Abstract
Pinholin S2168 is an essential part of the phage Φ21 lytic protein system to release the virus progeny at the end of the infection cycle. It is known as the simplest natural timing system for its precise control of hole formation in the inner cytoplasmic membrane. Pinholin S2168 is a 68 amino acid integral membrane protein consisting of two transmembrane domains (TMDs) called TMD1 and TMD2. Despite its biological importance, structural and dynamic information of the S2168 protein in a membrane environment is not well understood. Systematic site-directed spin labeling and continuous wave electron paramagnetic resonance (CW-EPR) spectroscopic studies of pinholin S2168 in 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) proteoliposomes are used to reveal the structural topology and dynamic properties in a native-like environment. CW-EPR spectral line-shape analysis of the R1 side chain for 39 residue positions of S2168 indicates that the TMDs have more restricted mobility when compared to the N- and C-termini. CW-EPR power saturation data indicate that TMD1 partially externalizes from the lipid bilayer and interacts with the membrane surface, whereas TMD2 remains buried in the lipid bilayer in the active conformation of pinholin S2168. A tentative structural topology model of pinholin S2168 is also suggested based on EPR spectroscopic data reported in this study.
INTRODUCTION
The most frequent cytocidal event in the biosphere is the bacteriophage infection cycle with the last stage of this cycle being bacterial cell lysis to release the mature neonate virus.1–3 Cell lysis is precisely controlled and synchronized by at least three groups of proteins.4–6 The first step of this process is the hole formation in the inner cytoplasmic membrane by the holin, followed by the murein layer (peptidoglycan) degradation by the endolysin and outer membrane disruption by the spanin complex.6–8 The entire process happens within seconds of holin triggering which occurs at an allele-specific time and concentration.1,9 The canonical holins form a nonspecific, microscale hole in the inner cytoplasmic membrane to allow the diffusion of functionally folded endolysin to the peptidoglycan for degradation.2,10–13 However, some phages (e.g. phage P1 or Φ21) represent a significantly different and alternative class of holin which make smaller holes in comparison to the canonical holins. These nanoscale holes are large enough for depolarization of membrane potential.2,13–15 This group of holin is responsible for the release of signal-anchor release endolysin and is known as pinholin for the small-scale pinholes it creates.13,14
Pinholin S21 is encoded by the S21 gene of phage Φ21. The S21 gene contains a dual start motif gene which encodes two proteins, the 68 amino acid long active pinholin (S2168) and the 71 amino acid long antiholin (S2171). Antiholin (S2171) is transcribed from the first codon of the S21 gene where active pinholin (S2168) is transcribed from the fourth codon with the first amino acid of S2168 being denoted as Met4. Pinholin S2168 has two transmembrane domains (TMDs) connected via a short periplasmic loop and cytoplasmic N and C-termini. Pinholin progressively accumulates in the bacterial inner cytoplasmic membrane as an inactive dimer with both TMDs remaining in the lipid bilayer. TMD1 of the active form of pinholin S2168 externalizes very quickly to the periplasm resulting in the active dimer.1,2,12 Within seconds of pinholin triggering, it forms heptametric holes by rapid oligomerization and reorientation of TMD2. A study of pinholin S2168 will describe the functionally and structurally unique group of holin which consists of ~900 proteins.16
The structure and functional model of pinholin S2168 has been reported by the Young group using biomolecular and functional studies.1,2,12,17–19 They have also used a computational approach to predict the number of monomers and the size of the pinhole using a truncated form of pinholin S2168 (TMD1 was deleted).1 However, dynamic information, as well as relative orientations and interactions of TMD1 and TMD2 of S2168 with lipid or other residues, were not extensively studied. For the confirmation of the proposed model, further biophysical studies were recommended.2
It has been widely recognized that the function and stability of proteins are interrelated with their structural topology and dynamic properties.20–24 Hence, it is important to know the structural topology and dynamic properties of membrane proteins in their native-like environment to understand their biological functions and mechanisms. Electron paramagnetic resonance (EPR) spectroscopy is a unique biophysical technique to study the structural topology and dynamic properties of proteins with high sensitivity in membrane mimic conditions.20,25–35 This study is focused on the structural topology and dynamic properties of full-length active pinholin (S2168) using EPR spectroscopy coupled with site-directed spin labeling (SDSL). Fmoc solid-phase peptide synthesis (Fmoc-SPPS) was used for sample preparation with high yield and flexibility of spin-label incorporation at any residue position of the peptide.36 In this EPR spectroscopic study of pinholin S2168, nitroxide spin label, MTSL (S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) was attached to a Cys side chain at specific sites using SDSL.37
This is the first biophysical study to probe the structural topology and dynamic properties of the full-length pinholin S2168. Using the combination of continuous wave (CW)-EPR spectroscopic line shape analysis and EPR power saturation data, we propose a tentative model of the active form of S2168, where TMD2 remains incorporated in lipid bilayers, while TMD1 partially externalizes or interacts with the head group of the lipid bilayers.
EXPERIMENTAL METHODS
Solid-Phase Peptide Synthesis.
All peptides were synthesized via optimized Fmoc SPPS.38 A 0.1 mmol peptide synthesis was conducted on an automated CEM Liberty Blue peptide synthesizer equipped with the Discovery Bio microwave system. The synthesis was started with a preloaded thermogravimetric analysis resin in a dimethylformamide (DMF) base solvent system where 20% (v/v) piperidine, 20% (v/v) N,N′-diisopropylcarbodiimide, and 10% (w/v) Oxyma in DMF were used as deprotecting, activator, and activator base, respectively. The cleavage reaction was run for at least 3 h under optimized cocktail conditions [trifluoroacetic acid (TFA) 94%, water 2.5%, 1,2-ethanedithiol 2.5%, triisoproylsilane (TIPS) 1%] or (TFA 85%, water 5%, anisole 5%, TIPS 5%) to remove the resin and side-chain protecting groups.38–41
Purification and Spin Labeling.
The crude peptide was purified by reverse-phase high-pressure liquid chromatography (HPLC) using a GE HPLC system. The sample was loaded onto a C4 (10 μm) preparative column (Vydac 214TP, 250 × 22 mm) and was run with a two solvent gradient system, where the polar solvent was 100% water and the nonpolar solvent was 90% acetonitrile with 10% water. 0.1% TFA was added to both solvents to acidify it. The gradient started from 5% nonpolar and increased up to 98% nonpolar by optimized run time.38 The purified pinholin peptide collected from HPLC elution was lyophilized. To attach the spin label, the lyophilized pure peptide was dissolved in dimethyl sulfoxide with 5-fold excess of MTSL (1:5 molar ratio) and stirred for 24 h in a dark environment. The spin-labeled (SL) peptide was lyophilized again and purified with a C4 semi-preparative column (Vydac 214TP, 250 × 10 mm), using the same solvent and gradient system to remove free MTSL and further purify the peptide sample. After each purification, the purity of the target peptide was confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Spin-labeling efficiency was calculated ~85–90% using CW-EPR.38
Peptide Incorporation into Proteoliposomes.
To mimic the membrane environment, SL pinholin peptides were incorporated into DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) proteoliposomes following the thin-film method.38 Pure SL peptides were dissolved in 2,2,2-trifluoroethanol and mixed with predissolved DMPC lipid solution in a pear-shaped flask. The organic solvent was gently evaporated by N2(g) purging to get a uniform thin film inside the pear-shaped flask. It was left under a vacuum desiccator overnight to remove any residual organic solvent. 20 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer (pH ≈ 7.0) was used to rehydrate the thin film to get the final concentration of 200 mM lipid and 200 μM peptide into proteoliposomes at a 1:1000 protein to lipid ratio to minimize the formation of oligomers. Before rehydration, both HEPES buffer and the sample flask were kept in a warm water bath for a short period of time to bring the temperature above the phase transition temperature of DMPC.42,43 The lipid film was dispersed from the side wall by several vortexes followed by warming in a water bath. Three more freeze–thaw cycles were performed before adding glycerol. 10% glycerol was added to the sample and mixed properly. Glycerol helps the sample to remain suspended for a longer duration at room temperature without phase separation. Sample homogeneity and size of the proteoliposomes were confirmed by dynamic light scattering (DLS) spectroscopy using ZETASIZER NANO Series (Malvern Instruments) at 25 °C in a disposable 40 μL microcuvette.
Circular Dichroism Spectroscopy.
Circular dichroism (CD) spectra for pinholin S2168 incorporated into proteoliposomes were collected using an Aviv CD spectrometer (model 435) in a quartz cuvette with a 1.0 mm path length. Pinholin S2168-incorporated proteoliposomes samples were diluted 10 times with 10 mM phosphate buffer to reduce the final HEPES buffer concentration (which gives absorbance at lower wavelength) as well as protein concentration (20 μM). Data were collected from 260 to 190 nm with an average of 10 scans per sample and 1 nm bandwidth at 25 °C.38
CW-EPR Spectroscopy.
CW-EPR spectra were collected at the X-band (~9.34 GHz) using a Bruker EMX spectrometer equipped with a ER 041xG microwave bridge and ER4119-HS cavity at the Ohio Advanced EPR Laboratory at Miami University. Each spectrum was acquired by the signal averaging of 10 scans with 3315 G central field, sweep width 150 G, 42 s field sweep, 100 kHz modulation frequency, 1 G modulation amplitude, and 10 mW microwave power. Each experiment was repeated at least three times. All experiments were carried out at room temperature.
Spin-Label Mobility Analysis.
The inverse line width (δ−1) of the first-derivative central resonance line (mI = 0) was calculated to determine the side-chain mobility. The mobility parameter, δ−1 was normalized to “scaled mobility” factor (Ms), using eq 1.20,31,44
| (1) |
where δi and δm are central line widths for the most immobilized and the most mobile side chains, respectively. The mean scaled mobility was calculated by taking the average of all studied SL residues.
To further explore the dynamic properties, the rotational correlation time (τ) was calculated using eq 2.32,33,45–47
| (2) |
where, K = 6.5 × 10−10 s, δ is the width of the center-line, and h0 and h−1 are the heights of the center and high field lines, respectively.46,47
CW-EPR Power Saturation Experiments.
CW-EPR power saturation experiments were performed on a Bruker EMX X-band spectrometer coupled with ER 041XG microwave bridge and ER 4123D CW-Resonator (Bruker BioSpin). Experimental setups were optimized following the published literature.28,34,35,45,48 Samples were loaded into gas-permeable TPX capillary tubes with a total volume of 3–4 μL at a concentration of 100–150 μM.48–51 EPR spectroscopic data were collected using a modulation amplitude of 1.0 G, a modulation frequency of 100 kHz, a field sweep of 42 s, and a sweep width of 90 G. Incident microwave powers were varied from 0.06 to 159 mW. Three to five scans were signal-averaged at each microwave power. For each SL site, the spectra were recorded under three equilibrium conditions. At first, the samples were equilibrated with a lipid-soluble paramagnetic relaxant (21% oxygen) followed by the equilibration with nitrogen gas (as control), and equilibration with a watersoluble paramagnetic relaxant (2 mM NiEDDA) with a continuous purge of nitrogen gas.51 Each set of experiment was repeated at least three times. NiEDDA was synthesized according to the published protocol.35,51 The samples were purged with nitrogen gas for at least 1 h, at a rate of 10 mL per min before starting nitrogen or NiEDDA data acquisition. The resonator was connected to the gas supply (air or nitrogen gas) during all measurements, and all the experiments were performed at room temperature. The peak-to-peak amplitudes (A) of the first derivative mI = 0 resonance lines were extracted and plotted against the square root of the incident microwave power. These data points were then fitted according to eq 3.35,50
| (3) |
where I is a scaling factor, P1/2 is the power where the first derivative amplitude is reduced to half of its unsaturated value, and ε is a measure of the homogeneity of saturation of the resonance line. The homogeneous and inhomogeneous saturation limits are ε = 1.5 and ε = 0.5, respectively.35 In eq 3, I, ε, and P1/2 are adjustable parameters and yield a characteristic P1/2 value for each equilibrium condition. Data analysis was performed using a MATLAB software script. The corresponding depth parameter (Φ) was calculated using eq 4.35
| (4) |
where ΔP1/2(O2) is the difference in the P1/2 values for oxygen and nitrogen equilibriums, and ΔP1/2(NiEDDA) is the difference in the P1/2 values for NiEDDA and nitrogen equilibriums.
RESULTS
Recently, the successful synthesis and spectroscopic characterization of the full-length active and inactive forms of pinholin S21 were reported by the Lorigan research group.38 For this study, the dynamic properties of pinholin S2168 were investigated by analyzing the CW-EPR spectra obtained from 39 SL positions of S2168 peptides incorporated into DMPC proteoliposomes at a mole ratio of 1:1000 to minimize the formation of oligomers. The representative DLS data are shown in Supporting Information, Figure S1, for pinholin S2168 G48R1 incorporated into DMPC proteoliposomes which confirmed the homogeneity of the proteoliposome samples.
The primary amino acid sequence of the full-length active pinholin S2168 is shown in Figure 1A. Residues studied with SDSL are indicated in blue. Predicted TMD1 and TMD2 are indicated by the two boxes. The predicted topology of pinholin S2168 is adapted from the literature and shown in Figure 1B.1,2,12,38 The R1 side-chain attachment to the protein via a disulfide bond is shown in Figure 1C.26
Figure 1.
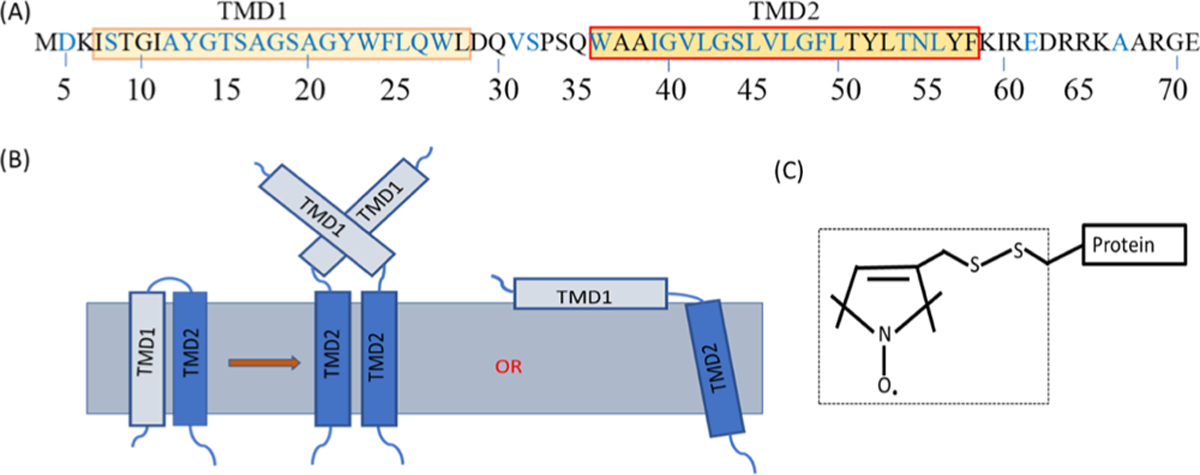
(A) Primary sequence of S2168, boxes indicate TMD1 and TMD2. The amino acid positions studied by EPR spectroscopy are shown in blue. (B) Predicted topology of S2168 is adapted from the literature.1,2,12,38 TMD1 completely externalizes from the lipid bilayer and remains in the periplasm or partially externalizes and stays on the surface of the lipid bilayer, where TMD2 remains in the lipid bilayer. (C) R1 side chain shown in the dotted box which is attached to the protein through the disulfide bond of a Cys residue.
Figure 2 shows the representative CD spectrum of pinholin S2168 G40R1 in DMPC proteoliposomes. Two minima around 222 and 208 nm, and a large positive peak close to 195 nm confirmed the α-helical secondary structure of pinholin S2168 with the R1 side chain.
Figure 2.
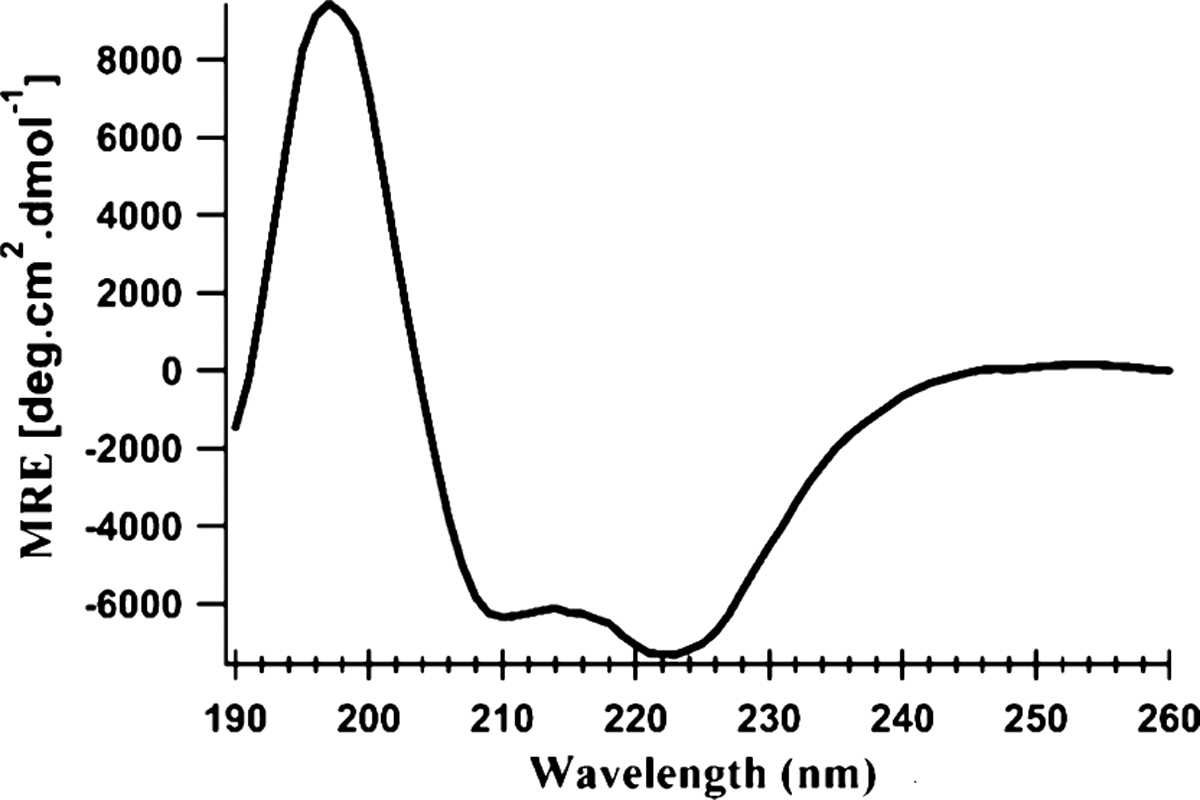
CD spectrum of pinholin S2168 G40R1 in DMPC proteoliposomes was collected at pH 7 and 298 K. Mean residue ellipticity is plotted against incident radiation wavelength.
CW-EPR Line-Shape Analysis of Pinholin S2168.
CW-EPR spectral analysis for a set of SL protein allows probing the structural and dynamic properties of the protein with a spatial resolution at the residue-specific level.45,52–56 Figure 3 shows 39 CW-EPR spectra collected for R1 side chains placed at strategic locations of S2168 incorporated into DMPC proteoliposomes.
Figure 3.
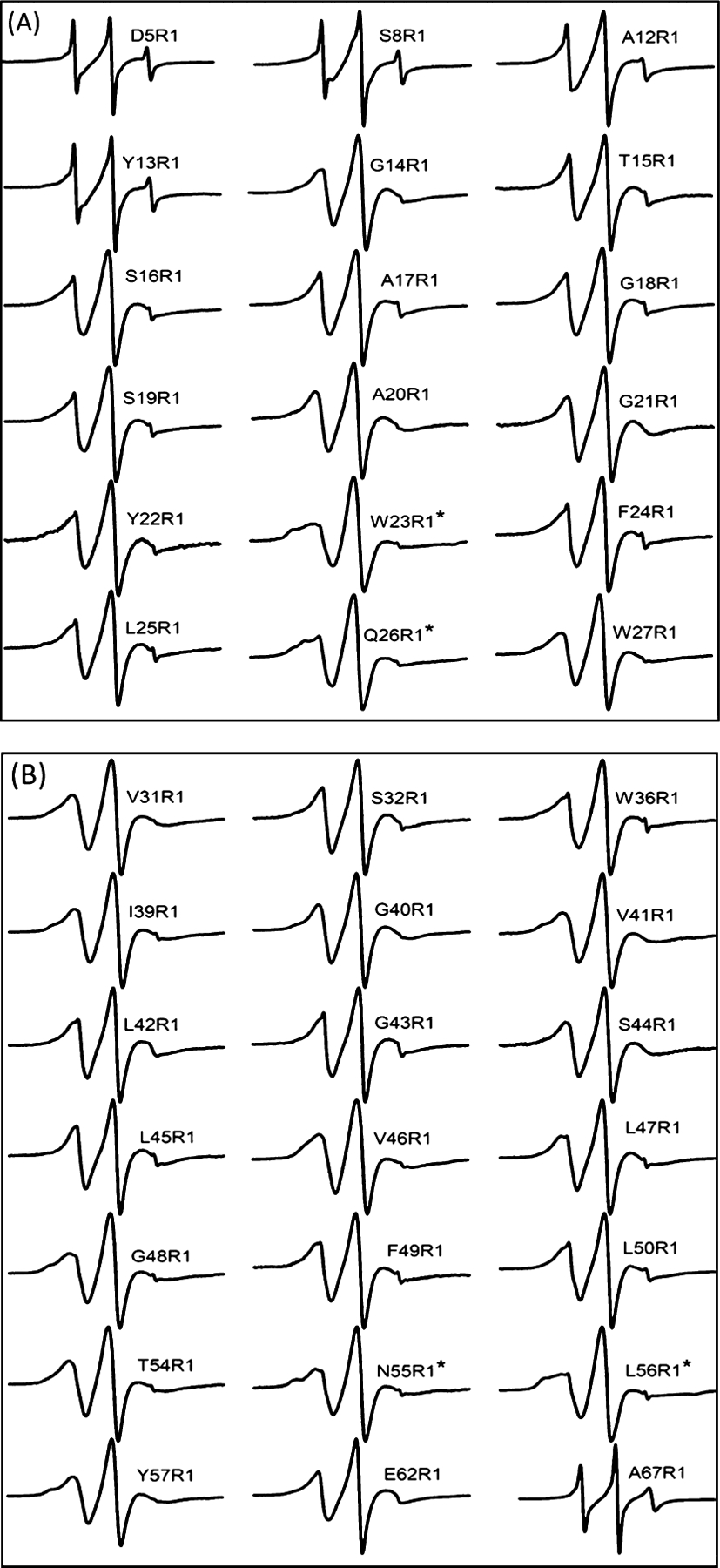
CW-EPR spectra of R1 side chains attached at indicated positions by replacing the native amino acid with Cys. All spectra were normalized to the highest spectral intensity. (A) EPR spectra from the N-terminal and TMD1 of pinholin S2168. (B) EPR spectra from the loop to the C-terminal of pinholin S2168 including TMD2. CW-EPR spectra composed of multiple components were marked with (*).
All CW-EPR spectra show the conventional three 14N hyperfine peaks of the R1 side chain attached to pinholin S2168. EPR spectra from the N- and C-termini show a relatively sharper central peak than the TMD regions which indicate higher mobility of the R1 side chain or protein backbone in the terminal regions when compared to the TMD residues. EPR spectra of residues within TMD1 and TMD2 show line broadening indicative of restricted mobility in those regions. Most of the spectra show a single motional component, which indicates monodispersion of conformational and motional properties of S2168. However, CW-EPR spectra for W23R1, Q26R1, N55R1, and L56R1 [indicated by (*) in Figure 3] show two spectral components (rigid and fast components), which are indicative of heterogeneous dynamics at those sites.52
Beside this qualitative information, CW-EPR line-shape analysis can be used to derive quantitative dynamic information. Inversion of the width of the central resonance line (δ−1) has been used as a semiquantitative measurement of nitroxide mobility.20,29,57,58 The mobility of individual residues is shown in Figure 4, by plotting δ−1 as a function of residue positions for pinholin S2168.
Figure 4.
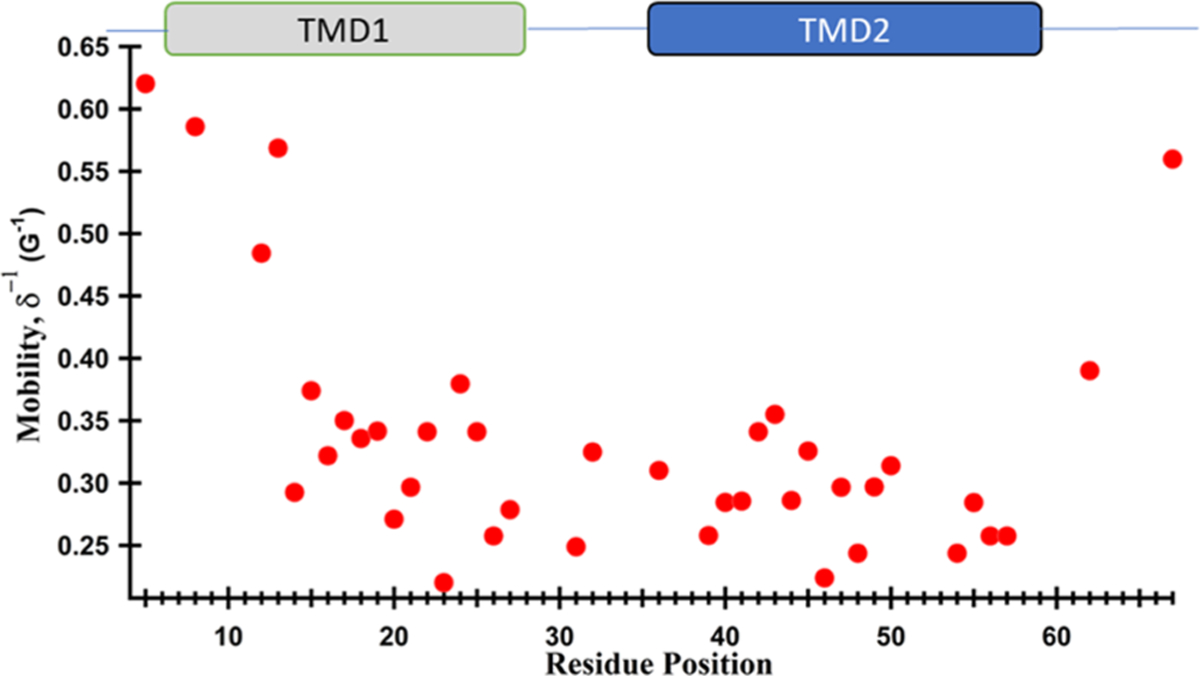
EPR mobility analysis of the R1 side chain of S2168, calculated from the inverse width of the central resonance line (δ−1). The larger δ−1 value indicates increasing motion.
Both N- and C-termini residues had higher mobility than residues inside the TMDs as speculated qualitatively from the line-shape analysis. The loop region had intermediate or restricted mobility which can be explained by the small loop size and the junction between the two slow motional segments (TMD1 and TMD2) of S2168. The highest mobility was observed for D5R1, present in the N-terminal with the δ−1 of 0.62 G−1 while the lowest mobility was 0.22 G−1 for W23R1 present in TMD1. Many more fluctuations of the δ−1 values were observed for the TMD1 residues, when compared to the δ−1 values of TMD2 residues. The δ−1 of the TMD1 varies from 0.22 to 0.59 G−1, while the δ−1 of the TMD2 varies between 0.22 and 0.36 G−1.
Scaled mobility (Ms) is another convenient method to compare the R1 dynamics for different sites relative to the most mobile and immobile sites. The Ms values were calculated using eq 1. The central line width was observed among the entire 39 residue set with the most mobile (D5R1) and immobile (W23R1) sites being 1.6 G (δm) and 4.5 G (δi), respectively. The individual Ms value as a function of the residue position is shown in Figure 5.
Figure 5.
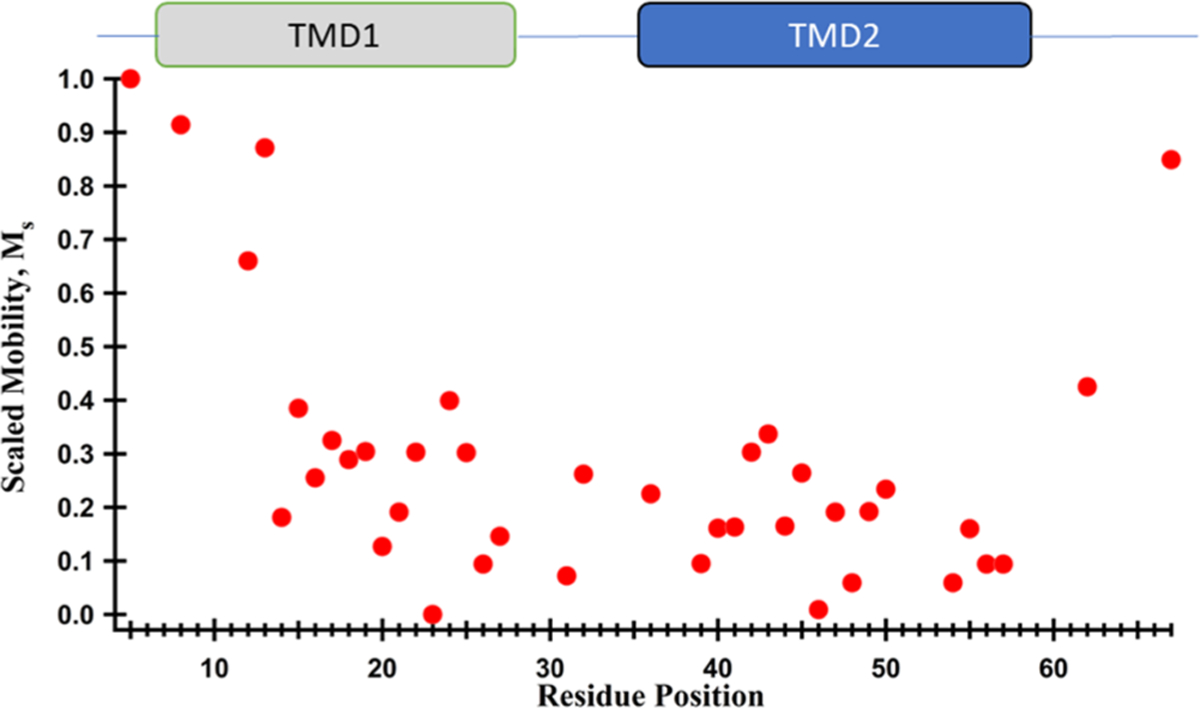
Scaled mobility as a function of the residue position for pinholin S2168.
From the individual Ms values, the average scaled mobility, , was calculated which reflect the compactness of the protein system, helical packing density, and local backbone motions of a protein or segment of a protein.20 An overall value of 0.27 was determined for pinholin S2168 which indicates moderate to tight helical packing.20 For the individual TMDs, values were 0.34 (TMD1; 8–27) and 0.17 (TMD2; 36–56), which indicates TMD2 has tighter helical packing when compared to TMD1.
To further explore the dynamic properties of pinholin S2168 in a membrane, the rotational correlation time (τ) was calculated. τ is the time required for the spin label to rotate one radian angle.45 The rotational correlation time of the R1 side chain is the result of three different rotational correlation times including the rotational correlation time of the entire protein (τR), local protein backbone fluctuations, and the flexibility of the SL side chain relative to the protein.20,29,59 For membrane proteins, τR value greater than 60 ns, which is out of EPR sensitivity, can be ignored.44,59 Therefore, the calculated τ values are the reflection of a combination of backbone fluctuations, side chain dynamics, and interactions with the surroundings. Figure 6 shows the calculated τ values for the corresponding residues of SL pinholin S2168.
Figure 6.
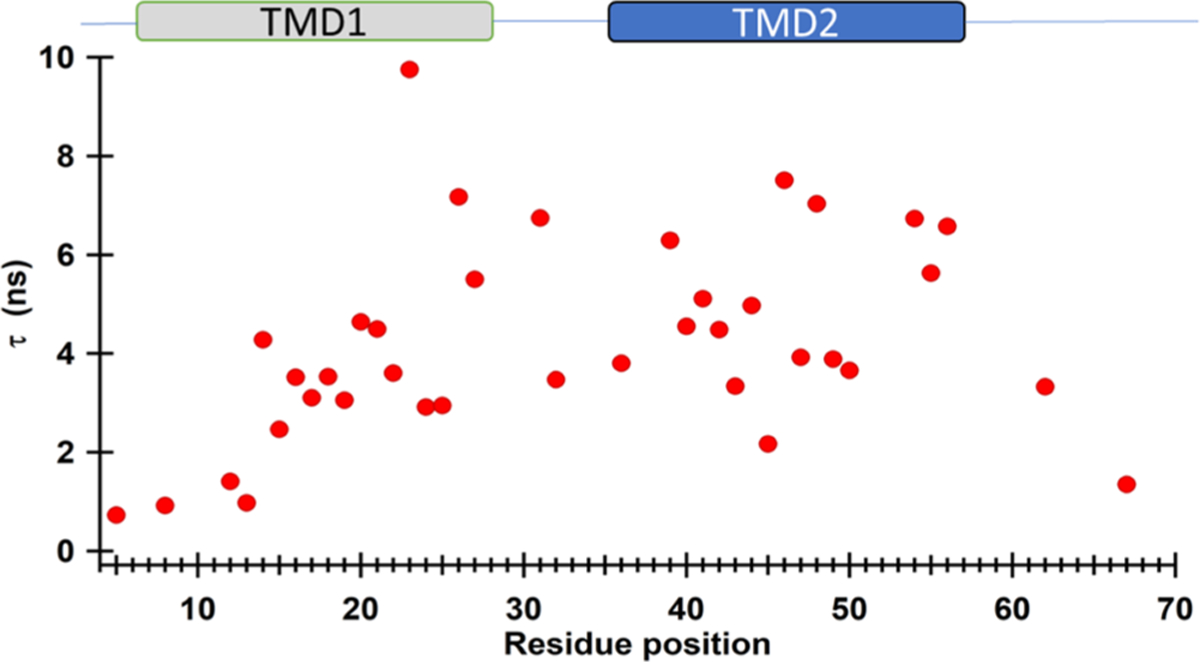
Rotational correlation time (τ) of SL pinholin S2168 in DMPC proteoliposomes as a function of the residue position.
The minimum and maximum τ values were calculated to be 0.73 ns (D5R1) and 9.75 ns (W23R1), respectively. Shorter τ values (below 2 ns) for N- and C-termini residues suggest higher spin label motion due to the lipid-free environments, where longer τ for TMD residues suggest their restricted motion due to lipid environment and/or restricted backbone motions. Dynamic patterns observed from the τ calculation are consistent with the side chain mobility data extracted from the central line broadening, which are shown in Figures 4 and 5.
Structural Topology with Respect to the Lipid Bilayer.
CW-EPR power saturation is a powerful and convenient biophysical technique to study the structural topology of proteins with respect to the lipid bilayer.35 In this study, CW-EPR power saturation experiments were performed on 31 SL pinholin S2168 samples incorporated into DMPC proteoliposomes. Representative CW-EPR power saturation curves are shown in Figure 7.
Figure 7.
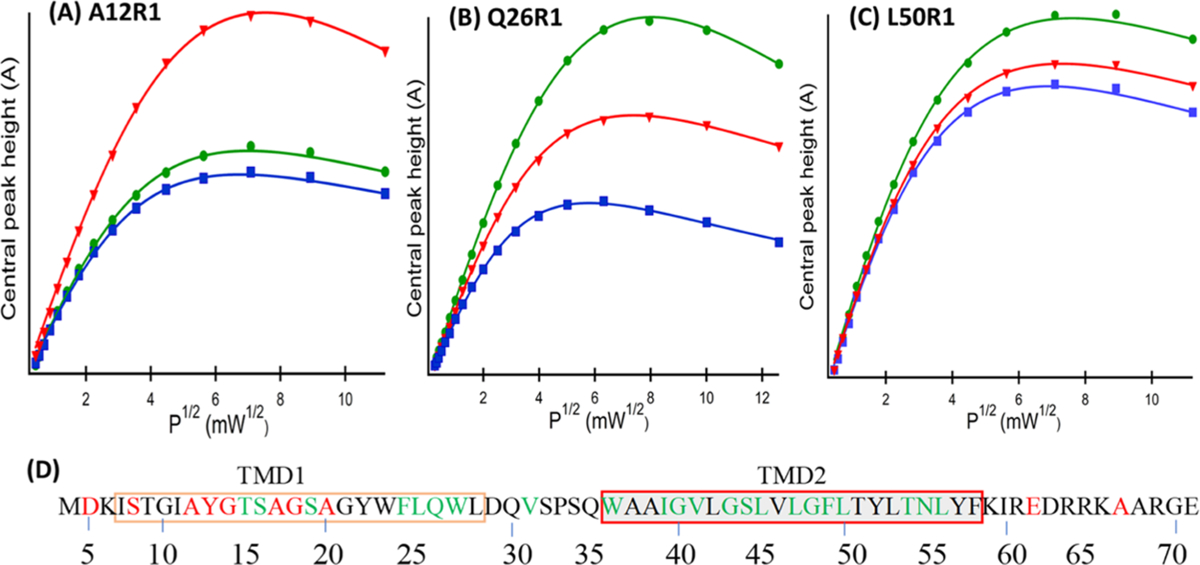
Representative CW-EPR power saturation curves of S2168 in DMPC proteoliposomes. (A) A12R1 and (B) Q26R1 are in TMD1 and (C) L50R1 is in TMD2. Red triangle represents NiEDDA, green circle represents oxygen, and blue square represents nitrogen spectra with their fitted line from eq 3. The amplitudes of first derivative mI = 0 peak were plotted against the square root of the incident microwave power. (D) Color-coded primary sequence of S2168, where green residues are buried in the lipid bilayer and red residues are solvent-exposed based on CW-EPR power saturation data. Black residues are not studied by the CW-EPR power saturation experiment.
The relative power saturation profile of the oxygen and NiEDDA spectra indicates that A12R1 was more accessible to the polar relaxing reagent NiEDDA, whereas Q26R1 and L50R1 were more accessible to nonpolar oxygen. This trend implies that residue A12R1 falls outside of the lipid bilayer, whereas Q26R1 and L50R1 are buried inside the lipid bilayer. For the quantitative analysis of membrane insertion, the depth parameter (Φ) was calculated for individual residues using eq 4. A positive Φ value indicates that the corresponding side chain is buried inside the lipid bilayer, whereas the negative Φ value indicates that the side chain is solvent-exposed or outside the lipid bilayer. Figure 7D shows residues buried in the lipid bilayer (green residues) or solvent-exposed (red residues) as suggested by Φ value measurements. The individual depth parameter (Φ) will be examined in detail in the Discussion section.
DISCUSSION
The qualitative and quantitative data reported in this study will provide a better understanding of side-chain dynamics and structural topology of pinholin S2168 in DMPC proteoliposomes. The prominent appearance of two motional components for W23R1, Q26R1, N55R1, L56R1 (indicated by an * in Figure 3) can be attributed to their positions in TMD1 or TMD2, and the bulky side chains on the same side of the helix.53,57 The R1 side chain shows different spectral components when its motion is restricted by a heterogenous interaction with the local environment.57 This heterogeneous interaction may arise from their position near the end of the helix or the hole opening.53 Interestingly, a similar trend was found for the bacterial K+ channel where residues at the beginning and the end of the helix (opening of the channel) show rigid and fast components.53 Another possible explanation for two components could be the bulky side chains on the same side of the helix (3 or 4 residues away from specific R1) which give characteristic anisotropic motion.57 W23 and Q26 are present in the vicinity of Y22, W23, Q26, and Q30, which can also contribute to the multicomponents observed in the W23R1 and Q26R1 spectra. Similar reasoning is applicable for the N55R1 and L56R1 EPR spectra, which are located at the interface of nonpolar lipid and polar solvent environments, and bulky side chains were present on the same side of the helix (Y52 and 59K).
In the active form of pinholin S2168, TMD1 was predicted to be externalized from the lipid bilayer, whereas TMD2 remains incorporated into the lipid bilayer.1,2,12,18 It was expected that the mobility of TMD1’s residues should be higher than the mobility of TMD2’s residues due to the protein segmental motion and lipid-free environment. The EPR data collected in this work indicated that only the N-terminal of TMD1 (S8 to Y13) had higher mobility when compared to TMD2, but residues G14 to W27 of TMD1 had no significant difference in mobility when compared to TMD2 residues. The average scaled mobility of TMD1 (0.34) was significantly higher than TMD2 (0.17). Again, the values allow us to compare the protein of interest with other protein systems studied independently.20,31,44 The value for TMD2 of pinholin S2168 is comparable with other channel-forming proteins, such as bacteriorhodopsin and annexin XII, which had tight helical packing with the values of 0.12 and 0.19, respectively, for the channel forming region.60 The value of TMD2 is harmonic with a predicted small pore size of S2168. This value implies tight, but somewhat flexible helical packing to allow interconversion of homotypic TMD2–TMD2 interaction to heterotypic TMD2–TMD2 interaction as reported by Young’s Lab.1,20
To resolve the relative orientation and interaction of TMD1 and TMD2 with respect to the lipid bilayer, we calculated the depth parameter (Φ) values and plotted them as a function of residue positions in Figure 8.
Figure 8.
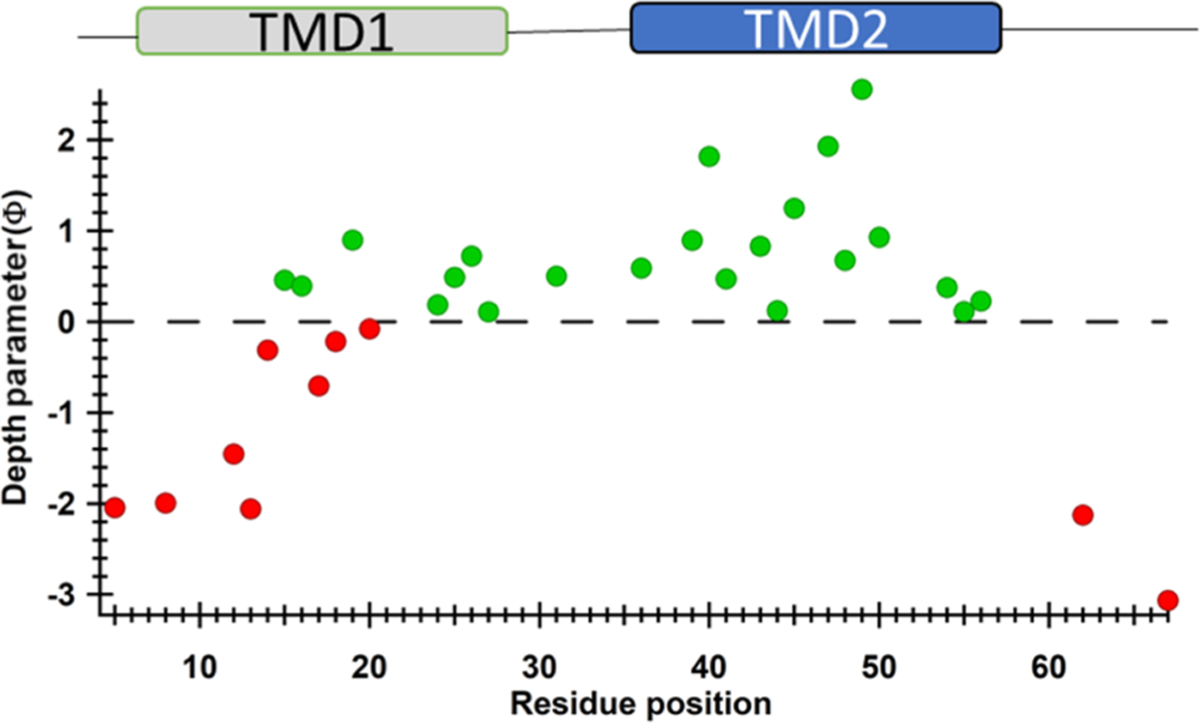
Membrane depth parameter (Φ) as a function of S2168 residue positions in DMPC proteoliposomes. Positive Φ values (green) indicate that the R1 side chains are embedded inside the lipid bilayer and negative Φ values (red) indicate that the R1 side chains are solvent-exposed.
The higher negative Φ values suggested that the N- and C-termini residues (e.g., D5R1, S8R1, E62R1, and A67R1) were highly solvent accessible and not buried in the lipid bilayer. It was predicted that all residues from TMD1 would show negative Φ values based on the assumption that TMD1 is externalized from the lipid bilayer as proposed by Park et al.2 Negative Φ values for residues 5–14 indicate that the N-terminal domain of TMD1 was solvent-exposed and externalized from the lipid bilayer. However, residues 24–27 of TMD1 showed positive Φ values, which implies that these residues were embedded in the lipid bilayer. Residues from the middle portion of TMD1 were partially solvent-exposed (G14R1, A17R1, G18R1, and A20R1) or buried in the lipid bilayer (T15R1, S16R1, and S19R1). It is worth mentioning here that residues T15, S16, and S19 are on the opposite side of the helix than G14, A17, G18, and A20.1 This indicates that the middle portion of TMD1 might be remained on the surface of the lipid bilayer, whereas some side chains were pointing out of the lipid bilayer and others were buried in the phospholipid membrane.
All residues of TMD2 (36–56) of pinholin had positive Φ values, which confirmed the membrane immersion of this segment. The first residue of TMD2 (W36) had a Φ value of 0.60, which gradually increased and reached to a maximum of 2.56 (F49) and then gradually decreased to the end of TMD2 (L56, Φ = 0.23). This is the characteristic pattern for a membrane-spanning segment of a protein or a membrane-embedded channel-forming segment of a protein.53 Although V41, S44, G48, and N55 are located inside of the lipid bilayer, they have shown relatively lower Φ values than their neighboring residues. This can be explained by the fact that all these residues have been identified as hole-lining residues or on the same side of the hole as proposed by the Young Lab.1
Based on the Φ values and mobility data, it can be inferred that instead of complete externalization of TMD1, this helical domain is partially externalized from the lipid bilayer where certain residues of TMD1 were exposed to solvent and others were buried in the lipid bilayer. This partial externalization can be explained by the presence of Trp (W27) at the C-terminal of TMD1. The smaller Φ value for W27R1 implies that it was in the lipid–water interface. The anchoring effect of the Trp residue has been widely recognized when it is located at the polar–nonpolar interface of a membrane.61,62
The partial externalization and orientation of TMD1 can be further explained by the amphipathic nature and presence of a glycine zipper in TMD1. Thermodynamically it may be more favorable for an amphipathic helix such as TMD1, initially buried in the membrane, to stay in a polar and nonpolar interface instead of in a fully polar environment. Again, partial externalization of TMD1 may be more favorable with less steric hindrance at the hole opening. In the active dimer of S2168, the hydrophilic faces of two TMD2s interact with each other by a homotypic TMD2–TMD2 interaction, which is facilitated by a homotypic TMD1–TMD1 interaction.2 However, the homotypic TMD2–TMD2 interaction is converted to heterotypic interaction in their heptameric form.1 This heterotypic interaction is a common phenomenon for channel proteins having a glycine zipper.1,63 A similar glycine zipper (G10xxxG14xxxG18) present in TMD1 may facilitate heterotypic interaction with other TMD1 via oligomerization. Heterotypic TMD1–TMD1 interactions will be more favorable if TMD1 is lying on the surface and interacting with neighboring TMD1(s). Park et al. proposed the complete externalization of TMD1 based on their disulfide-linked dimer and protease sensitivity for the S16C mutant which can be still possible at partial externalization conditions, where the S16 residue will be outside or on the surface of the lipid bilayer.2
Based on the EPR spectroscopic data presented in this study and considering the literature model for S2168, we are proposing a tentative structural topology model of pinholin S2168 as shown in Figure 9.
Figure 9.
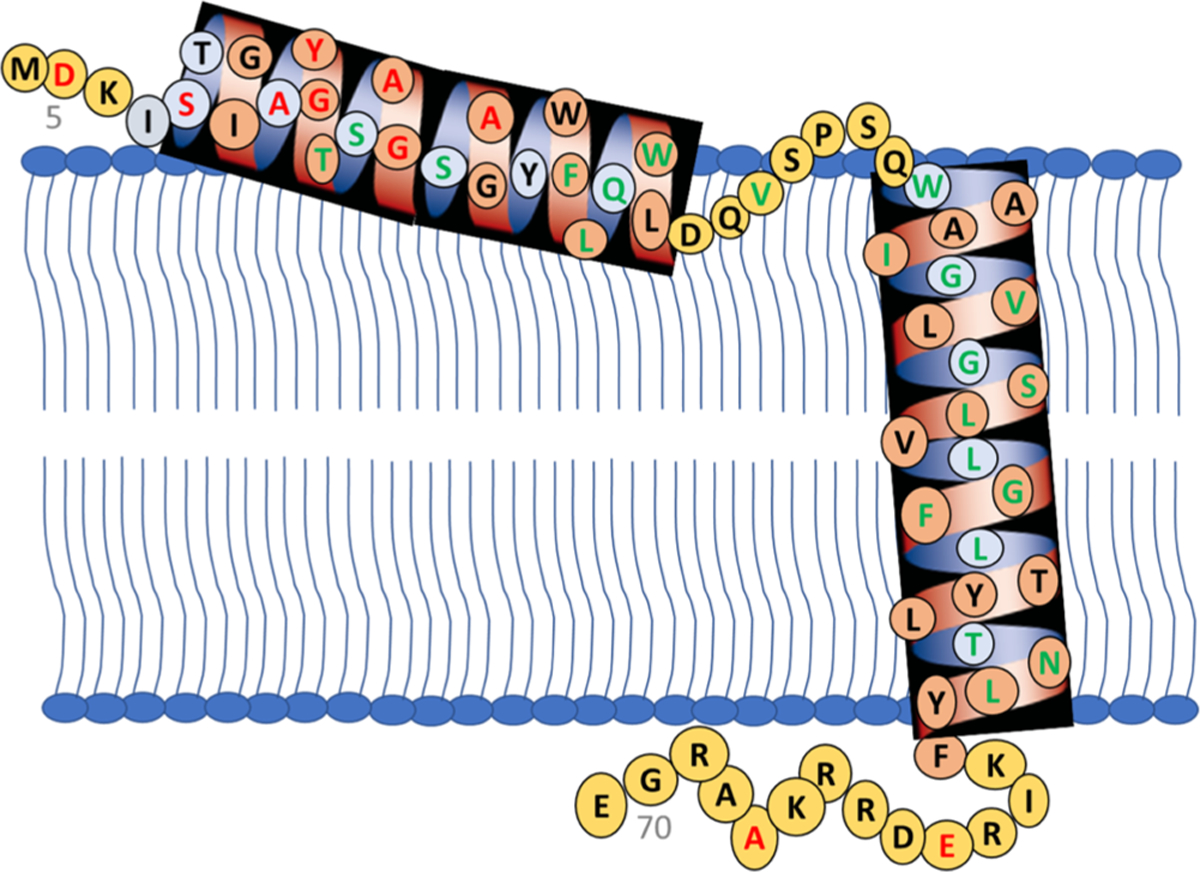
Proposed topology of active pinholin S2168 after partial externalization of TMD1 from the lipid bilayer. The red amino acids represent solvent-exposed and the green amino acids represent the lipid buried residues based on CW-EPR power saturation data. Black letters are for those residues which were not studied via EPR power saturation.
In the active conformation of S2168, the N-terminal remains in the periplasm, and TMD1 lays on the surface of the lipid bilayer with some residues pointing out of the lipid bilayer and other residues buried in the lipid environment. TMD2 remains incorporated in the lipid bilayer where one side of the helix is facing toward the pore and another side toward the lipid environment.1 The C-terminal of S2168 remains in the cytoplasm. Future biophysical experiments are needed to confirm the proposed topology model. The double electron–electron resonance EPR spectroscopic technique can be used to measure the distance between different segments of the S2168 to fine-tune the proposed topology.
CONCLUSIONS
This study reported on the structural topology and dynamic properties of the phage Φ21 lytic protein, pinholin S2168, in a lipid bilayer using EPR spectroscopic techniques. The CD data confirmed that pinholin S2168 maintains a predicted native α-helical secondary structure in DMPC proteoliposomes. R1 SDSL scanning of pinholin S2168 suggested that the N- and C-termini have higher mobility when compared to the two TMDs. The result for EPR power saturation indicates the partial externalization of TMD1 from the lipid bilayer, whereas TMD2 remains in the lipid bilayer. The structural topology model of S2168 presented in this study will be useful for future structural studies of pinholin as well as other holin systems using biophysical techniques.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Robert McCarrick for his continuous support for EPR instruments and data analysis. We are also grateful to the members of the Ry Young group at Texas A&M University for their experimental suggestions. This work was generously supported by the NIGMS/NIH Maximizing Investigator’s Research Award (MIRA) R35 GM126935, the NSF CHE-1807131 grant, the NSF (MRI-1725502) grant, the Ohio Board of Regents, and Miami University. G.A.L. would also like to acknowledge support from the John W. Steube Professorship.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.9b06480.
Representative DLS data (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Pang T; Savva CG; Fleming KG; Struck DK; Young R Structure of the lethal phage pinhole. Proc. Natl. Acad. Sci. U.S.A 2009, 106, 18966–18971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Park T; Struck DK; Deaton JF; Young R Topological dynamics of holins in programmed bacterial lysis. Proc. Natl. Acad. Sci. U.S.A 2006, 103, 19713–19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Abedon ST Phage Lysis In The Bacteriophages, 2nd ed.; Calendar R, Calendar RL, Abedon ST, Eds.; Oxford Univ Press: Oxford, 2006; pp 104–126. [Google Scholar]
- (4).Young R Phage lysis: three steps, three choices, one outcome. J. Microbiol 2014, 52, 243–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Berry J; Rajaure M; Pang T; Young R The spanin complex is essential for lambda lysis. J. Bacteriol 2012, 194, 5667–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Berry J; Summer EJ; Struck DK; Young R The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes. Mol. Microbiol 2008, 70, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Young R Phage lysis: do we have the hole story yet? Curr. Opin. Microbiol 2013, 16, 790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Young R Bacteriophage lysis - mechanism and regulation. Microbiol. Rev 1992, 56, 430–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Young R Bacteriophage holins: Deadly diversity. J. Mol. Microbiol. Biotechnol 2002, 4, 21–36. [PubMed] [Google Scholar]
- (10).Bläsi U; Young R Two beginnings for a single purpose: The dual-start holins in the regulation of phage lysis. Mol. Microbiol 1996, 21, 675–682. [DOI] [PubMed] [Google Scholar]
- (11).Wang I-N; Deaton J; Young R Sizing the holin lesion with an endolysin-beta-galactosidase fusion. J. Bacteriol 2003, 185, 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Pang T; Park T; Young R Mutational analysis of the S21 pinholin. Mol. Microbiol 2010, 76, 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Park T; Struck DK; Dankenbring CA; Young R The pinholin of lambdoid phage 21: Control of lysis by membrane depolarization. J. Bacteriol 2007, 189, 9135–9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Xu M; Struck DK; Deaton J; Wang I-N; Young R A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. U.S.A 2004, 101, 6415–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Xu M; Arulandu A; Struck DK; Swanson S; Sacchettini JC; Young R Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 2005, 307, 113–117. [DOI] [PubMed] [Google Scholar]
- (16).Kuppusamykrishnan H; Chau LM; Moreno-Hagelsieb G; Saier MH Jr. Analysis of 58 families of holins using a novel program, PhyST. J. Mol. Microbiol. Biotechnol 2016, 26, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Dewey JS; Savva CG; White RL; Vitha S; Holzenburg A; Young R Micron-scale holes terminate the phage infection cycle. Proc. Natl. Acad. Sci. U.S.A 2010, 107, 2219–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Pang T; Park T; Young R Mapping the pinhole formation pathway of S21. Mol. Microbiol 2010, 78, 710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).White R; Tran TAT; Dankenbring CA; Deaton J; Young R The N-terminal transmembrane domain of lambda S is required for holin but not antiholin function. J. Bacteriol 2010, 192, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Columbus L; Hubbell WL A new spin on protein dynamics. Trends Biochem. Sci 2002, 27, 288–295. [DOI] [PubMed] [Google Scholar]
- (21).Ishima R; Freedberg DI; Wang Y-X; Louis JM; Torchia DA Flap opening and dimer-interface flexibility in the free and inhibitor-bound HIV protease, and their implications for function. Structure 1999, 7, 1047–1055. [DOI] [PubMed] [Google Scholar]
- (22).Volkman BF; Lipson D; Wemmer DE; Kern D Two-state allosteric behavior in a single-domain signaling protein. Science 2001, 291, 2429–2433. [DOI] [PubMed] [Google Scholar]
- (23).Mulder FAA; Mittermaier A; Hon B; Dahlquist FW; Kay LE Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol 2001, 8, 932–935. [DOI] [PubMed] [Google Scholar]
- (24).Rozovsky S; Jogl G; Tong L; McDermott AE Solution-state NMR investigations of triosephosphate isomerase active site loop motion: Ligand release in relation to active site loop dynamics. J. Mol. Biol 2001, 310, 271–280. [DOI] [PubMed] [Google Scholar]
- (25).Sahu ID; McCarrick RM; Lorigan GA Use of electron paramagnetic resonance to solve biochemical problems. Biochemistry 2013, 52, 5967–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Altenbach C; Froncisz W; Hemker R; McHaourab H; Hubbell WL Accessibility of nitroxide side chains: Absolute Heisenberg exchange rates from power saturation EPR. Biophys. J 2005, 89, 2103–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pyka J; Ilnicki J; Altenbach C; Hubbell WL; Froncisz W Accessibility and dynamics of nitroxide side chains in T4 lysozyme measured by saturation recovery EPR. Biophys. J 2005, 89, 2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hubbell WL; Altenbach C Investigation of structure and dynamics in membrane-proteins using site-directed spin-labeling. Curr. Opin. Struct. Biol 1994, 4, 566–573. [Google Scholar]
- (29).Hubbell WL; McHaourab HS; Altenbach C; Lietzow MA Watching proteins move using site-directed spin labeling. Structure 1996, 4, 779–783. [DOI] [PubMed] [Google Scholar]
- (30).Hubbell WL; Gross A; Langen R; Lietzow MA Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol 1998, 8, 649–656. [DOI] [PubMed] [Google Scholar]
- (31).Hubbell WL; Cafiso DS; Altenbach C Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol 2000, 7, 735–739. [DOI] [PubMed] [Google Scholar]
- (32).Bates IR; Boggs JM; Feix JB; Harauz G Membrane-anchoring and charge effects in the interaction of myelin basic protein with lipid bilayers studied by site-directed spin labeling. J. Biol. Chem 2003, 278, 29041–29047. [DOI] [PubMed] [Google Scholar]
- (33).Klug CS; Feix JB Methods and applications of site-directed spin Labeling EPR Spectroscopy Biophysical Tools for Biologists: Volume 1 in Vitro Techniques; Elsevier, 2008; Vol. 84, pp 617–658. [DOI] [PubMed] [Google Scholar]
- (34).Altenbach C; Marti T; Khorana H; Hubbell W Transmembrane protein-structure - spin labeling of bacteriorhodopsin mutants. Science 1990, 248, 1088–1092. [DOI] [PubMed] [Google Scholar]
- (35).Altenbach C; Greenhalgh DA; Khorana HG; Hubbell WL A collision gradient-method to determine the immersion depth of nitroxides in lipid bilayers - application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A 1994, 91, 1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chandrudu S; Simerska P; Toth I Chemical methods for peptide and protein production. Molecules 2013, 18, 4373–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sahu ID; Lorigan GA Site-directed spin labeling EPR for studying membrane proteins. BioMed Res. Int 2018, 2018, 3248289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Drew DL Jr.; Ahammad T; Serafin RA; Butcher BJ; Clowes KR; Drake Z; Sahu ID; McCarrick RM; Lorigan GA Solid phase synthesis and spectroscopic characterization of the active and inactive forms of bacteriophage S21 pinholin protein. Anal. Biochem 2019, 567, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Lloyd-Williams P; Albericio F; Giralt E Chemical Approaches to the Synthesis of Peptides and Proteins; CRC Press, 1997. [Google Scholar]
- (40).Bottorf L; Sahu ID; McCarrick RM; Lorigan GA Utilization of C-13-labeled amino acids to probe the alpha-helical local secondary structure of a membrane peptide using electron spin echo envelope modulation (ESEEM) spectroscopy. Biochim. Biophys. Acta Biomembr 2018, 1860, 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Mayo DJ; Sahu ID; Lorigan GA Assessing topology and surface orientation of an antimicrobial peptide magainin 2 using mechanically aligned bilayers and electron paramagnetic resonance spectroscopy. Chem. Phys. Lipids 2018, 213, 124–130. [DOI] [PubMed] [Google Scholar]
- (42).Marsh D Thermodynamics of Phospholipid Self-Assembly. Biophys. J 2012, 102, 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Needham D; McIntosh TJ; Evans E Thermomechanical and transition properties of dimyristoylphosphatidylcholine cholesterol bilayers. Biochemistry 1988, 27, 4668–4673. [DOI] [PubMed] [Google Scholar]
- (44).White GF; Schermann SM; Bradley J; Roberts A; Greene NP; Berks BC; Thomson AJ Subunit organization in the TatA complex of the twin arginine protein translocase a site-directed EPR spin labeling study. J. Biol. Chem 2010, 285, 2294–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Sahu ID; Craig AF; Dunagan MM; Troxel KR; Zhang R; Meiberg AG; Harmon CN; McCarrick RM; Kroncke BM; Sanders CR; Lorigan GA Probing structural dynamics and topology of the KCNE1 membrane protein in lipid bilayers via site-directed spin labeling and electron paramagnetic resonance spectroscopy. Biochemistry 2015, 54, 6402–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Boggs JM; Moscarello MA Effect of basic-protein from human central nervous-system myelin on lipid bilayer structure. J. Membr. Biol 1978, 39, 75–96. [DOI] [PubMed] [Google Scholar]
- (47).Eletr S; Keith AD Spin-label studies of dynamics of lipid alkyl chains in biological-membranes - role of unsaturated sites. Proc. Natl. Acad. Sci. U.S.A 1972, 69, 1353–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Yu L; Wang W; Ling SL; Liu SL; Xiao L; Xin YL; Lai CH; Xiong Y; Zhang LH; Tian CL CW-EPR studies revealed different motional properties and oligomeric states of the integrin beta(1a) transmembrane domain in detergent micelles or liposomes. Sci. Rep 2015, 5, 7848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Popp CA; Hyde JS Effects of oxygen on electron-paramagnetic-res of nitroxide spin-label probes of model membranes. J. Magn. Reson 1981, 43, 249–258. [Google Scholar]
- (50).Klug CS; Su W; Feix JB Mapping of the residues involved in a proposed beta-strand located in the ferric enterobactin receptor FepA using site-directed spin-labeling. Biochemistry 1997, 36, 13027–13033. [DOI] [PubMed] [Google Scholar]
- (51).Oh KJ; Altenbach C; Collier RJ; Hubbell WL Site-directed spin labeling of proteins - applications to diphtheria toxin Bacterial Toxins: Methods and Protocols; Springer, 2000; Vol. 145, pp 147–169. [DOI] [PubMed] [Google Scholar]
- (52).Altenbach C; Flitsch SL; Khorana HG; Hubbell WL Structural studies on transmembrane proteins .2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry 1989, 28, 7806–7812. [DOI] [PubMed] [Google Scholar]
- (53).Perozo E; Cortes DM; Cuello LG Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat. Struct. Biol 1998, 5, 459–469. [DOI] [PubMed] [Google Scholar]
- (54).McHaourab HS; Perozo E Determination of protein folds and conformational dynamics using spin-labeling EPR spectroscopy Distance Measurements in Biological Systems by EPR; Springer, 2000; Vol. 19, pp 185–247. [Google Scholar]
- (55).Jeschke G; Bender A; Schweikardt T; Panek G; Decker H; Paulsen H Localization of the N-terminal domain in light-harvesting chlorophyll a/b protein by EPR measurements. J. Biol. Chem 2005, 280, 18623–18630. [DOI] [PubMed] [Google Scholar]
- (56).Vásquez V; Sotomayor M; Marien Cortes D; Roux B; Schulten K; Perozo E Three-dimensional architecture of membrane-embedded MscS in the closed conformation. J. Mol. Biol 2008, 378, 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).McHaourab HS; Lietzow MA; Hideg K; Hubbell WL Motion of spin-labeled side chains in T4 lysozyme, correlation with protein structure and dynamics. Biochemistry 1996, 35, 7692–7704. [DOI] [PubMed] [Google Scholar]
- (58).Isas JM; Langen R; Haigler HT; Hubbell WL Structure and dynamics of a helical hairpin and loop region in annexin 12: A site-directed spin labeling study. Biochemistry 2002, 41, 1464–1473. [DOI] [PubMed] [Google Scholar]
- (59).Bordignon E; Steinhoff H-J Membrane protein structure and dynamics studied by site-directed spin-labeling ESR Esr Spectroscopy in Membrane Biophysics; Springer, 2007; Vol. 27, pp 129–164. [Google Scholar]
- (60).Nicholls A; Sharp KA; Honig B Protein folding and association - insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Struct., Funct., Genet 1991, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- (61).Bortolus M; Dalzini A; Formaggio F; Toniolo C; Gobbo M; Maniero AL An EPR study of ampullosporin A, a medium-length peptaibiotic, in bicelles and vesicles. Phys. Chem. Chem. Phys 2016, 18, 749–760. [DOI] [PubMed] [Google Scholar]
- (62).de Planque MRR; Bonev BB; Demmers JAA; Greathouse DV; Koeppe RE; Separovic F; Watts A; Killian JA Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry 2003, 42, 5341–5348. [DOI] [PubMed] [Google Scholar]
- (63).Kim S; Jeon T-J; Oberai A; Yang D; Schmidt JJ; Bowie JU Transmembrane glycine zippers: Physiological and pathological roles in membrane proteins. Proc. Natl. Acad. Sci. U.S.A 2005, 102, 14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


