Abstract
Enterovirus D68 (EV-D68) is a member of the species Enterovirus D in the genus Enterovirus of the Picornaviridae family. EV-D68 was first isolated in the United States in 1962 and is primarily an agent of respiratory disease. Infections with EV-D68 have been rarely reported until recently, when reports of EV-D68 associated with respiratory disease increased notably worldwide. An outbreak in 2014 in the United States, for example, involved more than 1,000 cases of severe respiratory disease that occurred across almost all states. Phylogenetic analysis of all EV-D68 sequences indicates that the circulating strains of EV-D68 can be classified into two lineages, lineage 1 and lineage 2. In contrast to the prototype Fermon strain, all circulating strains have deletions in their genomes. Respiratory illness associated with EV-D68 infection ranges from mild illness that just needs outpatient service to severe illness requiring intensive care and mechanical ventilation. To date, there are no specific medicines and vaccines to treat or prevent EV-D68 infection. This review provides a detailed overview about our current understanding of EV-D68-related virology, epidemiology and clinical syndromes, pathogenesis, and laboratory diagnostics.
Keywords: enterovirus D68, respiratory infection, molecular epidemiology, lineage
A new picornavirus was isolated from the throat swab samples from children with acute lower respiratory tract illness in the United States during October–December 1962.1 As the properties of this virus, which was initially named “Fermon virus,” were similar to those of the enterovirus subgroup of the human picornavirus, the virus was designated as enterovirus 68. Based on the latest proposal by the International Committee of Taxonomy of Viruses (ICTV), this virus can be classified into species D of the genus Enterovirus of the Picornaviridae family and is therefore now designated as Enterovirus D68 (EV-D68). After a reevaluation of genetic, immunogenic, and receptor use properties, human rhinovirus 87 and EV-D68 were assigned to the same picornavirus serotype,2 3 indicating that EV-D68 has biological features of both enteroviruses and rhinoviruses.
Infections with EV-D68 have been rarely reported. Up until recently, during 36 years of EV surveillance, only 26 detections of EV-D68 had been reported in the United States.4 However, EV-D68 infections increased notably worldwide in recent years.5 6 7 8 9 10 11 12 13 14 15 16 Particularly, outbreaks caused by EV-D68 infection in 2014 in the United States have raised a great public health concern for a pandemic.17 18
In contrast to other EVs, EV-D68 is primarily an agent of respiratory disease,19 although neurological disease such as acute flaccid paralysis linked to EV-D68 infection was also noted in the 2014 outbreak.20 21 22 23 24 However, the virus' pathology and related characteristics are not fully understood. In this review, we focus on EV-D68-related respiratory infections, detailing virology, epidemiology and clinical syndromes, pathogenesis, and laboratory diagnostics.
Virology
Genome Structure
EV-D68 is a positive-sense, single-stranded RNA virus of approximately 7,300 nucleotides. The viral genome consists of the genome-linked protein VPg at the 5′ end, a 5′-untranslated region (UTR), a single open reading frame coding for a polyprotein, a 3′-UTR, and a poly (A) tail. The polyprotein is first cleaved into three parts: P1, P2, and P3 by its proteases 2A and 3C. P1 is cleaved into four viral capsid proteins VP1 to VP4, and P2 and P3 are cleaved into seven nonstructural proteins involved in protein processing and genome replication, including 2A-2C and 3A-3D25 (Fig. 1).
Fig. 1.
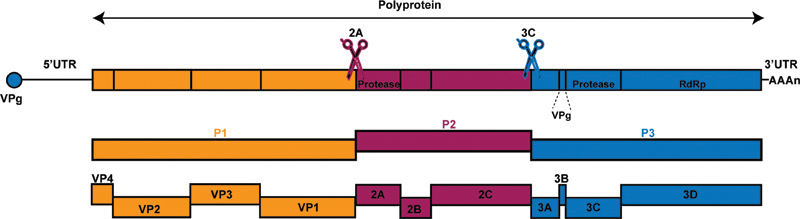
Schematic of the EV-D68 genome. The EV-D68 genome contains the genome-linked protein VPg at the 5′ end, a 5′ untranslated region (UTR), a single open reading frame codes a polyprotein, a 3′ UTR, and the poly (A) tail. The polyprotein is cleaved into 11 individual viral proteins by two viral proteases, 2Apro and 3Cpro, during translation. Proteases and RNA-dependent RNA polymerase (RDRP) are indicated in the schematic.
The VP1 gene is used to classify EVs into different serotypes or genotypes.26 Based on the phylogenetic tree generated by VP1 nucleotide sequences, the circulating strains of EV-D68 have been classified into two or three genetic groups by different researchers. These groups have been designated as clades A–C,10 27 28 29 clusters 1–3,9 lineages 1–3,30 31 or lineages 1–2.32 33 Based on all available partial sequences of VP1 and the complete genome sequences in each branch, the latest phylogenetic tree shows that the circulating strains of EV-D68 can be classified into two groups, that is, linages 1 and 2. The two lineages are cocirculating now. Accordingly, we labeled all available EV-D68 sequences as lineages 1 and 2 in this review (Fig. 2).
Fig. 2.
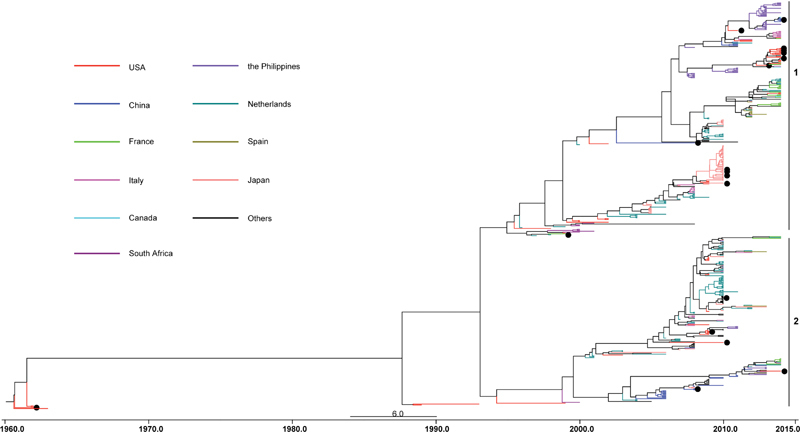
Maximum clade credibility tree based on Bayesian timescale phylogenetic analysis of EV-D68. The tree was inferred using partial VP1 sequences of EV-D68 strains worldwide available in GenBank as of March 23, 2015. These sequences are 339 nucleotides in length and correspond to nucleotide positions 2,521 to 2,859 of the EV-D68 prototype strain Fermon [AY426531]. The two lineages are marked by vertical lines. The strains that have complete genome sequences are indicated by black circles.
Due to the lack of proof-reading of RNA-dependent RNA polymerase during viral genome replication, mutations frequently occur and can accumulate during EV-D68 propagation, which leads to variations in the viral genome. Alignment analysis of all available complete sequences shows the occurrence of three distinct deletions in the viral genome among the circulating EV-D68 strains. Compared with the prototype Fermon strain, all circulating strains have a 23 nt deletion at position 681 to 703 in the 5′-UTR. The strains of lineage 1 have an additional 11 nt deletion at position 705 to 726 in the 5′-UTR, while strains of lineage 2 have an additional 3 nt deletion at position 2,806 to 2,808 in VP1 (Fig. 3A, B). These deletions were reported by Tokarz et al (as clade C and A)10 and Imamura and Oshitani (as lineages 1 and 3).31 One strain (CA/AFP/11–1767, GenBank accession number KM892501), which belongs to lineage 1, has neither the second deletion in the 5′-UTR nor the deletion in VP1, and is considered to be the intermediate in EV-D68 evolution (Fig. 3C).
Fig. 3.
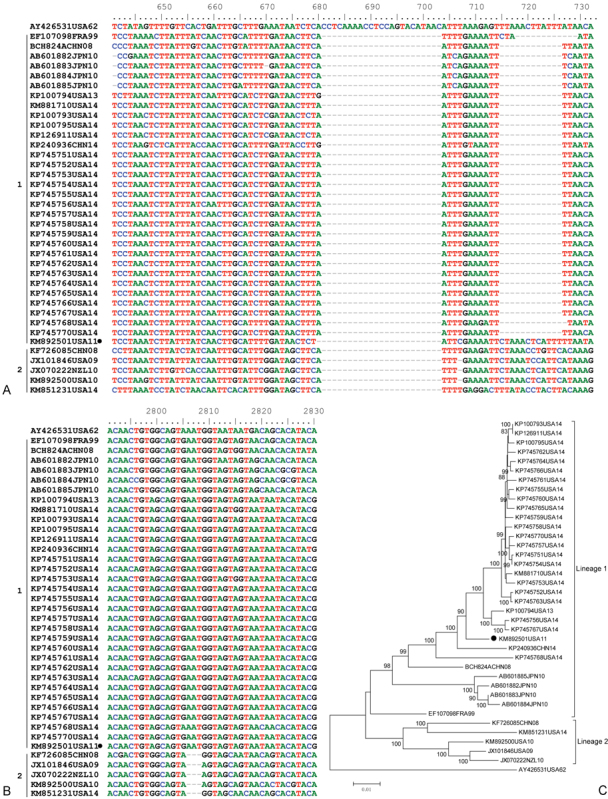
Nucletide alignment of all available complete EV-D68 genomes in GenBank. Nucletides are numbered relative to the start of the Fermon strain (AY426531) of EV-D68. The strains are indicated by Genbank accession numbers followed by state abbreviation and collection year. (A) Nucletide alignment of 5′UTR. Compared with the Fermon strain, all circulating strains have a 23 nt deletion at position 681–703. The strains of lineage 1 have an additional 11 nt deletion at position 705 to 726. (B) Nucletide alignment of VP1. Compared with the Fermon strain, strains of lineage 2 had 3 nt have a deletion at position 2,806 to 2,808. (C) Phylogenetic analysis of all available EV-D68 strains based on complete genomes. The trees, with 1,000 bootstrap replicates, were generated by using the neighbor-joining algorithm in MEGA 4.0. The strain has only one deletion as indicated by black circles.
Biological Properties
Although the prototype Fermon strain of EV-D68 was described as acid stable in the original report,1 Fermon and other EV-D68 strains were reported to be acid-labile afterward.2 19 Oberste et al demonstrated that EV-D68 strains grew better at 33°C than at 37°C.19 Acid lability and a lower optimum growth temperature indicate that EV-D68 shares similar biological features as human rhinoviruses.19 For example, EV-D68 can replicate in cells from different tissue origins susceptible to EVs and rhinoviruses, including rhesus primary kidney (RMK), human fetal diploid kidney (HFKD), human embryonic lung fibroblast (MRC5), human embryonic lung epithelium (A549), human rhabdomyosarcoma (RD), and some human leukocyte cell lines (MOLT, Jurkat, U937).19 34
The structure of EV-D68 has considerable similarities to those of rhinoviruses.35 The capsid of EV-D68 is composed of 60 copies of each of four structural proteins: VP1, VP2, VP3, and VP4. A deep depression (canyon) surrounds a pentagonal icosahedral vertex and is considered to be a receptor binding site. The canyon of EV-D68 is shallower and narrower than the canyon of other EVs, which indicates that EV-D68 may not use immunoglobulin-like receptors.35
Epidemiology
Although EV-D68 was identified around 50 years ago, it has been considered one of the most rarely reported serotypes until 2005.4 As part of respiratory infection surveillance during 2004–2005, Wang et al identified seven EV-D68 cases from throat-swab specimens at the Marine Corps Recruit Depot, San Diego.36 This was the first report suggesting that clonal EV-D68 was endemic in a single site. Nineteen further EV-D68 cases were isolated in October–November 2008 in France37 and 17 EV-D68 infections were detected from patients with acute respiratory tract infections (ARTIs) in Italy during the period of October 1, 2008, to September 30, 2009.38 39
EV-D68 infections attracted even more global attention when several clusters of severe respiratory illnesses occurred in Asia, Europe, and the United States during 2008–20105 6 7 8 40 (Fig. 4). In Asia, the first report about severe EV-D68 infections came from the Philippines in 2011,5 although 128 cases of EV-D68—the largest number of patients reported at that time6 30 40—were identified in 2010 in Japan. EV-D68 has been detected in Chinese adults with ARTIs since 2006.11 33 41 In Thailand, EV-D68 emerged after 2009.9 In Europe, EV-D68 infections were reported in the Netherlands,8 42 43 44 45 France,23 27 46 UK,32 and Italy.15 28 In the United States, EV-D68 cases were detected in Georgia, Pennsylvania, Arizona, and New York City during 2009–2010.7 10 Subsequent reports have documented EV-D68 detections in New Zealand47 and several African countries.10 29 The largest and most widespread EV-D68 outbreak occurred in the United States in 2014. This outbreak involved more than 1,000 cases of severe respiratory disease from almost all states in the United States.48
Fig. 4.
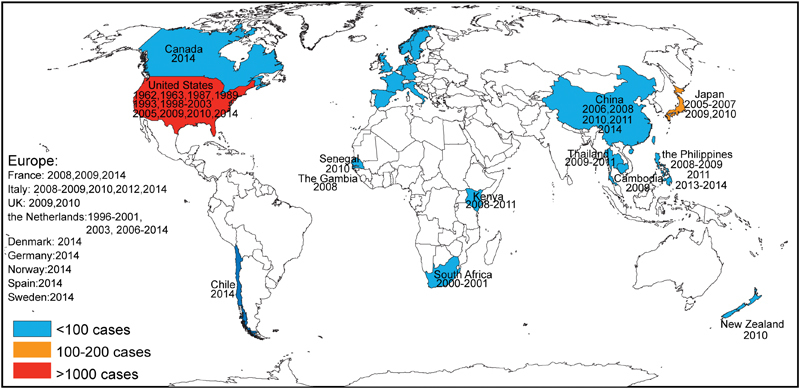
World map depicting EV-D68 prevalence (country and year). Red indicates countries in which more than 1,000 cases have been detected, orange indicates countries in which 100 to 200 cases have been detected, and blue indicates countries in which less than100 cases have been detected.
The outbreak in the United States initiated a new global climax of EV-D68 detection. In addition to the countries listed earlier, EV-D68 infections have been reported in many countries for the first time, including Germany,49 Spain,50 Sweden,51 Denmark,16 Norway,13 Cambodia,52 Canada,14 and Chile53 (Fig. 4).
EV-D68 infections involve all age groups and seem to have no sex predilection.8 42 Currently, most reports about EV-D68 infections derive from children. However, a study from the Netherlands involving all age groups found that the highest prevalence of EV68-positive patients was among persons aged 50 to 59 years.42
The seasonality of the EV-68 infections falls within or later than the typical enterovirus season (summer–fall).7 In the northern hemisphere, the peak of EV-D68 detections has occurred from August to October11 30 44 54 and infection has lasted till the winter.28 32 33 44 In the southern hemisphere, the peak incidence of EV-D68 was observed in June (winter) in New Zealand.47 In South Africa, EV-D68 was detected from April to July (autumn–winter).10 In Chile, the two EV-D68 positive cases were detected in September and October (spring).53 In Thailand, EV-D68 was detected in summer and rainy season,9 while in the Philippines, most of EV-D68 cases were detected in dry season,5 55 some were detected in rainy season in 2011.56
Clinical Features
As an agent of respiratory disease, EV-D68 infections can cause both upper respiratory tract infections (URTIs) and lower respiratory tract infections (LRTIs). An epidemic of EV-D68 usually coincides with an increased incidence of influenza-like illness42 or severe respiratory illness.54 In contrast to EV-D68-negative patients, EV-D68-positive patients have significantly more cough, dyspnea, and wheezing.5 7 42 57
The respiratory illness associated with EV-D68 infections ranges from relatively mild illness that just needs outpatient services to severe illness requiring intensive care and mechanical ventilation.7 EV-D68 has been associated with severe LRTIs in infants and young children, and frequently occurs in these patient population with complication.38 49 Some patients have been admitted to the pediatric intensive care units, some patients required positive airway pressure ventilation, and some patients required even mechanical ventilation.58 Up to now, at least 20 fatal cases of EV-D68 have been reported.7 32 59 60 Physicians should be alerted to acute, unexplained severe respiratory illness caused by EV-D68. Although EV-D68 usually causes URTIs in adults, some patients display mild symptoms.11 38 49 52 Pharyngeal congestion, headache, and myalgia were prominent symptoms in EV-D68-positive adults, but the incidence of these symptoms had no significantly difference from that of other EVs.11
EV-D68 infections have been significantly associated with asthma.32 For example, EV-D68 was detected in a cluster of asthma patients in 2010 in Japan.40 Vice versa, children with asthma are also more vulnerable to EV-D68 infections.12 In the initial reports about EV-D68 outbreak in Kansas City and Chicago in 2014, 68 and 73% of patients had a previous history of asthma or wheezing.54
Pathogenesis
Receptor
EV-D68 requires sialic acid (SA) as a cellular receptor for attachment and entry.61 Imamura et al investigated SA-binding specificities of EV-D68 and found that EV-D68 had a stronger affinity for α2,6-linked SAs than for α2,3-linked SA.62 As α2,6-linked SAs are predominant SA distributing in the human upper respiratory tract,63 these findings suggest that EV-D68 may have more affinity for the upper respiratory tract than for the lower respiratory tract. However, multiple reports about EV-D68 associating with LRTIs indicate that an additional receptor may be used by this virus.
Decay-accelerating factor (DAF) is a cellular receptor of EV-D70, which belongs to the same species as EV-D68.64 Blomqvist et al found that the cytopathic effects of EV-D68 were inhibited by monoclonal antibodies to DAF, which suggests that DAF might also be a cellular receptor of EV-D68.2
Viremia
Smura et al reported that EV-D68 had an unusually wide leukocyte tropism34 which provides a mechanism for EV-D68 to be transported to secondary target organs through viremia. Imamura et al detected EV-D68 using RT-PCR method in the serum of pediatric patients (1–4 years old) whose nasopharyngeal specimens were positive.65 These results suggest that after the primary infection of the respiratory tract, EV-D68 may cause systemic manifestations by viremia.
Immune Response
Although EV-D68 is a globally emerging pathogen, the molecular basis for EV-D68 pathogenesis is unclear. Similar to other enteroviruses and rhinoviruses, the innate and adaptive host immune responses play an important role in the pathogenesis of EV-D68 infection. On one hand, the innate immune system activates antiviral and immunomodulatory effects mediated by pattern recognition receptors, Toll-like receptors (TLRs), and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs).66 On the other hand, enteroviruses evolved various strategies to invade innate immune defenses for their replication.66 Our previous study demonstrated that 3Cpro of EV-D68 could cleave the TIR domain-containing adaptor inducing β interferon (TRIF) to inhibit TLR3-mediated innate immune responses.67 The interplay of EV-D68 and the TLR3-mediated pathway may represent an interface which determines the outcome of EV-D68 infection.
Neutralizing antibodies (NAbs) are pivotal to protecting the host against EV-D68 infection. Smura et al assessed NAbs against the Fermon strain in the Finnish population.34 They found that although Finnish people had high seroprevalence, the mean antibody levels against EV-68 showed a decreasing temporal trend from 1983 to 2002. As a rarely reported virus, children are less likely to build up specific immunity.12 The low levels of NAbs and limited cross-reactivity against the antisera of the circulating EV-D68 strains might be reasons for EV-D68 outbreaks in recent years.62 More studies are needed to comprehensively understand the pathogenesis of EV-D68.
Laboratory Diagnosis
At the initial stages of EV-D68 infections, patients usually present with influenza-like symptoms. Clinical manifestations alone cannot distinguish EV-D68 from other respiratory virus infections, and up to now, there are no commercially available diagnostic kits to identify EV-D68 from other enteroviruses or rhinoviruses. EV-D68 can only be diagnosed by specific tests on specimens collected from patients. The US Centers for Disease Control and Prevention (CDC) developed a new, faster real-time PCR tests for detecting EV-D68 viral RNA in October 2014.68 Wylie et al and Piralla et al subsequently developed other RT-PCR assays.59 69 These methods provide a way to rapidly test for EV-D68 infection, although final diagnosis still relies on partial sequencing of the structural protein gene VP4-VP2 or VP1 region.70
Antivirals
To date, there are no specific drugs and vaccines to treat and prevent EV-D68 infection. Liu et al reported that pleconaril, a capsid-binding compound that had activity against rhinoviruses and some enteroviruses, could bind to a hydrophobic drug-binding pocket in VP1 of the Fermon strain.35 The CDC tested pleconaril, pocapavir, and vapendavir which were demonstrated to have variable activities against some enteroviruses and found that they had no activity against circulating strains of EV-D68 at clinically relevant concentrations.57 71
The antiviral activities of several compounds, which target the nonstructural proteins of EV-D68, have been demonstrated in vitro. By screening the National Institutes of Health Clinical Collections 2 library of 281 compounds, Ulferts et al found that fluoxetine HCl, a selective serotonin reuptake inhibitor, could inhibit replication of EV-D68 in HeLa cells.71 72 Nonstructural protein 2C is the target of this compound.71 72 Gao et al screened the U.S. Drug Collection (1,040 compounds) and the International Drug Collection (240 compounds) and reported that itraconazole, a triazole antifungal agent, could inhibit replication of EV-D68 in RD cells by targeting nonstructural protein 3A.73 Wang et al found that peptidyl aldehyde NK-1.8k could suppress EV-D68 by targeting protease 3C.74 These compounds have broad-spectrum antiviral activity against enteroviruses. It is difficult to evaluate the in vivo activity of these compounds for lack of an animal model of EV-D68.
In conclusion, here we provide a detailed overview of virology, epidemiology and clinical syndromes, pathogenesis, and laboratory diagnostics of EV-D68, a globally emerging pathogen. Our review highlights that to date, most reports on EV-D68 have concentrated on its etiology. Little research has focused on the pathogenesis of EV-D68. For example, it is not known how EV-D68 causes other system symptoms such as neurological illness, as EV-D68 has not been detected in the cerebrospinal fluid of patients.57 It is also unclear how immune responses are induced at the site of infection. An animal model has not been developed. Therefore, more studies are needed to comprehensively understand the pathogenesis of EV-D68.
References
- 1.Schieble J H, Fox V L, Lennette E H. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol. 1967;85(2):297–310. doi: 10.1093/oxfordjournals.aje.a120693. [DOI] [PubMed] [Google Scholar]
- 2.Blomqvist S, Savolainen C, Råman L, Roivainen M, Hovi T. Human rhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J Clin Microbiol. 2002;40(11):4218–4223. doi: 10.1128/JCM.40.11.4218-4223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishiko H, Miura R, Shimada Y. et al. Human rhinovirus 87 identified as human enterovirus 68 by VP4-based molecular diagnosis. Intervirology. 2002;45(3):136–141. doi: 10.1159/000065866. [DOI] [PubMed] [Google Scholar]
- 4.Khetsuriani N Lamonte-Fowlkes A Oberst S Pallansch M A; Centers for Disease Control and Prevention. Enterovirus surveillance—United States, 1970-2005 MMWR Surveill Summ 20065581–20. [PubMed] [Google Scholar]
- 5.Imamura T, Fuji N, Suzuki A. et al. Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg Infect Dis. 2011;17(8):1430–1435. doi: 10.3201/eid1708.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaida A, Kubo H, Sekiguchi J. et al. Enterovirus 68 in children with acute respiratory tract infections, Osaka, Japan. Emerg Infect Dis. 2011;17(8):1494–1497. doi: 10.3201/eid1708.110028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (CDC) . Clusters of acute respiratory illness associated with human enterovirus 68—Asia, Europe, and United States, 2008-2010. MMWR Morb Mortal Wkly Rep. 2011;60(38):1301–1304. [PubMed] [Google Scholar]
- 8.Rahamat-Langendoen J, Riezebos-Brilman A, Borger R. et al. Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections. J Clin Virol. 2011;52(2):103–106. doi: 10.1016/j.jcv.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 9.Linsuwanon P, Puenpa J, Suwannakarn K. et al. Molecular epidemiology and evolution of human enterovirus serotype 68 in Thailand, 2006-2011. PLoS ONE. 2012;7(5):e35190. doi: 10.1371/journal.pone.0035190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tokarz R, Firth C, Madhi S A. et al. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol. 2012;93(Pt 9):1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Z, Gonzalez R, Wang Z. et al. Coxsackievirus A21, enterovirus 68, and acute respiratory tract infection, China. Emerg Infect Dis. 2012;18(5):821–824. doi: 10.3201/eid1805.111376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enterovirus D68: the unexpected guest. Lancet Infect Dis. 2014;14(11):1023. doi: 10.1016/S1473-3099(14)70968-5. [DOI] [PubMed] [Google Scholar]
- 13.Bragstad K, Jakobsen K, Rojahn A E. et al. High frequency of enterovirus D68 in children hospitalised with respiratory illness in Norway, autumn 2014. Influenza Other Respi Viruses. 2015;9(2):59–63. doi: 10.1111/irv.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drews S J, Simmonds K, Usman H R. et al. Characterization of enterovirus activity, including that of enterovirus D68, in pediatric patients in Alberta, Canada, in 2014. J Clin Microbiol. 2015;53(3):1042–1045. doi: 10.1128/JCM.02982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esposito S, Zampiero A, Ruggiero L, Madini B, Niesters H, Principi N. Enterovirus D68-associated community-acquired pneumonia in children living in Milan, Italy. J Clin Virol. 2015;68:94–96. doi: 10.1016/j.jcv.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 16.Midgley S E, Christiansen C B, Poulsen M W, Hansen C H, Fischer T K. Emergence of enterovirus D68 in Denmark, June 2014 to February 2015. Euro Surveill. 2015;20(17):21105. doi: 10.2807/1560-7917.es2015.20.17.21105. [DOI] [PubMed] [Google Scholar]
- 17.Brown B A Nix W A Sheth M Frace M Oberste M S Seven Strains of Enterovirus D68 Detected in the United States during the 2014 Severe Respiratory Disease Outbreak Genome Announc 201426. pii: e01201–e01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson R. Outbreaks of enterovirus D68 continue across the USA. Lancet Respir Med. 2014;2(10):791. doi: 10.1016/s2213-2600(14)70190-0. [DOI] [PubMed] [Google Scholar]
- 19.Oberste M S Maher K Schnurr D et al. Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses J Gen Virol 200485(Pt 9):2577–2584. [DOI] [PubMed] [Google Scholar]
- 20.Enterovirus D. Enterovirus D68: a new cause of acute paralysis? Arch Dis Child. 2015;100(6):541. doi: 10.1136/archdischild-2015-308763. [DOI] [PubMed] [Google Scholar]
- 21.Fyfe I. Infectious disease: neurological disease in children linked to enterovirus D68. Nat Rev Neurol. 2015;11(3):124. doi: 10.1038/nrneurol.2015.18. [DOI] [PubMed] [Google Scholar]
- 22.Greninger A L, Naccache S N, Messacar K. et al. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012-14): a retrospective cohort study. Lancet Infect Dis. 2015;15(6):671–682. doi: 10.1016/S1473-3099(15)70093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang M, Mirand A, Savy N. et al. Acute flaccid paralysis following enterovirus D68 associated pneumonia, France, 2014. Euro Surveill. 2014;19(44):20952. doi: 10.2807/1560-7917.es2014.19.44.20952. [DOI] [PubMed] [Google Scholar]
- 24.Messacar K, Schreiner T L, Maloney J A. et al. A cluster of acute flaccid paralysis and cranial nerve dysfunction temporally associated with an outbreak of enterovirus D68 in children in Colorado, USA. Lancet. 2015;385(9978):1662–1671. doi: 10.1016/S0140-6736(14)62457-0. [DOI] [PubMed] [Google Scholar]
- 25.Racaniello V R. Philadelphia, PA: Lippincott Williams & Wilkins; 2013. Picornarviridae: the viruses and their replication. [Google Scholar]
- 26.Oberste M S, Maher K, Kilpatrick D R, Pallansch M A. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73(3):1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renois F, Bouin A, Andreoletti L. Enterovirus 68 in pediatric patients hospitalized for acute airway diseases. J Clin Microbiol. 2013;51(2):640–643. doi: 10.1128/JCM.02640-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piralla A, Girello A, Grignani M. et al. Phylogenetic characterization of enterovirus 68 strains in patients with respiratory syndromes in Italy. J Med Virol. 2014;86(9):1590–1593. doi: 10.1002/jmv.23821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opanda S M, Wamunyokoli F, Khamadi S, Coldren R, Bulimo W D. Genetic diversity of human enterovirus 68 strains isolated in Kenya using the hypervariable 3′-end of VP1 gene. PLoS ONE. 2014;9(7):e102866. doi: 10.1371/journal.pone.0102866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda T, Mizuta K, Abiko C. et al. Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol Immunol. 2012;56(2):139–143. doi: 10.1111/j.1348-0421.2012.00411.x. [DOI] [PubMed] [Google Scholar]
- 31.Imamura T, Oshitani H. Global reemergence of enterovirus D68 as an important pathogen for acute respiratory infections. Rev Med Virol. 2015;25(2):102–114. doi: 10.1002/rmv.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauinger I L, Bible J M, Halligan E P, Aarons E J, MacMahon E, Tong C Y. Lineages, sub-lineages and variants of enterovirus 68 in recent outbreaks. PLoS ONE. 2012;7(4):e36005. doi: 10.1371/journal.pone.0036005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Q B Wo Y Wang H Y et al. Detection of enterovirus 68 as one of the commonest types of enterovirus found in patients with acute respiratory tract infection in China J Med Microbiol 201463(Pt 3):408–414. [DOI] [PubMed] [Google Scholar]
- 34.Smura T, Ylipaasto P, Klemola P. et al. Cellular tropism of human enterovirus D species serotypes EV-94, EV-70, and EV-68 in vitro: implications for pathogenesis. J Med Virol. 2010;82(11):1940–1949. doi: 10.1002/jmv.21894. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Sheng J, Fokine A. et al. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children. Science. 2015;347(6217):71–74. doi: 10.1126/science.1261962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Malanoski A P, Lin B. et al. Broad spectrum respiratory pathogen analysis of throat swabs from military recruits reveals interference between rhinoviruses and adenoviruses. Microb Ecol. 2010;59(4):623–634. doi: 10.1007/s00248-010-9636-3. [DOI] [PubMed] [Google Scholar]
- 37.Petitjean-Lecherbonnier J Dina J Nguyen E Gouarin S Lebigot E Vabret A Molecular diagnosis of respiratory enterovirus infections: Use of PCR and molecular identification for a best approach of the main circulating strains during 2008 Pathol Biol (Paris) 2011592113–121. [in French] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piralla A, Rovida F, Campanini G. et al. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J Clin Virol. 2009;45(4):311–317. doi: 10.1016/j.jcv.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Piralla A, Baldanti F, Gerna G. Phylogenetic patterns of human respiratory picornavirus species, including the newly identified group C rhinoviruses, during a 1-year surveillance of a hospitalized patient population in Italy. J Clin Microbiol. 2011;49(1):373–376. doi: 10.1128/JCM.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasegawa S, Hirano R, Okamoto-Nakagawa R, Ichiyama T, Shirabe K. Enterovirus 68 infection in children with asthma attacks: virus-induced asthma in Japanese children. Allergy. 2011;66(12):1618–1620. doi: 10.1111/j.1398-9995.2011.02725.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang T, Ren L, Luo M. et al. Enterovirus D68-associated severe pneumonia, China, 2014. Emerg Infect Dis. 2015;21(5):916–918. doi: 10.3201/eid2105.150036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meijer A, van der Sanden S, Snijders B E. et al. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology. 2012;423(1):49–57. doi: 10.1016/j.virol.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 43.Jaramillo-Gutierrez G, Benschop K S, Claas E C. et al. September through October 2010 multi-centre study in the Netherlands examining laboratory ability to detect enterovirus 68, an emerging respiratory pathogen. J Virol Methods. 2013;190(1–2):53–62. doi: 10.1016/j.jviromet.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Meijer A, Benschop K S, Donker G A, van der Avoort H G. Continued seasonal circulation of enterovirus D68 in the Netherlands, 2011-2014. Euro Surveill. 2014;19(42):20935. doi: 10.2807/1560-7917.es2014.19.42.20935. [DOI] [PubMed] [Google Scholar]
- 45.Poelman R, Schölvinck E H, Borger R, Niesters H G, van Leer-Buter C. The emergence of enterovirus D68 in a Dutch University Medical Center and the necessity for routinely screening for respiratory viruses. J Clin Virol. 2015;62:1–5. doi: 10.1016/j.jcv.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bal A, Schuffenecker I, Casalegno J S. et al. Enterovirus D68 nosocomial outbreak in elderly people, France, 2014. Clin Microbiol Infect. 2015;21(8):e61–e62. doi: 10.1016/j.cmi.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Todd A K, Hall R J, Wang J. et al. Detection and whole genome sequence analysis of an enterovirus 68 cluster. Virol J. 2013;10:103. doi: 10.1186/1743-422X-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norder H, Magnius L. Can sequence data predict enterovirus D68 infection outcome? Lancet Infect Dis. 2015;15(6):620–621. doi: 10.1016/S1473-3099(15)70107-6. [DOI] [PubMed] [Google Scholar]
- 49.Reiche J, Böttcher S, Diedrich S. et al. Low-level circulation of enterovirus D68-associated acute respiratory infections, Germany, 2014. Emerg Infect Dis. 2015;21(5):837–841. doi: 10.3201/eid2105.141900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gimferrer L, Campins M, Codina M G. et al. First Enterovirus D68 (EV-D68) cases detected in hospitalised patients in a tertiary care university hospital in Spain, October 2014. Enferm Infecc Microbiol Clin. 2015;33(9):585–589. doi: 10.1016/j.eimc.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dyrdak R, Rotzen-Ostlund M, Samuelson A, Eriksson M, Albert J. Coexistence of two clades of enterovirus D68 in pediatric Swedish patients in the summer and fall of 2014. Infect Dis (Lond) 2015;47(10):734–738. doi: 10.3109/23744235.2015.1047402. [DOI] [PubMed] [Google Scholar]
- 52.Ly N, Tokarz R, Mishra N. et al. Multiplex PCR analysis of clusters of unexplained viral respiratory tract infection in Cambodia. Virol J. 2014;11:224. doi: 10.1186/s12985-014-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torres J P, Farfan M J, Izquierdo G, Piemonte P, Henriquez J, O'Ryan M L. Enterovirus D68 infection, Chile, Spring 2014. Emerg Infect Dis. 2015;21(4):728–729. doi: 10.3201/eid2104.141766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Midgley C M, Jackson M A, Selvarangan R. et al. Severe respiratory illness associated with enterovirus D68 - Missouri and Illinois, 2014. MMWR Morb Mortal Wkly Rep. 2014;63(36):798–799. [PMC free article] [PubMed] [Google Scholar]
- 55.Furuse Y, Chaimongkol N, Okamoto M. et al. Molecular epidemiology of enterovirus d68 from 2013 to 2014 in Philippines. J Clin Microbiol. 2015;53(3):1015–1018. doi: 10.1128/JCM.03362-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imamura T, Suzuki A, Lupisan S. et al. Molecular evolution of enterovirus 68 detected in the Philippines. PLoS ONE. 2013;8(9):e74221. doi: 10.1371/journal.pone.0074221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oermann C M, Schuster J E, Conners G P, Newland J G, Selvarangan R, Jackson M A. Enterovirus d68. A focused review and clinical highlights from the 2014 U.S. Outbreak. Ann Am Thorac Soc. 2015;12(5):775–781. doi: 10.1513/AnnalsATS.201412-592FR. [DOI] [PubMed] [Google Scholar]
- 58.Stephenson J. CDC tracking enterovirus D-68 outbreak causing severe respiratory illness in children in the Midwest. JAMA. 2014;312(13):1290. doi: 10.1001/jama.2014.13256. [DOI] [PubMed] [Google Scholar]
- 59.Wylie T N, Wylie K M, Buller R S, Cannella M, Storch G A. Development and evaluation of an enterovirus D68 real-time reverse transcriptase polymerase chain reaction (RT-PCR) assay. J Clin Microbiol. 2015;53(8):2641–2647. doi: 10.1128/JCM.00923-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kreuter J D, Barnes A, McCarthy J E. et al. A fatal central nervous system enterovirus 68 infection. Arch Pathol Lab Med. 2011;135(6):793–796. doi: 10.5858/2010-0174-CR.1. [DOI] [PubMed] [Google Scholar]
- 61.Uncapher C R, DeWitt C M, Colonno R J. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology. 1991;180(2):814–817. doi: 10.1016/0042-6822(91)90098-v. [DOI] [PubMed] [Google Scholar]
- 62.Imamura T, Okamoto M, Nakakita S. et al. Antigenic and receptor binding properties of enterovirus 68. J Virol. 2014;88(5):2374–2384. doi: 10.1128/JVI.03070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nicholls J M, Bourne A J, Chen H, Guan Y, Peiris J S. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. 2007;8:73. doi: 10.1186/1465-9921-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karnauchow T M, Tolson D L, Harrison B A, Altman E, Lublin D M, Dimock K. The HeLa cell receptor for enterovirus 70 is decay-accelerating factor (CD55) J Virol. 1996;70(8):5143–5152. doi: 10.1128/jvi.70.8.5143-5152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Imamura T, Suzuki A, Lupisan S. et al. Detection of enterovirus 68 in serum from pediatric patients with pneumonia and their clinical outcomes. Influenza Other Respi Viruses. 2014;8(1):21–24. doi: 10.1111/irv.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harris K G, Coyne C B. Enter at your own risk: how enteroviruses navigate the dangerous world of pattern recognition receptor signaling. Cytokine. 2013;63(3):230–236. doi: 10.1016/j.cyto.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiang Z, Li L, Lei X. et al. Enterovirus 68 3C protease cleaves TRIF to attenuate antiviral responses mediated by Toll-like receptor 3. J Virol. 2014;88(12):6650–6659. doi: 10.1128/JVI.03138-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhuge J, Vail E, Bush J L. et al. Evaluation of a real-time reverse transcription-PCR assay for detection of enterovirus D68 in clinical samples from an outbreak in New York State in 2014. J Clin Microbiol. 2015;53(6):1915–1920. doi: 10.1128/JCM.00358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piralla A, Girello A, Premoli M, Baldanti F. A new real-time reverse transcription-PCR assay for detection of human enterovirus 68 in respiratory samples. J Clin Microbiol. 2015;53(5):1725–1726. doi: 10.1128/JCM.03691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaw J, Welch T R, Milstone A M. The role of syndromic surveillance in directing the public health response to the enterovirus D68 epidemic. JAMA Pediatr. 2014;168(11):981–982. doi: 10.1001/jamapediatrics.2014.2628. [DOI] [PubMed] [Google Scholar]
- 71.Tyler K L. Rationale for the evaluation of fluoxetine in the treatment of enterovirus D68-associated acute flaccid myelitis. JAMA Neurol. 2015;72(5):493–494. doi: 10.1001/jamaneurol.2014.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ulferts R, van der Linden L, Thibaut H J. et al. Selective serotonin reuptake inhibitor fluoxetine inhibits replication of human enteroviruses B and D by targeting viral protein 2C. Antimicrob Agents Chemother. 2013;57(4):1952–1956. doi: 10.1128/AAC.02084-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao Q, Yuan S, Zhang C. et al. Discovery of itraconazole with broad-spectrum in vitro antienterovirus activity that targets nonstructural protein 3A. Antimicrob Agents Chemother. 2015;59(5):2654–2665. doi: 10.1128/AAC.05108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Yang B, Zhai Y, Yin Z, Sun Y, Rao Z. Peptidyl aldehyde NK-1.8k suppresses enterovirus 71 and enterovirus 68 infection by targeting protease 3C. Antimicrob Agents Chemother. 2015;59(5):2636–2646. doi: 10.1128/AAC.00049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]


