Abstract

Xenobiotics are ubiquitous in the environment and modified in the human body by phase I and II metabolism. Liquid chromatography coupled to high resolution mass spectrometry is a powerful tool to investigate these biotransformation products. We present a workflow based on stable isotope-assisted metabolomics and the bioinformatics tool MetExtract II for deciphering xenobiotic metabolites produced by human cells. Its potential was demonstrated by the investigation of the metabolism of deoxynivalenol (DON), an abundant food contaminant, in a liver carcinoma cell line (HepG2) and a model for colon carcinoma (HT29). Detected known metabolites included DON-3-sulfate, DON-10-sulfonate 2, and DON-10-glutathione as well as DON-cysteine. Conjugation with amino acids and an antibiotic was confirmed for the first time. The approach allows the untargeted elucidation of human xenobiotic products in tissue culture. It may be applied to other fields of research including drug metabolism, personalized medicine, exposome research, and systems biology to better understand the relevance of in vitro experiments.
Introduction
Humans and other organisms are exposed to a multitude of xenobiotics during their lifetime through food and environment.1−3 The human organism utilizes different metabolic mechanisms to activate, detoxify, and excrete them.4 Phase I reactions cause an increase in polarity and thereby enhance the reactivity of the parent molecule, and phase II reactions form conjugates that can be excreted via urine. Phase II reactions include sulfation, glucuronidation, glutathione-conjugation, and acylation.5,6 The metabolic fate of xenobiotics in human systems is frequently incompletely understood, although crucial for the assessment of toxicity.
Liquid chromatography high resolution mass spectrometry (LC-HRMS) is the premier technique for the sensitive and selective detection and characterization of unknown metabolites and allows for the elucidation of xenobiotic biotransformation. However, the evaluation of LC-HRMS data sets, originating from untargeted metabolite profiling, is time-consuming and challenging and requires advanced bioinformatics tools.7 A powerful tool for the elucidation of novel metabolic pathways and respective data analysis is the recently developed MetExtract II algorithm.8 Concurrent analysis of stable isotopically labeled and native sample material allows the automated and comprehensive extraction of metabolic features relevant for the metabolism of practically any investigated small molecule. The practicability and power of this approach have already been proven in a number of plant-based experiments.8−12
Deoxynivalenol (DON), a secondary metabolite produced by Fusarium fungi, is a mycotoxin frequently found as a contaminant in wheat, corn, oat, and other grains as well as in products thereof.13 The toxin inhibits protein synthesis and subsequently also RNA and DNA synthesis.14 Additionally, ribotoxic stress causes stimulation of the production of cytokines, and induction of apoptosis is triggered by high concentrations of the mycotoxin.15 Acutely, it induces emesis and abdominal pain.16 The effects of chronic exposure to DON include, besides growth retardation and immunotoxicity, also impairment of reproduction and development in animals. The impact of low dose chronic exposure to humans remains elusive.17 The prevalence of the DON producing fungi in cereal based crops and the thermal stability of the toxin itself make the mycotoxin an issue for global food safety.16 Multiple human biomonitoring studies suggested that the established tolerable daily intake of DON (1 μg kg–1 body weight per day) is exceeded frequently in different populations, particularly by highly susceptible subgroups such as children and pregnant women.17−20 The metabolic fate of DON has been studied in humans in vivo,21,22 in animals,23−26 and also in the Fusarium host plant, wheat.9,27−29 DON-glucuronides (DON-GlcAs) were identified as the major urinary DON-metabolites in humans. While glucuronidation is the major detoxification pathway in humans, the formation of DON-GlcAs in human cell culture models has not been reported so far. The toxicological properties of DON biotransformation products significantly differ from the parent molecule. The ability of DON-sulfates to inhibit protein synthesis by inactivation of the ribosome was reduced,29 and consequently cytotoxicity was decreased compared to DON. However, an increase of cellular proliferation and activation of autophagy were described.30,31 Incubation of human cells with DON-glucuronide resulted in decreased cytotoxicity when compared to the parent toxin.32 In addition, conjugates with thiol or sulfhydryl containing compounds including DON-GSH showed no toxicity in human monocytes and did not enhance proinflammatory cytokines.33
In this study, an adapted workflow for the comprehensive elucidation of xenobiotic biotransformation through LC-HRMS, assisted by stable-isotope labeling and the bioinformatics tool MetExtract II, was established. Due to its potential for identifying known and unknown metabolic products, the in vitro metabolism of DON in human cell culture was studied with this technique. Two well-studied cell models of liver and colon, respectively, were tested and revealed unexpected insights into the in vitro metabolism of the model xenobiotic DON.
Results
Optimization of Tissue Culture and the Analytical Workflow
The selection of the most appropriate cell lines was based on a number of preliminary experiments in five different cell models (HepG2, Caco-2, HT29, HEK293, and T24). The uptake rate of DON from the extracellular medium into the cells after 24 h was lower for cells from the urinary tract (HEK293 and T24; below 0.3%) than for the other cell lines (approximately 1.5%). When screening for biotransformation products, DON-3-sulfates and DONS2 were confirmed by the correct m/z value and retention time with an authentic reference standard and were present in the supernatant of all cell lines after a 24 h incubation. Besides, the accurate masses corresponded to two further sulfur containing derivatives of DON, namely DON-GSH (Δppm −0.7) and DON-cysteine (Δppm −0.2). They were detected in the lysate of HepG2, HT29, and Caco-2 cells. In cell lines originating from liver (HepG2) and colon tumors (HT29, Caco-2) the number and relative concentrations of biotransformation products were higher than in kidney and bladder-derived cell lines. As the uptake of DON was slightly higher in HT29 than in Caco-2, HT29 was selected as an intestinal cell model besides HepG2 which served as a model for liver metabolism.
Initially, six-well plates were chosen for growing cells since formats with a lower volume resulted in low cell numbers and the inability to detect any DON biotransformation product by LC-HRMS. However, during the course of these pre-experiments we encountered that even six-well plates allowed only for the detection of the most abundant known biotransformation products but failed to enable the identification of low-abundance metabolites of potentially high biological impact. Therefore, we further up-scaled the approach to Petri dishes in order to maximize cell number and thus the concentrations of DON and its known and yet unknown metabolites in the cell lysate and in the extracellular medium. In addition, we increased the xenobiotic concentration from 1 μM to 10 μM (each 5 μM 12C-DON and 13C-DON). However, this required the incubation time to be reduced to 3 h since 10 μM DON induces cytotoxicity when cells are exposed to the mycotoxin for a longer period of time.16,30
The chromatographic separation was optimized in order to allow for the separation of DON conjugate isomers. The retention of DON on C-18 RP columns is known to be typically very limited due to its high polarity. Phase II metabolites are even more polar, and the separation of isomers constitutes a well-known separation issue.20,25 The selected column-eluent combination exhibited sufficient retention of the highly polar conjugates when an optimized, very flat gradient (increase from 5% to 40% eluent B between minutes 2 and 7) was applied. Different mobile phases were evaluated, and water and methanol, both containing 0.1% acetic acid, were deemed the most suitable eluents due to enhanced retention of DON conjugates. Despite the flat gradient, the total run time was below 12 min.
Stable Isotope-Assisted Data Processing and Targeted Evaluation
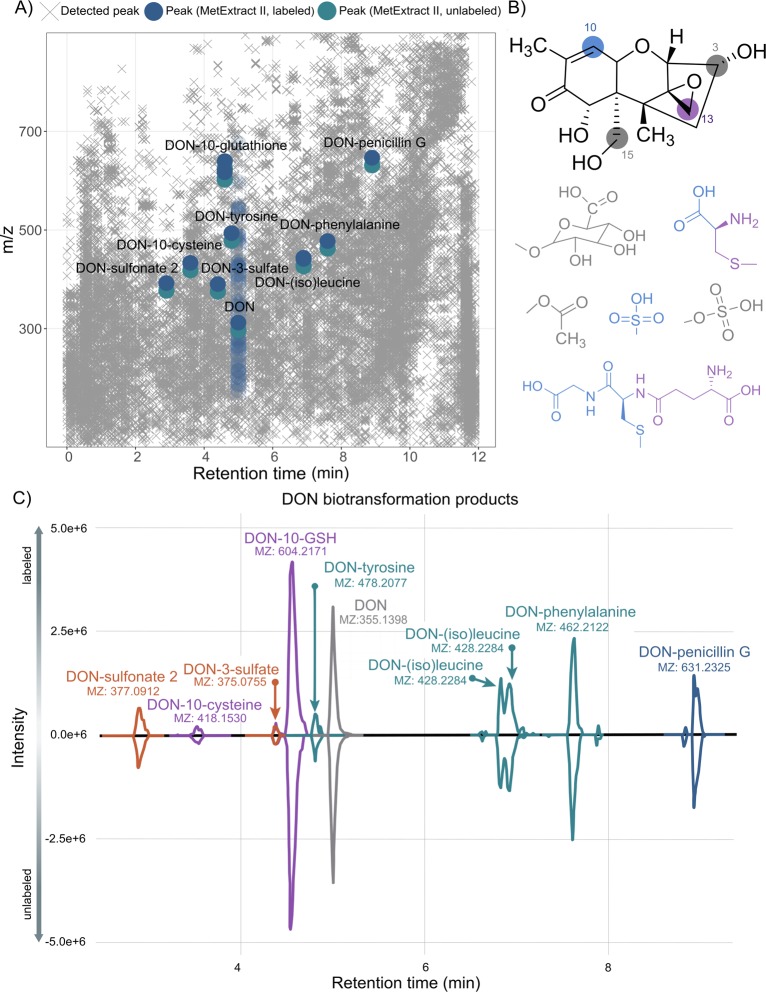
In order to capture all detectable biotransformation products of DON, the TracExtract approach, a module of MetExtract II,8 was utilized. As the exogenous DON was applied as a mixture of both native and uniformly 13C15-labeled toxin and similar metabolization of both forms can be assumed, all biotransformation products of DON formed in the cell cultures would also be present as a native and 13C-labeled form, while any other metabolite in the sample itself would only be present as a purely native form but not as a 13C-labeled form. Thus, this characteristic can be employed to automatically search for the native and 13C-labeled biotransformation products of DON with the MetExtract II software. The software tool is specifically designed to automatically search for coeluting ion pairs of native and 13C-labeled metabolite forms. In case of DON, which consists of 15 carbon atoms, a mass difference of 15.0503 m/z is expected if no carbon atom is cleaved. It only reports such metabolites but ignores any other chromatographic peaks in the LC-HRMS data thereby efficiently filtering out all other nontracer-derived metabolites from the data set. The reduction of peaks by the tool is visualized in Figure 2a. A detailed comparison was not conducted in this context.
Figure 2.
Overview of data reduction by MetExtract, chemical structure and potential reaction sites of the food toxin DON, and detected biotransformation products. (A) All detected features in the analyzed samples are illustrated by gray ×’s (×). Features of metabolic products of deoxynivalenol detected by the MetExtract II algorithm are highlighted in blue, and features related to the parent molecule DON (eluting at 5 min) are shown in faded blue symbols (except for the [M + H]+ adduct) for better visibility. (B) Possible reaction sites for conjugation of DON. Positions C-3 and C-15 are targets for sulfate-, glucuronide-, and acetyl-conjugation. GSH as well as cysteine conjugates are formed at positions C-10 and C-13. DON-sulfonates are preferably formed by Michael addition at position C-10. (C) XIC chromatograms of annotated DON metabolites. The XICs of the native metabolite forms are shown with negative intensities, while the XICs of the 13C-labeled metabolite forms are shown with positive intensity values. The XICs were extracted from different raw data files, and for each metabolite the dominant adduct/in-source fragment is shown.
For all masses of interest, agreement (retention time, accurate mass, MS2) with the available reference standards was checked. Known metabolites included in the multicomponent standard and other metabolites described in the literature like the plant metabolites DON-GSH, DON-cysteine, and DON-cysteinylglycine28,34,35 were annotated automatically by MetExtract II in case they were present. According to a study by Schwartz et al.,36 other potential metabolites, namely the DON-sulfonates, were included in the analysis.
Annotation of Novel Detoxification Products
The next step was the annotation of unknown biotransformation products. For ion pairs with a mass difference of 15.0503 between the 13C-labeled and the unlabeled compound an intact carbon scaffold of the DON-molecule was assumed. This applies to nearly all feature pairs not identified by authentic standards or reference MS2 spectra. Based on the known metabolites, thiols are conjugated to DON either via Michael addition at position C-10 or at position C-13, opening the epoxide moiety. Another possible reaction is the conjugation of DON at a hydroxy group, mainly at positions C-3 and C-15, by enzymatic reaction with activated cosubstrates (UDP-glucuronic acid, acetyl-CoA, or 3′-phosphoadenosine-5′-phosphosulfate) and elimination of water. The potential reaction sites of DON for glucuronidation (or glucosylation), acetylation, sulfation, and for reaction with thiol groups (like sulfonation) are highlighted in Figure 2b. With this knowledge the exact mass of molecules possibly conjugated to DON was calculated, and databases like the HMDB,37 METLIN,38 and MassBank39 were explored to see if matching molecules potentially conjugated to DON could be found. In order to further characterize annotated conjugates, MS2 spectra were recorded and compared to reference spectra from mzCloud. As no reference spectra for the intact conjugates were available, reference spectra of the binding molecules were considered. The comprehensive workflow established is shown schematically in Figure 1.
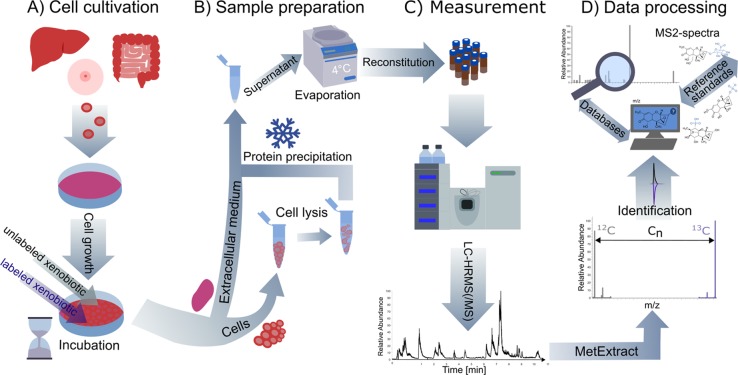
Figure 1.
Schematic representation of the developed workflow for the stable isotope-assisted elucidation of xenobiotic metabolism in human cells including (A) cell cultivation, (B) sample preparation, (C) LC-HRMS (/MS)-measurement, and (D) data processing and annotation. Metabolism of the model xenobiotic DON was investigated in an intestine and liver cell model.
In this study MetExtract II was applied to mammalian tissue culture, revealing ten ion pairs of interest. Authentic reference standards (commercially available or in-house synthesized), as well as MS2 reference spectra from the literature28,34,40 and the public databases HMDB,37 METLIN,38 and MassBank,39 served to annotate eight of the ten ion pairs of interest. In addition, one biotransformation product, DON-glutamylcysteine, was detected only manually. The identity of two metabolites, DON-3-sulfate and DONS2, was confirmed at level 1 by authentic reference standards. Other metabolites present in the standard mix such as the DON-glucuronides, acetyl-DONs, and acetyl-DON-sulfates were not detected. Table 1 reports all annotated and identified metabolites of DON. Most metabolites were annotated exclusively in the positive ionization mode, while DON-3-sulfate and DONS2 were solely detected in the negative ionization mode. The parent compound DON was found in both modes as well as DON-GSH and a putative DON-isoleucine/leucine conjugate. MS2 spectra were acquired for all compounds detected by MetExtract II, partly in additional experiments with inclusion lists and increased injection volume. DON-3-sulfate and DON-15-sulfate were distinguished by their fragmentation pattern. The fragment [M – CH2O – H]− with m/z 345.3425, which is formed when the CH2OH group at the C-6 position is cleaved off, is unique for DON-3-sulfate as for DON-15-sulfate the sulfate group is attached at this position.29 The described fragment was present at the earlier eluting peak at 4.37 min (standard compound). In the sample a peak at retention time 4.37 min, matching DON-3-sulfate was detected. Due to the low abundance of DON-sulfates in the sample, the fragment at m/z 345.3425 was not present in the MS2 spectrum of the sample (Supplementary Figure 1). Based on the availability of both DON-sulfate standards and measured retention times, the candidate was annotated as DON-3-sulfate.
Table 1. Metabolites of the Food Contaminant Deoxynivalenol Annotated by the Stable Isotope-Assisted Approach in HT29 and HepG2 Cells.
| DON metabolites | RT (min) | m/za | ion speciesb | mass accuracy (ppm) | polarityc | 13C-atomsd | sum formulae | levelf | MS2 |
|---|---|---|---|---|---|---|---|---|---|
| DON | 4.99 | 355.1402 | [M + Ac]− | 1.1 | ± | 15 | C15H20O6 | 1 | yes |
| DON-sulfonate 2 | 3.33 | 377.0907 | [M – H]− | –1.3 | – | 15 | C15H22O9S | 1 | yes |
| DON-10-cysteine | 3.50 | 418.1529 | [M + H]+ | –0.2 | + | 15 | C18H27NO8S | 2a | yes |
| DON-glutamylcysteineg | 4.25 | 547.1955 | [M + H]+ | –0.2 | + | 15 | C23H34N2O11S | 4 | no |
| DON-3-sulfate | 4.38 | 375.0748 | [M – H]− | –1.9 | – | 15 | C15H20O9S | 1 | yes |
| DON-10-glutathione | 4.56 | 604.2167 | [M + H]+ | –0.7 | ± | 15 | C25H37N3O12S | 2a | yes |
| DON-tyrosine | 4.81 | 478.2075 | [M + H]+ | –0.5 | + | 15 | C24H31O9N | 2b | yes |
| DON-(iso)leucine | 6.91 | 428.2280 | [M + H]+ | –1.0 | ± | 15 | C21H33O8N | 3 | yes |
| DON-phenylalanine | 7.59 | 462.2121 | [M + H]+ | –0.2 | + | 15 | C24H31O8N | 2b | yes |
| DON-penicillin G | 8.96 | 631.2313 | [M + H]+ | –1.9 | + | 15 | C31H38N2O10S | 2b | yes |
Accurate mass of most abundant ion species of each metabolite based on MS2 spectra from the second run.
Most abundant ion species.
Polarity where metabolite could be detected.
Number of 13C-atoms in the labeled conjugate.
Sum formula of the neutral metabolite.
Identification level based on Schymanski et al.41
Manually detected, not picked up by MetExtract.
The extracted ion chromatograms (XIC) at m/z 377.0916 of the sample containing DON-sulfonate [M – H]− was compared to single standards of three different DON-sulfonate isomers (Supplementary Figure 2). DONS1 (around 1.51 min) elutes earlier than DONS2 (around 3.40 min) and DONS3 (around 3.91 min). The peak in the experimental sample (extracellular medium, HT29) appeared at 3.33 min, which agrees with the retention time of the DONS2 standard. In the acquired MS2 spectrum, besides the parent ion, two further matching fragments (m/z 265.11 and m/z 138.03) were present in both the single standard of DONS2 and in the cell-derived sample. DON-10-GSH (Supplementary Figure 3) and DON-10-cysteine (Supplementary Figure 4) were characterized at identification level 2a by a successful match with MS2 spectra from the literature.34,40 In addition, a comparison of the MS2 spectrum and the retention time with a wheat sample in which the presence of DON-10-GSH was confirmed before35 further supported the identification. The remaining conjugates were annotated at level 2b (probable structure/diagnostic). The masses observed are consistent with the formation of adducts with different amino acids and an adduct with penicillin G. MS2 spectra were acquired and compared to available spectra of the amino acids and the antibiotic (penicillin G) in mzCloud (Figure 3). A good agreement of the obtained MS2 spectra and the ones from the literature was found despite the generally low abundance of these metabolites in our samples. For DON-(iso)leucine two peaks were present, originating from DON-leucine and its isomer DON-isoleucine. An assignment of the isomers was not conducted, and the peaks were not baseline separated. An overlay of the XICs of all annotated metabolites is illustrated in Figure 2c. All ion traces of the labeled metabolite ions are presented on the positive axis, while the overlaid XICs of the monoisotopic unlabeled compound ions appear on the negative axis. Most of the annotated metabolites were found in positive ionization mode; only DON-3-sulfate and DONS2 were present in negative mode. DON-(iso)leucine and DON-10-GSH were detected in both polarities. In addition to the automatically by MetExtract II derived DON derivatives, another conjugate, DON-γ-glutamylcysteine (m/z 547.1856, 4.25 min), was found manually at 4.25 min in the lysate of two out of three HepG2 samples. The retention time was, as expected, between those of DON-10-cysteine (3.56 min) and DON-10-GSH (4.57 min). However, the annotation of DON-γ-glutamylcysteine was somehow uncertain as the peak was of low abundance and no MS2 spectrum was triggered. The chemical structures of the newly annotated DON-metabolites remained ambiguous, and the reported conjugation sites can be regarded as preliminary as standards were lacking and the MS2 spectra did not contain enough information to determine their precise structure. Based on theoretical mass calculation of DON conjugates (i.e., addition without loss of water), the only possible reaction sites for conjugation were at position C-10 or at position C-13 (Figure 2b).
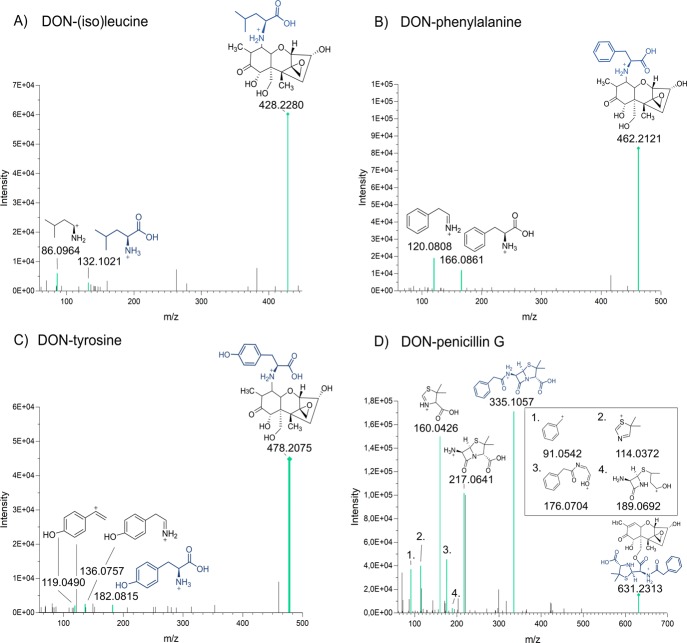
Figure 3.
MS2 spectra illustrating conjugates of DON with amino acids and an antibiotic in positive mode [M + H]+, where fragments present in the spectra of the pure compound found in mzCloud too are colored in turquoise. All spectra were acquired with a stepped normalized collision energy at 20 and 50 eV. (A) MS2 spectrum of DON-isoleucine/leucine of m/z 428.2292 at 6.82 min, (B) MS2 spectrum of DON-phenylalanine of m/z 462.2120 at 7.61 min, (C) MS2 spectrum of DON-tyrosine of m/z 478.2074 at 4.81 min, and (D) MS2 spectrum of DON-penicillin G of m/z 631 at 8.06 min.
Occurrence of Biotransformation Products
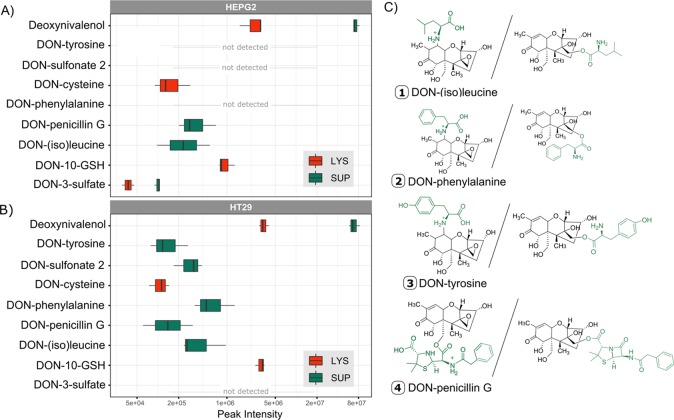
Not only the cell lysate samples but also the extracellular medium (supernatant) of the cells were screened for the presence of DON-metabolites excreted by the cells (Figure 4). In both cell lines, HT29 and HepG2, the abundance of the DON-metabolites was higher in the supernatant as compared to the lysate samples. Only DON-10-GSH and DON-10-cysteine were present exclusively in the lysate. Very low amounts of DON-3-sulfate were present in the lysate of HepG2 cells although the concentration was about three times lower than in the supernatant. All amino acid conjugates except the sulfur-containing cysteine and the penicillin conjugate were solely present in the supernatant. Interestingly, the sulfo-conjugation pattern was different in the supernatant of both cell lines. DON-3-sulfate was produced only in HepG2 cells, while DONS2 was excreted by HT29 cells. Amino acids and penicillin G were present in the medium; thus, a direct chemical reaction could theoretically be responsible for the conjugation of DON to these molecules. Consequently, an additional control experiment (#2) without cells but identical incubation conditions was performed. None of the DON biotransformation products were detected in the cell-free incubations, strongly supporting a cell mediated and enzymatic formation of the observed metabolites. Absolute quantification of the produced metabolites was not possible as no standards for the newly described compounds were available. However, in additional spiking experiments an average recovery of >90% in the cell lysates and >50% in the supernatants for the parent toxin in both positive and negative ionization mode was observed. The discrepancy originated most likely from matrix effects. Looking at the relative abundance, DON-10-GSH was clearly the most prevalent metabolite in both cell lines reaching values of about 1 × 106 cps for HepG2 and 3 × 106 cps for HT29 in the lysate samples. The DON-10-GSH was even in the same order of magnitude as the parent compound DON in the lysate. All other metabolites were of low abundance with a maximum signal intensity of 5 × 105 cps for DON-phenylalanine in HT29 and thereby close to the limit of detection.
Figure 4.
Peak intensities of annotated DON metabolites. (A) Peak intensities of unlabeled DON-metabolites in supernatant (SUP, green) and lysate (LYS, red) of HepG2 (top). Except for DON-3-sulfate and DONS2, measurements of the positive mode are shown. The average of all three biological replicates with a maximum n = 3 is plotted. The metabolite had to be present in at least two out of the three replicates. A logarithmic scale was used for the x-axis (abundance). (B) Abundance of unlabeled DON-metabolites in supernatant (green) and lysate (red) of HT29 (bottom). Except for DON-3-sulfate and DONS2 measurements of the positive mode are shown. The average of all three biological replicates with a maximum n = 3 was taken. The metabolite had to be present in at least two out of the three replicates. A logarithmic scale was used for the x-axis (abundance). (C) Putative structures of amino acids and xenobiotic conjugates of DON with both possible structures. For DON-penicillin G conjugation can occur either after epoxide opening at C-13 or at C-15-OH and C-3-OH (not shown) after lactam opening. For DON amino acid adducts the options include conjugation after epoxide opening at C-13 or at C-10 by Michael addition.
Discussion
The MetExtract II workflow8 was successfully adapted to cell culture experiments employing human cell models. The workflow provides an efficient way to detect novel biotransformation products of any xenobiotic with the only requirement that isotopically labeled substances need to be available. Our study revealed that various detoxification strategies were used by the tested cell models, and some metabolites were detected and further characterized for the first time. In humans, glucuronidation is normally by far the most prominent detoxification pathway in vivo.(20) Interestingly, the formation of DON glucuronides in human cell culture was not confirmed to date.19,42,43 In our tissue culture models DON-glucuronides were also absent despite the presence of uridine 5′-diphosphate (UDP)-glucuronosyltranferase transcripts involved in their formation.44−46 We assume that the concentrations of potentially formed DON glucuronides were most likely too low to be detected since the ionization efficiency of these conjugates was described as limited.30
DON-GSH and Related Metabolites
A common phase II reaction is glutathione conjugation. In plants this tripeptide conjugate and its processing products DON-cysteinylglycine and DON-cysteine have been described.9 However, glutathione-conjugate formation of DON has not been recognized as a mammalian detoxification mechanism so far. We are only aware of one previous study indicating that this pathway seems to exist for DON.47 In our experiment, we found two isomers of DON-10-GSH and DON-10-cysteine. The fragmentation pattern indicates that both were conjugated at position C-10 via Michael addition. The two isomers of DON-10-GSH had different fragmentation patterns,34 but both of them were likely to originate from DON-10-GSH as m/z 179.0482 (first peak, 4.36 min) and m/z 130.0499 (second peak, 4.54 min) were the most prominent peaks. Fragments like m/z 281.0836 and m/z 263.0733 characteristic for DON-13-GSH34 were absent. A comparison to a wheat sample in which this metabolite was previously found confirmed this annotation. Thus, they were probably derived from 9,10-diastereoisomers of the Michael adduct described in Stanic et al.34 which was formed in a nonenzymatic reaction with GSH after extended incubation at alkaline pH. The fragmentation pattern in the negative ionization mode supported this with a product ion at m/z 306.0772 being the base peak. To distinguish between a spontaneous reaction with glutathione and a glutathione-S-transferase (GST)-mediated conjugate formation, we performed incubations with the cell free medium. Neither DON-10-GSH nor other conjugates were detected in this control experiment (#2), indicating that the reaction was mediated by the cells. It is very unlikely that conditions exist in the cells that would allow for the nonenzymatic formation of Michael adducts. In the nonenzymatic synthesis of DON-10-GSH the reactants DON and GSH had to be incubated at pH 10.7 for 9 d.34 Formation in most alkaline compartments in the cells, such as the mitochondrial matrix (reported pH 8.5, Abad et al.48) or the peroxisomes (pH 8.2, Dansen et al.49), seems unlikely considering the short incubation time of 3 h. On the other hand, so far no mammalian GST catalyzing the DON-GSH adduct formation is known, and trichothecenes are generally believed to be unreactive with GSTs.50 However, plant GSTs catalyzing the formation of DON conjugates were recently identified.51 The level of glutathione S-transferases is lower in HepG2 compared to HT29 cells,52 which may explain the higher amount of formed DON-10-GSH in the colon carcinoma cell line compared to HepG2. DON-glutamylcysteine was detected in the lysate. During GSH-biosynthesis, γ-glutamylcysteine formation from glutamate and cysteine is the rate-limiting step catalyzed by the enzyme γ-glutamate-cysteine ligase.53 Again, a spontaneous reaction with the GSH precursor present in low amounts in the cell cannot be excluded, but the more likely explanation is that an enzymatically formed DON-GSH conjugate was processed by a peptidase cleaving glycine faster than the reaction occurred with γ-glutamyl-transpeptidase. Both activities are needed to generate DON-cysteine from DON-GSH. Cysteine might directly react nonenzymatically as well35 in an alkaline cell compartment, but this seems unlikely. A subsequent step in the detoxification through the mercapturic pathway would be the formation of N-acetyl-cysteine-DON.54 Neither our MetExtract II analysis nor a manual search of the LC-HRMS(/MS) raw data resulted in the detection of a metabolic feature corresponding to the m/z value of this structure, which might be explained by the short incubation time and the very low abundance already of the precursor DON-cysteine, which resulted in an MS2 spectrum with low intensity fragments. In one sample the fragment of the intact DON was present, indicating that the found substance was likely to be DON-S-10-cysteine. This was further supported by comparison with a literature spectrum.40 The positive mode MS2 spectrum of the epoxide adduct included prominent fragments at m/z 388.1427, m/z 281.0846, and m/z 263.0739. Neither of them was present in our spectrum. Moreover, product ions at m/z 122.0268 (Δ 4.9 ppm) and m/z 401.1268 (Δ 0.2 ppm) were detected in our study and were also reported in the published spectrum of DON-10-cysteine.40
DON-Sulfate and Sulfonate
Two sulfur conjugates were identified by comparison with reference standards, namely DON-3-sulfate and DON-sulfonate 2. DON-3-sulfate was identified before as a minor metabolite in human urine.30 However, DON-sulfonate was previously unknown to be produced by humans. It has only been described in rats and chickens so far.23,55 Several hypotheses have been formulated as to how DON-sulfonates are formed in animals by Wan et al.55 One possible pathway is formation via the DON-GSH conjugate. After a breakdown to DON-cysteine, enzymatic cleavage of the conjugate by cysteine-S-conjugate β-lyase, the hypothetical DON-SH could be formed. It is known that in proteins the SH group of cysteine can be further oxidized56 all the way to the sulfonate group. Yet, the oxidation of DON-SH would need high levels of H2O2 that are unlikely to exist in cells, potentially with the exception of peroxisomes. Another possibility would be the formation of sulfite from sulfate followed by nonenzymatic Michael addition to DON.23,55 The cell culture medium (DMEM) contains low amounts of magnesium sulfate but high levels of glutathione and cysteine as a sulfur source. We therefore propose as alternative, that cysteine is first converted by cysteine dioxygenase (KEGG R00893) to 3-sulfino-l-alanine. In the second step SO2 could be released either enzymatically by 3-sulfino-l-alanine 4-carboxy-lyase (KEGG R00863) or by spontaneous decay of this compound into alanine and SO2. The SO2 released is in an equilibrium with sulfite,57 which then might form nonenzymatically the Michael adduct.
Amino Acid and Penicillin Conjugates
The remaining ion pairs extracted by the MetExtract II algorithm were not identified by reference standards or respective literature spectra. When subtracting the mass of DON, the obtained masses were matching not only amino acids including phenylalanine, tyrosine, and (iso)leucine but also surprisingly the antibiotic penicillin G. As building blocks of proteins, amino acids are basal ingredients of cell culture, and higher concentrations can be found in DMEM (HT29) compared to RPMI 1640 (HepG2). In order to prevent bacterial contamination of the cell cultures, penicillin G was used as a supplement in both media. Conjugation with amino acids is a well-known metabolic reaction of xenobiotics in the human body. Typically, xenobiotics with a carboxyl group like salicylic acid or valproic acid are activated by the formation of CoA derivatives, which can react with the amino group of amino acids to build the corresponding N-substituted amides. A frequent reaction partner is glycine but also glutamine, glutamate, and taurine are involved. In most cases this is only a minor pathway in xenobiotic metabolism.58 DON does not have a carboxy group to be activated for conjugation. The only primary alcohol that could be converted to a carboxy group is C15-OH, but the subsequent amide formation with an amino acid would not lead to a mass consistent with the observed reaction product. The formation of an adduct which fits the mass of the found novel DON-amino acid conjugates can either occur at the epoxide moiety or by addition at the double bond without water loss. The putative structures of both options are presented in Figure 4c. In the first scenario, the epoxide would be opened (by an epoxide hydrolase, +H2O), and subsequently an ester with the carboxy group of the amino acid would be formed (−H2O). However, this is highly unlikely because so far no epoxide-hydrolase being able to act on the trichothecene’s epoxide has ever been characterized,59 and also the ester formation between the formed diol and the carboxy group is improbable to happen spontaneously. For the ester formation the amino acid would have to be activated to a thioester, which could occur by formation of an adenylate by tRNA synthetases or by a direct reaction with the charged tRNAs. Enzymes catalyzing ester formation with activated nonstandard amino acids have been described in secondary metabolite biosynthetic pathways (e.g., Lin et al.60). Alternatively, direct epoxide ring opening involving the amino group with amino acid esters has been described to occur without a catalyst at harsh conditions.61 The adenylates of amino acids or charged tRNA could potentially also undergo this reaction.
The second more likely option to form a DON-amino acid conjugate which is in line with the mass of the measured molecules is a Michael adduct formation by addition of the amino group of the amino acid with the double bond of the α,β unsaturated ketone structure in DON.62 Potentially, such reactions could occur with N-terminal amino groups of proteins rather than with free amino acids (no reaction was observed in the cell free control). Specific proteins might catalyze such a protein-adduct formation with DON, followed by proteolytic degradation leading to release of the respective amino acid conjugate.
In the supernatant of HT29 more amino acid conjugates including DON-phenylalanine, DON-(iso)leucine, and DON-tyrosine were detected compared to the other cell line, while for HepG2 cells only DON-(iso)leucine was found. A possible reason for higher biotransformation product concentrations might be the medium as in the basal medium of the HT29 cells (DMEM) higher concentrations of amino acids are present. However, the different metabolic activity, based on the varying conjugation products of both cell lines, might be another explanation. For the formation of the DON-penicillin conjugate, again two possibilities exist (Figure 4). The carboxylic acid of penicillin G (presumably after activation to a CoA derivative) forms an ester with the opened epoxide of DON. Alternatively,–in order to arrive at the observed mass–the lactam ring of penicillin is opened (+ H2O) to benzylpenicilloic acid, and one of the hydroxyl groups of DON (C-3 or C-15) forms an ester with one of the two (again presumably CoA-activated) carboxyl groups.
The presented isotope-assisted approach enables the global untargeted screening of DON conjugation products in mammalian cell cultures, and application to HepG2 and HT29 cell lines resulted in several known and novel DON conjugates. The most abundant conjugate, DON-10-GSH, was known as a detoxification mechanism of DON in plants.28 DON-sulfates and DON-sulfonates were also described to be less potent inhibitors of protein biosynthesis.30,63 The toxicity of the novel amino acid and penicillin conjugates is untested, but based on the structure of other known DON conjugates, an interaction with the ribosomal target site seems highly unlikely. However, as the majority of DON was not conjugated, the overall impact of metabolism on the toxicological potential is suspected to be minor in the specific case of DON. Only DON-10-GSH, formed in higher amounts, may have a significant effect. The pattern of metabolism for biotransformation products known in the past was clearly different from human in vivo experiments. Especially the lack of glucuronidation in the cells implies a different toxicological profile compared to the human in vivo metabolism.
Conclusion
The presented comprehensive workflow demonstrated its vast potential for deciphering novel human biotransformation products of xenobiotics. It will be a highly versatile tool for the untargeted detection and structural annotation of so far unknown metabolic products of potentially any small molecule in cell culture. These metabolites are not only interesting in the context of toxicology and pharmacology but are also an important contributor to the exposome. Our results further highlight the abundant and fast metabolism of the food contaminant DON in tissue culture and demonstrate the need to investigate xenobiotic metabolism thoroughly to understand its full biological impact.
Experimental Section
Chemicals and Standards
Reference standards were purchased from RomerLabs and Sigma-Aldrich Chemie GmbH or synthesized in-house.64,65 The 13C-DON was obtained from Romer Laboratories. A multistandard solution containing DON, de-epoxy-DON, DON-3-sulfate, DON-15-sulfate, DON-3-GlcA, DON-3,15-disulfate, 15-acetyl-DON-3-sulfate, 3,15-diacetyl-DON, 3-acetyl-DON, 3-acetyl-DON-15-sulfate, and 15-acetyl-DON was prepared in ACN/MeOH/H2O. In addition, individual standards of DON-sulfonate 1 (DONS1), DON-sulfonate 2 (DONS2), and DON-sulfonate 3 (DONS3), all conjugated at position C-10 and previously characterized by NMR,36 were available and diluted in 10% MeOH before the measurements.
Cell Culture
Pre-experiments without the cost-intensive isotopically labeled xenobiotic were performed to identify the most suitable model cell lines with sufficient metabolic activity. Five human cell models, all purchased from ATCC, were evaluated: HT29 and Caco-2 (both colorectal adenocarcinoma cells), HepG2 (hepatocellular adenocarcinoma cells), HEK293 (embryonic kidney cells), and T24 (bladder carcinoma cells). Cells were grown in six-well plates and incubated with 1 μM of unlabeled DON. Samples were harvested at two different time points (1 and 24 h).
For the main experiment employing native and uniformly 13C labeled DON, HT29 and HepG2 were selected. HT29 cells were cultivated in Dulbecco’s Modified Eagle Medium, and HepG2 cells were cultivated in RPMI 1640 Medium. Both basal media were supplemented with 10% heat inactivated fetal calf serum and 1% (v/v) of a solution containing penicillin G at 60 μg mL–1 and streptomycin at 100 μg mL–1 in the final medium. Cell culture media and supplements were purchased from GIBCO Invitrogen, Lonza Group Ltd., Sigma-Aldrich Chemie GmbH, and Sarstedt AG & Co. For cell cultivation and treatments humidified incubators at 37 °C and 5% CO2 were used. Cells were routinely tested for the absence of mycoplasma contamination and used for experiments at passages 10–14. Cells were seeded in 10 cm cell culture dishes (1.500.000 cells/Petri dish) from Sarstedt AG & Co with TC-treated surface, grown for 72 h, and incubated for 3 h after adding fresh medium containing 10 μM of DON, consisting of 5 μM of 12C-DON and 5 μM 13C-DON. Three individual biological experiments were performed. In addition, two control experiments were executed under identical incubation conditions as during the main experiment. For this purpose, medium without DON was added to the cells (#1) and medium containing 5 μM of 12C-DON and 5 μM 13C-DON was added to Petri dishes without cells (#2) (both in triplicates).
Sample Preparation
Following incubation, 4 mL of the extracellular medium was transferred into reaction tubes and diluted 1:5 with ice-cold ACN/MeOH (1/1; v/v). The tubes were vortexed, incubated for 1 h at −20 °C, and centrifuged for 15 min at 4 °C to precipitate the proteins. A volume of 10 mL supernatant was transferred into 15 mL tubes and evaporated to dryness at 4 °C using a vacuum concentrator. Samples were reconstituted in 0.4 mL of dilution solvent (MeOH/water; 10/90; v/v), centrifuged, transferred to HPLC vials with microinserts, and stored at −80 °C until LC-HRMS analysis.
To evaluate intracellular metabolites, cell extracts were prepared. Cells were rinsed twice with 2.5 mL of prewarmed PBS. Subsequently, 0.7 mL of ice-cold quenching solution (ACN/MeOH/water; 40/40/20; v/v/v) was added. Cells were scraped off and transferred into reaction tubes, before three cycles of freeze–thaw cycles and sonication were performed for cell lysis and metabolite extraction as follows: a) vortex for 30 s, b) liquid nitrogen for 1 min, c) thawing, and d) ultrasonication in ice bath for 10 min. To aid protein precipitation, the samples were incubated at −20 °C for 1 h after extraction, followed by centrifugation at 15.000 rpm and 4 °C. Supernatants were transferred into a new reaction tube and evaporated to dryness. Thereafter, the dried extracts were reconstituted in 240 μL of dilution solvent (MeOH/water; 10/90; v/v), sonicated for 10 min, and centrifuged for 15 min at 15.000 rpm and 4 °C. Samples were transferred to HPLC vials with microinserts and stored at −80 °C until LC-MS/MS analysis.
To verify that conjugation with media components was mediated by the cells and not an artifact or a chemical reaction, a cell-free and a DON-free incubation was repeated exactly as described above. Moreover, a spiking experiment was performed to estimate matrix effects during electrospray ionization. For this purpose, cell lysate samples and extracellular medium from HepG2 and HT29 incubations in which no DON was added to the medium (control experiment #1) were fortified in triplicate with native DON only.
LC-HRMS (/MS) Analysis
The samples were measured using a Vanquish UHPLC system coupled to a QExactive HF quadrupole-Orbitrap mass spectrometer via an ESI interface. A volume of 5 μL was injected onto an Acquity HSS T3, column (1.8 μm, 150 × 2.1 mm) maintained at 40 °C. The flow rate was set to 0.5 mL min–1. A gradient using water with 0.1% acetic acid (v/v) (eluent A) and methanol with 0.1% acetic acid (v/v) (eluent B) was applied. The gradient was optimized for separating highly polar DON conjugate isomers as follows: 0–2 min, 5% B; 2–7 min linear increase to 40% B; 7–9 min, linear increase to 100% B; 9–10.9 min, 100% B; 10.9–11 min, linearly decreased to 5% B; 11–11.9 min, equilibration at 5% B. The measurements were conducted consecutively in positive and negative mode. The settings of the ESI interface were as follows: sheath gas, 53 au; auxiliary gas, 14 au; capillary voltage, 3.5 kV (positive), 3.2 kV (negative); capillary temperature, 350 °C.
Full scan MS and data dependent MS2 spectra (ddMS2) for the ten most abundant ions in each full scan were acquired in a scan range of 60–900 m/z. The resolution (fwhm) was set to 60,000 (@ m/z 200) for full scan and 30,000 (@ m/z 200) for ddMS2 experiments. The automatic gain control (AGC) was set to 1 × 106 with a maximum injection time of 100 ms in full scan, whereas for the acquisition of ddMS2 2 × 105 with a maximum injection time of 60 ms was applied. The isolation window for the ddMS2 scans was 1 m/z. The ten most abundant ions (top 10) were selected for MS2 analysis. A stepped normalized collision energy (NCE) at 20 and 50 eV was applied. A minimum AGC of 1 × 103 and an intensity threshold of 1.7 × 104 were chosen. The apex trigger was set to 2–5 s. Dynamic exclusion was activated for 8 s. For generating MS2 spectra of suspected DON conjugates, the settings were adapted as follows: scan range, m/z 100–900; apex trigger, 3–8 s; dynamic exclusion, 10 s. Furthermore, an inclusion list consisting of all masses of interest was created (Supplementary Table 1). Specific m/z values for which MS2 spectra were already acquired in the initial run and the most abundant peaks in a solvent blank were suspended from MS2 triggering by an exclusion list. The injection volume was increased up to 20 μL in remeasurements in order to record spectra of higher quality for low abundance metabolites. The analysis of the samples obtained within the preliminary experiments was carried out according to Warth et al.30
Data Processing
To find all detectable biotransformation products of DON–known as well as putative novel ones–the MetExtract II software (module TracExtract) was employed.8 Briefly summarized, the software searched the LC-HRMS data for pairs of two chromatographic peaks from biotransformation products of DON that incorporated either the native or the uniformly 13C-labeled DON tracer. Such pairs showed complete chromatographic coelution as well as an m/z difference corresponding to the number of carbon atoms from the 13C-labeled DON. Any metabolite or chromatographic peak in the LC-HRMS data that was only present as a single form (i.e., native, non-13C-labeled form) was ignored by the software as it could be concluded that this peak was not derived from a biotransformation product of DON but rather an endogenous metabolite of the cells or any other unspecific compound (e.g., contamination, extraction artifact) present in the sample. Parameter settings for MetExtract II were as follows: module: TracExtract; intensity threshold (M and M′): 1.104 cps; ratio of native and 13C-labeled tracer form: 0.75–1.25; number of carbon atoms searched for: 10–15 and 30 (for dimer ions); number of isotopologs checked: 2; maximum isotopolog deviation from expected relative abundance: 25%; XIC (ppm): 5; maximum mass deviation (ppm): 5; Pearson correlation threshold (minimum): 0.85. Thermo Xcalibur Qual Browser and TraceFinder (Thermo Scientific) were used for further data evaluation. The graphs of the MS2 spectra were drawn using OriginPro 2018 or directly exported from Xcalibur. The preparation of pictures and graphical illustrations were done with Inkscape. The data processing for Figure 2a is described in the Supporting Information. Metabolite identification levels reporting follow the scheme proposed by Schymanski et al.41 For Figure 2a the raw data files from all samples were transformed to a mzML file with ProteoWizard (version 3). The xcms R-package was used to perform peak picking with the CentWave algorithm using the following setting: peakwidth: 5–40 s; noise: 1000; ppm: 5. The detected features are represented by gray ×’s in the figure.
Acknowledgments
We would like to express our gratitude towards M. Doppler for providing wheat samples for comparison purposes and L. Niederstätter and H. Schoeny for support during the LC-HRMS measurements. The measurements were performed at the Mass Spectrometry Centre (MSC) of the Faculty of Chemistry at the University of Vienna. Furthermore, we would like to thank the Austrian Science Fund (SFB Fusarium #F3702, #F3715, and #F3718) as well as the Provincial Government of Lower Austria (projects NoBiTUM, OMICS 4.0) for financial support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.9b01016.
Abbreviations, additional MS2 spectra and chromatograms of annotated metabolites, inclusion list for MS2 generation, and author contributions (PDF)
Author Contributions
⊥ M.F. and C.B. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Rappaport S. M.; Smith M. T. (2010) Environment and Disease Risks. Science 330, 460–461. 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecki M. M.; Walker D. I.; Vermeulen R.; Chadeau-Hyam M.; Jones D. P.; Miller G. W. (2019) The Exposome: Molecules to Populations. Annu. Rev. Pharmacol. Toxicol. 59, 107–127. 10.1146/annurev-pharmtox-010818-021315. [DOI] [PubMed] [Google Scholar]
- Vermeulen R.; Schymanski E. L.; Barabási A.-L.; Miller G. W. (2020) The exposome and health: Where chemistry meets biology. Science 367, 392–396. 10.1126/science.aay3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrap P.; Zheng G.; Wan Y.; Li T.; Hu W.; Li W.; Zhang H.; Zhang Z.; Hu J. (2017) Discovery of a widespread metabolic pathway within and among phenolic xenobiotics. Proc. Natl. Acad. Sci. U. S. A. 114, 6062–6067. 10.1073/pnas.1700558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croom E. (2012) Chapter Three - Metabolism of Xenobiotics of Human Environments, In Prog. Mol. Biol. Transl. Sci. (Hodgson E., Ed.), pp 31–88, Academic Press, 10.1016/B978-0-12-415813-9.00003-9. [DOI] [PubMed] [Google Scholar]
- Nicholson J. K.; Wilson I. D. (2003) Understanding ‘Global’ Systems Biology: Metabonomics and the Continuum of Metabolism. Nat. Rev. Drug Discovery 2, 668–676. 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- Gertsman I.; Barshop B. A. (2018) Promises and pitfalls of untargeted metabolomics. J. Inherited Metab. Dis. 41, 355–366. 10.1007/s10545-017-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueschl C.; Kluger B.; Neumann N. K. N.; Doppler M.; Maschietto V.; Thallinger G. G.; Meng-Reiterer J.; Krska R.; Schuhmacher R. (2017) MetExtract II: A Software Suite for Stable Isotope-Assisted Untargeted Metabolomics. Anal. Chem. 89, 9518–9526. 10.1021/acs.analchem.7b02518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluger B.; Bueschl C.; Lemmens M.; Berthiller F.; Häubl G.; Jaunecker G.; Adam G.; Krska R.; Schuhmacher R. (2013) Stable isotopic labelling-assisted untargeted metabolic profiling reveals novel conjugates of the mycotoxin deoxynivalenol in wheat. Anal. Bioanal. Chem. 405, 5031–5036. 10.1007/s00216-012-6483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng-Reiterer J.; Varga E.; Nathanail A. V.; Bueschl C.; Rechthaler J.; McCormick S. P.; Michlmayr H.; Malachová A.; Fruhmann P.; Adam G.; Berthiller F.; Lemmens M.; Schuhmacher R. (2015) Tracing the metabolism of HT-2 toxin and T-2 toxin in barley by isotope-assisted untargeted screening and quantitative LC-HRMS analysis. Anal. Bioanal. Chem. 407, 8019–8033. 10.1007/s00216-015-8975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassy A. W.; Bueschl C.; Lee H.; Lerno L.; Oberholster A.; Barile D.; Schuhmacher R.; Waterhouse A. L. (2015) Tracing flavonoid degradation in grapes by MS filtering with stable isotopes. Food Chem. 166, 448–455. 10.1016/j.foodchem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Nathanail A. V.; Varga E.; Meng-Reiterer J.; Bueschl C.; Michlmayr H.; Malachova A.; Fruhmann P.; Jestoi M.; Peltonen K.; Adam G.; Lemmens M.; Schuhmacher R.; Berthiller F. (2015) Metabolism of the Fusarium Mycotoxins T-2 Toxin and HT-2 Toxin in Wheat. J. Agric. Food Chem. 63, 7862–7872. 10.1021/acs.jafc.5b02697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2013) Deoxynivalenol in food and feed: occurrence and exposure. EFSA Journal 11, 3379. [Google Scholar]
- Pestka J. J. (2010) Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 84, 663–679. 10.1007/s00204-010-0579-8. [DOI] [PubMed] [Google Scholar]
- Pestka J. J.; Uzarski R. L.; Islam Z. (2005) Induction of apoptosis and cytokine production in the Jurkat human T cells by deoxynivalenol: role of mitogen-activated protein kinases and comparison to other 8-ketotrichothecenes. Toxicology 206, 207–219. 10.1016/j.tox.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Payros D.; Alassane-Kpembi I.; Pierron A.; Loiseau N.; Pinton P.; Oswald I. P. (2016) Toxicology of deoxynivalenol and its acetylated and modified forms. Arch. Toxicol. 90, 2931–2957. 10.1007/s00204-016-1826-4. [DOI] [PubMed] [Google Scholar]
- Turner P. C.; Rothwell J. A.; White K. L. M.; Gong Y.; Cade J. E.; Wild C. P. (2008) Urinary deoxynivalenol is correlated with cereal intake in individuals from the United kingdom. Environ. Health Perspect. 116, 21–25. 10.1289/ehp.10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C.; Li C.; Zhou S.; Wang X.; Xu H.; Wang D.; Gong Y. Y.; Routledge M. N.; Zhao Y.; Wu Y. (2018) Risk assessment of deoxynivalenol in high-risk area of China by human biomonitoring using an improved high throughput UPLC-MS/MS method. Sci. Rep. 8, 3901. 10.1038/s41598-018-22206-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šarkanj B.; Warth B.; Uhlig S.; Abia W. A.; Sulyok M.; Klapec T.; Krska R.; Banjari I. (2013) Urinary analysis reveals high deoxynivalenol exposure in pregnant women from Croatia. Food Chem. Toxicol. 62, 231–237. 10.1016/j.fct.2013.08.043. [DOI] [PubMed] [Google Scholar]
- Warth B.; Sulyok M.; Fruhmann P.; Berthiller F.; Schuhmacher R.; Hametner C.; Adam G.; Fröhlich J.; Krska R. (2012) Assessment of human deoxynivalenol exposure using an LC-MS/MS based biomarker method. Toxicol. Lett. 211, 85–90. 10.1016/j.toxlet.2012.02.023. [DOI] [PubMed] [Google Scholar]
- Vidal A.; Claeys L.; Mengelers M.; Vanhoorne V.; Vervaet C.; Huybrechts B.; De Saeger S.; De Boevre M. (2018) Humans significantly metabolize and excrete the mycotoxin deoxynivalenol and its modified form deoxynivalenol-3-glucoside within 24 h. Sci. Rep. 8, 5255. 10.1038/s41598-018-23526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth B.; Sulyok M.; Berthiller F.; Schuhmacher R.; Krska R. (2013) New insights into the human metabolism of the Fusarium mycotoxins deoxynivalenol and zearalenone. Toxicol. Lett. 220, 88–94. 10.1016/j.toxlet.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Schwartz-Zimmermann H. E.; Hametner C.; Nagl V.; Slavik V.; Moll W.-D.; Berthiller F. (2014) Deoxynivalenol (DON) sulfonates as major DON metabolites in rats: from identification to biomarker method development, validation and application. Anal. Bioanal. Chem. 406, 7911–7924. 10.1007/s00216-014-8252-3. [DOI] [PubMed] [Google Scholar]
- Schwartz-Zimmermann H. E.; Fruhmann P.; Dänicke S.; Wiesenberger G.; Caha S.; Weber J.; Berthiller F. (2015) Metabolism of Deoxynivalenol and Deepoxy-Deoxynivalenol in Broiler Chickens, Pullets, Roosters and Turkeys. Toxins 7, 4706–4729. 10.3390/toxins7114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz-Zimmermann H. E.; Hametner C.; Nagl V.; Fiby I.; Macheiner L.; Winkler J.; Dänicke S.; Clark E.; Pestka J. J.; Berthiller F. (2017) Glucuronidation of deoxynivalenol (DON) by different animal species: identification of iso-DON glucuronides and iso-deepoxy-DON glucuronides as novel DON metabolites in pigs, rats, mice, and cows. Arch. Toxicol. 91, 3857–3872. 10.1007/s00204-017-2012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka J.; Clark E.; Schwartz-Zimmermann H.; Berthiller F. (2017) Sex Is a Determinant for Deoxynivalenol Metabolism and Elimination in the Mouse. Toxins 9, 240. 10.3390/toxins9080240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeitzl C.; Warth B.; Fruhmann P.; Michlmayr H.; Malachová A.; Berthiller F.; Schuhmacher R.; Krska R.; Adam G. (2015) The Metabolic Fate of Deoxynivalenol and Its Acetylated Derivatives in a Wheat Suspension Culture: Identification and Detection of DON-15-O-Glucoside, 15-Acetyl-DON-3-O-Glucoside and 15-Acetyl-DON-3-Sulfate. Toxins 7, 3112. 10.3390/toxins7083112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluger B.; Bueschl C.; Lemmens M.; Michlmayr H.; Malachova A.; Koutnik A.; Maloku I.; Berthiller F.; Adam G.; Krska R.; Schuhmacher R. (2015) Biotransformation of the Mycotoxin Deoxynivalenol in Fusarium Resistant and Susceptible Near Isogenic Wheat Lines. PLoS One 10, e0119656 10.1371/journal.pone.0119656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth B.; Fruhmann P.; Wiesenberger G.; Kluger B.; Sarkanj B.; Lemmens M.; Hametner C.; Fröhlich J.; Adam G.; Krska R.; Schuhmacher R. (2015) Deoxynivalenol-sulfates: identification and quantification of novel conjugated (masked) mycotoxins in wheat. Anal. Bioanal. Chem. 407, 1033–1039. 10.1007/s00216-014-8340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth B.; Del Favero G.; Wiesenberger G.; Puntscher H.; Woelflingseder L.; Fruhmann P.; Sarkanj B.; Krska R.; Schuhmacher R.; Adam G.; Marko D. (2016) Identification of a novel human deoxynivalenol metabolite enhancing proliferation of intestinal and urinary bladder cells. Sci. Rep. 6, 33854. 10.1038/srep33854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Favero G.; Woelflingseder L.; Braun D.; Puntscher H.; Kütt M.-L.; Dellafiora L.; Warth B.; Pahlke G.; Dall’Asta C.; Adam G.; Marko D. (2018) Response of intestinal HT-29 cells to the trichothecene mycotoxin deoxynivalenol and its sulfated conjugates. Toxicol. Lett. 295, 424–437. 10.1016/j.toxlet.2018.07.007. [DOI] [PubMed] [Google Scholar]
- Wu X.; Murphy P.; Cunnick J.; Hendrich S. (2007) Synthesis and characterization of deoxynivalenol glucuronide: Its comparative immunotoxicity with deoxynivalenol. Food Chem. Toxicol. 45, 1846–1855. 10.1016/j.fct.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Stanic A.; Uhlig S.; Solhaug A.; Rise F.; Wilkins A. L.; Miles C. O. (2015) Nucleophilic Addition of Thiols to Deoxynivalenol. J. Agric. Food Chem. 63, 7556–7566. 10.1021/acs.jafc.5b02864. [DOI] [PubMed] [Google Scholar]
- Stanic A.; Uhlig S.; Sandvik M.; Rise F.; Wilkins A. L.; Miles C. O. (2016) Characterization of Deoxynivalenol-Glutathione Conjugates Using Nuclear Magnetic Resonance Spectroscopy and Liquid Chromatography-High-Resolution Mass Spectrometry. J. Agric. Food Chem. 64, 6903–6910. 10.1021/acs.jafc.6b02853. [DOI] [PubMed] [Google Scholar]
- Uhlig S.; Stanic A.; Hofgaard I.; Kluger B.; Schuhmacher R.; Miles C. (2016) Glutathione-Conjugates of Deoxynivalenol in Naturally Contaminated Grain Are Primarily Linked via the Epoxide Group. Toxins 8, 329. 10.3390/toxins8110329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz H. E.; Hametner C.; Slavik V.; Greitbauer O.; Bichl G.; Kunz-Vekiru E.; Schatzmayr D.; Berthiller F. (2013) Characterization of Three Deoxynivalenol Sulfonates Formed by Reaction of Deoxynivalenol with Sulfur Reagents. J. Agric. Food Chem. 61, 8941–8948. 10.1021/jf403438b. [DOI] [PubMed] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Marcu A.; Guo A. C.; Liang K.; Vázquez-Fresno R.; Sajed T.; Johnson D.; Li C.; Karu N.; Sayeeda Z.; Lo E.; Assempour N.; Berjanskii M.; Singhal S.; Arndt D.; Liang Y.; Badran H.; Grant J.; Serra-Cayuela A.; Liu Y.; Mandal R.; Neveu V.; Pon A.; Knox C.; Wilson M.; Manach C.; Scalbert A. (2018) HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 46, D608–D617. 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijas C.; Montenegro-Burke J. R.; Domingo-Almenara X.; Palermo A.; Warth B.; Hermann G.; Koellensperger G.; Huan T.; Uritboonthai W.; Aisporna A. E.; Wolan D. W.; Spilker M. E.; Benton H. P.; Siuzdak G. (2018) METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 90, 3156–3164. 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai H.; Arita M.; Kanaya S.; Nihei Y.; Ikeda T.; Suwa K.; Ojima Y.; Tanaka K.; Tanaka S.; Aoshima K.; Oda Y.; Kakazu Y.; Kusano M.; Tohge T.; Matsuda F.; Sawada Y.; Hirai M. Y.; Nakanishi H.; Ikeda K.; Akimoto N.; Maoka T.; Takahashi H.; Ara T.; Sakurai N.; Suzuki H.; Shibata D.; Neumann S.; Iida T.; Tanaka K.; Funatsu K.; Matsuura F.; Soga T.; Taguchi R.; Saito K.; Nishioka T. (2010) MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714. 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- Stanic A.; Uhlig S.; Solhaug A.; Rise F.; Wilkins A. L.; Miles C. O. (2016) Preparation and Characterization of Cysteine Adducts of Deoxynivalenol. J. Agric. Food Chem. 64, 4777–4785. 10.1021/acs.jafc.6b01158. [DOI] [PubMed] [Google Scholar]
- Schymanski E. L.; Jeon J.; Gulde R.; Fenner K.; Ruff M.; Singer H. P.; Hollender J. (2014) Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 48, 2097–2098. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- Gerding J.; Cramer B.; Humpf H.-U. (2014) Determination of mycotoxin exposure in Germany using an LC-MS/MS multibiomarker approach. Mol. Nutr. Food Res. 58, 2358–2368. 10.1002/mnfr.201400406. [DOI] [PubMed] [Google Scholar]
- Turner P. C.; Hopton R. P.; White K. L. M.; Fisher J.; Cade J. E.; Wild C. P. (2011) Assessment of deoxynivalenol metabolite profiles in UK adults. Food Chem. Toxicol. 49, 132–135. 10.1016/j.fct.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura H.; Iwaki M. (2011) In vitro and in vivo small intestinal metabolism of CYP3A and UGT substrates in preclinical animals species and humans: species differences. Drug Metab. Rev. 43, 476–498. 10.3109/03602532.2011.597401. [DOI] [PubMed] [Google Scholar]
- Woelflingseder L.; Warth B.; Vierheilig I.; Schwartz-Zimmermann H.; Hametner C.; Nagl V.; Novak B.; Šarkanj B.; Berthiller F.; Adam G.; Marko D. (2019) The Fusarium metabolite culmorin suppresses the in vitro glucuronidation of deoxynivalenol. Arch. Toxicol. 93, 1729–1743. 10.1007/s00204-019-02459-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama Y.; Sasaki Y.; Terasaki N.; Kawataki T.; Takekawa K.; Iwase Y.; Shimizu T.; Sanoh S.; Ohta S. (2018) Comparison of Drug Metabolism and Its Related Hepatotoxic Effects in HepaRG, Cryopreserved Human Hepatocytes, and HepG2 Cell Cultures. Biol. Pharm. Bull. 41, 722–732. 10.1248/bpb.b17-00913. [DOI] [PubMed] [Google Scholar]
- Juan-García A.; Juan C.; König S.; Ruiz M.-J. (2015) Cytotoxic effects and degradation products of three mycotoxins: Alternariol, 3-acetyl-deoxynivalenol and 15-acetyl-deoxynivalenol in liver hepatocellular carcinoma cells. Toxicol. Lett. 235, 8–16. 10.1016/j.toxlet.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Abad M. F.; Di Benedetto G.; Magalhães P. J.; Filippin L.; Pozzan T. (2004) Mitochondrial pH monitored by a new engineered GFP mutant. J. Biol. Chem. 279, 11521–11529. 10.1074/jbc.M306766200. [DOI] [PubMed] [Google Scholar]
- Dansen T. B.; Wirtz K. W. A.; Wanders R. J. A.; Pap E. H. W. (2000) Peroxisomes in human fibroblasts have a basic pH. Nat. Cell Biol. 2, 51–53. 10.1038/71375. [DOI] [PubMed] [Google Scholar]
- Nakamura Y.; Ohta M.; Ueno Y. (1977) Reactivity of 12, 13-Epoxytrichothecenes with Epoxide Hydrolase, Glutathione-S-Transferase and Glutathione. Chem. Pharm. Bull. 25, 3410–3414. 10.1248/cpb.25.3410. [DOI] [Google Scholar]
- Adam G. (2019) Method for biotransformation of trichothecenes, EP19181392.
- Lewis A. D.; Forrester L. M.; Hayes J. D.; Wareing C. J.; Carmichael J.; Harris A. L.; Mooghen M.; Wolf C. R. (1989) Glutathione S-transferase isoenzymes in human tumours and tumour derived cell lines. Br. J. Cancer 60, 327–331. 10.1038/bjc.1989.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. C. (2013) Glutathione synthesis. Biochim. Biophys. Acta, Gen. Subj. 1830, 3143–3153. 10.1016/j.bbagen.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. C. (2009) Regulation of glutathione synthesis. Mol. Aspects Med. 30, 42–59. 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan D.; Huang L.; Pan Y.; Wu Q.; Chen D.; Tao Y.; Wang X.; Liu Z.; Li J.; Wang L.; Yuan Z. (2014) Metabolism, Distribution, and Excretion of Deoxynivalenol with Combined Techniques of Radiotracing, High-Performance Liquid Chromatography Ion Trap Time-of-Flight Mass Spectrometry, and Online Radiometric Detection. J. Agric. Food Chem. 62, 288–296. 10.1021/jf4047946. [DOI] [PubMed] [Google Scholar]
- Rudyk O.; Eaton P. (2014) Biochemical methods for monitoring protein thiol redox states in biological systems. Redox Biol. 2, 803–813. 10.1016/j.redox.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbt M.; Monod M.; Yamada T.; Hertweck C.; Kunert J.; Staib P. (2013) Keratin Degradation by Dermatophytes Relies on Cysteine Dioxygenase and a Sulfite Efflux Pump. J. Invest. Dermatol. 133, 1550–1555. 10.1038/jid.2013.41. [DOI] [PubMed] [Google Scholar]
- Knights K. M.; Sykes M. J.; Miners J. O. (2007) Amino acid conjugation: contribution to the metabolism and toxicity of xenobiotic carboxylic acids. Expert Opin. Drug Metab. Toxicol. 3, 159–168. 10.1517/17425255.3.2.159. [DOI] [PubMed] [Google Scholar]
- Karlovsky P. (2011) Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives. Appl. Microbiol. Biotechnol. 91, 491–504. 10.1007/s00253-011-3401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Van Lanen S. G.; Shen B. (2009) A free-standing condensation enzyme catalyzing ester bond formation in C-1027 biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 106, 4183–4188. 10.1073/pnas.0808880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C.; Milcent T.; Crousse B.; Bonnet-Delpon D. (2009) Non Lewis acid catalysed epoxide ring opening with amino acid esters. Org. Biomol. Chem. 7, 2026–2028. 10.1039/b902081k. [DOI] [PubMed] [Google Scholar]
- Calow A. D. J.; Carbó J. J.; Cid J.; Fernández E.; Whiting A. (2014) Understanding α,β-Unsaturated Imine Formation from Amine Additions to α,β-Unsaturated Aldehydes and Ketones: An Analytical and Theoretical Investigation. J. Org. Chem. 79, 5163–5172. 10.1021/jo5007366. [DOI] [PubMed] [Google Scholar]
- Schwartz-Zimmermann H. E.; Wiesenberger G.; Unbekannt C.; Hessenberger S.; Schatzmayr G.; Berthiller F. (2014) Reaction of (conjugated) deoxynivalenol with sulphur reagents - novel metabolites, toxicity and application. World Mycotoxin J. 7, 187–197. 10.3920/WMJ2013.1632. [DOI] [Google Scholar]
- Fruhmann P.; Skrinjar P.; Weber J.; Mikula H.; Warth B.; Sulyok M.; Krska R.; Adam G.; Rosenberg E.; Hametner C.; Fröhlich J. (2014) Sulfation of deoxynivalenol, its acetylated derivatives, and T2-toxin. Tetrahedron 70, 5260–5266. 10.1016/j.tet.2014.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruhmann P.; Warth B.; Hametner C.; Berthiller F.; Horkel E.; Adam G.; Sulyok M.; Krska R.; Fröhlich J. (2012) Synthesis of deoxynivalenol-3-ß-D-O-glucuronide for its use as biomarker for dietary deoxynivalenol exposure. World Mycotoxin J. 5, 127–132. 10.3920/WMJ2011.1366. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.