Abstract
An automated, high-capacity, and high-throughput procedure for the rapid isolation and identification of biologically active natural products from a prefractionated library is presented. The semipreparative HPLC method uses 1 mg of the primary hit fraction and produces 22 subfractions in an assay-ready format. Following screening, all active fractions are analyzed by NMR, LCMS, and FTIR, and the active principle structural classes are elucidated. In the proof-of-concept study, we show the processes involved in generating the subfractions, the throughput of the structural elucidation work, as well as the ability to rapidly isolate and identify new and biologically active natural products. Overall, the rapid second-stage purification conserves extract mass, requires much less chemist time, and introduces knowledge of structure early in the isolation workflow.
Nature is an important resource for the discovery of new drugs and drug leads, with over half of all anticancer drugs based on chemical scaffolds isolated from plants, marine invertebrates, and microbes.1 However, despite their success as clinically used agents, compatibility issues that make natural product extracts challenging have reduced enthusiasm for high-throughput screening (HTS) of crude natural product libraries in targeted assay systems.2 In addition, after initial testing has been completed and activity confirmed, the process of compound isolation and identification is often slow and can increase the expense of screening support as (1) natural product extracts represent a mixture of compounds which can number up to hundreds of individual molecules and (2) bioassay-guided fractionation processes often include several iterations of fractionation and secondary screening. To improve the speed of hit identification, many natural product research laboratories are working with prefractionated natural product libraries.3 The use of prefractionated libraries reduces the complexity of individual test samples, simplifying the isolation and active principle identification effort and reducing screening support requirements. In addition, prefractionation can sequester nuisance compounds such as salts, tannins, and fatty acids and thus decrease the probability of false positive screening hits as well as improve the solubility of samples for liquid handling purposes.
The National Cancer Institute (NCI) Program for Natural Products Discovery (NPNPD) is a newly launched program for the NCI. The goals of the new initiative are to generate prefractionated samples (up to 1 000 000) for modern high-throughput screening technologies and to develop integrated analytical resources for the rapid isolation and structure elucidation of biologically active natural products. Recently, we published the development of chromatographic and automation procedures used in the generation of the NPNPD library of natural product fractions.4 The methodology is based on separation of the crude extract into seven fractions using solid-phase extraction (SPE) on C8 media and water/methanol step gradients. Proof-of-concept studies assessed the fraction library for a range of properties such as recovery and distribution of material and biological activity in cell-based and cell-free assays. In brief, we found good separation of mass across all seven fractions, enhanced activity of fractions compared to the crude extract, sequestration of common nuisance compounds, and demonstrated efficient identification of minor biologically active natural products. An ultraperformance liquid chromatography coupled to high-resolution mass spectrometry (UPLC–HRMS) metabolomic analysis of the pilot library with a minimum total ion current height (baseline) setting of 1 × 104 showed that each fraction can contain an average of 20 compounds. Thus, while the prefractionation process generates a simpler, less complicated natural product sample, each fraction is still a mixture of secondary metabolites and requires additional chromatography steps toward the identification of pure active compounds. Herein, we present a robust, rapid, and reproducible high-throughput HPLC-based second-stage method for further separation of the NPNPD fraction library and structural elucidation of the active principles.
The HPLC-based method chromatographically separates 1 mg of a primary SPE fraction on a C18 reversed-phase column into 22 subfractions, collected in 96 deep-well plates (Figure 1). This format can accommodate four sample injections to be collected in one plate with wells 1A–1D used as injection sites. The rest of column one is left empty for assay controls with fraction collection starting in well A2. Total run time, including gradient recovery, is 15 min per injection, and depending on the size of the collection bed used, the method enables high-throughput secondary separation workflow. For example, the large plate-based fraction collector used in this work is capable of processing 48 samples in a single run to generate 1056 subfractions in an assay-ready format in approximately 13.5 hours (Supporting Information (SI) Figure S1). In an HTS context, this workflow is able to process 500 primary hits and provide in excess of 10 000 wells containing dried-down pure and semipurified compounds back to the screening lab in a two-week window at a quantity that yields a sufficient amount of subfractions for multiple secondary assays.
Figure 1.
Summary of second-stage chromatography method used for the rapid, high-throughput isolation of natural products. Example depicts a representative HPLC run with fraction collection events and duty cycle analysis of a typical injection cycle shown. The gradient, represented as percentage acetonitrile, is shown in pink as well as UV absorbance traces at 254 nm (in blue), 280 nm (in green), and 320 nm (in orange).
Following HTS, for the purpose of active principle structural elucidation, second-stage HPLC chromatography is repeated, and the active wells are analyzed by 1H NMR, LCMS, and FTIR. To have confidence in the reproducibility of the system, during both the original screening and the repeat structural identification runs, chromatography standards covering a cLogP range of −1 to 4 are used every 9th injection. SI Figure S2 shows consistency of the separation of the standards on three identical HPLC systems in our laboratory as well as over time on a single column with over 250 injections. Precision was one of the crucial aspects of methods development in this work, as repeat HPLC separations used for structural elucidation may take place weeks or months after the original screening run and it was imperative to have the confidence in the reproducibility of the chromatography. The spectroscopic and spectrometric analytical data gathered on the active subfractions are then used to search in-house and commercially available databases of natural products to identify the structures responsible for the observed activity. This process is generally referred to as “dereplication” in the natural products community. While 1H NMR,5,6 2D NMR,7 MS,5 and MS/MS8 fingerprints are well-established and widely used techniques for the dereplication of natural products, we found FTIR to be equally valuable in the dereplication workflow. Modern IR instruments have a relatively small footprint, are sensitive (requiring only micrograms of sample), and provide a large data set across the spectroscopic range suitable for library generation and pattern matching. Overall, the active principle dereplication step combining different analytical techniques enables rapid identification of known compounds and introduces knowledge of active principle structural class early in the isolation workflow, notably in only two automated steps from the crude extract and consuming 1 mg of fraction material derived from the initial 250 mg of extract.
Results and Discussion
Throughput
To demonstrate proof-of-concept, 32 extracts that displayed activity against the NCI-60 human tumor cell line panel (NCI-60) were chosen from the NCI’s Natural Products Repository and prefractionated producing a total of 224 SPE fractions which were then tested in the NCI-60 one-dose assay at a concentration of 10 μg/mL. In total, 34 SPE fractions were significantly active defined as reaching the LC50 level against at least three different cell lines and were prioritized for second-stage HPLC work (Figure 2A). The active fractions were then chromatographed on a semipreparative HPLC column and 22 subfractions were collected from each run giving a total of 748 wells generated in approximately 10 h of instrument time. The mass in each well after second-stage HPLC was unknown, and the subfractions were tested at a nominal concentration of 1.1 μg/mL (see Materials and Methods for specific details on this calculation). Notably, 1 mg injection of the primary SPE hit fraction generated sufficient material to do follow-up one-dose testing on the NCI-60 panel, which is essentially 60 separate assays. The results showed that 75 subfractions reached the LC50 level against at least 3 different cell lines (Figure 2B). Four of the 34 fractions tested (11%) lost activity, which might be due to low testing concentration or decomposition of active principles during HPLC.
Figure 2.
HTS workflow summary. (A) Prefractionation and NCI-60 activity results. Rows in the table represent the 32 active extracts that were prefractionated, and columns represent 7 fractions. Green rectangles represent active fractions that had at least 3 cell lines reach LC50 at 10 μg/mL. (B) Second stage HPLC results. Rows in the table represent the 34 active fractions that were taken through the second-stage HPLC, and columns represent 22 subfractions. Green rectangles represent active subfractions that had at least 3 cell lines reach LC50.
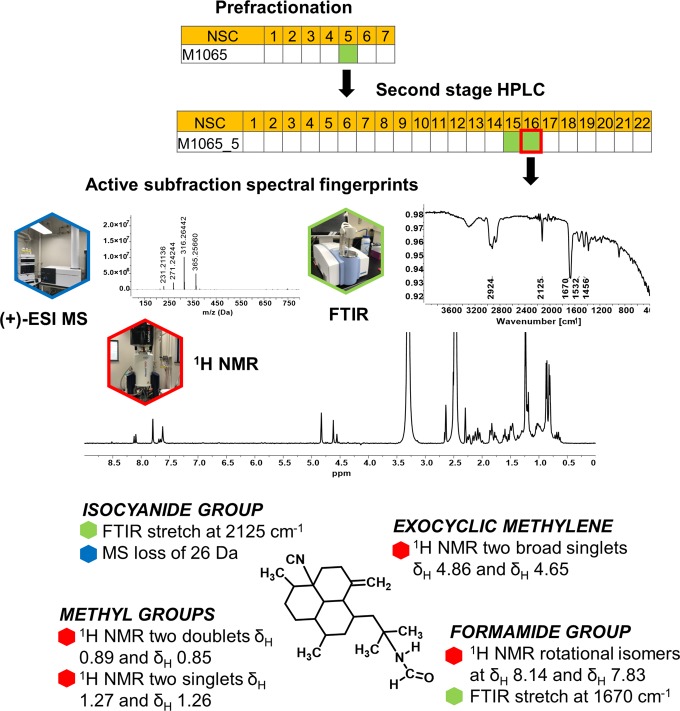
For active principle dereplication work, the entire HPLC run was repeated, and the 75 NCI-60 active subfractions were analyzed by 1H NMR (as well as 1H–1H COSY and 1H–13C HSQC if sufficient material was present), LCMS, and FTIR. Figure 3 shows the analytical spectra for the active subfraction sourced from the sponge Auletta sp. (NSC #C1065). Our prefractionation nomenclature designates the prefix M for prefractionated extracts sourced from marine organisms, followed by the numerical designation for the crude extract, the fraction, and the subfraction. Here, the active subfraction M1065_5_16, was sourced from the crude extract C1065, fraction 5, subfraction 16. The FTIR spectrum for M1065_5_16 showed a distinctive sharp band at 2125 cm–1 indicative of an isocyanide group and a broad carbonyl stretch at 1670 cm–1. The mass spectrometry data showed a sodium adduct ion [M + Na]+ at m/z 365.2566 as well as loss of HCN group at m/z 316.2644 and an additional loss of NH2CHO at m/z 271.2424. The 1H NMR spectrum showed two methyl doublets δH 0.89 (d, 3H, J = 6.3 Hz) and δH 0.85 (d, 3H, J = 6.1 Hz), two methyl singlets δH 1.27 (s, 3H) and δH 1.26 (s, 3H), and two exocyclic methylene broad singlets at δH 4.86 and δH 4.65 indicative of an amphilectene-type diterpene reported from haplosclerid sponges.9 In addition, the 1H NMR spectrum exhibited two downfield-shifted doublets in a 3:7 ratio δH 8.14 (0.3 H, d, J = 12.1 Hz) and δH 7.83 (0.7 H, d, J = 2.1 Hz) consistent with rotational isomers of a formamide group. Figure 3 depicts crucial spectroscopic and spectrometric features used to dereplicate the structure of 8-isocyano-15-formamido-11(20)-amphilectene (1)10 to be responsible for the activity of subfraction M1065_5_16. Using this approach on all of the hits, we found that most subfractions contained pure or semipurified compounds in quantities that enabled collection of high-quality analytical data and subsequent rapid dereplication of known compounds. In total, we dereplicated the structures of 28 known biologically active natural products as well as identified subfractions that contained new natural products (SI Figure S3). By standardizing the workflow and focusing on the active subfractions only, the approach presents a significant reduction in total instrument as well as dereplication time, resulting in a substantial decrease in hit identification timelines and introducing knowledge of structure early in the isolation workflow.
Figure 3.
Summary of screening results and spectral fingerprints of active subfraction for Auletta sp. C1065 extract. Crucial structural features on the 8-isocyano-15-formamido-11(20)-amphilectene molecule are highlighted with spectral features used to assign the structure.
Second Stage HPLC Scale-up Exemplified by an Amino Alkanol-Producing Haliclona chrysa Extract
The crude extract from the sponge Haliclona chrysa (NSC #C17747) showed moderate NCI-60 activity with a GI50 value of 13.6 μg/mL and selectivity for melanoma and colon cell line panels. A single SPE fraction 4 showed significant NCI-60 activity at 10 μg/mL and following second-stage HPLC, NCI-60 activity was concentrated in subfraction M17747_4_7, which contained a single major metabolite. Interpretation of the analytical data from the small-scale 1 mg HPLC injection depicted in SI Figure S4 revealed the active principle to be a long chain alkene-type compound with hydroxy and amino substituents and a mass that did not match any published structure. To obtain enough material for full structural elucidation as well as NCI-60 five-dose testing, SPE fraction 4 was subjected to a scale-up HPLC purification. Our scale-up procedure is designed to have 5 mg of the fraction material repeatedly injected onto the HPLC column using the same gradient and collection conditions as the original 1 mg small-scale run and instead of 96 deep-well plates, repeat injections are collected in 16 mL tubes. The 10 mm diameter semipreparative columns used for both the small-scale and scale-up work showed excellent reproducibility between the 1 and 5 mg injections without the need for method scaling. By collecting successive injections into the same tube, the large bed capacity liquid handlers shown in Figure S1C are capable of processing hundreds of milligrams of material in a single run. Thus, the automated, high-capacity, and high-throughput extract processing procedure yields pure or semipurified natural products in only two automated chromatographic steps, requiring much less chemist time and conserving extract mass. Figure 4 shows a comparison of the active subfraction M17747_4_7 sourced from small-scale (1 mg) and scale-up (5 mg) HPLC purifications by 1H NMR demonstrating repeatability and consistency of the method. Here, we isolated a pure natural product (>95%) with sufficient material for structural elucidation and biological activity assessment in two chromatographic steps.
Figure 4.
NCI-60 screening and scale-up summary for Haliclona chrysa extract. Fractions and subfractions that displayed significant NCI-60 activity are depicted in green. The 1H NMR spectra were acquired on a 600 MHz NMR spectrometer in DMSO-d6.
Subfraction M17747_4_7 yielded a pure new natural product, halaminol E (2), which was isolated as a weakly optically active amorphous solid with a molecular formula C16H29NO established by HRESIMS and 13C NMR measurements. The FTIR spectrum showed two distinctive sharp bands at 2925 and 2853 cm–1 indicative of a primary amine group and a broad hydroxyl group stretch at 3000 cm–1. The mass spectrometry data showed a protonated molecule [M + H]+ at m/z 252.23269 as well as loss of H2O from the precursor ion at m/z 234.2218. The 1H and 13C NMR spectra in DMSO-d6 showed resonances associated with a terminal conjugated diene, one additional olefin, an oxymethine, an aminomethine, two well-resolved methylenes, a methylene envelope, and a terminal methyl doublet resonance. The 2-amino-3-hydroxy terminus was established via 1H–1H COSY and 1H–13C HMBC correlations from the terminal methyl at C-1 (δH 1.04, d, J = 6.6 Hz, δC 15.9) to the amino group at C-2 (δH 2.86, q, J = 6.7 Hz, δC 51.2), hydroxy group at C-3 (δH 3.77, t, J = 7.5 Hz, δC 73.6), a trans olefin at C-4 and −5 (δH 5.64, dtd, J = 15.4, 6.8, 0.9 Hz, δC 133.0, C-4; and δH 5.38, ddt, J = 15.4, 7.2, 1.4 Hz, δC 130.0, C-5), and an allylic methylene resonance at C-6 (δH 1.99, m, δC 31.6). A contiguous sequence of resonances from C-7 to C-12 (δH 1.34–1.25, δC 28.7–28.5) suggested the presence of a central methylene envelope. A conjugated diene terminus was established via 1H–1H COSY and 1H–13C HMBC correlations from the terminal olefinic methylene at C-16 (δHa 5.09, ddt, J = 17.0, 2.0, 0.7 Hz, δHb 4.95, ddt, J = 10.1, 2.1, 0.7 Hz, δC 115.1), which continued onto a series of three sp2 hybridized methine resonances at C-15 (δH 6.29, dtd, J = 17.0, 10.1, 0.7 Hz, δC 137.2), C-14 (δH 6.04, m, δC 130.9), and C-13 (δH 5.72, dtd, J = 15.1, 7.0, 0.8 Hz, δC 135.3). The olefin at C-13 showed correlations to a methylene at C-12 (δH 2.05, m δC 31.9) and the spin system continued onto the methylene envelope to complete the planar structure of the new natural product 2. Geometry of the Δ4 and Δ13 double bonds was established as E based on the magnitude of the vicinal coupling constants (J4,5 = 15.4 Hz, J13,14 = 15.1 Hz) and the downfield chemical shift of the allylic methylenes at C-6 (δC 31.6) and C-12 (δC 31.9). Based on structural similarities to other 2-amino-3-alkanols isolated from Haliclona sp. sponges,11 the new natural product 2 was given the trivial name halaminol E.
Recently, it was proposed that the absolute configuration of 2-amino-3-alkanols can be determined upon comparison of the specific rotation and 13C NMR shifts of their N,O-diacetyl derivatives with the sign of the rotation being indicative of the absolute configuration at C-2 and the chemical shift for the C-1 methyl being indicative of the relative configuration of the threo and erythro diastereomers.12 The N,O-diacetyl halaminol E exhibited dextrorotatory specific rotation ([α]D +15, c 0.2, CHCl3;[α]D +10, c 0.2, MeOH) consistent with the (2R) configuration, while the 13C NMR chemical shift in chloroform-d of the C-1 methyl group (δC 17.9) was consistent with a threo configuration, suggesting the absolute configuration of 2 to be (2R,3R). However, the review exemplified only one (2R,3R) analogue13,14 with no 13C NMR shifts of its N,O-diacetyl derivative given for comparison, which prompted us to further examine the absolute configuration of halaminol E by semisynthetic methods. Reaction of 2 with 1,1′-carbonyldiimidazole afforded the oxazolidinone 3 in a good yield. The coupling constant between H-2 and H-3 of 7.4 Hz was consistent with a trans Δ4 oxazolidinone isomer,15 as was a selective 1D NOE irradiation of H-3 that showed an enhancement of the H-1 methyl resonance indicative of the threo configuration of the natural product. Ultimately, the configurational assignment was confirmed via exciton coupled circular dichroism (CD) spectrum of the bis-N-O-di(2-napthoyl) derivative 4. Compound 4 was obtained by hydrogenation of 2 (H2, Pd–C, MeOH), followed by a reaction with 2-napthoic acid16 (EDC HCl, 2-napthoic acid, DMAP, 5 equiv each, CH2Cl2, rt). The CD spectrum of 4 (MeCN) showed two strong Cotton Effects [λ 226 (Δε −62.4) and λ 243 (Δε −66.5)] consistent with threo-(2R,3R) configuration.17 Notably, semisynthetic studies confirmed the absolute configuration proposed by the specific rotation and 13C NMR shifts comparison model12 and halaminol E (2), is a rare naturally occurring (2R,3R) analogue in the 2-amino-3-alkanol series.
In NCI-60 activity testing, compound 2 exhibited low micromolar activity with a GI50 value of 6.76 μM (Table 1 and SI Figure S10), and selectivity for melanoma and colon cell line panels comparable to what was observed for the crude extract. Halaminol E bears structural resemblance to spisulosine (ES-285),18 a fully saturated C18 (2S,3R) 2-amino-3-alkanol analogue. Spisulosine showed promising early stage in vivo activity in animal models but had reported toxicity and limited tumor efficacy in several Phase I trials,19,20 with no further information on further clinical development available in the literature on this class of compound.
Table 1. NCI-60 Activity of the Active Principles of C17747 Haliclona chrysa Extract (2) and C15373 Hexabranchus sanguineus Extract (5–9)a.
| compound | GI50 (μM) | yield of crude (%) | yield of subfraction (%) |
|---|---|---|---|
| 2 | 6.76 | 3.7 | 100 |
| 5 | 6.76 | 0.12 | 23.33 |
| 6 | 4.37 | 0.21 | 19.35 |
| 7 | 0.40 | 0.09 | 17.78 |
| 8 | 0.18 | 0.15 | 12.90 |
| 9 | 0.03 | 0.017 | 1.67 |
GI50 is the concentration of the drug that causes 50% growth inhibition and values represent the average calculated across all 60 cell lines. Reported yields were calculated based on the recovery of the isolated compound from the two-step scale-up procedure.
Ability to Detect and Isolate Minor Biologically Active Natural Products Exemplified by the Nudibranch Hexabranchus sanguineus Extract
The extract of the nudibranch Hexabranchus sanguineus (NSC #C15373) showed potent NCI-60 activity with a GI50 value of 1.59 μg/mL. Of the seven SPE fractions generated, fraction M15373_6 was most active, and HPLC subfractions M15373_6_15, _16, and _20 were selected for active principle identification (Figure 5). In contrast to the two previous examples where the active subfractions contained pure compounds in a high yield, here they were comprised of a mixture of minor metabolites. To isolate the active principles, a scale-up study was conducted using 0.9 g of the crude extract. Following prefractionation, the active SPE fraction 6 was processed using the automated second-stage HPLC scale-up procedure (95 mg in 19 × 5 mg injections requiring 6 h of instrument time). For a comparison of small-scale and scale-up subfraction liquid chromatography UV–vis spectra, see SI Figure S17. The active fraction spectral fingerprints recorded for the small-scale (1 mg) injection were used to guide the isolation of pure compounds from subfraction M15373_6_15, which led to the identification of two natural products that were present in this subfraction, namely (−)-coelodiol (5) and 33-methyltetrahydrohalichondramide (7). A comparison of the active subfraction 1H NMR fingerprints shown in Figure 5 demonstrates the efficacy of the dereplication approach. Except for the resonances at δH 4.0–4.2 and 1.35–1.50, all other signals in the active subfraction spectrum could be assigned to the two pure compounds identified.
Figure 5.
NCI-60 screening and active principle identification summary for subfraction H. sanguineus extract. The active subfraction M15373_6_15 spectrum is compared to that of the two compounds isolated from this subfraction, namely (−)-coelodiol (5) and 33-methyltetrahydrohalichondramide (7). Resonances belonging to each of the two pure compounds 5 in red and 7 in blue are marked on the spectra of the subfraction mixture.
In total, H. sanguineus active subfractions yielded five compounds (5–9) with their activities, and isolated yields are presented in Table 1. The most active compound swinholide A (9) with an IC50 value of 30 nM was only present in 0.017% yield of the crude extract and 1.67% yield of the subfraction. Notably, while the 1H NMR fingerprint was not sensitive enough to detect this compound in the subfraction, the LCMS fingerprint did (SI Figure S18) and was used as a guiding principle for the scale-up isolation. The two-step automated procedure was able to concentrate the effective amount of compound close to 100 times in the subfraction compared to what was available in the crude extract with further purification requiring only one additional step of chromatography. In addition to the concentration of very potent minor natural products, we show ability to detect and isolate moderately active compounds that might have been undetected when screening crude extracts alone, or their activity masked by other more potent active principles. (−)-Coelodiol (5) and mycgranol (6) are orders of magnitude less potent than 33-methyltetrahydrohalichondramide (7), 33-methyldihydrohalichondramide (8), and swinholide A (9) and might have been missed if not for the drastic increase of the activity per unit mass going from the crude extract to the fraction shown in Table 1.
Conclusion
The NPNPD automated, high-capacity, high-throughput HPLC-based method for purification of natural product fraction mixtures is presented. The HPLC platform is capable of processing 48 samples in 14 h to generate 1056 subfractions in an assay-ready format. The resulting subfractions are pure or semipurified compounds, and for most the active principle structures could be elucidated using only 1 mg of the primary fraction. For prioritized hits of interest, we show an effective, automated, two-step scale-up procedure that is capable of processing hundreds of milligrams of material. A direct comparison of the new NPNPD methodology to more traditional natural products chemistry methods, used previously in the NCI Natural Products Branch, provided the following comparison of the isolation of an active agent from a marine organic extract. Using traditional methods to produce ∼50 mg of active compound took 4 purification steps and a total of 2–3 weeks. Using the NPNPD methods described here, purification of 45 mg to a similar level of purity (∼97%) required only 2 days. Overall, the rapid and repeatable second-stage purification conserves extract mass, requires much less chemist time, and introduces knowledge of structure early in the isolation workflow. As such, we believe this approach has the potential to significantly reduce natural-product-based active principle identification timelines, cost of screening, and enable faster outcomes for evaluating and identifying natural products in HTS.
Materials and Methods
Extraction
All marine biota used in this work was extracted according to the procedure outlined in McCloud.21 Briefly, frozen specimens were centrifuged in approximately 3 L of deionized water to generate the aqueous extract after which the solid biota material was freeze-dried and soaked in a mixture of methanol and dichloromethane (1:1) overnight to generate the organic extract. All work presented here was conducted on the organic solvent extracts.
Prefractionation
Automated prefractionation was carried out with a customized positive pressure solid phase extraction (ppSPE) workstation (Tecan Freedom Evo) with two robotic arms working together to process 88 extracts in parallel. A portion of the organic solvent extracts (250 mg) was weighed into barcoded tubes and dissolved in 4.5 mL of MeOH/EtOAc/MTBE (6:3:1). Dissolved samples were adsorbed onto cotton rolls (1.27 cm × 3.81 cm) contained within an empty SPE cartridge, followed by freeze-drying to remove solvents. Prefractionation was done on 2 g SPE cartridges (Thermo C8 SPE octyl, nonend-capped: 50 μm, 60 Å). Prior to prefractionation, each SPE cartridge was equilibrated with three column volumes of MeOH/H2O (5:95). Seven fractions were collected using the following elution: fraction 1 MeOH/H2O (5:95), fraction 2 MeOH/H2O (1:4), fraction 3 MeOH/H2O (2:3), fraction 4 MeOH/H2O (3:2), fraction 5 MeOH/H2O (4:1), fraction 6 MeOH, and fraction 7 MeCN/MeOH (1:1). A controlled rate of elution (<10 mL/min) was maintained, and the elution solvents (8 mL) were collected in preweighed 10 mL polypropylene FluidX 2D-barcoded tubes containing a linear barcoded protective jacket and dried at 20–35 °C using Genevac HT-12 high-performance centrifugal evaporation systems. The mass of each natural product fraction was determined using a BioMicroLab XL200 automated weighing station.
Second-Stage Semipreparative HPLC
Semipreparative HPLC was done on a Gilson HPLC purification system equipped with a GX-281 liquid handler, a 322-binary pump, and a 172-photodiode array detector. A mass of 1 mg of fraction was injected in 100 μL of DMSO. HPLC separations were performed on a Phenomenex Onyx Monolithic C18 [100 × 10 mm] column at a flow rate of 3.8 mL/min with the following conditions: an initial isocratic hold at 70% H2O (0.1% formic acid)/30% CH3CN (0.1% formic acid) from 0 to 1.5 min, followed by a linear gradient to CH3CN (0.1% formic acid) over 7.5 min, an isocratic hold at CH3CN (0.1% formic acid) for 3.5 min, followed by a linear gradient back to 70% H2O (0.1% formic acid)/30% CH3CN (0.1% formic acid) from 12.5 to 13.5 min and a final isocratic hold for 1.5 min. For each HPLC run, 22 fractions were collected in 30 s increments between 1.5 and 12.5 min in 96-deep well plates. A standard chromatography mix (30 μL) consisting of 2 mg mL–1 solution of uracil, acetanilide, acetophenone, propiophenone, valerophenone, and hexanophenone in DMSO was injected every 9th chromatography run.
Sample Preparation for NCI-60 Testing of Subfractions
Because the weights of each subfraction were unknown, a nominal amount of 45 μg per well was assumed (1 mg divided over 22 subfractions). A volume of 100 μL of DMSO was added to each well to give a nominal concentration of 450 μg/mL. For the NCI-60 one-dose testing, 75 μL of the solubilized subfraction was used and diluted 1:400 to give the final nominal testing concentration of 1.1 μg/mL.
Analytical Fingerprints from 1 mg Small-Scale HPLC Injection
NMR spectra were recorded at 25 °C on a 600 MHz Bruker Avance III spectrometer equipped with a Prodigy 5 mm cryo-probe. The subfraction samples were dissolved in 0.2 mL of DMSO-d6 and proton spectra were acquired in 3 mm NMR tubes over 128 scans using the standard zg30 Bruker pulse sequence. The 1H chemical shifts were referenced to the solvent peaks for dmso-d6 at δH 2.50 δC 39.50, and chloroform-d1 at δH 7.25 and δC 77.16. NMR FID processing and data interpretation was done using MestReNova software, version 11.0. For HRLCMS analysis subfraction samples were prepared in 200 μL of methanol and a 5 μL injection was done on a Phenomenex Kinetex C18 HPLC column [5 μm, 50 × 2.1 mm] at a flow rate of 1 mL/min and the following conditions: an initial isocratic hold at 95% H2O (0.1% formic acid)/5% CH3CN (0.1% formic acid) for 2 min, followed by a linear gradient to CH3CN (0.1% formic acid) over 20 min, and a final isocratic hold at CH3CN (0.1% formic acid) for 2 min. LCMS processing and data interpretation was done with MestReNova software, version 11.0. IR spectra were recorded as a thin dry film over 128 scans using a Bruker Alpha FTIR spectrometer. IR data processing and interpretation was done using OPUS 8.1 software package.
Haliclona chrysa: Collection, Extraction, and Isolation
The sponge H. chrysa was collected by SCUBA at a depth of 7 m in Tonga in November 1997 under contract with the Coral Reef Research Foundation for the National Cancer Institute. The specimen was taxonomically identified by P. L. Collin of the Coral Reef Research Foundation, and a voucher specimen (0CDN5438) was deposited at the Smithsonian Institution. The sponge (wet weight 111.45 g) was extracted in water, followed by a MeOH/DCM overnight soak according to the National Cancer Institute’s standard marine extraction procedure detailed in McCloud21 to give the organic solvent extract C17747 (6.64 g). A portion of the organic extract (250 mg) was prefractionated on a C8SPE (2 g) and fractions were collected using the standard prefractionation MeOH/H2O seven fraction elution scheme to yield fractions 1 m = 90.2 mg; 2 m = 6.4 mg; 3 m = 1.8 mg; 4 m = 46.8 mg; 5 m = 32.5 mg; 6 m = 19.4 mg; and 7 m = 4.3 mg. A portion of fraction 4 (m = 40.0 mg) was purified on a Phenomenex Onyx Monolithic C18 [100 × 10 mm] column using the standard second-stage semipreparative gradient conditions. For each of the eight HPLC runs, 5 mg was injected, and 22 fractions were combined from each of the 8 runs. Subfraction 7 yielded 8.1 mg of halaminol E (2) (3.7% crude extract yield).
Halaminol E (2)
Clear oil; [α]D = −7, [α]365 = −1.6, [α]405 = 0, [α]436 = −4, [α]546 = −4, [α]633 = −2.4, (c 0.5, MeOH); UV (MeOH) λmax (log ε) 227 (3.51) nm; IR (film) 3000, 2925, 2853, 1582, 1346, 1002, 969, 896 cm–1; 1H NMR (600 MHz, dmso-d6) data: δH 8.40 (1H, br s, OH-2), 6.29 (1H, dtd, J = 17.0, 10.1, 0.7 Hz, H-15), 6.04 (1H, m, H-14), 5.72 (1H, dtd, J = 15.1, 7.0, 0.8 Hz, H-13), 5.64 (1H, dtd, J = 15.4, 6.8, 0.9 Hz, H-5), 5.38 (1H, ddt, J = 15.4, 7.2, 1.4 Hz, H-4), 5.09 (1H, ddt, J = 17.0, 2.0, 0.7 Hz, H-16a), 4.95 (1H, ddt, J = 10.1, 2.1, 0.7 Hz, H-16b), 3.77 (1H, t, J = 7.5 Hz, H-3), 2.86 (1H, q, J = 6.7 Hz, H-2), 2.05 (2H, m, H-12), 1.99 (1H, m, H-6), 1.34 (4H, m, H-7/-11), 1.25 (6H, m, H-8/-9/-10), 1.04 (3H, d, J = 6.6 Hz, H-1); 13C NMR (151 MHz, dmso-d6) data: δC 137.2 (C-15), 135.3 (C-13), 133.0 (C-5), 130.9 (C-14), 130.0 (C-4), 115.1 (C-16), 73.6 (C-3), 51.2 (C-2), 31.9 (C-12), 31.6 (C-6), 28.7–28.5 (C-7/-8/-9/-10/-11), 15.9 (C-1); HRESIMS m/z [M + H]+ 252.23269 (calcd for C16H30NO+, 252.23219).
N-O-Diacetyl Halaminol E
A (1:1) solution of pyridine and acetic anhydride (0.3 mL) was added to halaminol E (2) (3.3 mg, 13.2 μM) and stirred at RT under N2 for 18 h. The reaction mixture was dried under vacuum and then dissolved in 6.0 mL of CH2Cl2, which was partitioned successively with 1 M aq. NaOH (3 × 3 mL), then brine (3 × 3 mL) and water (3 × 3 mL). The organic layer was dried over anhydrous Na2SO4 and evaporated to give a white solid. Final purification was done by HPLC using a Phenomenex Onyx C8 [150 × 22.5 mm] column at a flow rate of 15 mL and elution using a gradient solvent system from water (0.1% formic acid) to acetonitrile (0.1% formic acid) to give 1.8 mg of N-O-diacetyl halaminol E: white solid; [α]D = +15.2, [α]365 = +56.4, [α]405 = 36, [α]436 = +24, [α]546 = +12, [α]633 = +10, (c 0.5, CHCl3); [α]D = +10, [α]365 = +59, [α]405 = +38, [α]436 = +26, [α]546 = +12, [α]633 = +8, (c 0.5, MeOH); 1H NMR (600 MHz, chloroform-d1) data: δH 6.31 (1H, dt, J = 17.2, 10.3 Hz, H-15), 6.04 (1H, m, H-14), 5.74 (1H, m, H-5), 5.70 (1H, m, H-13), 5.47 (1H, br d, J = 9.0 Hz, NH-2) 5.37 (1H, ddt, J = 15.4, 7.3, 1.5 Hz, H-4), 5.19 (1H, m, H-3), 5.09 (1H, br d, J = 17.0, H-16a), 4.95 (1H, br d, J = 10.2 Hz, H-16b), 4.20 (1H, m, H-2), 2.08 (3H, s, OAc-3), 2.06 (2H, m, H-12), 2.02 (2H, m, H-6), 1.96 (3H, s, NAc-2), 1.37 (2H, m, H-11), 1.35 (2H, m, H-7), 1.23–1.30 (6H, m, H-8/-9/-10), 1.11 (3H, d, J = 6.8 Hz, H-1); 13C NMR (151 MHz, chloroform-d1) data: δC 170.0 (OCOCH3), 169.5 (NHCOCH3), 137.3 (C-15), 136.7 (C-5), 135.5 (C-13), 130.9 (C-14), 124.9 (C-4), 114.6 (C-16), 77.0 (C-3), 47.8 (C-2), 32.5 (C-12), 32.3 (C-6), 29.2–28.8 (C-7/-8/-9/-10/-11), 23.4 (OCOCH3), 21.3 (NHCOCH3), 17.9 (C-1); HRESIMS m/z [M + H]+ 336.25268 (calcd for C20H34NO3+, 336.25332).
Halaminol E Oxazolidinone (3)
Halaminol E(2) (3.8 mg, 15.1 μmol) was dissolved into 1.0 mL of dry CH2Cl2 and placed onto a magnetic stir pad. A solution of 1,1′-carbonyldiimidazole (2.9 mg, 18.2 μmol, 1.2 equiv) in 1.0 mL of dry CH2Cl2 was added into the reaction flask, and the reaction mixture was stirred for 2 h at RT. The reaction mixture was concentrated under vacuum, and the remaining residue was purified by HPLC on a Phenomenex Onyx C18 [100 × 10 mm] column at a flow rate of 3.8 mL and elution using a gradient solvent system from water (0.1% formic acid) to acetonitrile (0.1% formic acid) to give pure halaminol E oxazolidinone (3) (0.5 mg, 1.8 μmol): white solid; 1H NMR (600 MHz, DMSO-d6) data: δH 7.64 (1H, br s, NH-2), 6.29 (1H, dt, J = 17.0, 10.3 Hz, H-15), 6.04 (1H, dd, J = 15.3, 10.3 Hz, H-14), 5.81 (1H, m, H-5), 5.72 (1H, dt, J = 15.2, 7.0 Hz, H-13), 5.52 (1H, ddt, J = 15.3, 7.7, 1.5 Hz, H-4), 5.09 (1H, dd, J = 17.1, 1.9 Hz, H-16a), 4.95 (1H, dd, J = 10.2, 1.9 Hz, H-16b), 4.39 (1H, t, J = 7.4 Hz, H-3), 3.49 (1H, m, H-2), 2.04 (4H, br m, H-6/12), 1.34–1.25 (10H, m, H-7/8/9/10/11), 1.11 (3H, d, J = 6.1 Hz, H-1); 13C NMR (151 MHz, DMSO-d6) data: δC 157.6 (NHCOO), 137.2 (C-15), 136.1 (C-5), 135.3 (C-13), 130.9 (C-14), 126.6 (C-4), 115.1 (C-16), 83.4 (C-3), 53.3 (C-2), 31.9 (C-12), 31.4 (C-6), 28.6–28.2 (C-7/8/9/10/11), 19.0 (C-1); HRESIMS m/z [M + H]+ 278.21536 (calcd for C17H28NO2+, 278.21146).
N-O-Dinapthoyl Halaminol E (4)
Halaminol E (2) (15.0 mg, 59.7 μmol) was dissolved in dry MeOH (5 mL) and added into a 25 mL round-bottom flask containing a suspension of Pd/C (10%, 100.0 mg) in MeOH (10 mL). The air was evacuated and replaced with hydrogen gas three times. The reaction was attached to a hydrogen filled balloon and placed on a magnetic stir pad at RT and left stirring overnight (18 h), after which the reaction mixture was filtered and concentrated under vacuum to give 4,13,15-hexahydro-halaminol E (15.2 mg, ∼95%) as a white solid, HRESIMS m/z 258.279247 [M + H]+ (calcd for C16H36NO+ 258.279141). This material was carried onto the next reaction without further purification. A solution of 2-naphthoic acid (50.2 mg, 292 μmol, 5 equiv), EDC-HCl (61.5 mg, 321 μmol, 5.5 equiv) and DMAP (35.6 mg, 292 μmol) was dissolved into CH2Cl2 (4.0 mL) and placed in an ice water bath at 0 °C. The reaction mixture was stirred for 20 min under N2, then a solution of the 4,13,15-hexahydro-halaminol E (15 mg, 58 μmol) in CH2Cl2 (1.8 mL) was slowly added into the reaction flask. The reaction was allowed to slowly warm to RT and left stirring overnight (18 h). The reaction was quenched by adding CH2Cl2 (20 mL), and the mixture was washed successively with equal volumes of 10% HCl, water, NaHCO3 (satd.) and H2O. The organic layer was dried over anhydrous Na2SO4 and filtered, and the solvent was removed by rotary evaporation. The residue was purified by flash chromatography through a silica gel SPE cartridge, followed by HPLC using a Phenomenex Kinetex C8 [5 μm, 150 × 21.2 mm] column at a flow rate of 15 mL/min and elution using a gradient solvent system from water (0.1% formic acid) to acetonitrile (0.1% formic acid) to give N-O-dinapthoyl halaminol E (4) (0.6 mg, 1.1 μmol): white powder; ECD (CH3CN) λ 226 (Δε −62.4) and λ 243 (Δε −66.5); 1H NMR (600 MHz, DMSO-d6) data: δH 8.70 (1H, m), 8.55 (1H, d, J = 8.9 Hz), 8.38 (1H, t, J = 1.1 Hz), 8.10 (1H, dd, J = 8.3, 1.4 Hz), 8.07 (1H, dd, J = 8.5, 1.7 Hz), 8.04–7.92 (6H, m), 7.87 (1H, dd, J = 8.5, 1.8 Hz), 7.66 (1H, m), 7.61 (1H, m), 7.57 (1H, m), 5.25 (1H, ddd, J = 8.4, 5.8, 4.1 Hz), 4.54 (1H, m), 1.77 (1H, m), 1.34 (1H, m), 1.29 (3H, d, J = 6.9 Hz), 1.23 (4H, m), 1.17–1.12 (16H, m), 0.84 (t, J = 7.2 Hz, 3H); HRESIMS m/z [M + H]+ 566.36294 (calcd for C38H48NO3+, 566.36287).
Hexabranchus sanguineus: Collection, Extraction, and Isolation
Six H. sanguineus nudibranch specimens were collected in shallow waters on a reef top in Tanzania in January 1996 under contract with the Coral Reef Research Foundation for the National Cancer Institute. The animals were taxonomically identified by Lori J. Bell Collin of the Coral Reef Research Foundation, and a voucher specimen (0CDN3604) was deposited at the Smithsonian Institution. The combined nudibranchs (wet weight 84.24 g) were extracted in water, followed by a MeOH/DCM overnight soak according to the National Cancer Institute’s standard marine extraction procedure detailed in McCloud21 to give the organic solvent extract C15373 (2.36 g). A portion of the organic extract (4 × 250 mg) was prefractionated on a C8SPE cartridge (2 g), and fractions were collected using the standard prefractionation MeOH/H2O seven fraction elution scheme to yield fractions 1 m = 41.3 mg; 2 m = 86.9 mg; 3 m = 20.4 mg; 4 m = 10.2 mg; 5 m = 14.7 mg; 6 m = 159.2 mg; and 7 m = 209.3 mg. A portion of fraction 6 (m = 145 mg) was further purified on a Phenomenex Onyx Monolithic C18 [100 × 10 mm] column (29 × 5 mg injections) using the standard second-stage semipreparative gradient conditions. For each HPLC run, 5 mg was injected, and 22 fractions combined from each of the 29 runs. Subfractions M15373_6_15 (m = 4.5 mg) and _16 (m = 9.3 mg) were combined and subjected to preparative HPLC using a Phenomenex Kinetex C8 [5 μm, 150 × 21.2 mm] column at a flow rate of 15 mL/min with the following conditions: an initial isocratic hold at MeOH/H2O (4:6) for 5 min, followed by a linear gradient to MeOH/H2O (9:1) over 45 min, and a final hold at MeOH/H2O (9:1) for 10 min. A total of 120 fractions were collected in 30 s increments starting from time = 0 min. Fractions 75 to 80 were combined and then further purified on an identical column with the following conditions: an initial isocratic hold at MeOH/H2O (65:35) for 5 min, followed by a linear gradient to MeOH/H2O (85:15) over 45 min, and a final hold at MeOH/H2O (85:15) for 10 min, with fractions collected in 30 s increments starting from time = 0 min. Fraction 35 yielded 33-methyltetrahydrohalichondramide (7) (0.8 mg, 0.09% crude extract yield) and fraction 40 yielded 33-methyldihydrohalichondramide (8) (1.2 mg, 0.15% crude extract yield). Fractions 42–46 were combined and further purified on a Phenomenex Onyx Monolithic C18 [100 × 10 mm] with the following conditions: an initial isocratic hold at CH3CN/H2O with 0.1% formic acid (1:1) for 5 min, followed by a linear gradient to CH3CN over 45 min, and a final hold at CH3CN for 10 min, all at 10 mL/min and with fractions collected in 1 min increments starting from time = 0 min. Fraction 3 yielded coelodiol (5) (1.0 mg, 0.12% crude extract yield), and fraction 5 yielded mycgranol (6) (1.8 mg, 0.21% crude extract yield). Subfraction M15373_6_20 (m = 9 mg) was further purified on a Phenomenex Onyx Monolithic C18 [100 × 10 mm] with the following conditions: an initial isocratic hold at CH3CN/H2O with 0.1% formic acid (1:1) for 5 min, followed by a linear gradient to CH3CN over 45 min, and a final hold at CH3CN for 10 min, all at a 10 mL/min flow rate and with fractions collected in 1 min increments starting from time = 0 min. Fraction 15 yielded swinholide A (9) (0.15 mg, 0.017% crude extract yield).
(−)-Coelodiol (5)
Clear oil; [α]D = −6, (c 0.1, CHCl3); NMR spectroscopic data in agreement to those previously reported for (+)-coelodiol;22 HRESIMS m/z [M + H]+ 351.25322 (calcd for C21H35O4+, 351.25099).
Mycgranol (6)
Clear oil; chirooptical and NMR spectroscopic data in agreement to those previously reported;23 HRESIMS m/z [M + H]+ 349.23703 (calcd for C21H33O4+, 349.23734).
33-Methyltetrahydrohalichondramide (7)
Clear oil; NMR spectroscopic data in agreement to those previously reported;24 HRESIMS m/z [M + H]+ 855.47518 (calcd for C45H67N4O12+, 855.4750).
33-Methyldihydrohalichondramide (8)
Clear oil; chirooptical and NMR spectroscopic data in agreement to those previously reported;25 HRESIMS m/z [M + Na]+ 875.44086 (calcd for C45H64N4NaO12+, 875.44129).
Swinholide A (9)
Clear oil; 1H NMR spectroscopic data in agreement to those previously reported;26 HRESIMS m/z [M + Na]+ 1411.92072 (calcd for C78H132NaO20+, 1411.92042).
Acknowledgments
We thank the Molecular Pharmacology Branch, DTP, DCTD, NCI for performing the NCI 60-cell cytotoxicity assays in support of this study. We also M. Dyba and S. Tarasov (Biophysics Resource, SBL, NCI) for assistance with the ECD measurements. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E, by the National Cancer Institute’s Cancer MoonshotSM and by the Intramural Research Program of the National Cancer Institute. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.0c00139.
Additional figures, NMR spectra, and NCI-60 five-dose graphs (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Newman D. J.; Cragg G. M. (2016) Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661. 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- Henrich C. J.; Beutler J. A. (2013) Matching the power of high throughput screening to the chemical diversity of natural products. Nat. Prod. Rep. 30, 1284–1298. 10.1039/c3np70052f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey A. L.; Edrada-Ebel R.; Quinn R. J. (2015) The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discovery 14, 111–129. 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- Thornburg C. C.; Britt J. R.; Evans J. R.; Akee R. K.; Whitt J. A.; Trinh S. K.; Harris M. J.; Thompson J. R.; Ewing T. L.; Shipley S. M.; Grothaus P. G.; Newman D. J.; Schneider J. P.; Grkovic T.; O’Keefe B. R. (2018) NCI Program for Natural Product Discovery: A Publicly-Accessible Library of Natural Product Fractions for High-Throughput Screening. ACS Chem. Biol. 13, 2484–2497. 10.1021/acschembio.8b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G.; Mayhudin N. A.; Mitova M. I.; Sun L.; van der Sar S.; Blunt J. W.; Cole A. L. J.; Ellis G.; Laatsch H.; Munro M. H. G. (2008) Evolving Trends in the Dereplication of Natural Product Extracts: New Methodology for Rapid, Small-Scale Investigation of Natural Product Extracts. J. Nat. Prod. 71, 1595–1599. 10.1021/np8002222. [DOI] [PubMed] [Google Scholar]
- Grkovic T.; Pouwer R. H.; Vial M.-L.; Gambini L.; Noel A.; Hooper J. N. A.; Wood S. A.; Mellick G. D.; Quinn R. J. (2014) NMR Fingerprints of the Drug-like Natural-Product Space Identify Iotrochotazine A: A Chemical Probe to Study Parkinson’s Disease. Angew. Chem., Int. Ed. 53, 6070–6074. 10.1002/anie.201402239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Idelbayev Y.; Roberts N.; Tao Y.; Nannapaneni Y.; Duggan B. M.; Min J.; Lin E. C.; Gerwick E. C.; Cottrell G. W.; Gerwick W. H. (2017) Small Molecule Accurate Recognition Technology (SMART) to Enhance Natural Products Research. Sci. Rep. 7, 1–17. 10.1038/s41598-017-13923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; Porto C.; Bouslimani A.; Melnik A. V.; Meehan M. J.; Liu W.-T.; Crusemann M.; Boudreau P. D.; Esquenazi E.; Sandoval-Calderon M.; Kersten R. D.; Pace L. A.; Quinn R. A.; Duncan K. R.; Hsu C.-C.; Floros D. J.; Gavilan R. G.; Kleigrewe K.; Northen T.; Dutton R. J.; Parrot D.; Carlson E. E.; Aigle B.; Michelsen C. F.; Jelsbak L.; Sohlenkamp C.; Pevzner P.; Edlund A.; McLean J.; Piel J.; Murphy B. T.; Gerwick L.; Liaw C.-C.; Yang Y.-L.; Humpf H.-U.; Maansson M.; Keyzers R. A.; Sims A. C.; Johnson A. R.; Sidebottom A. M.; Sedio B. E.; Klitgaard A.; Larson C. B.; Boya P C. A.; Torres-Mendoza D.; Gonzalez D. J.; Silva D. B.; Marques L. M.; Demarque D. P.; Pociute E.; O’Neill E. C.; Briand E.; Helfrich E. J. N.; Granatosky E. A.; Glukhov E.; Ryffel F.; Houson H.; Mohimani H.; Kharbush J. J.; Zeng Y.; Vorholt J. A.; Kurita K. L.; Charusanti P.; McPhail K. L.; Nielsen K. F.; Vuong L.; Elfeki M.; Traxler M. F.; Engene N.; Koyama N.; Vining O. B.; Baric R.; Silva R. R.; Mascuch S. J.; Tomasi S.; Jenkins S.; Macherla V.; Hoffman T.; Agarwal V.; Williams P. G.; Dai J.; Neupane R.; Gurr J.; Rodriguez A. M. C.; Lamsa A.; Zhang C.; Dorrestein K.; Duggan B. M.; Almaliti J.; Allard P.-M.; Phapale P.; Nothias L.-F.; Alexandrov T.; Litaudon M.; Wolfender J.-L.; Kyle J. E.; Metz T. O.; Peryea T.; Nguyen D.-T.; Van Leer D.; Shinn P.; Jadhav A.; Muller R.; Waters K. M.; Shi W.; Liu X.; Zhang L.; Knight R.; Jensen P. R.; Palsson B. O.; Pogliano K.; Linington R. G.; Gutierrez M.; Lopes N. P.; Gerwick W. H.; Moore B. S.; Dorrestein P. C.; Bandeira N. (2016) Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavatta M. L.; Gavagnin M.; Manzo E.; Puliti R.; Mattia C. A.; Mazzarella L.; Cimino G.; Simpson J. S.; Garson M. J. (2005) Structural and stereochemical revision of isocyanide and isothiocyanate amphilectenes from the Caribbean marine sponge Cribochalina sp. Tetrahedron 61, 8049–8053. 10.1016/j.tet.2005.05.102. [DOI] [Google Scholar]
- Wratten S. J.; Faulkner D. J.; Hirotsu K.; Clardy J. (1978) Diterpenoid isocyanides from the marine sponge Hymeniacidon amphilecta. Tetrahedron Lett. 19, 4345–4348. 10.1016/S0040-4039(01)95220-7. [DOI] [Google Scholar]
- Clark R. J.; Garson M. J.; Hooper J. N. A. (2001) Antifungal alkyl amino alcohols from the tropical marine sponge Haliclona n. sp. J. Nat. Prod. 64, 1568–1571. 10.1021/np010246x. [DOI] [PubMed] [Google Scholar]
- Pearce A. N.; Hill C. A. E.; Page M. J.; Keyzers R. A.; Copp B. R. (2019) An Acetylenic Lipid from the New Zealand Ascidian Pseudodistoma cereum: Exemplification of an Improved Workflow for Determination of Absolute Configuration of Long-Chain 2-Amino-3-alkanols. J. Nat. Prod. 82, 2291–2298. 10.1021/acs.jnatprod.9b00504. [DOI] [PubMed] [Google Scholar]
- Gulavita N. K.; Scheuer P. J. (1989) Two epimeric aliphatic amino alcohols from a sponge. Xestospongia sp. J. Org. Chem. 54, 366–369. 10.1021/jo00263a019. [DOI] [Google Scholar]
- Mori K.; Matsuda H. (1992) Synthesis of sphingosine relatives. XII. Synthesis and absolute configuration of the two epimeric aliphatic amino alcohols [(5E,7E)-2-amino-5,7-tetradecadien-3-ols] isolated from a sponge, Xestospongia sp. Liebigs Ann. Chem. 2, 131–137. 10.1002/jlac.199219920125. [DOI] [Google Scholar]
- Polt R.; Sames D.; Chruma J. (1999) Glycosidase inhibitors: synthesis of enantiomerically pure aza-sugars from Schiff base amino esters via tandem reduction-alkenylation and osmylation. J. Org. Chem. 64 (17), 6147–6158. 10.1021/jo9820115. [DOI] [Google Scholar]
- Pearce A. N.; Copp B. R.; Molinski T. F. (2019) Enantiomeric variability of distaminolyne A. Refinement of ECD and NMR methods for determining optical purity of 1-amino-2-alkanols. Molecules 24 (1), 90–104. 10.3390/molecules24010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinski T. F.; Biegelmeyer R.; Stout E. P.; Wang X.; Frota M. L. C.; Henriques A. T. (2013) Halisphingosines A and B, Modified Sphingoid Bases from Haliclona tubifera. Assignment of Configuration by Circular Dichroism and van’t Hoff’s Principle of Optical Superposition. J. Nat. Prod. 76, 374–381. 10.1021/np300744y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros R.; Montejo de Garcini E.; Wandosell F.; Faircloth G.; Fernandez-Sousa J. M.; Avila J. (2000) The marine compound spisulosine, an inhibitor of cell proliferation, promotes the disassembly of actin stress fibers. Cancer Lett. (N. Y., NY, U. S.) 152, 23–29. 10.1016/S0304-3835(99)00428-0. [DOI] [PubMed] [Google Scholar]
- Baird R. D.; Kitzen J.; Clarke P. A.; Planting A.; Reade S.; Reid A.; Welsh L.; Lopez Lazaro L.; de las Heras B.; Judson I. R.; Kaye S. B.; Eskens F.; Workman P.; de Bono J. S.; Verweij J. (2009) Phase I safety, pharmacokinetic, and pharmacogenomic trial of ES-285, a novel marine cytotoxic agent, administered to adult patients with advanced solid tumors. Mol. Cancer Ther. 8, 1430–1437. 10.1158/1535-7163.MCT-08-1167. [DOI] [PubMed] [Google Scholar]
- Massard C.; Salazar R.; Armand J. P.; Majem M.; Deutsch E.; Garcia M.; Oaknin A.; Fernandez-Garcia E. M.; Soto A.; Soria J. C. (2012) Phase I dose-escalating study of ES-285 given as a three-hour intravenous infusion every three weeks in patients with advanced malignant solid tumors. Invest. New Drugs 30, 2318–2326. 10.1007/s10637-011-9772-8. [DOI] [PubMed] [Google Scholar]
- McCloud T. G. (2010) High throughput extraction of plant, marine and fungal specimens for preservation of biologically active molecules. Molecules 15, 4526–4563. 10.3390/molecules15074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattorusso E.; Romano A.; Taglialatela-Scafati O.; Bavestrello G.; Bonelli P.; Calcinai B. (2006) Coelodiol and coeloic acid, ent-isocopalane diterpenes from the Indonesian sponge Coelocarteria cfr. singaporensis. Tetrahedron Lett. 47, 2197–2200. 10.1016/j.tetlet.2006.01.109. [DOI] [Google Scholar]
- Rudi A.; Benayahu Y.; Kashman Y. (2005) Mycgranol, a new diterpene from the marine sponge Mycale aff. graveleyi. J. Nat. Prod. 68, 280–281. 10.1021/np0497550. [DOI] [PubMed] [Google Scholar]
- Dalisay D. S.; Rogers E. W.; Edison A. S.; Molinski T. F. (2009) Structure Elucidation at the Nanomole Scale. 1. Trisoxazole Macrolides and Thiazole-Containing Cyclic Peptides from the Nudibranch Hexabranchus sanguineus. J. Nat. Prod. 72, 732–738. 10.1021/np8007649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga S.; Fusetani N.; Hashimoto K.; Koseki K.; Noma M.; Noguchi H.; Sankawa U. (1989) Bioactive marine metabolites. 25. Further kabiramides and halichondramides, cytotoxic macrolides embracing trisoxazole, from the Hexabranchus egg masses. J. Org. Chem. 54, 1360–1363. 10.1021/jo00267a024. [DOI] [Google Scholar]
- Andrianasolo E. H.; Gross H.; Goeger D.; Musafija-Girt M.; McPhail K.; Leal R. M.; Mooberry S. L.; Gerwick W. H. (2005) Isolation of Swinholide A and Related Glycosylated Derivatives from Two Field Collections of Marine Cyanobacteria. Org. Lett. 7, 1375–1378. 10.1021/ol050188x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.