Programmed death-ligand 1 (PD-L1/B7-H1) serves as a cosignaling molecule in cell-mediated immune responses and contributes to chronicity of inflammation and the escape of tumor cells from immunosurveillance. Here, we investigated the molecular mechanisms leading to PD-L1 upregulation in human oral carcinoma cells and in primary human gingival keratinocytes in response to infection with Porphyromonas gingivalis (P. gingivalis), a keystone pathogen for the development of periodontitis.
KEYWORDS: PD-L1, B7-H1, Porphyromonas gingivalis, immune evasion, immune suppression, signaling pathway
ABSTRACT
Programmed death-ligand 1 (PD-L1/B7-H1) serves as a cosignaling molecule in cell-mediated immune responses and contributes to chronicity of inflammation and the escape of tumor cells from immunosurveillance. Here, we investigated the molecular mechanisms leading to PD-L1 upregulation in human oral carcinoma cells and in primary human gingival keratinocytes in response to infection with Porphyromonas gingivalis (P. gingivalis), a keystone pathogen for the development of periodontitis. The bacterial cell wall component peptidoglycan uses bacterial outer membrane vesicles to be taken up by cells. Internalized peptidoglycan triggers cytosolic receptors to induce PD-L1 expression in a myeloid differentiation primary response 88 (Myd88)-independent and receptor-interacting serine/threonine-protein kinase 2 (RIP2)-dependent fashion. Interference with the kinase activity of RIP2 or mitogen-activated protein (MAP) kinases interferes with inducible PD-L1 expression.
INTRODUCTION
The programmed death-ligand 1 (PD-L1/B7-H1) protein participates in cell-mediated immune responses and provides essential immune-regulatory functions in balancing immune reactions during infection and self-recognition. PD-L1 is expressed in different tissues and cell types, including the immune system. Constitutive expression of PD-L1 can be found on macrophages and antigen-presenting cells (APCs), while PD-L1 expression is induced in other cell types such as endothelial, epithelial, and B cells during inflammation (1–3). PD-L1 is recognized by the programmed death-1 (PD-1) receptor, which is expressed on activated T cells (2). PD-1 belongs to the CD28/CTLA-4 family of immunoglobulin proteins (4). Interaction between PD-1 on T cells and PD-L1 on APCs leads to the production of regulatory cytokines such as interleukin (IL)-10 and can induce apoptosis (5). These processes regulate the immune response, and sustained upregulation of PD-L1 supports chronic inflammation (6).
The expression of PD-L1 can be activated by cytokines, the complement system, and bacterial components which are sensed by pattern recognition receptors such as Toll-like receptors (TLRs) or nucleotide-binding and oligomerization domain receptors (NODs) (7).
Upregulation of PD-L1 has been reported after lipopolysaccharide (LPS) stimulation via the TLR4 receptor in macrophages, which led to the suggestion that this molecule may play a role in the immunosuppression observed in severe sepsis (8). In monocytes, the upregulation of PD-L1 is dependent on the NOD-like family member NOD2 (9). NOD2 and the NOD1 receptors contain a caspase recruiting domain (CARD) in the amino-terminal region and are involved in sensing of bacterial peptidoglycan (PDG) moieties (10).This process is important for recognition of pathogens such as Helicobacter pylori (H. pylori) (11).
Peptidoglycan (PDG) is one component of the protective barriers of the bacterial cell wall. PDG consists of glycan strands of alternating beta-1,4-linked N-acetylglucosamine (GlucNAc) and N-acetylmuramic acid (MurNAc), which are cross-linked by short peptide chains (12). The composition and architecture of the final peptidoglycan cell wall are diverse and differ in peptide composition between species (13). In H. pylori, acetyl residues were detected using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for analysis of PDG, and it was shown that acetylation as a mechanism for lysozyme resistance contributes to the survival of the germs in the host (14). Activation of NOD1 and NOD2 triggers downstream signaling by multiple proteins, including receptor-interacting protein kinase 2 (RIP2), which leads to the induction of proinflammatory transcription factors such as nuclear factor-kappa B (NF-κB) (15). These transcription factors, including signal transducer and activator of transcription (STAT) 1 and activator protein-1 (AP-1), jointly trigger inducible PD-L1 expression (16).
Periodontitis is a chronic inflammatory disease induced by bacteria in oral biofilms. Exogenous and/or endogenous factors can change the state of the biofilm from symbiotic to dysbiotic, which also affects microbial composition (17). Porphyromonas gingivalis (P. gingivalis), a keystone pathogen in periodontitis, possesses a variety of virulence factors, which may result in inadequate clearance of bacterial infections (18–20). P. gingivalis sheds outer membrane vesicles (OMVs), which deliver virulence factors into host cells to modulate the host immune response (21, 22). Periodontal infections are one of the most common bacterial infections, which can also lead to oral cancers (23, 24). Not only oral cancers but also malignancies from the breast, lung, pancreas, kidney, and the gastrointestinal tract frequently show high levels of PD-L1. The binding of PD-L1 to its receptor PD-1 on activated T helper cells may lead to anergy, apoptosis, or the development of regulatory T cells, which generates an immune-evasive microenvironment and prevents destruction of the cancer cell by cytotoxic T lymphocytes (25, 26). Thus, PD-L1 is currently being evaluated as a biomarker in cancer immunotherapy (27) and humanized anti-PD-1 or anti-PD-L1 antibodies are used therapeutically (28).
Here, we identified the bacterial components from P. gingivalis that lead to upregulation of PD-L1 in human SCC-25 oral squamous cell carcinoma cells and primary human gingival keratinocytes. The membrane fraction component PDG uses OMVs to enter SCC-25 cells and to trigger a cytosolic receptor in order to induce a RIP2- and mitogen-activated protein kinase (MAPK)-dependent signaling cascade resulting in inducible PD-L1 expression.
RESULTS
OMVs and PDG from P. gingivalis W83 trigger PD-L1 expression.
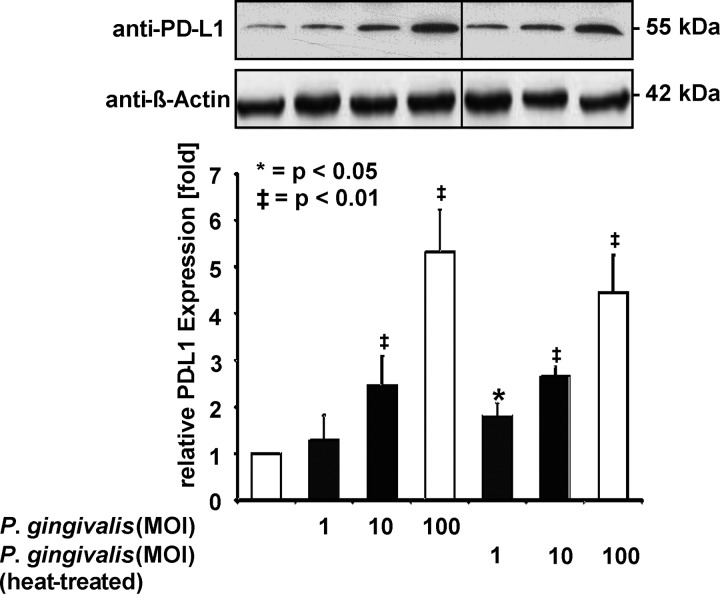
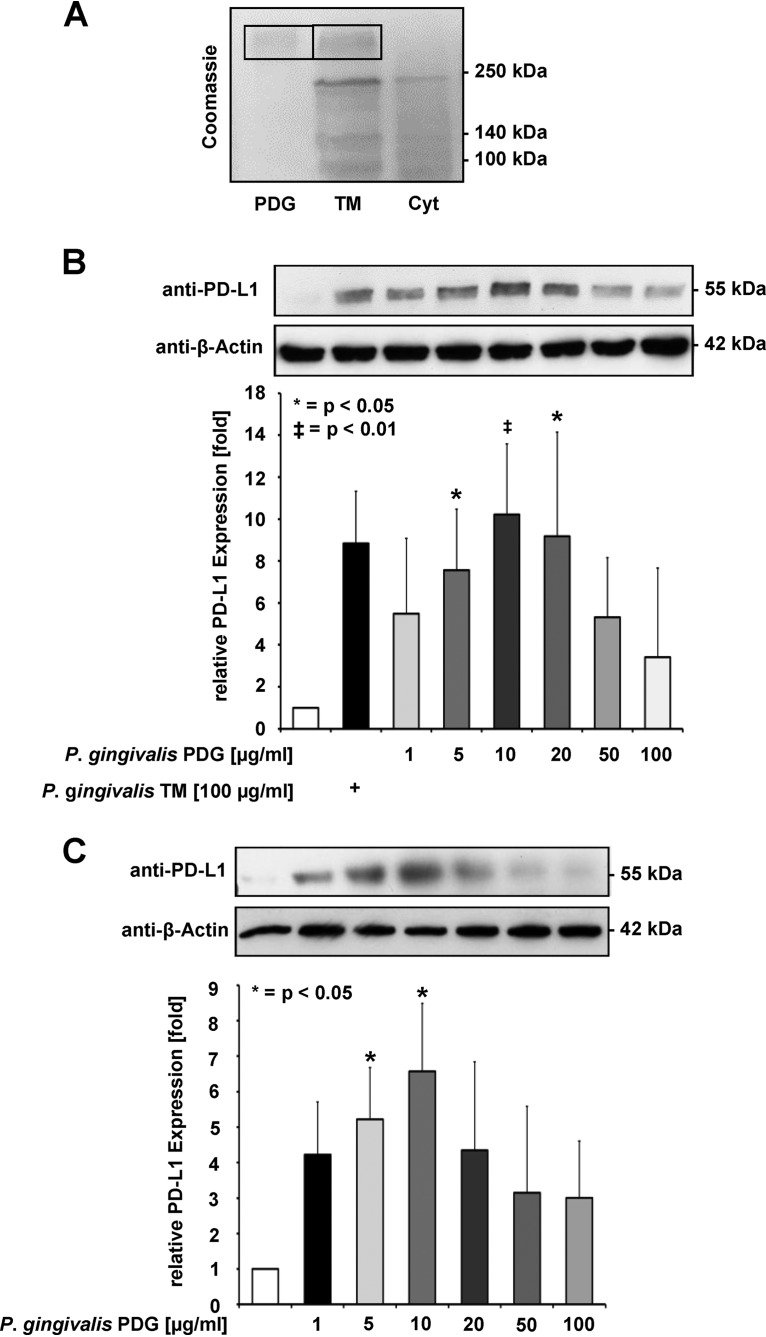
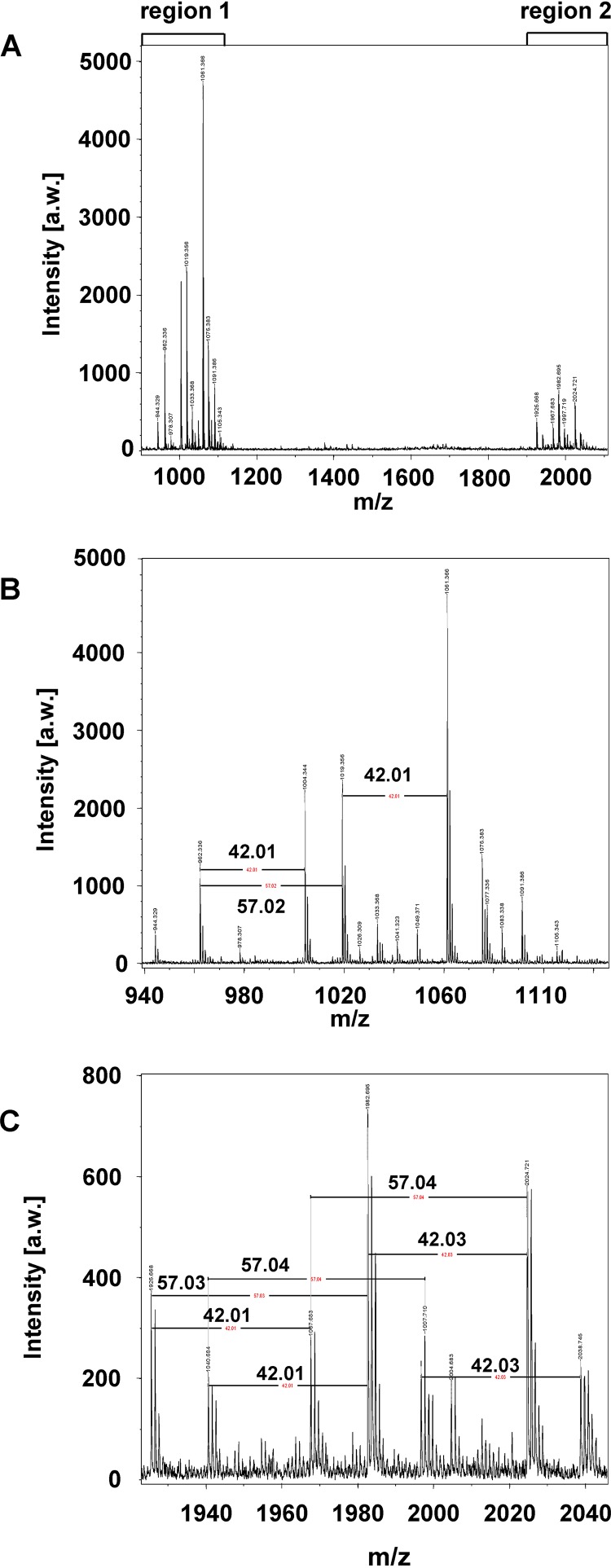
Infection of SCC-25 oral cancer cells with increasing multiplicities of infection (MOIs) of P. gingivalis strain W83 (referred to as P. gingivalis hereafter) triggered a dose-dependent increase of PD-L1 protein expression, as revealed by Western blotting and its quantitative analysis (Fig. 1). This increase was also seen after infection with heat-killed P. gingivalis (Fig. 1), suggesting that no heat-labile components are required for PD-L1 induction. PDG is an important bacterial component with the potential to trigger signaling in infected cells. Therefore, P. gingivalis extracts were fractionated into cytosolic and PDG-containing total membrane (TM) fractions. These fractions were analyzed by SDS-PAGE and Coomassie blue staining. Only the TM fraction contained a band comigrating with PDG, suggesting that this compound is contained in the TM fraction and not in the cytosolic fraction (Fig. 2A). Stimulation of SCC-25 cells with PDG prepared from P. gingivalis or with the TM fraction led to the induction of PD-L1 protein expression in a concentration-dependent manner, showing a peak of PD-L1 upregulation at around 10 μg/ml, as revealed by Western blotting (Fig. 2B). The PDG-induced upregulation was not suppressed by enzymatic digestion of the molecule (Fig. S1 in the supplemental material). Similar to SCC-25 cells, nontransformed primary human gingival keratinocytes (PHGKs) also exhibited this concentration-dependent PD-L1 upregulation, with a maximum at 10 μg/ml P. gingivalis PDG (Fig. 2C). The purity of our P. gingivalis-derived PDG preparations was tested by MALDI-TOF MS analysis after enzymatic treatment and revealed a survey spectrum (Fig. 3A) with two clusters of signals (regions 1 and 2; Fig. 3B and C). Besides these clusters, no other significant signals were detected, suggesting that the preparation was pure. Mass increments of 42 Da and 57 Da on PDG monomers (Fig. 3B and C) are typical for acetyl substituents and glycine-repeating residues, which is consistent with the structure of PDG.
FIG 1.
PD-L1 induction by P. gingivalis. Upper: viable and heat-killed bacteria were added to SCC-25 cells in the indicated MOIs. After 1 day, cells were harvested and equal amounts of protein contained in cell lysates were used for Western blotting using the indicated antibodies, including β-Actin as a loading control. Lower: triplicates of the Western blots were used for protein quantification using the Image J software. Protein amounts in untreated cells were arbitrarily set as 1; the error bars show standard deviations.
FIG 2.
Expression of PD-L1: P. gingivalis extracts were fractionated into cytosolic and PDG-containing total membrane (TM) fractions. (A) P. gingivalis was extracted to generate cytosolic (Cyt) and TM fractions. These fractions were separated together with a PDG control by SDS-PAGE and the gel was stained with Coomassie blue. The positions of molecular weight markers are shown; the position of PDG is indicated by a box. (B) The TM fraction and PDG in the indicated concentrations were used to treat SCC-25 cells for 1 day, followed by analysis of PD-L1 expression using Western blot (upper) and a quantitative analysis of three Western blots (lower). (C) Upper: primary human gingival keratinocytes (PHGKs) were incubated with P. gingivalis PDG at the indicated concentrations for 1 day. Cells were harvested and equal amounts of protein contained in cell lysates were used for Western blotting using PD-L1 antibodies. Lower: the results from three Western blots were quantified by Image J. PD-L1 protein expression from unstimulated cells was arbitrarily set as 1; the error bars show standard deviations; *, P < 0.05.
FIG 3.
Prepared muropeptides treated with mutanolysin from Streptomyces globisporus ATCC 21553 were used to analyze P. gingivalis-derived peptidoglycan using MALDI-TOF mass spectrometry. (A) Overview of the spectrum indicating two clusters of signals (regions 1 and 2). (B and C) Both clusters are composed of repeating motifs. The annotated mass differences of 42 Da show the presence of acetyl residues, whereas the mass differences of 57 Da indicate glycine-repeating units. Besides these clusters, no significant signals were detectable, indicating that the preparation was pure.
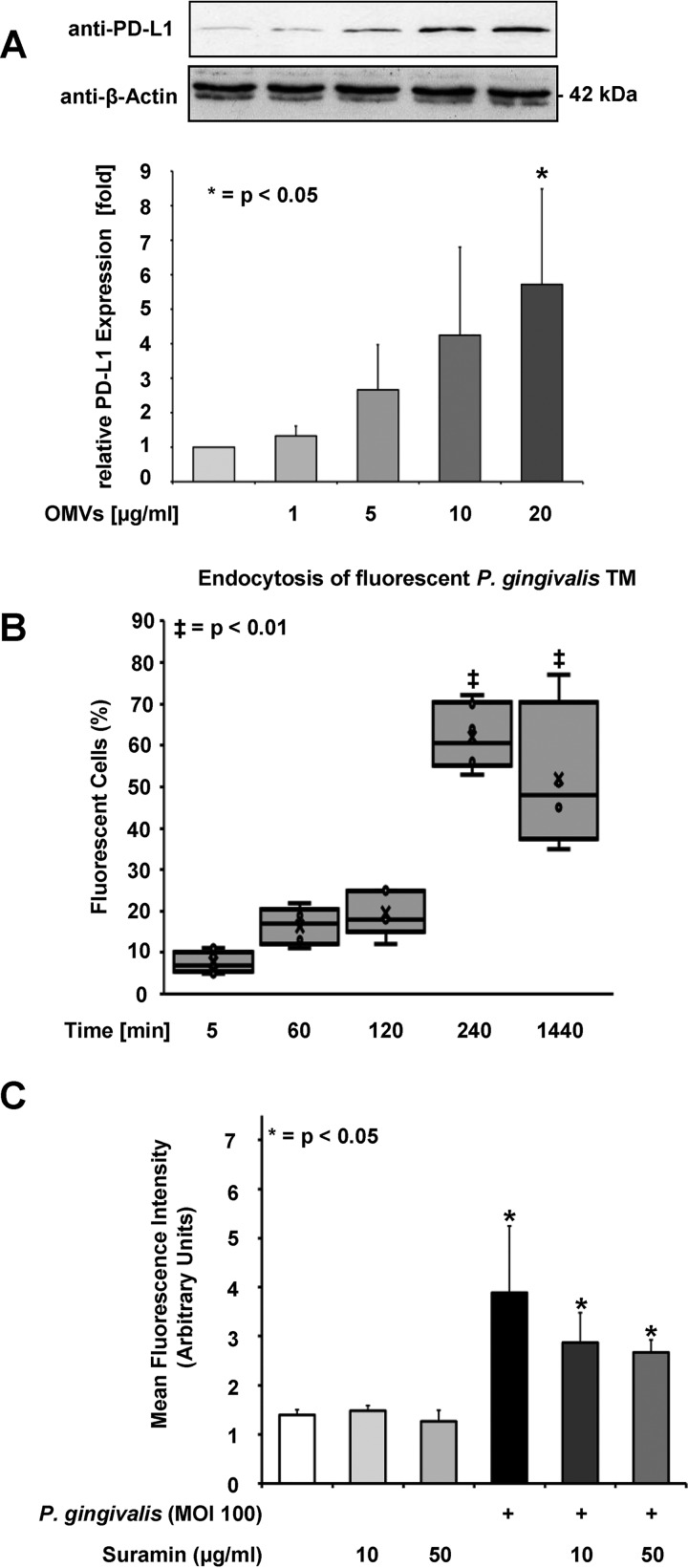
PDG and other ligands of pattern recognition receptors are often contained in OMVs that are shed by Gram-negative bacteria such as P. gingivalis (29). P. gingivalis-derived OMVs may enter the host cell via endocytosis to trigger cellular responses. OMVs derived from P. gingivalis strongly induced PD-L1 expression in SCC-25 cells at a concentration of 20 μg/ml (Fig. 4A). Further experiments showed the principal capacity of SCC-25 cells for endocytosis, and the time-dependent uptake of fluorescently labeled dextran (Fig. S2). The internalized total membrane (TM) fraction from P. gingivalis was labeled with a fluorochrome ester and internalized by SCC-25 cells in a time-dependent manner, reaching its maximum after 4 h and lasting until 24 h (Fig. 4B). Many pattern recognition receptors are contained in the cell membrane (30, 31). After incubation with different concentrations of suramin, an inhibitor of a broad range of membrane surface receptors (32, 33), PD-L1 expression remained unaffected (Fig. 4C), which excludes the involvement of specific surface receptors for the induction of PD-L1 expression. The biological activity of suramin was tested by its inhibitory effect on the viability of immortalized human gingival keratinocytes (IHGKs) (Fig. S3). Mitchen et al. (34) demonstrated the inhibitory effect of suramin in epithelial cells derived from normal, benign hyperplastic and cancerous human prostate tissue.
FIG 4.
OMVs and PDG from P. gingivalis strain W83 trigger PD-L1 expression. (A) SCC-25 cells were treated with the indicated concentrations of OMVs for 1 day and PD-L1 expression was determined by Western blotting (upper) and shown as the quantification of three independent experiments (lower). Triplicates of the Western blots were used for protein quantification using the Image J software. Protein amounts in untreated cells were arbitrarily set as 1; the error bars show standard deviations. (B) The P. gingivalis TM fraction was labeled with a fluorochrome ester and added to the cells for the indicated periods. Flow cytometry was used to determine the fluorescence uptake. Data represent the mean ± SD of 5 independent experiments. The results are shown as box plots. The boxes represent the 2nd and 3rd quartiles as the middle 50% of the scores. The line that divides the boxes displays the median. The lower and upper whiskers indicate the 1st and 4th quartiles. The crosses mark the mean values. (C) Cells were incubated with suramin at the indicated concentrations and infected with P. gingivalis (MOI 100) for 1 day and the PD-L1 expression was investigated using flow cytometry. The quantity of the PD-L1 expression is described as mean fluorescence intensity (MFI), provided in arbitrary units. All investigations were performed in three different independent experiments. The results (MFI of P. gingivalis infected against noninfected cells) were analyzed using independent two-sample Student’s t tests. The character of the evaluation was explorative. The probability of error was set to 5% and shown as P values; n = 3, *, P < 0.05; ‡, P < 0.01.
Cellular mechanisms mediating P. gingivalis-triggered PD-L1 expression.
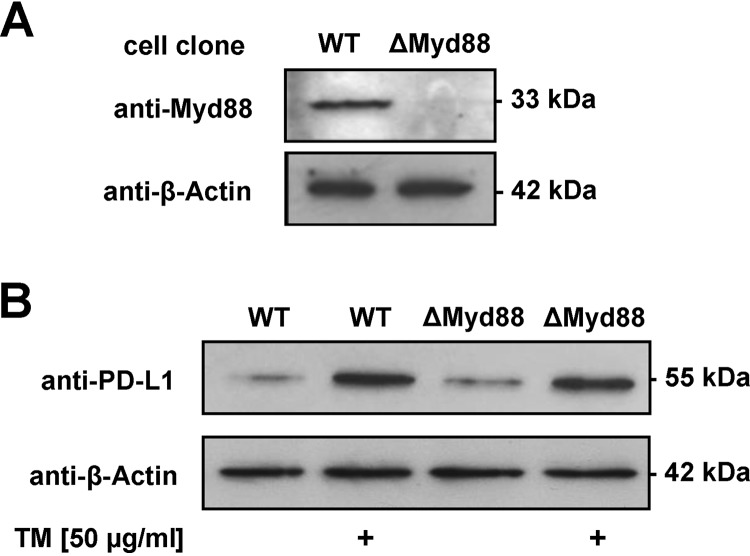
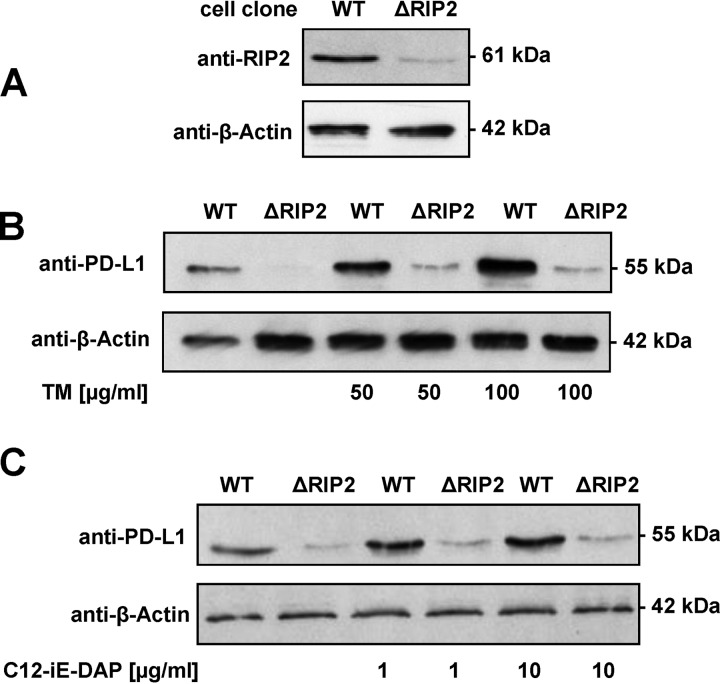
In colonic stromal and plasma cells, induction of PD-L1 expression has been shown to depend on myeloid differentiation primary response 88 (MyD88) (35, 36). To test whether induction of PD-L1 expression depends on MyD88, as has been suggested, MyD88 expression was altered by CRISPR-Cas9-mediated gene engineering (Fig. 5A). MyD88-deficient SCC-25 cells showed a largely unchanged induction of PD-L1 expression in response to stimulation with the TM fraction of P. gingivalis (Fig. 5B). Receptor-interacting protein kinase 2 (RIP2) plays an essential role in immune signaling (37, 38). The promoter region of RIP2 in SCC-25 cells was also mutated using CRISPR-Cas9, resulting in a strongly diminished RIP2 expression (Fig. 6A). In contrast to the wild-type (WT) cell line, the cells with impaired RIP2 expression showed only a very weak residual induction of PD-L1 protein levels after stimulation with P. gingivalis TM (Fig. 6B) or the NOD-agonistic PDG motif acetylated d-glutamyl-meso-diaminopimelic acid (C12-iE-DAP) (Fig. 6C).
FIG 5.
Myd88-independent PD-L1 induction by P. gingivalis. (A) SCC-25 cells were transfected with a plasmid encoding a Myd88-specific Cas9 enzyme cleaving in the first exon and individual cell clones were tested for expression of Myd88 and Cas9 by Western blotting as shown. (B) The indicated control and Myd88-deficient SCC-25 cells were stimulated with P. gingivalis TM and PD-L1 expression was analyzed by Western blotting.
FIG 6.
RIP2-dependent PD-L1 induction by P. gingivalis. (A) SCC-25 cells were transfected with a plasmid encoding a RIP2-specific Cas9 enzyme cleaving in the promoter just upstream from the transcription start site and individual cell clones were tested for expression of RIP2 by Western blotting as shown. (B) Cells with wild-type and reduced levels of RIP2 were treated for 1 day with the indicated concentrations of P. gingivalis TM and analyzed by Western blotting for PD-L1 expression as shown. (C) The experiment was done as described for panel B except that C12-iE-DAP was used as a stimulating agent.
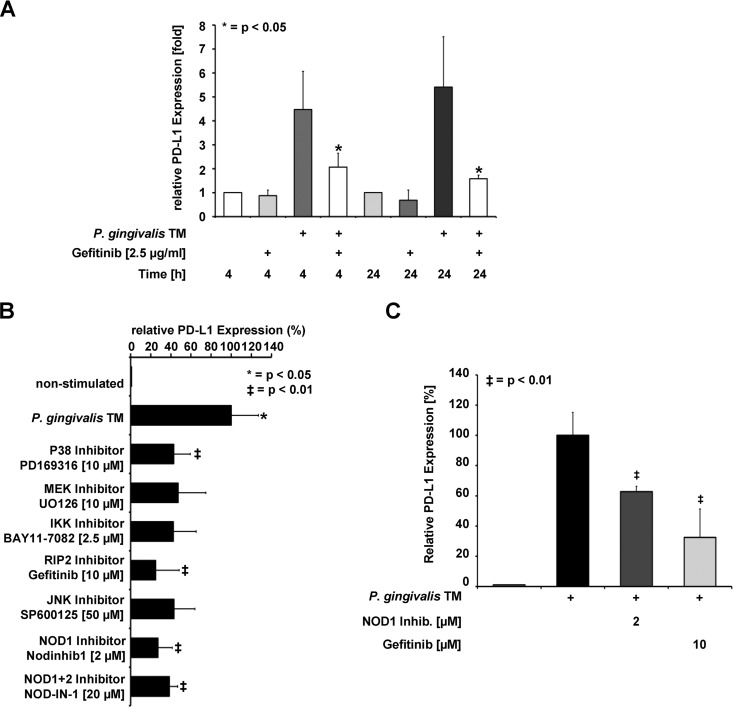
The impact of the RIP2 inhibitor gefitinib (39, 40) on inducible PD-L1 expression was tested by an independent experimental approach. Preincubation of cells with a low dose of gefitinib largely prevented PD-L1 expression triggered by TM extracts of P. gingivalis (Fig. 7A). Stimulation of pathogen recognition factors typically triggers MAPK signaling pathways that ultimately lead to the phosphorylation and activation of transcription factors. The MAP kinase 1/2 (MEK1/2) proteins are central signal integrators that relay upstream signals to downstream kinases, including extracellular signal-regulated kinases (ERKs) (41). Inhibition of the canonical NF-κB activation pathway by a specific IκB kinase (IKK) inhibitor caused a slight reduction of PD-L1 expression that was not statistically significant. Inhibition of all three major MAPK signaling modules (JNK, p38, and MEK1/2-ERK1/2) resulted in a significant, but incomplete, impairment of inducible PD-L1 expression (Fig. 7B), suggesting a significant, but not exclusive, contribution of MAPK signaling for this pathway. Directly targeting NOD1 or NOD1 together with NOD2 by specific inhibitors resulted in diminished PD-L1 upregulation (Fig. 7B). As this inhibition was not made stronger by an inhibitor targeting both NOD isoforms, we presume that NOD1 plays a critical role for this process. The importance of NOD1 for P. gingivalis-triggered PD-L1 upregulation was also evident in experiments with the primary human gingival keratinocytes (PHGKs) (Fig. 7C).
FIG 7.
P. gingivalis-induced expression of PD-L1 depends on the kinase activity of RIP2 and MAPKs. (A) SCC-25 cells were incubated with the RIP2 inhibitor gefitinib and stimulated with P. gingivalis TM for 4 and 24 h as shown, followed by the analysis of PD-L1 expression by Western blotting. (B) SCC-25 cells were incubated with the indicated MAPK and NOD inhibitors as shown and stimulated with the P. gingivalis TM fraction. PD-L1 protein expression was scored by immunoblotting and the results from three Western blots were quantified by Image J. Maximal PD-L1 protein expression from stimulated cells was arbitrarily set as 100%; the error bars show standard deviations. Triplicates of the Western blots were used for protein quantification using the Image J software; the error bars show standard deviations. (C) PHGKs were stimulated with the P. gingivalis TM fraction in the presence of the indicated inhibitors for 24 h. The analysis of PD-L1 expression was revealed by Western blotting; the results from three Western blots were quantified by Image J. Maximal PD-L1 protein expression from stimulated cells was arbitrarily set as 100%; the error bars show standard deviations.
DISCUSSION
Chronic inflammation can contribute to tumor development (42) and an association between periodontitis and the development of oral cancer has been described (43). Previous studies showed that P. gingivalis upregulates PD-L1 in oral cancer cells and in primary, as well as immortalized, human gingival keratinocytes (44). High constitutive levels of PD-L1 were detected in invasive oral squamous carcinoma cells (45) and in tissue samples of oral squamous cell carcinomas (46). PD-L1 expression is less of an oncogenic driver, but rather a coopted and maladaptive immune shield that protects the tumor from its immune microenvironment (27). Although upregulation of the PD-1/PD-L1 pathway may be important in limiting damage to the host, there is no doubt that impairing T cell responses may be beneficial for the invading pathogens (47). The relevance of PD-L1 expression levels for inflammation and eventually for development of cancer has put this protein at the forefront of many studies (48, 49).
The kinetics of P. gingivalis-induced PD-L1 expression is similar to that observed upon infection of gastric cells with H. pylori (50, 51). The present study shows that PD-L1 upregulation is caused by viable as well as heat-killed bacteria, indicating that PD-L1 expression is induced by heat-stable components and does not depend on viable P. gingivalis. Membrane components of P. gingivalis, such as PDG, are responsible for the upregulation of PD-L1. Consistent with published literature (52), these components are delivered to the infected cells in OMVs.
The rapid internalization of the labeled P. gingivalis TM fraction into SCC-25 cells suggests that these components are sensed by intracellular receptors such as NOD1 and NOD2, as has been shown for P. gingivalis-infected epithelial HEK-Blue cells (52). From a study addressing the development of Crohn’s disease, it is known that NOD2 serves as a sensor of peptidoglycan (53). Another study revealed that NOD1 detects peptidoglycan within early endosomes and promotes RIP2-dependent autophagy and inflammatory signaling in response to bacterial infection (54). Both NOD receptors are expressed in SCC-25 cells (Fig. S4), and NOD1 can be activated by its ligand C12-iE-DAP in these cells (see Fig. 6C). Direct inhibition of NOD1 leads to suppressed PD-L1 upregulation both in primary and in transformed cells (Fig. 7B and C), which underlines its relevance for this activation pathway. It was reported that NOD1 activation also plays a role in PD-L1 upregulation in liver sinusoidal endothelial cells (55). Consistent with the concept of an intracellular receptor for the induction of P. gingivalis-triggered signaling, interference with cell surface receptor function by suramin did not prevent inducible PD-L1 expression.
Downstream signaling seems to be Myd88-independent and to depend on RIP2, as revealed by knockout experiments and the use of the small molecule inhibitor gefitinib. RIP2 is a member of the RIP kinase family and contains an N-terminal kinase domain, a bridging intermediate domain, and a C-terminal CARD domain (56). Ligand binding triggers NOD2 oligomerization, which leads to the recruitment of RIP2 via CARD-CARD interactions, causing the formation of RIP2 filaments (57). This in turn leads to the activation of downstream effectors such as NF-κB and MAPKs (58). Inducible PD-L1 expression was only mildly affected by IKK inhibition, which indicates that at least the canonical NF-κB activation pathway is of limited relevance. Inhibition of MAPK signaling significantly interfered with PD-L1 expression, showing the functional relevance of these downstream effectors.
Upregulation of PD-L1 on tumor cells is one mechanism which protects these cells from host response and is of clinical relevance for the progression of the disease (65). The properties of P. gingivalis peptidoglycan in the upregulation of receptors on tumor cells make this Gram-negative anaerobic rod a pathogen comparable to H. pylori. With respect to the abundance of this mechanism in periodontitis and periodontal diseases, antimicrobial strategies may not only help to improve periodontal health but also have an impact on general health, especially in patients suffering from oral cancer.
MATERIALS AND METHODS
Cell culture.
The human oral tongue squamous cell carcinoma cell line SCC-25 was purchased from the DSMZ (number ACC 617; German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). Cells were cultured in a medium containing Dulbecco’s minimal essential medium (DMEM):Ham’s F12 (4:1 [vol/vol]), HEPES buffer, (Invitrogen, Karlsruhe, Germany), and 10% fetal calf serum (FCS) (Greiner, Frickenhausen, Germany). Primary human gingival keratinocytes (PHGKs) were obtained from gingival biopsy specimens of healthy volunteers, prepared and cultured in a serum-free medium containing DMEM:Ham’s F12 (4:1 [vol/vol]) and 10 mM HEPES (Invitrogen, Karlsruhe, Germany). The cells were seeded in 6-well plates at 1 × 106 cells per well and grown at 37°C in a humidified atmosphere with 5% CO2 to 80% confluence before stimulation.
Bacteria and growth conditions.
Porphyromonas gingivalis strain W83 was purchased from the American Type Culture Collection (ATCC) (LGC Standards GmbH, Wesel, Germany) and grown at 37°C in brain heart infusion broth (Difco, BD, Heidelberg, Germany) supplemented with hemine (5 μg/ml) and menadione (1 μg/ml) (Sigma-Aldrich, Munich, Germany) under anaerobic conditions using the Anaerocult A System (Merck, Darmstadt, Germany) until late log phase (OD = 1.0). Optical density (OD) was measured using a spectrophotometer at 600 nm with an optical density of 1.0 corresponding to 109 cells/ml. Heat inactivation of P. gingivalis cells was performed by centrifugation of cells followed by washing in phosphate-buffered saline (PBS), followed by incubation for 10 min at 70°C. Heat killing of bacteria was confirmed by plating cells on Brucella agar plates. Bacterial suspensions were adjusted to a concentration of 109 cells/ml using optical density measurements.
Infection and stimulation.
SCC-25 cells and primary human gingival keratinocytes (PHGKs) were seeded in 6-well plates (1 × 106 cells/well) and infection with P. gingivalis was performed in the indicated MOIs. Alternatively, cells were treated with OMVs, cell fractions, PDG, or C12-iE-DAP (tlrl-c12dap; InvivoGen) at the indicated concentrations. Inhibitors such as suramin or kinase inhibitors were added prior to infection. The biological activity of suramin was investigated by testing its inhibitory effect on the viability of epithelial cells using a cell proliferation kit I (tetrazolium salt, MTT) (Merck). Immortalized human gingival keratinocytes (IHGKs) were seeded at 1 × 104 cells per well in 96-well plates in 100 μl modified Lauer medium (59). After 24 h, medium was exchanged to a medium containing 10% FCS. Suramin was added freshly every day in the concentrations of 100 μM and 500 μM. The MTT test was performed following the manufacturer’s instructions after 24, 48, and 72 h.
Flow cytometric analysis.
For flow cytometric analysis, cells were harvested by scraping and stained with allophycocyanin-conjugated anti-human PD-L1 (eBioscience) in a dilution of 1:5 and incubated for 45 min at 4°C. Following two washing steps, cells were resuspended in Cytofix fixation buffer (BD Bioscience) and analyzed using a Cyan ADP flow cytometer (Dako). The quantity of the PD-L1 expression is described as mean fluorescence intensity (MFI), provided in arbitrary units. The investigations were performed in three different independent experiments. The results were analyzed using independent two-sample Student’s t tests. The character of the evaluation was explorative. Probability of error was set to 5% and shown as P values.
Membrane preparation.
P. gingivalis membrane preparations were produced as previously described (60). Briefly, harvested bacteria were disrupted by four passages through a high-pressure cell disruption device and membranes were sedimented from cleared lysate at 150,000 × g for 2 h at 4°C. After addition of DNase and RNase, the supernatant (cytosolic fraction) was isolated. Total membrane fractions were washed three times and stored in 10 mM HEPES, pH 7.4 at −20°C.
OMV isolation.
OMVs were prepared as published (61). P. gingivalis was grown in 500-ml cultures until late log phase (OD600 = 1.0) was reached, followed by pelleting of cells by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was filtered through 0.4 μm filters followed by filtration through 0.2 μm filters to remove the remaining bacterial cells. OMV were obtained by centrifugation at 100,000 × g for 60 min at 4°C. Pelleted vesicles were resuspended in 50 mM HEPES pH 6.8 and filtered through 0.22 μm Ultrafree spin filters (Millipore). Vesicles were again pelleted by centrifugation at 100,000 × g for 60 min at 4°C and finally resuspended in an adequate volume of 50 mM HEPES, pH 6.8, and stored at −80°C until use. The protein concentration was determined using the Pierce BCA protein assay kit (ThermoFisher).
Determination of endocytosis.
The agent pHrodo Green dextran (ThermoFisher) was used to test the occurrence of endocytosis in SCC-25 cells. Cells were seeded in 6-well plates at a density of 106 cells per well. The dextran was added at a concentration of 20 μg/ml and, after different time points, the cells were harvested by scraping. Cells were washed in PBS containing 2% (vol/vol) FCS and resuspended in fixation buffer (BD Bioscience). After filtration through a 50-μm filter unit, cells were analyzed in a Cyan ADP (Beckmann-Coulter) flow cytometer. Internalization of the P. gingivalis TM fraction by labeling with the fluorochrome pHrodo green STP ester (ThermoFisher) was performed according to the manufacturer’s protocol. The ester was diluted in dimethyl sulfoxide (DMSO) to generate a stock solution and further diluted in 100 mM sodium bicarbonate buffer, pH 8.5. After addition of the TM fraction/OMVs and incubation for 1 h at room temperature in the dark, the labeled TM was dialyzed for 24 h in 3 × 2 liters of 10 mM HEPES buffer. The labeled TM was added to the cells and the cells were harvested after the chosen time points as described above.
Cells were analyzed using a Cyan ADP flow cytometer (Dako). The quantities of fluorescent cells are provided in percentages. The investigations were performed in three different independent experiments. The results were analyzed using independent two-sample Student’s t tests. The character of the evaluation was explorative. The probability of error was set to 5% and shown as P values.
PDG isolation and characterization.
P. gingivalis PDG was isolated according to the protocol described by Desmarais (62) with modifications. Briefly, a late logarithmic culture of P. gingivalis was centrifuged at 8,000 × g for 10 min. The pellet was resuspended and trichloric acid (10% [vol/vol]) was added and the samples were incubated for 30 min at 4°C. Cells were washed three times in PBS and after the last wash, slowly pipetted into boiling SDS (final concentration 4% [wt/vol]). Cells were boiled for 3 h and stirred overnight without heat. Samples were centrifuged at 150,000 × g for 60 min at room temperature to pellet the PDG polymers. Pellets were resuspended in water and washed three times to remove the remaining SDS. After the last washing step, samples were resuspended in 10 mM Tris-HCl (Roth), Proteinase K (Sigma) was added, and samples were incubated overnight. Proteinase K digestion was stopped by incubating the samples at 70°C for 20 min. Samples were centrifuged 150,000 × g for 60 min at room temperature, weighed, and resuspended in water. To assess purity, the PDG was loaded on a 10% (wt/vol) SDS gel and low voltage (40 V) was applied for 2 h. Gels were fixed in 50% (vol/vol) methanol and 10% (vol/vol) glacial acetic acid overnight with gentle agitation. Afterward, gels were stained in 0.1% (wt/vol) Coomassie brilliant blue R-250 (Sigma) in 50% (vol/vol) methanol and 10% (vol/vol) glacial acetic acid for 20 min and then destained in 40% (vol/vol) methanol and 10% (vol/vol) glacial acetic acid.
To obtain a preparation with defined fractions of PDG for further characterization by mass spectrometry, an enzymatic digestion was performed by modified protocol of Glauner et al. (63). The PDG was digested using mutanolysin from Streptomyces globisporus ATCC 21553 (Sigma-Aldrich) in a concentration of 2,500 U/ml in Tris/EDTA (TE, 10 mM Tris/HCl, 1 mM EDTA) buffer (pH 6.3) at 37°C overnight. The enzymatic reaction was stopped by boiling for 3 min.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was performed on an Ultraflex TOF/TOF mass spectrometer (Bruker Daltonics, Bremen) equipped with a nitrogen laser and a LIFT-MS/MS facility. The instrument was operated in the positive-ion reflectron mode using 2.5-dihydroxybenzoic acid and methylendiphosphonic acid as matrix. Sum spectra consisting of 200 to 400 single spectra were acquired. For data processing and instrument control, the Compass 1.4 software package consisting of FlexControl 4.4, FlexAnalysis 3.4 4, Sequence Editor and BioTools 3.2 was used. External calibration was performed with a peptide standard (Bruker Daltonics).
CRISPR-Cas9 knockouts.
The plasmid pSpCas9(BB)-2A-Puro (pX459) (64) was used to target MyD88 with the sequence 5′-GTACTTGGAGATCCGGCAAC-3′ and RIP2 with the sequence 5′-CGTCCGCCCGCCACGCAGAC-3′. SCC-25 cells were transfected with the plasmids and selected by adding 0.6 μg/ml puromycin for 3 days. Survivors were picked and transferred to 96-well plates. Cell clones were analyzed for MyD88 and RIP2 expression using Western blotting.
Western blotting.
Cell lysis was done in radioimmunoprecipitation (RIPA)-buffer (ThermoFisher) and protein concentrations were determined by bicinchoninic acid (BCA) assays. Equal amounts of protein (20 μg) were loaded on an SDS gel and proteins were transferred to a nitrocellulose membrane by semidry transfer (Bio-Rad, Turboblotting). Successful protein transfer was tested by Ponceau S staining. For protein detection, membranes were blocked in 2% powdered milk for 1 h and then incubated in primary antibody (rabbit anti-human PD-L1, Thermo no. PA-5-20343; goat anti-human NOD1, Thermo no. PA-5-18027; goat anti-human NOD2, Thermo no. PA-5-18572; mouse anti-Flag antibody [M2], Sigma-Aldrich no. F3165; rabbit anti-human MyD88, Abcam no. ab2064; rabbit anti-human RIP2 Sigma-Aldrich no. HPA015273) overnight at 4°C. After washing, the membranes were incubated with secondary antibody (goat anti-Rabbit, Thermo no. 32460 and rabbit anti-goat IgG-HRP, Life Technologies no. 61-1820) (both 1:500 in 2% milk powder) for 1 h at room temperature. Antibody specificity for PD-L1 was checked using a blocking peptide (BP) (Thermo number PEP-0463) in a preadsorption assay. Following three washing steps, the blots were incubated with enhanced chemiluminescence (ECL) reagent (Bio-Rad). Chemiluminescence was detected using X-ray films (Amersham). β-Actin was used as loading control (1:5,000). X-ray films were scanned, and band intensity was measured by ImageJ software (SciJava). Triplicates of the Western blots were used for protein quantification using the Image J software. Protein amounts in untreated cells were arbitrarily set as 1; the error bars show standard deviations. The results (values of stimulated against nonstimulated cells) were analyzed using independent two-sample Student’s t tests. The probability of error was set to 5% and shown as P values, n = 3, * = P < 0.05, ‡= P < 0.01.
Statistical analysis.
All investigations were performed in three different independent experiments. The results were analyzed using independent two-sample Student’s t tests. The character of the evaluation was explorative. The probability of error was set to 5% and shown as P values, n = 3, * = P < 0.05, ‡ = P < 0.01.
Ethical considerations.
All experiments followed the guidelines of good clinical/laboratory practice (GCP/GLP) and the World Health Organization declaration, Helsinki 1964, latest update Seoul 2008 (59th WMA General Assembly, Seoul, October 2008).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the von-Behring-Röntgen-Stiftung. We thank Uwe Mamat, Manuel Hein, and Dörte Grella (Research Center Borstel) for the preparation of P. gingivalis membrane. The work of M.L.S. is supported by grants from the Deutsche Forschungsgemeinschaft SFB/TRR81, SFB1021, and SFB1213, and the GRK “Das inflammatorische Tumorsekretom: Vom grundlegenden Verständnis zu neuen Therapien.”
We declare no competing interests.
Author contribution statement: S. Groeger, M. L. Schmitz, and F. Denter wrote the manuscript and performed the experiments. G. Lochnit performed the MALDI-TOF analysis. M. L. Schmitz supported the knockout experiments. J. Meyle initiated the project and reviewed the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Chen Y, Zhang J, Guo G, Ruan Z, Jiang M, Wu S, Guo S, Fei L, Tang Y, Yang C, Jia Z, Wu Y. 2009. Induced B7-H1 expression on human renal tubular epithelial cells by the sublytic terminal complement complex C5b-9. Mol Immunol 46:375–383. doi: 10.1016/j.molimm.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, Ling V, Bowman MR, Carreno BM, Collins M, Wood CR, Honjo T. 2000. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamazaki T, Akiba H, Iwai H, Matsuda H, Aoki M, Tanno Y, Shin T, Tsuchiya H, Pardoll DM, Okumura K, Azuma M, Yagita H. 2002. Expression of programmed death 1 ligands by murine T cells and APC. J Immunol 169:5538–5545. doi: 10.4049/jimmunol.169.10.5538. [DOI] [PubMed] [Google Scholar]
- 4.Ishida Y, Agata Y, Shibahara K, Honjo T. 1992. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J 11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Chen L. 2004. T lymphocyte co-signaling pathways of the B7-CD28 family. Cell Mol Immunol 1:37–42. [PubMed] [Google Scholar]
- 6.Vakkila J, Lotze MT. 2004. Inflammation and necrosis promote tumour growth. Nat Rev Immunol 4:641–648. doi: 10.1038/nri1415. [DOI] [PubMed] [Google Scholar]
- 7.An LL, Gorman JV, Stephens G, Swerdlow B, Warrener P, Bonnell J, Mustelin T, Fung M, Kolbeck R. 2016. Complement C5a induces PD-L1 expression and acts in synergy with LPS through Erk1/2 and JNK signaling pathways. Sci Rep 6:33346. doi: 10.1038/srep33346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loke P, Allison JP. 2003. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc Natl Acad Sci U S A 100:5336–5341. doi: 10.1073/pnas.0931259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hewitt RE, Pele LC, Tremelling M, Metz A, Parkes M, Powell JJ. 2012. Immuno-inhibitory PD-L1 can be induced by a peptidoglycan/NOD2 mediated pathway in primary monocytic cells and is deficient in Crohn’s patients with homozygous NOD2 mutations. Clin Immunol 143:162–169. doi: 10.1016/j.clim.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Moreira LO, Zamboni DS. 2012. NOD1 and NOD2 signaling in infection and inflammation. Front Immunol 3:328. doi: 10.3389/fimmu.2012.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Memet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol 5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 12.Vollmer W. 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev 32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 13.Turner RD, Vollmer W, Foster SJ. 2014. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol 91:862–874. doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G, Lo LF, Forsberg LS, Maier RJ. 2012. Helicobacter pylori peptidoglycan modifications confer lysozyme resistance and contribute to survival in the host. mBio 3:e00409-12. doi: 10.1128/mBio.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy JV, Ni J, Dixit VM. 1998. RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J Biol Chem 273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 16.Ritprajak P, Azuma M. 2015. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol 51:221–228. doi: 10.1016/j.oraloncology.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Kumar PS, Mason MR, Brooker MR, O'Brien K. 2012. Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J Clin Periodontol 39:425–433. doi: 10.1111/j.1600-051X.2012.01856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat Rev Microbiol 10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamont RJ, Hajishengallis G. 2015. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med 21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie H. 2015. Biogenesis and function of Porphyromonas gingivalis outer membrane vesicles. Future Microbiol 10:1517–1527. doi: 10.2217/fmb.15.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veith PD, Chen Y-Y, Gorasia DG, Chen D, Glew MD, O'Brien-Simpson NM, Cecil JD, Holden JA, Reynolds EC. 2014. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res 13:2420–2432. doi: 10.1021/pr401227e. [DOI] [PubMed] [Google Scholar]
- 23.Tezal M, Grossi SG, Genco RJ. 2005. Is periodontitis associated with oral neoplasms? J Periodontol 76:406–410. doi: 10.1902/jop.2005.76.3.406. [DOI] [PubMed] [Google Scholar]
- 24.Tezal M, Sullivan MA, Reid ME, Marshall JR, Hyland A, Loree T, Lillis C, Hauck L, Wactawski-Wende J, Scannapieco FA. 2007. Chronic periodontitis and the risk of tongue cancer. Arch Otolaryngol Head Neck Surg 133:450–454. doi: 10.1001/archotol.133.5.450. [DOI] [PubMed] [Google Scholar]
- 25.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L. 2002. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 26.Wu CP, Jiang JT, Tan M, Zhu YB, Ji M, Xu KF, Zhao JM, Zhang GB, Zhang XG. 2006. Relationship between co-stimulatory molecule B7-H3 expression and gastric carcinoma histology and prognosis. World J Gastroenterol 12:457–459. doi: 10.3748/wjg.v12.i3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel SP, Kurzrock R. 2015. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther 14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 28.Zitvogel L, Kroemer G. 2012. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology 1:1223–1225. doi: 10.4161/onci.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medzhitov R, Janeway C Jr. 2000. The Toll receptor family and microbial recognition. Trends Microbiol 8:452–456. doi: 10.1016/S0966-842X(00)01845-X. [DOI] [PubMed] [Google Scholar]
- 31.Medzhitov R, Janeway C Jr. 2000. Innate immune recognition: mechanisms and pathways. Immunol Rev 173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 32.Hensey CE, Boscoboinik D, Azzi A. 1989. Suramin, an anti-cancer drug, inhibits protein kinase C and induces differentiation in neuroblastoma cell clone NB2A. FEBS Lett 258:156–158. doi: 10.1016/0014-5793(89)81639-4. [DOI] [PubMed] [Google Scholar]
- 33.Trieb K, Dorfinger K, Neuhold N, Selzer E, Wilfing A, Czernin S, Hermann M, Niederle B, Gessl A, Vierhapper H. 1992. Suramin affects differentiated and undifferentiated human thyroid epithelial cells in vitro. J Endocrinol 134:505–511. doi: 10.1677/joe.0.1340505. [DOI] [PubMed] [Google Scholar]
- 34.Mitchen J, Rago R, Wilding G. 1993. Effects of suramin on the proliferation of primary epithelial cell cultures derived from normal, benign hyperplastic and cancerous human prostates. Prostate 22:75–89. doi: 10.1002/pros.2990220110. [DOI] [PubMed] [Google Scholar]
- 35.Beswick EJ, Johnson JR, Saada JI, Humen M, House J, Dann S, Qiu S, Brasier AR, Powell DW, Reyes VE, Pinchuk IV. 2014. TLR4 activation enhances the PD-L1-mediated tolerogenic capacity of colonic CD90+ stromal cells. J Immunol 193:2218–2229. doi: 10.4049/jimmunol.1203441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Hamrouni A, Wolowiec D, Coiteux V, Kuliczkowski K, Hetuin D, Saudemont A, Quesnel B. 2007. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-{gamma} and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood 110:296–304. doi: 10.1182/blood-2006-10-051482. [DOI] [PubMed] [Google Scholar]
- 37.Park J-H, Kim Y-G, McDonald C, Kanneganti T-D, Hasegawa M, Body-Malapel M, Inohara N, Núñez G. 2007. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol 178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Jiang W, Duan N, Qian Y, Zhou Q, Ye P, Jiang H, Bai Y, Zhang W, Wang W. 2014. NOD1, RIP2 and Caspase12 are potentially novel biomarkers for oral squamous cell carcinoma development and progression. Int J Clin Exp Pathol 7:1677–1686. [PMC free article] [PubMed] [Google Scholar]
- 39.McKillop D, McCormick AD, Miles GS, Phillips PJ, Pickup KJ, Bushby N, Hutchison M. 2004. In vitro metabolism of gefitinib in human liver microsomes. Xenobiotica 34:983–1000. doi: 10.1080/02772240400015222. [DOI] [PubMed] [Google Scholar]
- 40.McKillop D, Partridge EA, Hutchison M, Rhead SA, Parry AC, Bardsley J, Woodman HM, Swaisland HC. 2004. Pharmacokinetics of gefitinib, an epidermal growth factor receptor tyrosine kinase inhibitor, in rat and dog. Xenobiotica 34:901–915. doi: 10.1080/00498250400009189. [DOI] [PubMed] [Google Scholar]
- 41.Roux PP, Blenis J. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feller L, Altini M, Lemmer J. 2013. Inflammation in the context of oral cancer. Oral Oncol 49:887–892. doi: 10.1016/j.oraloncology.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Whitmore SE, Lamont RJ. 2014. Oral bacteria and cancer. PLoS Pathog 10:e1003933. doi: 10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groeger S, Domann E, Gonzales JR, Chakraborty T, Meyle J. 2011. B7-H1 and B7-DC receptors of oral squamous carcinoma cells are upregulated by Porphyromonas gingivalis. Immunobiology 216:1302–1310. doi: 10.1016/j.imbio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Hirai M, Kitahara H, Kobayashi Y, Kato K, Bou-Gharios G, Nakamura H, Kawashiri S. 2017. Regulation of PD-L1 expression in a high-grade invasive human oral squamous cell carcinoma microenvironment. Int J Oncol 50:41–48. doi: 10.3892/ijo.2016.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groeger S, Howaldt HP, Raifer H, Gattenloehner S, Chakraborty T, Meyle J. 2017. Oral squamous carcinoma cells express B7-H1 and B7-DC receptors in vivo. Pathol Oncol Res 23:99–110. doi: 10.1007/s12253-016-0100-7. [DOI] [PubMed] [Google Scholar]
- 47.Fankhauser SC, Starnbach MN. 2014. PD-L1 limits the mucosal CD8+ T cell response to Chlamydia trachomatis. J Immunol 192:1079–1090. doi: 10.4049/jimmunol.1301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazanet MM, Hughes CC. 2002. B7-H1 is expressed by human endothelial cells and suppresses T cell cytokine synthesis. J Immunol 169:3581–3588. doi: 10.4049/jimmunol.169.7.3581. [DOI] [PubMed] [Google Scholar]
- 49.Youngnak-Piboonratanakit P, Tsushima F, Otsuki N, Igarashi H, Machida U, Iwai H, Takahashi Y, Omura K, Yokozeki H, Azuma M. 2004. The expression of B7-H1 on keratinocytes in chronic inflammatory mucocutaneous disease and its regulatory role. Immunol Lett 94:215–222. doi: 10.1016/j.imlet.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 50.Das S, Suarez G, Beswick EJ, Sierra JC, Graham DY, Reyes VE. 2006. Expression of B7-H1 on gastric epithelial cells: its potential role in regulating T cells during Helicobacter pylori infection. J Immunol 176:3000–3009. doi: 10.4049/jimmunol.176.5.3000. [DOI] [PubMed] [Google Scholar]
- 51.Beswick EJ, Pinchuk IV, Das S, Powell DW, Reyes VE. 2007. Expression of the programmed death ligand 1, B7-H1, on gastric epithelial cells after Helicobacter pylori exposure promotes development of CD4+ CD25+ FoxP3+ regulatory T cells. Infect Immun 75:4334–4341. doi: 10.1128/IAI.00553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cecil JD, O'Brien-Simpson NM, Lenzo JC, Holden JA, Chen Y-Y, Singleton W, Gause KT, Yan Y, Caruso F, Reynolds EC. 2016. Differential responses of pattern recognition receptors to outer membrane vesicles of three periodontal pathogens. PLoS One 11:e0151967. doi: 10.1371/journal.pone.0151967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. 2003. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 54.Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ, Thomas BJ, Malosse C, Gantier MP, Casillas LN, Votta BJ, Bertin J, Boneca IG, Sasakawa C, Philpott DJ, Ferrero RL, Kaparakis-Liaskos M. 2014. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe 15:623–635. doi: 10.1016/j.chom.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Huang S, Wu J, Gao X, Zou S, Chen L, Yang X, Sun C, Du Y, Zhu B, Li J, Yang X, Feng X, Wu C, Shi C, Wang B, Lu Y, Liu J, Zheng X, Gong F, Lu M, Yang D. 2018. LSECs express functional NOD1 receptors: a role for NOD1 in LSEC maturation-induced T cell immunity in vitro. Mol Immunol 101:167–175. doi: 10.1016/j.molimm.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 56.Humphries F, Yang S, Wang B, Moynagh PN. 2015. RIP kinases: key decision makers in cell death and innate immunity. Cell Death Differ 22:225–236. doi: 10.1038/cdd.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pellegrini E, Desfosses A, Wallmann A, Schulze WM, Rehbein K, Mas P, Signor L, Gaudon S, Zenkeviciute G, Hons M, Malet H, Gutsche I, Sachse C, Schoehn G, Oschkinat H, Cusack S. 2018. RIP2 filament formation is required for NOD2 dependent NF-kappaB signalling. Nat Commun 9:4043. doi: 10.1038/s41467-018-06451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA. 2002. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 59.Groeger S, Michel J, Meyle J. 2008. Establishment and characterization of immortalized human gingival keratinocyte cell lines. J Periodontal Res 43:604–614. doi: 10.1111/j.1600-0765.2007.01019.x. [DOI] [PubMed] [Google Scholar]
- 60.Groeger S, Jarzina F, Mamat U, Meyle J. 2017. Induction of B7-H1 receptor by bacterial cells fractions of Porphyromonas gingivalis on human oral epithelial cells: B7-H1 induction by Porphyromonas gingivalis fractions. Immunobiology 222:137–147. doi: 10.1016/j.imbio.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 61.Waller T, Kesper L, Hirschfeld J, Dommisch H, Kolpin J, Oldenburg J, Uebele J, Hoerauf A, Deschner J, Jepsen S, Bekeredjian-Ding I. 2016. Porphyromonas gingivalis outer membrane vesicles induce selective tumor necrosis factor tolerance in a Toll-like receptor 4- and mTOR-dependent manner. Infect Immun 84:1194–1204. doi: 10.1128/IAI.01390-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Desmarais SM, Cava F, de Pedro MA, Huang KC. 2014. Isolation and preparation of bacterial cell walls for compositional analysis by ultra performance liquid chromatography. J Vis Exp 83:e51183. doi: 10.3791/51183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glauner B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem 172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 64.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takahashi H, Sakakura K, Arisaka Y, Tokue A, Kaira K, Tada H, Higuchi T, Okamoto A, Tsushima Y, Chikamatsu K. 2019. Clinical and biological significance of PD-L1 expression within the tumor microenvironment of oral squamous cell carcinoma. Anticancer Res 39:3039–3046. doi: 10.21873/anticanres.13437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.