Glaesserella (Haemophilus) parasuis is a commensal bacterium of the upper respiratory tract in pigs and also the causative agent of Glässer’s disease, which causes significant morbidity and mortality in pigs worldwide. Isolates are characterized into 15 serovars by their capsular polysaccharide, which has shown a correlation with isolate pathogenicity. To investigate the role the capsule plays in G. parasuis virulence and host interaction, a capsule mutant of the serovar 5 strain HS069 was generated (HS069Δcap) through allelic exchange following natural transformation.
KEYWORDS: Glaesserella parasuis, Glässer’s disease, acapsular mutant, capsular polysaccharide
ABSTRACT
Glaesserella (Haemophilus) parasuis is a commensal bacterium of the upper respiratory tract in pigs and also the causative agent of Glässer’s disease, which causes significant morbidity and mortality in pigs worldwide. Isolates are characterized into 15 serovars by their capsular polysaccharide, which has shown a correlation with isolate pathogenicity. To investigate the role the capsule plays in G. parasuis virulence and host interaction, a capsule mutant of the serovar 5 strain HS069 was generated (HS069Δcap) through allelic exchange following natural transformation. HS069Δcap was unable to cause signs of systemic disease during a pig challenge study and had increased sensitivity to complement killing and phagocytosis by alveolar macrophages. Compared with the parent strain, HS069Δcap produced more robust biofilm and adhered equivalently to 3D4/31 cells; however, it was unable to persistently colonize the nasal cavity of inoculated pigs, with all pigs clearing HS069Δcap by 5 days postchallenge. Our results indicate the importance of the capsular polysaccharide to G. parasuis virulence as well as nasal colonization in pigs.
INTRODUCTION
Glaesserella (Haemophilus) parasuis is the etiologic agent of Glässer’s disease in pigs, which presents as a fibrinous polyserositis, arthritis, and meningitis (1, 2). In addition, it can be a contributor to swine respiratory disease and is found as a commensal bacterium in the nasal cavity of healthy swine (1). G. parasuis isolates are classified into 15 serovars, based on gene content and diversity at their capsular polysaccharide loci and via the Kielstein-Rapp-Gabrielson typing scheme (3, 4). While it appears that some serovars are more pathogenic and widespread, serotyping has shown an incomplete correlation with isolate virulence (3, 5, 6).
The importance of the capsular polysaccharide as a virulence factor for encapsulated bacteria has been investigated (7–10), which indicates the capsule-deficient counterparts to encapsulated bacteria are much less likely to cause invasive disease and are often reduced to avirulence. The reduction in virulence seen in capsular mutants has been attributed to the role that the capsule plays in adherence to host tissues and evasion of the host immune response, specifically inhibition of complement-mediated killing and phagocytosis (11). Additionally, there is evidence indicating that the capsular polysaccharide is essential for some encapsulated bacteria to colonize the mucous membranes of hosts (12).
The functions of the capsule have been partially evaluated in G. parasuis SH0165, a virulent serovar 5 strain (10). The capsular mutant (SH0165ΔcapD) was significantly less virulent than the wild-type bacteria and was unable to cause invasive disease in challenged pigs (10). SH0165Δcap was also highly susceptible to complement-mediated killing compared with the wild-type bacteria (10). This presented evidence for the importance of capsule in causing invasive disease; however, the characteristics of the capsular mutant were not fully elucidated, and the capacity of SH0165Δcap to colonize the swine nasal cavity was not evaluated. In this study, a capsule mutant of G. parasuis HS069 (HS069Δcap), a virulent serovar 5 strain, was used to examine the in vitro and in vivo characteristics of the capsule. We evaluated HS069Δcap for susceptibility to complement killing, biofilm formation, attachment to porcine macrophage 3D4/31 cells, and phagocytosis by porcine alveolar macrophages. In addition, swine were challenged with HS069 and HS069Δcap to evaluate virulence, capacity for nasal colonization, and stimulation of host immunity.
RESULTS
Development and confirmation of the HS069 capsule mutant.
The capsule locus of HS069 was removed by deleting the sequence from neuA_3 to etk (alternatively wzc or wzs) (Fig. 1A), which removed the biosynthesis and glycosyltransferase proteins contained within the serovar 5 capsule locus. The deleted sequence was confirmed with whole-genome sequencing using the PacBio sequencing platform. When the wild-type and HS069Δcap genomes were compared, no HS069Δcap reads mapped to the region of the capsule locus (Fig. 1B).
FIG 1.
Capsule locus deletion and verification. The entirety of the capsule locus was deleted from neuA_3 to etk as indicated (A). The loss of the capsule locus was verified using the Artemis compare tool (B). No sequence reads mapped to the region of the capsule locus.
Transmission electron microscopy (TEM) was also performed to phenotypically confirm the deletion of the capsule locus. In Fig. 2, the surface of the HS069 wild-type cells is irregular and thickened compared with that of HS069Δcap. This confirmed the absence of the capsular polysaccharide of the cell surface of HS069Δcap.
FIG 2.
Transmission electron microscopy visualization of capsule. The capsule layer of G. parasuis HS069 wild type (A) and HS069Δcap (B) were visualized using transmission electron microscopy to verify HS069Δcap was deficient in capsular polysaccharide production. The surface of HS069Δcap lacked a thickened, irregular surface associated with capsule production.
Growth characteristics and cellular morphology.
A comparison of the growth kinetics of G. parasuis HS069 and HS069Δcap grown in brain heart infusion (BHI+) broth indicated there was no alteration in cellular proliferation associated with the deletion of the capsule locus (see Fig. S1 in the supplemental material).
Biofilm formation.
Evaluation of static biofilm production indicated a similar capacity for G. parasuis isolates to produce biofilm at all starting cell densities tested (OD600 = 0.05, 0.125, and 0.25) (data not shown). Statistical evaluation of static biofilm production by HS069 and HS069Δcap grown from an initial cell density of OD600 = 0.05 indicated a significant enhancement in production associated with the loss of capsular polysaccharide production (P = 0.0193) (Fig. 3).
FIG 3.
Biofilm production by HS069 wild type and HS069Δcap. The capacity of G. parasuis HS069 and HS069Δcap to produce biofilm under static growth conditions was quantified using microtiter assays. Results here represent data from replicates with a starting OD600 of 0.05. The average absorbance at 538 nm is shown (column) with standard deviations indicated (error bars). Statistical significance is indicated by the asterisk (*) at a P value of <0.05.
Complement-mediated killing.
Sensitivity to complement-mediated killing (serum sensitivity) was evaluated using nontreated and heat-inactivated guinea pig serum (GPS). A significant increase in sensitivity to complement killing was noted for HS069Δcap compared with wild-type HS069 (P = 0.0207) (Fig. 4).
FIG 4.
Evaluation of sensitivity to complement killing. G. parasuis HS069 and HS069Δcap were screened for resistance to complement-mediated killing. Statistical analysis of the log10 reduction in CFU/ml was analyzed, and statistical significance is indicated by the asterisk (*) at a P value of <0.05.
Adherence capacity to the porcine alveolar macrophage cell line.
Confocal microscopy was used to evaluate the adherence capacity of HS069 and HS069Δcap to porcine alveolar macrophages (3D4/31 cells). A distinct difference in the pattern of attachment was visualized between HS069 and HS069Δcap (Fig. 5), with HS069Δcap producing aggregates of bacteria (Fig. 6B) that were not noted with the HS069 wild type (Fig. 6A). Because of the aggregation of HS069Δcap, adherence was evaluated both as a bacterial count per 3D4/31 cell and fluorescent intensity per 3D4/31 cell. There was no statistically significant difference in adherence capacity when HS069 and HS069Δcap were compared for bacterial counts (P = 0.0594) or compared for fluorescent intensity (P = 0.4296).
FIG 5.
Adherence capacity of HS069 (A) and HS069Δcap (B) to 3D4/31 cells. The capacity for HS069 and HS069Δcap to adhere to porcine alveolar macrophages was evaluated using the 3D4/31 cell line. The interaction between G. parasuis and 3D4/31 cells was evaluated by confocal microscopy in chamber slides. G. parasuis strains were stained using a monoclonal antibody to the outer membrane protein P5. Bacterial aggregates were noted when evaluating HS069Δcap (B) but were not produced by the wild-type HS069 isolate.
FIG 6.
Adherence capacity to porcine alveolar macrophages (3D4/31 cell line). The capacity of G. parasuis HS069 and HS069Δcap to adhere to porcine alveolar macrophages was evaluated using the 3D4/31 cell line. Due to the clusters of bacterial cells noted using confocal microscopy, adherence was evaluated both as bacterial cells detected per 3D4/31 cell (A) and fluorescent intensity per 3D4/31 cell (B). No statistical differences were noted in bacteria per cell or fluorescent intensity per cell.
Phagocytosis by primary porcine alveolar macrophages.
Changes in susceptibility to phagocytosis were assessed by incubation with isolated primary porcine alveolar macrophages. After 1 hour of incubation, there was no difference in phagocytosis between HS069 and HS069Δcap (P = 0.93); however, after 2 hours of incubation, significantly more HS069Δcap were phagocytosed than the HS069 wild type (P < 0.01) (Fig. 7).
FIG 7.
Evaluation of the susceptibility to phagocytosis. G. parasuis HS069 and HS069Δcap were screened for susceptibility to phagocytosis using porcine alveolar macrophages. Log fold reduction in G. parasuis [log10(CFU/ml)] is represented for both time points (1-hour and 2-hour incubation). The reduction of G. parasuis HS069 and HS069Δcap was not statistically different after 1 hour of incubation (P = 0.93); however, after 2 hours of incubation with PAMs, significantly more G. parasuis HS069Δcap was phagocytosed than HS069 wild type (P < 0.01).
Virulence assessment.
To assess the virulence of HS069Δcap compared with the parent strain, a total of 11 Caesarian-derived, colostrum-deprived (CDCD) pigs were challenged with HS069Δcap and 4 with wild-type HS069. The parent strain resulted in 100% mortality by day 2 postchallenge, while animals challenged with HS069Δcap showed no clinical signs of G. parasuis infection and survived until the end of the study (20 days postchallenge).
Colonization and immune stimulation.
A second study was conducted to evaluate nasal colonization of HS069Δcap in CDCD pigs. The presence of G. parasuis in nasal wash samples was assessed by PCR. Species-specific primers detected G. parasuis on days 1 (1 pig) and 3 (2 pigs) postchallenge; however, all pigs were negative by PCR by day 5 postchallenge.
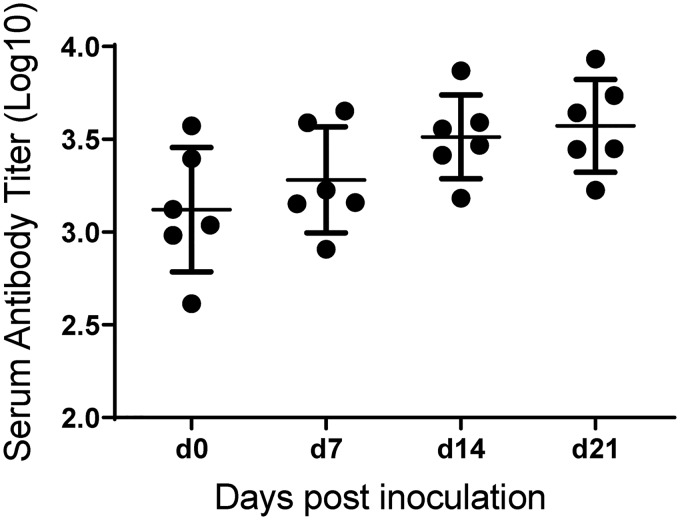
The serum antibody titer for animals challenged with HS069Δcap was determined at days 0, 7, 14, and 21 postchallenge (Fig. 8). A mild increase in serum antibody to HS069Δcap sonicate was seen over the study period, with the average log10 titer rising from 3.121 ± 0.336 to 3.572 ± 0.250 (mean ± standard deviation [SD]). However, upon intranasal challenge of HS069Δcap-inoculated animals with wild-type G. parasuis HS069, all animals succumbed to disease by 3 days postchallenge.
FIG 8.
Serum antibody titers for pigs inoculated with HS069Δcap. The serum antibody titer was detected using an ELISA to whole-cell sonicate. The data presented in this graph represent the animals in the second study investigating colonization with HS069Δcap.
DISCUSSION
Capsular polysaccharide is an important factor for the survival and virulence of many bacteria. It is known to function in adhesion and immune evasion through resistance to complement killing and phagocytosis (11). In this study, we sought to better evaluate the function of capsule for G. parasuis, the causative agent of Glässer’s disease in pigs. Capsule has been correlated with virulence, with some serovars associated with disease and others with nasal colonization (3, 5, 6). The importance of capsule for G. parasuis was previously investigated in the serovar 5 strain SH0165 (10). Wang and colleagues found SH0165ΔcapD to be less serum resistant and nonvirulent in a pig challenge than the highly virulent parent strain (10). To confirm and expand these findings and better understand how capsule contributes to G. parasuis disease, we generated a capsular mutant of the virulent serovar 5 strain HS069, followed by evaluation of sensitivity to complement killing, resistance to phagocytosis, and macrophage adhesion in vitro as well as virulence, colonization, and immune stimulation in vivo.
Here, the virulence of HS069Δcap was assessed with a combination of in vitro assays and an in vivo challenge. We found that, similar to previous reports, HS069Δcap was markedly less virulent in a pig challenge than the encapsulated parent strain. This confirmed findings by Wang and colleagues with SH0165Δcap (10). Reduced virulence of capsule-deficient isolates has been attributed, in part, to the marked increase in serum sensitivity in nonencapsulated G. parasuis strains, which was seen with both HS069Δcap (Fig. 4) and SH0165Δcap (10). Resistance to complement killing enables bacteria that penetrate mucosal defenses to better survive and disseminate to systemic sites, such as the central nervous system, joints, and serosal surfaces, as seen in G. parasuis infection.
Virulence has also been correlated with susceptibility to porcine alveolar macrophages (13), which serve as the first line of defense during pulmonary infection. To investigate the interaction between HS069Δcap and porcine alveolar macrophages, we examined the adherence capacity of HS069Δcap and the parent strain to 3D4/31 cells (Fig. 6). In this investigation, HS069 and HS069Δcap adhered equivalently, indicating the capsule is not playing a role in the interaction between G. parasuis HS069 and 3D4/31 cells. This analysis was made more complicated by the aggregation of HS069Δcap (Fig. 5); however, through analysis of fluorescent intensity, we were able to account for aggregated bacteria (Fig. 6B). We also evaluated HS069 and HS069Δcap for susceptibility to phagocytosis by primary PAMs, which revealed significantly more HS069Δcap phagocytosed over 2 hours than HS069 wild type. This indicates the importance of the G. parasuis capsule in protection against macrophage phagocytosis, which has not been shown previously in G. parasuis.
Additionally, we investigated the persistence of G. parasuis HS069Δcap in the nasal passageways, which indicated the importance of capsular polysaccharide not only in systemic infection but also in colonization. Although biofilm production was enhanced for HS069Δcap (Fig. 3), it did not contribute to persistent nasal colonization, and HS069Δcap was cleared from all pigs in the second study by 5 days postinoculation. This contrasts with colonization of the parent strain, which can persist in the nasal tract of vaccinated pigs (4/5 animals) through 11 days postchallenge (data not shown). Previous results assessing the role of capsule in colonization and adherence of other bacterial species have conflicted, with some studies reporting deficient colonization seen with capsular mutants and others indicating an increased capacity for adhesion to cell lines in vitro (12, 14–17). It has been hypothesized that the absence of capsule may increase the exposure of surface adhesins that contribute to adherence in vitro; however, most studies evaluated only in vitro adherence and have not investigated the effect of capsule in colonization in vivo. Our findings in this study indicate capsule may play a significant role in persistent colonization with G. parasuis in vivo, similar to that seen previously with Streptococcus pneumoniae (12).
The lack of virulence seen with G. parasuis capsular mutants could potentially make them a good candidate for an attenuated live vaccine; however, our investigation showed only a mild increase in G. parasuis serum antibody titer over the 21-day study period (Fig. 8) with an average log10 titer at 21 days postinoculation of 3.572 ± 0.250 (mean ± SD). The titers seen in animals inoculated with HS069Δcap are low compared with other studies in which animals were vaccinated with a single-dose HS069 bacterin (average log10 titer 21 days postvaccination, 4.221 ± 0.377) or after boost vaccination (average log10 day 42 titer, 4.752 ± 0.190), which has shown protection against homologous challenge (data not shown). The low titers associated with HS069Δcap are likely due to its rapid clearance from the nasal cavity, which would limit interaction with immune cells and development of an adaptive immune response. Furthermore, when pigs inoculated with HS069Δcap were challenged with the parent strain, we saw no protection from G. parasuis disease, indicating the antibody response generated was not protective.
Notably, this evaluation of the importance of capsule in G. parasuis colonization and infection involved the generation and use of a mutant deficient in all genes of the capsule locus. Because of this, we cannot eliminate the possibility that there are alternative functions for genes within the capsule locus that may be contributing to the phenotypes we observed in this study. Additionally, because of the size of the capsule locus, a mutant complemented with the deleted genes was not generated and comparisons can only be made between the wild type and the mutant.
This investigation of the G. parasuis capsule mutant HS069Δcap confirms the importance of capsule to a fully virulent phenotype in vivo that has been seen previously with SH0615ΔcapD. The capsular polysaccharide plays an important role in resistance to complement killing and phagocytosis, which may be a key factor in the dissemination of G. parasuis during infection to systemic sites. In this study, we also found capsule to be an essential factor in G. parasuis HS069 for persistent colonization of the swine nasal cavity. However, because of the rapid clearance of HS069Δcap from the nasal cavity, generation of antibody was minimal and no protection was provided against challenge with the parent strain making HS069Δcap a poor modified live vaccine candidate.
MATERIALS AND METHODS
Bacterial isolates.
The virulent serovar 5 G. parasuis strain HS069 was isolated from the lung of a pig with clinical signs of respiratory disease (18). Production and verification of HS069Δcap are described within the following mutant generation and results sections. G. parasuis strains were grown on brain heart infusion (BHI) plates or BHI broth (Becton, Dickinson and Company, Franklin Lakes, NJ) supplemented with 0.1 mg/ml NAD (NAD+) and 10% horse serum (referred to as BHI+). G. parasuis strains were also grown on chocolate agar, made with Columbia blood agar base (Thermo Fisher Scientific Inc., Waltham, MA) supplemented with 7% defibrinated horse blood (TCS Bioscience Ltd., Botolph Claydon, England), and lysed at 80°C for 10 minutes and 25 μg/ml NAD+. Media were supplemented with chloramphenicol (1 μg/ml) for selection purposes. All strains were grown at 37°C with 5% CO2.
Escherichia coli strain TOP10 (Invitrogen, Carlsbad, CA) was used as the cloning host. E. coli was grown in Luria-Bertani (LB) broth or agar (Oxoid). Media were supplemented with ampicillin (100 μg/ml) or chloramphenicol (50 μg/ml) where required for selection.
Mutant construction.
DNA manipulation. Genomic DNA extractions were performed using the DNeasy kit (Qiagen, Hilden, Germany), plasmid DNA extractions were performed using a plasmid miniprep kit (Qiagen), and PCRs were performed according to the manufacturers’ protocols with Phusion high-fidelity DNA polymerase mix (New England BioLabs, Ipswich, MA) for cloning and GoTaq green PCR mix (Promega, Madison, WI) for verification. PCR fragments were purified with the QIAquick PCR purification kit (Qiagen). Restriction enzymes were obtained from New England BioLabs. Plasmid DNA was extracted from the agarose gel using a gel extraction kit (Qiagen). DNA concentrations were measured using a NanoDrop instrument (Bio-Rad Laboratories, Hercules, CA).
Construction of plasmid pGEMT-neuAup-Cm-wzsdn. Plasmid pGEMT-neuAup-cat-wzsdn was constructed for deletion of the whole capsule gene cluster using a three-step cloning strategy. First, genomic DNA of G. parasuis isolate HS069 was used as a template to amplify the upstream and downstream regions flanking the 14-kb capsule locus (19, 20). The 647-bp upstream region of the neuA gene (neuAup) was amplified using primers P1 and P2, and the 731-bp downstream region of the wzs gene (wzsdn) was amplified using primers P3 and P4 (Table 1). In parallel, the chloramphenicol resistance cassette (Cm), containing the 9-bp DNA uptake signal sequence (USS) of 5′-ACCGCTTGT (21) was amplified from 50-ng linearized plasmid pUSScat (22) as the template with primers P5 and P6 (Table 1). Second, PCR products were digested and ligated into the pGEM-T vector. The PCR product of the upstream region of the neuA gene was digested with SacI/BamHI and the downstream region of the wzs gene was digested with SalI/BamHI. The pGEM-T vector was digested with SacI/SalI. Restricted products were gel purified and ligated. The constructed plasmid was transformed into E. coli TOP10, and transformants were confirmed using PCR. After purification, the plasmid pGEMT-neuAup-wzsdn was verified by Sanger sequencing. Finally, the purified plasmid pGEMT-neuAup-wzsdn and PCR product of the Cm were subject to BamHI restriction. The gel-purified fragments were mixed, ligated, and transformed into E. coli TOP10. The resulting plasmid pGEMT-neuAup-cat-wzsdn was extracted and confirmed by Sanger sequencing.
TABLE 1.
Primers utilized in the construction of HS069Δcap
| Primer | Sequence (5′–3′)a | Nucleotide positions in the SH0165 genomeb |
|---|---|---|
| P1 (neuAupFor1) | AAGACTGAGCTCTCGTTTTCCAGACAGCAATG | 49398–49417 |
| P2 (neuAupRev1) | AAGACTGGATCCCTCCTTTACATGCCCCCATC | 50044–50025 |
| P3 (wzsDnFor1) | AAGACTGGATCCTTGATGTAAGCGGTGGGATT | 64033–64052 |
| P4 (wzsDnRev1) | AAGCGAGTCGACAGTTGCGGCATAATCCAAAT | 64763–64744 |
| P5 (catFor) | GCGATGGATCCTGTGGAATTGTGAGCGGATA | n/ac |
| P6 (catRev) | GCGATGGATCCACAAGCGGTTTCAACTAACGG | n/a |
Restriction sites are underlined. GGATCC, BamHI; GTCGAC, SalI; GAGCTC, SacI.
From reference 20.
n/a, not available.
Construction of the whole capsule locus deletion mutant Δcap::Cm mutant of G. parasuis HS069. The plasmid pGEMT-neuAup-cat-wzsdn was linearized with SacI and used to transform G. parasuis HS069 using a natural transformation method, as described previously with some modifications (23). Briefly, HS069 was grown on chocolate agar overnight and suspended in BHI broth to achieve an optical density at 660 nm (OD660) of 2. Then, a 20-μl aliquot of a 1/10 dilution of the suspension was spread in a 10-mm area on prewarmed chocolate agar and 20 μl of 8 mM cAMP and 10 μl of donor DNA in Tris-EDTA (TE) buffer were added and mixed with the bacterial cells. The mixture was incubated at 37°C overnight. Bacterial cells were scraped up, suspended in 300 μl BHI broth, and plated onto chocolate agar with 1 μg/ml chloramphenicol. After incubation at 37°C for 2 days, suspected transformants were verified using PCR. For negative control, 10 μl of TE buffer without donor DNA was added to a bacterial spot. The deletion was also confirmed by whole-genome sequencing using the Illumina HiSeq 250 platform and PacBio RSII resequencing protocol of the single-molecule real-time (SMRT) analysis software v.2.3.0. Assembly of the PacBio-generated HS069 wild-type sequence was done using Hierarchical Genome Assembly Process (HGAP) (24), circularized using Circlator v.1.1.3 (25), and polished using Quiver v.1. The Illumina reads were subsequently mapped onto the PacBio assembly to correct small indels. A comparison of HS069Δcap against the wild type was made by mapping the Illumina and PacBio reads of HS069Δcap against the finished HS069 wild-type assembly.
TEM for capsule visualization.
Capsule was visualized via transmission electron microscopy (TEM) using previously described methods (26–28). Briefly, G. parasuis grown on BHI+ plates overnight was suspended in 0.1 M cacodylate buffer with 2.5% glutaraldehyde and 0.1% ruthenium red and incubated for 2 h at room temperature. Bacteria were pelleted and resuspended in 0.1 M cacodylate buffer with 2.5% glutaraldehyde and 1.0 mg/ml of polycationic ferritin and incubated for 30 min at room temperature. Bacteria were washed three times in 0.1 M cacodylate buffer. After being staining with ruthenium red and ferritin, samples were postfixed with 2% osmium tetroxide and rinsed three times in 0.1 M cacodylate buffer. The samples were processed through graded ethanols, propylene oxide, and embedded in Eponate 12 (Ted Pella Inc., Redding, CA). Following a 48-hour polymerization step, thin sections were taken and stained with 4% uranyl acetate and Reynolds’ lead stain. Sections were examined with a FEI Tecnai G2 BioTWIN electron microscope (FEI Co., Hillsboro, OR).
Growth kinetics.
Growth kinetics of G. parasuis HS069 and HS069Δcap were evaluated using a GeneQuant Pro spectrophotometer (Amersham PLC, Little Chalfont, United Kingdom). G. parasuis isolates were inoculated from a liquid overnight culture into BHI+ broth at an OD600 of 0.05. The cultures were incubated at 37°C with 5% CO2 and shaking at 200 rpm. OD600 readings were taken hourly for 8 h.
Biofilm assay using microtiter plate.
The microtiter plate assay for biofilm formation was adapted from a protocol described by Cassat et al. (29). Overnight cultures of G. parasuis were adjusted to an initial OD600 of 0.05, 0.125, or 0.25 in BHI+ broth. Diluted G. parasuis was plated in triplicate on a Nunc 96-well flat-bottom plate (Thermo Fisher Scientific Inc., Waltham, MA) and incubated statically for 48 hours at 37°C with 5% CO2. Cultures were gently removed from the plate and washed three times with sterile phosphate-buffered saline (PBS). The biofilm was fixed with 100% ethanol, allowed to dry for 10 minutes, and stained with 0.1% crystal violet. After incubating for 15 minutes at room temperature, plates were washed gently three times with PBS and allowed to dry overnight. Crystal violet was eluted from the biofilm with 150 μl of 100% ethanol for 10 minutes. The elution (120 μl) was transferred to a new 96-well plate and absorbance measured at 538 nm with a SpectraMax M5 instrument (Molecular Devices, LLC, Sunnyvale, CA). Three independent replicates were performed.
Complement-mediated killing.
To assess sensitivity to complement-mediated killing, G. parasuis cultures were treated with guinea pig serum (GPS) (Quidel, San Diego, CA). Heat-inactivated GPS (30 minutes at 56°C) was used as a negative control. G. parasuis cultures were grown on BHI+ plates and suspended in PBS to reach an OD600 of 0.42 (1 × 108 bacteria/ml). In a 96-well plate, 90 μl of GPS was added to 10 μl of G. parasuis (approximately 106 CFU). The G. parasuis and GPS incubated for 1 hour at 37°C, 5% CO2, and 100 rpm shaking. Serial dilutions were plated on BHI+ plates.
Adherence capacity to porcine alveolar macrophage cell line.
The interaction of G. parasuis HS069 and HS069Δcap with porcine alveolar macrophages was tested in vitro using the 3D4/31 cell line (ATCC, Manassas, VA). The 3D4/31 cells were maintained in complete advanced RPMI 1640 (Thermo Fisher Scientific Inc., Waltham, MA) as per ATCC recommendations. For the assay, 3D4/31 cells were plated into 4-well chamber slides (ibidi USA Inc., Madison, WI) at 5 × 105 cells/ml and allowed to adhere overnight. G. parasuis HS069 and HS069Δcap were added to the chambers to obtain an multiplicity of infection (MOI) of 10:1 and incubated for 1 hour at 37°C and 5% CO2. After incubating, 3D4/31 cells were washed to remove nonadherent bacteria, and chamber slides were placed on ice. G. parasuis cells were incubated with mouse monoclonal antibody to the outer membrane protein P5 (provided by M. Gottschalk) for 30 minutes at 4°C, followed by incubation with goat anti-mouse IgG3 (SouthernBiotech, Birmingham, AL) for 30 minutes at 4°C. Cells were fixed with ice-cold 50:50 methanol:acetone for 10 minutes and dried. Images were taken using the Nikon AR1+Si confocal microscope and evaluated using the NIS Elements software (Nikon Instruments Inc., Melville, NY). Bacterial intensity was evaluated using 10 random views.
Phagocytosis assessment using primary porcine alveolar macrophages.
Porcine alveolar macrophages (PAMs) were isolated as previously described (30, 31), with modifications. Briefly, the lungs of healthy pigs were flushed with PBS repeatedly until around 250 ml of fluid was collected. The lavage fluid was centrifuged at 1,000 rpm for 10 minutes. The pellet was washed twice and resuspended in complete RPMI 1640 medium (10% fetal bovine serum, 1 μg/ml fungizone, 100 U/ml penicillin, and 100 μg/ml streptomycin). PAMs were allowed to adhere to petri dishes for 2 hours at 37°C with 5% CO2. After 2 hours, media and nonadherent cells were aspirated, and adherent cells were washed with complete RPMI. Adherent cells were removed via cell scraping, washed twice with PBS, and resuspended in RPMI without antibiotics. Cells were counted and scored for viability using the Countess II automated cell counter (Invitrogen, Carlsbad, CA). PAMs were plated into 48-well plates with 5 × 105 PAMs per well and allowed to adhere for 20 minutes prior to starting the assay.
Bacterial stocks were generated by suspending agar-grown HS069 and HS069Δcap in PBS with 50% glycerol at an OD600 of 0.42. The stocks were quantified via serial dilution and frozen at –80°C until use. HS069 or HS069Δcap stocks were diluted to obtain an MOI of 10:1 in a 250-μl total volume per well (approximately 5 × 106 CFU/well).
To assess phagocytosis, the medium was aspirated, and RPMI containing G. parasuis was added to each well. The PAMs were incubated with the bacteria for 1 or 2 hours, and the supernatant was used to quantify nonphagocytosed bacteria. PAMs were isolated from four different animals, and 2 to 4 replicates were completed per animal for each isolate. The CFU/ml was quantified for HS069 and HS069Δcap inocula. The log fold reduction was calculated by subtracting the remaining bacteria at hour 1 or 2 from the initial inoculum.
G. parasuis challenge.
All experiments were approved by the National Animal Disease Center Institutional Animal Care and Use Committee. Caesarian-derived, colostrum-deprived (CDCD) pigs were derived at the National Animal Disease Center. At 4 weeks of age, pigs were intranasally challenged with 2 ml (1 ml per nostril) of 1 × 108 CFU/ml G. parasuis HS069 (4 pigs) or HS069Δcap (5 pigs) suspended in PBS. Pigs were monitored for clinical signs (lameness, respiratory distress, lethargy, and neurologic signs) and humanely euthanized when systemic signs of disease were noted. At necropsy, gross lesions were recorded and serum, nasal swabs, serosal swabs, joint fluid, lung lavage, and cerebral spinal fluid samples were obtained and plated for CFU counts. Serum and nasal swab samples were taken on day 0 and day 21 postchallenge. To investigate the vaccine potential of HS069Δcap, the pigs surviving challenge with HS069Δcap (5 pigs) were intranasally challenged with 2 ml (1 ml per nostril) of 1 × 108 of G. parasuis HS069 wild type on day 21 postchallenge with HS069Δcap. Pigs were monitored and treated as described above.
A follow-up study was conducted to evaluate nasal colonization of HS069Δcap in CDCD pigs. At 6 weeks of age, 6 pigs were inoculated intranasally with 2 ml (1 ml per nostril) of a 1 × 108-CFU/ml suspension of G. parasuis HS069Δcap in PBS. Nasal swabs were obtained on days 0, 1, 3, 5, 7, and 14 postchallenge for G. parasuis detection. G. parasuis species-specific PCR was run on these samples to detect bacterial colonization utilizing the primer set described by Howell et al. (32). Briefly, DNA was extracted from 50 μl of nasal swab samples using the MagMAX pathogen RNA/DNA kit (Thermo Fisher Scientific Inc., Waltham, MA). Extracted DNA was screened as previously described using the G. parasuis species-specific primers HPS_219690793-F (5′-ACAACCTGCAAGTACTTATCGGGAT-3′) and HPS_219690793-R (5′-TAGCCTCCTGTCTGATATTCCCACG-3′) (32).
ELISA for serum antibody titer.
Nunc MaxiSorp plates (Thermo Fisher Scientific Inc., Waltham, MA) were coated with 0.5 mg/ml of HS069Δcap sonicate in 100 mM carbonate-bicarbonate buffer (pH 9.6) at 4°C overnight. Plates were washed with 0.05% Tween 20 in PBS (PBST) followed by blocking for 1 hour at 37°C with 2% bovine serum albumin (BSA) in PBST. After being washed with PBST, serum samples were serially diluted in 1% BSA/PBST and applied to wells for 2 hours at 37°C. Plates were washed with PBST and horseradish peroxidase (HRP)-conjugated secondary goat anti-swine IgG antibody (SeraCare Life Sciences, Milford, MA) diluted 1:50,000 in 1% BSA/PBST was added and incubated for 1 hour at 37°C. Enzyme-linked immunosorbent assays (ELISAs) were developed using tetramethylbenzidine (TMB) substrate (Thermo Fisher Scientific Inc.). TMB was added to each well and incubated at room temperature for 15 minutes, and the reaction was halted with sulfuric acid (2 N). Absorbance at 450 nm was measured on a SpectraMax M5 instrument (Molecular Devices, LLC, Sunnyvale, CA).
Statistical analysis.
Statistical analysis was completed using GraphPad Prism 7.03 (GraphPad Software, La Jolla, CA). Biofilm formation was compared using an unpaired t test. Complement-mediated killing was evaluated as a log fold reduction in G. parasuis between heat-inactivated GPS and GPS and analyzed using an unpaired t test. Phagocytosis was evaluated using log fold reduction in G. parasuis between the inoculum and PAM-incubated wells. The difference in reduction between HS069 and HS069Δcap was compared using an ordinary one-way analysis of variance (ANOVA). A comparison of adherence to porcine alveolar macrophages was completed using unpaired t tests comparing both bacterial cells per 3D4/31 cell and fluorescent intensity per 3D4/31 cell. Welch’s corrections were used to account for differences in standard deviation when necessary. Results were considered significant at a P value of <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sarah Shore for sharing her expertise on microtiter biofilm assays.
This research was funded by the U.S. Department of Agriculture. This work was supported by a Longer and Larger (LoLa) grant from the Biotechnology and Biological Sciences Research Council (BBSRC grant numbers BB/G020744/1, BB/G019177/1, BB/G019274/1, and BB/G018553/1), the UK Department for Environment, Food and Rural Affairs, and Zoetis awarded to the Bacterial Respiratory Diseases of Pigs-1 Technology (BRaDP1T) consortium. L.A.W. is funded by a Sir Henry Dale Fellowship cofunded by the Royal Society and Wellcome Trust (grant number 109385/Z/15/Z). This project was additionally supported, in part, by an appointment to the Agricultural Research Service Postdoctoral Research Program administered by the Oak Ridge Institute for Science and Education (ORISE).
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. Funding sources had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Aragon V, Segales J, Oliveira S. 2012. Glasser’s disease, p 760–769. In Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW (ed), Diseases of swine, 10th ed John Wiley & Sons, Inc, West Sussex, United Kingdom. [Google Scholar]
- 2.Dickerman A, Bandara AB, Inzana TJ. 2019. Phylogenomic analysis of Haemophilus parasuis and proposed reclassification to Glaesserella parasuis, gen. nov., comb. nov. Int J Syst Evol Microbiol 70:180–186. doi: 10.1099/ijsem.0.003730. [DOI] [PubMed] [Google Scholar]
- 3.Kielstein P, Rapp-Gabrielson VJ. 1992. Designation of 15 serovars of Haemophilus parasuis on the basis of immunodiffusion using heat-stable antigen extracts. J Clin Microbiol 30:862–865. doi: 10.1128/JCM.30.4.862-865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell KJ, Peters SE, Wang J, Hernandez-Garcia J, Weinert LA, Luan SL, Chaudhuri RR, Angen O, Aragon V, Williamson SM, Parkhill J, Langford PR, Rycroft AN, Wren BW, Maskell DJ, Tucker AW; BRaDP1T Consortium. 2015. Development of a multiplex PCR assay for rapid molecular serotyping of Haemophilus parasuis. J Clin Microbiol 53:3812–3821. doi: 10.1128/JCM.01991-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aragon V, Cerdà-Cuéllar M, Fraile L, Mombarg M, Nofrarías M, Olvera A, Sibila M, Solanes D, Segalés J. 2010. Correlation between clinico-pathological outcome and typing of Haemophilus parasuis field strains. Vet Microbiol 142:387–393. doi: 10.1016/j.vetmic.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 6.Oliveira S, Blackall PJ, Pijoan C. 2003. Characterization of the diversity of Haemophilus parasuis field isolates by use of serotyping and genotyping. Am J Vet Res 64:435–442. doi: 10.2460/ajvr.2003.64.435. [DOI] [PubMed] [Google Scholar]
- 7.Jia Q, Lee BY, Bowen R, Dillon BJ, Som SM, Horwitz MA. 2010. A Francisella tularensis live vaccine strain (LVS) mutant with a deletion in capB, encoding a putative capsular biosynthesis protein, is significantly more attenuated than LVS yet induces potent protective immunity in mice against F. tularensis challenge. Infect Immun 78:4341–4355. doi: 10.1128/IAI.00192-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drysdale M, Heninger S, Hutt J, Chen Y, Lyons CR, Koehler TM. 2005. Capsule synthesis by Bacillus anthracis is required for dissemination in murine inhalation anthrax. EMBO J 24:221–227. doi: 10.1038/sj.emboj.7600495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith HE, Damman M, van der Velde J, Wagenaar F, Wisselink HJ, Stockhofe-Zurwieden N, Smits MA. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun 67:1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Xu X, Wu Y, Li L, Cao R, Cai X, Chen H. 2013. Polysaccharide biosynthesis protein CapD is a novel pathogenicity-associated determinant of Haemophilus parasuis involved in serum-resistance ability. Vet Microbiol 164:184–189. doi: 10.1016/j.vetmic.2013.01.037. [DOI] [PubMed] [Google Scholar]
- 11.Roberts IS. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol 50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 12.Magee AD, Yother J. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect Immun 69:3755–3761. doi: 10.1128/IAI.69.6.3755-3761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olvera A, Ballester M, Nofrarias M, Sibila M, Aragon V. 2009. Differences in phagocytosis susceptibility in Haemophilus parasuis strains. Vet Res 40:24. doi: 10.1051/vetres/2009007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaffner TO, Hinds J, Gould KA, Wuthrich D, Bruggmann R, Kuffer M, Muhlemann K, Hilty M, Hathaway LJ. 2014. A point mutation in cpsE renders Streptococcus pneumoniae nonencapsulated and enhances its growth, adherence and competence. BMC Microbiol 14:210. doi: 10.1186/s12866-014-0210-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu TY, Hsieh MK, Tan DH, Ou SC, Shien JH, Yen TY, Chang PC. 2015. Loss of the capsule increases the adherence activity but decreases the virulence of Avibacterium paragallinarum. Avian Dis 59:87–93. doi: 10.1637/10937-091414-reg. [DOI] [PubMed] [Google Scholar]
- 16.Xu B, Pei X, Su Y, Ma Z, Fan H. 2016. Capsule of Streptococcus equi subsp. zooepidemicus hampers the adherence and invasion of epithelial and endothelial cells and is attenuated during internalization. FEMS Microbiol Lett 363:fnw164. doi: 10.1093/femsle/fnw164. [DOI] [PubMed] [Google Scholar]
- 17.Rioux S, Galarneau C, Harel J, Kobisch M, Frey J, Gottschalk M, Jacques M. 2000. Isolation and characterization of a capsule-deficient mutant of Actinobacillus pleuropneumoniae serotype 1. Microb Pathog 28:279–289. doi: 10.1006/mpat.1999.0347. [DOI] [PubMed] [Google Scholar]
- 18.Howell KJ, Weinert LA, Chaudhuri RR, Luan SL, Peters SE, Corander J, Harris D, Angen O, Aragon V, Bensaid A, Williamson SM, Parkhill J, Langford PR, Rycroft AN, Wren BW, Holden MT, Tucker AW, Maskell DJ; BRaDP1T Consortium. 2014. The use of genome wide association methods to investigate pathogenicity, population structure and serovar in Haemophilus parasuis. BMC Genomics 15:1179. doi: 10.1186/1471-2164-15-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howell KJ, Weinert LA, Luan SL, Peters SE, Chaudhuri RR, Harris D, Angen O, Aragon V, Parkhill J, Langford PR, Rycroft AN, Wren BW, Tucker AW, Maskell DJ; BRaDP1T Consortium. 2013. Gene content and diversity of the loci encoding biosynthesis of capsular polysaccharides of the 15 serovar reference strains of Haemophilus parasuis. J Bacteriol 195:4264–4273. doi: 10.1128/JB.00471-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Z, Yue M, Zhou R, Jin Q, Fan Y, Bei W, Chen H. 2011. Genomic characterization of Haemophilus parasuis SH0165, a highly virulent strain of serovar 5 prevalent in China. PLoS One 6:e19631. doi: 10.1371/journal.pone.0019631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Li Y, Dai K, Wen Y, Wen X, Wu R, Huang X, Cao S. 2014. The confirmation of the DNA uptake signal sequence needed for genetic manipulation in Haemophilus parasuis. Vet Microbiol 173:395–396. doi: 10.1016/j.vetmic.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Bossé JT, Soares-Bazzolli DM, Li Y, Wren BW, Tucker AW, Maskell DJ, Rycroft AN, Langford PR; BRaDP1T Consortium. 2014. The generation of successive unmarked mutations and chromosomal insertion of heterologous genes in Actinobacillus pleuropneumoniae using natural transformation. PLoS One 9:e111252. doi: 10.1371/journal.pone.0111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bigas A, Garrido ME, de Rozas AM, Badiola I, Barbe J, Llagostera M. 2005. Development of a genetic manipulation system for Haemophilus parasuis. Vet Microbiol 105:223–228. doi: 10.1016/j.vetmic.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 25.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacques M, Foiry B. 1987. Electron microscopic visualization of capsular material of Pasteurella multocida types A and D labeled with polycationic ferritin. J Bacteriol 169:3470–3472. doi: 10.1128/jb.169.8.3470-3472.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borrathybay E, Sawada T, Kataoka Y, Okiyama E, Kawamoto E, Amao H. 2003. Capsule thickness and amounts of a 39 kDa capsular protein of avian Pasteurella multocida type A strains correlate with their pathogenicity for chickens. Vet Microbiol 97:215–227. doi: 10.1016/j.vetmic.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Kawamoto E, Okiyama E, Sasaki H, Sawada T, Mikazuki K, Ueshiba H. 2007. Ultrastructural characteristics of the external surfaces of Pasteurella pneumotropica from mice and Pasteurella multocida from rabbits. Lab Anim 41:285–291. doi: 10.1258/002367707780378087. [DOI] [PubMed] [Google Scholar]
- 29.Cassat JE, Lee CY, Smeltzer MS. 2007. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus. Methods Mol Biol 391:127–144. doi: 10.1007/978-1-59745-468-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cullen JM, Rycroft AN. 1994. Phagocytosis by pig alveolar macrophages of Actinobacillus pleuropneumoniae serotype 2 mutant strains defective in haemolysin II (ApxII) and pleurotoxin (ApxIII). Microbiology 140:237–244. doi: 10.1099/13500872-140-2-237. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Wang M, Zhang X, Wang P, Liu H, Wang Q. 2016. Heterogeneity of swine lung macrophages inoculated by porcine reproductive and respiratory syndrome virus. Food Agr Immunol 27:724–733. doi: 10.1080/09540105.2016.1160366. [DOI] [Google Scholar]
- 32.Howell KJ, Weinert LA, Peters SE, Wang J, Hernandez-Garcia J, Chaudhuri RR, Luan SL, Angen O, Aragon V, Williamson SM, Langford PR, Rycroft AN, Wren BW, Maskell DJ, Tucker AW. 2017. “Pathotyping” multiplex PCR assay for Haemophilus parasuis: a tool for prediction of virulence. J Clin Microbiol 55:2617–2628. doi: 10.1128/JCM.02464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.