LuxS/AI-2 is an important quorum sensing system which affects the growth, biofilm formation, virulence, and metabolism of bacteria. LuxS is encoded by the luxS gene, but how this gene is associated with a diverse array of physiological activities in Edwardsiella piscicida (E. piscicida) is not known. Here, we constructed an luxS gene mutant strain, the △luxS strain, to identify how LuxS/AI-2 affects pathogenicity.
KEYWORDS: Edwardsiella piscicida, LuxS/AI-2, quorum sensing, biofilm formation, virulence
ABSTRACT
LuxS/AI-2 is an important quorum sensing system which affects the growth, biofilm formation, virulence, and metabolism of bacteria. LuxS is encoded by the luxS gene, but how this gene is associated with a diverse array of physiological activities in Edwardsiella piscicida (E. piscicida) is not known. Here, we constructed an luxS gene mutant strain, the △luxS strain, to identify how LuxS/AI-2 affects pathogenicity. The results showed that LuxS was not found in the luxS gene mutant strain, and this gene deletion decreased E. piscicida growth compared to that of the wild-type strain. Meanwhile, the wild-type strain significantly increased penetration and motility in mucin compared to levels with the △luxS strain. The 50% lethal dose (LD50) of the E. piscicida △luxS strain for zebrafish was significantly higher than that of the wild-type strain, which suggested that the luxS gene deletion could attenuate the strain’s virulence. The AI-2 activities of EIB202 were 56-fold higher than those in the △luxS strain, suggesting that the luxS gene promotes AI-2 production. Transcriptome results demonstrated that between cells infected with the △luxS strain and those infected with the wild-type strain 46 genes were significantly differentially regulated, which included 34 upregulated genes and 12 downregulated genes. Among these genes, the largest number were closely related to cell immunity and signaling systems. In addition, the biofilm formation ability of EIB202 was significantly higher than that of the △luxS strain. The supernatant of EIB202 increased the biofilm formation ability of the △luxS strain, which suggested that the luxS gene and its product LuxS enhanced biofilm formation in E. piscicida. All results indicate that the LuxS/AI-2 quorum sensing system in E. piscicida promotes its pathogenicity through increasing a diverse array of physiological activities.
INTRODUCTION
A quorum sensing (QS) system was first discovered and described in Vibrio fischeri (V. fischeri) and Vibrio harveyi (V. harveyi), and it could regulate gene expression in response to increasing cell population density through autoinducer molecules (1). Based on the difference in autoinducer molecules, QS is classified into four types (2). One of these, autoinducer-2 (AI-2), is one of the universal languages for intraspecies and interspecies communication (3), which is a metabolic by-product of an luxS gene-encoded synthase, an enzyme involved primarily in the conversion of ribosylhomocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD), and the luxS gene is ubiquitous in the bacterial genomes (4–6). Except for the function of regulating the cell population density of bacteria, this system also plays a key role in the growth characteristics of bacteria and various physiological and biochemical functions, such as bioluminescence, sporulation, motility, conjugation, antibiotic production, biofilm formation, and secretion of virulence factors for infection or colonization (7–9).
Edwardsiella piscicida is an important zoonotic pathogen with a broad host range which particularly infects many farm-reared marine species and is responsible for large economic losses to aquaculture (10–12). Previous studies reported that LuxS of Edwardsiella spp. played a role in the antibacterial effect and its pathogenicity (13–16). However, the exact mechanism underlying the benefit of a functional LuxS system for physiological activities in the critical stage of E. piscicida has yet to be defined. But in some other bacteria, it was reported that the LuxS/AI-2 of Haemophilus parasuis (H. parasuis) affects its growth characteristics, biofilm formation, and virulence (2). LuxS of Borrelia burgdorferi performs a significant function during mammalian infection (17), and the LuxS/AI-2 of Streptococcus pneumoniae (S. pneumoniae) is necessary for its biofilm formation and colonization and regulates the expression of the genes involved in virulence and bacterial fitness during pneumococcal biofilm formation (18). LuxS of Salmonella enterica is considered a regulator of virulence and colonization. In Escherichia coli (E. coli), mutation rate plasticity is genetically switchable, dependent on the LuxS gene, and is socially mediated via cell-cell interactions (19). However, the luxS gene mutation in Campylobacter jejuni (C. jejuni) affected its motility through mucin and colonization under limited conditions (20). So the function of the QS-related gene luxS is very complicated, and the function of the luxS gene in the pathogenicity of bacteria needs further research.
Since luxS is involved in a very important regulating system, some luxS gene mutant strains were generated to study the function of this gene in E. coli, C. jejuni, and Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans) (20–22). However, luxS gene deletion mutants in E. piscicida have not been available, to our knowledge, to determine how this system is associated with a diverse array of physiological activities and abilities and to investigate if LuxS of E. piscicida plays a role similar to or different from that described for bacteria. Therefore, this study aimed to illustrate the function of the QS-related luxS gene of E. piscicida by comparing its role in pathogenicity to fish and host cells to that of to a luxS gene deletion mutant strain.
RESULTS
The luxS gene deletion mutant strain of E. piscicida was successfully constructed.
To characterize the luxS gene of E. piscicida EIB202, an in-frame deletion of the luxS gene was constructed, resulting in the E. piscicida ΔluxS strain (Fig. 1). The wild-type strain E. piscicida EIB202 and the mutant ΔluxS strain appeared as characteristic transparent colonies with black centers on deoxycholate-hydrogen sulfide-lactose (DHL) agar, reflecting hydrogen sulfide production, and had the same semitransparent off-white colonies on tryptic soy broth (TSB) agar.
FIG 1.
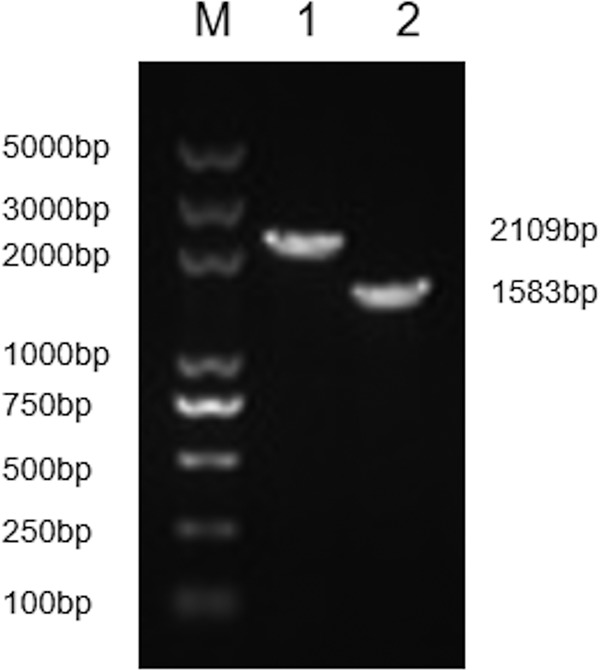
The luxS gene deletion in E. piscicida EIB202 was generated and identified. Lane 1, the wild-type strain EIB202; lane 2: the mutant ΔluxS strain; lane M, DNA marker DL5000.
Mutational analysis of LuxS.
Whole-cell proteins were analyzed with Western immunoblotting (Fig. 2). The results showed the amounts of LuxS detected in the wild-type strain EIB202; however, there was no LuxS production in the mutant ΔluxS strain. These results demonstrate that the luxS gene deletion significantly affected LuxS production and that this deleted gene is essential to the functioning of LuxS.
FIG 2.
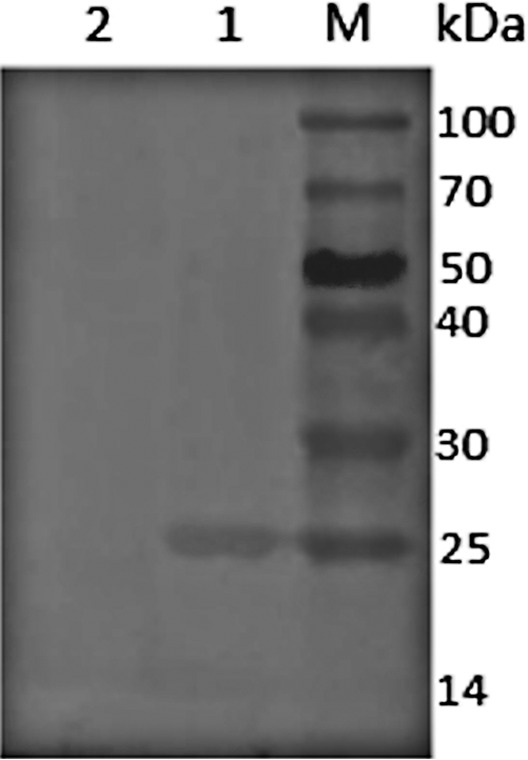
Western immunoblot analysis of luxS gene production in E. piscicida EIB202 and the mutant ΔluxS strain. Lane 1, E. piscicida EIB202; lane 2, ΔluxS mutant strain; lane M, protein marker.
The luxS gene deletion of E. piscicida attenuates virulence in fish.
After verifying that the zebrafish (Danio rerio) were disease free, it was found that E. piscicida EIB202 caused mortality in zebrafish 1 to 3 days after infection by intraperitoneal injection, whereas death occurred at 2 to 4 days after infection with the mutant strain E. piscicida ΔluxS. None of the fish in the control group died during the experiment. By extending the observation period to 15 days, infection with both strains was fully established. Both dead and moribund fish exhibited swelling and hemorrhaging in the abdomen. Fish infected with E. piscicida a ΔluxS exhibited a decrease in virulence compared to that of fish infected with E. piscicida EIB202 (50% lethal dose [LD50] of 4.28 × 104 CFU/fish and 9.59 × 105 CFU/fish, respectively). These results demonstrated that deletion of the luxS gene of E. piscicida attenuates virulence in zebrafish.
The luxS gene deletion decreases the motility of E. piscicida through mucin.
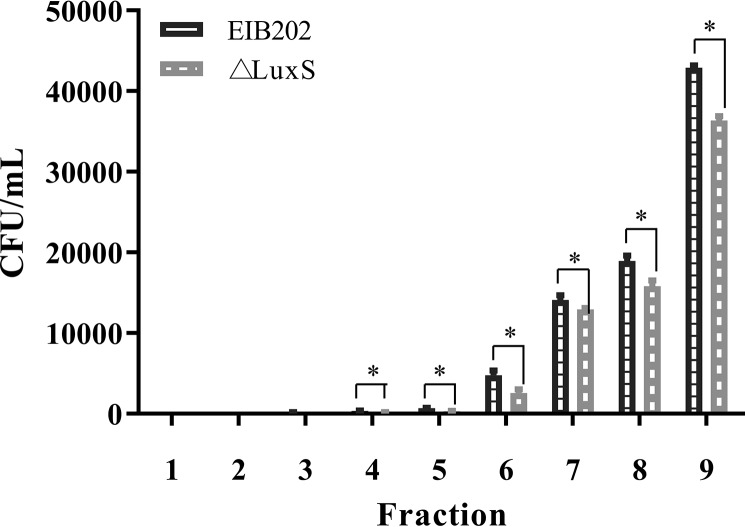
We simulated motility through mucin and compared the wild-type E. piscicida EIB202 and E. piscicida ΔluxS strains using a mucin column incubated at 28°C. It was found that the E. piscicida EIB202 strain could penetrate to fraction 3; however, the △luxS strain could only penetrate to fraction 4. When we compared the bacterial counts at each fraction of the mucin column, wild-type E. piscicida EIB202 counts were significantly higher between fractions 4 and 9 than those of E. piscicida ΔluxS (Fig. 3). In addition, counts of wild-type E. piscicida EIB202 were significantly higher than those of E. piscicida ΔluxS during the whole observation period. These results demonstrated that the luxS gene deletion has a significant effect on the mucin penetration ability (suspension ability) of E. piscicida.
FIG 3.
Penetration of E. piscicida EIB202 ΔluxS through mucus with incubation at 28°C. Columns of 30 mg/ml porcine gastric mucin in PBS were prepared in 1-ml syringes, with bacterium inoculate applied to the top of the column. Fraction 1 represents the bottom of the column, and fraction 9 represents the top. Graphs depict mean CFU counts (±SD) of each fraction in three independent experiments. *, P < 0.05, for significant differences between counts of E. piscicida.
The luxS gene promotes AI-2 production.
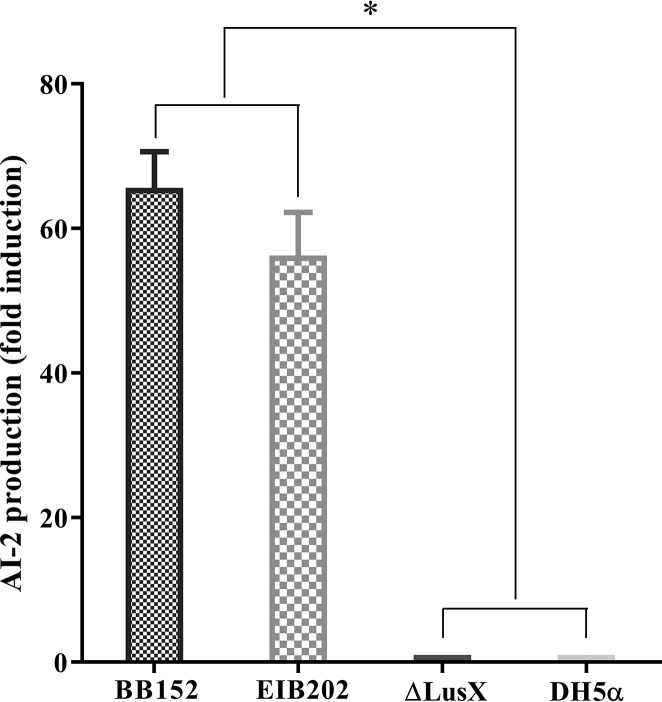
The luxS gene encodes an enzyme involved in the metabolism of S-ribosylhomocysteine, finally leading to the production of AI-2. AI-2 activities were measured in the BB152 luxS gene mutant, the △luxS strain, and the wild-type strain E. piscicida EIB202 by bioluminescence assay using the V. harveyi BB170 strain. As shown in Fig. 4, the AI-2 activities of the wild-type strain E. piscicida EIB202 were significantly increased compared to those of the luxS gene mutant strain. Also, the results demonstrated that the AI-2 activities of the BB152, EIB202, and △luxS strains were 65.63-fold, 56.23-fold, and 1.01-fold higher than those of the E. coli DH5ɑ strain (P < 0.05) (Fig. 4). These results suggested that the luxS gene was closely related to AI-2 in the E. piscicida EIB202 strain, that the luxS gene promoted AI-2 production, and that the luxS gene deletion led to an inability to synthesize AI-2.
FIG 4.
AI-2 activity in V. harveyi BB152, E. piscicida ΔluxS, and E. piscicida EIB202 using the V. harveyi bioluminescence assay. V. harveyi BB152 served as a positive control, and E. coli DH5α served as a negative control. Data are presented as means ± SD of three independent experiments. *, P < 0.05, for significant differences in levels of AI-2 production.
Biofilm formation in E. piscicida enhanced by LuxS.
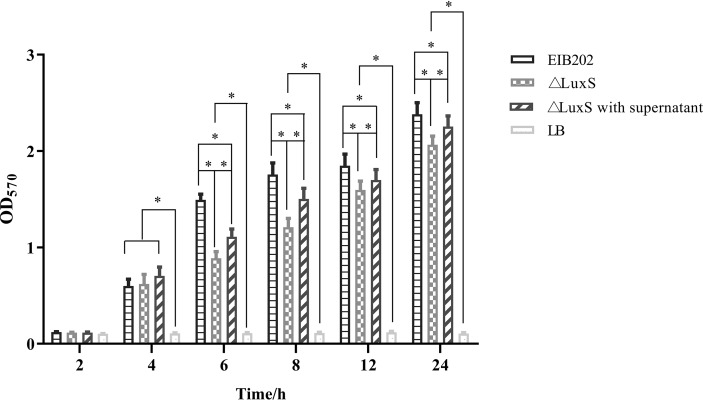
To investigate the differences in biofilm formation among the wild-type strain EIB202, the △luxS mutant strain, and the △luxS strain treated with supernatant of EIB202 under the same culture conditions, biofilm formation was quantitatively analyzed using a microtiter plate assay at different time points (Fig. 5). At the beginning of the first 4 h, biofilm formation was observed at the bottom of the microtiter plates, but there was no significant difference among the three experimental groups. However, after 6 h of cultivation, a significant difference in biofilm formation was found between the wild-type strain EIB202 and the △luxS strain treated with supernatant of EIB202; the same significant difference was observed between the △luxS strain treated with supernatant of EIB202 and the △luxS strain. Also, the biofilm formation ability of EIB202 was significantly higher than that of the △luxS strain treated with the EIB202 supernatant and significantly higher than that of the △luxS strain for the whole observation period. These data indicate that the luxS gene was involved in the formation of E. piscicida biofilms.
FIG 5.
Biofilm formation of E. piscicida ΔluxS and EIB202 strains. Sterile supernatant was added at 10% to overnight cultures of the wild-type strain E. piscicida EIB202 at the beginning of culture. Data are presented as means values of the OD570 (±SD) for three independent experiments. *, P < 0.0,5 for significant differences in the levels of the biofilm formation.
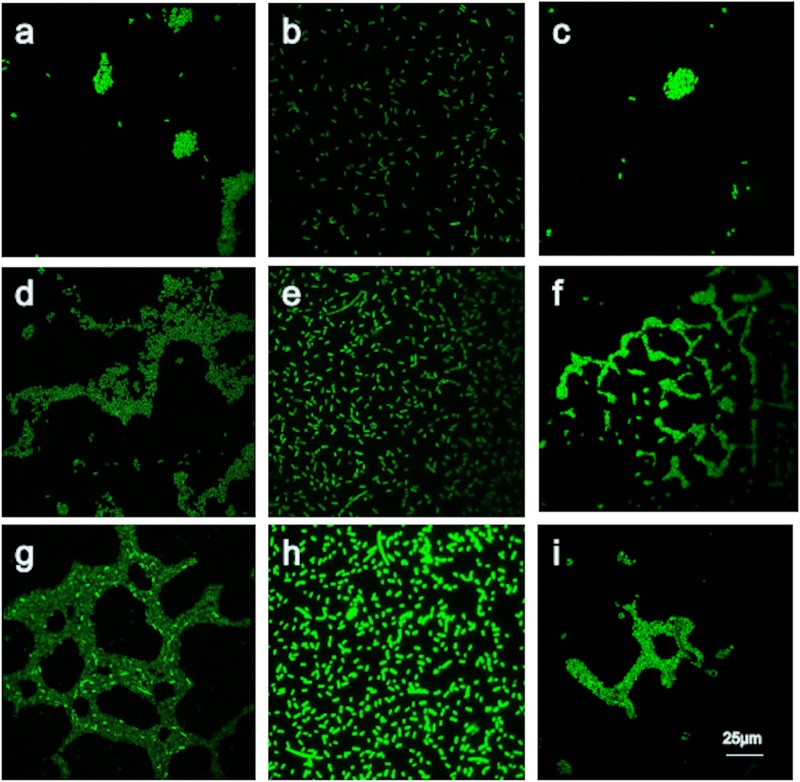
When we analyzed the biofilm structure and viability by confocal scanning laser microscopy (CLSM), an interesting result was found: even though the luxS gene deletion mutant was not able to effectively prevent biofilm formation, biofilm biomass was strongly reduced, as can be seen qualitatively in Fig. 6. The amount of biofilm biomass and the biofilm maximum depth for the luxS gene mutant strain were reduced compared to levels for the wild-type strain; however, the biofilm formation ability of the luxS gene mutant strain was partly restored to wild-type strain levels after addition of the supernatant of EIB202.
FIG 6.
Effects of the luxS gene deletion on the biofilm formation of E. piscicida EIB202. Sterile supernatant at 10% was added to overnight cultures of the wild-type strain EIB202 at the beginning of culture. a, EIB202 at 6 h; b, ΔluxS strain at 6 h; c, ΔluxS strain with supernatant at 6 h; d, EIB202 at 12 h; e, ΔluxS strain at 12 h; f, ΔluxS strain with supernatant at 12 h; g, EIB202 at 24 h; h, ΔluxS strain at 24 h; i, ΔluxS strain with supernatant at 24 h.
Annotation of the assembly and classification of unigenes.
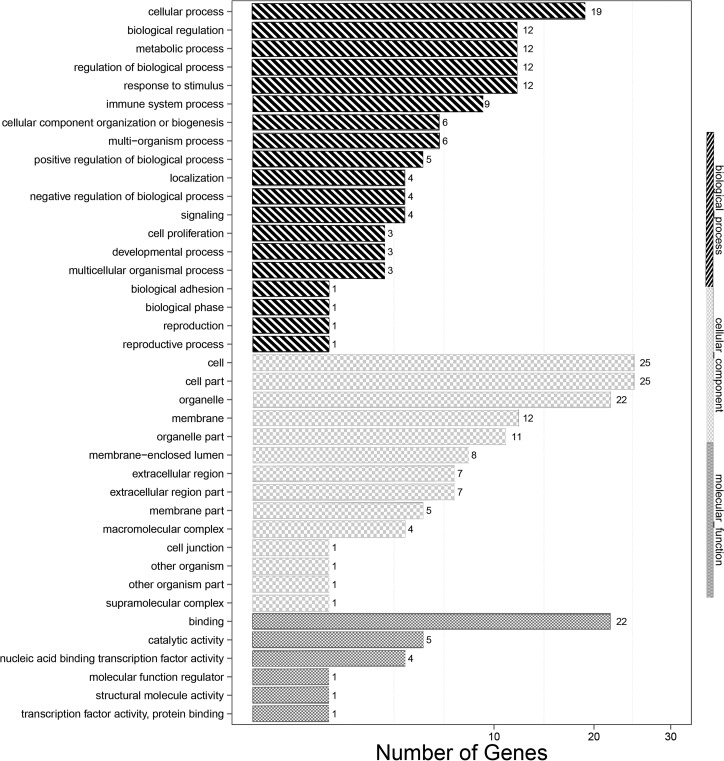
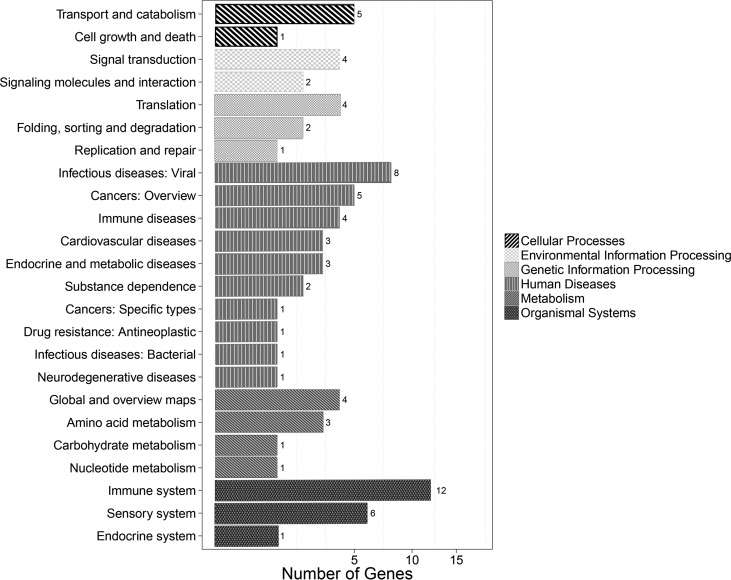
Gene Ontology (GO) classification describes the properties of genes by assigning these unigenes to biological processes, cellular components, and molecular functions. GO analysis was performed using the blast2 GO software. EIB202 versus △luxS GO analysis showed that the differentially expressed genes (DEGs) clustered in molecular functions, biological processes, and cellular components (Fig. 7). Cellular process (19 unigenes), biological regulation (12 unigenes), regulation of biological process (12 unigenes), and metabolic process (12 unigenes) represented the major categories of biology process. Cell (25 unigenes), cell part (25 unigenes), organelle (22 unigenes), and organelle part (11 unigenes) represented the majority of cellular components; binding (22 unigenes) and catalytic activity (5 unigenes) showed the highest percentages in the molecular function category. The unigenes were also annotated against the KEGG database and assigned to different pathways in six major groups, including genetic information processing, metabolism, cellular processes, organismal systems, human disease, and environmental information processing. In total, 46 transcripts had significant hits in the KEGG database for the EIB202 versus uhpA strain, and 12 genes were related to the immune system (Fig. 8).
FIG 7.
The transcripts of cells infected with EIB202 compared to those infected with the ΔluxS strain using the GO annotation method. The transcripts were assigned to different terms in three major categories of the GO database, namely, biological process, cellular component, and molecular function.
FIG 8.
The transcripts of cells infected with EIB202 compared to those infected with the ΔluxS strain using KEGG annotation. The transcripts were assigned to different pathways in six major categories of the KEGG database, including genetic information processing, metabolism, cellular processes, organismal systems, human disease, and environmental information processing.
Functional annotation and analysis of differentially expressed genes.
A total of 46 significantly differential genes were detected between the △luxS strain group and the EIB202 group, which included 34 upregulated genes and 12 downregulated genes. These up- and downregulated genes are presented in Table 1. Among those genes, the largest number were closely related to cell immunity and signaling systems.
TABLE 1.
Up- and downregulation in the cells infected with EIB202 compared to levels with ΔluxS strain infection
| Group and GeneID | Symbol | Description |
|---|---|---|
| Upregulated genes | ||
| 100041230 | Hist1h4m | Histone H4-like |
| 102639653 | LOC102639653 | KRAB box and zinc finger, C2H2-type domain containing |
| 68195 | Rnaset2b | Ribonuclease T2B precursor |
| 108167809 | LOC108167809 | Uncharacterized protein LOC105244343 isoform X1 |
| 105244999 | Gm40514 | Uncharacterized protein LOC105244999, partial |
| 100859931 | Gm20604 | AK010878-Moap1 protein |
| 100043125 | Cd300ld5 | Predicted gene 11710 precursor; predicted gene 11710 isoform X1 |
| 100040500 | Gm2808 | Tubby-related protein 4-like |
| 381308 | Mnda | Interferon-activated protein 205-B isoforms 1 and 2 |
| 236312 | Pyhin1 | Pyrin and HIN domain-containing protein 1 |
| 217310 | Hid1 | RIKEN cDNA C630004H02, isoform CRA_b and CRA_a |
| 108167415 | LOC108167415 | Uncharacterized protein LOC105246138 |
| 100039060 | 0610010B08Rik | KRAB box and zinc finger, C2H2 type domain containing |
| 280668 | Adam1a | Disintegrin and metalloproteinase domain-containing protein 1a precursor |
| 100042856 | Gm4070 | Interferon-induced very large GPTase 1 |
| 108167806 | LOC108167806 | Uncharacterized protein LOC105246138 |
| 328918 | Zscan30 | Zinc finger and SCAN domain-containing protein 30 isoform X1 and X3; zinc finger protein 397 opposite strand |
| 621823 | Psme2b | Proteasome activator complex subunit 2 isoform 1 |
| 381287 | A530032D15Rik | SP140 nuclear body protein family member isoform X3; SP140 nuclear body protein family member |
| 15061 | Ifi44l | Interferon-induced protein 44-like |
| 100040213 | Gm2666 | Proteinase-activated receptor 1-like |
| 219132 | Phf11d | PHD finger protein 11;11-like; 11D |
| 13549 | Dyrk1b | Dual specificity tyrosine phosphorylation-regulated kinase 1B isoform X1, X2, and X3, isoform CRA_b, isoform p65 |
| 665433 | Hist1h2ao | Histone H2A type 1-B/E |
| 15959 | Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3, isoform X1, isoform X2 |
| 209086 | Samd9l | Sterile alpha motif domain-containing protein 9-like |
| 94094 | Trim34a | Tripartite motif-containing protein 34A |
| 22169 | Cmpk2 | UMP-CMP kinase 2, mitochondrial precursor |
| 667370 | Ifit3b | Interferon-induced protein with tetratricopeptide repeats 3-like, isoform X1 |
| 15039 | H2-T22 | H2-T22 protein; histocompatibility 2, T region locus 22 precursor |
| 99899 | Ifi44 | Interferon-induced protein 44; interferon-induced protein 44, isoform X1 |
| 229900 | Gbp7 | mFLJ00316 protein |
| 100044068 | LOC100044068 | Interferon-inducible Ifi202b |
| 23962 | Oasl2 | 2′–5′ Oligoadenylate synthase-like protein 2 |
| Downregulated genes | ||
| 100043813 | Rps27rt | 40S ribosomal protein S27 |
| 15019 | H2-Q8 | H-2 class I histocompatibility antigen, Q8 alpha chain precursor |
| 432479 | 4930404N11Rik | Uncharacterized protein c19orf71 homolog |
| 105244844 | Gm40378 | Uncharacterized protein LOC105244844 |
| 101488212 | Gm21975 | Protein EVI2B precursor |
| 668605 | Gm9265 | Hippocalcin-like protein 1 |
| 100039672 | Msmp | Prostate-associated microseminoprotein precursor |
| 504193 | Npcd | Neuronal pentraxin with chromo domain isoform 1 and 2 |
| 71970 | Zbed5 | mCG145321, isoform CRA_a, partial; SCAN domain containing 3 |
| 22437 | Xirp1 | mCG14131, isoform CRA_c |
| 50708 | Hist1h1c | Histone H1.2 |
| 100328588 | Gm21948 | Nup62-Il4i1 protein precursor |
DISCUSSION
E. piscicida is a novel species within the genus Edwardsiella, which was first reported in 2013 (23). Genome sequence studies showed that the E. piscicida genome included the LuxS system (24). Much progress has been made over the last decade in understanding the role of LuxS, which is a metabolic enzyme with a key role in the production of the AI-2 molecule used for signaling in some species, including E. piscicida (5–7). However, a luxS gene deletion mutant of E. piscicida was first constructed, to our knowledge, in this study. In the present study, we investigated the effect of the E. piscicida luxS gene mutation on growth, penetration through mucin, biofilm formation capability, and pathogenicity in the zebrafish model. All results indicate that LuxS plays a key role in many physiological activities in E. piscicida.
A mutant ΔluxS strain was constructed, and a polarity effect also was verified to make sure that the change in the function of the ΔluxS strain was attributed only to the luxS gene rather than to other upstream or downstream genes. In this study, the result showed that there was no LuxS production in the mutant ΔluxS strain, which demonstrates that the luxS gene deletion significantly affected LuxS production and that this deleted gene is essential to the functioning of LuxS. In addition, this study found that the growth characteristics of the examined strains, the wild-type and the mutant strain, were similar except that the growth rate of the ΔluxS strain was lower than that of the wild-type strain. As with other studied bacterial species, E. piscicida maximally produces AI-2 during periods of high metabolic activity. The results demonstrated that the AI-2 activities of BB152 and EIB202 were significantly higher than those of the mutant strain. These results suggested that the luxS gene promoted AI-2 production and that the luxS gene deletion led to an inability to synthesize AI-2. A LuxS/AI-2 system was reported to exist in C. jejuni, Aeromonas hydrophila (A. hydrophila), E. coli, Actinobacillus pleuropneumoniae (A. pleuropneumoniae), Lactobacillus paraplantarum, A. actinomycetemcomitans, S. pneumoniae, and H. parasuis, etc., and LuxS inactivation affected the growth and/or virulence of the above-mentioned strains (2, 3, 8, 18, 21, 22, 25, 26).
The luxS gene was previously identified as a virulence determinant contributing to pathogenicity in bacteria, such as E. coli, A. pleuropneumoniae, etc. (21, 26). To further evaluate the role of the luxS gene in the pathogenesis of E. piscicida in vivo and in vitro, the LD50 in zebrafish and the motility of E. piscicida through mucin, respectively, were measured. The LD50 experiment showed that the virulence of the ΔluxS strain was attenuated about 23-fold. The result of the experiment measuring penetration through mucin found that the number of cells and the distance penetrated by the ΔluxS strain were sharply decreased compared to those of the wild-type strain for the whole experimental period, suggesting that the luxS gene deletion decreased the colonization ability and the virulence of E. piscicida. Although the effect of reducing the penetration ability of C. jejuni was not significant, the result is consistent with conclusions obtained from other bacteria, such as C. jejuni, H. parasuis, and S. pneumoniae (2, 18, 27). These data disprove prior conclusions and suggest that LuxS in E. piscicida does indeed perform a significant function(s) during fish infection.
To investigate the function of LuxS, the corresponding key genes and complex pathways were identified when the host cells were infected with the E. piscicida wild-type strain EIB202 and the luxS gene mutant strain. In this study, we used a transcriptome sequencing (RNA-Seq) platform and bioinformatics analysis to study the transcriptomic response of macrophages and to identify their potential functions and metabolic pathways using GO and KEGG annotations (28). The results with the EIB202 versus △luxS strain showed that a total of three GO terms were significantly enriched, and the top GO terms on the list included cellular response to interferon beta, response to interferon beta, and cellular response to interferon alpha. KEGG pathway analysis further showed that genes associated with 11 pathways were significantly enriched; these included genes associated with herpes simplex infection, ribosome biogenesis in eukaryotes, the NOD-like receptor signaling pathway, endocytosis, ribosome biogenesis in eukaryotes, viral myocarditis, Fc gamma R-mediated phagocytosis, antigen processing and presentation, viral carcinogenesis, the mRNA surveillance pathway, and graft-versus-host disease. Among these pathways, some were closely related to cell immunity and signaling systems. These data indicate that the luxS gene plays a key role in an E. piscicida-infected host.
The formation of a biofilm begins with the attachment of free-floating microorganisms to a surface, and it was reported that biofilm formation on biological surfaces in the host at the site of colonization played a key role in most infections because biofilm formation provides increased protection of bacteria from antibiotics and host defenses (29, 30). Therefore, some infections or diseases may effectively be prevented by inhibiting the formation of organisms by developing some materials or drugs based on control of biofilm formation or enhancing the host immune system (31–34). In recent years, some research has successfully evaluated methods to control unwanted biofilm formation (35, 36). In this study, the biofilm formation ability of EIB202 was significantly higher than that of the △luxS mutant treated with supernatant of EIB202 and significantly higher than that of the △luxS mutant for the whole observation period. These results suggested that the luxS gene could enhance the biofilm formation ability and that the product of the luxS gene could secrete outside the cell and could enhance biofilm formation ability, too. A similar study reported that inhibition of luxS gene expression could inhibit biofilm in E. coli and A. hydrophila (25, 37). It is well known that bacterial biofilms increase cell survival through the improved defense of the immune system, increased availability of nutrients, resistance to antibiotics, and better opportunities for cellular communication and transfer of genetic material.
In conclusion, the result of this study demonstrated that the deletion of the luxS gene in E. piscicida EIB202 reduced its ability for growth, mobility, biofilm formation, and production of AI-2 and for infection of zebrafish. These results suggested that the luxS gene of E. piscicida increased its growth and motility, promoted AI-2 production and biofilm formation, and enhanced its virulence to infect fish. All results indicate that the LuxS/AI-2 quorum sensing system in E. piscicida promotes its pathogenicity through an increase in a diverse array of physiological activities.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used include E. piscicida EIB202, E. coli DH5α λpir, E. coli S17-1 λpir, and V. harveyi BB152 and BB170 and plasmids pMD18-T and pDMK, which were from previous research (11, 21, 38). E. piscicida strains were grown at 28°C in tryptic soy broth (TSB; Becton, Dickinson, Sparks, MD) or tryptic soy agar (TSA; Becton, Dickinson) (39). E. coli strains were cultured in Luria-Bertani (LB; Becton, Dickinson) broth at 37°C. Antibiotics, when required, were added at the following final concentrations: ampicillin (Amp) 50 μg/ml; colistin (Col) 12.5 μg/ml; kanamycin (Km) 50 μg/ml; streptomycin (Sm) 100 μg/ml; and gentamicin (Gm) 25 μg/ml.
The macrophage RAW264.7 cell line was purchased from the type culture collection of the Chinese Academy of Sciences and cultured in Dulbecco’s modified Eagle’s minimum essential medium (DMEM) (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 incubator (40). All cells were passaged twice a week and maintained in exponential growth.
Construction of the luxS gene deletion mutants.
The construction of the gene luxS deletion in E. piscicida was performed as previously described (11). Briefly, PCR amplification was performed to prepare the upstream and downstream fragments of luxS with primer pairs (Table 2). The PCR products were then obtained with the next run of overlap PCR with the primer pair luxS-WF/luxS-WR after luxS-P1/luxS-P2 and luxS-P3/luxS-P4, generating a PCR product containing an in-frame deletion fragment of 515 bp in luxS. The overlap product was sequenced and cloned into the XbaI and SpeI sites of the suicide vector pDMK, which carried the R6K ori, sacB sucrose sensitivity gene, Cm resistance gene, and Km resistance gene. The resulting plasmid pDMK-fucP was transformed into E. coli DH5α and E. coli S17-1; then the plasmid was mated from E. coli SM10 λpir into E. piscicida EIB202 by conjugation (41). The unmarked in-frame deletion mutants were selected in two sequential homologous recombination processes on LB medium containing Col, Cm, and Km and then on LB with 10% (vol/vol) sucrose (11, 41). The targeted in-frame deletion mutants were confirmed by sequencing of the deleted region, and it was designated E. piscicida ΔluxS.
TABLE 2.
Primers used in this study for gene deletions
| Primer name | Primer sequence (5′–3′) |
|---|---|
| luxS-P1 | TGCTCTAGATCGCCGAGGTGCTGGATGGCATTAA |
| luxS-P2 | GTGAAGCCTACATTTCGTCACCTCCTCAAT |
| luxS-P3 | TGACGAAATGTAGGCTTCACCGATAGAAAAG |
| luxS-P4 | CGGACTAGTTTTACCACCTCCATGTCCCCCT |
| LuxS-WF | TGGACATTAACCCGTTCACTC |
| LuxS-WR | GCGGTAAGCAAAGAGGAGC |
| 16S-F | TAGGGAGGAAGGTGTGAA |
| 16S-R | CTCTAGCTTGCCAGTCTT |
| luxS-F | ATGCCGTTACTGGATAGCTTCA |
| luxS-R | CTAGGGGTGCAGACGGGC |
| pDMK-F | AAAGCTCTCATCAACCGTGGC |
| pDMK-R | TGCTCCAGTGGCTTCTGTTTC |
The analysis of luxS gene product by Western blotting.
Whole-cell proteins were prepared from cells grown in TSB medium to an optical density at 600 nm (OD600) of ~1 and resolved by electrophoresis in a 0.1% sodium dodecyl sulfate–12% polyacrylamide gel. After electrophoresis, the proteins were transferred to a nitrocellulose membrane and then analyzed by Western immunoblotting.
The Western blotting was performed as described previously (15). Briefly, bacterial cells were grown in LB medium to an OD600 of 1 and lysed with lysis buffer (100 mM NaH2PO4, 10 mM Tris-Cl, and 8 M urea, pH 8.0). The lysed cells were centrifuged at 4°C, and the supernatant was subjected to electrophoresis on 0.1% sodium dodecyl sulfate–12% polyacrylamide gels. After electrophoresis, the proteins were transferred to nitrocellulose membranes and then analyzed by Western immunoblotting with LuxS antibodies, as described previously (7).
AI-2 detection.
AI-2 detection was performed as previously described (3). The wild-type strain E. piscicida EIB202, the mutant ΔluxS strain, and DH5α were cultured in LB medium to an optical density of 1 at 600 nm. The cell-free culture fluid (CF) was obtained by filtering the supernatant through a 0.22-μm-pore-size filter (Millipore, Bedford, MA) and adjusting the pH to 7.0. The reporter strain V. harveyi BB170 was diluted 1:5,000 in autoinducer bioassay (AB) medium, and the LB medium was added to the diluted BB170 culture at 1:10 (vol/vol). The mixture was incubated at 28°C for 8 h. Aliquots (100-ml) were added to white, flat-bottomed, 96-well plates (Thermo Labsystems, Franklin, MA) to detect AI-2 activity. The CFs from BB152 and DH5α were used as positive and negative controls, respectively. Luminescence values were measured with a Tecan GENios Plus microplate reader.
Mucin penetration assay.
A mucin penetration assay was performed as described previously (20, 42). One-milliliter syringes containing 1 ml of porcine gastric mucin (Sigma-Aldrich) dissolved in phosphate-buffered saline (PBS) to a concentration of 30 mg/ml were used as mucin columns. The concentration of 30 mg/ml was selected as it was the highest concentration of mucin that could be used without the mucin viscosity interfering with the syringe function in the assay. A total of 100 ml of log-phase bacterial cultures was added to the top of the mucin columns and allowed to settle for 30 min at 28°C (11). Fractions of 0.1 ml (the first 0.1 ml collected is fraction 1, with subsequent fractions in chronological order until the last 0.1-ml fraction, or fraction 10) were collected from the bottom of the mucin columns, plated on TSB agar containing 12.5 mg/ml colistin, incubated overnight, and then enumerated for bacterial quantification (11). At least three independent experiments were performed for each strain at each temperature. Data were assessed for significant differences in the number of bacteria (CFU) compared to values for the wild type at each fraction and total CFU from fractions 1 to 9. Fraction 10 was not included in any of the analysis as this fraction included the very top of the mucin layer mixed with culture inoculum, thus making it impossible to determine if the bacteria in this sample had truly penetrated the mucin (20).
Test of bacterial virulence in a fish model.
The zebrafish (Dario rerio) used for virulence tests in this study were cultured animals, and all the experiments were designed in strict accordance with regulations of the local government (43). Fish from quarantined stocks were acclimatized for more than 1 week in the laboratory, and the study was the approved by the Animal Ethics Committee of Shandong Agricultural University (44, 45). The animals were determined to be disease free and were used as models to assess the virulence of E. piscicida strains (EIB202 and ΔluxS strains). These bacterial numbers were determined by the plate-counting method after estimations made using a hemocytometer (46). Each group of 15 fish was injected intraperitoneally with 20 μl of PBS containing bacterial cells at 104 CFU/ml, 105 CFU/ml, 106 CFU/ml, 107 CFU/ml, and 108 CFU/ml. The control groups were injected with 20 μl of PBS. The fish were maintained in static freshwater (50% of the volume was changed daily) at 28°C for 15 days. The LD50 values were calculated as described by Reed-Munch (47).
Biofilm formation.
Crystal violet (CV) staining was used to quantify biofilm formation of wild-type E. piscicida EIB202 and the mutant ΔluxS strain (3). Briefly, E. piscicida EIB202 was inoculated to an optical density of 1.0 at 600 nm; then the supernatant was obtained by centrifugation at 3,000 × g for 5 min, and the supernatant was filtered through a 0.22-μm-pore-size microporous filter to obtain a sterile supernatant. Overnight cultures were diluted to an optical density of 0.1 at 600 nm. Diluted culture (one culture of the ΔluxS strain group was treated with 10% sterile supernatant) was transferred to a 96-well plate (Corning, NY). After incubation at 28°C for 24 h, the wells were washed gently three times with phosphate-buffered saline (PBS), stained with 0.1% CV for 30 min at room temperature, rinsed with distilled water, and air dried. A total of 100 ml of 95% ethanol was added to each well to dissolve the CV. The optical density at 595 nm was determined using a Synergy 2 microplate reader (Biotek, Winooski, VT).
CLSM analysis of the biofilms.
Confocal scanning laser microscopy (CSLM) was performed as described previously (48, 49). Briefly, overnight cultures were diluted to an optical density of 0.1 at 600 nm (one culture of the ΔluxS strain group was treated with 10% sterile supernatant, as described above in “Biofilm formation”) and then transferred to a 24-well plate with a piece of cell slide placed at the bottom of each well before. All slides were washed twice with 0.9% NaCl after being cultured for 6 h, 12 h, and 24 h. After staining with SYBR green I, the biofilms were gently rinsed with 0.9% NaCl. The biofilm images were acquired in an Olympus FluoView FV1000 (Olympus, Lisbon, Portugal) confocal scanning laser microscope. For each condition, three independent biofilms were used. Images acquired from at least 20 different regions per surface were analyzed by determining the top and bottom layers of the biofilm and calculating the maximum thickness of each region.
Total RNA library construction of infected RAW26 macrophages.
Total RNA of six individuals was extracted from RAW26 macrophages infected with E. piscicida EIB202, the ΔuhpA strain, and control groups using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA quality and quantity were assessed and affirmed using NanoDrop, Qubit, and Bioanalyzer (50). The extracted RNA from RAW26 macrophages was used to isolate mRNA with an mRNA-Seq Sample Prep kit (Illumina) for constructing a nondirectional Illumina RNA-Seq library (28). The libraries were loaded onto flow cell channels for sequencing using an Illumina Genome Analyzer at Beijing Genomics Institute Co., Ltd. (Beijing, China). The cDNA library was sequenced on an Illumina Hi-Seq 2500 sequencing platform, and reads were used for gap filling of these scaffolds to generate final scaffold sequences (28). Sequence homology searches were performed using local BLAST programs against sequences in the NCBI nonredundant (nr/nt) protein/nucleotide database and the Swiss-Prot database (E value of <1e−5) (51).
Transcriptome annotation and gene ontology analysis.
All transcripts were compared to those in the NCBI databases, including the nonredundant (nr) protein database, GO database, Cluster of Orthologous Genes (COG) database, and KEGG database, for functional annotation using a BLAST search with an E value cutoff of 1e−5 (50). Functional annotation was performed with gene ontology (GO) terms (www.geneontology.org) that were analyzed using Blast2 GO software (https://www.blast2go.com/), and the COG and KEGG pathway annotations were performed using BLAST software against the COG and KEGG databases (28).
Statistical analysis.
Student's t test was used, with significance set at the 95% level. Error bars on graphs indicate the standard deviations (SD).
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 31402325), Natural Science Foundation of Shandong Province (ZR2019MC031), the fund earmarked for the Modern Agro-industry Technology Research System in Shandong Province (no. SDAIT-14-07), National Key R&D Program of China (2019YFD0900101), funds of the Shandong Double Tops Program, special funds from central finance to support the development of local universities and funds of the China Scholarship Council, and the Open Fund of Shandong Key Laboratory of Disease Control in Mariculture.
Y.S. and Y.L. performed the experiments, Y.S. and Q.L. analyzed the data, J.H. and J.C. helped collect data, and R.Z. helped edit references. X.W. designed and supervised the study and edited the manuscript.
REFERENCES
- 1.Nealson KH, Hastings JW. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol Res 43:496–518. doi: 10.1128/MMBR.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang B, Ku X, Zhang X, Zhang Y, Chen G, Chen F, Zeng W, Li J, Zhu L, He Q. 2019. The AI-2/luxS quorum sensing system affects the growth characteristics, biofilm formation, and virulence of Haemophilus parasuis. Front Cell Infect Microbiol 9:62. doi: 10.3389/fcimb.2019.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, Wu R, Zhang J, Li P. 2018. Overexpression of luxS promotes stress resistance and biofilm formation of Lactobacillus paraplantarum L-ZS9 by regulating the expression of multiple genes. Front Microbiol 9:2628. doi: 10.3389/fmicb.2018.02628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 5.Trappetti C, Gualdi L, Di Meola L, Jain P, Korir CC, Edmonds P, Iannelli F, Ricci S, Pozzi G, Oggioni MR. 2011. The impact of the competence quorum sensing system on Streptococcus pneumoniae biofilms varies depending on the experimental model. BMC Microbiol 11:75. doi: 10.1186/1471-2180-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao RM, Pasha SN, Sowdhamini R. 2016. Genome-wide survey and phylogeny of S-ribosylhomocysteinase (LuxS) enzyme in bacterial genomes. BMC Genomics 17:742. doi: 10.1186/s12864-016-3002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M, Li N, Yan M, Chang W, Hao J, Pang X, Wang X. 2015. Expression of Edwardsiella tarda luxS gene at different growth stage. Wei Sheng Wu Xue Bao 55:1201–1207. [PubMed] [Google Scholar]
- 8.Adler L, Alter T, Sharbati S, Golz G. 2014. Phenotypes of Campylobacter jejuni luxS mutants are depending on strain background, kind of mutation and experimental conditions. PLoS One 9:e104399. doi: 10.1371/journal.pone.0104399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Petri WA Jr, Haque R, Ahmed T, Gordon JI. 2014. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan M, Liu J, Li Y, Wang X, Jiang H, Fang H, Guo Z, Sun Y. 2018. Different concentrations of Edwardsiella tarda ghost vaccine induces immune responses in vivo and protects Sparus macrocephalus against a homologous challenge. Fish Shellfish Immunol 80:467–472. doi: 10.1016/j.fsi.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Liu G, Sun Y, Wang X, Fang H, Jiang H, Guo Z, Dong J. 2018. The role of regulator FucP in Edwardsiella tarda pathogenesis and the inflammatory cytokine response in tilapia. Fish Shellfish Immunol 80:624–630. doi: 10.1016/j.fsi.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Leung KY, Wang Q, Yang Z, Siame BA. 2019. Edwardsiella piscicida: a versatile emerging pathogen of fish. Virulence 10:555–567. doi: 10.1080/21505594.2019.1621648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu YH, Dang W, Liu CS, Sun L. 2010. Analysis of the effect of copper on the virulence of a pathogenic Edwardsiella tarda strain. Lett Appl Microbiol 50:97–103. doi: 10.1111/j.1472-765X.2009.02761.x. [DOI] [PubMed] [Google Scholar]
- 14.Sun B, Zhang M. 2016. Analysis of the antibacterial effect of an Edwardsiella tarda LuxS inhibitor. Springerplus 5:92. doi: 10.1186/s40064-016-1733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, Jiao X, Hu Y, Sun L. 2009. Attenuation of Edwardsiella tarda virulence by small peptides that interfere with LuxS/autoinducer type 2 quorum sensing. Appl Environ Microbiol 75:3882–3890. doi: 10.1128/AEM.02690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Sun L. 2012. Edwardsiella ictaluri LuxS: activity, expression, and involvement in pathogenicity. Pol J Microbiol 61:263–271. doi: 10.33073/pjm-2012-036. [DOI] [PubMed] [Google Scholar]
- 17.Arnold WK, Savage CR, Antonicello AD, Stevenson B. 2015. Apparent role for Borrelia burgdorferi LuxS during mammalian infection. Infect Immun 83:1347–1353. doi: 10.1128/IAI.00032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yadav MK, Vidal JE, Go YY, Kim SH, Chae SW, Song JJ. 2018. The LuxS/AI-2 quorum-sensing system of Streptococcus pneumoniae is required to cause disease, and to regulate virulence- and metabolism-related genes in a rat model of middle ear infection. Front Cell Infect Microbiol 8:138. doi: 10.3389/fcimb.2018.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krašovec R, Belavkin RV, Aston JA, Channon A, Aston E, Rash BM, Kadirvel M, Forbes S, Knight CG. 2014. Mutation rate plasticity in rifampicin resistance depends on Escherichia coli cell-cell interactions. Nat Commun 5:3742. doi: 10.1038/ncomms4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mou KT, Plummer PJ. 2016. The impact of the LuxS mutation on phenotypic expression of factors critical for Campylobacter jejuni colonization. Vet Microbiol 192:43–51. doi: 10.1016/j.vetmic.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Han X, Bai H, Liu L, Dong H, Liu R, Song J, Ding C, Qi K, Liu H, Yu S. 2013. The luxS gene functions in the pathogenesis of avian pathogenic Escherichia coli. Microb Pathog 55:21–27. doi: 10.1016/j.micpath.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Velusamy SK, Sampathkumar V, Godboley D, Fine DH. 2017. Survival of an Aggregatibacter actinomycetemcomitans quorum sensing luxS mutant in the mouths of Rhesus monkeys: insights into ecological adaptation. Mol Oral Microbiol 32:432–442. doi: 10.1111/omi.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abayneh T, Duncan JC, Henning S. 2013. Edwardsiella piscicida sp. nov.: a novel species pathogenic to fish. J Appl Microbiol 114:644–654. doi: 10.1111/jam.12080. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Yang M, Xiao J, Wu H, Wang X, Lv Y, Xu L, Zheng H, Wang S, Zhao G, Liu Q, Zhang Y. 2009. Genome sequence of the versatile fish pathogen Edwardsiella tarda provides insights into its adaptation to broad host ranges and intracellular niches. PLoS One 4:e7646. doi: 10.1371/journal.pone.0007646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali F, Yao Z, Li W, Sun L, Lin W, Lin X. 2018. In-silico prediction and modeling of the quorum sensing LuxS protein and inhibition of AI-2 biosynthesis in Aeromonas hydrophila. MOLECULES 23:2627. doi: 10.3390/molecules23102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Zhou R, Li T, Kang M, Wan Y, Xu Z, Chen H. 2008. Enhanced biofilm formation and reduced virulence of Actinobacillus pleuropneumoniae luxS mutant. Microb Pathog 45:192–200. doi: 10.1016/j.micpath.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Plummer PJ. 2012. LuxS and quorum-sensing in Campylobacter. Front Cell Infect Microbiol 2:22. doi: 10.3389/fcimb.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J, Ji X, Wang X, Li T, Wang H, Zeng Q. 2019. Identification and characterization of differentially expressed genes in hepatopancreas of oriental river prawn Macrobrachium nipponense under nitrite stress. Fish Shellfish Immunol 87:144–154. doi: 10.1016/j.fsi.2018.12.075. [DOI] [PubMed] [Google Scholar]
- 29.Chao Y, Bergenfelz C, Håkansson AP. 2017. In vitro and in vivo biofilm formation by pathogenic streptococci. Methods Mol Biol 1535:285–299. doi: 10.1007/978-1-4939-6673-8_19. [DOI] [PubMed] [Google Scholar]
- 30.Vasudevan R. 2014. Biofilms: microbial cities of scientific significance. J Microbiol Exp 1:84–98. doi: 10.15406/jmen.2014.01.00014. [DOI] [Google Scholar]
- 31.Wu H, Moser C, Wang H, Høiby N, Song Z. 2015. Strategies for combating bacterial biofilm infections. Int J Oral Sci 7:1–7. doi: 10.1038/ijos.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francolini I, Donelli G. 2010. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol Med Microbiol 593:227–238. doi: 10.1111/j.1574-695X.2010.00665.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Lu R, Ding L, Yan M, Chai X, Chen S, Xie Q. 2012. Polysaccharides, saponins and water decoction of Astragalus membranaceus significantly enhance the non-specific immune response in Spotted maigre (Nibea albifiora). Isr J Aquac 64:2012.743. [Google Scholar]
- 34.Pan T, Yan M, Chen S, Wang X. 2013. Effects of ten traditional Chinese herbs on immune response and disease resistance of Sciaenops ocellatus. Acta Ichthyol Piscat 43:41–49. doi: 10.3750/AIP2013.43.1.06. [DOI] [Google Scholar]
- 35.Giaouris EE, Simões MV. 2018. Pathogenic biofilm formation in the food industry and alternative control strategies, p 309–377. In Grumezescu A, Holban AM (ed), Foodborne diseases, handbook of food bioengineering. Academic Press, Cambridge, MA. [Google Scholar]
- 36.Koo H, Allan RN, Howlin RP, Hall-Stoodley L, Stoodley P. 2017. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol 15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuberi A, Misba L, Khan AU. 2017. CRISPR interference (CRISPRi) inhibition of luxS gene expression in E. coli: an approach to inhibit biofilm. Front Cell Infect Microbiol 7:214. doi: 10.3389/fcimb.2017.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao JF, Wang QY, Liu Q, Wang X, Liu H, Zhang YX. 2008. Isolation and identification of fish pathogen Edwardsiella tarda from mariculture in China. Aquac Res 40:13–17. doi: 10.1111/j.1365-2109.2008.02101.x. [DOI] [Google Scholar]
- 39.Wang X, Yan M, Hu W, Chen S, Zhang S, Xie Q. 2012. Visualization of Sparus macrocephalus Infection by GFP-Labeled Edwardsiella tarda. Isr J Aquac 64:2012.693. [Google Scholar]
- 40.Qin L, Sun Y, Zhao Y, Xu J, Bi K. 2017. In vitro model to estimate Edwardsiella tarda-macrophage interactions using RAW264.7 cells. Fish Shellfish Immunol 60:177–184. doi: 10.1016/j.fsi.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 41.Xiao J, Chen T, Liu B, Yang W, Wang Q, Qu J, Zhang Y. 2013. Edwardsiella tarda mutant disrupted in type III secretion system and chorismic acid synthesis and cured of a plasmid as a live attenuated vaccine in turbot. Fish Shellfish Immunol 35:632–641. doi: 10.1016/j.fsi.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 42.Naresh R, Hampson DJ. 2010. Attraction of Brachyspira pilosicoli to mucin. Microbiology 156:191–197. doi: 10.1099/mic.0.030262-0. [DOI] [PubMed] [Google Scholar]
- 43.He Y, Xu T, Fossheim LE, Zhang XH. 2012. FliC, a flagellin protein, is essential for the growth and virulence of fish pathogen Edwardsiella tarda. PLoS One 7:e45070. doi: 10.1371/journal.pone.0045070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Wang X, Wang X, Feng R, Luo Q, Huang J. 2018. Generation of a novel Streptococcus agalactiae ghost vaccine and examination of its immunogenicity against virulent challenge in tilapia. Fish Shellfish Immunol 81:49–56. doi: 10.1016/j.fsi.2018.06.055. [DOI] [PubMed] [Google Scholar]
- 45.Wang Q, Fu T, Li X, Luo Q, Huang J, Sun Y, Wang X. 2020. Cross-immunity in Nile tilapia vaccinated with Streptococcus agalactiae and Streptococcus iniae vaccines. Fish Shellfish Immunol 97:382–389. doi: 10.1016/j.fsi.2019.12.021. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Wang X, Zhang B, Li Z, Yang L, Li X, Ma H. 2020. A lysin motif-containing protein (SpLysMD3) functions as a PRR involved in the antibacterial responses of mud crab, Scylla paramamosain. Fish Shellfish Immunol 97:257–267. doi: 10.1016/j.fsi.2019.12.036. [DOI] [PubMed] [Google Scholar]
- 47.Reed LJ, Muench HA. 1938. Simple method of estimating fifty percent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 48.Gomes FIA, Teixeira P, Cerca N, Azeredo J, Oliveira R. 2011. Effect of farnesol on structure and composition of Staphylococcus epidermidis biofilm matrix. Curr Microbiol 63:354–359. doi: 10.1007/s00284-011-9984-3. [DOI] [PubMed] [Google Scholar]
- 49.Cerca N, Gomes F, Pereira S, Teixeira P, Oliveira R. 2012. Confocal laser scanning microscopy analysis of S. epidermidis biofilms exposed to farnesol, vancomycin and rifampicin. BMC Res Notes 5:244. doi: 10.1186/1756-0500-5-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sayadi A, Immonen E, Bayram H, Arnqvist G. 2016. The de novo transcriptome and its functional annotation in the seed beetle Callosobruchus maculatus. PLoS One 11:e0158565. doi: 10.1371/journal.pone.0158565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue S, Liu Y, Zhang Y, Sun Y, Geng X, Sun J. 2013. Sequencing and de novo analysis of the hemocytes transcriptome in Litopenaeus vannamei response to white spot syndrome virus infection. PLoS One 8:e76718. doi: 10.1371/journal.pone.0076718. [DOI] [PMC free article] [PubMed] [Google Scholar]