Abstract
The INO80 family of chromatin remodellers are multisubunit complexes that perform a variety of tasks on nucleosomes. Family members are built around a heterohexamer of RuvB-like protein, an ATP-dependent DNA translocase,nuclear actin and actin-related proteins, and a few complex-specific subunits. They modify chromatin in a number of ways including nucleosome sliding and exchange of variant histones. Recent structural information on INO80 and SWR1 complexes has revealed similarities in the basic architecture of the complexes. However, structural and biochemical data on the complexes bound to nucleosomes reveal these similarities to be somewhat superficial and their biochemical activities and mechanisms are very different. Consequently, the INO80 family displays a surprising diversity of function that is based upon a similar structural framework.
Current Opinion in Structural Biology 2020, 61:50–58
This review comes from a themed issue on Macromolecular assemblies
Edited by Xiaodong Zhang and Tom L Blundell
For a complete overview see the Issue and the Editorial
Available online 12th December 2019
https://doi.org/10.1016/j.sbi.2019.09.002
0959-440X/© 2019 The Author(s). Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Chromatin remodelling complexes facilitate access to nuclear DNA by altering the chromatin landscape, which is packaged into a repeating unit known as the nucleosome. Structures of the nucleosome first began to populate the Protein Data Bank in 1997 [1] and revealed how 145–147 base pairs (bp) of DNA wrap around the core histone octamer. This wrap can be divided into waypoints at superhelical locations across the surface of the histone octamer, which are mirrored on opposing faces. Of these waypoints, superhelical location 2 (SHL2), which is two DNA turns removed from the nucleosomal dyad, and SHL6, which is nearer the nucleosomal entry/exit site, are more readily distorted through protein:DNA interactions, making them the canonical sites for remodeller binding [2].
All remodelling enzymes, whether single or multi-subunit, utilise a superfamily II translocase motor [3] to mobilise nucleosomal DNA via ATP hydrolysis. This motor is the centrepiece of all remodellers and loss of its enzymatic activity results in a loss of remodelling in vivo and in vitro. High resolution cryo-electron microscopy (cryoEM) structures have been determined for members in each of the four remodelling families [4] in complex with a mononucleosome (Snf2 [5,6••], ISWI [7,8•], Chd1 [9••,10•], Chd4 [11], INO80 [12••,13••], SWR1 [14••], RSC [15, 16, 17] and SWI/SNF [18]), providing insight into how their motors interact with their substrate. Beyond the highly conserved motor, however, each remodeller carries unique accessory domains and variable subunit compositions that differentiate remodelling mechanisms from one another.
This is particularly evident in the INO80 family (Table 1). These large, multi-subunit complexes have crucial roles in double-strand break repair [19] and were first characterised in the early 2000s [20, 21, 22]. While the activity of other remodeller families appears to be limited to nucleosome sliding, members of the INO80 family perform additional functions with different outcomes on chromatin structure (Table 1). Consequently, whether these enzymes utilise a similar mechanism for remodelling or exhibit adaptations in line with these differences is currently unknown, but structural and biochemical studies have begun to address this question.
Table 1.
Subunit composition of INO80 family chromatin remodelling complexes
| Complex | INO80 | INO80 | SRCAP | SWR1 | TIP60 | NuA4 |
|---|---|---|---|---|---|---|
| Organism | Human | Yeast | Human | Yeast | Human | Yeast |
| Cellular role | DNA repair and Transcription | DNA repair and Transcription | DNA repair | DNA repair and Transcription | DNA repair | DNA repair |
| Biochemical activity | Nucleosome sliding | Histone exchange | Histone acetylation | Histone acetylation | ||
| Histone exchange | Histone exchange | |||||
| Function | Subunits | |||||
| Motor | Ino80 | Ino80 | SRCAP | Swr1 | TRRAP | Tra1 |
| Scaffolding | RuvBL1 | Rvb1 | RuvBL1 | Rvb1 | RuvBL1 | Eaf1 |
| RuvBL2 | Rvb2 | RuvBL2 | Rvb2 | RuvBL2 | Epl1 | |
| Epc1 | ||||||
| Regulation | Ino80B (Ies2) | Ies2 | Tip60 | Esa1 | ||
| Coupling | Actr5 (Arp5) | Arp5 | Actr6 (Arp6) | Arp6 | Actl6a (Arp4) | Arp4 |
| Ino80C (Ies6) | Ies6 | YL1 | Swc6 | Actin | Actin | |
| Actl6a (Arp4) | Arp4 | DMAP1 | Swc2 | |||
| Actr8 (Arp8) | Arp8 | Gas42 | Swc5 | |||
| Actin | Actin | Cfdp1 (Swc5) | Arp4 | |||
| Actl6a (Arp4) | Actin | |||||
| Actin | ||||||
| Undefined | Amida | Ies1 | Yaf9 | Eaf6 | Eaf6 | |
| Ino80D & E | Ies3 | Bdf1 | Gas41 | Yaf9 | ||
| MCRS1 | Ies4 | Swc3 | ING3 | Yng2 | ||
| NFRKB | Ies5 | Swc4 | Mrg15 | Eaf7 | ||
| UCH37 | Nhp10 | Swc7 | MrgBP | Eaf3 | ||
| YY1 | Taf14 | MrgX | Eaf5 | |||
| Eaf2 | Swc4 | |||||
| Brd8 | ||||||
| YL-1 | ||||||
| Available structural information | ||||||
| PDB IDs | 5OAF, 6HTS | 5NBN, 6FHS, 6FML | 6IGM | 6GEJ, 6GEN | 5J9Q, 5Y81, 5J9T, 5J9U, 5J9W, 5OJS, | |
| EMDB IDs (EMD-XXXX | 3954, 3772, 3773, 3774, 3775 | 2385, 2386, 4264, 4277, 4278, 4280, 6924, 8696 | 9668, 9669 | 4395, 4396, 3607, 5626, 5638 | ||
Structural similarities between INO80 and SWR1
INO80 family complexes share common core architectural features including several subunits of overlapping function (Table 1). A prominent unique and ubiquitous feature of INO80 family members is the hexameric ring of RuvB-like proteins (RuvBL1/2 or pontin/reptin in humans, Rvb1/2 in yeast) (Figure 1a). Even though ATPase activity of these AAA+ proteins is not required for remodelling by INO80 [12••] or SWR1 [14••], the hexameric structure acts as a linchpin for the motor subunit by enveloping a polypeptide insertion that extends from the second motor domain (Figure 1). Although the insertion is an INO80 family specific structural element, there is only ∼15% sequence identity (22% similarity) in this region between yeast INO80 and SWR1 (Figure 1b), and only 20% identity between human and yeast INO80. In turn, there are no obvious recurring sequence motifs that might drive the formation of the ‘spoked wheel’ architecture [23•]. By contrast, the flanking motor domain has significant sequence identity (about 50%) and structural overlap (RMSD 1.9 Å). The differences within the insert region impose a necessary asymmetry on the AAA+ ring, which enforces subunit interactions that are unique to each complex [23•]. For example, the asymmetry results in the association of only one Arp5 in INO80, despite there being two otherwise identical sites on the other RuvBL2/Rvb2 protomers. Therefore, the simplest description of the role of the hexameric ring is as an architectural scaffold upon which other subunits are assembled.
Figure 1.
A. Top view of yeast and human INO80, and yeast SWR1 core components. All motor subunits (green) of INO80 family members contain a large polypeptide insertion (blue) that is encapsulated by a heterohexamer of RuvBL1 and RuvBL2 subunits (Rvb1 and Rvb2 in yeast) (grey). This asymmetry facilitates the incorporation of some complex-specific subunits (red). B. Sequence alignment of yeast Swr1 and Ino80 motor domain insertion. The motor domain (flanking region, highlighted green) is highly conserved in all INO80 family members, but the interspersed insertion is highly variable. Red denotes sequence identity, yellow sequence similarity. Secondary structure of Swr1 and Ino80 are respectively shown above and below the alignment.
All members of the INO80 family also contain actin in combination with actin-related proteins (Arps) (Table 1). Two Arps (Arp7 and Arp9) are also found in yeast RSC [24] and SWI–SNF complexes [25]. Early studies showed that Arps are essential to remodelling by INO80 family enzymes [26] and are involved in histone recognition, with distinct preferences for certain histone types. Arp4, for example, interacts with unmodified [27] and phosphorylated H2A (γ-H2AX in humans) [28]. Arp8, in contrast, has a preference for H3/H4 tetramers [26,29,30], and Arp5 interacts with H2A:H2B histone dimers [12••,31].
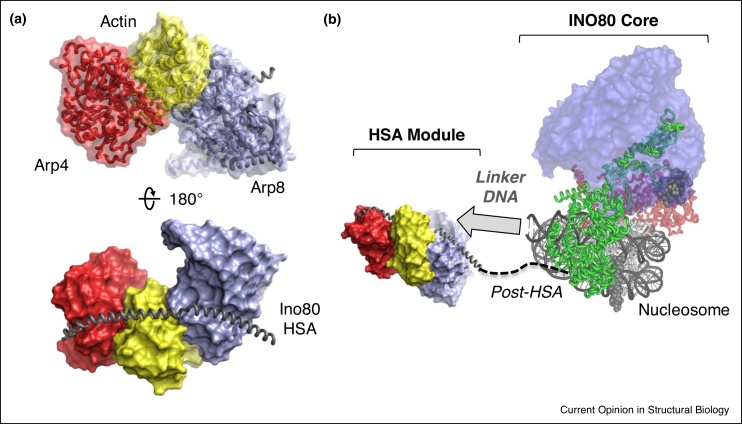
To accommodate some of these Arps, all Arp-containing remodellers have a helical region known as the HSA (helicase-SANT-associated) domain, which precedes the motor domain (Figure 2a). Structures of the HSA regions of a number of systems have been determined including INO80 bound to Arp4, Arp8 and actin (Figure 2a) [32••], RSC with Arp7 and Arp9 [33,34] and SWR1 complexed with Arp4 and actin [35]. These structures reveal that the HSA domain adopts an extended α-helical conformation with the actin/Arps sitting astride this in a staggered configuration via interactions with a hydrophobic groove at the base of the actin fold. In addition, the HSA domain has DNA-binding activity in both RSC [36] and INO80 [32••,37•].
Figure 2.
A. Cartoon representation of the INO80 HSA domain (grey) bound by Arp4 (red), actin (yellow) and Arp8 (light blue). The HSA domain forms an extended alpha-helical structure which accommodates Actin and Actin-related subunits in all INO80 family remodellers. B. Proposed location of the HSA-containing module relative to the motor and Rvb1/2 heterohexamer in Ino80. Based on biochemical work and 2D analysis of electron microscopy data, the HSA module is thought to reside on linker DNA projecting from the nucleosomal entry site. In this way it acts as a sensor of extranucleosomal DNA and can, presumably, regulate the function of the motor. For SWR1, the location of this module relative to the motor is still unclear.
Despite the contribution of actin and Arps to chromatin remodelling, a feature missing in all high resolution INO80 family remodeller cryoEM structures is the Arp-HSA module [12••,13••,14••]. Evidence suggests that this module resides on nucleosomal linker DNA in these structures [13••,32••] (Figure 2b), but it is unclear whether this is its only location. Low resolution structures of INO80 in the absence of a nucleosome [23•,38] show this module is tucked under the RuvBL1/2 ring and motor domain, implying that a dramatic conformational change occurs upon binding the substrate.
Mechanisms of translocation and coupling to chromatin remodelling
Recent structures have begun to determine how translocation is coupled to the nucleotide-binding and hydrolysis cycle of the main motor subunit of all remodellers [6••,8•], which share significant structural and sequence similarity even between families. This occurs in a 3’ to 5’ direction [39] via a cycle of discrete steps [40•] on the so called ‘tracking strand’. Binding of the motor creates a single base bulge in the direction of translocation. Nucleotide binding induces movement of a base on the opposing strand (the ‘guide strand’), thereby resetting the geometry of base pairing. Following hydrolysis and release of the nucleotide, the cycle repeats and produces a shuffle-like movement along the nucleosomal wrap.
This series of movements is self-contained, but can only result in productive movement of DNA relative to the histone core if the remodeller is physically tethered to one or multiple histones during translocation. Evidence from the structures of nucleosome-bound INO80 [12••,13••] and SWR1 [14••] suggests that Arp5:Ies6 and Arp6:Swc6, respectively, are in a prime position to fulfil this role. Support for this comes from recent biochemical interrogations of Arp5 and Ies6, which revealed a role for coupling the motor ATPase to productive nucleosome sliding [41, 42, 43]. In this way, Arp5 is not just important for recruitment [44,45] but directly involved in determining the extent of sliding in vivo, thereby explaining the dysfunction observed upon deletion of the gene [46, 47, 48]. In similar fashion, Snf2, the motor of SWI/SNF complexes, uses its SnAC (Snf2 ATP coupling) domain to remain tethered to the histone surface [49]. Deletion of Arp6 in SWR1 also leads to a loss of multiple subunits and histone exchange activity [50,51]. These tethers, therefore, support both structure and function of different chromatin remodellers.
Differences in a conserved bidentate interaction with the nucleosome
Even though INO80 and SWR1 share a common core architecture and features in line with other remodellers, their differences become apparent through their interaction with a nucleosome. Both INO80 [12••,13••] and SWR1 [14••] have a bidentate interaction with the nucleosomal wrap that is likely to be central to the mechanism of INO80 family enzymes (Figure 3). One contact is made at SHL6/7, where the entry/exit DNA is peeled away from the histone octamer, and a second occurs at SHL2/3. In both remodellers, this involves the motor and the aforementioned tether (Figure 3a). However, the motor–tether arrangement is inverted between INO80 and SWR1: while Ino80 and Arp5:Ies6 are bound at SHL6/7 and SHL3, respectively, the Swr1 motor binds at SHL2 with Arp6:Swc6 at SHL6 (Figure 3). The position of Swr1 at SHL2 is more in-line with that observed for nucleosome sliding enzymes such as Chd1 [9••], ISWI [7,8•] and Snf2h [52,53•], even though its biochemical activity is not nucleosome sliding, but histone exchange. Footprinting studies are consistent with the position of the INO80 and SWR1 motors within their respective structures [54,55•].
Figure 3.
A. (Left) Top view of the nucleosome, showing the locations of SHL2 and SHL6. The nucleosome view is the same in the middle and right panels. (Middle) Bidentate interaction of INO80 with the nucleosome. The motor (green) is bound at SHL6/7 and the tether (Arp5:Ies6, red) at SHL3. (Right) Bidentate interaction of SWR1 with the nucleosome. The motor (green) is bound at SHL2 (+1) and the tether (Arp5:Ies6, red) at SHL6. B. View of yeast INO80 complex down the dyad axis. The motor (green) is positioned at SHL6/7, while Arp5 and Ies6 (red) sit over the histone core and make contacts with SHL3. C. View of yeast SWR1 complex down the dyad axis. The motor (green) is engaged at SHL2 and has rotated by 35-degrees as a consequence of a 1 bp translocation. The Arp6:Swc6 heterodimer resides at SHL6 near the entry site, where the DNA has been peeled away.
This opposing arrangement of INO80 and SWR1 on the nucleosome means that they cannot employ the same mechanism for moving DNA across the octamer surface. In both cases ATP-dependent translocation likely occurs between SHL6/7 and SHL2/3, but the position of INO80 means that it would push entry side DNA against the Arp5:Ies6 anchor, while SWR1 would pull from Arp6:Swc6. As a result, the conformational state of the tether could play a crucial role in regulating DNA movement. Some evidence for this comes from a comparison of the nucleosome-bound human [12••] and yeast [13••] INO80 structures. The yeast complex shows a significant contact with the histone core via Arp5, which are absent in the human complex. These differences can be explained by the conditions used for the in vitro reconstitution of the nucleosome-bound complex: the human complex was formed in the presence of the non-hydrolysable ATP analogue ADP-beryllium fluoride (ADP-BeF3), whereas the yeast complex was not, suggesting that supplementation with ADP-BeF3 has favoured a conformation different to the yeast enzyme. Conversely, the SWR1-nucleosome complex shows a definitive engagement with the histone core via Swc6 (which forms a tight interaction with Arp6) in the presence of ADP-BeF3. Therefore, while the nucleotide-state of the tether is clearly important, identical states have opposite outcomes in INO80 and SWR1.
SWR1 and INO80 are distinguished further by their oligomeric states for remodelling. SWR1, like several other remodellers, acts as a monomer [51], whereas two human INO80 complexes are required for nucleosome sliding [56••]. This ‘functional dimerisation’ has also been observed for Snf2h [53•,57,58] and forms the basis for the proposed spacing mechanism of human ISWI complexes. For INO80, however, this finding is particularly surprising given the substantial size of a single complex relative to a nucleosome, let alone two complexes. Importantly, this means that neither structure of nucleosome-bound INO80 paints a complete picture of the remodelling mechanism.
The requirement for one or two complexes, as well as the opposing setup on the nucleosome, may ultimately relate to the outcome of chromatin remodelling by INO80 and SWR1. Continuous movement of DNA across the octamer surface is an integral part of nucleosome sliding by INO80 [59]. By contrast, SWR1 appears to perform histone exchange via limited translocation (six or seven base pairs) between the entry site and the motor with no net change in nucleosome position [54]. Consistent with these observations, a single base pair translocation is observed towards the dyad in the SWR1-nucleosome structure but only as far as the motor bound DNA segment [14••]. Therefore, the configuration of SWR1 is tightly linked to the region of DNA that wraps around the canonical H2A:H2B or the incoming replacement HTZ:H2B dimer.
Although histone exchange by yeast INO80 has been reported [60,61], it remains a contentious issue [62,63]. Nevertheless, recent experiments show that INO80 also exhibits a mode of limited translocation at the H2A:DNA interface in aid of histone exchange [55•]. High cooperativity of binding in vitro implies that INO80 interacts with the nucleosome almost exclusively in pairs [56••], but a condition may exist that favours a monomeric interaction and regulates a switch from continuous (sliding) to limited translocation (exchange). A significant proportion of ATPase activity is uncoupled from sliding and it may be that this has a role in histone exchange instead. Further work is needed to reveal a role for such a regulatory mechanism for INO80 in vivo.
Regulation of INO80 and SWR1
Nucleosomes undergo extensive post-translational modifications (PTMs) that can affect recruitment of effector proteins as well as the structure of chromatin itself [64]. Both TIP60 (another INO80 family member) and NuA4 complexes (Table 1), acetylate H4 and H2A.Z/HTZ histones [65], in an early stage of DNA repair [66]. Acetylation of H4 stimulates the incorporation of HTZ:H2B histone dimers by SWR1 although this appears to be an effect on affinity of binding rather than catalysis per se [67]. It is noteworthy that this mirrors the activating effects of H4 on nucleosome sliders Chd1 [68] and ISWI [7,69,70], which are also positioned at SHL2. However, despite the functional consequences on SWR1 activity, a direct interaction between SWR1 and the H4 N-terminal tail has not been demonstrated.
Histone tails also regulate the ATPase and nucleosome sliding activity of INO80 [71,72], with H3 tails inhibiting the complex [12••]. This is consistent with the location of the INO80 motor at SHL6/7 (Figure 3b), where the H3 tail trajectory follows the path of linker DNA, which is unpeeled in the nucleosome-bound structure. The H3 N-terminus is itself crucial to the stability of the nucleosomal entry/exit DNA and regulates release of the H2A:H2B dimer near the start of the wrap [73]. This suggests that INO80 overcomes the inherent stability conferred by the H3 N-terminal tail in order to loosen the DNA from the H2A:H2B dimer surface and perform its nucleosome sliding activity.
Furthermore, there are self-contained mechanisms of regulation in different remodellers, that act in addition to regulation by PTMs. For example, regions adjacent to the HSA domain are thought to regulate activities of both INO80 and SWR1, as well as RSC: under certain circumstances, a region C-terminal to the HSA (the post-HSA) interacts with the motor to control translocation [28••,74], while in Swr1 a region adjoining the HSA (referred to as the ‘Z-domain’ [75]) is responsible for binding the variant HTZ:H2B dimer, which in turn stimulates the ATPase activity of the motor [76]. As such, the HSA module and its complementary features regulate the motor depending on whether histones or DNA are engaged.
Future perspectives
Since their discovery, research on INO80 family enzymes over the last two decades has taken significant steps towards understanding the mechanistic basis for their biological functions, even though the role of accessory subunits remains largely unclear. In particular, the disorder of the essential HSA module in all current structures is a limitation that must be overcome to understand the remodelling mechanism in full. Furthermore, in contrast to SWR1 and all nucleosome sliders, the human INO80 complex functions as a dimer [56••] and the corresponding remodelling mechanism must also account for this observation.
One final note of caution is that these systems operate on chromatin and will never encounter isolated nucleosomes within the cell. This leads to the possibility that the evolution of accessory domains and multi-subunit architectures is in fact linked to higher order functions that cannot be described by studies centred on mono-nucleosome substrates alone. It would be prudent, therefore, to shift focus to poly nucleosome substrates that are more chromatin-like. In doing so, the field may reveal functionality for complexes beyond simple translocation.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was funded by grants from the Medical Research Council (MR/R009023/1), Cancer Research UK (C6913/A21608) and Wellcome Trust (209327/Z/17/Z).
References
- 1.Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Bowman G. Mechanisms of ATP-dependent nucleosome sliding. Curr Opin Struct Biol. 2010;20:73–81. doi: 10.1016/j.sbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 4.Clapier C.R., Cairns B.R. The biology of chromatin remodelling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 5.Xia X., Liu X., Li T., Fang X., Chen Z. Structure of chromatin remodeler Swi2/Snf2 in the resting state. Nat Struct Mol Biol. 2016;23:722–729. doi: 10.1038/nsmb.3259. [DOI] [PubMed] [Google Scholar]
- 6••.Li M., Xia X., Tian Y., Jia Q., Liu X., Lu Y., Li M., Li X., Chen Z. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature. 2019;567:409–413. doi: 10.1038/s41586-019-1029-2. [DOI] [PubMed] [Google Scholar]; This work proposes a mechanism for translocation of chromatin remodelling motors based on high resolution Snf2-nucleosome structures with ADP as well as the non-hydrolysable ATP analogue, ADP-BeF3.
- 7.Yan L., Wang L., Tian Y., Xia X., Chen Z. Structure and regulation of the chromatin remodeller ISWI. Nature. 2016;540:466–469. doi: 10.1038/nature20590. [DOI] [PubMed] [Google Scholar]
- 8•.Yan L., Wu H., Li X., Gao N., Chen Z. Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat Struct Mol Biol. 2019;26:258–266. doi: 10.1038/s41594-019-0199-9. [DOI] [PubMed] [Google Scholar]; This work corroborates the findings on translocation by Snf2, and proposes that all remodellers share a common mechanism for mobilising DNA around the histone core of the nucleosome.
- 9••.Farnung L., Vos S.M., Wigge C., Cramer P. Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature. 2017;550:539–542. doi: 10.1038/nature24046. [DOI] [PMC free article] [PubMed] [Google Scholar]; For decades, Chd1 has been the quintessential remodeller for biochemical work. The structure presented in this article begins to shed light on how different structural elements of Chd1 may contribute to its well characterised biochemistry.
- 10•.Sundaramoorthy R., Hughes A.L., El-Mkami H., Norman D.G., Ferreira H., Owen-Hughes T. Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. eLife. 2018 doi: 10.7554/eLife.35720. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ubiquitination is one of the many post-translational modifications that influence remodelling activities. Through a series of high-resolution cryoEM structures, the authors describe a possible mechanism for stimulation of Chd1 by ubiquitination of H2B.
- 11.Farnung L., Ochmann M., Cramer P. Nucleosome-CHD4 chromatin remodeller structures explains human disease mutations. BioRxiv. 2019 doi: 10.7554/eLife.56178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Ayala R., Willhoft O., Aramayo R.J., Wilkinson M., McCormack E.A., Ocloo L., Wigley D.B., Zhang X. Structure and regulation of human INO80-nucleosome complex. Nature. 2018;556:391–395. doi: 10.1038/s41586-018-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; The structure of the human INO80 remodeller bound to a nucleosome provides an initial glimpse at the collision complex between a large multi-subunit remodeller and its substrate. Supporting biochemistry further shows how INO80 is regulated by histone H3, not H4 as has been proposed for other remodellers.
- 13••.Eustermann S., Schall K., Kostrewa D., Lakomek K., Strauss M., Moldt M., Hopfner K.P. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018;556:386–390. doi: 10.1038/s41586-018-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Published at the same time as the human INO80-nucleosome structure, the high resolution structure of the yeast INO80 complex displays an altered conformation on the nucleosome compared to its counterpart. This demonstrates how nucleotide binding by non-motor subunits might modulate the interaction with the nucleosome.
- 14••.Willhoft O., Ghoneim M., Lin C.L., Chua E.Y.D., Wilkinson M., Chaban Y., Ayala R., McCormack E.A., Ocloo L., Rueda D.S., Wigley D.B. Structure and dynamics of the yeast SWR1-nucleosome complex. Science. 2018;362 doi: 10.1126/science.aat7716. [DOI] [PubMed] [Google Scholar]; Using a combination cryo-electron microscopy, biochemistry and single molecule microscopy, the authors demonstrated how the yeast SWR1 complex distorts the nucleosome in preparation for ATP-dependent histone exchange. Importantly, this work provides a structural basis for the differences within the INO80 family.
- 15.Wagner F.R., Dienemann C., Wang H., Stützer A., Tegunov D., Urlaub H., Cramer P. Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. BioRxiv. 2019 doi: 10.1038/s41586-020-2088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel A.B., Moore C.M., Greber B.J., Luo J., Ranish J., Nogales E. Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. BioRxiv. 2019 doi: 10.7554/eLife.54449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye Y., WuH, Chen K., Clapier C.R., Verma N., Zhang W., Deng H., Cairns B.R., Gao N., Chen Z. Structure of the RSC complex bound to the nucleosome. Science. 2019;366 doi: 10.1126/science.aay0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y., Reyes A.A., Malik S., He Y. Cryo-electron microscopy structure of a nucleosome-bound SWI/SNF chromatin remodeling complex. BioRxiv. 2019 [Google Scholar]
- 19.Poli J., Gasser S.M., Papamichos-Chronakis M. The INO80 remodeller in transcription, replication and repair. Philos Trans R Soc Lond B Biol Sci. 2017;372:1731. doi: 10.1098/rstb.2016.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen X., Mizuguchi G., Hamiche A., Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 21.Krogan N.J., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 22.Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodelling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 23•.Aramayo R.J., Willhoft O., Ayala R., Bythell-Douglas R., Wigley D.B., Zhang X. Cryo-EM structures of the human INO80 chromatin-remodeling complex. Nat Struct Mol Biol. 2018;25:37–44. doi: 10.1038/s41594-017-0003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article details the first high resolution structure of the INO80 insert region and how this interacts with the RuvBL1 and RuvBL2 (Rvb1/Rvb2 in yeast) heterohexamer. This hexamer is found in all INO80 family members and is also part of the Hsp90-dependent chaperone complex, R2TP.
- 24.Cairns B.R., Erdjument-Bromage H., Tempst P., Winston F., Kornberg R.D. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 25.Peterson C.L., Zhao Y., Chait B.T. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J Biol Chem. 1998;273:23641–23644. doi: 10.1074/jbc.273.37.23641. [DOI] [PubMed] [Google Scholar]
- 26.Shen X., Ranallo R., Choi E., Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 27.Harata M., Oma Y., Mizuno S., Jiang Y.W., Stillmann D.J., Wintersberger U. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol Biol Cell. 1999;10:2595–2605. doi: 10.1091/mbc.10.8.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Downs J.A., Allard S., Jobin-Robitaille O., Javaheri A., Auger A., Bouchard N., Kron S.J., Jackson S.P., Côté J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Gerhold C.B., Winkler D.D., Lakomek K., Seifert F.U., Fenn S., Kessler B., Witte G., Luger K., Hopfner K.P. Structure of actin-related protein 8 and its contribution to nucleosome binding. Nucleic Acids Res. 2012;40:11036–11046. doi: 10.1093/nar/gks842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saravanan M., Wuerges J., Bose D., McCormack E.A., Cook N.J., Zhang X., Wigley D.B. Interactions between the nucleosome histone core and Arp8 in the INO80 chromatin remodeling complex. Proc Natl Acad Sci U S A. 2012;109:20883–20888. doi: 10.1073/pnas.1214735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tosi A., Haas C., Herzog F., Gilmozzi A., Berninghausen O., Ungewickell C., Gerhold C.B., Lakomek K., Aebersold R., Beckmann R., Hopfner K.P. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154:1207–1219. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 32••.Knoll K.R., Eustermann S., Niebauer V., Oberbeckmann E., Stoehr G., Schall K., Tosi A., Schwarz M., Buchfellner A., Korber P., Hopfner K.P. The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling. Nat Struct Mol Biol. 2018;25:823–832. doi: 10.1038/s41594-018-0115-8. [DOI] [PubMed] [Google Scholar]; The HSA module is important for the structure and function of all INO80 remodellers. Using a combination of X-ray crystallography and biochemistry, the authors provide a molecular basis for the role of this subcomplex in nucleosome sliding and how this may be a conserved feature in other remodellers that contain similar elements.
- 33.Schubert H.L., Wittmeyer J., Kasten M.M., Hinata K., Rawling D.C., Héroux A., Cairns B.R., Hill C.P. Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler. Proc Natl Acad Sci U S A. 2013;110:3345–3350. doi: 10.1073/pnas.1215379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lobsiger J., Hunziker Y., Richmond T.J. Structure of the full-length yeast Arp7-Arp9 heterodimer. Acta Crystallogr D Biol Crystallogr. 2014;70:310–316. doi: 10.1107/S1399004713027417. [DOI] [PubMed] [Google Scholar]
- 35.Cao T., Sun L., Jiang Y., Huang S., Wang J., Chen Z. Crystal structure of a nuclear actin ternary complex. Proc Natl Acad Sci U S A. 2016;113:8985–8990. doi: 10.1073/pnas.1602818113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turegun B., Baker R.W., Leschziner A.E., Dominguez R. Actin-related proteins regulate the RSC chromatin remodeler by weakening intramolecular interactions of the Sth1 ATPase. Commun Biol. 2018;1 doi: 10.1038/s42003-017-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Brahma S., Ngubo M., Paul S., Udugama M., Bartholomew B. The Arp8 and Arp4 module acts as a DNA sensor controlling INO80 chromatin remodeling. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05710-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Footprinting analysis of the HSA-module of INO80 (containing Arp4, actin and Arp8) provides evidence for a role in extranucleosomal DNA length sensing, and corroborates the placement of the INO80 motor as observed in the high resolution cryoEM structures.
- 38.Zhang X., Wang X., Zhang Z., Cai G. Structure and functional interactions of INO80 actin/Arp module. J Mol Cell Biol. 2019;11:345–355. doi: 10.1093/jmcb/mjy062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha A., Wittmeyer J., Cairns B.R. Chromatin remodeling through directional DNA translocation from an internal nucleosome site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 40•.Sabantsev A., Levendovsky R.F., Zhuang X., Bowman G.D., Deindl S. Direct observation of coordinated DNA movements on the nucleosome during chromatin remodelling. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; A detailed single molecule study on Chd1 and Snf2h shows how ATP-dependent translocation leads to absorption of single base steps into the nucleosome from the entry site. In using a ‘buffering’ mechanism for remodelling, both remodellers are able to maintain nucleosome stability.
- 41.Watanabe S., Tan D., Lakshminarasimhan M., Washburn M.P., Hong E.J., Walz T., Peterson C.L. Structural analyses of the chromatin remodelling enzymes INO80-C and SWR-C. Nat Commun. 2015;6 doi: 10.1038/ncomms8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao W., Beckwith S.L., Zheng T., Young T., Dinh V.T., Ranjan A., Morrison A.J. Assembly of the Arp5 (actin-related protein) subunit involved in distinct INO80 chromatin remodeling activities. J Biol Chem. 2015;290:25700–25709. doi: 10.1074/jbc.M115.674887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willhoft O., Bythell-Douglas R., McCormack E.A., Wigley D.B. Synergy and antagonism in regulation of recombinant human INO80 chromatin remodeling complex. Nucleic Acids Res. 2016;44:8179–8188. doi: 10.1093/nar/gkw509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Attikum H., Fritsch O., Hohn B., Gasser S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y., Wang X., Bao S., Guo R., Johnson D.H., Shen X., Li L. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc Natl Acad Sci U S A. 2010;107:17274–17279. doi: 10.1073/pnas.1008388107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitayama K., Kamo M., Oma Y., Matsuda R., Uchida T., Ikura T., Tashiro S., Ohyama T., Winsor B., Harata M. The human actin-related protein hArp5: nucleo-cytoplasmic shuttling and involvement in DNA repair. Exp Cell Res. 2009;315:206–217. doi: 10.1016/j.yexcr.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 47.Kandasamy M.K., McKinney E.C., Deal R.B., Smith A.P., Meagher R.B. Arabidopsis actin-related protein ARP5 in multicellular development and DNA repair. Dev Biol. 2009;335:22–32. doi: 10.1016/j.ydbio.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao W., King D.A., Beckwith S.L., Gowans G.J., Yen K., Zhou C., Morrison A.J. The INO80 complex requires the Arp5-Ies6 subcomplex for chromatin remodeling and metabolic regulation. Mol Cell Biol. 2016;36:979–991. doi: 10.1128/MCB.00801-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sen P., Vivas P., Dechassa M.L., Mooney A.M., Poirier M.G., Bartholomew B. The SnAC domain of SWI/SNF is a histone anchor required for remodeling. Mol Cell Biol. 2013;33:360–370. doi: 10.1128/MCB.00922-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu W.H., Alami S., Luk E., Wu C.H., Sen S., Mizuguchi G., Wei D., Wu C. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- 51.Lin C.L., Chaban Y., Rees D.M., McCormack E.A., Ocloo L., Wigley D.B. Functional characterization and architecture of recombinant yeast SWR1 histone exchange complex. Nucleic Acids Res. 2017;45:7249–7260. doi: 10.1093/nar/gkx414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X., Li M., Xia X., Li X., Chen Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature. 2017;544:440–445. doi: 10.1038/nature22036. [DOI] [PubMed] [Google Scholar]
- 53•.Armache J.P., Gamarra N., Johnson S.L., Leonard J.D., Wu S., Narlikar G.J., Cheng Y. Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. eLife. 2019 doi: 10.7554/eLife.46057. [DOI] [PMC free article] [PubMed] [Google Scholar]; Similar to INO80, the SWI/SNF motor subunit, Snf2h, functions as a dimer to carry out ATP-dependent nucleosome sliding. This work presents a possible mechanism for allosteric control of two Snf2h protomers via the nucleosome acidic patch.
- 54.Ranjan A., Wang F., Mizuguchi G., Wei D., Huang Y., Wu C. H2A histone-fold and DNA elements in nucleosome activate SWR1-mediated H2A.Z replacement in budding yeast. eLife. 2015 doi: 10.7554/eLife.06845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Brahma S., Udugama M.I., Kim J., Hada A., Bhardwaj S.K., Hailu S.G., Lee T.H., Bartholomew B. INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun. 2017;8 doi: 10.1038/ncomms15616. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided the first hint that the INO80 complex is different to other remodellers due to its position near SHL6. The authors use this to propose a mechanism for the putative histone exchange activity of INO80.
- 56••.Willhoft O., McCormack E.A., Aramayo R.J., Bythell-Douglas R., Ocloo L., Zhang X., Wigley D.B. Crosstalk within a functional INO80 complex dimer regulates nucleosome sliding. eLife. 2017;6 doi: 10.7554/eLife.25782. pii: e25782. [DOI] [PMC free article] [PubMed] [Google Scholar]; Through a detailed biochemical study, the authors demonstrate the surprising finding that two INO80 complexes work together to slide nucleosomes.
- 57.Racki L.R., Yang J.G., Naber N., Partensky P.D., Acevedo A., Purcell T.J., Cooke R., Cheng Y., Narlikar G.J. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leonard J.D., Narlikar G.J. A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler. Mol Cell. 2015;57:850–859. doi: 10.1016/j.molcel.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou C.Y., Johnson S.L., Lee L.J., Longhurst A.D., Beckwith S.L., Johnson M.J., Morrison A.J., Narlikar G.J. The yeast INO80 complex operates as a tunable DNA length-sensitive switch to regulate nucleosome sliding. Mol Cell. 2018;69:677–688. doi: 10.1016/j.molcel.2018.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papamichos-Chronakis M., Watanabe S., Rando O.J., Peterson C.L. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe S., Radman-Livaja M., Rando O.J., Peterson C.L. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science. 2013;340:195–199. doi: 10.1126/science.1229758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang F., Ranjan A., Wei D., Wu C. Comment on “a histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme”. Science. 2016;353:358. doi: 10.1126/science.aad5921. [DOI] [PubMed] [Google Scholar]
- 63.Watanabe S., Peterson C.L. Response to comment on “a histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme”. Science. 2016;353:358. doi: 10.1126/science.aad6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keogh M.C., Mennella T.A., Sawa C., Berthelet S., Krogan N.J., Wolek A., Podolny V., Carpenter L.R., Greenblatt J.F., Baetz K., Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bird A.W., Yu D.Y., Pray-Grant M.G., Qiu Q., Harmon K.E., Megee P.C., Grant P.A., Smith M.M., Christman M.F. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 67.Altaf M., Auger A., Monnet-Saksouk J., Brodeur J., Piquet S., Cramet M., Bouchard N., Lacoste N., Utley R.T., Gaudreau L., Côté J. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hauk G., McKnight J.N., Nodelman I.M., Bowman G.D. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–723. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dang W., Kagalwala M.N., Bartholomew B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol. 2006;26:7388–7389. doi: 10.1128/MCB.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ludwigsen J., Pfennig S., Singh A.K., Schindler C., Harrer N., Forné I., Zacharias M., Mueller-Planitz F. Concerted regulation of ISWI by an autoinhibitory domain and the H4 N-terminal tail. eLife. 2017 doi: 10.7554/eLife.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Udugama M., Sabri A., Bartholomew B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol. 2011;31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwarz M., Schall K., Kallis E., Eustermann S., Guariento M., Moldt M., Hopfner K.P., Michaelis J. Single-molecule nucleosome remodeling by INO80 and effects of histone tails. FEBS Lett. 2018;592:318–331. doi: 10.1002/1873-3468.12973. [DOI] [PubMed] [Google Scholar]
- 73.Ferreira H., Somers J., Webster R., Flaus A., Owen-Hughes T. Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability. Mol Cell Biol. 2007;27:4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clapier C.R., Kasten M.M., Parnell T.J., Viswanathan R., Szerlong H., Sirinakis G., Zhang Y., Cairns B.R. Regulation of DNA translocation efficiency within the chromatin remodeler RSC/Sth1 potentiates nucleosome sliding and ejection. Mol Cell. 2016;62:452–461. doi: 10.1016/j.molcel.2016.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hong J., Feng H., Wang F., Ranjan A., Chen J., Jiang J., Ghirlando R., Xiao T.S., Wu C., Bai Y. The catalytic subunit of the SWR1 remodeler is a histone chaperone for the H2A.Z-H2B dimer. Mol Cell. 2014;53:498–505. doi: 10.1016/j.molcel.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luk E., Ranjan A., Fitzgerald P.C., Mizuguchi G., Huang Y., Wei D., Wu C. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell. 2010;143:725–736. doi: 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]