Abstract
While a growing body of modern phylogenetic research reveals that the Western Indochina represents a separate biogeographic subregion having a largely endemic freshwater fauna, the boundaries of this subregion are still unclear. We use freshwater mussels (Unionidae) as a model to reconstruct spatial patterns of freshwater biogeographic divides throughout Asia. Here, we present an updated freshwater biogeographic division of mainland Southeast Asia and describe 12 species and 4 genera of freshwater mussels new to science. We show that the Isthmus of Kra represents a significant southern biogeographic barrier between freshwater mussel faunas of the Western Indochina and Sundaland subregions, while the Indian and Western Indochina subregions are separated by the Naga Hills, Chin Hills, and Rakhine Yoma mountain ranges. Our findings highlight that the freshwater bivalve fauna of Southeast Asia primarily originated within three evolutionary hotspots (Western Indochina, Sundaland, and East Asian) supplemented by ancient immigrants from the Indian Subcontinent.
Subject terms: Biodiversity, Biogeography, Taxonomy
Introduction
Freshwater mussels (Unionida) are an economically and environmentally important group of aquatic animals having a broad distribution on all continents except Antarctica1,2. Southeast Asia houses one of the richest endemic faunas of freshwater mussels globally3–7. Unfortunately, freshwater mussels are among the most endangered animal groups at the global scale, with numerous local extinctions triggered by multiple anthropogenic impacts and climate changes8–11. Human-mediated degradation of natural habitats, e.g. water pollution, river damming, and irrigation practices, appears to be the most influential factor causing the decline and local extinctions of freshwater mussels11–13. It was shown that even a prehistoric decline in freshwater mussels corresponds to the early development of agricultural techniques14. Alien species may represent a significant threat to native freshwater mussel assemblages in Southeast Asia13 and other regions15. For example, the tropical lineage of Sinanodonta woodiana (Lea, 1834) is widely spread throughout Malaysia, the Indonesian Archipelago, and the Philippines16–18, while the temperate lineage of this taxon was found in Myanmar19.
Recent advances in mitogenomic20 and multi-locus nuclear21 phylogenetic modeling reveal that two widespread Southeast Asian subfamilies of the Unionidae, i.e. Pseudodontinae22 and Rectidentinae22,23, represent tribes within the monophyletic Gonideinae. The genus- and species-level taxonomy of freshwater mussels in Southeast Asia is still poorly known3, but several integrative studies performed in Myanmar, Thailand, Laos, and Malaysia have recently improved our knowledge about the diversity and biogeographic patterns in the region. These studies found that freshwater mussel faunas of Myanmar, from the Ayeyarwady to Salween and Dawei basins, could be considered as a separate and distinct freshwater biogeographic subregion from the Indian and Sundaland subregions4–7,22,24. This subregion harbors a species-rich, largely endemic fauna of freshwater mussels, with one endemic tribe, Leoparreysiini Vikhrev, Bolotov & Kondakov, 20174,6. Broad-scale phylogenetic research revealed that the Unionidae fauna of Malaysia has 9 native species16, all of which are representatives of Sundaland genera4,22.
Our knowledge of freshwater mussels from the Mekong Basin and coastal rivers of Thailand and Cambodia is also far from being complete. However, there have been recent advances in the research on the freshwater mussel diversity in this region. It is now known that the genus Contradens Haas, 1911 in Thailand contains at least three allopatric species, one of which is widespread throughout the Chao Phraya, Mae Klong, and Bang Pakong basins corresponding to the former Siam Paleo-River system25. In contrast, two other species share restricted ranges in the northeastern part of the Khorat Plateau25. Two monotypic genera from the Mekong Basin, i.e. Unionetta Haas, 1955 and Harmandia Rochebrune, 1881, were found to be members of the tribe Indochinellini Bolotov et al., 201826. It was revealed recently that Scabies Haas, 1911, another member of the Indochinellini, contains at least eight valid species from Thailand, while S. songkramensis Kongim & Panha, 2015 takes a distant phylogenetic position leading to non-monophyly of this genus in its current understanding25,27. The genus Ensidens Frierson, 1911 in Thailand includes two large clades, one of which contains four species and corresponds to tributaries of the Middle Mekong drainage28. The second clade comprises two species: one species primarily from the Siam Paleo-River system, and another species from the Lower Mekong and Bang Pakong River basins.
The present study aims to update the freshwater biogeographic divisions of mainland Southeast Asia using freshwater mussels (Unionidae) as a model group. Based on the results of a broad-scale field survey throughout Myanmar, Thailand, and northern Laos, we clarify the western and southern boundaries of the Western Indochina Subregion. During this extensive assessment, we discovered several novel genera and species of freshwater mussels that are described here to improve our current understanding of the Unionidae systematics in Southeast Asia. Finally, we show that the Isthmus of Kra is a significant biogeographic barrier separating freshwater mussel faunas of the Western Indochina and Sundaland subregions.
Results
New Unionidae genera and species from Southeast Asia
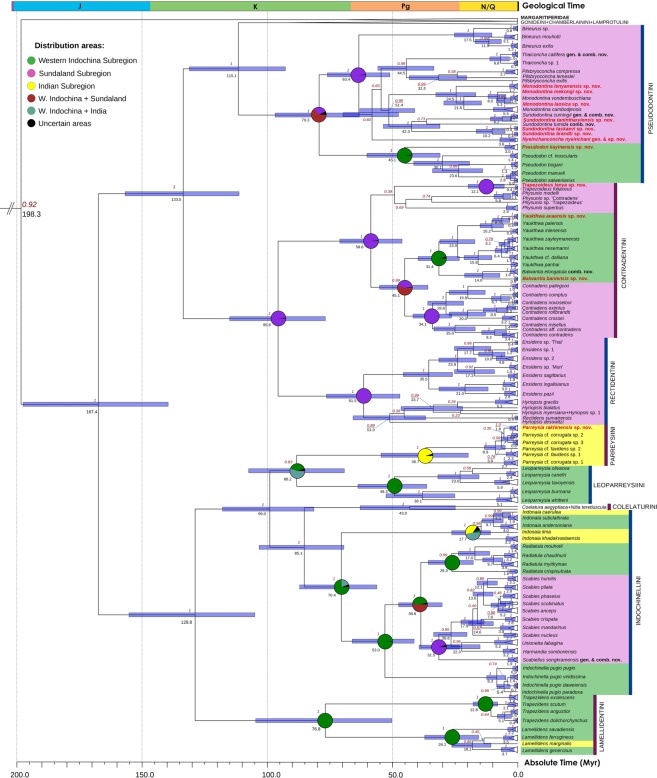
Our multi-locus phylogenies were constructed using BEAST v2.6.1, MrBayes v3.2.6 and IQ-TREE v1.6.11 based on the mitochondrial cytochrome c oxidase subunit I (COI), small ribosomal RNA (16 S rRNA), and the nuclear large ribosomal RNA (28 S rRNA) gene fragments. These analyses returned well-resolved consensus phylogenies having a similar topology (Fig. 1 and Supplementary Fig. 1). We found that available freshwater mussel taxa from Southeast Asia cluster to at least 25 genera, four of which are new to science and are described here. The novel genera represent distant monotypic lineages (Scabiellus gen. nov. and Nyeinchanconcha gen. nov.) and well-supported clades with several species (Sundadontina gen. nov. and Thaiconcha gen. nov.). An integrative species delimitation analysis indicates that our dataset contains 12 species that do not have available names and can be considered new to science (Figs. 1–5, Tables 1–3, Supplementary Tables 1–2). Each novel species can be distinguished from its sister taxa by conchological and molecular characters. A description of each new species is presented below. Mean shell parameters for the type series of new species are presented in Table 1. Two more unnamed species-level lineages, i.e. Ensidens sp. ‘Mun’ and Ensidens sp. ‘Thai’ (Fig. 1), appear to represent cryptic species and require separate research.
Figure 1.
Time-calibrated multi-locus phylogeny of the Unionidae based on the complete data set of mitochondrial and nuclear sequences (five partitions: three codons of COI + 16 S rRNA + 28 S rRNA). Red numbers near nodes are BPP of BEAST v2.6.1. Black numbers near nodes are the node ages. Node bars are 95% HPD of the divergence time. Age reconstructions for weakly supported nodes (BPP < 0.75) are omitted. Pie charts at nodes indicate the probabilities of certain ancestral areas for clades of interest with respect to combined results under two different statistical modeling approaches (S-DIVA and Bayesian MCMC analysis). New generic and species names are colored red. Outgroup and non-target clades are collapsed. Stratigraphic chart according to the International Commission on Stratigraphy, 2019.
Figure 5.
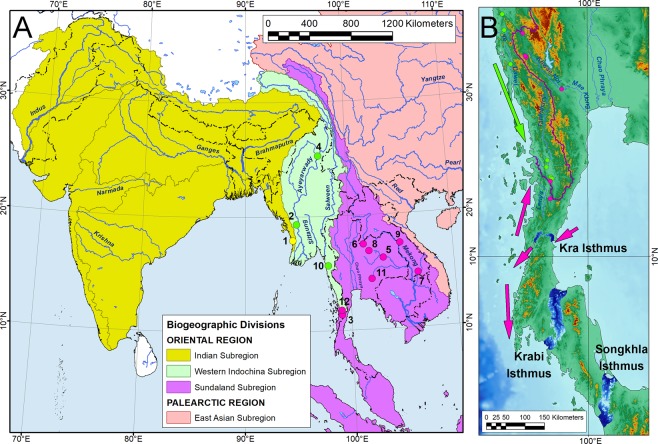
Updated freshwater biogeographic division of the mainland Southeast Asia based on freshwater mussel phylogenetics. (A) Freshwater biogeographic division of the mainland Southeast Asia. Color circles indicate the localities of taxa having biogeographic affinities to the Indian (yellow), Western Indochina (green), and Sundaland (pink) faunas. Type localities of new freshwater mussel species and occurrences of two cryptic Ensidens lineages are numbered as follows: Parreysia rakhinensis sp. nov. (1), Balwantia baniensis sp. nov. (2), Trapezoideus lenya sp. nov. and Monodontina lenyanensis sp. nov. (3), Yaukthwa avaensis sp. nov. (4), Ensidens sp. ‘Mun’ (5), Ensidens sp. ‘Thai’ (6), Monodontina laosica sp. nov. (7), M. mekongi sp. nov. (8), Nyeinchanconcha nyeinchani gen. & sp. nov. (9), Pseudodon kayinensis sp. nov. (10), Sundadontina brandti sp. nov. and S. taskaevi sp. nov. (11), and S. tanintharyiensis sp. nov. (12). (B) Boundary between the Western Indochina and Sundaland freshwater subregions at the southern margin of the Kra Isthmus (Tanintharyi – Lenya drainage divide). Violet line indicates the boundary between freshwater subregions based on drainage divides of the corresponding river basins. Green circles indicate records of the Western Indochina fauna representatives: Leoparreysia tavoyensis, Trapezidens scutum, T. exolescens4,24, Indochinella pugio daweiensis6. Pink circles indicate records of the Sundaland fauna representatives: Trapezoideus foliaceus7, Trapezoideus lenya sp. nov., Monodontina lenyanensis sp. nov., and Sundadontina tanintharyiensis sp. nov. All freshwater mussel taxa in Malaysia are members of the Sundaland fauna16,22. Arrows indicate putative ancient (pre-Pleistocene) dispersal routes of the Western Indochina (green) and Sundaland (pink) Unionidae species around the Isthmus of Kra and surrounding areas inferred from our statistical biogeographic analyses and distribution data (Supplementary Tables 1 and 3). Blue gradient shading indicates the putative ancient seaways crossing the Thai-Malay Peninsula based on the ArcGIS modeling (hydrologically conditioned DEM with elevation levels < 120 m) and published data36. The map was created using ESRI ArcGIS 10 software (https://www.esri.com/arcgis); the topographic base of the map was created with Natural Earth Free Vector and Raster Map Data (https://www.naturalearthdata.com), Global Self-consistent Hierarchical High-resolution Geography, GSHHG v2.3.7 (https://www.soest.hawaii.edu/wessel/gshhg), HydroSHEDS (https://www.hydrosheds.org)81, The General Bathymetric Chart of the Oceans, GEBCO (https://www.gebco.net), and Vector Map (VMap) Level 0 (http://gis-lab.info/qa/vmap0-eng.html) (Maps: Mikhail Yu. Gofarov).
Table 1.
Mean shell parameters (mm) for the type series of new freshwater mussel species (Unionidae) from Southeast Asia.
| Species | Shell Length (SL) | Shell Height (SH) | Shell Width (SW) | |||
|---|---|---|---|---|---|---|
| Mean ± s.e.m. | Min-max | Mean ± s.e.m. | Min-max | Mean ± s.e.m. | Min-max | |
| Parreysia rakhinensis sp. nov. (N = 26) | 45.9 ± 1.7 | 29.3–62.3 | 28.8 ± 1.1 | 18.5–41.7 | 17.3 ± 0.7 | 10.3–24.2 |
| Balwantia baniensis sp. nov. (N = 5) | 54.1 ± 2.9 | 46.1–61.4 | 22.7 ± 1.2 | 19.5–26.3 | 14.2 ± 0.9 | 12.7–16.7 |
| Trapezoideus lenya sp. nov. (N = 4) | 35.2 ± 0.8 | 33.1–36.3 | 20.6 ± 0.6 | 19.1–21.4 | 11.3 ± 0.6 | 10.2–12.6 |
| Yaukthwa avaensis sp. nov. (N = 10) | 37.8 ± 2.8 | 24.7–45.6 | 21.0 ± 1.4 | 14.1–25.1 | 14.4 ± 1.2 | 8.7–17.0 |
| Monodontina laosica sp. nov. (N = 1) | n/a | 61.4 | n/a | 41.4 | n/a | 19.0 |
| M. lenyanensis sp. nov. (N = 10) | 51.1 ± 3.6 | 31.2–63.4 | 32.4 ± 2.2 | 20.3–40.1 | 17.5 ± 1.4 | 11.0–23.7 |
| M. mekongi sp. nov. (N = 1) | n/a | 65.7 | n/a | 42.2 | n/a | 20.3 |
| Pseudodon kayinensis sp. nov. (N = 11) | 52.6 ± 3.4 | 37.8–71.0 | 30.9 ± 1.9 | 22.4–42.5 | 15.9 ± 1.1 | 10.3–21.2 |
| Sundadontina brandti sp. nov. (N = 3) | 78.4 ± 5.0 | 71.3–85.4 | 49.3 ± 3.6 | 43.8–53.7 | 26.3 ± 1.4 | 24.1–27.7 |
| S. tanintharyiensis sp. nov. (N = 3) | 52.5 ± 8.3 | 39.2–61.2 | 37.7 ± 6.6 | 27.2–45.1 | 20.4 ± 3.5 | 14.7–23.2 |
| S. taskaevi sp. nov. (N = 2) | 71.4 ± 11.4 | 60.0–82.7 | 47.5 ± 4.7 | 42.8–52.2 | 26.0 ± 3.3 | 22.6–29.3 |
| Nyeinchanconcha nyeinchani gen. & sp. nov. (N = 3) | 48.2 ± 9.5 | 33.7–59.5 | 26.6 ± 7.8 | 15.1–37.1 | 15.0 ± 4.8 | 7.9–21.5 |
n/a – not available.
Table 3.
Taxonomic review of freshwater mussel genera under discussion within the boundaries of Southeast Asia (Myanmar, Thailand, Cambodia, Laos, and the Lower Mekong in Vietnam).
| Genus and species | Type locality | Distribution |
|---|---|---|
| Parreysia Conrad, 1853 | ||
| P. rakhinensis sp. nov. | Kyeintali Stream upstream of Ohtein village [17.9193°N, 94.5946°E], Myanmar | Coastal rivers of the Rakhine State, Myanmar (from Kyeintali to Ann) |
| Scabiellus gen. nov. | ||
| S. songkramensis (Kongim & Panha, 2015) gen. & comb. nov. [ = Scabies songkramensis Kongim & Panha, 2015] | Houy Plahang Stream [17.4061°N, 103.8336°E], Songkram River Basin, Thailand30 | Songkhram and Kam river basins, and the corresponding section of the Mekong River, Thailand30 |
| Balwantia Prashad, 1919 stat. rev. | ||
| B. elongatula (Bolotov et al., 2019) comb. nov. [ = Yaukthwa elongatula Bolotov et al., 2019] | Chindwin River [23.1918ºN, 94.3217ºE], Ayeyarwady Basin, Myanmar6 | Chindwin River, Ayeyarwady Basin, Myanmar6 |
| B. baniensis sp. nov. | Bani River at Bangong village [19.3247°N, 94.9839°E], Ayeyarwady Basin, Myanmar | Bani River, Myanmar |
| Trapezoideus Simpson, 1900 | ||
| T. foliaceus (Gould, 1843) [ = Unio foliaceus Gould, 1843] | Tavoy [Dawei River], British Burma7 | Mae Klong River, Thailand, and Dawei River, Myanmar7 |
| T. lenya sp. nov. | 14 Mile Stream [11.3508°N, 99.1093°E], Lenya Basin, Myanmar | Lenya Basin, Myanmar |
| Yaukthwa Konopleva et al., 2019 | ||
| Y. avaensis sp. nov. | Tarkat Stream [25.2758°N, 97.2722°E], tributary of Ayeyarwady River, Myanmar | Ayeyarwady River, Myanmar |
| Y. dalliana (Frierson, 1913) [ = Parreysia dalliana Frierson, 1913] | Lashio River near Lashio [approx. 22.9946°N, 97.7650°E], Ayeyarwady Basin, Myanmar7 | Upper Ayeyarwady Basin, Myanmar7 |
| Y. inlenensis Konopleva et al., 2019 | Mway Stream [19.7266°N, 97.0992°E], a tributary of Nam Pilu River, Salween Basin, Myanmar7 | Lake Inle Area, Salween Basin, Myanmar7 |
| Y. nesemanni (Konopleva, Vikhrev & Bolotov, 2017) [ = Trapezoideus nesemanni Konopleva, Vikhrev & Bolotov, 2017] | Thauk Ye Kupt River [19.3075°N, 96.7219°E], Sittaung Basin, Myanmar4,7 | Sittaung Basin, Myanmar4,7 |
| Y. paiensis Konopleva et al., 2019 | Khong River [19.4246°N, 98.4013°E], tributary of the Pai River, Salween Basin, Thailand7 | Pai River, Salween Basin, Thailand7 |
| Y. panhai (Konopleva, Bolotov & Kondakov, 2017) [ = Trapezoideus panhai Konopleva, Bolotov & Kondakov, 2017] | Kyan Hone River [19.5059°N, 96.8280°E], Sittaung Basin, Myanmar4,7 | Sittaung Basin, Myanmar4,7 |
| *Y. peguensis (Anthony, 1865) [ = Unio peguensis Anthony, 1865] | Pegu, British Burmah [Bago River, Myanmar]7 | Bago River, Myanmar7 |
| Y. zayleymanensis (Preston, 1912) [ = Trapezoideus foliaceus var. zayleymanensis Preston, 1912] | Bhamo, Ayeyarwady River [approx. 24.2669°N, 97.2210°E]7 | Ayeyarwady River, Myanmar7 |
| Monodontina Conrad, 1853 | ||
| *M. aeneola (Drouet & Chaper, 1892) comb. nov. [ = Pseudodon aeneolus Drouet & Chaper, 1892] | Sebruang River [approx. 0.4937°N, 111.8931°E], Kapuas Basin, western Borneo82 | Kapuas Basin, western Borneo82 |
| M. cambodjensis (Petit de la Saussaye, 1865) [ = Monocondylea cambodjensis Petit de la Saussaye, 1865; = *Monocondylus orbicularis Morelet, 1866; = *Unio subtrigonus Sowerby, 1867; = *U. vagulus Fischer, 1891; = *Pseudodon cambodjensis tenerrimus Brandt, 1974] | Battambang [approx. 13.0929°N, 103.2001°E], Mekong Basin, Cambodia83 | Mekong Basin in Thailand and Cambodia, few rivers in Malaysia16,31 |
| M. laosica sp. nov. | Houai Pin, about 300 m upstream of the mouth [14.7944°N, 106.4842°E], Mekong Basin, Laos | Mekong Basin in Laos |
| M. lenyanensis sp. nov. | 14 mile stream [11.3508°N, 99.1093°E], Lenya River basin, Myanmar | Lenya Basin, Myanmar |
| M. mekongi sp. nov. | Headwater of the Phong River [16.8616°N, 101.9105°E], Mekong Basin, Thailand | Phong River, Mekong Basin, Thailand |
| M. vondembuschiana (Lea, 1840) [ = Margaritana vondembuschiana Lea, 1840; = *Alasmodonta crispata Mousson, 1849; = *A. zollingeri Mousson, 1849; = *Monodontina buschiana Conrad, 1853; = *Monocondyloea planulata Lea, 1859; = *M. hageni Strubell, 1897 syn. nov.] | Java84 | Malaysia, Sumatra and Java |
| *M. walpolei (Hanley, 1871) comb. nov. [ = Monocondylaea walpolei Hanley, 1871; = *Pseudodon crassus Drouet & Chaper, 1892 syn. nov.] | Sarawak, Borneo (by lectotype designation)85 | Northern Borneo85,86 |
| Nyeinchanconcha gen. nov. | ||
| N. nyeinchani gen. & sp. nov. | Small stream arising at a cave near Ban Kouanphavang [17.4578°N, 104.9263°E], Nam Done River drainage, Mekong Basin, Laos | Mekong Basin in Laos |
| Pseudodon Gould, 1844 | ||
| P. avae (Theobald, 1873) [ = Monocondylaea avae Theobald, 1873] | Mandalay, Burmah4 | Ayeyarwady Basin, Myanmar4 |
| P. bogani Bolotov, Kondakov & Konopleva, 2017 | Kanni River [19.0545°N, 96.5131°E], Sittaung Basin, Myanmar4 | Sittaung Basin, Myanmar4 |
| *P. crebristriatus (Anthony, 1865) [ = Monocondyloea crebristriata Anthony, 1865; = *Pseudodon (Trigonodon) crebristriatus var. curvata Preston, 1912] | Pegu, British Burmah4 | Bago Basin, Myanmar4 |
| P. inoscularis (Gould, 1844) [ = Anodon inoscularis Gould, 1844] | River Salwen, Tavoy, Brit. Burmah4 | ?Dawei River, Myanmar |
| P. kayinensis sp. nov. | Winyaw River [15.6685°N, 97.9496°E], Ataran River basin, Myanmar | Salween Basin, Myanmar |
| P. manueli Konopleva, Kondakov & Vikrev, 2017 | Pyowne Stream [18.9694°N, 96.5309°E], Sittaung Basin, Myanmar4 | Sittaung Basin, Myanmar4 |
| *P. peguensis (Anthony, 1865) [ = Monocondyloea peguensis Anthony, 1865] | Pegu, British Burmah4 | Bago Basin, Myanmar4 |
| P. salwenianus (Gould, 1844) [ = Anodon salweniana Gould, 1844] | Salwen River, British Burmah4 | Salween Basin, Myanmar4 |
| Sundadontina gen. nov. | ||
| S. brandti sp. nov. | Headwater of the Mun River [14.4138°N, 102.0821°E], Mekong Basin, Thailand | Mun River, Mekong Basin, Thailand |
| S. cumingii (Lea, 1850) gen. & comb. nov. [ = Anodonta cumingii Lea, 1850; *Pseudodus chaperi Morgan, 1885 syn. nov.] | Malacca87 | Malaysia16 |
| *S. harmandi (Crosse & Fischer, 1876) comb. nov. [ = Pseudodon harmandi Crosse & Fischer, 1876] | Cambodia88 | Lower Mekong Basin in Cambodia |
| *S. mabilli (Rochebrune, 1881) comb. nov. [ = Pseudodon mabilli Rochebrune, 1881] | Mekong, Shigloni Breithon, Cochinchina89 | Lower Mekong Basin in southern Vietnam |
| *S. moreleti Crosse & Fischer, 1876 [ = Pseudodon moreleti Crosse & Fischer, 1876] | Mekong, Kompang Cham Province, Cambodia88 | Lower Mekong Basin in Cambodia |
| S. tanintharyiensis sp. nov. | Chaung Nauk Pyan stream [11.7620°N, 99.1124°E], Lenya River basin, Myanmar | Lenya Basin, Myanmar |
| *S. ponderosa (Preston, 1909) comb. nov. [ = Pseudodon ponderosa Preston, 1909] | Nan-ko, Siam [Nan River, Chao Phraya Basin, Thailand]90 | Chao Phraya Basin, Thailand |
| *S. sulcatum (Rochebrune, 1881) comb. nov. [ = Pseudodon sulcatum Rochebrune, 1881] | Mouth of the Mekong River, Cochinchina89 | Mekong Delta in southern Vietnam |
| S. taskaevi sp. nov. | Headwater of the Mun River [14.4138°N, 102.0821°E], Mekong Basin, Thailand | Mun River, Mekong Basin, Thailand |
| S. tumida (Morelet, 1866) comb. nov. [ = Monocondylus tumidus Morelet, 1866] | Cambodia91 | Lower Mekong Basin in Cambodia and southern Vietnam |
| Thaiconcha gen. nov. | ||
| T. callifera (Martens, 1860) gen. & comb. nov. [ = Anodonta callifera Martens, 1860; Pseudodon ellipticum Conrad, 1865 syn. nov.; P. thomsoni Morlet, 1884 syn. nov.] | Siam [Thailand]92 | Mekong Basin in Cambodia and Thailand |
| *T. ovalis (Morlet, 1889) comb. nov. [ = Pseudodon ovalis Morlet, 1889] | Srakeo River, Siam [Thailand]93 | Bang Pakong Basin, Thailand |
*These nominal taxa were provisionally placed in the corresponding genera or in the synonymy on the basis of conchological features alone, and they are in need of future molecular study and subsequent rearrangements.
New freshwater mussel species distribution and their biogeographic affinities in Southeast Asia
Here, we examined freshwater mussels from several poorly known, remote basins such as the rivers of the Rakhine Coast (western Myanmar), the Tanintharyi (former Tenasserim) River, and the Lenya River (southeastern Myanmar) (Fig. 5A). We found that the rivers of the Rakhine Coast are inhabited by one species, Parreysia rakhinensis sp. nov. This is the first record of a Parreysiini member in Myanmar. The fauna of the Tanintharyi Basin includes several species belonging to endemic genera of the Western Indochina Subregion, i.e. Trapezidens scutum and Leoparreysia tavoyensis. In contrast, Trapezoideus lenya sp. nov., Monodontina lenyanensis sp. nov., and Sundadontina tanintharyiensis sp. nov. were discovered from the Lenya River basin, which is located just south of the Tanintharyi Basin (Fig. 5B). These species are members of the genera widely distributed throughout the Sundaland Subregion.
Evolutionary biogeography and time-calibrated phylogeny
Our statistical biogeographic analyses (the combined results of S-DIVA and Bayesian MCMC runs) and time-calibrated Bayesian phylogeny reveal the high levels of endemism of freshwater mussel fauna within each subregion (Fig. 1 and Supplementary Table 3). The fauna of the Sundaland Subregion contains several endemic radiations of freshwater mussels belonging to two subfamilies, i.e. the subtribe Pilsbryoconchina, the tribe Rectidentini, the genera Trapezoideus, Physunio, Contradens (Gonideinae), and the so-called Mekong’s Indochinellini group (Harmandia, Scabies, Scabiellus gen. nov., and Unionetta) (Parreysiinae). The subtribe Pseudodontina, the genus Yaukthwa (Gonideinae), tribe Leoparreysiini, the genera Radiatula, Indochinella, and Trapezidens (Parreysiinae) are endemic clades to the Western Indochina Subregion. The fauna of the Indian Subregion contains the endemic clade Parreysiini, and shares some genus-level subclades with the Western Indochina, i.e. Indonaia, Lamellidens, and, most probably, Balwantia.
Our time-calibrated Bayesian phylogenetic model reveals that there were several splits between the Western Indochinese and Sundaland clades (Fig. 1). The oldest split between such groups occurred between the subtribes Pseudodontina and Pilsbryoconchina in the Late Cretaceous (mean age = 79.3 Myr, 95% HPD = 63.2–96.8 Myr). The Pseudodontina most likely diversified in the Western Indochinese Region and the Pilsbryoconchina evolved in the Sundaland Subregion (probability = 97.8% and 98.7%, respectively) (Supplementary Table 3). The Rectidentini + Contradentini clade (the former subfamily Rectidentinae21) appears to have evolved within the Sundaland Subregion since the Late Cretaceous (probability = 98.4%; mean age = 95.6 Myr, 95% HPD = 76.8–115.0 Myr). A colonization event of the Yaukthwa + Balwantia clade to Western Indochina occurred in the Early Eocene (mean age = 45.6 Myr, 95% HPD = 35.8–55.1 Myr) followed by an intra-area radiation (probability = 95.8%). Conversely, the Indochinellini seems to be a primary Western Indochinese clade evolving in situ since the Late Cretaceous (probability = 75.0%; mean age = 70.4 Myr, 95% HPD = 56.1–87.0 Myr). Divergence of Mekong’s Indochinellini clade from Western Indochinese Radiatula placed in the Late Eocene (probability = 50.9%; mean age = 38.6 Myr, 95% HPD = 30.1–47.5 Myr). After the vicariance event separating these taxa, each group diversified in isolation within a corresponding subregion (probability = 95–100%). Our analyses indicate that the Parreysiini clade is a group evolved within the Indian Subregion (probability = 95.2%), whereas the Leoparreysiini diversified within the Western Indochina (probability = 99.6%). The Indian and Indochinese Indonaia species groups most likely diverged in the mid-Miocene (mean age = 14.6 Myr, 95% HPD = 9.4–21.3 Myr) via a dispersal event from Western Indochina to India but with a rather low probability (47.0%).
Three novel species from the Lenya Basin have sister taxa in the Mekong River and smaller basins emptying into the Gulf of Thailand. These species likely separated by a series of splits occurred from the Oligocene – Miocene boundary to the Late Miocene as follows: (1) Monodontina lenyanensis sp. nov. vs M. mekongi sp. nov. + M. vondembuschiana + M. laosica sp. nov. + M. cambodjensis (mean age = 24.5 Myr, 95% HPD = 16.7–32.6 Myr); (2) Trapezoideus lenya sp. nov. vs T. foliaceus (mean age = 12.1 Myr, 95% HPD = 5.3–19.7 Myr); and (3) Sundadontina tanintharyiensis sp. nov. vs S. cumingii (mean age = 6.2 Myr, 95% HPD = 2.3–10.8 Myr). The split between Parreysia rakhinensis sp. nov. from western Myanmar and several species from India occurred in the Late Miocene (mean age = 5.5 Myr, 95% HPD = 2.9–8.1 Myr).
Taxonomic account. Family Unionidae Rafinesque, 1820
Subfamily Parreysiinae Henderson, 1935
Tribe Parreysiini Henderson, 1935
Type genus: Parreysia Conrad, 1853 (by original designation)
Comments: A monotypic Indian tribe4.
Distribution: Indian Subregion from the Indus Basin29 to the coastal basins of the Rakhine State of Myanmar.
GenusParreysiaConrad, 1853
Type species: Unio multidentatus Philippi, 1847 (by original designation)
Comments: A diverse Indian genus, in which the modern systematics and number of species are still uncertain, due to the lack of available molecular data. Several species from Western Indochina originally assigned to Parreysia were recently transferred to another genus, Leoparreysia Vikhrev, Bolotov & Aksenova, 2017, belonging to the tribe Leoparreysiini Vikhrev, Bolotov & Kondakov, 20174. Parreysia rakhinensis sp. nov. is the first member of the true Parreysiini discovered in Myanmar (Table 3).
Distribution: As for the tribe.
Parreysia rakhinensissp. nov
Figures 2A,B, 5A, Tables 1–2, Supplementary Table 2
Figure 2.
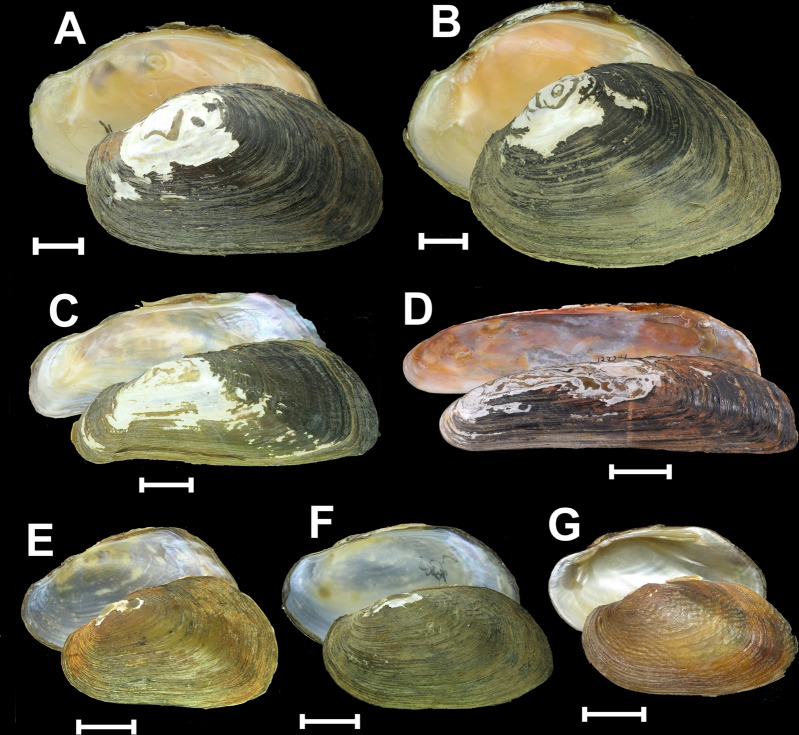
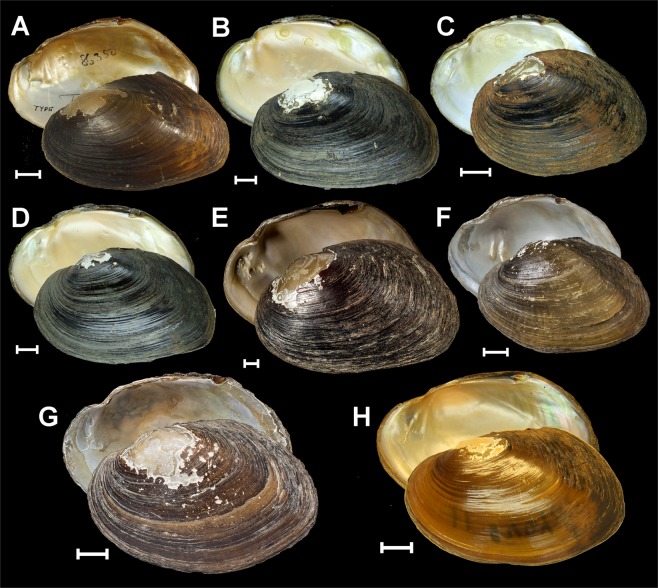
Shells of the Parreysiini, Indochinellini, Contradentini, and Rectidentini from Southeast Asia. (A) Parreysia rakhinensis sp. nov., Kyeintali Stream, Rakhine Coast, Myanmar (holotype RMBH biv652_1). (B) Parreysia rakhinensis sp. nov., Ann River, Rakhine Coast, Myanmar (paratype RMBH biv659_3). (C) Balwantia baniensis sp. nov., Bani River, Ayeyarwady Basin, Myanmar (holotype RMBH 666_2). (D) Balwantia soleniformis (Benson, 1836) comb. rev., Brahmaputra River, India (specimen USNM 127246). (E) Trapezoideus lenya sp. nov., 14th Mile Stream, Lenya Basin, southeastern Myanmar (holotype RMBH biv629_2). (F) Yaukthwa avaensis sp. nov., unnamed small stream, a tributary of the Ayeyarwady River, Myanmar (holotype RMBH biv680_3). (G) Scabiellus songkramensis (Kongim & Panha, 2015) gen. & comb. nov., Songkhram River, Mekong Basin, Thailand (topotype, collection of S. Tumpeesuwan, Mahasarakham University). Scale bars = 1 cm [A-C, E-G] and 3 cm [D]. Photos: Ekaterina S. Konopleva [A-C, E, F], Ellen Strong [D], and Benchawan Nahok [G].
Table 2.
Molecular diagnoses of the new freshwater mussel species (Unionidae) from Southeast Asia.
| New species | Mean COI p-distance from the most closely related species (%) | Most closely related species | Fixed unique nucleotide differences based on the sequence alignment of congeners | ||
|---|---|---|---|---|---|
| COI | 16 S rRNA | 28 S rRNA | |||
| Parreysia rakhinensis sp. nov. | 2.12 | Parreysia cf. corrugata sp.2 | 29 G, 429 C, 557 A | n/a | n/a |
| Balwantia baniensis sp. nov. | 5.05 | Balwantia elongatula comb. nov. | 15 T, 17 G, 35 C, 101 G, 104 C, 159 C, 167 G, 170 A, 173 A, 182 G, 194 T, 207 T, 236 C, 242 A, 243 C, 248 T, 263 A, 290 A, 296 G, 338 A, 347 A, 353 G, 404 A, 443 T, 470 T, 506 T, 537 C, 542 G, 579 C, 587 T, 626 T, 635 C | 192 T, 253 C, 263 G, 313 T, 464 T, 469 T | 212 C, 497 T, 609 C |
| Trapezoideus lenya sp. nov. | 4.46 | Trapezoideus foliaceus | 6 T, 14 G, 35 G, 38 G, 86 A, 92 G, 93 T, 98 C, 170 T, 209 A, 317 G, 353 T, 365 G, 383 T, 392 C, 401 A, 402 T, 404 A, 413 T, 461 G, 479 A, 527 G, 557 A, 584 C, 599 G, 608 T, 617 C, 629 A, 647 T, 654 C | 155 T, 234 C, 252 T, 299 C, 316 A, 344 A, 355 G | n/a |
| Yaukthwa avaensis sp. nov. | 2.90 | Yaukthwa paiensis | 365 A, 626 A | n/a | n/a |
| Monodontina laosica sp. nov. | 2.40 | Monodontina mekongi sp. nov. | 89 A, 164 G, 347 A, 500 C, 539 A, 608 G | 18 G, 48 C, 191 T | n/a |
| Monodontina lenyanensis sp. nov. | 7.59 | Monodontina vondembuschiana | 86 G, 96 C, 146 A, 149 G, 197 A, 200 C, 206 A, 284 C, 287 T, 290 A, 389 A, 479 A, 480 T, 500 A, 512 A, 518 A, 521 A, 531 A, 611 C | 258 C, 295 T, 334 T | 528 G, 609 T, 738 C, 755 A |
| Monodontina mekongi sp. nov. | 2.40 | Monodontina laosica sp. nov. | 56 A, 89 T, 257 A, 515 C, 527 G, 596 C | 18 A, 48 T, 155 G | 638 A, 768 A |
| Nyeinchanconcha nyeinchani gen. & sp. nov. | 9.42 | Sundadontina brandti sp. nov. | 134 T, 194 C, 278 C, 299 C, 575 A | 19 C, 150 C, 185 C, 196 A, 234 A, 236 C, 242 G, 296 A, 320 T, 329 T, 335 T, 338 G, 343 A, 457 T, 465 T | n/a |
| Pseudodon kayinensis sp. nov. | 11.10 | Pseudodon cf. inoscularis | 14 A, 23 T, 53 A, 62 G, 65 C, 68 A, 83 A, 101 T, 104 C, 110 A, 131 A, 179 C, 203 C, 213 G, 224 T, 266 G, 272 A, 291 C, 311 A, 314 T, 326 A, 338 G, 344 G, 347 G, 371 A, 380 A, 437 C. 464 C, 470 G, 482 A, 491 A, 521 A, 524 A, 569 C, 587 C, 605 A, 623 T | 7 C, 15 T, 25 G, 47 T, 48 C, 127 G, 147 C, 154 T, 159 G, 234 C, 240 C, 243 C, 247 T, 253 T, 293 C, 310 T, 311 C, 319 A, 320 C, 323 A, 329 T, 331 C | 489 C |
| Sundadontina brandti sp. nov. | 3.95 | Sundadontina taskaevi sp. nov. | 116 C, 398 C, 483 G, 518 C, 531 G, 581 A | 18 C, 49 C, 371 C, 445 T | n/a |
| Sundadontina tanintharyiensis sp. nov. | 2.46 | Sundadontina cumingii gen. & comb. nov. | 86 T, 206 G, 287 A, 338 G, 485 C | 12 C, 14 T, 15 T, 17 T, 20 C, 47 T, 48 C, 162 T, 168 A, 172 G, 193 C, 194 A, 234 A, 243 G, 248 C, 253 G, 263 T, 318 G, 322 A, 323 T, 329 A, 335 T, 344 T, 375 T, 440 C, 475 A | n/a |
| Sundadontina taskaevi sp. nov. | 3.95 | Sundadontina brandti sp. nov. | 149 A, 269 C, 380 A, 401 A, 440 C, 518 T, 536 A | 159 T, 265 T, 342 A | n/a |
n/a – not available. Del – deletion mutation.
Holotype: RMBH biv0652_1, MYANMAR: Kyeintali Stream upstream of Ohtein village, 17.9193°N, 94.5946°E, 04.xii.2018, Bogan, Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg. Reference sequence numbers of the holotype are as follows: MN275091 (COI), MN307275 (16 S rRNA), and MN307218 (28 S rRNA). Shell measurements of the holotype are as follows: shell length (SL) 58.7 mm, shell height (SH) 33.7 mm, and shell width (SW) 19.6 mm.
Paratypes: MYANMAR: type locality, same collecting date, and collectors, 4 specimens (RMBH biv0652_2, biv0652_3, biv0652_4, biv0652_6), Sa Lu Stream, 18.1631°N, 94.4997°E, 04.xii.2018, 5 specimens (RMBH biv0653_1, biv0653_2, biv0653_3, biv0654_1, biv0654_2) and 24 specimens (NCSM 113365), Thandwe River near Ywar Shai village, 18.3741°N, 94.4952°E, 04.xii.2018, 4 specimens (RMBH biv0655_1, biv0655_2, biv0655_3, biv0656), Shwehle Stream, 18.6174°N, 94.3508°E, 05.xii.2018, 3 specimens (RMBH biv0657_1, biv0657_2, biv0657_3) and 22 specimens (NCSM 113366), Toungup River, 18.8439°N, 94.3447°E, 06.xii.2018, 3 specimens (RMBH biv0658_1, biv0658_2, biv0658_3) and 13 specimens (NCSM 113367), Ann River near Ann town, 19.8026°N, 94.0449°E, 07.xii.2018, 3 specimens (RMBH biv0659_1, biv0659_2, biv0659_3), tributary of the Ann River, 19.8035°N, 94.0460°E, 07.xii.2018, 3 specimens (RMBH biv0660_1, biv0660_2, biv0660_3) and 10 specimens (NCSM 113360), Bogan, Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg.
Etymology: The new species name is derived from the Rakhine State of Myanmar, in which it is widely distributed.
Diagnosis: The new species is conchologically and genetically close to a group of Parreysia species from India with affinity to several nominal taxa such as P. corrugata and P. favidens. Its shell varies from ovate-rounded to ovate-elongated, rather thick, umbo slightly elevated, pseudocardinal teeth massive and usually indented, lateral teeth curved and strong, muscle attachment scars deep. The new species differs from all the congeners by fixed nucleotide substitutions in the COI gene fragment, while other genes from Indian Parreysia taxa are not available (Table 2).
Description: Medium-sized mussel: SL 29.3–62.3 mm, SH 18.5–41.7 mm, SW 10.3–24.2 mm. Shell shape variable, from ovate and strongly inflated to ovate-elongated and sub-compressed; inequilateral, rather solid. Umbo usually slightly elevated but may be much more developed at some specimens, with v-shaped sculpture visible only in small mussels due to strong erosion in old mussels. Periostracum from olive-green to brown. Nacre whitish, with bright peach or orange spot near the umbo cavity area, shining. Right valve with one curved lateral tooth and two pseudocardinal teeth, anterior tooth small and somewhat lamellar, posterior tooth massive, very indented. Left valve with two curved lateral teeth and two strongly indented pseudocardinal teeth, the anterior tooth usually higher than the posterior tooth. Anterior adductor scar ovate and deep, posterior adductor scar rounded and well-visible.
Distribution: Rivers and streams of the Rakhine Coast of Myanmar emptying into the Bay of Bengal.
Tribe Indochinellini Bolotov, Pfeiffer, Vikhrev & Konopleva, 2018
Type genus: Indochinella Bolotov, Pfeiffer, Vikhrev & Konopleva, 2018 (by original designation)
Comments: A large Oriental tribe, which contains seven genera: Harmandia Rochebrune, 1882, Indochinella Bolotov, Pfeiffer, Vikhrev & Konopleva, 2018, Indonaia Prashad, 1918, Radiatula Simpson, 1900, Scabies Haas, 1911, Scabiellus gen. nov., and Unionetta Haas, 19555,26,27.
Distribution: This group is widespread throughout the Indian, Western Indochina and Sundaland subregions5,26,27.
GenusScabiellusgen. nov
Figure 2G Type species: Scabies songkramensis Kongim & Panha, 2015.
Comments: Monotypic genus (Table 3) representing a distinct phylogenetic lineage that is not sister to the other members of Scabies, including its type species, S. scobinatus (Lea, 1856) (Fig. 1). Although Scabiellus gen. nov. is conchologically similar to Scabies and several taxa of Indochinella by having a v-shaped shell sculpture30, this external similarity appears to be only a result of convergence.
Diagnosis: Small mussels, shell length up to 33 mm. Shell thick, rather short, сuneiform, with a broad and elevated umbo, broad anterior margin and narrower posterior margin. Periostracum brown. Dark brown v-shaped sculpture covers the entire shell disc.
Etymology: The name of this genus derived from the genus Scabies, in which its type species was described initially.
Distribution: Scabiellus songkramensis gen. & comb. nov. is recorded from rivers of the Khorat Plateau in Thailand, i.e. the Songkhram and Kam river basins, and the corresponding section of the Mekong River26,27,30.
Subfamily Gonideinae Ortmann, 1916
Comments: Here we use this subfamily in a broader sense, with the former subfamilies Pseudodontinae22 and Rectidentinae22,23 being tribes within the monophyletic Gonideinae, as suggested based on the mitogenomic20 and multi-locus nuclear21 phylogenies.
Tribe Contradentini Modell, 1942
Type genus: Contradens Haas, 1911 (by original designation)
Comments: This tribe includes six valid genera: Balwantia Prashad, 1919, Contradens Haas, 1911, Trapezoideus Simpson 1900, Physunio Simpson, 1900, Solenaia Conrad, 1869, and Yaukthwa Konopleva et al., 20197,21.
Distribution: Northeastern India (Brahmaputra Basin) and Southeast Asia, including the Greater Sunda Islands2,3,7,29,31.
GenusBalwantiaPrashad, 1919 stat. rev
Type species: Anodonta soleniformis Benson, 1836 (by original designation)
Comments: This genus contains ultra-elongated mussels externally resembling members of Solenaia32 (Fig. 2D) but is distantly related to Yaukthwa phylogenetically (Fig. 1). Previously, it was considered a synonym of Solenaia29 and was not used as a valid genus name since the last monograph of Haas33. Two species were recorded from Myanmar, one of which is new to science and described here (Table 3). They were collected from deep burrows which they dig in hard clay and soft sandstone substrate as does Balwantia soleniformis, their putative Indian relative from the upper Brahmaputra River34,35.
Distribution: Ayeyarwady and upper Brahmaputra basins29,32,34.
Balwantia baniensissp. nov
Figure 2C, 5A, Tables 1–2, Supplementary Table 2
Holotype: RMBH biv0666_2, MYANMAR: Bani River near Bangong village, 19.3247°N, 94.9839°E, Ayeyarwady Basin, 09.xii.2018, Bogan, Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg. Reference sequence numbers of the holotype are as follows: MN275077 (COI), MN307264 (16 S rRNA), and MN307206 (28 S rRNA). Shell measurements of the holotype are as follows: SL 57.0 mm, SH 23.2 mm, and SW 15.3 mm.
Paratypes: MYANMAR: type locality, same collecting date, and collectors, 4 specimens (RMBH biv0666_1, biv0666_3, biv0666_4, and biv0666_5), 9 specimens (NCSM 113369).
Etymology: The new species name is dedicated to Bani River, a tributary of the Ayeyarwady River, in which it was collected.
Diagnosis: The new species can be distinguished from Balwantia elongatula comb. nov. by having a rostrate anterior margin (vs. rounded), a more inflated shell (vs. flattened), and by the presence of bars from umbo along the dorsal margin. The new species also differs from Balwantia elongatula comb. nov. by fixed nucleotide substitutions in the COI, 16 S rRNA and 28 S rRNA gene fragments (Table 2).
Description: Medium-sized mussel: SL 46.1–61.4 mm, SH 19.5–26.3 mm, SW 12.7–16.7 mm. Shell somewhat trapezoidal, elongated, not very inequilateral, thin, and rather inflated, narrow and rostrate anteriorly, broad and truncated posteriorly, ventral margin slightly curved. Posterior slope covered by elongated, slightly curved bars. Umbo eroded, slightly elevated, without clear sculpture. Periostracum olive-yellow or brownish, the central part of the shell usually lighter than the posterior side. Nacre blue-whitish, sometimes with yellow spots, shining. Lateral teeth very thin, almost straight, by one on each valve. Pseudocardinal teeth reduced. Anterior adductor scars somewhat ovate or drop-like, not deep. Posterior adductor scars ovate or rounded shape, shallow.
Distribution: Bani River, Ayeyarwady Basin, central Myanmar.
GenusTrapezoideusSimpson, 1900
Type species: Unio foliaceus Gould, 1843 (by original designation)
Comments: Small genus with a restricted range that was previously considered a monotypic entity7. However, we found one additional congeneric species (Table 3), which is described here.
Distribution: Southern Myanmar (Lenya and Dawei basins) and southwestern Thailand (Mae Klong Basin)7.
Trapezoideus lenyasp. nov
Figures 2E, 5A, Tables 1–2, Supplementary Table 2
Holotype: RMBH biv0629_2, MYANMAR: 14 Mile Stream, 11.3508°N, 99.1093°E, Lenya River basin, 24.xi.2018, Bogan, Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg. Reference sequence numbers of the holotype are as follows: MN275068 (COI), MN307257 (16 S rRNA), and MN307198 (28 S rRNA). Shell measurements of the holotype are as follows: SL 36.3 mm, SH 21.4 mm, and SW 11.3 mm.
Paratypes: MYANMAR: type locality, same collecting date, and collectors, 3 specimens (RMBH biv0629_1, biv0629_3, and biv0629_5), 3 specimens (NCSM 113368).
Etymology: The name of new species is derived from the Lenya River, a coastal freshwater basin in southern Myanmar, from which this species was collected.
Diagnosis: The new species is morphologically similar to Trapezoideus foliaceus but differs in shell shape, being higher posteriorly and narrower anteriorly, with a straight ventral margin. The new species also differs from its congener by fixed nucleotide substitutions in the COI and 16 S rRNA gene fragments (Table 2).
Description: Small mussel: SL 33.1–36.3 mm, SH 19.1–21.4 mm, SW 10.2–11.3 mm. Shell rounded, somewhat trapezoidal, inequilateral, rather thin and compressed, anterior margin rounded and narrow, posterior margin broad and somewhat truncated, dorsal margin high with minute bars extended from the umbo, ventral margin straight. Umbo small, slightly projected, eroded. Periostracum olive-brown. Nacre bluish gray. Pseudocardinal teeth thin and lamellar, two teeth in right valve and one tooth in left valve. Lateral teeth slender, elongated, slightly curved, one in right valve and two in left valve. Anterior adductor scar somewhat drop-like, shallow, posterior adductor scar almost reduced.
Distribution: Lenya River basin, southern Myanmar.
GenusYaukthwaKonoplevaet al., 2019
Type species: Trapezoideus nesemanni Konopleva, Vikhrev & Bolotov, 2017 (by original designation)
Comments: A large genus with at least eight species7, including a species newly described here (Table 3).
Distribution: Endemic to the Western Indochina Subregion7.
Yaukthwa avaensissp. nov
Figure 2F, 5A, Tables 1–2, Supplementary Table 2
Holotype: RMBH biv0680_3, MYANMAR: Tarkat Stream, 25.2758°N, 97.2722°E, tributary of the Ayeyarwady River, 23.iii.2018, Nyein Chan leg. Reference sequence numbers of the holotype are as follows: MN275071 (COI), MN307259 (16 S rRNA), and MN307200 (28 S rRNA). Shell measurements of the holotype are as follows: SL 38.3 mm, SH 20.8 mm, and SW 14.1 mm.
Paratypes: MYANMAR: type locality, same collecting date, and collector, 9 specimens (RMBH biv0680_1, biv0680_5, biv0680_2, biv0680_4, biv0680_6, biv0680_7, biv0680_8, biv0680_9, and biv0680_10).
Etymology: The name of the new species is derived from the ancient Ava Kingdom in central Myanmar.
Diagnosis: The new species can be distinguished from its sister species Yaukthwa paiensis by having a more curved ventral margin and stronger inflation of the shell. The new species also differs from its congeners by fixed nucleotide substitutions in the COI gene fragment (Table 2).
Description: Small mussel: SL 24.7–46.6 mm, SH 14.1–25.1 mm, SW 8.7–17.0 mm. Shell subtrapezoidal, inequilateral, moderately thick and rather inflated. Anterior margin rounded, dorsal margin elevated posteriorly, ventral outline slightly curved, posterior slope truncated and usually covered by small striae. Umbo slightly elevated and strongly eroded at some specimens. Periostracum from light to dark brown. Nacre bluish, with yellow spots. Pseudocardinal teeth thin, lamellar, one tooth in the left valve and two teeth in the right valve. Lateral teeth elongated, slightly curved, one in the right valve and two in the left valve. Adductor muscle scars shallow.
Distribution: Middle section of the Ayeyarwady River, central Myanmar.
Tribe Pseudodontini Frierson, 1927
Type genus: Pseudodon Gould, 1844 (by original designation)
Comments: This tribe includes seven valid genera: Bineurus Simpson, 1900, Monodontina Conrad, 1853, Nyeinchanconcha gen. nov., Pilsbryoconcha Simpson, 1900, Sundadontina gen. nov., Thaiconcha gen. nov. (subtribe Pilsbryoconchina Bolotov, Vikhrev & Tumpeesuwan, 2017), and Pseudodon Gould, 1844 (subtribe Pseudodontina s. str.).
Distribution: Southeast Asia from the Ayeyarwady River to the Mekong Basin, Malaysia and the Greater Sunda Islands2–4,22,31.
Subtribe Pilsbryoconchina Bolotov, Vikhrev & Tumpeesuwan, 2017
Type genus: Pilsbryoconcha Simpson, 1900 (by original designation)
GenusMonodontinaConrad, 1853
=Suborbiculus Simpson, 1900
Type species: Margaritana vondembuschiana Lea, 1840 (by original designation)
Comments: This genus contains seven species (Table 3), three of which are new to science and described here.
Distribution: Sundaland Subregion (Lenya Basin in Myanmar, Mekong Basin, Malaysia, and the Greater Sunda Islands)4.
Monodontina laosicasp. nov
Figure 3C, 5A, Tables 1–2, Supplementary Table 2
Figure 3.
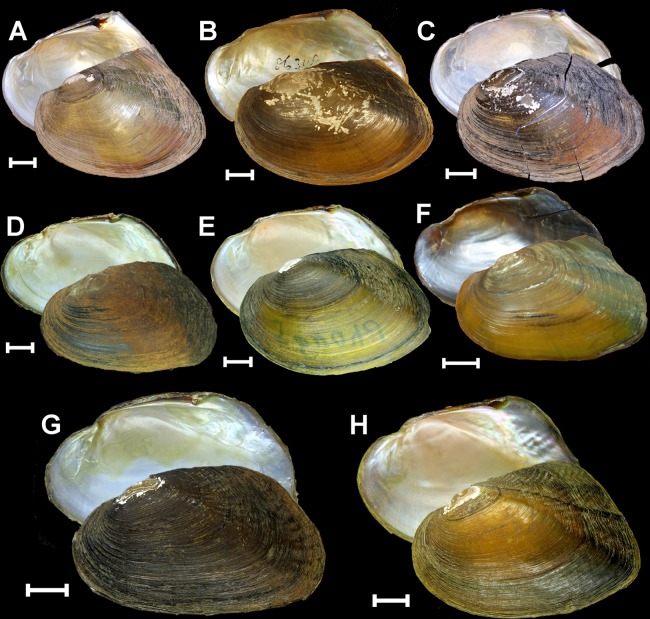
Shells of the Pseudodontini from Southeast Asia. (A) Monodontina cambodjensis (Petit de la Saussaye, 1865), Pursat River, Mekong Basin, Cambodia (specimen UMMZ 304350). (B) Monodontina vondembuschiana (Lea, 1840), Java (holotype USNM 86348). (C) Monodontina laosica sp. nov., Houai Pin Stream, a tributary of the Vang Ngao River, Mekong Basin, southern Laos (holotype UMMZ 304650). (D) Monodontina lenyanensis sp. nov., 14 Mile Stream, Lenya Basin, Myanmar (holotype RMBH biv628_2). (E) Monodontina mekongi sp. nov., headwater of the Phong River, Mekong Basin, Thailand (holotype RMBH biv122). (F) Nyeinchanconcha nyeinchani gen. & sp. nov., small stream arising at cave near Ban Kouanphavang, Mekong Basin, central Laos (holotype NCSM 84884). (G) Pseudodon kayinensis sp. nov., Winyaw River, Ataran Basin, southeastern Myanmar (holotype RMBH biv618_1). (H) Pseudodon salwenianus (Gould, 1844), unnamed stream, Salween Basin, Myanmar (a topotype specimen RMBH biv639_3). Scale bars = 1 cm. Photos: Taehwan Lee [A, C], Ilya V. Vikhrev [B], Ekaterina S. Konopleva [D, E, G, H], and Jamie M. Smith [F].
Holotype: UMMZ 304650, LAOS: ca. 300 m upstream of the mouth of Houai Pin Stream, 14.7944°N, 106.4842°E, a tributary of the Vang Ngao River, Mekong Basin, 21.v.2009, Kottelat et al. leg. Reference sequence numbers of the holotype are as follows: KP795029 (COI) and KP795052 (16 S rRNA). Shell measurements of the holotype are as follows: SL 61.4 mm, SH 41.4 mm, and SW 19.0 mm.
Etymology: The name of the new species is derived from the country of Laos, in which it was recorded.
Diagnosis: This species can be distinguished from its sister taxa by having a higher dorsal margin and reduced pseudocardinal teeth. The new species also differs from its congeners by fixed nucleotide substitutions in the COI and 16 S rRNA gene fragments (Table 2).
Description: Medium-sized mussel. Shell ovate, inequilateral, rather inflated, with high dorsal margin, creating a wing, rounded anteriorly, truncated posteriorly, ventral margin curved. Umbo not prominent, eroded. Periostracum brownish with yellow and rusty sites. Nacre blue-whitish with cream tint near the umbo. Pseudocardinal teeth weak. Both muscle scars shallow.
Distribution: Vang Ngao River, Mekong Basin, southern Laos.
Monodontina lenyanensissp. nov
Figures 3D, 5A, Tables 1–2, Supplementary Table 2
Holotype: RMBH biv0628_2, MYANMAR: 14 Mile Stream, 11.3508°N, 99.1092°E, Lenya River basin, 24.xi.2018, Bogan, Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg. Reference sequence numbers of the holotype are as follows: MN275055 (COI), MN307246 (16 S rRNA), and MN307187 (28 S rRNA). Shell measurements of the holotype are as follows: SL 63.4 mm, SH 40.1 mm, and SW 23.7 mm.
Paratypes: MYANMAR: type locality, same collecting date, and collectors, 9 specimens (RMBH biv0628_1, biv0628_3, biv0628_4, biv0628_5, biv0628_6, biv0628_7, biv0628_8, biv0628_9, and biv0628_10), 9 specimens (NCSM 104012).
Etymology: This new species is dedicated to the Lenya River, its type locality.
Diagnosis: This species can be distinguished from its sister taxa by presenting an ovate, elongated, rather solid and inflated shell, not elevated umbo, tubercular or pyramidal pseudocardinal teeth, and rather reduced muscle scars. The new species also differs from its congeners by fixed nucleotide substitutions in the COI and 16 S rRNA gene fragments (Table 2).
Description: Medium-sized mussel: SL 31.2–63.4 mm, SH 20.3–40.1 mm, SW 11.0–23.7 mm. Shell ovate, elongated, inequilateral, rather solid and inflated, rounded anteriorly, broad and truncated posteriorly, dorsal margin high, ventral margin slightly curved. Umbo small, slightly elevated, eroded. Periostracum rusty-brown, smooth. Nacre white-yellowish. One pseudocardinal tooth in each valve, which is tubercular-like or more pyramidal and sharper, rather high and strong, smooth or slightly ribbed. Lateral teeth reduced. Anterior adductor scar ovate, rather prominent; posterior adductor scar reduced, weakly developed.
Distribution: Lenya River basin, southern Myanmar.
Monodontina mekongisp. nov
Figures 3E, 5A, Tables 1–2, Supplementary Table 2
Holotype: RMBH biv0122, THAILAND: clay bottom, headwaters of the Phong River, 16.8616°N, 101.9105°E, Mekong Basin, Loei Province, 09.iv.2014, Bolotov, Vikhrev, Spitsyn and Gofarov leg. Reference sequence numbers of the holotype are as follows: KX865861 (COI), KX865632 (16 S rRNA), and KX865733 (28 S rRNA). Shell measurements of the holotype are as follows: SL 65.7 mm, SH 42.2 mm, and SW 20.3 mm.
Etymology: The name of this species is derived from the Greater Mekong Basin, its type locality.
Diagnosis: This species is conchologically and genetically related with Monodontina vondembuschiana and M. laosica sp. nov. but it can be distinguished from these species by presenting an uninflated, weak pseudocardinal teeth (vs. stouter), and a curved and lower dorsal margin (vs. strait and higher). The new species also differs from its congeners by fixed nucleotide substitutions in the COI, 16 S rRNA and 28 S rRNA gene fragments (Table 2).
Description: Medium-sized mussel. Shell obovate, slightly higher posteriorly, inequilateral, thin, semitransparent, not inflated; anterior margin rounded, posterior margin angulate, ventral margin curved. Umbo slightly elevated, eroded, without clear sculpture. Periostracum olive yellow. Nacre whitish. Pseudocardinal teeth weak, flatten, more developed in the right valve than in the left one. Both muscle scars shallow, anterior scar irregular; posterior scar somewhat drop-like, almost invisible.
Distribution: Phong River, Mekong Basin, northern Thailand.
GenusNyeinchanconchagen. nov
Type species: Nyeinchanconcha nyeinchani gen. & sp. nov.
Comments: Remarkable monotypic genus (Table 3).
Diagnosis: Shell elliptical, resembling that of the genus Lamellidens Simpson, 1900 (Parreysiinae: Lamellidentini), slightly elevated posteriorly, moderately thick and inflated, umbo not elevated, pseudocardinal teeth strong and somewhat pyramidal in each valve; anterior adductor scar drop-like, developed, usually contiguous with pedal retractor scar; the posterior muscle scar shallow.
Etymology: This genus is dedicated to our friend Mr. Nyein Chan, an enthusiastic conservation biologist from FFI – Myanmar Program, Yangon, Myanmar, for his valuable contribution to the conservation of freshwater ecosystems in Southeast Asia. This genus name means “Shell of Nyein Chan” (“concha” being shell in Latin).
Distribution: Mekong Basin in Laos.
Nyeinchanconcha nyeinchanigen. & sp. nov
Figure 3F, 5A, Tables 1–2, Supplementary Table 2
Holotype: NCSM 84884, LAOS: small stream arising at a cave near Ban Kouanphavang, 17.4578°N, 104.9263°E, Nam Done River drainage, Mekong Basin, Khammouane Province, 17.v.2012, M. Kottelat et al. leg. Reference sequence numbers of the holotype are as follows: KX822662 (COI) and KX822618 (28 S rRNA). Shell measurements of the holotype are as follows: SL 50.9 mm, SH 27.8 mm, and SW 15.6 mm.
Paratypes: LAOS: type locality, same date, and collectors, 1 specimen (NCSM 113351); Nam Phiat River near Phon Bong village, 18.0839°N, 104.9781°E, ca. 2 km from confluence with the Namkading River, Mekong Basin, Bolikhamsai Province, 12.v.2009, 1 specimen (UMMZ 304648), M. Kottelat et al. leg.
Etymology: This species is dedicated to our friend Mr. Nyein Chan, a conservation biologist from FFI – Myanmar Program, Yangon, Myanmar.
Diagnosis: The species is morphologically and genetically more similar to Sundadontina brandti sp. nov. but differs from it by a more elliptical shell without marked elevation of the dorsal margin, and by pyramidal and weaker pseudocardinal teeth. The new species also differs from other Pseudodontini taxa by fixed nucleotide substitutions in the COI and 16 S rRNA gene fragments (Table 2).
Description: Medium-sized mussel: SL 33.7–59.9 mm, SH 15.1–37.0 mm, SW 7.9–15.6 mm. Shell elliptical, inequilateral, rounded anteriorly, truncated posteriorly, dorsal margin elevated, ventral margin slightly curved. Umbo not prominent, eroded. Periostracum dark brown. Nacre whitish with cream tint near the umbo. Pseudocardinal teeth somewhat pyramidal, stout. Anterior adductor scar pronounced; posterior adductor scar weak.
Distribution: Nam Done and Nam Phiat rivers, Mekong Basin, Laos.
GenusSundadontinagen. nov
Type species: Anodonta cumingii Lea, 1850.
Comments: This genus contains at least 10 species, three of which are new to science and described here (Table 3).
Diagnosis: Shell ovate or elongate-ovate, rather thick and strong, umbo not projected, pseudocardinal teeth stout and tubercular-like; anterior muscle scar ovate and well-developed, usually contiguous with pedal retractor scar; the posterior muscle scar shallow.
Etymology: The name of this genus is derived from that of the genus Monodontina, but with another prefix highlighting its broad distribution across the ancient Sundaland.
Distribution: Sundaland Subregion: Lenya Basin in Myanmar, Mekong Basin in Thailand, Cambodia, and southern Vietnam, Chao Phraya Basin in Thailand, Malaysia.
Sundadontina brandtisp. nov
Figures 4B, 5A, Tables 1–2, Supplementary Table 2
Figure 4.
Shells of the Pseudodontini from Southeast Asia. (A) Sundadontina cumingii (Lea, 1850) gen. & comb. nov., Malacca (holotype USNM 86350). (B) Sundadontina brandti sp. nov., headwater of the Mun River, Mekong Basin, Thailand (holotype RMBH biv475_2). (C) Sundadontina tanintharyiensis sp. nov., Chaung Nauk Pyan Stream, Lenya Basin, Myanmar (holotype RMBH biv643_4). (D) Sundadontina taskaevi sp. nov., headwater of the Mun River, Mekong Basin, Thailand (holotype RMBH biv475_1). (E) Sundadontina moreleti (Crosse & Fischer, 1876) comb. nov., Mekong Basin, Cambodia (syntype MNHN-IM-2000–34623). (F) Sundadontina tumida (Morelet, 1866) comb. nov., Cambodia (holotype NHMUK 93-2-4-1734). (G) Thaiconcha callifera (Martens, 1860) gen. & comb. nov., Siam (holotype NHMUK 1859-8-1-20). (H) Thaiconcha callifera (Martens, 1860) gen. & comb. nov., (a topotype specimen RMBH biv120_11). Scale bars = 1 cm. Photos: Ilya V. Vikhrev [A], Ekaterina S. Konopleva [B-D, H], Kevin Webb (NHMUK Photographic Unit) [F, G], and Manuel Caballer (2018 MNHN Project: RECOLNAT No. ANR-11-INBS-0004) [E].
Holotype: RMBH biv0475_2, THAILAND: headwater of the Mun River, 14.4138°N, 102.0821°E, Mekong Basin, Khorat Plateau, Nakhon Ratchasima Province, 12.iii.2018, Bolotov, Vikhrev, and local villagers leg. Reference sequence numbers of the holotype are as follows: MN275058 (COI), MN307249 (16 S rRNA), and MN307190 (28 S rRNA). Shell measurements of the holotype are as follows: SL 85.4 mm, SH 53.7 mm, and SW 27.7 mm.
Paratypes: THAILAND: type locality, same collecting date, and collectors, 2 specimens (RMBH biv0475_3, biv0475_4).
Etymology: This species is named in the memory of Dr. Rolf Arthur Max Brandt (1917–1989), one of the most influential freshwater malacologists of the last century. This prominent scientist worked in Southeast Asia on freshwater mollusks and authored the freshwater mollusks of Thailand31.
Diagnosis: The species is similar to Sundadontina taskaevi sp. nov. but can be distinguished from it by having a more slender and higher pseudocardinal tooth on the right valve. The new species also differs from its congeners by fixed nucleotide substitutions in the COI and 16 S rRNA gene fragments (Table 2).
Description: Large mussel: SL 71.3–85.4 mm, SH 43.8–53.7 mm, SW 24.1–27.7 mm. Shell ovate, very inequilateral, solid, not very inflated, rounded anteriorly, truncated posteriorly, dorsal margin curved, ventral margin slightly rounded. Periostracum brownish-black. Nacre creamy. Umbo very small, not developed, eroded. Left valve with one tubercle-like pseudocardinal tooth, right valve with one rectangular and high pseudocardinal tooth. Anterior muscle scar rather well-developed, ovate; posterior muscle scar slightly visible.
Distribution: Mun Basin, Thailand.
Sundadontina tanintharyiensissp. nov
Figure 4C, 5A, Tables 1–2, Supplementary Table 2
Holotype: RMBH biv0643_4, MYANMAR: Chaung Nauk Pyan Stream, 11.7620°N, 99.1124°E, Lenya River basin, 16.vi.2018, Nyein Chan leg. Reference sequence numbers of the holotype are as follows: MN275057 (COI), MN307248 (16 S rRNA), and MN307189 (28 S rRNA). Shell measurements of the holotype are as follows: SL 57.1 mm, SH 40.8 mm, and SW mm.
Paratypes: MYANMAR: type locality, same collecting date, and collectors, 2 specimens (RMBH biv0643_1, biv0643_6) and 3 specimens (NCSM 113364).
Etymology: The new species name is derived from the Tanintharyi Region of Myanmar, in which its type locality is situated.
Diagnosis: The new species is similar to Sundadontina cumingii gen. & comb. nov. but can be distinguished from it by having a more rounded and inflated shell. The new species also differs from its congeners by fixed nucleotide substitutions in the COI and 16 S rRNA gene fragments (Table 2).
Description: Medium-sized mussel: SL 39.2–61.2 mm, SH 27.2–45.1 mm, SW 14.7–23.2 mm. Shell ovate or slightly elongated, inequilateral, moderately solid and rather inflated. Anterior margin rounded, dorsal and ventral margin curved, posterior margin subangular. Umbo not elevated, eroded. Periostracum rusty-brown, smooth. Nacre yellowish white. Each valve with one tubercle-like, smooth pseudocardinal tooth. Lateral teeth reduced. Anterior muscle scar ovate or drop-like, rather prominent; posterior muscle scar drop-like and very shallow.
Distribution: Lenya River basin, southern Myanmar.
Sundadontina taskaevisp. nov
Figures 4D, 5A, Tables 1–2, Supplementary Table 2
Holotype: RMBH biv0475_1, THAILAND: headwater of the Mun River, 14.4138°N, 102.0821°E, Mekong Basin, Khorat Plateau, Nakhon Ratchasima Province, 12.iii.2018, Bolotov and Vikhrev leg. Reference sequence numbers of the holotype are as follows: MN275061 (COI), MN307251 (16 S rRNA), and MN307192 (28 S rRNA). Shell measurements of the holotype are as follows: SL 82.7 mm, SH 52.2 mm, and SW 29.3 mm.
Paratypes: THAILAND: type locality, same collecting date, and collectors, 1 specimen (RMBH biv0475_5).
Etymology: This species is named in memory of the late Dr. Anatoly Ivanovich Taskaev (1944–2010), a well-known Russian biologist.
Diagnosis: The new species is similar to Sundadontina brandti sp. nov. but can be distinguished from it by having broader, stronger, tubercle-like pseudocardinal teeth. The new species also differs from its congeners by fixed nucleotide substitutions in the COI and 16 S rRNA gene fragments (Table 2).
Description: Large mussel: SL 60.0–82.7 mm, SH 42.8–52.2 mm, SW 22.6–29.3 mm. Shell elongate-ovate, inequilateral, rather solid and inflated, rounded anteriorly, dorsal margin convex, ventral margin slightly curved. Periostracum blackish, with brown lines. Nacre creamy. Umbo slightly elevated, eroded. Each valve with one tubercle-like, smooth pseudocardinal tooth. The tooth in right valve more developed, rather trapezoidal, with broader base. Anterior muscle scar ovate, rather well-developed; posterior muscle scar shallow.
Distribution: Mun River, Thailand.
GenusThaiconchagen. nov
Type species: Anodonta callifera Martens, 1860.
Comments: This genus contains at least two valid species (Table 3).
Diagnosis: Shell large, thick, elliptical or rounded, moderately inflated. Pseudocardinal teeth rather well developed, muscle attachment scars deep.
Etymology: The name of this genus means “a shell from Thailand”.
Distribution: Mekong Basin in Thailand and Cambodia.
SubtribePseudodontina Frierson, 1927
Type genus: Pseudodon Gould, 1844 (by original designation)
GenusPseudodonGould, 1844
Type species: Anodon inoscularis Gould, 1844 (by original designation)
Comments: This genus contains eight species, one of which is new to science and described here (Table 3).
Distribution: Endemic clade to the Western Indochina Subregion4.
Pseudodon kayinensissp. nov
Figure 3G, 5A, Tables 1–2, Supplementary Table 2
Holotype: RMBH biv0618_1, MYANMAR: Winyaw River, 15.6685°N, 97.9496°E, Ataran River basin, 20.xi.2018, Vikhrev, Bogan, Lopes-Lima, and local villagers leg. Reference sequence numbers of the holotype are as follows: MN275043 (COI). Shell measurements of the holotype are as follows: SL 59.6 mm, SH 34.9 mm, and SW 17.7 mm.
Paratypes: MYANMAR: type locality, same collecting date, and collectors, 4 specimens (RMBH biv0618_2, biv0618_3, biv0618_4, biv0618_5) and 4 specimens (NCSM 104014); Ko Du Kwe Stream, 15.6132°N, 98.2363°E, Zami River, Ataran River basin, 26.ii.2018, 5 specimens (RMBH biv0637_1, biv0637_2, biv0637_3, biv0637_4, biv0637_5) and 5 specimens (NCSM 113362), Than Win leg.; unnamed stream, 17.0292°N, 97.8100°E, Hlaingbwe River basin, 17.xi.2018, 3 specimens (RMBH biv0638_1, biv0638_2, biv0638_3) and 5 specimens (NCSM 104015), Than Win leg.
Etymology: The name of new species is derived from its distribution range, i.e. the Kayin State in Myanmar.
Diagnosis: The new species is conchologically more similar to Pseudodon bogani but can be distinguished from it by having a more curved dorsal margin and clearly ornamented posterior side. The new species also differs from its congeners by fixed nucleotide substitutions in the COI, 16 S rRNA and 28 S rRNA gene fragments (Table 2).
Description: Rather large mussel: SL 37.8–71.0 mm, SH 22.4–42.5 mm, SW 10.3–21.2 mm. Shell from ovate to elliptical, elongated, inequilateral, moderately inflated and thick. Anterior margin rounded, posterior margin somewhat truncated, dorsal margin curved and rather high, ventral margin straight or slightly curved. Umbo not elevated, eroded. Periostracum olive-brown to dark brown, the surface from umbo to posterior margin clearly ribbed, having curved bars covering the entire dorsal margin and then radiate along the posterior slope. Nacre whitish, sometimes with yellow sites. Pseudocardinal teeth high, tubercular-like, in each valve. Anterior adductor scar rather well developed, ovate; posterior adductor scar drop-like, more or less visible.
Distribution: Hlaingbwe and Ataran River basins in southern Myanmar.
Discussion
The Isthmus of Kra as a significant biogeographic barrier for the Unionidae
It was shown that freshwater basins of the Malacca Peninsula represent a part of the Sundaland Subregion16,17 and that the Dawei and Tanintharyi river basins belong to the Western Indochina Subregion4,5,7,24. However, the location of the southern boundary of the Western Indochina Subregion was until now unclear, because freshwater mussel faunas of the southern edge of Myanmar south of the Tanintharyi Basin were almost unknown. It was assumed that the Western Indochinese fauna could spread throughout the western coastal rivers of southern Thailand as far south as the Kangar-Pattani Line (7°N latitude along the Thai-Malay border)5 that corresponds to a putative ancient seaway36,37. In this study, however, we found that freshwater mussel species inhabiting the Lenya River basin such as Trapezoideus lenya sp. nov., Monodontina lenyanensis sp. nov., and Sundadontina tanintharyiensis sp. nov. belong to the Sundaland fauna, and these species were separated from their sister taxa inhabiting the Mekong River and smaller basins of the Gulf of Thailand drainage during the Miocene, with the last split occurring ca. 6 Myr ago. Based on these results, we can conclude that the boundary between freshwater mussel faunas of the Western Indochina and Sundaland subregions is located along the Tanintharyi – Lenya drainage divide just north of the Isthmus of Kra (Fig. 5B). While freshwater mussel faunas between the Lenya Basin and the Malay Peninsula are poorly known31, the fauna of Malaysia contains only typical Sundaland unionid taxa supporting our conclusion3,16. There is an admixture of Sundaland's taxa, i.e. Trapezoideus foliaceus, in the Dawei River7,24 that can reflect an ancient river capture.
This isthmus is a major biogeographic barrier corresponding to a putative ancient seaway36,37 that influenced distribution ranges of a plethora of animal and plant taxa38 and corresponds to the separation of the Oriental (Indo-Burmese) and Sundaland biotas39. A growing body of phylogeographic and phylogenetic research indicates that this barrier is reflected through abrupt changes in bird40, frog41, snake42, lizard43, giant centipede44 and spider45 assemblages around the Isthmus of Kra area. However, such examples are still poorly known among freshwater animals. Tarebia granifera (Lamarck, 1816) (Thiaridae), a freshwater snail species, shares two distant species-level lineages probably diverged due to marine transgressions through the Isthmus of Kra approximately 5 Myr ago46. The high level of genetic divergence between populations of the giant freshwater prawn Macrobrachium rosenbergii (de Man, 1879) (Palaemonidae) clearly supports the existence of a hypothetical seaway north of the Isthmus of Kra37. Specimens of the Blue Panchax killifish Aplocheilus panchax (Hamilton, 1822) (Aplocheilidae) collected just north of the Isthmus of Kra share clear mtDNA affinities to the Indian clade, while those from localities south of this isthmus represent a separate Sundaic clade47. In summary, our novel findings agree with available data on other freshwater and terrestrial taxa revealing the presence of a significant biogeographic barrier at the Isthmus of Kra area. There are two more putative connections via lowlands and river valleys at the Krabi and Songkhla isthmuses (Fig. 5B). Freshwater mussel assemblages of these areas are still unknown, but they undoubtedly belong to the Sundaland fauna.
Eastern edge of the Indian Subregion
The biogeographic boundary between the Indian and Western Indochina freshwater subregions is located along the Naga Hills, Chin Hills, and Rakhine Yoma ranges separating rivers of the Rakhine Coast from the Ayeyarwady Basin. The eastern edge of the Indian Subregion covers the entire Rakhine Coast of Myanmar with numerous coastal freshwater basins. Freshwater animal species inhabiting this area have clear affinities to the Indian fauna, e.g. the sponge Corvospongilla ultima (Annandale, 1910), the polychaete worm Namalycastis indica (Southern, 1921), and the bivalves Novaculina gangetica Benson, 1830, Lamellidens marginalis (Lamarck, 1819)48,49, and Parreysia rakhinensis sp. nov. The freshwater fish fauna of the Rakhine Coast contains numerous local endemic species that are also related to the fauna of the Indian Subcontinent50–52, being this area considered a regional hotspot of freshwater fish diversity53. Our time-calibrated phylogeny indicates that Parreysia rakhinensis sp. nov. has close affinities to Indian taxa, and the split between these lineages occurred ca. 5 Myr ago.
Insights into the genus-level taxonomy of the Southeast Asian Unionidae
Based on our comprehensive multi-locus phylogeny, we introduce four new genera and twelve new species of freshwater mussels from Southeast Asia. Scabiellus gen. nov. represents a remarkable example of convergent evolution of the shell patterns in freshwater bivalves as it is conchologically similar to members of two other genera of the tribe Indochinellini, i.e. Scabies and Indochinella. The range of this monotypic genus corresponds to the Khorat Plateau, a putative evolutionary hotspot at the Middle Mekong Basin, that presents high levels of endemism in freshwater animals, e.g. bivalves26–28, fish54, and softshell turtles55,56. In summary, the Indochinellini contains three genera (Indochinella, Indonaia, and Radiatula) west of the Salween – Mekong drainage divide and four genera (Harmandia, Scabies, Scabiellus gen. nov., and Unionetta) to the east of the same boundary.
The tribe Pseudodontini shares one of the largest monophyletic radiations of freshwater mussels in Southeast Asia, with numerous genus- and species level clades4,22 that were traditionally placed into two genera, Pseudodon and Pilsbryoconcha29,31. Based on our multi-locus data set, we had previously resurrected two more valid genera, Monodontina and Bineurus, and indicated the presence of at least three genus-level clades new to science4. To establish an updated taxonomy of the Pseudodontini, in the present study we describe three new genera: Nyeinchanconcha gen. nov., Sundadontina gen. nov., and Thaiconcha gen. nov. The first monotypic genus from the Middle Mekong Basin in central Laos represents another example of shell convergence as it conchologically resembles members of the tribe Lamellidentini by having a rather thin, ovate shell with a strongly reduced hinge plate. Sundadontina seems to be a large and conchologically variable genus widespread from the Mekong Basin to Thailand, southern Myanmar, and Malaysia. Thaiconcha is a remarkable genus that is phylogenetically sister to Bineurus but conchologically differs by having a less elongated, ovate-shaped shell.
Ancient faunal exchanges between freshwater mussel faunas
The time-calibrated phylogeny suggests that there were several ancient exchanges between faunas of the Sundaland and Western Indochina subregions starting as early as the Late Cretaceous (ca. 80 Myr ago), when the subtribes Pseudodontina and Pilsbryoconchina were separated. The Rectidentini + Contradentini clade most likely evolved within the Sundaland Subregion, with an expansion of a single clade to Western Indochina, while the Indochinellini clade shows an opposite pattern. Both the colonization events were placed in the Late Eocene (ca. 40–46 Myr ago) and were probably triggered by a wet and warm climatic episode during this period57. We suggest that these splits may reflect ancient river captures/splits with subsequent colonization/vicariance events in freshwater mussels22. The recent geological study suggests that the paleo-Mekong River was established as a large river in the Middle Miocene due to increased erosion during a period of high monsoon precipitation58. Other research assumes that the paleo-Ayeyarwady probably originated sometime between the Late Eocene and Early Oligocene59. However, our results indicate that these ages might be underestimated, and the paleo-Mekong and paleo-Ayeyarwady rivers could have been initiated since the Late Cretaceous as did the paleo-Yangtze System60. Two ancient monophyletic mussel radiations (age 51–55 Myr) were previously discovered within the putative paleo-Mekong catchment22 also suggesting at least the Early Eocene age of this freshwater system. In summary, our findings support the hypothesis that Southeast Asian freshwater bivalve fauna primarily originated within three evolutionary hotspots (Western Indochina, Sundaland, and East Asian)5,6 supplemented by ancient (Late Miocene) immigrants that colonized freshwater systems of the western coast of Myanmar from the Indian Subcontinent.
Methods
Data sampling
Mussel specimens were collected from various water bodies throughout Myanmar, Thailand and northern Laos from 2012 to 2018. A foot tissue snip from each specimen was preserved in 96% ethanol immediately after collection. To find the boundaries between biogeographic subregions, we collected freshwater mussels throughout small and medium-sized freshwater basins of the Rakhine Coast and the southern edge of Myanmar in 2018 under a National Geographic Society grant No. NGS-274R-18.
Studied museum collections
The freshwater mussel shell lots were studied in the malacological collections of the Russian Museum of Biodiversity Hotspots [RMBH], Federal Center for Integrated Arctic Research, Russian Academy of Sciences, Arkhangelsk, Russia, National Museum of Natural History [NMNH], Smithsonian Institution, Washington, DC, United States of America, British Museum of Natural History [NHMUK], London, United Kingdom, Muséum National d’Histoire Naturelle [MNHN], Paris, France, Museo Civico di Storia Naturale di Genova [MSNG], Genoa, Italy, California Academy of Natural Sciences, San Francisco, United States of America [CAS], North Carolina Museum of Natural Sciences [NCSM], Raleigh, United States of America, and the University of Michigan Museum of Zoology [UMMZ], Ann Arbor, United States of America.
Molecular data and phylogenetic analyses
Multi-locus phylogeny (3 codons of COI + 16 S rRNA + 28 S rRNA) was reconstructed using 271 haplotypes of the Parreysiinae and Gonideinae members from Southeast Asia, East Asia, India, and Africa (Supplementary Table 1). Representatives of the Margaritiferidae, Iridinidae, Etheriidae, Mycetopodidae, Hyriidae, and Trigoniidae were used as outgroup. We used IQ-TREE v1.6.1161 and MrBayes v3.2.662 as described in our previous work6. Bayesian calculations were performed at the San Diego Supercomputer Center through the CIPRES Science Gateway63. The best-fit evolutionary models applied to each partition in the IQ-TREE and MrBayes runs based on Bayesian Information Criterion (BIC) of Model Finder implemented in the IQ-TREE web server61 were as follows: F81 + G (1st codon of COI); GTR + G (2nd codon of COI); TN + I + G (3rd codon of COI); GTR + I + G (16 S rRNA); and TIM2 + I + G (28 S rRNA).
Time-calibrated phylogeny
The time-calibrated phylogeny was reconstructed in BEAST v2.6.164 based on an external COI evolutionary rate (0.265 ± 0.06% substitutions per site per million years) estimated for the Unionidae65. This rate can be considered a reliable estimate as it is largely congruent with the data inferred from a mitogenomic reconstruction20. The same multi-locus dataset as for the IQ-TREE and MrBayes phylogenetic analyses (3 codons of COI + 16 S rRNA + 28 S rRNA) was estimated. The evolutionary rate was implemented only to the COI partition. The HKY + G model was applied for each gene partition. The analyses were run using a lognormal relaxed clock algorithm with the Yule speciation process as the tree priors66,67. Calculations were performed at the San Diego Supercomputer Center through the CIPRES Science Gateway63. We conducted four searches, each with 5 × 107 generations and tree sampling every 1000th generation. The log files were checked visually with Tracer v. 1.768. Most of ESS values were recorded as > 300, a few of them were registered > 100. All runs were compiled with LogCombiner v1.8.467 using an additional re-sampling every 10,000th generation and 25% burn-in. The maximum clade credibility tree was obtained using TreeAnnotator v1.8.467.
Statistical biogeographic analyses
To reconstruct ancestral areas with RASP v3.269, we used the set of 15,004 time-calibrated binary trees that were combined from the four runs of BEAST v2.6.1 (see above). As a condensed tree, we used the user-specified consensus tree, which was calculated based on this set of trees with TreeAnnotator v1.8.4 (see above). Non-target sequences (outgroup taxa and species, the ranges of which are situated beyond Southeast Asia and the Indian Subcontinent) were removed from the tree set using the appropriate option of the software. We used only one haplotype per species. Ancestral area patterns were reconstructed using two probabilistic algorithms: Statistical Dispersal-Vicariance Analysis (S-DIVA) and Bayesian MCMC analysis. Three possible distribution areas were assigned as follows: (A) Western Indochina, (B) Sundaland, and (C) Indian subregions. The S-DIVA analyses were calculated with the following parameters: max areas = 3; allow reconstruction with max reconstructions = 100; max reconstructions for final tree = 1,000; and allowing extinctions. The MCMC analysis was performed with default settings and 500,000 generations. In addition to the reconstructions obtained from each analysis separately, we used summary results of the two kinds of analyses, which were combined with RASP v3.269.
Species delimitation and diagnostics of new taxa
To delimit and diagnose species in our dataset, we used an integrative approach4–7,70–73 based on the phylogenetic and morphological analyses. First, we applied an automatic species delimitation approach to delimit the Molecular Operational Taxonomic Units (MOTUs) that may correspond to biological species. The maximum likelihood COI phylogeny of each tribe inferred from IQ-TREE v1.6.1161 was used as an input tree for the Poisson Tree Process (PTP) modeling through the PTP web-service (http://mptp.h-its.org)74. An uncorrected COI mean p-distance to the nearest neighbor of each species-level lineage was calculated in MEGA775. Second, each MOTU within the clades of interest was studied using morphological criteria (shell shape, umbo position, structure of pseudocardinal and lateral teeth, shape of muscle attachment scars), and was compared with the original descriptions of nominal taxa to link each clade to a biological species. Three shell dimensions of each specimen, included in the type series of new taxa, i.e., the length, height, and width of the shell (all at the maximum diameter), were measured using calipers ( ± 0.1 mm) (Table 1 and Supplementary Table 2). The molecular diagnosis of every new species was designed using fixed nucleotide substitutions, which were estimated for each gene separately using a Toggle Conserved Sites tool of MEGA775 at a 50% level. For the diagnoses, an alignment of congeneric haplotype sequences (tribe-level alignment for Nyeinchanconcha nyeinchani gen. & sp. nov.) was performed using the Muscle algorithm implemented in MEGA775. All deleterious mutations were retained for the analyses. While numerous recent studies reveal that using an integrative approach for freshwater mussel taxonomic research is rather straightforward4–7,70–73, its application to freshwater gastropods is more difficult due to several shortcomings such as a possible incongruence in a mitochondrial phylogeny76,77 and often higher DNA barcoding thresholds between species46,78,79. At first glance, the differences between these groups can be explained by slower evolutionary rates of freshwater mussels20,65 compared with those of freshwater gastropods80.
Nomenclatural acts
The electronic edition of this article conforms to the requirements of the amended International Code of Zoological Nomenclature (ICZN), and hence the new names contained herein are available under that Code from the electronic edition of this article. This published work and the nomenclatural acts it contains have been registered in ZooBank (http://zoobank.org), the online registration system for the ICZN. The LSID for this publication is: urn:lsid:zoobank.org:pub:C6AF4F5B-8526–4FF6-BF08-D6697BE24E66. The electronic edition of this paper was published in a journal with an ISSN and has been archived and is available from PubMed Central.
Supplementary information
Acknowledgements
We are grateful to Prof. Dr. Matthias Glaubrecht and an anonymous reviewer for their valuable comments on the earlier version of this paper. This study was supported by the Ministry of Science and Higher Education of Russia, the Ministry of Europe and Foreign Affairs of France (MEAE), and the Ministry of Higher Education, Research and Innovation of France (MESRI) under project No. 05.616.21.0114 of the Hubert Curien Partnership (PHC) for the Franco-Russian Cooperation for Science and Technology (PHC Kolmogorov 2019). We are grateful to the late Dr. Tony Whitten (Fauna & Flora International – Asia-Pacific, UK), Mr. Frank Momberg (Director for Program Development and Asia-Pacific Program Director of Fauna & Flora International, UK) and Mr. Mark Grindley (Country Director of Fauna & Flora International – Myanmar Program, Myanmar), and the staff of the Department of Fisheries of the Ministry of Agriculture, Livestock and Irrigation of Myanmar for their great help during this study. Our research in Myanmar was performed under the survey permission No. 5/6000/LFR(210/2018) dated on 23 January 2018 issued by the Ministry of Agriculture, Livestock and Irrigation of Myanmar and the export permission No. NWCD/CITES/9/5666/2018 dated on 28 June 2018 issued by the Forest Department of the Ministry of Environmental Conservation and Forestry of Myanmar. Our samples from Thailand were taken under the export permission No. 11501110316100766 dated on 15 March 2018 issued by the Suvarnabhumi Airport Fish Inspection Office. We are thankful to Dr. Adam J. Baldinger (Museum of Comparative Zoology [MCZ], Harvard University, Cambridge, United States of America), Dr. Virginie Héros (Muséum National d’Histoire Naturelle [MNHN], Paris, France), Dr. Taehwan Lee (University of Michigan Museum of Zoology [UMMZ], Ann Arbor, United States of America), Ms. Jamie M. Smith (North Carolina Museum of Natural Sciences [NCSM], Raleigh, United States of America), Dr. Ellen E Strong (National Museum of Natural History [NMNH], Smithsonian Institution, Washington, DC, United States of America), Dr. Tom S. White and Kevin Webb (British Museum of Natural History [NHMUK], London, United Kingdom) for providing images of shells from museum collections.
Author contributions
I.N.B. developed the concept of the study. I.N.B., I.V.V., A.E.B., M.L.-L., S.T., K.T., Z.L., N.C., and T.W. collected samples. A.V.K. and E.S.K. designed and processed molecular analyses. E.S.K. performed morphological research and phylogenetic modeling. M.Y.G. created the maps. I.N.B. and E.S.K. wrote the paper, with input from I.V.V., M.L.-L., A.E.B., Z.L., N.C., A.V.K., M.Y.G., O.V.A., and T.W. All authors discussed the final version of the manuscript.
Data availability
The type series of the new taxa are deposited in the Russian Museum of Biodiversity Hotspots [RMBH], Federal Center for Integrated Arctic Research, Russian Academy of Sciences, Arkhangelsk, Russia; the North Carolina Museum of Natural Sciences [NCSM], Raleigh, United States of America; and the University of Michigan Museum of Zoology [UMMZ], Ann Arbor, United States of America. The molecular sequences obtained in this study are available in GenBank. Sequence accession numbers and collecting locality for each specimen are presented in Supplementary Tables 1–2. Shell measurements for the type series of the new species are given in Table 1 and Supplementary Table 2.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-63612-5.
References
- 1.Bogan AE. Global diversity of freshwater mussels (Mollusca, Bivalvia) in freshwater. Hydrobiologia. 2008;595:139–147. doi: 10.1007/s10750-007-9011-7. [DOI] [Google Scholar]
- 2.Graf DL, Cummings KS. Review of the systematics and global diversity of freshwater mussel species (Bivalvia: Unionoida) Journal of Molluscan Studies. 2007;73:291–314. doi: 10.1093/mollus/eym029. [DOI] [Google Scholar]
- 3.Zieritz A, et al. Diversity, biogeography and conservation of freshwater mussels (Bivalvia: Unionida) in East and Southeast Asia. Hydrobiologia. 2018;810:29–44. doi: 10.1007/s10750-017-3104-8. [DOI] [Google Scholar]
- 4.Bolotov IN, et al. New taxa of freshwater mussels (Unionidae) from a species-rich but overlooked evolutionary hotspot in Southeast Asia. Scientific Reports. 2017;7:1–18. doi: 10.1038/s41598-017-11957-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolotov IN, et al. A new genus and tribe of freshwater mussel (Unionidae) from Southeast Asia. Scientific Reports. 2018;8:1–12. doi: 10.1038/s41598-018-28385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotov IN, et al. Eight new freshwater mussels (Unionidae) from tropical Asia. Scientific Reports. 2019;9:1–15. doi: 10.1038/s41598-019-48528-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konopleva ES, et al. A new genus and two new species of freshwater mussels (Unionidae) from western Indochina. Scientific Reports. 2019;9:1–14. doi: 10.1038/s41598-019-39365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lydeard C, et al. The global decline of nonmarine mollusks. BioScience. 2004;54:321–330. doi: 10.1641/0006-3568(2004)054[0321:TGDONM]2.0.CO;2. [DOI] [Google Scholar]
- 9.Bolotov IN, et al. Climate warming as a possible trigger of keystone mussel population decline in oligotrophic rivers at the continental scale. Scientific Reports. 2018;8:1–9. doi: 10.1038/s41598-017-18873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopes-Lima M, et al. Biology and conservation of freshwater bivalves: past, present and future perspectives. Hydrobiologia. 2014;735:1–13. doi: 10.1007/s10750-014-1902-9. [DOI] [Google Scholar]
- 11.Lopes-Lima M, et al. Conservation of freshwater bivalves at the global scale: diversity, threats and research needs. Hydrobiologia. 2018;810:1–14. doi: 10.1007/s10750-017-3486-7. [DOI] [Google Scholar]
- 12.Bogan AE. Freshwater bivalve extinctions (Mollusca: Unionoida): a search for causes. Integrative and Comparative Biology. 1993;33:599–609. doi: 10.1093/icb/33.6.599. [DOI] [Google Scholar]
- 13.Ferreira-Rodríguez N, et al. Research priorities for freshwater mussel conservation assessment. Biological Conservation. 2019;231:77–87. doi: 10.1016/j.biocon.2019.01.002. [DOI] [Google Scholar]
- 14.Peacock E, Haag WR, Warren ML., Jr. Prehistoric decline in freshwater mussels coincident with the advent of maize agriculture. Conservation Biology. 2005;19:547–551. doi: 10.1111/j.1523-1739.2005.00036.x. [DOI] [Google Scholar]
- 15.Ricciardi A, Neves RJ, Rasmussen JB. Impending extinctions of North American freshwater mussels (Unionoida) following the zebra mussel (Dreissena polymorpha) invasion. Journal of Animal Ecology. 1998;67:613–619. doi: 10.1046/j.1365-2656.1998.00220.x. [DOI] [Google Scholar]
- 16.Zieritz A, et al. Factors driving changes in freshwater mussel (Bivalvia, Unionida) diversity and distribution in Peninsular Malaysia. Science of the Total Environment. 2016;571:1069–1078. doi: 10.1016/j.scitotenv.2016.07.098. [DOI] [PubMed] [Google Scholar]
- 17.Zieritz A, et al. Changes and drivers of freshwater mussel diversity and distribution in northern Borneo. Biological Conservation. 2018;219:126–137. doi: 10.1016/j.biocon.2018.01.012. [DOI] [Google Scholar]
- 18.Bolotov IN, et al. Spreading of the Chinese pond mussel, Sinanodonta woodiana, across Wallacea: One or more lineages invade tropical islands and Europe. Biochemical Systematics and Ecology. 2016;67:58–64. doi: 10.1016/j.bse.2016.05.018. [DOI] [Google Scholar]
- 19.Vikhrev IV, et al. A tropical biodiversity hotspot under the new threat: Discovery and DNA barcoding of the invasive Chinese pond mussel Sinanodonta woodiana in Myanmar. Tropical Conservation Science. 2017;10:1–11. doi: 10.1177/1940082917738151. [DOI] [Google Scholar]
- 20.Froufe E, et al. Mesozoic mitogenome rearrangements and freshwater mussel (Bivalvia: Unionoidea) macroevolution. Heredity. 2020;124:182–196. doi: 10.1038/s41437-019-0242-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeiffer JM, Breinholt JW, Page LM. Unioverse: A phylogenomic resource for reconstructing the evolution of freshwater mussels (Bivalvia, Unionoida) Molecular Phylogenetics and Evolution. 2019;137:114–126. doi: 10.1016/j.ympev.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 22.Bolotov IN, et al. Ancient river inference explains exceptional Oriental freshwater mussel radiations. Scientific Reports. 2017;7:1–14. doi: 10.1038/s41598-017-02312-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopes-Lima M, et al. Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): Defining modern subfamilies and tribes. Molecular Phylogenetics and Evolution. 2017;106:174–191. doi: 10.1016/j.ympev.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 24.Konopleva ES, Bolotov IN, Vikhrev IV, Gofarov MY, Kondakov AV. An integrative approach underscores the taxonomic status of Lamellidens exolescens, a freshwater mussel from the Oriental tropics (Bivalvia: Unionidae) Systematics and Biodiversity. 2017;15:204–217. doi: 10.1080/14772000.2016.1249530. [DOI] [Google Scholar]
- 25.Jeratthitikul E, et al. Integrative taxonomy reveals phenotypic plasticity in the freshwater mussel Contradens contradens (Bivalvia: Unionidae) in Thailand, with a description of a new species. Systematics and Biodiversity. 2019;17:134–147. doi: 10.1080/14772000.2018.1554607. [DOI] [Google Scholar]
- 26.Pfeiffer JM, Graf DL, Cummings KS, Page LM. Molecular phylogeny and taxonomic revision of two enigmatic freshwater mussel genera (Bivalvia: Unionidae incertae sedis: Harmandia and Unionetta) reveals a diverse clade of Southeast Asian Parreysiinae. Journal of Molluscan Studies. 2018;84:404–416. doi: 10.1093/mollus/eyy028. [DOI] [Google Scholar]
- 27.Jeratthitikul E, Sucharit C, Prasankok P. Molecular phylogeny of the Indochinese freshwater mussel genus Scabies Haas, 1911 (Bivalvia: Unionidae) Tropical Natural History. 2019;19:21–36. [Google Scholar]
- 28.Muanta S, Jeratthitikul E, Panha S, Prasankok P. Phylogeography of the freshwater bivalve genus Ensidens (Unionidae) in Thailand. Journal of Molluscan Studies. 2019;85:224–231. doi: 10.1093/mollus/eyz013. [DOI] [Google Scholar]
- 29.Subba Rao, N. V. Handbook of freshwater molluscs of India (Calcutta, 1989).
- 30.Kongim B, Sutcharit C, Panha S. Cytotaxonomy of unionid freshwater mussels (Unionoida, Unionidae) from northeastern Thailand with description of a new species. ZooKeys. 2015;514:93–110. doi: 10.3897/zookeys.514.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandt RAM. The non-marine aquatic mollusca of Thailand. Archiv für Mollusckenkunde. 1974;105:1–423. [Google Scholar]
- 32.Prashad B. Studies on the anatomy of Indian Mollusca. The soft parts of some Indian Unionidae. Records of the Indian Museum. 1919;16:289–296. doi: 10.5962/bhl.part.25924. [DOI] [Google Scholar]
- 33.Haas F. Superfamilia Unionacea. Das Tierreich. 1969;88:1–663. [Google Scholar]
- 34.Godwin–Austen HH. Description of a new species of Margaritanopsis (Unionidae) from the Southern Shan States, with notes on Solenaia soleniformis. Records of the Indian Museum. 1919;16:203–205. [Google Scholar]
- 35.Annandale, N. Addendum. Further note on the burrows of Solenaia soleniformis. Records of the Indian Museum16, 205–206 (1919).
- 36.Woodruff DS. Neogene marine transgressions, palaeogeography and biogeographic transitions on the Thai–Malay Peninsula. Journal of Biogeography. 2003;30:551–567. doi: 10.1046/j.1365-2699.2003.00846.x. [DOI] [Google Scholar]
- 37.De Bruyn M, Nugroho E, Hossain MM, Wilson JC, Mather PB. Phylogeographic evidence for the existence of an ancient biogeographic barrier: the Isthmus of Kra Seaway. Heredity. 2005;94:370–378. doi: 10.1038/sj.hdy.6800613. [DOI] [PubMed] [Google Scholar]
- 38.Parnell J. The biogeography of the Isthmus of Kra region: a review. Nordic Journal of Botany. 2013;31:001–015. doi: 10.1111/j.1756-1051.2012.00121.x. [DOI] [Google Scholar]
- 39.Woodruff DS. Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today’s patterns and the future of the remaining refugial-phase biodiversity. Biodiversity and Conservation. 2010;19:919–941. doi: 10.1007/s10531-010-9783-3. [DOI] [Google Scholar]
- 40.Dejtaradol A, et al. Indochinese-Sundaic faunal transition and phylogeographical divides north of the Isthmus of Kra in Southeast Asian Bulbuls (Aves: Pycnonotidae) Journal of Biogeography. 2016;43:471–483. doi: 10.1111/jbi.12662. [DOI] [Google Scholar]
- 41.Buddhachat K, Suwannapoom C. Phylogenetic relationships and genetic diversity of the Polypedates leucomystax complex in Thailand. PeerJ. 2018;6:1–13. doi: 10.7717/peerj.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulcahy DG, Lee JL, Miller AH, Zug GR. Troublesome Times: Potential cryptic speciation of the Trimeresurus (Popeia) popeiorum complex (Serpentes: Crotalidae) around the Isthmus of Kra (Myanmar and Thailand) Zootaxa. 2017;4347:301–315. doi: 10.11646/zootaxa. [DOI] [PubMed] [Google Scholar]
- 43.Zug GR, Mulcahy DG, Vindum JV. Resurrection of Bronchocela burmana Blanford, 1878 for the Green Crested Lizard (Squamata, Agamidae) of southern Myanmar. ZooKeys. 2017;657:141–156. doi: 10.3897/zookeys.657.11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siriwut W, Edgecombe GD, Sutcharit C, Panha S. The centipede genus Scolopendra in mainland Southeast Asia: molecular phylogenetics, geometric morphometrics and external morphology as tools for species delimitation. PLoS ONE. 2015;10:1–37. doi: 10.1371/journal.pone.0135355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F, Li S. Paleocene–Eocene and Plio–Pleistocene sea-level changes as “species pumps” in Southeast Asia: Evidence from Althepus spiders. Molecular Phylogenetics and Evolution. 2018;127:545–555. doi: 10.1016/j.ympev.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 46.Veeravechsukij N, et al. Molecular phylogeography and reproductive biology of the freshwater snail Tarebia granifera in Thailand and Timor (Cerithioidea, Thiaridae): morphological disparity versus genetic diversity. Zoosystematics and Evolution. 2018;94:461–493. doi: 10.3897/zse.94.28981. [DOI] [Google Scholar]
- 47.Beck SV, et al. Plio-Pleistocene phylogeography of the Southeast Asian Blue Panchax killifish, Aplocheilus panchax. PloS ONE. 2017;12:1–17. doi: 10.1371/journal.pone.0179557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolotov IN, et al. Discovery of a silicate rock-boring organism and macrobioerosion in fresh water. Nature Communications. 2018;9:1–11. doi: 10.1038/s41467-018-05133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolotov IN, et al. Discovery of Novaculina myanmarensis sp. nov. (Bivalvia: Pharidae: Pharellinae) closes the freshwater razor clams range disjunction in Southeast Asia. Scientific Reports. 2018;8:1–12. doi: 10.1038/s41598-018-34491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bohlen J, Šlechtová V, Udomritthiruj K. Schistura hypsiura, a new species of loach (Cobitoidea: Nemacheilidae) from South-West Myanmar. Raffles Bulletin of Zoology. 2014;62:21–27. [Google Scholar]
- 51.Kullander SO. Taxonomy of chain Danio, an Indo-Myanmar species assemblage, with descriptions of four new species (Teleostei: Cyprinidae) Ichthyological Exploration of Freshwaters. 2015;25:357–380. [Google Scholar]
- 52.Barman AS, Singh M, Pandey PK. DNA barcoding and genetic diversity analyses of fishes of Kaladan River of Indo-Myanmar biodiversity hotspot. Mitochondrial DNA Part A. 2018;29:367–378. doi: 10.1080/24701394.2017.1285290. [DOI] [PubMed] [Google Scholar]
- 53.Kullander SO, Britz R. Description of Danio absconditus, new species, and redescription of Danio feegradei (Teleostei: Cyprinidae), from the Rakhine Yoma hotspot in south-western Myanmar. Zootaxa. 2015;3948:233–247. doi: 10.11646/zootaxa.3948.2.5. [DOI] [PubMed] [Google Scholar]
- 54.Sriwattanarothai N, Steinke D, Ruenwongsa P, Hanner R, Panijpan B. Molecular and morphological evidence supports the species status of the Mahachai fighter Betta sp. Mahachai and reveals new species of Betta from Thailand. Journal of Fish Biology. 2010;77:414–424. doi: 10.1111/j.1095-8649.2010.02715.x.. [DOI] [PubMed] [Google Scholar]
- 55.Fritz U, Gemel R, Kehlmaier C, Vamberger M, Praschag P. Phylogeography of the Asian softshell turtle Amyda cartilaginea (Boddaert, 1770): evidence for a species complex. Vertebrate Zoology. 2014;64:229–243. [Google Scholar]
- 56.Ihlow F, et al. Integrative taxonomy of Southeast Asian snail-eating turtles (Geoemydidae: Malayemys) reveals a new species and mitochondrial introgression. PloS ONE. 2016;11:e0153108. doi: 10.1371/journal.pone.0153108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klaus S, Morley R, Plath M, Zhang Y-P, Li J-T. Biotic interchange between the Indian subcontinent and mainland Asia through time. Nature Communications. 2016;7:1–6. doi: 10.1038/ncomms12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nie J, et al. Rapid incision of the Mekong River in the middle Miocene linked to monsoonal precipitation. Nature Geoscience. 2018;11:944–948. doi: 10.1038/s41561-018-0244-z. [DOI] [Google Scholar]
- 59.Zhang P, et al. Palaeodrainage evolution of the large rivers of East Asia, and Himalayan-Tibet tectonics. Earth-Science Reviews. 2019;192:601–630. doi: 10.1016/j.earscirev.2019.02.003. [DOI] [Google Scholar]
- 60.Wang P, Zheng H, Liu S, Hoke G. Late Cretaceous drainage reorganization of the Middle Yangtze River. Lithosphere. 2018;10:392–405. doi: 10.1130/L695.1. [DOI] [Google Scholar]
- 61.Nguyen L-T, Schmidt HA, Haeseler, von A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller, M., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE). 1–8 (IEEE, 2010).
- 64.Bouckaert R, et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology. 2019;15:1–28. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Froufe E, et al. Who lives where? Molecular and morphometric analyses clarify which Unio species (Unionida, Mollusca) inhabit the southwestern Palearctic. Organisms Diversity & Evolution. 2016;16:597–611. doi: 10.1007/s13127-016-0262-x. [DOI] [Google Scholar]
- 66.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biology. 2006;4:1–12. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rambaut A, et al. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology. 2018;67:901–904. doi: 10.1093/sysbio/syy032.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu Y, Harris AJ, Blair C, He XJ. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Molecular Phylogenetics and Evolution. 2015;87:46–49. doi: 10.1016/j.ympev.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 70.Keogh SM, Simons AM. Molecules and morphology reveal ‘new’ widespread North American freshwater mussel species (Bivalvia: Unionidae) Molecular Phylogenetics and Evolution. 2019;138:182–192. doi: 10.1016/j.ympev.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 71.Pfeiffer JM, et al. Phylogeny of Mesoamerican freshwater mussels and a revised tribe‐level classification of the Ambleminae. Zoologica Scripta. 2019;48:106–117. doi: 10.1111/zsc.12322. [DOI] [Google Scholar]
- 72.Inoue K, Harris JL, Robertson CR, Johnson NA, Randklev CR. A comprehensive approach uncovers hidden diversity in freshwater mussels (Bivalvia: Unionidae) with the description of a novel species. Cladistics. 2020;36:88–113. doi: 10.1111/cla.12386. [DOI] [PubMed] [Google Scholar]
- 73.Smith CH, Johnson NA, Inoue K, Doyle RD, Randklev CR. Integrative taxonomy reveals a new species of freshwater mussel, Potamilus streckersoni sp. nov.(Bivalvia: Unionidae): implications for conservation and management. Systematics and Biodiversity. 2019;17:331–348. doi: 10.1080/14772000.2019.1607615. [DOI] [Google Scholar]
- 74.Kapli P, et al. Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics. 2017;33:1630–1638. doi: 10.1093/bioinformatics/btx025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Köhler F. Rampant taxonomic incongruence in a mitochondrial phylogeny of Semisulcospira freshwater snails from Japan (Cerithioidea: Semisulcospiridae) Journal of Molluscan Studies. 2016;82:268–281. doi: 10.1093/mollus/eyv057. [DOI] [Google Scholar]
- 77.Schniebs K, Gloeer P, Vinarski MV, Hundsdoerfer AK. A barcode pitfall in Palaearctic Stagnicola specimens (Mollusca: Lymnaeidae): Incongruence of mitochondrial genes, a nuclear marker and morphology. North-Western Journal of Zoology. 2016;12:239–354. [Google Scholar]
- 78.Köhler F, Glaubrecht M. Toward a systematic revision of the Southeast Asian freshwater gastropod Brotia H. Adams, 1866 (Cerithioidea: Pachychilidae): an account of species from around the South China Sea. Journal of Molluscan Studies. 2001;67:281–318. doi: 10.1093/mollus/67.3.281. [DOI] [Google Scholar]
- 79.Aksenova OV, et al. Species richness, molecular taxonomy and biogeography of the radicine pond snails (Gastropoda: Lymnaeidae) in the Old World. Scientific Reports. 2018;8:1–17. doi: 10.1038/s41598-018-29451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.von Rintelen T, Stelbrink B, Marwoto RM, Glaubrecht M. A snail perspective on the biogeography of Sulawesi, Indonesia: origin and intra-island dispersal of the viviparous freshwater gastropod Tylomelania. PLoS ONE. 2014;9:1–11. doi: 10.1371/journal.pone.0098917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lehner B, Verdin K, Jarvis A. New global hydrography derived from spaceborne elevation data. Eos. 2008;89:93–942. doi: 10.1029/2008EO100001. [DOI] [Google Scholar]
- 82.Drouët H, Chaper M, Voyage de M. Chaper a Bornéo. Unionidae. Mémoires de la Société zoologique de France. 1892;5:145–154. [Google Scholar]
- 83.Petit de la Saussaye S. Note sur le genre Monocondylea de d’Orbigny, et description d’une espèce nouvelle. Journal de Conchyliologie. 1865;13:15–19. [Google Scholar]
- 84.Lea I. Descriptions of new fresh water and land shells. Proceedings of the American Philosophical Society. 1840;1:284–289. [Google Scholar]
- 85.Johnston RI. Lectotypes for two species of Asiatic Unionidae in the genus Pseudodon. The Nautilus. 1948;62:48–51. [Google Scholar]
- 86.Hanley S. Descriptions of a new species of Monocondylaea. Proceedings of the Zoological Society of London. 1871;2:587–588. [Google Scholar]
- 87.Lea I. Description of five new species of Anodontae, collected by H. Cuming, Esq., in the East Indies. Proceedings of the Zoological Society of London. 1850;18:197–199. [Google Scholar]
- 88.Crosse H, Fischer P. Mollusques fluviatiles, recueillis au Cambodge, par la mission scientifique française de 1873. Journal de Conchyliologie. 1876;24:313–334. [Google Scholar]
- 89.Rochebrune A-T. Documents sur la faune malacologique de la Cochinchine et du Cambodge. Bulletin de la Société philomathique de Paris. 1881;6:35–74. [Google Scholar]
- 90.Preston HB. Descriptions of new species of Macrochlamys and Pseudodon from Siam. Proceedings of the Malacological Society of London. 1909;8:202. [Google Scholar]
- 91.Morelet A. Description d’espèces appartenant à la faune malacologique de l’Indo-Chine. Journal de Conchyliologie. 1866;14:62–64. [Google Scholar]
- 92.Martens EV. On the Mollusca of Siam. Proceedings of the Zoological Society of London. 1860;1860:6–36. [Google Scholar]
- 93.Morlet L. Catalogue des Coquilles recueillies, par M. Pavie, dans le Cambodge et le Royaume de Siam, et description d’espèce nouvelles. Journal de Conchyliologie. 1889;37:121–199. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The type series of the new taxa are deposited in the Russian Museum of Biodiversity Hotspots [RMBH], Federal Center for Integrated Arctic Research, Russian Academy of Sciences, Arkhangelsk, Russia; the North Carolina Museum of Natural Sciences [NCSM], Raleigh, United States of America; and the University of Michigan Museum of Zoology [UMMZ], Ann Arbor, United States of America. The molecular sequences obtained in this study are available in GenBank. Sequence accession numbers and collecting locality for each specimen are presented in Supplementary Tables 1–2. Shell measurements for the type series of the new species are given in Table 1 and Supplementary Table 2.