Abstract
Background
The aim of this study was to analyze the microbiological characteristics of nasopharyngeal carriage Haemophilus influenzae isolates collected from children with respiratory infections in Beijing hospital and Youyang Hospital of China.
Methods
The serotypes of all isolates were determined using latex agglutinated antisera (a‐f). The minimum inhibitory concentrations (MICs) of 11 antibiotics were determined using E‐test strips. For the beta‐lactamase‐negative ampicillin‐resistant (BLNAR) isolates, ftsI gene was sequenced based on fragments amplified by PCR. STs of H influenzae isolates were determined by multi‐locus sequence typing.
Results
The overall carriage rate of H influenzae in the study population was 9.1% (362/3984). One hundred and ninety H influenzae isolates which were selected in our study were non‐typeable (NTHi) and 44 (23.2%) of them were positive for β‐lactamase. All isolates were susceptible to ceftriaxone and levofloxacin. Susceptibility rates to erythromycin and sulfamethoxazole‐trimethoprim in Beijing were significantly higher than Youyang (P < .05). Thirty‐six BLNAR isolates were identified. The MLST analysis showed 108 STs in 190 isolates, the most common of which were ST408 (11, 5.8%), ST914 (10, 5.3%), ST57 (9, 4.7%), and ST834 (6, 3.2%). Twelve STs were detected in both of the study sites, which covered 63 isolates.
Conclusions
All isolates in the present study were NTHi, which suggested widespread of this type in China. The BLNAR isolates were detected more frequently than before. Because high genetic diversity of NTHi isolates of H influenzae exists worldwide, it is important to continuously monitor these bacteria in the future.
Keywords: antimicrobial susceptibility, children, Haemophilus influenzae, respiratory infection
Abbreviations
- BLNAR
beta‐lactamase‐negative ampicillin‐resistant
- CC
clonal complex
- MIC
minimal inhibitory concentration
- MLST
multiple locus sequence types
- NTHi
non‐typeable Haemophilus influenzae
- PCR
polymerase chain reaction
1. INTRODUCTION
Haemophilus influenzae (H influenzae), one of the most common bacterial pathogens causing respiratory tract infections in humans, can also cause bacterial meningitis and sepsis in children.1 It is usually colonized in the nasopharynx, especially in children, which is the first step leading to a severe infectious diseases. Previous study has shown that the nasopharyngeal carriage rate of H influenzae in children under 5 years old with acute upper respiratory tract infections is as high as 26.3%.2 It is also the second most common bacterial pathogen causing pneumonia in Chinese children.3, 4
In the past several decades, its resistance of H influenzae has increased in many countries, but there is a significant difference between geographic sites and monitoring time.5 A previous study in Beijing showed that the percentage of β‐lactamase positive was 4% in 2000, 13% in 2002, 27% in 2010, and 31% in 2012, and few β‐lactamase‐negative ampicillin‐resistant (BLNAR) isolates were detected.2 Variations in penicillin‐binding protein 3 (PBP3), which is encoded by the ftsI gene, have infrequently been analyzed in H influenzae isolates in China.
In the present study, H influenzae isolates collected from Chinese children with respiratory infections was monitored in Beijing and discrepancies in the epidemiology between two areas located far apart in China (Beijing City and Youyang County) were identified. The carriage rates, distribution of serotypes, drug resistance, multi‐locus sequence types (MLSTs), and PBP3 of H influenzae isolates collected from children with respiratory infections in two hospitals were determined and compared.
2. MATERIALS AND METHODS
2.1. Study sites
A total of 3984 children in Beijing Children's Hospital affiliated with Capital Medical University [Beijing Children's Hospital] and People's Hospital of Youyang County [Youyang Hospital] were involved in this study. Beijing Children's Hospital is a specialized children's hospital located in northern China and is the largest children's hospital in China. The team at Beijing Children's Hospital has been monitoring the epidemiology of H influenzae for about 20 years. This study analyzed the data collected in Beijing Children's Hospital in 2014. Youyang Hospital is a general hospital in southern China. Since 2015, the Department of Pediatrics of Youyang Hospital has been conducting the pathogens investigation involving H influenzae. This study analyzed the data collected in Youyang Hospital in 2015.
2.2. Study population and sample storage
Nasopharyngeal swabs were collected from outpatients with acute respiratory infections evaluated at Beijing Children's Hospital from January to December 2014. The swab samples were immediately placed into transfer tubes with culture medium composed of skimmed milk, glucose, and glycerin (skimmed milk‐tryptone glucose‐glycerol [STGG], Qingdao Tianqi Biotechnology Co., Ltd.), as previously reported.6, 7 Samples were sent to the Microbiology Laboratory at Beijing Children's Hospital for culture within 2 hours after collection. Hospitalized patients with acute respiratory infections who were evaluated at Youyang Hospital were enrolled from January to December 2015. Because there is no chocolate culture substrate in Youyang Hospital, the STGG transfer tubes were incubated at 37°C for about 2 hours and then stored in a −60°C freezer immediately. The samples were transferred to Beijing Children's Hospital every 6 months. Finally, 362 H influenzae were isolated.
Inclusion criteria were as follows: (a) children of both sexes, aged 3 months to 14 years and (b) children with diagnosis of community‐acquired upper or lower respiratory infections. Exclusion criteria were as follows: (a) children had received any immunotherapeutic agents within 2 months before enrollment; (b) children had received antibiotic treatment within 2 months before enrollment and were hypersensitive to the antibiotic prescribed; (c) children had severe underlying disease, such as leukemia; and (d) children had previously been included in the study.
Before enrollment and implementation of any study procedure, the parents and/or legal guardians of each participant signed a written informed consent document. This study was approved by the Ethics Committees of the two hospitals. No ethical issues were encountered in this study.
2.3. Bacterial culture and identification
After incubated in the culture media at 37°C for 1 hour, 30 μL of transfer medium was added to culture samples from Beijing Children's Hospital, which was cultured on chocolate medium plates (Guangdong HuanKai Microbial Technology Co., Ltd.). The samples from Youyang Hospital were transferred to Beijing for bacterial culture. The plates were incubated in a 37°C and 5% CO2 incubator for 1 hour and examined after 24 and 48 hours the same methods as for the samples from Beijing Children's Hospital.
Bacterial identification proceeded according to standard procedures, including colony morphologic features, gram staining, and requirement for both X and V factors (Oxoid, Basingstoke, UK). Detection of β‐lactamase activity was determined using the chromogenic cephalosporin nitrocefin (BR66A; Oxoid) method with a known β‐lactamase‐positive isolate as a control.
2.4. Molecular identification
All of the isolates were determined not only by regular microbiologic procedures, but also by present molecular identification methods. All H influenzae isolates were confirmed by genotype identification of the outer membrane protein P6 and 16S RNA genes, which are specific for H influenzae. A polymerase chain reaction (PCR) was performed to identify the genotype by using primers and the procedure as previous described with some modification.8, 9
2.5. Serotyping
Serotypes of all isolates were determined using latex agglutination antisera (a‐f; Statens Serum Institut). Non‐typeable isolates (NTHis) were confirmed not only by negative reactions of antisera, but also by negative PCR amplification of the bexA gene, as described in previous reports.10, 11
2.6. Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of all isolates for ampicillin, amoxicillin‐clavulanic acid, cefuroxime, ceftriaxone, cefepime, meropenem, levofloxacin, erythromycin, tetracycline, sulfamethoxazole‐trimethoprim, and chloramphenicol were determined using E‐test strips (AB Biodisk). MIC50/MIC90 was defined as the MIC of antibiotics that inhibit the growth of 50%/90% of isolates. H influenzae ATCC10211, ATCC49247, and ATCC49766 were used as quality control strains in each test batch. Breakpoints were determined in accordance with the Clinical and Laboratory Standards Institute (CLSI) 2016 criterion.12
2.7. ftsI mutation in BLNAR isolates
The ftsI gene was sequenced based on fragments of BLNAR isolates amplified by PCR. The PCR procedure and the sequencing primers were the same as previously reported.13 Sequences were analyzed using Lasergene software (DNASTAR) and compared with the ftsI gene sequence of the Rd KW 20 H influenzae strain to detect nucleotide substitutions. As previously described, different molecular groups were subsequently classified.14, 15 According to the amino acid transposition point of pbp3, it can be divided into types I, II, III, and III‐like.16 Group II isolates were typed based on other amino acid substitution patterns in the transpeptidase region of PBP3 (338‐573).13, 16 The other 13 substitutions used for typing in group II have been previously reported.16 Unreported types were defined as “Ch” plus capital letters to distinguish the unreported types from types reported in previous studies.
2.8. MLST
The STs of H influenzae isolates were determined by multi‐locus sequence typing as described in previous reports.17, 18 Briefly, seven housekeeping genes (adk, atpG, frdB, fucK, mdh, pgi, and recA) were amplified from chromosomal DNA, and the products were sent to BGI Company for sequencing on both strands. The STs were determined by comparing the allelic profiles with recognized STs on the MLST website (https://pubmlst.org/hinfluenzae/). New alleles and allelic profiles identified in the present study were submitted to the MLST database for name assignment. eBURST software (version 3; available at http://haemophilus.mlst.net/eburst/) was used to estimate the relationships between the isolates and to assign strains to a clonal complex (CC) using the stringent group definition of six of seven shared alleles.
2.9. Statistical analysis
The antimicrobial susceptibility data and STs were analyzed with WHONET 5.6 software, as recommended by the World Health Organization. The chi‐squared test and Fisher's exact test were used for statistical comparisons and performed with SPSS software (version 16.0; SPSS, Inc). A two‐tailed cutoff of P < .05 was considered as statistical significance.
3. RESULTS
3.1. H influenzae strain carriage rates
A total of 362 isolates of H influenzae were identified. The proportion of H influenzae positive culture in the study population was 9.1% (362/3984). The carriage rate of H influenzae by age and study site was shown in Table 1. The results showed that the carriage rates in different age groups ranged from 7.5% to 15.4%, and the highest carriage rate was in “3‐4” group. In addition, there was no significant difference in the carriage rates of any age group between the two study sites.
Table 1.
Carriage rates of Haemophilus influenzae isolates in different age group in two sites
| Age (years) | Total (n = 362) | Study sites | |||
|---|---|---|---|---|---|
| Beijing Children' Hospital (n = 271) | Youyang Hospital (n = 91) | χ 2 | P | ||
| <1 | 34 (9.3%) | 84 (9.6%) | 55 (8.9%) | 0.22 | .17 |
| 1‐2 | 27 (7.5%) | 44 (7.2%) | 13 (8.8%) | 0.45 | .24 |
| 2‐3 | 30 (8.2%) | 30 (7.9%) | 9 (9.6%) | 0.28 | .19 |
| 3‐4 | 56 (15.4%) | 47 (13.4%) | 9 (12.7%) | 0.02 | .22 |
| 4‐5 | 38 (10.6%) | 23 (11.2%) | 2 (6.5%) | 0.65 | .35 |
| ≥5 | 29 (7.7%) | 43 (8.5%) | 3 (3.3%) | 3.02 | .29 |
| Total | 362 (9.1%) | 271 (9.2%) | 91 (8.6%) | 0.36 | .16 |
3.2. Serotype distribution of H influenzae isolates
All of the H influenzae isolates tested in the present study could not be serotyped with latex agglutination antisera. In fact, no isolates of bexA and type b genes were successfully amplified.
3.3. β‐lactamase determination and antibiotic susceptibility pattern of H influenzae isolates
A total of 99 isolates were randomly chosen based on age from Beijing Children's Hospital, and a total of 91 isolates were randomly chosen from Youyang Hospital. All 190 isolates were tested for β‐lactamase activity and antibiotic susceptibility patterns. The test results were shown in Table 2. It was showed that among the 190 isolates, 44 (23.2%) were positive for β‐lactamase, 42 (22.1%) were positive for TEM, and none were positive for ROB. Although the susceptibility rates of isolates to ampicillin in the two study sites approached statistical significance, the susceptibility rates to other β‐lactamase antibiotics were close. All isolates were susceptible to ceftriaxone and levofloxacin. Moreover, susceptibility rates of isolates to erythromycin and sulfamethoxazole‐trimethoprim in Beijing Children's Hospital were significantly higher than susceptibility rates in Youyang Hospital (P < .05).
Table 2.
The results of β‐lactamase activity and antibiotic susceptibility pattern of H influenzae isolates
| β‐lactamase/Antimicrobials | Total (n = 190) | Study sites | |||
|---|---|---|---|---|---|
| Beijing (n = 99) |
Youyang (n = 91) |
χ 2 | P | ||
| β‐lactamase‐positive [n(%)] | 44 (23%) | 22 (22.2%) | 22 (24.1%) | 0.102 | .750 |
| TEM [n (%)] | 42 (95%) | 22 (100%) | 20 (90.9%) | ||
| ROB [n (%)] | 0 (0%) | 0 (0%) | 0 (0%) | ||
| Ampicillin | 3.845 | .050 | |||
| Susceptible [n (%)] | 112 (58.9%) | 65 (65.7%) | 47 (51.6%) | ||
| Intermediate [n (%)] | 25 (13.2%) | 10 (10.1%) | 15 (16.5%) | ||
| Resistant [n (%)] | 53 (27.9%) | 24 (24.2%) | 29 (31.9%) | ||
| MIC50 (mg/L) | 1 | 0.75 | 1 | ||
| MIC90 (mg/L) | ≥256 | 12 | ≥256 | ||
| MIC range (mg/L) | 0.094 to ≥256 | 0.094 to ≥256 | 0.125 to ≥256 | ||
| Amoxicillin‐clavulanic acid | 1.640 | .200 | |||
| Susceptible [n (%)] | 177 (93.2%) | 90 (90.9%) | 87 (95.6%) | ||
| Resistant [n (%)] | 13 (6.8%) | 9 (9.1%) | 4 (4.4%) | ||
| MIC50 (mg/L) | 1 | 0.5 | 1 | ||
| MIC90 (mg/L) | 3 | 4 | 3 | ||
| MIC range (mg/L) | 0.023 to ≥256 | 0.023 to ≥256 | 0.25 to ≥256 | ||
| Cefuroxime | 0.696 | .404 | |||
| Susceptible [n (%)] | 151 (79.5%) | 81 (81.8%) | 70 (76.9%) | ||
| Intermediate [n (%)] | 16 (8.4%) | 7 (7.1%) | 9 (9.9%) | ||
| Resistant [n (%)] | 23 (12.1%) | 11 (11.1%) | 12 (13.2%) | ||
| MIC50 (mg/L) | 1 | 1 | 1 | ||
| MIC90 (mg/L) | 16 | 16 | 24 | ||
| MIC range (mg/L) | 0.064 to ≥256 | 0.064 to ≥256 | 0.19 to ≥256 | ||
| Ceftriaxone | — | — | |||
| Susceptible [n (%)] | 190 (100%) | 99 (100%) | 91 (100%) | ||
| MIC50 (mg/L) | 0.016 | 0.016 | 0.016 | ||
| MIC90 (mg/L) | 0.19 | 0.19 | 0.19 | ||
| MIC range (mg/L) | 0.002‐5 | 0.002‐0.5 | 0.002‐0.25 | ||
| Levofloxacin | — | — | |||
| Susceptible [n (%)] | 190 (100%) | 99 (100%) | 91 (100%) | ||
| MIC50 (mg/L) | 0.016 | 0.016 | 0.016 | ||
| MIC90 (mg/L) | 0.25‐0.75 | 0.25 | 0.75 | ||
| MIC range (mg/L) | 0.004‐1 | 0.004‐0.5 | 0.008‐1 | ||
| Erythromycin | 13.970 | <.001 | |||
| Susceptible [n (%)] | 104 (54.7%) | 67 (67.7%) | 37 (40.7%) | ||
| Intermediate [n (%)] | — | — | — | ||
| Resistant [n (%)] | — | — | — | ||
| MIC50 (mg/L) | 4 | 4 | 6 | ||
| MIC90 (mg/L) | 256 | 64 | ≥256 | ||
| MIC range (mg/L) | 0.38 to ≥256 | 0.38 to ≥256 | 2 to ≥256 | ||
| Tetracycline | 2.885 | .089 | |||
| Susceptible [n (%)] | 179 (94.2%) | 96 (97%) | 83 (91.2%) | ||
| Intermediate [n (%)] | 4 (2.1%) | 2 (2%) | 2 (2.2%) | ||
| Resistant [n (%)] | 7 (3.7%) | 1 (1%) | 6 (6.6%) | ||
| MIC50 (mg/L) | 0.25 | 0.25 | 0.38 | ||
| MIC90 (mg/L) | 0.5 | 0.38 | 0.5 | ||
| MIC range (mg/L) | 0.047‐12 | 0.047‐12 | 0.19‐8 | ||
| Sulfamethoxazole‐trimethoprim | 4.322 | .038 | |||
| Susceptible [n (%)] | 73 (38.4%) | 45 (45.5%) | 28 (30.8%) | ||
| Intermediate [(%)] | 4.7% | 7.1% | 2.2% | ||
| Resistant [(%)] | 56.8% | 47.5% | 67% | ||
| MIC50 (mg/L) | ≥32 | 2 | ≥32 | ||
| MIC90 (mg/L) | ≥32 | ≥32 | ≥32 | ||
| MIC range (mg/L) | 0.008 to ≥32 | 0.008 to ≥32 | 0.047 to ≥32 | ||
| Chloramphenicol | 2.885 | .089 | |||
| Susceptible [n (%)] | 179 (94.2%) | 96 (96%) | 83 (91.2%) | ||
| Intermediate [n (%)] | 1 (0.5%) | 0 | 1 (1.1%) | ||
| Resistant [n (%)] | 10 (5.3%) | 4 (4%) | 6 (6.6%) | ||
| MIC50 (mg/L) | 0.5 | 0.38 | 0.5 | ||
| MIC90 (mg/L) | 0.75 | 0.5 | 1 | ||
| MIC range (mg/L) | 0.094 to ≥256 | 0.094‐8 | 0.25 to ≥256 | ||
3.4. Genotypes of BLNAR isolates
None of the TEM‐positive isolates were susceptible to ampicillin. The 36 isolates without β‐lactamase genes were not susceptible to ampicillin (ie, BLNAR strains), of which 14 (14.1%) were from Beijing Children's Hospital and 22 (24.2%) from Youyang Hospital. Among the 14 isolates of Beijing Children's Hospital, 10 isolates were in group III, and the others were in group II. Among the 22 isolates of Youyang Hospital, three isolates were in group III, four isolates were in group III‐like, one isolate was in group I, and the others were in group II (Table 3).
Table 3.
Categorization of PBP3 substitution types according to amino acid substitution pattern in the transpeptidase region of PBP3 (338‐573)
| Patterns | Groups | Types | No. | PBP3 substitutions for typing according to previous references 13 and 16 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D350 | S357 | M377 | S385 | I449 | G490 | A502 | R517 | N526 | A530 | T532 | V547 | N569 | ||||
| 01 | II | F | 1 | K | ||||||||||||
| 02 | II | O | 2 | T | K | I | S | |||||||||
| 03 | II | ChC | 1 | E | K | S | ||||||||||
| 04 | II | ChA | 6 | E | T | K | I | S | ||||||||
| 05 | II | ChD | 1 | V | K | |||||||||||
| 06 | II | B | 2 | V | K | I | S | |||||||||
| 07 | III‐like | 1 | I | T | H | S | I | S | ||||||||
| 08 | II | ChE | 1 | N | T | K | ||||||||||
| 09 | II | D | 3 | N | E | K | S | |||||||||
| 10 | II | ChB | 2 | N | N | K | ||||||||||
| 11 | I | 1 | N | N | I | H | I | S | ||||||||
| 12 | III | 8 | N | N | I | T | K | I | S | |||||||
| 13 | III | 1 | N | N | I | T | K | S | I | S | ||||||
| 14 | III‐like | 1 | N | N | I | T | H | I | ||||||||
| 15 | III‐like | 1 | N | N | I | T | H | I | S | |||||||
| 16 | III‐like | 1 | N | N | I | T | H | S | I | S | ||||||
| 17 | III | 1 | N | N | I | T | T | K | I | S | ||||||
| 18 | III | 1 | N | N | I | T | V | K | ||||||||
| 19 | III | 1 | N | N | I | T | E | K | S | |||||||
| Patterns | Groups | Types | No. | Other PBP3 substitutions found only in the present study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L389 | G489 | T512 | A520 | G555 | Y557 | Y558 | V562 | N568 | A586 | A587 | ||||
| 01 | II | F | 1 | L | ||||||||||
| 02 | II | O | 1 | E | ||||||||||
| 07 | III‐like | 1 | F | X | H | |||||||||
| 08 | II | ChE | 1 | T | ||||||||||
| 10 | II | ChB | 2 | L | ||||||||||
| 11 | I | 1 | F | S | H | |||||||||
| 12‐1 | III | 7 | F | L | ||||||||||
| 12‐2 | III | 1 | F | L | S | S | ||||||||
| 13 | III | 1 | F | E | H | S | ||||||||
| 14 | III‐like | 1 | F | E | H | |||||||||
| 15 | III‐like | 1 | F | H | ||||||||||
| 16 | III‐like | 1 | F | T | ||||||||||
| 17 | III | 1 | F | |||||||||||
| 18 | III | 1 | F | |||||||||||
| 19 | III | 1 | F | |||||||||||
The sequence analysis results revealed that 24 amino acid substitutions at 22 positions between amino acids 338 and 573 in PBP3 were associated with reference to H influenzae Rd KW20 (Table 3). Previously reported substitutions (A368T, A437S, and A554D) were not detected in present BLNAR isolates. Four types which were previously referred to as B, D, F, and O were detected in present study, and five new types were determined in group II isolates. Finally, 19 patterns were identified in the present research (Table 4). Although amino acid substitutions at the other nine positions between amino acids 338 and 573 in PBP3 were also detected, no additional patterns were revealed. Two additional substitutions at amino acids 586 and 587 were existed in an isolate of pattern 12. In group II, type ChA, identified in six isolates from Youyang Hospital, was the most prevalent type. In group III, pattern 12 was the most common type, and this pattern was found in six isolates from Beijing Children's Hospital and in two isolates from Youyang Hospital. Other patterns found in both Beijing Children's Hospital and Youyang Hospital were pattern 09 (type D) and pattern 10 (type ChB).
Table 4.
The MICs (mg/L) distribution of BLNAR isolates by different PBP3 groups and substitution types
| Patterns | Groups | Types | No. | Ampicillin | Amoxicillin/clavulanate | Cefuroxime | Ceftriaxone |
|---|---|---|---|---|---|---|---|
| 01 | II | F | 1 | 1.5 | 2 | 2 | 0.064 |
| 02 | II | O | 2 | 1.5 | 3 | 4‐8 | 0.016‐0.032 |
| 03 | II | ChC | 1 | 1.5 | 1.5 | 1.5 | 0.047 |
| 04 | II | ChA | 6 | 1.5‐3 | 2‐4 | 4‐12 | 0.016‐0.023 |
| 05 | II | ChD | 1 | 1.5 | 1.5 | 8 | 0.023 |
| 06 | II | B | 2 | 1.5 | 2 | 2‐4 | 0.016 |
| 07 | III‐like | 1 | 16 | 8 | 64 | 0.25 | |
| 08 | II | ChE | 1 | 1.5 | 6 | 6 | 0.016 |
| 09 | II | D | 3 | 1.5 | 2‐4 | 3‐6 | 0.032‐0.064 |
| 10 | II | ChB | 2 | 1.5‐2 | 3‐12 | 4‐24 | 0.064‐0.125 |
| 11 | I | 1 | 3 | 3 | 1.5 | 0.25 | |
| 12‐1 | III | 7 | 1‐6 | 0.125‐48 | 6‐>256 | 0.19‐0.5 | |
| 12‐2 | III | 1 | 1.5 | 8 | 24 | 0.38 | |
| 13 | III | 1 | 1.5 | 4 | 64 | 0.094 | |
| 14 | III‐like | 1 | 12 | 4 | >256 | 0.25 | |
| 15 | III‐like | 1 | 2 | 2 | >256 | 0.125 | |
| 16 | III‐like | 1 | 6 | 4 | >256 | 0.25 | |
| 17 | III | 1 | 1.5 | >256 | >256 | 0.38 | |
| 18 | III | 1 | 0.75 | 8 | 32 | 0.125 | |
| 19 | III | 1 | 6 | 4 | >256 | 0.19 |
The MIC50s of group II type ChB against ampicillin, amoxicillin/clavulanate, cefuroxime, and ceftriaxone were 2, 3, 6, and 0.023 mg/L, respectively.
The MIC50s against ampicillin, amoxicillin/clavulanate, cefuroxime, and ceftriaxone were 2, 6, >256, and 0.25 mg/L, respectively.
3.5. Multi‐locus sequence typing of H influenzae isolates
The multi‐locus sequence typing analysis revealed that 108 STs, the most common of which were ST408 (11, 5.8%) and ST914 (10, 5.3%), followed by ST57 (9, 4.7%) and ST834 (6, 3.2%). eBURST analysis identified 13 CCs and 68 singletons (Figure 1). Four CCs, each included 10 or more isolates, were as follows: CC1497 (21 isolates), CC259 (15 isolates), CC57 (11 isolates), and CC107 (10 isolates). Each of the following CC/STs contained five isolates: CC11, ST113, ST474, ST486, ST1556, and ST1558. Twelve STs were determined in both of the study sites, which covered 63 isolates.
Figure 1.
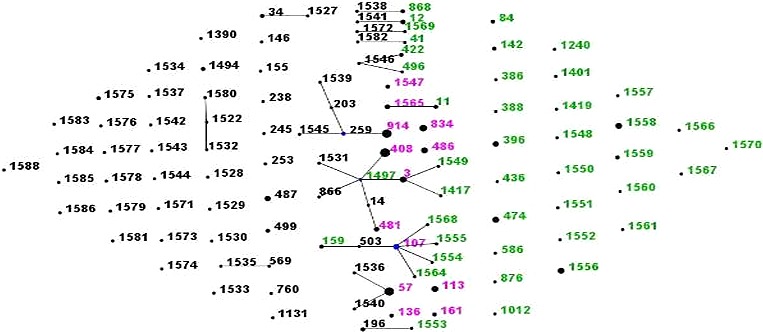
Population snapshot of 190 H influenzae strains revealed by eBURST analysis. The lines indicated the presence of single locus variant links in particular sequence types (STs), which were indicated by circles. The size of the circle corresponded to the number of isolates belonging to the ST. The green ST numbers indicated isolates from Youyang Hospital, the black numbers indicated isolates from Beijing Children's Hospital, and the pink numbers indicates isolates from Youyang Hospital and Beijing Children's Hospital
4. DISCUSSION
The present study revealed that the overall H influenzae carriage rate in children with respiratory tract infections was 9.1%, and the most common age was 3‐4 years old, which was similar to a report from Shanghai, China.19
In our study, none of the isolates could be typed by antisera, which was further confirmed by negative nucleic acid amplification of bexA and type b genes. This result was different from a study from Shanghai, China,19 in which all isolates were identified as type b. In most recent epidemiologic studies, isolates of H influenzae type b have been rarely detected. Jiang et al20 reported that Hib (2.7% of H influenzae isolates) was rare among Chinese children in Beijing, Shanghai, and Guangzhou. The proportion of Hib in H influenzae isolates was also very low in other studies (0%‐4.7%).21, 22, 23, 24 Since the typing method was not described in the Shanghai study,19 which reported 100% of Hib, it was possible that all H influenzae isolates were mistaken for type b.
In China, the Hib vaccines have been on the market in the private sector for nearly 20 years, and the coverage rate of vaccines are close to 50% in infants,25 which plays an important role in preventing Hib infection and transmission in China. After nearly 20 years of Hib immunization, NTHis have been identified more and more frequently. Hu26 and Hu27 reported that NTHis are responsible for 6.17% and 10.08% of pediatric lower respiratory tract infections, respectively. NTHis represent the major proportion of H influenzae isolates cultured in nasopharyngeal specimens from healthy individuals, and the proportion is usually more than 90% except the Hua study (80.7%).22 One study in Italy reported that after 15 years of introduction of Hib vaccine, 97.0% (98/101) of the isolates were NTHis in the oropharyngeal carriage rate of H influenzae in young children in two Italian cities.28 A study from Spain showed that with the introduction of the Hib vaccine, NTHis have become more and more common in invasive isolates.29 The NTHis can even cause infectious outbreaks in nursing homes with the spread of some clones. Andersson et al30 reported that an outbreak of the beta‐lactam‐resistant NTHi and ST14 was associated with severe clinical outcomes in Sweden. A corollary study showed that ST14 clone was associated with increased clinical virulence and resistance to several antimicrobial agents.31 ST14 has also been identified in one of the present isolates, which was included the most common CC1497. Therefore, CC1497 should be monitored in the future.
The β‐lactamase‐positive rate and the antimicrobial resistance pattern of the 190 isolates tested were similar to our previous report.3 BLNAR isolates were previously considered rare in China (0%‐1.3%).32, 33 However, in the current study, 36 isolates of BLNAR were identified from 190 test isolates (18.9%). The BLNAR proportion in Beijing Children's Hospital (14.1%) was clearly higher than that in previous investigation, and the BLNAR proportion in Youyang Hospital (24.2%) was more impressive.3 In Japan, where Hib vaccine had not been introduced in 2006, the BLNAR was determined in more than 50% of the H influenzae isolates.34 The increase in BLNAR isolates resulted in more discrepancies between the β‐lactamase‐positive and ampicillin‐resistant rates than before, and the β‐lactamase‐positive rate no longer represented the ampicillin‐resistant rate. Therefore, the resistant rates of amoxicillin/clavulanate, cefuroxime, and ceftriaxone were increased to varying degrees.
Compared with previous reports, known substitutions of PBP3 amino acid for grouping could be determined in each of the tested BLNAR isolate. Groups I, II, III, and III‐like were identified, and group III was the most common. Isolates in group III showed higher maximum MICs against ampicillin, amoxicillin/clavulanate, cefuroxime, and ceftriaxone than that in groups I and II. Characteristic alterations of PBP3 in group III usually cause cephalosporin resistance. The BLNAR isolates in groups III and III‐like were all determined as the third‐stage L389F substitution (first stage was R517H or N526K substitution, and second stage was S385T substitution).35 Interestingly, one isolate in group I was shown to have L389F and R517H substitutions, without S385T substitution. Most of the isolates in group II in the present study could not be shown to have any reported types based on the amino acid substitutions from 338 to 573 of the transpeptidase region of PBP3. A few isolates in group II showed MICs as high as group III, and some isolates in group III showed low MICs. It suggested that other amino acid substitutions might play a role in the resistance pattern. The variations in amino acid substitution and MIC values showed the complexity of resistance mechanism of BLNAR isolates.
The homology of present isolates was very weak. Among the typed 190 isolates, 108 STs were determined. Although some shared CCs/STs were identified in both Beijing Children's Hospital and Youyang Hospital, most singletons were found in only one site. High heterogeneity was also found in BLNAR isolates, which was in accordance with the variations in the ftsI amino acid substitutions. High heterogeneity of NTHi or BLNAR was also revealed in other cities or countries. Tian et al36 typed 273 isolates of H influenzae from pediatric pneumonia patients in Chengdu, China, and 50 different STs (including 39 novel STs) were identified by multi‐locus sequence typing. No ST was correlated with isolates from Korea, which is geographically adjacent to China. A study involving 316 invasive NTHi isolates from children in England and Wales during 2003‐2010 revealed that a genetically heterogeneous population (155 STs) had different biotypes.37 A report from Italy showed that 98 of 101 were NTHi isolates, and 76 STs were identified among the 98 NTHi strains, of which only four STs (ST12, ST57, ST238, and ST1238) comprised of more than three isolates.28 Among 18 BLNAR NTHis collected in Spain, 15 different genetically diverse STs were identified.29 Unlike high genetic variability of NTHis, encapsulated isolates express widely clonal characteristics.38 The high heterogeneity of NTHis is a block to the development of vaccines and may also be the reason for rare outbreaks of NTHi infections worldwide.
However, there were also some limitations in this study. Firstly, the sample from Youyang Hospital was stored at −60°C for 6 months. We assumed that the bacteria would not change in this process. Secondly, the sample size was too small for two hospitals to represent all hospitals in China. In future study, more hospitals located in different regions of China should be involved. It will be better to culture the bacteria in local laboratories than to do it after samples storage and transport.
5. CONCLUSION
In conclusion, all isolates in the present study were NTHi, which suggested widespread of this type in China. The BLNAR isolates were detected more frequently than before. Because high genetic diversity of NTHi isolates of H influenzae exist worldwide, it is important to continuously monitor these bacteria in the future.
AUTHORS' CONTRIBUTIONS
QD, WS, XC, CC, QM, KY, and SQ designed the study. XC and CC collected the specimens from Youyang. QD, WS, and QM identified the bacteria and performed the antimicrobial susceptibility tests. QD, WS, and KY performed the molecular studies. QD, WS, KY, and SQ collected the data, analyzed them, interpreted the results, and drafted the manuscript. All authors reviewed and revised the manuscript and approved the final version.
ETHICAL APPROVAL
Before enrollment and implementation of any study procedure, the parents and/or legal guardians of each participant signed the written informed consent document. This study was approved by the Ethics Committee of the two hospitals. No ethical problems were encountered in this study.
Dong Q, Shi W, Cheng X, et al. Widespread of non‐typeable Haemophilus influenzae with high genetic diversity after two decades use of Hib vaccine in China. J Clin Lab Anal. 2020;34:e23145 10.1002/jcla.23145
Funding information
This study was supported by a national clinical center project from the Ministry of Science and Technology of the People's Republic of China (2013BAI09B11) and the Beijing Talents Fund (2015000021469G209).
Contributor Information
Kaihu Yao, Email: jiuhu2655@sina.com.
Suyun Qian, Email: syqian1211@163.com.
DATA AVAILABILITY STATEMENT
The datasets analyzed during the present study are available from the corresponding authors Kaihu Yao (email address: jiuhu2655@sina.com) and Suyun Qian (email address: syqian1211@163.com) on reasonable request.
REFERENCES
- 1. Jacobs MR, Johnson CE. Macrolide resistance: an increasing concern for treatment failure in children. Pediatr Infect Dis J. 2003;22(Suppl):S131‐S138. [DOI] [PubMed] [Google Scholar]
- 2. Zhu H, Wang A, Tong J, et al. Nasopharyngeal carriage and antimicrobial susceptibility of Haemophilus influenzae among children younger than 5 years of age in Beijing, China. BMC Microbiol. 2015;15:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang YJ, Vuori‐Holopainen E, Yang Y, et al. Relative frequency of Haemophilus influenzae type b pneumonia in Chinese children as evidenced by serology. Pediatr Infect Dis J. 2002;21(4):271‐277. [DOI] [PubMed] [Google Scholar]
- 4. Wang A, Yu S, Yao K, et al. Antimicrobial susceptibility of Haemophilus influenzae strains and antibiotics usage patterns in pediatric outpatients: results from a children's hospital in China (2000–2004). Pediatric Pulmonol. 2008;43(5):457‐462. [DOI] [PubMed] [Google Scholar]
- 5. Alpuche C, Garau J, Lim V. Global and local variations in antimicrobial susceptibilities and resistance development in the major respiratory pathogens. Int J Antimicrob Agents. 2007;30(Suppl 2):135‐138. [DOI] [PubMed] [Google Scholar]
- 6. Kaijalainen T, Ruokokoski E, Ukkonen P, Herva E. Survival of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis frozen in skim milk‐tryptone‐glucose‐glycerol medium. J Clin Microbiol. 2004;42(1):412‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O'Brien KL, Bronsdon MA, Dagan R, et al. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J Clin Microbiol. 2001;39(3):1021‐1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strålin K, Bäckman A, Holmberg H, Fredlund H, Olcén P. Design of a multiplex PCR for Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae and Chlamydophila pneumoniae to be used on sputum samples. APMIS. 2005;113(2):99‐111. [DOI] [PubMed] [Google Scholar]
- 9. Strålin K, Korsgaard J, Olcén P. Evaluation of a multiplex PCR for bacterial pathogens applied to bronchoalveolar lavage. Eur Respir J. 2006;28(3):568‐575. [DOI] [PubMed] [Google Scholar]
- 10. Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. PCR for capsular typing of Haemophilus influenzae . J Clin Microbiol. 1994;32(10):2382‐2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davis GS, Sandstedt SA, Patel M, Marrs CF, Gilsdorf JR. Use of bexB to detect the capsule locus in Haemophilus influenzae . J Clin Microbiol. 2011;49(7):2594‐2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clinical and Laboratory Standards Institute (CLSI) . Performance standards for antimicrobial susceptibility testing. In: Twenty‐sixth informational supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2016; M100‐S26. [Google Scholar]
- 13. Skaare D, Allum AG, Anthonisen IL, et al. Mutant ftsI genes in the emergence of penicillin‐binding protein‐mediated beta‐lactam resistance in Haemophilus influenzae in Norway. Clin Microbiol Infect. 2010;16(8):1117‐1124. [DOI] [PubMed] [Google Scholar]
- 14. Dabernat H, Delmas C, Seguy M, et al. Diversity of beta‐lactam resistance‐conferring amino acid substitutions in penicillin‐binding protein 3 of Haemophilus influenzae . Antimicrob Agents Chemother. 2002;46(7):2208‐2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ubukata K, Shibasaki Y, Yamamoto K, et al. Association of amino acid substitutions in penicillin‐binding protein 3 with beta‐lactam resistance in beta‐lactamase‐negative ampicillin‐resistant Haemophilus influenzae . Antimicrob Agents Chemother. 2001;45(6):1693‐1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skaare D, Anthonisen IL, Caugant DA, et al. Multilocus sequence typing and ftsI sequencing: a powerful tool for surveillance of penicillin‐binding protein 3‐mediated beta‐lactam resistance in nontypeable Haemophilus influenzae . BMC Microbiol. 2014;14:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144(Pt 11):3049‐3060. [DOI] [PubMed] [Google Scholar]
- 18. Meats E, Feil EJ, Stringer S, et al. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol. 2003;41(4):1623‐1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu J, Sun X, Huang Z, et al. Streptococcus pneumoniae and Haemophilus influenzae type b carriage in Chinese children aged 12–18 months in Shanghai, China: a cross‐sectional study. BMC Infect Dis. 2016;16:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang M, Wang YJ, Gao W, et al. Genotyping of ampicillin‐resistant Haemophilus influenzae . Chin J Pediatrics. 2005;43(9):685‐689. (in Chinese). [PubMed] [Google Scholar]
- 21. Hu YY, Yu SJ, Liu G, Gao W, Yang YH. Antimicrobial susceptibility of Haemophilus influenzae among children in Beijing, China, 1999–2000. Acta Paediatr. 2002;91(2):136‐140. [DOI] [PubMed] [Google Scholar]
- 22. Hua CZ, Sun LY, Li JP, Zhao LY, Chen ZM, Yu HM. Carriage and serotypes of Haemophilus influenza in young children attending a kindergarten in Hangzhou. Chin J Lab Med. 2005;28(4):389‐391. (in Chinese). [Google Scholar]
- 23. Deng QL, Deng L, Xie YQ, et al. Study on serotype and the resistance of Haemophilus influenzae from children with respiratory tract infection in Guangzhou. Chin J Pract Pediatrics. 2009;24(5):362‐365. (in Chinese). [Google Scholar]
- 24. Ye JL, Mei LL, Luo Y, et al. Study on carriage status and typing of Haemophilus influenzae among children in Zhejiang, 2007–2008. Disease Surveillance. 2009;24(10):782‐785. (in Chinese). [Google Scholar]
- 25. Zheng JS, Cao L, Cao LS, Yuan P, Wang HQ. Analysis on reported data of immunization monitoring system for category II vaccine in China, 2012. Zhonghua Yu Fang Yi Xue Za Zhi. 2013;47(10):928‐932. (in Chinese). [PubMed] [Google Scholar]
- 26. Hu Y, Xia XL, Huang HL, et al. Study on serotype and drug resistance of Haemophilus influenzae isolates from children with respiratory tract infection in Kunming. Exp Lab Med. 2013;31(2):118‐120. (in Chinese). [Google Scholar]
- 27. Hu J, Wang X, Ai T, et al. Multicenter prospective epidemiological studies on Haemophilus influenzae infection among hospitalized children with lower respiratory tract infections. Zhonghua Er Ke Za Zhi. 2016;54(2):119‐125. (in Chinese). [DOI] [PubMed] [Google Scholar]
- 28. Giufrè M, Daprai L, Cardines R, et al. Carriage of Haemophilus influenzae in the oropharynx of young children and molecular epidemiology of the isolates after fifteen years of H influenzae type b vaccination in Italy. Vaccine. 2015;33(46):6227‐6234. [DOI] [PubMed] [Google Scholar]
- 29. García‐Cobos S, Arroyo M, Pérez‐Vázquez M, et al. Isolates of β‐lactamase‐negative ampicillin‐resistant Haemophilus influenzae causing invasive infections in Spain remain susceptible to cefotaxime and imipenem. J Antimicrob Chemother. 2014;69(1):111‐116. [DOI] [PubMed] [Google Scholar]
- 30. Andersson M, Resman F, Eitrem R, et al. Outbreak of a beta‐lactam resistant non‐typeable Haemophilus influenzae sequence type 14 associated with severe clinical outcomes. BMC Infect Dis. 2015;15:581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Månsson V, Skaare D, Riesbeck K, Resman F. The spread and clinical impact of ST14CC‐PBP3 type IIb/A, a clonal group of non‐typeable Haemophilus influenzae with chromosomally mediated β‐lactam resistance‐a prospective observational study. Clin Microbiol Infect. 2017;23(3):209. [DOI] [PubMed] [Google Scholar]
- 32. Gong SD, Hua CZ, Li JP, Chen XJ, Shang SQ, Chen ZM. Analysis on antibiotic resistance patterns and epidemiological features of 546 Haemophilus influenzae strains isolated from children during 2007–2014 in Children's Hospital, Zhejiang University School of Medicine. Chin J Pract Pediatrics. 2016;31(12):915‐919. (in Chinese). [Google Scholar]
- 33. Wang H, Ning YZ, Chen HB, et al. Guideline and consensus: standardization of antibacterial susceptibility test report. Chin J Lab Med. 2016;39(01):18‐22. [Google Scholar]
- 34. Hasegawa K, Kobayashi R, Takada E, et al. Nationwide Surveillance for Bacterial Meningitis. High prevalence of type b beta‐lactamase‐non‐producing ampicillin‐resistant Haemophilus influenzae in meningitis: the situation in Japan where Hib vaccine has not been introduced. J Antimicrob Chemother. 2006;57(6):1077‐1082. [DOI] [PubMed] [Google Scholar]
- 35. Skaare D, Anthonisen IL, Kahlmeter G, et al. Emergence of clonally related multidrug resistant Haemophilus influenzae with penicillin‐binding protein 3‐mediated resistance to extended‐spectrum cephalosporins, Norway, 2006 to 2013. Euro Surveill. 2014;19(49). 10.2807/1560-7917.es2014.19.49.20986 [DOI] [PubMed] [Google Scholar]
- 36. Tian G, Zhang L, Li M, et al. Genotypic characteristics of Haemophilus influenzae isolates from pediatric pneumonia patients in Chengdu city, Sichuan, China. J Microbiol. 2009;47(4):494‐497. [DOI] [PubMed] [Google Scholar]
- 37. Collins S, Vickers A, Ladhani SN, et al. Clinical and molecular epidemiology of childhood invasive nontypeable Haemophilus influenzae disease in England and Wales. Pediatr Infect Dis J. 2016;35(3):e76‐e84. [DOI] [PubMed] [Google Scholar]
- 38. Staples M, Graham RMA, Jennison AV. Characterisation of invasive clinical Haemophilus influenzae isolates in Queensland, Australia using whole‐genome sequencing. Epidemiol Infect. 2017;145(8):1727‐1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the present study are available from the corresponding authors Kaihu Yao (email address: jiuhu2655@sina.com) and Suyun Qian (email address: syqian1211@163.com) on reasonable request.


