Abstract
Background
This study aimed to explore the predictive value of integrin α7 (ITGA7) for acute myeloid leukemia (AML) risk and subsequently investigate its correlation with risk stratification and prognosis in AML patients.
Methods
Bone marrow samples were obtained from 196 de novo AML patients prior to initiation of treatment and from 50 subjects underwent bone marrow donation or bone marrow biopsy for non‐hematologic malignant disease (as controls). ITGA7 mRNA and protein expressions were detected by real‐time quantitative polymerase chain reaction and Western blot assays, respectively. In AML patients, the risk stratification was assessed, and complete remission (CR), event‐free survival (EFS), and overall survival (OS) were evaluated.
Results
Both ITGA7 mRNA and protein expressions were increased in AML patients compared with controls, and their expressions were correlated with poorer risk stratification. For prognosis, ITGA7 mRNA expression and protein expression were declined in CR patients compared to non‐CR patients. Meanwhile, both EFS and OS were shorter in ITGA7 mRNA high expression patients compared to ITGA7 mRNA low expression patients, as well as ITGA7 protein high expression patients compared to ITGA7 protein low expression patients.
Conclusion
Integrin α7 might serve as a potential biomarker for predicting increased AML risk and worse prognosis in AML patients.
Keywords: acute myeloid leukemia, complete remission, event‐free survival, integrin α7, overall survival
1. INTRODUCTION
Acute myeloid leukemia (AML), a kind of medullary blood cell cancer, is featured by the clonal disorder of hemopoietic stem cells characterized by differentiation suppression and the cellular accumulation at different stages of incomplete maturation, as well as decreased production of healthy hemopoietic elements.1 Clinically, a larger number of AML patients present with several clinical symptoms including feeling tired, shortness of breath, easy bruising and bleeding, as well as increased risk of infection.1 Despite progress in AML treatment in recent decades, there has been little progress in improving the prognosis for AML patients. According to previous studies, AML could be cured in just 35%‐40% patients who are 60 years of age or younger, and only 5%‐15% patients who are older than 60 years of age.2 In order to solve the problem of poor prognosis in AML, investigation of candidate biomarkers is essential to early diagnosis and disease progress monitoring to improve AML prognosis.
Integrins are one family of cell adhesion molecules composing of heterodimeric α and β subunits connected by noncovalent interactions, which could mediate cell‐cell or cell‐matrix adhesion to devoting into multiple physiological and pathological processes.3, 4 Integrin α7 (ITGA7), belonging to the integrin family of adhesion molecules, is a member of the extracellular matrix binding proteins localized on chromosome 12p13 and consists of over 27 exons spanning a region of about 22.5 kb, which participants in a broad spectrum of cellular processes (such as survival, proliferation, migration and invasion), and has been identified as tumor promoter in various solid carcinomas (including oral squamous cell carcinoma (OSCC)).5, 6 Meanwhile, ITGA7 has been considered as a functional cancer stem cells (CSCs) marker involved in the moderation of stem cell‐like properties in several cancer cells (including OSCC and glioblastoma).6, 7 Although the oncogenic role of ITGA7 has been described in different solid carcinomas, no data have been reported so far on its role in hematologic malignancies, including AML. Considering the information mentioned above about ITGA7 as the tumor promoter in solid carcinomas, and the oncogenic influence of integrins on the pathology of hematologic malignancies (particularly AML), we hypothesized that ITGA7 might also have deteriorated influence on disease progression and prognosis in AML patients.8, 9 In the present study, our aim was to explore the predictive value of ITGA7 for AML risk and subsequently investigate its correlation with risk stratification, complete remission (CR), event‐free survival (EFS), and overall survival (OS) in AML patients.
2. METHODS
2.1. Participants
From January 2016 to December 2018, 196 de novo AML patients treated in Guizhou Provincial People's Hospital were consecutively recruited in this study. Patients were included if they met the following criteria: (a) confirmed diagnosis of primary AML in accordance with World Health Organization (WHO) Classification of Tumors of Hematopoietic and Lymphoid Tissues (2008); (b) age above 18 years; (c) life expectancy more than 12 months, which was assessed by the experienced physician according to the clinical status and examinations; (d) able to be followed up regularly. The exclusion criteria included the following: (a) secondary or relapsed AML; (b) received treatment for AML before recruitment; (c) presenting with severe infection; (d) complicated with other malignancies; and (e) pregnant or breastfeeding women. In addition, 50 subjects who underwent bone marrow donation or bone marrow biopsy for non‐hematologic malignant disease during the same period were enrolled in the study as controls. The present study protocol was approved by the Institutional Review Board of Guizhou Provincial People's Hospital and was conducted in accordance with the provisions of the Declaration of Helsinki. All participants signed the informed consents before recruitment.
2.2. Clinical data collection
After enrollment, baseline clinical characteristics of AML patients were documented, which consisted of age, gender, French‐American‐British (FAB) classification, cytogenetics, molecular genetics, white blood cell (WBC) count, and so on. The risk stratification was assessed based on the cytogenetics and molecular genetics according to the National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology of AML (Version 1.2015).
2.3. Bone marrow sample collection and detection
After the collection of written informed consents from the participants, bone marrow samples were obtained from all participants (for AML patients, collection of bone marrow sample was performed prior to initiation of treatment). Then, real‐time quantitative polymerase chain reaction (RT‐qPCR) assay was conducted to detect the ITGA7 mRNA relative expression in the bone marrow, and the Western blot was applied to determine the ITGA7 protein expression in the bone marrow.
2.4. RT‐qPCR for ITGA7 mRNA expression
The extraction of total RNA was performed by using RNeasy Protect Mini Kit (Qiagen). Subsequently, 1ug RNA was reversely transcribed to cDNA through using ReverTra Ace® qPCR RT Kit (Toyobo). qPCR was carried out by THUNDERBIRD® SYBR® qPCR Mix (Toyobo). The qPCR amplification was performed in the following condition: 95°C for 5 minutes, followed by 40 cycles of 95°C for 5 seconds, 61°C for 30 seconds. The primers of ITGA7 were as follows: forward (5′‐>3′): GCCACTCTGCCTGTCCAATG, reverse (5′‐>3′): CGGAGGTGCTAAGGATGAGGTA. The calculation was performed by using 2‐△△Ct. Glyceraldehyde‐phosphate dehydrogenase (GAPDH) was used as the internal reference. The primers of GAPDH were as follows: forward (5′‐>3′): TGACCACAGTCCATGCCATCAC, reverse (5′‐>3′): GCCTGCTTCACCACCTTCTTGA.
2.5. Western blot for ITGA7 protein expression
Total protein extraction was carried out using RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific), and the measurement of protein concentration was measured by using Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). The 20 μg protein sample was separated by NuPAGE™ 4%‐12% Bis‐Tris Protein Gel (Invitrogen) and then transferred to nitrocellulose filter membrane (Millipore, USA). After blocked with bull serum albumin (BSA) (Sigma), incubation was performed with the primary antibody (Rabbit Anti‐ITGA7 Antibody [1:200 dilution, Abcam]) at 4°C overnight, and then the secondary antibody (horseradish peroxidase‐conjugated goat anti‐rabbit immunoglobulin G antibody [1:1000 dilution, Abcam]) was used for incubation at room temperature for 1h. After exposed by the enhanced chemiluminescence (ECL) and X‐ray film (Kodak), the membrane was photographed by using ImageQuant LAS‐4000 (GE Healthcare). The density of immunoblotting results was determined by ImageJ software (Java), and the relative density of target protein was normalized by GAPDH density as a ratio.
2.6. Treatment and follow‐up
Appropriate induction chemotherapy was administered to patient according to the clinical status, and the chemotherapy regimens were formulated by the treating physician referring to NCCN Clinical Practice Guidelines in Oncology of AML (Version 1.2015). After induction chemotherapy, remission status was assessed. CR was defined as bone marrow (BM) with at least 20% cellularity and BM blasts below 5% at steady state after treatment, without cytological evidence of leukemia, no transfusion requirement, leukocyte count above 1 × 109/L and platelet count above 100 × 109/L. Surveillance and follow‐up were performed as the Guidelines recommended. The last follow‐up date was December 31st, 2018, and follow‐up duration was ranging from 2.0 to 36.0 months with a median follow‐up duration of 16.0 months. EFS was defined as the time interval from initiation of treatment to disease recurrence, progression, or death; OS was defined as the time interval from initiation of treatment to death.
2.7. Statistical analysis
Data analyses were performed using SPSS 24.0 (SPSS Inc), and figures were plotted using GraphPad Prism 7.01 (GraphPad Software Inc). Continuous variables were displayed as mean value ± standard deviation (SD) or median and interquartile range (IQR), and categorical variables were displayed as number (percentage). Comparisons between groups were determined by Wilcoxon rank‐sum test, and correlation analyses were determined by Spearman's rank correlation test. OS and EFS were illustrated by Kaplan‐Meier curve, and the difference of OS and EFS between groups was determined by the log‐rank test. P value <.05 was considered as significant.
3. RESULTS
3.1. Baseline characteristics in AML patients
The mean age of these 196 de novo AML patients was 45.8 ± 15.0 years, and there were 112 (57.1%) male and 84 (42.9%) female (Table 1). For FAB classification, the number of AML patients with M1, M2, M4, M5, and M6 was 2 (1.0%), 64 (32.7%), 55 (28.1%), 59 (30.0%), and 16 (8.2%), respectively. The number of AML patients with monosomal karyotype, FLT3‐ITD mutation, isolated biallelic CEBPA mutation, and NPM1 mutation was 15 (7.7%), 48 (24.5%), 17 (8.7%), and 68 (34.7%), respectively. For risk stratification, there were 52 (26.5%) patients with favorable‐risk stratification, 75 (38.3%) patients with intermediate‐risk stratification, and 69 (35.2%) patients with poor‐risk stratification. Detailed information of other baseline characteristics was shown in Table 1.
Table 1.
Baseline characteristics of patients
| Items | AML patients (N = 196) |
|---|---|
| Age (years), mean ± SD | 45.8 ± 15.0 |
| Gender, No. (%) | |
| Male | 112 (57.1) |
| Female | 84 (42.9) |
| FAB classification, No. (%) | |
| M1 | 2 (1.0) |
| M2 | 64 (32.7) |
| M4 | 55 (28.1) |
| M5 | 59 (30.0) |
| M6 | 16 (8.2) |
| Cytogenetics, No. (%) | |
| −5 or −5q | 1 (0.5) |
| t(9; 11) | 4 (2.0) |
| t(9; 22) | 5 (2.6) |
| −7 or −7q | 5 (2.6) |
| 11q23 | 7 (3.6) |
| +8 | 8 (4.1) |
| t(8; 21) | 9 (4.6) |
| inv(16) or t(16; 16) | 19 (9.7) |
| CK | 22 (11.2) |
| NK | 95 (48.4) |
| Others (not included in better or poor risk) | 21 (10.7) |
| Monosomal karyotype, No. (%) | 15 (7.7) |
| FLT3‐ITD mutation, No. (%) | 48 (24.5) |
| Isolated biallelic CEBPA mutation, No. (%) | 17 (8.7) |
| NPM1 mutation, No. (%) | 68 (34.7) |
| Risk stratification, No. (%) | |
| Favorable risk | 52 (26.5) |
| Intermediate risk | 75 (38.3) |
| Poor risk | 69 (35.2) |
| WBC (×109/L), median (IQR) | 16.8 (8.1‐27.8) |
Abbreviations: AML, acute myeloid leukemia; CEBPA, CCAAT/enhancer‐binding protein α; CK, complex karyotype; FAB classification, French‐American‐Britain classification systems; FLT3‐ITD, internal tandem duplications in the FMS‐like tyrosine kinase 3; IQR, interquartile range; NK, normal karyotype; NPM1, nucleophosmin 1; SD, standard deviation; WBC, white blood cell.
3.2. The ITGA7 mRNA and protein expressions in AML patients
The median value of ITGA7 mRNA expression was 1.615 (0.680‐2.918) in AML patients, while was 1.036 (0.593‐1.668) in controls, and it was higher in AML patients compared to controls (P < .001; Figure 1A). Western blot assay was performed to detect the ITGA7 protein expression, and the examples of ITGA7 protein expressions in AML patient and control were shown in Figure 1B, and the median value of ITGA7 protein expression was higher in AML patients (0.704 [0.486‐1.180]) than that in controls (0.578 [0.373‐0.730]; P < .001; Figure 1C).
Figure 1.
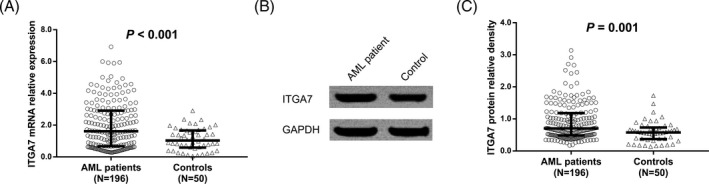
Integrin α7 expression in AML patients and controls. A, Comparison of ITGA7 mRNA expression between AML patients and controls; B, the examples of ITGA7 protein expressions in AML patient and control; C, comparison of ITGA7 protein expression between AML patients and controls. P value <.05 was considered as significant. Comparisons between groups were determined by Wilcoxon rank‐sum test. ITGA7, integrin α7; AML, acute myeloid leukemia
3.3. The ITGA7 mRNA and protein expressions in AML patients with different risk stratification
The median value of ITGA7 mRNA expression in AML patients with favorable risk, intermediate risk, and poor risk was 0.978 (0.428‐2.467), 1.644 (0.677‐2.882), and 2.022 (1.285‐3.422), respectively, and ITGA7 high mRNA expression was correlated with poorer risk stratification in AML patients (P < .001; Figure 2A). In addition, Western blot assay was carried out to detect ITGA7 protein expression, and the examples of ITGA7 protein expressions in AML patients with favorable risk, intermediate risk, and poor risk were shown in Figure 2B. The median value of ITGA7 protein expression in AML patients with favorable risk, intermediate risk, and poor risk was 0.607 (0.399‐1.015), 0.612 (0.483‐1.444), and 0.903 (0.666‐1.221), respectively, and ITGA7 high protein expression was associated with poorer risk stratification in AML patients (P = .002) as well (Figure 2C).
Figure 2.
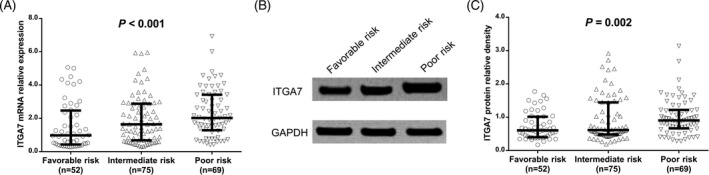
Integrin α7 expressions in AML patients with different risk stratification. A, comparison of ITGA7 mRNA expression among AML patients with favorable‐risk stratification, AML patients with intermediate‐risk stratification and AML patients with poor‐risk stratification; B, the examples of ITGA7 protein expression in AML patients with favorable risk, intermediate risk, and poor risk; C, comparison of ITGA7 protein expression among AML patients with favorable‐risk stratification, AML patients with intermediate‐risk stratification and AML patients with poor‐risk stratification. Correlation analyses were determined by Spearman's rank correlation test. P value <.05 was considered as significant. ITGA7, integrin α7; AML, acute myeloid leukemia
3.4. The ITGA7 mRNA and protein expressions in CR patients and non‐CR patients
There were 155(79.1%) CR patients and 41 (20.9%) non‐CR patients (Figure 3A). The median value of ITGA7 mRNA expression was 1.554 (0.637‐2.678) in CR patients and 2.597 (0.862‐4.015) in non‐CR patients, and it was reduced in CR patients compared to non‐CR patients (P = .012; Figure 3B). Also, the median value of ITGA7 protein expression was decreased in CR patients (0.686 [0.483‐1.021]) than that in non‐CR patients (1.098 [0.500‐1.606]; P = .028; Figure 3C).
Figure 3.
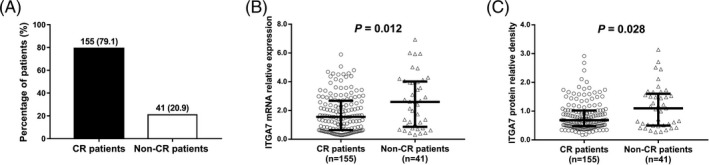
Comparison of ITGA7 expression between CR patients and non‐CR patients. A, The percentage of patients achieved CR and patients who did not achieve CR; B, comparison of ITGA7 mRNA expression between CR patients and non‐CR patients; C, comparison of ITGA7 protein expression between CR patients and non‐CR patients. Comparisons between groups were determined by Wilcoxon rank‐sum test. P value <.05 was considered as significant. CR, complete remission; ITGA7, integrin α7; AML, acute myeloid leukemia
3.5. Correlations of ITGA7 mRNA and protein expressions with EFS and OS in AML patients
According to the median value, all AML patients were divided into ITGA7 mRNA high expression group and ITGA7 mRNA low expression group, and EFS was shorter in ITGA7 mRNA high expression patients (median value: 8.0 [6.2‐9.8] months) compared to ITGA7 mRNA low expression patients (median value: 24.0 [17.4‐30.6] months; P < .001; Figure 4A). Meanwhile, OS was worse in ITGA7 mRNA high expression patients (median value: 22.0 [19.6‐24.5] months) compared to ITGA7 mRNA low expression patients (median value: 30.2 [28.3‐32.2] months; P < .001) as well (Figure 4B). In addition, we also classified all AML patients into ITGA7 protein high expression patients and ITGA7 protein low expression patients and we discovered that EFS in ITGA7 protein high expression patients (median value: 9.0 [6.7‐11.3] months) was shorter compared to ITGA7 protein low expression patients (median value: 19.0 [13.0‐25.0] months; P = .001; Figure 4C), and OS was also worse in ITGA7 protein high expression patients (median value: 23.4 [20.6‐26.2] months) than that in ITGA7 protein low expression patients (median value: 29.5 [27.4‐31.5] months; P = .001) as well (Figure 4D).
Figure 4.
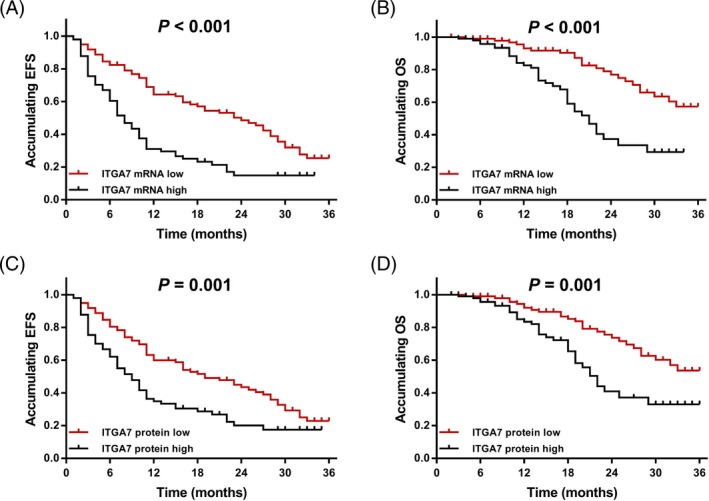
The EFS and OS in AML patients. A, The correlation of ITGA7 mRNA expression with EFS in AML patients; B the correlation of ITGA7 mRNA expression with OS in AML patients; C, the correlation of ITGA7 protein expression with EFS in AML patients; and D, the correlation of ITGA7 protein expression with OS in AML patients. OS and EFS were illustrated by Kaplan‐Meier curve, and the difference of OS and EFS between groups was determined by the log‐rank test. P value <.05 was considered as significant. EFS, event‐free survival; OS, overall survival; ITGA7, integrin α7; AML, acute myeloid leukemia
4. DISCUSSION
In this study, we found three interesting results as follows: firstly, ITGA7 expression was higher in AML patients compared to controls. Secondly, ITGA7 high expression was correlated with poorer risk stratification in AML patients. Thirdly, ITGA7 high expression was correlated with lower CR achievement, worse EFS and shorter OS in AML patients.
Integrin, large and complex transmembrane glycoproteins belonging to adhesion receptors, modulates various cell functions upon ligand binding, which serve as mechanosensors and mechanotransducers of the extracellular environment.3, 10 As one of common members of the integrin family, ITGA7 is a kind of primary extracellular matrix receptor and could use its β1 chain to form heterodimers, subsequently transducing signals from the matrix to cells, which has been discovered to promote tumor progression of different carcinomas via multiple pathways.11 For instance, an appealing study shows that ITGA7 binds with S100P to activate focal adhesion kinase (FAK)/serine protein kinase (AKT)‐ zinc finger E‐box Binding Homeobox 1 (ZEB1) signaling way, which thereby promotes cell migration and cell invasion in lung cancer.12 Another recent report provides strong in vitro and in vivo evidence that ITGA7 interacts with laminin‐induced outside‐in signaling to participant in the growth and invasion of glioblastoma stem‐like cells.10 Meanwhile, an interesting in vitro experiment reveals that ITGA7 knockdown reduces cell proliferation and cell invasion, while enhances cell apoptosis rate in breast cancer cells.13 Additionally, a previous study highlights a surprising function of ITGA7 as a tumor promoter in OSCC malignancy that ITGA7 not only upregulates stemness‐associated genes expressions (such as octamer‐binding transcription factor 4 [OCT4], SRY [sex‐determining region Y]‐box 2 [SOX2], Nanog homeobox [NANOG] and CD90), but also enhances abilities to self‐renew of OSCC cells via activating the FAK‐mediated signaling pathways.6 Together with these previous studies, ITGA7 contributes to the development and progression processes of different types of solid carcinomas.
Although several studies have been performed for investigating the underlying mechanism of ITGA7 in different solid carcinomas, there is still no study investigating the role of ITGA7 in hematologic malignancies, including AML. However, several previous studies have been performed for investigation the roles of integrins in hematologic malignancies, particular in AML. For example, β2 integrins (including Mac‐1) have been reported to activate spleen tyrosine kinase (Syk)/signal transduction and transcription activator (SATA) signaling axis to promote AML cell survival and proliferation.8 Another study reveals that AlphavBeta3 integrin interacts with the fibroblast growth factor receptor to enhances AML cell proliferation.9 Further, alpha4beta1 integrin binding and CXCR4 chemokine receptors (CD184) activation are prerequisites for the migration of CD34+ haematopoietic progenitors and AML cells beneath marrow stromal cells.14 Hence, various integrins might participate in the pathology of AML. Based on the above information that ITGA7 presents with oncogenic effects on the pathology of different solid carcinomas, and several integrins have potential to promote development and progression of hematologic malignancies, particularly AML, we hypothesized that ITGA7 might also contribute to the deterioration in AML patients. In order to investigate the role of ITGA7 in AML patients, we herein enrolled 196 de novo AML patients as well as 50 controls, and we discovered that both ITGA7 mRNA and protein expressions were higher in AML patients compared to controls. One possible reason was that ITGA7 interacted with FAK/Akt signaling and Syk/SATA signaling axis to promote AML cell survival and proliferation, which subsequently caused high risk of AML.8, 12 Another reason was that ITGA7 could increase leukemia stem cells (LSC) properties including self‐renew through mediating various pathways (such as activating the FAK‐mediated signaling pathways and interacting with FMS‐like tyrosine kinase‐3), thereby contributed to the increased AML risk.12, 15 Although there are no data about the role of ITGA7 in hematologic malignancies, including AML, the results in this study were in line with these previous studies that disclose the increased expression of ITGA7 in patients with different solid carcinomas (including glioblastoma, OSCC, and prostate cancer).6, 10, 16
Risk stratification has been considered as the golden standard for predicting the prognosis in AML patients.17 For the sake of exploration whether ITGA7 has an impact on the prognosis in AML patients, we investigated the correlation of ITGA7 with risk stratification of AML, and we discovered that ITGA7 high expression was associated with poorer risk stratification in AML patients. Therefore, we further investigated the direct influence of ITGA7 on prognosis, and the results showed that ITGA7 high expression was correlated with unfavorable CR achievement, worse EFS, and shorter OS in AML patients, which might due to that (a) ITGA7 could regulate multiple genes or pathways to not only promote cell survival and proliferation (such as activating FAK/AKT‐ZEB1 signaling way), but also induce LSC properties including self‐renew (such as promoting FAK‐mediated signaling pathways and interacting with laminin‐induced outside‐in signaling), subsequently aggravated disease severity and lead to worse prognosis in AML patients.6, 10, 12 (b) ITGA7 promoted chemoresistance via the FAK/Akt/Caspase‐9/Caspase‐3 pathway, thereby decreased the efficacy of chemotherapy and caused lower CR achievement as well as worse survival in AML patients.6 However, the detailed mechanism of ITGA7 in AML and the clear reasons for the predictive value of ITGA7 for survival in AML are still unclear, further study is necessary. These results suggested that ITGA7 may represent a potentially valuable target for anti‐tumor therapy for AML, which might open a new avenue for future studies on the nature of ITGA7 as potential biomarker for development in AML patients.
Three impressive results were discovered in this study, whereas several limitations still existed. The detailed information was as follows: (a) All patients were just from our hospital, selected bias might exist. Thus, further multi‐center study is necessary. (b) 196 de novo AML patients were enrolled in this study, while there were just 50 subjects underwent bone marrow donation or bone marrow biopsy for non‐hematologic malignant disease as controls recruited. A different number of patients and controls might cause lower statistical power. (c) The underlying mechanism of ITGA7 in AML is still unclear, further study is needed. (d) we performed RT‐qPCR to detect ITGA7 mRNA expression, and Western blot assays to determine ITGA7 protein expression, while further validation with IHC assays for detection of ITGA7 expression is necessary. (e) In this study, all subjects who had to undergo bone marrow donation or bone marrow biopsy for non‐hematologic malignant disease during the same period could be enrolled as controls, while bone marrow donation and bone marrow biopsy were invasive operation, which would cause bleeding, pain, and infection risk, and thus, there were just several subjects who were willing to undergo bone marrow donation or bone marrow biopsy for non‐hematologic malignant disease. Therefore, the sample size of controls in this study was less (of 50 subjects), which was far less compared to AML patients. Further study with the same number of AML patients and controls is greatly needed.
In summary, ITGA7 high expression correlates with increased disease risk, poorer risk stratification, and worse prognosis in AML, which might serve as a potential biomarker for AML in the future.
CONFLICTS OF INTEREST
The authors of this work have nothing to disclose.
ACKNOWLEDGMENTS
None.
Zeng M, Ding S, Zhang H, Huang Q, Ren W, Guo P. Predictive value of integrin α7 for acute myeloid leukemia risk and its correlation with prognosis in acute myeloid leukemia patients. J Clin Lab Anal. 2020;34:e23151 10.1002/jcla.23151
Zeng and Ding contributed equally to this work.
REFERENCES
- 1. Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894‐1907. [DOI] [PubMed] [Google Scholar]
- 2. Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136‐1152. [DOI] [PubMed] [Google Scholar]
- 3. Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673‐687. [DOI] [PubMed] [Google Scholar]
- 4. Zhang X, Li L, Li N, et al. Salt bridge interactions within the beta2 integrin alpha7 helix mediate force‐induced binding and shear resistance ability. FEBS J. 2018;285(2):261‐274. [DOI] [PubMed] [Google Scholar]
- 5. Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10(1):9‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ming XY, Fu L, Zhang LY, et al. Integrin alpha7 is a functional cancer stem cell surface marker in oesophageal squamous cell carcinoma. Nat Commun. 2016;7:13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carrasco‐Garcia E, Auzmendi‐Iriarte J, Matheu A. Integrin alpha7: a novel promising target in glioblastoma stem cells. Stem Cell Investig. 2018;5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oellerich T, Oellerich MF, Engelke M, et al. Beta2 integrin‐derived signals induce cell survival and proliferation of AML blasts by activating a Syk/STAT signaling axis. Blood. 2013;121(19):3889‐3899, S3881‐S3866. [DOI] [PubMed] [Google Scholar]
- 9. Shah CA, Bei L, Wang H, Altman JK, Platanias LC, Eklund EA. Cooperation between alphavbeta3 integrin and the fibroblast growth factor receptor enhances proliferation of Hox‐overexpressing acute myeloid leukemia cells. Oncotarget. 2016;7(34):54782‐54794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haas TL, Sciuto MR, Brunetto L, et al. Integrin alpha7 Is a functional marker and potential therapeutic target in glioblastoma. Cell Stem Cell. 2017;21(1): 35‐50 e39. [DOI] [PubMed] [Google Scholar]
- 11. Tan LZ, Song Y, Nelson J, Yu YP, Luo JH. Integrin alpha7 binds tissue inhibitor of metalloproteinase 3 to suppress growth of prostate cancer cells. Am J Pathol. 2013;183(3):831‐840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsu YL, Hung JY, Liang YY, et al. S100P interacts with integrin alpha7 and increases cancer cell migration and invasion in lung cancer. Oncotarget. 2015;6(30):29585‐29598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai X, Gao C, Zhang L, Yang S. Integrin alpha7 high expression correlates with deteriorative tumor features and worse overall survival, and its knockdown inhibits cell proliferation and invasion but increases apoptosis in breast cancer. J Clin Lab Anal. 2019;33:e22979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burger JA, Spoo A, Dwenger A, Burger M, Behringer D. CXCR4 chemokine receptors (CD184) and alpha4beta1 integrins mediate spontaneous migration of human CD34+ progenitors and acute myeloid leukaemia cells beneath marrow stromal cells (pseudoemperipolesis). Br J Haematol. 2003;122(4):579‐589. [DOI] [PubMed] [Google Scholar]
- 15. Muranyi AL, Dedhar S, Hogge DE. Targeting integrin linked kinase and FMS‐like tyrosine kinase‐3 is cytotoxic to acute myeloid leukemia stem cells but spares normal progenitors. Leuk Res. 2010;34(10):1358‐1365. [DOI] [PubMed] [Google Scholar]
- 16. Ren B, Yu YP, Tseng GC, et al. Analysis of integrin alpha7 mutations in prostate cancer, liver cancer, glioblastoma multiforme, and leiomyosarcoma. J Natl Cancer Inst. 2007;99(11):868‐880. [DOI] [PubMed] [Google Scholar]
- 17. Estey EH. Acute myeloid leukemia: 2013 update on risk‐stratification and management. Am J Hematol. 2013;88(4):318‐327. [DOI] [PubMed] [Google Scholar]


