Abstract
Since the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Wuhan, China, the number of globally confirmed cases according to World Health Organization statistics reached 292 124 in 189 countries by 22 March 2020. The number of deaths reached 12 784, with estimated case-fatality rates ranging from 0.5% to 5.7%. Children population seems to be the least affected by the disease, while the highest rate of death is among the elderly and people with comorbidities. Most infected individuals are asymptomatic or only exhibit mild symptoms. After the incubation period, the most common symptoms are fever, cough and fatigue. Asymptomatic carrier state is of paramount importance because of carriers' ability to spread the infection and to shed the virus into the air and surroundings. Although much is still unknown about SARS-CoV-2, the scientific research is moving at an unprecedented pace towards understanding the nature, effective control, prevention and treatment of SARS-CoV-2. Various reports have suggested an in vivo evolution of the virus, which may explain the rapid spread and changing epidemiology of SARS-CoV-2, but further evidence is needed. Unfortunately, no effective treatment or therapeutic drug is available for the disease; only supportive treatment and classical intervention measures are available for confronting the SARS-CoV-2 pandemic.
Keywords: 2109-nCoV, Asymptomatic, Coronavirus, COVID-19, Epidemic, Europe, Pneumonia, Quarantine, Transmission
Introduction
Emerging viral diseases are frequent public health threats as a result of their potential to develop from small outbreaks into epidemics and pandemics. On 31 December 2019 Chinese health authorities announced dozens of pneumonia infections in Wuhan city (Hubei province) without a recognized aetiology [1]. The first reported patients with pneumonia in Wuhan had a history of visiting or association with a local seafood market where wild animals are sold. The infectious agent was subsequently identified on 7 January 2020 as a novel coronavirus (2019-nCoV) thought to have originated from the Huanan seafood market [2]. Because of its marked similarity in terms of clinical symptoms and biological nature with the causative agent of severe acute respiratory syndrome (SARS), the novel coronavirus was named SARS-CoV-2 by the International Committee on Taxonomy of Viruses. After rapid isolation of the virus, its genome sequence became publically available and was submitted on 12 January 2020 to GenBank (accession no. MN908947.2; https://www.ncbi.nlm.nih.gov/nuccore/MN908947.2). Phylogenetic analyses showed that SARS-CoV-2 is closely related to two SARS coronaviruses of bat origin, bat-SL-CoVZC45 and bat-SL-CoVZXC21, but is distant from human SARS-CoV (79% sequence homology) and Middle East respiratory syndrome coronavirus (50%) [3,4]. Epidemiologic investigations showed that different animals (bats, pangolins, snakes) could have been intermediate hosts that facilitated the spillover of SARS-CoV-2 as a distinct human Betacoronavirus from bats to the human population [[4], [5], [6]].
Since 26 February 2020 reported new cases outside China have surpassed cases being reported from mainland China [7]. Currently the numbers are increasing rapidly, especially in Europe, with overwhelming deaths in Italy. As of 22 March 2020 the total number of cases of infection was 292 142 in 189 countries and territories around the globe, with approximately 12 784 cases of death. Reports have shown that adults and the elderly are the most infected by SARS-CoV-2, with a slight predominance in men; only a low proportion of children have contracted the infection [[8], [9], [10]]. Moreover, the presence of comorbidities such as diabetes or cardiovascular or respiratory disorders greatly affects outcome. Indeed, being elderly or having a delay in diagnosis were found to substantially increase the case-fatality rate [10,11].
In this review, we aim to concisely discuss what is currently known about SARS-CoV-2 and the resulting pandemic.
Virology and genome of SARS-CoV-2
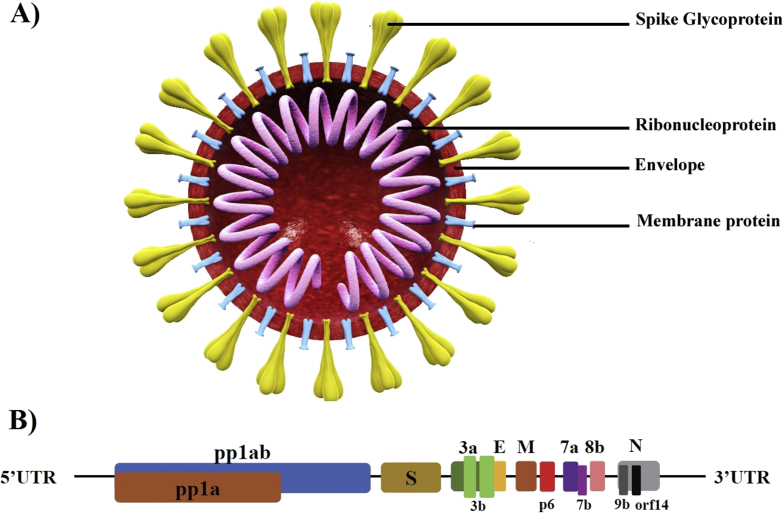
SARS-CoV-2 is an enveloped positive-sense unsegmented single-strand RNA virus that belong to the genus Betacoronavirus (Fig. 1(A)) [12]. The whole-genome sequences of SARS-CoV-2 isolated from patients living in or visiting Wuhan showed a genome 29 844 to 29 891 nt in size, encoding approximately 9860 aa and lacking the haemagglutinin-esterase gene [4,12]. The SARS-CoV-2 genome has great sequence similarity (89–96.3%) with two bat coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21, and 79% to 82% with that of human SARS-CoV [4,12,13]. The genome contains 14 open reading frames (ORFs) encoding 27 proteins (Fig. 1(B)). The longest ORF is located at the 5ʹ terminus encoding for 15 nonstructural proteins collectively involved in virus replication and possibly in immune evasion. The 3ʹ terminus of the genome encodes for structural and accessory proteins [3]. Hypervariable genomic hot spots have been detected in the spike gene and in other ORFs for nonstructural proteins [14].
Fig. 1.
Structure and genome organization of SARS-CoV-2. (A) General structure of SARS-CoV-2 virion. (B) Genome composition of SARS-CoV-2 shows that 14 ORFs exist. First two ORFs at 5ʹ untranslated region are coding for polyprotein (pp1a/ab) required for virus replication, followed by structural proteins for spike (S), membrane (M) and nucleoprotein (N). At 3ʹ terminus, accessory genes (3a, 3b, p6, 7a, 7b, 8b, 9b and orf14) are located with flanking ORFs. Accessory proteins are not required for virus replication or other known functions. Adapted with permission from Wu et al. [3]. ORF, open reading frame; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Interestingly, the unique aspects of SARS-CoV-2 were found in genes of spike glycoprotein, orf8 and orf3b. The spike protein is composed of two subunits, the S1 domain of a single polypeptide containing the receptor-binding domain and the S2 domain, composed of highly conserved polypeptides associated with the envelope. The external subdomain of SARS-CoV-2 spike globular head S1 has only 40% similarity with its counterparts in the bat and human SARS-CoV virion [12]. The outer portion of the external subdomain responsible for direct contact with the human receptor (angiotensin-converting enzyme 2, ACE-2) has the most diversity in amino acids. Such variations are believed to have evolved from homologous recombination between a bat coronavirus and another coronavirus of unknown origin [5]. Orf3b is a putative novel protein that seems to play a pivotal role in pathogenesis of SARS-CoV-2, while Orf8 is an accessory protein shorter than its counterparts found in other Betacoronavirus with unknown function.
Epidemiology and transmission of SARS-CoV-2
During the past two decades China has witnessed the emergence of three respiratory virus outbreaks that turned into epidemics: avian influenza H5N1 in 1997 [15], SARS-CoV in 2003 [16] and the ongoing SARS-CoV-2 (2019–2020). Only the first cases of SARS-CoV-2 had associations with the Huanan seafood market; subsequent sources of infection were infected individuals. Human-to-human transmission is believed to be the major route for worldwide disease spread [17]. The intermediate host in Wuhan was suspected to be snakes and/or pangolins [5], but this remains to be confirmed. Travel and importation played a critical role in the delivery of SARS-CoV-2 to Korea, Japan, the Middle East and Europe [18,19]. During the early weeks of the epidemic, the basic reproductive values (R0) were calculated to be between 2 and 3.5, which is higher than SARS [20,21]. The R0 is defined as the expected number of secondary subjects infected by a single infectious person in a susceptible population.
The World Health Organization (WHO) declared SARS-CoV-2 to be a public health emergency of international concern on 30 January 2020 and as a controllable pandemic on 11 March 2020. According to WHO statistics (as of 22 March 2020) most reported cases were in China (81 498), Italy (53 578), Spain (24 926), Germany (21 463), Iran (20 610), France (14 296) Korea (8897) and the United States (15 219). Populations of about 54.6% of reporting countries are undergoing local transmission of the infection [22]. Shockingly, more than 160 000 confirmed cases were reported within the last 12 days (10–22 March) [7]. On the basis of reports from the Chinese Centre for Disease Control and Prevention (CCDC), human-to-human transmission via respiratory droplets and contact are the major routes by which people acquire the infection. So far no reliable evidence of vertical (intrauterine) transmission has been described in the literature. Transmission of infection to healthcare workers resulted in more than 2000 cases of disease in China and 1423 in Italy (as of 17 March 2020) [23,24].
Early March 2020 saw a rapid increase in the number of cases in Italy, Spain, France, Germany and Iran. Currently all European countries are affected, and thousands of new cases are being reported daily, without known dynamics. Perhaps there is no preexisting immunity in the population; all individuals are considered susceptible. The transmission chain might go unnoticed during the incubation period of infected or asymptomatic individuals, who actively and unwittingly transfer the infection. An as yet unknown new mutant type may be a major factor for the current overwhelming increase in cases despite the preemptively implemented precautions and countermeasures taken by at-risk countries. Indeed, analysis of 103 genomes derived from SARS-CoV-2 showed that genomes fall into one of two types: S (the ancestor) and L [25]. The L type was prevalent during early January 2020 but decreased in later weeks. Furthermore, recent studies support the evolution of the virus by successive mutations and recombination processes [26,27]. Comparative studies of genomes sequenced from cases of disease occurring in European subjects compared to previously sequenced genomes in China are needed to verify the previous findings.
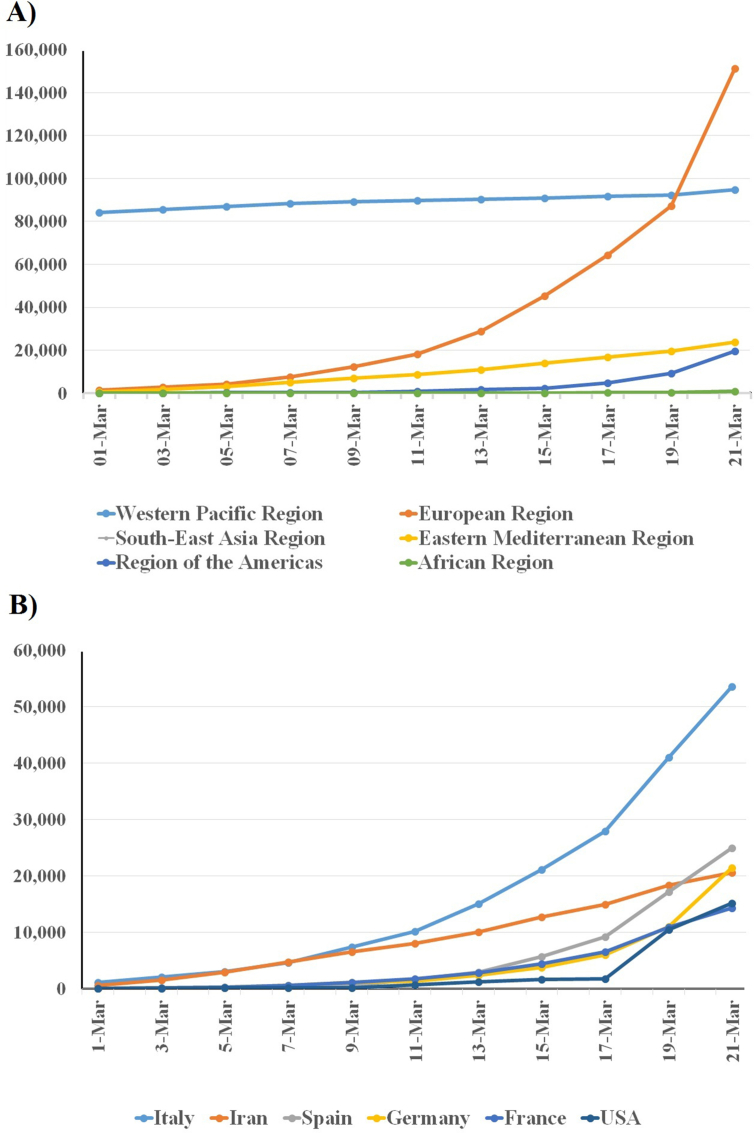
By mid-March 2020 the number of new cases in China was decreasing with time; Italy and Iran are now the new hot spots from which new infections are disseminated to nearby countries. In the Middle East and central Europe the number of cases reported to the WHO are growing with time (Fig. 2). In the European continent the total confirmed cases reached 141 858 with 7319 cases of death until 22 March 2020 (10:00 CET) [28]. According to the epidemiologic data reported by the Italian Higher Institute of Health, the mortality rate in Italy (5.4%) is higher than China's (2.3%); total deaths in Italy exceeded the number documented in China. This may be attributed to the fact that most infected people are elderly, in addition to the incapacity of Italy's healthcare system to cope with the rapid increase of new cases [23].
Fig. 2.
Reported cases of SARS-CoV-2 from 1 to 21 March 2020. (A) Total confirmed cases in all regions of the world. (B) Newly reported cases of SARS-CoV-2 in most affected countries (with >8000 cases). Numbers were obtained from World Health Organization situation reports (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports). SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Turning into a pandemic
Unlike the precedent SARS outbreak, SARS-CoV-2 had unique characteristics and chances to become a pandemic. Firstly, the initial case definition set by the CCDC was narrow and was based solely on signs of pneumonia. Subsequent analysis of the first cases showed that about 81% of patients experience only mild disease, which undoubtedly increased transmission within the community by asymptomatic carriers [10]. The WHO estimated the mortality rate to be between 3% and 4%, with increased mortality in the elderly or in individuals with comorbidities. Because of the lack of data regarding the true number of infected individuals worldwide, it is still too early to calculate exact mortality rates. Secondly, the 22 March 2020 definition of a suspected case set by WHO occurs in a patient with any acute respiratory illness who has been in contact with a person with a case of confirmed or probable SARS-CoV-2 in the last 14 days before onset of symptoms. This definition is based on reports that calculated the incubation period to range from 1 to 14 days [29]. However, recent reports have found that the incubation period might be as long as 24 days [9,30]. This period is much longer than the 14 days used by the WHO to guide quarantine policies, so truly infected persons may be missed during the disease's long incubation period, and they may contribute to the spread of SARS-CoV-2. Additionally, international surveillance estimated that up to two thirds of exported cases from China went undetected [31].
Thirdly, containment measures were challenging in the first phase of the outbreak in Wuhan because of its high population, immense trade and wide travel connections within China and the rest of the world. Furthermore, the industry, commerce and travel of China with the rest of the world has doubled since 2000, which aided the rapid spread of SARS-CoV-2 [32,33]. The Spring Festival and Lunar Year events are also believed to have played a critical role in the spread of SARS-CoV-2 within China, as millions of Wuhan dwellers travelled out of the city before the implementation of lockdown measures just after the outbreak. The same situation applies to Europe, with intense international travel and business, as well as free travel within the eurozone. The exact underlying reasons for the ongoing rapid spread of SARS-CoV-2 in Europe remain unknown. The suspected in vivo evolution of the virus and its transmission by asymptomatic carriers may be major factors [27,34,35].
Fourthly, in SARS-CoV-2 infections, shedding of virions starts earlier than clinical manifestations, and the shedding peak is still unknown [36]. Isolation of individuals after onset of symptoms will be too late, and transmission to others would already have occurred during the incubation period [30,37]. Person-to-person transmission of SARS-CoV-2 from asymptomatic patients was documented in first clusters in Germany and Vietnam during early February 2020 [38,39]. Dependence on detection of high body temperature (fever) for ruling out asymptomatic carriage, as was used during SARS containment, has been deemed inefficient in SARS-CoV-2 [40]. Additionally, the R0 value of SARS-CoV-2 is higher than of SARS (3.28 vs. 2.2) [20,21]. Indeed, the high transmissibility of SARS-CoV-2 is obvious by a global increase of more than 72 000 cases in 3 days according to WHO reports (from 18 to 20 March 2020) [7].
Pathology and clinical features
SARS-CoV-2 exploits the ACE-2 receptor, the same receptor as SARS, in the human lower respiratory tract for entry into lung cells [41,42]. Cellular surface serine protease TMPRSS2 is also used by SARS-CoV-2 for priming of spike protein S to facilitate membrane fusion with host cells [41]. ACE-2 is also prevalent on other cells outside lung [43]. The exact pathophysiology of SARS-CoV-2 and its closely related outbreak-associated coronaviruses remains elusive. Cytokine storm and virus evasion of cell-mediated immunoresponse play important roles in disease pathogenesis and severity [44]. Higher levels of inflammatory chemokines and cytokines (GCSF, IP-10, MCP-1, MIP-1α, TNF-α and other interleukins) result in lung injury that requires urgent admission to intensive care units [8].
Children comprise the population least affected by the disease; however, this observation might be biased by the small number of children sampled in early reports. Moreover, children have less contact with potential sources because they engage in fewer outdoor activities and have parental protection. Some researchers believe that the immune systems of children develop less intense cytokine storms or that their respiratory tracts are healthier because they have not been exposed to pollutants as adults; however, evidence supporting this is still unavailable.
After an incubation period of 5 to 14 days, SARS-CoV-2–infected people commonly manifest features of pneumonia, including fever, dry cough, dyspnoea, myalgia and fatigue. Acute respiratory distress syndrome (ARDS) is a feature in severe cases. Other documented symptoms include productive cough, headache, haemoptysis and diarrhoea [8]. A report from the CCDC estimated that only 5% of patients were critically ill (e.g. shock, respiratory failure requiring mechanical ventilation, multiple organ dysfunction), and 14% of patients had severe pneumonia (e.g. shortness of breath, low oxygen saturation, >50% of lung parenchyma involved at chest imaging within 1–2 days). Meanwhile, most patients (81%) showed no or mild pneumonia [10]. The overall case-fatality rate was 2.3%; however, case-fatality rates of between 5.8% in Wuhan to 0.7% in the rest of China were reported. Fortunately, no cases of death were reported among non–critically ill subjects [45].
Leukocyte count in most cases shows lymphopenia; nonetheless, total white cell count may be lower or higher than normal. d-Dimer, prothrombin time, hepatic transaminases and procalcitonin are high among critically ill patients. Abnormal chest radiographic features were observed in almost all patients [8]. Bilateral patchy shadows were frequently noted on chest X-ray, while ground-glass opacifications are found in patients with ARDS. Typical computed tomographic (CT) findings include multiple ground-glass opacifications with or without reticular pattern, and parenchymal consolidations involving both lungs [46,47]. Other common CT features include microvascular dilatation, thick interlobar septa and air bronchogram [47]. Interestingly, CT changes have also been noted in patients before the onset of clinical symptoms and before virus RNA detection in upper respiratory specimens [48]. Time required for recovery ranges from 2 weeks in mild infections to 3 to 6 weeks in severe disease. Extrapulmonary complications include acute injuries in heart, liver and kidneys [9,49].
Diagnosis
Travel history is no longer valid as a criterion to build a diagnosis because local transmission accounts for most cases of acquisition of infection in locations where no cases have been previously identified. Clinical manifestations accompanied by radiographic evaluation and laboratory diagnosis (detection of virus RNA) are the only possible approach for definitive diagnosis [50]. Generalizations regarding symptoms and laboratory indices still cannot be made because SARS-CoV-2 pathology and its clinical picture are still not completely understood. Because of variations in epidemiology and clinical features of SARS-CoV-2 infections, physicians and specialists are strongly recommended to continually update their management strategies on the basis of WHO interim guidelines for diagnosis and cases definitions (suspected, probable and confirmed).
The most convenient laboratory tests for SARS-CoV-2 diagnosis is real-time reverse transcriptase PCR of nasopharyngeal specimens. A regularly updated source for various protocols that are based on real-time reverse transcriptase PCR assay is available online via the WHO [51]. As in any diagnostic test, false-positive and false-negative results have been reported, but at a very low frequency [30]. According to recommendations of the US Centers for Disease Control and Prevention, accepted clinical specimens are bronchoalveolar lavage fluid, nasopharyngeal swabs (but not throat swabs) and blood [52]. Bronchoalveolar lavage fluid samples were found to be better than other respiratory specimens (positive rate 93%), followed by sputum samples (72%), nasal swab (63%), fibrobronchoscope brush biopsy samples (46%), pharyngeal swab (32%) and faeces (29%) [53]. As a result of the increasing number of healthcare-associated infections, strict adherence to use of personal protective equipment and precautions against airborne pathogens is highly recommended. A concise guide for healthcare staff, researcher and public health workers has been published [54].
Confronting the pandemic
In the absence of specific therapy and anti–SARS-CoV-2 drugs, the urgent implementation of classical public health measures is the only reliable action that can be taken to mitigate the pandemic and to gradually control the spread of infection. A recent small-size clinical trial has reported promising results for SARS-CoV-2 treatment in France. The administration of 600 mg of hydroxychloroquine plus azithromycin daily resulted in clearance of the virus from the nasopharynx within 6 ix days [55]. The mechanism of action is the alkalinization of epithelial phagolysosome containing the virus. These results are the basis of the current therapeutic protocol at IHU Méditerranée Infection, Marseille, France. Further large-size ongoing randomized clinical trials (including hydroxychloroquine, among other drugs) will provide further evidence for treatment options.
China's rigorous and monumental efforts in implementing public health measures have obviously been effective, as the number of new cases is sharply declining. Indeed, China has applied the largest quarantine in history to contain the disease. China's efforts have cut the person-to-person transmission chain and have demonstrated that isolation, quarantine, social distancing and community containment are the only promising measures to fight SARS-CoV-2. As a respiratory virus of high transmissibility, applying social distancing to reduce interactions between people on a mass scale is beneficial, especially when detection of cases is swift [56]. If community transmission continued to expand, social distancing would be insufficient and community containment would be needed. The efficiency of community containment is apparent in China, where normal daily life is gradually being resumed in Wuhan after a marked decline in cases. Nonetheless, such measures are challenging and require partnership between local and state officials in different fields. The media should also tap into their ability to communicate trusted information about containment benefits and to preempt false rumours and panic.
As the world witnesses a marked decline in the number of new cases in China, the will of political and healthcare authorities should be engaged to implement a lockdown of hot spot zones and to perform a comprehensive tracing of contacts associated with patients with confirmed disease. The cost of wide tracking and community containment cannot be outweighed by long-term economic losses and disease burden if transmission continues. Additionally, rapid responses and decision making alongside free (or at least affordable) laboratory testing will facilitate the delineation of hot spot zones. Certain community containment procedures, such as cancellation of public events and closure of institutions, are evident in many countries. These precautions will not be fruitful unless parallel thorough surveillance accompanied by medical observation and legal actions are taken if quarantine is violated. Recently Italy, France, Spain and the Philippines initiated implementation of lockdown measures, with law enforcement in some cases, to control the overwhelming increase in cases. In third world countries, lockdown measures are extremely challenging as a result of the high proportion of low-wage jobs where workers seek employment opportunities on a daily basis. Finally, efficient deactivation of different coronaviruses (SARS-CoV-2 was not evaluated) has been reported with disinfectants such as 62% to 71% ethanol, 0.5% hydrogen peroxide or 0.1% sodium hypochlorite within 1 minute; this efficacy is highly likely to apply to SARS-CoV-2 [57].
Issues to be investigated
Despite intense research and clinical investigations, certain issues of the SARS-CoV-2 pandemic await exploration. Firstly, the origin of the virus and the nature of its spillover from its natural reservoir to humans are still unknown. It is known that bats and pangolins are hunted for food in China, but so far these animals have not been confirmed to be the origin of the disease [6]. Moreover, the mode of virus replication and the human immunoresponse to SARS-CoV-2 are still largely unknown. Secondly, the virus was detected in faecal samples from infected patients, but the disease's potential for faecal–oral transmissibility remains unaddressed by researchers [53,58,59]. Thirdly, phylogenetic analyses have shown genomic variations in SARS-CoV-2 genomes sequenced in different countries through different periods. Whether in vivo evolution occurs in humans or in another intermediate animal host is still unproven. Although the evidence suggests the existence and identity of an intermediate host, this has not been experimentally proven. Fourthly, the period during which the SARS-CoV-2 virion retains its infectivity when shed and deposited on surfaces and in the environment is also unclear. Indeed, the efficacy of handwashing in deactivating the SARS-CoV-2 virion has not been tested. Fifthly, the reason SARS-CoV-2 infections predominate in men is unknown. A smoking habit in Asian men was expected to be the underlying cause, but no solid evidence is yet available [60]. Sixthly, the initial administration of corticosteroids during the early stage of the SARS epidemic was justified because corticosteroids mitigated the inflammatory response, but it was subsequently halted as a result of the delay in virus clearance [61]. Lately a small-size (n = 201) observational study reported that methylprednisolone treatment was associated with a decrease in the risk of death in patients with ARDS [62]. Whether corticosteroids will have the same effect in SARS-CoV-2 infection is still unclear.
Conclusion
The number of global epicentres of SARS-CoV-2 is expected to multiply, especially in poor nations. Genomic, virologic and medical analyses are progressing at an unprecedented speed. Such analyses are expected to contribute to understanding of the disease's epidemiology, pathogenesis, treatment and prevention. The definition of a suspected case is expected to be revised at any moment, so medical staff and epidemiologists should keep themselves updated for better control of the pandemic. Moreover, they are also strongly advised to use protective measures because they comprise an at-risk population as a result of nosocomial exposure. The disease's clinical manifestations vary widely; patients with mild or asymptomatic disease may not seek medical care, so their disease may remain undetected, thereby exacerbating transmission. The exact routes of SARS-CoV-2 pathogenesis and transmission, as well as the dynamics of the pandemic, are under intense investigation. Right now, classical public health measures and control interventions are the only way to fight the SARS-CoV-2 pandemic.
Conflict of interest
None declared.
References
- 1.World Health Organization . World Health Organization; Geneva: 2020. Novel coronavirus (2019-nCoV) situation report—1. [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cyranoski D. Mystery deepens over animal source of coronavirus. Nature. 2020;579:18–19. doi: 10.1038/d41586-020-00548-w. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization Situation reports. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Available at:
- 8.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 11.Yang S., Cao P., Du P., Wu Z., Zhuang Z., Yang L. Early estimation of the case fatality rate of COVID-19 in mainland China: a data-driven analysis. Ann Transl Med. 2020;8:128. doi: 10.21037/atm.2020.02.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen F., Yu H., Guo J., Li Y., Luo K., Huang S. Identification of the hyper-variable genomic hotspot for the novel coronavirus SARS-CoV-2. J Infect. 2020 doi: 10.1016/j.jinf.2020.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander D.J., Brown I.H. History of highly pathogenic avian influenza. Rev Sci Tech. 2009;28:19–38. doi: 10.20506/rst.28.1.1856. [DOI] [PubMed] [Google Scholar]
- 16.Chan-Yeung M., Seto W.H., Sung J.J.Y. Severe acute respiratory syndrome: patients were epidemiologically linked. BMJ. 2003;326:1393. doi: 10.1136/bmj.326.7403.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giovanetti M., Benvenuto D., Angeletti S., Ciccozzi M. The first two cases of 2019-nCoV in Italy: where they come from? J Med Virol. 2020;92:518–521. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernard Stoecklin S., Rolland P., Silue Y., Mailles A., Campese C., Simondon A. First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.6.2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipsitch M., Cohen T., Cooper B., Robins J.M., Ma S., James L. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organization . World Health Organization; Geneva: 2020. Coronavirus disease 2019 (COVID-19) situation report—51. [Google Scholar]
- 23.L’epidemiologia per la sanità pubblica; Istituto Superiore di Sanità. Sorveglianza integrata COVID-19: i principali dati nazionali. https://www.epicentro.iss.it/coronavirus/sars-cov-2-sorveglianza-dati Available at:
- 24.Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. [Google Scholar]
- 25.Tang X., Wu C., Li X., Song Y., Yao X., Wu X. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi H. 2019 Novel coronavirus is undergoing active recombination. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L. Genomic diversity of SARS-CoV-2 in coronavirus disease 2019 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.European Centre for Disease Prevention and Control Situation update for the EU/EEA and the UK. https://www.ecdc.europa.eu/en/cases-2019-ncov-eueea as of 17 March 2020 08:00 2020. Available at:
- 29.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhatia S., Imai N., Cuomo-Dannenburg G., Baguelin M., Boonyasiri A., Cori A. 2020. Report 6: relative sensitivity of international surveillance.https://www.researchgate.net/publication/339677490_Report_6_Relative_sensitivity_of_international_surveillance [preprint] [Google Scholar]
- 32.Wilson M.E., Chen L.H. Travellers give wings to novel coronavirus (2019-nCoV) J Travel Med. 2020 doi: 10.1093/jtm/taaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogoch, Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Potential for global spread of a novel coronavirus from China. J Travel Med. 2020 doi: 10.1093/jtm/taaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L., Shen F., Chen F., Lin Z. Origin and evolution of the 2019 novel coronavirus. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angeletti S., Benvenuto D., Bianchi M., Giovanetti M., Pascarella S., Ciccozzi M. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020 doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilder-Smith A., Chiew C.J., Lee V.J. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu P., Zhu J., Zhang Z., Han Y. A familial cluster of infection associated with the 2019 novel coronavirus indicating possible person-to-person transmission during the incubation period. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382:872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020 doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quilty B.J., Clifford S., Flasche S., Eggo R.M. Effectiveness of airport screening at detecting travellers infected with novel coronavirus (2019-nCoV) Eurosurveillance. 2020;25:2000080. doi: 10.2807/1560-7917.ES.2020.25.5.2000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.World Health Organization . World Health Organization; Geneva: 2020. Report of the WHO–China Joint Mission on Coronavirus Disease 2019 (COVID-19) [Google Scholar]
- 46.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou S., Wang Y., Zhu T., Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. AJR Am J Roentgenol. 2020 doi: 10.2214/AJR.20.22975. [DOI] [PubMed] [Google Scholar]
- 48.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zu Z.Y., Di Jiang M., Xu P.P., Chen W., Ni Q.Q., Lu G.M. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020 doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.World Health Organization . 2020. Coronavirus disease (COVID-19) technical guidance: laboratory testing for 2019-nCoV in humans.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance Available at: [Google Scholar]
- 52.US Centers for Disease Control and Prevention . 2020. Interim guidelines for collecting, handling, and testing clinical specimens from persons for coronavirus disease 2019 (COVID-19)https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html Available at: [Google Scholar]
- 53.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin Y.H., Cai L., Cheng Z.S., Cheng H., Deng T., Fan Y.P. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil Med Res. 2020;7:4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Wilder-Smith A., Freedman D.O. Isolation, quarantine, social distancing and community containment: pivotal role for old-style public health measures in the novel coronavirus (2019-nCoV) outbreak. J Travel Med. 2020 doi: 10.1093/jtm/taaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J Med Virol. 2020 doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burki T. Outbreak of coronavirus disease, 2019. Lancet Infect Dis. 2020;20:292–293. doi: 10.1016/S1473-3099(20)30076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]