Abstract
Antibody-mediated humoral immunity is thought to play a central role in mediating the immunopathogenesis of acute DENV infection, but limited data are available on the diversity, specificity, and functionality of the antibody response at the molecular level elicited by primary or secondary DENV infection. In order to close this functional gap in our understanding of DENV-specific humoral immunity, we utilized high-throughput single cell RNA sequencing to investigate B cells circulating in both primary and secondary natural DENV infections. We captured full-length paired immunoglobulin receptor sequence data from 9,027 B cells from a total of 6 subjects, including 2,717 plasmablasts. In addition to IgG and IgM class-switched cells, we unexpectedly found a high proportion of the DENV-elicited plasmablasts expressing IgA, principally in individuals with primary DENV infections. These IgA class-switched cells were extensively hypermutated even in individuals with a serologically confirmed primary DENV infection. Utilizing a combination of conventional biochemical assays and high-throughput shotgun mutagenesis, we determined that DENV-reactive IgA class-switched antibodies represent a significant fraction of DENV-reactive Igs generated in response to DENV infection, and that they exhibit a comparable epitope specificity to DENV-reactive IgG antibodies. These results provide insight into the molecular-level diversity of DENV-elicited humoral immunity and identify a heretofore unappreciated IgA plasmablast response to DENV infection.
Keywords: Dengue, Plasmablast, Single-cell RNA sequencing, IgA, Monoclonal antibody
Research in context.
Evidence before this study
Antibody-mediated humoral immunity is thought to play a central role in mediating the immunopathogenesis of acute DENV infection. However, limited data are available on the molecular-level diversity, specificity, and functionality of the antibody response elicited by primary or secondary DENV infection. At the onset of this study in 2018 we queried Pubmed for articles either directly comparing/contrasting the plasmablast immunoglobulin repertoires elicited by either primary or secondary DENV infection, or for articles that assessed the DENV-elicited plasmablast repertoires in an unbiased fashion without up-front immunoglobulin isotype selection. No previously published report satisfied either of these criteria.
Added value of this study
Our study is the first to examine the molecular level diversity and antigen specificity of the plasmablast antibody repertoire elicited by both primary and secondary dengue in an unbiased and high-throughput fashion. This approached revealed a previously unappreciated role for DENV-reactive IgA in both primary and secondary dengue, with IgA the dominant plasmablast-restricted isotype observed following primary DENV infection.
Implications of all the available evidence
These results identify a heretofore unappreciated role for DENV-reactive IgA in the humoral response to DENV infection, especially in the setting of primary DENV infection.
Alt-text: Unlabelled box
1. Introduction
DENV is a positive-sense single-stranded RNA virus of the family Flaviviridae, genus Flavivirus, that is maintained in a primarily anthroponotic cycle between the Aedes aegypti mosquito and humans [1]. Consisting of four genetically and immunologically distinct serotypes (DENV-1, −2, −3, and −4), DENV is thought to infect between 280 and 550 million people worldwide every year, with as many as 100 million infections resulting in some degree of clinical presentation [2,3]. While DENV infection is subclinical in the majority of cases, in susceptible individuals it can cause a debilitating flu-like illness known as dengue fever. The majority of individuals suffering from dengue fever recover without the need for extensive medical intervention, but approximately 500,000 individuals per year develop severe dengue, dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS), which has a mortality rate of up to 20% [4], [5], [6], [7].
A unique epidemiological feature of DENV infection is that severe immunopathological symptoms are more likely to occur in individuals previously infected with a heterologous viral serotype compared to individuals without any preexisting DENV immunity [8]. While the mechanisms behind the multifaceted immunopathogenesis of dengue are incompletely understood and may involve some degree of genetic predisposition [9,10], waning antibody-mediated cross-recognition of heterotypic DENV is one potential explanation for the increased frequency of severe disease in individuals experiencing a secondary DENV infection [5,11]. Antibody-dependent enhancement (ADE) of DENV infection has been observed in various in vitro experimental models and in in vivo adoptive transfer models [5,[12], [13], [14], [15]], and is thought to be primarily facilitated by Fc-receptor mediated endocytosis of IgG1-opsonized DENV particles [16], [17], [18]. Even though discrete DENV E protein antibody epitopes have been identified as particularly amenable to antibody-mediated immune enhancement of infection [19], any DENV-reactive IgG1 antibody with a low IC50/EC50 ratio is theoretically capable of enhancing DENV infection when present at an appropriate concentration [20], [21], [22]. However, while significant correlative data exist, definitive in vivo evidence of ADE in humans has been elusive. Therefore, understanding the molecular diversity of DENV-elicited humoral immunity is critical for extending our understanding of risk factors associated with severe dengue, especially the relative abundance of antibody subclasses with the potential for promoting or inhibiting ADE.
The primary source of circulating DENV-reactive antibodies persisting after the resolution of infection are non-dividing, terminally differentiated, bone-marrow resident, plasma cells [23], [24], [25]. However, while plasma cell-derived antibodies take several weeks to peak and stabilize after initial antigen exposure, B cell plasmablasts can be found in circulation just days after an initial pathogen exposure [23,24,26]. The majority of plasmablasts generated in response to infection rapidly undergo apoptosis following the resolution of inflammation, but a fraction of these cells terminally differentiate into long-lived plasma cells and take-up residency in the bone marrow [25,[27], [28], [29]]. A direct precursor/progeny relationship has been demonstrated for plasmablasts and plasma cells in animal studies, and the frequency of plasmablast-phenotype B cells circulating 5–10 days after vaccination has been shown to positively correlate with serum antibody titers achieved several weeks after DENV antigen exposure [30], [31], [32], 33, [34].
The functional relationship between DENV-elicited plasmablasts and the durable humoral immune profile present after the resolution of infection is further reinforced by work assessing the clonal diversity and antigen specificity of the plasmablast population generated in response to DENV infection. Previous work has demonstrated that the plasmablasts elicited by a secondary DENV infection primarily express broadly cross-reactive, moderately neutralizing, and extensively hypermutated immunoglobulins [32,[35], [36], [37]]. This antigen specificity is reminiscent of the humoral immune profile observed in most individuals following resolution of a secondary DENV infection [33]. In all of these studies, 70%–90% of the immunoglobulins expressed by circulating plasmablasts were observed to bind DENV, and the overwhelming majority of the plasmablasts appear to be derived from memory B cells, as indicated by both the relatively restricted clonal diversity of the population, as well as the extensive pre-existing SHM burden of the antibodies [32,[35], [36], [37]].
In contrast to the abundance of data available on the plasmablast phenotype associated with secondary DENV infection, relatively little data are available on the immunoglobulin receptor specificity and functionality of plasmablasts circulating in response to a primary DENV infection. The most well characterized dataset addressing this question is derived from a human primary dengue serotype 2 infection model [38]. As in secondary DENV infection, abundant plasmablast expansion was observed following infection. However, in contrast to the specificity and functional profile of immunoglobulins generated in response to secondary DENV infection, only ~40% of plasmablast-derived antibodies reacted with DENV, with the majority of antibodies exhibiting serotype-specific reactivity and generally poor neutralization potential. Again, this profile closely mirrors the functional profile of serum following primary DENV infection [39,40].
Although these previous studies provide valuable insight into the molecular complexity of the humoral immune response elicited by DENV infection, there are several significant limitations inherent within the current publication record. Firstly, no data are available on the plasmablast-derived immunoglobulin diversity and functionality following natural primary DENV infection. Secondly, the aforementioned studies restricted their immunoglobulin receptor analysis to cells that expressed IgG or IgM, either by up-front selection during the cell isolation procedure or by virtue of the primers used to amplify the immunoglobulin receptor sequences [32,[35], [36], [37]]. This lack of an unbiased approach to the analysis of plasmablast and immunoglobulin receptor sequence diversity leaves open the possibility that significant features of the humoral immune profile associated with either primary or secondary infection may have been overlooked. Filling this potential knowledge gap may generate new tools for the rapid clinical assessment of previous DENV exposure, and may also provide some insight into the divergent clinical outcomes observed following either primary or secondary DENV infection.
To fill this knowledge gap, we utilized single cell RNA sequencing technology to assess the clonal and transcriptional diversity of plasmablasts elicited in response to primary and secondary DENV infections in an unbiased and high-throughput fashion. Using this approach, we were able to capture full-length paired immunoglobulin receptor sequence data from 9,027 B cells circulating in response to both primary and secondary DENV infection, including 2,717 DENV-elicited plasmablasts. We found an unexpectedly high proportion of plasmablasts expressing IgA in PBMC from patients with primary infection, in addition to the expected IgG and IgM class-switched plasmablasts. These IgA class-switched cells were extensively hypermutated, even in individuals with serologically confirmed primary DENV infection. IgA class-switched Igs expressed by plasmablasts in both primary and secondary DENV infections neutralized DENV and reacted with canonical epitopes on the DENV E protein. Our data demonstrate a previously unappreciated role for IgA class-switched B cells during acute DENV infection - especially during a primary DENV infection - and highlight the utility of unbiased scRNAseq analysis in identifying novel immunological consequences of viral infection.
2. Materials and methods
2.1. Sample collection
Peripheral blood mononuclear cells (PBMC) and plasma were isolated from whole blood specimens obtained from children enrolled in a hospital-based acute illness study in Bangkok, Thailand, the design of which has been previously described [41,42]. Blood samples were obtained serially during acute infection and at early and late convalescent time points; the term ‘fever day’ is used to report acute illness time points relative to Day 0, defined as the day of defervescence. The infecting virus type (DENV-1-4) was determined by RT-PCR and/or virus isolation as previously described [43], and serology (EIA and HAI assays) was used to distinguish primary and secondary DENV infections [44]. Written informed consent was obtained from each subject and/or his/her parent or guardian. The study protocol was approved by the Institutional Review Boards of the Thai Ministry of Public Health, the Office of the U.S. Army Surgeon General, the University of Massachusetts Medical School, and the University of Rhode Island. PBMC and plasma samples were cryopreserved for later analysis.
2.2. Flow cytometry and cell sorting
Cryopreserved PBMC were thawed and placed in RPMI 1640 medium supplemented with 10% heat-inactivated normal human serum (100–318, Gemini Bio-Products), L-glutamine, penicillin, and streptomycin prior to analysis. Cell viability was assessed using CTL-LDC Dye (Cellular Tecnology Limited [CTL], Shaker Heights, OH) and a CTL-ImmunoSpot S6 Ultimate-V Analyzer (CTL). Surface staining for flow cytometry analysis and cell sorting was performed in PBS supplemented with 2% FBS at room temperature. Aqua Live/Dead (ThermoFisher, L34957) was used to exclude dead cells in all experiments. Antibodies and dilutions used for flow cytometry analysis are listed in Supplemental Table 7. Cell sorting was performed on a BD FACSAria Fusion instrument, and data analyzed using FlowJo v10.2 software (Treestar).
2.3. Single-cell RNA sequencing library generation
Flow-sorted viable B and T cell suspensions were prepared for single-cell RNA sequencing using the Chromium Single-Cell 5′ Reagent version 2 kit and Chromium Single-Cell Controller (10x Genomics, CA) [45]. Approximately 2000–8000 cells per reaction suspended at a density of 50–500 cells/μL in PBS plus 0.5% FBS were loaded for gel bead-in-emulsion (GEM) generation and barcoding. Reverse transcription, RT-cleanup, and cDNA amplification were performed to isolate and amplify cDNA for downstream 5′ gene or enriched V(D)J library construction according to the manufacturer's protocol. Libraries were constructed using the Chromium Single-Cell 5′ reagent kit, V(D)J Human B Cell Enrichment Kit, 3′/5′ Library Construction Kit, and i7 Multiplex Kit (10x Genomics, CA) according to the manufacturer's protocol.
2.4. Sequencing
scRNAseq 5′ gene expression libraries and BCR V(D)J enriched libraries were sequenced on an Illumina NovaSeq 6000 instrument using the S1, S2, or S4 reagent kits (300 cycles). Libraries were balanced to allow for ~150,000 reads/cell for 5′ gene expression libraries, and ~20,000 reads/cell for BCR V(D)J enriched libraries. Sequencing parameters were set for 150 cycles for Read1, 8 cycles for Index1, and 150 cycles for Read2. Prior to sequencing, library quality and concentration were assessed using an Agilent 4200 TapeStation with High Sensitivity D5000 ScreenTape Assay and Qubit Fluorometer (Thermo Fisher Scientific) with dsDNA BR assay kit according to the manufacturer's recommendations.
2.5. 5′ gene expression analysis/visualization
The 5′ gene expression alignment from sorted PBMC was performed using the 10x Genomics Cell Ranger pipeline [45]. Sample demultiplexing, alignment, barcode/UMI filtering, and duplicate compression was performed using the Cell Ranger software package (10x Genomics, CA, v2.1.0) and bcl2fastq2 (Illumina, CA, v2.20) according to the manufacturer's recommendations, using the default settings and mkfastq/count commands, respectively. Transcript alignment was performed against a human reference library generated using the Cell Ranger mkref command and the Ensembl GRCh38 v87 top-level genome FASTA and the corresponding Ensembl v87 gene GTF.
Multi-sample integration, data normalization, visualization, and differential gene expression was performed using the R package Seurat (v3.0) [46,47]. Non-viable cells and doublets were filtered from the dataset by removing cells with >10% mitochondrial RNA content, and those cells expressing fewer than 200 or more than 6,000 unique genes. Differential gene expression analysis was performed using a Wilcoxon rank sum test with Bonferoni correction to control for False Discovery Rate (FDR).
2.6. Immunoglobulin sequence analysis
Sorted B cell immunoglobulin clonotype identification, alignment, and annotation was performed using the 10x Genomics Cell Ranger pipeline. Sample demultiplexing and clonotype alignment was performed using the Cell Ranger software package (10x Genomics, CA, v2.1.0) and bcl2fastq2 (Illumina, CA, v2.20) according to the manufacturer's recommendations, using the default settings and mkfastq/vdj commands, respectively. Immunoglobulin clonotype alignment was performed against a filtered human V(D)J reference library generated using the Cell Ranger mkvdjref command and the Ensembl GRCh38 v87 top-level genome FASTA and the corresponding Ensembl v87 gene GTF. Immunoglobulin clonotype visualization, diversity assessment, and analysis were performed using either the Loupe VDJ Browser (10x Genomics, CA, v2.0.0) or custom R code. Paired immunoglobulin clonotype identity, hypermutation burden, and clonal lineage was assessed using the software package BRILIA, with a 15% sequence similarity threshold for clonal lineage assignment [48]. Heavy chain restricted framework and CDR hypermutation burdens were calculated using the IMGT/HighV-QUEST server [49].
2.7. Recombinant monoclonal antibody synthesis
The variable regions from the heavy and light chains of targeted immunoglobulin sequences were codon optimized, synthesized in vitro and subcloned into a pcDNA3.4 vector containing the human IgG1 Fc region by a commercial partner (Genscript). Transfection grade plasmids were purified by maxiprep and transfected into a 293-6E expression system. Cells were grown in serum-free FreeStyle 293 Expression Medium (Thermo Fisher), and the cell supernatants collected on day 6 for antibody purification. Following centrifugation and filtration, the cell culture supernatant was loaded onto an affinity purification column, washed, eluted, and buffer exchanged to the final formulation buffer (PBS). Antibody lot purity was assessed by SDS-PAGE, and the final concentration determined by 280 nm absorption. The clonotype information for all monoclonal antibodies generated as part of this study is listed in Supplemental Table 2.
2.8. Viruses
DENV-1-4 (strains Nauru/West Pac/1974, S16803, CH53489, and TVP-360, respectively) propagated in C6/36 mosquito cells were utilized for ELISA, FlowNT50, and ADE assays. Virus for ELISA was purified by ultracentrifugation through a 30% sucrose solution and the virus pellet was resuspended in PBS.
2.9. Monoclonal antibody DENV-capture ELISA
Monoclonal antibody DENV reactivity was assessed using a 4G2 DENV capture ELISA protocol. In short, 96 well NUNC MaxSorb flat-bottom plates were coated with 2 μg/ml flavivirus group-reactive mouse monoclonal antibody 4G2 (Envigo Bioproducts, Inc.) diluted in borate saline buffer. Plates were washed and blocked with 0.25% BSA + 1% Normal Goat Serum in PBS after overnight incubation. DENV-1, -2, -3 or -4 (strains Nauru/West Pac/1974, S16803, CH53489, and TVP-360) were captured for 2 h in the appropriate wells, followed by extensive washing. Serially diluted monoclonal antibody samples were incubated for 1 h at RT on the captured virus, and DENV-specific antibody binding quantified using anti-human IgG HRP (Sigma-Aldrich, SAB3701362). Secondary antibody binding was quantified using the TMB Microwell Peroxidase Substrate System (KPL, cat. #50-76-00) and Synergy HT plate reader (BioTek, Winooski, VT). Antibody data were analyzed by nonlinear regression (One site total binding) to determine EC50 titers in GraphPad Prism 8 (GraphPad Software, La Jolla, CA).
2.10. Flow cytometry-based 50% neutralization (FlowNT50) assay
Neutralizing antibody titers of monoclonal antibodies and heat-inactivated sera were determined using a flow cytometry-based neutralization assay in U937 cells expressing DC-SIGN as previously described [50,51]. Serial dilutions of antibody or sera were mixed with an equal volume of virus, diluted to achieve 10%–15% infection of cells/well, and incubated for 1 h at 37 °C. After 1 h of incubation, an equal volume of medium (RPMI-1640 supplemented with 10% FBS, 1% penicillin/streptomycin, 1% L-glutamine (200 mM), and 1% nonessential amino acids (10 mM)) containing 5 × 104 U937-DC-SIGN cells were added to each serum-virus mixture and incubated 18–20 h overnight in a 37 °C, 5% CO2, humidified incubator. Following overnight incubation, the cells were fixed, permeabilized and immunostained with flavivirus group-reactive mouse monoclonal antibody 4G2 (Envigo Bioproducts, Inc.), and secondary polyclonal goat anti-mouse IgG PE-conjugated antibody (#550589, BD Biosciences). The percentage of infected cells were quantified on a BD Accuri C6 Plus flow cytometer (BD Biosciences). Data were analyzed by nonlinear regression to determine 50% neutralization titers in GraphPad Prism 8 (GraphPad Software, La Jolla, CA).
2.11. Antibody-dependent enhancement (ADE) assay
In vitro antibody-dependent enhancement of DENV infection was quantified as previously described [52]. Two-fold serial dilutions of antibody or heat-inactivated sera were incubated with virus (in sufficient amounts to infect 10%–15% of U937-DC-SIGN cells) at 1:1 for 1 h at 37 °C. This mixture was then added to a 96-well plate containing 5 × 104 cells (K562) per well in duplicate. Cells were infected 18–20 h overnight in a 37 °C, 5% CO2, humidified incubator. Processing and quantification continued as outlined in the FlowNT50 methods. Fold-infection relative to control serum was reported.
2.12. Serum IgG/IgM/IgA ELISA
DENV-reactive serum IgM/IgG/IgA levels were assessed using a modified 4G2 DENV capture ELISA protocol. 96 well NUNC MaxSorb flat-bottom plates were coated with 2 μg/ml flavivirus group-reactive mouse monoclonal antibody 4G2 (Envigo Bioproducts, Inc.) diluted in borate saline buffer. Plates were washed and blocked with 0.25% BSA +1% Normal Goat Serum in PBS after overnight incubation. DENV-1 (strain Nauru/West Pac/1974) was captured for 2 h in separate wells, followed by extensive washing. Serially diluted serum samples were plated in duplicate and incubated for 1 h at RT on the captured virus. DENV-specific IgM/IgG/IgA levels were quantified using anti-human IgM HRP (Seracare, 5220-0328), anti-human IgG HRP (Sigma-Aldrich, SAB3701362), and anti-human IgA HRP (Biolegend, 411,002). Secondary antibody binding was quantified using the TMB Microwell Peroxidase Substrate System (KPL, cat. #50-76-00) and Synergy HT plate reader. Single-dilution ELISA was performed using a 1/500 serum dilution for IgM and IgA assays, and a 1/20,000 serum dilution for IgG assays.
2.13. Shotgun mutagenesis epitope mapping
Epitope mapping was performed as outlined previously [53]. Expression constructs for prM/E from DENV-1 (strain Nauru/West Pac/1974) and DENV-2 (strain 16681) were subjected to high-throughput ‘Shotgun Mutagenesis’ to generate comprehensive mutation libraries, with each prM/E polyprotein residue mutated to alanine (and alanine residues to serine), which were arrayed into 384-well plates (one mutation per well). For MAb library screening, plasmids encoding the DENV protein variants were transfected individually into human HEK-293T cells and allowed to express for 22 h before fixing cells in 4% paraformaldehyde (Electron Microscopy Sciences), and permeabilizing with 0.1% (w/v) saponin (Sigma-Aldrich) in PBS plus calcium and magnesium (PBS++). Cells were incubated with purified mAbs (0.1–2.0 µg/mL) diluted in 10% NGS (Sigma)/0.1% saponin, pH 9.0. Before screening, the optimal concentration was determined for each antibody, using an independent immunofluorescence titration curve against wild-type prM/E to ensure that signals were within the linear range of detection and that signal exceeded background by at least 5-fold. Antibodies were detected using 3.75 µg/mL Alexa Fluor 488-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) in 10% NGS/0.1% saponin. Cells were washed three times with PBS++/0.1% saponin followed by 2 washes in PBS. Mean cellular fluorescence was detected using an Intellicyt high throughput flow cytometer (HTFC, Intellicyt). Antibody reactivity against each mutant protein clone was calculated relative to reactivity with wild-type prM/E, by subtracting the signal from mock-transfected controls and normalizing to the signal from wild-type protein-transfected controls. The entire library data for each mAb were compared to the equivalent data from control mAbs. Mutations within clones were identified as critical to the MAb epitope if they did not support reactivity of the test MAb, but supported reactivity of other, control antibodies. This counter-screen strategy facilitates the exclusion of DENV prM/E protein mutants that are mis-folded or have an expression defect.
2.14. Statistical analysis
All statistical analysis was performed using GraphPad Prism 8 Software (GraphPad Software, La Jolla, CA). A P-value < 0.05 was considered significant.
3. Results
3.1. Robust B cell plasmablast circulation occurs following primary and secondary DENV infection
Following introduction into a human host by an infected mosquito during blood meal acquisition, DENV asymptomatically replicates for 3–14 days prior to the onset of measurable viremia or any clinical manifestation of infection (Fig. 1a) [54]. As the precise interval between when an individual is infected with DENV and when viremia/symptoms manifest is highly variable, the day of defervescence (referred here as “fever day 0”) is used as a reference point to standardize the kinetics of infection-associated immunopathology. While viremia generally resolves by fever day 0, lymphocyte activation in the periphery is generally not detectable until just before fever abatement, with maximal T and B cell activation occurring in the subsequent 3–5 days (“early convalescence”; Fig. 1a) [34,[55], [56], [57]]. While this kinetic profile has been described in previous studies of secondary DENV infection, limited data are available regarding the functional diversity of plasmablasts activated in response to both primary and secondary DENV infection. Therefore, to determine if there are any differences in the phenotype and immunoglobulin repertoire of plasmablasts activated in response to either primary or secondary DENV infection, we analyzed PBMC from 6 pediatric patients with serologically confirmed primary or secondary DENV infection.
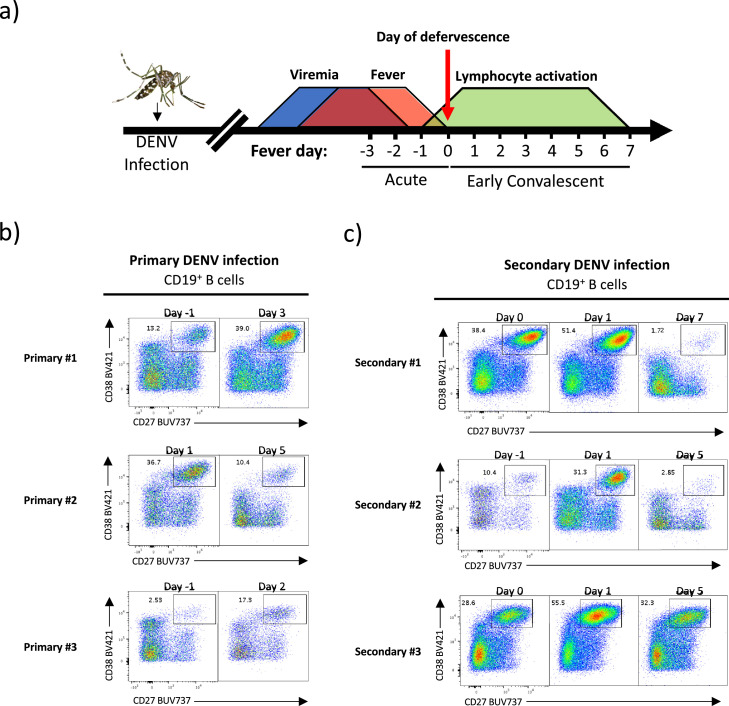
Fig. 1.
Kinetics of B cell plasmablast circulation following natural primary and secondary DENV infection. Plasmablast phenotype B cells (CD19+CD27+CD38+) are evident in circulation following both primary and secondary natural DENV infection. (A) Schematic representation of dengue kinetics and PBMC sample collection. (B) Quantification of plasmablast phenotype B cell frequency in subjects with serologically confirmed primary DENV infections by flow cytometry. (C) Quantification of plasmablast phenotype B cell frequency in subjects with serologically confirmed secondary DENV infection by flow cytometry.
We first analyzed the frequency of circulating plasmablasts during the acute (febrile) and early convalescent period by flow cytometry. Consistent with other previously published reports [34,[55], [56], [57]], robust plasmablast expansion was observed in PBMC of subjects with primary and secondary DENV infections. For individuals with serologically confirmed primary DENV infections, a significant population of plasmablasts were observed at two timepoints per subject (1 acute, early convalescent) (Fig. 1b and Supplemental Fig. 1), and individuals with serologically confirmed secondary DENV infections exhibited significant plasmablast expansion over three time points (2 acute, 1 early convalescent) (Fig. 1c and Supplemental Fig. 1).
3.2. Identification and transcriptional characterization of DENV-elicited B cell plasmablasts by scRNAseq following primary or secondary DENV infection
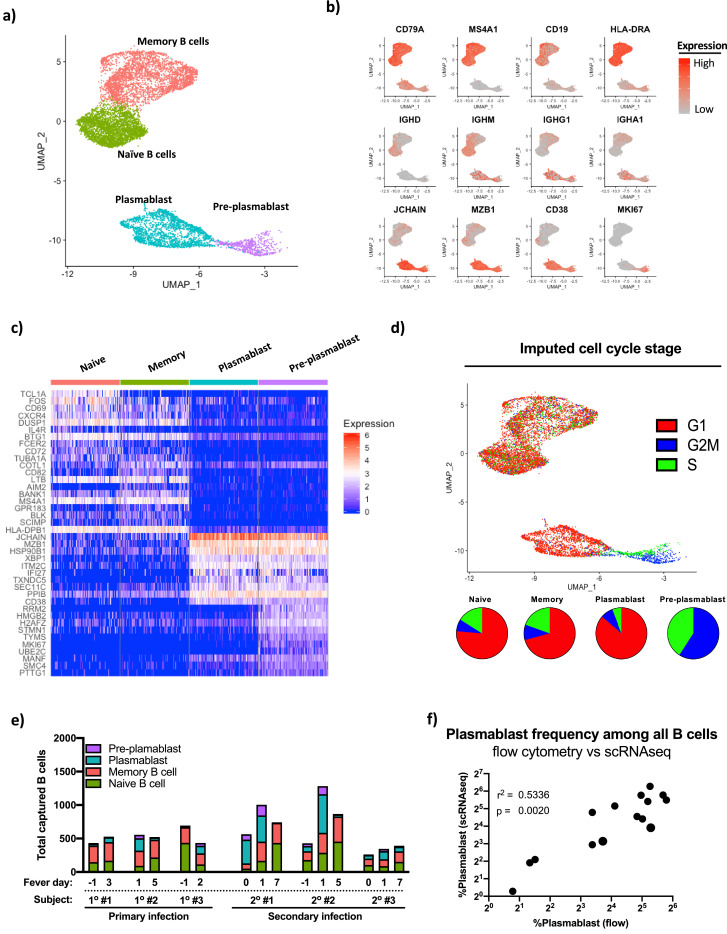
To better understand the clonal and transcriptional diversity of the plasmablast repertoire elicited by primary and secondary DENV infections, we subjected sorted total CD3+ T cells and CD19+ B cells from the samples highlighted in Fig. 1 to single cell RNA sequencing (scRNAseq) analysis using the 10x Genomics platform (Supplemental Fig. 2). Cells were processed so as to recover both the unbiased 5′ gene expression profile from each cell, as well as the paired full-length immunoglobulin sequences utilizing targeted nested PCR. B cells were identified within the parental dataset and extracted for subsequent targeted analysis. Using this approach, we recovered a total of 9,027 B cells with high-quality transcriptomic and immunoreceptor data from the 6 subjects included in our analysis (range, 260–1281 per subject-time point). Four transcriptionally distinct populations of B cells were identified within the total captured B cell pool by unsupervised hierarchical clustering of the cells based on their complete gene expression profile (Fig. 2a). Differential gene expression analysis of these four statistically distinct B cell populations determined that these clusters correspond to canonical naïve B cells, memory B cells, plasmablasts, and pre-plasmablasts (Fig. 2b and c, Supplemental Table 1). Consistent with their ascribed functional potential, transcriptional imputation of the cell-cycle stage of the B cells captured in this dataset revealed that the naïve B cells, memory B cells, and plasmablasts were primarily in the G1 stage of the cell cycle, while pre-plasmablasts were exclusively in G2M/S (Fig. 2d). All four distinct populations of B cells were identified within each subject at each timepoint, with significant population expansion/contraction observed in each subject (Table 1, Fig. 2e). The frequency of plasmablasts as assessed by scRNAseq analysis of sorted CD19+ B cells correlated with the results obtained from flow-cytometry analysis of the same samples (Fig. 2f).
Fig. 2.
Identification, quantification, and transcriptional characterization of B cell subsets by scRNAseq. Conventional B cell subset identification by unsupervised hieratical clustering of scRNAseq data from individuals with primary and secondary DENV infections. (A) UMAP projection of transcriptionally defined B cell populations from all subjects/time points included in this analysis with key populations demarcated. (B) Expression of canonical key B cell lineage genes across all cells in the dataset. (C) Heat map of most differentially expressed gene products as defined by Wilcoxon Log Rank test distinguishing naïve B cells, memory B cells, plasmablasts, and pre-plasmablasts across all subjects and time points. Data are down-sampled to 200 cells/population for visualization. (D) Imputation of cell cycle stage based on the expression of canonical cell-cycle stage associated gene products within naïve B cells, memory B cells, plasmablasts and pre-plasmablasts in the integrated dataset. (E) Abundance of naïve B cells, memory B cells, plasmablasts, and pre-plasmablasts longitudinally within each subject in the study as defined by scRNAseq. (F) Correlation analysis of the frequency of B cell plasmablasts as defined by flow cytometry and scRNAseq for all subjects and time points within the study. r2 and p value calculated by linear regression.
Table 1.
Subject, timepoint, and cell-type information.
| Subject | Age / Sex | Infection | Serotype | Severity | Time point (fever day) | Total B cells | Cell type breakdown |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Naïve | Memory | Plasmablast | Pre-plasmablast | |||||||
| Primary #1 | 9/M | Primary | DENV1 | DF | −1 | 430 | 146 (33.9%) | 246 (57.2%) | 28 (6.5%) | 10 (2.3%) |
| 3 | 525 | 168 (32%) | 278 (52.9%) | 71 (13.5%) | 8 (1.5%) | |||||
| Primary #2 | 9/M | Primary | DENV1 | DF | 1 | 554 | 90 (16.2%) | 228 (41.1%) | 185 (33.4%) | 51 (9.2%) |
| 5 | 521 | 215 (41.2%) | 266 (51.1%) | 33 (6.3%) | 7 (1.3%) | |||||
| Primary #3 | 5/M | Primary | DENV1 | DF | −1 | 693 | 434 (62.6%) | 233 (33.6%) | 22 (3.2%) | 4 (0.5%) |
| 2 | 432 | 112 (25.9%) | 166 (38.4%) | 112 (25.9%) | 42 (9.7%) | |||||
| Secondary #1 | 12/M | Secondary | DENV3 | DHF | 0 | 562 | 49 (8.7%) | 78 (13.9%) | 355 (63.2%) | 80 (14.2%) |
| 1 | 1002 | 165 (16.5%) | 288 (28.7%) | 391 (39%) | 158 (15.8%) | |||||
| 7 | 741 | 432 (58.3%) | 300 (40.5%) | 8 (1.1%) | 1 (0.01%) | |||||
| Secondary #2 | 13/F | Secondary | DENV1 | DHF | −1 | 427 | 178 (41.7%) | 131 (30.7%) | 79 (18.5%) | 39 (9.1%) |
| 1 | 1281 | 285 (22.2%) | 299 (23.3%) | 576 (45%) | 121 (9.4%) | |||||
| 5 | 865 | 452 (52.2%) | 376 (43.5%) | 31 (3.6%) | 6 (0.7%) | |||||
| Secondary #3 | 11/F | Secondary | DENV1 | DF | 0 | 260 | 103 (39.6%) | 96 (36.9%) | 53 (20.4%) | 8 (3.1%) |
| 1 | 344 | 85 (24.7%) | 104 (30.2%) | 121 (35.2%) | 34 (9.9%) | |||||
| 5 | 390 | 150 (38.5%) | 157 (40.2%) | 68 (17.4%) | 15 (3.8%) | |||||
3.3. Isotype distribution and somatic hypermutation burden of immunoglobulins expressed by circulating B cells during primary and secondary natural DENV infection
Using the population identifiers assigned by the transcriptional profile of the B cells captured in our scRNAseq analysis, we assessed the antibody isotype diversity and immunoglobulin somatic hypermutation (SHM) burden of naïve B cells, memory B cells, and plasmablasts/pre-plasmablasts present in individuals experiencing primary or secondary DENV infections. As pre-plasmablasts and plasmablasts are direct clonal relatives and only differ in their stage of the cell cycle, (Fig. 1c and d, Supplemental Table 1) [24,58], these two populations were merged in all subsequent immunoglobulin receptor analysis. To maximize data quality in downstream evaluation, immunoglobulin sequence analysis was restricted to those cells from which a single paired full-length heavy chain and full-length light chain sequence was obtained.
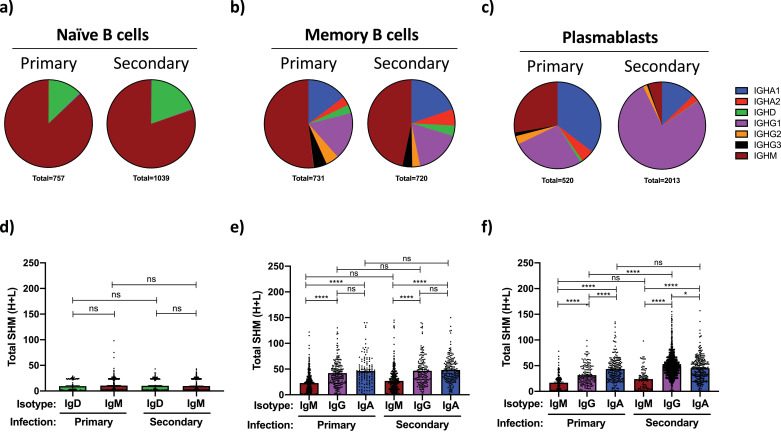
Consistent with their canonical profile, the Ig sequences expressed by transcriptionally-demarcated naïve B cells were observed to be IgD or IgM in isotype with little-or-no SHM away from their germline configuration (Fig. 3a and d). No difference in the isotype distribution or SHM burden was observed within the naïve B cell compartment between individuals with primary versus secondary DENV infection. In contrast, transcriptionally-defined memory B cells were observed to express Igs with a diverse array of isotypes, including IgA1, IgA2, IgG1, IgG2, IgG3, IgM, and to a lesser extent IgD (Fig. 3b). However, no difference in the isotype distribution of the Igs expressed by memory B cells was observed in PBMC from individuals with primary versus secondary DENV infection. The SHM burden of the IgM, IgG, and IgA immunoglobulins expressed by memory B cells were higher than those observed in naïve B cells, with IgG and IgA class-switched memory B cells having a significantly higher SHM burden than their IgM expressing counterparts (Fig. 3e). No difference in the SHM burden was observed in the IgM, IgG, and IgA expressing memory B cell compartment between individuals with primary versus secondary DENV infection.
Fig. 3.
Isotype distribution and somatic hypermutation burden of immunoglobulins expressed by circulating B cells during primary and secondary natural DENV infections. Immunoglobulin sequence analysis was restricted to cells from which a full-length heavy/light chain pair was successfully isolated and annotated by scRNAseq. (A) Isotype distribution of immunoglobulins expressed by transcriptionally defined naïve B cells within all subjects at all time points captured in the dataset. (B) Isotype distribution of immunoglobulins expressed by transcriptionally defined memory B cells within all subjects in all time points captured in the dataset. (C) Isotype distribution of immunoglobulins expressed by transcriptionally defined plasmablast phenotype B cells within all subjects at all time points captured in the dataset. Plasmablasts and pre-plasmablasts were merged for this analysis. (D) Total somatic hypermutation (SHM) burden of immunoglobulins expressed by naïve B cells from all subjects at all time points captured in this analysis. Analysis split by isotype. (E) Total SHM burden of immunoglobulins expressed by memory B cells from all subjects at all time points captured in this analysis. Analysis split by isotype. (F) Total SHM burden of immunoglobulins expressed by plasmablast phenotype B cells from all subjects at all time points captured in this analysis. Analysis split by isotype. Error bars +/- SEM. * p < 0.05, **** p < 0.0001, one-way ANOVA.
In contrast to the modest differences in isotype distribution and SHM burden observed in naïve and memory B cells identified in individuals with primary versus secondary DENV infections, significant differences were observed within the corresponding plasmablast compartments. Plasmablasts identified in individuals with primary DENV infections had nearly equal ratios of IgA, IgG and IgM expressing cells, while individuals experiencing a secondary DENV infection exhibited a significant IgG1 bias within the plasmablast compartment, with only modest IgA or IgM representation (Fig. 3c and Supplemental Fig. 3, Table 2). No difference was observed in the SHM burden of the IgM expressing plasmablasts between individuals with either primary or secondary DENV infections, while IgG expressing plasmablasts from individuals with a secondary DENV infection exhibited a significantly higher SHM burden than their counterparts from individuals with primary DENV infections (Fig. 3f and Supplemental Fig. 3). Surprisingly, the IgA class-switched plasmablasts in both primary and secondary DENV infections exhibited an equally high SHM burden, with the accumulation of mutations following similar patterns in both the framework regions and CDRs of the antibodies (Fig. 3f and Supplemental Figs. 3 and 4). This is despite the fact that by all other metrics the individuals classified as experiencing a primary DENV infection have never encountered DENV antigen previously. Therefore, in addition to the anticipated differences in IgM and IgG expressing plasmablasts, we contend that primary DENV infection can be characterized by a high relative proportion of extensively hypermutated IgA expressing plasmablasts. Clonal expansion was evident in the both the IgG and IgA expressing plasmablast compartments from subjects with either primary or secondary DENV infection (Supplemental Fig. 5), but no discernable trend or enrichment in Ig gene segment usage was evident (Supplemental Fig. 6).
Table 2.
Plasmablast isotype breakdown.
| Subject | Time point (fever day) | Total number of plasmablast | # paired heavy/light | % recovery | Plasmablast isotype distribution (fully annotated) |
||
|---|---|---|---|---|---|---|---|
| IgM | IgG | IgA | |||||
| Primary #1 | −1 | 38 | 35 | 92.11% | 9 (26.5%) | 4 (11.8%) | 21 (61.8%) |
| 3 | 79 | 73 | 92.41% | 6 (8.3%) | 22 (30.6%) | 44 (61.1%) | |
| Primary #2 | 1 | 236 | 222 | 94.07% | 82 (36.9%) | 55 (24.8%) | 85 (38.3%) |
| 5 | 40 | 36 | 90.00% | 5 (13.9%) | 24 (66.7%) | 7 (19.4%) | |
| Primary #3 | −1 | 26 | 21 | 80.77% | 2 (10.5%) | 4 (21.1%) | 13 (68.4%) |
| 2 | 154 | 133 | 86.36% | 39 (29.8% | 53 (40.4%) | 39 (29.8%) | |
| Secondary #1 | 0 | 435 | 396 | 91.03% | 12 (3%) | 352 (89.1%) | 31 (7.8%) |
| 1 | 549 | 494 | 89.98% | 10 (2%) | 428 (86.8%) | 55 (11.2%) | |
| 7 | 9 | 8 | 88.89% | 3 (37.5%) | 5 (62.5%) | 0 (0%) | |
| Secondary #2 | −1 | 118 | 104 | 88.14% | 3 (2.9%) | 61 (59.2%) | 39 (37.9%) |
| 1 | 697 | 642 | 92.11% | 17 (2.7%) | 560 (87.4%) | 64 (10%) | |
| 5 | 37 | 33 | 89.19% | 6 (18.8%) | 6 (18.8%) | 20 (62.5%) | |
| Secondary #3 | 0 | 61 | 54 | 88.52% | 14 (25.9%) | 22 (40.7%) | 18 (33.3%) |
| 1 | 155 | 141 | 90.97% | 18 (12.8%) | 82 (58.2%) | 41 (29.1%) | |
| 5 | 83 | 75 | 90.36% | 11 (14.7%) | 40 (53.3%) | 24 (32%) | |
3.4. Functional characterization of DENV-elicited plasmablast immunoglobulins
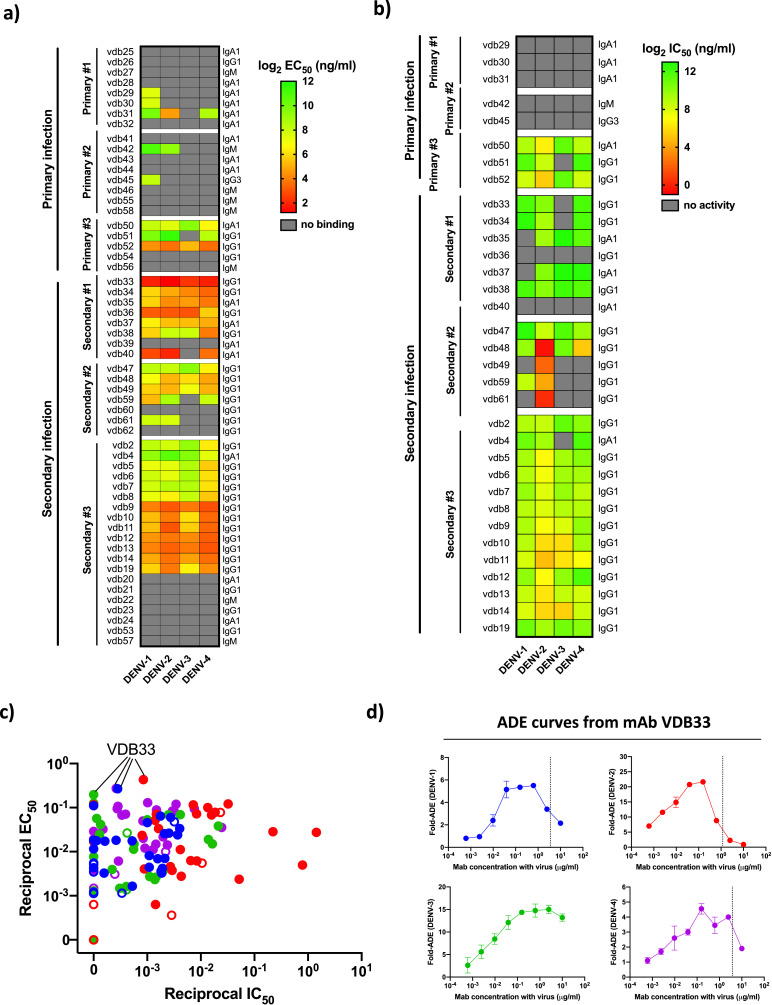
To determine the antigen specificity and functional potential of the Igs expressed by DENV-elicited plasmablasts captured in our scRNAseq dataset – especially IgA class-switched cells - we synthesized 5–20 monoclonal antibodies (mAb) from each subject included in our analysis. We selected Ig sequences based on the clonal abundance of the sequence and the amount of the Ig transcript expressed within the corresponding plasmablast (> 1000 copies/cell). The only exception to these criteria were VDB-5, -6,- 7, and -8, which were selected based on their clonal relationship to VDB-2. Using these benchmarks, we selected and synthesized a total of 56 mAbs from DENV-elicited plasmablasts: 21 from individuals with primary DENV infection and 35 from individuals with secondary DENV infection (Supplemental Table 2). Within the selected pool of mAb candidates were antibodies with parental IgM, IgG1, IgG3, and IgA1 isotypes. The mAbs were screened for the ability to bind DENV-1, -2, -3 and -4 by virus-capture ELISA. In agreement with previous results obtained from individuals with experimental primary DENV infection and subjects with natural secondary infections, 38% (8/21) of the mAbs generated from individuals with a primary DENV infection exhibited DENV reactivity, while 71% (25/35) of mAbs from secondary infection plasmablasts exhibited DENV reactivity (Fig. 4a, Supplemental Table 3, Supplemental Fig. 7). The mAbs from secondary DENV infection exhibited significant serotype cross-reactivity, while moderate bias towards the infecting serotype was observed in mAbs from primary infections (Fig. 4a). Notably, 47% (8/17) of the mAbs synthesized from antibodies with a parental IgA isotype were DENV reactive. This is a slightly lower rate than what was observed for IgG1 mAbs (76.6%, 23/30), but indicates that a significant fraction of the IgA expressing plasmablasts found in circulation following acute DENV infection are indeed virus-reactive.
Fig. 4.
DENV-elicited plasmablast-derived monoclonal antibody specificity and functional activity. (A) Heat map summary of the specificity and avidity of 61 mAbs derived from immunoglobulin sequences extracted from the scRNAseq analysis of plasmablast-phenotype B cells from individuals experiencing primary or secondary natural DENV infections. (B) Heat map summary of the neutralization potential (IC50) of 33 mAbs derived from immunoglobulin sequences extracted from the scRNAseq analysis of plasmablast-phenotype B cells from individuals experiencing primary or secondary natural DENV infections. Only mAbs exhibiting DENV specificity by ELISA (EC50 < 5μg/ml) were assessed by FlowNT. (C) Scatter plot of reciprocal DENV-1, −2, −3, and −4 IC50/EC50 values calculated for all plasmablast-derived mAbs generated in this study which exhibited an EC50 value of <5 μg/ml for at least one viral serotype. Symbol color indicates the virus serotype tested (DENV-1 in blue, DENV-2 in red, DENV-3 in green, DENV-4 in purple). Symbol fill indicates if the antibody was isolated from an individual with a primary (open) or secondary (closed) DENV infection. (D) ADE potential of mAb VDB33 against DENV-1, −2, −3, and −4. All values are shown as fold-increased infection compared to no antibody. Dashed line indicates the mAb IC50 as determined by FlowNT using the same viral preparation as used in the ADE assay. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Any mAb exhibiting DENV binding ability was subsequently tested for DENV neutralization potential using a flow-cytometry based microneutralization assay (Fig. 4b, Supplemental Table 3, Supplemental Fig. 7). mAbs from individuals experiencing primary DENV infections exhibited relatively poor neutralization potential, with only 37.5% (3/8) DENV reactive mAbs exhibiting any neutralization activity. In contrast, 92% (23/25) of the DENV-reactive mAbs generated from individuals experiencing a secondary DENV infection exhibited viral neutralization, with 60% (15/25) capable of neutralizing all 4 DENV serotypes to some degree. Of note, mAbs generated from individuals experiencing secondary DENV infections often exhibited better binding and neutralization activity towards serotypes other than the infecting virus at the time of sample collection, potentially providing insight into the virologic exposure history of the individual.
While the neutralization potential of a DENV-reactive mAb provides some indication of its theoretical in vivo efficacy, the ratio between neutralization potential and avidity is an important secondary consideration. Antibodies requiring high viral occupancy in order to neutralize viral infectivity – reflected in a high EC50/IC50 ratio – are potentially capable of facilitating ADE in situations where they are present at a concentration where they can bind a virus but not neutralize it if they possess an Fc domain capable of interacting with Fc receptors expressed by DENV-susceptible leukocytes. Indeed, while some relationship is apparent between the EC50 and IC50 values of the mAbs analyzed (Fig. 4c), many mAbs sit above a theoretical linear line of identity for some DENV serotypes, and therefore may be capable of facilitating ADE if they possessed an appropriate Fc domain. Of particular note, mAb VDB33 demonstrated a uniquely high EC50/IC50 ratio, as it potently binds all four DENV serotypes (EC50 2.32–5.07 ng/ml) while showing poor neutralization potential (IC50 1181 to >10,000 ng/ml). Shotgun mutagenesis epitope mapping revealed that this mAb bound the fusion loop of the DENV E protein, the canonical binding site for antibodies thought to facilitate ADE (Fig. 6a). Indeed, when tested in an in vitro ADE assay, VDB33 exhibited dramatic ADE potential, especially against DENV3 (Fig. 4d). This is notable, as the individual from which VDB33 was obtained was experiencing a secondary DENV-3 infection (resulting in DHF) at the time of sample collection. A contemporaneous serum sample from this individual exhibited a very similar ADE profile as VDB33 (Supplemental Fig. 8). While it is not possible to draw a direct causal connection between this plasmablast-derived antibody (or its relatives) and the clinical manifestation of DENV infection observed in the individual from which it was isolated using these data, these findings underscore the utility of assessing the molecular level detail of DENV-elicited humoral immunity in future studies.
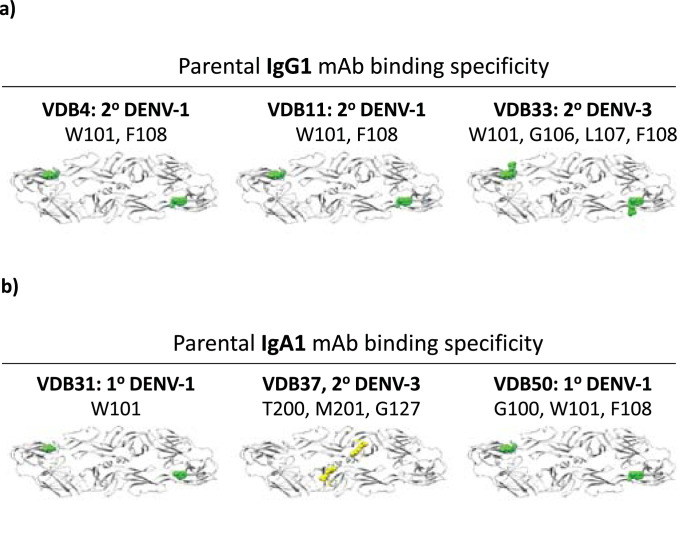
Fig. 6.
Binding specificity of plasmablast-derived/DENV-reactive monoclonal antibodies. (A) Shotgun mutagenesis epitope mapping data from IgG1-derived mAbs. (B) Shotgun mutagenesis epitope mapping data from IgA1-derived mAbs. Color scheme for indicated residues: green = fusion loop, yellow = domain II hinge region. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. DENV-reactive serum immunoglobulin isotype diversity in primary and secondary DENV infection
To determine if the DENV-elicited immunoglobulin isotype diversity observed in the plasmablast compartment was reflected in the contemporaneous antibodies in circulation, we performed DENV-capture ELISA on serum samples from individuals with either primary or secondary DENV infection and quantified the relative proportions of DENV-reactive IgM, IgG, and IgA present therein. To provide temporal resolution to the analysis, four time-points per subject were analyzed: two samples during acute viral infection (acute 1, acute 2), one sample obtained 3–7 days following fever abatement (early-convalescent), and one sample collected 6 months post infection (Fig. 5, Supplemental Table 4).
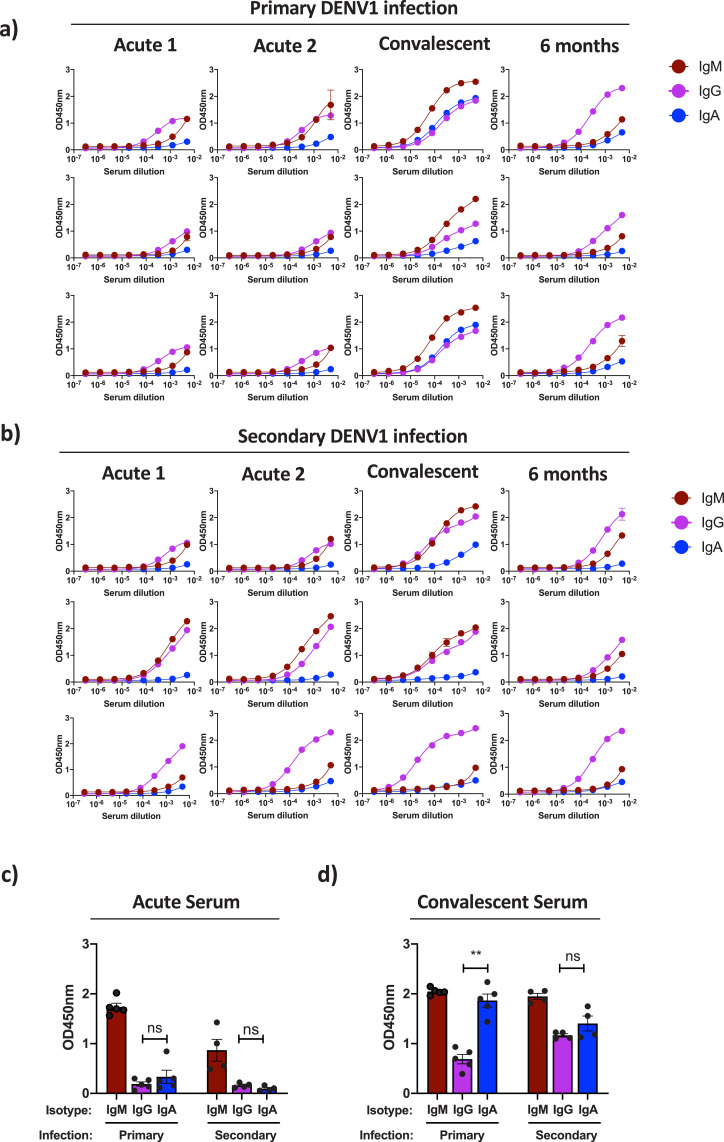
Fig. 5.
Kinetics and magnitude of DENV-reactive IgM/IgG/IgA in circulation following primary or secondary DENV infection. (A) Limiting dilution DENV-1 capture ELISA analysis of relative virus-reactive IgG, IgA, IgM levels present in serum obtained from three subjects with serologically- and PCR-confirmed primary DENV-1 infections. (B) Limiting dilution DENV-1 capture ELISA analysis of relative virus-reactive IgG, IgA, IgM levels present in serum obtained from three subjects with serologically- and PCR-confirmed secondary DENV-1 infections. (C) Single-dilution DENV-1 capture ELISA analysis of the relative virus-reactive IgG, IgA, IgM levels present in acute infection serum samples obtained from subjects with serologically and PCR confirmed primary (n = 5) and secondary (n = 4) DENV-1 infections. (D) Single-dilution DENV-1 capture ELISA analysis of the relative virus-reactive IgG, IgA, IgM levels present in early-convalescent serum samples obtained from subjects with serologically and PCR confirmed primary and secondary DENV-1 infections. Error bars indicate +/- SEM. ** p < 0.01, one-way ANOVA.
Relatively modest DENV-reactive Ig levels were observed in acute serum samples obtained from individuals with either primary (Fig. 5a) or secondary (Fig. 5b) DENV-1 infections. However, a dramatic increase in circulating DENV-reactive Igs was observed in early-convalescent serum samples from both primary and secondary DENV infections (Fig. 5a and b). Notably, while DENV-reactive IgA was detectable in all subjects at this time point, IgA represented a much higher fraction of the total DENV-reactive Ig pool in individuals experiencing a primary DENV infection, while IgG production dominated in secondary DENV-1 infections (Fig. 5a and b). Total DENV-reactive Ig titers returned to more modest levels in all subjects by 6 months post infection, with little DENV-reactive IgA observed (Fig. 5a and b). Therefore, despite the fact that DENV-reactive IgA represent an appreciable fraction of the total DENV-reactive Ig in circulation just following the resolution of infection – especially in individuals experiencing a primary DENV infection - DENV-reactive IgA does not appear to persist in circulation.
To further validate these observations, single-dilution ELISA analysis was performed on acute and convalescent serum samples from an additional 9 individuals experiencing either a primary or secondary DENV-1 infection (Supplemental Table 5). As was observed in the limiting-dilution ELISA analysis, relatively little DENV-reactive IgG or IgA was observed in acute infection time points from subjects with either primary or secondary infection (Fig. 5c). However, the DENV-reactive IgG and IgA signals increased dramatically in the corresponding early convalescent serum samples, with IgA comprising a much higher fraction of the total DENV-reactive Ig signal in samples from primary DENV-1 infections than from samples from secondary infections (Fig. 5d). These data confirm that DENV-reactive IgA represents a significant fraction of the total DENV-reactive Ig pool following the resolution of acute infection, especially in individuals experiencing a primary DENV infection.
3.6. Epitope specificity of plasmablast-derived/DENV-reactive mAbs
In light of the unexpectedly high frequency of DENV-reactive IgA clones observed in our dataset and the pervasiveness of DENV-reactive IgA in contemporaneous serum samples– especially in individuals experiencing a primary DENV infection - we endeavored to determine if DENV-reactive IgA Igs recognize an unconventional epitope on DENV or if their antigen specificity mirrored that of other “conventional” DENV-reactive antibody isotypes. To this end, we utilized shotgun mutagenesis epitope mapping of prM/E from DENV-1 (strain Nauru/West Pac/1974) and DENV-2 (strain 16681) to determine the binding specificity of six parentally IgG1 and IgA1 mAbs identified in subjects with either primary or secondary DENV infections.
The three parentally IgG1 DENV-reactive mAbs picked for epitope mapping were found to exclusively interact with the fusion loop region of the DENV E protein (Fig. 6a, Supplemental Table 6). Three parentally IgA mAbs generated from subjects with primary or secondary DENV infections were observed to either bind the DENV E protein fusion loop (VDB31/VDB50) or the DENV E protein domain II hinge region (VDB37) (Fig. 6b, Supplemental Table 6). These results indicate that DENV-reactive IgA antibodies are capable of recognizing multiple distinct epitopes on the DENV E protein even in individuals who have not previously encountered DENV, and that DENV-reactive IgG and IgA isotype antibodies can overlap in their epitope specificity.
4. Discussion
In this study, we utilized single cell RNA sequencing technology to assess the clonal and transcriptional diversity of plasmablasts elicited in response to primary and secondary DENV infections in an unbiased and high-throughput fashion. We found that an unexpectedly high proportion of DENV-elicited plasmablasts expressed IgA Igs, notably in individuals experiencing a primary DENV infection. These IgA immunoglobulins were extensively hypermutated, even in individuals with serologically confirmed primary DENV infection. Utilizing a combination of conventional biochemical assays and high-throughput shotgun mutagenesis, we determined that DENV-reactive IgA antibodies represented a significant fraction of DENV-reactive Igs generated in response to DENV infection, and that they have a overlapping epitope specificity to “conventional” DENV-reactive IgM and IgG class-switched antibodies reported in the literature [36,38,59,60]. These results provide insight into the molecular-level diversity of DENV-elicited humoral immunity, and suggest a hitherto unappreciated role for DENV-reactive IgA in the humoral response to DENV infection, especially in the setting of primary DENV infection.
In addition to revealing a potential role for DENV-reactive IgA in the setting of acute DENV infection, the data presented in this study provide molecular-level resolution and reinforcement to several fundamental features of DENV-elicited humoral immunity. This includes the observations that the majority of the individual antibody clones elicited by a primary DENV infection exhibit relatively poor neutralization activity, and the serotype cross-reactive humoral immune profile associated with secondary DENV infections extends to the level of individual antibody clones. Our findings must be considered in light of several limitations, however. These data were generated by the analysis of samples obtained from a small number of children, most of whom were experiencing DENV-1 infections. Although viral tropism and clinical manifestations of the different DENV serotypes are thought to be similar, some caution must be used in extrapolating these observations to other age groups and to other DENV serotypes. Immunological history and host/pathogen genetics both play significant roles in determining the pathogenesis and cellular/molecular level response to DENV infection [61]. However, tools such as scRNAseq and other high-throughput/high-resolution modalities can now reliably capture many of these complex variables, hopefully providing better insight and more actionable information in the setting of acute DENV infection.
The induction of virus specific IgA in response to acute infection is not an uncommon phenomenon, especially for viruses that exhibit significant mucosal tropism [62]. The unique cytokine environment found in mucosal tissues is thought to provide the necessary immunological cues to nascently activated B cells to facilitate the IgA1 antibody class-switch event [63]. However, DENV has no known mucosal involvement either during transmission or replication. DENV-reactive IgA generated in response to acute infection is not without precedent, although much of the previous work on IgA in the context of DENV infection has focused on the feasibility of utilizing urine/saliva restricted DENV-reactive IgA as a non-invasive diagnostic [64], [65], [66], [67]. A single paper has compared directly the titers of IgA in primary and secondary DENV infections [67], and no previous study has assessed the prevalence of IgA class-switched plasmablasts present following acute DENV infection. The paucity of previous data on the abundance and specificity of DENV-specific IgA following acute viral infection is partially attributable to the relatively brief window during which significant levels of these DENV-reactive IgA antibodies are present in serum following primary DENV infection (only in early convalescent serum samples in our study). In addition, the majority of previous studies assessing the molecular diversity of DENV-elicited humoral immunity also intentionally limited their analysis to include IgG and IgM antibodies proteins and/or genes [32,[35], [36], [37]].
Although the precise role – if any - that DENV-elicited IgA plays in influencing the immunopathogenesis of DENV is still unclear, it is noteworthy that while we have demonstrated that the antigen-binding domains of DENV-elicted IgA clones can effectively neutralize DENV infectivity, the restricted expression profile of IgA Fc receptors implies that DENV-reactive IgA clones cannot easily facilitate classical antibody-dependent immune enhancement of DENV infection. Therefore, much like IgM, DENV-reactive IgA might contribute neutralizing activity to DENV-reactive serum, while antagonizing IgG1 mediated ADE. As the average neutralization potential of the DENV-reactive IgA clones tested in this study varied widely, the ability of these clones to neutralize DENV in a complex humoral environment may be limited, even if these antibodies were present at an equimolar ratio as their IgG counterparts during the resolution phase of a DENV infection. However, the epitope mapping data presented herein suggests that at least some DENV-reactive IgA clones are capable of binding the same epitopes as their IgG1 counterparts. Therefore, it is possible that DENV-specific IgA may limit intra-host viral propagation during the resolution phase of an acute DENV infection by competing with infection-enhancing DENV-specific IgG and preventing internalization of infectious virions through IgG Fc receptors. This scenario does not necessitate that an IgA opsonized virus is less infectious than a un-opsonized virus in the context of Fc-receptor independent infection, but it does suggest that an IgA opsonized virus might be less infectious than an IgG1 opsonized virus from the standpoint of a cell expressing an IgG1-reactive Fc receptor.
In light of the consistently high SHM burden observed in IgA expressing plasmablasts circulating in response to primary DENV infection, we additionally propose that the majority of these cells are reactivated memory B cells that were initially stimulated by an antigen other than DENV. Memory B cells have both a numerical and functional advantage over naïve B cells upon antigen engagement, and a relatively small population of memory B cells that happen to cross-react with DENV in addition to their “cognate” antigen could certainly be expected to contribute to a primary response to infection. Furthermore, it is perhaps unsurprising that a small number of IgA class-switched memory B cells with some degree of DENV cross reactivity exist in any individual prior to a primary DENV infection, as it has been suggested that more than 70% of the total antibody production in humans is IgA in isotype, albeit mostly at mucosal surfaces [68]. Therefore, it is possible that all primary pathogen infections include a significant IgA component, as the largest pool of pre-existing memory B cells that is subsequently capable of responding/cross-reacting with a “novel” antigen would be IgA in isotype. This interpretation follows the concept of “original antigen sin”, wherein a previous infection has a direct impact on the specificity and efficacy of immunity generated in response to a subsequent infection/challenge, but extends the concept to primary DENV infection [69]. This indelible feature of pre-existing immunity can also be observed in our data from secondary DENV infections, wherein antibody specificity is occasionally biased towards DENV serotypes other than the infecting type. These data suggest that immunological memory generated in response to completely different stimuli may impact future immune responses simply because of putative antigenic similarity.
Declaration of Competing Interest
A.T.W reports grants from Military Infectious Disease Research Program, during the conduct of the study.
B.J.D, E.D., M.E.F, A.G., J.L., and A.L.R. reports grants from National Institute of Allergy and Infectious Diseases, during the conduct of the study.
B.J.D. reports other support from Integral Molecular, outside the submitted work.
S.J.T reports other support from US DoD, other support from GSK, during the conduct of the study; personal fees and other support from GSK Vaccines, personal fees and other support from Takeda, personal fees and other support from Merck, personal fees and other support from PrimeVax, personal fees and other support from Themisbio, personal fees and other support from Chugai Pharma, personal fees and other support from Cormac Life Sciences, personal fees and other support from HHS NVPO / Tunnel Govt Services, personal fees and other support from Janssen, other support from GreenMark Partners, personal fees from Tremeau Pharma, outside the submitted work; In addition, Dr. Thomas has a patent US10086061B2 (combined flavivirus vaccines) issued.
All other authors have nothing to disclose.
Acknowledgments
Acknowledgements
None.
Funding
This work was supported by the Military Infectious Disease Research Program (MIDRP), the Congressionally Directed Medical Research Program (CDMRP), and the National Institutes of Allergy and Infectious Disease (NIAID, P01AI034533, Rothman). The epitope mapping analysis was supported by the NIH contract HHSN272201400058C (BJD). These funders had any no role in study design, data collection, data analysis, interpretation, or writing of this report.
Data availability
The authors declare that all data supporting the findings of this study are available within this article and its Supplementary Information files, or from the corresponding author upon reasonable request. Single-cell RNAseq gene expression data have been deposited in the Gene Expression Omnibus database (GSE145307).
Disclaimer
The opinions or assertions contained herein are the private views of the authors and are not to be construed as reflecting the official views of the US Army or the US Department of Defense, or the National Institutes of Health. Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70–25.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102733.
Appendix. Supplementary materials
References
- 1.Gubler D.J. Aedes aegypti and aedes aegypti-borne disease control in the 1990s: top down or bottom up. Charles Franklin Craig lecture. Am J Trop Med Hyg. 1989;40(6):571–578. doi: 10.4269/ajtmh.1989.40.571. [DOI] [PubMed] [Google Scholar]
- 2.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepard D.S., Coudeville L., Halasa Y.A., Zambrano B., Dayan G.H. Economic impact of dengue illness in the Americas. Am J Trop Med Hyg. 2011;84(2):200–207. doi: 10.4269/ajtmh.2011.10-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guzman M.G., Harris E. Dengue. Lancet. 2015;385(9966):453–465. doi: 10.1016/S0140-6736(14)60572-9. [DOI] [PubMed] [Google Scholar]
- 5.Sangkawibha N., Rojanasuphot S., Ahandrik S., Viriyapongse S., Jatanasen S., Salitul V. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol. 1984;120(5):653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 6.Thein S., Aung M.M., Shwe T.N., Aye M., Zaw A., Aye K. Risk factors in dengue shock syndrome. Am J Trop Med Hyg. 1997;56(5):566–572. doi: 10.4269/ajtmh.1997.56.566. [DOI] [PubMed] [Google Scholar]
- 7.Yoon I.K., Srikiatkhachorn A., Hermann L., Buddhari D., Scott T.W., Jarman R.G. Characteristics of mild dengue virus infection in Thai children. Am J Trop Med Hyg. 2013;89(6):1081–1087. doi: 10.4269/ajtmh.13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke D.S., Nisalak A., Johnson D.E., Scott R.M. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38(1):172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 9.Rothman A.L. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. 2011;11(8):532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 10.Lan N.T., Hirayama K. Host genetic susceptibility to severe dengue infection. Trop Med Health. 2011;39(4 Suppl):73–81. doi: 10.2149/tmh.2011-S08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katzelnick L.C., Gresh L., Halloran M.E., Mercado J.C., Kuan G., Gordon A. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358(6365):929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halstead S.B. Immune enhancement of viral infection. Prog Allergy. 1982;31:301–364. [PubMed] [Google Scholar]
- 13.Halstead S.B. Dengue: hematologic aspects. Semin Hematol. 1982;19(2):116–131. [PubMed] [Google Scholar]
- 14.Kliks S.C., Nisalak A., Brandt W.E., Wahl L., Burke D.S. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. Am J Trop Med Hyg. 1989;40(4):444–451. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- 15.Zellweger R.M., Prestwood T.R., Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe. 2010;7(2):128–139. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang T.T., Sewatanon J., Memoli M.J., Wrammert J., Bournazos S., Bhaumik S.K. IgG antibodies to dengue enhanced for FcgammaRIIIA binding determine disease severity. Science. 2017;355(6323):395–398. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kou Z., Lim J.Y., Beltramello M., Quinn M., Chen H., Liu S. Human antibodies against dengue enhance dengue viral infectivity without suppressing type i interferon secretion in primary human monocytes. Virology. 2011;410(1):240–247. doi: 10.1016/j.virol.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Blackley S., Kou Z., Chen H., Quinn M., Rose R.C., Schlesinger J.J. Primary human splenic macrophages, but not t or b cells, are the principal target cells for dengue virus infection in vitro. J Virol. 2007;81(24):13325–13334. doi: 10.1128/JVI.01568-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodenhuis-Zybert I.A., Moesker B., da Silva Voorham J.M., van der Ende-Metselaar H., Diamond M.S., Wilschut J. A fusion-loop antibody enhances the infectious properties of immature flavivirus particles. J Virol. 2011;85(22):11800–11808. doi: 10.1128/JVI.05237-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halstead S.B. Dengue antibody-dependent enhancement: knowns and unknowns. Microbiol Spectr. 2014;2(6):1–18. doi: 10.1128/microbiolspec.AID-0022-2014. [DOI] [PubMed] [Google Scholar]
- 21.Halstead S.B., Venkateshan C.N., Gentry M.K., Larsen L.K. Heterogeneity of infection enhancement of dengue 2 strains by monoclonal antibodies. J Immunol. 1984;132(3):1529–1532. [PubMed] [Google Scholar]
- 22.Dejnirattisai W., Jumnainsong A., Onsirisakul N., Fitton P., Vasanawathana S., Limpitikul W. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328(5979):745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shlomchik M.J., Weisel F. Germinal center selection and the development of memory b and plasma cells. Immunol Rev. 2012;247(1):52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 24.Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15(3):160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 25.Radbruch A., Muehlinghaus G., Luger E.O., Inamine A., Smith K.G., Dorner T. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6(10):741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida T., Mei H., Dorner T., Hiepe F., Radbruch A., Fillatreau S. Memory b and memory plasma cells. Immunol Rev. 2010;237(1):117–139. doi: 10.1111/j.1600-065X.2010.00938.x. [DOI] [PubMed] [Google Scholar]
- 27.Benner R., Hijmans W., Haaijman J.J. The bone marrow: the major source of serum immunoglobulins, but still a neglected site of antibody formation. Clin Exp Immunol. 1981;46(1):1–8. [PMC free article] [PubMed] [Google Scholar]
- 28.Slifka M.K., Matloubian M., Ahmed R. Bone marrow is a major site of long-term antibody production after acute viral infection. J Virol. 1995;69(3):1895–1902. doi: 10.1128/jvi.69.3.1895-1902.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu V.T., Berek C. The establishment of the plasma cell survival niche in the bone marrow. Immunol Rev. 2013;251(1):177–188. doi: 10.1111/imr.12011. [DOI] [PubMed] [Google Scholar]
- 30.Nakaya H.I., Wrammert J., Lee E.K., Racioppi L., Marie-Kunze S., Haining W.N. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson K.J., Liu Y., Roskin K.M., Glanville J., Hoh R.A., Seo K. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16(1):105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wrammert J., Onlamoon N., Akondy R.S., Perng G.C., Polsrila K., Chandele A. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J Virol. 2012;86(6):2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.] Balakrishnan T., Bela-Ong D.B., Toh Y.X., Flamand M., Devi S., Koh M.B. Dengue virus activates polyreactive, natural IgG B cells after primary and secondary infection. PLoS ONE. 2011;6(12):e29430. doi: 10.1371/journal.pone.0029430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haltaufderhyde K., Srikiatkhachorn A., Green S., Macareo L., Park S., Kalayanarooj S. Activation of peripheral T follicular helper cells during acute dengue virus infection. J Infect Dis. 2018;218(10):1675–1685. doi: 10.1093/infdis/jiy360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Appanna R., Kg S., Xu M.H., Toh Y.X., Velumani S., Carbajo D. Plasmablasts during acute dengue infection represent a small subset of a broader virus-specific memory B cell pool. EBioMedicine. 2016;12:178–188. doi: 10.1016/j.ebiom.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu M., Hadinoto V., Appanna R., Joensson K., Toh Y.X., Balakrishnan T. Plasmablasts generated during repeated dengue infection are virus glycoprotein-specific and bind to multiple virus serotypes. J Immunol. 2012;189(12):5877–5885. doi: 10.4049/jimmunol.1201688. [DOI] [PubMed] [Google Scholar]
- 37.Priyamvada L., Cho A., Onlamoon N., Zheng N.Y., Huang M., Kovalenkov Y. B cell responses during secondary dengue virus infection are dominated by highly cross-reactive, memory-derived plasmablasts. J Virol. 2016;90(12):5574–5585. doi: 10.1128/JVI.03203-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nivarthi U.K., Tu H.A., Delacruz M.J., Swanstrom J., Patel B., Durbin A.P. Longitudinal analysis of acute and convalescent b cell responses in a human primary dengue serotype 2 infection model. EBioMedicine. 2019;41:465–478. doi: 10.1016/j.ebiom.2019.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Alwis R., Beltramello M., Messer W.B., Sukupolvi-Petty S., Wahala W.M., Kraus A. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl Trop Dis. 2011;5(6):e1188. doi: 10.1371/journal.pntd.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabin A.B. Research on dengue during World War II. Am J Trop Med Hyg. 1952;1(1):30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- 41.Vaughn D.W., Green S., Kalayanarooj S., Innis B.L., Nimmannitya S., Suntayakorn S. Dengue in the early febrile phase: viremia and antibody responses. J Infect Dis. 1997;176(2):322–330. doi: 10.1086/514048. [DOI] [PubMed] [Google Scholar]
- 42.Kalayanarooj S., Vaughn D.W., Nimmannitya S., Green S., Suntayakorn S., Kunentrasai N. Early clinical and laboratory indicators of acute dengue illness. J Infect Dis. 1997;176(2):313–321. doi: 10.1086/514047. [DOI] [PubMed] [Google Scholar]
- 43.Libraty D.H., Endy T.P., Houng H.S., Green S., Kalayanarooj S., Suntayakorn S. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J Infect Dis. 2002;185(9):1213–1221. doi: 10.1086/340365. [DOI] [PubMed] [Google Scholar]
- 44.Innis B.L., Nisalak A., Nimmannitya S., Kusalerdchariya S., Chongswasdi V., Suntayakorn S. An enzyme-linked immunosorbent assay to characterize dengue infections where dengue and Japanese encephalitis co-circulate. Am J Trop Med Hyg. 1989;40(4):418–427. doi: 10.4269/ajtmh.1989.40.418. [DOI] [PubMed] [Google Scholar]
- 45.Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee D.W., Khavrutskii I.V., Wallqvist A., Bavari S., Cooper C.L., Chaudhury S. BRILIA: integrated tool for high-throughput annotation and lineage tree assembly of B-cell repertoires. Front Immunol. 2016;7:681. doi: 10.3389/fimmu.2016.00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aouinti S., Giudicelli V., Duroux P., Malouche D., Kossida S., Lefranc M.P. IMGT/StatClonotype for pairwise evaluation and visualization of NGS IG and TR IMGT clonotype (AA) diversity or expression from IMGT/HighV-quest. Front Immunol. 2016;7:339. doi: 10.3389/fimmu.2016.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraus A.A., Messer W., Haymore L.B., de Silva A.M. Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J Clin Microbiol. 2007;45(11):3777–3780. doi: 10.1128/JCM.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Alwis R., Smith S.A., Olivarez N.P., Messer W.B., Huynh J.P., Wahala W.M. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A. 2012;109(19):7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCracken M.K., Gromowski G.D., Friberg H.L., Lin X., Abbink P., De La Barrera R. Impact of prior flavivirus immunity on Zika virus infection in rhesus macaques. PLoS Pathog. 2017;13(8) doi: 10.1371/journal.ppat.1006487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davidson E., Doranz B.J. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014;143(1):13–20. doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodenhuis-Zybert I.A., Wilschut J., Smit J.M. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Mol Life Sci. 2010;67(16):2773–2786. doi: 10.1007/s00018-010-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurane I., Innis B.L., Nimmannitya S., Nisalak A., Meager A., Janus J. Activation of t lymphocytes in dengue virus infections. High levels of soluble interleukin 2 receptor, soluble CD4, soluble CD8, interleukin 2, and interferon-gamma in sera of children with dengue. J Clin Invest. 1991;88(5):1473–1480. doi: 10.1172/JCI115457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boonpucknavig S., Lohachitranond C., Nimmanitya S. The pattern and nature of the lymphocyte population response in dengue hemorrhagic fever. Am J Trop Med Hyg. 1979;28(5):885–889. [PubMed] [Google Scholar]
- 57.Friberg H., Bashyam H., Toyosaki-Maeda T., Potts J.A., Greenough T., Kalayanarooj S. Cross-reactivity and expansion of dengue-specific t cells during acute primary and secondary infections in humans. Sci Rep. 2011;1:51. doi: 10.1038/srep00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jourdan M., Caraux A., Caron G., Robert N., Fiol G., Reme T. Characterization of a transitional preplasmablast population in the process of human b cell to plasma cell differentiation. J Immunol. 2011;187(8):3931–3941. doi: 10.4049/jimmunol.1101230. [DOI] [PubMed] [Google Scholar]
- 59.Dejnirattisai W., Wongwiwat W., Supasa S., Zhang X., Dai X., Rouvinski A. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. 2015;16(2):170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lok S.M., Ng M.L., Aaskov J. Amino acid and phenotypic changes in dengue 2 virus associated with escape from neutralisation by IgM antibody. J Med Virol. 2001;65(2):315–323. doi: 10.1002/jmv.2036. [DOI] [PubMed] [Google Scholar]
- 61.Martina B.E., Koraka P., Osterhaus A.D. Dengue virus pathogenesis: an integrated view. Clin Microbiol Rev. 2009;22(4):564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corthesy B. Multi-faceted functions of secretory IgA at mucosal surfaces. Front Immunol. 2013;4:185. doi: 10.3389/fimmu.2013.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8(6):421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nawa M., Takasaki T., Ito M., Inoue S., Morita K., Kurane I. Immunoglobulin A antibody responses in dengue patients: a useful marker for serodiagnosis of dengue virus infection. Clin Diagn Lab Immunol. 2005;12(10):1235–1237. doi: 10.1128/CDLI.12.10.1235-1237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balmaseda A., Guzman M.G., Hammond S., Robleto G., Flores C., Tellez Y. Diagnosis of dengue virus infection by detection of specific immunoglobulin M (IgM) and IgA antibodies in serum and saliva. Clin Diagn Lab Immunol. 2003;10(2):317–322. doi: 10.1128/CDLI.10.2.317-322.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao H., Qiu S., Hong W.X., Song K.Y., Wang J., Yang H.Q. Dengue specific immunoglobulin a antibody is present in urine and associated with disease severity. Sci Rep. 2016;6:27298. doi: 10.1038/srep27298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vazquez S., Cabezas S., Perez A.B., Pupo M., Ruiz D., Calzada N. Kinetics of antibodies in sera, saliva, and urine samples from adult patients with primary or secondary dengue 3 virus infections. Int J Infect Dis. 2007;11(3):256–262. doi: 10.1016/j.ijid.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 68.Kerr M.A. The structure and function of human IgA. Biochem J. 1990;271(2):285–296. doi: 10.1042/bj2710285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Halstead S.B., Rojanasuphot S., Sangkawibha N. Original antigenic sin in dengue. Am J Trop Med Hyg. 1983;32(1):154–156. doi: 10.4269/ajtmh.1983.32.154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within this article and its Supplementary Information files, or from the corresponding author upon reasonable request. Single-cell RNAseq gene expression data have been deposited in the Gene Expression Omnibus database (GSE145307).