Abstract
Purpose
We aimed to estimate the carrier frequency of Zellweger spectrum disorder (ZSD), a rare autosomal-recessive disease, and the associated disease incidence based on data from the Exome Aggregation Consortium (ExAC) of approximately 60,000 individuals.
Methods
We obtained variants from ExAC in 13 PEX genes associated with ZSD. Potentially pathogenic missense variants were identified with computational variant analysis tools according to three stringency levels. Using variants classified as potentially pathogenic, we estimated the carrier frequency and the associated incidence for the entire ExAC population and its sub-populations. We also evaluated variants based on pathogenicity criteria for sequence variant interpretation outlined by the American College of Medical Genetics and Genomics (ACMG) and calculated the carrier frequency and incidence based on those variants.
Results
The bioinformatically estimated incidence rate of ZSD in the ExAC population is 1 in 83,841 using our least stringent pathogenicity cutoff. Under clinical guidelines outlined by ACMG, the estimated incidence is 1 in 3,275,751 births.
Conclusion
We outlined a process for estimating the ZSD disease carrier frequency and incidence in a large consortium using bioinformatics tools. Our results are close to current newborn screening estimates in New York of 1 in 90,000 births, estimated from 1.08 million screenings.
Keywords: Zellweger spectrum disorder, peroxisome biogenesis, carrier frequency, incidence rate, X-linked adrenoleukodystrophy
INTRODUCTION
Peroxisome biogenesis disorders (PBDs) represent a spectrum of conditions associated with faulty peroxisome assembly and function in this organelle.1 There are two subtypes of PBDs which are distinguished by measurements of plasma very long chain fatty acid (VLCFA) levels and erythrocyte membrane plasmalogens. The first, rhizomelic chondroplasia punctate (RCDP), results from variants in the PEX7 gene. The second, Zellweger spectrum disorder (ZSD), is autosomal recessive and results from variants in 13 genes: PEX1, PEX2, PEX3, PEX5, PEX6, PEX10, PEX11β, PEX12, PEX13, PEX14, PEX16, PEX19, and PEX26.1,2 ZSD has different severities that used to be described as separate disorders: Zellweger syndrome (ZS, severe), neonatal adrenoleukodystrophy (NALD, intermediate), and infantile Refsum disease (IRD, mild).1 Symptoms are present at birth in severe cases or can manifest later in childhood in less severe cases. Since peroxisomes are tied to many processes in the body, ZSD has a wide range of symptoms. They include craniofacial abnormalities, hypotonia, seizures, blindness, deafness, enlarged liver, renal cysts, and myelin degradation.1–3
A related condition, X-linked adrenoleukodystrophy (X-ALD), was recently added to the recommended uniform newborn screening panel.4 Because of their similar biochemical features, screening for X-ALD might also identify some ZSD cases through elevated VLCFAs, the markers used for X-ALD screening. Increased detection of ZSD will probably lead to demand for more comprehensive information about the condition, including carrier rate for recurrence risk assessment. A better understanding of the carrier frequency will be helpful to extended family members’ genetic counseling following positive newborn screening results.
Approximately 1 in 50,000 births is thought to be affected by PBDs.1,2,5–7 This estimate is based on a combination of observations, and often it is unclear which level of cases (ZS, ZSD, or PBD) is included in the incidence. In 1975, Danks et al. reported on eight cases of ZS in the state of Victoria, Australia, that occurred over the course of 13 years.8 They used these cases and the number of births in Victoria (882,765) over the same period to estimate the incidence of ZS at 1 in 100,000. In a 1987 review of ZS, Hans Zellweger stated that he believed the incidence to be higher than that reported by Danks et al. because in the Netherlands the disease “is more likely to affect one in 25,000 to 50,000 newborns,”7 but no data were reported to support this claim. Lazarow and Moser noticed that the Kennedy Krieger Institute accounted for 0.79 in 100,000 (1 in 126,582) incident ZS cases in the US between 1985 and 1995.9 In certain sub-populations, the incidence is much higher, such as the French-Canadian population in the Saguenay-Lac-St-Jean region, where the estimated frequency of ZS is 1 in 12,191.10 Subsequent literature regarding ZSD has cited a middle figure of 1 in 50,000.1,5,6 The range of estimates reported in the literature indicates a need for a systematic approach to obtain a more accurate carrier frequency of ZSD in a large population that relies on a genetic definition of ZSD.
To date, there are few reports on genetic disease carrier frequency assessment using a combination of large population-based data and variant analysis tools to assess pathogenicity.11,12 In this study, we aimed to demonstrate the process of estimating the carrier frequency and associated incidence rate of ZSD using the Exome Aggregation Consortium (ExAC) database and several bioinformatics tools that assess pathogenicity of genetic variants. ExAC is a compilation of high-quality exome data from approximately 60,000 individuals, which have been filtered to contain unrelated adults without a history of severe childhood disease.13 Bioinformatics tools used include Sorting Intolerant from Tolerant (SIFT), Polymorphism Phenotyping v2 (PolyPhen-2), Combined Annotation-Dependent Depletion (CADD), and Deleterious Annotation of genetic variants using Neural Networks (DANN), all of which evaluate missense variants based on evolutionary conservation, protein structure, or a combination of both.14–17 Also used was the conservation-based PHylogenetic Analysis with Space/Time models tool (PhastCons), which evaluates nucleotides based on conservation.18 Of the tools listed here, SIFT and PolyPhen-2 are among the most used for clinical assessment of missense variants.19 We also assessed these variants according to the American College of Medical Genetics and Genomics (ACMG) criteria to see how they would be evaluated in the current clinical practice setting.19 To our knowledge, this study is the first to report estimates of ZSD carrier frequency and incidence rates based on a large consortium database and a genetic definition of the disease.
MATERIALS AND METHODS
Databases and Genetic Variants
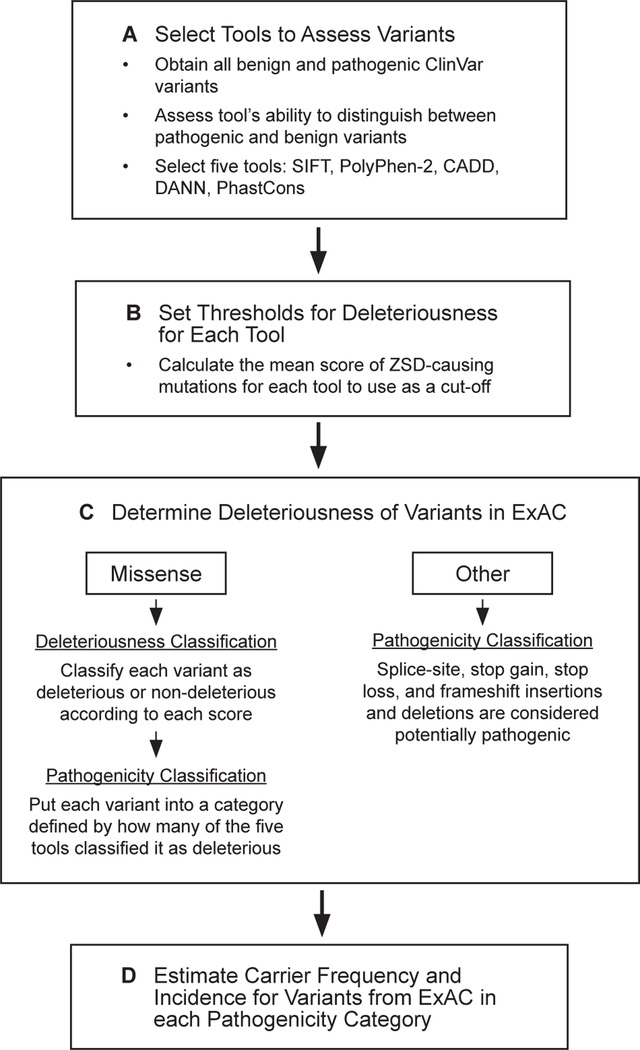
As described below, our study used three lists of variants from different databases in the process of (1) selecting the variant analysis tools that we would use to assess variant deleteriousness, (2) establishing a threshold for each of those tools, and (3) evaluating the carrier frequency of ZSD (Figure 1).
Figure 1: Analysis outline.
Workflow to estimate ZSD carrier frequency and incidence.
To evaluate which variant analysis tools are most informative in assessing variant deleteriousness, we used genome-wide missense variants (ie, not limited to variants in the thirteen PEX genes) reported as either pathogenic (15,406 variants) or benign (3,932 variants) in ClinVar.20 This is a repository of human variants with a reported clinical significance such as benign or pathogenic.
To establish a threshold for deleteriousness for variant analysis tools, we used known ZSD-causing missense variants from OMIM (Online Mendelian Inheritance of Man)21 and dbPEX (PEX Gene Database).22 OMIM provides information on publications that report pathogenic variants, and dbPEX is a PBD-specific database of variants. Thresholds can vary depending on the disease, and setting a disease-specific deleteriousness threshold was the method of choice in a previous study.11,16 The following inclusion criteria were applied to ZSD variants: first, they were present in the ExAC population; second, they had a deleteriousness score for each variant analysis tool that we used (scores were not available for some variants); and third, they met the baseline deleteriousness thresholds of each variant analysis tool. From the over 200 variants in dbPEX and OMIM, 34 were present in the ExAC database. Of those, 15 were non-synonymous variants and therefore had scores for each variant analysis tool. Of these, 6 did not meet the standard deleteriousness cutoffs provided by the variant analysis tools. This resulted in 9 variants used to establish deleteriousness thresholds (Table 1).
Table 1:
Variants known to cause Zellweger spectrum disorder found in OMIM or dbPEX databases.
| rsID | Gene | Chromosome | RefSeq Accession Number | cDNA | Protein |
|---|---|---|---|---|---|
| rs61750420 | PEX1 | 7 | NM_001282677 | c.G2357A | p.G786D |
| NM_000466 | c.G2528A | p.G843D | |||
| NM_001282678 | c.G1904A | p.G635D | |||
| rs61750427 | PEX1 | 7 | NM_001282677 | c.T2795C | p.I932T |
| NM_000466 | c.T2966C | p.I989T | |||
| NM_001282678 | c.T2342C | p.I781T | |||
| rs61750425 | PEX1 | 7 | NM_001282677 | c.G2675A | p.R892Q |
| NM_000466 | c.G2846A | p.R949Q | |||
| NM_001282678 | c.G2222A | p.R741Q | |||
| rs61753231 | PEX6 | 6 | NM_000287 | c.G2579A | p.R860Q |
| rs61753226 | PEX6 | 6 | NM_000287 | c.T2534C | p.I845T |
| rs61753229 | PEX6 | 6 | NM_000287 | c.G2435A | p.R812Q |
| - | PEX10 | 1 | NM_002617 | c.G932A | p.R311Q |
| NM_153818 | c.G992A | p.R331Q | |||
| rs62641228 | PEX26 | 22 | NM_001127649 | c.C292T | p.R98W |
| NM_001199319 | c.C292T | p.R98W | |||
| NM_017929 | c.C292T | p.R98W | |||
| rs61752134 | PEX26 | 22 | NM_001127649 | c.C350T | p.P117L |
| NM_001199319 | c.C350T | p.P117L | |||
| NM_017929 | c.C350T | p.P117L | |||
To assess the carrier frequency of ZSD, we extracted allele frequencies from the ExAC database in the following 13 genes: PEX1, PEX2, PEX3, PEX5, PEX6, PEX10, PEX11β, PEX12, PEX13, PEX14, PEX16, PEX19, and PEX26. We then analyzed whether these variants were potentially pathogenic using the deleteriousness scores from the selected tools and our criteria for pathogenicity, as described below. Variants were excluded if at least one individual in the ExAC population was homozygous for it. There were six homozygous variants. In addition to the nine variants used to establish a deleteriousness threshold, a total of 2,104 variants from ExAC were assessed for the carrier frequency calculation.
Variant Analysis Tool Selection and Bioinformatics Variant Pathogenicity Assessment
ANNOVAR23 was used to annotate all variants, including ClinVar variants used for the variant analysis tool selection, known ZSD-causing variants from OMIM and dbPEX used to establish the threshold for deleteriousness, and the ExAC variants included in the carrier frequency calculation. ANNOVAR also extracted allele frequencies from the ExAC database. Annotations used the GRCh37 human reference sequence and RefSeq gene definitions. Variant deleteriousness and frequency in the ExAC population were analyzed with SAS (v. 9.4) and R (v. 3.2.1). We use the term deleteriousness when referring to one bioinformatics tool’s assessment of a variant and pathogenicity when referring to a composite score of all five bioinformatics tools.
To select the most informative variant analysis tools, we imported all ClinVar variants identified as pathogenic (15,406) and benign (3,932) into ANNOVAR, which provided scores for 16 different variant analysis tools. Then we conducted further evaluation as described below.
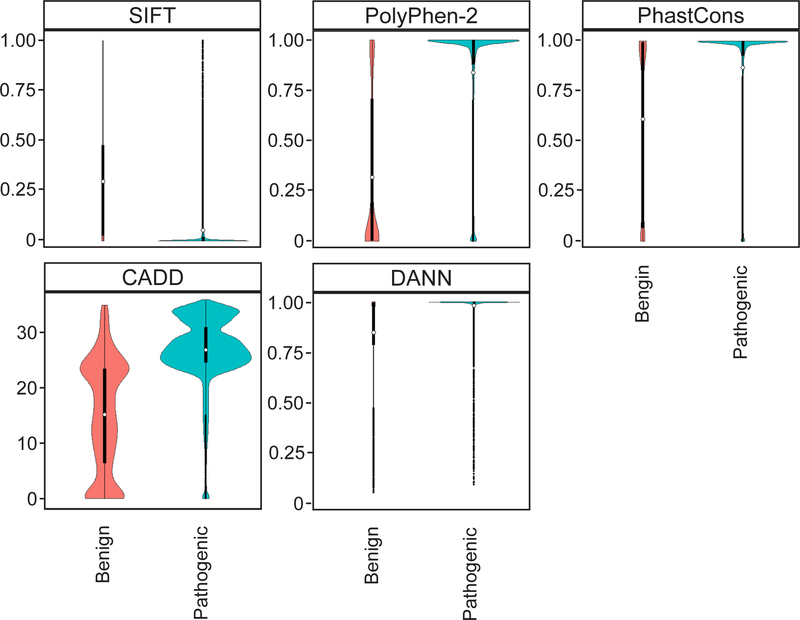
First, we qualitatively examined the spread, shape, and overlap of the distributions of pathogenic and benign ClinVar variant scores from each tool. We preferred tools that had narrow distributions of scores, especially for pathogenic variants, and tools where the overlap between the benign and pathogenic variant distributions was minimized. Second, we selected tools that represent various approaches in determining the deleteriousness of variants, as stated in the ACMG guidelines for variant assessment.19 These categories include evolutionary conservation of amino acids, protein structure and function, and nucleotide conservation. Third, we conducted a literature search to select tools that are widely-cited in peer-reviewed journals. We chose SIFT, PolyPhen-2, and PhastCons because they each represented at least one of the three mentioned approaches to determining deleteriousness, they were widely cited, and the comparison of benign and pathogenic variant distributions had the qualities we described above (Figure 2). We chose CADD and DANN because they represented a fourth approach in determining deleteriousness, which is based on a comparison of variants that survived natural selection and simulated variants. The distributions of pathogenic and benign variants for the two scores met our outlined criteria. CADD was widely referenced in the literature while DANN is a relatively new tool.
Figure 2: Violin plots comparing scores for 15,406 pathogenic and 3,932 benign variants from the ClinVar database for the five variant analysis tools used to assess deleteriousness.
In each plot, benign variants are on the left and pathogenic variants are on the right. The deleteriousness scores are along the y-axis. Higher values indicated a higher probability that the variant is damaging in all scores except for SIFT, where a low score is associated with deleteriousness. The y-axis for CADD is a logarithmically transformed score, and the rest are linear probabilities. The x-axis represents the probability density of variants along the range of scores. The CADD plot appears different because its y-axis is on a logarithmic instead of linear scale.
The SIFT algorithm predicts whether an amino acid substitution is deleterious using evolutionary conservation of amino acids.14 SIFT generates a probability for every amino acid in the protein based on how often that amino acid is observed in alignments with homologous sequences. The lower the probability of an amino acid substitution, the higher the likelihood is for that substitution to be deleterious. PolyPhen-2 uses a combination of 11 tools based on amino acid sequence and protein structure to predict if an amino acid substitution is deleterious.15 PolyPhen-2 (HumVar-trained model) generates a score that estimates the probability of a variant being damaging. High scores indicate variants that are more likely to be damaging. CADD compares derived alleles to simulated de novo variants and ranks each one relative to the rest based on how likely it is that the allele is derived or simulated.16 It is based on the principle that there are fewer derived than simulated deleterious variants because of natural selection. We used a Phred-like scaled version of this C-score, which is equivalent to −10log10 (rank/total number of substitutions). A variant with a scaled C-score between 20 and 29 means that the variant is in the 1st percentile of the “most deleterious substitutions that you can do to the human genome.”24 A score between 30 and 39 means that it is in the 0.1th percentile of the most deleterious substitutions. DANN is similar to CADD except that it uses a deep neural network instead of a linear kernel support vector machine to compare derived and simulated alleles.17 The higher the score, the more likely a variant is to be damaging. PhastCons (20-way mammalian score) is based on phylogenetic hidden Markov models and generates a conservation score for each variant using a cross-species alignment.18 A high score indicates that the variant has a higher probability of being in an evolutionarily conserved element and that changing it would be deleterious.
After selecting these five tools, we established a threshold of deleteriousness for each one using the nine missense PEX gene variants that were reported in OMIN and dbPEX as disease-causing. We calculated the mean scores of this set of variants for SIFT (mean = 0.007, standard deviation [SD] = 0.017), PolyPhen-2 (mean = 0.999, SD = 0.002), CADD (mean = 33.167, SD = 2.677), DANN (mean = 0.999, SD = 0.000), and PhastCons (mean = 0.988, SD = 0.014) and used that as the cutoff between deleterious and non-deleterious for each tool when evaluating variants in the ExAC database. If a missense variant in ExAC had a score equal to or above (below for SIFT) the mean, it was considered deleterious.
To evaluate which missense variants in the 13 PEX genes from the ExAC database were potentially pathogenic, we categorized them by the number of variant analysis tools that classified them as deleterious. An allele was classified as pathogenic under three levels of stringency: pathogenic if deemed deleterious by at least three out of the five tools (3/5), at least four out of the five tools (4/5), and all of the tools (5/5). Each tool carried the same weight in the composite scores.
Insertions and deletions (indels) causing frameshift, stop-loss, stop-gain, and splice site variants were all considered as potentially pathogenic. The splice site variants that ANNOVAR annotates as “splicing” are by default in the +1, +2, −1, and −2 positions (personal communication with Dr. Wang of ANNOVAR). These are well-conserved positions, and changes to these nucleotides are recognized to affect protein splicing. We included these variants in the “other variants” category. The frequencies of the known ZSD-causing missense variants are also included in this category.
Variant Assessment with ACMG Criteria
Variants were categorized with clinical interpretation software, Cartagenia (Allisa Interpret), to evaluate each variant using a series of databases, allele frequency information, and functional predictions. Pathogenicity was categorized according to the standards and guidelines set forth by ACMG.19 Evidence for and against pathogenicity were weighted as strong (previously described function, loss of function), moderate (loss of initiation, premature stop codon, disruption of stop codon, whole-gene deletion, frame shifting indel, and disruption of splicing), or supporting (nonsynonymous substitution, in-frame indel, support from multiple functional prediction algorithms). Each variant was interpreted based on the cumulative evidence supporting its categorization as pathogenic or likely pathogenic.
Carrier Frequency and Disease Incidence Estimation
To date, variants in 13 PEX genes have been linked to ZSD. We calculated the ExAC population carrier frequency for every PEX gene in each of the three pathogenicity categories described above. We summed the allele frequencies from the ExAC population within those categories along with the frequencies of the frameshift indels, stop loss, stop gain, splice site, and known ZSD-causing variants. We then estimated the incidence rates for each gene based on those frequencies and the Hardy-Weinberg Equilibrium principle: 1 = p2 + 2pq + q2. The p represents the frequency of the major (non-disease) allele, which we assume to be approximately 1. The q represents the minor allele frequency, q2 the frequency of affected individuals (including compound heterozygotes), and 2pq the carrier frequency. Then, we summed the estimated carrier frequencies and incidence rates across all genes. As an example, the carrier frequency (2pq) for PEX1 in the 3/5 pathogenicity category is 0.00635682. To solve for q, we divide that number by 2 (assuming p is 1) and get 0.00317841. We then square that to get a gene-level incidence (q2) of 1.01 × 10−5. We also estimated the carrier frequency and incidence for the African, non-Finnish European, Finnish, admixed American, South Asian, and East Asian genetic ancestry groups within the consortium. The same process for carrier frequency and incidence estimates was followed for the variants categorized according to ACMG criteria as pathogenic or likely pathogenic.
RESULTS
Bioinformatics Assessment of Carrier Frequency and Incidence
To estimate ZSD carrier frequency, we assessed 1,953 missense variants and 151 additional variants, which include frameshift indels, stop gain or loss variants, and splice-site variants. There were 9 known ZSD-causing variants available, for a total of 160 variants in the “other” category that were counted as pathogenic. There were 231, 82, and 24 missense variants in the 3/5, 4/5, and 5/5 pathogenicity categories. The 3/5 category is inclusive of the 4/5 and 5/5 categories, and the 4/5 category is inclusive of the 5/5 category. In the sub-population assessments, the same variants were assessed in each population except for one variant in the other category, which was not available for the Finnish population. Overall, the top three genes that had the most pathogenic variants across all composite scores were PEX1, PEX6 and PEX12, in descending order. According to dbPEX, the top three genes with the highest number of unique variants associated with ZSD are PEX6, PEX1, and PEX12, in descending order. However, the number of recorded cases with PEX1 variants is high relative to cases with variants in other genes.
The estimated incidence of ZSD in the entire ExAC population using the 3/5, 4/5, and 5/5 thresholds is 1 in 83,841, 1 in 121,749, and 1 in 139,557 births, respectively (Table 2). In the ExAC sub-populations, using the lowest stringency level of 3/5 (at least three out of five variant analysis tools classified variant as deleterious) the incidence ranges from 1 in 31,165 births in the East Asian population to 1 in 263,531 births in the admixed American population (Table 2). At the highest stringency level where all five variant analysis tools classified a variant as deleterious, the incidence ranged from 1 in 76,630 births in the Non-Finnish European population to 1 in 2,702,703 births in the Finnish population. No individuals in this latter sub-population had missense variants that were in the 5/5 category, which could be due to the low probability of observing these variants in the small population of only 3,307 people, the smallest of the ExAC populations. The incidence estimated in the entire ExAC population is mainly reflective of the Non-Finnish European sub-population because 33,370 people out of the total 60,706 are in this population (Table 2).
Table 2:
Carrier frequency and estimated incidence of Zellweger spectrum disorder (ZSD).
| Threshold (at least N deleterious scores out of 5) | Missense Variantsa (frequency) | Other Variantsb (frequency) | Carrier Frequency (1 in N People) | Incidencec (1 in N Births) |
|---|---|---|---|---|
| ExAC All (60,706 people) | ||||
| 3/5 | 0.007120 | 0.006623 | 1 in 73 | 1 in 83,841 |
| 4/5 | 0.003376 | 0.006623 | 1 in 100 | 1 in 121,749 |
| 5/5 | 0.001125 | 0.006623 | 1 in 129 | 1 in 139,557 |
| ExAC Non-Finnish European (33,370 people) | ||||
| 3/5 | 0.005248 | 0.008724 | 1 in 72 | 1 in 62,993 |
| 4/5 | 0.002572 | 0.008724 | 1 in 89 | 1 in 71,074 |
| 5/5 | 0.000681 | 0.008724 | 1 in 106 | 1 in 76,630 |
| ExAC South Asian (8,256 people) | ||||
| 3/5 | 0.010096 | 0.002739 | 1 in 78 | 1 in 135,223 |
| 4/5 | 0.006292 | 0.002739 | 1 in 111 | 1 in 181,312 |
| 5/5 | 0.004743 | 0.002739 | 1 in 134 | 1 in 211,340 |
| ExAC admixed American (5,789 people) | ||||
| 3/5 | 0.006420 | 0.003603 | 1 in 100 | 1 in 263,531 |
| 4/5 | 0.002293 | 0.003603 | 1 in 170 | 1 in 382,865 |
| 5/5 | 0.000960 | 0.003603 | 1 in 219 | 1 in 473,094 |
| ExAC African (5,203 people) | ||||
| 3/5 | 0.007412 | 0.006650 | 1 in 71 | 1 in 107,356 |
| 4/5 | 0.002479 | 0.006650 | 1 in 110 | 1 in 242,098 |
| 5/5 | 0.000296 | 0.006650 | 1 in 144 | 1 in 309,959 |
| ExAC East Asian (4,327 people) | ||||
| 3/5 | 0.012900 | 0.005000 | 1 in 56 | 1 in 31,165 |
| 4/5 | 0.002300 | 0.005000 | 1 in 137 | 1 in 244,349 |
| 5/5 | 0.000300 | 0.005000 | 1 in 189 | 1 in 298,285 |
| ExAC Finnish (3,307 people) | ||||
| 3/5 | 0.010800 | 0.001400 | 1 in 82 | 1 in 36,456 |
| 4/5 | 0.010400 | 0.001400 | 1 in 85 | 1 in 36,483 |
| 5/5 | 0.000000 | 0.001400 | 1 in 714 | 1 in 2,702,703 |
There are 231, 82, and 24 missense variants in the 3/5, 4/5, and 5/5 categories, respectively.
Other variants category includes the known ZSD-causing, stop loss, stop gain, frameshift insertions, frameshift deletions, and splice-site variants, all of which are considered deleterious. There are 161 other variants in each population except for the Finnish population where there are 160 variants.
Total incidence is calculated by summing gene-level incidence rates.
ACMG Assessment of Carrier Frequency and Incidence
The same variants assessed using the bioinformatics criteria discussed above were also assessed using ACMG criteria.19 Of the variants extracted from ExAC, 11 were classified as pathogenic, and 33 were classified as likely pathogenic for a total of 44 variants that factored into the carrier frequency estimate. There are four missense variants in the likely pathogenic category and none in the pathogenic category. The remaining 40 variants are frameshift indels, stop gain or loss variants, and splice-site variants.
The estimated incidence of ZSD in the entire ExAC population including variants classified as pathogenic and likely pathogenic according to ACMG criteria is 1 in 3,275,751 births (Table 3). The estimate decreases to 1 in 10,413,631 births if variants classified as pathogenic are the only ones included. For ExAC sub-populations, the total incidence ranged from 1 in 1,230,228 births in the non-Finnish European group to 1 in 94,886,541 births in the South Asian group (Table 3).
Table 3:
Carrier frequency and estimated incidence of Zellweger spectrum disorder estimated with variants that pass ACMG criteria to classify sequence variants.
| ACMG Pathogenicity Ratinga | Carrier Frequency (1 in N People) | Incidenceb (1 in N Births) |
|---|---|---|
| ExAC All (60,706 people) | ||
| Pathogenic and Likely Pathogenic | 1 in 531 | 1 in 3,275,751 |
| Pathogenic Only | 1 in 1,198 | 1 in 10,413,631 |
| Likely Pathogenic Only | 1 in 953 | 1 in 14,426,436 |
| ExAC Non-Finnish European (33,370 people) | ||
| Pathogenic and Likely Pathogenic | 1 in 347 | 1 in 1,230,228 |
| Pathogenic Only | 1 in 690 | 1 in 3,103,913 |
| Likely Pathogenic Only | 1 in 696 | 1 in 6,688,356 |
| ExAC South Asian (8,256 people) | ||
| Pathogenic and Likely Pathogenic | 1 in 2,258 | 1 in 94,886,541 |
| Pathogenic Only | 1 in 6,211 | 1 in 291,523,941 |
| Likely Pathogenic Only | 1 in 3,547 | 1 in 190,211,615 |
| ExAC admixed American (5,789 people) | ||
| Pathogenic and Likely Pathogenic | 1 in 1,145 | 1 in 15,407,026 |
| Pathogenic Only | 1 in 3,333 | 1 in 80,000,000 |
| Likely Pathogenic Only | 1 in 1,745 | 1 in 30,859,004 |
| ExAC African (5,203 people) | ||
| Pathogenic and Likely Pathogenic | 1 in 781 | 1 in 9,040,685 |
| Pathogenic Only | 1 in 2,533 | 1 in 67,826,224 |
| Likely Pathogenic Only | 1 in 1,129 | 1 in 13,072,318 |
| ExAC East Asian (4,327 people) | ||
| Pathogenic and Likely Pathogenic | 1 in 2,000 | 1 in 57,142,857 |
| Pathogenic Only | 1 in 5,000 | 1 in 200,000,000 |
| Likely Pathogenic Only | 1 in 3,333 | 1 in 133,333,333 |
| ExAC Finnish (3,307 people) | ||
| Pathogenic and Likely Pathogenic | 1 in 714 | 1 in 2,702,703 |
| Pathogenic Only | 1 in 5,000 | 1 in 100,000,000 |
| Likely Pathogenic Only | 1 in 833 | 1 in 2,777,778 |
There are 11 pathogenic and 33 likely pathogenic variants included in analysis.
Total incidence is calculated by summing gene-level incidence rates.
ACMG American College of Medical Genetics and Genomics.
DISCUSSION
Our study estimates the ZSD carrier frequency and incidence rates using a large consortium database. Recent advancements in bioinformatics tools for variant assessment, and efforts to create large databases of human genomic information, have generated possibilities to estimate carrier frequencies based on large population data. One challenge with these bioinformatics tools is to develop a procedure for their use that represents biological or pathogenic processes. We outline an approach for selecting informative variant analysis tools that uses the entire ClinVar repository of “benign” and “pathogenic” non-synonymous variants. These variants are then leveraged to select tools that discern between reported benign and pathogenic variants. Combining this with our other described criteria, we have more confidence in the reliability of the tools we use for variant evaluation than if we had selected tools based on convenience or familiarity. Instead of using default deleteriousness thresholds, we calibrated each tool with ZSD-causing variants. Then we evaluated missense variants based on a combination of the five tools, setting three thresholds to determine whether variants were pathogenic. In addition to other variants assumed to be pathogenic, we calculated the carrier frequency and estimated the associated incidence.
Our bioinformatically estimated incidence of ZSD in the whole ExAC population of 1 in 83,841 births is similar to recent estimates from newborn screening in New York of approximately 1 in 90,000 births (calculated from 12 ZSD cases in 1.08 million births, personal communication with Dr. Joseph Orsini, August 2018), and lower than the figure of 1 in 50,000 births that is often cited.1,2,6 Our analysis was limited to PEX variants present in the ExAC database, which did not include all variants, such as large indels that are known to cause ZSD. Therefore, the frequency of those could not be included in the estimate. In addition, if ANNOVAR did not provide SIFT, PolyPhen-2, CADD, DANN, or PhastCons scores for a missense variant in ExAC, it could not be analyzed. There were 102 missense variants that did not have these scores. The combination of these limitations means that some pathogenic variants may not have been included in the estimates, which means the incidence in the ExAC population could be higher than what we estimated. Another factor that could lead to an underestimate of the incidence is that we may have excluded hypomorphic alleles that result in the disease only when paired with a more deleterious allele. For example, we excluded at least one: PEX6-R601Q, a homozygous variant in ExAC. When this allele is paired with a null allele it results in ZSD.25 Conversely, we may have overestimated the carrier frequency because in the absence of clinical information about the variants, we may have falsely classified some variants as pathogenic.
Under the current clinical setting, using ACMG guidelines, the whole ExAC population incidence would be estimated at 1 in 3,275,751 births, which is much lower than the bioinformatics assessment or the observed estimate. The design of ACMG criteria aims to greatly limit the possibility of falsely assessing a non-pathogenic variant as pathogenic. The criteria focus on combining clinical, bioinformatics, variant type, and other lines of evidence about a variant to make a definitive judgement about its pathogenicity. If a variant’s pathogenicity has not been assessed in multiple ways, it cannot be classified as pathogenic even if it is potentially pathogenic. The low incidence estimate using ACMG criteria likely reflects this lack of information. ACMG criteria are necessary for clinical diagnosis but may not be suitable for estimating the disease incidence.
Our bioinformatically estimated incidence supports that ZSD is a rare disease. Besides the constraints associated with data availability, the incidence is dependent on the deleteriousness cutoffs for each score and the pathogenicity thresholds we set. We took a conservative approach to estimating the carrier frequency and set deleteriousness thresholds using the mean scores of known ZSD-causing variants. Our thresholds were more stringent than the default thresholds for each variant analysis tool. For example, the default threshold for SIFT is 0.05, and our threshold was more stringent at 0.007. SIFT has a 20% false positive rate at the 0.05 level.26 Another limitation is that variants we used to set the deleteriousness threshold did not represent all PEX genes. We also had the added stringency of a compound score for pathogenicity. The five tools we selected in our compound score provided results that were similar to what obtained from a large newborn screening cohort. Additional replicable studies in other disorders are needed to evaluate if the utility of the same five tools can be generalized. Currently, there are no agreed-upon guidelines for computationally evaluating variant pathogenicity.
Despite the described limitations, one interesting finding from the ExAC sub-populations suggests that the incidence of ZSD varies by population composition. This range in ZSD incidence highlights that it would be important to investigate whether different sub-groups are more heavily impacted by ZSD.
Our study, collectively with other recent research, provides a starting point for calibrating bioinformatics approaches of disease carrier frequency estimation.11,12 An opportunity for refinement of this method comes with the recent expansion of the recommended newborn screening panel to include X-ALD, which can also detect ZSD. Our estimates appeared close to currently available newborn screening data for ZSD in New York, and these estimates will be further verified as new newborn screening data emerge. However, newborn screening is ongoing, and at any given point, the obtained incidence rate may not be as similar as we estimated with our method. In addition, future analyses could work with the larger gnomAD dataset, which is in early beta mode but includes 126,216 exomes and 15,136 genomes.27 Bioinformatics approaches to carrier frequency estimations are an important resource when other methods for assessing variant pathogenicity are limited and when population-based gene variant testing is implausible.
ACKNOWLEDGMENTS
This work was supported by the Wynne Grace Mateffy Professorship and the Center for Demography of Health and Aging at the University of Wisconsin-Madison (Center Grant: P30 AG017266; NIA Training Grant T32 AG00129). We appreciate the advice given by Dr. Nancy Braverman, Dr. Joseph Hacia, Ms. Ann Moser, Dr. Joseph Orsini, Dr. Steven Steinberg, and Dr. Michael Wangler. We thank Kallie Grassinger and Katy Penland for their assistance in data organization and manuscript preparation. Computational resources were supported by a core grant to the Center for Demography and Ecology at the University of Wisconsin-Madison (P2C HD047873).
Footnotes
The authors declare no conflict of interest.
REFERENCES
- 1.Braverman NE, Raymond GV, Rizzo WB, et al. Peroxisome biogenesis disorders in the Zellweger spectrum: An overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab. 2016;117(3):313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinberg S, Chen L, Wei L, et al. The PEX Gene Screen: molecular diagnosis of peroxisome biogenesis disorders in the Zellweger syndrome spectrum. Mol Genet Metab. 2004;83(3):252–263. [DOI] [PubMed] [Google Scholar]
- 3.Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochim Biophys Acta. 2006;1763(12):1733–1748. [DOI] [PubMed] [Google Scholar]
- 4.Burwell SM, Secretary of Health and Human Services. Letter to Joseph A. Bocchini, Committee Chairperson of the Advisory Committee on Heritable Disorders in Newborns and Children. February 16, 2016. [Google Scholar]
- 5.Steinberg SJ, Raymond GV, Braverman NE, Moser AB. Peroxisome Biogenesis Disorders, Zellweger Syndrome Spectrum In: Pagon RA, Adam MP, Ardinger HH, et al. , eds. GeneReviews(R). Seattle (WA): University of Washington, Seattle.; 2003. [Updated 2012]. [Google Scholar]
- 6.Gould SJ, Raymond GV, Valle D. The Peroxisome Biogenesis Disorders In: Scriver CR, Beaudet AL, Sly WS, Valle D, eds. The Metabolic and Molecular Bases of Inherited Disease. Vol 2 8 ed. New York, NY: McGraw-Hill; 2001:3181–3218. [Google Scholar]
- 7.Zellweger H. The cerebro-hepato-renal (Zellweger) syndrome and other peroxisomal disorders. Dev Med Child Neurol. 1987;29(6):821–829. [DOI] [PubMed] [Google Scholar]
- 8.Danks DM, Tippett P, Adams C, Campbell P. Cerebro-hepato-renal syndrome of Zellweger. The Journal of Pediatrics. 1975;86(3):382–387. [DOI] [PubMed] [Google Scholar]
- 9.Lazarow PB, Moser HW. Disorders of Peroxisome Biogenesis In: Scriver CR, Beaudet AL, Sly WS, Valle D, eds. The Metabolic and Molecular Bases of Inherited Disease. Vol 2 8 ed. New York, NY: McGraw-Hill; 1995:2287–2314. [Google Scholar]
- 10.Levesque S, Morin C, Guay SP, et al. A founder mutation in the PEX6 gene is responsible for increased incidence of Zellweger syndrome in a French Canadian population. BMC Med Genet. 2012;13:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Appadurai V, DeBarber A, Chiang PW, et al. Apparent underdiagnosis of Cerebrotendinous Xanthomatosis revealed by analysis of ~60,000 human exomes. Mol Genet Metab. 2015;116(4):298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sleat DE, Gedvilaite E, Zhang Y, Lobel P, Xing J. Analysis of large-scale whole exome sequencing data to determine the prevalence of genetically-distinct forms of neuronal ceroid lipofuscinosis. Gene. 2016;593(2):284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. [DOI] [PubMed] [Google Scholar]
- 15.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quang D, Chen Y, Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31(5):761–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; (Baltimore, MD: ). https://omim.org/. Accessed June 6, 2016. [Google Scholar]
- 22.dbPEX, PEX Gene Database. http://www.dbpex.org/home.php. Accessed February 19, 2016.
- 23.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Combined Annotation Dependent Depletion (CADD). Information Page. http://cadd.gs.washington.edu/info. Accessed June, 2017.
- 25.Ratbi I, Falkenberg KD, Sommen M, et al. Heimler Syndrome Is Caused by Hypomorphic Mutations in the Peroxisome-Biogenesis Genes PEX1 and PEX6. Am J Hum Genet. 2015;97(4):535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.gnomAD. Home Page. http://gnomad.broadinstitute.org/. Accessed October 17, 2018.