Abstract
RNA splicing, the process through which intervening segments of noncoding RNA (introns) are excised from pre-mRNAs to allow for the formation of a mature mRNA product, has long been appreciated for its capacity to add complexity to eukaryotic proteomes. However, evidence suggests that the utility of this process extends beyond protein output and provides cells with a dynamic tool for gene regulation. In this review, we aim to highlight the role that intronic RNA plays in mediating specific splicing outcomes in pre-mRNA processing, as well as explore an emerging class of stable intronic sequences that have been observed to act in gene expression control. Building from underlying flexibility in both sequence and structure, intronic RNA provides mechanisms for post-transcriptional gene regulation that are amenable to the tissue and condition specific needs of eukaryotic cells. This article is part of a Special Issue entitled: RNA structure and splicing regulation edited by Francisco Baralle, Ravindra Singh and Stefan Stamm.
1. Introduction
Pre-mRNA splicing is a key post-transcriptional process in which non-coding introns are excised from transcripts allowing for coding exons to be ligated together to generate a mature mRNA product [1]. Through alternative usage of splice sites, this process can lead to the inclusion or exclusion of exons, termed alternative splicing, or potential redefinitions of the boundaries between introns and exons in mRNA isoforms. In addition to dramatically diversifying the proteome [2–5], these differences have the potential to affect mRNA stability, localization, and translation, offering a unique regulatory mechanism to eukaryotic cells [6,7]. Since their initial identification [8,9], introns have emerged as distinctive features of eukaryotic genomes, with the overall abundance and length of introns varying greatly between organisms. Yet, their potential functions, as well as the evolutionary constraints that drive their maintenance in genomes remains poorly understood.
Splicing is regulated through an extensive network of protein-RNA interactions involving the recognition of cis elements within the pre-mRNA by trans-acting factors [10,11]. Among these cis elements, the most conserved sequences are the 5′ and 3′ splice sites (5ss and 3ss), which define the intron boundaries, as well as the branch point site (bps) and polypyrimidine tract which typically reside within a confined distance upstream of the 3ss [10,11] (Fig. 1A). Additionally, auxiliary cis elements within the intron or flanking exons can either act as enhancer or silencer elements to promote or repress exon splicing through interactions with trans-acting splicing regulators. The precise recognition of splice sites and splicing reaction is catalyzed by a massive ribonucleoprotein (RNP) complex, termed the spliceosome, which is de novo assembled onto each intron [12,13] cotranscriptionally [14–16].
Fig. 1.
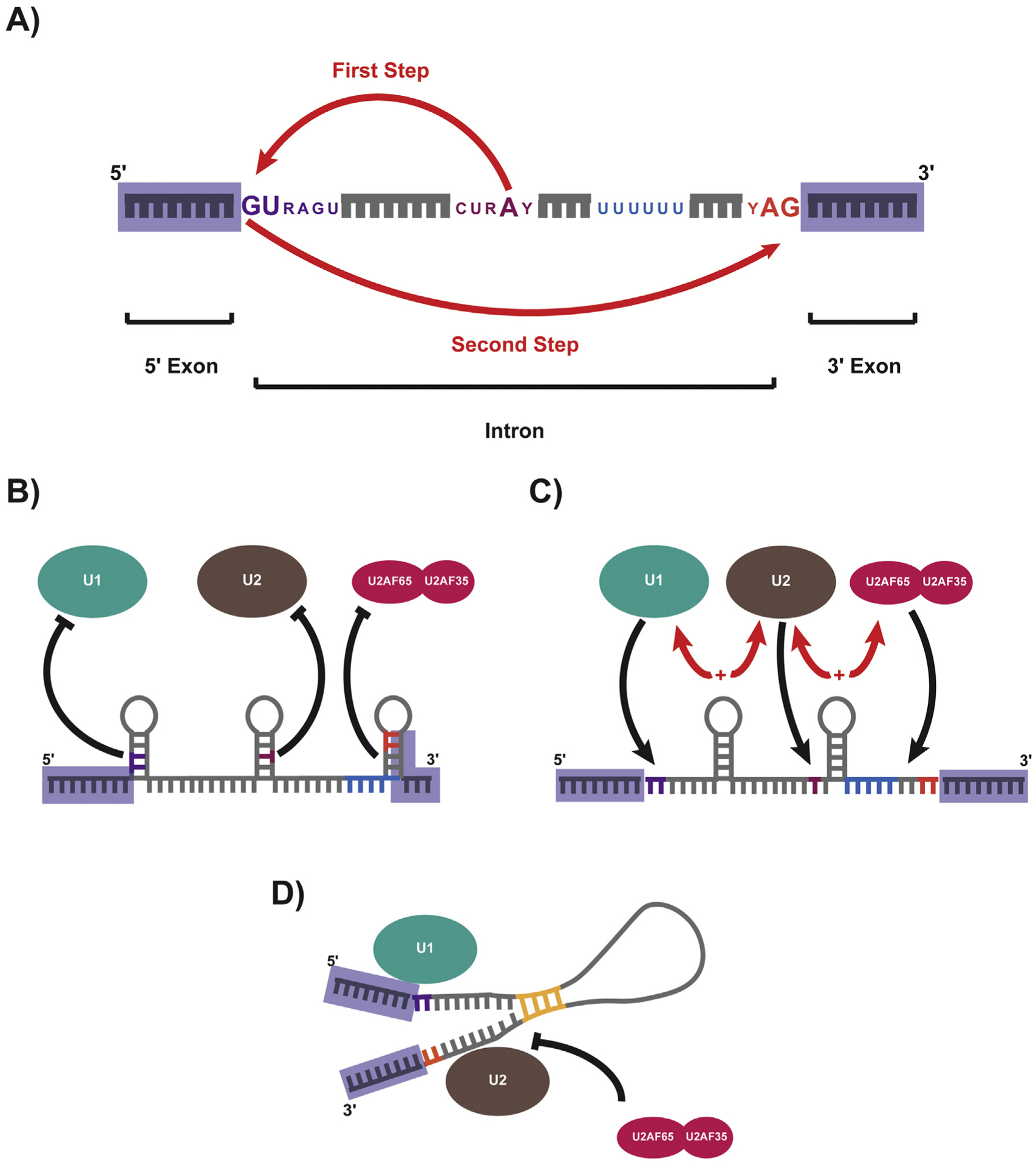
The role of RNA structure in pre-mRNA splicing. A) Canonical consensus sequences with a pre-mRNA that mediate splicing: The 5ss (purple), Branchpoint sequence (maroon), polypyrimidine tract (blue), and 3ss (orange). Splicing proceeds through a two-step transesterification reaction. In the first step the 2′OH of the branchpoint carries out a nucleophilic attack (represented by red arrow) on the 5ss, while in the second step the 3′OH of the 5′ exon attacks the first nucleotide downstream of the 3ss. B) Diagram of structures with inhibitory effects on splicing. Local stem loops repress the binding of U1 and U2 snRNPs at the 5ss and 3ss respectively, by pairing cis-elements within the double stranded structures. C) Diagram of structures that promote efficient splicing. Local stem loops serve to bring important cis-elements (such as the 5ss and branchpoint, as well as the branchpoint and 3ss) into closer proximity to one another, which promotes the interaction between snRNPs. D) Diagram of long range intra-intronic repeat elements (yellow) which pair flanking ends of intron to promote efficient splicing. Such pairings are suggested to obviate the need for essential splicing elements (such as the U2AF heterodimer). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The core of the spliceosome consists of five U-rich small nuclear RNPs (snRNPS), which are named for their small nuclear RNA (snRNA) components: U1, U2, U4, U5, and U6. The snRNPs coordinate dynamic base pairing between the different snRNAs and between snRNAs and the pre-mRNA to obtain secondary and tertiary structures that define the catalytic center of the spliceosome [13,17]. Hundreds of additional proteins are required for spliceosomal function [13,18]. Pre-mRNA splicing takes place through a two-step transesterification reaction, in which the 5ss, the branchpoint, and the 3ss are the substrates for splicing catalysis [19–21] (Fig. 1A). In the first step, the 2′OH of the branchpoint carries out a nucleophilic attack on the 5ss, generating a 5′ exon with a free 3′OH and a branched intron lariat that is attached to the 3′ exon. In the second step, the 3′OH of the 5′ exon attacks the first nucleotide downstream of the 3ss, releasing a ligated 5′ exon-3′ exon product as well as the excised intron lariat. Following splicing, the intronic lariat is debranched and rapidly degraded, which has led to the perception that they are largely composed of superfluous sequence. However, emerging research suggests that intronic RNA may serve as a dynamic mediator of gene regulation; operating to both coordinate specific splicing outcomes and provide post-transcriptional feedback in response to environmental and cellular cues. This new role for introns will be the subject of this review.
2. Introns as regulators of alternative splicing
2.1. The cis-acting splice code
Genome-wide studies estimate that 90–95% of human genes undergo some level of alternative splicing [2,3]. Differences in isoformspecific read densities indicate that the majority of alternative splicing events vary between tissues, whereas variation between individuals is significantly less common [3]. The relative abundance of these different isoforms is thought to be dependent on splice site strength, the influence of cis-acting enhancer and repressor elements, and the expression level of trans-acting factors (such as RNA binding proteins (RBPs) and splicing factors). Regardless of the expression of trans-acting factors, combinations of RNA features associated with the regulation of splicing have been proposed to make up a ‘splicing code’ which dictates outcomes in a cell and condition specific manner [11,22]. While strong splice sites contain sequences that are reliably recognized by the splicing machinery leading to ‘constitutive splicing’ in of exons, weak splice sites are dependent on the presence of additional cis-acting sequence elements and recognition of splicing-factors for their use. Extensive coordination of splicing outcomes are observed throughout development and cell differentiation, often relying on tight regulation of RBP expression rather than alterations in the overall transcription rates of alternatively spliced genes [23]. In this way, underlying flexibility in cis sequences can be exploited by the cell to generate global shifts in gene expression.
While extensive emphasis has been placed on the sequence features that define intron-exon boundaries (including splice sites, the polypyrimidine tract, and accessory silencers and enhancers), the identification, selection and regulation of branchpoints has remained largely unexplored. This is in large part due to the technical challenges posed in identifying branch points. Excised lariats, which represent the most direct record of branch point selection, are quickly degraded following splicing and are low abundance RNAs in sequencing data sets. Additionally, computational prediction of branchpoints is complicated by the low information content of the human branchpoint consensus sequence [24–27]. Beyond these technical challenges, the essential nature of branchpoints in splicing catalysis has led to the assumption that they occupy a basal, rather than regulatory, role in splicing. However, studies within specific introns suggest that branchpoint selection may contribute to the regulation of splice site recognition. Both alternative and constitutive introns can possess multiple branchpoints, which allows competing branchpoints to influence 3ss recognition [28–30], 5ss recognition [31], cassette exons [25,27], and mutually exclusive exons [32,33]. Moving beyond studies of isolated introns, Taggert et al. (2012) took advantage of the unusual inverted alignment of intronic reads generated when reverse transcriptase traverses the branch point nucleotide in cDNA libraries to identify de novo branch points [34]. Such a genome-wide approach was subsequently scaled up to both enrich for lariat cDNA using RNase R digestion [35] and include more extensive sequencing data sets encompassing diverse human tissues [36]. In this way, it was shown that the vast majority of human introns (95%) have multiple branchpoints which display tissue-specific usage in almost three-quarters of constitutive introns [36]. Such findings suggest that branchpoint recognition represents a major source of gene regulation, extending the role of intronic cis-elements in mediating splicing outcomes.
2.2. RNA structure – adding layers to cis-acting splice regulation
Beyond the direct sequence of cis-acting splicing elements, RNA structure within pre-mRNAs can also play a major role in mediating splicing outcomes (Fig. 1). The ability of RNA to form transient secondary structures allows it to influence splicing through both regulation of access to splicing signals and by altering the physical spacing among sequence elements [37]. Splicing enhancers and silencers that reside within double-stranded sequences have been shown to exert reduced influence over splicing outcomes, as many sequence-specific RBPs are unable to recognize their corresponding binding sites within double-stranded structures [38]. Additionally, a systematic analysis of regional variations in GC composition demonstrated an association between increased RNA secondary structure and alternatively spliced exons [39], suggesting that control of splice site accessibility may be a mechanism for regulating alternative splicing.
RNA structures often work to inhibit splicing by disrupting the recognition of pre-mRNA by snRNPs, an initiating event in spliceosome assembly and function (Fig. 1B). Stem structures at splice sites weaken exon inclusion [40–47], often in a dosage-dependent manner [48,49]. Such pairings at either the 5ss or the branchpoint obstruct recruitment of the U1 and U2 snRNPs [44,46,47,50]. Similarly, structures that accumulate within the polypyrimidine tract and 3ss can disrupt binding of the accessory U2AF heterodimer (composed of U2AF65 and U2AF35, which bind the polypyrimidine tract and 3ss, respectively), which in turn prevents recruitment of the U2 snRNP to the branchpoint [51] (Fig. 1B). In vitro splicing assays demonstrate limited accumulation of free upstream exon or lariat RNA for pre-mRNA containing structured 3ss, suggesting that splicing inhibition through this method may not solely disrupt the second catalytic step of splicing, but rather may function to disrupt earlier steps in splicing [29,41]. Beyond core cis sequences, RNA secondary structure has the potential to obscure additional splicing enhancers and prevent the accumulation of key accessory factors. In this way, RNA secondary structure provides a sensitive mechanism to regulate alternative splicing by repressing exon usage.
RNA structure also holds the potential to positively effect splicing, often by bringing important cis-acting sequences into closer proximity to one another (Fig. 1C). In the yeast Saccharomyces cerevisiae, secondary structure between the 5ss and branchpoint facilitate splicing by minimizing the physical distance between the U1 and U2 complexes [52–54]. Similarly, branch points, which are often restricted in their distance from the 3ss, can circumvent these physical limitations through structured regions that minimize the effective distance between more distal branchpoints (in some cases extending hundreds of nucleotides away from the 3ss) in order to reinforce efficient splicing [55–57] (Fig. 1C). Interestingly, work in yeast has shown that the spliceosome uses all available 3ss within a defined window from the branchpoint, and that ~70% of all possible 3ss selections are mediated by pre-mRNA structure [58]. One such stem structure was shown to be sensitive to changes in temperature, effectively serving as a thermosensor for alternative 3ss selection; suggesting a more direct role for pre-mRNA in mediating splicing outcomes in response to environmental cues [58].
Work by Lin, Taggart, and Lim et al. (2016) further expanded the role of RNA structure in splicing to include the influence of longer range base-pairings [59] (Fig. 1D). Long, evolutionarily conserved stretches of AC and GT repeats were identified co-occurring at the ends of introns in both fish and lamprey. Pairing of these terminal elements greatly reduces the distance between the 5ss and 3ss and greatly enhances splicing. AC and GT repeat-containing introns contain significantly weaker polypyrimidine tracts and are insensitive to U2AF2 disruption, suggesting that these long range interactions overcome the need for essential splicing factors [59] (Fig. 1D). While efforts to identify a comparable mechanisms in humans is ongoing, pairing of G and C triplets at the 5′ and 3′ ends of introns have been observed [59]. Additionally, extended stretches of G and C rich intronic sequence appearing across conditionally skipped exons has been proposed as a secondary structure-based mechanism for regulating exon skipping [60] and further efforts to identify conserved RNA structures that extend beyond individual introns has suggested that such long range interactions could play regulatory roles in alternative splicing [61].
Sequence variations that disrupt secondary structure have the potential to be deleterious, potentially causing structural changes that can alter a host of post-transcriptional processing events [62,63]. In some instances, such mutations can lead to the expression of disease-specific mRNA isoforms [64,65]. Recent efforts to profile the landscape and variation of RNA secondary structure in human transcriptomes identified as much as 15% of all transcribed single nucleotide variants alter local RNA structure [66]. These variants were shown to be more likely to result in splicing changes relative to variants that do not alter secondary structure, with a unique RNA secondary structure signature at exon-exon junctions appearing to favor more accessible AG dinucleo-tides at the end of the 5′ exon and more structured nucleotides at the start of the 3′ exon [66]. Additionally, computational analyses of genomic variants determined that intronic disease mutations within 30 nucleotides of a splice site are significantly more likely to disrupt splicing outcomes than common variants [67]. These findings suggest that variants that alter RNA secondary structure may play a more prominent role in human disease than has been previously appreciated. Work by Soemidi et al. (2017) using a Massively Parallel Splicing Assay (MaPSy), which allows for direct comparison of splicing outcomes between thousands of mutant and wildtype substrates in vivo [68], demonstrated that mutations that cause changes in RNA secondary structure can significantly alter splicing outcomes [69]. While MaPSy originally focused on exonic variants, expansion into intronic variants holds the potential to greatly expand our understanding of the role of intronic RNA and RNA structure in mediating splicing outcomes under a host of physiological and disease states.
3. Functional roles for excised introns beyond splicing
In general, lariat RNAs derived from excised introns are rapidly recognized and linearized by the debranching enzyme (DBR) and degraded by exonucleases [70–73] (Fig. 2A). This turnover is thought to both free up nucleotides to be used during subsequent rounds of transcription, as well as recycle splicing factors that remain bound to the spliced intronic lariat [74]. Failure to process intronic lariats causes a dramatic reduction in fitness leading to impaired growth in the yeast Schizosaccharomyces pombe [75] and embryonic lethality in both plants [76] and animals [77]. While the vast majority of intronic RNA is rapidly degraded, a number of exceptions have emerged which hint at an extended function for introns in post-transcriptional gene regulation. The observation that many families of non-coding RNAs (such as small nucleolar RNAs (snoRNAs), microRNAs (miRNAs), small interfering RNAs (siRNAs), and long non-coding RNAs (lncRNAs)) are preferentially associated with introns in humans suggests that introns may undergo additional post-splicing processing to generate functional RNA species [78]. And indeed such processing of excised lariats has been observed [79–83]. Additionally, a growing number of lariats have been shown to accumulate under physiological conditions [84–88], hinting that such accumulation is evolutionarily conserved and may potentially hold as yet unknown functions in the cell. Moreover, two landmark studies in yeast have demonstrated that introns promote cell survival in yeast under saturated growth or stress conditions, by selectively stabilizing a subset of excised intronic RNAs [89,90]. Given that lariats represent a direct readout of transcription, they have the potential to serve as a highly responsive layer in gene regulation and represent an emerging class of functional RNA species.
Fig. 2.
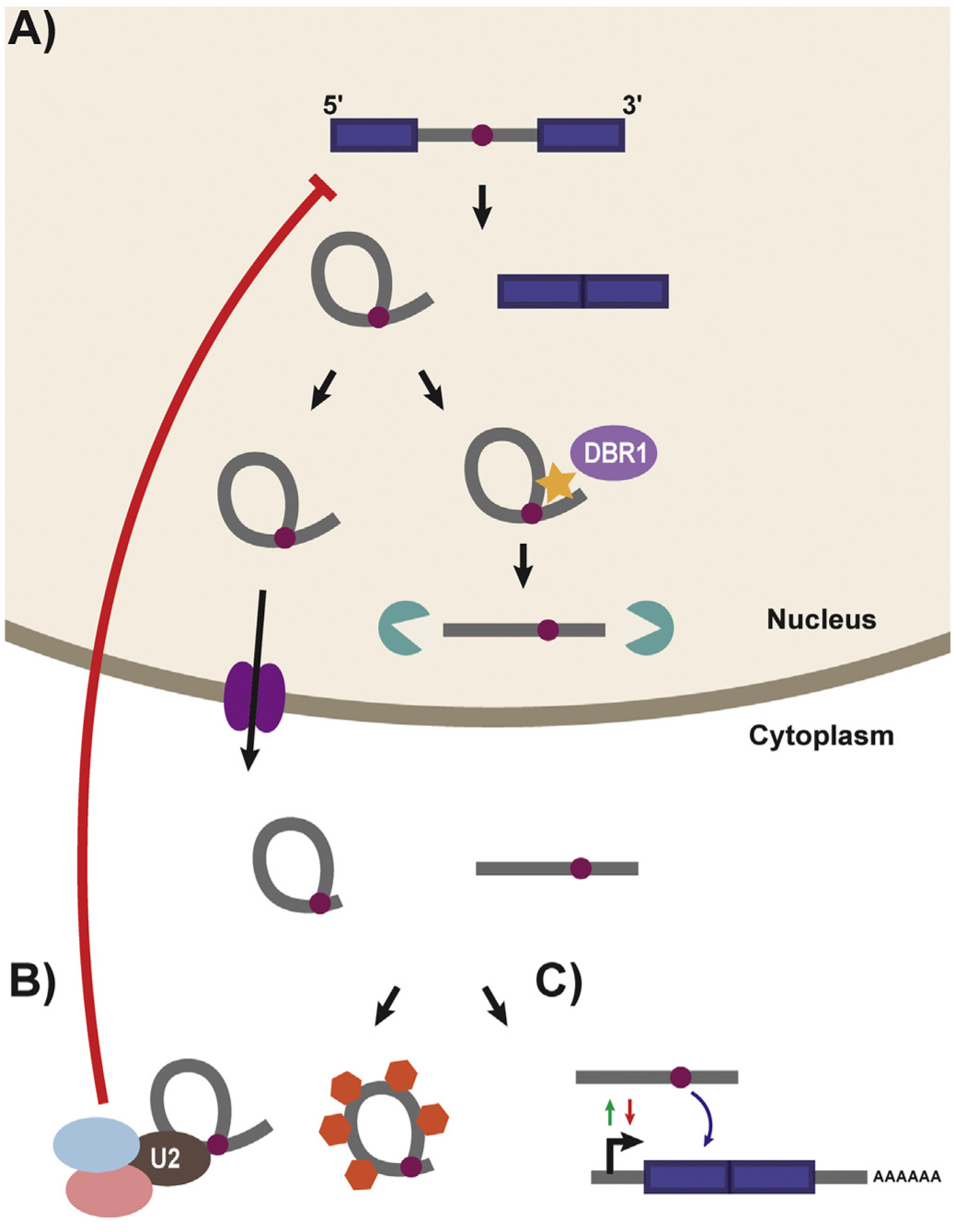
Schematic of Lariat Processing and sisRNA Accumulation/Function. A) Following splicing both a ligated exon product and intronic lariat are produced. The lariat is typically debranched by the debranching enzyme (DBR1:light purple) and processed for degradation in the nucleus by exonucleases (green). However, some stable lariats persist in both the nuclease and cytoplasm. Export to the cytoplasm proceeds through the NXF1/NXT1 system (dark purple) and leads to the accumulation of both stable lariat RNAs with trimmed tails and linearized intronic segments. B) sisRNAs have been show to work in trans (left hand portion of the panel) to control post-transcriptional processing by acting as molecular sponges, or sinks, for proteins (orange hexagons) including splice and RNA processing factors. Additionally the accumulation of snRNPs on stable lariats is suggested to cause global changes in RNA splicing efficiency. sisRNAs are also known to function in cis (right hand portion of the panel) to both positively and negatively mediate the expression of their host genes through feedback loops. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. Stable intronic RNAs
While early detection of lariat RNAs relied on the ability of reverse transcriptase to read through branchpoints in targeted RT-PCR reactions [91], advancements in high-throughput sequencing technologies and bioinformatic analyses have allowed for genome-wide screens for stable intronic lariats [34]. Using this approach, work out of the laboratory of Joseph Gall reported the first genome-wide identification of intronic RNAs in the extracted oocyte germinal vesicle (GV or nucleus) of Xenopus tropicalis [92] (Fig. 2A). These ‘intronic sequences’ represent more than 90% of actively transcribed genes during oogenesis and are highly stable; still detected two days post transcriptional inhibition [92]. In many cases these ‘intronic sequences’ are significantly shorter than the full-length introns from which they are derived [92]. Follow up work demonstrated that similar stable intronic sequence RNAs (sisRNAs) are also present in the cytoplasm of Xenopus tropicalis (Fig. 2A) and in fact are concentrated at a higher molar ratio than their nuclear counterparts [85]. These cytoplasmic sisRNAs are resistant to RNase R treatment and are believed to be stable excised lariat molecules. Contrary to nuclear sisRNAs, cytoplasmic sisRNAs represent only a subset of genes expressed in the Xenopus transcriptome, but similarly encompass both coding and noncoding genes related to virtually all known cellular functions [85]. Further examination of the host genes from which cytoplasmic sisRNAs are generated revealed a bias towards their excision from smaller introns [85]. Despite their differences, both nuclear and cytoplasmic sisRNAs persist in mature oocytes from early embryogenesis through the start of zygotic transcription at the blastula stage [85]. This persistence throughout early development strongly suggests that sisRNAs may play a regulatory role in gene expression.
Additional studies identified cytoplasmic lariats in human, mouse, chicken, and zebrafish cells [86]. These lariats continued to be restricted in size (mostly 100–500 nucleotides in length) and tended to use an unusual cytosine branchpoint [86]. Cytosine branchpoints are an unfavorable target for DBR1 recognition and debranching [93], which provides a possible mechanism through which cytoplasmic sisRNAs escape debranching. Interestingly, stable lariats are suggested to be selectively exported from the nucleus to the cytoplasm by the NXF1/NXT1 system, suggesting that their accumulation is not due to simple leakage from the nucleus [86] (Fig. 2A). One possible explanation for their behavior centers around the idea that they remain stably associated to the mRNA from which they are derived, acting in a regulatory capacity to control translation in the cytoplasm. However, such a stable association between mRNA and cognate sisRNAs is not observed [86].
Several examples of sisRNAs assuming a direct role in gene regulation have been identified in Drosophila melanogaster, where both circular and linear sisRNAs are found to persist throughout early development and into adult tissues [94] (Fig. 2A). One such sisRNAs, sisR-1, was shown to regulate the expression of its host gene regena, by binding to a cis-natural antisense transcript, ASTR. While association between ASTR and regena promotes regena expression, disruption of this interaction by sisR-1 binding to ASTR generates a negative feedback loop which results in down regulation of host gene expression. This repression is developmentally regulated and continues throughout embryogenesis [94]. Additional sisRNAs with inhibitory roles, including sisR-2 and sisR-3 (which repress dFAR1 and the lncRNA CR44148, respectively) were also identified in Drosophila [94,95], and seem to possess a common predicted secondary structure. In each case, these linear sisRNAs are highly paired and possess an exposed 3′ tail which has been hypothesized to play a role in both mediating intronic stability and function [96]. Also acting in cis, expression of the stable lariat RNA sisR-4, has been shown to upregulate expression of its host gene, deadpan [87], suggesting that sisRNAs do not act exclusively to down-regulate gene expression. sisR-4 activates an enhancer present within a common host intron, which in turn promotes transcription of the deadpan locus in a positive feedback loop [87]. Such cis-acting means of regulation, in which introns are utilized to self-regulate their parental genes, allows for direct pairing of transcriptional rates with feedback loops in a particularly efficient and responsive manner (Fig. 2B).
In addition to operating in cis, some sisRNAs have been observed to function as molecular sinks or sponges, affecting post-transcriptional regulation of broader gene regulatory networks by sequestering transacting factors from their normal functions (Fig. 2B). In Arabidopsis, stable intronic lariat RNAs compete with pre-miRNAs for binding with the DCL1/HYL1 dicing complex [97]. Over expression of nuclear sisRNAs effectively inhibits miRNA processing, with potentially broad implication for gene regulation [97]. Similarly, in human cell lines it was shown that linear sisRNAs derived from the 15q11-q13 genomic region (distinctive for their snoRNA terminal sequences) accumulate in the nucleus (outside of snoRNA associated nuclear bodies), where they sequester the alternative splicing factor Fox2 [83]. This interaction is shown to specifically regulate the splicing profile of Fox2 sensitive genes [83]. Lastly, the over accumulation of intronic lariat RNAs in the cytoplasm of yeast was shown to sequester the RNA binding protein TDP-43 [98]. This inhibition disrupts the potentially toxic effects of TDP-43 mutations associated with Amyotrophic Lateral Sclerosis (ALS) [98], suggesting that the role of sisRNAs as a molecular sink could be an attractive point for therapeutic intervention. However, this provocative finding is yet to be supported by follow up experiment confirming an influence of lariat levels in ALS.
In light of the emergence of sisRNAs as regulators of gene expression, recent landmark papers examining the function of introns in yeast cells [89,90] stand out as particularly exciting. Parallel work by the Bartel and Elela laboratories identified a subset of excised introns, which selectively accumulate as linear RNAs during periods of TORC1-mediated stress. These sisRNAs function to reduce growth rates, promoting cell survival under starvation conditions and offering significant growth advantages to yeast cells upon reentry into log-phase growth [89,90]. This accumulation is independent of host gene expression and leads to a shift in global splicing patterns [89,90]. As ribosomal mRNAs are the substrate of a disproportionate number of splicing events in yeast cells, this shift leads to a corresponding repression of ribosomal protein genes [89,90]. Interestingly, the predicted structure of starvation responsive introns features extensive base-pairing between the intron (typically encompassing either the 5ss or branchpoint) and the 5′ UTR [90]. Mutations that disrupt these structures were found to impact endogenous control over growth rates, suggesting that gene context, and in particular host 5′ UTR, may influence intron stability and function [90]. Regulation of such conserved structures represents an attractive mechanism for mediating conditional intron stability. The capacity to regulate the stability of excised introns in response to dynamic environmental cues in order to mediate global gene expression patterns, suggests that intronic RNA may hold tremendous utility in post-transcriptional gene regulation.
Understanding sisRNAs as conditionally regulated opens up exciting possibilities for gene regulation. However, efforts to shift focus from yeast to other, more intron-rich, eukaryotic species presents challenges. An attractive approach to gaining insights into potential roles for stabilized lariats is to characterize cells in which ‘normal’ lariat processing has been disrupted. By promoting the overaccumulation of intronic lariat molecules, cells lose the capacity to selectively enrich for sisRNAs. One such study in patients harboring mutations in DBR1 revealed tissue specific and cell intrinsic susceptibility to the herpes simplex virus 1 (HSV1) [99]. Over accumulation of lariats lead to increased vulnerability to viral infection of the brainstem in patients harboring the HSV1 virus [99]. Such an outcome suggests that control over lariat accumulation impairs virus recognition by host cells, potentially damaging cellular defenses against viral invasion. This is in fitting with the observation that HSV-1 infection itself increases lariat accumulation. Alternatively, control over viral RNA lariats may be disrupted, which have been shown to mediate the switch between lytic and latent infection [100]. In either case, control over lariat accumulation underlies a shift in cellular homeostasis that implies a function for sisRNAs and furthers the broadening field of intronic RNA function.
4. Concluding remarks
Moving beyond the image of intronic RNA sequence as ‘junk’ material whose primary function is to add complexity to the protein coding content of the genome, eukaryotic introns are emerging as regulators of cellular function. Both through mediation of specific RNA splicing events and through the regulated stability of functional intronic RNAs, introns serve a key role in post-transcriptional gene regulation. Research aimed at better understanding intronic RNA structure as a dynamic component of gene expression seem destined to provide insights into how cells are able to respond to a diversity of external stimuli and further our understanding of the coding potential inherent to eukaryotic gene architecture.
Acknowledgements
This work was supported by the National Institutes of Health under award numbers R01GM127472 and R01GM105681.
Footnotes
This article is part of a Special Issue entitled: RNA structure and splicing regulation edited by Francisco Baralle, Ravindra Singh and Stefan Stamm.
Transparency document
The Transparency document associated with this article can be found, in online version.
References
- [1].Lee Y, Rio DC, Mechanisms and regulation of alternative Pre-mRNA splicing. (2015) Annu. Rev. Biochem 84: 291–323. doi: 10.1146/annurevbiochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. (2008) Nat. Genet 40: 1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- [3].Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. (2008) Nature 456: 470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. (2010) Nature 463: 457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LDN, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang TC, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TSK, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. (2014) Nature 509: 575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. (2013) Cell 152: 1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Heyn P, Kalinka AT, Tomancak P, Neugebauer KM. Introns and gene expression: Cellular constraints, transcriptional regulation, and evolutionary consequences. (2015) BioEssays. 37: 148–154. doi: 10.1002/bies.201400138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. (1977) Proc. Natl. Acad. Sci 74: 3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. (1977) Cell 12: 1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- [10].Wang Z, Burge CB. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. (2008) RNA 14: 802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. (2010) Nature 465: 53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- [12].Hoskins AA, Moore MJ. The spliceosome: A flexible, reversible macromolecular machine. (2012) Trends Biochem. Sci 37: 179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. (2017) Nat. Rev. Mol. Cell Biol 18: 655–670. doi: 10.1038/nrm.2017.86. [DOI] [PubMed] [Google Scholar]
- [14].Khodor YL, Rodriguez J, Abruzzi KC, Tang C-HA, Marr MT, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. (2011) Genes Dev. 25: 2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schmidt U, Basyuk E, Robert M-C, Yoshida M, Villemin J-P, Auboeuf D, Aitken S, Bertrand E. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. (2011) J. Cell Biol 193: 819–829. doi: 10.1083/jcb.201009012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SAE, Schedl P, Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. (2011) Cell 147: 1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Herzel L, Ottoz DSM, Alpert T, Neugebauer KM. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. (2017) Nat. Rev. Mol. Cell Biol 18:637–650. doi: 10.1038/nrm.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Matlin AJ, Moore MJ. Spliceosome assembly and composition. (2007) Adv. Exp. Med. Biol 623:14–35. Doi: 10.1007/978-0-387-77374-2_2. [DOI] [PubMed] [Google Scholar]
- [19].Reed R, Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. (1985) Cell 41: 95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- [20].Frendewey D, Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. (1985) Cell 42: 355–367. doi: 10.1016/S0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- [21].Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. (1986) Cell 47: 555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- [22].Fu XD. Towards a splicing code. (2004) Cell 119: 736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- [23].Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. (2017) Nat. Rev. Mol. Cell Biol 18: 437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhuang Y, Goldstein AM, Weiner AM. UACUAAC is the preferred branch site for mammalian mRNA splicing. (1989) Proc. Natl. Acad. Sci 86:2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kol G, Lev-Maor G, Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. (2005) Hum. Mol. Genet 14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- [26].Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. (2008) Nucleic Acids Res. 36: 2257–2267. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Corvelo A, Hallegger M, Smith CWJ, Eyras E. Genome-wide association between branch point properties and alternative splicing. (2010) PLoS Comput. Biol 6: e1001016. doi: 10.1371/journal.pcbi.1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Reed R, Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. (1988) Genes Dev. 2: 1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- [29].Smith CW, Chu TT, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. (1993) Mol. Cell. Biol 13: 4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bradley RK, Merkin J, Lambert NJ, Burge CB. Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. (2012) PLoS Biol. 10: e1001229. doi: 10.1371/journal.pbio.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Noble JC, Prives C, Manley JL. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. (1988) Genes Dev. 2: 1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- [32].Mullen MP, Smith CWJ, Patton JG, Nadal-Ginard B. α-Tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. (1991) Genes Dev. 5: 642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- [33].Southby J, Gooding C, Smith CWJ. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutually exclusive exons. (1999) Mol. Cell. Biol 19: 2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Taggart AJ, Desimone AM, Shih JS, Filloux ME, Fairbrother WG. Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo. (2012) Nat. Struct. Mol. Biol 19: 719–721. doi: 10.1038/nsmb.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mercer TR, Clark MB, Andersen SB, Brunck ME, Haerty W, Crawford J, Taft RJ, Nielsen LK, Dinger ME, Mattick JS. Genome-wide discovery of human splicing branchpoints. (2015) Genome Res. 25: 290–303. doi: 10.1101/gr.182899.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pineda JMB, Bradley RK. Most human introns are recognized via multiple and tissue-specific branchpoints. (2018) Genes Dev. 32: 577–591. doi: 10.1101/gad.312058.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. (2004) Mol. Cell. Biol 24: 10505–10514. doi: 10.1128/mcb.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. (2013) PLoS Genet. 3: e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang J, Kuo CCJ, Chen L. GC content around splice sites affects splicing through pre-mRNA secondary structures. (2011) BMC Genomics 12: 90. doi: 10.1186/1471-2164-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Solnick D. Alternative splicing caused by RNA secondary structure. (1985) Cell 43: 667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- [41].Watakabe A, Inoue K, Sakamoto H, Shimura Y. A secondary structure at the 3′ splice site affects the in vitro splicing reaction of mouse immunoglobulin μ chain pre-mRNAs. (1989) Nucleic Acids Res. 17: 8159–8169. doi: 10.1093/nar/17.20.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Estes PA, Cooke NE, Liebhaber SA. A native RNA secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. (1992) J. Biol. Chem 267: 14902–14908. [PubMed] [Google Scholar]
- [43].Liu HX, Goodall GJ, Kole R, Filipowicz W. Effects of secondary structure on pre-mRNA splicing: hairpins sequestering the 5′ but not the 3′ splice site inhibit intron processing in Nicotiana plumbaginifolia. (1995) EMBO J. 14: 377–388. doi: 10.1002/j.1460-2075.1995.tb07012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sirand-Pugnet P, Durosay P, D’Orval BC, Brody E, Marie J. β-tropomyosin pre-mRNA folding around a muscle-specific exon interferes with several steps of spliceosome assembly. (1995) J. Mol. Biol 251: 591–602. doi: 10.1006/jmbi.1995.0458. [DOI] [PubMed] [Google Scholar]
- [45].Varani L, Hasegawa M, Spillantini MG, Smith MJ, Murrell JR, Ghetti B, Klug A, Goedert M, Varani G. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. (1999) Proc. Natl. Acad. Sci 96: 8229–8234. doi: 10.1073/pnas.96.14.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. (1997) RNA 3: 405–419. [PMC free article] [PubMed] [Google Scholar]
- [47].Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. (2007) Nucleic Acids Res. 35: 371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Goguel V, Wang Y, Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. (1993) Mol. Cell. Biol 13: 6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Plass M, Codony-Servat C, Ferreira PG, Vilardell J, Eyras E. RNA secondary structure mediates alternative 3’ss selection in Saccharomyces cerevisiae. (2012) RNA. 18: 1103–1115. doi: 10.1261/rna.030767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jiang Z, Cote J, Kwon JM, Goate AM, Wu JY. Aberrant Splicing of tau Pre-mRNA Caused by Intronic Mutations Associated with the Inherited Dementia Frontotemporal Dementia with Parkinsonism Linked to Chromosome 17. (2000) Mol. Cell. Biol 20: 5360–5363. doi: 10.1128/mcb.20.14.5360-5360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Warf MB, Diegel JV, Von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. (2009) Proc. Natl. Acad. Sci 106: 9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Goguel V, Rosbash M. Splice site choice and splicing efficiency are positively in-fluenced by pre-mRNA intramolecular base pairing in yeast. (1993) Cell 72: 893–901. doi: 10.1016/0092-8674(93)90578-E. [DOI] [PubMed] [Google Scholar]
- [53].Libri D, Stutz F, McCarthy T, Rosbash M. RNA structural patterns and splicing: molecular basis for an RNA-based enhancer. (1995) RNA 1: 425–436. [PMC free article] [PubMed] [Google Scholar]
- [54].Howe KJ, Ares M. Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. (1997) Proc. Natl. Acad. Sci 94: 12467–12472. doi: 10.1073/pnas.94.23.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chebli K, Gattoni R, Schmitt P, Hildwein G, Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. (1989) Mol. Cell. Biol 9: 4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Deshler JO, Rossi JJ. Unexpected point mutations activate cryptic 3′ splice sites by perturbing a natural secondary structure within a yeast intron. (1991) Genes Dev. 5: 1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- [57].Rogic S, Montpetit B, Hoos HH, Mackworth AK, Ouellette BFF, Hieter P. Correlation between the secondary structure of pre-mRNA introns and the efficiency of splicing in Saccharomyces cerevisiae. (2008) BMC Genomics 9: 355–357. doi: 10.1186/1471-2164-9-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Meyer M, Plass M, Pérez-Valle J, Eyras E, Vilardell J. Deciphering 3’ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. (2011) Mol. Cell 43: 1033–1039. doi: 10.1016/j.molcel.2011.07.030. [DOI] [PubMed] [Google Scholar]
- [59].Lin CL, Taggart AJ, Lim KH, Cygan KJ, Ferraris L, Creton R, Huang YT, Fairbrother WG. RNA structure replaces the need for U2AF2 in splicing. (2016) Genome Res. 26:12–23. doi: 10.1101/gr.181008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Miriami E, Margalit H, Sperling R. Conserved sequence elements associated with exon skipping. (2003) Nucleic Acids Res. 31:1974–1983. doi: 10.1093/nar/gkg279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pervouchine DD, Khrameeva EE, Pichugina MY, Nikolaienko OV, Gelfand MS, Rubtsov PM, Mironov AA. (2012) Evidence for widespread association of mammalian splicing and conserved long-range RNA structures. (2013) RNA 18:1–15. doi: 10.1261/rna.029249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Shabalina SA, Spiridonov NA, Kashina A. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. (2013) Nucleic Acids Res. 41: 2073–2094. doi: 10.1093/nar/gks1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sabarinathan R, Wenzel A, Novotny P, Tang X, Kalari KR, Gorodkin J. Transcriptome-wide analysis of UTRs in non-small cell lung cancer reveals cancer-related genes with SNV-induced changes on RNA secondary structure and miRNA target sites. (2014) PLoS One 9: e82699. doi: 10.1371/journal.pone.0082699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Clouet d’Orval B, d’Aubenton Carafa Y, Sirand-Pugnet P, Gallego M, Brody E, Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. (1991) Science 252: 1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- [65].Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden HH, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevena M, De Graaff E, Wauters E, Van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JBJ, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Costra BA, Hardy J, Goate A, Van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. (1998) Nature 393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- [66].Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, Chang HY. Landscape and variation of RNA secondary structure across the human transcriptome. (2014) Nature 505: 706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RKC, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, Morris Q, Barash Y, Krainer AR, Jojic N, Scherer SW, Blencowe BJ, Frey BJ, The human splicing code reveals new insights into the genetic determinants of disease. (2015), Science 347 (2015) 1254806, 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Soemedi R, Cygan KJ, Rhine CL, Wang J, Bulacan C, Yang J, Bayrak-Toydemir P, McDonald J, Fairbrother WG. Pathogenic variants that alter protein code often disrupt splicing. (2017) Nat. Genet 49: 848–855. doi: 10.1038/ng.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Soemedi R, Cygan KJ, Rhine CL, Glidden DT, Taggart AJ, Lin CL, Fredericks AM, Fairbrother WG. The effects of structure on pre-mRNA processing and stability. (2017) Methods 125: 36–44. doi: 10.1016/j.ymeth.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ruskin B, Krainer AR, Maniatis T, Green MR. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. (1984) Cell 38: 317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- [71].Ruskin B, Green MR. An RNA processing activity that debranches RNA lariats. (1985) Science 229: 135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- [72].Clement JQ, Qian L, Kaplinsky N, Wilkinson MF. The stability and fate of a spliced intron from vertebrate cells. (1999) RNA 5: 206–220. doi: 10.1017/S1355838299981190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. (2003) Mol. Cell 12: 1453–1465. doi: 10.1016/S1097-2765(03)00488-X. [DOI] [PubMed] [Google Scholar]
- [74].Han B, Park HK, Ching T, Panneerselvam J, Wang H, Shen Y, Zhang J, Li L, Che R, Garmire L, Fei P. Human DBR1 modulates the recycling of snRNPs to affect alternative RNA splicing and contributes to the suppression of cancer development. (2017) Oncogene 36: 5382–5391. doi: 10.1038/onc.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Khalid MF, Damha MJ, Shuman S, Schwer B. Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. (2005) Nucleic Acids Res. 33: 6349–6360. doi: 10.1093/nar/gki934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang H, Hill K, Perry SE. An Arabidopsis RNA Lariat Debranching Enzyme Is Essential for Embryogenesis. (2004) J. Biol. Chem 279: 1468–1473. doi: 10.1074/jbc.M309106200. [DOI] [PubMed] [Google Scholar]
- [77].Zheng S, Vuong BQ, Vaidyanathan B, Lin JY, Huang FT, Chaudhuri J. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. (2015) Cell 161: 762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A. Critical association of ncRNA with introns. (2011) Nucleic Acids Res. 39: 2357–2366. doi: 10.1093/nar/gkq1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. (2007) Cell 130: 89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. (2007) Nature 448: 83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Dumesic PA, Natarajan P, Chen C, Drinnenberg IA, Schiller BJ, Thompson J, Moresco JJ, Yates JR, Bartel DP, Madhani HD. Stalled spliceosomes are a signal for RNAi-mediated genome defense. (2013) Cell 152: 957–968. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ooi SL, Samarsky DA, Fournier MJ, Boeke JD. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. (1998) RNA 4: 1096–1110. doi: 10.1017/S1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. (2012) Mol. Cell 48: 219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- [84].Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular Intronic Long Noncoding RNAs. (2013) Mol. Cell 51: 792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- [85].Talhouarne GJS, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. (2014) RNA 20: 1476–1487. doi: 10.1261/rna.045781.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Talhouarne GJS, Gall JG. Lariat intronic RNAs in the cytoplasm of vertebrate cells. (2018) Proc. Natl. Acad. Sci 115: 7970–7977. doi: 10.1073/pnas.1808816115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Tay MLI, Pek JW. Maternally inherited stable intronic sequence RNA triggers a self-reinforcing feedback loop during development. (2017) Curr. Biol 27: 1062–1067. doi: 10.1016/j.cub.2017.02.040. [DOI] [PubMed] [Google Scholar]
- [88].Zhang X, Zhang Y, Wang T, Li Z, Cheng J, Ge H, Tang Q, Chen K, Liu L, Lu C, Guo J, Zheng B, Zheng Y. A comprehensive map of intron branchpoints and lariat RNAs in plants. (2019) Plant Cell 31: 956–973. doi: 10.1105/tpc.18.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Morgan JT, Fink GR, Bartel DP. Excised linear introns regulate growth in yeast. (2019) Nature 565: 606–611. doi: 10.1038/s41586-018-0828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Parenteau J, Maignon L, Berthoumieux M, Catala M, Gagnon V, Abou Elela S. Introns are mediators of cell response to starvation. (2019) Nature 565: 612–617. doi: 10.1038/s41586-018-0859-7. [DOI] [PubMed] [Google Scholar]
- [91].Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. (2006) Nucleic Acids Res. 34: e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gardner EJ, Nizami ZF, Conover Talbot J, Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. (2012) Genes Dev. 26:2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Montemayor EJ, Katolik A, Clark NE, Taylor AB, Schuermann JP, Combs DJ, Johnsson R, Holloway SP, Stevens SW, Damha MJ, Hart PJ. Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1. 2014. Nucleic Acids Res. 42: 10845–10855. doi: 10.1093/nar/gku725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Pek JW, Osman I, Tay MLI, Zheng RT. Stable intronic sequence RNAs have possible regulatory roles in Drosophila melanogaster. (2015) J. Cell Biol 211: 243–251. doi: 10.1083/jcb.201507065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Osman I, Pek JW. A sisRNA/miRNA axis prevents loss of germline stem cells during starvation in Drosophila. (2018) Stem Cell Reports 11: 4–12. doi: 10.1016/j.stemcr.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chan SN, Pek JW. Stable intronic sequence RNAs (sisRNAs): an expanding universe. (2019) Trends Biochem. Sci 44: 258–272. doi: 10.1016/j.tibs.2018.09.016. [DOI] [PubMed] [Google Scholar]
- [97].Li Z, Wang S, Cheng J, Su C, Zhong S, Liu Q, Fang Y, Yu Y, Lv H, Zheng Y, Zheng B. Intron Lariat RNA inhibits microRNA biogenesis by sequestering the dicing complex in Arabidopsis. (2016) PLoS Genet. 12: e1006422. doi: 10.1371/journal.pgen.1006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z, Fang X, Shorter J, Krogan NJ, Finkbeiner S, Farese RV, Gitler AD. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. (2012) Nat. Genet 44: 1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhang SY, Clark NE, Freije CA, Pauwels E, Taggart AJ, Okada S, Mandel H, Garcia P, Ciancanelli MJ, Biran A, Lafaille FG, Tsumura M, Cobat A, Luo J, Volpi S, Zimmer B, Sakata S, Dinis A, Ohara O, Garcia Reino EJ, Dobbs K, Hasek M, Holloway SP, McCammon K, Hussong SA, DeRosa N, Van Skike CE, Katolik A, Lorenzo L, Hyodo M, Faria E, Halwani R, Fukuhara R, Smith GA, Galvan V, Damha MJ, Al-Muhsen S, Itan Y, Boeke JD, Notarangelo LD, Studer L, Kobayashi M, Diogo L, Fairbrother WG, Abel L, Rosenberg BR, Hart PJ, Etzioni A, Casanova JL. Inborn errors of RNA lariat metabolism in humans with brainstem viral infection. (2018) Cell. 172: 952–965. doi: 10.1016/j.cell.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Rivas HG, Schmaling SK, Gaglia MM. Shutoff of host gene expression in in-fluenza a virus and herpesviruses: similar mechanisms and common themes. (2016) Viruses 8: 102 [DOI] [PMC free article] [PubMed] [Google Scholar]


