Abstract
Although efficient human-to-human transmission of avian influenza virus has yet to be seen, in the past two decades avian-to-human transmission of influenza A viruses has been reported. Influenza A/H5N1, in particular, has repeatedly caused human infections associated with high mortality, and since 1998 the virus has evolved into many clades of variants with significant antigenic diversity. In 2013, three (A/H7N9, A/H6N1, and A/H10N8) novel avian influenza viruses (AIVs) breached the animal-human host species barrier in Asia. In humans, roughly 35% of A/H7N9-infected patients succumbed to the zoonotic infection, and two of three A/H10N8 human infections were also lethal; however, neither of these viruses cause influenza-like symptoms in poultry. While most of these cases were associated with direct contact with infected poultry, some involved sustained human-to-human transmission. Thus, these events elicited concern regarding potential AIV pandemics. This article reviews the human incursions associated with AIV variants and the potential role of pigs as an intermediate host that may hasten AIV evolution. In addition, we discuss the known influenza A virus virulence and transmission factors and their evaluation in animal models. With the growing number of human AIV infections, constant vigilance for the emergence of novel viruses is of utmost importance. In addition, careful characterization and pathobiological assessment of these novel variants will help to identify strains of particular concern for future pandemics.
Keywords: avian influenza A virus, humans, pathogenicity, transmission, pandemic, influenza vaccines
Influenza A viruses are RNA viruses of the family Orthomyxoviridae. Each possesses an eight-segmented RNA viral genome of negative polarity, which encodes 10 to 12 functional viral proteins depending on the virus strain.1 The segmented nature of the viral RNA genome combined with its error-prone polymerase result in a virus that can rapidly evolve and adapt in new host species. This can produce novel strains with the potential to cause influenza pandemics.2 Moreover, coinfection of a single host with two or more viruses facilitates genetic reassortment, the rearrangement of genetic material through the exchange of gene segments, creating progeny viruses with a novel gene constellation that may confer the ability to infect new hosts.2
Although wild waterfowl (Anseriformes) and shorebirds (Charadriiformes) are generally regarded as the natural hosts, these viruses have been found in many other avian species as well as in mammalian hosts including humans.2 Influenza viruses are further classified into different subtypes based on the antigenic variation of their surface glycoproteins, hemagglutinin (HA), and neuraminidase (NA). To date, there are ∼16 HA and 9 NA subtypes, most of which have been found in aquatic birds.2 New strains (H17–18 and N10–11) have also been identified in bat species.3 4
Despite their asymptomatic infection of wild birds, introduction of influenza A viruses into a new host such as terrestrial poultry species can cause severe illness often leading to high mortality. Thus, avian influenza viruses (AIVs) can be separated into two pathotypes, namely, high pathogenicity (HP) or low pathogenicity (LP) AIVs.5 Some HPAI strains of the H5 (e.g., H5N1) and H7 (e.g., H7N1, H7N3, and H7N7) subtypes have demonstrated high lethality in chickens (up to 100% mortality) with wide tissue (extrapulmonary) distribution of viral replication. In contrast, LPAI strains are mainly limited to target organs of the intestinal and/or respiratory tracts.2 In humans, infection with HPAI H5N1 viruses results in a 60% mortality rate worldwide6 Furthermore, in recent years, the number of influenza A viruses crossing the animal-human host species barrier has increased. Here, we provide a general overview of these zoonotic infections and discuss their pandemic risk potential.
Factors Affecting Interspecies Transmission of Influenza A Viruses
Despite the potential to infect a wide variety of animal species, influenza A viruses are generally host-specific and are not readily transmissible from one species to another. Several factors (e.g., environmental, viral, and host factors) can affect or determine the ability of influenza viruses to transmit efficiently from one host to another (Fig. 1). While the molecular basis for host range restriction has not yet been clearly defined, the factors governing this function have frequently been linked to the viral surface glycoproteins, particularly HA, and their interaction with sialic acid receptors on host epithelial cells.7 8 9 Influenza virus infection is initiated via HA, which binds to sialic acid-containing glycans that are associated with glycoproteins and glycolipids on the surface of epithelial cells. The HAs of human influenza virus strains preferentially bind to oligosaccharides that terminate with sialic acid linked to galactose by α(2,6) linkages (Sia [α2,6]Gal), whereas the HAs of avian influenza virus strains prefer oligosaccharides that terminate with a sialic acid linked to galactose by α(2,3) linkages (Sia [α2,3]Gal).10 11 The receptor-binding domain (RBD) of HA is located in a globular head domain formed by the 190 helix at the top of HA, the 220 loop at the edge of the globular head, and the 130 loop at the other edge of the globular head.12 Amino acid changes in and around the RBD dramatically alter the receptor binding preference of influenza viruses, which help define virus transmission. However, the specific amino acids that determine receptor-binding specificity vary among the different HA subtypes. Regardless, mutations in the 220 loop may alter the orientation of this loop and thus, optimize the contact with specific receptor types13 to facilitate the release of virus particles from infected host cells. In addition, the NA protein is also likely to contribute to viral transmissibility13 14 and it was recently reported that an optimal balance between HA and NA activities is critical for efficient respiratory droplet transmission of a pandemic 2009 H1N1 virus in ferrets.15 Beyond HA and NA, additional viral proteins have been examined for their contributing roles in influenza virus transmission. For example, amino acid residues in the polymerase basic protein 2 (PB2) protein are associated with mammalian adaptation, efficient transmission via respiratory droplets between ferrets, and replication in human cells.7 16 17 It is also worth mentioning that along with other segments, the PB1 genes of the human pandemic viruses in 1957 (Asian flu, H2N2) and 1968 (Hong Kong flu, H3N2) came from avian virus sources.18 Therefore, acquisition of the avian PB1 gene was speculated to be a crucial factor in the emergence of reassortants with a competitive advantage over seasonal influenza viruses with regard to replication and virulence.19 Aside from differences in the genetic makeup of viruses, the immunological history of the host can also impact an individual's susceptibility to infection, as well as the potential for immune control of virus infection and subsequent disease outcome.20 Collectively, these features account for the virulence, expansion of host range, and interspecies transmission of AIVs.
Fig. 1.
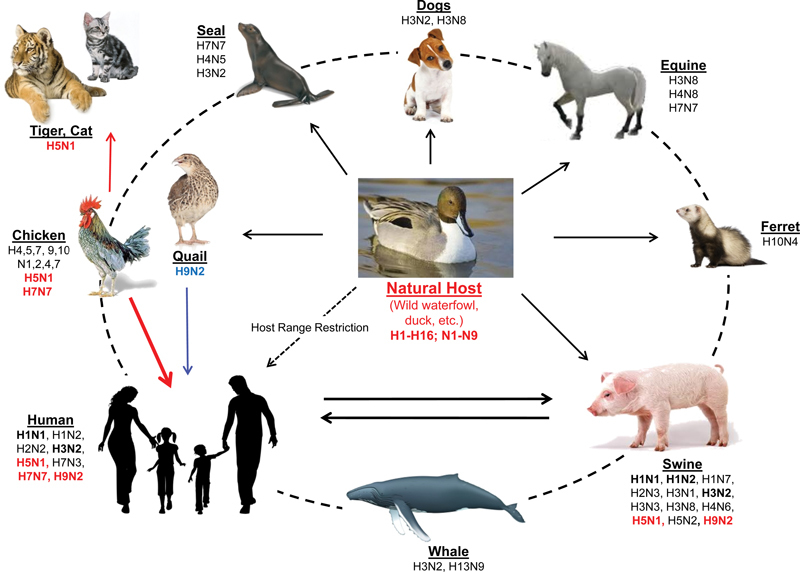
Reservoir of influenza viruses. It is known that wild aquatic birds are the source of all influenza viruses in other species. Although transmission between humans and pigs had already been demonstrated and confirmed, direct transmission of avian-to-humans have been less frequent (such as those with H9N2 and H5N1 subtypes) but sometimes with fatal outcome.
Emergence of the Animal-Human Interface Avian Influenza A Viruses
Various AIV subtypes have occasionally crossed the animal–human interface, most commonly after close human contact with infected birds21 (Table 1). Of note, humans also possess AIV-susceptible cells in the lower respiratory tract (e.g., lungs), which express receptors that are not expressed by upper respiratory tract tissues (e.g., nasal passages and trachea).22 Thus, it is not surprising that extensive or prolonged exposure to affected poultry increases the chances of interspecies transmission.
Table 1. Reported cases of human infections with avian influenza A viruses.
| Subtype | Nation and year | Symptoms | Confirmed cases/ fatalities |
|---|---|---|---|
| H4N8 | US (1991) | Acute respiratory symptoms | 1/0 |
| H5N1 | HK (1997), AZ, BD, CN, DJ, EG, HK, ID, IQ, KH, LA, MM, NG, PK, TH, TR, VN (2003–2013) | Fever, headache, muscle aches, cough, sore throat, eye infections, pneumonia, acute respiratory distress | 826/440 |
| H6N1 | US (1991), TW (2013) | Acute respiratory symptoms and Influenza-like illness |
3/0 |
| H7N2 | US (2002–2003), UK (2007) | Conjunctivitis, mild influenza-like illness | 4/0 |
| H7N3 | IT (1999–2003), CA (2004), UK (2006), MX (2012) | Conjunctivitis, no fever, respiratory symptoms | 12/0 |
| H7N7 | US (1959,1979), AU (1977), UK (1996), NL (2003) | Conjunctivitis, influenza-like illness, fever, pneumonia | 90/1 |
| H7N9 | CN (2013–2015), HK (2013), TW (2013), CA (2015), US (2015) | Acute respiratory distress syndrome and multi-organ failure | 678/271 |
| H9N2 | CN, HK (1998–1999, 2003, 2007, 2009), BD (2011) | Low fever, headache, sneezing, runny nose, vomiting, influenza-like illness | 16/0 |
| H10N7 | US (1991), EG (2004), AT (2010) | Acute respiratory symptoms | 10/0 |
| H10N8 | CN (2013–2014) | Severe pneumonia | 3/0 |
Abbreviations: AZ, Azerbaijan; AT, Austria; BD, Bangladesh; KH, Cambodia; CA, Canada; CN, China; DJ, Djibouti; EG, Egypt; HK, Hong Kong Special Administrative Region of China; ID, Indonesia; IQ, Iraq; IT, Italy; LA, Laos; MX, Mexico; MM, Myanmar; NL, Netherland; NG, Nigeria; PK, Pakistan; TH, Thailand; TR, Turkey; TW, Taiwan; UK, United Kingdom; US, United States; VN, Vietnam.
Distinct features of the HA protein control not only the species specificity of the virus, but also its tissue specificity and therefore its ability to spread within a host. Different lineages of viruses have become stably adapted to avian and mammalian species through diverse receptor preferences, for example, AIVs recognize specific sialic acid receptors of susceptible cells lining the intestinal gut of avian hosts.23 However, specific amino acid substitutions in HA generated through repeated adaptation can shift receptor recognition, which, in turn, effectively switches binding preferences to receptors expressed by mammalian hosts including humans. Further genetic modification of the HA cleavage motif of some subtypes (e.g., H5 and H7) can result in an HA that is more readily cleaved by ubiquitous cellular proteases.24 It should be noted that these genetic alterations were observed during novel host adaption. For example, during consecutive passages in chickens, a highly virulent AIV was isolated from a virulent-swan virus that normally replicates poorly in chickens. The more virulent chicken strain was associated with accumulation of polybasic residues in HA cleavage sites,7 which is typically seen among highly pathogenic avian influenza (HPAI) H5 and H7 viruses and promotes efficient biological activation facilitating growth in various organs outside the respiratory tract.24 25
Among the vast pool of AIVs in nature, the HPAI A/H5N1 virus is believed to represent the greatest threat for the next flu pandemic (Table 1). Since its first appearance in Guangdong, China in 1996, this virus has already spawned various clusters of antigenically divergent HPAI H5 lineages that spread widely in Asia toward Europe and Africa26 causing devastating poultry outbreaks. In humans, more than 640 HPAI A/H5N1 infections have been reported by the World Health Organization (WHO) since 1997, of which ∼60% were fatal.6 This high mortality rate may be related to aberrant innate immune responses, which were found to correlate with the atypical virulence of the high mortality HPAI A/H5N1.6 27 28 29 In 2014, new cases were reported in Asia with Vietnam and Cambodia recording fatalities during the year.6 The first imported case in North America was also recently confirmed in a resident of Alberta, Canada who returned from Beijing, China in December 27, 2013, was hospitalized, and died within a few days due to viral infection.30 At present, there are no imminent indications that contemporary HPAI A/H5N1 viruses have the capacity to support efficient, sustained viral transmission in humans. However, two independent studies have simultaneously demonstrated that a low number of changes in the HPAI A/H5N1 viral genome can confer airborne transmission between mammals.12 31
Human infection with A/H9N2 AIVs was first documented in Hong Kong in 1999.32 Retrospectively, it was postulated that the HPAI A/H5N1 virus that caused the 1997 human outbreaks in Hong Kong acquired most of its gene segments from avian A/H9N2 subtypes that were circulating in 1997.33 Sporadic cases of AIV A/H9N2 zoonoses have since occurred with at least 16 human infections as shown in the influenza virus sequence database34 (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html), including a recent case in Hong Kong in late December 2013, the first in 4 years.35 Serologic studies also showed that some poultry workers in China and India were positive for A/H9N2 virus infection.36 37 38 Notably, avian A/H9N2 infections induced relatively mild clinical disease with no mortality in contrast to the more virulent HPAI A/H5N1.
Prior to 1996, human infections with the H7 AIV subtype were primarily due to laboratory or occupational exposures.39 Direct avian-to-human transmission of a low pathogenic avian influenza (LPAI) A/H7N7 was first confirmed in 1996 in an English woman who tended to infected pet ducks.40 In 2002, LPAI A/H7N2 virus was detected in Virginia41 and isolated from an immunocompromised New York resident42; this subtype was also found among poultry workers in Wales, England.43 Similarly, an LPAI A/H7N3 virus was recovered from a poultry worker with conjunctivitis in the United Kingdom,44 and retrospective serologic analysis of workers who responded during the 2002–2003 H7N3 Italian poultry outbreak identified seven seropositive individuals. In contrast to these LPAI H7 strains,45 HPAI A/H7N3 variants were responsible for zoonotic infections among poultry workers in British Columbia, Canada,46 and Jalisco, Mexico.47 Before 2013, the largest human H7 outbreak occurred in 2003 in the Netherlands48 and was caused by an HPAI A/H7N7 virus, involved over 80 human cases including one fatality, and was associated with the destruction of more than 30 million birds. Although the number of human cases during this outbreak was quite high, only three cases were individuals who had not been in contact with infected poultry, suggesting that human-to-human transmission of HPAI A/H7N7 was quite limited. Furthermore, with the exception of the cases in the United States, most H7 patients have generally presented with mild respiratory disease commonly accompanied by conjunctivitis.39
However, the pathobiology of H7 AIVs changed when human cases caused by A/H7N9 viruses emerged after late March 2013.49 As of March 9th, 2015, there have been 631 laboratory-confirmed cases of human infection with the A/H7N9 virus reported to the WHO with more than 36% mortality including 4 cases in Taipei, 13 cases in Hong Kong SAR, 1 case in a Chinese traveler reported from Malaysia, and 2 cases in Canadian citizens who traveled to China.50 In contrast to the A/H5N1 and A/H9N2 AIVs, A/H7N9 replicates efficiently in poultry without inducing remarkable influenza-like disease.51 52 The product of multiple reassortment events, this virus includes an HA gene that may have originated from AIVs of duck origin and an NA gene that may have transferred from migratory birds, while the internal genes originated from prevailing A/H9N2 viruses in Chinese poultry53 (Fig. 2). While the A/H7N9 AIV does not contain a pathotypic HA cleavage motif, suggesting that these viruses are of low pathogenicity in avian species, it can infect and replicate well in mammalian species due to receptor binding site mutations.51 52 This has important public health implications since viruses with low pathogenicity are able to spread in domestic poultry undetected. Thus, in contrast to HPAI A/H5 and A/H7 AIVs, which kill infected birds rapidly, A/H7N9 has the potential to cause silent epidemics and outbreaks resulting in sporadic infections in humans who come into direct contact with infected birds. Notably, human infections with A/H7N9 viruses have been characterized by rapid progression to severe pulmonary disease and acute respiratory distress syndrome, reminiscent of human HPAI A/H5N1 disease.51 52 53 Although there is not yet any evidence of sustained human-to-human transmission,52 54 55 the scale of these zoonotic infections highlights the need for enhanced surveillance. Moreover, the geographic location of many Asian countries puts them well within the East Asian flyway of migratory waterfowls, enhancing the likelihood of spread. In addition, an increase in Chinese tourism due to improved air travel could also contribute to global spread of viruses. Thus, the potential exists for A/H7N9 to be carried by infected migratory wild birds and/or human travelers as in cases reported in Taiwan, Malaysia, and Canada.56 57
Fig. 2.
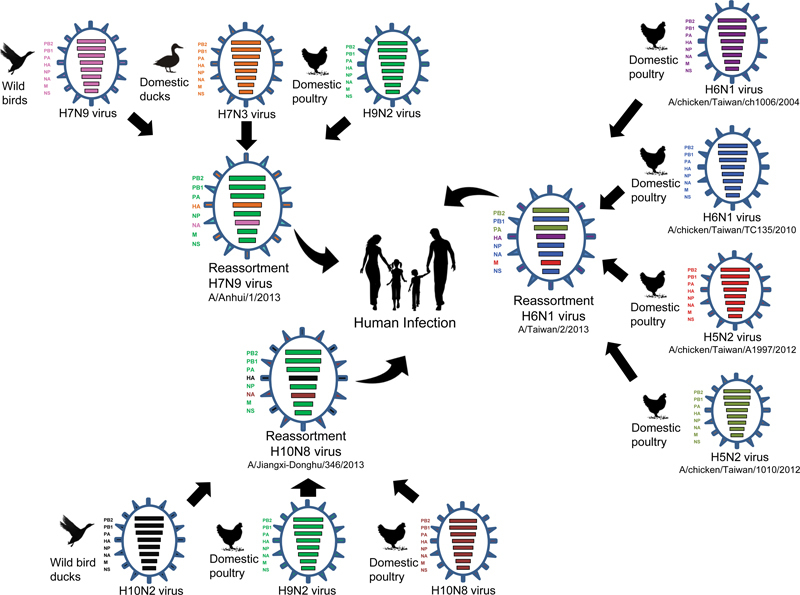
Schematic diagram model of multiple reassortment events of human infected novel influenza viruses from avian (H7N9, H10N8 and H6N1). The colors of the eight gene segments in the ovals represent origin viruses. As the diagram show, same colors is same linage (H9N2) and different colors is different viruses.
In addition to the more frequently isolated strains mentioned above, H6 and H10 AIV human infections have also been recorded. In May 2013, the first human case of A/H6N1 AIV infection was reported in Taiwan.58 Genetic analyses showed that the virus was highly homologous to chicken H6N1 viruses of Taiwanese origin (Fig. 2), except with a G228S substitution in the HA protein that might increase the affinity for the human α(2–6) linked sialic acid receptor.58 The patient was hospitalized after developing a high fever, cough, and shortness of breath, but received early treatment and fully recovered. Occurrences of A/H10N7 infections were also independently recorded in Egypt59 and Australia60 with both patients having only minor symptoms and eventually recovering. In contrast, two of the three A/H10N8 AIV infections reported in China since December 2013 resulted in death.61 62 It is also noteworthy that the internal gene constellations of the newly emergent A/H10N8 viruses contain corresponding segments from A/H9N2 virus subtypes prevailing in Chinese poultry and it does not cause remarkable disease in birds as like it's A/H7N9 predecessor49 52 61 (Fig. 2).
The persistent and sporadic outbreaks of various AIVs in poultry and humans, respectively, underscore the likelihood of AIVs becoming the next influenza pandemic strain. Therefore, the pandemic potential of any subtypes of AIVs should be considered and the continued need to monitor domestic and wild bird populations to better understand interspecies transmission. Such monitoring would also lead to a better understanding of the importance of avian hosts in the ecology of influenza viruses.
Role of Pigs as Intermediate Hosts and Mediators of Rapid Virus Evolution
With the inherent capacity of influenza A viruses to evolve and adapt, it is only a matter of time before novel AIV variants capable of sustaining person-to-person transmission arise. Influenza infection of pigs is highly regarded as an important turning point in the evolution and ecology of influenza A viruses due to the dual susceptibility of pigs to human and animal influenza viruses.11 23 This susceptibility is due to sialyl oligosaccharide receptors lining the respiratory tract, which possess both N-acetylneuraminic acid α(2,3)-galactose (preferred by AIVs) and α(2,6)-galactose (preferred by mammalian influenza viruses). In addition to serving as sources for direct interspecies transmission of virus to people, swine are also known as genetic mixing vessels due to the frequent coinfection and recombination of influenza viruses from various sources and lineages63 64(Fig. 3).
Fig. 3.
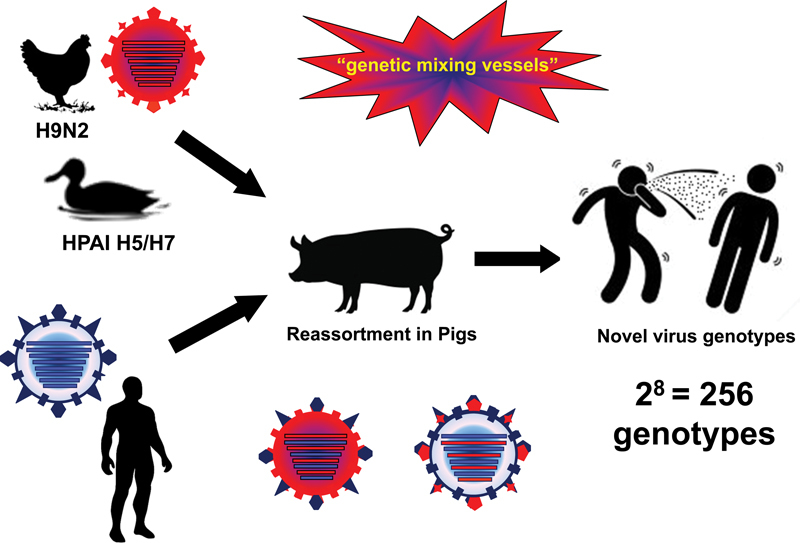
Pigs promote the generation of novel viruses with pandemic potential. Pigs are potential reservoir of old virus strains for subsequent infection of human populations. Avian influenza viruses that infect humans could undergo adaptation altering its receptor-binding affinity to those for human epithelial receptors. Lastly, coinfection of an avian and mammalian virus in a single host cell could generate novel virus genotypes (∼256 recombinant viruses could be generated from two parental viruses), one of which could cause the next pandemic.
Some of the AIVs mentioned previously are known to have the capacity to infect and become adapted in swine populations under natural and experimental conditions. Although domestic pigs appear to have low susceptibility,65 HPAI A/H5N1 viruses have been isolated from wild pig herds in China,66 67 68 69 Indonesia,70 71 and Vietnam.72 Since 1998, several A/H9N2 viruses have been also been isolated from pigs in Hong Kong and mainland China.72 73 74 75 Furthermore, variants of the A/H7N9 virus have been shown to infect pigs in experimental settings, although they were not transmissible to naïve pigs or other animals.76
Successful cross-species transmission of influenza virus is dependent on both host and virus genetic factors, and subsequent spread within a new host population typically requires a period of adaptation of the virus to the new host.2 However, simultaneous infection of a pig with different viruses could promote genetic reassortment that may significantly alter viral evolution and result in the rapid production of reassortant viruses with pandemic potential (Fig. 1). Therefore, with the significant role of pigs in the ecology of influenza viruses, these transmission events should be closely monitored and minimized to prevent the risk of generating viruses with greater human health concerns.
Determinants of Viral Pathogenicity
For influenza virus infections pathogenicity is defined as the ability of an influenza virus to cause disease in the host.77 The pathogenesis of influenza viruses is a polygenic trait with molecular determinants that may differ among animal species (Table 2).78 79 Although factors that define virulence and host range have been commonly linked to the viral surface glycoproteins, the internal gene segments also contribute to virus adaptation, pathogenesis, transmission, and immune evasion. Nonstructural protein 1 appears to contribute to the efficient replication of human influenza A viruses (i.e., 1918 pandemic virus) and is a virulence determinant of HPAI H5N1 viruses that acts by restricting the induction of the host interferon response.80 Additionally, the viral polymerase genes also play significant roles in adaptation and pathogenesis. Of the identified molecular markers of virulence in the viral polymerases, a glutamic acid (E)-to-lysine (K) alteration at position 627 in PB2 is perhaps most commonly observed, particularly among HPAI H5N1 and H7 viruses.7 81 In the absence of 627K, the aspartic acid (D)-to-asparagine (N) alteration at residue 701 of the PB2 protein has been shown to similarly enhance growth and transmission in guinea pigs17 and to expand the host range of avian H5N1 viruses to mice and humans.16 17 In addition, the expression of the PB1-F2 protein, which is associated with mitochondrial targeting, membrane disruption, and subsequent apoptosis, was suggested to contribute to virulence.82 83 Mutations in the polymerase genes of highly pathogenic viruses, such as the E627K mutation of the PB2 protein and the N66S mutation of the PB1-F2 protein, have been associated with an impaired adaptive immune response and increased host cell apoptosis, which may contribute to enhanced viral replication and extrapulmonary spread. Beside the viral virulence factor of described previously, host factors are also responsible for the pathogenesis of AIV infection. Therefore, comprehensive understanding of how virus pathogenesis is mediated by various viral or host factors could propose the clue for rapid detection of next pandemic potentials AIVs and the development of new therapies.
Table 2. Common determinants of viral pathogenicity.
| Protein | Position | Pathogenicity | Function | Reference (s) | |
|---|---|---|---|---|---|
| Low | High | ||||
| PB2 | 591 | Gln (Q) | Lys (K) | Increased virulence in mammals | Yamada et al 103 |
| 627 | Glu (G) | Lys (K) | Replicative ability in some mammals, including humans | Subbarao et al81 | |
| Hatta et al7 | |||||
| 701 | Asp (D) | Asn (N) | Nuclear import kinetics affecting replicative ability in mice | Li et al16 | |
| Gabriel et al104 | |||||
| PB1-F2 | 66 | Asn (N) | Ser (S) | Induction of apoptosis | Conenello et al82 |
| PA | 97 | Thr (T) | Ile (I) | Enhanced virulence | Song et al105 |
|
HA
(H5 numbering) |
Cleavage site | Single basic amino acid | Polybasic amino acid | HA cleavability, tropism | Kawaoka et al106 |
| 222 | Gln (Q) | Leu (L) | Increased virus binding to 2,6, airborne transmissibility in mammals | Yen et al107 | |
| 223 | Ser (S) | Asn (N) | |||
| 224 | Glu (G) | Ser (S) | |||
|
NA
(N1 numbering) |
223 | Thr (T) | Ile (I) | Increased virulence in mammals | LeGoff et al108 |
| 275 | His (H) | Tyr (Y) | de Vries et al109 | ||
| NS1 | 92 | Asp (D) | Glu (E) | Evasion/suppression of interferon response | Seo et al80 |
Abbreviations: HA, hemagglutinin; NA, neuraminidase; NS1, nonstructural protein 1; PA, polymerase acidic; PB1-F2, alternate open reading frame near the 5' end of the polymerase basic 1 gene; PB2, polymerase basic protein 2.
Animal Models for Influenza Virus Studies
The use of mammalian models of influenza virus infection has allowed for the extrapolation of experimental results into the context of human infection. Furthermore, the use of a single model to evaluate both pathogenicity and transmissibility has resulted in a fuller understanding of the capacity of influenza viruses to cause disease. Mice are among the most commonly used mammalian models for evaluating influenza infection and offer many advantages over other laboratory species including low cost and the broad availability of reagents and of transgenic mice with targeted gene disruptions39 77 (1 2 3). Moreover, cells in the respiratory tract of commonly used laboratory strains of mice express both SAa2,6 and SAa2,3 receptors, making them susceptible to both human and avian influenza viruses. The parameter most commonly used for measuring influenza virus disease in mice is weight loss (20–25% decrease in body weight generally requires euthanasia). In addition, blood oxygen saturation (Sa02) can be used as a metric of disease severity, as decreases correlate with increases in viral replication.84 In addition, measuring viral titers in the respiratory tissues is critical since even modest reductions in viral load can be associated with profound differences in lethality. Unfortunately, most human influenza strains do not replicate efficiently in mice without prior virus adaptation, thereby limiting the utility of the mouse model. Furthermore, many clinical signs of influenza virus infection in humans are not present in mice, and they are a poor model for virus transmission.85
Table 3. General animal models for influenza virus.
| Animal model | Advantages | Disadvantages |
|---|---|---|
| Mice | Low cost (purchase, maintenance, and reproduction) |
Not for natural host of influenza virus |
| Well-characterized genetics; microarray and knockouts | Anatomy and histology of respiratory tract and pattern of influenza virus attachment dissimilar to humans | |
| Minimal host variability and background pathology of inbred SPF strains | Most strains demonstrate hypothermia, but not real fever | |
| Availability of molecular virology/Immunology reagent | ||
| Unsuitable for live-attenuated vaccines | ||
| Unsuitable for transmission experiments | ||
| Ferrets | Pathology of influenza viral pneumonia comparable to humans | Variable outcome results depend on the age, inoculum titer, and volume |
| Anatomy and histology of respiratory tract moderately similar to humans and similar pattern of influenza virus attachment | Very limited ferret specific immunological reagents | |
| Suitable for transmission experiments | Require special caging; No SPF animals, so need to confirm Aleutian disease and initial influenza seronegative status, | |
| Apt animal size for blood and tissue sampling | Outbred, Relatively expensive | |
| Few molecular biological reagents available | ||
| Guinea Pigs | Human and Avian influenza virus isolates replicate without prior adaptation | Variable host responses, Need to confirm initial influenza seronegative status |
| Suitable for transmission experiments | Pathology of influenza viral pneumonia dissimilar to humans | |
| Usually no clinical symptoms after virus challenge, | ||
| Non-human primates | Pathology of influenza viral pneumonia comparable to humans | Expensive, Need of animal handling experience |
| May display similar clinical symptoms to humans | No SPF animals, thus need to confirm initial influenza seronegative status | |
| Anatomy and histology of respiratory tract and immune response similar to humans | May different susceptible to human influenza viruses | |
| Many available molecular biological reagents and cross-reaction with human reagents | May different disease outcome and clinical signs dependent on species, virus strain, and inoculation routes | |
| Similar immune responses as humans | Need to confirm the sialic acid receptor distribution and pattern of viral attachment in respiratory tracts |
Abbreviation: SPF, specific pathogen free.
Guinea pigs have emerged as an alternative small mammalian model for Influenza.39 77 Unlike mice, guinea pigs support replication of human influenza viruses without prior adaptation and can be used to model virus transmission to susceptible contact animals.86 However, guinea pigs are an inferior model for viral pathogenesis and also lack many clinical signs of infection that are observed in humans.87 Although not widely used for the study of influenza A viruses, the cotton rat model supports the replication of human influenza viruses without prior adaptation and is suitable for the study of both innate and adaptive immune responses.88 89 However, cotton rats lack a sneeze reflex and are not well suited to study virus transmissibility, similar to the mouse model.88
In contrast to these small mammalian models, nonhuman primates offer the ability to study virus infection in an animal that is similarly complex to humans; however, size, cost, and ethical considerations limit the use of this species in most settings.90 91
Apart from nonhuman primates, the ferret is currently the only small mammalian model available that is equally well suited to studying these aspects of influenza pathobiology. Human and avian influenza viruses replicate efficiently in the respiratory tract of ferrets without prior adaptation, and following infection with some highly pathogenic viruses, extrapulmonary spread of virus can result.92 Unlike other small mammalian models, numerous clinical signs found in humans following seasonal or avian influenza virus infection are present in the ferret following experimental inoculation with these strains through the intranasal route (Table 3). Elevated body temperatures are detected as early as 1 day postinoculation, and more virulent viruses generally elicit high fevers that can persist for several days.93 94 Nasal discharge and sneezing are also frequently observed in infected ferrets up to 1 week postinoculation with sneezing detected more frequently following seasonal influenza virus infection.95 Additionally, transmission models have been established using the ferret to measure the capacity of influenza viruses to spread to naïve contact ferrets in the presence of direct contact (i.e., pair-housing an infected and a naïve ferret) or by the spread of respiratory droplets in the absence of direct contact (i.e., separating infected and naïve ferrets with a perforated wall to allow air exchange only).95 96 97 Successful virus transmission is detected by testing contact ferrets for the presence of virus in nasal secretions and seroconversion at the end of the observation period. Daily clinical assessment of inoculated and contact ferrets further provides additional means to evaluate the effects of virus infection and transmission effectiveness. The most commonly used animal species for viral pathogenesis and antiviral drug screening of AIVs are the laboratory mouse, the ferret, and the cynomolgus macaque (Macaca fascicularis).22 In addition, ferrets and guinea pigs are used in influenza transmission studies. However, each animal model has its advantages and disadvantages in a particular purpose. Therefore, we should be considered in relation to the research question concerned to select the proper animal model.
Concluding Remarks
To date, none of the AIVs described previously have displayed efficient human-to-human dissemination beyond index cases. However, the growing number of human infections by AIVs raises concerns that one or more might eventually evolve into a variant capable of causing a global pandemic. Although factors that define virulence and host range have been commonly linked to the viral surface glycoproteins, pathogenesis and transmission of AIVs also relies on additional genes, and molecular determinants may differ among animal species.78 79 Therefore, continuous efforts are needed to define the viral virulence markers and host restriction factors responsible for zoonotic transmission.
Among the AIVs, the A/H7N9 virus is considered a likely candidate to emerge as a pandemic strain in humans. It is noteworthy that following the initial outbreaks, A(H7N9) underwent further reassortment with local avian A/H9N2 strains in mainland China, an event which may have caused the sudden increase in human cases seen in early 2014.98 As of March 2015, there have been 631 laboratory-confirmed cases of human infection with A/H7N9. In contrast, HPAI A/H5N1 viruses took more than 5 years to attain a similar number of cases. In addition, unlike HPAI A/H5N1 viruses, which cause almost 100% mortality in affected poultry, A/H7N9 and H10N6 do not cause obvious symptoms in poultry thereby increasing the difficulty of virus tracking and surveillance. Moreover, the enzootic cocirculation of A/H5N1, A/H9N2, and A/H7N9 viruses has given rise to many novel reassortants with other viruses such as H10N8,61 H10N6,99 H5N8,100 H5N6,101 and H7N6.102 The possibility that the continued circulation of these viruses will result in the generation of more virulent and human-transmissible variants, however remote at the moment, is likely under the right conditions and with appropriate hosts. Collectively, these concerns highlight the importance of continued worldwide virus monitoring and surveillance in human and animal populations alike. It should be noted that AIV infections differ from human influenza infections in many ways, including method of viral transmission, viral dissemination, clinical features, pathogenesis, and host response. Understanding the molecular determinants of these differences through careful in vitro experiments and the use of appropriate animal models may help to identify the reassortant viruses with the highest potential to cause future pandemics.
Funding
This work was partly supported by a grant from the Korea Healthcare Technology R&D project, Ministry of Health & Welfare, Republic of Korea (Grant No.: A103001), and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science (NRF-2007–0054930).
References
- 1.Palese P Shaw M L Orthomyxoviridae: the virus and their replication Philadelphia, PA: Lippincott Williams & Wilkins; 2007:p.1647–1690 [Google Scholar]
- 2.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong S, Zhu X, Li Y. et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10):e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong S, Li Y, Rivailler P. et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109(11):4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swayne D E, Radin M J, Hoepf T M, Slemons R D. Acute renal failure as the cause of death in chickens following intravenous inoculation with avian influenza virus A/chicken/Alabama/7395/75 (H4N8) Avian Dis. 1994;38(1):151–157. [PubMed] [Google Scholar]
- 6.World Health Organization (WHO) Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO, 2003–2014 Available at http://www.who.int/influenza/human_animal_interface/EN_GIP_20140124CumulativeNumberH5N1cases.pdf. Accessed on February 28, 2014.
- 7.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293(5536):1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 8.Kawaoka Y, Chambers T M, Sladen W L, Webster R G. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology. 1988;163(1):247–250. doi: 10.1016/0042-6822(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 9.Tumpey T M, Maines T R, Van Hoeven N. et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315(5812):655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 10.Matrosovich M, Suzuki T, Hirabayashi Y, Garten W, Webster R G, Klenk H D. Gangliosides are not essential for influenza virus infection. Glycoconj J. 2006;23(1-2):107–113. doi: 10.1007/s10719-006-5443-y. [DOI] [PubMed] [Google Scholar]
- 11.Rogers G N, Paulson J C, Daniels R S, Skehel J J, Wilson I A, Wiley D C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 12.Imai M, Watanabe T, Hatta M. et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486(7403):420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol. 2012;2(2):160–167. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, Zhong Y, Qin Y, Sun S, Li Z. The evolutionary pattern of glycosylation sites in influenza virus (H5N1) hemagglutinin and neuraminidase. PLoS ONE. 2012;7(11):e49224. doi: 10.1371/journal.pone.0049224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yen H L, Liang C H, Wu C Y. et al. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A. 2011;108(34):14264–14269. doi: 10.1073/pnas.1111000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Chen H, Jiao P. et al. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol. 2005;79(18):12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steel J, Lowen A C, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5(1):e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaoka Y, Krauss S, Webster R G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63(11):4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L M, Davis C T, Zhou H, Cox N J, Donis R O. Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PLoS Pathog. 2008;4(5):e1000072. doi: 10.1371/journal.ppat.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belser J A, Wadford D A, Pappas C. et al. Pathogenesis of pandemic influenza A (H1N1) and triple-reassortant swine influenza A (H1) viruses in mice. J Virol. 2010;84(9):4194–4203. doi: 10.1128/JVI.02742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina R A, García-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9(8):590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440(7083):435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, Couceiro J NS, Kelm S. et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72(9):7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garten W Klenk H Cleavage activation of the influenza virus hemagglutinin and its role in pathogenesis Basel: Karger; 2008:p.156–167 [Google Scholar]
- 25.Stieneke-Gröber A, Vey M, Angliker H. et al. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11(7):2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonnberg S, Webby R J, Webster R G. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 2013;178(1):63–77. doi: 10.1016/j.virusres.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Jong M D, Simmons C P, Thanh T T. et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrone L A, Plowden J K, García-Sastre A, Katz J M, Tumpey T M. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4(8):e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szretter K J, Gangappa S, Lu X. et al. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol. 2007;81(6):2736–2744. doi: 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Center for Infectious Disease Research and Policy (CIDRAP) Fatal H5N1 case in Canada is North America's first Minneapolis: CIDRAP; c2014 Available at http://www.cidrap.umn.edu/news-perspective/2014/01/fatal-h5n1-case-canada-northamericas-first. Accessed on February 28, 2014.
- 31.Herfst S, Schrauwen E J, Linster M. et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336(6088):1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peiris M, Yuen K Y, Leung C W. et al. Human infection with influenza H9N2. Lancet. 1999;354(9182):916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 33.Guan Y, Shortridge K F, Krauss S, Webster R G. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci U S A. 1999;96(16):9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao Y, Bolotov P, Dernovoy D. et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82(2):596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei L Kao E Hong Kong sees first case of H9N2 avian flu in four years Hong Kong: South China Morning Post; c2013. 2015. Available at http://www.scmp.com/news/hong-kong/article/1393266/86-year-old-man-infected-h9n2-avian-flu. Accessed on February 28, 2013. [Google Scholar]
- 36.Huang R, Wang A R, Liu Z H. et al. Seroprevalence of avian influenza H9N2 among poultry workers in Shandong Province, China. Eur J Clin Microbiol Infect Dis. 2013;32(10):1347–1351. doi: 10.1007/s10096-013-1888-7. [DOI] [PubMed] [Google Scholar]
- 37.Pawar S D, Tandale B V, Raut C G. et al. Avian influenza H9N2 seroprevalence among poultry workers in Pune, India, 2010. PLoS ONE. 2012;7(5):e36374. doi: 10.1371/journal.pone.0036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou P, Zhu W, Gu H. et al. Avian influenza H9N2 seroprevalence among swine farm residents in China. J Med Virol. 2014;86(4):597–600. doi: 10.1002/jmv.23869. [DOI] [PubMed] [Google Scholar]
- 39.Belser J A, Bridges C B, Katz J M, Tumpey T M. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis. 2009;15(6):859–865. doi: 10.3201/eid1506.090072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurtz J, Manvell R J, Banks J. Avian influenza virus isolated from a woman with conjunctivitis. Lancet. 1996;348(9031):901–902. doi: 10.1016/S0140-6736(05)64783-6. [DOI] [PubMed] [Google Scholar]
- 41.Centers for Disease Control and Prevention (CDC) Update: influenza activity—United States and worldwide, 2003-04 season, and composition of the 2004-05 influenza vaccine MMWR Morb Mortal Wkly Rep 20045325547–552., a1 [PubMed] [Google Scholar]
- 42.Ostrowsky B, Huang A, Terry W. et al. Low pathogenic avian influenza A (H7N2) virus infection in immunocompromised adult, New York, USA, 2003. Emerg Infect Dis. 2012;18(7):1128–1131. doi: 10.3201/eid1807.111913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avian influenza A/(H7N2) outbreak in the United Kingdom. Euro Surveill. 2007;12(5):E070531.2. [PubMed] [Google Scholar]
- 44.Nguyen-Van-Tam J S, Nair P, Acheson P. et al. Outbreak of low pathogenicity H7N3 avian influenza in UK, including associated case of human conjunctivitis. Euro Surveill. 2006;11(5):E060504.2. doi: 10.2807/esw.11.18.02952-en. [DOI] [PubMed] [Google Scholar]
- 45.Puzelli S, Di Trani L, Fabiani C. et al. Serological analysis of serum samples from humans exposed to avian H7 influenza viruses in Italy between 1999 and 2003. J Infect Dis. 2005;192(8):1318–1322. doi: 10.1086/444390. [DOI] [PubMed] [Google Scholar]
- 46.Tweed S A, Skowronski D M, David S T. et al. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis. 2004;10(12):2196–2199. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Centers for Disease Control and Prevention (CDC) . Notes from the field: highly pathogenic avian influenza A (H7N3) virus infection in two poultry workers—Jalisco, Mexico, July 2012. MMWR Morb Mortal Wkly Rep. 2012;61(36):726–727. [PubMed] [Google Scholar]
- 48.Fouchier R A, Schneeberger P M, Rozendaal F W. et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101(5):1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao R, Cao B, Hu Y. et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 50.World Health Organization(WHO) Human Infection with Avian Influenza A(H7N9) Available at http://www.wpro.who.int/outbreaks_emergencies/H7N9/en/. Accessed on March 10, 2015.
- 51.Zhang Q, Shi J, Deng G. et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 2013;341(6144):410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe T, Kiso M, Fukuyama S. et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501(7468):551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu D, Shi W, Shi Y. et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381(9881):1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 54.Rudge J W, Coker R. Human to human transmission of H7N9. BMJ. 2013;347:f4730. doi: 10.1136/bmj.f4730. [DOI] [PubMed] [Google Scholar]
- 55.Yi L, Guan D, Kang M. et al. Family clusters of avian influenza A H7N9 virus infection in Guangdong Province, China. J Clin Microbiol. 2015;53(1):22–28. doi: 10.1128/JCM.02322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.World Health Organization(WHO) WHO risk assessment of human infections with avian influenza A(H7N9) virus Available at http://www.who.int/influenza/human_animal_interface/influenza_h7n9/RiskAssessment_H7N9_23Feb20115.pdf. Accessed on February 23, 2015.
- 57.Centers for Disease Control and Prevention H7N9 case detected in Malaysia Available at http://www.cdc.gov/flu/news/h7n9-case-malaysia.htm. Accessed on February 13, 2014.
- 58.Yuan J, Zhang L, Kan X. et al. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis. 2013;57(9):1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 59.Pan American Health Organization (PAHO) (2014) Avian influenza virus A (H10N7) circulating among humans in Egypt c2007. Available at http://www1.paho.org/hq/dmdocuments/2010/Avian_Influenza_Egypt_070503.pdf. Accessed on February 28, 2014.
- 60.Arzey G G, Kirkland P D, Arzey K E. et al. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis. 2012;18(5):814–816. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen H, Yuan H, Gao R. et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383(9918):714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 62.Health and Family Planning Commision of Jiangxi Province Jiangxi 1 new confirmed cases of human infection with avian influenza H10N8 Available at http://www.jxwst.gov.cn/gzdt/201402/t20140213_308109.htm. Accessed on February 28, 2014.
- 63.Castrucci M R, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster R G. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193(1):503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 64.Ma W, Kahn R E, Richt J A. The pig as a mixing vessel for influenza viruses: Human and veterinary implications. J Mol Genet Med. 2008;3(1):158–166. [PMC free article] [PubMed] [Google Scholar]
- 65.Lipatov A S, Kwon Y K, Sarmento L V. et al. Domestic pigs have low susceptibility to H5N1 highly pathogenic avian influenza viruses. PLoS Pathog. 2008;4(7):e1000102. doi: 10.1371/journal.ppat.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He L, Zhao G, Zhong L. et al. Isolation and characterization of two H5N1 influenza viruses from swine in Jiangsu Province of China. Arch Virol. 2013;158(12):2531–2541. doi: 10.1007/s00705-013-1771-y. [DOI] [PubMed] [Google Scholar]
- 67.Li H, Yu K, Yang H. et al. Isolation and characterization of H5N1 and H9N2 influenza viruses from pigs in China. Chin J Prev Vet Med. 2003;26:1–6. [Google Scholar]
- 68.Shi W F, Gibbs M J, Zhang Y Z. et al. Genetic analysis of four porcine avian influenza viruses isolated from Shandong, China. Arch Virol. 2008;153(1):211–217. doi: 10.1007/s00705-007-1083-1. [DOI] [PubMed] [Google Scholar]
- 69.Zhu Q, Yang H, Chen W. et al. A naturally occurring deletion in its NS gene contributes to the attenuation of an H5N1 swine influenza virus in chickens. J Virol. 2008;82(1):220–228. doi: 10.1128/JVI.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takano R, Nidom C A, Kiso M. et al. A comparison of the pathogenicity of avian and swine H5N1 influenza viruses in Indonesia. Arch Virol. 2009;154(4):677–681. doi: 10.1007/s00705-009-0353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nidom C A, Takano R, Yamada S. et al. Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis. 2010;16(10):1515–1523. doi: 10.3201/eid1610.100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi Y K, Nguyen T D, Ozaki H. et al. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J Virol. 2005;79(16):10821–10825. doi: 10.1128/JVI.79.16.10821-10825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peiris J SM, Guan Y, Markwell D, Ghose P, Webster R G, Shortridge K F. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol. 2001;75(20):9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cong Y L Pu J Liu Q F et al. Antigenic and genetic characterization of H9N2 swine influenza viruses in China J Gen Virol 200788(Pt 7):2035–2041. [DOI] [PubMed] [Google Scholar]
- 75.Yu H, Zhou Y J, Li G X. et al. Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Vet Microbiol. 2011;149(1–2):254–261. doi: 10.1016/j.vetmic.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 76.Nichol K L, Treanor J J. Vaccines for seasonal and pandemic influenza. J Infect Dis. 2006;194 02:S111–S118. doi: 10.1086/507544. [DOI] [PubMed] [Google Scholar]
- 77.O'Donnell C D, Subbarao K. The contribution of animal models to the understanding of the host range and virulence of influenza A viruses. Microbes Infect. 2011;13(5):502–515. doi: 10.1016/j.micinf.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459(7249):931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen H, Bright R A, Subbarao K. et al. Polygenic virulence factors involved in pathogenesis of 1997 Hong Kong H5N1 influenza viruses in mice. Virus Res. 2007;128(1-2):159–163. doi: 10.1016/j.virusres.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 80.Seo S H, Hoffmann E, Webster R G. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8(9):950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 81.Subbarao E K, London W, Murphy B R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67(4):1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conenello G M, Zamarin D, Perrone L A, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3(10):1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zamarin D, García-Sastre A, Xiao X, Wang R, Palese P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005;1(1):e4. doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sidwell R W, Huffman J H, Gilbert J. et al. Utilization of pulse oximetry for the study of the inhibitory effects of antiviral agents on influenza virus in mice. Antimicrob Agents Chemother. 1992;36(2):473–476. doi: 10.1128/aac.36.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schulman J L, Kilbourne E D. Experimental transmission of influenza virus infection in mice. i. the period of transmissibility. J Exp Med. 1963;118:257–266. doi: 10.1084/jem.118.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lowen A C, Mubareka S, Tumpey T M, García-Sastre A, Palese P. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A. 2006;103(26):9988–9992. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Hoeven N, Belser J A, Szretter K J. et al. Pathogenesis of 1918 pandemic and H5N1 influenza virus infections in a guinea pig model: antiviral potential of exogenous alpha interferon to reduce virus shedding. J Virol. 2009;83(7):2851–2861. doi: 10.1128/JVI.02174-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eichelberger M C. The cotton rat as a model to study influenza pathogenesis and immunity. Viral Immunol. 2007;20(2):243–249. doi: 10.1089/vim.2007.0017. [DOI] [PubMed] [Google Scholar]
- 89.Sadowski W, Semkow R, Wilczyński J, Kruś S, Kańtoch M. The cotton rat (Sigmodon hispidus) as an experimental model for studying viruses in human respiratory tract infections. I. Para-influenza virus type 1, 2 and 3, adenovirus type 5 and RS virus [article in Polish]. Med Dosw Mikrobiol. 1987;39(1):33–42. [PubMed] [Google Scholar]
- 90.Kobasa D, Jones S M, Shinya K. et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445(7125):319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 91.Rimmelzwaan G F Kuiken T van Amerongen G Bestebroer T M Fouchier R A Osterhaus A D A primate model to study the pathogenesis of influenza A (H5N1) virus infection Avian Dis 200347(3, Suppl):931–933. [DOI] [PubMed] [Google Scholar]
- 92.Belser J A, Szretter K J, Katz J M, Tumpey T M. Use of animal models to understand the pandemic potential of highly pathogenic avian influenza viruses. Adv Virus Res. 2009;73:55–97. doi: 10.1016/S0065-3527(09)73002-7. [DOI] [PubMed] [Google Scholar]
- 93.Sweet C, Bird R A, Cavanagh D, Toms G L, Collie M H, Smith H. The local origin of the febrile response induced in ferrets during respiratory infection with a virulent influenza virus. Br J Exp Pathol. 1979;60(3):300–308. [PMC free article] [PubMed] [Google Scholar]
- 94.Zitzow L A, Rowe T, Morken T, Shieh W J, Zaki S, Katz J M. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J Virol. 2002;76(9):4420–4429. doi: 10.1128/JVI.76.9.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maines T R, Chen L M, Matsuoka Y. et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A. 2006;103(32):12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herlocher M L, Elias S, Truscon R. et al. Ferrets as a transmission model for influenza: sequence changes in HA1 of type A (H3N2) virus. J Infect Dis. 2001;184(5):542–546. doi: 10.1086/322801. [DOI] [PubMed] [Google Scholar]
- 97.Yen H L, Lipatov A S, Ilyushina N A. et al. Inefficient transmission of H5N1 influenza viruses in a ferret contact model. J Virol. 2007;81(13):6890–6898. doi: 10.1128/JVI.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Meng Z, Han R, Hu Y. et al. Possible pandemic threat from new reassortment of influenza A(H7N9) virus in China. Euro Surveill. 2014;19(6):pii 20699. doi: 10.2807/1560-7917.es2014.19.6.20699. [DOI] [PubMed] [Google Scholar]
- 99.Ma C, Lam T T-Y, Chai Y. et al. Emergence and evolution of H10 subtype influenza viruses in poultry in China. J Virol. 2015;89(7):3534–3541. doi: 10.1128/JVI.03167-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao K, Gu M, Zhong L. et al. Characterization of three H5N5 and one H5N8 highly pathogenic avian influenza viruses in China. Vet Microbiol. 2013;163(3–4):351–357. doi: 10.1016/j.vetmic.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 101.Qi X, Cui L, Yu H, Ge Y, Tang F. Whole-genome sequence of a reassortant H5N6 avian influenza virus isolated from a live poultry market in China, 2013. Genome Announc. 2014;2(5):706–714. doi: 10.1128/genomeA.00706-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lam T T-Y, Zhou B, Wang J. et al. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature. 2015;522(7554):102–105. doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 103.Yamada S, Hatta M, Staker B L. et al. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010;6(8):e1001034. doi: 10.1371/journal.ppat.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gabriel G, Herwig A, Klenk H D. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008;4(2):e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Song M S, Pascua P N, Lee J H. et al. The polymerase acidic protein gene of influenza a virus contributes to pathogenicity in a mouse model. J Virol. 2009;83(23):12325–12335. doi: 10.1128/JVI.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kawaoka Y, Webster R G. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc Natl Acad Sci U S A. 1988;85(2):324–328. doi: 10.1073/pnas.85.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yen H L, Peiris J S. Mapping antibody epitopes of the avian H5N1 influenza virus. PLoS Med. 2009;6(4):e1000064. doi: 10.1371/journal.pmed.1000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.LeGoff J, Rousset D, Abou-Jaoudé G. et al. I223R mutation in influenza A(H1N1)pdm09 neuraminidase confers reduced susceptibility to oseltamivir and zanamivir and enhanced resistance with H275Y. PLoS ONE. 2012;7(8):e37095. doi: 10.1371/journal.pone.0037095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Vries E, de Vries R P, Wienholts M J. et al. Influenza A virus entry into cells lacking sialylated N-glycans. Proc Natl Acad Sci U S A. 2012;109(19):7457–7462. doi: 10.1073/pnas.1200987109. [DOI] [PMC free article] [PubMed] [Google Scholar]


