Abstract
Background
Yellow-feathered chickens (YFCs) have a long history in China. They are well-known for the nutritional and commercial importance attributable to their yellow color phenotype. Currently, there is a huge paucity in knowledge of the genetic determinants responsible for phenotypic and biochemical properties of these iconic chickens. This study aimed to uncover the genetic structure and the molecular underpinnings of the YFCs trademark coloration.
Results
The whole-genomes of 100 YFCs from 10 major traditional breeds and 10 Huaibei partridge chickens from China were re-sequenced. Comparative population genomics based on autosomal single nucleotide polymorphisms (SNPs) revealed three geographically based clusters among the YFCs. Compared to other Chinese indigenous chicken genomes incorporated from previous studies, a closer genetic proximity within YFC breeds than between YFC breeds and other chicken populations is evident. Through genome-wide scans for selective sweeps, we identified RALY heterogeneous nuclear ribonucleoprotein (RALY), leucine rich repeat containing G protein-coupled receptor 4 (LGR4), solute carrier family 23 member 2 (SLC23A2), and solute carrier family 2 member 14 (SLC2A14), besides the classical beta-carotene dioxygenase 2 (BCDO2), as major candidates pigment determining genes in the YFCs.
Conclusion
We provide the first comprehensive genomic data of the YFCs. Our analyses show phylogeographical patterns among the YFCs and potential candidate genes giving rise to the yellow color trait of the YFCs. This study lays the foundation for further research on the genome-phenotype cross-talks that define important poultry traits and for formulating genetic breeding and conservation strategies for the YFCs.
Keywords: Yellow, Chicken, Genome, BCDO2, Breeding, Color, Genetic diversity
Background
Different cultures and ethnicities around the globe have developed unique cuisines, into which chickens are incorporated in diverse ways. Chicken consumption is popular globally, with the preference for chicken meat eclipsing that of red meat [1, 2]. Yellow-feathered chickens, otherwise known as “three-yellow chickens” because of their characteristic yellow beak, feathers, and feet [1], and herein abbreviated as YFCs, are a favorite choice for traditional broths and soups in Asian countries, particularly in Korea and southern China. YFCs have been described in the ancient Chinese agricultural text “Qimin Yaoshu” written around 540 C.E [3], and their importance is evidenced by the incredible leap in their demand. For instance, the production of YFC meat in China reached 4445 kt in 2015, representing 31.8% of the national broiler meat yields [4]. The YFCs’ unique meat flavor and color appeal are important factors driving this strong consumer preference. In addition to serving as a traditional nutritional and commercial mainstay for millions of people living in China and its purlieus, YFCs are reported to have contributed to the recent breeding of European chickens [5], indicating a broadening utility of the YFCs. At the present, more than 15 traditional breeds of YFCs are dispersed widely in China [6]. However, these attributes are threatened by the aggressive genetic selection for rapid growth and high feed conversion efficiency in China and other Asian countries [7]. Previous research on YFCs primarily focused on understanding the chemical properties of meat and soups [2, 8–10], or their genetic diversity compared to commercial breeds using low-density markers [1, 11, 12]. Hardly any genome-wide investigations of the population structure and genetic basis of the unique YFC phenotypic traits have been conducted, a major drawback in rational improvement and conservation of these chickens.
In this study, we sought to accomplish an extensive sequencing of YFC populations across China to support their in-depth studies into their evolutionary biology. We also aimed to implement comparative population genetic analyses to determine the genetic structure of the YFCs and retrieve the footprints of selection for their unique color property. This study provides vital resources and insights to facilitate effective avicultural strategies.
Results
Characteristics of the genome datasets
We performed an initial in-depth characterization of the genomes of the 100 YFCs from 10 different breeds and 10 Huaibei (HB) partridge chickens (used for comparisons) sequenced in this study (Fig. 1a; Additional file 1). An average of 86,155,900 clean reads per genome are obtained after quality control protocols, which were then aligned to the reference genome, yielding a mean mapping rate at 87.12% (Additional files 2 and 3, Additional file 4: Fig. S1). The total average base coverage across the genome is 96.35% at a sequencing depth target of 1X, 86.81% at 4X, 41.87% at 10X, and down to 0.36% at 30X (Additional file 4: Fig. S2). The average number of nucleotides in each genome is 11,916,810,290 after filtration, with an average GC content at 44.53% (Additional file 5).
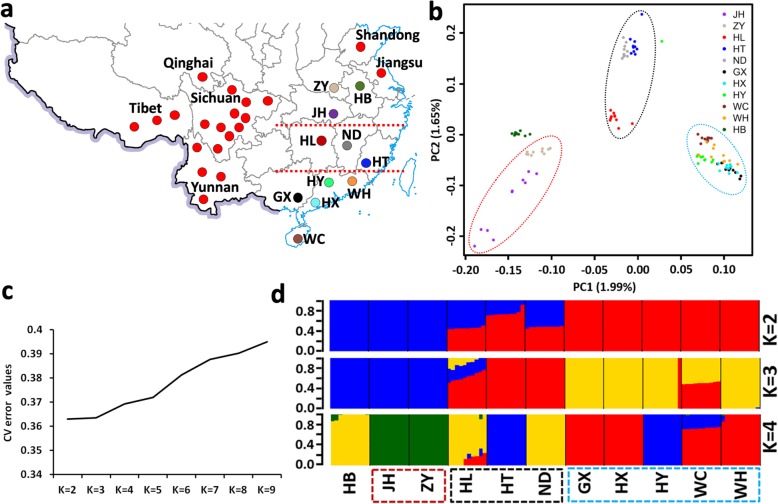
Fig. 1.
Population genomic analysis of the YFCs. a Sampling map (adapted from http://bzdt.ch.mnr.gov.cn/) showing the geographical locations of all chicken breeds/populations. The newly described chicken breeds are noted by their respective population’s IDs, while red cycles indicate chicken populations retrieved from previous studies [13, 14]. The two populations from Shandong and Jiangsu provinces are Yuanbao bantams. The dotted horizontal lines demarcate the three population clusters. b Principal component analysis (PCA) of all 110 chickens sequenced in this study. YFCs’ clustering patterns are highlighted by dotted red (northern cluster), black (central cluster), and blue (southern cluster) circles. Breed code: HB, Huaibei partridge; ZY, Zhengyang Yellow; JH, Jianghan; ND, Ningdu Yellow; HL, Huanglang; HT, Hetian; WH, Wuhua Yellow; HY, Huiyang bearded; HX, Huaixiang; GX, Guangxi Yellow; WC, Wenchang. c-d ADMIXTURE analysis for K = 2, K = 3, and K = 4
For comparative analyses, we merged the 100 YFC and 10 Huaibei partridge chicken genomes with 104, 10, and 1 previously published Chinese chicken, red junglefowl (Gallus gallus; RJF), and green junglefowl (Gallus varius; GVF) genomes, respectively, retaining a total of 3,065,814 common autosomal single nucleotide polymorphisms (SNPs).
Genome variants in the yellow-feathered chickens
After filtration, 16,817,111 single nucleotide polymorphisms (SNPs) and 1,289,024 InDels (insertion or deletion of bases) (≤ 50 bp) were retained. The structural variations (SVs) and the increase or decrease of the copy number of large (> 1 kb) genomic fragments were analyzed. All these genomic variants in the newly generated dataset are summarized in Additional file 4: Fig. S3. Briefly, most of the SNPs are located in intergenic followed by intronic genomic regions (Additional file 4: Fig. S4a). Those located within coding sequences are mainly associated with synonymous or nonsynonymous coding attributes (Additional file 4: Fig. S4b). There are more transitions (11,943,736; 71.02%) than transversions (4,873,375; 28.98%) in the dataset. G- > A and C- > T substitutions are the common transitions at 28% while A- > G and T- > C substitutions are around 21% (Additional file 4: Fig. S5). Different transversions show a low but relatively uniform distribution rate in the dataset. The total average ratio of transitions to transversions is 2.53 (Additional file 6). Analysis of the heterogeneity of clean SNPs shows that about 2,033,275 and 2,259,628 SNPs per genome are homogenous and heterozygous hybrids, respectively (Additional file 7).
Among the high quality InDels, there are more deletions than insertions (785,806 (60.96%) versus 503,218 (39.04%)). The genomic locations of these InDels are summarized in Additional file 4: Fig. S6a, where most InDels are located in non-coding (i.e. intergenic and intronic) regions. Additionally, the four most common genomic consequences of the InDels include frameshift or non-frameshift insertions and deletions (Additional file 4: Fig. S6b).
Besides SNPs and InDels, SVs, which represent a large range of chromosomal variations encompassing large genomic regions have been characterized. These include large fragment deletions (DEL), insertions (INS), inversions (INV), translocations, and duplications [15, 16]. Intrachromosomal translocations (65%) and deletions (26%) are predominant in the dataset, while inversions and interchromosomal translocations are present in lower proportions (Additional file 4: Fig. S7a, Additional file 8). Analysis of copy number variations (CNVs; 95,918 in total), divided into deletions and duplications, reveals an overall higher proportion of deletions (56.5%) than duplications (43.5%) (Additional file 4: Fig. S7b, Additional file 9).
Population structure
Principal component Analysis (PCA) was performed for all the 100 YFC genomes, revealing a general separation of YFCs from Henan (Zhengyang, ZY) and Hubei (Jianghan, JH) (Fig. 1b) into a northern cluster. The YFCs from Guangxi (Guangxi Yellow, GX), Guangdong (Huaixiang, HX, Huiyang bearded, HY, and Wuhua Yellow, WH), and Hainan (Wenchang, WC) form a southern cluster, while those from Hunan (Huanglang, HL), Jiangxi (Ningdu Yellow, ND), and Fujian (Hetian, HT) group into a central cluster. This finding is supported by ADMIXTURE analysis (Fig. 1c and d). At the lowest cross validation error value, corresponding to K = 2, the northern (blue) and southern (red) clusters show a complete separation, whereas the central cluster exhibits a signal of admixture with the northern and southern clusters. These three clusters were verified when K = 3, with HL and WC showing admixed ancestries. When K = 4, both HT and HY harbor the same ancestry component, which also contribute to WC.
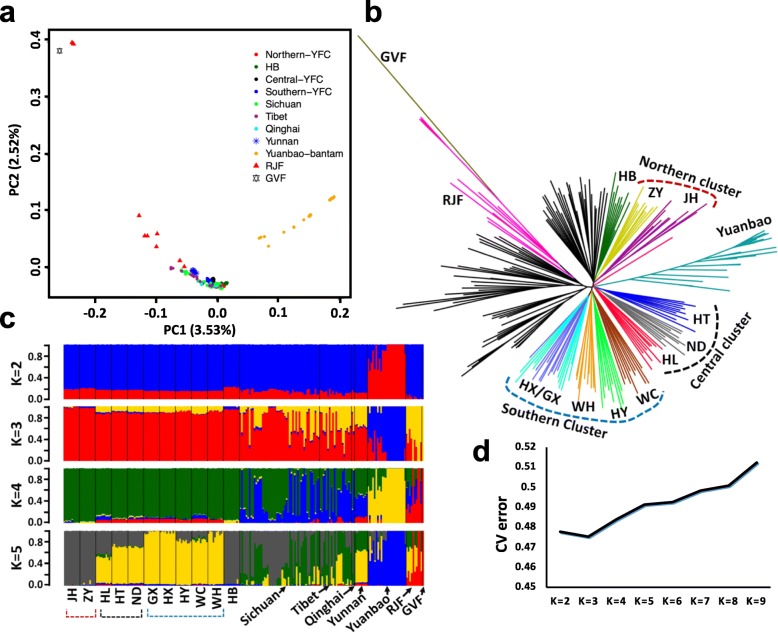
We implemented comparative population genetic analyses the YFCs against other indigenous chickens from China, RJF, and GVF. In the PCA (Fig. 2a), YFCs tend to cluster together and appear to be in close proximity to HB partridge chickens and a few indigenous chickens from Sichuan and Tibet. These patterns imply a close congruity in the total genomic architecture of the YFCs. Yuanbao bantams form a distant cluster from the YFCs and other chickens, underscoring the genomic effects of differential breeding trajectories [13, 17]. Neighbor joining (NJ) phylogeny (Fig. 2b) and ADMIXTURE (Fig. 2c and d) corroborates the findings of the PCA and further clarifies the northern, central, and southern YFC clustering pattern inferred from Fig. 1b-d.
Fig. 2.
Population genomic analysis of YFCs in the context of other Chinese indigenous chickens. a PCA showing the evolutionary relationships among YFCs, other Chinese indigenous chickens, red junglefowl (RJF), and green junglefowl (GVF). b Neighbor joining tree including all chickens and RJF (root). The tree was viewed and edited using FigTree software (v1.4.3). c-d ADMIXTURE analysis for K = 2 up to K = 5. The lowest cross validation error is observed when K = 2
Detection of selective sweeps
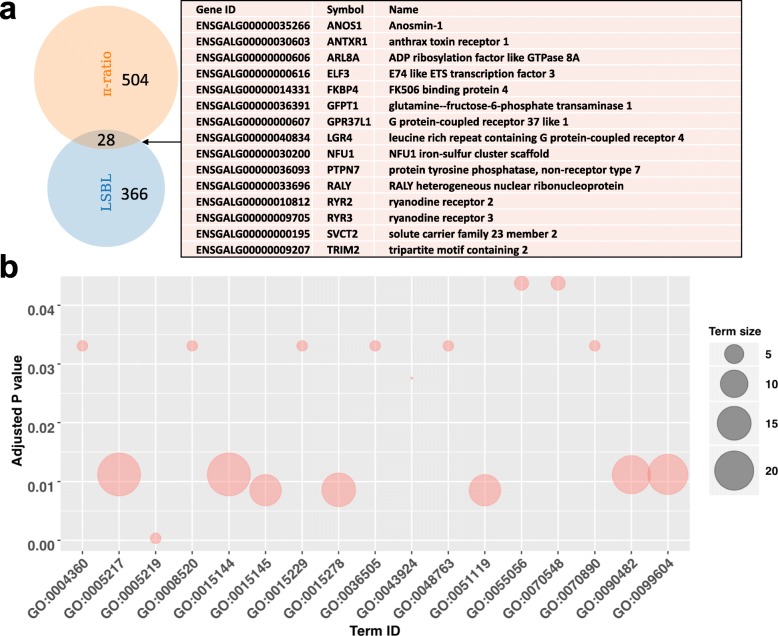
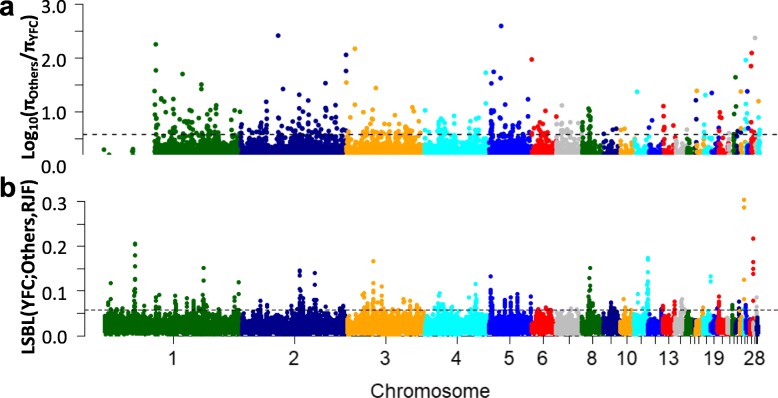
Genome-wide scans for signals of selection attributable to the YFCs phenotype identified 268 analytical windows within the top 1% of the Locus-specific branch length (LSBL) test, and 370 windows in the π-ratio test (Additional files 10 and 11). These correspond to 366 and 504 positively selected genes (PSGs), respectively. A total of 28 PSGs were concurrently identified in the top 1% by the two selection tests (Fig. 3a). This is a relatively small overlap, possibly owing to the differences in the selection tests. Among the 28 genes are genes that are associated with pigmentation including: RALY heterogeneous nuclear ribonucleoprotein (RALY), leucine rich repeat containing G protein-coupled receptor 4 (LGR4), ryanodine receptor 2 (RYR2), RYR3, solute carrier family 23 member 2 (SLC23A2), and SLC2A14. Functional enrichment assessment showed significant gene ontology (GO) terms including vitamin transport activity (GO:0090482; Fig. 3b), intersecting with SLC23A2 and SLC2A14, which play roles in pigmentation. There are additional genes above the top 1% significance threshold in either of the selection tests. These genes are important for understanding the color trait and other properties of interest like meat quality of the YFCs. They include BCDO2, IL-18, FBXO5, COL1A2, COL4A2, COL6A1, COL6A2 in LSBL; and GDF8, HSPA5, SHISA9, COL4A1, and COL23A1 in the π-ratio test (Fig. 4).
Fig. 3.
Candidate positively selected genes (PSGs). a PSGs in the top 1% windows of LSBL and π-ratio genome selection tests. The identities of annotated genes overlapping in both tests are shown. b Summary of the functional enrichment analysis of the 28 overlapping PSGs. Only terms with adjusted P values less than 0.05 are shown
Fig. 4.
Genome-wide distributions of selection signals. a π-ratio contrasting Yellow-feathered chickens (YFC) against chicken with non-yellow (black) phenotypes, denoted as Others. b Locus-specific branch length (LSBL) analysis contrasting YFC against non-yellow chicken with red junglefowl (RJF) outgroup. The horizontal dotted lines represent the top 1% cut-off
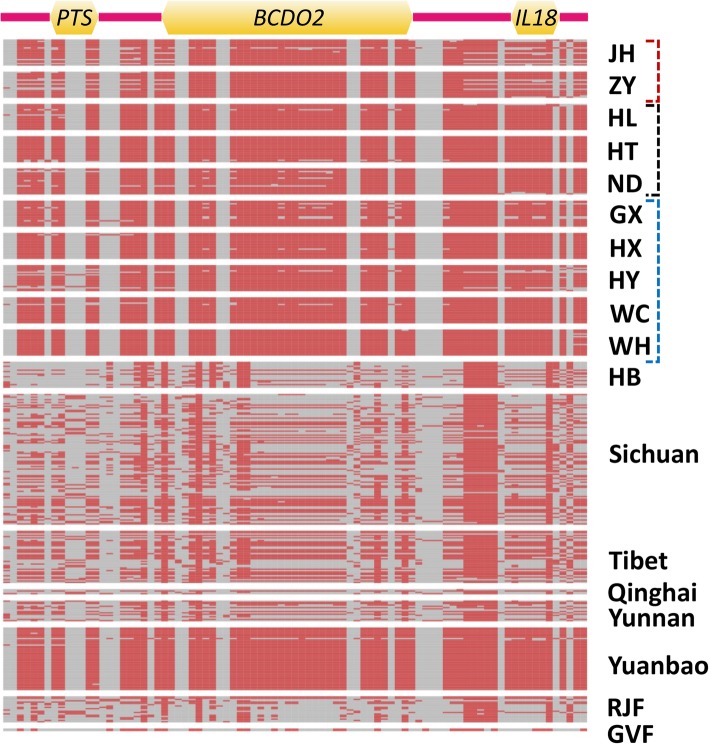
BCDO2 haplotype differentiation
BCDO2 gene is a classical yellow color gene in chicken. We investigated its haplotype structure, also encompassing the proximal flanking genes. BCDO2 showed a homogenous haplotype pattern across the 10 YFC breeds (Fig. 5). Interestingly, Yuanbao breed also bears the same pattern as the YFCs. On the other hand, HB partridge chickens, which initially showed a close genomic proximity to the northern cluster YFCs (Fig. 1), clearly exhibits a synonymous BCDO2 haplotype pattern to the other Chinese indigenous chickens rather than to the YFCs (Fig. 5). Overall, the haplotype differentiation pattern of BCDO2 and its flanking genes (IL18 and PTS) is consistent with the selection of these genes as candidate PSGs for the yellow pigmentation phenotype.
Fig. 5.
Genomic diversity and BCDO2 gene structure of YFCs and other chickens. Haplotype pattern analysis of the yellow skin gene, BCDO2 across different populations separated by white gaps. Immediate flanking genes are indicated. Grey color denotes reference alleles while Indian red indicates alternative alleles. Initials represent chicken breeds/population names as defined in the methods. The three YFC clusters are indicated by red, black, and blue boxes representing northern, central, and southern clusters, respectively
Discussion
We provide the first comprehensive whole-genome sequencing data and genomic variants for the YFCs. We also describe the genetic structure and molecular background of the distinguished color phenotype of these chickens. YFCs are a traditional nutritional and commercial mainstay for millions of people living in China and its purlieus, and are believed to have contributed to the recent breeding of European chickens [5]. Next generation sequencing has augmented scientific research into the molecular foundations of various complex phenotypic poultry traits such as body size in chicken [13], body size and plumage color in ducks [18], as well as maturation and plumage color in domestic quails [19], among others. In this study, we characterize not only the SNPs in the genomes of the YFCs but also other variants including InDels, structural variations (SV), and copy number variations (CNV) to facilitate research of these chickens. Particularly, SVs are increasingly gaining research interest as they can lead to the birth of new genes, change gene copy number as well as their expression profiles, eventually affecting phenotypic evolution and adaptation of organisms to local environments [20–23], hence will be an important resource to extend the SNP-based genetic analyses [24, 25]. Similarly, CNVs are linked with phenotypic evolution and have supported high-impact evolutionary investigations on complex diseases and economically important traits [26, 27]. For instance, in chicken, sequence duplication near the first intron of SOX5 gene is linked with the chicken pea-comb trait [28], an inverted duplication covering EDN3 gene leads to dermal hyperpigmentation [29], and a partial duplication of PRLR gene is associated with late feathering [30].
Our current comparative population genomics analysis was anchored on genome-wide SNPs of the YFCs, other indigenous chickens, and wild ancestors. Population structure analysis revealed an overall distinctive genomic architecture of the YFCs from other Chinese indigenous chickens (PCA and NJ phylogenetic tree). Interestingly, a three-way sub-clustering pattern is consistent in PCA, ADMIXTURE, and NJ phylogenetic tree and amazingly mirrors the geographical distributions of the YFCs. The 10 YFC populations divide into northern, central and southern clusters, agreeing with the trends earlier proposed by microsatellite-based studies of chickens from these regions of China [1, 11, 12]. This sub-structuring may be reflective of some extent of differential exchange of genetic materials in neighboring locations, breeding histories, or natural and artificial selection drivers as described in several chicken populations [17, 27]. This explains the existence of genomic grouping among populations with close phenotypic appearances such as the YFCs. A crucial point to note is the signals of admixture at K = 3 and 4 in the ADMIXTURE analysis. Hetian (HT) and Huiyang bearded (HY) YFCs are historically ascribed to the Hakka Chinese [6] who are thought to have immigrated from northern China, and have preserved their distinguished cultures, languages [31], and even genetic attributes [32]. Wenchang (WC) chickens are reported to have originated from crossbreeding of chickens brought into Hainan Province by people (including the Hakka) from Guangdong and Fujian Provinces [6]. The results of PCA and ADMIXTURE (K = 2 and 3) suggest that the Huaibei (HB) partridge chickens have a close relationship with YFCs of the northern cluster, consistent with their geographical proximity. Nevertheless, it is incomprehensible that HB, Huanglang (HL), and Ningdu Yellow (ND) shared dominant ancestry component at K = 4. Compared to other indigenous Chinese chickens, the YFCs tend have a closer genetic semblance among themselves than with other chickens, inferring a possible overriding effect of selection for the outstanding phenotypic traits of the YFCs.
Fundamental to the genomic selection scans in this study is the identification of RALY, LGR4, RYR2, RYR3, and SLC23A2 as well as its related homologue, SLC2A14. These genes stood out as candidate genes under selection in the YFCs, having significant signals both in LSBL and π-ratio scans. There is a known epistatic relationship bringing together RALY, ASIP, and MC1R [33]. ASIP gene codes for agouti-signaling protein, which antagonizes the α-MSH hormone (melanocyte stimulating hormone) for the melanocyte-1 receptor (MC1R) counteracting the production of eumelanin (black/brown melanin) and favoring the synthesis of pheomelanin (yellow/red melanin) [34]. Both ASIP and MC1R are genes which continue to be synonymous with nearly all studies on pigmentation in mammalian and avian species [35–39]. Interestingly, it has been demonstrated that a > 90 kb deletion upstream of avian ASIP, encompassing portions of the RALY locus, places ASIP under the regulatory control of RALY promoter [40]. The resulting up-regulation of ASIP underlies the yellow feather phenotype in quails and is interestingly associated with down-regulation of SLC24A5 [40]. SLC24A5 is an important gene in pigmentation whose roles in eumelanogenesis has been clearly demonstrated in both human and zebrafish [41]. We detected two members of the solute carrier family (SLC), SLC23A2 and SLC2A14. SLC23A2 is a major mediator of the transport of ascorbic acid, an indispensable metabolite that is fundamental for survival [42]. Anomalies in the availability of this vitamin have been associated with neonatal jaundice and yellow chromophore in eye lenses of human and humanized mouse model [43, 44]. It is key to note that although the selection of neither ASIP nor MC1R did not reach significance in our analyses, genes with which they are closely related, particularly RALY and SLC family homologues such as SLC23A2 and SLC2A14 point to the possibility of a gene network encompassing the PSGs identified in this study, working in conjunction with ASIP and MC1R in the determination of the yellow color trait of the YFCs.
From the selection sweep analysis, it was not a surprise to detect a strong selection for the BCDO2 gene and a common genetic architecture of the gene among all the YFCs. Even the HB chickens which are phylogenetically and geographically close to the YFCs in the northern cluster were clearly distinguishable based on the BCDO2 haplotype structure, depicting a possible marked differentiation of indigenous chickens at trait-linked genome compartment under selection pressure, despite likely closeness at the total genome level. Besides the YFCs, the BCDO2 haplotype for yellow skin is also observed in Yuanbao chicken, which also have yellow skin, and at a low frequency in some indigenous chicken, consistent with the reporting of related haplotypes of this gene in southern China chickens [45].
Moreover, our results show some clues for meat quality which is a major economic feature in chicken production. RYR2 and RYR3 [46–48]; IL-18 [49]; FBXO5 [50]; COL1A2, COL4A2, COL6A1; COL6A2; COL4A1, and COL23A1 [51]; GDF8 [52]; HSPA5 [49]; SHISA9 [53]; all bearing strong selection signals in the YFCs, are important determinants of meat quality in domestic animals. These genes provide a foundation for understanding the meat properties of the YFCs, which would attract more concerns to investigate the detailed function roles in future studies.
Conclusions
In summary, this study provides an invaluable resource for further research on the molecular mechanisms conferring complex traits that are of high economic and nutritional value. Through genomic insights regarding key genes behind the unique traits of YFCs and a comprehensive data resource, this study paves way for reconstructing the breeding history and formulating future conservation and breed improvement strategies for YFCs.
Methods
Samplings and sequencing of the yellow-feathered chickens
Unrelated chickens were identified with the help of pedigree records. Wing-vein blood samples were then collected by trained local veterinary personnel, from 100 birds of 10 YFC breeds, 10 chickens per breed. These breeds include Guangxi Yellow (GX), Hetian (HT), Huaixiang (HX), Huanglang (HL), Huiyang bearded (HY), Jianghan (JH), Ningdu Yellow (ND), Wenchang (WC), Wuhua Yellow (WH), and Zhengyang Yellow (ZY) (Additional file 1). Ten Huaibei partridge (HB) chickens were also sampled for comparison. Animal handling and experimentation was conducted according to the guidelines approved by the Animal Ethics Committee of Jiaying University and Kunming Institute of Zoology, Chinese Academy of Sciences. Genomic DNA was extracted using the standard phenol-chloroform method and checked for quantity and quality using agarose gel electrophoresis and NanoDrop spectrophotometer 2000 (NanoDrop, Wilmington, DE, USA). Whole genome sequencing for these 110 samples was carried out to a median depth of 12.22X (ranging from 9.34 to 18.66) (Additional file 1) using the Illumina HiSeq X10 platform at the Genedenovo Biotechnology Co., Ltd. (Guangzhou, China).
Identification and annotation of whole-genome variants
To obtain high quality clean reads, stringent quality filtering procedures were applied as follows: 1) removing reads with ≥ 10% unidentified nucleotides (N); 2) removing reads with > 50% bases having phred scores of ≤ 20; and 3) removing reads aligned to the barcode adapter. The clean reads were then aligned to the chicken reference genome (Galgal5) [54] using the BWA-MEM alignment algorithm [55] implemented in the BWA [56] with options ‘mem 4 -k 32 -M’. Further quality control processes were performed using the SortSam and MarkDuplicates tools in the Picards package (picard-tools-1.56) to sort and remove possible duplicates in the aligned BAM files, and the RealignerTargetCreator, IndelRealigner, and BaseRecalibrator tools in the Genome Analysis Toolkit (GATK 2.6–4) [57] for local realignment and base quality recalibration. The bedtools software (v.2.25.0) [58] was employed to generate sequencing coverage statistics.
Variant calling was performed using the GATK’s Unified Genotyper. SNPs and InDels were filtered by the GATK’s VariantFiltration with options “-Window 4, -filter “QD < 2.0 || FS > 60.0 || MQ < 40.0 “, -G_filter “GQ < 20″”, excluding those exhibiting segregation distortions or sequencing errors. The ANNOVAR [59] was used to assign putative genomic positions of SNPs and InDels against the chicken gene database in ENSEMBL (release 92.5). The structural variations (SVs) in these 110 chicken genomes were assessed using the BreakDancer package (Max1.1.2.) [60, 61], and the CNVnator program (v.0.3.2) [62] was employed to classify copy number variations (CNVs).
Analysis of population genetic structure
The evolutionary interactions among the 100 YFCs were examined using principal component analysis (PCA) following the GCTA approach [63] and maximum-likelihood-based ADMIXTURE [64] at K = 2 to 9. The PLINK package (v.1.90) was used to obtain pruned data with parameters “--indep-pairwise 50 10 0.1” [65] for the PCA and ADMIXTURE analyses. To perform a comparative analysis of the YFC genomes generated in this study against those of other chicken populations, 104 previously published whole genomes [13, 14] of Chinese indigenous chickens (Sichuan, n = 50; Tibet, n = 20; Qinghai, n = 2; Yunnan, n = 8; and Yuanbao bantams, n = 24), as well as 10 RJF and 1 GVF genomes were included. After merging our dataset of the 110 chicken genomes with the additional 115 genomes, 3,065,814 common SNPs were retained for subsequent analyses. The PCA was performed as stated above and a neighbor joining (NJ) tree rooted to RJF was constructed using the RapidNJ program [66] with 100 bootstrap replications.
Genomic targets of selection in yellow-feathered chickens
To retrieve the genetic foundation for the outstanding phenotypic properties of the YFCs, we performed genome-wide scans for signals of selection using locus-specific branch length (LSBL) statistics [67] and π-ratio. Use of multiple statistical approaches helps to manage inherent differences of individual tests and increase the reliability of the selective sweep detection [68]. The comparative genomic analysis approach involved genomes of the YFCs against 24 chickens with contrasting non-yellow phenotypes (non-YFCs), i.e. black-phenotype Chinese chickens (five Emei black fowl, four Miyi fowl, five Muchuan black-bone fowl, and five Tuanfu black-bone fowl; Additional file 1) and RJF. In the LSBL, we computed LSBL(A;B,C) = (FST(AB) + FST(AC)–FST(BC))/2 to assess the population differentiation between YFCs and other chickens, set as ‘YFCs;non-YFCs,RJF’. FST values were calculated as described elsewhere [69] with a 50-kb sliding window and 25-kb stepwise increments. π-ratio was performed by first calculating the genetic diversity (π) for YFCs and the 24 non-YFCs populations using VCFtools [70] in 50-kb windows with 25-kb stepwise increments, then computing π-ratio (πnon-YFCs/πYFCs). An empirical cutoff of 99th percentile was used to retrieve candidate selective sweeps, which were then annotated using variant effect predictor (VEP) to identify the putative positively selected genes (PSGs) [71].
We performed functional enrichment analysis using g:Profiler [72] to obtain a global overview on the biological functions of the candidate PSGs with concordantly significant selection signals in the two genomic selection scans employed. A Benjamini-Hochberg false discovery rate (FDR) significance threshold was set at 0.05.
Assessment of the classical yellow skin gene, BCDO2
BCDO2 gene, located in chromosome 24: 6,110,301-6,130,965 reverse strand (Galgal5, [54]), is believed to be substantially associated with yellow skin pigmentation in chickens following a possible introgression from grey junglefowl (Gallus sonneratii) in South Asia [73]. We evaluated the haplotype variability of BCDO2 gene and its flanking genes, a stretch of the genome from chromosome 24:6,105,000 - 6,145,000. Haplotypes for were phased using BEAGLE software (v.3.3.2) with the default parameters [74] and viewed as heatmaps. Gene structure representation was done using the Gene Structure Display Server (GSDS v.2) [75].
Supplementary information
Additional file 1. Individual sample characteristics of all chickens sequenced in this study.
Additional file 2. Reads filtering statistics for the chicken sequenced in this study.
Additional file 3. Genome alignment statistics for all chickens sequenced in this study.
Additional file 4: Fig. S1 Constitution of clean sequencing reads of all 110 chicken genomes produced in this study. Fig. S2 Summary of the average sequencing coverage of all 110 chicken genomes generated in this study. Fig. S3 Circos plot depicting the genomic variants landscape in each chromosome. Fig. S4 Annotation of the clean genomic SNPs of all 110 chickens sequenced in this study. Fig. S5 Transition-transversion analysis of the clean SNPs of all 110 chicken genomes sequenced in this study. Fig. S6 Annotation of the clean InDels of all 110 chicken genomes sequenced in this study. Fig. S7 Summary of the structural variations (SVs) and copy number variations (CNVs) in all 110 chicken genomes generated in this study.
Additional file 5. Base information statistics before and after quality filtering of all chicken genomes sequenced in this study.
Additional file 6. Individual transitional and transversional SNP statistics of all chickens sequenced in this study.
Additional file 7. Annotation of the hybrid status of SNPs in each chicken genome sequenced in this study.
Additional file 8. Annotation of structural variations in the chicken genomes sequenced in this study.
Additional file 9. Annotation of copy number variations in the chicken genomes sequenced in this study.
Additional file 10. The top 1% genomic windows in the selection scan by LSBL test.
Additional file 11. The top 1% genomic windows in the selection scan by π-ratio test.
Acknowledgements
We would like to recognize the important contribution of chicken farms which made this study possible. N.O.O. is grateful for the support from CAS-TWAS President’s Fellowship Program for Doctoral Candidates. We are grateful to Guangzhou Genedenovo Biotechnology Co., Ltd. for assisting in sequencing and part of bioinformatics analysis.
Abbreviations
- BCDO2
Beta-carotene dioxygenase 2
- CNV
Copy number variation
- FDR
False discovery rate
- GO
Gene ontology
- GVF
Green junglefowl
- InDel
Insertion or deletion of bases
- INS
Insertions
- INV
Inversions
- LGR4
Leucine rich repeat containing G protein-coupled receptor 4
- LSBL
Locus-specific branch length
- NJ
Neighbor joining
- PCA
Principal component analysis
- PSGs
Positively selected genes
- RALY
RALY heterogeneous nuclear ribonucleoprotein
- RJF
Red junglefow
- RYR2
Ryanodine receptor 2
- SLC23A2
Solute carrier family 23 member 2
- SLC2A14
Solute carrier family 2 member 14
- SNP
Single nucleotide polymorphism
- SV
Structural variation
- VEP
Variant effect predictor
- YFC
Yellow-feathered chicken
Authors’ contributions
XH and BD supervised the project. XH, ZW, WL, JC, MZ, FZ, SJ, ZG, WL and DH collected samples and performed DNA preparation. YZ provided genomic data for comparison. NO, MP, ZW and XH performed data analysis, interpretation of results, and writing of the manuscript. JH, CM, SO, YZ, XZ, BD, WL, JC, MZ, FZ, SJ, ZG and DH participated in writing and revising the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Science & Technology Planning Project of Guangdong Province (2016A030303068), Natural Science Foundation of Guangdong (2014A030307018), Provincial and municipal key construction projects in Jiaying University, and the Bureau of Science and Technology of Yunnan Province. The funding bodies did not participate in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All new sequencing data generated in this study have been deposited in the NCBI sequence read archive (SRA) under accession number SRP155577. Further details are provided in Additional file 1. Additional requests can be channeled to the corresponding authors.
Ethics approval and consent to participate
Verbal informed consent for sampling was obtained from the conservation farms. Standard techniques were used to collect blood samples from each animal, by trained local veterinary personnel, and this procedure was approved by the ethics committee of Jiaying University and Kunming Institute of Zoology, Chinese Academy of Sciences. The study was approved by Jiaying University and the Internal Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences (SMKX2017005).
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xunhe Huang, Newton O. Otecko and Minsheng Peng contributed equally to this work.
Contributor Information
Yaping Zhang, Email: zhangyp@mail.kiz.ac.cn.
Xiquan Zhang, Email: xqzhang@scau.edu.cn.
Bingwang Du, Email: dudu903@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-6736-4.
References
- 1.Huang XH, Zhang JF, He DL, Zhang XQ, Zhong FS, Li WN, Zheng QM, Chen JB, Du BW. Genetic diversity and population structure of indigenous chicken breeds in South China. Front Agr Sci Eng. 2016;3(2):97–101. doi: 10.15302/J-FASE-2016102. [DOI] [Google Scholar]
- 2.Qi J, Li X, Zhang W, Wang H, Zhou G, Xu X. Influence of stewing time on the texture, ultrastructure and in vitro digestibility of meat from the yellow-feathered chicken breed. Anim Sci J. 2018;89(2):474–482. doi: 10.1111/asj.12929. [DOI] [PubMed] [Google Scholar]
- 3.Meng XB, Xu TS, Du BW, Teng XH. Chicken art – ancient Chinese wisdom on chicken-raising with an appendix of the Chinese art of calligraphy. Beijing: China Agriculture Press; 2017. [Google Scholar]
- 4.Zheng MQ, Gong GF, Gao HJ, Yao WY, Lv SY, Tian LJ, Wen J. Report of Chinese broiler industry development in 2015. China Poult. 2016;38:67–70. [Google Scholar]
- 5.Loog L, Thomas MG, Barnett R, Allen R, Sykes N, Paxinos PD, Lebrasseur O, Dobney K, Peters J, Manica A, et al. Inferring allele frequency trajectories from ancient DNA indicates that selection on a chicken gene coincided with changes in medieval husbandry practices. Mol Biol Evol. 2017;34(8):1981–1990. doi: 10.1093/molbev/msx142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.China National Commission of Animal Genetic Resources . Animal genetic resources in china – poultry. Beijing: China Agriculture Press; 2011. [Google Scholar]
- 7.Wang Y, Hui X, Wang H, Kurban T, Hang C, Chen Y, Xing J, Wang J. Association of H-FABP gene polymorphisms with intramuscular fat content in three-yellow chickens and Hetian-black chickens. J Anim Sci Biotechnol. 2016;7:9. [DOI] [PMC free article] [PubMed]
- 8.Chiang PD, Yen CT, Mau JL. Non-volatile taste components of various broth cubes. Food Chem. 2007;101(3):932–937. doi: 10.1016/j.foodchem.2006.02.041. [DOI] [Google Scholar]
- 9.Duan Y, Zheng FP, Chen HT, Huang MQ, Xie JC, Chen F, Sun BG. Analysis of volatiles in Dezhou braised chicken by comprehensive two-dimensional gas chromatography/high resolution-time of flight mass spectrometry. LWT Food Sci Technol. 2015;60:1235–1242. doi: 10.1016/j.lwt.2014.09.006. [DOI] [Google Scholar]
- 10.Jaturasitha S, Srikanchai T, Kreuzer M, Wicke M. Differences in carcass and meat characteristics between chicken indigenous to northern Thailand (black-boned and Thai native) and imported extensive breeds (Bresse and Rhode Island red) Poult Sci. 2008;87(1):160–169. doi: 10.3382/ps.2006-00398. [DOI] [PubMed] [Google Scholar]
- 11.Qu LJ, Li XY, Xu GF, Chen KW, Yang HJ, Zhang LC, Wu GQ, Hou ZC, Xu GY, Yang N. Evaluation of genetic diversity in Chinese indigenous chicken breeds using microsatellite markers. Sci China C Life Sci. 2006;49(4):332–341. doi: 10.1007/s11427-006-2001-6. [DOI] [PubMed] [Google Scholar]
- 12.Yu YB, Wang JY, Mekki DM, Tang QP, Li HF, Gu R, Ge QL, Zhu WQ, Chen KW. Evaluation of genetic diversity and genetic distance between twelve Chinese indigenous chicken breeds based on microsatellite markers. Int J Poult Sci. 2006;5(6):550–556. doi: 10.3923/ijps.2006.550.556. [DOI] [Google Scholar]
- 13.Wang MS, Huo YX, Li Y, Otecko NO, Su LY, Xu HB, Wu SF, Peng MS, Liu HQ, Zeng L, et al. Comparative population genomics reveals genetic basis underlying body size of domestic chickens. J Mol Cell Biol. 2016;8(6):542–552. doi: 10.1093/jmcb/mjw044. [DOI] [PubMed] [Google Scholar]
- 14.Li D, Che T, Chen B, Tian S, Zhou X, Zhang G, Li M, Gaur U, Li Y, Luo M, et al. Genomic data for 78 chickens from 14 populations. Gigascience. 2017;6(6):1–5. doi: 10.1093/gigascience/gix026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker M. Structural variation: the genome's hidden architecture. Nat Methods. 2012;9(2):133–137. doi: 10.1038/nmeth.1858. [DOI] [PubMed] [Google Scholar]
- 16.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12(5):363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang MS, Otecko NO, Wang S, Wu DD, Yang MM, Xu YL, Murphy RW, Peng MS, Zhang YP. An evolutionary genomic perspective on the breeding of dwarf chickens. Mol Biol Evol. 2017;34(12):3081–3088. doi: 10.1093/molbev/msx227. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z, Li M, Cheng H, Fan W, Yuan Z, Gao Q, Xu Y, Guo Z, Zhang Y, Hu J, et al. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat Commun. 2018;9(1):2648. doi: 10.1038/s41467-018-04868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y, Zhang Y, Hou Z, Fan G, Pi J, Sun S, Chen J, Liu H, Du X, Shen J, et al. Population genomic data reveal genes related to important traits of quail. Gigascience. 2018;7(5):giy049. doi: 10.1093/gigascience/giy049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen NF, Teague B, Alkan C, Antonacci F. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paudel Y, Madsen O, Megens H, Frantz LAF, Bosse M, Bastiaansen JWM, Crooijmans RPMA, Groenen MAM. Evolutionary dynamics of copy number variation in pig genomes in the context of adaptation and domestication. BMC Genomics. 2013;14(1):449. doi: 10.1186/1471-2164-14-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu SH, He ZW, Zhang Z, Guo ZX, Guo WX, Lyu HM, Li JF, Yang M, Du ZL, Huang YL, et al. The origin, diversification and adaptation of a major mangrove clade (Rhizophoreae) revealed by whole-genome sequencing. Natl Sci Rev. 2017;4(5):721–734. doi: 10.1093/nsr/nwx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Wang K, Wang L, Yang Y, Ni Z, Xie X, Shao X, Han J, Wan D, Qiu Q. Genome-wide patterns of copy number variation in the Chinese yak genome. BMC Genomics. 2016;17(1):379. doi: 10.1186/s12864-016-2702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu R, Xing S, Wang J, Zheng M, Cui H, Crooijmans R, Li Q, Zhao G, Wen J. A new chicken 55K SNP genotyping array. BMC Genomics. 2019;20(1):410. doi: 10.1186/s12864-019-5736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye S, Gao N, Zheng R, Chen Z, Teng J, Yuan X, Zhang H, Chen Z, Zhang X, Li J, et al. Strategies for obtaining and pruning imputed whole-genome sequence data for genomic prediction. Front Genet. 2019;10:673. doi: 10.3389/fgene.2019.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S, Lin X, Zhang Z, Jiang M, Rao Y, Nie Q, Zhang X. Copy number variation in SOX6 contributes to chicken muscle development. Genes. 2018;9(1):42. [DOI] [PMC free article] [PubMed]
- 27.Yi G, Qu L, Liu J, Yan Y, Xu G, Yang N. Genome-wide patterns of copy number variation in the diversified chicken genomes using next-generation sequencing. BMC Genomics. 2014;15:962. doi: 10.1186/1471-2164-15-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright D, Boije H, Meadows JR, Bed'hom B, Gourichon D, Vieaud A, Tixier-Boichard M, Rubin CJ, Imsland F, Hallbook F, et al. Copy number variation in intron 1 of SOX5 causes the pea-comb phenotype in chickens. PLoS Genet. 2009;5(6):e1000512. [DOI] [PMC free article] [PubMed]
- 29.Dorshorst B, Molin AM, Rubin CJ, Johansson AM, Stromstedt L, Pham MH, Chen CF, Hallbook F, Ashwell C, Andersson L. A complex genomic rearrangement involving the endothelin 3 locus causes dermal hyperpigmentation in the chicken. PLoS Genet. 2011;7(12):e1002412. doi: 10.1371/journal.pgen.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elferink MG, Vallee AA, Jungerius AP, Crooijmans RP, Groenen MA. Partial duplication of the PRLR and SPEF2 genes at the late feathering locus in chicken. BMC Genomics. 2008;9:391. [DOI] [PMC free article] [PubMed]
- 31.Hsieh T. Origin and migrations of the Hakkas. Chin Soc Pol Sci Rev (Beijing) 1929;13:202–227. [Google Scholar]
- 32.Chen J, Zheng H, Bei JX, Sun L, Jia WH, Li T, Zhang F, Seielstad M, Zeng YX, Zhang X, et al. Genetic structure of the Han Chinese population revealed by genome-wide SNP variation. Am J Hum Genet. 2009;85(6):775–785. doi: 10.1016/j.ajhg.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nazari-Ghadikolaei A, Mehrabani-Yeganeh H, Miarei-Aashtiani SR, Staiger EA, Rashidi A, Huson HJ. Genome-wide association studies identify candidate genes for coat color and mohair traits in the Iranian Markhoz goat. Front Genet. 2018;9:105. doi: 10.3389/fgene.2018.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97(3):222–34. [DOI] [PubMed]
- 35.Alshanbari F, Castaneda C, Juras R, Hillhouse A, Mendoza MN, Gutierrez GA, Ponce de Leon FA, Raudsepp T. Comparative FISH-Mapping of MC1R, ASIP, and TYRP1 in new and old world camelids and association analysis with coat color phenotypes in the dromedary (Camelus dromedarius). Front Genet. 2019;10:340. [DOI] [PMC free article] [PubMed]
- 36.Nakamura K, Tozaki T, Kakoi H, Owada S, Takasu M. Variation in the MC1R, ASIP, and MATP genes responsible for coat color in Kiso horse as determined by SNaPshot genotyping. J Vet Med Sci. 2019;81(1):100–2. [DOI] [PMC free article] [PubMed]
- 37.Shang S, Yu Y, Zhao Y, Dang W, Zhang J, Qin X, Irwin DM, Wang Q, Liu F, Wang Z, et al. Synergy between MC1R and ASIP for coat color in horses (Equus caballus). J Anim Sci. 2019;97(4):1578–85. [DOI] [PMC free article] [PubMed]
- 38.Yang CW, Ran JS, Yu CL, Qiu MH, Zhang ZR, Du HR, Li QY, Xiong X, Song XY, Xia B, et al. Polymorphism in MC1R, TYR and ASIP genes in different colored feather chickens. 3 Biotech. 2019;9(5):203. [DOI] [PMC free article] [PubMed]
- 39.Zhang J, Liu F, Cao J, Liu X. Skin transcriptome profiles associated with skin color in chickens. PLoS One. 2015;10(6):e0127301. doi: 10.1371/journal.pone.0127301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nadeau NJ, Minvielle F, Ito S, Inoue-Murayama M, Gourichon D, Follett SA, Burke T, Mundy NI. Characterization of Japanese quail yellow as a genomic deletion upstream of the avian homolog of the mammalian ASIP (agouti) gene. Genetics. 2008;178(2):777–86. [DOI] [PMC free article] [PubMed]
- 41.Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310(5755):1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 42.Eck PK, Corpe C, Levine MA. Temporo-spacial microanatomical distribution of the murine sodium-dependent ascorbic acid transporters Slc23a1 and Slc23a2 in the kidney throughout development. Biochem Cell Biol. 2017;95(3):421–7. [DOI] [PMC free article] [PubMed]
- 43.Ayyappan S, Philip S, Bharathy N, Ramesh V, Kumar CN, Swathi S, Kumar AA. Antioxidant status in neonatal jaundice before and after phototherapy. J Pharm Bioallied Sci. 2015;7(Suppl 1):S16–S21. doi: 10.4103/0975-7406.155766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan X, Reneker LW, Obrenovich ME, Strauch C, Cheng R, Jarvis SM, Ortwerth BJ, Monnier VM. Vitamin C mediates chemical aging of lens crystallins by the Maillard reaction in a humanized mouse model. Proc Natl Acad Sci U S A. 2006;103(45):16912–16917. doi: 10.1073/pnas.0605101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao YS, Jia XX, Tang XJ, Fan YF, Lu JX, Huang SH, Tang MJ. The genetic diversity of chicken breeds from Jiangxi, assessed with BCDO2 and the complete mitochondrial DNA D-loop region. PLoS One. 2017;12(3):e0173192. [DOI] [PMC free article] [PubMed]
- 46.Droval AA, Binneck E, Marin SR, Paiao FG, Oba A, Nepomuceno AL, Shimokomaki M. A new single nucleotide polymorphism in the ryanodine gene of chicken skeletal muscle. Genet Mol Res. 2012;11(2):821–829. doi: 10.4238/2012.April.3.4. [DOI] [PubMed] [Google Scholar]
- 47.Paiao FG, Ferracin LM, Pedrao M, Kato T, Shimokomaki M. Skeletal muscle calcium channel ryanodine and the development of pale, soft, and exudative meat in poultry. Genet Mol Res. 2013;12(3):3017–3027. doi: 10.4238/2013.August.20.3. [DOI] [PubMed] [Google Scholar]
- 48.Strasburg GM, Chiang W. Pale, soft, exudative Turkey—the role of ryanodine receptor variation in meat quality. Poult Sci. 2009;88(7):1497–1505. doi: 10.3382/ps.2009-00181. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Kim SW, Do KT, Ha YK, Lee YM, Yoon SH, Kim HB, Kim JJ, Choi BH, Kim KS. Analyses of porcine public SNPs in coding-gene regions by re-sequencing and phenotypic association studies. Mol Biol Rep. 2011;38(6):3805–3820. doi: 10.1007/s11033-010-0496-1. [DOI] [PubMed] [Google Scholar]
- 50.Edea Z, Hong JK, Jung JH, Kim DW, Kim YM, Kim ES, Shin SS, Jung YC, Kim KS. Detecting selection signatures between Duroc and Duroc synthetic pig populations using high-density SNP chip. Anim Genet. 2017;48(4):473–477. doi: 10.1111/age.12559. [DOI] [PubMed] [Google Scholar]
- 51.Sun Y, Zhao G, Liu R, Zheng M, Hu Y, Wu D, Zhang L, Li P, Wen J. The identification of 14 new genes for meat quality traits in chicken using a genome-wide association study. BMC Genomics. 2013;14:458. doi: 10.1186/1471-2164-14-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu PA, Lo LL, Chen YC, Hsu CC, Shiau JW, Lin EC, Lin RS, Wang PH. Polymorphisms in the promoter region of myostatin gene are associated with carcass traits in pigs. J Anim Breed Genet. 2014;131(2):116–122. doi: 10.1111/jbg.12053. [DOI] [PubMed] [Google Scholar]
- 53.Wang H, Zhang L, Cao J, Wu M, Ma X, Liu Z, Liu R, Zhao F, Wei C, Du L. Genome-wide specific selection in three domestic sheep breeds. PLoS One. 2015;10(6):e0128688. doi: 10.1371/journal.pone.0128688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warren WC, Hillier LW, Tomlinson C, Minx P, Kremitzki M, Graves T, Markovic C, Bouk N, Pruitt KD, Thibaud-Nissen F, et al. A new chicken genome assembly provides insight into avian genome structure. G3 (Bethesda) 2017;7(1):109–117. doi: 10.1534/g3.116.035923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30(20):2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinlan AR. BEDTools. The Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:1121–34. [DOI] [PMC free article] [PubMed]
- 59.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6(9):677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan X, Abbott TE, Larson D, Chen K. BreakDancer: identification of genomic structural variation from paired-end read mapping. Curr Protoc Bioinformatics. 2014;45:15.6.1–15.611. doi: 10.1002/0471250953.bi1506s45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21(6):974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simonsen M, Mailund T, Pedersen CNS. In: Rapid neighbor-joining. In: proceedings of the eighth international workshop on algorithms in bioinformatics: 15–19 September 2008. Crandall KA, Lagergren J, editors. Karlsruhe: Springer Berlin Heidelberg; 2008. [Google Scholar]
- 67.Shriver MD, Kennedy GC, Parra EJ, Lawson HA, Sonpar V, Huang J, Akey JM, Jones KW. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genomics. 2004;1(4):274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma Y, Ding X, Qanbari S, Weigend S, Zhang Q, Simianer H. Properties of different selection signature statistics and a new strategy for combining them. Heredity. 2015;115(5):426–36. [DOI] [PMC free article] [PubMed]
- 69.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12(12):1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47(W1):W191–W1W8. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eriksson J, Larson G, Gunnarsson U, Bed'hom B, Tixier-Boichard M, Stromstedt L, Wright D, Jungerius A, Vereijken A, Randi E, et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008;4(2):e1000010. doi: 10.1371/journal.pgen.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Individual sample characteristics of all chickens sequenced in this study.
Additional file 2. Reads filtering statistics for the chicken sequenced in this study.
Additional file 3. Genome alignment statistics for all chickens sequenced in this study.
Additional file 4: Fig. S1 Constitution of clean sequencing reads of all 110 chicken genomes produced in this study. Fig. S2 Summary of the average sequencing coverage of all 110 chicken genomes generated in this study. Fig. S3 Circos plot depicting the genomic variants landscape in each chromosome. Fig. S4 Annotation of the clean genomic SNPs of all 110 chickens sequenced in this study. Fig. S5 Transition-transversion analysis of the clean SNPs of all 110 chicken genomes sequenced in this study. Fig. S6 Annotation of the clean InDels of all 110 chicken genomes sequenced in this study. Fig. S7 Summary of the structural variations (SVs) and copy number variations (CNVs) in all 110 chicken genomes generated in this study.
Additional file 5. Base information statistics before and after quality filtering of all chicken genomes sequenced in this study.
Additional file 6. Individual transitional and transversional SNP statistics of all chickens sequenced in this study.
Additional file 7. Annotation of the hybrid status of SNPs in each chicken genome sequenced in this study.
Additional file 8. Annotation of structural variations in the chicken genomes sequenced in this study.
Additional file 9. Annotation of copy number variations in the chicken genomes sequenced in this study.
Additional file 10. The top 1% genomic windows in the selection scan by LSBL test.
Additional file 11. The top 1% genomic windows in the selection scan by π-ratio test.
Data Availability Statement
All new sequencing data generated in this study have been deposited in the NCBI sequence read archive (SRA) under accession number SRP155577. Further details are provided in Additional file 1. Additional requests can be channeled to the corresponding authors.