Abstract
Meyerozyma guilliermondii, a non-conventional yeast that naturally assimilates xylose, is considered as a candidate for biotechnological production of the sugar alternative xylitol. Because the genes of the xylose metabolism were yet unknown, all efforts published so far to increase the xylitol yield of this yeast are limited to fermentation optimization. Hence, this study aimed to genetically engineer this organism for the first time with the objective to increase xylitol production. Therefore, the previously uncharacterized genes of M. guilliermondii ATCC 6260 encoding for xylose reductase (XR) and xylitol dehydrogenase (XDH) were identified by pathway investigations and sequence similarity analysis. Cloning and overexpression of the putative XR as well as knockout of the putative XDH genes generated strains with about threefold increased xylitol yield. Strains that combined both genetic modifications displayed fivefold increase in overall xylitol yield. Enzymatic activity assays with lysates of XR overexpressing and XDH knockout strains underlined the presumed functions of the respective genes. Furthermore, growth evaluation of the engineered strains on xylose as sole carbon source provides insights into xylose metabolism and its utilization for cell growth.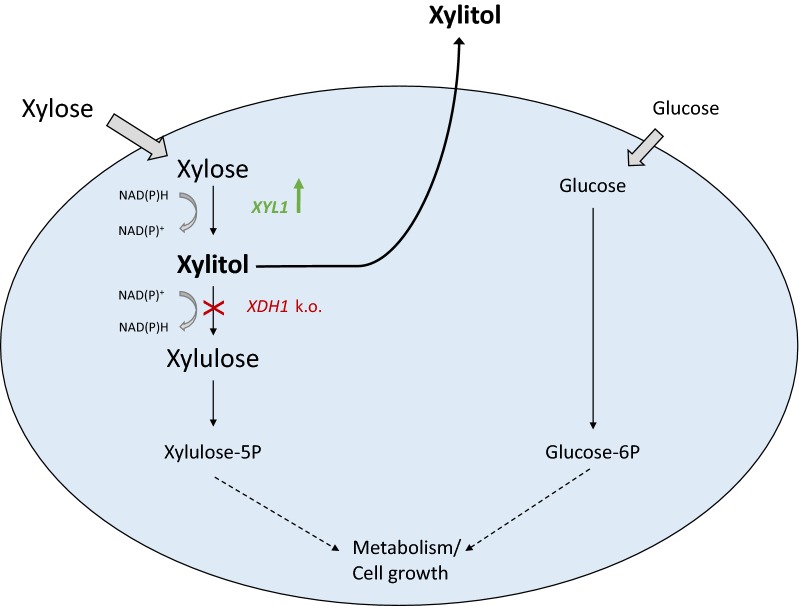
Keywords: Meyerozyma guilliermondii, Xylose, Xylitol, Xylitol dehydrogenase, Xylose reductase
Keypoints
New xylose reductase (XYL1) and xylitol dehydrogenase (XDH1) genes were identified.
M. guilliermondii was genetically modified to increase production of xylitol.
Homologous overexpression of XYL1 and knockout of XDH1 promote xylitol production.
Introduction
The interest on the five-carbon sugar alcohol xylitol has been constantly rising over the last years. This is not only because it is metabolized in an insulin independent manner and, therefore, is suitable for people suffering from diabetes, but also because of the rising interest in low-calorie diets (Ylikahri 1979). Xylitol tastes as sweet as sucrose whilst harboring about 40% less calories (Zacharis 2012). Furthermore, its anti-cariogenic properties make xylitol an interesting agent for the pharmaceutical industry (Mäkinen 1979).
Currently xylitol is produced chemically, which has some disadvantages in terms of energy demand, wastewater pollution and extensive purification requirements. The need of pure d-xylose as raw material for a nickel-catalyzed hydrogenation is one of the main cost rising factors, as this d-xylose has to be purified from mainly lignocellulosic material in a laborious and expensive way (Granström et al. 2007a; Rafiqul and Sakinah 2013).
In contrast, two biotechnological approaches are evolving over the last years: the enzymatic approach and the microbial process. For the enzymatic attempt, d-xylose is converted by purified xylose reductase (XR) from yeast. Herein the conversion is very efficient, but also has the drawback of using pure d-xylose as starting material (Kitpreechavanich et al. 1984; Neuhauser et al. 1998; Nidetzky et al. 2000). For the microbial xylitol production, the lignocellulosic starting material has to be pre-treated in certain ways, depending on the organism used, but the xylose does not have to be purified (Granström et al. 2007a; Rafiqul and Sakinah 2013; Albuquerque et al. 2014; Venkateswar Rao et al. 2015). This approach uses the fact that some microorganisms, bacteria as well as yeasts, are capable of producing xylitol in a fermentative way. Although some bacteria as Corynebacterium sp., Enterobacter liquefaciens or Mycobacterium smegmatis have been reported to produce xylitol (Winkelhausen and Kuzmanova 1998; Chen et al. 2010; Rafiqul and Sakinah 2013), yeasts are the more investigated candidates for microbial xylitol production (reviewed e.g. in (Granström et al. 2007b) and (Jeffries and Jin 2004)). Whilst there are several attempts of genetic modification of Saccharomyces cerevisiae to produce xylitol (Jeppsson et al. 2006; Dasgupta et al. 2017), other, non-conventional yeasts, like amongst others Candida sp., Kluyveromyces sp. or Trichoderma reesei, are natural producers of this sugar alcohol (Winkelhausen and Kuzmanova 1998; Chen et al. 2010; Venkateswar Rao et al. 2015), and are therefore probably more promising candidates for biotechnological production of xylitol.
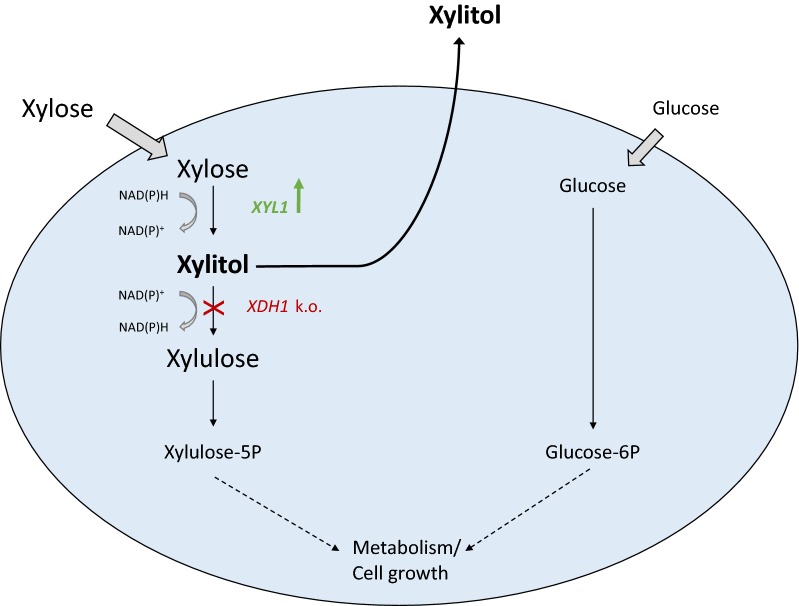
Generally, there are two ways microbes metabolize xylose. Bacteria use a one step process, catalysed by xylose isomerase (XI), for the direct conversion from xylose to xylulose (Karhumaa et al. 2007) whilst yeast and fungi use a two-step process (Fig. 1). The first step is the xylose reductase (XR; EC 1.1.1.307) or aldose reductase (EC 1.1.1.21) catalyzed conversion of xylose into xylitol, which then is further converted by xylitol dehydrogenase (XDH; EC 1.1.1.9) or l-iditol 2-dehydrogenase (EC 1.1.1.14) to form d-xylulose, which can be used in the pentose phosphate pathway (Granström et al. 2007b; Karhumaa et al. 2007; Rafiqul and Sakinah 2013), or by l-xylulose reductase (EC 1.1.1.10) or d-iditol 2-dehydrogenase (EC1.1.1.15) to form l-xylulose.
Fig. 1.
Illustration of xylose metabolism pathway. Xylose metabolism (part) of xylose-utilizing non-conventional yeasts (suggested in (da Silva et al. 2005)) Xylose metabolizing enzymes covered in this study are highlighted in green (XYL1 = xylose reductase) and red (XDH1 = xylitol dehydrogenase)
Meyerozyma guilliermondii (anamorph: Candida guilliermondii) is a non-conventional yeast, which produces xylitol naturally, and therefore has been under intense investigation for many years. Its relatively high tolerance to inhibitors which typically occur in hydrolysates of lignocellulosic material, which is a low-cost substrate of great interest, makes it an interesting organism for the use in biorefineries (Pereira et al. 2011). So far, most of the past work was focused on the optimization of the fermentation conditions or the substrate preparation (reviewed e.g. in (Mohamad et al. 2015)) but no genetic optimization regarding xylitol production in M. guilliermondii is available in the literature. One reason for the lack of research in this field may be, that the prerequisite for this intention, the availability of genetic tools for the organism, was just fulfilled by the work of different scientists in the last decade (Millerioux et al. 2011; Foureau et al. 2013; Defosse et al. 2014).
The encouraging potential of enhancing xylitol production genetically has been previously shown with other yeasts like Candida tropicalis or Trichoderma reesei (Ko et al. 2006b; Hong et al. 2014). Ko et al. (2006a, b) could reach a 97% xylose to xylitol conversion in their xylitol dehydrogenase gene disrupted C. tropicalis strain, compared to 67.2% of the wildtype (Ko et al. 2006a). Likewise, a knockout of XDH in T. reesei resulted in an increase from undetectable to 8.6 mM xylitol (Hong et al. 2014). In general, xylose metabolism in CTG clade family members, like C. tropicalis, P. stipitis or M. caribbica has been quite intensively studied (Amore et al. 1991; Byoung et al. 2006; Sukpipat et al. 2017). However, only limited data are available for M. guilliermondii: Handumrongkul et al. (1998) overexpressed a XYL1 gene of M. guilliermondii ATCC 20118 in P. pastoris (Handumrongkul et al. 1998). Yet, genes responsible for xylose metabolism in M. guilliermondii have not been investigated within the organism itself. Consequently, the aim of this study was to identify and manipulate XR (XYL1) and XDH (XDH1) genes in M. guilliermondii and evaluate the impact on xylitol production.
Materials and methods
Strains and culture conditions
The M. guilliermondii strains used and generated in this study are listed in Table 1. KU141F1 was kindly provided by N. Papon (Foureau et al. 2013) and was used as parental strain for all genetic modifications in this study. KU141F1 and hence all strains derived thereof are originated from the reference strain M. guilliermondii ATCC 6260 (American Type Culture Collection, ATCC Manassas, USA). The strains were maintained on YNB(S)-agar (Yeast Nitrogen Base without amino acids 6.7 g l−1, Lactan; sucrose 20 g l−1; agar 20 g l−1) or YPS-agar (yeast extract 10 g l−1; peptone from casein 20 g l−1; sucrose 20 g l−1; agar 20 g l−1). Liquid cultures were prepared in YNB(S)- or YPS-media and incubated at 30 °C and 170 rpm in an orbital shaker. Escherichia coli XL1-blue was used as host for cloning. The wild-type and recombinant E. coli were cultured in Luria–Bertani (LB, Roth) liquid medium or on LB-agar plates, with or without ampicillin (100 mg l−1) as required, at 37 °C.
Table 1.
M. guilliermondii strains generated and used in this study
| Strain | Genotype | Plasmid | Refs. |
|---|---|---|---|
| ATCC 6260 | wild type | – | – |
| KU141F1* | ura5, ku70Δ | – | (Foureau et al. 2013) |
| Ctrl (KU [EV])* | ura5, ku70Δ | [pG-URA5-PACT1-TTRP1] | (Foureau et al. 2013)/This study |
| xdh∆::URA5* | ura5, ku70Δ, xdh1∆::URA5 | – | This study |
| xdhΔ* | ura5, ku70Δ, xdh1Δ (xdh1∆::REP) | [pG-URA5-PACT1-TTRP1] | This study |
| KU [XR]* | ura5, ku70Δ | [pU-XR] | This study |
| xdhΔ [XR]* | ura5, ku70Δ, xdh1Δ (xdh1∆::REP) | [pU-XR] | This study |
| xdh∆::XR* | ura5, ku70Δ, xdh1∆::XYL1 | – | This study |
* KU141F1 and hence all strains derived thereof are originated from the reference strain M. guilliermondii ATCC 6260
Genetic modifications
The used primers are listed in Additional file 1: Table S1, plasmids in Table 2. For the knockout of the putative XDH gene, XDH1 (PGUG_05726), 1000 bp upstream (primer P1 and P2) and 1000 bp downstream (primer P3 and P4) of XDH1 were amplified by PCR and cloned up- and down-stream of the URA5-selection cassette of pGRU5R to generate pU-XDH-KO. The knockout cassette was amplified from the plasmid (primer P1 and P4) and transformed into M. guilliermondii KU141F1 to generate xdh∆::URA5. The successful homologous recombination was verified by PCR (primer S1, S2 and S3, shown in Additional file 1: Fig. S2). To eliminate the URA5-selection marker from the genome again, URA-blaster was performed as described by Foreau et al. (Foureau et al. 2012) by plating the mutants on media containing 1 mg ml−1 5-FOA (5-Fluoroorotic acid) and 300 ng ml−1 Uridine, resulting in the xdhΔ strain.
Table 2.
plasmids used and generated in this study
| Plasmid | Purpose | Source/Refs. |
|---|---|---|
| pG-RU5R | Backbone for knockout plasmids, URA5 marker | (Millerioux et al. 2011) |
| pG-URA5-PACT1-TTRP1 | Expression plasmid backbone for protein expression in M. guilliermondii (= empty vector control [pU-EV]) | (Defosse et al. 2014) |
| pU-XDH-KO | Plasmid for genomic knockout of XDH1 (PGUG_05726) with URA5 | This study |
| pU-XR | Plasmid for the expression of XYL1 (PGUG_00922) in M. guilliermondii (= [pU-XR]) | This study |
| pU-XDH-XR-KO | Plasmid for genomic replacement of XDH1 (PGUG_01218) by XYL1 (PGUG_00922) | This study |
In order to overexpress the putative XR-gene, XYL1 (PGUG_00922), it was amplified by PCR from the genome of M. guilliermondii ATCC6260 (primer P5 and P6) and cloned under the control of PAct1, a constitutively active promoter into pG-URA5-PACT1-TTRP1 to generate pU-XR. This was transformed into M. guilliermondii KU141F1 as described below and selected on YNB(S) agar plates to generate the strain KU [XR]. Further, pU-XR was transformed into the xdhΔ strain to generate the xdhΔ [XR] strain.
Moreover, the open reading frame of XDH1 was replaced in the genome of M. guilliermondii KU141F1 by XYL1. Therefore, XYL1 was amplified by PCR from the genome of M. guilliermondii KU141F1 (primer P9 and P10) and cloned into pU-XDH1-KO, generating pU-XDH1-XR-KO. The knockout cassette was amplified from the plasmid (primer P1 and P4) and transformed into M. guilliermondii KU141F1 to generate the strain xdh1∆::XR-URA5. The successful homologous recombination was verified by PCR (primer S1, S2 and S3). The URA5 selection marker was released by URA-blaster as described above, resulting in the xdh1∆::XR strain.
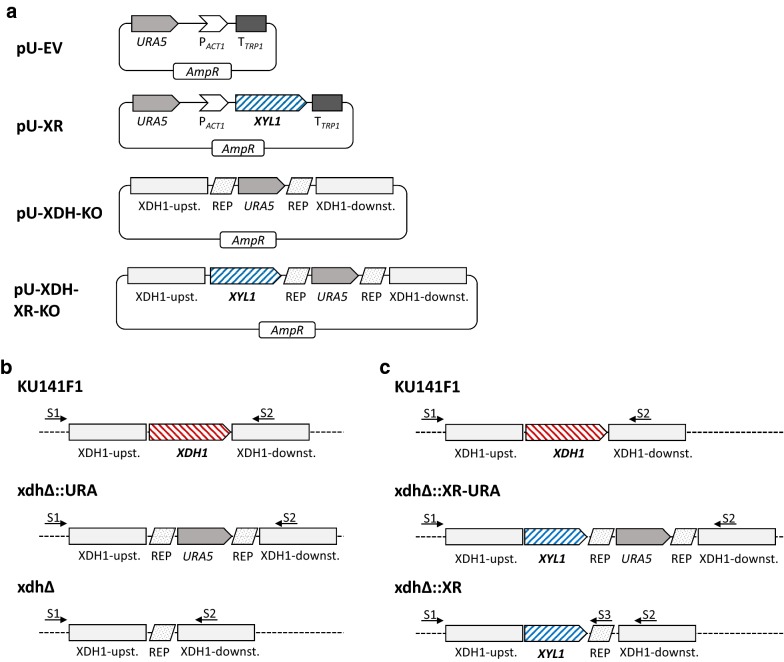
Additionally, pG-URA5-PACT1-TTRP1 without insert was transformed into KU141F1 (KU [EV]), xdhΔ (xdhΔ [EV]) and xdh∆::XR (xdh∆::XR [EV]) to serve as control strains. All the plasmids and resulting genetic modifications are illustrated in Fig. 2.
Fig. 2.
Schematic representation of the plasmids used and generated in this study, and the resulting genetic modifications. a shows the vectors used in this study: pU-EV is the empty vector used as control, pU-XR is used for the overexpression of the xylose reductase gene XYL1, pU-XDH-KO for the knockout of the xylitol dehydrogenase gene XDH1, and pU-XDH-XR-KO for genomic replacement of XDH1 by XYL1. b Deletion of XDH1 by homologous recombination of the selection cassette (xdh∆::URA) and after release of URA5 by Ura-Blaster (xdhΔ) in the parental strain M. guilliermondii KU141F1. c Replacement of XDH1 by XYL1 (xdh∆::XR-URA) and after release of URA5 by Ura-Blaster (xdh∆::XR) in the parental strain KU141F1; AmpR: Ampicillin resistance gene; URA5: selection marker for uridine-prototrophy, PACT1: ACT1 promotor for constitutive expression of XYL1, TTRP1: Terminator of XYL1 expression; REP: repetitive sequences for the use of the Ura-Blaster system to release URA5 selection marker; XDH1-upstr.: genomic sequence 1000 bp upstream of XDH1; XDH1-downst.: genomic sequence 1000 bp downstream of XDH1; S1, S2 and S3 indicate sequencing primer binding sites
Yeast transformation
The method for the transformation of M. guilliermondii was adapted from Boretsky et al. (2007).
Preparation of competent cells: The main culture was inoculated in 50 ml YPS + xylose (10 g l−1) and grown to an OD600 of approximately 0.5. The whole culture was harvested, washed and suspended in LiAc/TE-Buffer (lithium acetate 100 mM; TRIS–HCl 10 mM; EDTA 1 mM; pH 7.5). After incubation for 1 h at 30 °C and shaking, cells were harvested and re-suspended in LiAc/TE-Buffer to a concentration of approximately 5 × 109 cfu ml−1 (OD 0.5 equals 2.4 × 107 cfu ml−1). The suspension was spread into 50 µl aliquots for the transformation.
Transformation: 2–10 µg plasmid DNA or 2–2.5 µg linear DNA, 250 µl 50% PEG (polyethylene glycol) in LiAc/TE buffer and 5 µl herring sperm DNA (freshly denaturated at 95 °C for 10 min, chilled on ice) were added to the competent cells, mixed carefully and incubated at 30 °C for 30 min. After a heat shock (42 °C, 15 min) cells were centrifuged at 4 °C, re-suspended in 1 ml pre-warmed (30 °C) YPS and incubated for 3 to 5 h at 30 °C with shaking. Cells ware harvested, re-suspended in 100 µl 1 M sucrose, plated on selection plates and incubated at 30 °C for 2–3 days.
Enzyme activity assay
The enzymatic activities of XDH and XR were analyzed after a modified protocol from Veras et al. (2017). Briefly, 10 OD units of an exponentially growing culture (cultivated in YNB(S) + 10 g l−1 xylose for 8 h) are harvested and washed with distilled water. Cells were lysed using Y-PER® (Thermo Scientific, Vienna, Austria) according to the manufacturers protocol. The total protein in the cell free lysate was quantified by Roti®-Quant (Lactan, Graz, Austria) as described by the manufacturer.
The XR-reaction mix consisted of triethanolamine buffer (100 mM, pH 7.0), NADPH (0.2 mM) and xylose (350 mM). The XDH-reaction mix consisted of triethanolamine buffer (100 mM, pH 9.0), NAD+ (0.3 mM), xylitol (300 mM). The reactions were started by adding 1:10 crude cell lysate to the respective reaction mix and incubated at room temperature. Change of the absorbance at 340 nm, corresponding to co-factor conversion, was followed over 10 min. Enzyme activities (U mg−1protein) were calculated by using 6.22 ml (µmol cm)−1 as the molar absorption coefficient and 1 enzyme unit is defined as 1 μmol of cofactor reduction or oxidation per minute.
Sugar and xylitol quantification
To analyze saccharides and xylitol, the culture samples were centrifuged at the indicated time points, and the supernatant was used for the quantification by HPLC, using a Jasco HPLC 2000 plus series (Biolab, Vienna, Austria) with an Aminex hpx 87 h column at 65 °C. H2SO4 (c = 5 mmol l−1) was used as eluent at an isocratic flow rate of 0.8 ml min−1. Data acquisition was performed with a refractive index detection. Data were analyzed with ChromPass (Version 1.8.6.1, Jasco Europe, Italy).
Statistical analysis
All experiments were performed at least three times, each time in triplicates. Statistical analysis was performed with Prism v8.0.2—GraphPad Software, San Diego California, USA. Outliers were detected by a Grubbs outlier test and statistical significance was calculated via one-way ANOVA followed by a Tukey HSD test (xylitol yield) or students t test (enzymatic activity assays). Values are displayed as means with standard deviation of the mean. p < 0.05 = *, p < 0.005 = ** and p < 0.005 = ***.
Results
To find the putative genes for XR and XDH in M. guilliermondii, investigations of the pentose and glucuronate interconversions pathway of M. guilliermondii 6260 (NCBI ref. gen. seq. AAFM00000000.1, assembly: GCF_000149425.1 from whole genome shotgun sequencing (Butler et al. 2009), were performed on the KEGG website (https://www.genome.jp/kegg-bin/show_pathway?pgu00040). This analysis revealed the enzyme activity EC 1.1.1.307 (d-xylose reductase) to PGUG_00922 of the M. guilliermondii ATCC6260 genomic scaffold 1 (NCBI ref. seq. NW_001809800.1). BLAST analysis revealed 94.32% sequence identity to the previously described XR of M. guilliermondii ATCC 20118 (alignment in Additional file 1: Fig. S1), which was 70.6% identical to XR of S. stipitis (Handumrongkul et al. 1998). Furthermore, activity EC 1.1.1.9 (d-xylulose reductase or NAD + -dependent xylitol dehydrogenase), which catalyzes the reaction from d-xylulose to xylitol and vice versa, was linked to PGUG_05726 of the M. guilliermondii ATCC6260 genomic scaffold 8 (NCBI ref. seq. NW_001809793.1). BLAST analysis revealed a 97.24% sequence identity to a previously described XDH of M. caribbica (Sukpipat et al. 2017).
Cloning of XYL1 and knockout of XDH1
PCR-amplification of the XYL1 gene from M. guilliermondii ATCC 6260 genomic DNA with primers P5 and P6 yielded a 955 bp fragment, which was subsequently cloned into the expression vector pG-URA5-PACT1-TTRP1 (Defosse et al. 2014). The successful cloning of the resulting expression plasmid (pU-XR) was confirmed by sequencing, then the plasmid was transformed into M. guilliermondii strains KU141F1 and xdh∆ (see Fig. 2 for a schematic representation of the plasmids and resulting genetic modifications). Transformants grown on selection medium without uridine allowed for selection of pU-XR carrying transformants.
To disrupt the putative XDH1 gene, two approaches were applied: in a first attempt, the genomic site of PGUG_5726 (XDH1) was disrupted by the URA5 selection cassette (pU-XDH-KO). In the second approach, the XDH1 gene was disrupted by the XYL1 genomic sequence (pU-XDH-XR-KO). Thereby, a stable integration of an additional XYL1 gene copy into the genomic DNA with a simultaneous disruption of XDH1 was generated. The knockout of XDH1 by URA5 and XYL1, respectively, yielding the strains xdh∆ and xdh∆::XR, was screened by PCR (Fig. S2) and confirmed by sequencing.
Enzymatic activity assessment of XYL1 overexpressing and XDH1 knockout strains
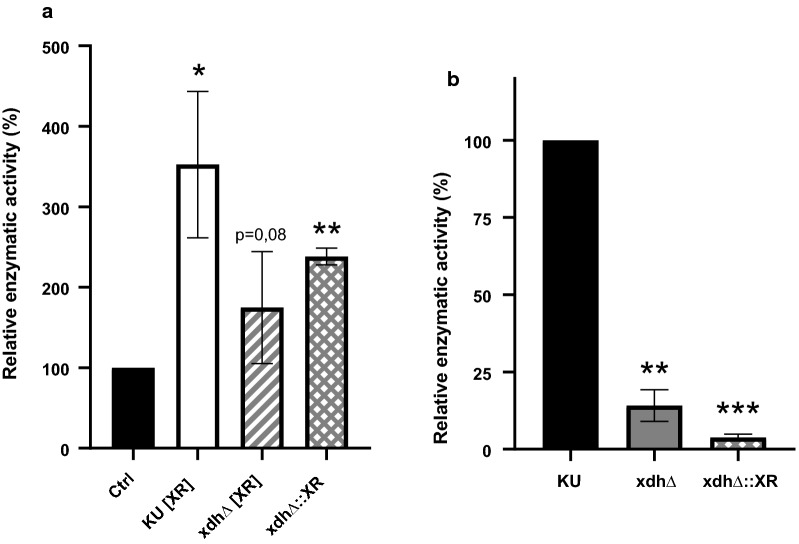
In order to characterize the putative XYL1 and XDH1 genes, enzymatic activity assays with crude cell lysates of strains overexpressing XYL1 and strains with disrupted XDH1 genes were performed. When xylose and NADPH were provided as substrates, lysates of KU [XR] indeed displayed 350% increased NADPH to NADP + conversion, see Fig. 3a. Lysates of xdh∆ [XR] displayed 175% and the xdh∆::XR strain 240% increased XR activity. Vice versa, lysates of xdh∆ and xdh∆::XR exhibited strongly decreased NAD + to NADH conversion when xylitol and NAD + were provided, see Fig. 3b. In ∆xdh only 14% and in xdh∆::XR only 4% of the parental strain’s XDH activity were remaining. This provides clear evidence that both genes actually encoded for the predicted enzymes, which converted the expected substrates and co-factors (xylose and NADPH for XR, and xylitol and NAD + for XDH).
Fig. 3.
Enzymatic activity analysis of M. guilliermondii cell lysates. Measurement of co-factor conversion rates is displayed as a relative enzymatic XR-activity of lysates of KU [EV] (Ctrl), KU [XR], xdhΔ [XR] and xdh∆::XR; b relative enzymatic XDH-activity of lysates of KU, xdhΔ and xdh∆::XR; Displayed are mean values ± SD of at least 3 replicate experiments, each performed in triplicates. The control and parental strains were set to 100%. Asterisks indicate p-values (* = p < 0.05; ** = p < 0.005 and *** = p<0.001)
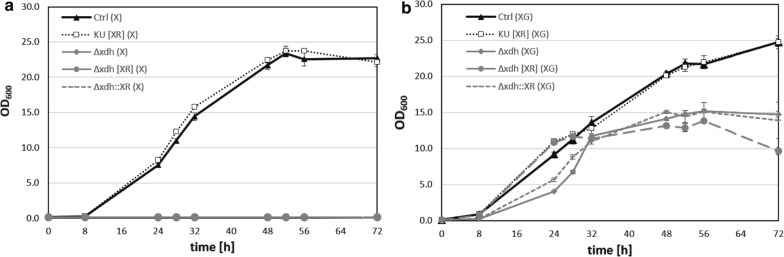
Growth evaluation of mutant strains
To get an insight in growth behavior and xylose metabolism of the genetically engineered strains, growth in xylose containing media was assessed. As can be seen in Fig. 4, overexpression of XYL1 did not markedly affect growth characteristics of M. guilliermondii. However, mutants with disrupted XDH1 genes were not able to grow at all on media with xylose as the sole carbon source, see Fig. 4a. Supplementation with glucose as an additional carbon sources allowed growth of the various XDH1 knockout strains (Fig. 4b). However, they grew notably slower and to considerably lower final densities compared to the control strain.
Fig. 4.
Growth evaluation of M. guilliermondii strains with xylose only or xylose + glucose as carbon source. Growth comparison of parental strain Ctrl (KU [EV]) (filled triangles) with mutant strains KU [XR] (open square), xdhΔ (filled diamond), xdhΔ [XR] (filled circle) and xdh∆::XR (dashed lines) in YNB media with a 20 g l−1 xylose (X) or b 20 g l−1 xylose and 20 g l−1 glucose (XG); Displayed are mean values ± SD of triplicates
Xylitol production of genetically engineered strains
Finally, the xylitol production of all strains generated during this project was analyzed in shake flask fermentations (shown in Fig. 5). Overexpression of XYL1 resulted in a threefold increase in xylitol accumulation after 72 h, yielding 3.89 ± 0.47 g l−1 for KU [XR], compared to 1.29 ± 0.35 g l−1 for the control strain KU [EV]. Similarly, xdhΔ showed increased xylitol accumulation to comparable amounts, 3.41 ± 0.73 g l−1. Although overall growth of double mutant strains xdhΔ [XR] and xdh∆::XR was substantially lower than that of KU [XR], xylitol yield after 72 h was the highest of all the strains investigated in this study. xdhΔ [XR] produced 5.31 ± 0.69 g l−1 after 72 h of fermentation, resulting in a volumetric productivity of 0.074 g l−1 h−1 and a specific productivity of 0.27 g xylitol per g xylose. The knockout of XDH1 by XYL1 (xdh∆::XR) yielded 4.28 ± 1.30 g l−1 after 72 h of fermentation, resulting in a volumetric productivity of 0.060 g l−1 h−1 and a specific productivity of 0.21 g xylitol per g xylose. The yields and productivities of the above mentioned strains are summarized in Table 3.
Fig. 5.
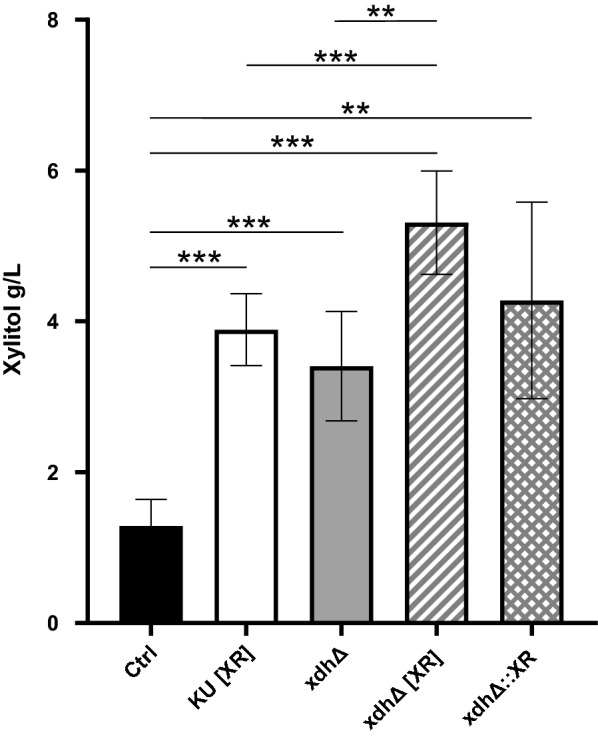
Xylitol production of M. guilliermondii mutant strains. Xylitol yield (in g l−1 as determined by HPLC) of KU [EV] (Ctrl); KU [XR]; xdhΔ, xdhΔ [XR] and xdh∆::XR after 72 h cultivation in YNB with 20 g l−1 xylose and 20 g l−1 glucose; Displayed are mean values ± SD of at least 3 replicate experiments, each performed in triplicates. Asterisks indicate p-values (** = p < 0,005 and *** = p<0,001)
Table 3.
Yields (after 72 h growth on 20 g l−1 glucose and 20 g l−1 xylose) and productivities of the strains used in this study
| Strain name | Yield (g l−1) | Vol. productivity (g l−1 h−1) | Spec. productivity (g xylitol per g xylose) |
|---|---|---|---|
| Ctrl (KU [EV]) | 1.29 ± 0.35 | 0.018 | 0.06 |
| xdhΔ | 3.41 ± 0.73 | 0.047 | 0.17 |
| KU [XR] | 3.89 ± 0.47 | 0.054 | 0.19 |
| xdhΔ [XR] | 5.31 ± 0.69 | 0.074 | 0.27 |
| xdh∆::XR | 4.28 ± 1.30 | 0.060 | 0.21 |
Discussion
In this study, we provide data that allow interpretation of the function of two previously undescribed M. guilliermondii genes involved in xylose metabolism. On the one hand, we cloned and homologously overexpressed XYL1 (PGUG_0922), which encodes for a putative XR. Indeed, overexpression of XYL1 (KU [XR], xdhΔ [XR], xdh∆::XR) enabled higher xylitol accumulation in the medium, see Fig. 5. Moreover, lysates of XYL1 overexpressing strains KU [XR] and xdh∆::XR showed significantly higher enzymatic activity on xylose with NAPDH provided as substrate (shown in Fig. 3a). Although the increase of co-factor conversion rates can also be observed in ∆xdh [XR] lysates, here the effect was not as outspoken as in the other XR overexpressing strains. It has been demonstrated that xylose reductases preferentially use NADPH as co-factor and that this specificity is determined by a conserved IPKS amino acid motif (Silva et al. 1996; Kostrzynska et al. 1998; Petschacher et al. 2005). PGUG_00922 of M. guilliermondii encodes for this sequence motif, highlighted in Additional file 1: Fig. S1, as well. Together, these data strongly support our hypothesis that M. guilliermondii PGUG_00922 is indeed a XYL1 gene encoding for a XR that uses NADPH as a cofactor.
On the other hand, lysates of XDH1 disrupted mutants (xdhΔ, xdh∆::XR) exhibited significantly lower enzymatic activity on xylitol with NAD + provided as a cofactor compared to lysates of the parental strain (KU), as shown in Fig. 3b, thereby providing evidence that the putative XDH1 gene of M. guilliermondii PGUG_05726 is indeed a XDH that uses NAD + as a cofactor.
Mutants with disrupted XDH1 genes (xdhΔ, xdh∆::XR) were not able to grow on xylose as sole carbon source (Fig. 4a). Conversely, the parental strain (Ctrl) grew equally well on xylose as a sole carbon source (Fig. 4a) compared to when glucose is present (Fig. 4b). These data strongly indicate that xylose metabolism is completely blocked in XDH1 knockout strains. Obviously, its function (converting xylitol to d- or l-xylulose) could not be compensated by other enzymes, at least not in the experimental conditions applied. Hence, XDH1 is probably the only enzyme in M. guilliermondii ATCC 6260 capable of processing xylitol and thus providing carbon for cell growth. Growth of M. guilliermondii XDH1 knockout strains could be restored to a certain extent by supplementation with glucose (Fig. 4b). Correspondingly, the necessity of co-substrates for adequate growth of XDH-defective mutants has been demonstrated for xylose assimilating yeasts, like C. tropicalis, P. stipitis, K. marxianus and D. hansenii (Kim et al. 2001; Ko et al. 2006a; Pal et al. 2013; Zhang et al. 2014). Contrasting to our observations in M. guilliermondii, knockout of an XDH gene in C. tropicalis actually resulted in lower xylitol yield compared to the parental strain, due to growth impairment of the knockout strain (Ko et al. 2006a). Although, addition of glucose enabled growth of the XDH1 knockout strains we generated, the parental strain still grew significantly better under these conditions. The major differences in the overall growth rate between wild type and XDH1 defective strains can most probably be attributed to differences in the ability to utilize xylose. However, the parental strain also grew faster than xdhΔ and xdh∆::XR within the first 24–32 h. In this early growth phase only glucose is supposed to be used (shown by (Gurgel et al. 1998) and observed by us, data not shown). This might indicate that an intact xylose metabolic pathway seems to be required for realization of the full growth potential.
Overexpression of XYL1 in addition to knockout of XDH1 resulted in significantly increased xylitol levels (Fig. 5). Conversely, homologous overexpression of XR in an XDH-disrupted mutant of T. reesei only slightly increased total xylitol yield (Hong et al. 2014). However, in our hands xylitol production improves significantly only when overexpressed from a plasmid (xdhΔ [XR]). Simply supplying another genetic copy of XYL1 at the XDH1 locus (xdh∆::XR) only slightly increased the total xylitol yield. This might be due to different gene copy numbers or promotor activities of the expression vector and the XDH1 locus.
The overall xylitol yields of the M. guilliermondii ATCC 6260 strain are intrinsically rather low compared to other xylitol producing yeasts or fungi (Pal et al. 2016; Dasgupta et al. 2017). Although total xylitol yield remains within the single-digit gram per liter range, it could be substantially enhanced by overexpression of XR and knockout of XDH in M. guilliermondii ATCC 6260. With the remarkable relative increase of about 500% of the double mutants in mind, it would be highly interesting to expand the strategy applied here, to other strains, like M. guilliermondii FTI 20037, that produce higher amounts of xylitol per se (Barbosa et al. 1988; Winkelhausen and Kuzmanova 1998; Das and De 2005). As shown for other Candida spp., optimization of media and fermentation conditions could certainly enhance this strains xylitol production rate (Horitsu et al. 1992). Further studies examining the underlying mechanisms of this relatively low xylitol productivity of M. guilliermondii ATCC 6260 and addressing the respective rate-limiting steps could provide further insights into xylitol production and xylitol metabolism in general. It would be interesting to see whether expression of heterologous XRs, improving xylose uptake or co-factor balancing could enhance this strains productivity.
Meyerozyma guilliermondii is a promising potential candidate that can grow on alternative substrates as carbon sources in biotechnological production processes (Carvalho et al. 2002; Mussatto et al. 2006; Pereira et al. 2011; Cassabarbosa et al. 2015; Hernández-Pérez et al. 2016; Martini et al. 2016; López-Linares et al. 2018). Insight into the genetic mechanisms underlying its xylose metabolism and its growth behavior will facilitate its future use as cell factory. Moreover, XRs of various species have been expressed in S. cerevisiae in order to enable fermentation of xylose and thereby improve biotechnological production of e.g. ethanol (Amore et al. 1991; Karhumaa et al. 2007; Watanabe et al. 2007; Petschacher and Nidetzky 2008; Bengtsson et al. 2009). The characterization of additional XRs will allow greater variability and fine-tuning of xylose conversion, also in heterologous expression systems.
Supplementary information
Additional file 1. Additional figures and table.
Acknowledgements
We would like to thank Daniel Sulzenbacher and Peter Preiner for technical assistance, and Manuel Selg for critically reviewing the manuscript.
Code availability
Not applicable.
Authors’ contributions
DA and AZ conceived and designed research. DA and NU conducted experiments. All authors analyzed data. DA and AZ wrote the manuscript. All authors read and approved the manuscript.
Funding
The project was funded by the European Regional Development Fund under the Grant Agreement Number 2015-132711.
Availability of data and materials
All relevant data are supplied within the manuscript and additional file.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors read the manuscript and expressed their consent for publication.
Consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13568-020-01012-8.
References
- Amore R, Kötter P, Küster C, Ciriacy M, Hollenberg CP. Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (XYL1) from the xylose-assimilating yeast Pichia stipitis. Gene. 1991;109:89–97. doi: 10.1016/0378-1119(91)90592-Y. [DOI] [PubMed] [Google Scholar]
- Barbosa MFS, de Medeiros MB, de Mancilha IM, Schneider H, Lee H. Screening of yeasts for production of xylitol from d-xylose and some factors which affect xylitol yield in Candida guilliermondii. J Ind Microbiol. 1988;3:241–251. doi: 10.1007/BF01569582. [DOI] [Google Scholar]
- Bengtsson O, Hahn-Hägerdal B, Gorwa-Grauslund MF. Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol Biofuels. 2009 doi: 10.1186/1754-6834-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boretsky YR, Pynyaha YV, Boretsky VY, Kutsyaba VI, Protchenko OV, Philpott CC, Sibirny AA. Development of a transformation system for gene knock-out in the flavinogenic yeast Pichia guilliermondii. J Microbiol Methods. 2007;70:13–19. doi: 10.1016/j.mimet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJP, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PWJ, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KAT, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MPH, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NAR, Lorenz MC, Birren BW, Kellis M, Cuomo CA. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459(7247):657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byoung SK, Heung CJ, Jung HK. Molecular cloning and characterization of NAD + -dependent xylitol dehydrogenase from Candida tropicalis ATCC 20913. Biotechnol Prog. 2006;22:1708–1714. doi: 10.1021/bp060263i. [DOI] [PubMed] [Google Scholar]
- Carvalho W, Silva SS, Converti A, Vitolo M. Metabolic behavior of immobilized Candida guilliermondii cells during batch xylitol production from sugarcane bagasse acid hydrolyzate. Biotechnol Bioeng. 2002;79:165–169. doi: 10.1002/bit.10319. [DOI] [PubMed] [Google Scholar]
- Cassabarbosa LA, Procópio REL, Matos ITSR, Filho SA. Isolation and characterization of yeasts capable of efficient utilization of hemicellulosic hydrolyzate as the carbon source. Genet Mol Res. 2015;14:11605–11612. doi: 10.4238/2015.September.28.12. [DOI] [PubMed] [Google Scholar]
- Chen X, Jiang ZH, Chen S, Qin W. Microbial and bioconversion production of d-xylitol and its detection and application. Int J Biol Sci. 2010;6:834–844. doi: 10.7150/ijbs.6.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva DDV, de das Almeida Felipe MG, de Mancilha IM, Silva SS. Evaluation of inoculum of Candida guilliermondii grown in presence of glucose on xylose reductase and xylitol dehydrogenase activities and xylitol production during batch fermentation of sugarcane bagasse hydrolysate. Appl Biochem Biotechnol. 2005;121–124:427–437. doi: 10.1385/ABAB:121:1-3:0427. [DOI] [PubMed] [Google Scholar]
- Das A, De A. Evaluation of inoculum of Candida guilliermondii grown in presence of glucose on xylose reductase and xylitol dehydrogenase activities and xylitol production during batch fermentation of sugarcane bagasse hydrolysate. Biotechnol Fuels Chem. 2005;121:427–437. doi: 10.1385/abab:121:1-3:0427. [DOI] [PubMed] [Google Scholar]
- Dasgupta D, Bandhu S, Adhikari DK, Ghosh D. Challenges and prospects of xylitol production with whole cell bio-catalysis: a review. Microbiol Res. 2017;197:9–21. doi: 10.1016/j.micres.2016.12.012. [DOI] [PubMed] [Google Scholar]
- De Albuquerque TL, Da Silva IJ, De MacEdo GR, Rocha MVP. Biotechnological production of xylitol from lignocellulosic wastes: a review. Process Biochem. 2014;49:1779–1789. doi: 10.1016/j.procbio.2014.07.010. [DOI] [Google Scholar]
- Defosse TA, Melin C, Obando Montoya EJ, Lanoue A, Foureau E, Glévarec G, Oudin A, Simkin AJ, Crèche J, Atehortùa L, Giglioli-Guivarc’h N, Clastre M, Courdavault V, Papon N. A new series of vectors for constitutive, inducible or repressible gene expression in Candida guilliermondii. J Biotechnol. 2014;180:37–42. doi: 10.1016/j.jbiotec.2014.03.034. [DOI] [PubMed] [Google Scholar]
- Foureau E, Courdavault V, Simkin AJ, Pichon O, Crèche J, Giglioli-Guivarc’h N, Clastre M, Papon N. Optimization of the URA-blaster disruption system in Candida guilliermondii: efficient gene targeting using the URA3 marker. J Microbiol Methods. 2012;91:117–120. doi: 10.1016/j.mimet.2012.07.020. [DOI] [PubMed] [Google Scholar]
- Foureau E, Courdavault V, Rojas LF, Dutilleul C, Simkin AJ, Crèche J, Atehortùa L, Giglioli-Guivarc’h N, Clastre M, Papon N. Efficient gene targeting in a Candida guilliermondii non-homologous end-joining pathway-deficient strain. Biotechnol Lett. 2013;35:1035–1043. doi: 10.1007/s10529-013-1169-7. [DOI] [PubMed] [Google Scholar]
- Granström TB, Izumori K, Leisola M. A rare sugar xylitol. Part II: biotechnological production and future applications of xylitol. Appl Microbiol Biotechnol. 2007;74:273–276. doi: 10.1007/s00253-006-0760-4. [DOI] [PubMed] [Google Scholar]
- Granström TB, Izumori K, Leisola M. A rare sugar xylitol. Part I: the biochemistry and biosynthesis of xylitol. Appl Microbiol Biotechnol. 2007;74:277–281. doi: 10.1007/s00253-006-0761-3. [DOI] [PubMed] [Google Scholar]
- Gurgel PV, Vieira DC, Furlan SA, de Mancilha IM. Growth of Candida guilliermondii FTI 20037 on mixed substrate. Braz Arch Biol Technol. 1998;41(3):335–338. doi: 10.1590/S1516-89131998000300010. [DOI] [Google Scholar]
- Handumrongkul C, Ma DP, Silva JL. Cloning and expression of Candida guilliermondii xylose reductase gene (xyl1) in Pichia pastoris. Appl Microbiol Biotechnol. 1998;49:399–404. doi: 10.1007/s002530051189. [DOI] [PubMed] [Google Scholar]
- Hernández-Pérez AF, de Arruda PV, das de Felipe MGA. Sugarcane straw as a feedstock for xylitol production by Candida guilliermondii FTI 20037. Brazilian J Microbiol. 2016;47:489–496. doi: 10.1016/j.bjm.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Dashtban M, Kepka G, Chen S, Qin W. Overexpression of d-xylose reductase (xyl 1) gene and antisense inhibition of d-xylulokinase (xyiH) gene increase xylitol production in Trichoderma reesei. Biomed Res Int. 2014 doi: 10.1155/2014/169705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horitsu H, Yahashi Y, Takamizawa K, Kawai K, Suzuki T, Watanabe N. Production of xylitol from d-xylose by Candida tropicalis: optimization of production rate. Biotechnol Bioeng. 1992;40:1085–1091. doi: 10.1002/bit.260400912. [DOI] [PubMed] [Google Scholar]
- https://www.genome.jp/kegg-bin/show_pathway?pgu00040 KEGG—Pentose and glucuronate inter conversions—Meyerozyma guilliermondii. https://www.genome.jp/kegg-bin/show_organism?org=pgu. Accessed 15 June 2017
- Jeffries TW, Jin YS. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biotechnol. 2004;63:495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hägerdal B, Gorwa-Grauslund MF. The expression of a Pichia stipitis xylose reductase mutant with higher KM for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93:665–673. doi: 10.1002/bit.20737. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Sanchez RG, Hahn-Hägerdal B, Gorwa-Grauslund MF. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb Cell Fact. 2007;6:1–10. doi: 10.1186/1475-2859-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Chung YS, Seo JH, Jo DH, Park YH, Ryu YW. High-yield production of xylitol from xylose by a xylitol dehydrogenase defective mutant of Pichia stipitis. J Microbiol Biotechnol. 2001;11:564–569. [Google Scholar]
- Kitpreechavanich V, Hayashi M, Nishio N, Nagai S. Conversion of d-xylose into xylitol by xylose reductase from Candida pelliculosa coupled with oxidoreductase system of methanogen strain HU. Biotechnol Lett. 1984;6:651–656. doi: 10.1007/BF00133831. [DOI] [Google Scholar]
- Ko BS, Kim J, Kim JH. Production of xylitol from d-xylose by a xylitol dehydrogenase gene-disrupted mutant of Candida tropicalis. Appl Environ Microbiol. 2006;72:4207–4213. doi: 10.1128/AEM.02699-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko BS, Rhee CH, Kim JH. Enhancement of xylitol productivity and yield using a xylitol dehydrogenase gene-disrupted mutant of Candida tropicalis under fully aerobic conditions. Biotechnol Lett. 2006;28:1159–1162. doi: 10.1007/s10529-006-9068-9. [DOI] [PubMed] [Google Scholar]
- Kostrzynska M, Sopher CR, Lee H. Mutational analysis of the role of the conserved lysine-270 in the Pichia stipitis xylose reductase. FEMS Microbiol Lett. 1998 doi: 10.1016/S0378-1097(97)00555-7. [DOI] [PubMed] [Google Scholar]
- López-Linares JC, Romero I, Cara C, Castro E, Mussatto SI. Xylitol production by Debaryomyces hansenii and Candida guilliermondii from rapeseed straw hemicellulosic hydrolysate. Bioresour Technol. 2018;247:736–743. doi: 10.1016/j.biortech.2017.09.139. [DOI] [PubMed] [Google Scholar]
- Mäkinen K. Xylitol and oral health. Adv Food Res. 1979;25:137–158. doi: 10.1016/S0065-2628(08)60236-0. [DOI] [PubMed] [Google Scholar]
- Martini C, Tauk-Tornisielo SM, Codato CB, Bastos RG, Ceccato-Antonini SR. A strain of Meyerozyma guilliermondii isolated from sugarcane juice is able to grow and ferment pentoses in synthetic and bagasse hydrolysate media. World J Microbiol Biotechnol. 2016 doi: 10.1007/s11274-016-2036-1. [DOI] [PubMed] [Google Scholar]
- Millerioux Y, Clastre M, Simkin AJ, Marais E, Sibirny AA, Noël T, Crèche J, Giglioli-Guivarc’h N, Papon N. Development of a URA5 integrative cassette for gene disruption in the Candida guilliermondii ATCC 6260 strain. J Microbiol Methods. 2011;84:355–358. doi: 10.1016/j.mimet.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Mohamad NL, Mustapa Kamal SM, Mokhtar MN. Xylitol biological production: a review of recent studies. Food Rev Int. 2015;31:74–89. doi: 10.1080/87559129.2014.961077. [DOI] [Google Scholar]
- Mussatto SI, Silva CJSM, Roberto IC. Fermentation performance of Candida guilliermondii for xylitol production on single and mixed substrate media. Appl Microbiol Biotechnol. 2006;72:681–686. doi: 10.1007/s00253-006-0372-z. [DOI] [PubMed] [Google Scholar]
- Neuhauser W, Steininger M, Haltrich D, Kulbe KD, Nidetzky B. A pH-controlled fed-batch process can overcome inhibition by formate in NADH-dependent enzymatic reduction using formate dehydrogenase-catalyzed coenzyme regeneration. Biotechnol Bioeng. 1998;60:277–282. doi: 10.1002/(SICI)1097-0290(19981105)60:3<277::AID-BIT2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Nidetzky B, Neuhauser W, Haltrich D, Kulbe KD. Continuous enzymatic production of xylitol with simultaneous coenzyme regeneration in a charged membrane reactor. Biotechnol Bioeng. 2000;52:387–396. doi: 10.1002/(SICI)1097-0290(19961105)52:3<387::AID-BIT4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Pal S, Choudhary V, Kumar A, Biswas D, Mondal AK, Sahoo DK. Studies on xylitol production by metabolic pathway engineered Debaryomyces hansenii. Bioresour Technol. 2013;147:449–455. doi: 10.1016/j.biortech.2013.08.065. [DOI] [PubMed] [Google Scholar]
- Pal S, Mondal AK, Sahoo DK. Molecular strategies for enhancing microbial production of xylitol. Process Biochem. 2016;51:809–819. doi: 10.1016/j.procbio.2016.03.017. [DOI] [Google Scholar]
- Pereira RS, Mussatto SI, Roberto IC. Inhibitory action of toxic compounds present in lignocellulosic hydrolysates on xylose to xylitol bioconversion by Candida guilliermondii. J Ind Microbiol Biotechnol. 2011;38:71–78. doi: 10.1007/s10295-010-0830-6. [DOI] [PubMed] [Google Scholar]
- Petschacher B, Nidetzky B. Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb Cell Fact. 2008 doi: 10.1186/1475-2859-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschacher B, Leitgeb S, Kavanagh KL, Wilson DK, Nidetzky B. The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography. Biochem J. 2005;385:75–83. doi: 10.1042/BJ20040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqul ISM, Sakinah AMM. Processes for the production of xylitol—a review. Food Rev Int. 2013;29:127–156. doi: 10.1080/87559129.2012.714434. [DOI] [Google Scholar]
- Silva DSS, Vitolo M, Jr, Felipe MGA. Xylose reductase and xylitol dehydrogenase activities of d-xylose-xylitol-fermenting Candida guilliermondii. J Basic Microbiol. 1996;36:187–191. doi: 10.1002/jobm.3620360305. [DOI] [Google Scholar]
- Sukpipat W, Komeda H, Prasertsan P, Asano Y. Purification and characterization of xylitol dehydrogenase with l-arabitol dehydrogenase activity from the newly isolated pentose-fermenting yeast Meyerozyma caribbica 5XY2. J Biosci Bioeng. 2017;123:20–27. doi: 10.1016/j.jbiosc.2016.07.011. [DOI] [PubMed] [Google Scholar]
- Venkateswar Rao L, Goli JK, Gentela J, Koti S. Bioconversion of lignocellulosic biomass to xylitol: an overview. Bioresour Technol. 2015;213:299–310. doi: 10.1016/j.biortech.2016.04.092. [DOI] [PubMed] [Google Scholar]
- Veras HCT, Parachin NS, Almeida JRM. Comparative assessment of fermentative capacity of different xylose-consuming yeasts. Microb Cell Fact. 2017;16:1–8. doi: 10.1186/s12934-017-0766-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Saleh AA, Pack SP, Annaluru N, Kodaki T, Makino K. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology. 2007;153:3044–3054. doi: 10.1099/mic.0.2007/007856-0. [DOI] [PubMed] [Google Scholar]
- Winkelhausen E, Kuzmanova S. Microbial conversion of d-xylose to xylitol. J Ferment Bioeng. 1998;86:1–14. doi: 10.1016/S0922-338X(98)80026-3. [DOI] [Google Scholar]
- Ylikahri R. Metabolic and nutritional aspects of xylitol. In: Chichester CO, editor. Advances in food research. New York: Academic Press; 1979. pp. 159–180. [DOI] [PubMed] [Google Scholar]
- Zacharis C (2012) Xylitol. In: Molinary SV, Quinlan ME (eds) Sweeteners and sugar alternatives in food technology. pp 347–382
- Zhang J, Zhang B, Wang D, Gao X, Hong J. Xylitol production at high temperature by engineered Kluyveromyces marxianus. Bioresour Technol. 2014;152:192–201. doi: 10.1016/j.biortech.2013.10.109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional figures and table.
Data Availability Statement
All relevant data are supplied within the manuscript and additional file.