Abstract
Background
This study aimed to investigate the correlations of long non‐coding RNA maternally expressed gene 3 (lnc‐MEG3), microRNA (miR)‐21, and lnc‐MEG3/miR‐21 axis with disease risk, inflammation, disease severity, and 28‐day mortality of sepsis.
Methods
Totally, 219 sepsis patients and 219 health controls (HCs) were enrolled. Plasma samples were obtained from sepsis patients within 24 hours after admission and from HCs on enrollment to detect lnc‐MEG3 and miR‐21 expressions by real‐time quantitative polymerase chain reaction.
Results
The lnc‐MEG3 expression and lnc‐MEG3/miR‐21 axis were increased, while miR‐21 expression was decreased in sepsis patients compared with HCs. Lnc‐MEG3 (area under the curve (AUC): 0.887, 95% confidence interval (CI): 0.856‐0.917) and lnc‐MEG3/miR‐21 axis (AUC: 0.934, 95% CI: 0.909‐0.958) had good values for predicting elevated sepsis risk, while miR‐21 (AUC: 0.801, 95% CI: 0.758‐0.844) presented a good predictive value for reduced sepsis risk. Furthermore, lnc‐MEG3 expression and lnc‐MEG3/miR‐21 axis positively correlated with, whereas miR‐21 expression negatively correlated with acute pathologic and chronic health evaluation II, sequential organ failure assessment score, serum creatinine, C‐reactive protein, tumor necrosis factor‐α, interleukin (IL)‐1β, IL‐6, and IL‐17 in sepsis patients. Additionally, lnc‐MEG3 (AUC: 0.704, 95% CI: 0.626‐0.783) and lnc‐MEG3/miR‐21 axis (AUC: 0.669, 95% CI: 0.589‐0.750) exhibited acceptable values in predicting higher 28‐day mortality risk, while miR‐21 (AUC: 0.588, 95% CI: 0.505‐0.672) presented a poor predictive value for lower 28‐day mortality risk in sepsis patients.
Conclusion
Lnc‐MEG3 might serve as a potential biomarker for the development, progression, and prognosis prediction of sepsis via interacting with miR‐21.
Keywords: 28‐day mortality, disease severity, Lnc‐MEG3, miR‐21, sepsis
1. INTRODUCTION
Sepsis, a devastating and life‐threatening syndrome, is caused by an aberrant systemic host response to infections, resulting in excessive inflammatory responses and multiple organ failures.1 Sepsis has emerged as a major public health problem with an increasing incidence worldwide, being the main contributor of death among patient admitted to the intensive care units.2 Despite the improvements in antibiotic treatments, ventilator management, and resuscitative strategies, the prognosis of sepsis is still unsatisfied due to under‐recognition of sepsis, delayed assessment and development of antibiotic resistance.1, 3 Currently, there is still a lack of sensible and efficient disease monitoring and prognostic biomarkers for sepsis in clinical practice. Therefore, it is of great need to explore novel and rapidly measurable biomarkers for assisting on the identification of sepsis and prognosis prediction in sepsis patients.
Long non‐coding RNAs (LncRNAs), a class of non‐coding RNA, are characterized by non‐coding transcripts longer than 200 nucleotides in length with no open reading frame, which function as decoys, scaffolds, guides, and enhancers to regulate DNA, RNA and proteins.4 Among the identified lncRNAs, long non‐coding RNA maternally expressed gene 3 (lnc‐MEG3) is reported to be positively associated with inflammation and organ injury.5, 6 For instance, lnc‐MEG3 enhance inflammatory response via upregulating the expressions of early growth response protein 1 and Toll‐like receptor 4.5 Another study illuminates that overexpression of lnc‐MEG3 upregulates the creatine kinase and lactate dehydrogenase activities, inducing cell apoptosis and myocardial ischemia‐reperfusion injury.6 Furthermore, it is reported that lnc‐MEG3 acts as a molecular sponge for several microRNAs such as microRNA (miR)‐21, miR‐181a, and miR‐7‐5p to inhibit their downstream signaling pathways.5, 7, 8 Among these target miRNAs, miR‐21 functions as an important contributor for preventing inflammation and organ dysfunctions such as liver, kidney, and lung in sepsis.9, 10, 11 Based on these previous studies, we hypothesized that lnc‐MEG3 might be involved in the development and progression of sepsis via interacting with miR‐21. Therefore, this study aimed to investigate the correlations of lnc‐MEG3, miR‐21 and lnc‐MEG3/miR‐21 axis with disease risk, inflammation, disease severity, and 28‐day mortality of sepsis.
2. MATERIALS AND METHODS
2.1. Participants
This study consecutively recruited 219 sepsis patients between July 2016 and June 2019. All patients included in this study must met the following criteria: (a) diagnosed as sepsis based on the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis‐3)12; (b) age above 18 years; (c) not complicated with hematological malignancies or solid tumors; (d) seronegative for human immunodeficiency virus (HIV); (e) willing to participate in the study. The exclusion criteria were as follows: (a) received immunosuppressant within 3 months before enrollment; (b) died within 24 hours after admission; (c) history of stem cell transplant; (d) complicated with autoimmune disorders (eg, systemic lupus erythematosus, rheumatoid arthritis); and (e) pregnant or lactating woman. In addition, 219 healthy subjects who had no history of malignancies or severe infection, or no obvious abnormalities in biochemical indexes were enrolled as healthy controls (HCs), during the same period. The study was approved by the Ethics Committee of our hospital. All participants or their guardians signed the informed consents.
2.2. Data and sample collection
After enrollment, all clinical features of sepsis patients were recorded. Demographic characteristics included age, gender, body mass index (BMI), and smoke status. Chronic comorbidities included chronic obstructive pulmonary disease (COPD), cardiomyopathy, chronic kidney failure, and cirrhosis. Biochemical indexes included serum creatinine (Scr), albumin, white blood cell (WBC), C‐reactive protein (CRP), tumor necrosis factor‐α(TNF‐α), interleukin‐1β (IL‐1β), IL‐6, and IL‐17. Besides, peripheral blood samples of sepsis patients were collected within 24 hours after admission, and peripheral blood samples of HCs were collected on enrollment. Then, the peripheral blood samples were immediately separated for plasma at the condition of 3000 g for 25 minutes (4°C). The levels of TNF‐α, IL‐1β, IL‐6, and IL‐17 in plasma were measured by enzyme‐linked immunosorbent assay (ELISA), and the levels of lnc‐MEG3 and miR‐21 in plasma were detected by real‐time quantitative polymerase chain reaction (qPCR).
2.3. Severity assessment
The severity of sepsis was assessed using acute pathologic and chronic health evaluation II (APACHE II) score, which included acute physiology score, age score, and chronic health score. The total score of APACHE II ranges from 0 to 71, and the higher score represented more severe disease condition.13 The severity of organ dysfunction of sepsis patients was evaluated using sequential organ failure assessment (SOFA) score, which included respiration score, coagulation score, liver score, cardiovascular score, central nervous system score, and renal score. The total SOFA score was calculated by summing up each item scores (range 0‐24), and higher score represented more severe organ dysfunction.14
2.4. ELISA
Commercial human ELISA Kits (Thermo Fisher Scientific) were used to measure the level of TNF‐α, IL‐1β, IL‐6, and IL‐17 in plasma, and all procedures were performed according to the manufacturer's instruction. In brief, plasma samples were added to the wells of target‐specific antibody pre‐coated microplate. After incubation, the wells were washed to remove unbound material. The sandwich was formed by the addition of the second antibody, and a tetramethylbenzidine substrate was added to produce measurable signal. After stop solution was added, the optical density was measured at 450 nm wavelengths on microplate reader (BioTek).
2.5. Real‐time quantitative polymerase chain reaction (qPCR)
Firstly, total RNA was extracted from plasma using QIAamp RNA Blood Mini Kit (Qiagen). Then, cDNA was synthesized using iScript™ Reverse Transcription Supermix (Bio‐Rad). Subsequently, qPCR was performed to quantify lnc‐MEG3 and miR‐21 relative expressions using QuantiNova SYBR Green PCR Kit (Qiagen). Lnc‐MEG3 relative expression was calculated by 2−△△Ct method using GAPDH as internal reference, and miR‐21 relative expression was calculated by 2−△△Ct method using U6 as internal reference. Primers (designed based on a previous study) applied were as follows15: lnc‐MEG3, forward: 5′ CTCCCCTTCTAGCGCTCACG 3′, reverse: 5′ CTAGCCGCCGTCTATACTACCGGCT 3′; GAPDH, forward: 5′ GAGTCCACTGGCGTCTTCAC 3′, reverse: 5′ ATCTTGAGGCTGTTGTCATACTTCT 3′; miR‐21, forward: 5′ ACACTCCAGCTGGGTAGCTTATCAGACTGA 3’, reverse: 5′ TGTCGTGGAGTCGGCAATTC 3′; U6 forward: 5′ CTCGCTTCGGCAGCACATATACTA 3′, reverse: 5′ ACGAATTTGCGTGTCATCCTTGC 3′.
2.6. Treatment and follow‐up
Appropriate treatment regimens were given to the sepsis patients according to guideline of sepsis.16 All patients were followed up to 28 days after enrollment, and during follow‐up, the number of deaths was recorded to calculate 28‐day mortality. And based on the survival status in 28‐day follow‐up, patients were categorized into survivors and deaths.
2.7. Statistical analysis
Statistical analysis was performed with the use of SPSS 24.0 software (IBM), and figure was made using GraphPad Prism 7.01 software (GraphPad Software). Continuous variables were expressed as mean ± standard deviation (SD) or median and interquartile range (IQR). Categorical variables were expressed as count (percentage). Comparison of lnc‐MEG3 relative expression, miR‐21 relative expression, or lnc‐MEG3/miR‐21 axis between two groups was determined by Wilcoxon rank‐sum test. Correlation of lnc‐MEG3 relative expression, miR‐21 relative expression, or lnc‐MEG3/miR‐21 axis with APACHE II score, SOFA score, and biochemical indexes was analyzed by Spearman's rank correlation test. The performances of continuous variables in predicting sepsis risk and 28‐day mortality risk were performed using receiver operating characteristic (ROC) curve and the area under the curve (AUC) with 95% confidence interval (CI). P value < 0.05 was considered significant.
3. RESULTS
3.1. Patients’ clinical characteristics
The mean age was 56.5 ± 10.3 years in sepsis patients. And there were 76 (34.7%) females and 143 (65.3) males. Besides, smoke was found in 81 (37.0%) sepsis patients. As for chronic comorbidities, 32 (14.6%), 73 (33.3%), 23 (10.5%), and 43 (19.6%) sepsis patients were with COPD, cardiomyopathy, chronic kidney failure, and cirrhosis, respectively. Furthermore, the mean APACHE II score and mean SOFA score were 13.7 ± 6.1 and 6.2 ± 2.8, respectively, in sepsis patients. The detailed information of other characteristics was listed in Table 1.
Table 1.
Clinical characteristics of sepsis patients
| Items | Sepsis patients (N = 219) |
|---|---|
| Age (y), Mean ± SD | 56.5 ± 10.3 |
| Gender, No. (%) | |
| Female | 76 (34.7) |
| Male | 143 (65.3) |
| BMI (kg/m2), Mean ± SD | 23.3 ± 5.0 |
| Smoke, No. (%) | 81 (37.0) |
| Chronic comorbidities, No. (%) | |
| COPD | 32 (14.6) |
| Cardiomyopathy | 73 (33.3) |
| Chronic kidney failure | 23 (10.5) |
| Cirrhosis | 43 (19.6) |
| APACHE II score, Mean ± SD | 13.7 ± 6.1 |
| SOFA score, Mean ± SD | 6.2 ± 2.8 |
| Scr (mg/dL), median (IQR) | 1.6 (1.2‐2.3) |
| Albumin (g/L), median (IQR) | 26.8 (21.6‐35.6) |
| WBC (×109/L), median (IQR) | 12.6 (2.9‐26.8) |
| CRP (mg/L), median (IQR) | 106.1 (61.4‐154.7) |
| TNF‐α (pg/mL), median (IQR) | 212.5 (131.4‐315.3) |
| IL‐1β (pg/mL), median (IQR) | 9.7 (4.3‐19.5) |
| IL‐6 (pg/mL), median (IQR) | 90.5 (48.3‐171.3) |
| IL‐17 (pg/mL), median (IQR) | 192.1 (95.0‐288.3) |
Abbreviations: APACHE II, acute pathologic and chronic health evaluation II; BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRP, C‐reactive protein; IL, interleukin; IQR, interquartile range; Scr, serum creatinine; SD, standard deviation; SOFA, sequential organ failure assessment; TNF‐α, tumor necrosis factor‐α; WBC, white blood cell.
3.2. The values of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis for predicting sepsis risk
The lnc‐MEG3 relative expression was elevated in sepsis patients (2.325 [1.703‐3.912]) compared with HCs (0.994 [0.662‐1.362]; P < .001; Figure 1A). While, the miR‐21 relative expression was reduced in sepsis patients (0.277 [0.193‐0.451]) compared with HCs (0.967 [0.400‐1.630]; P < .001; Figure 1B). Besides, the lnc‐MEG3/miR‐21 axis was increased in sepsis patients (8.362 [4.011‐16.874]) compared with HCs (1.058 [0.729‐1.720]; Figure 1C). Further ROC curve analysis displayed that lnc‐MEG3 (AUC: 0.887, 95% CI: 0.856‐0.917), miR‐21 (AUC: 0.801, 95% CI: 0.758‐0.844), and lnc‐MEG3/miR‐21 axis (AUC: 0.934, 95% CI: 0.909‐0.958) exhibited good values for predicting sepsis risk, among which lnc‐MEG3/miR‐21 axis had a numerically superior predictive value for sepsis risk (Figure 1D).
Figure 1.
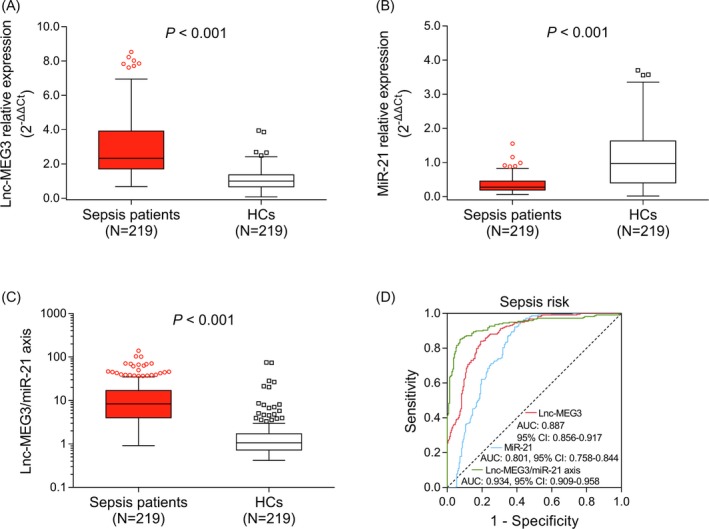
The predictive values of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis for sepsis risk. The comparisons of lnc‐MEG3 relative expression (A), miR‐21 relative expression (B), and lnc‐MEG3/miR‐21 axis (C) between sepsis patients and HCs. And the performances of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis in predicting sepsis risk (D). The comparisons of lnc‐MEG3 relative expression, miR‐21 relative expression and lnc‐MEG3/miR‐21 axis between sepsis patients and HCs were assessed by Wilcoxon rank‐sum test. P < .05 was considered significant. The abilities of lnc‐MEG3 relative expression, miR‐21 relative expression, and lnc‐MEG3/miR‐21 axis in predicting sepsis risk were identified by ROC curve and AUC with 95% CI. Lnc‐MEG3, long non‐coding RNA maternally expressed gene 3; AUC, area under the curve; CI, confidence interval; HCs, healthy controls; miR‐21, microRNA‐21; and ROC, receiver operating characteristic
3.3. Associations of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis with APACHE II and SOFA score in sepsis patients
Lnc‐MEG3 relative expression (P < .001, r = .398) and lnc‐MEG3/miR‐21 axis (P < .001, r = .494) were positively associated with APACHE II score, while miR‐21 relative expression was negatively associated with APACHE II score in sepsis patients (P < .001, r = −.469; Figure 2A‐C). Regarding SOFA score, lnc‐MEG3 relative expression (P < .001, r = .345) and lnc‐MEG3/miR‐21 axis (P < .001, r = .428) were positively associated with SOFA score, while miR‐21 relative expression was negatively associated with SOFA score in sepsis patients (P < .001, r = −.404; Figure 2D‐F).
Figure 2.
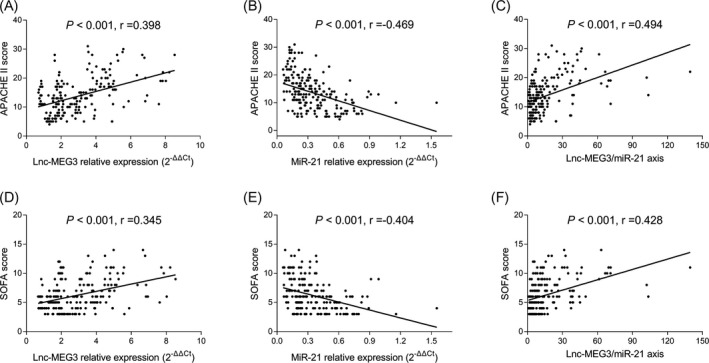
Correlations of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis with disease severity in sepsis patients. The associations of lnc‐MEG3 relative expression (A), miR‐21 relative expression (B), and lnc‐MEG3/miR‐21 axis (C) with APACHE II score in sepsis patients. Besides, the associations of lnc‐MEG3 relative expression (D), miR‐21 expression (E) and lnc‐MEG3/miR‐21 axis (F) with SOFA score in sepsis patients. The associations of lnc‐MEG3 relative expression, miR‐21 expression and lnc‐MEG3/miR‐21 axis with APACHE II score and SOFA score in sepsis patients were evaluated by Spearman's rank correlation test. P < .05 was considered significant. APACHE II, acute pathologic and chronic health evaluation II; Lnc‐MEG3, long non‐coding RNA maternally expressed gene 3; miR‐21, microRNA‐21; SOFA, sequential organ failure assessment
3.4. Associations of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis with biochemical and inflammatory indexes in sepsis patients
The lnc‐MEG3 relative expression was positively correlated with Scr (P = .001, r = .230), CRP (P < .001, r = .476), TNF‐α (P < .001, r = .305), IL‐1β (P < .001, r = .312), IL‐6 (P < .001, r = .354), and IL‐17 (P < .001, r = .375), while negatively correlated with albumin in sepsis patients (P = .044, r = −.136; Table 2). No correlation of lnc‐MEG3 relative expression with WBC was observed in sepsis patients (P = .587, r = .037). As for miR‐21 relative expression, it was negatively correlated with Scr (P = .003, r = −.202), CRP (P < .001, r = −.280), TNF‐α (P = .026, r = −.150), IL‐1β (P = .001, r = −.231), IL‐6 (P = .008, r = −.180), and IL‐17 (P = .002, r = −.212), while positively correlated with albumin in sepsis patients (P < .001, r = 0.403; Table 2). And there was no correlation of miR‐21 relative expression with WBC in sepsis patients (P = .225, r = −.082). In terms of lnc‐MEG3/miR‐21 axis, it was positively correlated with Scr (P < .001, r = .255), CRP (P < .001, r = .440), TNF‐α (P < .001, r = .268), IL‐1β (P < .001, r = .322), IL‐6 (P < .001, r = .307), and IL‐17 (P < .001, r = .338), while negatively correlated with albumin in sepsis patients (P < .001, r = −.299; Table 2). And no correlation of lnc‐MEG3/miR‐21 axis with WBC was observed in sepsis patients (P = .359, r = .062).
Table 2.
Correlation of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis with biochemical indexes in sepsis patients
| Items | Lnc‐MEG3 relative expression | MiR‐21 relative expression | Lnc‐MEG3/miR‐21 axis | |||
|---|---|---|---|---|---|---|
| P value | Correlation coefficient (r) | P value | Correlation coefficient (r) | P value | Correlation coefficient (r) | |
| Scr | .001 | 0.230 | .003 | −0.202 | <0.001 | .255 |
| Albumin | .044 | −0.136 | <.001 | 0.403 | <0.001 | ‐.299 |
| WBC | .587 | 0.037 | .225 | −0.082 | 0.359 | .062 |
| CRP | <.001 | 0.476 | <.001 | −0.280 | <0.001 | .440 |
| TNF‐α | <.001 | 0.305 | .026 | −0.150 | <0.001 | .268 |
| IL‐1β | <.001 | 0.312 | .001 | −0.231 | <0.001 | .322 |
| IL‐6 | <.001 | 0.354 | .008 | −0.180 | <0.001 | .307 |
| IL‐17 | <.001 | 0.375 | .002 | −0.212 | <0.001 | .338 |
Correlation was determined by spearman rank correlation test.
Abbreviations: CRP, C‐reactive protein; IL, interleukin; Scr, serum creatinine; TNF, tumor necrosis factor; WBC, white blood cell.
3.5. Associations of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis with comorbidities in sepsis patients
The lnc‐MEG3 relative expression was associated with cardiomyopathy (P < .014), while there was no association of lnc‐MEG3 relative expression with COPD (P = .854), chronic kidney failure (P = .851), or cirrhosis (P = .448) in sepsis patients (Table 3). As for miR‐21 relative expression, no correlation of miR‐21 relative expression with COPD (P = .945), cardiomyopathy (P = .859), chronic kidney failure (P = .916), or cirrhosis (P = .805) was observed in sepsis patients (Table 3). Regarding lnc‐MEG3/miR‐21 axis, there was no correlation of lnc‐MEG3/miR‐21 axis with COPD (P = .800), cardiomyopathy (P = .141), chronic kidney failure (P = .792), or cirrhosis (P = .359) in sepsis patients (Table 3).
Table 3.
Correlation of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis with comorbidities in sepsis patients
| Items | Lnc‐MEG3, median (IQR) | MiR‐21, median (IQR) | Lnc‐MEG3/miR‐21 axis, median (IQR) |
|---|---|---|---|
| COPD | |||
| No | 2.400 (1.648‐3.912) | 0.284 (0.193‐0.432) | 7.684 (4.155‐17.033) |
| Yes | 2.004 (1.777‐3.904) | 0.239 (0.184‐0.505) | 8.763 (3.311‐14.516) |
| P value | .854 | .945 | .800 |
| Cardiomyopathy | |||
| No | 2.092 (1.643‐3.446) | 0.265 (0.188‐0.460) | 7.609 (4.004‐15.278) |
| Yes | 3.065 (1.788‐4.624) | 0.305 (0.199‐0.424) | 10.314 (4.056‐22.540) |
| P value | .014 | .859 | .141 |
| Chronic kidney failure | |||
| No | 2.225 (1.707‐3.822) | 0.281 (0.188‐0.457) | 8.051 (4.120‐16.723) |
| Yes | 3.432 (1.169‐4.779) | 0.259 (0.217‐0.432) | 9.534 (2.888‐23.252) |
| P value | .851 | .916 | .792 |
| Cirrhosis | |||
| No | 2.759 (1.594‐4.010) | 0.277 (0.193‐0.431) | 8.598 (4.176‐17.919) |
| Yes | 2.035 (1.874‐3.055) | 0.277 (0.183‐0.475) | 7.477 (3.376‐12.695) |
| P value | .448 | .805 | .359 |
Comparison was determined by Wilcoxon rank‐sum test.
Abbreviations: COPD, chronic obstructive pulmonary disease; IQR, interquartile range.
3.6. The comparisons of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis between survivors and deaths in sepsis patients
Sepsis patients were categorized into survivors and deaths based on survival status in 28‐day follow‐up. The lnc‐MEG3 relative expression was higher in deaths (3.469 [1.898‐5.126]) than that in survivors (2.080 [1.536‐3.507]; P < .001; Figure 3A). And the miR‐21 relative expression was lower in deaths (0.251 [0.171‐0.360]) than that in survivors (0.287 [0.193‐0.479]; P = .049; Figure 3B). Besides, the lnc‐MEG3/miR‐21 axis was raised in deaths (13.572 [6.032‐29.089]) compared with survivors (7.195 [3.289‐13.197]; P < .001; Figure 3C).
Figure 3.
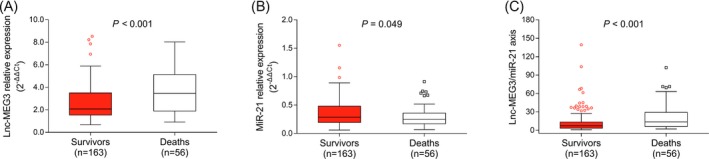
The lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis in survivors and deaths. The comparisons of lnc‐MEG3 relative expression (A), miR‐21 relative expression (B), and lnc‐MEG3/miR‐21 axis (C) between survivors and deaths, which were conducted by Wilcoxon rank‐sum test. P < .05 was considered significant. Lnc‐MEG3, long non‐coding RNA maternally expressed gene 3; miR‐21, microRNA‐21
3.7. The predictive values of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis for 28‐day mortality risk in sepsis patients
Lnc‐MEG3 (AUC: 0.704, 95% CI: 0.626‐0.783) and lnc‐MEG3/miR‐21 axis (AUC: 0.669 95% CI: 0.589‐0.750) presented acceptable values for predicting 28‐day mortality risk, while miR‐21 had a poor value for predicting 28‐day mortality risk in sepsis patients (AUC: 0.588, 95% CI: 0.505‐0.672; Figure 4A). Besides, APACHE II score (AUC: 0.793, 95% CI: 0.729‐0.857) and SOFA score (AUC: 0.758, 95% CI: 0.687‐0.830) were with relatively good predictive values for 28‐day mortality risk in sepsis patients (Figure 4B). Furthermore, Scr (AUC: 0.724, 95% CI: 0.648‐0.800), WBC (AUC: 0.624, 95% CI: 0.547‐0.701), and CRP (AUC: 0.711, 95% CI: 0.631‐0.791) presented acceptable values in predicting 28‐day mortality risk, while albumin failed to predict 28‐day mortality risk in sepsis patients (AUC: 0.556, 95% CI: 0.464‐0.647; Figure 4C). Additionally, TNF‐α (AUC: 0.629, 95% CI: 0.548‐0.710), IL‐1β (AUC: 0.709, 95% CI: 0.636‐0.783), and IL‐17 (AUC: 0.616, 95% CI: 0.531‐0.700) had fair predictive values for 28‐day mortality risk, whereas IL‐6 could not predict 28‐day mortality risk in sepsis patients (AUC: 0.576, 95% CI: 0.492‐0.659; Figure 4D). These suggested that lnc‐MEG3 and lnc‐MEG3/miR‐21 axis exhibited relatively good values for predicting 28‐day mortality risk in sepsis patients, which were similar with commonly applied prognostic factors such as APACHE II score, and SOFA score. Whereas, miR‐21 had a poor predictive value for 28‐day mortality risk in sepsis patients, which was much less valuable compared with those common prognostic factors.
Figure 4.
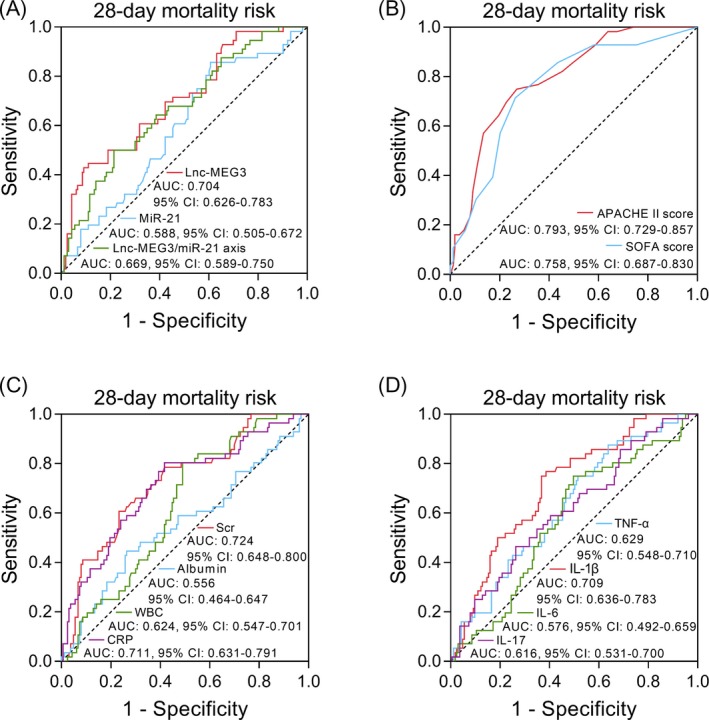
ROC curve analyses of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis for predicting 28‐day mortality risk in sepsis patients. The performances of lnc‐MEG3, miR‐21, lnc‐MEG3/miR‐21 axis (A), APACHE II, SOFA score (B), Scr, albumin, WBC, CRP (C), TNF‐α, IL‐1β, IL‐6, and IL‐17 (D) in predicting 28‐day mortality risk in sepsis patients were assessed by ROC curve and AUC with 95% CI. APACHE II, acute pathologic and chronic health evaluation II; AUC, area under the curve; CI, confidence interval; CRP, C‐reactive protein; IL, interleukin; Lnc‐MEG3, long non‐coding RNA maternally expressed gene 3; miR‐21, microRNA‐21; ROC, receiver operating characteristic; Scr, serum creatinine; SOFA, sequential organ failure assessment; TNF‐α, tumor necrosis factor‐α; WBC, white blood cell
4. DISCUSSION
In the present study, we have discovered that: (a) Lnc‐MEG3 and lnc‐MEG3/miR‐21 axis had good values for predicting higher sepsis risk, while miR‐21 was of good predictive value for lower sepsis risk, among which lnc‐MEG3/miR‐21 axis presented a numerically superior predictive value for sepsis risk. (b) Lnc‐MEG3 and lnc‐MEG3/miR‐21 axis were positively associated with, but miR‐21 was negatively associated with APACHE II score, SOFA score, Scr, CRP, TNF‐α, IL‐1β, IL‐6, and IL‐17 in sepsis patients. And lnc‐MEG3 as well as lnc‐MEG3/miR‐21 axis negatively correlated with, whereas miR‐21 positively correlated with albumin in sepsis patients. (c) Lnc‐MEG3 and lnc‐MEG3/miR‐21 axis exhibited acceptable values for predicting elevated 28‐day mortality risk, while miR‐21 presented a poor predictive value for reduced 28‐day mortality risk in sepsis patients.
Lnc‐MEG3, an imprinted gene, is transcribed from human chromosome 14q32.3 that is implied in the induction of inflammatory response and multiple organ dysfunction such as heart.8, 17, 18 For instance, the overexpression of lnc‐MEG3 enhances the expressions and secretions of inflammatory cytokines such as IL‐1β, IL‐6, IL‐8, and TNF‐α in osteoarthritis.17 Another study discloses that lnc‐MEG3 promotes apoptosis in cardiomyocytes via direct binding with fused in sarcoma protein in myocardial infarction.18 Besides, it is evident that lnc‐MEG3 directly binds to miR‐21 and subsequently inhibits its downstream signaling pathways.7, 8 As for miR‐21, it has been proposed as a regulator that elicits effect on inhibiting inflammatory responses and protecting several vital organs from damage in sepsis.10, 11 Based on the effects of lnc‐MEG3 and miR‐21 on inflammation and organ injury as well as the interaction between lnc‐MEG3 and miR‐21, we speculated that lnc‐MEG3 might contribute to the development of sepsis via interacting with miR‐21. However, no related study has been published yet. Our study discovered that lnc‐MEG3 relative expression and lnc‐MEG3/miR‐21 axis were elevated in sepsis patients compared with HCs, while miR‐21 relative expression was attenuated in sepsis patients compared with HCs, and subsequent ROC curve analysis exhibited that lnc‐MEG3 (AUC: 0.887, 95% CI: 0.856‐0.917) and lnc‐MEG3/miR‐21 axis (AUC: 0.934, 95% CI: 0.909‐0.958) had good values for predicting higher, while miR‐21 (AUC: 0.801, 95% CI: 0.758‐0.844) was of good predictive value for lower sepsis risk, among which lnc‐MEG3/miR‐21 axis presented a numerically better predictive value for sepsis risk. The possible explanations were as follows: (a) Lnc‐MEG3 intensified the inductions and release of inflammatory cytokines (such as IL‐1β, IL‐6, IL‐8, and TNF‐α), which enhanced the inflammatory responses and induced the damage as well as necrosis of tissues in several vital organs (such as lung, kidney, and heart.), thereby, resulting in accelerated organ injury, multiple organ failure, and the development of sepsis. (b) Lnc‐MEG3 might decrease the expression of miR‐21, which subsequently attenuated the inhibitory and protective effect of miR‐21 on inflammatory responses and organ injuries respectively, thereby, leading to a higher sepsis risk.10
Furthermore, our study analyzed the correlations of lnc‐MEG3, miR‐21, and lnc‐MEG3/miR‐21 axis with inflammation, severity of organ dysfunction, and disease severity in sepsis patients and disclosed that lnc‐MEG3 and lnc‐MEG3/miR‐21 axis were positively associated, while miR‐21 was negatively associated with inflammation (CRP, TNF‐α, IL‐1β, IL‐6, IL‐17) and disease severity (Scr, SOFA score and APACHE II score) in sepsis patients. These might be explained by that: (a) Lnc‐MEG3 amplified the release and secretion of pro‐inflammatory cytokines and induced cell apoptosis, necrosis of tissues as well as subsequent organ injuries, thus, resulting in elevated inflammation and disease severity in sepsis patients. (b) MiR‐21 decreased the production of pro‐inflammatory mediators via its downstream pathways (such as nuclear factor kappa‐light‐chain‐enhancer of activated B cells) and alleviated tissue damage as well as organ dysfunctions, thereby, leading to lower levels of inflammation and attenuated disease severity in sepsis patients.11 Notably, lnc‐MEG3/miR‐21 axis exhibited relatively higher correlation coefficients with disease severity (APACHE II score and SOFA score) compared with lnc‐MEG3 and miR‐21 individually, which implied that lnc‐MEG3/miR‐21 axis was with better clinical value in sepsis progression.
Additionally, our study also revealed that lnc‐MEG3 (AUC: 0.704, 95% CI: 0.626‐0.783) and lnc‐MEG3/miR‐21 axis (AUC: 0.669, 95% CI: 0.589‐0.750) had acceptable predictive values for higher 28‐day mortality risk, while miR‐21 was of a poor predictive value for lower 28‐day mortality risk in sepsis patients (AUC: 0.588, 95% CI: 0.505‐0.672). The possible reasons were as follows: (a) Lnc‐MEG3 and lnc‐MEG3/miR‐21 axis were associated with increased inflammation, multiple organ dysfunction and accelerated disease severity, thus, resulting in raised 28‐day mortality risk in sepsis patients. (b) MiR‐21 exhibited a dual effect in the regulation of inflammation and organ injuries. From a review, miR‐21 suppressed inflammatory responses and alleviated organ injuries in some cases, whereas it promoted inflammatory responses and accelerated organ injuries in other cases.19 Thus, the value of miR‐21 for predicting 28‐day mortality risk was poor in sepsis patients. Interestedly, compared with common prognostic factors (such as APACHE II score, SOFA score et al), lnc‐MEG3/miR‐21 axis exhibited a numerically inferior predictive value for 28‐day mortality risk in sepsis. However, the predictive value of lnc‐MEG3/miR‐21 axis still implied its potential as a biomarker that could be used to assist with disease monitoring and prognosis in sepsis.
There were certain limitations in the present study. Firstly, the sample size was relatively small, which might reduce the statistic power. Secondly, sepsis patients who died within 24 hours after admission were excluded in order to eliminate the extremely severe disease conditions (confounding factor). However, at the same time, the exclusion of sepsis patients who died within 24 hours after admission might cause selection bias and limit the generalizability of the findings. Lastly, the detailed mechanisms of lnc‐MEG3 and miR‐21 on the development and progression of sepsis were not investigated.
In conclusion, lnc‐MEG3/miR‐21 axis predicts higher sepsis risk and correlates with elevated systemic inflammation, disease severity as well as 28‐day mortality risk of sepsis.
Na L, Ding H, Xing E, et al. Lnc‐MEG3 acts as a potential biomarker for predicting increased disease risk, systemic inflammation, disease severity, and poor prognosis of sepsis via interacting with miR‐21. J Clin Lab Anal. 2020;34:e23123 10.1002/jcla.23123
Lei Na and Huajie Ding contributed equally to this work.
Contributor Information
Jian Yu, Email: yiyou3068682472@163.com.
Changyu Yu, Email: xchang28426@sina.com.
REFERENCES
- 1. Szilagyi B, Fejes Z, Pocsi M, Kappelmayer J, Nagy B Jr. Role of sepsis modulated circulating microRNAs. EJIFCC. 2019;30(2):128‐145. [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang TN, Li D, Xia J, et al. Non‐coding RNA: a potential biomarker and therapeutic target for sepsis. Oncotarget. 2017;8(53):91765‐91778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sinha M, Jupe J, Mack H, Coleman TP, Lawrence SM, Fraley SI. Emerging technologies for molecular diagnosis of sepsis. Clin Microbiol Rev. 2018;31(2):e00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang L, Wang H, Shen Q, Feng L, Jin H. Long non‐coding RNAs involved in autophagy regulation. Cell Death Dis. 2017;8(10):e3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zha F, Qu X, Tang B, et al. Long non‐coding RNA MEG3 promotes fibrosis and inflammatory response in diabetic nephropathy via miR‐181a/Egr‐1/TLR4 axis. Aging. 2019;11(11):3716‐3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zou L, Ma X, Lin S, Wu B, Chen Y, Peng C. Long noncoding RNA‐MEG3 contributes to myocardial ischemia‐reperfusion injury through suppression of miR‐7‐5p expression. Biosci Rep. 2019;39(8):BSR20190210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu M, Wang X, Gu Y, Wang F, Li L, Qiu X. MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR‐21 through the PI3K/Akt pathway. Arch Biochem Biophys. 2019;661:22‐30. [DOI] [PubMed] [Google Scholar]
- 8. Huang P, Huang FZ, Liu HZ, Zhang TY, Yang MS, Sun CZ. LncRNA MEG3 functions as a ceRNA in regulating hepatic lipogenesis by competitively binding to miR‐21 with LRP6. Metab Clin Exp. 2019;94:1‐8. [DOI] [PubMed] [Google Scholar]
- 9. Pan T, Jia P, Chen N, et al. Delayed remote ischemic preconditioning confersrenoprotection against septic acute kidney injury via exosomal miR‐21. Theranostics. 2019;9(2):405‐423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu WD, Xu J, Zhang M, Zhu TM, Zhang YH, Sun K. MicroRNA‐21 inhibits lipopolysaccharide‐induced acute lung injury by targeting nuclear factor‐kappaB. Exp Ther Med. 2018;16(6):4616‐4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jia P, Wu X, Dai Y, et al. MicroRNA‐21 Is required for local and remote ischemic preconditioning in multiple organ protection against sepsis. Crit Care Med. 2017;45(7):e703‐e710. [DOI] [PubMed] [Google Scholar]
- 12. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis‐3). JAMA. 2016;315(8):801‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13(10):818‐829. [PubMed] [Google Scholar]
- 14. Vincent JL, Moreno R, Takala J, et al. The SOFA (sepsis‐related organ failure assessment) score to describe organ dysfunction/failure. On behalf of the working group on sepsis‐related problems of the European society of intensive care medicine. Intensive Care Med. 1996;22(7):707‐710. [DOI] [PubMed] [Google Scholar]
- 15. Li ZY, Yang L, Liu XJ, Wang XZ, Pan YX, Luo JM. The long noncoding RNA MEG3 and its target miR‐147 regulate JAK/STAT pathway in advanced chronic myeloid leukemia. EBioMedicine. 2018;34:61‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dellinger RP, Levy MM, Rhodes A, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39(2):165‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z, Chi X, Liu L, et al. Long noncoding RNA maternally expressed gene 3 knockdown alleviates lipopolysaccharide‐induced inflammatory injury by up‐regulation of miR‐203 in ATDC5 cells. Biomed Pharmacother. 2018;100:240‐249. [DOI] [PubMed] [Google Scholar]
- 18. Wu H, Zhao ZA, Liu J, et al. Long noncoding RNA Meg3 regulates cardiomyocyte apoptosis in myocardial infarction. Gene Ther. 2018;25(8):511‐523. [DOI] [PubMed] [Google Scholar]
- 19. Sheedy FJ. Turning 21: Induction of miR‐21 as a key switch in the inflammatory response. Front Immunol. 2015;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]


