Abstract

The biocatalytic synthesis of amides from carboxylic acids and primary amines in aqueous media can be achieved using the ATP-dependent amide bond synthetase McbA, via an adenylate intermediate, using only 1.5 equiv of the amine nucleophile. Following earlier studies that characterized the broad carboxylic acid specificity of McbA, we now show that, in addition to the natural amine substrate 2-phenylethylamine, a range of simple aliphatic amines, including methylamine, butylamine, and hexylamine, and propargylamine are coupled efficiently to the native carboxylic acid substrate 1-acetyl-9H-β-carboline-3-carboxylic acid by the enzyme, to give amide products with up to >99% conversion. The structure of wild-type McbA in its amidation conformation, coupled with modeling and mutational studies, reveal an amine access tunnel and a possible role for residue D201 in amine activation. Amide couplings were slower with anilines and alicyclic secondary amines such as pyrrolidine and piperidine. The broader substrate specificity of McbA was exploited in the synthesis of the monoamine oxidase A inhibitor moclobemide, through the reaction of 4-chlorobenzoic acid with 1.5 equiv of 4-(2-aminoethyl)morpholine, and utilizing polyphosphate kinases SmPPK and AjPPK in the presence of polyphosphoric acid and 0.1 equiv of ATP, required for recycling of the cofactor.
Keywords: biocatalysis, amides, ATP, McbA, cofactor recycling
There is renewed interest in biocatalytic methods for the formation of amide bonds, which is the number one employed reaction within the pharmaceutical sector, because enzymes can offer attractive alternatives to standard abiotic methods of amide synthesis.1 Biocatalytic alternatives may eliminate the need for stoichiometric coupling reagents and also confer enantioselectivity in the case of chiral amine or acid components.2−5 Lipases, for example, catalyze the aminolysis of esters in organic solvents6 to form chiral amides, and penicillin acylases (PACs) catalyze the amidation of phenylacetic acid derivatives to form semisynthetic penicillins.7 In addition, Li and co-workers have recently described an intracellular lipase SpL from a Sphingomonas sp., which catalyzes enantioselective amidation of both esters and carboxylic acids with high selectivity in organic solvents.8 However, complementary routes to amides that permit the coupling of carboxylic acids and amides in aqueous media are also of interest. One natural strategy for amide bond formation is the adenylation–thiolation–condensation cascade of reactions observed in, for example, the biosynthesis of thiomarinols in Pseudoalteromonas,9 in which the adenylation of a carboxylic acid is followed by thioesterification to Coenzyme A and then aminolysis of the thioester to form an amide bond. These reactions inspired the construction of libraries of Coenzyme A ligases (CLs) and N-acyltransferases (NATs) by Lovelock and co-workers,10 who then used the best-performing enzymes to construct a whole-cell system for the synthesis of the kinase inhibitor losmapimod from 6-chloronicotinic acid and neopentylamine. Similar chemistry is observed in nonribosomal peptide synthases (NRPSs),11 which first catalyze the adenylation of a carboxylic acid, followed by the formation of thioester linkage to a phosphopantetheine on an acyl carrier protein, after which attack of an amine nucleophile catalyzed by a ligase domain forms the amide bond. Recent experiments by Kobayashi and co-workers,12−15 Kino,16 and Campopiano17 have demonstrated that members of the adenylate-forming enzyme superfamily can be exploited for amide bond formation reactions in which the adenylate intermediate is attacked directly by an amine nucleophile. However, the role of the domain in catalyzing amide bond formation itself has yet to be conclusively demonstrated. In one example, Flitsch and co-workers showed that the adenylate formed by a carboxylic acid reductase (CAR) adenylation domain could be recruited for the milligram-scale synthesis of the anticonvulsant compound ilepcimide, when presented with a large excess of the amine precursor, piperidine.18
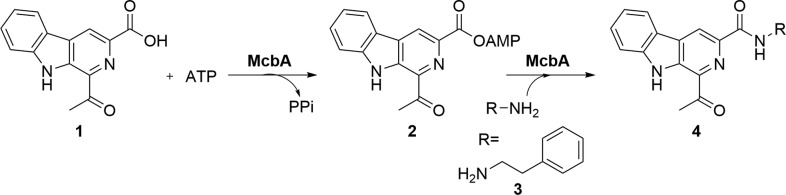
Enzymes that actively catalyze both the adenylation of carboxylic acid precursors and amide bond formation within one polypeptide chain also exist. These “amide bond synthetases” (ABSs) are exemplified by the biosynthetic enzyme McbA from Marinactinospora thermotolerans, which was described by Ju and co-workers.19 McbA catalyzes the adenylation of 1-acetyl-9H-β-carboline-3-carboxylic acid 1 (Scheme 1) to give intermediate 2, followed by amidation with 2-phenylethylamine 3, provided in equimolar amounts, to give the amide 4 as part of the biosynthetic pathway toward the marinacarboline antibiotics.19
Scheme 1. Synthesis of β-Carboline Amide 4via Adenylate Intermediate 2 by McbA from Marinactinospora thermotolerans.
The same group investigated the substrate specificity of McbA for the amine component of the coupling reaction, and found that, while tryptamine, and halogenated derivatives of 3 were accepted as substrates, the specificity of the enzyme appeared to be rather narrow.20 In further experiments, we have shown that the carboxylic acid specificity of McbA extends to bicyclic aromatic and heteroaromatic acids, in addition to benzoic acid,21 suggesting scope for the application of McbA to the synthesis of a wider range of amides than was previously envisaged. We also reported the three-dimensional structure of McbA, which revealed the possible basis for relaxed carboxylic acid specificity within the active site.21 In this report, in the interests of further expanding the applicability of the enzyme, we have revisited the amine specificity of McbA and, following a new screen of acid-amine partners, applied the enzyme to the synthesis of the amide pharmaceutical moclobemide. With a view to further application, we have incorporated an ATP recycling system in these reactions, using a 0.1 mol equiv of the cofactor to effect up to 63% conversions in the reactions.
Having established a broader spectrum of carboxylic acid substrates for the enzyme,21 we turned our attention to the amine specificity of McbA, with a view to targeting the synthesis of known pharmaceutical amide products. In the first instance, we tested a library of amines, including aliphatic, aromatic, and alicyclic representatives (Scheme 2) against 1 for amide coupling, using only 1.5 equiv of amine for screening, and 5 equiv of ATP. Conversions were monitored by HPLC against chemically synthesized amide product reference standards. The Supporting Information (SI) details protocols for enzyme production (Section 2 in the SI), amide standard synthesis and characterization (Sections 3 and 4 in the SI), analytical procedures (Section 5 in the SI), biotransformations (Section 6 in the SI), and representative NMR spectra (Section 13 in the SI). A graph, showing the time course of these reactions, is also provided (see Figure S1 in the SI). No amide products were observed in control reactions that did not contain either ATP or McbA. Scheme 2 shows that a wider range of amines is accepted for coupling to 1 using McbA than previously evaluated.20 Small or aliphatic primary amines, such as methylamine 5, amylamine 7, and propargylamine 9 are particularly well-tolerated. In addition, 4-(2-aminoethyl)morpholine 11, which is structurally similar to the natural substrate 2-phenylethylamine 3, was also converted. A longer aliphatic chain on the amine appeared to be a characteristic of better amine substrates, as the conversion with benzylamine 15 was rapid but those with aromatic amine nucleophiles such as aniline 14 and 3,5-dimethylalanine 12 were slower. However, more activated anilines, including 5-aminoindazole 18 and 5-amino-1,3-benzodioxole 17, were converted, if more slowly. Secondary alicyclic amines, such as pyrrolidine 19 and piperidine 20 were converted very slowly at the level of amine equivalents (1.5 equiv) used in the screen. The results suggest that steric factors, but also electronic factors that determine the nucleophilicity of the amine, have a role in the constraints on amine acceptance by McbA.
Scheme 2. Amine Substrates Used in This Study.
McbA-catalyzed coupling of β-carboline acid 1 with amines 5–20 to give amides (21–36), with conversions after 1 and 16 h determined by HPLC. Reactions contained 1 (0.4 mM); amine (1.5 equiv); McbA (1 mg mL–1); ATP (5 equiv) in 50 mM KPi buffer (pH 7.5) at 37 °C.
The amine specificity of McbA was evaluated in the context of the structure of the enzyme. We have previously solved the structure of a mutant of McbA in which an active site lysine residue K483 has been mutated to alanine, in complex with 1 and AMP.21 In common with other members of the adenylase superfamily,22 McbA has a two-domain structure and adopts different conformations suitable for the two half-reactions that lead from carboxylic acid to amide. Hence, the adenylation conformer McbAAd catalyzes the synthesis of the adenylate 2 and then undergoes a domain rotation, to create a channel for amine access in the amidation conformer McbAAm. The rotation, and the formation of the channel, are analogous to the change from adenylation to thiolation conformations observed with other adenylases, such as 4-chlorobenzoyl-CoA ligase (4-CBCL),23 and the subsequent accommodation of the phosphopantetheinyl nucleophile in the new access channel in that enzyme. We have now determined a higher resolution structure, of wild-type (wt) McbA, in complex with 1 and AMP (crystallization protocols and data collection and refinement statistics are provided in Section 7 of the SI). Interestingly, the asymmetric unit of the structure contained four molecules of the McbAAd and one molecule of the McbAAm conformer, as obtained for the K483 mutant.
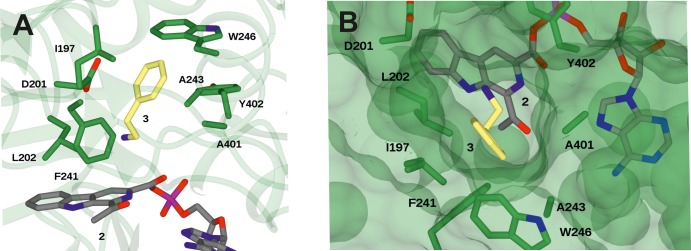
The structure of wt-McbA reveals the position of active site residues, including K483, in the adenylation conformer of the enzyme McbAAd (see Figure S2 in the SI). To provide further insight into the determinants of amine recognition in McbA, adenylate 2 was first modeled into the active site of the amidation conformer McbAAm, followed by 2-phenylethylamine 3, using AutoDock Vina.24 The model (Figure 1) illustrates the accommodation of the amine within the entry channel created by the domain rotation that creates McbAAm and suggests a hydrophobic binding site for the amine formed by residues including I197, F241, A243, and Y246, from the large domain, and A401 from the rotated smaller domain. The model also places the amine in proximity to D201, which is structurally homologous to histidine H207 in 4-CBCL that is thought to be important in the catalytic mechanism of that enzyme.23 We suggest that the role of D201 may be to activate amines for attack at the adenylate intermediate. In order to examine this hypothesis, mutant D201A was constructed (Section 8 in the SI), expressed, purified, and assayed against the wt-McbA enzyme in the coupling of β-carboline acid 1 with propargylamine 9. A comparison of the time courses of each reaction showed that D201A was significantly less active than the wt-McbA, showing just 2% residual activity (see Figure S3 in the Supporting Information).
Figure 1.
(A) Structure of wt-McbAAm modeled with the adenylate intermediate 2 and 2-phenylethylamine 3. (B) Surface view of wt-McbAAm–2–3 rotated 90° relative to the view in panel (A), showing access channel for the amine.
The broader amine specificity of McbA, particularly including amines 10 and 11, in conjunction with our previous observation that McbA accepts benzoic acid as a substrate,21 led us to evaluate the enzyme for small-scale syntheses of pharmaceutical amide compounds. First, we envisaged a synthesis of the monoamine oxidase A inhibitor moclobemide 38, through the coupling of para-chlorobenzoic acid 37 with 4-(2-aminoethyl)morpholine 11 (see Scheme 3). Initial experiments were encouraging, with conversions of 63% for 37 → 38 observed for reactions in which 4 mM of 37 was incubated with 6 mM of 11 and 20 mM ATP. However, further optimization failed to increase these conversions, and we hypothesized that inhibition of McbA by AMP, inorganic pyrophosphate (PPi), or ATP, which has been observed previously with ATP-dependent ligases,25 may be a factor. Reactions performed in the presence of 1 mM or 4 mM AMP gave similar conversions (see Section 9 and Figure S4 in the SI); however, conversions in the presence of 1 mM or 4 mM Pi were significantly lower than in standard reactions. In order to overcome the apparent Pi inhibition, 0.3 U inorganic pyrophosphatase (IPase) was added to the McbA-catalyzed synthesis of 38.
Scheme 3. Synthesis of Moclobemide 38 from 4-Chlorobenzoic Acid 37 and 4-(2-Aminoethyl)morpholine 11 Using McbA.
This scheme also illustrates the ATP recycling system comprising the polyphosphate kinases AjPPK2-II, SmPPK2-I and polyphosphate (PolyPn).
The conversion after 24 h was increased to >99%, compared to 60% with no IPase added (Section 10 in the SI). These conditions were applied to the transformation of 10 mg of 37 with 1.5 equiv of 11, giving a conversion of 70%, as determined by NMR (Section 11 and Figure S5 in the SI), and an isolated yield of 38 of 11 mg (64%).
The amidation of the native substrate beta-carboline 1 with propargylamine 9 was used as a model reaction to study inhibition by ATP. The activity of McbA was highest at 4 mM ATP but decreased thereafter to approximately half the optimum value value at 16 mM ATP (Section 12 and Figure S6 in the SI). One possible solution to this problem is the use of an ATP recycling system that would permit lower concentrations of ATP within the reaction. This would also be advantageous, with respect to process costs in amidation reactions using McbA and related enzymes. We envisaged, as a suitable system, the polyphosphate kinases SmPPK2-I from Sinorhizobium meliloti and AjPPK2-II from Acinetobacter johnsonii described by Andexer and co-workers,26,27 which use inexpensive polyphophoric acid (PolyPn) for the phosphorylation of AMP and ADP to ADP and ATP, respectively (see Scheme 3). Equivalent enzymes have recently been employed for the regeneration of ATP in the reduction of carboxylic acids using CARs by Winkler and co-workers.28 The efficiency of the recycling system, with respect to the reaction between 11 and 37, was tested in reactions consisting of McbA, AjPPK2-II, SmPPK2-I, and polyphosphate, using increasing equivalents of ATP (Figure 2). At 0.1 equiv of ATP, the reaction in the absence of the recycling system achieved, as expected, only the theoretical maximum of ∼10% conversion after 48 h.
Figure 2.
Comparison of 0.1–0.8 equiv (eq) of ATP with and without ATP recycling system. Reactions for the (−) recycling system (black) contained McbA (1 mg mL–1), acid 37 (4 mM), amine 11 (1.5 equiv), and ATP (x eq) in 50 mM KPi buffer (pH 7.5) at 37 °C. Reactions for the (+) recycling system (white) also contained polyphosphate (20 mM), AjPPK2-II, and SmPPK2-I (each at 0.1 mg mL–1, at 280 nm).
However, the inclusion of the polyphosphate kinase system increased the conversion to 36% after 48 h. Further optimization of the system, in which the concentration of AjPPK2-II and SmPPK2 was doubled to 0.2 mg mL–1 and a further 1 mg mL–1 of McbA was added after 6 h of reaction time, resulted in a conversion of 63% at 0.1 equiv of ATP.
The need for more selective and sustainable routes to amides suggests that biocatalytic routes may form part of the selection of greener methods for amide synthesis in the future. While a range of enzymes is available, each has its advantages and disadvantages, so the development of complementary systems will be beneficial. The ATP-dependent amide bond synthetases are attractive because they permit, where required, the coupling of carboxylic acid and amine partners, supplied in equimolar amounts, and in an aqueous medium, to form pharmaceutical-type amide products. In addition to improvements in the system gained through the optimization of process-specific technologies, such as ATP recycling, the potential of these enzymes may also be realized through directed evolution methods to increase stability and turnover, and broadened substrate specificity for wider application.
Acknowledgments
M.P. was funded by an iCASE studentship award from GSK in collaboration with the British Biotechnology and Biological Sciences Research Council (BBSRC). We are grateful to Prof. Jennifer Andexer of the University of Freiburg for the gift of plasmids containing genes encoding the polyphosphate kinases AjPPK2-II and SmPPK2-I.
Glossary
Abbreviations
- ATP
adenosine triphosphate
- NRPS
nonribosomal peptide synthase
- ABS
amide bond synthetase
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscatal.0c00929.
Synthetic protocols and analytical data on substrates/products; biotransformation procedures; X-ray crystallographic methods and data (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Brown D. G.; Boström J. Analysis of Past and Present Synthetic Methodologies on Medicinal Chemistry: Where Have all the New Reactions Gone?. J. Med. Chem. 2016, 59 (10), 4443–4458. 10.1021/acs.jmedchem.5b01409. [DOI] [PubMed] [Google Scholar]
- Goswami A.; van Lanen S. G. Enzymatic Strategies and Biocatalysts for Amide Bond Formation: Tricks of the Trade Outside of the Ribosome. Mol. BioSyst. 2015, 11, 338–353. 10.1039/C4MB00627E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzer J.; Steiner K. Amides in Nature and Biocatalysis. J. Biotechnol. 2016, 235, 32–46. 10.1016/j.jbiotec.2016.03.023. [DOI] [PubMed] [Google Scholar]
- Dorr B. M.; Fuerst D. E. Enzymatic Amidation for Industrial Applications. Curr. Opin. Chem. Biol. 2018, 43, 127–133. 10.1016/j.cbpa.2018.01.008. [DOI] [PubMed] [Google Scholar]
- Petchey M. R.; Grogan G. Enzyme-Catalysed Synthesis of Secondary and Tertiary Amides. Adv. Synth. Catal. 2019, 361, 3895–3914. 10.1002/adsc.201900694. [DOI] [Google Scholar]
- Lima R. N.; dos Anjos C. S.; Orozco E. V. M.; Porto A. L. M. Veratility of Candida antarctica Lipase in the Amide Bond Formation Applied in Organic Synthesis and Biotechnological Processes. Mol. Catal. 2019, 466, 75–105. 10.1016/j.mcat.2019.01.007. [DOI] [Google Scholar]
- Marešová H.; Plačková M.; Grulich M.; Kyslík P. Current State and Perspectives of Penicillin G Acylase-Based Biocatalyses. Appl. Microbiol. Biotechnol. 2014, 98, 2867–2879. 10.1007/s00253-013-5492-7. [DOI] [PubMed] [Google Scholar]
- Zeng S.; Liu J.; Anankanbil S.; Chen M.; Guo Z.; Adams J. P.; Snajdrova R.; Li Z. Amide Synthesis via Aminolysis of Ester or Acid with an Intracellular Lipase. ACS Catal. 2018, 8, 8856–8865. 10.1021/acscatal.8b02713. [DOI] [Google Scholar]
- Dunn Z. D.; Wever W. J.; Economou N. J.; Bowers S. S.; Li B. Enzymatic Basis of ‘Hybridity’ in Thiomarinol Biosynthesis. Angew. Chem., Int. Ed. 2015, 54, 5137–5141. 10.1002/anie.201411667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott H. K.; Thomas P. J.; Tew D.; Fuerst D. E.; Lovelock S. L. A Versatile Biosynthetic Approach to Amide Bond Formation. Green Chem. 2018, 20, 3426–3431. 10.1039/C8GC01697F. [DOI] [Google Scholar]
- Winn M.; Fyans J. K.; Zhuo Y.; Micklefield J. Micklefield. Recent Advances in Engineering Nonribosomal Peptide Assembly Lines. Nat. Prod. Rep. 2016, 33, 317–347. 10.1039/C5NP00099H. [DOI] [PubMed] [Google Scholar]
- Abe T.; Hashimoto Y.; Sugimoto S.; Kobayashi K.; Kumano T.; Kobayashi M. Amide Compound Synthesis by Adenylation Domain of Bacillibactin Synthetase. J. Antibiot. 2017, 70, 435–442. 10.1038/ja.2016.117. [DOI] [PubMed] [Google Scholar]
- Abe T.; Kobayashi K.; Kawamura S.; Sakaguchi T.; Shiiba K.; Kobayashi M. Dipeptide Synthesis by Internal Adenylation Domains of a Multidomain Enzyme Involved in Nonribosomal Peptide Synthesis. J. Gen. Appl. Microbiol. 2019, 65, 1–10. 10.2323/jgam.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Abe T.; Hashimoto Y.; Hosaka H.; Tomita-Yokotani K.; Kobayashi M. Discovery of Amide (Peptide) Bond Synthetic Activity in Acyl-CoA Synthetase. J. Biol. Chem. 2008, 283, 11312–11321. 10.1074/jbc.M709654200. [DOI] [PubMed] [Google Scholar]
- Abe T.; Hashimoto Y.; Zhuang Y.; Ge Y.; Kumano T.; Kobayashi M. Peptide Bond Synthesis by a Mechanism Involving an Enzymatic Reaction and a Subsequent Chemical Reaction. J. Biol. Chem. 2016, 291, 1735–1750. 10.1074/jbc.M115.700989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R.; Hirai K.; Suzuki S.; Kino K. A Chemoenzymatic Process for Amide Bond Formation by an Adenylating Enzyme-Mediated Mechanism. Sci. Rep. 2018, 8, 2950. 10.1038/s41598-018-21408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti P. M.; Richardson S. M.; Kariem N. M.; Campopiano D. J. Synthesis of N-Acyl Amide Natural Products Using a Versatile Adenylating Biocatalyst. MedChemComm 2019, 10, 1192–1196. 10.1039/C9MD00063A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. J. L.; Weise N. J.; Frampton J. D.; Dunstan M. S.; Hollas M. A.; Derrington S. R.; Lloyd R. C.; Quaglia D.; Parmeggiani F.; Leys D.; Turner N. J.; Flitsch S. L. Adenylation Activity of Carboxylic Acid Reductases Enables the Synthesis of Amides. Angew. Chem., Int. Ed. 2017, 56, 14498–14501. 10.1002/anie.201707918. [DOI] [PubMed] [Google Scholar]
- Chen Q.; Ji C.; Song Y.; Huang H.; Ma J.; Tian X.; Ju J. Discovery of McbB, an Enzyme Catalyzing the c-Carboline Skeleton Construction in the Marinacarboline Biosynthetic Pathway. Angew. Chem., Int. Ed. 2013, 52, 9980–9984. 10.1002/anie.201303449. [DOI] [PubMed] [Google Scholar]
- Ji C.; Chen Q.; Li Q.; Huang H.; Song Y.; Ma J.; Ju J. Chemoenzymatic Synthesis of -Carboline Derivatives Using McbA, a New ATP-Dependent Amide Synthetase. Tetrahedron Lett. 2014, 55, 4901–4904. 10.1016/j.tetlet.2014.07.004. [DOI] [Google Scholar]
- Petchey M.; Cuetos A.; Rowlinson B.; Dannevald S.; Frese A.; Sutton P. W.; Lovelock S.; Lloyd R. C.; Fairlamb I. J. S.; Grogan G. The Broad Aryl Acid Specificity of the Amide Bond Synthetase McbA Suggesst Potential for the Biocatalytic Synthesis of Amides. Angew. Chem., Int. Ed. 2018, 57, 11584–11588. 10.1002/anie.201804592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick A. M. Conformational Dynamics in the Acyl-CoA Synthetases, Adenylation Domains of Non-Ribosomal Peptide Synthetases, and Firefly Luciferase. ACS Chem. Biol. 2009, 4, 811–827. 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reger A. S.; Wu R.; Dunaway-Mariano D.; Gulick A. M. Structural Characterization of a 140° Domain Movement in the Two-Step Reaction Catalyzed by 4-Chlorobenzoate: CoA Ligase. Biochemistry 2008, 47, 8016–8025. 10.1021/bi800696y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2009, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C.; Lloyd A.; Sanschagrin F.; Maaroufi H.; Clarke T.; Blewett A.; Dowson C.; Roper D. I.; Bugg T. D. H.; Levesque R. C. Pseudomonas aeruginosa MurE Amide Ligase: Enzyme Kinetics and Peptide Inhibitor. Biochem. J. 2009, 421, 263–272. 10.1042/BJ20081395. [DOI] [PubMed] [Google Scholar]
- Andexer J. N.; Richter M. Emerging Enzymes for ATP Regeneration in Biocatalytic Processes. ChemBioChem 2015, 16, 380–386. 10.1002/cbic.201402550. [DOI] [PubMed] [Google Scholar]
- Mordhorst S.; Maurer A.; Popadić D.; Brech J.; Andexer J. N. A Flexible Polyphosphate-Driven Regeneration System for Coenzyme A Dependent Catalysis. ChemCatChem 2017, 9, 4164–4168. 10.1002/cctc.201700848. [DOI] [Google Scholar]
- Strohmeier G. A.; Eiteljörg I. C.; Schwarz A.; Winkler M. Enzymatic One-Step Reduction of Carboxylates to Aldehydes with Cell-Free Regeneration of ATP and NADPH. Chem. - Eur. J. 2019, 25, 6119–6123. 10.1002/chem.201901147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.