Graphical abstract

Keywords: Pancreatic ductal adenocarcinoma, Immune cell infiltration, M0 macrophages, Prognosis
Abstract
The details of the immunological microenvironment and its clinical implications for pancreatic cancer are still unclear. In this study, we obtained data from public databases, such as the Gene Expression Omnibus, the Cancer Genome Atlas Program, the International Cancer Genome Consortium Data Portal, the ArrayExpress Data Warehouse, and the cBioPortal for Cancer Genomics. We used these data to evaluate the pattern of immune cells infiltration in pancreatic ductal adenocarcinoma (PDAC) tissues. We observed that the levels of M0 macrophages and activated dendritic cells in tumor tissues were significantly higher than that in para-tumor tissues. M0 macrophages, gamma delta T cells and naive CD4 T cells were independent predictive factors of a poor outcome for PDAC patients. An immune score determined by M0 macrophages, gamma delta T cells and naive CD4 T cells could predict the survival of patients. The results of this study suggest that the infiltration of immune cells, such as M0 macrophages, may be a possible target for the treatment of PDAC. However, these findings need to be confirmed by additional studies.
Introduction
Pancreatic cancer is the fourth leading cause of cancer-related death in the USA. In contrast to the declining mortality from breast and lung cancer, the mortality from pancreatic cancer increased 0.3% per year from 2011 to 2015 [1], [2], and the 5-year survival rate is only 9% [1]. The poor outcomes of pancreatic cancer have been attributed to that this disease is resistant to chemoradiation [3]. Thus, the exploration of novel targets for the treatment of pancreatic cancer is urgently needed.
Immune cells are essential cell types involved in the pancreatic tumor, and several studies demonstrate that targeting immune cells is a promising treatment for cancers [4], [5]. For example, previous studies suggested that the immune checkpoint inhibitor drugs, which target programmed cell death-1 (PD1) or its ligand PD-L1, have successfully improved the survival of patients with hematological malignancies [6], [7], [8] or patients with solid cancers [9], [10], [11]. However, the PD-L1 antibody, BMS-956559, failed to result in an objective response in pancreatic cancer patients [12]. This suggests that targeting the immune checkpoint may not always benefit patients, and the identification of novel therapeutic targets for immunotherapy of pancreatic cancer is necessary.
Thus, in this study, we evaluated the levels of immune cells in pancreatic ductal adenocarcinoma (PDAC) and para-PDAC tissues using the Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT), a robust algorithm that can accurately calculate the levels of 22 human immune cell phenotypes [13], and determined the immune cells which affect the survival of patients. In addition, we evaluated whether the classical signaling pathways of immune reactions were involved in the infiltration of immune cells. This could help us to understand the details of the immunological microenvironment and provide potential targets for the diagnosis and treatment of pancreatic cancer.
Materials and methods
Gene expression profiles of PDAC
A systematic search was performed to obtain the gene expression profiles of PDAC. The search, “pancreatic ductal adenocarcinoma OR PDAC”, was conducted in several public databases, such as the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), The Cancer Genome Atlas Program (TCGA, https://portal.gdc.cancer.gov/), the International Cancer Genome Consortium (ICGC, https://icgc.org/) Data Portal, the ArrayExpress Data Warehouse (https://www.ebi.ac.uk/arrayexpress/), and the cBioPortal for Cancer Genomics (http://www.cbioportal.org/). As indicated in Fig. 1, 364 series were excluded according to the following exclusion criteria: (1) the sample sizes of the series were 40 or fewer; (2) data were obtained from cells, not tissues; and (3) the data were related to microRNA, lncRNA, or DNA, not mRNA. In addition, series for which the survival information of the patients was unavailable were also excluded from further analysis.
Fig. 1.
Flowchart of the study. GEO: Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). TCGA: The Cancer Genome Atlas Program (https://portal.gdc.cancer.gov/). ICGC: International Cancer Genome Consortium Data Portal (https://icgc.org/). ArrayExpress: ArrayExpress Data Warehouse (https://www.ebi.ac.uk/arrayexpress/). cBioPortal: cBioPortal for Cancer Genomics (http://www.cbioportal.org/).
Evaluating immune cell infiltration by CIBERSORT
Eight series from GEO (GSE102238, GSE21501, GSE28735, GSE57495, GSE62452, GSE71729, GSE78229, and GSE85916), two series from ICGC (ICGC-AU and ICGC-CA), two series from ArrayExpress (E-MTAB-6134 and MTAB 2780), one series from cBioportal-qcmg and one series from TCGA were included in this study. Finally, immune cell infiltrations of 1700 patients were estimated by CIBERSORT (https://cibersort.stanford.edu/) [13], a free and robust algorithm for calculating the cellular composition of a tissue. The LM22 (22 immune cell types) was used as a reference gene expression signature. The immune cell composition analyses were performed with 100 permutations using the default parameters. A total of 662 cases were excluded from further analysis because the P-value determined by CIBERSORT was greater than 0.05 [14]. Subsequently, duplicated samples (N = 41) in the GSE78229 and GSE62452 series, as well as samples (N = 118) that failed to provide survival information, were excluded, and thus a total of 879 samples were included for further analysis.
We used 45 paired samples of a para-tumor and a tumor to evaluate the predictive value of immune cell infiltration, and 830 samples (41 paired tumors and 789 non-paired tumors) were used to investigate the prognostic significance of immune cell infiltration (Fig. 1). These samples were randomly enrolled in either the training cohort or the validation cohort (Fig. 1) using the R Project for Statistical Computing (R version 3.6.1) and the ‘sampling’ package.
Statistical analysis
The percentage of immune cells in each tissue (Fig. 2A) was presented in histograms using R project and the Package ‘ggplot2′. In addition, a box plot and a Wilcoxon test (Fig. 2B) were used to determine the statistical significance of the differences in immune cells between para-tumor and tumor tissues.
Fig. 2.
The levels of immune cells in PDAC tissues and para-PDAC tissues. Two GEO series, GSE62452 (N = 22 pairs) and GSE28735 (N = 23 pairs), were used to evaluate the pattern of immune cell infiltration between PDAC tissue and para-tumor tissue (A). The levels of M0 macrophages (M0) and activated dendritic cells (ADCs) in tumor tissues were significantly high than that in para-tumor tissues. However, the level of naive B cells was significantly reduced (B). The multiple logistic regression demonstrated M0 and ADC were the independent predictors of PDAC and a predictive score was determined by M0 and ADC (C). The ROC curve suggested that M0 in combination with ADC could significantly distinct PDAC from non-PDAC (D). Wilcoxon test and P < 0.05 indicates significantly difference for Fig. 2B.
To determine the markers for predicting PDAC, 31 paired samples were randomly split and assigned to the training cohort, and binary logistic regression was performed (Fig. 2C). The results were internally validated by 1000-fold bootstrapping with the help of SPSS 19.0 (IBM, New York, USA) [15]. Subsequently, the predictive score for each individual was calculated by the coefficients of each variable (Fig. 2C), and the following formula was used to determine the score: Probability = exp (predictive score) / [1 + exp (predictive score)] [16]. The receiver operating characteristic (ROC) curve and the area under the curve (AUC) were drawn to evaluate the predictive performance of this score (Fig. 2D) [17]. In addition, to validate the predictive performance of the immune cells, a ROC curve was developed for the validation cohort (Fig. 2E).
To determine the prognostic significance of immune cells, a univariate Cox proportional-hazards model was constructed (Fig. 3), and the variables that significantly influenced the survival of patients were then used to develop a multivariable Cox proportional-hazards model. The Schoenfeld residual test was performed to evaluate the assumptions of the multivariable Cox proportional-hazards model (Fig. 4 and Fig. 5A) [18].
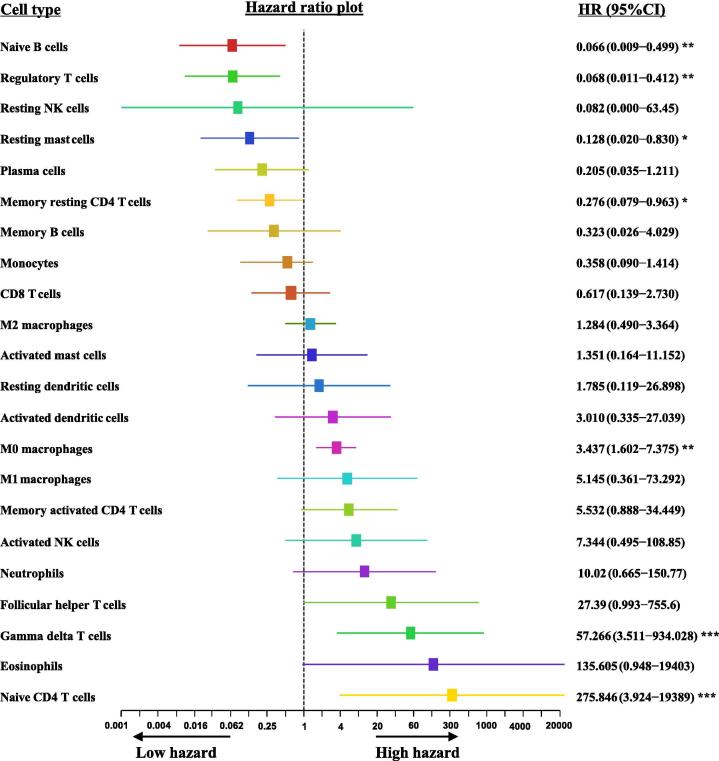
Fig. 3.
The univariate cox proportional hazards regression model of immune cell infiltration. In the training cohort, we observed that Naive B cells, regulatory T cells, resting mast cells, and memory resting CD4 T cells significantly decreased the hazard ratio for death. However, M0 macrophages, gamma delta T cells and naive CD4 T cells significantly increased the hazard ratio for death. * indicates P < 0.05, ** indicates P < 0.01. ***indicates P < 0.001.
Fig. 4.
Evaluating the proportional hazards assumption of multiple Cox regression. The schoenfeld residual of naive B cells (A), regulatory T cells (B), resting mast cells (C), memory resting CD4 T cells (D), M0 macrophages (E), gamma delta T cells (F), and naive CD4 T cells (G) were not dependent on the time. This suggests that the assumption of multiple Cox regression is satisfied.
Fig. 5.
Development and validation of immune score for diagnosis of PDAC. The multiple cox proportional hazards regression suggested that M0 macrophages, naive CD4 T cells and gamma delta T cells were independent risk factors of survival, and an immune score was developed based on these variables (A). The optimal cut-off of this index was 0.4, which was determined by X-title (B and C). The Kaplan-Meier curve and log-rank test suggested that the survival of patients with an immune score no greater than 0.4 was significantly longer than the survival of those with an immune score greater than 0.4 in training cohort (D), validation cohort (E), and the entire cohort (F). In addition, and the relapse-free survival (RFS) time of patients whose immune score was no greater than 0.4 was longer than that of patients with an immune score greater than 0.4 (G). The prognostic power of immune score was significantly superior to the TNM stage in both the training cohort and the validation cohort (H).
The optimal cutoff of the immune score was determined with the help of X-title [19]. The X-tile program divided the patients into a training set (upper-left quartile of Fig. 5B) and a validation set (the small long strip on the bottom of Fig. 5B), and the optimal cut-point (black dot) occurs at the brightest pixel (red) in the region of the validation set [19]. In addition, a plot of χ2 log-rank indicates the correlation between the cutoff point and survival (Fig. 5B). Red coloration suggests an inverse correlation between the cutoff and survival, while green coloration indicates a direct association. The histogram (Fig. 5C) shows that the optimal cutoff was used to divide patients into a short and a long survival group.
To evaluate the prognostic performance of the immune cell infiltration, we calculated Kaplan-Meier curves and log-rank tests (Fig. 5D-5G). Harrell’s concordance index (C-index) was used to investigate if the immune score was superior to the TNM stage in predicting the survival of patients (Fig. 5H). In addition, in order to explore the functional biomarkers that might be related to the changes in the immunological tumor microenvironment between patients with higher and lower immune scores, gene set enrichment analysis (GSEA) was performed with the GSEA Desktop v4.0.3 (1,000 permutations) using the TCGA samples [20]. The functional gene set files “c5.all.v6.2.symbols.gmt” were used to summarize and elucidate specific and well-defined biological processes or molecular functions.
Results
Immune cell infiltration between PDAC tissues and para-PDAC tissues
We observed that the levels of M0 macrophages and activated dendritic cells in PDAC were significantly (P = 0.010 and P < 0.0001, respectively) higher than in para-PDAC. However, compared with para-PDAC, the level of naive B cells in PDAC was significantly (P < 0.0001, Fig. 2B) decreased. There were no significant differences between PDAC and para-PDAC in regard to the levels of other immune cells.
To evaluate if M0 macrophages, activated dendritic cells and naive B cells were independent predictors of PDAC, we performed logistic regression (enter method), and it was internally validated by 1000-fold bootstrapping. We observed that M0 macrophages and activated dendritic cells were both independent factors that could be used to distinguish PDAC from para-PDAC (Fig. 2C).
In order to evaluate the discriminatory ability of M0 macrophages and activated dendritic cells for PDAC, a predictive score was determined by the following formula: Predictive score = 18.477 × M0 macrophages + 22.467 × activated dendritic cells – 2.498, and ROC curves were generated for the training (Fig. 2D) and the validation cohort (Fig. 2E). We observed that the AUCs were 0.865 (95% CI: 0.775–0.955, P < 0.001) and 0.837 (95% CI: 0.685–0.988, P = 0.002), respectively. This suggests that the M0 macrophages and the activated dendritic cells have an acceptable discriminatory ability for predicting PDAC.
Prognostic significance of immune cells for PDAC
To evaluate the prognostic significance of tumor infiltrating immune cells, 830 PDAC samples were randomly divided into a training cohort (N = 581) and a validation cohort (N = 249). Subsequently, a univariate Cox proportional-hazards model was constructed for the training cohort (Fig. 3). We observed that the presence of naive B cells (P = 0.008), regulatory T cells (P = 0.003), resting mast cells (P = 0.003), and memory resting CD4 T cells (P = 0.043) were significantly correlated with a decreased risk of death. However, the presence of M0 macrophages (P = 0.002), gamma delta T cells (P < 0.001), and naive CD4 T cells (P < 0.001) were significantly correlated with an increased risk of death.
The Schoenfeld residual test indicated that these variables are independent of time (Fig. 4). This suggests that the assumptions of the multivariate Cox proportional-hazards model are satisfied. We constructed a multivariate Cox regression model (Forward: LR) and determined that only M0 macrophages, gamma delta T cells, and naive CD4 T cells were independent predictors of survival (Fig. 5A). The immune score of each patient was determined by the following formula: Immune score = 1.400 × M0 macrophages + 4.007 × gamma delta T cells + 5.426 × naive CD4 T cells. The optimal cutoff of the immune score (cutoff value = 0.4) was determined by X-tile (Fig. 5B and 5C). To evaluate the performance of the immune score, Kaplan-Meier curves were constructed for the training cohort (Fig. 5D), the validation cohort (Fig. 5E), and the entire cohort (Fig. 5F). We observed that the Kaplan-Meier curves were significantly distinct, and the survival of patients with an immune score no greater than 0.4 was significantly longer than the survival of those with an immune score greater than 0.4 (P < 0.05, Fig. 5D-F). In addition, we obtained the relapse-free survival time from the TCGA series (N = 104) and constructed Kaplan-Meier curves. Again, we observed that the Kaplan-Meier curves were clearly separated, and the relapse-free survival time of patients whose immune score was no greater than 0.4 was longer than that of patients with an immune score greater than 0.4 (Fig. 5G). Moreover, in order to compare the prognostic significance of the TNM stage and the immune score, we calculated Harrell’s concordance index. We observed that the immune score was significantly superior to the TNM stage in both the training cohort and the validation cohort (Fig. 5H).
Utilizing GSEA to identify potential targets for regulating immune cells
To identify the genes that might be involved in regulation of the immunological microenvironment, the individuals from the TCGA database were divided into two groups, the immune score ≤ 0.4 group (N = 96) and the immune score >0.4 group (N = 26), and GSEA was performed. We observed that the biological processes related to cell chemotaxis (Fig. 6A), leukocyte chemotaxis (Fig. 6B), and chemokine mediated signaling pathways (Fig. 6C) were inactivated in patients with immune score >0.4. We, therefore, evaluated the expression of chemotactic factors at the transcription level. We observed that the expression of chemokine (C-X-C motif) ligand 9 (CXCL9), CXCL10, CXCL11, CXCL13, chemokine (C-C motif) ligand 15 (CCL15), CCL17, chemokine (C-X-C motif) receptor 2 (CXCR2), and CXCR6 were significantly decreased in patients with an immune score >0.4 (Fig. 6D). In addition, these patients also had a low enrichment score for the following biological processes, such as activation of immune response (Fig. 6E), immune response regulating cell surface receptor signaling pathway (Fig. 6F), antigen receptor mediated signaling pathway (Fig. 6G), natural killer cell activation (Fig. 6H), and dendritic cell migration (Fig. 6I). In addition, the molecular function of cytokine receptor activity (Fig. 6J) was also deficient in these patients.
Fig. 6.
Gene set enrichment analysis (GSEA) of PDAC with different immune score. 122 samples from TCGA were divided into two groups, the immune score ≤ 0.4 group (N = 96) and the immune score greater than 0.4 group (N = 26). PDAC patients with immune score >0.4 have a low enrichment score for the following biological processes of cell chemotaxis (A), leukocyte chemotaxis (B) and chemokine mediated signaling pathways (C). The expression levels of chemokine (C-X-C motif) ligand 9 (CXCL9), CXCL10, CXCL11, CXCL13, chemokine (C-C motif) ligand 15 (CCL15), CCL17, chemokine (C-X-C motif) receptor 2 (CXCR2), and CXCR6 were significantly decreased in patients with an immune score >0.4, * indicates P < 0.05 , ** indicates P < 0.01. ***indicates P < 0.001 (D). PDAC patients with immune score >0.4 have a low enrichment score for the following biological processes of activation of immune response (E), immune response regulating cell surface receptor signaling pathway (F), antigen receptor mediated signaling pathway (G), natural killer cell activation (H), dendritic cell migration (I) and the molecular function of cytokine receptor activity (J).
Discussion
It is well known that pancreatic cancer cells are surrounded by an abundant stromal microenvironment, which is composed of several non-cancer cells, such as immune cells, endothelial cells, and cancer-associated fibroblasts [21], [22]. Notably, the tumor-associated macrophages (TAMs), recruited by pancreatic carcinoma cells via the CCL2-CCR2 chemokine axis, are the most frequent infiltrated immune cells. Based on the polarization states, TAMs can be divided into three types: inactivated macrophages (M0 macrophage), classically (M1) or alternatively (M2) activated macrophages. The results of most studies have suggested that macrophages are promoters of tumors and this pro-tumor effect is mediated by the M2 macrophage [23]. This concept is supported by the fact that M2 macrophages can cause dysregulation of the T cell receptor signaling pathway and activate cytotoxic CD8 T cell activity by secreting immunosuppressive factors, such as arginase-1, TGF-β, and IL-10 [24], [25], [26], [27]. Additionally, Ye et al. demonstrated that TAMs could promote the progression of PDAC by facilitating the Warburg effect, in which both cytokines and the microenvironment are involved [28]. Some studies have determined that M1 macrophages can active inflammatory responses and induce the death of tumors by secreting pro-inflammatory cytokines, such as IL-12, IL-16, and INF-γ [29], [30]. However, in contrast to M1 and M2 macrophages, as far as we know, no studies have evaluated the interaction between M0 macrophages, the precursors of M1 and M2, and pancreatic cancer cells. The results of the present study suggest that M0 macrophages accumulate in the tumor tissues and their presence can be used to predict a poor patient outcome. Thus, eliminating the M0 macrophages might be a promising strategy to fight against PDAC. However, additional studies are necessary to evaluate the mechanism of how M0 macrophages decrease the survival of patients. Is this dependent on the conversion of M0 to M2 or is there a direct interaction between M0 macrophages and tumor cells?
Naive CD4 T cells might be another promising target for the treatment of PDAC. This is supported by the present study and previous studies [31], [32], [33]. For example, Pan et al. reported that naive T cells could convert into tumor-specific Tregs cells, in the presence of myeloid-derived suppressor cells, and support the survival of tumor cells in stressful tumor environments [32].
This study suggested that activated dendritic cells (DCs), the antigen-presenting cells in the innate immune system, were also infiltrating the tumor tissues. Notably, the role of DCs in pancreatic cancer is still contradictive. Leone et al. suggested that DCs can present antigens to CD8 T cells and stimulate the CD8 effector memory population to secrete IFN-γ, which exerts an antitumor activity [34]. However, DCs can also promote immune evasion of tumors cells. For example, Argentiero et al. reported that the level of DCs is significantly higher in PDAC patients with metastatic lymph nodes, and these DCs can upregulate the immunosuppressive WNT pathway [35]. This might be the reason why dendritic cells can promote tumor metastasis and immune evasion of cancer cells [35], [36], [37], [38].
The role of gamma delta T cells in cancer in cancer is also contradictive. It has been reported that gamma delta T cells could secret IFN-γ, which inhibits tumor progression [39]. Interestingly, the results of the present study suggest that gamma delta T cells are associated with a poor prognosis of pancreatic cancer. This is supported by many studies, which reported that gamma delta T cells can promote angiogenesis and tumor cell growth [40], [41], [42]. The contradictive role of dendritic cells and gamma delta T cells suggest that targeting these immune cells might not always benefit the patients. Additional studies are needed to explore the interaction between these cells and PDAC cells. This will help to provide a basis for novel therapeutics of PDAC.
Notably, our findings have two clinical implications: First, measuring the levels of infiltrating M0 and activated dendritic cells might improve the diagnosis of PDAC. Second, stratifying patients according to their immune score, which is determined from the levels of M0 macrophages, gamma delta T cells and naive CD4 T cells, might help clinicians determine which patients can benefit from immune therapy. However, even though the other immune cells were excluded from the Cox proportional-hazards model, and the immune score showed a promising performance for predicting the survival of patients. The interaction among tumor cells and the immune system which is consists of various innate and adaptive immune cells, are complex and other factors may be also involved in these interaction. For example, Leone et al. demonstrated that endothelial cells could act as antigen presenting cells to stimulate the central memory CD8 T cell population, which exhibits pro-tumor activity via the production of IL-10 and TGF-β [34]. Also, the GSEA suggested that some genes involved in chemotaxis, such as CXCL9, CXCL10, CXCL11, CXCL13, CCL15, CCL17, CXCR2, and CXCR6, are involved in the regulation of tumor progression. This suggests that targeting one type of immune cell or chemotaxis may not be sufficient for treating PDAC.
When interpreting the results of this study, it should be kept in mind that although CIBERSORT is one of the best in silico approaches to date, CIBERSORT evaluates the immune cell infiltration into tissues and assumes that these cells have the same gene expression profile as the immune cells in peripheral blood [43]. Besides, the limitations of the TCGA database should also be taken into account: First, samples in which the cell nuclei were less than 60% were excluded by the pathologist [43]. This might lead to the removal of many immune-infiltrated tumors from the analysis. Second, although we have tried our best to review the gene expression profiles systematically, this study is restricted since the analyses did not include data from genome-wide molecular assays. In addition, this is a retrospective study, and therefore, the results might be influenced by reporting bias [43].
In conclusion, this study suggested that the levels of M0 macrophages and activated dendritic cells in the tissue of PDAC were significantly higher than in para-tumor tissues, while the levels of naive B cells in the PDAC tissue was significantly decreased. We showed that the percentage of M0 macrophages and activated dendritic cells could distinguish PDAC from non-PDAC. This implies that M0 macrophages and activated dendritic cells may be valuable markers for the diagnosis of PDAC. However, this finding needs further investigation. In addition, we observed that the presence of M0 macrophages, gamma delta T cells and naive CD4 T cells were independent prognostic factors of PDAC patients. An immune score, which was based on M0 macrophages, gamma delta T cells and naive CD4 T cells, could successfully stratify patients by survival time. This might help clinicians in choosing an optimal individualized treatment for PDAC patients. However, the diagnostic and prognostic utility of immune cells should be investigated by a further study, in which the scientists compare the amount of peripheral blood immune cells in pancreatic cancer patients and pancreatic or diabetes.
Author contributions
Conceptualization: XZ, HC, CX, SS. Analysis and acquisition of data: CX, SS, XZ, GZ, YS, PG, ZY, MH, MN. Data Curation: CX, SS. Writing - Original Draft: CX, XZ. Writing - Review & Editing: All authors. Funding acquisition: HC, XZ.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects
Declaration of Competing Interest
The authors have declared no conflict of interest
Acknowledgement
This study was supported by the National Natural Science Foundation of China (No. 81573751 & 81973646).
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Peng Gong, Email: doctorgongpeng@szu.edu.cn.
Hailong Chen, Email: chenhailong@dmu.edu.cn.
Xianbin Zhang, Email: zhangxianbin@hotmail.com.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X., Kumstel S., Jiang K., Meng S., Gong P., Vollmar B. LW6 enhances chemosensitivity to gemcitabine and inhibits autophagic flux in pancreatic cancer. J Adv Res. 2019;20:9–21. doi: 10.1016/j.jare.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamisawa T., Wood L.D., Itoi T., Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 4.Watt J., Kocher H.M. The desmoplastic stroma of pancreatic cancer is a barrier to immune cell infiltration. Oncoimmunology. 2013;2(12):1–3. doi: 10.4161/onci.26788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan J.Q., Wang M.F., Chen H.L., Shang D., Das J.K., Song J. Current advances and outlooks in immunotherapy for pancreatic ductal adenocarcinoma. Mol Cancer. 2020;19(1):1–22. doi: 10.1186/s12943-020-01151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ansell S.M., Lesokhin A.M., Borrello I., Halwani A., Scott E.C., Gutierrez M. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2015;372(4):311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsirigotis P., Savani B.N., Nagler A. Programmed death-1 immune checkpoint blockade in the treatment of hematological malignancies. Ann Med. 2016;48(6):428–439. doi: 10.1080/07853890.2016.1186827. [DOI] [PubMed] [Google Scholar]
- 8.Lesokhin A.M., Ansell S.M., Armand P., Scott E.C., Halwani A., Gutierrez M. Nivolumab in patients with relapsed or refractory hematologic malignancy: preliminary results of a phase Ib study. J Clin Oncol. 2016;34(23):2698–2704. doi: 10.1200/JCO.2015.65.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Topalian S.L., Sznol M., McDermott D.F., Kluger H.M., Carvajal R.D., Sharfman W.H. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32(10):1020–1030. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang Y.K., Boku N., Satoh T., Ryu M.H., Chao Y., Kato K. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10111):2461–2471. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 11.Thind K., Padrnos L.J., Ramanathan R.K., Borad M.J. Immunotherapy in pancreatic cancer treatment: a new frontier. Therap Adv Gastroenterol. 2017;10(1):168–194. doi: 10.1177/1756283X16667909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brahmer J.R., Tykodi S.S., Chow L.Q., Hwu W.J., Topalian S.L., Hwu P. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou R., Zhang J., Zeng D., Sun H., Rong X., Shi M. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer. Cancer Immunol Immunother. 2019;68(3):433–442. doi: 10.1007/s00262-018-2289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moons K.G., Altman D.G., Reitsma J.B., Ioannidis J.P., Macaskill P., Steyerberg E.W. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162(1):W1–73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 16.Wilks D.S. Extending logistic regression to provide full-probability-distribution MOS forecasts. Meteorol Appl. 2009;16(3):361–368. [Google Scholar]
- 17.Bewick V., Cheek L., Ball J. Statistics review 13: receiver operating characteristic curves. Crit Care. 2004;8(6):508–512. doi: 10.1186/cc3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hess K.R. Graphical methods for assessing violations of the proportional hazards assumption in Cox regression. Stat Med. 1995;14(15):1707–1723. doi: 10.1002/sim.4780141510. [DOI] [PubMed] [Google Scholar]
- 19.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 20.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J.P. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23(23):3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 21.Ren B., Cui M., Yang G., Wang H., Feng M., You L. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol Cancer. 2018;17(1):1–15. doi: 10.1186/s12943-018-0858-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng J., Sun B.F., Chen C.Y., Zhou J.Y., Chen Y.S., Chen H. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sica A., Larghi P., Mancino A., Rubino L., Porta C., Totaro M.G. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18(5):349–355. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F., Wang H., Wang X., Jiang G., Liu H., Zhang G. TGF-beta induces M2-like macrophage polarization via SNAIL-mediated suppression of a pro-inflammatory phenotype. Oncotarget. 2016;7(32):52294–52306. doi: 10.18632/oncotarget.10561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Binnemars-Postma K., Storm G., Prakash J. Nanomedicine strategies to target tumor-associated macrophages. Int J Mol Sci. 2017;18(5):1–27. doi: 10.3390/ijms18050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon S., Martinez F.O. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32(5):593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Mantovani A., Sozzani S., Locati M., Allavena P., Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 28.Ye H., Zhou Q., Zheng S., Li G., Lin Q., Wei L. Tumor-associated macrophages promote progression and the Warburg effect via CCL18/NF-kB/VCAM-1 pathway in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9(5):1–19. doi: 10.1038/s41419-018-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubil E., Caskey L., Holtzhausen A., Hunter D., Story C., Earp H.S. Tumor-secreted Pros1 inhibits macrophage M1 polarization to reduce antitumor immune response. J Clin Invest. 2018;128(6):2356–2369. doi: 10.1172/JCI97354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goswami K.K., Ghosh T., Ghosh S., Sarkar M., Bose A., Baral R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell Immunol. 2017;316:1–10. doi: 10.1016/j.cellimm.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Su S., Liao J., Liu J., Huang D., He C., Chen F. Blocking the recruitment of naive CD4(+) T cells reverses immunosuppression in breast cancer. Cell Res. 2017;27(4):461–482. doi: 10.1038/cr.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan P.Y., Ma G., Weber K.J., Ozao-Choy J., Wang G., Yin B. Immune stimulatory receptor CD40 is required for T-cell suppression and T regulatory cell activation mediated by myeloid-derived suppressor cells in cancer. Cancer Res. 2010;70(1):99–108. doi: 10.1158/0008-5472.CAN-09-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin B., Auffray C., Delpoux A., Pommier A., Durand A., Charvet C. Highly self-reactive naive CD4 T cells are prone to differentiate into regulatory T cells. Nat Commun. 2013;4:1–12. doi: 10.1038/ncomms3209. [DOI] [PubMed] [Google Scholar]
- 34.Leone P., Di Lernia G., Solimando A.G., Cicco S., Saltarella I., Lamanuzzi A. Bone marrow endothelial cells sustain a tumor-specific CD8(+) T cell subset with suppressive function in myeloma patients. Oncoimmunology. 2019;8(1):1–12. doi: 10.1080/2162402X.2018.1486949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Argentiero A., De Summa S., Di Fonte R., Iacobazzi R.M., Porcelli L., Da Via M. Gene expression comparison between the lymph node-positive and -negative reveals a peculiar immune microenvironment signature and a theranostic role for WNT targeting in pancreatic ductal adenocarcinoma: a pilot study. Cancers (Basel) 2019;11(7):1–19. doi: 10.3390/cancers11070942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marti L.C., Pavon L., Severino P., Sibov T., Guilhen D., Moreira-Filho C.A. Vascular endothelial growth factor-A enhances indoleamine 2,3-dioxygenase expression by dendritic cells and subsequently impacts lymphocyte proliferation. Mem Inst Oswaldo Cruz. 2014;109(1):70–79. doi: 10.1590/0074-0276130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao F., Xiao C., Evans K.S., Theivanthiran T., DeVito N., Holtzhausen A. Paracrine Wnt5a-beta-Catenin Signaling Triggers a Metabolic Program that Drives Dendritic Cell Tolerization. Immunity. 2018;48(1) doi: 10.1016/j.immuni.2017.12.004. pp. 147–160 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fainaru O., Almog N., Yung C.W., Nakai K., Montoya-Zavala M., Abdollahi A. Tumor growth and angiogenesis are dependent on the presence of immature dendritic cells. FASEB J. 2010;24(5):1411–1418. doi: 10.1096/fj.09-147025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva-Santos B., Serre K., Norell H. gammadelta T cells in cancer. Nat Rev Immunol. 2015;15(11):683–691. doi: 10.1038/nri3904. [DOI] [PubMed] [Google Scholar]
- 40.Wu P., Wu D., Ni C., Ye J., Chen W., Hu G. gammadeltaT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity. 2014;40(5):785–800. doi: 10.1016/j.immuni.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Hede D., Polese B., Humblet C., Wilharm A., Renoux V., Dortu E. Human papillomavirus oncoproteins induce a reorganization of epithelial-associated gammadelta T cells promoting tumor formation. Proc Natl Acad Sci USA. 2017;114(43):E9056–E9065. doi: 10.1073/pnas.1712883114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin C., Lagoudas G.K., Zhao C., Bullman S., Bhutkar A., Hu B. Commensal microbiota promote lung cancer development via gammadelta T cells. Cell. 2019;176(5) doi: 10.1016/j.cell.2018.12.040. pp. 998–1013 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H. Immunity. 2018;48(4) pp. 812–830 e14. [Google Scholar]