Abstract
In this study, we investigated the diversity of AAB from fermenting cocoa and the production of acetic acid in response to various environmental conditions. Ribosomal 16S gene sequence analysis and PCR-RFLP showed a restricted microbiota mainly composed of Acetobacter pasteurianus, Acetobacter tropicalis and Acetobacter okinawensis sp., consistently found in all six regions studied. Meanwhile Acetobacter malorum, Acetobacter ghanensis and Gluconobacter oxydans were isolated as minor species in specific regions. The dominant species were mainly isolated in the first 72 h period of natural cocoa fermentation while the minor species were present toward the later stages. Acetobacter okinawensis, a newly isolated species, was able to yield an unusually high quantity, up to 62 g/L of acetic acid at 30 °C. However, a shift of temperature to 35 °C severely impaired acid production in most strains of this species. While acetic acid production increases for up to 6 days in Acetobacter okinawensis and Acetobacter pasteurianus, it decreases beyond 4 days in Acetobacter tropicalis strains. The production of acetic acid was strongly dependent on environmental conditions, with optimal production between pH 4 and 5, under ethanol concentration below 8% and temperatures above 35–40 °C, corresponding to conditions prevailing in the first half of fermentation process. Acetobacter tropicalis was more productive at higher ethanol concentration and Acetobacter okinawensis at low pH. Species diversity and different behavior of strains highlight the importance of valuable starter selection for well-controlled cocoa fermentation.
Electronic supplementary material
The online version of this article (10.1007/s13197-019-04226-2) contains supplementary material, which is available to authorized users.
Keywords: Acetic acid bacteria, Diversity, Cocoa fermentation, Phenotypic properties, Acid production, Ivory Coast
Introduction
Fermentation of cocoa beans is the first step in the chocolate-making process. It involves a 5- to 7-days fermentation on the farm during which microorganisms grow within the pulp material and induce biochemical changes deep inside the beans (Afoakwa et al. 2007; De Vuyst and Weckx 2016; Nielsen et al. 2007; Schwan 1998; Schwan and Wheals 2004). The microbiota responsible for cocoa fermentation is mostly dominated by yeasts, Lactic Acid Bacteria (LAB), Acetic Acid Bacteria (AAB), and various species of Bacillus whose growth follows a well-defined microbial time-dependent succession (De Vuyst and Weckx 2016; Nielsen et al. 2007; Schwan and Wheals 2004). At the onset of cocoa fermentation, in anaerobic process, yeasts first produce ethanol from free sugars and degrade the pulp by enzymatic activity of pectinolytic enzymes (Camu et al. 2008a; Papalexandratou et al. 2011, 2013; Schwan and Fleet 2014). Further degradation of the viscous and sticky pulp by Bacillus strains takes place during the advanced stage of the process (Ouattara et al. 2008). The disappearance of the pulp allows air to percolate through the fermenting cocoa bean heaps, resulting in aerobic conditions favorable to AAB growth (Schwan and Wheals 2004). At this stage, ethanol is oxidized into acetic acid by the AAB and this diffuses deep into the cotyledons, increasing the inner acidity of the beans. This increase in acidity leads to the activation of pH-dependent hydrolytic enzymes which, in turn, results in the generation of free amino acids, from bean storage proteins, identified as being specific precursors of chocolate flavors and aromas (Biehl et al. 1993; De Vuyst and Weckx 2016; Voigt et al. 1994). Two well-studied enzymes, aspartic endoprotease and serine carboxypeptidase, both require an acidic pH (4–5) for their activity (Camu et al. 2008a; Jinap et al. 2008; Voigt et al. 1994; Ziegleder, 2009). The acidification of cocoa beans also promotes the oxidation of polyphenols, resulting in the loss of cocoa astringency and bitterness (Camu et al. 2008b; Misnawi et al. 2003).
The physiological pH of the bean is almost pH 7 and the acidic pH required for hydrolytic enzyme activities is provided by acetic acid diffusion into the beans (Biehl et al. 1993; Lefeber et al. 2010; Nielsen et al. 2007). Thus, acetic acid production from AAB activities plays a central role in the development of the desirable traits characteristic of certain chocolate flavors. It is now established that an efficient cocoa fermentation process should include an acidification step (De Vuyst et al. 2010; Schwan and Wheals 2004; Nielsen et al. 2007). However, cocoa fermentation is still an empirical and traditional process that is difficult to control, very often giving a variable quality product (Schwan, 1998; Kongor et al. 2016; Ouattara et al. 2017a). Recently, we reported the isolation of valuable AAB strains that have the ability to produce a high concentration of acetic acid (Soumahoro et al. 2015). Such AAB could be selected for use as starter strains to improve the fermentation process of cocoa. Thus, a study of AAB strains is crucial for solving the problem of variable cocoa fermentation quality. Until now, limited knowledge has been available regarding the AAB microbiota diversity involved in Ivorian cocoa fermentation as previous studies have mainly focused on the response of AAB to fermentation stress (Soumahoro et al. 2015; Yao et al. 2014).
In this paper, we first describe the diversity of the AAB involved in cocoa fermentation from six different Ivory Coast producing regions. Ivory Coast is the leading country in term of cocoa production in the world and these regions are among the most important producing region in this country. Secondly, we characterize the high acid-producing bacteria selected with regard to the conditions encountered during cocoa fermentation. A better understanding of the cocoa microbiota may help screening performant strains as starter for further control of cocoa fermentation, and this may in turn deliver a consistent quality of fermented and dried beans.
Materials and methods
Fermentation, culture conditions and isolation of bacteria
Cocoa pods were harvested from six cocoa producing regions, namely Agneby-Tiassa (geographic coordinates 5° 59′ North 4° 28′ West), Guemon (6° 29′ 41″ North 6° 57′ 59″ West), Indénié-Djuablin (6° 44′ North 3° 29′ West), Loh-Djiboua (5° 55′ North 5° 37′ West), Nawa (5° 47′ 00″ North 6° 36′ 00″ West), Sud-Comoé (5° 28′ 06′ North 3° 12′ 25′ West), in Ivory Coast. Heap fermentations, of about 50 kg of cocoa beans, were conducted for six days using banana leaves, as described by Samagaci et al. (2016). Beans were laid out on, and then covered with, banana leaves. During fermentation, a total of 100 g of beans were collected from the top, the middle and the bottom layers of the cocoa mass at 12 h intervals. Each sample was used for the numeration and isolation of AAB. For this purpose, a subset of 25 g of each sample were added to 225 mL of 0.1% (w/v) buffered (pH 7.2) peptone water (DIFCO, Abidjan, Ivory Coast), contained in a 500 mL sterile flask, and shaken for 2 to 5 min to disperse the bacteria in the liquid (initial dilution). Next, this suspension was used to prepare a serial dilution, up to 10–6, using trypton saline (DIFCO, Abidjan, Ivory Coast). 0.1 mL of each dilution was spread onto a potato medium containing 0.5% (w/v) d-glucose, 1% yeast extract (w/v), 1% peptone (w/v), 2% glycerol (v/v), 1.5% potato extract (w/v) and 4% ethanol (v/v), supplemented with 0.0016% bromocresol green (w/v) to monitor pH variation and nystatin (50 μg/mL) to inhibit fungal growth (Duthatai and Pathom-Aree 2007).
After a 48 h-incubation at 30 °C, colonies with yellow areas, due to acidification of the green colored agar, were selected for biochemical identification. The colonies that were oxidase negative, showing Gram negative staining and absolute aerobic metabolism were presumed to be AAB (De Ley et al. 1984). These isolated strains were stored in Eppendorf tubes, at − 80 °C, in Luria Bertani medium supplemented with 20% glycerol, for further studies.
Ribosomal 16S RNA gene amplification and RFLP procedures
To get insight into the diversity of the AAB microorganisms isolated, a genetic approach that consisted in sequencing and realizing RFLP (Random Fragment Length Polymorphism) from ribosomal 16S gene (16S rRNA), was performed. This technique is generally well-accepted as the best target for studying phylogenetic relationships (Vandamme et al. 1996; Rossello-Mora and Amann 2001). For this purpose, a multiple alignment (https://npsaprabi.ibcp.fr/NPSA/npsa_clustalw.html) of 16S rRNA genes, from various species of AAB, enabled us to design the forward (5′-AGTGGCGGACGGGTGAGTA-3′) and reverse (5′-CCAACTCCCATGGTGTGACG-3′) primers. These specific primers anneal to the most highly conserved 5′ and 3′ regions of the 16S rRNA gene, respectively, and after PCR they generate an amplicon of approximately 1300 bp. To perform the PCR reactions, bacteria grown for 24 h on agar plates were suspended in 100 µL of sterile distilled water and the resulting suspensions were used as DNA templates (Ouattara et al. 2017b). PCR amplification was carried out in a Sensoquest Labcycler, as described previously (Ouattara et al. 2011). Reactions were performed in a final volume of 50 µL containing 1 µl of bacterial suspension, 1.25 U of Taq DNA polymerase (Biolabs, Lyon, France), 5 µL of 10X standard buffer; 1 µL deoxynucleoside triphosphate (10 mM), 2 µL of each primer (10 µM) (Eurofins Genomics, Allemagne) and 38.75 µL of water. After an initial denaturation, at 95 °C for 4 min, reactions were run for 35 cycles, each cycle comprising: denaturation at 95 °C for 1 min, annealing at 56 °C for 1 min and extension at 72 °C for 1 min. Finally, a 10 min extension at 72 °C was carried out. The presence and yield of specific PCR products were monitored using agarose 0.8% (w/v) gel electrophoresis at 70 V, for 2 h, in 1X Tris Borate EDTA buffer and visualized with ethidium bromide staining and UV transillumination.
For RFLP analysis, the 1300 bp PCR products were digested with AluI, HaeIII and TaqI (Thermo Scientific, France) in separate reactions. The digestions were carried out in 20 µL reaction volumes containing 12 µL of the PCR product solution, 2 µL of the commercial buffer, 5 µL of sterilized distilled water and 1 µL (10 U) of restriction enzyme (Thermo Scientific, France). Enzymatic reactions were performed for 2 h, at 37 °C for AluI and HaeIII and at 65 °C for TaqI. The digestion products were then subjected to electrophoresis on 2% agarose gels in 1 × Tris-Borate-EDTA buffer, at 35 V, overnight. The number and size of the fragments obtained forming a restriction profile or fingerprint were visualized and digitalized with a gel print system. Strains were grouped based on their restriction profiles.
16S rRNA gene partial sequencing and sequence analysis
The hyper variable (HV) sections of the ribosomal gene of 16S RNA, including the fragments V1, V2 and V6, were assessed to include specific sequences that allow for the identification of the bacterial species (Gray et al. 1984; Woese et al. 1983). For sequencing and identification purposes, this section of about 500 bp was targeted for PCR amplification using the primers F27 (5′-AGAGTTTGATCCTGGCTCAG-3′) and R520 (5′-ACCGCGGCTGCTGGC-3′) (Anzai et al. 2000), complementary to the positions 531–517 of conserved sequences of the E. coli reference 16S gene. PCR reactions were performed in the same conditions as described above. The yield of amplification products was verified using 1% agarose gel electrophoresis. Next, PCR amplicons were purified using PCR Clean-up (Macherey Nagel, Germany) and sequenced by EurofinR, Genomics, (Germany) using the primer F27. The basic local alignment search tool (BLAST, blastN) from the NCBI database site (blast.ncbi.nlm.nih.gov/) was used to find the closest sequences relative to the amplified 16S rRNA genes in order to identify our AAB strains.
Screening of high acetic acid producers
Strains identified as AAB were subjected to screening in order to target the high acetic acid producers. The method of screening was developed in two steps. In the first step, we used a solid medium composed of 0.05% glucose (w/v), 0.3% peptone (w/v), 0.5% yeast extract (w/v), 1.5% CaCO3 (calcium carbonate) (w/v), 1.2% agar (w/v), and 4% ethanol (v/v) (Andelib and Nuran 2009). The calcium carbonate imparts a white color to the medium. This medium was inoculated with a drop from a pure 18–24 h culture of the AAB strains and incubated, at 30 °C, for six days. During incubation, calcium carbonate is neutralized by the acetic acid produced by the bacterial strains, forming a soluble salt of calcium acetate and CO2. The disappearance of calcium carbonate, due to acid production, is evident from the clear halo that develops around the drop of culture and the size of this halo is relative to the amount of acid produced.
In the second step, strains presenting the largest haloes were used for a second screening. These strains were grown in liquid medium with the same composition as that described above but without agar and calcium carbonate. To this medium, 0.01% MgSO4 (w/v) and 0.27% NaHPO4 (w/v) were added to promote acid production. For inoculation purposes, a culture was prepared by growing the bacteria in the liquid medium to an absorbance of OD600 = 0.5 and 1 mL of this culture was used to inoculate 150 mL of liquid medium, contained in a 500 mL flask. The cultures were incubated for 6 days at 30 °C, under agitation at 160 rpm. During incubation, a 10 mL daily sample of liquid medium was aseptically withdrawn and analyzed to determine bacterial growth, the pH of the medium, and to quantify acetic acid production.
The quantification of total acetic acid yielded in the medium was measured by titration of a cell free supernatant with 0.1 N NaOH, in the presence of phenolphthalein as a pH indicator. Acetic acid concentration (Pa) was calculated, as described previously (Nanda et al. 2001).
Quantification of acetic acid by HPLC
An aliquot of cell free supernatant was used to further quantify acetic acid with a new method of separation using High Performance Liquid Chromatography (HPLC) as we have previously described (Ouattara et al. 2017b).
Influence of culture conditions on acetic acid production
Variations in pH and temperature, together with alcohol and acid accumulation, are the main environmental factors encountered during cocoa fermentation. The effects of these individual factors on acetic acid production were analyzed using the selected high acetic acid-producing strains as models. The standard inoculum of these strains was calibrated as described above (see “Screening of high acetic acid producers” section) and the OD600 = 0.5 allowed using approximately the same size of microorganisms. Strains were grown, for 6 days, in a standard liquid medium containing 0.5% d-glucose (w/v), 1% yeast extract (w/v), 1% peptone (w/v), 2% glycerol (v/v), 1.5% potato extract (w/v) and 4% ethanol (v/v), pH 6. To assess the influence of temperature on acid production, bacterial cultures were incubated at different temperatures ranging from 30 to 50 °C, at pH 6. The pH effect was analyzed by incubating the cultures using the same medium buffered alternatively with HCl or NaOH at different pH levels, from 3 to 6, at 30 °C. Concerning the effects of alcohol, ethanol was added to the culture medium described above at concentrations ranging from 4 to 15%. The same type of experiments was performed individually with organic acids (acetic acid, lactic acid and citric acid) added individually to the medium at concentrations ranging from to 0.1 to 3% and maintaining the culture pH medium at pH 6 using NaOH. Bacterial growth and acid production in the medium were monitored, respectively, by measuring the turbidity (OD600) and by HPLC quantification of acetic acid.
Results
Diversity of AAB isolated from fermenting beans
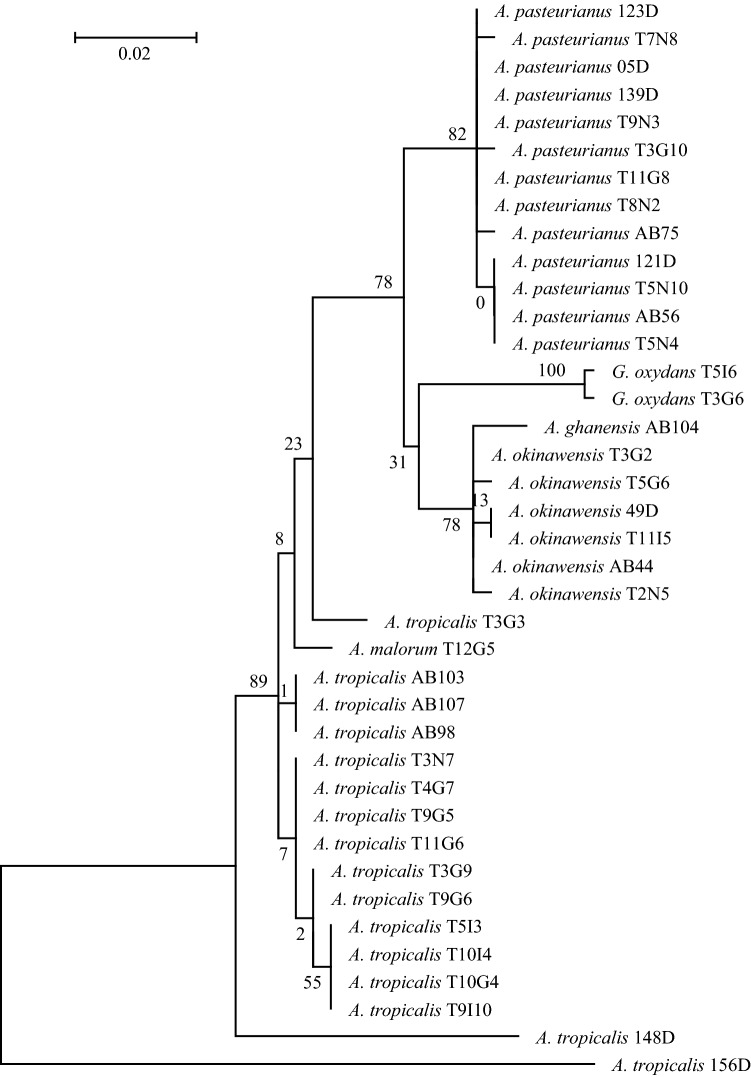
In the six different regions analyzed, a total of 672 AAB strains were isolated from fermenting beans in this study. The analysis of RFLP showed that these strains can be classified into six distinct groups (Table 1). TaqI displayed the highest discriminatory power and raised five distinct profiles while HaeIII and AluI generated only two profiles (Table 1). A total of six species, namely Acetobacter pasteurianus, Acetobacter tropicalis, Acetobacter okinawensis, Acetobacter ghanensis, Acetobacter malorum and Gluconobacter oxydans, were characterized (Table 1). The RFLP group 1, corresponding to Acetobacter pasteurianus, accounts for 394 isolates and 58.63% of all characterized AAB isolates (Table 1). The RFLP groups 2 and 3, corresponding to Acetobacter tropicalis and Acetobacter okinawensis, account for 134 and 130 isolates, respectively (20% for each species) (Table 1). The groups 4, 5 and 6, corresponding to Acetobacter malorum, Gluconobacter oxydans and Acetobacter ghanensis are minority species (Table 1). Furthermore, the AAB 16S rRNA gene sequence analysis revealed very weak intraspecies variation (less than 2%) within these different restriction groups isolated from the six regions (Fig. 1). Although, beside the identification methods used in ours experiments, a typing method (Random Amplified Polymorphic DNA -RAPD- or Pulsed Field Gel Electrophoresis -PFGE-) should provide additional accuracy to differentiate the strains into each species.
Table 1.
AAB species isolated and their RFLP group
| Species | 16S rRNA gene length (bp) | AluI restriction fragments (bp) | HaeIII restriction fragments (bp) | TaqI restriction fragments (bp) | Number of strains | RFLP group |
|---|---|---|---|---|---|---|
| Acetobacter pasterianus | 1300 | (450 + 320 + 200 + 280) | (550 + 300 + 180 + 150) | (500 + 380 + 300 + 120) | 394 | 1 |
| Acetobacter ghanensis | 1300 | (350 + 220 + 200 + 180 + 150 + 120) | (1000 + 250) | (500 + 380 + 300 + 120) | 2 | 2 |
| Acetobacter tropicalis | 1300 | (350 + 220 + 200 + 180 + 150 + 120) | (550 + 300 + 180 + 150) | (500 + 380 + 150 + 130 + 140) | 134 | 3 |
| Acetobacter okinawensis | 1300 | (350 + 220 + 200 + 180 + 150 + 120) | (1000 + 250) | (650 + 380 + 270) | 130 | 4 |
| Acetobacter malorun | 1300 | – | (550 + 300 + 180 + 150) | (800 + 380 + 120) | 8 | 5 |
| Gluconobacter oxydans | 1300 | – | (550 + 300 + 180 + 150) | (380 + 200 + 160 + 130 + 120 + 110 + 100 + 100) | 4 | 6 |
Fig. 1.
Phylogenetic tree of AAB strains isolated from Ivorian cocoa fermentation. The partial sequences of the hyper variable region of the 16S rRNA genes were aligned using ClustalW and the evolutionary history was inferred using the Maximum Likelihood method based on the General Time Reversible model. The tree with the highest log likelihood (− 1023.9028) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying the Maximum Parsimony method. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. The analysis involved 39 nucleotide sequences. All positions with less than 95% site coverage were eliminated, i.e. fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 358 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al. 2013)
Distribution of AAB strains in the six cocoa producing regions
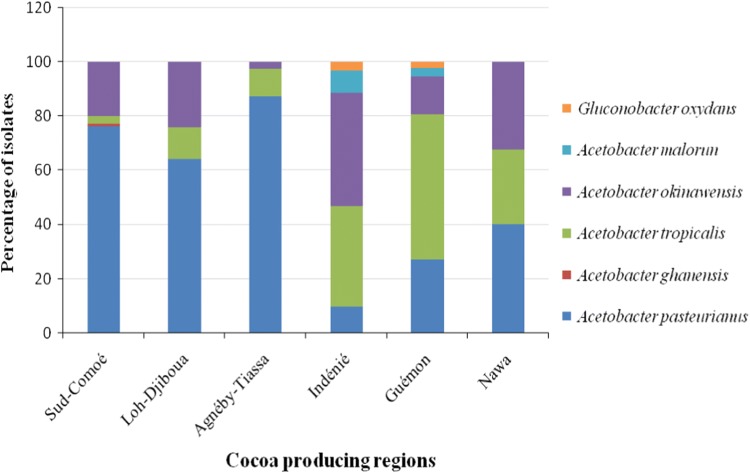
The six AAB species are not homogenously distributed throughout the six cocoa producing regions (Fig. 2). The regions of Guemon and Indenié recorded the greatest diversity of AAB, with five distinct species. The most prevalent AAB species isolated in these two regions were Acetobacter okinawensis (41.94%) and Acetobacter tropicalis (37.09%) in Indenié, whilst Acetobacter pasteurianus (28.88%) and Acetobacter tropicalis (53.76%) dominated in the Guemon region (Fig. 2). The minor species in these regions were Acetobacter malorum (3 to 8%) and Gluconobacter oxydans (2 to 3%). In the region of Sud-Comoé, four AAB species were present with Acetobacter pasteurianus (76.19%) and Acetobacter okinawensis (20%) as the dominant species. The weakest biodiversity was found in the Lôh-Djiboua, Agneby-Tiassa and Nawa regions, where only three distinct species were present, namely A. pasteurianus, A. okinawensis and A. tropicalis, with varying proportions from one region to another (Fig. 2). Globally, A. pasteurianus, A. tropicalis and A. okinawensis are the most prevalent species in all the selected regions, while the minority species, A. malorum, A. ghanensis and G. oxydans, are restricted to specific regions.
Fig. 2.
Distribution of AAB isolates in the different cocoa producing regions
Evolution of the AAB population during cocoa fermentation
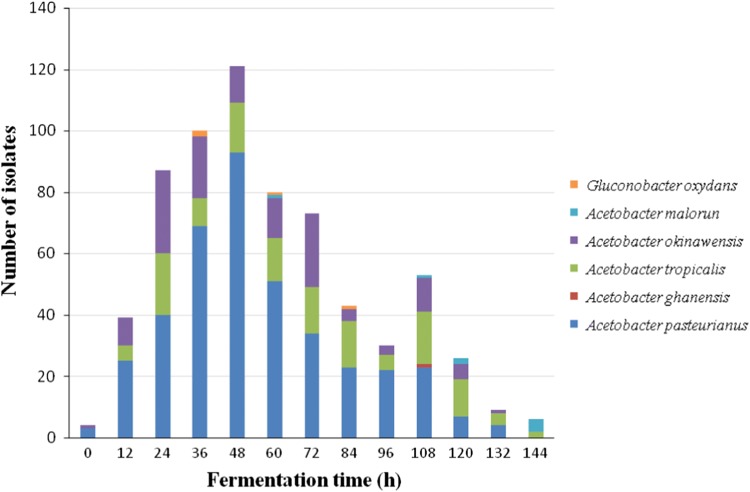
Regardless their regions of origin, all the 672 strains were analyzed for the time at which they could be isolated during cocoa fermentation. This allowed monitoring the general trend of dynamic changes in AAB species during fermentation (Fig. 3). Overall, the AAB population size increased gradually up to 48 h and then declined. The clear reduction in the AAB population size at the end of fermentation indicates that this medium is extremely deleterious for AAB and very few of them survive. The dominant species, A. pasteurianus, A. tropicalis and A. okinawensis, were isolated during almost the entire fermentation process, with the exception of the very late phase. However, 54% of the A. pasteurianus strains were isolated between 36 and 72 h of fermentation, with the highest number at 48 h post-initiation of the process. Likewise, most of the A. okinawensis strains (73.84%) were isolated during a fermentation time spanning from 24 to 72 h. While A. pasteurianus and A. okinawensis were the only species isolated at the beginning of the fermentations, they were absent at the end of the process (144 h). Hence, A. pasteurianus and A. okinawensis appear to be the first AAB colonizing species during cocoa fermentation. In contrast, the population of A. tropicalis remained relatively constant between 24 and 120 h of fermentation. This species was absent at the beginning of fermentation but was present at the end of the process. As regards the minority species, G. oxydans was isolated in the middle period of fermentation, between 36 and 84 h, while A. malorum and A. ghanensis were mostly found in the advanced and final steps of the fermentation process, between 60 and 144 h. Therefore, all the species were not found at the same time points during the fermentation process. A. pasteurianus and A. okinawensis completely disappeared at the end of the fermentation, suggesting that they cannot survive under the hostile conditions resulting from advanced fermentation. Minority species emerged when the overall AAB population size declined, indicating a higher tolerance to harsh fermentation conditions in these bacteria.
Fig. 3.
Isolation time of the different AAB species during natural cocoa fermentation
Effects of environmental conditions on acetic acid production
Acetic acid production from ethanol is a key property for an efficient fermentation of cocoa. Therefore, all the isolates were screened for their potential for acetic acid production. We observed that AAB strains produced acetic acid on solid medium containing 4% of ethanol to form a halo with a diameter of 3.4 cm. Only strains producing a halo diameter superior to 1.7 cm, equivalent to 50% of the maximum halo diameter, were arbitrarily considered as the best producers. On this basis, 135 strains out of the 672 isolates were retained for further analysis in liquid medium at 30 °C. Of these, 35 strains produced more than 25 g/L of acetic acid, as determined by titration with sodium hydroxide in the presence of phenolphthalein or with HPLC (Table 2). Thus, less than 5% (35/ 672) of the total AAB strains could be considered as high acetic acid producers.
Table 2.
Different patterns of acid production by AAB regarding growth temperature (acetic acid by NaOH titration or HPLC)
| Strains | Temperatures (°C) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 30 | 35 | 40 | 45 | 50 | ||||||
| Acid production (g/L) | Bacterial growth (OD600) | Acid production (g/L) | Bacterial growth (OD600) | Acid production (g/L) | Bacterial growth (OD600) | Acid production (g/L) | Bacterial growth (OD600) | Acid production (g/L) | Bacterial growth (OD600) | |
| Acetobacter pasteurianus T11G8 | 48.61 | 0.74 | 0 | 0.69 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter okinawensis T5G6 | 55.31 | 1.04 | 0 | 0.71 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianusT6D121 | 45.38 | 0.94 | 0 | 0.91 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianus T3G10 | 35.23 | 0.952 | 0 | 0.79 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter okinawensisT2N6 | 38.18 | 1.045 | 0 | 1.67 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianus T4N5 | 34.26 | 1.028 | 0 | 1.95 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter okinawensis T5N8 | 32.41 | 1.025 | 0 | 1.49 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T9I10 | 31.53 | 1.235 | 0 | 1.65 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianusT3G3 | 30.50 | 1.913 | 0 | 1.10 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter okinawensisT10I6 | 28.23 | 1.040 | 0 | 1.07 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter okinawensisT10I4 | 38.64 | 1.043 | 0 | 1.46 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter okinawensisT2N5 | 30.20 | 1.045 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T12G4 | 26.60 | 1.242 | 0 | 2.66 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianus T9N3 | 32.30 | 0.989 | 0 | 1.24 | 0 | 1.27 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T4N3 | 32.00 | 0.596 | 0 | 0.76 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianus T9N3 | 34.10 | 1.940 | 0 | 8.68 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianus T9N5 | 29.50 | 2.695 | 0 | 3.93 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianus T5N4 | 31.00 | 4.745 | 0 | 4.09 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter okinawensisT11I5 | 62.20 | 0.978 | 35.12 | 1.60 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianusT11G3 | 53.22 | 1.045 | 34.89 | 1.16 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T12G5 | 28.07 | 0.982 | 30.81 | 1.42 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T3G9 | 30.00 | 1.350 | 29 | 1.52 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianusT7N8 | 26.50 | 1.208 | 23.45 | 1.56 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T9G5 | 47.25 | 0.965 | 31.25 | 0.63 | 23.47 | 0.87 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianusT0N5 | 44.36 | 1.025 | 30.98 | 1.30 | 4.65 | 0.90 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T4G7 | 39.69 | 0.960 | 25.55 | 0.88 | 4.70 | 0.64 | 0 | 0 | 0 | 0 |
| Acetobacter okinawensis T3G2 | 29.88 | 0.670 | 38.31 | 0.66 | 10.79 | 0.42 | 0 | 0 | 0 | 0 |
| Acetobacter pasteurianusT5N10 | 31.17 | 0.885 | 25.26 | 0.59 | 22.30 | 0.60 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T9G6 | 29.39 | 0.930 | 31.56 | 0.80 | 33.33 | 0.72 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T10G4 | 28.60 | 0.987 | 28.25 | 1.25 | 9.64 | 1.04 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T10G3 | 27.04 | 1.242 | 29.59 | 1.92 | 4.87 | 1.87 | 0 | 0 | 0 | 0 |
| Acetobacter tropicalis T7I1 | 45.18 | 1.103 | 36.37 | 1.31 | 6.65 | 1.20 | 5.44 | 2.05 | 0 | 0 |
| Acetobacter tropicalis T3N7 | 28.51 | 1.004 | 28.68 | 0.96 | 21.46 | 0.84 | 2.76 | 0.43 | 0 | 0 |
| Acetobacter pasteurianusT9N4 | 37.10 | 3.450 | 7.15 | 3.10 | 1.71 | 2.08 | 1.63 | 0.56 | 0 | 0 |
| Acetobacter tropicalis T11G6 | 37.98 | 0.930 | 29.43 | 0.74 | 21.17 | 0.79 | 3.54 | 0.32 | 1.15 | 0.126 |
These 35 Acetobacter strains belonged to the species A. pasteurianus (14 strains), A. tropicalis (12 strains), A. okinawensis (8 strains) and A. malorum (1 strain). Table 2 also presents the different patterns of acetic acid production in AAB strains in response to changes in temperature. Maximum acid production, at 30 °C, was 62.20 g/L of acetic acid in the A. okinawensis strain T11I5. However, acid production in this strain was severely reduced to 35.12 g/L when the growth temperature was increased to 35 °C (Table 2). Furthermore, at 35 °C, half of the isolates failed to produce acetic acid although the growth capacity remained high. Remarkably, only a few A. tropicalis strains are able to produce acetic acid at temperatures up to 45 °C (Table 2). Six AAB strains, presenting the ability to significantly produce acetic acid at 40 °C (highlighted in Table 2), were retained as model strains for evaluating the effect of pH, alcoholic and acidic conditions on acetic acid production. These are the Acetobacter species tropicalis (T9G5, T9G6, T3N7, T11G6), okinawensis (T3G2) and pasteurianus (T5N10).
Two tendencies in acetic acid production were observed when changing the pH from 3 to 6. All the A. tropicalis strains demonstrated acetic acid production with a peak at pH 5, while the optimum pH was 4 for A. okinawensis, and A. pasteurianus (data not shown).
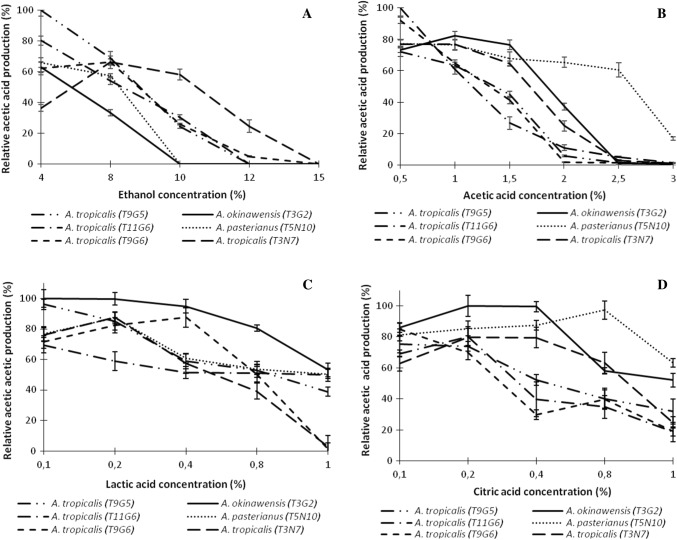
Likewise, the major metabolites produced during cocoa fermentation, notably ethanol, lactic acid and acetic acid (Ouattara et al. 2011; Schwan and Wheals 2004), were tested for their impact on acetic acid production in the AAB strains. When the ethanol content in the growth medium was increased from 4 to 15%, changes in the behavior of the strains were observed. Acetic acid production in A. pasteurianus and A. okinawensis appeared to be relatively dependent on ethanol while the A. tropicalis T3N7 strain retained approximately 25% of its potential of acid production in the presence of 12% ethanol. The other A. tropicalis strains displayed an intermediate behavior (Fig. 4a). When considering the impact of acetic acid, the reaction product, on the behavior of the strains (Fig. 4b), a constant decrease in acetic acid production was observed in the presence of 0.5% to 2% acetic acid with the A. tropicalis strains T9G5, T11G6 and T9G6, while A. tropicalis strain T3N7 still retained 25% of its potential acetic acid production in the presence of 2% acetic acid. In the same conditions, A. okinawensis expressed up to 37.33% of its acid production capacity whereas A. pasteurianus was the most productive at high acetic acid concentrations (2.5%), retaining 60.49% of its potential acid production (Fig. 4b). When varying the lactic or citric acid concentration from 0.1 to 1%, a decrease in acetic acid production was observed in all AAB strains (Fig. 4c, d). However, A. okinawensis and A. pasteurianus retained 50% of their acetic acid producing capacity in the presence of 1% lactic or citric acid while, at the same lactic acid concentration (1%), A. tropicalis strains failed to produce any acetic acid,with the exception of the strain T9G5 which retained 38% of its production capacity (Fig. 4c, d). These strains retained approximately 20% of their potential acid production in 1% citric acid (Fig. 4d).
Fig. 4.
Influence of different compounds on acetic acid production in AAB isolates. Strains were grown in standard medium containing different concentrations of the compounds. Error bars indicate standard deviation between two biological replicates
Kinetics of acetic acid production during AAB growth
Acetic acid production in AAB isolates was monitored for six days, corresponding to the duration of natural cocoa fermentation. The dynamics of acid production, reported in supplemental material (Fig S1), indicate two groups of strains. In the first group, composed of A. tropicalis strains, a progressive increase in acid production was measured, reaching 30 to 40 g/L on the fourth day of culture. There was a moderate decrease in acid production during the remaining growth period. Interestingly, the second group, including A. okinawensis and A. pasteurianus strains, was characterized by a lag phase on the first day of culture with very low acid production (less than 3 g/L). Thereafter, the quantity of acetic acid produced by these AAB strains steadily increased, reaching up to 45–52 g/L at the end of the culture period. The increase in acetic acid production was particularly evident between 24 (1 day) and 96 h (4 days) in the A. okinawensis strain, compared with the other strains. Thus, the data obtained with the six AAB strains studied highlights their importance complementary roles in an efficient fermentation of cocoa.
Discussion
AAB are the main microbial actor reported to have a direct impact on the formation of the chocolate flavor precursors during cocoa fermentation (Schwan and Wheals 2004; Camu et al. 2008a). This study is the first investigation into the diversity and the physiology of AAB isolated from fermenting cocoa in six main cocoa producing regions from Ivory Coast. Three species, Acetobacter pasteurianus, Acetobacter tropicalis and Acetobacter okinawensis, representing more than 80% of total isolates, were consistently present in all of the six selected Ivory Coast regions. Three minor AAB species, Acetobacter ghanensis, Acetobacter malorum and Gluconobacter oxydans, representing less than 2% of the total isolates, were only found in specific regions. Some of the dominant AAB species, particularly Acetobacter pasteurianus, have worldwide distribution since they have been isolated from fermenting cocoa beans in Malaysia, Brazil, Ecuador and Ghana (Nielsen et al. 2007; Papalexandratou et al. 2011, 2013). Another dominant AAB species, Acetobacter tropicalis, seems to have a more restricted distribution since it has only been isolated in Ghana and Brazil (Nielsen et al. 2007; Pereira et al. 2012). In addition, Visintin et al. (2016) have isolated Acetobacter pasteurianus as a dominant AAB species from cocoa fermentation in Ivory Coast. However, the role of Acetobacter suzygii as an important species in Ivorian cocoa fermentation reported previously (Visintin et al. 2016) could not be confirmed in this study. With the exception of Acetobacter okinawensis, the AAB species identified in this study have been previously identified in Ghanaian cocoa fermentation (Nielsen et al. 2007). The resemblance in the general composition of the AAB flora involved in Ivorian and Ghanaian cocoa fermentation may be due, in part, to the geographic proximity of these neighboring countries. However, this microflora is unequally distributed in the six Ivorian regions, indicating the variability of the spontaneous microbial community that ferments cocoa from one region to another. These observations led to the hypothesis that this diversity could be the main determining factor in the variability of cocoa quality.
Cocoa fermentation is a complex process, characterized by the successive growth of different species of microorganisms that have a strong impact on the final quality of the products (Camu et al. 2008b; Schwan, 1998). In this study, the dominant species were mostly isolated during the 24–72 h time period, while the minor species were more generally isolated in the more advanced stages of the fermentation process. As the prevailing conditions during cocoa fermentation (pH, temperature, chemical composition, oxygen, etc.) were dynamic, each microbial species grew when the conditions become favorable, resulting in different times of emergence. Papalexandratou et al. (2013) reported that A. pasteurianus and A. senegalensis dominated the initial phase of Malaysian cocoa fermentation while the growth of A. ghanensis occurred during the middle phase. However, in Ecuador, A. pasteurianus was rather isolated in the later stages of fermentation.
In an efficient natural cocoa fermentation, the maximum AAB population size occurred in the 48–72 h time period (Lefeber et al. 2010; Schwan and Wheals 2004), which is in parallel with the timing of intensive acetic acid production (Biehl et al. 1993; Lefeber et al. 2010). Thus, AAB strains growing in the early stage of cocoa fermentation, prior to 72 h, may play an important and/or different role from that of the species growing during the later stages.
Moreover the peak of temperature of the fermenting cocoa (40–43 °C) was generally reached in the period 48–72 h (data not shown). The dominant species particularly A. pasteurianus was found with high rate of isolation during this period, suggesting that the growth of this species may be related to the raise of temperature considered as an indicator of successful fermentation process (Barel 2013). This indicates the relevance of screening the starter strains among these dominant species.
Additionally, the strains, screened from the AAB microbiota isolated from fermenting cocoa, were able to produce high levels of acetic acid, up to 60 g/L, as revealed by HPLC analysis and NaOH titration. The strains characterized in this study showed a higher level of activity than most of the strains isolated in previous studies (Konate et al. 2014; Romero-Cortes et al. 2012; Soumahoro et al. 2015). However, high acetic acid production in AAB could reach up to 100 g/L in optimized specific processes (Sharafi et al. 2010). Our screening for AAB strains was performed particularly based on their high acetic acid production. Of the 672 isolated AAB strains, only 35 strains produced more than 25 g/L of acetic acid. Thus, less than 5% of the total AAB strains could be considered as being high acetic acid producers, which may have a significant impact on chocolate aroma development when using them as starter strains.
Ivorian cocoa fermentation was dominated by more than 99% of AAB belonging to the Acetobacter genus. This is an important feature since this genus is characterized by its capacity to over-oxidize acetic acid which can, at a high concentration, dissimulate the cocoa aroma (Raspor and Goranovic 2016). An over-oxidation of excess acetic acid (CO2 + H2O) should have a role in homeostasis, modulating the concentration of this acid in the fermenting cocoa, as was observed in this study. Several features were highlighted in our study, especially regarding AAB behavior with respect to the environmental conditions. For instance, some strains failed to produce acetic acid at temperatures above 35 °C, while others were still actively producing acetic acid at 45–50 °C. Likewise, acetic acid production was affected by acidic conditions induced by 1% lactic acid or citric acid to a differing extent depending on the AAB strain. This indicates some diversity in the physiological properties of various AAB strains that may help to maintain a high level of acetic acid production as the bacteria can function in relay throughout the changes in the fermentation conditions.
Conclusion
This study reveals a limited diversity of AAB species in Ivorian cocoa fermentation, dominated by A. pasteurianus, A. tropicalis and A. okinawensis although the AAB microbiota varies from one region to another. The AAB microbiota studied is characterized by a significant variability in its phenotypic properties, with a different level and kinetics of acetic acid production and different behavior related to the environmental conditions. Furthermore, the occurrence of Acetobacter okinawensis, which shows particular characteristics, demonstrated the complexity of the cocoa fermentation ecosystem. This, in turn, indicates the possibility of finding strains that exhibit particular biological properties, which could be exploited within a controlled cocoa fermentation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by an ASCAD Grant (Ascad 2ed.P1/2014) and Souleyman Soumahoro was supported by a PhD fellowship from the Embassy of France in Ivory Coast, under the AMRUGE/C2D program. The authors gratefully acknowledge Julien Wawrzyniak, Alexandre Duprey and Xuejiao Jiang for their technical assistance.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Andelib AY, Nuran DA (2009) Isolation of cellulose producing bacteria from wastes of vinegar fermentation. In: Proceedings of the world congress on engineering and computer science, vol I, San Francisco, USA, pp 978–988
- Afoakwa EO, Paterson A, Fowler M. Factors influencing rheological and textural qualities in chocolate—a review. Trends Food Sci Tech. 2007;18:290–298. doi: 10.1016/j.tifs.2007.02.002. [DOI] [Google Scholar]
- Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H. Phylogenetic affiliation of the pseudomonas based on 16S rRNA sequence. Int J Syst Evol Microbiol. 2000;50:1563–1589. doi: 10.1099/00207713-50-4-1563. [DOI] [PubMed] [Google Scholar]
- Barel M (2013) Qualité du cacao: L'impact du traitement post-récolte. Editions Quae. Livre electronique. pp 31–38
- Biehl B, Voigt J, Heinrichs H, Senjuk V, Bytof G. pH-dependent enzymatic formation of oligopeptides and amino acids, the aroma precursors in raw cocoa beans. In: Lafforest J, editor. Proceedings in XIth international cocoa research conference. Yamassoukro, Ivory Coast: Cocoa Producers’ Alliance; 1993. pp. 717–722. [Google Scholar]
- Camu N, De Winter T, Addo SK, Takrama JS, Bernaert H, De Vuyst L. Fermentation of cocoa beans: influence of microbial activities and polyphenol concentrations on the flavour of chocolate. J Sci Food Agric. 2008;88:2288–2297. doi: 10.1002/jsfa.3349. [DOI] [Google Scholar]
- Camu N, Gonzalez A, De Winter T, Van Schoor A, De Bruyne K, Vandamme P, Takrama JS, Addo SK, De Vuyst L. Influence of turning and environmental contamination on the dynamics of populations of lactic acid and acetic acid bacteria involved in spontaneous cocoa bean heap fermentation in Ghana. Appl Environ Microbiol. 2008;74:86–98. doi: 10.1128/AEM.01512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ley J, Gillis M, Swings J. Family VI. Acetobacteraceae. In: Krieg NR, Holt JG, editors. Bergey’s manual of systematic bacteriology. 1. Baltimore: Williams and Wilkins Co; 1984. pp. 267–278. [Google Scholar]
- De Vuyst L, Weckx S. The cocoa bean fermentation process: from ecosystem analysis to starter culture development. J Appl Microbiol. 2016;121:5–17. doi: 10.1111/jam.13045. [DOI] [PubMed] [Google Scholar]
- De Vuyst L, Lefeber T, Papalexandratou Z, Camu N. The functional role of lactic acid bacteria in cocoa bean fermentation. In: Mozzi F, Raya RR, Vignolo GM, editors. Biotechnology of lactic acid bacteria: novel applications. Ames: Wiley-Blackwell; 2010. pp. 301–326. [Google Scholar]
- Duthatai F, Pathom-aree W. Application of chemical dyes as colour indicator for selective isolation of acetic acid bacteria. Res J Microbiol. 2007;2:885–888. doi: 10.3923/jm.2007.885.888. [DOI] [Google Scholar]
- Gray MW, Sankoff D, Cedergren RJ. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucl Acids Res. 1984;12:5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinap S, Ikrawan Y, Bakar J, Saari N, Lioe HN. Aroma precursors and methylpyrazines in underfermented cocoa beans induced by endogenous carboxypeptidase. J Food Sci. 2008;73:141–147. doi: 10.1111/j.1750-3841.2008.00858.x. [DOI] [PubMed] [Google Scholar]
- Konate M, Akpa EE, Koffi LB, Kra KAS, Megnanou RM, Niamke S. Isolation of thermotolerant and high acetic acid-producing Acetobacter pasteurianus from Ivorian palm wine. Emir. J Food Agric. 2014;26:773–785. doi: 10.9755/ejfa.v26i9.18122. [DOI] [Google Scholar]
- Kongor JE, Hinneh M, Van de Walle D, Afoakwa EO, Boeckx P, Dewettinck K. Factors influencing quality variation in cocoa (Theobroma cacao) bean flavour profile—a review. Food Res Int. 2016;82:44–52. doi: 10.1016/j.foodres.2016.01.012. [DOI] [Google Scholar]
- Lefeber T, Janssens M, Camu N, De Vuyst L. Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media to compose a starter culture for cocoa bean fermentation. Appl Environ Microbiol. 2010;76:7708–7716. doi: 10.1128/AEM.01206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misnawi SJ, Jamilah B, Nazamid S. Effects of incubation and polyphenol oxidase enrichment on colour, fermentation index, procyanidins and astringency of unfermented and partly fermented cocoa beans. Int J Food Sci Technol. 2003;38:285–295. doi: 10.1046/j.1365-2621.2003.00674.x. [DOI] [Google Scholar]
- Nanda K, Taniguchi M, Ujike S, Ishihara N, Mori H, Ono H, Murooka Y. Characterization of acetic acid bacteria in traditional acetic acid fermentation of rice vinegar (komesu) and unpolished rice vinegar (kurosu) produced in Japan. Appl Environ Microbiol. 2001;67:986–990. doi: 10.1128/AEM.67.2.986-990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Syst Evol Microbiol. 2007;114:168–186. doi: 10.1016/j.ijfoodmicro.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Ouattara HG, Ban-Koffi L, Karou GT, Sangare A, Niamke LS, Diopoh JK. Implication of Bacillus sp. in the production of pectinolytic enzymes during cocoa fermentation. World J Microbiol Biotechnol. 2008;24:1753–1760. doi: 10.1007/s11274-008-9683-9. [DOI] [Google Scholar]
- Ouattara HG, Reverchon S, Niamke SL, Nasser W. Molecular identification and pectate lyase production by Bacillus strains involved in cocoa fermentation. Food Microbiol. 2011;28:1–8. doi: 10.1016/j.fm.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Ouattara HG, Reverchon S, Niamke SL, Nasser W. Regulation of the synthesis of pulp degrading enzymes in Bacillus isolated from cocoa fermentation. Food Microbiol. 2017;63:255–262. doi: 10.1016/j.fm.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Ouattara HD, Ouattara HG, Droux M, Reverchon S, Nasser W, Niamke SL. Lactic acid bacteria involved in cocoa beans fermentation from Ivory Coast: Species diversity and citrate lyase production. Int J Food Microbiol. 2017;256:11–19. doi: 10.1016/j.ijfoodmicro.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Papalexandratou Z, Camu N, Falony G, De Vuyst L. Comparison of the bacterial species diversity of spontaneous cocoa bean fermentations carried out at selected farms in Ivory Coast and Brazil. Food Microbiol. 2011;28:964–973. doi: 10.1016/j.fm.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Papalexandratou Z, Lefeber T, Bahrim B, Lee OS, Daniel HM, De Vuyst L. Hanseniaspora opuntiae, Saccharomyces cerevisiae, Lactobacillus fermentum, and Acetobacter pasteurianus predominate during well-performed Malaysian cocoa bean box fermentations, underlining the importance of these microbial species for a successful cocoa bean fermentation process. Food Microbiol. 2013;35:73–85. doi: 10.1016/j.fm.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Pereira GVM, Migue MGCP, Ramos CL, Schwan RF. Microbiological and physicochemical characterization of small-scale cocoa fermentations and screening of yeast and bacterial strains to develop a defined starter culture. Appl Environ Microbiol. 2012;78:5395–5405. doi: 10.1128/AEM.01144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspor P, Goranovic D (2016) Biotechnological application of acetic acid bacteria in food fermentations. Biotechnology. Vol VII. Encyclopedia of life support system
- Rossello-Mora R, Amann R. The species concept of prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- Romero-Cortes T, Robles-Olvera V, Rodriguez-Jimenes G, Ramírez-Lepe M. Isolation and characterization of acetic acid bacteria in cocoa fermentation. Afr J Microbiol Res. 2012;6:339–347. doi: 10.5897/AJMR11.986. [DOI] [Google Scholar]
- Samagaci L, Ouattara H, Niamké S, Lemaire M. Pichia kudrazevii and Candida nitrativorans are the most well-adapted and relevant yeast species fermenting cocoa in Agneby-Tiassa, a local Ivorian cocoa producing region. Food Res Int. 2016;89:773–780. doi: 10.1016/j.foodres.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Schwan RF. Cocoa fermentation conducted with a defined microbial cocktail inoculum. Appl Environ Microbiol. 1998;64:1477–1483. doi: 10.1128/AEM.64.4.1477-1483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan RF, Wheals AE. The microbiolgy of cocoa fermentation and its role in chocolate quality. Crit Rev Food Sci Nutr. 2004;44:205–221. doi: 10.1080/10408690490464104. [DOI] [PubMed] [Google Scholar]
- Schwan RF, Fleet GH. Cocoa and coffee fermentations. New York: CRC Press Taylor & Francis Group; 2014. [Google Scholar]
- Sharafi SM, Rasooli I, Beheshti MK. Isolation, characterization and optimization of indigenous acetic acid bacteria and evaluation of their preservation method. Iran J Microbiol. 2010;2:41–48. [PMC free article] [PubMed] [Google Scholar]
- Soumahoro S, Ouattara HG, Goualie BG, Koua G, Doue G, Niamké SL. Occurrence of high acetic acid-producing bacteria in Ivorian cocoa fermentation and analysis of their response to fermentative stress. Am J Biosci. 2015;3:70–79. doi: 10.11648/j.ajbio.20150303.12. [DOI] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P, Pot B, Gillis M, De Vos P, Kesters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/MMBR.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin S, Alessandria V, Valente A, Dolci P, Cocolin L. Molecular identification and physiological characterization of yeasts, lactic acid bacteria and acetic acid bacteria isolated from heap and box cocoa bean fermentations in West Africa. Int J Food Microbiol. 2016;216:69–78. doi: 10.1016/j.ijfoodmicro.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Voigt J, Biehl B, Heinrichs H, Kamaruddin S, Marsoner GG, Hugi A. In-vitro formation of cocoa-specific aroma precursors—aroma-related peptides generated from cocoa-seed protein by cooperation of an aspartic endoprotease and A carboxypeptidase. Food Chem. 1994;49:173–180. doi: 10.1016/0308-8146(94)90155-4. [DOI] [Google Scholar]
- Woese CR, Gutell R, Gupta R, Noller HF. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983;47:621–669. doi: 10.1128/MMBR.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Ouattara HG, Goualie B, Soumahoro S, Niamke S. Analysis of some functional properties of acetic acid bacteria involved in Ivorian cocoa fermentation. J Appl Biosci. 2014;75:6282–6290. doi: 10.4314/jab.v75i1.13. [DOI] [Google Scholar]
- Ziegleder G. Flavour development in cocoa and chocolate. In: Beckett ST, editor. Industrial chocolate manufacture and use. 4. New York: Blackwell Publishing Ltd.; 2009. pp. 169–191. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.