Abstract
Background
Climate change, including higher temperatures (HT) has a detrimental impact on wheat productivity and modeling studies predict more frequent heat waves in the future. Wheat growth can be impaired by high daytime and nighttime temperature at any developmental stage, especially during the grain filling stage. Leaf chlorophyll content, leaf greenness, cell membrane thermostability, and canopy temperature have been proposed as candidate traits to improve crop adaptation and yield potential of wheat under HT. Nonetheless, a significant gap exists in knowledge of genetic backgrounds associated with these physiological traits. Identifying genetic loci associated with these traits can facilitate physiological breeding for increased yield potential under high temperature stress condition in wheat.
Results
We conducted genome-wide association study (GWAS) on a 236 elite soft wheat association mapping panel using 27,466 high quality single nucleotide polymorphism markers. The panel was phenotyped for three years in two locations where heat shock was common. GWAS identified 500 significant marker-trait associations (MTAs) (p ≤ 9.99 × 10− 4). Ten MTAs with pleiotropic effects detected on chromosomes 1D, 2B, 3A, 3B, 6A, 7B, and 7D are potentially important targets for selection. Five MTAs associated with physiological traits had pleiotropic effects on grain yield and yield-related traits. Seventy-five MTAs were consistently expressed over several environments indicating stability and more than half of these stable MTAs were found in genes encoding different types of proteins associated with heat stress.
Conclusions
We identified 500 significant MTAs in soft winter wheat under HT stress. We found several stable loci across environments and pleiotropic markers controlling physiological and agronomic traits. After further validation, these MTAs can be used in marker-assisted selection and breeding to develop varieties with high stability for grain yield under high temperature.
Keywords: Single nucleotide polymorphisms1, Genotyping-by-sequencing2, Marker-trait associations3, Quantitative trait loci4, Genome-wide association study5, Physiological traits6, Marker-assisted breeding7
Background
Worldwide, wheat is grown on more than 218 million hectares of land and provides approximately 20% of dietary calorie needs [1]. Although, there has been a substantial increase in yield since the Green Revolution, the pace of increase in yield production is not predicted to match demand resulting from increase in human population and changing weather pattern [2, 3]. High temperature (HT) stress is one of the major consequences of climate change and poses a serious threat to wheat production [4]. Global temperature has increased by 0.5 °C in the twentieth century [5] and this warming trend is expected to continue up to 1.5–4.5 °C by the end of twenty-first century, resulting in elevated daytime maximum (HDT) and nighttime minimum temperatures. (HNT) [6]. Post-anthesis heat stress is very common in wheat growing areas and can cause large reductions in grain yield [7]. Although, some researchers suggest that HDT and HNT cause damage of a similar magnitude to winter wheat [8], others report a stronger negative impact on yield of HNT compared to HDT [9].
Genetic improvement of yield under HT via direct selection is hindered by the quantitative nature of grain yield and large genotype by environment interaction. Optimizing carbohydrate partitioning using traits such as spike fertility (SF), internode partitioning, and spike organ partitioning is essential to overcome sink limitations and increase harvest index (HI) [10, 11]. Although higher grain yield potential of wheat has been largely a result of increased HI under HT, increased biomass and crop adaptation are equally important. Physiological trait (PT) selection insures development of stress resilient genotypes with functioning metabolic activities including photosynthesis and respiration under HT [12]. Moreover, previous studies have reported strong correlations of these traits with grain yield. Therefore, PTs can serve as indirect selection tools to select superior genotypes from large numbers of breeding lines for stress environments and to compensate for a large genotype by environment interaction. This is important to wheat breeders since it can save substantial amounts of labor, time, and money and permits rapid screening of a large number of genotypes in relatively short time [13, 14]. Selection of desirable physiological trait (PTs) associated with heat adaptation and combinations is essential for future improvement of crops and provides opportunities for optimizing genetic yield gain. A model proposed to improve yield in wheat under heat stress includes partitioning of assimilates, radiation use efficiency (RUE), and light interception (LI) [15]. Some of the candidate PTs associated with these components are well documented as being heat adaptive traits, including higher leaf chlorophyll content measured as SPAD (soil-plant analyses development) value, intact leaf greenness measured as normalized difference vegetation index (NDVI), membrane thermostability (MT), and canopy temperature (CT). While these PTs are good candidates for improving heat tolerance and yield potential in wheat, limited knowledge on their genetic basis prevent full exploitation [11, 16]. Use of these indirect selection tools is limited in breeding programs due to their complex evaluation procedure [17]. Therefore, identifying novel genetic loci (QTLs) associated with PTs under heat stress and using them as selection tools can results in a cumulative genetic effect on yield, which is the basis of maker assisted physiological trait breeding [18].
Association mapping is a powerful approach that utilizes genetic diversity and historical recombination events to provide a high resolution map of trait-linked loci [19]. Although genome-wide association studies (GWAS) have been used to identify quantitative trait loci (QTL) in wheat for various simply inherited traits like disease resistance [20], currently, limited information are available for GWAS of complex PTs, particularly under HT. Recently, the International Wheat Genome Sequencing Consortium (IWGSC) published a full chromosome-anchored reference genome which allows more precise curation of marker trait associations (MTAs) identified by GWAS. In this study, GWAS was performed on 236 advanced soft red winter wheat accessions using 27,466 SNPs generated by GBS. The panel was phenotyped at two heat stress locations over three years. The objectives of this study were: i) to identify novel MTAs linked to NDVI, CT, SPAD and MT under HT and ii) to identify candidate genes for these MTAs and investigate their underlying function.
Results
Phenotypic analyses
There was significant genotypic variation (P < 0.001) for all measured traits (Additional file 3), as expected given the diverse genetic backgrounds of the SWAMP. Environments (growing years and locations) and their interaction were all significant (P < 0.05) determinants of phenotypic traits except for MT (Additional file 3). Trait means and a summary in response to each environment are provided (Table 1). All traits had moderate heritabilities, ranging from 60% for MT to 35% for CT (Table 1).
Table 1.
Summary of adjusted means of physiological traits for the SWAMP
| Traits | Citra | Quincy | Combined | H2 | |||
|---|---|---|---|---|---|---|---|
| Mean | Range | Mean | Range | Mean | Range | ||
| SPAD | 51.11 | 25.68–61.16 | 48.57 | 13.28–52.55 | 50.25 | 31.56–61.36 | 0.49 |
| MT | 56.72 | 22.27–77.21 | 59.47 | 23.68–61.95 | 57.63 | 26.29–74.26 | 0.60 |
| CT | 26.38 | 24.87–28.23 | 23.65 | 21.47–25.41 | 25.28 | 23.91–26.72 | 0.35 |
| NDVIa | 0.73 | 0.58–0.80 | 0.74 | 0.59–0.86 | 0.73 | 0.62–0.81 | 0.56 |
| NDVIg | 0.59 | 0.37–0.77 | 0.62 | 0.41–0.76 | 0.61 | 0.43–0.75 | 0.40 |
SPAD, soil-plant analyses development; MT, cell membrane thermostability (%); CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling
Pearson correlation coefficients (r) among PTs and their relationship with GY, GN, SF, HI, SHI and TGW were calculated using the combined dataset. SPAD was positively correlated with MT (0.31***), GY (0.50***), SF (0.25***), GN (0.30***), HI (0.46***), SHI (0.37***) and TGW (0.26***) (Additional file 4). Similarly, MT was positively correlated with NDVIa (0.22***), NDVIg (0.31***), GY (0.60***), SF (0.29***), GN (0.33***), HI (0.58***), SHI (0.44***) and TGW (0.40***). Pearson correlation coefficients results were further supported by principal components (PC) analysis that showed SPAD and MT were closely associated with GY, SF, GN, HI and SHI (Additional file 5). CT was negatively correlated with GY (− 0.18**), GN (− 0.17*), MT (− 0.25***), and SPAD (− 0.25***). NDVIa and NDVIg were positively correlated with GY, GN, HI and TGW (Additional file 4).
Genetic data, LD decay, and population structure
Population structure analysis of the SWAMP was performed in our previous study using 27,466 high quality GBS-derived SNP markers (minor allele frequency; MAF > 0.05 and missing data < 20%) [10]. Briefly, these SNP markers were distributed throughout the A (9958, ~ 36%), B (9,968, ~ 36%) and D (6,954, ~ 25%) genomes. A total of 686 SNPs where found on unplaced scaffolds and thus were classified as unmapped SNPs. Chromosome 2B had the highest number of SNPs (1960) and chromosome 4D had the lowest (571). The population structure analysis grouped the 236 SWAMP lines into three genetic demes containing 49, 144, and 43 lines respectively (Additional file 6). PC analysis revealed substantial admixture among lines in the SWAMP, with the first and second PC explaining only 4.7 and 3.1% of the total genotypic variance, respectively (Additional file 6).
LD was computed using the “LDcorSV” package in R to determine the approximate marker density required for GWAS [10]. The LD decay below the line of critical value (r2 = 0.2) was estimated at 1182, 1920 and 2916 bp for ranges of 30,000, 40,000, and 50,000 bp, respectively, across the whole genome (Additional file 7). The magnitude of change in LD decay between a sample range of 30,000–50,000 bp was 1734 bp. Population structure was investigated to avoid false positive associations in GWAS (Additional file 6) [19].
Marker-trait association
The GWAS identified novel MTAs for all measured PTs and explained a large portion of phenotypic variances from 5 to 23% (Additional file 8). The FarmCPU model with kinship and PC scores was used to identify MTAs for each trait using 27,466 GBS-derived SNPs. The SNP markers were uniformly distributed throughout the chromosomes of each genome (Additional file 8). GWAS was conducted on three datasets: BLUEC (Citra), BLUEQ (Quincy) and BLUEA (combined). We identified 500 significant MTAs for PTs distributed across 21 chromosomes (Additional file 8). The highest number of MTAs was detected in BLUEA (192) followed by BLUEQ (177) and BLUEC (131) (Table 2). The highest number of MTAs were identified in the B genome (225 MTAs), compared to A (139 MTAs) and D (136 MTAs) genomes. We identified 94 MTAs for SPAD across three datasets on chromosomes with phenotypic variance explained (PVE) ranging from 13 to 20% (Table 2, Additional file 8). For MT, 95 MTAs were identified with PVEs ranging from 5 to 15%. We detected the highest number of MTAs for CT (110) with PVEs ranging from 8 to 13%. For NDVIa, and NDVIg we detected 102 and 99 significant MTAs respectively with PVEs ranging from 5 to 23%. (Table 2, Additional file 8).
Table 2.
Summary of significant marker–trait associations for physiological traits
| Traits | BLUEC | BLUEQ | BLUEA | Total |
|---|---|---|---|---|
| SPAD | 28 | 33 | 33 | 94 |
| MT | 25 | 35 | 35 | 95 |
| CT | 24 | 49 | 37 | 110 |
| NDVIa | 22 | 32 | 48 | 102 |
| NDVIg | 32 | 28 | 39 | 99 |
| Total | 131 | 177 | 192 | 500 |
SPAD, soil-plant analyses development; MT, cell membrane thermostability (%); CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling. BLUEC, BLUEs values derived across Citra; BLUEQ, BLUEs values derived across Quincy, and BLUEA: BLUEs values derived across all environment
Co-localized MTAs controlling multiple PTs were detected in the study. Ten pleiotropic SNP markers on chromosomes 1D, 2B, 3A, 3B, 5A, 6A, 7B, and 7D were detected across different environments (Table 3). Seven of them were associated with NDVIa and NDVIg indicating common MTAs for NDVI expressed during anthesis and grain filling period. SNP S7D_635578722 had a positive allelic effect for SPAD, NDVIa and NDVIg. SNP S2B_466014434 was associated with SPAD and MT and had a negative allelic effect on both traits. S3A_609909640 was associated with SPAD and CT and had a negative allelic effect on CT and a positive allelic effect on SPAD (Table 3). Interestingly, we detected five significant MTAs on chromosome 3A (S3A_12554694 and S3A_12554700), 5A (S5A_590056740), 6B (S6B_149148874), and 7D (S7D_18808932) for PTs, which were associated with GY and other yield related traits in our previous study [10].
Table 3.
List of significant markers associated with multiple phenotypic traits (pleiotropy) in the SWAMP
| SNP | Trait | Dataset | -log10(p) | Effect | PVE |
|---|---|---|---|---|---|
| S1D_479711964 | NDVIa | BLUEA | 5.26 | 0.00 | 0.20 |
| NDVIa | BLUEC | 5.52 | 0.01 | 0.23 | |
| NDVIg | BLUEA | 3.40 | 0.01 | 0.16 | |
| S2B_466014434 | MT | BLUEC | 3.31 | −4.85 | 0.06 |
| SPAD | BLUEA | 3.83 | −1.89 | 0.20 | |
| SPAD | BLUEC | 3.34 | −1.91 | 0.16 | |
| S3A_609909640 | CT | BLUEC | 4.11 | −0.20 | 0.11 |
| SPAD | BLUEQ | 4.51 | 1.78 | 0.16 | |
| S3B_785311769 | NDVIa | BLUEC | 6.02 | 0.00 | 0.22 |
| NDVIg | BLUEA | 3.01 | 0.01 | 0.13 | |
| NDVIg | BLUEQ | 3.34 | 0.01 | 0.07 | |
| S5A_356222133 | NDVIa | BLUEQ | 3.26 | 0.01 | 0.11 |
| NDVIg | BLUEQ | 3.02 | 0.02 | 0.08 | |
| S5A_356222163 | NDVIa | BLUEQ | 3.26 | 0.01 | 0.11 |
| NDVIg | BLUEQ | 3.02 | 0.02 | 0.08 | |
| S5A_590056740 | NDVIa | BLUEA | 5.79 | −0.01 | 0.18 |
| NDVIg | BLUEA | 3.10 | −0.02 | 0.13 | |
| S6A_39961388 | NDVIa | BLUEC | 3.12 | 0.01 | 0.22 |
| NDVIg | BLUEA | 3.66 | 0.02 | 0.17 | |
| S7B_701649275 | NDVIa | BLUEA | 5.82 | 0.00 | 0.20 |
| NDVIg | BLUEA | 3.78 | 0.01 | 0.15 | |
| S7D_635578722 | NDVIa | BLUEA | 3.49 | 0.01 | 0.18 |
| NDVIa | BLUEQ | 3.34 | 0.02 | 0.09 | |
| NDVIg | BLUEA | 3.15 | 0.03 | 0.15 | |
| NDVIg | BLUEC | 4.17 | 0.02 | 0.11 | |
| SPAD | BLUEQ | 3.41 | −3.40 | 0.15 |
SPAD, soil-plant analyses development; MT, cell membrane thermostability (%); CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling. BLUEC, BLUEs values derived across Citra; BLUEQ, BLUEs values derived across Quincy, and BLUEA: BLUEs values derived across all environment
Seventy-five out of 500 MTAs were expressed in multiple environments and were considered as stable MTAs (Table 4). We identified 23 stable markers for SPAD on chromosomes 1B, 1D, 2B, 2D, 3B, 4A, 5B, 5D, 6B, 6D, 7A, and 7D with PVEs ranging from 13 to 19% (Table 4, Additional file 8). For MT, we identified 20 stable MTAs on chromosomes 1B, 1D, 2A, 2D, 3A, 3D, 5B, 5D, 6A, 6B, and 7B with PVEs ranging from 5 to 14%. Similarly, 17 stable MTAs were detected for CT with PVE ranging from 8 to 13%. For NDVIa and NDVIg, we identified 3 and 12 stable markers respectively that were unique to corresponding growth stages (Table 4).
Table 4.
List of significant markers expressed in multiple environments (stable) in the SWAMP
| SPAD | MT | CT | NDVIa | NDVIg |
|---|---|---|---|---|
| S1B_4335636AC | S1B_602752201AQ | S2B_693094464AQ | S2B_717098540AC | S3A_699988530AQ |
| S1B_204428462AC | S1B_602752209AQ | S2B_693094466AQ | S3A_108025984AC | S3A_699991338AQ |
| S1D_907114AQ | S1B_602752224AQ | S5B_601343966AQ | S6D_201817286AC | S3A_732890228AQ |
| S1D_907133AQ | S1B_602752226AQ | S5B_602833771AQ | S3A_736970882AQ | |
| S2B_466014437AC | S1D_262475151AC | S5B_606014586AQ | S3A_737114441AQ | |
| S2D_574118879AQ | S2A_70446757AC | S5B_607207678AQ | S2D_570960728AQ | |
| S2D_11171031AC | S2D_602734684AC | S5B_607207704AQ | S3B_785311773AQ | |
| S3B_792189571AQ | S2D_634968398AQ | S5B_608350950AQ | S5B_583295527AC | |
| S4A_625244392AC | S3A_12554694AQ | S5B_610295429AQ | S1B_677572998AQ | |
| S5B_592791824AQ | S3A_12554700AQ | S5B_617291841AQ | S7B_426882778AQ | |
| S5D_220760001AC | S3B_509660072AC | S5B_621237427AQ | S2A_93482025AQ | |
| S6B_131285725AC | S3D_590224603ACQ | S5B_622494564AQ | S3B_784466451AQ | |
| S6B_150497646AC | S3D_590224620AC | S5B_622494601AQ | ||
| S6B_462165779AC | S5B_487440465AQ | S5B_622494604AQ | ||
| S6D_16178496AC | S5B_509105168AQ | S5B_643470598AQ | ||
| S6D_16178499AC | S5B_509105191AQ | S6B_149148874AC | ||
| S6D_16178505AC | S5D_184110184AC | S7B_265453929ACQ | ||
| S7A_563391742AQ | S6A_58259479AQ | |||
| S7A_565347529AQ | S6B_42215716AQ | |||
| S7A_579648980AQ | S7B_464657928AQ | |||
| S7A_644864716AC | ||||
| S7A_644864763AC | ||||
| S7D_37142233AC |
ACombined; CCitra; QQuincy
SPAD, soil-plant analyses development; MT, cell membrane thermostability (%); CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling. BLUEC, BLUEs values derived across Citra; BLUEQ, BLUEs values derived across Quincy, and BLUEA: BLUEs values derived across all environment
Gene annotation
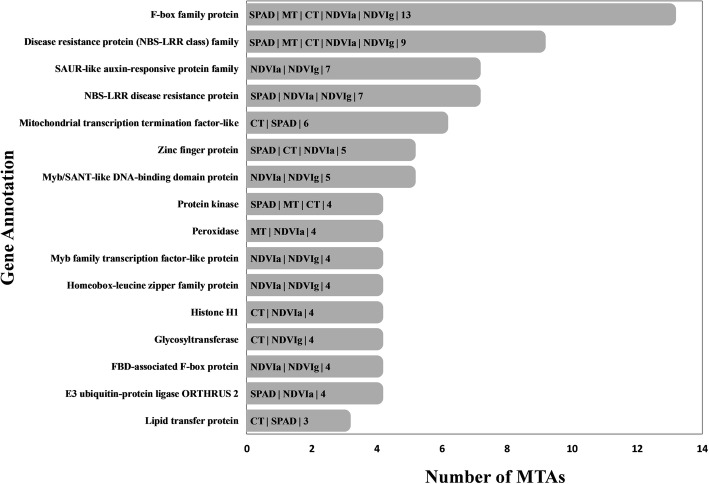
Functional annotation of all stable MTAs was carried out using the IWGSC v1.0 reference genome sequence assembly. Forty-one out of 75 stable MTAs associated with PTs were anchored within functional genes. These MTAs had a wide range of functional annotations and are potential candidate genes for QTLs of interest (Table 5). Candidate genes were further investigated using past literatures to understand their possible functions. We discovered that these candidate genes encode different classes of proteins including F-box family proteins, RNA-binding proteins, disease resistance protein and protein kinase that have suggestive roles in response to biotic and abiotic stresses (Table 5). In addition, gene annotation was also carried out for all the significant MTAs (Fig. 2). Interestingly, we identified several MTAs (linked to different traits) in different chromosomes that had common genes with exact same annotation (Fig. 2 and Additional file 8).
Table 5.
List of potential candidate genes and anchoring markers associated with physiological traits
| SNP | SNP | Dataset | -log10(p) | Effect | PVE | Alleles | Gene-ID | Annotation |
|---|---|---|---|---|---|---|---|---|
| S1B_4335636 | SPAD | BLUEA | 3.46 | 1.19 | 0.17 | G/A | TraesCS1B01G007900 | E3 ubiquitin-protein ligase ORTHRUS 2 |
| BLUEC | 3.01 | 1.20 | 0.14 | |||||
| S1D_907114 | SPAD | BLUEA | 3.65 | 1.74 | 0.18 | G/C | TraesCS1D01G003300 | SNF1-related protein kinase regulatory subunit beta-2 |
| BLUEQ | 3.13 | 2.33 | 0.14 | |||||
| S1D_907133 | SPAD | BLUEA | 3.65 | 1.74 | 0.18 | G/C | TraesCS1D01G003300 | SNF1-related protein kinase regulatory subunit beta-2 |
| BLUEQ | 3.13 | 2.33 | 0.14 | |||||
| S2D_574118879 | SPAD | BLUEA | 3.98 | 2.78 | 0.19 | A/G | TraesCS2D01G469000 | GDSL-like Lipase/Acylhydrolase superfamily protein |
| BLUEQ | 3.09 | 3.53 | 0.14 | |||||
| S2D_11171031 | SPAD | BLUEC | 3.48 | 1.42 | 0.15 | T/C | TraesCS2D01G026300 | Glutamyl-tRNA (Gln) amidotransferase subunit A |
| BLUEA | 3.07 | 1.21 | 0.17 | |||||
| S4A_625244392 | SPAD | BLUEA | 3.35 | 1.63 | 0.17 | C/T | TraesCS4A01G346600 | F-box protein |
| BLUEC | 3.22 | 1.75 | 0.14 | |||||
| S5D_220760001 | SPAD | BLUEC | 3.34 | 1.64 | 0.14 | G/A | TraesCS5D01G138700 | disease resistance protein (TIR-NBS-LRR class) family |
| BLUEA | 3.18 | 1.46 | 0.17 | |||||
| S6D_16178496 | SPAD | BLUEA | 3.59 | −1.66 | 0.19 | C/T | TraesCS6D01G038900 | 2-oxoglutarate (2OG) and Fe (II)-dependent oxygenase superfamily protein |
| BLUEC | 3.55 | −1.81 | 0.16 | |||||
| S6D_16178499 | SPAD | BLUEA | 3.59 | −1.66 | 0.19 | A/T | TraesCS6D01G038900 | 2-oxoglutarate (2OG) and Fe (II)-dependent oxygenase superfamily protein |
| BLUEC | 3.55 | −1.81 | 0.16 | |||||
| S6D_16178505 | SPAD | BLUEA | 3.59 | −1.66 | 0.19 | A/C | TraesCS6D01G038900 | 2-oxoglutarate (2OG) and Fe (II)-dependent oxygenase superfamily protein |
| BLUEC | 3.55 | −1.81 | 0.16 | |||||
| S6B_462165779 | SPAD | BLUEA | 4.15 | 1.34 | 0.18 | C/G | TraesCS6B01G257900 | Adenosine kinase-like protein |
| BLUEC | 3.19 | 1.27 | 0.14 | |||||
| S7A_565347529 | SPAD | BLUEA | 4.54 | 1.38 | 0.19 | T/G | TraesCS7A01G389100 | TBC1 domain family member |
| BLUEQ | 4.38 | 1.98 | 0.15 | |||||
| S7A_579648980 | SPAD | BLUEQ | 3.63 | −1.78 | 0.15 | C/G | TraesCS7A01G399700 | Telomere repeat-binding factor like-protein |
| BLUEA | 3.48 | −1.19 | 0.17 | |||||
| S1D_262475151 | MT | BLUEC | 3.53 | 4.15 | 0.07 | T/C | TraesCS1D01G190700 | Heavy metal transport/detoxification superfamily protein |
| BLUEA | 3.52 | 4.14 | 0.10 | |||||
| S3D_590224603 | MT | BLUEA | 4.89 | 4.60 | 0.12 | T/C | TraesCS3D01G501200 | Protein kinase |
| BLUEC | 4.80 | 4.57 | 0.09 | |||||
| BLUEQ | 3.20 | 4.98 | 0.12 | |||||
| S3D_590224620 | MT | BLUEC | 3.61 | 4.21 | 0.06 | G/A | TraesCS3D01G501200 | Protein kinase |
| BLUEA | 3.12 | 3.87 | 0.09 | |||||
| S5B_487440465 | MT | BLUEQ | 3.36 | 4.36 | 0.11 | A/C | TraesCS5B01G302900 | Plant/T31B5–30 protein |
| BLUEA | 3.23 | 3.11 | 0.09 | |||||
| S5B_509105168 | MT | BLUEQ | 4.25 | −6.33 | 0.14 | A/G | TraesCS5B01G325000 | F-box family protein |
| BLUEA | 3.78 | −4.32 | 0.11 | |||||
| S5B_509105191 | MT | BLUEQ | 4.25 | 6.33 | 0.14 | T/C | TraesCS5B01G325000 | F-box family protein |
| BLUEA | 3.78 | 4.32 | 0.11 | |||||
| S6B_42215716 | MT | BLUEC | 3.46 | 6.04 | 0.07 | A/G | TraesCS6B01G063500 | Peroxidase |
| BLUEA | 3.08 | 5.64 | 0.09 | |||||
| S2B_693094464 | CT | BLUEC | 3.83 | −0.24 | 0.10 | G/A | TraesCS2B01G496300 | BTB/POZ and MATH domain-containing protein 2 |
| BLUEA | 3.10 | −0.16 | 0.08 | |||||
| S2B_693094466 | CT | BLUEC | 3.83 | −0.24 | 0.10 | G/A | TraesCS2B01G496300 | BTB/POZ and MATH domain-containing protein 2 |
| BLUEA | 3.10 | −0.16 | 0.08 | |||||
| S5B_610295429 | CT | BLUEA | 4.41 | 0.29 | 0.12 | T/G | TraesCS5B01G436700 | Lipid transfer protein |
| BLUEQ | 4.27 | 0.34 | 0.11 | |||||
| S5B_608350950 | CT | BLUEQ | 4.85 | −0.34 | 0.12 | C/T | TraesCS5B01G433700 | Maintenance of telomere capping protein 2 |
| BLUEA | 3.19 | −0.23 | 0.09 | |||||
| S5B_621237427 | CT | BLUEQ | 4.37 | −0.34 | 0.11 | C/T | TraesCS5B01G448700 | Mitochondrial transcription termination factor-like |
| BLUEA | 3.22 | −0.24 | 0.10 | |||||
| S5B_606014586 | CT | BLUEQ | 3.37 | 0.30 | 0.10 | T/C | TraesCS5B01G431300 | Peptidase M50 family protein |
| BLUEA | 3.03 | 0.23 | 0.10 | |||||
| S5B_602833771 | CT | BLUEA | 3.70 | −0.24 | 0.11 | C/T | TraesCS5B01G426900 | DNA topoisomerase |
| BLUEQ | 3.64 | −0.28 | 0.10 | |||||
| S5B_617291841 | CT | BLUEA | 4.79 | −0.30 | 0.13 | A/G | TraesCS5B01G445300 | Endo-1,4-beta-xylanase |
| BLUEQ | 3.51 | −0.30 | 0.10 | |||||
| S5B_643470598 | CT | BLUEA | 3.16 | 0.18 | 0.10 | G/A | TraesCS5B01G470200 | Protein phosphatase 2C |
| BLUEQ | 3.07 | 0.22 | 0.08 | |||||
| S5B_601343966 | CT | BLUEA | 3.17 | −0.24 | 0.11 | C/T | TraesCS5B01G425500 | Zinc finger CCCH domain-containing protein 16 |
| BLUEQ | 3.14 | −0.29 | 0.10 | |||||
| S2B_717098540 | NDVIa | BLUEC | 8.41 | −0.02 | 0.22 | C/T | TraesCS2B01G522200 | SAUR-like auxin-responsive protein family |
| BLUEA | 4.17 | −0.01 | 0.18 | |||||
| S1B_677572998 | NDVIg | BLUEA | 3.48 | 0.02 | 0.12 | C/G | TraesCS1B01G467900 | Methyltransferase |
| BLUEQ | 3.32 | 0.03 | 0.07 | |||||
| S2D_570960728 | NDVIg | BLUEQ | 3.79 | −0.03 | 0.07 | A/C | TraesCS2D01G464800 | Multidrug resistance protein ABC transporter family protein |
| BLUEA | 3.13 | −0.02 | 0.13 | |||||
| S3A_737114441 | NDVIg | BLUEQ | 3.49 | 0.02 | 0.07 | T/C | TraesCS3A01G519100 | rRNA N-glycosidase |
| BLUEA | 3.41 | 0.02 | 0.14 | |||||
| S3B_785311773 | NDVIg | BLUEQ | 3.34 | 0.01 | 0.07 | A/G | TraesCS3B01G550500 | Myb/SANT-like DNA-binding domain protein |
| BLUEA | 3.01 | 0.01 | 0.13 | |||||
| S5B_583295527 | NDVIg | BLUEC | 6.60 | 0.02 | 0.12 | A/G | TraesCS5B01G407600 | Myb family transcription factor-like protein |
| BLUEA | 5.00 | 0.02 | 0.16 | |||||
| S7B_426882778 | NDVIg | BLUEQ | 3.28 | 0.03 | 0.08 | A/G | TraesCS7B01G226400 | RING finger protein 13 |
| BLUEA | 3.01 | 0.02 | 0.14 | |||||
| S3B_785311769 | NDVIa | BLUEC | 6.02 | 0.00 | 0.22 | A/G | TraesCS3B01G550500 | Myb/SANT-like DNA-binding domain protein |
| NDVIg | BLUEQ | 3.34 | 0.01 | 0.07 | ||||
| NDVIg | BLUEA | 3.01 | 0.01 | 0.13 | ||||
| S7B_701649275 | NDVIa | BLUEA | 5.82 | 0.00 | 0.20 | C/T | TraesCS7B01G434600 | FBD-associated F-box protein |
| NDVIg | BLUEA | 3.78 | 0.01 | 0.15 | ||||
| S3A_699988530 | NDVIg | BLUEQ | 4.46 | −0.04 | 0.12 | A/G | TraesCS3A01G466000 | F-box family protein |
| BLUEA | 3.48 | −0.03 | 0.16 | |||||
| S7D_635578722 | NDVIg | BLUEC | 4.17 | 0.02 | 0.11 | T/C | TraesCS7D01G552700 | NBS-LRR disease resistance protein |
| NDVIa | BLUEA | 3.49 | 0.01 | 0.18 | ||||
| SPAD | BLUEQ | 3.41 | −3.40 | 0.15 | ||||
| NDVIa | BLUEQ | 3.34 | 0.02 | 0.09 | ||||
| NDVIg | BLUEA | 3.15 | 0.03 | 0.15 |
SPAD, soil-plant analyses development; MT, cell membrane thermostability; CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling. BLUEC, BLUEs values derived across Citra; BLUEQ, BLUEs values derived across Quincy, and BLUEA: BLUEs values derived across all environment; PVE, phenotypic variance explained
Fig. 2.
Potential candidate gene functions harboring SNPs affecting physiological traits under heat stress. The traits and count of marker–trait associations (for two or more traits) located within genes that have the same gene annotation is shown inside a bar. SPAD, soil-plant analyses development; MT, cell membrane thermostability; CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling
Discussion
Growth and development of wheat is very sensitive to HT during anthesis and grain filling [7]. The impact of HDT and HNT on wheat growth and grain yield is well documented in many studies [7, 9, 21]. In this study, the SWAMP was evaluated in two heat prone environments of southeast USA with a goal to identify significant MTAs for use in breeding to improve adaptability and optimize yield potential. Short episodes of HDT (> 30 °C) and HNT (> 21 °C) were common during from anthesis to grain filling period in both environments (Additional file 1), thus, the MTAs identified in this study can provide useful information to understand the genetic bases of PTs under HDT and HNT.
HT reduces leaf area index and increases senescence rate which subsequently impairs photosynthesis and reduces grain yield [15, 22]. Measuring chlorophyll content using SPAD, as a proxy for the entire photosynthetic complex, indicates photosynthetic potential. Higher expression of SPAD values during reproductive stages in wheat have been associated with heat tolerance resulting in higher grain yield potential [23–25]. In this study, genotypes showed significant genotypic variation in SPAD values with moderate broad-sense heritability (0.49) (Table 1). Previous studies reported similar heritability for SPAD values [20]. Pearson’s correlation showed strong positive relationship of SPAD with GY, SF, GN, HI, SHI, TGW, and MT. The result was supported by PC biplot analysis where SPAD was clustered with GY, SF, GN, HI, SHI, and MT (Additional file 5). We identified 94 MTAs for SPAD with PVEs ranging from 13 to 20% (Table 2, Fig. 1b) out of which 24 MTAs were expressed in multiple environments suggesting the genetic stability of these MTAs under different environments (Table 4). Stable MTAs were located in chromosomes 1B, 1D, 2B, 2D, 3B, 4A, 5B, 5D, 6B, 6D, 7A, and 7D (Fig. 1a). In this study, SPAD shared common MTAs with MT (S2B_466014434), CT (S3A_609909640), and NDVIa and NDVIg (S7D_635578722) (Table 3). These are potential new targets for multi-trait improvement and marker assisted breeding. Thirteen of these stable MTAs had functional annotation suggesting their involvement in abiotic stress including heat stress (Table 5). Three markers within 9 bp range on chromosome 6D (S6D_16178496, S6D_16178499, S6D_16178505) were within gene, TraesCS6D01G038900, with functional annotation of 2-oxoglutarate (2OG) and Fe (II)-dependent oxygenase superfamily protein. This gene has been reported to increase oxidative stress and reduce flower and pod number in canola when affected by heat stress [26]. One MTA on chromosome 4A (S4A_625244392) within gene TraesCS4A01G346600 had functional annotation as F-box protein. A gene (TaFBA1) encoding homologous F-box protein was reported to regulate gene expression and improve enzymatic antioxidant levels in response to heat stress in tobacco [27]. These proteins area involved in regulating many other biological processes, including biotic and abiotic stresses, floral development, embryogenesis, hormonal responses, and senescence [28]. Another MTA (S2D_574118879) within gene TraesCS2D01G469000 (GDSL-like Lipase/Acylhydrolase superfamily protein) has a predicted functional role in response to thermal stress in sorghum [29]. We also found MTAs associated with drought stress. Moreover, we found several other MTAs whose annotations suggest different roles including response to drought stress (TBC1 domain family member, SNF1-related protein kinase regulatory subunit beta-2) plant senescence (E3 ubiquitin-protein ligase ORTHRUS 2), salt stress (Glutamyl-tRNA (Gln) amidotransferase subunit A), disease resistance (Disease resistance protein (NBS-LRR class) family), metabolism (Adenosine kinase-like protein) and antioxidant defense (Telomere repeat-binding factor like-protein) [30–36]. Our study identified novel MTAs associated with SPAD measurements in US soft wheat under HT conditions which contributes to a better understanding of the genetic basis of SPAD traits in wheat. Upon further validation, these MTAs can be used in future marker assisted breeding programs to overcome sink limitations, improve HI and ultimately increase yield potential.
Fig. 1.
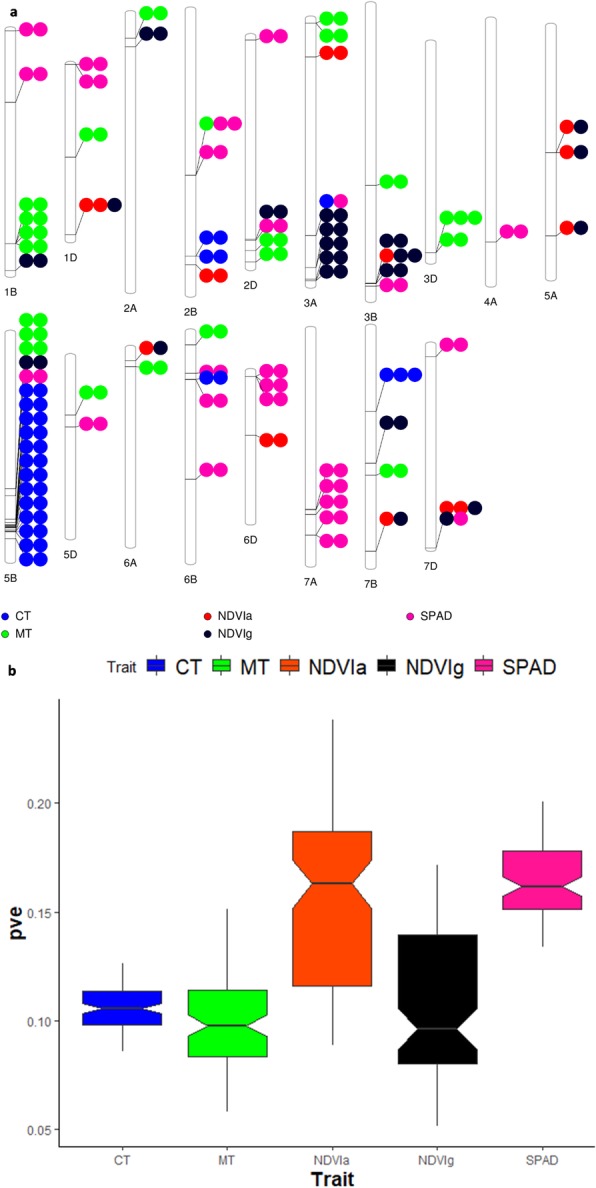
Summary of GWAS. a genome-wide distribution of stable markers trait associations and b) range of percentage of variation explained for physiological traits in SWAMP. SPAD, soil-plant analyses development; MT, cell membrane thermostability; CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling
Another important trait that provides rapid measurement of crops to characterize the canopy for LAI and leaf greenness is NDVI. High leaf chlorophyll content at anthesis and the ability to retain greenness (delayed senescence) during grain filling stages is associated with higher heat tolerance [37]. Studies have confirmed that NDVI could be used to predict grain yield in wheat [14]. We found significant genotypic variation among SWAMP genotypes for NDVI at anthesis and grain filling period. NDVIa and NDVIg showed moderate broad sense heritability of 0.56 and 0.40 respectively (Table 1) which aligned with the results from previous studies [38]. NDVI showed significant positive correlation with GY, GN, HI, TGW, and MT at anthesis and grain filling stages. Some studies have reported a strong correlation of NDVI with wheat grain yield at any growth stages [14]. Others have reported a varied relationship of NDVI with grain yield, depending on growth conditions [39]. Here, we found several unique as well as common MTAs associated with NDVIa and NDVIg. We identified total 102 MTAs for NDVIa with PVEs ranging from 8 to 23% (Table 2, Fig. 1b). Six out of 95 MTAs were expressed in several environments indicating stability of these markers under different environments (Table 4). For NDVIg, 99 significant MTAs were detected with PVE ranging from 5 to 17% (Table 2, Fig. 1b) with 14 stable MTAs (Table 2, Table 4, Fig. 1a). One MTA for NDVIa on chromosome 2B (S2B_717098540) is annotated as SAUR-like auxin-responsive protein family (TraesCS2B01G522200) and has been reported to be upregulated under heat stress in Arabidopsis [40]. For NDVIg, MTA on chromosome 1B (S1B_677572998) within gene TraesCS1B01G467900 is annotated as Methyltransferase which has a predicted role of genetic or epigenetic regulation of heat responses in plants [41]. Another MTA (S5B_583295527) within gene TraesCS5B01G407600 had functional annotation for Myb family transcription factor-like protein. Myb transcription factor were reported to be significantly induced by heat treatment in rice and wheat and thus play important roles in response to high temperature stress [42, 43]. Moreover, we found several other MTAs whose annotations suggest different roles including response to abiotic stress including drought stress (Multidrug resistance protein ABC transporter family protein, Myb/SANT-like DNA-binding domain protein, RING finger protein 13) [44–47]. Eight common MTAs were detected for NDVI at anthesis and grain filing period on chromosomes 1D, 3B, 5A, 6A, 7B, and 7D which can be potentially important targets for marker assisted selection. Three MTAs (S3A_699988530, S7B_701649275, and S7D_635578722) have annotations suggesting their role in heat tolerance [27, 28, 48]. Our study identified novel MTAs associated with NDVI which provides insight into the genetic basis of this trait in wheat at different growth stages in US soft wheat under HDT and HNT conditions.
The selective permeability of plasma membrane is highly sensitive to heat stress affecting growth and development of a plant [49]. Therefore, MT is another important PT for understanding heat tolerance in wheat where lower expression of solute leakage leaf tissue indicates stability (tolerance) of cell membrane to elevated temperature [24]. We measured solute leakage from heat stressed plant tissue to estimate damage to cell membrane [50]. Overall, we found significant genotypic variation in SWAMP for MT with moderate broad sense heritability (0.60) (Table 1) in agreement with previous studies [51]. MT was positively correlated with GY, SF, GN, HI, SHI, TGW, NDVIa, NDVIg and SPAD indicating that MT can be used as an additive component trait to improve yield potential in wheat under HT (Additional file 4). We identified 95 MTAs for MT with PVEs ranging from 5 to 15% (Table 2, Fig. 1b) indicating its quantitative nature. Twenty out of 95 MTAs were expressed in multiple environments indicating stability of these markers under different environments (Table 4). Twenty stable MTAs were located in chromosomes 1B, 1D, 2A, 2D, 3A, 3B, 3D, 5B, 5D, 6A, 6B, and 7B (Fig. 1a). Seven of these stable MTAs had functional annotation suggesting their involvement in abiotic stress including heat stress (Table 5). Two MTAs (S3D_590224603, and S3D_590224620) within 17 bp of each other were detected within gene TraesCS3D01G501200 with a functional annotation of protein kinase (Table 5). Protein kinases have been found to play a role in plant defense and adaptation responses and were reported to be upregulated by heat stress in durum wheat [52]. Another MTA (S1D_262475151) within gene TraesCS1D01G190700 (Heavy metal transport/detoxification superfamily protein) was reported to have an important role in growth and development of canola under heat stress conditions [53]. An MTA on chromosome 6B (S6B_42215716) within gene TraesCS6B01G063500 is annotated as peroxidase. Heat stress triggers the production and accumulation of harmful reactive oxygen species like hydrogen peroxide and their detoxification by antioxidant systems is important for protecting plants against heat stress [54, 55]. A significant increase in the activity of peroxidase (antioxidant) under short term heat stress has been reported in heat tolerant genotypes indicating efficient antioxidative defense system in wheat [56]. Two MTAs within 23 bp on chromosome 2B (S5B_509105168 and S5B_509105191) were in gene TraesCS5B01G325000 with annotation as F-box family protein.
Another parameter that has been frequently used to estimate heat tolerance in wheat is CT [57]. Lower canopy temperature indicates water status and transpiration rate in controlling temperature to avoid dehydration under stress [37, 58]. In this study, there was significant genotypic variation in CT with moderate broad-sense heritability (0.35) (Table 1). CT showed a significant negative correlation with GY, GN, SPAD, and MT. Genotypes with cooler canopies are presumed to have better root systems and retain chlorophyll content and membrane stability resulting in higher yield under high temperature. We identified 110 MTAs for CT with PVEs ranging from 8 to 13% (Table 2, Fig. 1b). Seventeen out of 110 MTAs were expressed in multiple environments indicating stability of these markers under different environments (Table 4). Stable MTAs were located in chromosomes 2B, 5B, 5D, 6B, and 7B (Fig. 1a). Ten of the stable MTAs were located in five genes with annotated functions. An MTA on chromosome 5B (S5B_610295429) is within gene TraesCS5B01G436700 which is annotated as a lipid transfer protein. Lipid transfer proteins are low molecular weight proteins that are involved in many biological roles, such as anther development, different signaling pathways and heat stress both at the seedling and the grain-filling stages [59]. One MTA (S5B_621237427) on chromosome 5B was found in gene (TraesCS5B01G448700). This gene is annotated as mitochondrial transcription termination factor-like protein and is reported to be involved in heat tolerance in Arabidopsis [60]. Another MTA detected on chromosomes 5B (S5B_601343966), has gene annotation for Zinc finger CCCH domain-containing protein 16 (TraesCS5B01G425500). This protein was found to be over expressed upon high temperature stress in bread wheat [61]. We found several other MTAs whose annotations suggest different roles including abiotic stress response (BTB/POZ and MATH domain-containing protein 2), senescence (Maintenance of telomere capping protein 2, Protein Phosphatase 2C) and salinity stress (Peptidase M50 family protein) [62–64].
In summary, we detected 500 MTAs located in different chromosomes out of which 81 MTAs were linked to the same trait in multiple environments (suggesting stability) and ten MTAs linked to multiple traits (suggesting pleiotropy) (Table 3-4). Notably, MTAs associated with multiple PTs within different genomic regions had the same functional annotation (Fig. 2; and Additional file 8). For instance, 13 MTAs for SPAD, MT, CT, NDVIa and NDVIg were annotated as F-box family proteins (Fig. 2). Similarly, the genes annotated as zinc finger proteins harbored MTAs for SPAD, CT and NDVIa. This result indicated the likely gene families that are important for physiological traits to improve yield potential under heat stress.
Some MTAs associated with PTs had pleiotropic effects with GY and other yield related traits (Additional file 9). These MTAs have been described in our previous studies. In a previous study [10], we found two MTAs (S3A_12554694 and S3A_12554700) within 6 bp were associated with TGW and GY. In this study, we found the same MTA associated with MT indicating that plasma membrane thermostability may contribute to increased TGW and GY under heat stress condition. Another MTA on chromosome 7D (S7D_18808932) also had a pleiotropic effect on MT and TGW. MTAs with pleiotropic effect on CT and TGW were detected on chromosome 6B (S6B_149148874) under multiple HT environments. One MTA (S5A_590056740), associated with HI in our previous study [10] was also linked to NDVIa and NDVIg. The co-localization of observed MTAs for HI with NDVIa and NDVIg suggests that HI is highly dependent upon greenness at anthesis as well as grain filling period under heat stress. This might indicate that higher photosynthetic reserves at anthesis can later be translocated to the developing grain.
In recent years, rapid progress in ground-based and unmanned aerial high-throughput field phenotyping have resulted in a variety of non-invasive imaging techniques which can lead to significant improvements in precision and speed of phenotyping for PTs in large plant populations with high resolution and high precision. These platforms can have large impact on validating as well as finding new genetic loci that are relevant to heat tolerance.
Conclusion
A large number of MTAs were identified in soft red winter wheat under the environments with HDT and HNT conditions. The MTAs were detected for four PTs: SPAD, MT, CT, NDVI for which there is reasonable evidence of being heat adaptive. We found several stable loci across environments and pleiotropic markers controlling multiple traits among PTs and also associated with yield-related traits under hot environments. We identified candidate genes affecting several important biological processes in plants including response to heat stress. Identifying regulatory loci associated with traits like PTs and yield-related traits can assist in developing ideotypes that can maximize the amount of assimilates and conversion of enhanced carbon capture and biomass growth for improving yield potential. Further validation of these MTAs in different controlled environmental conditions is required before they can be used extensively in marker assisted selection and breeding for heat tolerance in wheat.
Methods
Plant materials and experimental design
Field evaluations were conducted on 236 advanced genotypes of a soft red winter wheat association mapping panel (SWAMP) that are well adapted to the warm and humid south and southeastern regions of the USA. These lines were developed by public and private soft wheat breeding programs in the south and southeastern USA and the list is available in the NCBI database with accession number PRJNA578088 (https://www.ncbi.nlm.nih.gov//bioproject/PRJNA578088). The seeds were collected from different soft wheat breeding programs (University of Arkansas, University of Georgia and Louisiana State University) from in the south and southeastern USA. The SWAMP was evaluated for PTs in five trials at the two heat stressed locations in Florida: Citra and Quincy. Citra had more frequent episodes of HT (> 30 °C) during the grain filling period than Quincy (Additional file 1). In Citra, the SWAMP was phenotyped for three growing seasons: 2015–2016 (29.407215 °N, − 82.1876 °W, Elevation = 23 m), 2016–2017 (29.405701 °N, − 82.175818 °W, Elevation = 23 m), and 2017–2018 (29.403853 °N, − 82.17429 °W, Elevation = 23 m). In Quincy, SWAMP was evaluated in two growing seasons: 2015–2016 (30.5546202 °N, − 84.59533 °W, Elevation = 76 m) and 2016–2017 (30.549658 °N, − 84.59835 °W, Elevation = 76 m). Traits assessed in each year and used in GWAS can be found in (Additional file 2). All yield trials were planted in six row plots (3 m length × 1.5 m width) at the seeding rate of 100 kgh− 1. The SWAMP was planted in randomized augmented block design [65] in all trials with 236 un-replicated entries and three repeated check varieties (SS8641, AGS2000 and Jamestown).
Trait measurement
Four physiological traits (PTs) including CT, SPAD, NDVI, and MT were measured at different time points. CT was measured three times during grain filling period (Zadoks stages 67, 72, and 77) using a handheld infrared thermometer (Fluke 572–2 IR thermometer, Fluke Corporation, Everett WA) on sunny days when the temperature reached the daily high between 1300 and 1500 h [66]. CT data were taken from the same side of each plot at 50 distance from the edge and approximately 50 cm above the canopy at an angle of 30o to the horizontal. The average of three time point readings was used for the association analysis. Chlorophyll content was measured on flag leaves from eight random main stems for each plot at anthesis plus seven days (Zadoks stage 72) using a handheld self-calibrated SPAD chlorophyll meter (Minolta SPAD-502 Spectrum Technologies Inc., Plainfield, IL, US). The SPAD-502 instrument provides a convenient means of assessing relative leaf chlorophyll concentration. The chlorophyll content was measured on intact flag leaves one third of the way from the base of the abaxial surface. The average of eight readings was used for further statistical analysis.
SPAD Chlorophyll meter data were taken on the same day or the closest possible day coinciding with CT and MT. NDVI was measured at anthesis (NDVIa; Zadoks stage 65) and grain filling stage (NDVIg; Zadoks stage 77). A GreenSeeker handheld crop sensor (Trimble Navigation Limited 935 Stewart Drive Sunnyvale, California 94,085) was used for collecting NDVI readings. The GreenSeeker handheld crop sensor was hold 50 cm above the canopy facing the center of the bed. A 30–40 NDVI readings were recorded/plot and the mean value of those readings represented the NDVI value of the respective plot. To determine MT, flag leaves were collected from ten random main stems at anthesis plus seven days (Zadoks stage 72) from each plot. One cm diameter leaf disks from each leaf were extracted from midway between the base and the tip of the leaf blade using a leaf disc puncher and placed in glass vials containing 20 ml deionized water. The vials were placed in shaker for 24 h at room temperature to ensure diffusion of electrolytes. After 24 h, electrolyte leakage was measured using a conductometer (Thermo Scientific Orion Star A212) followed by autoclaving the vials (0.10 MPa pressure, 121 °C for 15 min) to release all the electrolytes from plant tissue. The vials were placed in shaker for 24 h and electrolyte leakage was measured again. MT was expressed in percentage units as the reciprocal of relative leakage [50].
where T1 is the conductivity reading after heat treatment, and T2 is the conductivity reading after autoclaving. Grain yield (GY) and yield-related traits including grain number (GN), harvest index (HI), thousand grain weight (TGW), spike fertility (SF), and spike harvest index (SHI) were also calculated to determine correlation among traits. The details of the calculation of these traits is described in previous study [10].
Phenotypic data analysis
Combined analysis of variance (ANOVA) was conducted assuming a mixed linear model. The ‘lme4’ package [67] and the R software program (v3.5.1, R Development Core Team) were used to calculate best linear unbiased estimates (BLUEs) assuming a fixed genotypic effect (all other effects were random):
where the phenotypic response (Yijk) is a function of the overall mean (μ), ith genotype (Gi), jth environment, genotype-environment interaction (GEij), kth block (Bk) nested within the jth environment (Ej), and the residual error (εijk).
BLUEs for combined as well as individual locations were also calculated and therefore will be discussed hereafter as BLUEC (BLUE values estimated from Citra), BLUEQ (BLUE values estimated from Quincy) and BLUEA (BLUE values estimated from all environments). Broad sense heritability was calculated assuming random genotypic effect (all other effects were random) and was obtained by:
where H2, broad-sense heritability estimate; σ2G, genetic variance; σ2G × E, genotype-by-environmental variance; σ2e, residual variance; n, number of environments; and r, number of replications.
Pearson’s correlations were calculated from BLUEs in R using the “corrplot” package (v3.5.1, R Development Core Team) and used to determine the direction and magnitude of measured trait associations. Associations between traits were also explored in principle component (PC) biplot analysis using the package “factoextra” in R [68].
Genotyping
The detail of genotyping process, SNP discovery and filtering criteria has been described in previous study [10]. In brief, DNA was isolated from fresh, young leaves using a modified Cetyl trimethylammonium bromide (CTAB) protocol [69]. The GBS libraries were prepared using MspI and PstI-HF restriction enzymes and pooled together in 96-plex and sequenced in an Ion Torrent Proton sequencer (Thermo Fisher Scientific, Waltham, MA, USA) at the USDA Central Small Grain Genotyping Lab, Kansas State University, Manhattan, KS, USA. Prior to analysis, 80 poly-A bases were appended to the 3′ end of all sequencing reads so that TASSEL 5.0 would attempt to use reads shorter than 64 bases rather than discarding short reads. SNP calling was performed in TASSEL v5.0 GBS v2.0 discovery pipeline [70] and aligned to the Chinese Spring wheat (RefSeq v1) genome sequence [71] using the default settings of BWA (version 0.6.1). The markers were filtered based on the criteria of minor allele frequency (MAF > 5%) and missing values (< 20%).
Linkage disequilibrium, population structure and GWAS analysis
Linkage disequilibrium (LD) and population structure analysis of the SWAMP has been described in detail previously [10]. Briefly, “LDcorSV” package [72] in R (v3.5.1, R Development Core Team) was used to estimate LOESS (Locally weighted scatterplot smoothing) regressions of mean r2 (coefficient of LD) between pairs of SNPs sampled at the range of 30,000, 40,000, and 50,000 bp. The intersection between critical value (r2 = 0.2) and LOESS line was considered as the distance beyond which LD starts to decay. Population structure was observed using Bayesian information criterion (BIC) score provided by discriminant analysis of principal components (DAPC, “adegenet” package, R Development Core Team 2013) [73] to determine the optimum number of demes supported by the results. The principal component analysis was performed using “prcomp” (“stats” package) to investigate the genetic differentiation among and within demes.
GWAS was performed in three BLUE datasets (BLUEC, BLUEQ, BLUPA) for each trait to identify significant MTAs in SWAMP using Fixed and random model Circulating Probability Unification (FarmCPU) model [74, 75] executed in the Genome Association Prediction Integrated Tool (GAPIT) package in R software package [76]. The first three principal components were used as covariates by observing model fit in Q-Q (quantile-quantile) plots, and kinship was determined from FarmCPU [74]. A uniform value of -log10(p) = 4.00 (p = 9.99 × 10− 4) was used as the cut-off to define significant MTAs based on Q-Q plots [77, 78]. Candidate genes associated with significant MTAs and their annotation were identified using Chinese Spring wheat reference genome (IWGSC RefSeq v1.0) [71]. The putative genes were further investigated in past literature to determine their association with phenotypic traits under heat stress.
Supplementary information
Additional file 1. Weather table showing number of hours in daytime (> 24 °C) and nighttime (> 15 °C) temperature during grain filling stages (Mar 15 - Apr 30). The soft wheat association mapping panel (SWAMP) was planted in three seasons in Citra (2015/2016, 2016/2017, 2017, 2018) and two seasons in Quincy (2015/2016, 2016/2017).
Additional file 2. Physiological traits for the SWAMP assessed in each year and used in GWAS.
Additional file 3. Summary of ANOVA results testing the effects of genotype (G), environment (E), and genotype-by-environment interaction (G × E). The table includes mean square values and significance level of each term. SPAD, soil-plant analyses development; MT, cell membrane thermostability; CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling.
Additional file 4. Pearson’s correlation coefficient (r) between physiological traits in SWAMP. SF, spike fertility (grains g-1 chaff weight); GY, grain yield (kg h-1); GN, grain number m-2; TGW, thousand grain weight (g); SHI, spike harvest index; HI, harvest index; SPAD, soil-plant analyses development; MT, cell membrane thermostability; CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling.
Additional file 5. Principal component bi-plot analysis of measured traits for the SWAMP. SF, spike fertility (grains g− 1 chaff weight); GY, grain yield (kg h− 1); GN, grain number m− 2; TGW, thousand grain weight (g); SHI, spike harvest index; HI, harvest index; SPAD, soil-plant analyses development; MT, cell membrane thermostability; CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling.
Additional file 6. Population structure of the SWAMP based on 27,466 SNPs. (A) bar charts showing posterior probabilities of assignment to three groups based on algorithms of discriminant analysis of principal components (DAPC). (B) Population structure among demes inferred from PC analysis. The populations were colored based on the posterior of probability assigned to three genetic groups inferred from DAPC.
Additional file 7. Linkage disequilibrium represented by the r2 against physical distance (in bp) showing LD decay. LOESS regressions of mean r2 between pairs of SNPs vs. physical distance were sampled at 30,000 (red), 40,000 (blue), and 50,000 (green) bp. Grey line represents the critical value beyond which LD is likely caused by physical linkage.
Additional file 8. Summary of all significant markers and their functional annotations associated with eight traits in SWAMP.
Additional file 9. Summary of significant pleiotropic MTAs associated with PTs, GY and other yield related traits.
Acknowledgements
The research work was conducted through the financial support of UF/IFAS early career award program. The authors acknowledge the contribution of Agronomy department of UF to provide field and lab facilities to conduct the research.
Abbreviations
- HT
high temperature
- MTAs: GWAS
genome-wide association study
- MTAs
marker-trait associations
- HDT
daytime maximum temperature
- HNT
night-time minimum temperatures
- HI
harvest index
- PTs
physiological traits
- RUE
radiation use efficiency
- LI
light interception
- SPAD
soil-plant analyses development
- MT
membrane thermostability
- CT
canopy temperature
- NDVI
normalized difference vegetation index
- NDVIa
normalized difference vegetation index at anthesis
- NDVIg
normalized difference vegetation index at grain filling
- QTL
quantitative trait loci
- SWAMP
soft winter wheat association mapping panel
- GY
grain yield
- TGW
thousand grain weight
- SF
spike fertility
- SHI
spike harvest index
- BLUEs
best linear unbiased estimates
- BLUEC
BLUE values estimated from Citra
- BLUEQ
BLUE values estimated from Quincy
- BLUEA
BLUE values estimated from all environments
- LD
linkage disequilibrium
- FarmCPU
Fixed and random model Circulating Probability Unification
- PVE
phenotypic variance explained
Authors’ contributions
MAB and SP conceived and designed the study. SP completed the study and data analysis and wrote the manuscript. SP, JG, JK, DS, MA, JM, SK and SS collected the phenotypic data on association panel and edited the manuscript. GB and PA performed marker analysis and SNP calling. SA, SG, BB, AB, and SH edited the manuscript. All Authors read and approved the manuscript.
Funding
This research was funded by UF/IFAS early career award. The funding sources had no influence on the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The phenotypic datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The genotypic datasets generated and/or analyzed during the current study are available in the NCBI using accession number PRJNA578088 (https://www.ncbi.nlm.nih.gov//bioproject/PRJNA578088).
Ethics approval and consent to participate
The field trail experiments in the current study were permitted by University of Florida.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-6717-7.
References
- 1.FAOSTAT. Statistical databases and datasets of the Food and Agriculture Organization of the United Nations. http://www.faostatfaoorg/. 2016.
- 2.Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. Food security: the challenge of feeding 9 billion people. science. 2010;327(5967):812–818. [DOI] [PubMed]
- 3.Lobell D, Schlenker W, Costa-Roberts J. Climate trends and global crop production since 1980. Science. 2011;333(6042):616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- 4.Pradhan GP, Prasad PV, Fritz AK, Kirkham MB, Gill BS. Effects of drought and high temperature stress on synthetic hexaploid wheat. Funct Plant Biol. 2012;39(3):190–198. doi: 10.1071/FP11245. [DOI] [PubMed] [Google Scholar]
- 5.IPCC. Climate change 2007: the physical science basis: summary for policymakers. http://www.userstelenetbe/jjanssens/CommentsSPM4webpdf. 2007.
- 6.IPCC. Climate change 2001: the scientific basis. http://www.keneamazonnet/Documents/Publications/Virtual-Library/Impacto/9pdf. 2013:881.
- 7.Farooq M, Bramley H, Palta JA, Siddique KH. Heat stress in wheat during reproductive and grain-filling phases. Crit Rev Plant Sci. 2011;30(6):491–507. doi: 10.1080/07352689.2011.615687. [DOI] [Google Scholar]
- 8.Narayanan S, Prasad P, Fritz A, Boyle D, Gill B. Impact of high night-time and high daytime temperature stress on winter wheat. J Agron Crop Sci. 2015;201(3):206–218. doi: 10.1111/jac.12101. [DOI] [Google Scholar]
- 9.Lobell DB, Ortiz-Monasterio JI. Impacts of day versus night temperatures on spring wheat yields. Agron J. 2007;99(2):469–477. doi: 10.2134/agronj2006.0209. [DOI] [Google Scholar]
- 10.Pradhan S, Babar M, Robbins K, Bai G, Mason RE, Khan J, Shahi D, Avci M, Guo J, Bhatta M, et al. Understanding the genetic basis of spike fertility to improve grain number, harvest index, and grain yield in wheat under high temperature stress environments. Front Plant Sci. 2019;10:1481. doi: 10.3389/fpls.2019.01481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foulkes MJ, Slafer GA, Davies WJ, Berry PM, Sylvester-Bradley R, Martre P, Calderini DF, Griffiths S, Reynolds MP. Raising yield potential of wheat. III. Optimizing partitioning to grain while maintaining lodging resistance. J Exp Bot. 2011;62(2):469–486. doi: 10.1093/jxb/erq300. [DOI] [PubMed] [Google Scholar]
- 12.Xu Q, Paulsen AQ, Guikema JA, Paulsen GM. Functional and ultrastructural injury to photosynthesis in wheat by high temperature during maturation. Environ Exp Bot. 1995;35(1):43–54. doi: 10.1016/0098-8472(94)00030-9. [DOI] [Google Scholar]
- 13.Reynolds M, Rajaram S, Sayre K. Physiological and genetic changes of irrigated wheat in the post–green revolution period and approaches for meeting projected global demand. Crop Sci. 1999;39(6):1611–1621. doi: 10.2135/cropsci1999.3961611x. [DOI] [Google Scholar]
- 14.Babar M, Reynolds M, Van Ginkel M, Klatt A, Raun W, Stone M. Spectral reflectance indices as a potential indirect selection criteria for wheat yield under irrigation. Crop Sci. 2006;46(2):578–588. doi: 10.2135/cropsci2005.0059. [DOI] [Google Scholar]
- 15.Cossani CM, Reynolds MP. Physiological traits for improving heat tolerance in wheat. Plant Physiol. 2012;160(4):1710–1718. doi: 10.1104/pp.112.207753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chenu K, Deihimfard R, Chapman SC. Large-scale characterization of drought pattern: a continent-wide modelling approach applied to the Australian wheatbelt–spatial and temporal trends. New Phytol. 2013;198(3):801–820. doi: 10.1111/nph.12192. [DOI] [PubMed] [Google Scholar]
- 17.Loss SP, Siddique K. Morphological and physiological traits associated with wheat yield increases in Mediterranean environments. Adv Agron. 1994;52:229–276. doi: 10.1016/S0065-2113(08)60625-2. [DOI] [Google Scholar]
- 18.Reynolds M, Langridge P. Physiological breeding. Curr Opin Plant Biol. 2016;31:162–171. doi: 10.1016/j.pbi.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Sukumaran S, Yu J. Association mapping of genetic resources: achievements and future perspectives. Genomics of plant genetic resources. 2014:207–35.
- 20.Ogbonnaya FC, Rasheed A, Okechukwu EC, Jighly A, Makdis F, Wuletaw T, Hagras A, Uguru MI, Agbo CU. Genome-wide association study for agronomic and physiological traits in spring wheat evaluated in a range of heat prone environments. Theor Appl Genet. 2017:1–17. [DOI] [PubMed]
- 21.Prasad P, Pisipati S, Ristic Z, Bukovnik U, Fritz A. Impact of nighttime temperature on physiology and growth of spring wheat. Crop Sci. 2008;48(6):2372–2380. doi: 10.2135/cropsci2007.12.0717. [DOI] [Google Scholar]
- 22.Sharma DK, Andersen SB, Ottosen C-O, Rosenqvist E. Phenotyping of wheat cultivars for heat tolerance using chlorophyll a fluorescence. Funct Plant Biol. 2012;39(11):936–947. doi: 10.1071/FP12100. [DOI] [PubMed] [Google Scholar]
- 23.Rosyara UR, Subedi S, Duveiller E, Sharma RC. Photochemical efficiency and SPAD value as indirect selection criteria for combined selection of spot blotch and terminal heat stress in wheat. J Phytopathol. 2010;158(11–12):813–821. doi: 10.1111/j.1439-0434.2010.01703.x. [DOI] [Google Scholar]
- 24.Reynolds M, Balota M, Delgado M, Amani I, Fischer R. Physiological and morphological traits associated with spring wheat yield under hot, irrigated conditions. Funct Plant Biol. 1994;21(6):717–730. doi: 10.1071/PP9940717. [DOI] [Google Scholar]
- 25.Reynolds M, Singh R, Ibrahim A, Ageeb O, Larque-Saavedra A, Quick J. Evaluating physiological traits to complement empirical selection for wheat in warm environments. Euphytica. 1998;100(1–3):85–94. doi: 10.1023/A:1018355906553. [DOI] [Google Scholar]
- 26.Rahaman MM. Genome-Wide Association Study of Heat Tolerance in Rapeseed/Canola (Brassica napus L.). North Dakota State University. 2016.
- 27.Li Q, Wang W, Wang W, Zhang G, Liu Y, Wang Y, Wang W. Wheat F-box protein gene TaFBA1 is involved in plant tolerance to heat stress. Front Plant Sci. 2018;9:521. doi: 10.3389/fpls.2018.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. F-box proteins everywhere. Curr Opin Plant Biol. 2006;9(6):631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Chopra R, Burow G, Burke JJ, Gladman N, Xin Z. Genome-wide association analysis of seedling traits in diverse Sorghum germplasm under thermal stress. BMC Plant Biol. 2017;17(1):12. doi: 10.1186/s12870-016-0966-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belknap WR, Garbarino JE. The role of ubiquitin in plant senescence and stress responses. Trends Plant Sci. 1996;1(10):331–335. doi: 10.1016/S1360-1385(96)82593-0. [DOI] [Google Scholar]
- 31.Oyiga BC. Genetic variation of traits related to salt stress response in Wheat (Triticum aestivum L.). Universitäts-und Landesbibliothek Bonn; 2016.
- 32.Bhatta M, Morgounov A, Belamkar V, Baenziger P. Genome-wide association study reveals novel genomic regions for grain yield and yield-related traits in drought-stressed synthetic hexaploid wheat. Int J Mol Sci. 2018;19(10):3011. doi: 10.3390/ijms19103011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moffatt BA, Wang L, Allen MS, Stevens YY, Qin W, Snider J, von Schwartzenberg K. Adenosine kinase of Arabidopsis. Kinetic properties and gene expression. Plant Physiol. 2000;124(4):1775–1785. doi: 10.1104/pp.124.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woldesemayat AA, Ntwasa M. Pathways and Network Based Analysis of Candidate Genes to Reveal Cross-Talk and Specificity in the Sorghum (Sorghum bicolor (L.) Moench) Responses to Drought and It's Co-occurring Stresses. Front Genet. 2018;9:557–557. [DOI] [PMC free article] [PubMed]
- 35.Deshmukh AB, Datir SS, Bhonde Y, Kelkar N, Samdani P, Tamhane VA. De novo root transcriptome of a medicinally important rare tree Oroxylum indicum for characterization of the flavonoid biosynthesis pathway. Phytochemistry. 2018;156:201–213. doi: 10.1016/j.phytochem.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 36.Katam R, Sakata K, Suravajhala P, Pechan T, Kambiranda DM, Naik KS, Guo B, Basha SM. Comparative leaf proteomics of drought-tolerant and -susceptible peanut in response to water stress. J Proteome. 2016;143:209–226. doi: 10.1016/j.jprot.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds M, Trethowan R. Physiological interventions in breeding for adaptation to abiotic stress. Frontis. 2007:127–44.
- 38.Sukumaran S, Reynolds MP, Sansaloni C. Genome-Wide Association Analyses Identify QTL Hotspots for Yield and Component Traits in Durum Wheat Grown under Yield Potential, Drought, and Heat Stress Environments. Frontiers in Plant Science. 2018;9(81). [DOI] [PMC free article] [PubMed]
- 39.Freeman K, Raun W, Johnson G, Mullen R, Stone M, Solie J. Late-season prediction of wheat grain yield and grain protein. Commun Soil Sci Plant Anal. 2003;34(13–14):1837–1852. doi: 10.1081/CSS-120023219. [DOI] [Google Scholar]
- 40.Kim E. How does light affect the heat stress response in Arabidopsis? 2018. [Google Scholar]
- 41.Liu J, Feng L, Li J, He Z. Genetic and epigenetic control of plant heat responses. Frontiers in Plant Science. 2015;6(267). [DOI] [PMC free article] [PubMed]
- 42.Zhang X, Rerksiri W, Liu A, Zhou X, Xiong H, Xiang J, Chen X, Xiong X. Transcriptome profile reveals heat response mechanism at molecular and metabolic levels in rice flag leaf. Gene. 2013;530(2):185–192. doi: 10.1016/j.gene.2013.08.048. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y, Tian X, Wang F, Zhang L, Xin M, Hu Z, Yao Y, Ni Z, Sun Q, Peng H. Characterization of wheat MYB genes responsive to high temperatures. BMC Plant Biol. 2017;17(1):208. doi: 10.1186/s12870-017-1158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang J, Park J, Choi H, Burla B, Kretzschmar T, Lee Y, Martinoia E. Plant ABC transporters. Arabidopsis Book. 2011;9:–e0153. [DOI] [PMC free article] [PubMed]
- 45.Puri A. Quantitative proteome analysis of alfalfa in drought stress under the influence of miR156. 2019. [Google Scholar]
- 46.Song W-Y, Chen X, Huang X. Drought Tolerant Plants. US Patent Application No 16/186,954. 2019.
- 47.Toueni M, Ben C, Le Ru A, Gentzbittel L, Rickauer M. Quantitative Resistance to Verticillium Wilt in Medicago truncatula Involves Eradication of the Fungus from Roots and Is Associated with Transcriptional Responses Related to Innate Immunity. Frontiers in Plant Science. 2016;7(1431). [DOI] [PMC free article] [PubMed]
- 48.Zhang X, Xiong H, Liu A, Zhou X, Peng Y, Li Z, Luo G, Tian X, Chen X. Microarray data uncover the genome-wide gene expression patterns in response to heat stress in rice post-meiosis panicle. Journal of Plant Biology. 2014;57(6):327–336. doi: 10.1007/s12374-014-0177-z. [DOI] [Google Scholar]
- 49.Bajji M, Kinet J-M, Lutts S. The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 2002;36(1):61–70. doi: 10.1023/A:1014732714549. [DOI] [Google Scholar]
- 50.Ibrahim AM, Quick JS. Genetic control of high temperature tolerance in wheat as measured by membrane thermal stability. Crop Sci. 2001;41(5):1405–1407. doi: 10.2135/cropsci2001.4151405x. [DOI] [Google Scholar]
- 51.Fokar M, Nguyen HT, Blum A. Heat tolerance in spring wheat. I. Estimating cellular thermotolerance and its heritability. Euphytica. 1998;104(1):1–8. doi: 10.1023/A:1018346901363. [DOI] [Google Scholar]
- 52.Rampino P, Mita G, Fasano P, Borrelli GM, Aprile A, Dalessandro G, De Bellis L, Perrotta C. Novel durum wheat genes up-regulated in response to a combination of heat and drought stress. Plant Physiol Biochem. 2012;56:72–78. doi: 10.1016/j.plaphy.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 53.Rahaman M, Mamidi S, Rahman M. Association mapping of agronomic traits of canola ('Brassica napus' L.) subject to heat stress under field conditions. Australian Journal of Crop Science. 2017;11(9):1094.
- 54.Esfandiari E, Shekari F, Shekari F, Esfandiari M. The effect of salt stress on antioxidant enzymes activity and lipid peroxidation on wheat seedling. Notulae Botanicae Horti Agrobotanici Cluj-Napoca. 2007;35(1):48–56. [Google Scholar]
- 55.Almeselmani M, Deshmukh P, Sairam R. High temperature stress tolerance in wheat genotypes: role of antioxidant defence enzymes. Acta Agronomica Hungarica. 2009;57(1):1–14. doi: 10.1556/AAgr.57.2009.1.1. [DOI] [Google Scholar]
- 56.Gupta NK, Agarwal S, Agarwal VP, Nathawat NS, Gupta S, Singh G. Effect of short-term heat stress on growth, physiology and antioxidative defence system in wheat seedlings. Acta Physiol Plant. 2013;35(6):1837–1842. doi: 10.1007/s11738-013-1221-1. [DOI] [Google Scholar]
- 57.Amani I, Fischer R, Reynolds M. Canopy temperature depression association with yield of irrigated spring wheat cultivars in a hot climate. J Agron Crop Sci. 1996;176(2):119–129. doi: 10.1111/j.1439-037X.1996.tb00454.x. [DOI] [Google Scholar]
- 58.Blum A. Plant breeding for stress environments. CRC Press. 1988.
- 59.Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9(5):244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 60.Kim M, Lee U, Small I. Des francs-Small CC, Vierling E. mutations in an Arabidopsis mitochondrial transcription termination factor–related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell. 2012;24(8):3349–3365. doi: 10.1105/tpc.112.101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chauhan H, Khurana N, Tyagi AK, Khurana JP, Khurana P. Identification and characterization of high temperature stress responsive genes in bread wheat (Triticum aestivum L.) and their regulation at various stages of development. Plant Mol Biol. 2011;75(1–2):35–51. doi: 10.1007/s11103-010-9702-8. [DOI] [PubMed] [Google Scholar]
- 62.Kushwaha HR, Joshi R, Pareek A, Singla-Pareek SL. MATH-domain family shows response toward abiotic stress in Arabidopsis and Rice. Front Plant Sci. 2016;7:923. doi: 10.3389/fpls.2016.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Brien M, Grogan H, Kavanagh K. Proteomic response of Trichoderma aggressivum f. europaeum to Agaricus bisporus tissue and mushroom compost. Fungal biology. 2014;118(9–10):785–791. doi: 10.1016/j.funbio.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 64.Renaud AL. Genetic regulation of maize and sorghum under abiotic stress. Purdue University. 2015.
- 65.Federer WT, Raghavarao D. On augmented designs. Biometrics. 1975:29–35.
- 66.Pask A, Pietragalla J, Mullan D, Reynolds M. Physiological breeding II: a field guide to wheat phenotyping. CIMMYT. 2012.
- 67.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. https://www.apdf/pdf/14065823pdf. 2014:1–51.
- 68.Kassambara A, Mundt F. Factoextra: extract and visualize the results of multivariate data analyses. https://mranmicrosoftcom/snapshot/2016-06-09/web/packages/factoextra/factoextrapdf. 2016.
- 69.Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard R. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci. 1984;81(24):8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 71.Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Choulet F, Distelfeld A, Poland J. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361(6403). [DOI] [PubMed]
- 72.Desrousseaux D, Sandron F, Siberchicot A, Cierco-Ayrolles C, Mangin B, Siberchicot MA. Package 'LDcorSV'. https://www.cranstatunipdit/web/packages/LDcorSV/LDcorSVpdf. 2017.
- 73.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11(1):94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu X, Huang M, Fan B, Buckler ES, Zhang Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016;12(2):e1005767. doi: 10.1371/journal.pgen.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arora S, Singh N, Kaur S, Bains NS, Uauy C, Poland J, Chhuneja P. Genome-wide association study of grain architecture in wild wheat Aegilops tauschii. Front Plant Sci. 2017;8:886. doi: 10.3389/fpls.2017.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28(18):2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 77.Sukumaran S, Xiang W, Bean SR, Pedersen JF, Kresovich S, Tuinstra MR, Tesso TT, Hamblin MT, Yu J. Association mapping for grain quality in a diverse sorghum collection. The Plant Genome. 2012;5(3):126–135. doi: 10.3835/plantgenome2012.07.0016. [DOI] [Google Scholar]
- 78.Sukumaran S, Dreisigacker S, Lopes M, Chavez P, Reynolds MP. Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor Appl Genet. 2015;128(2):353–363. doi: 10.1007/s00122-014-2435-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Weather table showing number of hours in daytime (> 24 °C) and nighttime (> 15 °C) temperature during grain filling stages (Mar 15 - Apr 30). The soft wheat association mapping panel (SWAMP) was planted in three seasons in Citra (2015/2016, 2016/2017, 2017, 2018) and two seasons in Quincy (2015/2016, 2016/2017).
Additional file 2. Physiological traits for the SWAMP assessed in each year and used in GWAS.
Additional file 3. Summary of ANOVA results testing the effects of genotype (G), environment (E), and genotype-by-environment interaction (G × E). The table includes mean square values and significance level of each term. SPAD, soil-plant analyses development; MT, cell membrane thermostability; CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling.
Additional file 4. Pearson’s correlation coefficient (r) between physiological traits in SWAMP. SF, spike fertility (grains g-1 chaff weight); GY, grain yield (kg h-1); GN, grain number m-2; TGW, thousand grain weight (g); SHI, spike harvest index; HI, harvest index; SPAD, soil-plant analyses development; MT, cell membrane thermostability; CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling.
Additional file 5. Principal component bi-plot analysis of measured traits for the SWAMP. SF, spike fertility (grains g− 1 chaff weight); GY, grain yield (kg h− 1); GN, grain number m− 2; TGW, thousand grain weight (g); SHI, spike harvest index; HI, harvest index; SPAD, soil-plant analyses development; MT, cell membrane thermostability; CT, canopy temperature (°C); NDVIa, normalized difference vegetation index at GS65; NDVIg, normalized difference vegetation index at grain filling.
Additional file 6. Population structure of the SWAMP based on 27,466 SNPs. (A) bar charts showing posterior probabilities of assignment to three groups based on algorithms of discriminant analysis of principal components (DAPC). (B) Population structure among demes inferred from PC analysis. The populations were colored based on the posterior of probability assigned to three genetic groups inferred from DAPC.
Additional file 7. Linkage disequilibrium represented by the r2 against physical distance (in bp) showing LD decay. LOESS regressions of mean r2 between pairs of SNPs vs. physical distance were sampled at 30,000 (red), 40,000 (blue), and 50,000 (green) bp. Grey line represents the critical value beyond which LD is likely caused by physical linkage.
Additional file 8. Summary of all significant markers and their functional annotations associated with eight traits in SWAMP.
Additional file 9. Summary of significant pleiotropic MTAs associated with PTs, GY and other yield related traits.
Data Availability Statement
The phenotypic datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The genotypic datasets generated and/or analyzed during the current study are available in the NCBI using accession number PRJNA578088 (https://www.ncbi.nlm.nih.gov//bioproject/PRJNA578088).