Key Points
Question
What are the genetic risk loci for Parkinson disease in an Asian population?
Findings
In this genetic-association study of 31 575 individuals, 1 novel locus SV2C showed robust replication in European and Japanese samples; 11 genome-wide significant loci were identified in an Asian-only meta–genome-wide association study in 6724 individuals with Parkinson disease and 24 851 controls. Substantial overlap in genetic risk factors appeared to exist between Asian and European individuals, with 63 of 78 polymorphic European risk variants displaying a similar direction of association.
Meaning
Risk prediction models constructed based on Asian and European risk variants appear to be associated with improvements in stratification of Parkinson disease risk in the worldwide population of Asian individuals.
Abstract
Importance
Large-scale genome-wide association studies in the European population have identified 90 risk variants associated with Parkinson disease (PD); however, there are limited studies in the largest population worldwide (ie, Asian).
Objectives
To identify novel genome-wide significant loci for PD in Asian individuals and to compare genetic risk between Asian and European cohorts.
Design Setting, and Participants
Genome-wide association data generated from PD cases and controls in an Asian population (ie, Singapore/Malaysia, Hong Kong, Taiwan, mainland China, and South Korea) were collected from January 1, 2016, to December 31, 2018, as part of an ongoing study. Results were combined with inverse variance meta-analysis, and replication of top loci in European and Japanese samples was performed. Discovery samples of 31 575 individuals passing quality control of 35 994 recruited were used, with a greater than 90% participation rate. A replication cohort of 1 926 361 European-ancestry and 3509 Japanese samples was analyzed. Parkinson disease was diagnosed using UK Parkinson’s Disease Society Brain Bank Criteria.
Main Outcomes and Measures
Genotypes of common variants, association with disease status, and polygenic risk scores.
Results
Of 31 575 samples identified, 6724 PD cases (mean [SD] age, 64.3 [10] years; age at onset, 58.8 [10.6] years; 3472 [53.2%] men) and 24 851 controls (age, 59.4 [11.4] years; 11 030 [45.0%] men) were analyzed in the discovery study. Eleven genome-wide significant loci were identified; 2 of these loci were novel (SV2C and WBSCR17) and 9 were previously found in Europeans. Replication in European-ancestry and Japanese samples showed robust association for SV2C (rs246814; odds ratio, 1.16; 95% CI, 1.11-1.21; P = 1.17 × 10−10 in meta-analysis of discovery and replication samples) but showed potential genetic heterogeneity at WBSCR17 (rs9638616; I2=67.1%; P = 3.40 × 10−3 for hetereogeneity). Polygenic risk score models including variants at these 11 loci were associated with a significant improvement in area under the curve over the model based on 78 European loci alone (63.1% vs 60.2%; P = 6.81 × 10−12).
Conclusions and Relevance
This study identified 2 apparently novel gene loci and found 9 previously identified European loci to be associated with PD in this large, meta–genome-wide association study in a worldwide population of Asian individuals and reports similarities and differences in genetic risk factors between Asian and European individuals in the risk for PD. These findings may lead to improved stratification of Asian patients and controls based on polygenic risk scores. Our findings have potential academic and clinical importance for risk stratification and precision medicine in Asia.
This genetic association study compares gene loci for Parkinson Disease in cohorts of Asian and European individuals with Parkinson disease.
Introduction
Parkinson disease (PD) (gene: OMIM 168600) is one of the most common age-related neurodegenerative diseases worldwide and has been a factor in more than 200 000 deaths and 3.2 million disability-adjusted life-years worldwide in 2016.1,2 Parkinson disease presents as a hypokinetic movement disorder characterized by bradykinesia, postural instability, rigidity, and resting tremors resulting from loss of nigrostriatal dopaminergic neurons and other nondopaminergic structures. Several genes containing rare pathogenic variants have been identified in familial PD, suggesting that, although genetic factors play a role in PD pathogenesis, the disease is heterogeneous and associated with multiple genes and pathways.
Large-scale meta-analyses of genome-wide association studies (GWASs) of approximately 40 000 PD cases and 20 000 proxy cases in the European population have since identified several dozen loci that implicate lysosomal function and other pathways in PD pathogenesis and confirmed the involvement of familial PD genes, such as SNCA (OMIM 163890) and LRRK2 (OMIM 609007), in sporadic PD as well. The Asian population is the largest worldwide and thus makes up a significant fraction of patients with PD globally.1 Studies have demonstrated both similarities and differences in genetic risk factors underlying PD in Asian and European individuals,3,4 such as the absence of the MAPT (OMIM 157140) H2 protective haplotype and the LRRK2 G2019S risk variant in Asian individuals and the identification of the Asian-specific LRRK2 R1628P and G2385R variants. However, the numbers of samples analyzed have been relatively small to date.
To identify novel, potentially Asian-specific loci and conduct a robust comparison between genetic risk for PD in Asian and European individuals, we conducted what was, to our knowledge, the first East Asian meta-GWAS in 31 575 individuals (6724 patients and 24 851 controls) and replicated the top single-nucleotide variants (SNVs) (formerly SNPs) in up to 56 306 cases and 1 417 791 controls from the meta-analyses by the International Parkinson’s Disease Genomics Consortium (IPDGC),5 1239 cases and 451 025 controls from the UK Biobank,6 and 988 cases and 2521 controls from Japan.4
Methods
Patients and ethnically and regionally matched controls were recruited by 13 independent centers and study groups from 6 regions across East Asia (eMethods, eTable 1 in the Supplement). Patients were diagnosed with PD using the UK Parkinson’s Disease Society Brain Bank criteria. Written informed consent was obtained according to the Declaration of Helsinki.7 This study was approved by the SingHealth Centralized Institutional Review Board and Nanyang Technological University Institutional Review Board. The study was conducted from January 1, 2016, to December 31, 2018. This study followed the Strengthening the Reporting of Genetic Association Studies (STREGA) reporting guideline.
We analyzed SNVs within the 2 novel loci in 988 cases and 2521 controls from Japan.4 We also analyzed our top SNVs in the largest European-ancestry PD GWAS from the IPDGC (56 306 cases and 1 417 791 controls).5,8 Summary statistics of SNVs within the loci of interest were assessed in the UK Biobank study on G20: Parkinson disease (1239 PD cases and 451 025 controls, eMethods in the Supplement).6
Statistical Analysis
A total of 34 162 Han Chinese and South Korean samples were genotyped, followed by sample filtering (eMethods, eFigure 1 in the Supplement). Imputation of untyped SNVs and prephasing was conducted using the multiethnic 1000 Genomes Project phase 3 reference panel (released in October 2014), followed by stringent quality-control filtering at the SNV level (eTable 2, eFigure 2 in the Supplement). Logistic regression analyses were conducted on genotype dosages. Results were combined by fixed-effects inverse variance meta-analysis (eMethods in the Supplement).
Polygenic risk scores were calculated in 2536 PD cases and 21 840 population-based controls from Singapore/Malaysia based on 11 genome-wide significant Asian SNVs, 78 SNVs from European-ancestry studies,5,8,9 or a combination of Asian and European SNVs (80 SNVs) (eMethods in the Supplement).
The percentage of the total variance explained was estimated via Nagelkerke pseudo R2 analysis. Area under the curve (AUC) estimates were conducted to assess receiver operating characteristic curve differences (eMethods in the Supplement). Significance was assessed at genome-wide significance level P < 5 × 10−8 for GWAS and P < .05 for other comparisons.
Results
A total of 31 575 samples remained after quality-control filtering, consisting of 6724 PD cases (mean [SD] age, 64.3 [10] years; age at onset, 58.8 [10.6] years; 3472 [53.2%] men) and 24 851 controls (age, 59.4 [11.4] years; 11 030 [45.0%] men) from China (2279 cases, 2021 controls), Taiwan (216 cases, 225 controls), Hong Kong (199 cases, 166 controls), South Korea (1494 cases, 599 controls), and Chinese participants from Singapore/Malaysia (2536 cases, 21 840 controls) (eTable 1B in the Supplement). Association statistics were combined using fixed-effects meta-analysis at a total of 5 843 213 SNVs (minor allele frequency ≥1%; λ genomic control (genomic inflation factor) = 1.082; λ1000 = 1.0077; λ genomic control (genomic inflation factor) for minor allele frequency ≥5% = 1.092; λ1000 = 1.0087; linkage disequilibrium score intercept = 1.02) that were genotyped or successfully imputed at high quality across all 5 data sets (eFigures 1-3 in the Supplement). Sensitivity analyses using leave-one-out meta-analyses suggested that the effect size estimates were not driven by any single study (eFigure 4, eTable 3 in the Supplement).
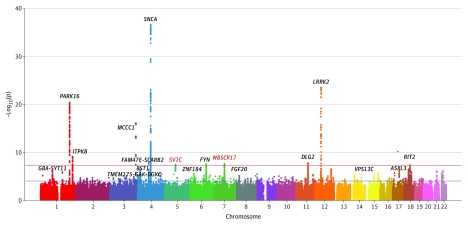
Our meta-analysis revealed 11 genome-wide significant loci, of which 9 were previously described (PARK16 [OMIM 613164], ITPKB [OMIM 147522], MCCC1 [OMIM 609010], SNCA [OMIM 163890], FAM47E-SCARB2 [OMIM 602257], DLG2 [OMIM 603583], LRRK2 [OMIM 609007)], RIT2 [OMIM 609592], and FYN [OMIM 137025]) (Figure 1). We identified 2 new associations at SV2C (OMIM 610291) and WBSCR17 (OMIM 615137). We also observed a strong association (P < 1 × 10−5) at 7 other loci that have previously (GBA [OMIM 606463]-SYT11 [OMIM 163890], BST1 [OMIM 600387], TMEM175 [OMIM 616660]-GAK [OMIM 602052)]-DGKQ [OMIM 601207], ZNF184 [OMIM 602277], FGF20 [OMIM 605558], VPS13C [OMIM 608879], and ASXL3 [OMIM 615115]) been reported to be associated with PD in European individuals (Figure 1). Of the 16 previously reported loci with P < 1 × 10−5, the top-associated SNV was in high linkage disequilibrium (r2>0.75) with the reported European SNV within 7 loci. We observed allelic heterogeneity at LRRK2, ITPKB, ZNF184, FAM47E-SCARB2 and GBA/SYT11 in which the top Asian SNV was independent of the reported European SNV, and LD differences at SNCA, FYN, VPS13C, and ASXL3 (eTable 4 in the Supplement), thus demonstrating differences in the underlying genetic architecture between Asian and European individuals at overlapping loci.
Figure 1. Genome-Wide Association Study of Parkinson Disease in East Asian Individuals.
Meta–genome-wide association studies of 5 East Asian sample collections, with novel loci labeled in red, previously reported loci in black, and genome-wide significant loci in bold font. Horizontal lines indicate the thresholds for genome-wide significant association (5 × 10−8) (red) and suggestive association (1 × 10−4) (blue).
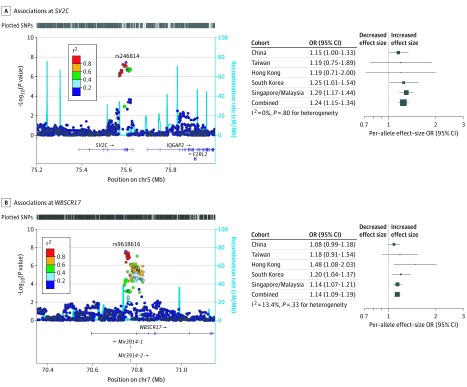
We observed genome-wide significant association at rs246814 (odds ratio [OR], 1.24; 95% CI, 1.15-1.34; P = 3.48 × 10−8) located within an intron of the SV2C gene (Figure 2A, Table 1). Consistent association was observed across all 5 East Asian data sets (I2 = 0, P = .80 for heterogeneity). This SNV is in complete LD (r2 = 1 in 1000 Genomes Project data and >0.96 in our samples) with a missense variant p.Asp543Asn (rs31244) within SV2C (OR, 1.24; 95% CI, 1.14-1.33; P = 6.22 × 10−8). Although this nonsynonymous change is predicted by SIFT and PolyPhen to be tolerated and benign, respectively, the change occurs within an extracellular or luminal domain of SV2C and may affect N-linked glycosylation of this domain10 via the creation of a new glycosylation site (Asn543-Asp544-Thr545). The SNV rs246814 also tags SNVs located within potential transcription factor binding motifs and DNase hypersensitivity sites (eTables 5 and 6 in the Supplement). SV2C is expressed in the basal ganglia and dopaminergic neurons and has previously been evaluated as a functional PD candidate gene because of its restricted expression in brain regions relevant to PD.11
Figure 2. Two Novel Parkinson Disease Risk Loci.
Associations at SV2C (A) and WBSCR17 (B) in the Asian meta–genome-wide association studies. chr Indicates chromosome; cM/Mb, centimorgan/megabase (genomic location in reference build 37 [Hg19]); OR, odds ratio; and SNV, single-nucleotide variant (formerly SNP).
Table 1. Association and Meta-analysis Results at SV2C and WBSCR17.
| Study | MAF, % | OR (95% CI) | P value | I2, % | P value for heterogeneity | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| SV2C rs246814: C | ||||||
| China | 10.32 | 9.13 | 1.15 (1.00-1.33) | .054 | ||
| Taiwan | 10.51 | 8.74 | 1.19 (0.75-1.89) | .45 | ||
| Hong Kong | 9.79 | 8.67 | 1.19 (0.71-2.00) | .51 | ||
| Korea | 11.92 | 9.63 | 1.25 (1.01-1.54) | .04 | ||
| Singapore/Malaysia | 10.45 | 8.29 | 1.29 (1.17-1.44) | 1.44 × 10−6 | ||
| Combined discovery | 1.24 (1.15-1.34) | 3.48 × 10−8 | 0 | .80 | ||
| Japana | 11.08 | 10.13 | 1.11 (0.94-1.31) | .2 | ||
| UK Biobank | 7.75b | 1.09 (0.94-1.26) | .25 | |||
| IPDGC | ||||||
| All | 8.23b | 1.07 (1.04-1.11) | 3.62 × 10−5 | |||
| Clinical | 8.42b | 1.13 (1.06-1.21) | 2.95 × 10−4 | |||
| Combined replication (IPDGC all)c | 1.07 (1.04-1.11) | 9.74 × 10−6 | 0 | .92 | ||
| Combined discovery plus replication (all) | 1.11 (1.07-1.13) | 6.02 × 10−10 | 48 | .06 | ||
| Combined replication (IPDGC clinical) | 1.12 (1.06-1.19) | 7.80 × 10−5 | 0 | .90 | ||
| Combined discovery plus replication (clinical) | 1.16 (1.11-1.21) | 1.17 × 10−10 | 0 | .50 | ||
| WBSCR17 rs9638616:T | ||||||
| China | 49.24 | 47.22 | 1.08 (0.99-1.18) | .08 | ||
| Taiwan | 48.13 | 44.17 | 1.18 (0.91-1.54) | .21 | ||
| Hong Kong | 47.26 | 38.39 | 1.48 (1.08-2.03) | .01 | ||
| Korea | 56.68 | 52.29 | 1.20 (1.04-1.37) | 9.50 × 10−3 | ||
| Singapore/Malaysia | 47.06 | 43.27 | 1.14 (1.07-1.21) | 1.93 × 10−5 | ||
| Combined discovery | 1.14 (1.09-1.19) | 2.53 × 10−8 | 13.4 | .33 | ||
| Japana | 41.19 | 40.16 | 1.04 (0.94-1.16) | .43 | ||
| UK Biobank | 31.43b | 0.97 (0.89-1.06) | .53 | |||
| IPDGC | ||||||
| All | 32.44b | 1.00 (0.98-1.02) | .76 | |||
| Clinical | 31.81b | 1.01 (0.95-1.06) | .85 | |||
| Combined replication (IPDGC all)c | 1.00 (0.98-1.02) | .77 | 0 | .59 | ||
| Combined discovery plus replication (all) | 1.02 (1.00-1.04) | .04 | 78.5 | 3.16 × 10−5 | ||
| Combined replication (IPDGC clinical) | 1.00 (0.96-1.05) | .89 | 0 | .591 | ||
| Combined discovery plus replication (clinical) | 1.06 (1.03-1.10) | 8.37 × 10−5 | 67.1 | 3.40 × 10−3 | ||
Abbreviations: IPDGC, International Parkinson’s Disease Genomics Consortium; MAF, minor allele frequency; OR, odds ratio.
rs246813 was used as a proxy for rs246814 (r2 = 0.99) and rs1317290 was used as a proxy for rs9638616 (r2 = 0.90) in data from Japan.4
Number represents the mean minor allele frequency in percentage across combined cases and controls.
Replication was performed using either the full IPDGC data set of 56 306 cases, 1 417 791 controls (all), or the IPDGC clinically diagnosed subset of 15 056 cases and 12 637 controls (clinical) in which there is no overlap with the UK Biobank samples. The Japan and UK Biobank data sets were included in both analyses.
Genome-wide significant association was also observed at a second novel locus tagged by rs9638616 (OR, 1.14; 95% CI, 1.09-1.19; P = 2.53 × 10−8) (Figure 2B, Table 1). This SNV is located within an intron of the WBSCR17 gene and near genes encoding microRNAs mir-3914-1 and mir3914-2. Similarly, consistent association was observed across the 5 data sets (I2 = 13.4%, P = .33 for heterogeneity). To our knowledge, neither of these 2 genes has previously been implicated in PD.
We then evaluated the association evidence at SNVs and loci previously reported to show genome-wide significant association with PD in European populations5,8,9 in our GWAS meta-analysis results (Table 2; eTable 7 in the Supplement). Of the 78 SNVs polymorphic in Asian samples, only 3 showed genome-wide significant association in Asian individuals, and another 6 SNVs were associated at P < 1 × 10−5 (Table 2). A total of 63 SNVs had ORs in same direction (38 with P < .05), 15 had ORs in the opposite direction (all P > .05 except MEX3C [OMIM: 611005]) (eFigure 5 in the Supplement). We recognize that our Asian sample set is smaller than the largest European GWAS and has limited statistical power to validate these loci. However, the fraction of polymorphic SNVs showing same direction of association (63/78 = 80.8%) and the strong enrichment for significant SNVs (38/78 = 48.7% at P < .05; median P = .055, λ = 8.08) (eFigure 5 in the Supplement) suggest a substantial but incomplete overlap in genetic risk between Asian and European populations. At the locus level, SNVs with P < 1 × 10−5 were observed in 16 of the previously reported loci (eTable 4, eFigure 6 in the Supplement), while there was no evidence of linked or independent signals crossing P < 1 × 10−5 at the remaining loci (eFigure 6 in the Supplement).
Table 2. Variants at Reported PD Risk Loci With P < .01 in Asian Discovery Samples5,8,9,a.
| P value | No. of variants | Locus names |
|---|---|---|
| P < 5 × 10−8 | 3 | SNCA, MCCC1, PARK16 |
| P < 1 × 10−5 | 9 | BST1, GAK, ITPKB, RIT2, DLG2, FYN |
| P < 1 × 10−4 | 12 | ASXL3, VPS13C, FGF20 |
| P < 1 × 10−3 | 13 | RPS12 |
| P < .01 | 25 | ZNF184, SH3GL2, CCDC62, RCORL, RIMS1, UBAP2, RNF141, SCAF11, FBRSL1, RPS6KL1, UBTF, STK39 |
| P < .05 | 39 | 38 In same direction, 1 in opposite direction (MEX3C) |
Abbreviation: PD, Parkinson disease.
Full single-nucleotide variant (formerly SNP) identifiers and association statistics are listed in eTable 7 in the Supplement.
To determine whether the 2 novel SNVs are associated with PD risk in other populations, we evaluated summary statistics from the largest European-ancestry data sets available online: the UK Biobank (1239 cases, 451 025 controls) and the most recent meta-GWAS by the IPDGC5 (up to 56 306 cases, 1 417 791 controls). Given that the IPDGC data set includes proxy cases and web-based diagnosed cases and controls, we also analyzed only the subset of clinically diagnosed PD cases consisting of 15 056 cases and 12 637 controls (Table 1). In addition, we analyzed SNVs within these 2 loci in 988 cases and 2521 controls from Japan.4 Both risk variants were present at lower frequencies in European populations compared with Asian populations (Table 1).
We observed consistent association at SV2C in samples of Japanese (OR, 1.11; 95% CI, 0.94-1.31; P = .24)4 and European ancestry populations, including IPDGC full (OR, 1.07; 95% CI, 1.04-1.11; P = 3.62 × 10−5)5,8 and IPDGC clinically diagnosed sub-data set (OR,1.13; 95% CI, 1.06-1.21; P = 2.95 × 10−4)5 and UK Biobank data (OR, 1.09; 95% CI, 0.94-1.26; P = .25).6 Based on the full replication data sets, significant replication was observed at the SV2C locus (replication meta-analysis: OR, 1.07; 95% CI, 1.04-1.11; P = 9.74 × 10−6; I2 = 0%; P = .92 for heterogeneity; combined meta-analysis: OR, 1.11; 95% CI, 1.07-1.13; P = 6.02 × 10−10; I2 = 48%; P = .06 for heterogeneity) (Table 1). Meta-analysis of Asian consortium discovery samples with the European and Japanese clinically diagnosed PD replication samples5 provided support for the association at both the lead SNV SV2C rs246814 (OR, 1.16; 95% CI, 1.11-1.21; P = 1.17 × 10−10; I2 = 0%; P = .50 for heterogeneity) (Table 1) and the missense variant p.Asp543Asn rs31244 (OR, 1.16; 95% CI, 1.11-1.21; P = 1.80 × 10−10; I2 = 0%; P = .53 for heterogeneity) with low intercohort and interethnic heterogeneity.
The WBSCR17 SNV rs9638616 did not appear to be associated with PD risk in European data, IPDGC full (OR, 1.00; 95% CI, 0.98-1.02; P = .76) and clinically diagnosed data sets (OR, 1.01; 95% CI, 0.95-1.06; P = .85), UK BioBank (OR, 0.97; 95% CI, 0.89-1.06; P = .53), or Japan (OR, 1.04; 95% CI, 0.94-1.16; P = .43) PD GWAS. This SNV (OR, 1.06; 95% CI, 1.03-1.10; P = 8.37 × 10−5; I2 = 67.1%; P = 3.40 × 10−3 for heterogeneity) and locus did not reach genome-wide significance in a meta-analysis between the discovery, Japanese,4 and European clinically diagnosed PD samples (Table 1).5
Functional annotation and gene set analyses confirmed the enrichment of brain-expressed genes among the 22 genes mapped to the 11 genome-wide significant loci (eFigures 7-9 in the Supplement). Most of the 22 genes are expressed in the human brain throughout childhood and adulthood (eFigure 9 in the Supplement). Higher SV2C expression was found in the substantia nigra, striatum, and hypothalamus compared with other brain regions based on GTEX v7 data.12 WBSCR17 showed increased expression throughout multiple regions of the brain, similar to other known PD GWAS genes (eFigures 7-9 in the Supplement). There was no evidence for an association of the 2 novel SNVs (rs246814 and rs9638616) with gene expression levels in the brain or other relevant tissues.12
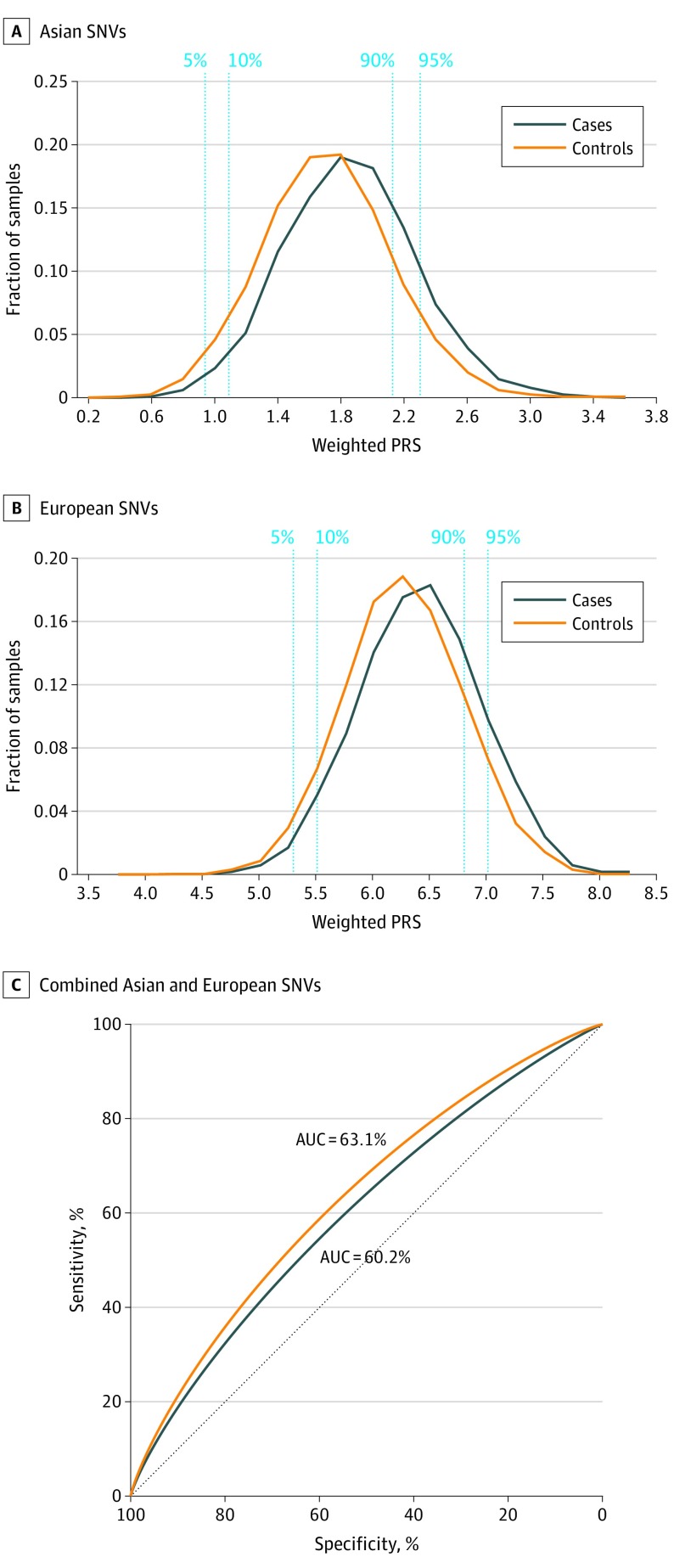
We calculated polygenic risk scores based on the 11 genome-wide significant SNVs identified in this Asian PD study. To evaluate the utility of SNVs identified by European GWASs in estimating risk in the Asian population, we calculated separate scores using 90 risk variants (78 polymorphic) from previously reported European loci using effect sizes derived from the GWAS in which they were first reported. We then evaluated the polygenic risk score distribution in the largest Asian subset of 2536 PD cases and 21 840 controls from Singapore/Malaysia (Figure 3).
Figure 3. Polygenic Risk Score (PRS) Analysis in Asian Samples.
Polygenic risk score distribution using 11 genome-wide significant Asian single-nucleotide variant (SNVs) (formerly, SNP) (A) and 90 known Parkinson disease SNVs (78 polymorphic) (B) identified in European samples and receiver operator curve based on polygenic risk prediction of Parkinson disease with previously reported SNVs (black) (area under the curve [AUC], 60.2%) vs combined European and Asian SNVs (orange) (AUC, 63.1%) (C).
In the weighted polygenic risk score distribution based on the 11 Asian SNVs, we observed a 4.0- and 3.5-fold difference in risk between the top and bottom 5% and 10% of the polygenic risk score distribution in controls (Figure 3A), respectively. We also observed that higher polygenic risk scores are significantly correlated with a younger age at onset in patients with PD (β = −1.784, P = 5.17 × 10−4), consistent with previous observations.13 In contrast, there was no correlation between the age of the controls and polygenic risk scores (β = 0.16, P = .21). We estimated a 0.29-year decrease in age at onset for every additional copy of risk allele present among the 11 loci. Assessment within our Asian PD data set of weighted PRS scores based on the 78 European SNVs revealed a 2.9- and 2.2-fold difference in risk between the top and bottom 5% and 10% of polygenic risk score distribution in controls, respectively (Figure 3B).
These 11 Asian SNVs were estimated to account for 2.61% of the variance in PD risk in this data set (AUC, 60.4%; 95% CI, 59.5%-61.8%), while the 78 polymorphic European SNVs explained 2.57% of the variance in the same data set (AUC, 60.2%; 95% CI, 59.0%-61.2%). The AUCs were not significantly different between the 2 models (P = .83). Although the European PD SNVs are still able to discriminate Asian cases and controls, their utility is limited by allelic heterogeneity, LD differences, and variability in effect sizes because of gene-gene or gene-environment interactions. Combining the European and Asian loci, we observed a significant improvement in AUC (63.1%; 95% CI, 62.1%-64.4%) over the model based on European loci alone (P = 6.81 × 10−12) (Figure 3C), and similar to that in European samples (AUC, 65.1%).5 Similar improvements were observed in the China (66.2% vs 64.7%, P = .005) and South Korea (69.5% vs 68.0%; P = .04) data sets. These analyses suggest that the data resolution conferred by polygenic risk score modeling will progressively improve as further research in Asian samples reveal additional PD risk loci.
Discussion
To our knowledge, we have conducted the largest multicenter Asian GWAS on PD to date, analyzing 31 575 individuals (6724 cases and 24 851 controls) from 6 regions across East Asia. We observed genome-wide significant association signals at 11 loci and consistent association at nominal significance (P < .05) at 51 other previously reported loci. Of the 2 novel loci we identified, we observed replication of the association at SV2C across 3 independent sample collections from European-ancestry and Japanese populations.
Strengths and Limitations
Our study had both strengths and limitations. The top-associated haplotype at SV2C is consistent between Asian and European-ancestry samples (eFigure 10A in the Supplement). Despite differences in LD patterns, the top SNV rs246814 is in near perfect LD with p.Asp543Asn (rs31244) and 2 other flanking SNVs (rs246813 and rs246815) in both Asian and European populations (eFigure 10B in the Supplement), suggesting that the functional variant likely resides on this common haplotype. The lack of significant replication at WBSCR17 in the Japanese data set may be attributed to the small effect sizes observed at this locus (68.5% power to detect an association at α = .05). There is no significant genetic heterogeneity between the Japanese replication samples and our East Asian discovery GWAS samples (P = .24 for heterogeneity, I2 = 25.6%). Future validation in larger collections of Asian samples will be needed to determine whether WBSCR17 is an Asian-specific PD risk variant.
The Asian population is the largest worldwide and also one of the most ethnically diverse.1 Our study aimed to identify loci common to 2 major East Asian populations (Han Chinese and South Koreans), with a significant proportion of our GWAS samples (1494 cases and 599 controls) being South Korean. We note that ethnic differences in allele frequencies and effect sizes are known to exist between Han Chinese, South Korean, and Japanese individuals that our study may not have identified. Further differences may exist between these and other unrepresented Asian populations in this study (eg, South Asian).1 Future collections should include substantial numbers of these samples that will allow further evaluation of the differences in genetic risk between various ethnic Asian subgroups.
Although we have analyzed what is, to our knowledge, the largest Asian sample collection to date, the study still has limited statistical power to analyze rare alleles (minor allele frequency <5%) or common alleles with smaller effects (OR, <1.15) (eFigure 11 in the Supplement). Future GWASs of larger collections (>20 000) of PD cases in the Asian population are expected to lead to identification of multiple novel PD loci, some of which are likely to be population-specific. While the polygenic risk score calculated using common GWAS SNVs appear to be applicable, albeit with limited predictive value, across different populations, we expect more accurate estimates when population-specific risk loci and effect sizes along with rarer alleles are taken into consideration.
Conclusions
We believe our study is notable in several aspects. First, we provide evidence for the association of genetic variants, including a nonsynonymous variant, in SV2C with PD risk in humans. SV2C has been evaluated for a role in synaptic function and neurotransmitter release in the basal ganglia. SV2C and other synaptic vesicle 2 proteins, SV2A and SV2B, localize to the surface of synaptic vesicles of neurons. Unlike the other 2 proteins, SV2C showed restricted expression, primarily to phylogenetically old brain regions, such as the pallidum, brainstem, substantia nigra, and olfactory bulb, which are regions directly affected by PD pathologic changes.14 Dardou et al15 estimated that over 70% of dopaminergic neurons in the substantia nigra and ventral tegmentum and about 45% of cholinergic striatal interneurons in mice express SV2C. The investigators subsequently demonstrated increased SV2C expression in dopamine-depleted mouse models and increased tyrosine hydroxylase expression in SV2C knockout mice.16 Dunn et al11 later observed a significant reduction in synaptic striatal dopamine release in SV2C knockout mice, which causes mild motor deficits in the animals. The investigators further demonstrated altered expression of SV2C in postmortem brain tissue from human patients with PD with an SNCA mutation, but not from patients with other neurodegenerative conditions.11 The SV2C protein appears to mediate dopamine homeostasis and motor function and may therefore serve as a therapeutic candidate for such deficits in PD. The missense variant Asp543Asn (rs31244) occurs within an extracellular/luminal domain containing multiple N-linked glycosylation sites. The presence of an additional asparagine residue on the risk haplotype may affect glycosylation of SV2C within this domain through introduction of a new glycosylation site matching the consensus sequence Asn-X-Ser/Thr and therefore alter its function and interaction with other proteins.10 The association we now report between this naturally occurring SV2C missense allele and increased risk of PD lends further credence to SV2C being a potential therapeutic target, although further fine-mapping efforts and functional work will be needed to elucidate the biologic mechanisms underlying the association at this locus.
In addition, although our results demonstrate similarities in PD genetic risk between European and East Asian individuals with clear overlap in risk loci and consistency in effect sizes, there are some differences in the overall underlying genetic architecture involving allele frequency and LD patterns and allelic heterogeneity, leading to an improvement in the polygenic risk score model on inclusion of SNVs identified in Asian individuals. The newly observed association at SV2C underpins this point and suggests that future, larger studies focused on the Asian population will be important to reveal novel loci that are Asian specific or Han Chinese specific. Furthermore, future trans-ethnic meta-analysis with the European samples will help to fine-map these shared loci and further contribute to our understanding of similarities and differences in the genetic risk underlying PD.
eMethods. Detailed Procedures
eTable 1. Sample Information
eTable 2. Imputev22 info scores
eTable 3. Sensitivity Analyses Using Leave-One-Out Meta-Analysis Using Correlation Between Beta Estimated Across All 5 843 213 snps Using All 5 Datasets and Beta Estimated When One Dataset Is Left Out
eTable 4. Allele frequency and pairwise linkage disequilibrium7 between top-associated SNPs identified in this study vs reported SNPs at overlapping loci with P < 10−5
eTable 5. RegulomeDB Annotation of SNPs in Novel Loci
eTable 6. HaploReg Annotation of SNPs in Novel Loci
eTable 7. Lookup of 78 Polymorphic SNPs in Previously Reported PD Loci
eFigure 1. PCA Plots by Affection Status (Case or Control) and Region in Singapore/Malaysia, China, South Korea, Taiwan and Hong Kong
eFigure 2. Examples of Genotyping Cluster Plots for rs31244 and rs6679073 Showing Good Clustering of Samples in Three Different Genotype Groups
eFigure 3. QQ Plot Before and After Excluding Snps in Known Loci With P < 1e-5
eFigure 4. Sensitivity Analyses Using Leave-One-Out Meta-analysis Showing Beta Estimates at Each of the 11 Genome-Wide Significant Loci From Meta-analysis (Fixed Effects) Using All Five Datasets and Five Other Meta-analyses When One Dataset Is Left Out inTurn
eFigure 5. QQ Plot Showing P value Distribution of 78 Polymorphic SNPs Identified to be PD-Associated in European POPULATIONS (Median P = .060, λ = 7.74) in Our Discovery GWAS on Asian-Samples and Beta-Beta Plot Showing the Correlation of Effect Sizes Estimated in Europeanvs Asian Populations
eFigure 6. Regional Association Plots at Previously-Reported European Loci Where At Least One SNP Is Associated With PD at P < 10−5 and None of the SNPs Within the Locus Is Associated With PD at P < 10−5.
eFigure 7. FUMA GENE2FUNC Analysis of 22 SNPs in 11 Asian Genome-Wide Significant Loci
eFigure 8. GTEX v7 Expression of Novel Genes SV2C and WBSCR17 Showing Enriched Expression in Brain Tissue
eFigure 9. Gene Expression in Brainspan 29 Different Ages of Brain Samples
eFigure 10. Regional Association Plots of Associations at the SV2C Locus in Asian PD Genetics Consortium Discovery Dataset and the European-Ancestry IPDGC Dataset; Pairwise LD (r2) Between the SV2C Lead SNP rs246814 and Flanking SNPs Plotted by Distance (Base Pairs) From the Lead SNP
eFigure 11. Estimated Statistical Power to Detect Loci of a Given Effect Size (X-Axis) at a Given Minor Allele Frequency
eReferences
References
- 1.Lim SY, Tan AH, Ahmad-Annuar A, et al. Parkinson’s disease in the Western Pacific Region. Lancet Neurol. 2019;18(9):865-879. doi: 10.1016/S1474-4422(19)30195-4 [DOI] [PubMed] [Google Scholar]
- 2.Dorsey ER, Elbaz A, Nichols E, et al. ; GBD 2016 Parkinson’s Disease Collaborators . Global, regional, and national burden of Parkinson’s disease, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018;17(11):939-953. doi: 10.1016/S1474-4422(18)30295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foo JN, Tan LC, Irwan ID, et al. Genome-wide association study of Parkinson’s disease in East Asians. Hum Mol Genet. 2017;26(1):226-232. [DOI] [PubMed] [Google Scholar]
- 4.Satake W, Nakabayashi Y, Mizuta I, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41(12):1303-1307. doi: 10.1038/ng.485 [DOI] [PubMed] [Google Scholar]
- 5.Nalls MA, Blauwendraat C, Vallerga CL, et al. ; 23andMe Research Team; System Genomics of Parkinson’s Disease Consortium; International Parkinson’s Disease Genomics Consortium . Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18(12):1091-1102. doi: 10.1016/S1474-4422(19)30320-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50(11):1593-1599. doi: 10.1038/s41588-018-0248-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Medical Association . World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. doi: 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 8.Nalls MA, Pankratz N, Lill CM, et al. ; International Parkinson’s Disease Genomics Consortium (IPDGC); Parkinson’s Study Group (PSG) Parkinson’s Research: the Organized GENetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson’s Disease Consortium; Alzheimer Genetic Analysis Group . Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46(9):989-993. doi: 10.1038/ng.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang D, Nalls MA, Hallgrímsdóttir IB, et al. ; International Parkinson’s Disease Genomics Consortium; 23andMe Research Team . A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49(10):1511-1516. doi: 10.1038/ng.3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao G, Zhang S, Mahrhold S, et al. N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A. Nat Struct Mol Biol. 2016;23(7):656-662. doi: 10.1038/nsmb.3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn AR, Stout KA, Ozawa M, et al. Synaptic vesicle glycoprotein 2C (SV2C) modulates dopamine release and is disrupted in Parkinson disease. Proc Natl Acad Sci U S A. 2017;114(11):E2253-E2262. doi: 10.1073/pnas.1616892114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The GTEx Consortium . Human genomics: the Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648-660. doi: 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blauwendraat C, Heilbron K, Vallerga CL, et al. ; 23andMe Research Team; International Parkinson’s Disease Genomics Consortium (IPDGC) . Parkinson’s disease age at onset genome-wide association study: defining heritability, genetic loci, and α-synuclein mechanisms. Mov Disord. 2019;34(6):866-875. doi: 10.1002/mds.27659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janz R, Südhof TC. SV2C is a synaptic vesicle protein with an unusually restricted localization: anatomy of a synaptic vesicle protein family. Neuroscience. 1999;94(4):1279-1290. doi: 10.1016/S0306-4522(99)00370-X [DOI] [PubMed] [Google Scholar]
- 15.Dardou D, Dassesse D, Cuvelier L, Deprez T, De Ryck M, Schiffmann SN. Distribution of SV2C mRNA and protein expression in the mouse brain with a particular emphasis on the basal ganglia system. Brain Res. 2011;1367:130-145. doi: 10.1016/j.brainres.2010.09.063 [DOI] [PubMed] [Google Scholar]
- 16.Dardou D, Monlezun S, Foerch P, et al. A role for Sv2c in basal ganglia functions. Brain Res. 2013;1507:61-73. doi: 10.1016/j.brainres.2013.02.041 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Detailed Procedures
eTable 1. Sample Information
eTable 2. Imputev22 info scores
eTable 3. Sensitivity Analyses Using Leave-One-Out Meta-Analysis Using Correlation Between Beta Estimated Across All 5 843 213 snps Using All 5 Datasets and Beta Estimated When One Dataset Is Left Out
eTable 4. Allele frequency and pairwise linkage disequilibrium7 between top-associated SNPs identified in this study vs reported SNPs at overlapping loci with P < 10−5
eTable 5. RegulomeDB Annotation of SNPs in Novel Loci
eTable 6. HaploReg Annotation of SNPs in Novel Loci
eTable 7. Lookup of 78 Polymorphic SNPs in Previously Reported PD Loci
eFigure 1. PCA Plots by Affection Status (Case or Control) and Region in Singapore/Malaysia, China, South Korea, Taiwan and Hong Kong
eFigure 2. Examples of Genotyping Cluster Plots for rs31244 and rs6679073 Showing Good Clustering of Samples in Three Different Genotype Groups
eFigure 3. QQ Plot Before and After Excluding Snps in Known Loci With P < 1e-5
eFigure 4. Sensitivity Analyses Using Leave-One-Out Meta-analysis Showing Beta Estimates at Each of the 11 Genome-Wide Significant Loci From Meta-analysis (Fixed Effects) Using All Five Datasets and Five Other Meta-analyses When One Dataset Is Left Out inTurn
eFigure 5. QQ Plot Showing P value Distribution of 78 Polymorphic SNPs Identified to be PD-Associated in European POPULATIONS (Median P = .060, λ = 7.74) in Our Discovery GWAS on Asian-Samples and Beta-Beta Plot Showing the Correlation of Effect Sizes Estimated in Europeanvs Asian Populations
eFigure 6. Regional Association Plots at Previously-Reported European Loci Where At Least One SNP Is Associated With PD at P < 10−5 and None of the SNPs Within the Locus Is Associated With PD at P < 10−5.
eFigure 7. FUMA GENE2FUNC Analysis of 22 SNPs in 11 Asian Genome-Wide Significant Loci
eFigure 8. GTEX v7 Expression of Novel Genes SV2C and WBSCR17 Showing Enriched Expression in Brain Tissue
eFigure 9. Gene Expression in Brainspan 29 Different Ages of Brain Samples
eFigure 10. Regional Association Plots of Associations at the SV2C Locus in Asian PD Genetics Consortium Discovery Dataset and the European-Ancestry IPDGC Dataset; Pairwise LD (r2) Between the SV2C Lead SNP rs246814 and Flanking SNPs Plotted by Distance (Base Pairs) From the Lead SNP
eFigure 11. Estimated Statistical Power to Detect Loci of a Given Effect Size (X-Axis) at a Given Minor Allele Frequency
eReferences