown increased the colony-forming capacity of CT26 cells as measured by a colony-formation assay. n The graph shows the statistical results of the colonies (from left to right: P = 0.0003; P = 0.0006). o FDFT1 overexpression inhibited CT26 cell invasion as measured by a Transwell assay. Scale bar: 100 µm. p The graph shows the number of invaded cells. q FDFT1 knockdown increased CT26 cell invasion as measured by a Transwell assay. Scale bar: 100 µm. r The graph shows the number of invaded cells. Error bars, mean ± SD, the data are from three independent experiments. Two-sided t tests. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, compared with the control group.
To further investigate the effect of FDFT1 on cell proliferation, CCK8, EdU, and colony-formation assays were conducted. FDFT1 overexpression inhibited CRC cell proliferation (Fig. 3e; Supplementary Fig. 15d), whereas FDFT1 knockdown promoted it (Fig. 3f; Supplementary Fig. 16b). In EdU immunofluorescence staining assay, FDFT1 overexpression resulted in a lower relative fold fraction of EdU-positive cells (Fig. 3g, h; Supplementary Fig. 15g, h), yet FDFT1 knockdown resulted in a greater relative fold fraction of EdU-positive cells (Fig. 3i, j; Supplementary Fig. 16f, g). In addition, FDFT1 overexpression decreased the colony-forming capacity of CRC cells (Fig. 3k, l; Supplementary Fig. 15e, f), whereas FDFT1 knockdown enhanced it (Fig. 3m, n; Supplementary Fig. 16e, h). To further investigate the effect of FDFT1 on cell invasion, a Transwell assay was performed. FDFT1 overexpression inhibited cell invasion (Fig. 3o, p), whereas FDFT1 knockdown increased it (Fig. 3q, r). In addition, FDFT1 overexpression induced G0/G1 phase cell cycle arrest and apoptosis (Supplementary Figs. 13b, c and 15i, j), whereas FDFT1 knockdown induced S phase cell cycle arrest and attenuated apoptosis (Supplementary Figs. 14b, c and 16i, j). In summary, FDFT1 overexpression inhibited cell proliferation and induced G0/G1 phase cell cycle arrest and apoptosis, while FDFT1 knockdown promoted cell proliferation, induced S phase cell cycle arrest, and attenuated apoptosis in CRC cells.
We also extend our experiments to PIK3CA-Mut cell line (HCT116 and HT29) and the human colorectal mucosal epithelial cell line NCM460 to see whether fasting or FDFT1 have effects on cell proliferation. The results showed that the effect of fasting and FDFT1 on PIK3CA-Mut cell lines (HCT116 and HT29) proliferation were similar to that in the previous PIK3CA-WT cell lines (CT26 and SW620) (Supplementary Fig. 17a–p), but FDFT1 had no effect on NCM460 proliferation (Supplementary Fig. 18a–d).
FDFT1 is an important downstream target of fasting in CRC
We further evaluated whether the inhibitory effect of fasting on CRC increases when combined with FDFT1 overexpression. Our data demonstrated that compared with either treatment alone, FDFT1 overexpression combined with fasting had the greatest inhibitory effect on CRC cell proliferation (Fig. 4a; Supplementary Fig. 19c). Moreover, an EdU assay showed that FDFT1 overexpression combined with fasting yielded the most marked reduction in the proliferative capacity of CRC cells (Supplementary Fig. 19a, b, d, e). Furthermore, we studied FDFT1 protein levels in four groups in vitro. Western blotting indicated that fasting and FDFT1 overexpression increased the protein level of FDFT1 in CRC cells. Fasting exerted an additive effect on FDFT1 expression level in cells overexpressing FDFT1 in the suppression of CRC cell proliferation (Fig. 4b).
Fig. 4. FDFT1 is a downstream target of fasting in suppressing CRC proliferation.
a FDFT1 overexpression and fasting for 48 h inhibited CT26 cell proliferation as measured by a CCK8 assay. Compared with either treatment alone, FDFT1 overexpression combined with fasting for 48 h had the most obvious inhibitory effect on CT26 cell proliferation (from left to right: P = 0.0021; P = 0.0005; P = 0.0003). b Western blotting indicated that fasting 48 h and FDFT1 overexpression increased the protein level of FDFT1 in CRC cells. Fasting exerted an additive effect on FDFT1 expression level in cells overexpressing FDFT1 in the suppression of CRC cell proliferation. c, d Photograph of dissected tumors (first line: CT26 cells + normal diet; second line: CT26 cells + FMD; third line: FDFT1-overexpressing CT26 cells + normal diet; fourth line: FDFT1-overexpressing CT26 cells + FMD; n = 4; both P < 0.0001). Both the FMD and FDFT1 overexpression inhibited tumor growth in the mice. The FMD combined with the implantation of FDFT1-overexpressing CT26 cells had the most obvious inhibitory effect on tumor growth in the mice. e Representative 18F-FDG microPET/CT imaging of tumor-bearing mice. f The ratio of the tumor SUVmax in the four groups. The SUVmax was decreased most significantly in the FDFT1-overexpressing CT26 cells + FMD group (n = 3; from left to right: P = 0.0018; P = 0.0018; P = 0.0003). g The protein expression of FDFT1 in dissected tumor samples was evaluated by IHC. Scale bar: 100 µm. h Graph shows the quantitative analysis of FDFT1 staining (n = 3). i The effect of FDFT1 knockdown, fasting 48 h and FDFT1 knockdown combined with fasting 48 h on CT26 cell proliferation was evaluated by CCK8 (upper: P = 0.005; lower: P = 0.0045). j, k Photograph of dissected tumors (first line: CT26 cells + normal diet; second line: CT26 cells + FMD; third line: shFDFT1 CT26 cells + normal diet; fourth line: shFDFT1 CT26 cells + FMD; n = 4; P = 0.0006; P = 0.0002; P = 0.0003). The FMD inhibited tumor growth in mice. FDFT1 knockdown promoted tumor growth in mice. The FMD combined with shFDFT1 CT26 cells did not inhibit tumor growth in the mice. Error bars, mean ± SD, the data are from three independent experiments. Two-sided t tests. *P < 0.05, **P < 0.01, ***P < 0.001, compared with the control group (or normal diet group). #P < 0.05, ##P < 0.01.
To further confirm the effect of combining FDFT1 overexpression with fasting on cell proliferation observed in vitro, we subcutaneously injected CT26 cells and FDFT1-overexpressing CT26 cells into BALB/c mice. Our results indicated that both the FMD and FDFT1 overexpression inhibited tumor growth in the mice. Compared with FMD alone or FDFT1-overexpressing alone, FMD combined with FDFT1 overexpressing had the most dramatic inhibitory effect on tumor growth in the mice (Fig. 4c, d). Consistent with these results, FMD combined with FDFT1 overexpressing had the most significant inhibitory effect on 18F-FDG uptake in the in the xenograft model (Fig. 4e, f). In addition, the protein expression of FDFT1 in dissected tumor samples evaluated by IHC indicated that either FMD or FDFT1 overexpression with a normal diet increased the protein level of FDFT1 in vivo. (Fig. 4g, h).
Our previous data showed that fasting upregulated the expression of FDFT1 and FDFT1 acted as a tumor suppressor in CRC. Our data also suggested that fasting synergizes with FDFT1 overexpression in the inhibition of CRC proliferation in vitro and in vivo. To evaluate whether the inhibitory effect of fasting on CRC is mediated by FDFT1, we assessed whether FDFT1 knockdown would reverse the inhibitory effect of fasting on CRC. Our data demonstrated that FDFT1 knockdown combined with fasting reversed the inhibitory effect of fasting on CRC cell proliferation (Fig. 4i; Supplementary Fig. 20c). Moreover, an EdU assay showed that FDFT1 knockdown combined with fasting abrogated the fasting inhibition of CRC cell proliferation (Supplementary Fig. 20a, b, d, e).
To further confirm our in vitro observation, we subcutaneously injected CT26 cells and FDFT1-knockdown CT26 cells into BALB/c mice. When the FMD was combined with the implantation of FDFT1 knockdown CT26 cells in vivo, the inhibitory effect of fasting on CRC tumor growth was reversed (Fig. 4j, k). Overall, these results demonstrated that FDFT1 was an important downstream target of fasting that mediates the inhibition of CRC cell proliferation.
The mTOR level is inversely correlated with FDFT1 level in CRC
To identify the core signaling pathway underlying the inhibitory effect of FDFT1 on the proliferation of CRC cells, the 8-plex iTRAQ proteomic technique and bioinformatics analysis were performed again. The most enriched pathways involving DEGs between FDFT1 knockdown and control CT26 cells included the “mTOR signaling” and “Glycolysis” pathways in Ingenuity Pathway Analysis (IPA) (Supplementary Fig. 21a). The most enriched pathways involving DEGs between FDFT1-overexpressing and control CT26 cells included the “mTOR signaling” and “Oxidative phosphorylation” pathways in IPA (Supplementary Fig. 21b). We speculated that FDFT1 inhibited the mTOR-related pathway, thereby suppressing glycolysis in CRC cells. First, we investigated the correlation between FDFT1 and mTOR. mTOR protein levels decreased following FDFT1 overexpression, but increased following FDFT1 knockdown (Fig. 5a). Our results suggested that FDFT1 expression is inversely correlated with mTOR expression. To further confirm our observation, the expression of FDFT1 and mTOR was examined by IHC staining in parallel human CRC specimens from FUSCC (Fig. 5b). Patients with high FDFT1 expression had low mTOR expression, which increased the validity of our previous results. Therefore, our data implied that FDFT1 expression was negatively correlated with mTOR expression, and that mTOR was a potential downstream target of FDFT1 in CRC.
Fig. 5. mTOR expression is inversely correlated with FDFT1 expression.
a FDFT1 overexpression inhibited the protein level of mTOR, whereas FDFT1 knockdown increased the protein level of mTOR in CT26 and SW620 cells. b FDFT1 expression and mTOR expression were negatively correlated in CRC patient samples. Scale bar: 200 µm. c The mTOR silencing efficiency of the siRNA in CT26 and SW620 cells was validated by western blotting. d The effect of mTOR silencing on FDFT1 expression level in CT26 was evaluated by western blotting. e The protein level of mTOR when FDFT1 knockdown combined with or without mTOR inhibitor in CT26. f The protein level of pS6k, S6k, pS6, and S6 under the effect of fasting and FDFT1 overexpression in CRC cells. g, h CCK8 proliferation assays showed that the silencing of mTOR decreased the proliferation of CT26 and SW620 cells. k, n The silencing of mTOR reduced glucose uptake and lactate production in CT26 and SW620 cells. i, j, l, o The silencing of mTOR decreased the ECAR and increased the OCR in CT26 and SW620 cells. m The silencing of mTOR decreased the expression of AKT, HIF1α, and proteins encoded by relevant glycolytic genes, such as GLUT1, HK2, LDHA, GPI, PGK1, in CT26 and SW620 cells. p–t Based on TCGA data set analysis, mTOR expression was positively correlated with AKT1, HIF1α, GLUT1, HK2, and LDHA expression. Error bars, mean ± SD, the data are from three independent experiments. Two-sided t tests. *P < 0.05, **P < 0.01, ***P < 0.001, compared with the control group.
To make the link between FDFT1 and mTOR stronger, we evaluated the effect of mTOR silencing on FDFT1 expression level and whether treatment with an mTOR inhibitor would reverse the effects of FDFT1 knockdown. First, we transfected CT26 and SW620 cells with siRNA targeting mTOR (Fig. 5c). Our results indicated that mTOR silencing increased the FDFT1 expression level in CT26 cell (Fig. 5d). An mTOR inhibitor prevents an increase in mTOR protein levels in FDFT1 knockdown cells (Fig. 5e). Moreover, an mTOR activator prevents an increase in FDFT1 protein levels in FDFT1 overexpression cells, identifying an inverse relationship between FDFT1 and mTOR (Supplementary Fig. 22a). Meanwhile, phosphorylation of two mTOR downstream target proteins, S6k and S6, were decreased under fasting or FDFT1 overexpression in colorectal cells (Fig. 5f; Supplementary Fig. 22b).
To explore the function of mTOR in CRC, we examined the influence of mTOR on CRC cell proliferation by CCK8 and EdU assays and found that the silencing of mTOR decreased cell proliferation (Fig. 5g, h). In the EdU immunofluorescence staining assay, the silencing of mTOR resulted in a lower relative fold fraction of EdU-positive cells (Supplementary Fig. 22c–f). These data showed that mTOR was a positive regulator of CRC cell proliferation.
Next, the impact of mTOR on glycolysis in CRC cells was assessed. Silencing mTOR reduced glucose uptake and lactate production in CRC cells (Fig. 5k, n). Furthermore, the silencing of mTOR decreased the ECAR, but increased the OCR of CT26 and SW620 cells (Fig. 5i, j, l, o). In addition, we examined the protein expression of AKT, HIF1α, and relevant glycolytic genes by western blotting. As expected, the silencing of mTOR decreased the expression of AKT, HIF1α, and proteins encoded by relevant glycolytic genes, such as GLUT1, HK2, LDHA, GPI, and PGK1 (Fig. 5m). Finally, we analyzed the correlation between mTOR expression and AKT, HIF1α, GLUT1, HK2, and GPI expression in TCGA data sets. Consistent with the results of previous studies, a strong correlation was observed between mTOR expression and AKT1, HIF1α, GLUT1, HK2, and GPI expression (Fig. 5p–t). Therefore, these observations supported the hypothesis that mTOR positively regulates the aerobic glycolysis involved in CRC cell proliferation.
FDFT1 inhibits the AKT-mTOR-HIF1α pathway in CRC glycolysis
Our iTRAQ proteomics analysis indicated that FDFT1 is probably a negative regulator of glucose metabolism in CRC. Our results showed that FDFT1 overexpression reduced glucose uptake and lactate production in CT26 and SW620 cells (Fig. 6a, b; Supplementary Fig. 23a, b). Subsequent analysis indicated that in cells overexpressing FDFT1, ECAR was reduced, but OCR was increased (Fig. 6c, d; Supplementary Fig. 23c, d). In contrast, FDFT1 knockdown increased glucose uptake and lactate production (Fig. 6e, f; Supplementary Fig. 23e, f). Moreover, in FDFT1 knockdown, ECAR was increased, but OCR was decreased (Fig. 6g, h; Supplementary Fig. 23g, h). Furthermore, FDFT1 overexpression inhibited the protein and mRNA expression of mTOR-targeted glycolytic enzymes, including GLUT1, HK2, PGK1, GPI, and LDHA (Fig. 6i, m; Supplementary Fig. 23i, m). In contrast, FDFT1 knockdown increased the protein and mRNA expression of mTOR-targeted glycolytic enzymes, including GLUT1, HK2, PGK1, GPI, and LDHA (Fig. 6j, m; Supplementary Fig. 23j, m). These results confirmed that FDFT1 was a negative regulator of glucose metabolism in CT26 and SW620 cells.
Fig. 6. FDFT1 inhibits glycolysis through suppressing the AKT-mTOR-HIF1α pathway in CRC.
a FDFT1 overexpression reduced glucose uptake in CT26 cells. b FDFT1 overexpression decreased lactate production via glycolysis in CT26 cells. c ECAR was reduced when FDFT1 was overexpressed in CT26 cells. d OCR was increased when FDFT1 was overexpressed in CT26 cells. e FDFT1 knockdown increased glucose uptake in CT26 cells. f FDFT1 knockdown increased lactate production via glycolysis in CT26 cells. g ECAR was increased when FDFT1 was knocked down in CT26 cells. h OCR was decreased when FDFT1 was knocked down in CT26 cells. i, m FDFT1 overexpression inhibited the protein and mRNA expression of mTOR-targeted glycolytic enzymes, including GLUT1, HK2, PGK1, GPI, and LDHA, in CT26 cells. j, m FDFT1 knockdown increased the protein and mRNA expression of mTOR-targeted glycolytic enzymes, including GLUT1, HK2, PGK1, GPI, and LDHA, in CT26 cells. k, n FDFT1 overexpression decreased the protein and mRNA expression of AKT, mTOR, and HIF1α. l, n FDFT1 knockdown increased the protein and mRNA expression of AKT, mTOR, and HIF1α. o Photograph of dissected tumors (the first line: normal diet, the second line: FMD + glucose, the third line: FMD, n = 5). p The tumor volumes were measured every 2 days after the 13th day. The FMD + glucose group can reverse the tumor growth inhibition induced by the FMD (n = 5; ns: P = 0.1838; P = 0.0001). q The protein level of FDFT1 and mTOR in dissected tumor samples from normal diet group, FMD group and FMD + glucose group was measured by western blotting. r The glucose level in these three groups. Error bars, mean ± SD, the data are from three independent experiments. Two-sided t tests. *P < 0.05, **P < 0.01, ***P < 0.001, compared with the control group (or normal diet group). #P < 0.05, ##P < 0.01.
Because the AKT-mTOR-HIF1α pathway is an essential pathway that governs glycolysis and proliferation in CRC and FDFT1 expression is inversely correlated with mTOR expression, we decided to determine whether FDFT1 is negatively correlated with the AKT-mTOR-HIF1α pathway. Notably, FDFT1 overexpression decreased the protein and mRNA expression of AKT, mTOR, and HIF1α in CRC cells (Fig. 6k, n; Supplementary Fig. 16k, n). In contrast, FDFT1 knockdown increased the protein and mRNA expression of AKT, mTOR, and HIF1α (Fig. 6l, n; Supplementary Fig. 23l, n). These results suggested that FDFT1 inhibited glucose metabolism through suppressing the AKT-mTOR-HIF1α pathway in CRC.
Because fasting and FDFT1 upregulation reduced mTOR, AKT, and HIF1α expression in vitro, we hypothesized that the effect of fasting may be due to a strong glucose reduction during FMD, so we added a group of FMD + glucose in vivo tumor formation experiment. In this group, glucose was added to drinking water in correspondence with the FMD. This group can reverse the tumor growth inhibition induced by the FMD (Fig. 6o, p). Compared with FMD group, the protein level of FDFT1 and mTOR was reversed in FMD + glucose group (Fig. 6q). The glucose level in FMD + glucose mice was significantly higher than that of the FMD group (Fig. 6r).
We also examined the effect of fasting, FDFT1 overexpression and fasting combined with FDFT1 overexpression on the glycolysis and total cholesterol production in CRC cell lines and found that overexpression of FDFT1 and fasting reduced glycolysis and total cholesterol levels. Meanwhile, overexpression of FDFT1 combined with fasting had additive effects in reducing glycolysis and total cholesterol levels in CRC cell lines (Supplementary Fig. 24a–n). The mRNA level of key genes in cholesterol biosynthesis and efflux pathway were activated after fasting or FDFT1 upregulation (Supplementary Fig. 25a–c).
Fasting and mTOR inhibitor synergize in suppressing CRC proliferation
We explored whether rapamycin, a pharmacological inhibitor of mTOR, can synergize with FMD in inhibiting the proliferation of CRC. The results indicated that both FMD and 1 mg/kg dose of rapamycin can robustly inhibit the proliferation of CRC and drastically improve survival (Fig. 7a–c). In addition, we found that rapamycin therapy could increase the levels of FDFT1 protein and mRNA like fasting (Fig. 7d, e). Moreover, we also found that fasting and rapamycin synergize in delaying CRC progression, improving survival, and upregulating FDFT1 (Fig. 7a–e). Weight profile in four groups is shown in Supplementary Fig. 26. Overall, these results demonstrated that fasting and rapamycin can be potential therapeutic implications to inhibit the proliferation of CRC.
Fig. 7. Fasting and mTOR inhibitor synergize in suppressing CRC proliferation and clinical significance of the FDFT1/AKT-mTOR-HIF1α pathway in CRC patients.
a CT26 cells were injected into BALB/c mice. When the tumors were palpable, the mice were randomly assigned to the normal diet group, FMD group, the rapamycin 1 mg/kg group and FMD + rapamycin 1 mg/kg group. Photograph of dissected tumors (the first line: normal diet, the second line: FMD, the third line: rapamycin 1 mg/kg, the fourth line: FMD + rapamycin 1 mg/kg, n = 5). b The tumor volumes were measured every 3 days after the 9th day (n = 30; ***P = 0.0008, P = 0.0003; #P = 0.0133). On day 9 after inoculation, all the tumor were palpable. c Kaplan–Meier analysis of the overall survival of mice after the inoculation in normal diet group, FMD group, normal diet mice treated with rapamycin 1 mg/kg group and FMD + rapamycin 1 mg/kg group (n = 30; log-rank score: P = 0.0049 for FMD group, P = 0.0058 for rapamycin 1 mg/kg group; P = 0.00069 for FMD + rapamycin 1 mg/kg group.) d, e The expression level of FDFT1 in four groups was evaluated by western blotting and qRT-PCR (**P = 0.0025, P = 0.0097; ***P = 0.0008; #P = 0.0133, P = 0.0351). f–j Survival analysis stratified by combining FDFT1 levels with AKT1, mTOR, HIF1α, GLUT1, and HK2 levels from CRC patients in the TCGA cohort. k Proposed model of the mechanism underlying the fasting-mediated regulation of glucose metabolism via the FDFT1/AKT-mTOR-HIF1α axis in colorectal cancer. Fasting upregulates the expression of FDFT1 during the inhibition of colorectal cancer cell aerobic glycolysis and proliferation. FDFT1, whose downregulation is correlated with malignant progression and poor prognosis in CRC, acts as a critical tumor suppressor in CRC. We then observed that FDFT1 is an important downstream target of fasting that mediates the inhibition of CRC cell proliferation. Mechanistically, FDFT1 inhibits the AKT-mTOR-HIF1α pathway, impairing aerobic glycolysis, and thereby suppressing the proliferation of CRC cells. There is also a reverse regulation of FDFT1 by mTOR. Error bars, mean ± SD, the data are from three independent experiments. Two-sided t tests. Kaplan–Meier analysis and log-rank tests were used in panel c. *P < 0.05, **P < 0.01, ***P < 0.001, compared with normal diet group. #P < 0.05, ##P < 0.01.
Clinical significance of the FDFT1/AKT-mTOR-HIF1α pathway in CRC patients
To further validate our observation, we examined the clinical significance of AKT1, mTOR, HIF1α, GLUT1, and HK2 expression with FDFT1 in CRC by analyzing the TCGA data set. Because we speculated FDFT1 as a tumor suppressor in CRC, we assessed the expression of FDFT1 with that of AKT1, mTOR, HIF1α, GLUT1, and HK2. The patients with a high expression of FDFT1 and a low expression of AKT1, mTOR, HIF1α, GLUT1, and HK2 exhibited longer survival than those with a low expression of FDFT1 and a high expression of the AKT-mTOR-HIF1α pathway and glycolytic genes (Fig. 7f–j; Supplementary Fig. 27).
Discussion
Mounting evidence indicates that fasting exerts extensive antitumor effects in various cancers8,29,36,37. However, the mechanism by which fasting inhibits CRC remains unclear. Our results showed that fasting inhibits the malignant progression of CRC by impairing aerobic glycolysis. In particular, our in vivo and in vitro experiments showed that fasting could dramatically elevate the expression of the cholesterogenic gene FDFT1. Moreover, we showed that FDFT1 played an important tumor-suppressive role with clinical significance and function in CRC, and was also an important downstream target of fasting. Mechanistically, FDFT1 performs its tumor-inhibitory function on glucose metabolism by negatively regulating AKT/mTOR/HIF1α signaling. Furthermore, the pharmacological mTOR inhibitor rapamycin can synergize with FMD in inhibiting the proliferation of CRC. We conclude that fasting negatively regulates glucose metabolism and cell proliferation via the FDFT1/AKT-mTOR-HIF1α axis in CRC (Fig. 7k).
Cancer cells usually exhibit aberrant metabolism resulting from metabolic reprogramming. The most prominent metabolic reprogramming occurring in cancer results in aerobic glycolysis in preference to mitochondrial oxidative phosphorylation, which provides continuous energy and nutrients to support uncontrolled proliferation; this reprogramming is termed the Warburg effect8,23,31,38. Previous studies have demonstrated that caloric restriction or fasting can alter the overall metabolic state of cancer cells, including the dependence on aerobic glycolysis8,11,39,40. In this study, we provided evidence that fasting inhibits the proliferation of CRC cells. Proteomics analysis showed that the glycolysis pathway was highly downregulated during fasting. A series of aerobic glycolysis-related assays indicated that fasting plays a vital role in inhibiting glycolysis in CRC cells. The proteasome pathway also was downregulated during fasting, which can be another promising direction to explore41–43. We confirmed these results in an in vivo xenograft model. Fasting dramatically inhibited 18F-FDG uptake and attenuated tumor growth in this xenograft model. These results suggested that, similar to traditional cancer therapy, fasting can be a potential therapeutic approach in CRC. Given the critical role of fasting in CRC glucose metabolism, we further explored the potential underlying mechanism and downstream effectors.
Some studies have suggested that the antitumor effect of fasting is mediated through the modulation of insulin-like growth factor (IGF)-1 or insulin, or through the enhancement of antitumor immunity15,16,37,44. To investigate the downstream target of fasting that mediates the inhibition of CRC cell proliferation, we analyzed the GSE6065329 data set (from a study on fasting-induced anti-Warburg effects in CRC) to identify DEGs between the control and fasting groups. Intriguingly, the results indicated that fasting upregulates the “Steroid biosynthesis” pathway, and that FDFT1 was dramatically upregulated. FDFT1 acts at a branch point in the mevalonate pathway, and is the first specific enzyme in cholesterol synthesis, catalyzing the reaction that produces squalene synthase34,45. Clinically, high FDFT1 expression in CRC is associated with better prognosis in The Cancer Genome Atlas (TCGA) data sets. However, FDFT1 has seldom been studied in the field of cancer research. Limited studies addressed FDFT1 in prostate cancer35, ovarian cancer46, and lung cancer47, but there is no information on the function of FDFT1 in CRC. Therefore, we selected FDFT1 as the target gene. Our study validated that fasting upregulates the expression of FDFT1 in CRC cells both in vitro and in vivo. Then, we observed that the downregulation of FDFT1 was correlated with malignant progression and poor prognosis in CRC in both the FUSCC cohort and TCGA data sets.
To our knowledge, the biological function of FDFT1 in CRC has not been studied. Our study showed that FDFT1 negatively regulates the proliferation of CRC cells, and acts as a tumor suppressor in CRC. Furthermore our data suggested that fasting synergizes with FDFT1 overexpression in the inhibition of CRC cell proliferation in vitro and in vivo. However, when fasting was combined with FDFT1 knockdown in vivo, the inhibitory effect of fasting on CRC was reversed, indicating that FDFT1 was an important downstream target of fasting that mediated the inhibition of CRC cell proliferation.
To further identify the core signaling pathway underlying the inhibitory effect of FDFT1 on the proliferation of CRC cells, another proteomics analysis was performed. Based on the bioinformatics results, we speculated that FDFT1 inhibited the mTOR-related pathway, thereby suppressing glycolysis in CRC cells. mTOR is a conserved serine/threonine kinase that plays a key role in integrating multiple physiological stimuli to regulate cell growth and metabolic pathways27,28,40,48. mTOR deregulation occurs in many human pathologies, including cancer, metabolic diseases, nervous system diseases, and inflammation49,50. There are already many mTOR inhibitors for the treatment of human cancer, and many more have been evaluated in clinical trials51–55. mTOR activates glycolysis through modulating the expression of the transcription factor HIF-1α, indirectly upregulating the transcription of almost all glycolytic genes in tumor cell5,28,56. Our results indicated that FDFT1 expression is negatively correlated with mTOR expression. Consistent with the data in previous reports27,57–60, our data showed that mTOR is a positive regulator of aerobic glycolysis and proliferation in CRC cells. HIF1α is a key regulator of the Warburg effect and regulates the expression of a variety of metabolism-related proteins61. The correlation between mTOR and HIF1α prompted us to validate that FDFT1 could regulate glucose metabolism through the AKT-mTOR-HIF1α pathway. Through the effect of fasting on glucose metabolism as well as on the connection among mTOR signaling, fasting and glucose metabolism have been reported. To our knowledge, the impact of FDFT1 on inhibiting the Warburg effect has not been reported during fasting; thus, our results innovatively validated that FDFT1 is a negative regulator of glucose metabolism through suppressing the AKT-mTOR-HIF1α pathway during fasting in CRC. As the clinical application of FDFT1 will be challenging, we explored whether a pharmacological inhibitor of mTOR could be an effective therapy for colorectal cancer. Consistent with our in vitro observations, mTOR inhibitor can synergize with FMD in supressing the proliferation of CRC. Moreover, patients with high FDFT1 expression and low expression of the AKT-mTOR-HIF1α pathway and glycolytic genes exhibited longer survival, adding validation to our hypothesis. The survival data also show a worse prognosis of FDFT1-low tumors regardless of the expression of other mTOR pathway genes, which suggest an alternative pathway for FDFT1 to effect cancer proliferation potential. Several studies have found that cholesterol deprivation can inhibit tumor growth, decrease the phosphorylation of AKT, promote apoptosis62, decrease Bcl-xl, downregulate caspase 3 activation63, or hinder entry of cells into the S phase64. Our study showed that fasting and FDFT1 overexpression reduced the total cholesterol production in CRC cell lines. So we agree there may be other pathways for FDFT1 affect tumor growth. The mechanism by which FDFT1 affects mTOR needs to be further explored. We will focus on studying these problems in the future.
In conclusion, we demonstrated that fasting was a negative regulator of glucose metabolism and proliferation via the FDFT1/AKT-mTOR-HIF1α axis in CRC cells. A therapy based on mTOR inhibition could be a promising approach against CRC. These results indicated a good function of the cholesterogenic gene FDFT1 in CRC glucose metabolism and suggest that FDFT1 is a potential marker in CRC. Our results also elucidate potential therapeutic implications (involving fasting and mTOR) for CRC and indicate potential crosstalk between a cholesterogenic gene and glycolysis.
Methods
Cells and reagents
CRC cell lines CT26, SW620, HCT116, HT29, and human colorectal cancer mucosal epithelial cell line NCM460 were obtained from the American Type Culture Collection (ATCC; Shanghai, China) and cultured in DMEM (Biological Industries, USA) supplemented with 10% FBS (Gibco, USA). Isogenic PIK3CA mutant (HCT116 and HT29) were generated in advance65,66. All culture media contained 100 U/ml penicillin and 100 mg/ml streptomycin and maintained at 37 °C in a humidified atmosphere containing 5% CO2. The fasting mimic medium comprised of glucose-free DMEM (Gibco, USA) supplemented with 0.5 g/L glucose and 1% FBS; fasting was mimicked by incubating cells in this medium for 24 h or 48 h67,68.To determine if mTOR can reverse FDFT1 overexpression, pretreat CT26 and SW620 with mTOR activator MHY1485 (10 µmM) for 24 h.
Lentivirus production and stable cell line selection
The PGMLV-CMV-FDFT1-EF1-ZsGreen1-T2A-Puro (Genomeditech, Shanghai, China) plasmid was used to generate the FDFT1 overexpression constructs. The PGMLV-hU6-FDFT1-CMV-ZsGreen1-PGK-Puro (Genomeditech) plasmid was used to generate the FDFT1 shRNA constructs. The 21-bp sequences targeting FDFT1 were GTGTTTAACTTCTGTGCTATT, GCAGGTATTCAAAGGAGTAGT, and GCCGTCAAAGCTATCATATAC. The PGMLV-CMV-SREBP2-EF1-ZsGreen1-T2A-Puro (Genomeditech) plasmid was used to generate the SREBP2 overexpression constructs. The PGMLV-hU6-SREBP2-CMV-ZsGreen1-PGK-Puro (Genomeditech) plasmid was used to generate the SREBP2 shRNA constructs. The sequences targeting SREBP2 were ATGATGCGAGGCTGAGTTGTC and CCCTGGCTGTCCTGTGTAATAC. Lentiviral particles were produced by transfecting the psPAX2 and pMD2.G plasmids into HEK293T cells using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocol. Stable cell lines were obtained by infection with the lentiviral particles followed by puromycin selection. The efficiency of transfection was examined using real-time PCR and western blotting.
The siRNA transfection
We silenced mTOR expression in CRC cells by using siRNA-mediated silencing. The siRNAs targeting mTOR were 5′-CGAUCCAGUUGUCAUGGAAdTdT-3′/5′-UUCCAUGACAACUGGAUCGdTdT-3′, 5′-GAGACUUGAUGGAAGAGAAdTdT-3′/5′-UUCUCUUCCAUCAAGUCUCdTdT-3′, and 5′-GGUCGGAGUUUAAGGUCUAdTdT-3′/5′-UAGACCUUAAACUCCGACCdTdT-3′ (Proteintech, Shanghai, China). siRNA duplexes targeting mTOR were transfected into colorectal cancer cells using Lipofectamine 2000 (Invitrogen, USA) in serum-free medium according to the manufacturer’s instructions.
Cell proliferation assay
Cell proliferation was examined using a Cell Counting Kit-8 (CCK8; Dojindo Laboratories, Japan). Briefly, cell suspensions (3 × 103/well) were seeded in 96-well culture plates and incubated for 5 days. CCK8 solution (10 µL) was added to each well, and the cells were cultured for another 4 h. The optical density was measured at 450 nm using a microplate reader.
Colony-formation assay
Cells were seeded in triplicate in six-well plates at a density of 500 cells/well. After 14 days, most single colonies contained more than 50 cells. The cells were fixed with 4% paraformaldehyde for 20 min at room temperature, stained with 4 mg/mL crystal violet for 20 min, and counted under a light microscope (Leica DM IL, Germany).
EdU assay cells
In total, 1 × 105 cells from each group were seeded on coverslips and cultured for 24 h. Click-iT EdU Imaging Kits (#C10339, Invitrogen, USA) were used to label and detect the incorporated EdU according to the manufacturer’s protocol. The staining results were observed under an inverted fluorescence microscope (IX51; Olympus, Tokyo, Japan).
Transwell migration assay
The Transwell migration assay was performed using 24-well cell culture inserts containing a transparent PET membrane (8.0-µm pore size, #353097; BD Biosciences, USA). A total of 2 × 105 cells in 200 µL serum-free DMEM were added to the upper chamber, and 800 µL DMEM supplemented with 10% FBS was added to the lower chamber. Following 24 h of incubation, the migrated cells on the bottom of the membrane were fixed with 4% PFA for 20 min and stained with 0.1% Crystal violet for further analysis.
Cell cycle and apoptosis analysis
Cell cycle analysis was performed via propidium iodide (PI) staining following the manufacturer’s protocols (Signalway Antibody, #CA002). To identify apoptotic cells, a PE Annexin V Apoptosis Detection Kit I (BD Biosciences, #559763) was used according to the manufacturer’s protocol. The cells were analyzed by flow cytometry (Cell Lab Quanta, Beckman Coulter, USA).
Glycolysis analysis
Glucose Uptake Fluorometric Assay Kits (Biovision, #K666-100) and Lactate Colorimetric Assay Kits (Biovision, #K627-100) were purchased, and glycolysis was detected in CRC cells according to the manufacturer’s protocols.
OCR and ECAR
Cellular mitochondrial function and cellular glycolytic capacity were measured by using a Seahorse Bioscience XF96 Extracellular Flux Analyzer according to the manufacturer’s instructions for the Seahorse XF Glycolysis Stress Test Kit and Cell Mito Stress Test Kit (Seahorse Bioscience, USA). Briefly, 2 × 104 cells were seeded into 96-well cell culture XF microplates and incubated overnight for further testing. The ECAR and OCR values were calculated after normalization to the total cell number and are plotted as the mean ± SD.
Protein extraction and western blot analysis
Briefly, total protein was extracted, qualified by BCA protein assay reagent, separated by SDS-PAGE, and detected by immunoblotting with specific antibodies. Antibodies against FDFT1 (Abcam, ab195046), mTOR (Abcam, ab10268), AKT (Cell Signaling, #9272), vinculin (Cell Signaling, #4650), HIF1α (Novus, NB100-105), GLUT1 (HIF-Proteintech, 66290-1-lg), LDHA (Proteintech, 19987-1-AP), HK2 (Proteintech, 22029-1-AP), PGK1 (Proteintech, 17811-1-AP), and GPI (Abcam, ab66340) were purchased from the designated manufacturers. Vinculin was used as the loading control.
RNA isolation and quantitative real-time PCR
The total RNA was extracted using TRIzol reagent (Invitrogen). A Takara PrimeScript RT reagent kit was used for reverse transcription to obtain cDNA. The expression of the candidate genes and GAPDH was assessed by quantitative real-time PCR using an ABI 7900HT real-time PCR system (Applied Biosystems). The primer sequences are listed in Supplementary Table 1.
Proteomics
After samples were lysed in 1 × protease inhibitor cocktail (Roche Ltd., Switzerland), the protein was digested with sequence-grade modified trypsin (Promega, WI) and lyophilized. The resultant peptide mixture was labeled with an iTRAQ 8-plex labeling kit (Sciex) following the manufacturer’s instructions. The peptide mixture was fractionated by high pH separation using an Ultimate 3000 system (ThermoFisher Scientific, USA). The peptides were analyzed by on-line nanospray LC-MS/MS. Tandem mass spectra were processed by PEAKS Studio version 8.5 (Bioinformatics Solutions Inc., Canada). Differentially expressed proteins were filtered if their fold change was greater than 1.5 and they contained at least two unique peptides with a significance greater than 13. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD012029 and 10.6019/PXD012029.
Clinical samples
CRC and paired noncancerous tissues were obtained from 81 patients who underwent surgical resection without preoperative chemotherapy or radiotherapy at FUSCC from 2012 to 2013. Prior patient consent and approval from the Institutional Research Ethics Committee were obtained. The patients were followed up after surgery every 3 months. All tissues were frozen at −125 °C until use.
IHC staining
IHC staining of paraffin-embedded tissues with antibodies against FDFT1 (Abcam, ab195046) was performed and scored according to standard procedures. The staining score was determined by two independent pathologists at our center.
Animal model
BALB/c mice (female, 4–6 weeks of age, 18–20 g; Shanghai SLAC Laboratory Animal Co., Ltd) were housed in a specific pathogen-free (SPF) environment. In total, 2 × 104 cells (CT26, CT26 pcDNA-FDFT1, or CT26 shFDFT1) were injected subcutaneously into the right flank of the mice. When the tumors were palpable, the mice were randomly assigned to the control group or the FMD group. In the control group, food was provided ad libitum with TD.7912 rodent chow. The average daily consumption of mice was 14.9 kJ in the control group. The FMD consisted of three components11,67–70, designated as the day 1 diet, day 2–3 diet, and day 4–7 diet, fed in this order. The day 1 diet contained only 50% of the calories of the normal intake, which consists of a variety of vegetable powders, low-calorie broth powders, essential fatty acids, extra virgin olive oil, minerals, and vitamins, containing 7.67 kJ/g; carbohydrate 2.2k J/g, protein 0.46k J/g, fat 5.00 kJ/g. The day 2–3 diet contained only 10% of the normal calories intake, which consists of glycerol and low-calorie broth powders, containing 1.48 kJ/g, carbohydrates 1.47 kJ/g, and protein/fat 0.01 kJ/g. For the day 4–7 diet, the mice were fed their normal intake; this progression was followed by another cycle of the FMD. The animals had free access to water. At the end of the study, the mice underwent microPET/CT scanning. After scanning, the tumors were surgically dissected. Fasting mimic group + glucose group were provided with 10 mg/kg glucose supplementation in the drinking water during the FMD. All animal experiments were performed according to the procedures approved by the Institutional Animal Care and Use Committee of Fudan University.
MicroPET/CT imaging
MicroPET/CT scanning and image analysis were performed using an Inveon microPET/CT system and the manufacturer-supplied display software (Inveon Research Workplace, Siemens Medical Solutions, USA). Each tumor-bearing mouse was injected with 5.55 MBq (150 μCi) of 18F-fluorodeoxyglucose (18F-FDG) via the tail vein. Ten-min static scans were acquired at 1.0 h post injection, and the animals were maintained under isoflurane anesthesia during the scanning period. The images were reconstructed using a three-dimensional ordered-subset expectation-maximization (OSEM3D)/maximum algorithm. Inveon Research Workplace was used to obtain the maximum standard uptake value (SUVmax).
In vivo rapamycin treatment
Rapamycin (1 mg/kg/d, CAS 53123-88-9, Santa Cruz Biotechnology, USA) was injected into mice intraperitoneally when the tumors were palpable. FMD also was started when the tumors were palpable.
Total cholesterol levels
The total cell cholesterol levels were measured by a cholesterol/cholesteryl ester quantitation kit (Abcam, ab65359) according to the manufacturers’ instructions.
Statistical analysis
The experiments were repeated at least three times. All data are presented as the means ± SDs. SPSS 17.0 software (IBM, USA) and GraphPad Prism 7.0 software were used for data analysis. Two-sided unpaired Student’s t tests, one-way analysis of variance, chi-square tests, Kaplan–Meier analysis and log-rank tests were used to evaluate the data. Differences were considered significant at *P < 0.05; **P < 0.01; ***P < 0.01; ****P < 0.0001; #P < 0.05; and ##P < 0.01.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Ping Zhang for providing flow cytometry analyses and Jianping Zhang for providing microPET/CT imaging. We also thank Zhenhuan Zhao, Zhenhua Jiang, Jun Li, Ping Wei, and Yi Qin for valuable comments and suggestions. This study was supported by, the National Key Research and Development Program of China (2018YFC2001904), National Natural Science Foundation of China (no. 81372101, no. 81873948, no. 81871591 and no. 81572626), State Key Basic Research Program (973) project (2015CB553404), the Shanghai Anticancer Association EYAS project (SACA-CY1B07), the Key Technology and Development Program of Shanghai (no.17411963400), Shanghai Municipal Commission of Health and Family Planning, Key Developing Disciplines (2015ZB0104), the Shanghai Shenkang Hospital Development Center Clinical Science and Technology Innovation Project (SHDC12018105) and the 2018 Xin Chen Cup Young Anesthesiologist Training Grant.
Author contributions
M.L.W., D.M., and C.H.M. designed the study; M.L.W., X.Y.C., H.L., Z.R.S., M.M.Z., and P.F.S. performed the experiments; Q.Y., Y.J.X., and N.J. analyzed the data; J.P.Z. and J.Z. provided reagents; M.L.W. and W.K.C. wrote the paper; M.L.W. and X.P.Z. revised the paper.
Data availability
Raw mass spectrometry proteomics data files have been deposited in the ProteomeXchange Consortium with the data set identifier PXD012029 and 10.6019/PXD012029. All the other data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file.
Competing interests
The authors declare no competing financial interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41467-025-65107-1"
Peer review information Nature Communications thanks Marios Giannakis, Valter Longo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mei-lin Weng, Wan-kun Chen, Xiang-yuan Chen.
Change history
10/20/2025
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41467-025-65107-1
Contributor Information
Duan Ma, Email: duanma@fudan.edu.cn.
Xiao-ping Zhang, Email: zxpsibs@163.com.
Chang-hong Miao, Email: whitedolphin2007@hotmail.com.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-15795-8.
References
- 1.Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Brenner, H., Kloor, M. & Pox, C. P. Colorectal cancer. Lancet383, 1490–1502 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Global Burden of Disease Cancer C et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol.3, 524–548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker, E. & Rex, D. K. Advances in CRC prevention: screening and surveillance. Gastroenterology154, 1970–1984 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Schreuders, E. H. et al. Colorectal cancer screening: a global overview of existing programmes. Gut64, 1637–1649 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Fontana, L. & Partridge, L. Promoting health and longevity through diet: from model organisms to humans. Cell161, 106–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Longo, V. D. & Panda, S. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab.23, 1048–1059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nencioni, A., Caffa, I., Cortellino, S. & Longo, V. D. Fasting and cancer: molecular mechanisms and clinical application. Nat. Rev. Cancer18, 707–719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luigi, F., Meyer, T. E., Samuel, K. & Holloszy, J. O. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc. Natl Acad. Sci. USA101, 6659–6663 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eitan, E. et al. In a randomized trial in prostate cancer patients, dietary protein restriction modifies markers of leptin and insulin signaling in plasma extracellular vesicles. Aging cell16, 1430–1433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei, M. et al. Fasting-mimicking diet and markers/risk factors for aging, diabetes, cancer, and cardiovascular disease. Science Transl. Med.9, eaai8700 (2017). [DOI] [PMC free article] [PubMed]
- 12.Robertson, L. T. et al. Protein and calorie restriction contribute additively to protection from renal ischemia reperfusion injury partly via leptin reduction in male mice. J. Nutr.145, 1717–1727 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauro, C. R. et al. Preoperative dietary restriction reduces intimal hyperplasia and protects from ischemia-reperfusion injury. J. Vasc. Surg.63, 500–509 e501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton, E. F. et al. Early time-restricted feeding improves insulin sensitivity, blood pressure, and oxidative stress even without weight loss in men with prediabetes. Cell Metab.27, 1212–1221 (2018). [DOI] [PMC free article] [PubMed]
- 15.Efeyan, A., Comb, W. C. & Sabatini, D. M. Nutrient-sensing mechanisms and pathways. Nature517, 302–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buono, R. & Longo, V. D. Starvation, stress resistance, and cancer. Trends Endocrinol. Metab.29, 271–280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tulsian, R., Velingkaar, N. & Kondratov, R. Caloric restriction effects on liver mTOR signaling are time-of-day dependent. Aging10, 1640–1648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blagosklonny, M. V. Fasting and rapamycin: diabetes versus benevolent glucose intolerance. Cell Death Dis.10, 607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Pavlova, N. N. & Thompson, C. B. The emerging hallmarks of cancer metabolism. Cell Metab.23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warburg, O. On the origin of cancer cells. Science123, 309–314 (1956). [DOI] [PubMed] [Google Scholar]
- 22.Cairns, R. A., Harris, I. S. & Mak, T. W. Regulation of cancer cell metabolism. Nat. Rev. Cancer11, 85–95 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Liberti, M. V. & Locasale, J. W. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci.41, 211–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polivka, J. Jr. & Janku, F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharm. Ther.142, 164–175 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Xiao, Y. et al. PDGF promotes the Warburg effect in pulmonary arterial smooth muscle cells via activation of the PI3K/AKT/mTOR/HIF-1 alpha signaling pathway. Cell Physiol. Biochem.42, 1603–1613 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Charbonnier, L. M. et al. Functional reprogramming of regulatory T cells in the absence of Foxp3. Nat. Immunol.20, 1208–1219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mossmann, D., Park, S. & Hall, M. N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer18, 744–757 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Shih-Chin, C. et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science345, 1250684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bianchi, G., Martella, R., Ravera, S., Marini, C. & Longo, V. D. Fasting induces anti-Warburg effect that increases respiration but reduces ATP-synthesis to promote apoptosis in colon cancer models. Oncotarget6, 11806 (2015). [DOI] [PMC free article] [PubMed]
- 30.Patterson, R. E. & Sears, D. D. Metabolic effects of intermittent fasting. Annu. Rev. Nutr.37, 371–393 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Lien, E. C. & Vander Heiden, M. G. A framework for examining how diet impacts tumour metabolism. Nat. Rev. Cancer19, 651–661 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Brusselmans, K. et al. Squalene synthase, a determinant of Raft-associated cholesterol and modulator of cancer cell proliferation. J. Biol. Chem.282, 18777–18785 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Do, R., Kiss, R. S., Gaudet, D. & Engert, J. C. Squalene synthase: a critical enzyme in the cholesterol biosynthesis pathway. Clin. Genet.75, 19–29 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Coman, D. et al. Squalene synthase deficiency: clinical, biochemical, and molecular characterization of a defect in cholesterol biosynthesis. Am. J. Hum. Genet.103, 125–130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukuma, Y. et al. Role of squalene synthase in prostate cancer risk and the biological aggressiveness of human prostate cancer. Prostate Cancer Prostatic Dis.15, 339–345 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Marinac, C. R. et al. Prolonged nightly fasting and breast cancer prognosis. JAMA Oncol2, 1049–1055 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Flanagan, C. H., Smith, L. A., Mcdonell, S. B. & Hursting, S. D. When less may be more: calorie restriction and response to cancer therapy. BMC Med.15, 106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Upadhyay, M., Samal, J., Kandpal, M., Singh, O. V. & Vivekanandan, P. The Warburg effect: insights from the past decade. Pharmacol. Therapeut.137, 318–330 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Vernieri, C. et al. Targeting cancer metabolism: dietary and pharmacologic interventions. Cancer Discov.6, 1315–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masui, K. et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab.18, 726–739 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams, J. The proteasome: a suitable antineoplastic target. Nat. Rev. Cancer4, 349–360 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Arlt, A. et al. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2). Oncogene28, 3983–3996 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Fang, Y. et al. CD36 inhibits beta-catenin/c-myc-mediated glycolysis through ubiquitination of GPC4 to repress colorectal tumorigenesis. Nat. Commun.10, 3981 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun, P. et al. Fasting inhibits colorectal cancer growth by reducing M2 polarization of tumor-associated macrophages. Oncotarget8, 74649–74660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shechter, I. et al. Localization of the squalene synthase gene (FDFT1) to human chromosome 8p22-p23.1. Genomics20, 116–118 (1994). [DOI] [PubMed] [Google Scholar]
- 46.Zheng, L., Li, L., Lu, Y., Jiang, F. & Yang, X. A. SREBP2 contributes to cisplatin resistance in ovarian cancer cells. Exp. Biol. Med.243, 655–662 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dehghani, M. et al. Relationship of SNP rs2645429 in farnesyl-diphosphate farnesyltransferase 1 gene promoter with susceptibility to lung cancer. Int J. Genomics2018, 4863757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salmond, R. J. mTOR regulation of glycolytic metabolism in T cells. Front. Cell Develop. Biol.6, 122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabatini, D. M. Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc. Natl Acad. Sci. USA114, 11818–11825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hua, H. et al. Targeting mTOR for cancer therapy. J. Hematol. Oncol.12, 71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baselga, J. et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med.366, 520–529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franz, D. N. et al. Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet381, 125–132 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Yao, J. C. et al. Everolimus for the treatment of advanced, non-functional neuroendocrine tumours of the lung or gastrointestinal tract (RADIANT-4): a randomised, placebo-controlled, phase 3 study. Lancet387, 968–977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janku, F., Yap, T. A. & Meric-Bernstam, F. Targeting the PI3K pathway in cancer: are we making headway? Nat. Rev. Clin. Oncol.15, 273–291 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Masoud, G. N. & Li, W. HIF-1 α pathway: role, regulation and intervention for cancer therapy. Acta Pharmaceutica Sin. B5, 378–389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosinsky, R. L. et al. USP22 exerts tumor-suppressive functions in colorectal cancer by decreasing mTOR activity. Cell Death Differ.27, 1328–1340 (2019). [DOI] [PMC free article] [PubMed]
- 58.Wang, H. et al. Targeting mTOR suppressed colon cancer growth through 4EBP1/eIF4E/PUMA pathway. Cancer Gene Ther. 1–13. 10.1038/s41417-019-0117-7 (2019). [DOI] [PubMed]
- 59.Zhong, Z. et al. PORCN inhibition synergizes with PI3K/mTOR inhibition in Wnt-addicted cancers. Oncogene38, 6662–6677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen, X. et al. DEPTOR is an in vivo tumor suppressor that inhibits prostate tumorigenesis via the inactivation of mTORC1/2 signals. Oncogene39, 1557–1571 (2019). [DOI] [PMC free article] [PubMed]
- 61.Semenza, G. L. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol. Sci.33, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhuang, L., Kim, J., Adam, R. M., Solomon, K. R. & Freeman, M. R. Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. J. Clin. Investig.115, 959–968 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li, Y. C., Park, M. J., Ye, S. K., Kim, C. W. & Kim, Y. N. Elevated levels of cholesterol-rich lipid rafts in cancer cells are correlated with apoptosis sensitivity induced by cholesterol-depleting agents. Am. J. Pathol.168, 1107–1118 (2006). [DOI] [PMC free article] [PubMed]
- 64.Duncan, R. E., El-Sohemy, A. & Archer, M. C. Mevalonate promotes the growth of tumors derived from human cancer cells in vivo and stimulates proliferation in vitro with enhanced cyclin-dependent kinase-2 activity. J. Biol. Chem.279, 33079–33084 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Song, B. L. et al. Human acyl-CoA:cholesterol acyltransferase 2 gene expression in intestinal Caco-2 cells and in hepatocellular carcinoma. Biochem. J.394, 617–626 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samuels, Y. et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell7, 561–573 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Brandhorst, S. et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab.22, 86–99 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Biase, S. et al. Fasting-mimicking diet reduces HO-1 to promote T cell-mediated tumor cytotoxicity. Cancer Cell30, 136–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng, C. W. et al. Fasting-mimicking diet promotes Ngn3-driven beta-Cell regeneration to reverse diabetes. Cell168, 775–788.e712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rangan, P. et al. Fasting-mimicking diet modulates microbiota and promotes intestinal regeneration to reduce inflammatory bowel disease pathology. Cell Rep.26, 2704–2719.e2706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry proteomics data files have been deposited in the ProteomeXchange Consortium with the data set identifier PXD012029 and 10.6019/PXD012029. All the other data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file.