Abstract
The intestinal reclamation of bile acids is crucial for the maintenance of their enterohepatic circulation. The majority of bile acids are actively absorbed via specific transport proteins that are highly expressed in the distal ileum. The uptake of bile acids by intestinal epithelial cells modulates the activation of cytosolic and membrane receptors such as the farnesoid X receptor (FXR) and G protein-coupled bile acid receptor 1 (GPBAR1), which has a profound effect on hepatic synthesis of bile acids as well as glucose and lipid metabolism. Extensive research has focused on delineating the processes of bile acid absorption and determining the contribution of dysregulated ileal signaling in the development of intestinal and hepatic disorders. For example, a decrease in the levels of the bile acid-induced ileal hormone FGF15/19 is implicated in bile acid-induced diarrhea (BAD). Conversely, the increase in bile acid absorption with subsequent overload of bile acids could be involved in the pathophysiology of liver and metabolic disorders such as fatty liver diseases and type 2 diabetes mellitus. This review article will attempt to provide a comprehensive overview of the mechanisms involved in the intestinal handling of bile acids, the pathological implications of disrupted intestinal bile acid homeostasis, and the potential therapeutic targets for the treatment of bile acid-related disorders.
Introduction
Bile acids, the major organic solutes in bile, are avidly absorbed in the distal small intestine and returned to the liver for resecretion, resulting in continuous circulation between the liver and the intestine. The intestinal absorption processes are robust and efficient, resulting in only 5% of the total secreted bile acids being lost in the feces. Bile acids are synthesized from cholesterol in the liver and modified by the intestinal bacteria producing a group of molecules that have common structure but differ in the number and spatial location of the hydroxy groups on the steroid rings. These differences in the structural features among bile acids have a significant impact on their functions. In the intestinal lumen, bile acids function as lipid solubilizers that are crucial for lipid absorption. Beyond solubilizing lipids, bile acids play other important physiological roles, modulating a number of hepatic and intestinal functions and exerting systemic effects such as increasing energy expenditure and improving insulin sensitivity. The decrease in intestinal absorption reduces the circulating pool of bile acids, disrupting bile acid homeostasis and altering bile acid signaling processes. For example, certain types of bile acid malabsorption (BAM) are associated with an increase in hepatic bile acid synthesis and an elevation in their intestinal luminal concentrations. These alterations may explain the mechanisms underlying some cases of bile acid diarrhea (BAD). On the other hand, the increase in bile acid absorption may contribute to the development of severe liver damage, and inhibition of bile acid absorption should be considered in the treatment of hepatic disorders such as nonalcoholic liver disease (NAFLD) and its severe form, nonalcoholic steatohepatitis (NASH). In light of the critical roles of intestinal absorption of bile acids in the maintenance of their homeostasis, extensive research during the past two decades has focused on understanding the molecular mechanisms underlying these processes. The following sections will highlight the current knowledge about the intestinal responses to bile acids with an emphasis on the processes involved in their absorption and their regulation under physiological and pathophysiological conditions.
Bile Acid Structure
Bile acids are one of the most structurally diverse classes of small biomolecules. Remarkably, at least 84 distinct unconjugated bile acids and 25 bile alcohols have been identified in vertebrates (162). It is important to note that subtle structural differences between bile acids dramatically affect their biological roles. During the past 100 years, much effort has been dedicated to identifying bile acid structures and their composition in humans. Because of this important basic research, we now know the structures, the relative abundance, and the basis of the structure-function relationship of these molecules in humans and other animals.
Structural features of bile acids
Despite their wide structural diversity, bile acids do have several defining structural features. All bile acids possess a lipophilic steroid body that is hydroxylated at one or more positions and a side chain that terminates as a carboxylic acid (162). The bile acid pool of mammals and most vertebrates is composed entirely of compounds with 24 carbon atoms, while more primitive animals may produce bile alcohols with up to 27 carbons. The presence of both a lipophilic ring and hydroxyl and carboxyl moieties makes bile acids amphipathic molecules, which is necessary for their function as physiological detergents. Bile acids are hydroxylated at the 3α position (3α-OH) on steroid ring A, but many are hydroxylated at other sites as well. Indeed, the majority of bile acid diversity is the result of differential hydroxylation of the steroid ring. In addition to their hydroxylation, bile acids have a side chain extending from C-17 on steroid ring D. The carboxylic acid moiety at the end of the side chain is conjugated to either taurine or glycine in the liver, which lowers the pKa and enhances water solubility at physiological pH. Conjugation of bile acids to either taurine or glycine also increases their resistance to precipitation by high concentration of Ca2+ in the gallbladder where they are being concentrated (160).
Mammalian bile acid species
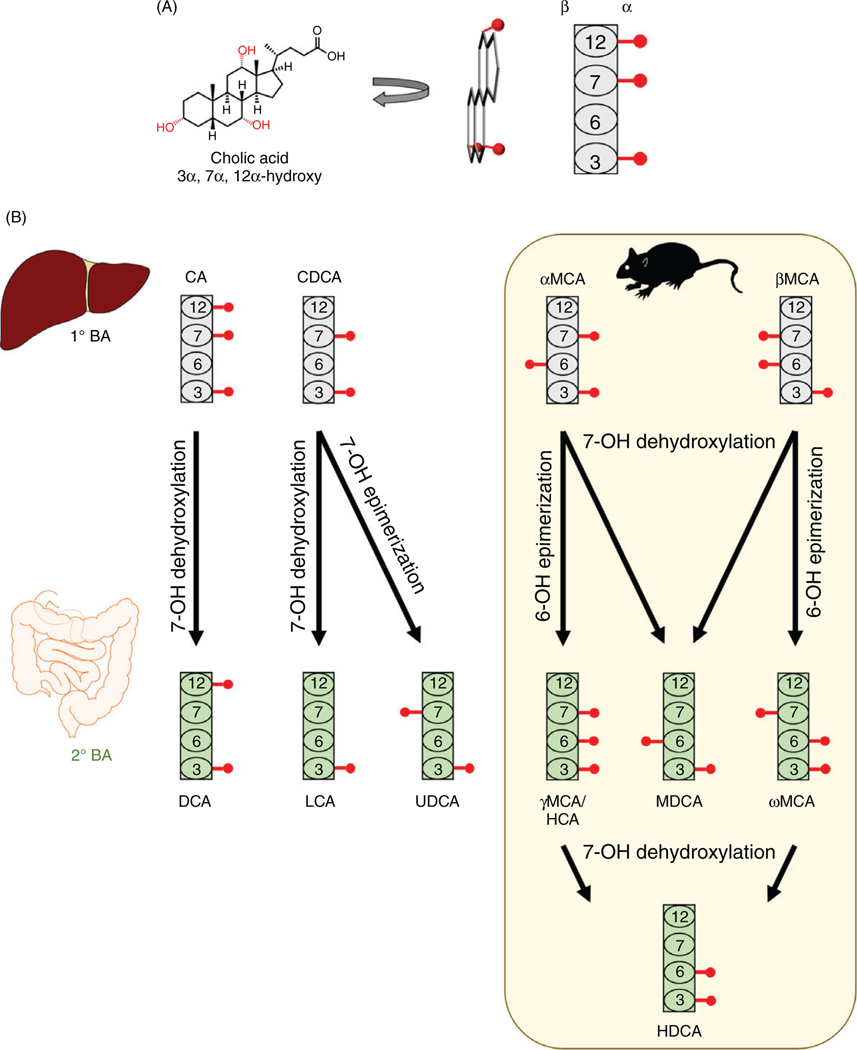
Of all the bile acid species that have been identified, only a few are of physiologic relevance in mammals. The chemical structures of physiologically important bile acids are shown in Figure 1. Humans synthesize just two bile acids: cholic acid (CA) and chenodeoxycholic acid (CDCA). Both are produced from cholesterol in the liver and are found in all mammals, as well as many lower species. The structural difference between CA and CDCA is slight. CA is 3α-, 7α -, and 12α-hydroxylated, while CDCA is only 3α- and 7α-hydroxylated. CA and CDCA, along with the bacterial metabolite deoxycholic acid (DCA; 3α - and 12α-OH), are the major bile acid species in humans. Combined, these three bile acids and their conjugated forms account for greater than 90% of biliary bile acids in humans (160). Additionally, several minor bile acids in humans have been identified, including lithocholic acid (LCA; 3α-OH) and ursodeoxycholic acid (UDCA; 3α - and 7β-OH). DCA and LCA are generated in the intestine by bacteria via 7β-dehydroxylation of CA and CDCA, respectively. Like CA and CDCA, DCA and LCA are found in all mammals and numerous other species. Similarly, UDCA is produced from CDCA by intestinal bacteria via 7-OH epimerization, although it can be formed in the hepatocytes of several species of bears (107, 145). Low levels of tauroursodeoxycholic acid (TUDCA) were detected in germ-free mice, indicating that it is also a primary bile acid in mice (331).
Figure 1.
Structure of common mammalian bile acids. (A) The structure of bile acids can be visualized by orienting the plane of the molecule perpendicular to the page, such that α-hydroxyl moieties are on the right side of the steroid nucleus and β-hydroxyl moieties are on the left side. This can be represented by a block diagram, where the rectangles represent the steroid nucleus with selected carbon atoms labeled by number and the red circles represent the hydroxyl group. (B) Structures of primary (gray) and secondary (green) bile acids. Bacterial production of secondary bile acids occurs via the reactions depicted by black arrows. Muricholic acids and their derivatives, which are predominantly found in rodents, are shown in the yellow box.
Because much research in the field is conducted in mice, it is critical to understand the differences between human and mouse bile acid pools. In addition to the bile acids noted above, mice produce a group of bile acids termed muricholic acids (MCAs), which are hydroxylated at the 6-C position. The two major MCAs in mice are α-muricholic acid (a-MCA; 3α-, 6β-, and 7α-OH) and β-muricholic acid (β-MCA; 3α -, 6β-, and 7β-OH). Interestingly, the MCAs constitute a majority of the mouse bile acid pool. The consequences of these different bile acid profiles are just beginning to be uncovered and will be discussed in detail later. However, it is clear that bile acid signaling differs greatly between human and mice because of these chemical differences in the bile acid pool.
Bile Acid Synthesis
The primary bile acids CA and CDCA are synthesized in the liver from cholesterol at a rate of approximately 500 mg daily in humans (79, 321). The secondary bile acids, which include DCA, LCA, and UDCA, are produced by bacteria from bile acids in the intestinal lumen (160, 402). The process of primary bile acid synthesis requires at least 17 enzymes, which are located in the endoplasmic reticulum, mitochondria, peroxisomes, and cytosol. The liver is the sole organ that contains all of the enzymes required for BA biosynthesis. However, many other tissues possess enzymes that synthesize intermediate compounds, which can then be circulated to the liver for completion of the synthesis. There are two parallel pathways by which BA synthesis occurs: the classical (neutral) pathway, which accounts for up to 70% of human BA synthesis, and the alternative (acidic) pathway, which is a relatively minor route for bile acid synthesis under physiological conditions (46). Hepatic bile acids (either de novo synthesized or recycled from the intestine) are conjugated to the amino acids taurine or glycine in a two-step process mediated by the bile acid-coenzyme A ligase (BAL) and the bile acid-coenzyme A:amino acid N-acyltransferase (BAT) (362).
The classical pathway occurs exclusively in hepatocytes and leads to production of both CA and CDCA in most mammals. The first reaction in the classical pathway is catalyzed by cytochrome P450 7A1 (cholesterol 7α-hydroxylase; CYP7A1), localized to the endoplasmic reticulum. CYP7A1 hydroxylates the 7α position of cholesterol to produce 7-hydroxy cholesterol. This enzyme is rate limiting and is critical for efficient synthesis of bile acids. Indeed, disruption of CYP7A1 by homozygous knockout of the Cyp7a1 gene in mice reduces bile acid synthesis by approximately 60% and bile acid pool size by approximately 75%, which results in increased fecal lipids, hypercholesterolemia, and poor survival (109, 181,337,338).
The other enzyme of the classical pathway is cytochrome P450 8B1 (sterol 12α-hydroxylase; CYP8B1), which is also located in the ER. CYP8B1 is responsible for hydroxylating the 12α carbon of the steroid ring. Thus, CYP8B1 is the enzymatic branch point for the synthetic routes of CA, which is 12α-hydroxylated, and CDCA, which is not. As expected, Cyp8b1 knockout mice have a markedly different bile acid profile compared to wild-type animals. While wild-type mice have bile acid profiles rich with CA, Cyp8b1 knockout mice are deficient in CA and DCA and have increased levels of CDCA and its derivatives, including UDCA and MCAs (240). Interestingly, a recent study showed that CYP8B1 is inactivated in certain species such as naked-mole rats and elephants in which CA is missing (341).
The alternative pathway for bile acid synthesis is a minor contributor to the total bile acid pool. In adult humans, it accounts for no more than approximately 30% of total bile acid synthesis (46). The main product of the alternative pathway is CDCA. Recent studies showed that the expression of enzymes involved in the alternative pathway was upregulated in CYP7A1 knockout mice on a near-pure C57BL/6J background, leading to a decrease in CA/CDCA due to an increase in CDCA synthesis (118). It is noted in these studies that CA is detected in the pool of bile acids in CYP7A1-deficient mice, indicating that CA could also be synthesized from the products of the alternative pathways. The first and the rate-limiting step of the alternative pathway is the entry of cholesterol into the mitochondria, which is mediated by the steroidogenic acute regulatory protein (StAR) (292). Then, the mitochondrial sterol 27-hydroxylase (CYP27A1) oxidizes the side chain of cholesterol. CYP27A1 is expressed in hepatocytes, as well as a variety of extrahepatic sites, including macrophages and endothelial cells (13, 308, 312). Thus, the alternative pathway can be initiated outside of the liver. Interestingly, hepatic CYP27A1 is also critical in the classical pathway, where it oxidizes intermediates of BA synthesis. Expectedly, mice deficient in Cyp27a1 display bile acid pools approximately 1/3 the size of the wild-type mice (315). The second step in the alternative pathway involves the hydroxylation of 27-hydroxycholesterol to 3β,7α-dihydroxy-5-cholestenoic acid mediated by the CYB7B1 enzyme. The product of this reaction is then converted to CDCA (236).
In addition to CYP7A1, CYP8B1, and CYP27A1, which are key enzymes in bile acid synthesis, Cyp2c70 is essential for bile acid synthesis in mice. Takahashi et al. recently identified this enzyme as the bile acid 6β-hydroxylase and demonstrated that Cyp2c70 is required for the synthesis of MCAs (373). Mice with the Cyp2c cluster knocked out (Cyp2c ko) and Cyp2c ko mice with a human CYP2C9 transgene (hCYP2C9) were unable to synthesize MCA species and displayed a corresponding increase in CDCA. The bile acid profile in these mice more closely resembles that of humans, who do not produce MCAs. Although the implications of the different bile acid pools in mice and humans have not been fully investigated, it is clear that bile acid structure underlies the molecular basis of bile acid functions, as will be discussed in the following sections.
Enterohepatic Circulation of Bile Acids
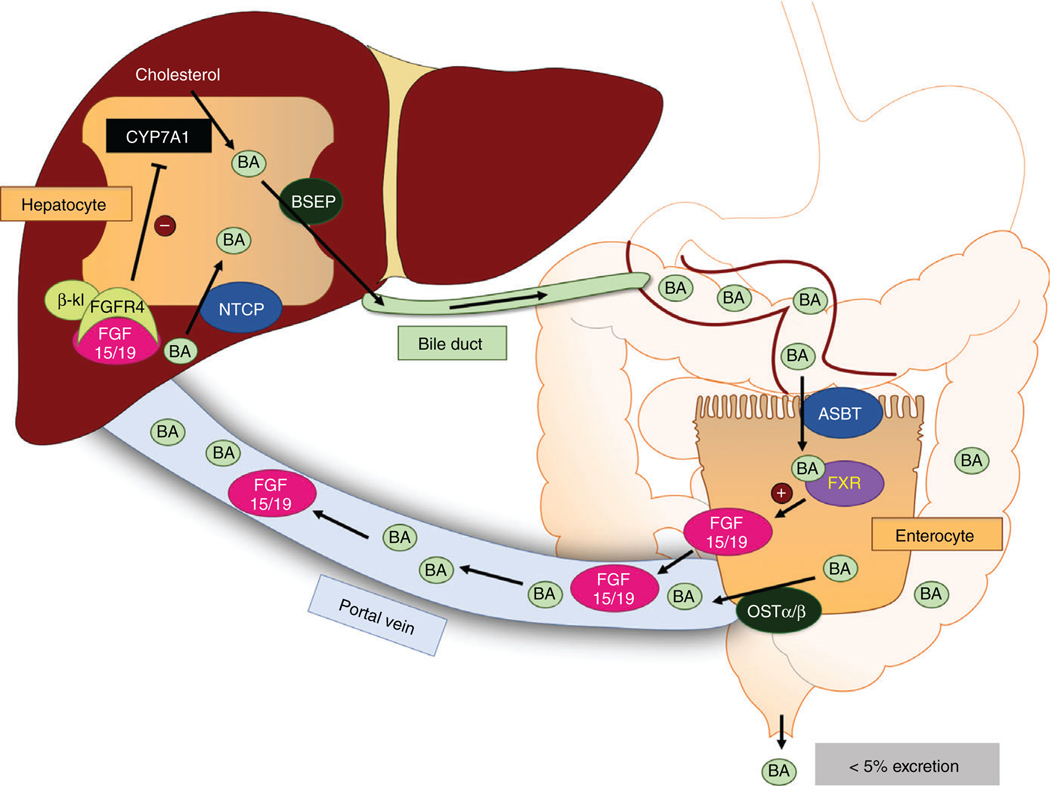
Following the synthesis in hepatocytes, bile acids are excreted across the canalicular membrane by the bile salt export pump (BSEP). Biliary BAs are stored in the gallbladder and secreted into the small intestine following the intake of food, where they act as detergents and signaling molecules. Luminal BAs travel through the small intestine and are actively reabsorbed in the distal ileum by the apical sodium-dependent bile acid transporter (ASBT). Reabsorbed BAs are shuttled from the apical to the basolateral membrane of the enterocyte and are transported into the portal circulation by the organic solute transporter α/β heterodimer (OSTα/β). These BAs are returned to the liver via the portal vein and enter hepatocytes by the sinusoidal sodium-taurocholate cotransporting polypeptide (NTCP), establishing the enterohepatic circulation of bile acids (Figure 2). We discuss in detail the intestinal BA transporters in the following sections.
Figure 2.
Enterohepatic circulation of bile acids. Bile acids (BAs) are synthesized from cholesterol in the liver via the rate-limiting enzyme CYP7A1 and secreted into the bile duct by the bile salt export pump (BSEP). These BAs are then deposited into the small intestine, where they emulsify lipids and serve as signaling molecules. Luminal BAs are taken up into ileal enterocytes by the apical sodium-dependent bile acid transporter (ASBT) and exported into the portal blood by the organic solute transporter α/β heterodimer (OSTα/β). These BAs return to the liver, where they are taken into hepatocytes (mainly in the periportal region of the hepatic acinus) by the Na+-taurocholate cotransporting polypeptide (NTCP) and resecreted by BSEP. Within the enterocyte, BAs activate the farnesoid X receptor (FXR), which induces fibroblast growth factor 15/19 (FGF15/19) expression and secretion into the portal blood. FGF15/19 travels to the liver, where it binds to the FGF receptor 4 (FGFR4)/β-klotho (β-kl) complex. Activation of this complex reduces CYP7A1 expression, thus decreasing bile acid synthesis.
Functions of Bile Acids
In addition to their vast structural diversity, bile acids perform a wide variety of physiological roles in the body. Some of their functions, particularly their role as detergents, have been known for more than 100 years. However, more recent research has demonstrated that bile acids also have critical signaling roles affecting different functions such as immune response and maintenance of glucose and lipid homeostasis.
Dietary lipid absorption
The physiological role of bile acid as detergents is historically the best recognized of BA functions. Indeed, bile acids are of critical importance for proper lipid absorption. Bile acids are amphipathic and are present at high concentration in the intestinal lumen, allowing them to form micelles and emulsify dietary fats and cholesterol. In the gut lumen, these micelles increase lipid surface area, promoting digestion by lipases and enhancing absorption at the brush border membrane (164). In some pathological states, such as BAM, an insufficient bile acid pool size limits the ability to aid lipid absorption and leads to lipid malabsorption (55).
Importantly, bile acid structure greatly influences the extent to which they aid lipid absorption. Amino acid conjugation maintains high BA concentration in the lumen by preventing passive diffusion. Furthermore, bile acids in which all hydroxyl moieties are facing one side of the molecule are more hydrophobic and are thus better lipid emulsifiers. Studies performed by Wang et al. (406) demonstrated that when mice were supplemented with CA and CDCA, cholesterol absorption was enhanced relative to nonsupplemented diets. However, when supplemented with more hydrophilic bile acids such as MCAs, cholesterol absorption was reduced. These studies clearly illustrate the relationship between bile acid structure and function as digestive emulsifiers.
Bile acids as signaling molecules
While the role of bile acids in facilitating the absorption of lipids has been known for over 100 years, the effects of bile acids on other aspects of metabolism have only recently been recognized. By the early 1990s, a link between bile acid and cholesterol homeostasis was clear, but the molecular mechanisms of this interaction were not well known (108). The molecular basis underlying bile acids’ importance in metabolism has started to clarify over the past 20 years. A number of researchers reported bile acid interactions with cellular receptors, including the nuclear receptors farnesoid X receptor (FXR), pregnane X receptor (PXR), and vitamin D receptor (VDR), as well as the membrane G protein-coupled bile acid receptor (G protein-coupled bile acid receptor 1, GPBAR1; GPCR19; and TGR5) and sphingosine-1-phosphate receptor (S1PR) (378). These seminal findings identified new molecular targets for bile acid-based therapies and helped renew interest in the field of bile acid physiology. Indeed, bile acids are now understood as complex signaling molecules that exert a wide array of effects on bile acid, cholesterol, glucose, and lipid homeostasis.
Farnesoid X Receptor
The FXR (NR1H4) is a nuclear receptor that was originally identified in 1995 (125). In 1999, several groups simultaneously reported that FXR is a receptor for bile acids (255, 293, 407). When activated by BAs, FXR binds the response elements on DNA to activate transcription of target genes primarily in the liver and intestine. Since its identification as a bile acid receptor, FXR has received great attention as a metabolic regulator and therapeutic target.
Bile acids as FXR ligands
It is important to note that slight differences in bile acid structure result in large functional differences, and this is particularly apparent when interactions with FXR are studied. Early reports suggested that CDCA was the most potent FXR activator among endogenous bile acids, followed by DCA and LCA (130, 255, 293). Some of these studies suggested that CA and conjugated BAs were not FXR agonists. (255) However, when the bile acid transporters NTCP or ASBT were coexpressed, these bile acids were shown to activate FXR (293, 407). Furthermore, tauro- and glyco-conjugated bile acids activated FXR as strongly as the respective unconjugated BAs in cells that expressed NTCP. These studies indicated that FXR is activated by both conjugated and unconjugated bile acids, provided that these compounds are able to reach the cytoplasm of the cell.
Interestingly, not all bile acids are able to activate FXR. LCA, for example, does not appear to strongly activate this receptor. Furthermore, some bile acids have actually been shown to antagonize FXR. These FXR antagonists appear to have one defining structural feature: a β-OH moiety located on the B ring of the steroid nucleus. UDCA (3α- and 7β-OH), which is present at low concentrations in humans, acts as an FXR antagonist under some conditions (272). Similarly, aMCA (3α-, 6β, and 7α-OH) and β-MCA (3α-, 6β, and 7β-OH), two of the major rodent bile acid species, were recently identified as FXR antagonists (170, 331). The finding that mice have higher concentrations of FXR antagonist bile acids has complicated the interpretation of bile acid studies conducted in these animals, as the basal FXR signaling is likely to be considerably lower in mice. The Gonzalez Laboratory recently generated a mouse model that lacks the enzyme Cyp2c70 and does not synthesize MCAs, leading to a bile acid profile that more closely resembles humans (373). Although the consequences of this humanized bile acid pool have yet to be investigated, it is expected that basal FXR activation is increased in humanized Cyp2c mice, as FXR antagonist MCAs are replaced with FXR agonist BA species. Studies conducted in mice with humanized bile acid profiles could allow for the reconciliation of conflicting studies with respect to the physiological and therapeutic effects of bile acid signaling.
FXR regulation of bile acid synthesis and transport
Because bile acids are relatively strong detergents, they are potentially toxic to cells. Therefore, cellular levels of bile acids must be tightly controlled. Shortly after it was identified as a bile acid receptor, it was discovered that FXR is a major regulator of bile acid homeostasis and may function to protect cells against bile acid toxicity.
Initially, studies focused on the role of hepatic FXR in mediating a negative feedback mechanism that inhibits bile acid synthesis. Several groups identified a negative feedback mechanism by which hepatic FXR inhibits bile acid synthesis (74, 140, 247). These groups showed that FXR transactivates small heterodimer partner (SHP; NR0B2) expression. SHP is a member of the nuclear receptor gene family that lacks a DNA-binding domain. Instead, SHP interacts with other transcription factors to repress their activity. In the liver, hepatic SHP was shown to impair CYP7A1 expression, and therefore bile acid synthesis, by inhibiting liver receptor homolog 1 (LRH-1) and hepatic nuclear factor 4α (HNF4α) activity (1, 140, 206). Similarly, hepatic SHP represses CYB8B1 through inhibition of LRH-1 and HNF4α (195, 206). The inhibition of bile acid synthesis by FXR may also occur via SHP-independent pathways via stimulating the expression of the RNA-binding protein ZEP36L1 with a subsequent increase in CYP7A1 mRNA degradation (374, 408).
Hepatic FXR regulates the expression of bile acid transporters to reduce cellular bile acids. Hepatic FXR inhibits the expression of the NTCP through an SHP-dependent process, decreasing the uptake of bile acids from the portal circulation (100). Conversely, hepatic FXR transactivates the expression of the BSEP, enhancing the excretion of intracellular BAs into the bile canaliculi (11). Thus, one of the major functions of hepatic FXR is to reduce the bile acid synthesis and modulate the transporter expression to decrease the bile acid concentration within the hepatocyte.
It was later discovered that intestinal farnesoid X receptor (iFXR) was also an important mediator of feedback inhibition of bile acid synthesis (179). Activation of FXR in the intestinal epithelium promotes transcription of fibroblast growth factor 19 (FGF19; FGF15 in rodents). FGF15/19 is secreted into the portal blood and circulates to the liver, where it binds to the FGF receptor 4 (FGFR4)/β-klotho complex on hepatocytes. The downstream c-Jun-N-terminal kinase (JNK) pathways repress CYP7A1 transcription in an SHP-dependent manner, reducing BA synthesis (200). The iFXR-FGF15/19-CYP7A1 axis was shown to be essential for feedback inhibition of BA synthesis. When FXR is knocked out specifically in the intestine, the total bile acid pool increased in size (360). Unlike hepatic FXR, iFXR activation only appears to regulate CYP7A1 expression and not CYP8B1 (198). This suggests that iFXR inhibits bile acid synthesis, while hepatic FXR is able to modulate primarily the composition of the BA pool.
iFXR has also been shown to regulate the expression of epithelial bile acid transporters. Landrier et al. (223) demonstrated that iFXR binds to the response elements in the OSTα/β promoter, enhancing the OSTα/β expression. This results in increased export of BAs into the portal circulation. Additionally, iFXR promotes the expression of the ileal bile acid-binding protein (IBABP) (142, 178), which binds and sequesters BAs within the cytosol of enterocytes.
Like in the liver, iFXR activation promotes SHP expression. Intestinal SHP inhibits retinoic acid receptor (RAR)-dependent transcription of the apical sodium-dependent bile acid transporter (ASBT). This leads to lower ASBT expression in enterocytes and decreased bile acid uptake from the intestinal lumen. Thus, through both direct and indirect mechanisms, FXR acts as a bile acid sensor to protect hepatocytes and enterocytes from excessive bile acid accumulation and associated toxicity.
Regulation of lipid metabolism
In addition to regulating bile acid homeostasis, FXR has been shown to regulate several other metabolic pathways, including hepatic lipid metabolism. In the liver, FXR activation lowers the triglyceride levels through SHP repression of the lipogenic transcription factor sterol regulatory element-binding protein 1c (SREBP1c) (412). Additionally, FXR activation by bile acids induces expression of peroxisome proliferator-activated receptor α (PPARα; NR1C1), a nuclear receptor that is a key regulator of lipolysis (301). Furthermore, the FXR/SHP pathway was shown to inhibit the transcription factor SREBP2, which has the effect of reducing de novo cholesterol synthesis (199). Thus, it seems likely that FXR activation impairs lipogenesis through SREBP1c inhibition, enhances fatty acid β-oxidation through a PPARα-dependent mechanism, and may reduce cholesterol synthesis through SREBP2 inhibition.
Interestingly, recent evidence has suggested that activation of iFXR is able to promote browning of adipocytes throughout the body. Work by Fang et al. demonstrated that fexaramine, a gut-restricted FXR agonist, enhances lipolysis in adipocytes and protects against diet-induced obesity (DIO) in mice (113). Though the precise mechanism is unclear, it may be related to alterations in the systemic bile acid profile in fexaramine-treated mice that promote signaling via other receptors, such as GPBAR1 (discussed later).
Regulation of glucose metabolism
It has also been demonstrated that bile acid signaling is an important regulator of glucose metabolism, although the exact role of FXR is still to be precisely defined. Several studies have shown that FXR activity promotes glucose tolerance and insulin sensitivity. FXR−/− mice display high fasting glucose and poor insulin sensitivity (62,251). Additionally, FXR agonists restore both hepatic and peripheral insulin sensitivity in diabetic (db/db) and obese (ob/ob and DIO) mice (62, 253, 439). However, the interpretation of these results is controversial, as other studies generated opposite results showing that FXR deficiency is protective against insulin resistance in ob/ob and high fat diet (HFD)-fed mice (47, 306, 438). Interestingly, recent evidence suggests that different FXR splice variants may be induced by various bioenergetic states, with each splice variant producing somewhat different effects on glucose and lipid metabolism (91). However, the differential roles of FXR splice variants remain largely unknown. Furthermore, there is evidence of large sex differences in the effects of FXR metabolic signaling, but this is an area that requires further study (438).
Because of the central role of the liver in maintaining glucose homeostasis, much research in this field has focused on hepatic FXR signaling. Nevertheless, FXR in other organs also regulates glucose homeostasis. For example, FXR in pancreatic β cells is important for efficient insulin production and glucose-stimulated insulin transcription and secretion (106, 303, 313).
Interestingly, the role of iFXR in maintaining glucose homeostasis is more controversial. iFXR activation using the gut-restricted FXR agonist fexaramine improves insulin sensitivity and reduces hepatic gluconeogenesis in HFD-fed mice (113). However, recent evidence has suggested that iFXR inhibition also promotes glucose homeostasis. Jiang et al. recently showed that inhibition of iFXR via glycine-conjugated β-muricholic acid (Gly-MCA) or intestine-specific knockout of FXR reduces hepatic gluconeogenesis, improves glucose tolerance, and enhances insulin sensitivity (183). In addition, these studies demonstrated that iFXR inhibition reduces hepatic triglycerides and protects against DIO. It was further shown that iFXR antagonism reduces intestinal ceramide production, which may be responsible for the observed effects on metabolic homeostasis (429).
Clearly, FXR is a crucial regulator of several metabolic pathways. However, elucidating the differences in FXR signaling between organ systems and FXR isoforms will be critical for delineating the complicated effects on metabolism and designing therapies against this target (Table 1).
Table 1.
Functions of Hepatic FXR (hFXR) and Intestinal FXR (iFXR)
| hFXR | iFXR | |
|---|---|---|
| Hepatic bile acid synthesis | SHP-dependent reduction in CYP7A1 | Reduction via FGF15/19-FGFR4-CYP7A1 axis |
| Bile acid transporters | ↑ Export (BSEP) | ↑ Export (OSTα/β) |
| ↓ Import (NTCP) | ↓ Import (ASBT) | |
| Lipid metabolism | ↑ Lipolysis | ↑ Ceramide synthesis |
| ↓ Lipogenesis | Promotes adipose tissue browning | |
| Glucose homeostasis | ↑ Glucose tolerance and insulin sensitivity | Controversial — both activation and inhibition may ↑ glucose tolerance and insulin sensitivity and ↓ hepatic gluconeogenesis |
Regulation of immune response
The roles of bile acids as signaling molecules have recently extended to the modulation of immune response. Indeed, the effects of bile acids on the immune response were observed in earlier studies showing that incubation with CDCA suppressed the LPS-induced secretion of TNFα in monocytes (54). More recent investigations provided evidence for FXR expression in human CD4+ T cells, CD8+ T cells, and CD14 monocyte subpopulations of immune cells (336), suggesting a role for this nuclear receptor in bile acid-mediated immune modulation. This notion was further supported by studies showing that the activation of FXR by the synthetic agonist INT-747 (obeticholic acid, OCA) also decreased the secretion of TNFα from activated human PBMCs and CD14+ monocytes (132). The findings of those studies implied potential therapeutic effects of FXR agonists in the treatment of inflammatory disorders such as inflammatory bowel disease (IBD). In fact, the experiments of Gadaleta et al. (132) demonstrated that administration of OCA protected against dextran sulfate sodium (DSS) and 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis in mice. It is also evident that the lack of FXR in knockout mice increased the severity of experimentally induced intestinal inflammation (394). These data along with findings showing a decrease in FXR expression in IBD patients (394) indicate that changes in bile acids and/or the expression of FXR may be involved in the dysregulation of the immune response and the development of inflammatory disorders including IBD. More research is warranted to determine the roles of bile acids in the development of inflammatory disorders.
Antimicrobial effects
Multiple lines of evidence indicated the antimicrobial effects of bile acids. First, the small intestine where conjugated bile acids are usually present at high concentration (~10mM) has less bacterial flora as compared to the colon (161). Second, intestinal bacterial overgrowth occurs in cirrhotic patients in whom bile acid synthesis and secretion are decreased (36). Also, the administration of conjugated bile acids to rats with CCl4-induced cirrhosis resulted in a decrease in the associated bacterial overgrowth (245). Few reports have shown a direct inhibition of bacterial growth in vitro by unconjugated bile acids (44). However, compelling evidence was provided by Inagaki et al. (180), demonstrating the involvement of FXR in mediating the antimicrobial effects of bile acids. Those investigators showed that the bile duct ligation (BDL) and blocking bile flow to the intestine in mice caused bacterial overgrowth and mucosal damage that was augmented in the absence of FXR. Conversely, the same authors showed that the administration of the FXR agonist GW4064 reduced the mucosal damage and the bacterial overgrowth and translocation in the BDL mouse model. The activation of FXR was shown to induce the expression of genes with antibacterial activities such as the inducible nitric oxide synthase in the intestine (180). It is becoming more evident that the relationship between gut bacteria and bile acids is complex. It is clear that bile acids profoundly affect the bacterial composition of gut microflora, and that gut microbiome is one of the major determinants of the bile acid profile. The reciprocal relationship between bile acids and gut bacteria is discussed in detail in other reviews (317, 355).
Pregnane X Receptor
The PXR (NR1I2) is a nuclear receptor that is highly expressed in the liver and the intestine (202). PXR activation promotes the transcription of detoxifying enzymes, such as CYP3A4 and other cytochrome P450 enzymes, which is a critical function of the intestine and the liver (231). In 2001, it was shown that bile acids, particularly the highly hepatotoxic LCA, activate PXR to promote CYP3A4 expression, which then metabolizes and detoxifies bile acids (356, 430). Additionally, PXR activation inhibits bile acid synthesis by repressing transcription of CYP7A1 (238, 297). In this manner, hepatic PXR appears to aid FXR in protecting against bile acid-induced toxicity. However, it has been shown that PXR knockout mice are actually resistant to LCA hepatotoxicity (237). Thus, additional research is required to better understand the effects of bile acid-dependent PXR activation.
Vitamin D Receptor
The VDR (NR1I1) is expressed in many tissues, including osteoblasts, renal cells, and enterocytes. In 2002, intestinal VDR was identified as a nuclear receptor that is activated by the secondary bile acid LCA (254). Much like PXR, binding of LCA to VDR promotes transcription of CYP3A enzymes, which metabolizes bile acids and other toxins. Additionally, recent evidence suggests that VDR activation promotes FGF15 synthesis, which represses CYP7A1 transcription in the liver (334). Thus, VDR acts to reduce bile acid synthesis and promote bile acid degradation. Although intestinal VDR activation by BAs certainly may suppress bile acid synthesis genes through the FGF15 pathway, the functional importance is likely minor, given that disruption of the FXR-FGF15/19-CYP7A1 axis is sufficient to relieve feedback inhibition of BA synthesis. It seems more likely that the primary role of intestinal VDR in bile acid homeostasis is through promotion of bile acid detoxification.
G protein-coupled bile acid receptor 1
Even before bile acids had been shown to activate nuclear receptors, there was a large body of evidence that they were involved in mediating some signaling pathways. In 1976, Conley et al. showed that DCA induced adenylyl cyclase activity and impaired phosphodiesterase E activity in rabbit colon, leading to an increase in cAMP (89). They speculated that this pathway was responsible for the secretory effects of colonic bile acids. Indeed, later studies confirmed that bile acid-induced Cl− secretion in the colon was a result of increased cAMP (304). In 2002, the Tanaka Laboratory identified and cloned a Gs-coupled membrane receptor for bile acids that they named the membrane receptor for bile acids (M-BAR) (258). They demonstrated by Northern blot that M-BAR was expressed in many tissue types, including the kidney, spleen, placenta, heart, leukocytes, and the GI tract. The following year, researchers at Takeda Chemical Industries identified a GPCR they named the Takeda G protein-coupled receptor 5 (TGR5), which bound bile acids and induced cAMP in monocytes (192). Indeed, these groups had identified the same protein, which is now also known as the GPBAR1.
Bile acids are the only known endogenous ligands for this receptor, and secondary bile acids have a higher affinity than do primary bile acids. LCA is the strongest endogenous activator of GPBAR1 (EC50 < 1 μM), followed by DCA, CDCA, and CA (192). In addition, numerous synthetic GPBAR1 agonists have been developed, with some displaying high potency in the nanomolar range (144). Importantly, GPBAR1 is able to bind conjugated bile acids, meaning that these BA species can exert signaling effects without needing to enter the cell. This allows conjugated BAs to signal in a vast array of cell types that do not express BA transporters.
Since its discovery, GPBAR1 has been recognized as having important functions throughout the body. As it relates to this article, it is important to note that GPBAR1 is an important regulator of bile acid homeostasis, immune response, energy expenditure, and glucose homeostasis. Global GPBAR1 knockout results in a smaller bile acid pool size (~20% decrease) and a shift from hydrophilic MCAs to CA (102,259). In addition, female mice but not males display increased weight gain relative to wild-type littermates (259). GPBAR1 knockout mice were also shown to be more susceptible to TNBS-induced colitis, suggesting anti-inflammatory roles for the receptor (88). This conclusion was later supported by studies from Biagioli et al. showing that activating GPBAR1 by the synthetic agonist BAR501 caused a shift in the M1/M2 phenotype of intestinal macrophages along with a decrease in the levels of pro-inflammatory cytokines (43). Evidence also suggests that GPBAR1 activation by bile acids induces thyroid hormone release, promoting adipocyte browning and enhancing global energy expenditure in mice, suggesting a possible mechanism for weight gain in GPBAR1 knockouts (411). Interestingly, in humans, supplementation with CDCA stimulated GPBAR1 and induced adipose browning and increased energy expenditure, suggesting that this phenomenon may translate to the clinic (50).
In addition, many studies have pointed to the central role of gastrointestinal GPBAR1 activation in mediating glucose homeostasis. Activation of GPBAR1 in the L cells of the duodenum induces release of the insulinotropic hormone glucagon-like peptide-1 (GLP-1) (191, 375). Indeed, when TCA is administered rectally in humans, GLP-1 secretion is induced (425). Additionally, GPBAR1 activation has been shown to have a direct positive effect on insulin release at the level of pancreatic β cells (217). Clearly, GPBAR1 is critical for a variety of cellular processes both within and outside of the intestine. It is not surprising, therefore, that this receptor is an attractive therapeutic target with several agonists currently in clinical trials. It will be interesting to determine the clinical efficacy of these GPBAR1 agonists.
Sphingosine-1-phosphate receptor 2
The S1PR is a GPCR that binds the lipid sphingosine-1-phosphate (319). Studies in the Hylemon Laboratory demonstrated that conjugated bile acids activate sphingosine-1-phosphate receptor type 2 (SIPR2) to trigger ERK1/2 and AKT intracellular pathways via the coupling of the receptor to Gi protein (361). These pathways have a direct effect on hepatic bile acid, glucose, and lipid metabolism, making S1PR2 an interesting drug target for the treatment of several liver diseases (218).
Although the majority of these studies were performed in primary hepatocytes, S1PR2 is also highly expressed in the intestine and cholangiocytes (72, 73, 409). The activation of S1PR2 receptor was shown to mediate bile acid-induced cholangiocytes proliferation. Furthermore, the lack of S1PR2 resulted in less cholangiocyte proliferation and liver injury in a BDL model of cholestasis in mice (410). The precise role of S1PR2 in the intestine is an area of active research. Recent evidence suggests that S1PR2 signaling enhances proliferation of intestinal epithelia and is important for maintaining the epithelial barrier (72, 73). However, the physiological and/or pathophysiological roles of BAs in these intestinal processes are still unclear.
Molecular Mechanisms of Bile acid Absorption
The enterohepatic circulation of the bile was first postulated by Moritz Schiff in 1870 (316). In 1936, it was reported that when concentrated bile acid solutions were injected into the small intestine of animals, bile acids were absorbed by the intestine and recovered in portal and systemic blood (185). Over the next decades, the concept of enterohepatic circulation of bile acids became well accepted, as evidence for both passive and active intestinal absorption continued to emerge (101, 209). However, the molecular mechanisms of this absorptive process remained elusive until relatively recently, when bile acid transporters were discovered.
It had long been known that at physiological pH, conjugated BAs are ionized and membrane impermeable. Therefore, their transport across epithelial cells requires membrane-transport proteins. The hepatic NTCP (SLC10A1) facilitates bile acid uptake at the sinusoidal membrane of the hepatocyte, and the BSEP secretes bile acids into the bile canaliculi. In the ileum, this transepithelial transport is mediated by the apical sodium-dependent bile acid transporter (ASBT; SLC10A2) at the apical membrane and the OSTα/β heterodimer at the basolateral membrane (Figure 3). This process of intestinal bile acid absorption is crucial for maintaining bile acid homeostasis and enterohepatic circulation.
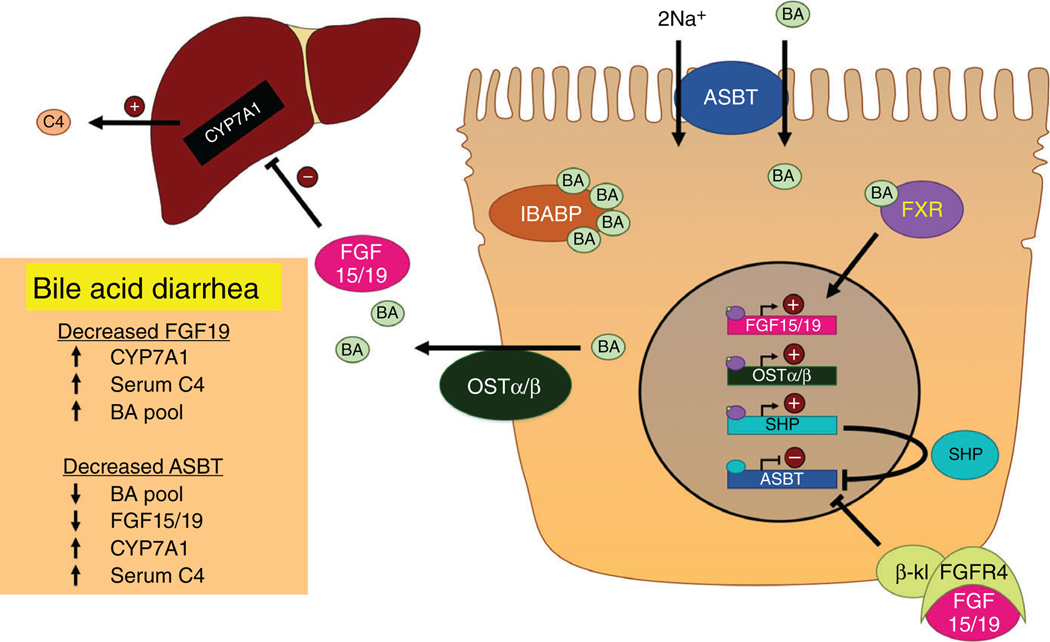
Figure 3.
Bile acid transport and signaling in the ileum. Luminal BAs are taken up into ileal enterocytes by the apical sodium-dependent bile acid transporter (ASBT), where they are bound by the ileal bile acid-binding protein (IBABP) and transported to the basolateral membrane. BAs are exported basolaterally by the organic solute transporter α/β heterodimer (OSTα/β) and returned to the liver. Intracellular BAs activate the farnesoid X receptor (FXR), which heterodimerizes with the retinoid X receptor (RXR) and translocates to the nucleus. There, FXR transactivates fibroblast growth factor 15/19 (FGF15/19), OSTα/β, and the small heterodimeric partner (SHP). ASBT expression is negatively regulated by SHP within the cell and by an autocrine/paracrine function of FGF15/19, which binds to the FGF receptor 4 (FGFR4)/β-klotho (β-kl) complex and activates signaling processes that repress ASBT. Bile acid diarrhea can be caused by disruptions in these processes. Notably, reduced FGF19 disrupts the negative feedback of bile acid synthesis resulting in bile acid overproduction, while reduced ASBT impairs bile acid absorption.
In addition to their active uptake by ASBT, BAs with a high enough pKa are able to passively diffuse through membranes. Unlike active absorption, passive absorption can occur in all regions of the small and large intestines. This primarily relates to unconjugated BAs, which have a high enough pKa(~6) that they are able to undergo nonionic passive diffusion through membranes (101, 209). Hepatic conjugation to taurine and glycine decreases the pKa of BAs, rendering them charged and approximately 10-fold less permeable to membranes (209, 333). On the other hand, colonic bacteria deconjugate and dehydroxylate BAs, increasing the pKa and allowing bile acids to passively diffuse into the portal circulation (257). Several lines of evidence, however, clearly demonstrate that ASBT and OSTα/β are critical for the intestinal absorption and enterohepatic circulation of bile acids.
Apical sodium-dependent bile acid transporter (ASBT)
The apical sodium-dependent bile acid transporter (ASBT) is responsible for the uptake of BAs from the intestinal lumen into enterocytes and is the rate-limiting enzyme for intestinal BA absorption. Like NTCP, ASBT is a sodium-dependent bile acid cotransporter in the SLC10A gene family.
The presence of an Na+-dependent bile acid transporter in the ileum was confirmed in 1992 in brush border membrane vesicles from pig (273) and rabbit (215) ileum. Much of the early work uncovering the role of ASBT was performed in the laboratory of Paul Dawson, who first cloned ASBT in 1994 from a hamster ileum cDNA library (422). ASBT was later identified in the rat (87, 343, 344), human (92, 285, 423, 424), mouse (219, 325), and chicken (277) ileum. These early studies also showed ASBT expression in the cells of the proximal renal tubule (87, 92, 344) as well as cholangiocytes (9, 81, 226). Recently, ASBT expression in the brain has been shown, although the functional relevance is as yet unknown (281). Within the adult intestine of humans and rodents, ASBT is most highly expressed in villus cells of the distal ileum, with negligible expression in the colon (12, 168, 357, 415). The distribution of ASBT expression along the intestine mirrors the conjugated bile acid uptake capacity, highlighting the essential role of this transporter in bile acid absorption (357).
A great deal of evidence has supported the importance of ASBT in intestinal bile acid absorption. For instance, inactivating mutations in ASBT have been linked to some cases of primary bile acid malabsorption (PBAM), a serious condition that presents with steatorrhea and loss of bile acids in the stool (285). Additionally, mice with targeted deletion of the Slc10a2 gene display similar symptoms, with steatorrhea, loss of bile acids in the stool, decreased bile acid pool size, and enhanced bile acid synthesis (95).
ASBT and cholesterol metabolism
Because bile acid synthesis from cholesterol is responsible for approximately 50% of cholesterol elimination, ASBT-dependent recycling of bile acids is intimately related to cholesterol homeostasis. It has been known for decades that disrupting the intestinal absorption of bile acids leads to loss of bile acids in the stool and a concomitant increase in hepatic uptake of cholesterol to increase the synthesis of bile acids (108, 134). In this regard, it is interesting to note that patients with PBAM have low plasma low-density lipoprotein (LDL) cholesterol levels, suggesting that loss of bile acids in the stool is protective against hypercholesterolemia (285). Indeed, ASBT knockout mice also show resistance to diet-induced and genetic models of hypercholesterolemia (94, 220).
Given that genetic ablation of ASBT reduces serum cholesterol, it is not surprising that small molecule inhibitors of ASBT have been shown to have similar antihyperlipidemic effects. Indeed, several ASBT inhibitors have been developed by pharmaceutical companies (42, 78, 201, 318). Although most are in trials for other indications (see below), they have been shown to enhance bile acid synthesis and reduce plasma cholesterol levels in several rodents (201, 235, 318, 418), ApoE knockout mice (42), pigs (171), monkeys (201), and humans (78, 348).
Gene structure and variants
The SLC10A2 gene is located on chromosome 13 in humans (NCBI Gene ID #6555) and chromosome 8 in mice (NCBI Gene ID #20494) (219, 424). In humans, mature ASBT mRNA consists of 6 exons comprising a 1044-nucleotide (nt) coding region, a 597-nt 5′-untranslated region, and a 2135-nt 3′-untranslated region. One splice variant has been reported, coding for a 154-aa bile acid efflux protein of unknown physiological significance that has been called t-ASBT (227).
Several genetic variants of ASBT have been reported, including some that result in altered protein function (216). The first reported variant was a single-nucleotide polymorphism (SNP) resulting in a P290S mutant that abolished BA transport activity (423). Later, L243P and T262M variants that abolished transport activity were reported in a family with PBAM, along with a A171S variant that did not affect transport (285). Screening approaches have since been used to identify and characterize other variants in Korean (291), German (314), and American (158) populations.
Protein structure and function
ASBT is a 348-aa transmembrane (TM) protein with an extracellular glycosylated N-terminus and a cytoplasmic C-terminus. The presence of a bile acid-binding protein with an apparent molecular weight of 43 kDa, which would later be identified as ASBT, was first discovered in 1983 by photoaffinity labeling of ileal BBMVs (210). Studies in the early 1990s similarly found a bile acid-binding protein of similar molecular weight (or a ~90 kDa dimer) and noted that it was responsible for the Na+-dependent bile acids (213–215, 242, 243, 343, 347). Once ASBT was cloned, it was determined that the protein has a molecular weight of approximately 38 kDa, but a higher apparent molecular weight as a result of its glycosylation (423). Electrophysiological studies showed that ASBT is electrogenic with a 2:1 Na+: BA stoichiometry (416,417).
With respect to the structure of ASBT, the precise topology has been a matter of debate, in large part because the crystal structure of vertebrate ASBT has yet to be determined. Although initial predictions suggested that ASBT comprised seven TM domains (422), several lines of evidence supported a 9-TM model. For instance, membrane insertion scanning provided support for a 9-TM model (147), and a recent crystal structure of bacterial ASBT homologues revealed a structure consistent with 9-TM ASBT model (169,441). However, strong evidence from dual label epitope insertion scanning mutagenesis and other methods refutes the 9-TM model in favor of a 7-TM model (33, 436).
Several laboratories, particularly that of Peter Swaan, have devoted a great deal of effort into elucidating not only the membrane topology of ASBT, but also the functional importance of the individual domains of this protein. The functions of individual domains discussed below assume a 7-TM model of ASBT (Figure 4). Several of the TM domains are crucial for ASBT transporter function. TM domain 1 is important for bile acid and Na+ binding and translocation and also confers stability to the protein (93). TM domain 2 appears to be most important for Na+ translocation via coordination with the Na+ ion (322). The portion of TM domain 3 closest to the cytosol forms the route for bile acids as they exit ASBT and enter the cytosol (174). Additionally, the cytosolic half of TM domain 4 is another important part of the translocation pathway (197). TM domains 5 and 6 possess GxxxG motifs that are important for ASBT helix-helix interaction, conferring stability to the proteins without being directly involved with substrate translocation (270). TM domain 6 also appears important for Na+-binding and translocation, while TM domain 7 is crucial for bile acid binding and translocation, as both of these domains line the translocation pathway (139, 173, 176, 212).
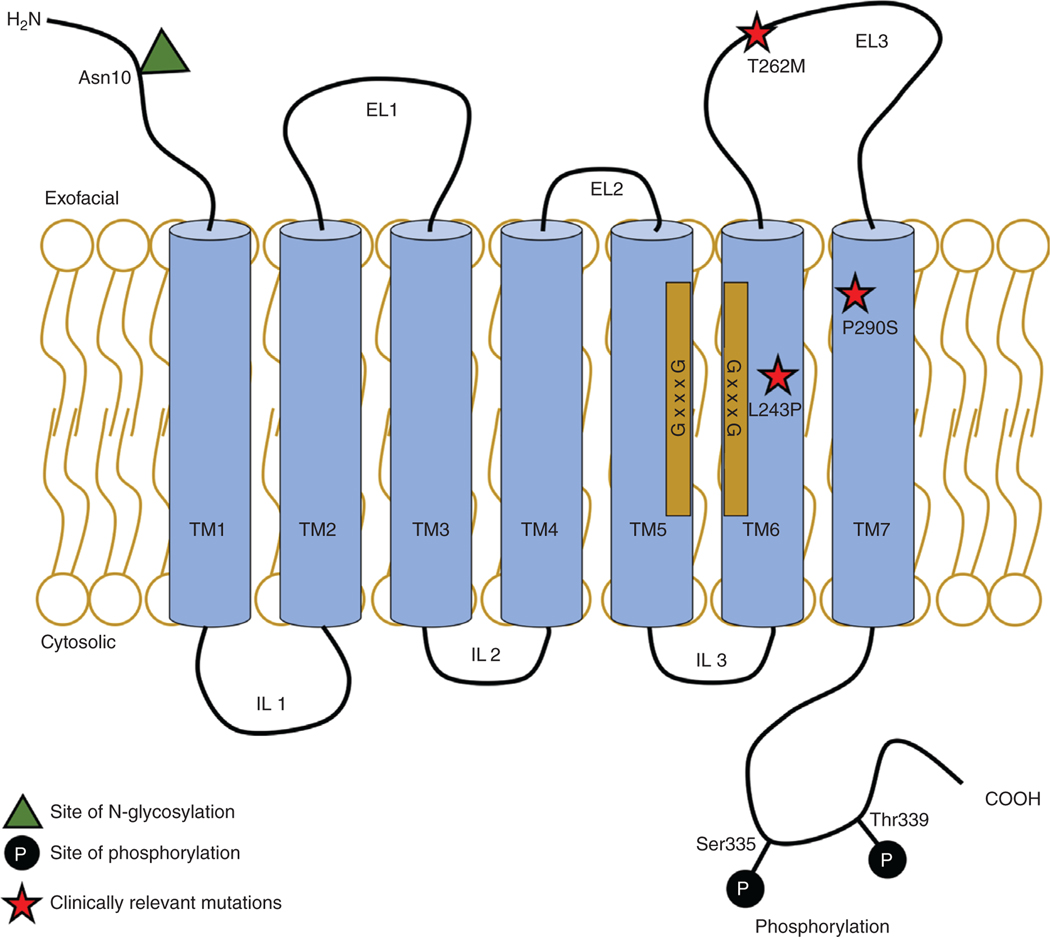
Figure 4.
Topology of ASBT. A cartoon of the 7-transmembrane ASBT topology with transmembrane (TM), extracellular loop (EL), and intracellular loop (IL) domains. Sites of posttranslational modifications (glycosylation and phosphorylation), clinically relevant missense mutations, and GxxxG motifs are shown.
The soluble loops of ASBT have also been shown to be critical for protein function. A pair of aspartic acid residues (Asp120 and Asp122) on extracellular loop (EL) 1 are critical for Na+ -binding, and Asp124 appears to be important for binding to the 7α-OH of many bile acids (175). In addition, several other residues in this highly conserved loop are crucial for protein expression, localization, and substrate affinity (326, 328). EL 3 is also important for transporter activity via its electrostatic interactions with bile acids, which may represent the initial bile acid-binding interaction site (31). Furthermore, a truncated mutant of ASBT revealed the importance of 14 amino acids in the C-terminal cytoplasmic tail in apical localization of the protein (367, 370).
Of particular interest is the important role of ASBT cysteine residues. Several studies have probed the 13 cysteine residues of ASBT and noted a number of these residues that are both highly conserved between ASBT and NTCP, and are crucial for transport function, trafficking, and stability (32, 82, 324, 327, 368). Notably, mutation of the highly conserved Cys51, Cys105, Cys132, and Cys255 to Ala or Thr nearly abolishes [3H]-TC uptake (32). Mutations at other sites, such as Cys69 or Cys314, somewhat reduce [3H]-TC uptake. In an effort to understand the mechanisms underlying the role of cysteine residues, a recent study concluded that cysteine residues in ASBT are likely not important for its oligomerization (82). However, the precise role of many of these important residues requires further study.
Substrate specificity of ASBT
Bile acids are the only known endogenous substrates for ASBT. While ASBT is known to have a narrower substrate specificity than NTCP, it is clear that ASBT is able to efficiently transport a wide variety of bile acids, both conjugated and unconjugated (92, 205, 244). ASBT does not appear to have a strong preference for conjugated compared to unconjugated bile acids (92). However, CA and its conjugated derivatives appear to have a lower affinity (~ 15–35 μM) than CDCA and its derivatives (~3–10 μM) (92, 244, 325, 327, 377, 422). TLCA appears to be a high-affinity substrate of ASBT (244), while DCA species have an intermediate affinity between CA and CDCA (175, 422). Thus, the structural differences in bile acids determine their affinity for ASBT.
Because ASBT is an intestinal transporter that absorbs relatively large molecules, a great deal of work has been devoted to using this transport system to aid in the absorption of drugs via bile acid-conjugated prodrugs (151, 346). Several groups have demonstrated that bile acids conjugated to synthetic molecules are substrates for ASBT and other bile acid transporters, such as NTCP (25, 246, 346, 377). For example, studies have shown that the antiviral acyclovir conjugate to CDCA (acyclovir valylchenodeoxycholate) has a favorable affinity to ASBT and enhanced the oral availability of acyclovir by twofold in rats (385). Indeed, the precise structural requirements of ASBT have been meticulously elucidated. 3D-quantitative structure-activity relationship studies demonstrated that anionic side chains at the 24-C position are preferred to neutral side chains (34, 440). Additionally, 7-C conjugates are not efficiently transported by ASBT, although the presence of a 7α-OH moiety is not necessary for transport (205). Conversely, conjugates at the 3-C position (such as to naproxen and chlorambucil) are actively transported by ASBT (34, 205). These studies provided important details with respect to the ASBT substrate-binding pocket and the substrate specificity of the transporter.
Interestingly, recent evidence has indicated an alternative transport process that ASBT may use to transport large molecules into the cell (6,7). When bile acids were conjugated to a macromolecule and applied to cells expressing ASBT, vesicular endocytosis was induced and both ASBT and the macromolecule were internalized. This alternative, receptor-mediated ASBT uptake could be useful for designing drug delivery systems that target bile acid absorption pathways.
ASBT inhibitors as therapeutics
Because bile acid signaling is closely tied to lipid and glucose homeostasis, inflammatory pathways, and colonic electrolyte secretion, it is not surprising that inhibitors of ASBT are of interest in a wide variety of intestinal and liver diseases (4). Currently, there are several ASBT inhibitors in development, though none have been approved by the FDA as of this writing. In addition to their potential for lowering plasma LDL cholesterol (see above), these small molecule inhibitors have shown promise in preclinical and clinical studies for the treatment of chronic constipation (65, 78, 276, 348), NASH (289, 310), type 2 diabetes mellitus (71, 249, 379, 426), and cholestatic pruritus (24,153, 264). It will be interesting to see how these promising drug candidates fare in future clinical trials.
Given that modulating ASBT may be useful for the treatment of several disorders, it is important to understand the cellular mechanisms underlying its regulation. In the following sections, the regulation of ASBT at transcriptional, post-transcriptional, and posttranslational levels will be discussed.
Transcriptional regulation of ASBT
Transcription of ASBT mRNA is tightly regulated to maintain proper expression of this important transporter (97). The basal mRNA expression of ASBT is controlled by several important transcription factors, most notably the hepatocyte nuclear factor 1α (HNF1α). Mice with deletion of the Tcf1 gene, which codes for HNF1α, completely lack ASBT expression in the ileum and kidney (342). The minimal ASBT promoter contains three HNF1α-binding sites, which have been shown to bind the ASBT promoter and stimulate its activity as judged by electrophoretic mobility shift assays (EMSA) and luciferase reporter assays, respectively (187). Furthermore, intestinal deletion of the sirtuin 1 (SIRT1) metabolic sensor decreases HNF1α dimerization and function, with a concomitant reduction in ASBT mRNA expression (193).
The ASBT promoter also possesses two activated protein-1 (AP-1) response elements that are responsive to the c-Jun and c-Fos transcription factors (67). The upstream AP-1 site was shown to bind a c-Jun homodimer, and the downstream site was shown to bind a c-Jun/c-Fos heterodimer (69). Promoter studies revealed that c-Jun activates the ASBT promoter, while c-Fos leads to repression of the promoter (278). In agreement with this model, c-Fos knockout mice display enhanced ileal ASBT expression (278). An unidentified component of human serum was also shown to stimulate ASBT mRNA expression in a manner dependent on the AP-1 response elements (104).
The expression of ASBT is thought to be regulated through a negative feedback mechanism whereby the presence of bile acids leads to a reduction in ASBT expression. This process occurs by several distinct mechanisms. The first mechanism appears to be an autocrine/paracrine mechanism that is initiated by FGF15/19, which is a major gene target of FXR (105, 135, 350). Much like the signaling pathway in hepatocytes, FGF15/19 binds to the FGFR4/β-klotho receptor complex. This leads to MEK1/2 activation, c-Jun and c-Fos phosphorylation, and translocation to the nucleus, which represses ASBT transcription. FXR-dependent feedback inhibition also involves three other transcription factors: the LRH-1, the fetoprotein transcription factor (FTF), and the retinoic acid receptor/retinoid X receptor heterodimer (RAR/RXR). LRH-1 is an important positive regulator of ASBT basal expression, as evidenced by the finding that LRH-1 knockout mice show reduced ASBT expression (230). Similarly, the LRH-1 orthologue FTF induces ASBT expression via direct binding to the ASBT promoter in rabbits and human Caco2 cells (233, 290). It appears that LRH-1 may be more important for ASBT expression in mice, while FTF may serve the same role in humans (290). Additionally, RAR/RXR contributes to ASBT basal expression through binding to retinoic acid response elements (279). These transcription factors appear to be responsible for a negative feedback regulation of ASBT, whereby intracellular bile acids in the epithelium reduce ASBT expression. This important regulatory mechanism involves activation of FXR by cytosolic bile acids, which induces expression of the SHP, an inhibitory factor. SHP impairs the activity of LRH-1 and FTF, reducing ASBT transcription (68, 233, 290). Similarly, SHP impairs RAR/RXR activity, also leading to a decrease in ASBT mRNA expression (279).
It is important to note that this negative feedback regulatory mechanism is a species-dependent phenomenon. As described above, it appears that in mice, rabbits, and humans, iFXR activation reduces ASBT expression. In rats, however, it appears that CA induces ASBT mRNA and protein expression (359). It is possible that this relates to the lack of a gall bladder in rats. Still, it will be important to further illuminate the mechanisms by which bile acids regulate ASBT transcription.
In addition to bile acids, ASBT transcription is affected by other steroid-like molecules, including cholesterol, glucocorticoids, and vitamin D. It has been shown that cholesterol downregulates ASBT expression and function in several model systems. In Caco2 cells and mouse models, cholesterol or 25-hydroxycholesterol administration led to a reduction in ASBT mRNA via a mechanism dependent on HNF1α and the sterol regulatory binding element protein 2 (SREBP2) and independent of FXR (10, 265, 376). SREBP2, which translocates to the nucleus in low cholesterol conditions, was shown to transactivate ASBT (376). In agreement with these data, administration of the cholesterol synthesis inhibitor atorvastatin in mice led to a reduction in ileal ASBT mRNA expression (401). The glucocorticoid receptor (GR) has also been shown to bind and activate the ASBT promoter. In vitro promoter studies, in vivo studies in mice and rats, and clinical work showed that glucocorticoids induce ASBT transcription through GR binding to glucocorticoid response elements in the ASBT promoter, and increase intestinal bile acid absorption (90, 186, 283, 287, 428). Similarly, the activation of the VDR was shown to transactivate ASBT (75, 84, 85).
In addition to regulation by endogenous and exogenous steroids and sterols, ASBT transcription is affected by pathological processes such as inflammation. Inflammatory processes have been shown to cause a significant reduction in ASBT expression, in part through a transcriptional mechanism. In rats with indomethacin-induced ileitis and in human Crohn’s disease (CD) patients, ASBT mRNA expression has been shown to be reduced. Further studies showed that the pro-inflammatory cytokines interleukin-1β (IL-1β) and tumor necrosis factor (TNF) increase phosphorylation and nuclear translocation of c-Fos, leading to repression of the ASBT promoter (69, 278).
There are several other transcription factors that have been shown to regulate ASBT, though these are less well studied than those discussed above. Positive regulators include the PPARα (187), the nuclear factor-E2-related factor 2 (NRF2) (414), caudal-type homeobox proteins CDX1 and CDX2 (252), and the constitutive androstane receptor (CAR) (77). Additionally, the GATA4 transcription factor was found to be a negative regulator of ASBT expression, as GATA4 knockout resulted in increased ASBT mRNA expression (41). Interestingly, it has been shown that intestinal bacteria reduce ASBT expression partially via a mechanism dependent on GATA4 (288). It has also been shown that insulin is a negative regulator of the ASBT promoter, although the molecular mechanisms are not fully understood (15). These transcriptional regulators of ASBT are crucial for maintaining proper ASBT expression and bile acid homeostasis (Table 2).
Table 2.
Transcriptional Regulators of ASBT
| Transcription factor | ASBT transcription | References |
|---|---|---|
| HNF1 α | ↑ | (187, 193, 342) |
| c-Jun | ↑ | (67, 278) |
| c-Fos | ↓ | (67, 278) |
| FXR | ↓ (Indirect) | (105, 135, 350) |
| LRH-1 | ↑ | (230) |
| FTF | ↑ | (233, 290) |
| RAR/RXR | ↑ | (279) |
| SHP | ↓ | (68, 233, 279, 290) |
| SREBP2 | ↑ | (10, 265, 376) |
| GR | ↑ | (90, 186, 283, 287, 428) |
| VDR | ↑ | (75, 84, 85) |
| PPARα | ↑ | (187) |
| NRF2 | ↑ | (414) |
| CDX1, CDX2 | ↑ | (252) |
| CAR | ↑ | (77) |
| GATA4 | ↓ | (41, 288) |
Posttranscriptional regulation of ASBT
Although much is known about the transcriptional regulation of ASBT, relatively little research has investigated its post-transcriptional regulation. Of the few studies that have been performed in this area, most have related to the role of the 3′-untranslated region of ASBT mRNA. The Shneider Laboratory showed that a metastasis-associated gene 1 (MTA1) element within the 3′-UTR interacts with the RNA-binding proteins Hu antigen R (HuR) and tristetraprolin (70, 352). They showed that HuR stabilized reporter mRNA, while tristetraprolin decreased reporter mRNA half-life. Interestingly, the overall effect of the 3′-UTR appeared to destabilize mRNA in two intestinal epithelial cell lines, which may be important for the rapid regulation of ASBT. This important posttranscriptional regulation appears to be critical in the ontogenesis of ASBT expression, as discussed in the section titled “ASBT ontogeny and aging.”
Posttranslational regulation of ASBT
ASBT has been shown to be subject to rapid, posttranslational modifications (PTMs). It is possible that these acute regulatory mechanisms represent adaptive responses to a rapidly changing intestinal milieu. ASBT protein stability, subcellular localization, and activity have all been shown to be modulated by PTMs such as ubiquitination, glycosylation, and phosphorylation, as well as by interactions with lipids and plasma membrane microdomains.
One of the key posttranslational mechanisms by which ASBT is regulated is through modulation of protein stability and degradation. It has been shown that ASBT is degraded by the ubiquitin-proteasome pathway in both cholangiocytes and ileocytes (267, 427). Interestingly, ASBT turnover is fairly rapid, as the protein has a relatively short half-life of approximately 6 h (274, 427). Several mechanisms have been shown to modulate the rate of ASBT proteasomal degradation. For instance, the pro-inflammatory cytokine IL-1β enhances ASBT ubiquitination and degradation via a mechanism that involves JNK-dependent phosphorylation of ASBT at Ser335 and Thr339 (427). Furthermore, feeding with CA, but not conjugated TC or TDC, decreased ASBT protein expression via a ubiquitin-dependent mechanism (267). Similarly, the SIRT1 activator resveratrol was shown to promote ASBT ubiquitination and proteasomal degradation, independent of Ser335/Thr339 phosphorylation (83).
ASBT is also subject to N-glycosylation at its extracellular N-terminal domain (274, 370). This glycosylation was shown to enhance ASBT protein stability, as inhibition of N-glycosylation by tunicamycin or mutation of the Asn10 glycosylation site resulted in shorter protein half-life (274). It has been found that ASBT protein expression is higher in rats with streptozotocin-induced diabetes (15). This may be partially the result of enhanced glycosylation, as in vitro glucose treatment promoted mature glycosylation and increased protein stability.
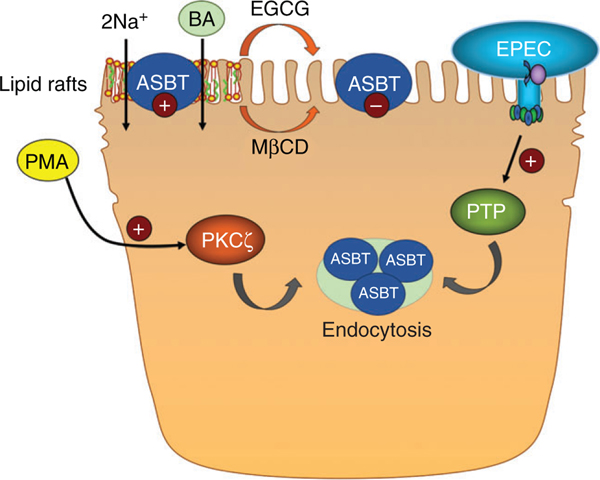
Phosphorylation of ASBT has also been shown to rapidly modulate ASBT function. As discussed above, Ser335/Thr339 phosphorylation was shown to promote ASBT degradation (427). In addition, activation of the protein kinase C ζ (PKCζ) pathway significantly inhibits [3H]-TC uptake within 60 min of treatment with the PKC activator phorbol 12-myristate 13-acetate (PMA) (329). These studies demonstrated that treatment with PMA led to internalization of ASBT and decreased surface expression. Conversely, tyrosine phosphorylation of ASBT was shown to enhance transporter activity, although the kinase and phosphorylation site have yet to be determined (16, 377). Interestingly, enteropathogenic Escherichia coli (EPEC) was shown to rapidly reduce ASBT function via an effector molecule that led to tyrosine dephosphorylation and internalization of ASBT (16). Future studies should focus on further clarifying the role of phosphorylation in the regulation of ASBT function. With respect to the modulation of ASBT by enteric bacteria, an additional report showed that reducing Enterobacteria in the gut by treatment with ampicillin resulted in a significant increase in ASBT protein levels concomitant with an increase in the levels of bile acids in the portal blood (266). The increase in ASBT was not associated with changes in the mRNA. Since the increase occurred in response to eliminating the bacteria by antibiotics, the authors concluded that Enterobacteria suppress ASBT expression via posttranscriptional mechanisms. The studies with EPEC and Enterobacteria provide strong evidence showing that ASBT protein expression and its level on the plasma membrane is sensitive to the enteric bacteria. Further investigations are warranted to determine the significance of these findings in the relation to the development of infections and to understand the interactions between gut microbiota and intestinal absorption of bile acids.
Another critical posttranslational regulatory mechanism involves the association of ASBT with plasma membrane lipid raft microdomains. Studies have demonstrated that ASBT is associated with lipid rafts, and disruption of this association reduces ASBT function (14, 17). For instance, depletion of plasma membrane cholesterol by methyl-β-cyclodextrin led to a shift of ASBT to nonraft domains of plasma membrane and was associated with reduced ASBT function without a change in its surface expression (17). Similarly, the green tea catechin (−)-epigallocatechin-3-gallate (EGCG) rapidly reduces ASBT function and is associated with a shift in ASBT into the nonraft domain of plasma membrane (14). It is clear that these rapid posttranslational regulatory mechanisms are crucial to allow ASBT to respond to environmental changes, and additional studies in this area are warranted.
Given that the apical sodium-dependent bile acid transporter is rapidly regulated by a variety of posttranslational mechanisms, it is important to recognize the techniques used to study these acute functional changes. Conventional methods to assess the function of ASBT, which rely on measuring the uptake of radiolabeled bile acids, provide a measurement of transporter function only at a single time point. These methods are likely insufficient to study dynamic, posttranslational changes in ASBT activity, necessitating the development of real-time methods. Several recent approaches have been developed to quantify the aspects of bile acid physiology in -real time (76, 115, 263, 311, 389, 392, 397–399). Additionally, a recent bioluminescence-based approach relying on a CA-luciferin-conjugated probe has shown promising results for measuring real-time ASBT activity in vitro and in isolated intestinal epithelial cells (377). Further development of these crucial methods will allow for a more comprehensive understanding of the acute regulation of ASBT and other bile acid transporters. Such in vivo real-time measurement of ASBT function could also be useful to examine the potential recruitment of ASBT to the proximal part of the ileum as a mechanism to increase bile acid absorption suggested by previous studies (159) rather than an increase in the amount of ASBT protein in the distal ileum.
ASBT ontogeny and aging
The expression of ASBT shows distinct changes throughout an individual’s lifespan. The newborn ileum shows minimal expression of ASBT, followed by a dramatic increase in expression after weaning. It appears that hormonal changes are responsible for ASBT ontogenic development. It has been demonstrated that the thyroxine hormone is critical for the induction of ASBT mRNA expression after weaning (177, 269). Additionally, the ontogeny of ASBT may be at least partially explained by its posttranscriptional regulation by RNA-binding proteins. HuR, which has been shown to stabilize ASBT, shows a pattern of ontogenic expression similar to that of ASBT. Conversely, the destabilizing RNA-binding protein tristetraprolin shows the inverse ontogenic pattern (70, 352). Thus, the enhanced expression of ASBT after weaning could be in part the result of altered RNA-binding protein expression. Additionally, the intestinal development of ASBT expression in early life mirrors the expression of the transcription factors c-Jun and c-Fos, which are poorly expressed in mice during the neonatal period and sharply increased after weaning (67). The fact that ASBT expression is low prior to weaning is crucial for intestinal health, as the neonatal intestine is not equipped to respond to intracellular bile acids. Indeed, aberrantly high ASBT expression has been linked to neonatal necrotizing enterocolitis (NEC) (150).
While it is clear that ASBT expression is enhanced after weaning, there is also evidence that aging decreases intestinal bile acid absorption. Studies by Gao et al. demonstrated that 3H-TC uptake across ileum in Ussing chambers was reduced approximately 50% in a cohort of aged ~2-year-old) C57BL/6 mice relative to a younger (<1-year-old) cohort (133). There was a corresponding approximately 50% decrease in ASBT mRNA expression in the older cohort, suggesting that decreased ASBT expression in old age may be responsible for the reduced bile acid absorption in these mice. On this note, a 300-participant clinical study found that serum levels of bile acids decrease with age in men, but not women (129). Although the physiological implications of these findings were not directly investigated in either of these studies, it is reasonable to speculate that reduced intestinal bile acid absorption in older individuals may result in blunted bile acid signaling through FXR and other receptors. Future studies should focus on uncovering these functional consequences as well as the mechanisms underlying this age- and sex-related reduction in ASBT expression.
Besides age, ASBT expression also seems to be enhanced in postpartum individuals. ASBT protein expression and transport function were increased in postpartum rats corresponding to an increase in food intake (271). Interestingly, ASBT mRNA expression was not affected, suggesting a possible posttranscriptional mechanism. Although the mechanism underlying this phenomenon has not been elucidated, it is likely related to the increased energy demand during lactation. It is clear that the changes in ASBT expression that occur throughout the lifespan are physiologically important and warrant further study.
Ileal bile acid-binding protein (IBABP)
The ileal bile acid-binding protein (IBABP; FABP6) is a 14 kDa member of the fatty acid binding protein family of intracellular lipid binding proteins (188). Initially, this protein was identified as gastrotropin, given its ability to promote gastric acid and pepsinogen secretion (98, 388,405). In 1990, a soluble bile acid-binding protein with a molecular weight of 14 kDa was identified in rat ileum by photoaffinity labeling (242). This protein was shown to be the major intracellular carrier for ileal bile acids and was soon shown to be identical to gastrotropin (241, 400). IBABP was cloned from rat ileum in 1994 (136) and was later cloned in human (131, 284) and rabbit (358). The longitudinal expression and ontogenic pattern of IBABP in the intestine mirrors that of ASBT, with high expression in the ileum after weaning and low expression in other regions (137, 146, 177).
While it is clear that IBABP binds the intracellular bile acids in the ileum, the importance of this protein in the absorption of bile acids is a matter of current study. There is evidence that IBABP binds to ASBT, which enhances bile acid transport activity. FABP6 (IBABP) knockout mice do show mild abnormalities in bile acid absorption (305). Intestinal tissues from these mice display approximately 60% reduced transport of [3H]-TC across ileal gut sacs, suggesting a role for IBABP in bile acid absorption. In addition, female mice had a significant increase and males had a trend toward an increase in fecal bile acid excretion, with both sexes showing a trend toward decreased bile acid pool size. Hepatic bile acid synthesis genes Cyp7a1 and Cyp8b1 were significantly lower than wild-type littermates, indicating alterations in bile acid signaling. Furthermore, Li et al. recently demonstrated that feeding with medium-chain fatty acids (MCFAs) reduced IBABP protein expression but not intestinal bile acid transporter expression (234). These mice showed decreased plasma LDL cholesterol levels and a decrease in bile acid absorption, suggesting that the loss of IBABP may contribute to impaired bile acid absorption. However, ASBT function was not measured in these mice, leaving open the possibility that posttranslational mechanisms are responsible for decreased bile acid absorption. In contrast, both whole-body and intestine-specific FXR knockout mice, which also lack appreciable expression of IBABP, do not show defects in bile acid absorption (204, 360). The fact that these mice have a normal bile acid pool size and intestinal reabsorption suggests that while IBABP may enhance intestinal bile acid absorption, it is not essential for the process and even minimal expression may recover the absorptive phenotype.
Other studies have identified a role for IBABP in bile acid sensing and signaling. It could be expected that IBABP might sequester cytosolic bile acids, preventing them from binding receptors such as FXR. However, it appears that IBABP is actually important for enhancing FXR signaling. Two reports have demonstrated that IBABP was necessary for full activation of FXR (112, 275). Additionally, this work provided evidence that IBABP forms a complex with FXR that translocates to the nucleus upon addition of bile acids (275). Thus, while the role of IBABP in intestinal absorption of bile acids is still unclear, it might play a crucial role in bile acid signaling.
Gene structure and variants
The IBABP gene is coded by seven exons on chromosome 5 in humans (Gene ID 2172) (284) and four exons on chromosome 11 in mice (Gene ID 16204) (45). The major IBABP mRNA transcript is approximately 600 bp in length and is spliced from four exons (131, 136, 284). There are two additional transcripts that code for a longer IBABP protein. However, this variant is primarily associated with colorectal cancer and is regulated differently from the major IBABP isoform (103, 111). In this article, only the major isoform of IBABP will be further discussed.
There is only one IBABP mutant that has received clinical attention. A Thr79Met mutation is associated with protection against type 2 diabetes mellitus among obese individual in a case-control study and two cohort studies, although the molecular mechanisms underlying this protection have yet to be investigated (121, 122). While no other clinically significant mutations of IBABP have been reported, it has been demonstrated that gallstone carriers have approximately 65% lower IBABP mRNA and protein expression compared to healthy control patients (40).
Protein structure and function
IBABP is a 128-aa protein with a predicted molecular weight of approximately 14 kDa. Several solution and crystal structures of IBABP have been determined, showing a protein with a β-barrel and two α-helices (114). One of the key structural features of IBABP is that it possesses two internal bile acid-binding sites that act in a positively cooperative manner (114). This phenomenon was first recognized in 1998 when it was found that bile acid photoaffinity labeling was enhanced, rather than reduced, by the presence of natural bile acids (211). Further study in the Cistola Laboratory confirmed that IBABP binds two bile acid molecules with “low intrinsic affinity but an extraordinarily high degree of cooperativity” and proposed mechanistic models of ligand binding (380–384). Similarly, studies in the Molinari Laboratory have helped to define the structural determinants of bile acid binding to IBABP, as well as the liver bile acid-binding protein FABP1 (114, 143, 434). These studies exquisitely describe IBABP cooperativity and ligand selectivity using several complimentary approaches and account for structural features of both the protein and bile acid ligands, proposing a 2:1 bile acid/protein ratio with a multistep binding mechanism. Additional work has identified a putative third bile acid-binding site on the exterior of the protein (387).
Substrate specificity of IBABP
In addition to bile acids, IBABP has been shown to bind some other steroids including progesterone, but has low affinity for fatty acids, suggesting that bile acids are the primary substrates for this protein (136, 442). Structural studies using solution NMR suggest that IBABP is more flexible than the other members of the FABP family, allowing for relatively large bile acid molecules to enter the binding pockets (248). Among bile acids, human IBABP has lower affinity for unconjugated bile acids compared to conjugated and prefers taurine-conjugated BAs to glycine-conjugated, with apparent Km values in the range of 50 to 400 μM (442). Furthermore, the presence of a 7α-OH moiety decreases affinity, while a 12α-OH increases affinity (442). Recent work has demonstrated that human and mouse IBABP bind β-MCAs with low affinity, indicating that the 6β-hydroxylation is not preferred by IBABP. Interestingly, two-dimensional NMR methods helped to identify a preference of certain bile acids for each of the two IBABP binding sites (381). When either GC or GCDC is present individually, both sites are fully occupied. However, in equimolar concentrations, one site is preferentially occupied by GC, while the other is occupied by GCDC (167, 381). This was confirmed with the taurine conjugates TC and TCDC (23). These findings suggest that in addition to allowing for cooperative binding, the two binding pockets of IBABP may have evolved to efficiently bind the wide variety of bile acids that exist in nature. Future studies could further elucidate the importance of these distinct binding sites as it relates to substrate specificity.
Regulation of IBABP
Not surprisingly, the expression of IBABP is regulated by bile acids. This phenomenon has been known since the late 1990s, when it was shown that IBABP expression was induced by human bile and bile acids (61, 189, 190). An FXR response element in the FABP6 promoter was soon identified, supporting the role of FXR signaling in the induction of IBABP (142, 178). Indeed, FXR was shown to be critical for the expression of IBABP, as FXR knockout mice do not express IBABP (204, 349, 360). In humans as well, administration of bile acids induced IBABP mRNA expression (437).
In addition to bile acids, cholesterol can also affect the transcription of IBABP. In Caco2 cells, it was shown that cholesterol depletion resulted in binding of SREBP-1c to a sterol response element located in the FABP6 promoter, enhancing its transcription (433). Similarly, it was shown that the liver X receptor (LXR), which can be activated by oxysterols, binds to the FABP6 FXR response element to activate transcription (224).
Other transcription factors have also been shown to regulate IBABP transcription. A peroxisome proliferator-activated receptor (PPAR) response element was identified within the FABP6 promoter, and treatment with the PPARα-PPARβ/δ agonist bezafibrate induced promoter activity, suggesting a role for these receptors in mediating IBABP expression (225). Additionally, the CDX2 transcription factor, which is highly expressed in the ileum, was shown to bind and activate the FABP6 promoter (35).
Organic solute transporter α/β heterodimer (OSTα/β)
The OSTα/β heterodimer (SLC51A/B) is a plasma membrane glycoprotein that is the basolateral exporter of bile acids out of enterocytes and toward the portal circulation. Prior to the discovery of OSTα/β, several other candidate basolateral bile acid transporters were explored, including t-ASBT (a splice variant of ASBT) (227) and the multidrug-resistance protein 3 (MRP3) (435). OSTα/β was first cloned in 2001 from little skate Raja erinacea liver (409). This work identified an organic anion transporter comprising a predicted 7-TM protein as well as a 1-TM ancillary protein, which would later be identified as OSTα and OSTβ, respectively. After its identification in the skate, OSTα/β was cloned from human and mouse liver, although the mRNA expression for these transporters was found in several other tissues including the small intestine and kidney (339). Further study demonstrated conclusively that OSTα/β localized to the basolateral membrane of ileocytes, renal proximal tubular cells, and cholangiocytes, where it functions as a bile acid export protein (27, 96). In general, it has been shown that OSTα/β expression mirrors that of ASBT, highlighting the intertwined function of these two transporters (27, 96).
The first in vivo studies characterizing OSTα/β were published in 2008 from Paul Dawson’s laboratory (309). These studies reported that OSTα knockout mice had a decreased bile acid pool size secondary to impaired intestinal bile acid absorption, similar to ASBT knockout mice. OSTα knockout mice also display reduced renal and biliary bile acid absorption, further supporting the crucial role of OSTα/β in bile acid absorption (29). However, unlike ASBT knockout mice, OSTα knockout mice displayed a paradoxical decrease in hepatic bile acid synthesis genes such as Cyp7a1. This finding is particularly instructive, as it highlights the role of FGF15/19 in maintaining bile acid homeostasis. Unlike in the case of impaired ASBT, apical bile acid uptake is largely functional in OSTα knockout mice. Thus, luminal bile acids are taken into enterocytes, where they bind to FXR and induce FGF15/19, decreasing bile acid synthesis (309). However, bile acid absorption is still impaired as basolateral export is dysfunctional. This results in a phenotype of decreased bile acid synthesis and approximately 60% decreased pool size, with fecal bile acids in the normal range. As would be expected with decreased bile acid pool size, OSTα knockout mice displayed lipid malabsorption. They were also protected against age-related weight gain and insulin resistance (419). However, some maladaptive changes were seen in the small intestine of OSTα knockout mice. As a result of bile acid accumulation within enterocytes, these mice showed bile acid toxicity, oxidative stress, and morphological changes including decreased villus height and increased proliferation of ileal epithelial cells (117). Further studies showed that OSTα knockout did not protect against atherosclerosis, primarily because of suppressed bile acid synthesis and reduced hepatic cholesterol uptake (94, 220). Expectedly, when OSTα knockout was superimposed on an FXR knockout background, FGF15 feedback was not present and bile acid synthesis, pool size, and fecal excretion increased dramatically (221, 222). These data demonstrate conclusively that OSTα/β is indispensable for intestinal bile acid absorption and maintenance of bile acid homeostasis.
Gene structure and variants
In humans, the OSTα gene (SLC51A) is located on chromosome 3, while the OSTβ gene (SLC51B) is present on chromosome 15. In mice, these genes are located on chromosome 16 and 9 for OSTα and OSTβ, respectively. In both species, OSTα is coded by nine exons and OSTβ is coded by four exons (339).
Until very recently, no deleterious OSTα/β variants had been identified. However, a novel OSTβ frameshift mutation was reported in two related patients with congenital diarrhea and cholestasis (366). This mutation renders the OSTβ nonfunctional and was associated with a dramatic decrease in OSTα protein expression and [3H]-TC uptake activity.
Protein structure and function
OSTα is a 340-aa polypeptide with seven predicted TM domains, an extracellular N-terminus, and a cytosolic C-terminus. In contrast, OSTβ is a 128-aa protein with an extracellular N-terminus, a single TM domain, and a large cytosolic C-terminal domain (339). Unlike ASBT, OSTα/β functions independent of ion gradients (Na+, K+, H+, or Cl−) or ATP, but rather relies on a facilitated diffusion mechanism for transporting its substrates (27, 28, 339).
Dimerization is crucial for the proper expression, localization, and function of OSTα/β. When either OSTα or OSTβ is expressed independently, no transport function is observed for either [3H]-estrone-3-sulfate or [3H]-TC (96, 339, 409). The OSTβ TM domain was shown to be necessary for both dimer formation and transport function (86). Interestingly, deletion of 27 aa from the OSTβ extracellular N-terminal domain or the majority of the cytosolic C-terminal domain (save for two Arg residues near the TM domain) did not dramatically affect [3H]-TC transport (86, 369). However, mutation of the highly conserved TM residues Trp34 and Asn35 to Ala resulted in nearly complete abolishment of transporter activity (86). Additionally, further N-terminal deletion of 35 amino acids did abolish dimerization, which prevented proper membrane trafficking and protein function (371). There is also recent evidence that Leu20 and Leu21 are important for protein trafficking, as LL20/21AA mutation resulted in poor plasma membrane expression (431). It is not yet known why deletion of this region is tolerated while mutation is not, but it may be the result of altered lipophilicity in the LL20/21AA mutant that prevents proper interaction among the OSTβ subunit, OSTα, and the membrane itself.
Substrate specificity of OST
The substrate specificity of OSTα/β is relatively wide, as it is able to transport a range of organic anions and steroid molecules. OSTα/β shows robust transport of bile acids such as TC, as well as conjugated steroids such as estrone-3-sulfate (339). Additionally, this protein has been shown to transport digoxin and prostaglandin E2, supporting its wide substrate specificity. However, some other steroids, including estradiol, are not substrates for OSTα/β (339).
Regulation of OSTα/β
OSTα and OSTβ appear to be regulated in concert, as neither subunit is functional on its own. A large body of evidence supports that FXR is the primary regulator of OSTα/β expression. Endogenous and exogenous FXR agonists transactivate both genes, and FXR response elements are present in the promoters of both subunits (223, 229), Additionally, FXR binding to these promoters has been confirmed by EMSA (16251721). There is important evidence supporting the role of bile acids in regulating OSTα/β expression via FXR. First, bile acid feeding suppresses OSTα/β expression in a manner dependent on FXR, as this effect is not seen in FXR knockout mice (443). Additional studies in human ileal explants confirmed that bile acid treatment induces OSTα/β mRNA and protein expression (437). Second, bile acids were also shown to have a negative regulatory role via FXR-SHP-dependent repression of LRH1, which is another transactivator of OSTα/β genes (127). Thus, bile acids and other FXR agonists both positively and negatively regulate OSTα/β, allowing for careful titration of protein expression. It appears that in most cases, the positive induction predominates, which is expected as OSTα/β serves to protect epithelial cells against bile acid-induced toxicity.
Other transcription factors have been shown to regulate OSTα/β, though they may be less critical than FXR and LRH-1. The LXR was shown to transactivate OSTα/β via binding to response elements shared with FXR (286). Similarly, GR was also shown to transactivate OSTα/β in human and rat ileal tissue (196). Additionally, retinoic acid was shown to transactivate OSTβ via RAR/CAR binding to its promoter. Interestingly, hypoxic conditions were also shown to induce OSTα/β expression in hepatocytes via hypoxia-inducible-factor-1α (HIF-1α) binding to hypoxia response elements within the OSTα and OSTβ promoters (332). While these regulatory mechanisms may be important for OSTα/β expression, the in vivo relevance must be investigated further to better elucidate the roles of these transcription factors.
Pharmacological inhibition of OSTα/β
Pharmacological inhibitors of OSTα/β have not generated as much interest as those for ASBT. A recent report identified clofazimine, a rarely used antibiotic, as a dose-dependent inhibitor of OSTα/β (390). These studies showed that OSTα/β inhibition resulted in increased iFXR activity in vivo, indicating intestine-specific activation of FXR.
Disorders of Bile Acid Absorption
Numerous diseases have elements of dysfunctional intestinal bile acid absorption. In some cases, disruption of intestinal BA absorption is the primary defect, while in other cases BA absorption is not involved in pathogenesis, but its inhibition may be useful for treatment. Here, we discuss several of these disorders, including BAM, NEC, cholestasis, type 2 diabetes mellitus (T2DM), and nonalcoholic liver disease/nonalcoholic steatohepatitis (NAFLD/NASH).
Bile acid diarrhea (BAD)
Interruption of intestinal bile acid absorption results in bile acid accumulation in the colon, leading to watery diarrhea and bile acid loss in the stool. BAD is classified into three types depending on the underlying mechanisms of BAM (55):
Type 1: Secondary to ileal resection, inflammation, or radiation
Type 2: Primary (idiopathic)
Type 3: Secondary to gastrointestinal diseases that affect absorption, such as small intestinal bacterial overgrowth or celiac disease
Clinically, BAD can be diagnosed by several tests that have been extensively discussed elsewhere (48, 396). The 75selenium homotaurocholic acid test (75SeHCAT) uses a synthetic 75Se-labeled BA that is actively absorbed by ASBT in the ileum and is not passively absorbed or metabolized by bacteria. The patient ingests a 75SeHCA capsule, and 75Se gamma-decay is measured in either the abdomen or the stool by a gamma camera. Patients with BAD display decreased 75SeHCA retention and increased excretion. The 75SeHCAT is the definitive test for diagnosing BAD and used in European countries but is not approved in the United States. This has necessitated the development of other clinical tests for the diagnosis of BAM.
Measurement of fecal bile acids, both total and individual species, can also provide insight into bile acid absorption. For instance, many patients with chronic functional diarrhea and diarrhea-prominent irritable bowel syndrome (IBS-D) have altered fecal bile acid composition [31, 32, 396]. In this test, 48 h of stool is collected and homogenized, followed by deconjugation and extraction of bile acids. Total bile acids can be measured by enzymatic assay, and individual species can be quantified by GC-MS or LC-MS. This procedure is technically challenging, making fecal bile acid measurement difficult to perform and largely unavailable in the clinical setting.
Markers of bile acid synthesis can be used as surrogates for bile acid absorption. Serum concentration of 7α-hydroxy-4-cholesten-3-one (C4), an intermediate in the classical bile acid synthesis pathway, is indicative of CYP7A1 activity and bile acid synthesis (330). When bile acids are lost in the stool, serum C4 rapidly increases as cholesterol is converted to intermediates of BA synthesis. Indeed, serum C4 measured by LC-MS is positively correlated with total fecal bile acids, indicating that C4 may be used as a marker for BA loss (58, 395).
Serum FGF19 may be another useful marker for BAM, with the advantage that it can be measured using ELISA-based tests. Impaired BA absorption is expected to decrease FXR activation and FGF19 synthesis. Indeed, serum FGF19 is inversely correlated with serum C4 (232) and positively correlated with SeHCAT retention (295). It is important to note that fasted serum FGF19 appears to be a less reliable metric than the other clinical tests discussed. It has been reported that fasting FGF19 has positive and negative predictive values of just 61% and 82%. However, FGF19 levels following the intake of food supplemented with CDCA may have better predictive power (48). These studies suggest that reduced serum FGF19 is a clinical feature of BAM and may be another appropriate clinical test for diagnosis of BAM. However, more studies will be required to determine the proper testing conditions. Indeed, it may be necessary to measure both C4 and FGF19 to accurately determine the degree of BAM. While FGF19 could be easily measured in humans, the measurement of FGF15 in rodents was found to be challenging for unclear reason (268). This limits the use of preclinical models to evaluate the levels of FGF15 in different physiological and pathological conditions.
Depending on the severity of malabsorption, a large quantity of BAs can pass through the ileum to enter the colon. Supraphysiological concentrations of colonic BAs have several pathogenic effects. Bile acids, particularly hydrophobic BAs, are cytotoxic as a result of their detergent nature (21, 335). Physiologic concentrations of colonic BAs have been shown to stimulate propagating motor waves, accelerating colonic transit and enhancing the urge to defecate (30). Many bile acid species also induce Cl− secretion from colonocytes, resulting in secretory diarrhea, the predominant feature of BAD (55, 262). In cultured T84 colonic cells, unconjugated CDCA and DCA were shown to be to the most potent bile acids in stimulating Cl− secretion (18, 194). The stimulation was much stronger when cells were exposed to bile acids from the basolateral membrane. This observation suggests that luminal bile acids reach the basolateral membrane of colonocytes after a disruption in the epithelial barrier to induce chloride secretion. The sequence of events involved in bile acid-mediated chloride secretion require further investigations.
Type 1 BAD
Type 1 BAD is BAM secondary to gross ileal dysfunction, such as in ileal resection, radiation enteritis, or IBD (particularly CD, as this frequently affects the distal ileum). BA malabsorption in these disorders may be caused by one or a combination of several factors. First, removal or destruction of absorptive cells in the distal ileum results in the loss of ASBT and prevents BA reabsorption. Furthermore, ileal dysfunction may cause impaired FGF19 production and disinhibition of BA synthesis, exacerbating the condition by enhancing bile acid load in the intestine (163). In addition, intestinal transit time decreases in patients with ileal resection (110), potentially leading to insufficient time to adequately absorb BAs. Interestingly, SI transit time is lengthened in many IBD patients who have not had resection (120), suggesting that transit time is unlikely to underlie BAM in these patients.
Type 1 BAD was first recognized in the 1960s when it was discovered that the terminal ileum was responsible for BA reabsorption. It became clear that following ileal resection or ileitis, patients with diarrhea excreted bile acids at a faster rate than individuals with intact ileum (165, 261).
This increased excretion of bile acids can be measured using markers of BAM. Recently, Camilleri et al. (58) demonstrated that 50% of patients with ileal resection or CD in the distal ileum have elevated serum C4, indicating BAM. Similarly, approximately 25% of children in a German cohort of pediatric CD patients had elevated serum C4 (141). In this cohort, 70% with chronic diarrhea had elevated C4, compared to only 15% of those without diarrhea, indicating that diarrhea in these patients may be related to disruptions in BA homeostasis. Interestingly, nearly all CD patients with diarrhea but low disease activity scores displayed elevated serum C4, suggesting that BAM unrelated to inflammation may be the underlying cause of diarrhea in these patients.
In the case of ileal resection, it is easy to understand the mechanism of BAM. Resection reduces or eliminates the absorptive area for bile acids, leading to BAM. However, in the case of inflammatory disease, the molecular mechanisms are considerably more complex. Over the past 15 years, it has become apparent that in the case of CD-associated BAD, reduced ASBT expression is a likely culprit in the pathogenesis.
Numerous studies have demonstrated that ASBT mRNA and protein expression is reduced in the ileum of patients with CD relative to healthy subjects (166, 182, 186, 420). Furthermore, these studies showed that glucocorticoids, which are often used in the treatment of CD, induced ASBT protein expression and promoter activity. Additional work showed that inflammatory cytokines, including IL-1β, decreased ASBT expression dramatically (69, 278). This reduction in ASBT protein expression by inflammatory cytokines was shown to be the result of both reduced ASBT mRNA transcription and enhanced ASBT protein degradation by the ubiquitin-proteasome pathway (427).
Interestingly, ASBT mRNA expression is reduced in both active ileitis and in CD patients in remission, suggesting a potential mechanism for the BAM and diarrhea seen in patients without active disease (182). It will be of particular interest to determine the molecular mechanisms underlying this reduction in ASBT expression among patients in remission, as this may yield new therapeutic targets for the disease.
Another interesting observation was made by Dekaney and colleagues showing the appearance of ASBT, IBABP, and OSTα/β in the colon after ileocecal resection in mice (99). This induction in the expression was absent in germ-free mice indicating dependency on intestinal bacteria. Also, the increase in the expression of these transporters was significantly attenuated in FXR knockout, suggesting a role for FXR transcription factor. The authors suggested that the abnormal expression of bile acid transporters may play an adaptive function protecting the colonic mucosa from the high levels of luminal bile acids as a result of removing the distal ileum. Importantly, these adaptations may partially compensate for the lack of ileal bile acid absorption. These findings underscore the interplay among the intestinal microflora, FXR, and bile acid transporters and warrant detail investigations to determine the molecular mechanism underling these interactions.
Type 2 BAD
Type 2 BAD, also referred to as primary bile acid diarrhea (PBAD), includes a collection of disorders that present as BA malabsorption and diarrhea not attributable to another disease. Remarkably, 25% to 50% of diarrhea-predominant IBS-D display some degree of BAM (22, 351,413). Although the molecular mechanisms underlying most cases of PBAD are still a matter of active investigation, the general causes can be separated into two groups: disorders of impaired BA absorption and disorders of enhanced BA synthesis (Figure 3).
Type 2 BAD can be caused by two mechanisms: impaired bile acid absorption or enhanced bile acid synthesis. As early as the 1970s, it was recognized in case reports that defective ileal bile acid absorption was a likely cause of some cases of congenital diarrhea (155, 156). After the discovery of ASBT, several inactivating mutations in this transporter were reported, including some of clinical relevance (285). Although some patients with inactivating ASBT mutations present with mild chronic diarrhea and no nutritional deficiency, many present in infancy with severe chronic diarrhea, steatorrhea, and fat-soluble vitamin malabsorption. Because of the large amount of bile acids lost in the stool, these patients have low FXR signaling in the gut. Thus, FGF19 production is reduced and bile acid synthesis is greatly increased, as indicated by elevated CYP7A1 expression and serum C4 in these patients (55, 296). Because of the compensatory overproduction of bile acids, plasma cholesterol levels remain low throughout their entire life. These patients respond well to bile acid sequestrants (BASs) such as cholestyramine, which bind bile acids in the colon and limit the severity of the diarrhea.
It has been postulated that accelerated small intestinal transit may allow bile acids to escape ileal absorption via ASBT and enhance BA leakage into the colon. It has been shown that patients with PBAD have accelerated small bowel transit time (323). This seems unlikely to underlie BAD pathogenesis given that ASBT is a high-affinity transporter with Km values in the low micromolar range. It seems more likely that accelerated transit time is an effect of BAD, rather than a cause. It should be stated that several recent studies have identified SNPs in the bile acid receptor GBPAR1 that reduce responsiveness to BAs and are associated with altered small and large bowel transit times among IBS-D patients (59, 60). However, these polymorphisms were found among IBS-D patients both with and without BA malabsorption, indicating that there may be no direct relationship between GPBAR1 SNPs and type 2 BAD.
It is also important to consider the potential of other ileal BA transporters, particularly OSTα/β, to contribute to PBAD. Although intestinal OSTα/β is essential for absorption of bile acids, this transporter did not appear to be of clinical relevance in BAM until quite recently. Just one instance of a clinically important OSTβ mutation has been reported in humans (366). This mutation resulted in a truncated OSTβ mutant associated with low OSTα protein expression. The patients (two family members) with this mutation presented with PBAM associated with mild cholestatic liver disease. However, many questions remain unanswered with respect to the clinical relevance of OSTα/β polymorphisms. Interestingly, OSTα knockout mice display reduced bile acid absorption that is counterintuitively accompanied by reduced BA synthesis (309). In these mice, bile acids are transported from the gut lumen into enterocytes by ASBT, where they can activate FXR-FGF15 signaling. Thus, bile acid synthesis is reduced in these animals despite BA malabsorption. It is possible that this reduction in BA synthesis is protective against the severe phenotype associated with BAM in the ASBT knockout model, a finding that may be of interest in the development of therapeutics. Nevertheless, alterations in OSTα/β expression and function have also been implicated in several other diseases of the enterohepatic system. For instance, OSTα/β is induced in cholestatic disease (49), likely as a result of FXR dysregulation. Additionally, OSTα/β is suppressed in CD (282), although the cause or importance of this finding has yet to be uncovered.
Although the most severe cases of type 2 BAD are typically the result of mutations in ASBT, the vast majority of patients with PBAD do not have defects in ileal BA absorption (393). Although the precise mechanisms remain unclear, it has recently been recognized that hepatic overproduction of BAs is likely responsible for many of these cases. Despite having no appreciable defect in ileal BA absorption, many PBAD patients have reduced serum FGF19 and enhanced BA synthesis (increased serum C4) (184,404, 421). It is possible that defective FGF19 synthesis or secretion in these patients leads to disinhibition of hepatic BA synthesis in the liver and increased BA secretion into the intestine. The resulting increase in ileal BA load overcomes the absorptive capacity of ASBT, allowing BAs to leak into the colon.
It is also possible that defects in FGF19 signaling play a role in this disorder. Recent studies from the Camilleri laboratory have provided extensive knowledge regarding potential genetic causes of enhanced BA synthesis in type 2 BAD. This group has identified gene variants that predispose patients to PBAD in FGFR4 and β-klotho, the FGF19 hepatic receptor and coreceptor, respectively (57, 59, 184).
Treatment of BAD
The staple of treatment for BAD has been cholestyramine, a bile acid-binding resin. These resins bind and sequester bile acids in the gut lumen, preventing them from stimulating secretion in the colon. In 1969, Alan Hofmann first reported that cholestyramine treatment relieved diarrhea secondary to ileal resection (now known as type 1 BAD) (165). This treatment modality is similarly effective at relieving BAM symptoms in other types of BAD (124, 320, 413). A systematic meta-analysis of 15 adult-onset type 2 BAD studies demonstrated cholestyramine response rates of 96% in severe disease (75SeHCAT 7-day retention <5%), 80% in moderate disease (75SeHCAT <10%), and 70% in mild disease (75SeHCAT <15%) (413). Nevertheless, cholestyramine is often poorly tolerated by patients. Indeed, in a small trial, nearly half of patients discontinued cholestyramine due to side effects or poor compliance, despite the efficacy of the treatment (320).
Colesevelam is a newer generation bile acid-binding resin that is generally much better tolerated than cholestyramine and may be useful for treating BAD (307). In a randomized, placebo-controlled trial of CD patients with BAD, approximately 65% of colesevelam-treated patients showed a greater than 30% reduction in liquid stools/day (the primary endpoint of this study) compared to on approximately 20% of patients receiving a placebo (p = 0.036) (37). Of the 15 patients receiving colesevelam, 5 had adverse effects potentially related to the drug. However, these events resolved without stopping treatment in all but 1 patient, who eventually discontinued the drug. This study indicates that colesevelam may be an effective treatment option with fewer side effects and improved compliance compared to cholestyramine.
It is critical to note that while bile acid-binding resins can effectively relieve the symptoms of BAD, they do not address the underlying causes of malabsorption. In fact, because sequestered bile acids are inaccessible to ASBT, there is an overall increase in bile acids lost in the stool (56). This results in decreased iFXR activation, leading to further reduction in FGF19 expression and a concomitant increase in hepatic bile acid synthesis from cholesterol (this is the fundamental reason for their usefulness in treating hypercholesterolemia). Indeed, cholestyramine has been shown in humans to reduce serum FGF19 and increase serum C4 (56, 207, 250). It is somewhat concerning that bile acids are likely unable to participate in their vast intestinal signaling functions in the presence of binding resins. Thus, it may be useful to develop new treatments for BAD that do not interfere with BA signaling.
In this regard, three novel potential BAD therapies have been identified. First, a novel colonic-release formulation of cholestyramine was recently tested in a small trial (12 healthy individuals and 19 patients with BAD) (19). In this study, the colonic-release formulation improved clinical symptoms but did not decrease serum FGF19 or increase serum C4, in contrast to the traditional formulation. Larger trials are necessary, but this is a promising improved formulation of a drug that has been used successfully for decades in the treatment of BAD.
Another recent clinical study used hydroxypropyl cellulose (HPC), a common excipient and food additive, as a placebo control in a trial for the use of cholestyramine in type 2 BAD (116). However, it became evident that HPC was also somewhat effective in treating BAD, although not as effective as cholestyramine. HPC has previously been shown to adsorb bile acids, suggesting a possible mechanism for this effect (256, 302, 386). Interestingly, HPC does not appear to increase serum C4, indicating that it may allow for some retention of bile acid signaling while also limiting their effect on colonic secretion (53).
Another promising potential therapy for BAD is the semisynthetic FXR agonist OCA (6α-ethyl CDCA). A recent proof of concept study was conducted with 10 patients with type 1, 10 patients with type 2 BAM, and 7 control patients with idiopathic chronic diarrhea (403). Patients were treated with OCA for 2 weeks. Following treatment with OCA, fasting serum FGF19 was significantly increased in patients with type 2 BAD and trended toward an increase in type 1 and control patients. Similarly, serum C4 was decreased in type 2 patients and control patients and trended toward an increase in type 1 BAD patients. Clinically, type 2 BAD patients showed improvements in weekly stool number and stool consistency. Interestingly, while type 1 patients did not display the same magnitude of improvement in these measures, a subgroup with ileal resection did respond quite well to OCA therapy. Control idiopathic diarrhea patients did not show improvement in clinical outcomes with OCA, indicating that this treatment is specific for disorders of BAM. It appears likely that OCA acts to decrease BA synthesis via the FXR-FGF19-CYP7A1 axis, reducing BA secretion into the intestine. Because the bile acids that are present in the lumen are presumably able to engage in their signaling roles, this treatment option may compare favorably to bile acid-binding resins with respect to side effects and patient compliance. Notably, there were only a few minor adverse events reported in this study, and none that resulted in discontinuation of OCA. Future studies will be valuable in establishing the efficacy and safety of this promising drug for the treatment of BAD.
It is crucial to note that a serious barrier currently exists in the treatment of BAD, particularly in the United States. Because there are no approved diagnostic tests for BAD available in the United States, it is difficult to establish a set of concrete diagnostic criteria for these diseases. It is likely that this has contributed to a great number of missed diagnoses and has limited the available treatment options. Thus, the development and approval of new diagnostic tests will ultimately be critical for effective treatment of BAD.
Necrotizing enterocolitis (NEC)
NEC is a common disease among premature infants that is characterized by severe hemorrhage and inflammatory necrosis in the distal ileum and proximal colon (149). It was noted that increased levels of hepatic pro-inflammatory cytokines were associated with exacerbated intestinal inflammation in a rat model of NEC (148). Given that the liver is the site for bile acid synthesis and the intestinal segment expressing ASBT (distal ileum) is the site predominantly affected by NEC, Halpern and colleagues postulated that disturbances in bile acid homeostasis may play a critical role in the development of NEC (149). In the initial studies, this group showed that the luminal level of bile acids and the expression of ASBT were increased in an animal model of NEC (149). The sequestration of bile acids in this NEC model caused a significant reduction in the severity of NEC. In subsequent studies, Halpern and colleagues provided convincing evidence demonstrating that ASBT-deficient mice were protected against the development of NEC and that ASBT inhibitors significantly reduced the severity of the damage in a rat model of NEC (150). Since the proteins involved in the key processes involved in bile acid homeostasis are not mature in newborns (26, 157, 177, 344, 363–365), the increase in ASBT leads to intracellular accumulation and toxic levels of bile acids in the ileal enterocytes, causing intestinal damage and contributing to the pathology of NEC. These studies also provide evidence for the therapeutic effects of ASBT inhibitors as potential intervention to treat NEC. Beside the treatment of NEC, it is also preferable to identify a marker that predicts the development of NEC in premature neonates. A recent study has shown that fecal levels of unconjugated bile acids were significantly higher before the development of NEC as compared to their levels in neonates that did not develop NEC (172). Collectively, these studies underscore the central role of bile acids in general and ileal ASBT in particular in the pathogenesis of NEC. Understanding the mechanisms by which disturbances in bile acid homeostasis lead to NEC will unravel therapeutic targets and establish biomarkers for predicting the occurrence of this disease in premature infants.
Cholestasis
The obstruction of bile flow from the liver, known as cholestasis, leads to the accumulation of toxic biliary compounds, causing liver damage. The clinical features of cholestatic liver diseases are mainly jaundice and pruritus (353). Cholestasis can be categorized as either intrahepatic or posthepatic, depending on the location of the obstruction. Cholangiopathies, including primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC), represent examples of posthepatic cholestasis. Since bile acid accumulation in hepatocytes is associated with bile acid-induced toxicity, FXR agonists that reduce bile acid synthesis have proven beneficial for the treatment of these diseases. Recent studies showed that OCA significantly reduced cholestatic liver injury in animal models of cholestasis and in human patients (119, 208, 280, 299). Indeed, OCA was recently approved by the FDA for use in PBC (to be used for PBC patients who are not responding to UDCA) and is currently in phase II trials for PSC. However, side effects including increased pruritus and decreased levels of serum HDL cholesterol may pose a limitation for this approach. Blocking intestinal bile acid absorption to reduce bile acid load into the liver has also shown promising results in the treatment of cholangiopathies. Studies showed that ASBT inhibitors reduced liver injury in the multidrug-resistance protein 2 (MDR2)-deficient mouse model of cholestasis (24, 264). These inhibitors have been shown to significantly reduce pruritus among PBC patients (5, 154, 345). It is possible that combination therapies that include ASBT inhibitors will provide additional benefits while minimizing side effects such as pruritus.
Progressive familial intrahepatic cholestasis (PFIC) is a group of inherited disorders with clinical manifestations which often appear in late adolescence but may be recognized as early as infancy (353). Among these disorders, PFIC type 1 has been shown to occur due to mutations in the ATP8B1 gene, which encodes for P-type adenosine triphosphatase (FIC1 protein) that functions as aminophospholoipid flippase (126, 203). Mice harboring a homozygous, PFIC1-causing mutation exhibited an increase in intestinal bile acid absorption, suggesting that bile acid overload from the intestine may contribute to the associated cholestasis (298, 340). More extensive investigations showed that FIC1 loss-of-function leads to a decrease in FXR activity (66). Since FXR is a negative regulator of ASBT, this reduction in FXR activity causes an increase in ASBT expression. Using an siRNA-mediated approach, Chen et al. showed that the knockout of ATP8B1 in intestinal epithelial Caco2 cells caused an increase in ASBT expression and promoter activity (66). They concluded that in the presence of loss-of-function mutations in ATP8B1, ASBT expression is increased. Recent studies, however, have challenged this conclusion. In this regard, van der Mark and colleagues showed that the loss of ATP8B1 in intestinal Caco2 cells did not alter ASBT expression but rather reduced its levels on the apical membrane (391). In contrast to the studies of Chen et al., these studies were performed using stable transfection of shRNA targeting FIC1, providing a more robust and chronic knockout. The decrease in ASBT surface expression implies a reduction in bile acid absorption that may explain the diarrhea associated with PFIC1. One important conclusion from these seemingly contradictory results is that cellular ASBT expression and membrane localization is sensitive to the loss of ATP8B1 function and may contribute to pathophysiology of cholestasis or diarrhea associated with PFIC1. Further careful in vivo studies are warranted to examine these regulatory mechanisms and to determine the exact roles of ASBT in PFIC.
Metabolic disorders
Disruptions in intestinal bile acid absorption have also been implicated in metabolic disorders, such as diabetes mellitus and nonalcoholic liver disease (NAFLD). As has been described earlier, the postprandial increase in luminal bile acids results in enhanced iFXR signaling, which controls many aspects of glucose and lipid homeostasis. As this signaling process requires cytosolic bile acids to bind to FXR, BASs and small molecule inhibitors of ASBT have emerged for their potential to modulate iFXR signaling.
Diabetes mellitus
In the United States, diabetes mellitus affects nearly 10% of the population, costing $245 billion and causing more than 250,000 deaths annually (64). While there are a large number of therapies available, the health burden of this disease warrants the investigation of novel therapeutics.
In this regard, BASs were first developed as LDL-cholesterol-lowering agents, but have recently gained attention for their ability to improve hyperglycemia in T2DM patients. It has been shown that T2DM patients have slightly altered bile acid homeostasis compared to healthy individuals (2, 39, 51, 52, 239). While it is not clear whether changes in bile acid homeostasis are important for the pathogenesis of T2DM, it is clear that targeting BA pathways are useful for the treatment of this disease (354). The BAS colesevelam was FDA approved in 2008 for the treatment of diabetes, and in 2011, a meta-analysis confirmed that the use of colesevelam improved glycemic control in T2DM patients (152). However, the mechanisms of colesevelam’s effects of glucose homeostasis are not yet fully understood. It may be dependent on alterations in iFXR signaling, hepatic FXR signaling, GPBAR1 signaling, or a combination of these. It has been demonstrated that colesevelam and other BASs increase plasma levels of the incretin hormone GLP-1, whose release is positively regulated by the GPBAR1 receptor (354, 372). This could be attributed to colonic GPBAR1 activation by bile acids, despite still being bound to BASs.
Changes in bile acid homeostasis were suggested to contribute to the beneficial effects of bile acids in improving glucose metabolism in obese patients with T2DM (3). In this regard, the levels of bile acids were shown to be increased in individuals who had undergone a Roux-en-Y gastric bypass surgery (294). In light of their roles in stimulating energy expenditure in the body, this increase indicates a possible role for bile acids in decreasing body weight by the surgery. Additional interesting findings also supported the roles of bile acids in mediating the effects of bariatric surgery including the observation that bile diversion operations in mice were associated with increased insulin sensitivity, weight loss, and remarkable decrease in hepatic steatosis (123). Future studies are needed to determine the mechanisms involved in mediating the effects of bile acids on glucose metabolism in the preclinical models of bariatric surgery before extrapolating the results to humans.
Inhibitors of ASBT have also gained interest for the treatment of T2DM (71, 379,426). ASBT inhibition by luminally restricted inhibitors has proved effective in lowering plasma glucose in preclinical models, likely via a mechanism similar to that seen with BASs (71, 426). In rodent models, plasma GLP-1 and insulin were increased, which correlated with improved glycemic control. A phase I trial showed modest benefits for T2DM even at relatively low dosage, suggesting that ASBT inhibitors may be of clinical benefit in this disease. In future studies it will be important not only to show clinical benefit, but also to delineate the mechanisms underlying the improved glycemic control when ileal bile acid absorption is inhibited.
Nonalcoholic liver disease (NAFLD)
NAFLD is a disorder that is highly prevalent in individuals with obesity and T2DM and is thus of growing concern worldwide (300). In fact, more than 25% of the world’s population is affected by this disease. Our knowledge with respect to pathogenesis of NAFLD is constantly evolving, but it is thought to represent a progressive spectrum of liver disease beginning with simple steatosis. In a subset of this population (~10%–15%), steatosis progresses to NASH, which is characterized by inflammatory infiltration and can progress to liver fibrosis, cirrhosis, and hepatocellular carcinoma (38, 128, 432). As of this writing, there are no FDA approved agents for the treatment of NAFLD or NASH, though there are a number of them being tested in II and III clinical trials.
Because bile acids are important modulators of lipid metabolism, glucose homeostasis, and energy expenditure, it is understandable that targeting bile acid pathways might be attractive for treating NALFD. Indeed, several experimental drugs for NAFLD and NASH target these pathways. The majority of BA-targeting drugs being explored for NAFLD/NASH are FXR agonists, although there is some preclinical evidence that intestine-specific FXR antagonists may have some benefit in these disorders (138, 183). Other experimental medications target the GPBAR1 or a dual FXR/GPBAR1 agonists. These classes of drugs have been extensively reviewed elsewhere and are beyond the scope of this article (8, 20, 63, 80). However, we briefly discuss the potential for inhibitors of bile acid absorption in the treatment of the NAFLD/NASH. Unlike diabetes, it appears that BASs may be of limited utility for NASH. Preclinical studies in rodent models have shown that colesevelam improves steatosis and hepatic inflammation (260). However, a small clinical study in NASH patients showed that colesevelam had little effect on relevant outcomes compared to placebo and may even increase hepatic steatosis (228). Finally, it should be noted that ASBT knockout mice were shown to be protected against development of diet-induced hepatic steatosis (310). Since the lack of ASBT is expected to reduce the activation of iFXR, this observation is consistent with the notion that intestine-specific FXR antagonism is beneficial in treating NASH. Indeed, ASBT inhibitors may represent a promising therapeutic as they have been shown to be successful in a preclinical NASH model (310) and a phase I trial (379).
Conclusion
The field of bile acids has been flourishing as researchers have begun to unravel the roles of bile acids as signaling molecules. The discovery of the involvement of bile acid receptors TGR5 and FXR in regulating energy expenditure, lipid metabolism, and glucose homeostasis ignited an interest in understanding the roles of bile acids in metabolic disease. It is clear that receptors in the GI tract are central to the systemic effects of bile acids. There is also convincing evidence that bile acid absorption is crucial in dictating the intestinal, hepatic, and extra-enterohepatic effects of bile acid. Interventions targeting intestinal bile acid absorption and receptors have shown promising therapeutic effects for common diseases such as fatty liver disorders and diabetes. The complex nature of the processes involved in bile acid handling and absorption warrants additional detailed investigations to better understand the roles of the intestine in controlling bile acid homeostasis and signaling. Further investigation is still required, for example, to delineate the detailed interactions between gut microbiota and intestinal bile acid absorption. Understanding the exact roles of intestine-specific agonists and antagonists of bile acid receptors in altering bile acid homeostasis and other functions remains a fruitful area of investigation. Also, a more complete understanding of the posttranscriptional modifications of proteins responsible for bile acid absorption such as ASBT is needed. Utilizing system-based approaches such as profiling of gut microbial communities, as well as proteomics and metabolomics approaches, should enrich our understanding of the intestinal handing of bile acids. Such knowledge will increase our ability to design intestine-specific strategies to alter bile acid homeostasis and treat diseases such as BAM, diabetes mellitus, fatty liver diseases, and hypercholesterolemia.
Figure 5.
Acute, posttranslational regulation of ASBT. Functional ASBT, which transports two Na+ ions along with one bile acid, is present in lipid rafts. Movement of ASBT out of rafts, either by the green tea catechin (−)-epigallocatechin-3-gallate (EGCG) or methyl-β-cyclodextrin (MβCD), decreases ASBT function. Activation of protein kinase C ζ (PKCζ) by phorbol 12-myristate 13-acetate (PMA) results in endocytosis and reduced function of ASBT. Similarly, activation of protein tyrosine phosphatases (PTP) by enteropathogenic Escherichia coli (EPEC) causes endocytosis of ASBT.
Didactic Synopsis.
Major Teaching Points
The enterohepatic circulation of bile acids entails synthesis and secretion from the liver, reabsorption from the intestine, recirculation back to liver via the portal blood, and resecretion from the liver.
Bile acids are efficiently reabsorbed in the intestine with only 5% lost in the feces.
Bile acid absorption is critical for the negative feedback inhibition of their synthesis in the liver via the secretion of ileal fibroblast growth factor FGF19/15 hormone.
Bile acids are crucial for lipid digestion and absorption due to their roles as fat solubilizers. Bile acids also activate specific receptors and signaling pathways to modulate a number of hepatic and intestinal functions and exert systemic effects such as increasing energy expenditure and improving insulin sensitivity.
Disturbances in the enterohepatic circulation of bile acids contribute to the pathophysiology of hepatic and intestinal disorders such as cholestatic liver diseases and inflammatory bowel disorders.
Acknowledgements
The work in the authors’ laboratories is supported by merit review grants from the Department of Veterans Affairs (VA): BX000152 (WAA) and BX002011 (PKD); F30 grant DK117535 (ALT); and R01 grants: DK109709 (WAA), DK54016 (PKD), DK81858 (PKD), DK92441 (PKD), and DK98170 (RKG), from NIDDK/NIH. WAA is supported by a VA Research Career Scientist Award. PKD is also supported by a VA Senior Research Career Scientist Award.
Footnotes
Related Articles
Intestinal transport of bile acids
Bile acid metabolism and signaling
Bile formation and secretion
Bile proteome in health and disease
References
- 1.Abrahamsson A, Gustafsson U, Ellis E, Nilsson L-M, Sahlin S, Björkhem I, Einarsson C. Feedback regulation of bile acid synthesis in human liver: Importance of HNF-4alpha for regulation of CYP7A1. Biochem Biophys Res Commun 330: 395–399, 2005. [DOI] [PubMed] [Google Scholar]
- 2.Abrams JJ, Ginsberg H, Grundy SM. Metabolism of cholesterol and plasma triglycerides in nonketotic diabetes mellitus. Diabetes 31: 903–910, 1982. [DOI] [PubMed] [Google Scholar]
- 3.Albaugh VL, Banan B, Ajouz H, Abumrad NN, Flynn CR. Bile acids and bariatric surgery. Mol Aspects Med 56: 75–89, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Dury S, Marschall H-U. Ileal bile acid transporter inhibition for the treatment of chronic constipation, cholestatic pruritus, and NASH. Front Pharmacol 9:931,2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Dury S, Wahlström A, Wahlin S, Langedijk J, Elferink RO, Ståhlman M, Marschall H-U. Pilot study with IBAT inhibitor A4250 for the treatment of cholestatic pruritus in primary biliary cholangitis. Sci Rep 8:6658,2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Hilal TA, Chung SW, Alam F, Park J, Lee KE, Jeon H, Kim K, Kwon IC, Kim I-S, Kim SY, Byun Y. Functional transformations of bile acid transporters induced by high-affinity macromolecules. Sci Reep 4:4163,2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hilal TA, Park J, Alam F, Chung SW, Park JW, Kim K, Kwon IC, Kim I-S, Kim SY, Byun Y. Oligomeric bile acid-mediated oral delivery of low molecular weight heparin. J Control Release 175: 17–24, 2014. [DOI] [PubMed] [Google Scholar]
- 8.Ali AH, Carey EJ, Lindor KD. Recent advances in the development of farnesoid X receptor agonists. Ann Transl Med 3: 5, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alpini G, Glaser SS, Rodgers R, Phinizy JL, Robertson WE, Lasater J, Caligiuri A, Tretjak Z, LeSage GD. Functional expression of the apical Na+-dependent bile acid transporter in large but not small rat cholangiocytes. Gastroenterology 113: 1734–1740, 1997. [DOI] [PubMed] [Google Scholar]
- 10.Alrefai WA, Sarwar Z, Tyagi S, Saksena S, Dudeja PK, Gill RK. Cholesterol modulates human intestinal sodium-dependent bile acid transporter. Am J Physiol Gastrointest Liver Physiol 288: G978–G985, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem 276: 28857–28865, 2001. [DOI] [PubMed] [Google Scholar]
- 12.Anderle P, Sengstag T, Mutch DM, Rumbo M, Praz V, Mansourian R, Delorenzi M, Williamson G, Roberts M-A. Changes in the transcriptional profile of transporters in the intestine along the anterior-posterior and crypt-villus axes. BMC Genomics 6: 69, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson S, Davis DL, Dahlbäck H, Jörnvall H, Russell DW. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem 264: 8222–8229, 1989. [PubMed] [Google Scholar]
- 14.Annaba F, Kumar P, Dudeja AK, Saksena S, Gill RK, Alrefai WA. Green tea catechin EGCG inhibits ileal apical sodium bile acid transporter ASBT. Am J Physiol Gastrointest Liver Physiol 298: G467–G473, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Annaba F, Ma K, Kumar P, Dudeja AK, Kineman RD, Shneider BL, Saksena S, Gill RK, Alrefai WA. Ileal apical Na+-dependent bile acid transporter ASBT is upregulated in rats with diabetes mellitus induced by low doses of streptozotocin. Am J Physiol Gastrointest Liver Physiol 299: G898–G906, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Annaba F, Sarwar Z, Gill RK, Ghosh A, Saksena S, Borthakur A, Hecht GA, Dudeja PK, Alrefai WA. Enteropathogenic Escherichia coli inhibits ileal sodium-dependent bile acid transporter ASBT. Am J Physiol Gastrointest Liver Physiol 302: G1216–G1222, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Annaba F, Sarwar Z, Kumar P, Saksena S, Turner JR, Dudeja PK, Gill RK, Alrefai WA. Modulation of ileal bile acid transporter (ASBT) activity by depletion of plasma membrane cholesterol: Association with lipid rafts. Am J Physiol Gastrointest Liver Physiol 294: G489–G497, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ao M, Sarathy J, Domingue J, Alrefai WA, Rao MC. Chenodeoxycholic acid stimulates Cl(–) secretion via cAMP signaling and increases cystic fibrosis transmembrane conductance regulator phosphorylation in T84 cells. Am J Physiol Cell Physiol 305: C447–C456, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appleby RN, Bajor A, Gillberg P-G, Graffner H, Simrén M, Ung KA, Walters J. Effects of conventional and a novel colonic-release bile acid sequestrant, A3384, on fibroblast growth factor 19 and bile acid metabolism in healthy volunteers and patients with bile acid diarrhoea. United European Gastroenterol J 5: 380–388, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arab JP, Karpen SJ, Dawson PA, Arrese M, Trauner M. Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology 65: 350–362, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araki Y, Fujiyama Y, Andoh A, Nakamura F, Shimada M, Takaya H, Bamba T. Hydrophilic and hydrophobic bile acids exhibit different cytotoxicities through cytolysis, interleukin-8 synthesis and apoptosis in the intestinal epithelial cell lines. IEC-6 and Caco-2 cells. Scand J Gastroenterol 36: 533–539, 2001. [DOI] [PubMed] [Google Scholar]
- 22.Aziz I, Mumtaz S, Bholah H, Chowdhury FU, Sanders DS, Ford AC. High prevalence of idiopathic bile acid diarrhea among patients with diarrhea-predominant irritable Bowel syndrome based on Rome III criteria. Clin Gastroenterol Hepatol 13: 1650.e2–1655.e2, 2015. [DOI] [PubMed] [Google Scholar]
- 23.Badiee M, Tochtrop GP. Bile acid recognition by mouse ileal bile acid binding protein. ACS Chem Biol 12: 3049–3056, 2017. [DOI] [PubMed] [Google Scholar]
- 24.Baghdasaryan A, Fuchs CD, Österreicher CH, Lemberger UJ, Halil-basic E, Påhlman I, Graffner H, Krones E, Fickert P, Wahlström A, Ståhlman M, Paumgartner G, Marschall H-U, Trauner M. Inhibition of intestinal bile acid absorption improves cholestatic liver and bile duct injury in a mouse model of sclerosing cholangitis. J Hepatol 64: 674–681,2016. [DOI] [PubMed] [Google Scholar]
- 25.Balakrishnan A, Polli JE. Apical sodium dependent bile acid transporter (ASBT, SLC10A2): Apotential prodrug target. Mol Pharm 3:223–230, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balistreri WF, Heubi JE, Suchy FJ. Immaturity of the enterohepatic circulation in early life: Factors predisposing to “physiologic” maldigestion and cholestasis. J Pediatr Gastroenterol Nutr 2: 346–354, 1983. [PubMed] [Google Scholar]
- 27.Ballatori N, Christian WV, Lee JY, Dawson PA, Soroka CJ, Boyer JL, Madejczyk MS, Li N. OSTalpha-OSTbeta: A major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology 42: 1270–1279, 2005. [DOI] [PubMed] [Google Scholar]
- 28.Ballatori N, Christian WV, Wheeler SG, Hammond CL. The heteromeric organic solute transporter, OSTα-OSTβ/SLC51: A transporter for steroid-derived molecules. Mol Aspects Med 34: 683–692, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballatori N, Fang F, Christian WV, Li N, Hammond CL. Ostalpha-Ostbeta is required for bile acid and conjugated steroid disposition in the intestine, kidney, and liver. Am J Physiol Gastrointest Liver Physiol 295: G179–G186, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bampton PA, Dinning PG, Kennedy ML, Lubowski DZ, Cook IJ. The proximal colonic motor response to rectal mechanical and chemical stimulation. Am J Physiol Gastrointest Liver Physiol 282: G443–G449, 2002. [DOI] [PubMed] [Google Scholar]
- 31.Banerjee A, Hussainzada N, Khandelwal A, Swaan PW. Electrostatic and potential cation-pi forces may guide the interaction of extracellular loop III with Na+ and bile acids for human apical Na+-dependent bile acid transporter. Biochem J 410: 391–400, 2008. [DOI] [PubMed] [Google Scholar]
- 32.Banerjee A, Ray A, Chang C, Swaan PW. Site-directed mutagenesis and use of bile acid-MTS conjugates to probe the role of cysteines in the human apical sodium-dependent bile acid transporter (SLC10A2). Biochemistry 44: 8908–8917, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Banerjee A, Swaan PW. Membrane topology of human ASBT (SLC10A2) determined by dual label epitope insertion scanning mutagenesis. New evidence for seven transmembrane domains. Biochemistry 45: 943–953, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baringhaus KH, Matter H, Stengelin S, Kramer W. Substrate specificity of the ileal and the hepatic Na(+)/bile acid cotransporters of the rabbit. II. A reliable 3D QSAR pharmacophore model for the ileal Na(+)/bile acid cotransporter. J Lipid Res 40: 2158–2168, 1999. [PubMed] [Google Scholar]
- 35.Barley NF, Taylor V, Shaw-Smith CJ, Chakravarty P, Howard A, Legon S, Walters JRF. Human ileal bile acid-binding protein promoter and the effects of CDX2. Biochim Biophys Acta 1630: 138–143, 2003. [DOI] [PubMed] [Google Scholar]
- 36.Bauer TM, Steinbrückner B, Brinkmann FE, Ditzen AK, Schwacha H, Aponte JJ, Pelz K, Kist M, Blum HE. Small intestinal bacterial overgrowth in patients with cirrhosis: Prevalence and relation with spontaneous bacterial peritonitis. Am J Gastroenterol 96: 2962–2967, 2001. [DOI] [PubMed] [Google Scholar]
- 37.Beigel F, Teich N, Howaldt S, Lammert F, Maul J, Breiteneicher S, Rust C, Göke B, Brand S, Ochsenkühn T. Colesevelam for the treatment of bile acid malabsorption-associated diarrhea in patients with Crohn’s disease: A randomized, double-blind, placebo-controlled study. J Crohns Colitis 8: 1471–1479, 2014. [DOI] [PubMed] [Google Scholar]
- 38.Benedict M, Zhang X. Non-alcoholic fatty liver disease: An expanded review. World J Hepatol 9: 715–732, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennion LJ, Grundy SM. Effects of diabetes mellitus on cholesterol metabolism in man. N Engl J Med 296: 1365–1371, 1977. [DOI] [PubMed] [Google Scholar]
- 40.Bergheim I. Apical sodium bile acid transporter and ileal lipid binding protein in gallstone carriers. J Lipid Res 47: 42–50, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Beuling E, Kerkhof IM, Nicksa GA, Giuffrida MJ, Haywood J, aan de Kerk DJ, Piaseckyj CM, Pu WT, Buchmiller TL, Dawson PA, Krasinski SD. Conditional Gata4 deletion in mice induces bile acid absorption in the proximal small intestine. Gut 59: 888–895, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhat BG, Rapp SR, Beaudry JA, Napawan N, Butteiger DN, Hall KA, Null CL, Luo Y, Keller BT. Inhibition of ileal bile acid transport and reduced atherosclerosis in apoE−/− mice by SC-435. J Lipid Res 44: 1614–1621,2003. [DOI] [PubMed] [Google Scholar]
- 43.Biagioli M, Carino A, Cipriani S, Francisci D, Marchianò S, Scarpelli P, Sorcini D, Zampella A, Fiorucci S. The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J Immunol 199: 718–733, 2017. [DOI] [PubMed] [Google Scholar]
- 44.Binder HJ, Filburn B, Floch M. Bile acid inhibition of intestinal anaerobic organisms. Am J Clin Nutr 28: 119–125, 1975. [DOI] [PubMed] [Google Scholar]
- 45.Birkenmeier EH, Rowe LB, Crossman MW, Gordon JI. Ileal lipid-binding protein (Illbp) gene maps to mouse chromosome 11. Mamm Genome 5: 805–806, 1994. [DOI] [PubMed] [Google Scholar]
- 46.Björkhem I, Araya Z, Rudling M, Angelin B, Einarsson C, Wikvall K. Differences in the regulation of the classical and the alternative pathway for bile acid synthesis in human liver. No coordinate regulation of CYP7A1 and CYP27A1. J Biol Chem 277: 26804–26807, 2002. [DOI] [PubMed] [Google Scholar]
- 47.Bjursell M, Wedin M, Admyre T, Hermansson M, Böttcher G, Görans-son M, Lindén D, Bamberg K, Oscarsson J, Bohlooly-Y M. Ageing Fxr deficient mice develop increased energy expenditure, improved glucose control and liver damage resembling NASH. PLoS One 8: e64721, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borup C, Wildt S, Rumessen JJ, Bouchelouche PN, Graff J, Damgaard M, McQuitty C, Rainteau D, Munck LK. Chenodeoxycholic acid stimulated fibroblast growth factor 19 response - a potential biochemical test for bile acid diarrhoea. Aliment Pharmacol Ther 45: 1433–1442, 2017. [DOI] [PubMed] [Google Scholar]
- 49.Boyer JL, Trauner M, Mennone A, Soroka CJ, Cai S-Y, Moustafa T, Zollner G, Lee JY, Ballatori N. Upregulation of a basolateral FXR-dependent bile acid efflux transporter OSTalpha-OSTbeta in cholestasis in humans and rodents. Am J Physiol Gastrointest Liver Physiol 290: G1124–G1130, 2006. [DOI] [PubMed] [Google Scholar]
- 50.Broeders EPM, Nascimento EBM, Havekes B, Brans B, Roumans KHM, Tailleux A, Schaart G, Kouach M, Charton J, Deprez B, Bouvy ND, Mottaghy F, Staels B, van Marken Lichtenbelt WD, Schrauwen P. The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab 22: 418–426, 2015. [DOI] [PubMed] [Google Scholar]
- 51.Brufau G, Bahr MJ, Staels B, Claudel T, Ockenga J, Böker KH, Murphy EJ, Prado K, Stellaard F, Manns MP, Kuipers F, Tietge UJ. Plasma bile acids are not associated with energy metabolism in humans. Nutr Metab (Lond) 7:73,2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brufau G, Stellaard F, Prado K, Bloks VW, Jonkers E, Boverhof R, Kuipers F, Murphy EJ. Improved glycemic control with colesevelam treatment in patients with type 2 diabetes is not directly associated with changes in bile acid metabolism. Hepatology 52: 1455–1464, 2010. [DOI] [PubMed] [Google Scholar]
- 53.Brydon WG, Walters JRF, Ghosh S, Culbert P. Letter: Hydroxypropyl cellulose as therapy for chronic diarrhoea in patients with bile acid malabsorption - possible mechanisms. Aliment Pharmacol Ther 44: 306–307, 2016. [DOI] [PubMed] [Google Scholar]
- 54.Calmus Y, Guechot J, Podevin P, Bonnefis MT, Giboudeau J, Poupon R. Differential effects of chenodeoxycholic and ursodeoxycholic acids on interleukin 1, interleukin 6 and tumor necrosis factor-alpha production by monocytes. Hepatology 16: 719–723, 1992. [DOI] [PubMed] [Google Scholar]
- 55.Camilleri M. Bile acid diarrhea: Prevalence, pathogenesis, and therapy. Gut Liver 9: 332–339, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Camilleri M, Acosta A, Busciglio I, Boldingh A, Dyer RB, Zinsmeister AR, Lueke A, Gray A, Donato LJ. Effect of colesevelam on faecal bile acids and bowel functions in diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther 41: 438–448, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camilleri M, Klee EW, Shin A, Carlson P, Li Y, Grover M, Zinsmeister AR. Irritable bowel syndrome-diarrhea: Characterization of genotype by exome sequencing, and phenotypes of bile acid synthesis and colonic transit. Am J Physiol Gastrointest Liver Physiol 306: G13–G26, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camilleri M, Nadeau A, Tremaine WJ, Lamsam J, Burton D, Odunsi S, Sweetser S, Singh R. Measurement of serum 7alpha-hydroxy-4-cholesten-3-one (or 7alphaC4), a surrogate test for bile acid malabsorption in health, ileal disease and irritable bowel syndrome using liquid chromatography-tandem mass spectrometry. Neurogastroenterol Motil 21: 734–e43, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Camilleri M, Shin A, Busciglio I, Carlson P, Acosta A, Bharucha AE, Burton D, Lamsam J, Lueke A, Donato LJ, Zinsmeister AR. Genetic variation in GPBAR1 predisposes to quantitative changes in colonic transit and bile acid excretion. Am J Physiol Gastrointest Liver Physiol 307: G508–G516, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camilleri M, Vazquez-Roque MI, Carlson P, Burton D, Wong BS, Zinsmeister AR. Association of bile acid receptor TGR5 variation and transit in health and lower functional gastrointestinal disorders. Neurogastroenterol Motil 23: 995–999, e458, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campana G, Pasini P, Roda A, Spampinato S. Regulation of ileal bile acid-binding protein expression in Caco-2 cells by ursodeoxycholic acid: Role of the farnesoid X receptor. Biochem Pharmacol 69: 1755–1763,2005. [DOI] [PubMed] [Google Scholar]
- 62.Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk TH, Grefhorst A, Abdelkarim M, Caron S, Torpier G, Fruchart J-C, Gonzalez FJ, Kuipers F, Staels B. The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J Biol Chem 281: 11039–11049,2006. [DOI] [PubMed] [Google Scholar]
- 63.Carr RM, Reid AE. FXR agonists as therapeutic agents for nonalcoholic fatty liver disease. Curr Atheroscler Rep 17: 500, 2015. [DOI] [PubMed] [Google Scholar]
- 64.Centers for Disease Control and Prevention. National diabetes statistics report. Atlanta, GA: Centers for Disease Control and Prevention, 2017. [Google Scholar]
- 65.Chedid V, Vijayvargiya P, Camilleri M. Elobixibat for the treatment of constipation. Expert Rev Gastroenterol Hepatol 12: 951–960, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen F, Ananthanarayanan M, Emre S, Neimark E, Bull LN, Knisely AS, Strautnieks SS, Thompson RJ, Magid MS, Gordon R, Balasub-ramanian N, Suchy FJ, Shneider BL. Progressive familial intrahepatic cholestasis, type 1, is associated with decreased farnesoid X receptor activity. Gastroenterology 126: 756–764, 2004. [DOI] [PubMed] [Google Scholar]
- 67.Chen F, Ma L, Al-Ansari N, Shneider B. The role of AP-1 in the transcriptional regulation of the rat apical sodium-dependent bile acid transporter. JBiol Chem 276: 38703–38714, 2001. [DOI] [PubMed] [Google Scholar]
- 68.Chen F, Ma L, Dawson PA, Sinal CJ, Sehayek E, Gonzalez FJ, Bres-low J, Ananthanarayanan M, Shneider BL. Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J Biol Chem 278: 19909–19916, 2003. [DOI] [PubMed] [Google Scholar]
- 69.Chen F, Ma L, Sartor RB, Li F, Xiong H, Sun A-Q, Shneider B. Inflammatory-mediated repression of the rat ileal sodium-dependent bile acid transporter by c-fos nuclear translocation. Gastroenterology 123:2005–2016,2002. [DOI] [PubMed] [Google Scholar]
- 70.Chen F, Shyu A-B, Shneider BL. Hu antigen R and tristetraprolin: Counter-regulators of rat apical sodium-dependent bile acid transporter by way of effects on messenger RNA stability. Hepatology 54: 1371–1378, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen L, Yao X, Young A, McNulty J, Anderson D, Liu Y, Nystrom C, Croom D, Ross S, Collins J, Rajpal D, Hamlet K, Smith C, Gedulin B. Inhibition of apical sodium-dependent bile acid transporter as a novel treatment for diabetes. Am J Physiol Endocrinol Metab 302: E68–E76, 2012. [DOI] [PubMed] [Google Scholar]
- 72.Chen T, Huang Z, Liu R, Yang J, Hylemon PB, Zhou H. Sphingosine-1 phosphate promotes intestinal epithelial cell proliferation via S1PR2. Front Biosci (Landmark Ed) 22: 596–608, 2017. [DOI] [PubMed] [Google Scholar]
- 73.Chen T, Xue H, Lin R, Huang Z. MiR-126 impairs the intestinal barrier function via inhibiting S1PR2 mediated activation of PI3K/AKT signaling pathway. Biochem Biophys Res Commun, 2017. DOI: 10.1016/j.bbrc.2017.03.043. [DOI] [PubMed] [Google Scholar]
- 74.Chen W, Owsley E, Yang Y, Stroup D, Chiang JY. Nuclear receptor-mediated repression of human cholesterol 7alpha-hydroxylase gene transcription by bile acids. J Lipid Res 42: 1402–1412, 2001. [PubMed] [Google Scholar]
- 75.Chen X, Chen F, Liu S, Glaeser H, Dawson PA, Hofmann AF, Kim RB, Shneider BL, Pang KS. Transactivation of rat apical sodium-dependent bile acid transporter and increased bile acid transport by 1alpha,25-dihydroxyvitamin D3 via the vitamin D receptor. Mol Pharmacol 69: 1913–1923, 2006. [DOI] [PubMed] [Google Scholar]
- 76.Cheng K, Metry M, Felton J, Shang AC, Drachenberg CB, Xu S, Zhan M, Schumacher J, Guo GL, Polli JE, Raufman J-P. Diminished gallbladder filling, increased fecal bile acids, and promotion of colon epithelial cell proliferation and neoplasia in fibroblast growth factor 15-deficient mice. Oncotarget 9: 25572–25585, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng S, Zou M, Liu Q, Kuang J, Shen J, Pu S, Chen L, Li H, Wu T, Li R, Li Y, Jiang W, Zhang Z, He J. Activation of constitutive androstane receptor prevents cholesterol gallstone formation. Am J Pathol 187: 808–818, 2017. [DOI] [PubMed] [Google Scholar]
- 78.Chey WD, Camilleri M, Chang L, Rikner L, Graffner H. A randomized placebo-controlled phase IIb trial of a3309, a bile acid transporter inhibitor, for chronic idiopathic constipation. Am J Gastroenterol 106: 1803–1812, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chiang JYL. Bile acids: Regulation of synthesis. J Lipid Res 50: 1955–1966, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chiang JYL, Pathak P, Liu H, Donepudi A, Ferrell J, Boehme S. Intestinal farnesoid X receptor and Takeda G protein couple receptor 5 signaling in metabolic regulation. Dig Dis 35: 241–245, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chignard N, Mergey M, Veissière D, Parc R, Capeau J, Poupon R, Paul A, Housset C. Bile acid transport and regulating functions in the human biliary epithelium. Hepatology 33: 496–503, 2001. [DOI] [PubMed] [Google Scholar]
- 82.Chothe PP, Czuba LC, Moore RH, Swaan PW. Human bile acid transporter ASBT (SLC10A2) forms functional non-covalent homodimers and higher order oligomers. Biochim Biophys Acta 1860: 645–653, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chothe PP, Swaan PW. Resveratrol promotes degradation of the human bile acid transporter ASBT (SLC10A2). Biochem J 459:301–312,2014. [DOI] [PubMed] [Google Scholar]
- 84.Chow ECY, Maeng H-J, Liu S, Khan AA, Groothuis GMM, Pang KS. 1alpha,25-Dihydroxyvitamin D(3) triggered vitamin D receptor and farnesoid X receptor-like effects in rat intestine and liver in vivo. Biopharm Drug Dispos 30: 457–475, 2009. [DOI] [PubMed] [Google Scholar]
- 85.Chow ECY, Sondervan M, Jin C, Groothuis GMM, Pang KS. Comparative effects of doxercalciferol (1α-hydroxyvitamin D2) versus calcitriol (1α,25-dihydroxyvitamin D3) on the expression of transporters and enzymes in the rat in vivo. J Pharm Sci 100: 1594–1604, 2011. [DOI] [PubMed] [Google Scholar]
- 86.Christian WV, Li N, Hinkle PM, Ballatori N. β-Subunit of the OSTα-OSTβ organic solute transporter is required not only for heterodimerization and trafficking but also for function. J Biol Chem 287: 21233–21243, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Christie DM, Dawson PA, Thevananther S, Shneider BL. Comparative analysis of the ontogeny of a sodium-dependent bile acid transporter in rat kidney and ileum. Am J Physiol 271: G377–G385, 1996. [DOI] [PubMed] [Google Scholar]
- 88.Cipriani S, Mencarelli A, Chini MG, Distrutti E, Renga B, Bifulco G, Baldelli F, Donini A, Fiorucci S. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS ONE 6: e25637, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Conley DR, Coyne MJ, Bonorris GG, Chung A, Schoenfield LJ. Bile acid stimulation of colonic adenylate cyclase and secretion in the rabbit. Am J Dig Dis 21:453–458, 1976. [DOI] [PubMed] [Google Scholar]
- 90.Coon S, Kekuda R, Saha P, Sundaram U. Glucocorticoids differentially regulate Na-bile acid cotransport in normal and chronically inflamed rabbit ileal villus cells. Am J Physiol Gastrointest Liver Physiol 298: G675–G682, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Correia JC, Massart J, de Boer JF, Porsmyr-Palmertz M, Martínez-Redondo V, Agudelo LZ, Sinha I, Meierhofer D, Ribeiro V, Björnholm M, Sauer S, Dahlman-Wright K, Zierath JR, Groen AK, Ruas JL. Bioenergetic cues shift FXR splicing towards FXRα2 to modulate hepatic lipolysis and fatty acid metabolism. Mol Metab 4: 891–902, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Craddock AL, Love MW, Daniel RW, Kirby LC, Walters HC, Wong MH, Dawson PA. Expression and transport properties of the human ileal and renal sodium-dependent bile acid transporter. Am J Physiol 274: G157–G169, 1998. [DOI] [PubMed] [Google Scholar]
- 93.da Silva TC, Hussainzada N, Khantwal CM, Polli JE, Swaan PW. Transmembrane helix 1 contributes to substrate translocation and protein stability of bile acid transporter SLC10A2. J Biol Chem 286: 27322–27332, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dawson PA. Impact of inhibiting ileal apical versus basolateral bile acid transport on cholesterol metabolism and atherosclerosis in mice. Dig Dis 33:382–387, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dawson PA, Haywood J, Craddock AL, Wilson M, Tietjen M, Kluckman K, Maeda N, Parks JS. Targeted deletion of the ileal bile acid transporter eliminates enterohepatic cycling of bile acids in mice. J Biol Chem 278: 33920–33927, 2003. [DOI] [PubMed] [Google Scholar]
- 96.Dawson PA, Hubbert M, Haywood J, Craddock AL, Zerangue N, Christian WV, Ballatori N. The heteromeric organic solute transporter alphabeta, OSTalpha-Ostbeta, is an ileal basolateral bile acid transporter. J Biol Chem 280: 6960–6968, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dawson PA, Lan T, Rao A. Bile acid transporters. J Lipid Res 50:2340–2357, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Defize J, Wider MD, Walz D, Hunt RH. Isolation and partial characterization of gastrotropin from canine ileum: Further studies of the parietal and chief cell response. Endocrinology 123: 2578–2584, 1988. [DOI] [PubMed] [Google Scholar]
- 99.Dekaney CM, von Allmen DC, Garrison AP, Rigby RJ, Lund PK, Henning SJ, Helmrath MA. Bacterial-dependent up-regulation of intestinal bile acid binding protein and transport is FXR-mediated following ileocecal resection. Surgery 144: 174–181, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Denson LA, Sturm E, Echevarria W, Zimmerman TL, Makishima M, Mangelsdorf DJ, Karpen SJ. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology 121: 140–147, 2001. [DOI] [PubMed] [Google Scholar]
- 101.Dietschy JM. Mechanisms for the intestinal absorption of bile acids. J Lipid Res 9: 297–309, 1968. [PubMed] [Google Scholar]
- 102.Donepudi AC, Boehme S, Li F, Chiang JYL. G-protein-coupled bile acid receptor plays a key role in bile acid metabolism and fasting-induced hepatic steatosis in mice. Hepatology 65: 813–827, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.D’Onofrio M, Zanzoni S, Munari F, Monaco HL, Assfalg M, Capaldi S. The long variant of human ileal bile acid-binding protein associated with colorectal cancer exhibits sub-cellular localization and lipid binding behaviour distinct from those of the common isoform. Biochim Biophys Acta Gen Subj 1861: 2315–2324, 2017. [DOI] [PubMed] [Google Scholar]
- 104.Duane WC, Xiong W, Lofgren J. Transactivation of the human apical sodium-dependent bile acid transporter gene by human serum. J Steroid Biochem Mol Biol 108: 137–148, 2008. [DOI] [PubMed] [Google Scholar]
- 105.Duane WC, Xiong W, Wolvers J. Effects of bile acids on expression of the human apical sodium dependent bile acid transporter gene. Biochim Biophys Acta 1771: 1380–1388, 2007. [DOI] [PubMed] [Google Scholar]
- 106.Düfer M, Hörth K, Wagner R, Schittenhelm B, Prowald S, Wagner TFJ, Oberwinkler J, Lukowski R, Gonzalez FJ, Krippeit-Drews P, Drews G. Bile acids acutely stimulate insulin secretion of mouse β-cells via farnesoid X receptor activation and K(ATP) channel inhibition. Diabetes 61: 1479–1489,2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eggert T, Bakonyi D, Hummel W. Enzymatic routes for the synthesis of ursodeoxycholic acid. J Biotechnol 191: 11–21, 2014. [DOI] [PubMed] [Google Scholar]
- 108.Einarsson K, Ericsson S, Ewerth S, Reihnér E, Rudling M, Ståhlberg D, Angelin B. Bile acid sequestrants: Mechanisms of action on bile acid and cholesterol metabolism. Eur J Clin Pharmacol 40 (Suppl 1): S53–S58, 1991. [PubMed] [Google Scholar]
- 109.Erickson SK, Lear SR, Deane S, Dubrac S, Huling SL, Nguyen L, Bollineni JS, Shefer S, Hyogo H, Cohen DE, Shneider B, Sehayek E, Ananthanarayanan M, Balasubramaniyan N, Suchy FJ, Batta AK, Salen G. Hypercholesterolemia and changes in lipid and bile acid metabolism in male and female cyp7A1-deficient mice. J Lipid Res 44: 1001–1009, 2003. [DOI] [PubMed] [Google Scholar]
- 110.Fallingborg J, Pedersen P, Jacobsen BA. Small intestinal transit time and intraluminal pH in ileocecal resected patients with Crohn’s disease. Dig Dis Sci 43: 702–705, 1998. [DOI] [PubMed] [Google Scholar]
- 111.Fang C, Dean J, Smith JW. A novel variant of ileal bile acid binding protein is up-regulated through nuclear factor-kappaB activation in colorectal adenocarcinoma. Cancer Res 67: 9039–9046, 2007. [DOI] [PubMed] [Google Scholar]
- 112.Fang C, Filipp FV, Smith JW. Unusual binding of ursodeoxycholic acid to ileal bile acid binding protein: Role in activation of FXR. J Lipid Res 53:664–673, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fang S, Suh JM, Reilly SM, Yu E, Osborn O, Lackey D, Yoshihara E, Perino A, Jacinto S, Lukasheva Y, Atkins AR, Khvat A, Schnabl B, Yu RT, Brenner DA, Coulter S, Liddle C, Schoonjans K, Olefsky JM, Saltiel AR, Downes M, Evans RM. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med 21: 159–165, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Favretto F, Ceccon A, Zanzoni S, D’Onofrio M, Ragona L, Molinari H, Assfalg M. The unique ligand binding features of subfamily-II iLBPs with respect to bile salts and related drugs. Prostaglandins Leukot Essent Fatty Acids 95: 1–10, 2015. [DOI] [PubMed] [Google Scholar]
- 115.Felton J, Cheng K, Said A, Shang AC, Xu S, Vivian D, Metry M, Polli JE, Raufman J-P. Using multi-fluorinated bile acids and in vivo magnetic resonance imaging to measure bile acid transport. J Vis Exp, 2016. DOI: 10.3791/54597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fernández-Bañares F, Rosinach M, Piqueras M, Ruiz-Cerulla A, Modolell I, Zabana Y, Guardiola J, Esteve M. Randomised clinical trial: Colestyramine vs. hydroxypropyl cellulose in patients with functional chronic watery diarrhoea. Aliment Pharmacol Ther 41: 1132–1140, 2015. [DOI] [PubMed] [Google Scholar]
- 117.Ferrebee CB, Li J, Haywood J, Pachura K, Robinson BS, Hinrichs BH, Jones RM, Rao A, Dawson PA. Organic solute transporter α-β protects ileal enterocytes from bile acid-induced injury. Cell Mol Gastroenterol Hepatol 5: 499–522, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ferrell JM, Boehme S, Li F, Chiang JYL. Cholesterol 7α-hydroxylase-deficient mice are protected from high-fat/high-cholesterol diet-induced metabolic disorders. J Lipid Res 57: 1144–1154, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fiorucci S, Clerici C, Antonelli E, Orlandi S, Goodwin B, Sadeghpour BM, Sabatino G, Russo G, Castellani D, Willson TM, Pruzanski M, Pellicciari R, Morelli A. Protective effects of 6-ethyl chenodeoxycholic acid, a farnesoid X receptor ligand, in estrogen-induced cholestasis. J Pharmacol Exp Ther 313: 604–612, 2005. [DOI] [PubMed] [Google Scholar]
- 120.Fischer M, Siva S, Wo JM, Fadda HM. Assessment of small intestinal transit times in ulcerative colitis and Crohn’s disease patients with different disease activity using video capsule endoscopy. AAPS Pharm-SciTech 18: 404–409, 2017. [DOI] [PubMed] [Google Scholar]
- 121.Fisher E, Grallert H, Klapper M, Pfäfflin A, Schrezenmeir J, Illig T, Boeing H, Döring F. Evidence for the Thr79Met polymorphism of the ileal fatty acid binding protein (FABP6) to be associated with type 2 diabetes in obese individuals. Mol Genet Metab 98: 400–405, 2009. [DOI] [PubMed] [Google Scholar]
- 122.Fisher E, Nitz I, Lindner I, Rubin D, Boeing H, Möhlig M, Hampe J, Schreiber S, Schrezenmeir J, Döring F. Candidate gene association study of type 2 diabetes in a nested case-control study of the EPIC-Potsdam cohort - role of fat assimilation. Mol Nutr Food Res 51: 185–191,2007. [DOI] [PubMed] [Google Scholar]
- 123.Flynn CR, Albaugh VL, Cai S, Cheung-Flynn J, Williams PE, Brucker RM, Bordenstein SR, Guo Y, Wasserman DH, Abumrad NN. Bile diversion to the distal small intestine has comparable metabolic benefits to bariatric surgery. Nat Commun 6: 7715, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ford GA, Preece JD, Davies IH, Wilkinson SP. Use of the SeHCAT test in the investigation of diarrhoea. Postgrad Med J 68: 272–276, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81:687–693, 1995. [DOI] [PubMed] [Google Scholar]
- 126.Frankenberg T, Miloh T, Chen FY, Ananthanarayanan M, Sun A-Q, Balasubramaniyan N, Arias I, Setchell KDR, Suchy FJ, Shneider BL. The membrane protein ATPase class I type 8B member 1 signals through protein kinase C zeta to activate the farnesoid X receptor. Hepatology 48: 1896–1905, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Frankenberg T, Rao A, Chen F, Haywood J, Shneider BL, Dawson PA. Regulation of the mouse organic solute transporter alpha-beta, OSTalpha-Ostbeta, by bile acids. Am J Physiol Gastrointest Liver Physiol 290: G912–G922, 2006. [DOI] [PubMed] [Google Scholar]
- 128.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 24: 908–922, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Frommherz L, Bub A, Hummel E, Rist MJ, Roth A, Watzl B, Kulling SE. Age-related changes of plasma bile acid concentrations in healthy adults—results from the cross-sectional KarMeN study. PLoS ONE 11: e0153959, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fujino T, Une M, Imanaka T, Inoue K, Nishimaki-Mogami T. Structure-activity relationship of bile acids and bile acid analogs in regard to FXR activation. J Lipid Res 45: 132–138, 2004. [DOI] [PubMed] [Google Scholar]
- 131.Fujita M, Fujii H, Kanda T, Sato E, Hatakeyama K, Ono T. Molecular cloning, expression, and characterization of a human intestinal 15-kDa protein. Eur J Biochem 233: 406–413, 1995. [DOI] [PubMed] [Google Scholar]
- 132.Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen ECL, Renooij W, Murzilli S, Klomp LWJ, Siersema PD, Schipper MEI, Danese S, Penna G, Laverny G, Adorini L, Moschetta A, van Mil SWC. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 60: 463–472, 2011. [DOI] [PubMed] [Google Scholar]
- 133.Gao T, Feridooni HA, Howlett SE, Pelis RM. Influence of age on intestinal bile acid transport in C57BL/6 mice. Pharmacol Res Perspect 5: e00287, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Garbutt JT, Kenney TJ. Effect of cholestyramine on bile acid metabolism in normal man. J Clin Invest 51: 2781–2789, 1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ghosh A, Chen F, Banerjee S, Xu M, Shneider BL. c-Fos mediates repression of the apical sodium-dependent bile acid transporter by fibroblast growth factor-19 in mice. Am J Physiol Gastrointest Liver Physiol 306: G163–G171, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gong YZ, Everett ET, Schwartz DA, Norris JS, Wilson FA. Molecular cloning, tissue distribution, and expression of a 14-kDa bile acid-binding protein from rat ileal cytosol. Proc Natl Acad Sci U S A 91: 4741–4745, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gong YZ, Kato T, Schwartz DA, Norris JS, Wilson FA. Ontogenic and glucocorticoid-accelerated expression of rat 14 kDa bile acid-binding protein. Anat Rec 245: 532–538, 1996. [DOI] [PubMed] [Google Scholar]
- 138.Gonzalez FJ, Jiang C, Xie C, Patterson AD. Intestinal farnesoid X receptor signaling modulates metabolic disease. Dig Dis 35: 178–184,2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.González PM, Hussainzada N, Swaan PW, MacKerell AD, Polli JE. Putative irreversible inhibitors of the human sodium-dependent bile acid transporter (hASBT; SLC10A2) support the role of transmembrane domain 7 in substrate binding/translocation. Pharm Res 29: 1821–1831,2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 6: 517–526, 2000. [DOI] [PubMed] [Google Scholar]
- 141.Gothe F, Beigel F, Rust C, Hajji M, Koletzko S, Freudenberg F. Bile acid malabsorption assessed by 7 alpha-hydroxy-4-cholesten-3-one in pediatric inflammatory bowel disease: Correlation to clinical and laboratory findings. J Crohns Colitis 8: 1072–1078, 2014. [DOI] [PubMed] [Google Scholar]
- 142.Grober J, Zaghini I, Fujii H, Jones SA, Kliewer SA, Willson TM, Ono T, Besnard P. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene: Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J Biol Chem 274: 29749–29754, 1999. [DOI] [PubMed] [Google Scholar]
- 143.Guariento M, Assfalg M, Zanzoni S, Fessas D, Longhi R, Molinari H. Chicken ileal bile-acid-binding protein: A promising target of investigation to understand binding co-operativity across the protein family. Biochem J 425: 413–424, 2009. [DOI] [PubMed] [Google Scholar]
- 144.Guo C, Chen W-D, Wang Y-D. TGR5, not only a metabolic regulator. Front Physiol 7: 646, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hagey LR, Crombie DL, Espinosa E, Carey MC, Igimi H, Hofmann AF. Ursodeoxycholic acid in the Ursidae: Biliary bile acids of bears, pandas, and related carnivores. J Lipid Res 34: 1911–1917, 1993. [PubMed] [Google Scholar]
- 146.Håkansson P, Andersson I, Nyström S, Löfgren L, Amrot LF, Li H. Ontogenetic development and spatial distribution of the ileal apical sodium-dependent bile acid transporter and the ileal lipid-binding protein in apoE knockout and C57BL/6 mice. Scand J Gastroenterol 37: 1089–1096, 2002. [DOI] [PubMed] [Google Scholar]
- 147.Hallén S, Brändén M, Dawson PA, Sachs G. Membrane insertion scanning of the human ileal sodium/bile acid co-transporter. Biochemistry 38: 11379–11388, 1999. [DOI] [PubMed] [Google Scholar]
- 148.Halpern MD, Holubec H, Dominguez JA, Meza YG, Williams CS, Ruth MC, McCuskey RS, Dvorak B. Hepatic inflammatory mediators contribute to intestinal damage in necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol 284: G695–G702, 2003. [DOI] [PubMed] [Google Scholar]
- 149.Halpern MD, Holubec H, Saunders TA, Dvorak K, Clark JA, Doelle SM, Ballatori N, Dvorak B. Bile acids induce ileal damage during experimental necrotizing enterocolitis. Gastroenterology 130: 359–372, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Halpern MD, Weitkamp J-H, Mount Patrick SK, Dobrenen HJ, Khailova L, Correa H, Dvorak B. Apical sodium-dependent bile acid transporter upregulation is associated with necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol 299: G623–G631, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Han X, Sun J, Wang Y, He Z. PepTl, ASBT-linked prodrug strategy to improve oral bioavailability and tissue targeting distribution. Curr Drug Metab 16: 71–83, 2015. [DOI] [PubMed] [Google Scholar]
- 152.Handelsman Y. Role of bile acid sequestrants in the treatment of type 2 diabetes. Diabetes Care 34 (Suppl 2): S244–S250, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hegade VS, Kendrick SFW, Dobbins RL, Miller SR, Richards D, Storey J, Dukes G, Gilchrist K, Vallow S, Alexander GJ, Corrigan M, Hirschfield GM, Jones DEJ. BAT117213: Ileal bile acid transporter (IBAT) inhibition as a treatment for pruritus in primary biliary cirrhosis: Study protocol for a randomised controlled trial. BMC Gastroenterol 16: 71, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hegade VS, Kendrick SFW, Dobbins RL, Miller SR, Thompson D, Richards D, Storey J, Dukes GE, Corrigan M, Oude Elferink RPJ, Beuers U, Hirschfield GM, Jones DE. Effect of ileal bile acid transporter inhibitor GSK2330672 on pruritus in primary biliary cholangitis: A double-blind, randomised, placebo-controlled, crossover, phase 2a study. Lancet 389: 1114–1123, 2017. [DOI] [PubMed] [Google Scholar]
- 155.Heubi JE, Balistreri WF, Fondacaro JD, Partin JC, Schubert WK. Primary bile acid malabsorption: Defective in vitro ileal active bile acid transport. Gastroenterology 83: 804–811, 1982. [PubMed] [Google Scholar]
- 156.Heubi JE, Balistreri WF, Partin JC, Schubert WK, McGraw CA. Refractory infantile diarrhea due to primary bile acid malabsorption. J Pediatr 94:546–551, 1979. [DOI] [PubMed] [Google Scholar]
- 157.Heubi JE, Balistreri WF, Suchy FJ. Bile salt metabolism in the first year of life. J Lab Clin Med 100: 127–136, 1982. [PubMed] [Google Scholar]
- 158.Ho RH, Leake BF, Urquhart BL, Gregor JC, Dawson PA, Kim RB. Functional characterization of genetic variants in the apical sodium-dependent bile acid transporter (ASBT; SLC10A2). J Gastroenterol Hepatol 26: 1740–1748, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hofmann AF. Regulation of ileal bile acid transport: Desirability of measuring transport function as well as transporter activity. Hepatology 29: 1335–1337, 1999. [DOI] [PubMed] [Google Scholar]
- 160.Hofmann AF. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med 159: 2647–2658, 1999. [DOI] [PubMed] [Google Scholar]
- 161.Hofmann AF, Eckmann L. How bile acids confer gut mucosal protection against bacteria. Proc Natl Acad Sci U S A 103: 4333–4334, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: Structural variation and possible evolutionary significance. J Lipid Res 51: 226–246, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hofmann AF, Mangelsdorf DJ, Kliewer SA. Chronic diarrhea due to excessive bile acid synthesis and not defective ileal transport: A new syndrome of defective fibroblast growth factor 19 release. Clin Gastroenterol Hepatol 7: 1151–1154, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hofmann AF, Mysels KJ. Bile salts as biological surfactants. Colloids Surf 30: 145–173, 1987. [Google Scholar]
- 165.Hofmann AF, Poley JR. Cholestyramine treatment of diarrhea associated with ileal resection. N Engl J Med 281: 397–402, 1969. [DOI] [PubMed] [Google Scholar]
- 166.Holzer A, Harsch S, Renner O, Strohmeyer A, Schimmel S, Wehkamp J, Fritz P, Stange EF. Diminished expression of apical sodium-dependent bile acid transporter in gallstone disease is independent of ileal inflammation. Digestion 78: 52–59, 2008. [DOI] [PubMed] [Google Scholar]
- 167.Horváth G, Bencsura Á, Simon Á, Tochtrop GP, DeKoster GT, Covey DF, Cistola DP, Toke O. Structural determinants of ligand binding in the ternary complex of human ileal bile acid binding protein with glycocholate and glycochenodeoxycholate obtained from solution NMR. FEBS J 283: 541–555, 2016. [DOI] [PubMed] [Google Scholar]
- 168.Hruz P, Zimmermann C, Gutmann H, Degen L, Beuers U, Terracciano L, Drewe J, Beglinger C. Adaptive regulation of the ileal apical sodium dependent bile acid transporter (ASBT) in patients with obstructive cholestasis. Gut 55: 395–402, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hu N-J, Iwata S, Cameron AD, Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478: 408–411, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hu X, Bonde Y, Eggertsen G, Rudling M. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J Intern Med 275: 27–38, 2014. [DOI] [PubMed] [Google Scholar]
- 171.Huff MW, Telford DE, Edwards JY, Burnett JR, Barrett PHR, Rapp SR, Napawan N, Keller BT. Inhibition of the apical sodium-dependent bile acid transporter reduces LDL cholesterol and apoB by enhanced plasma clearance of LDL apoB. Arterioscler Thromb Vasc Biol 22: 1884–1891, 2002. [DOI] [PubMed] [Google Scholar]
- 172.Hulzebos CV, van Zoonen AGJF, Hulscher JBF, Schat TE, Kooi EMW, Koehorst M, Boverhof R, Krabbe PFM, Groen AK, Verkade HJ. Fecal bile salts and the development of necrotizing enterocolitis in preterm infants. PLoS ONE 12: e0168633, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hussainzada N, Banerjee A, Swaan PW. Transmembrane domain VII of the human apical sodium-dependent bile acid transporter ASBT (SLC10A2) lines the substrate translocation pathway. Mol Pharmacol 70: 1565–1574,2006. [DOI] [PubMed] [Google Scholar]
- 174.Hussainzada N, Claro Da Silva T, Swaan PW. The cytosolic half of helix III forms the substrate exit route during permeation events of the sodium/bile acid cotransporter ASBT. Biochemistry 48: 8528–8539, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hussainzada N, da Silva TC, Zhang EY, Swaan PW. Conserved aspartic acid residues lining the extracellular loop 1 of sodium-coupled bile acid transporter ASBT Interact with Na+ and 7alpha-OH moieties on the ligand cholestane skeleton. J Biol Chem 283: 20653–20663, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hussainzada N, Khandewal A, Swaan PW. Conformational flexibility of helix VI is essential for substrate permeation of the human apical sodium-dependent bile acid transporter. Mol Pharmacol 73: 305–313, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hwang ST, Henning SJ. Ontogenic regulation of components of ileal bile acid absorption. Exp Biol Med (Maywood) 226: 674–680, 2001. [DOI] [PubMed] [Google Scholar]
- 178.Hwang ST, Urizar NL, Moore DD, Henning SJ. Bile acids regulate the ontogenic expression of ileal bile acid binding protein in the rat via the farnesoid X receptor. Gastroenterology 122: 1483–1492, 2002. [DOI] [PubMed] [Google Scholar]
- 179.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2:217–225, 2005. [DOI] [PubMed] [Google Scholar]
- 180.Inagaki T, Moschetta A, Lee Y-K, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A 103: 3920–3925, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ishibashi S, Schwarz M, Frykman PK, Herz J, Russell DW. Disruption of cholesterol 7alpha-hydroxylase gene in mice. I. Postnatal lethality reversed by bile acid and vitamin supplementation. J Biol Chem 271: 18017–18023, 1996. [DOI] [PubMed] [Google Scholar]
- 182.Jahnel J, Fickert P, Hauer AC, Hogenauer C, Avian A, Trauner M. Inflammatory bowel disease alters intestinal bile acid transporter expression. Drug Metab Dispos 42: 1423–1431, 2014. [DOI] [PubMed] [Google Scholar]
- 183.Jiang C, Xie C, Lv Y, Li J, Krausz KW, Shi J, Brocker CN, Desai D, Amin SG, Bisson WH, Liu Y, Gavrilova O, Patterson AD, Gonzalez FJ. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun 6: 10166, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Johnston IM, Nolan JD, Pattni SS, Appleby RN, Zhang JH, Kennie SL, Madhan GK, Jameie-Oskooei S, Pathmasrirengam S, Lin J, Hong A, Dixon PH, Williamson C, Walters JRF. Characterizing factors associated with differences in FGF19 blood levels and synthesis in patients with primary bile acid diarrhea. Am J Gastroenterol 111: 423–432, 2016. [DOI] [PubMed] [Google Scholar]
- 185.Josephson B, Rydin A. The resorption of the bile acids from the intestines. Biochem J 30: 2224–2228, 1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jung D, Fantin AC, Scheurer U, Fried M, Kullak-Ublick GA. Human ileal bile acid transporter gene ASBT (SLC10A2) is transactivated by the glucocorticoid receptor. Gut 53: 78–84, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jung D, Fried M, Kullak-Ublick GA. Human apical sodium-dependent bile salt transporter gene (SLC10A2) is regulated by the peroxisome proliferator-activated receptor alpha. J Biol Chem 277: 30559–30566, 2002. [DOI] [PubMed] [Google Scholar]
- 188.Kaikaus RM, Bass NM, Ockner RK. Functions of fatty acid binding proteins. Experientia 46: 617–630, 1990. [DOI] [PubMed] [Google Scholar]
- 189.Kanda T, Foucand L, Nakamura Y, Niot I, Besnard P, Fujita M, Sakai Y, Hatakeyama K, Ono T, Fujii H. Regulation of expression of human intestinal bile acid-binding protein in Caco-2 cells. Biochem J 330 (Pt 1): 261–265, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kanda T, Niot I, Foucaud L, Fujii H, Bernard A, Ono T, Besnard P. Effect of bile on the intestinal bile-acid binding protein (I-BABP) expression. In vitro and in vivo studies. FEBS Lett 384: 131–134, 1996. [DOI] [PubMed] [Google Scholar]
- 191.Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagonlike peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun 329: 386–390, 2005. [DOI] [PubMed] [Google Scholar]
- 192.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem 278: 9435–9440, 2003. [DOI] [PubMed] [Google Scholar]
- 193.Kazgan N, Metukuri MR, Purushotham A, Lu J, Rao A, Lee S, Pratt-Hyatt M, Lickteig A, Csanaky IL, Zhao Y, Dawson PA, Li X. Intestine-specific deletion of SIRT1 in mice impairs DCoH2-HNF-1α-FXR signaling and alters systemic bile acid homeostasis. Gastroenterology 146: 1006–1016, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Keely SJ, Scharl MM, Bertelsen LS, Hagey LR, Barrett KE, Hofmann AF. Bile acid-induced secretion in polarized monolayers of T84 colonic epithelial cells: Structure-activity relationships. Am J Physiol Gastrointest Liver Physiol 292: G290–G297, 2007. [DOI] [PubMed] [Google Scholar]
- 195.Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell 2:713–720, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Khan AA, Chow ECY, Porte RJ, Pang KS, Groothuis GMM. Expression and regulation of the bile acid transporter, OSTalpha-OSTbeta in rat and human intestine and liver. Biopharm Drug Dispos 30: 241–258,2009. [DOI] [PubMed] [Google Scholar]
- 197.Khantwal CM, Swaan PW. Cytosolic half of transmembrane domain IV of the human bile acid transporter hASBT (SLC10A2) forms part of the substrate translocation pathway. Biochemistry 47: 3606–3614, 2008. [DOI] [PubMed] [Google Scholar]
- 198.Kim I, Ahn S-H, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, Gonzalez FJ. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res 48: 2664–2672, 2007. [DOI] [PubMed] [Google Scholar]
- 199.Kim Y-C, Byun S, Zhang Y, Seok S, Kemper B, Ma J, Kemper JK. Liver ChIP-seq analysis in FGF19-treated mice reveals SHP as a global transcriptional partner of SREBP-2. Genome Biol 16: 268, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kir S, Zhang Y, Gerard RD, Kliewer SA, Mangelsdorf DJ. Nuclear receptors HNF4α and LRH-1 cooperate in regulating Cyp7a1 in vivo. J Biol Chem 287: 41334–41341, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kitayama K, Nakai D, Kono K, van der Hoop AG, Kurata H, de Wit EC, Cohen LH, Inaba T, Kohama T. Novel non-systemic inhibitor of ileal apical Na+-dependent bile acid transporter reduces serum cholesterol levels in hamsters and monkeys. Eur J Pharmacol 539: 89–98, 2006. [DOI] [PubMed] [Google Scholar]
- 202.Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92: 73–82, 1998. [DOI] [PubMed] [Google Scholar]
- 203.Klomp LWJ, Vargas JC, van Mil SwC, Pawlikowska L, Strautnieks SS, van Eijk MJT, Juijn JA, Pabón-Peña C, Smith LB, DeYoung JA, Byrne JA, Gombert J, van der Brugge G, Berger R, Jankowska I, Pawlowska J, Villa E, Knisely AS, Thompson RJ, Freimer NB, Houwen RHJ, Bull LN. Characterization of mutations in ATP8B1 associated with hereditary cholestasis. Hepatology 40: 27–38, 2004. [DOI] [PubMed] [Google Scholar]
- 204.Kok T, Hulzebos Cv, Wolters H, Havinga R, Agellon LB, Stellaard F, Shan B, Schwarz M, Kuipers F. Enterohepatic circulation of bile salts in farnesoid X receptor-deficient mice: Efficient intestinal bile salt absorption in the absence of ileal bile acid-binding protein. J Biol Chem 278:41930–41937, 2003. [DOI] [PubMed] [Google Scholar]
- 205.Kolhatkar V, Polli JE. Structural requirements of bile acid transporters: C-3 and C-7 modifications of steroidal hydroxyl groups. Eur J Pharm Sci 46: 86–99,2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kong B, Wang L, Chiang JYL, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 56:1034–1043,2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kovár J, Lenícek M, Zimolová M, Vítek L, Jirsa M, Pitha J. Regulation of diurnal variation of cholesterol 7alpha-hydroxylase (CYP7A1) activity in healthy subjects. Physiol Res 59: 233–238, 2010. [DOI] [PubMed] [Google Scholar]
- 208.Kowdley KV, Luketic V, Chapman R, Hirschfield GM, Poupon R, Schramm C, Vincent C, Rust C, Parés A, Mason A, Marschall H-U, Shapiro D, Adorini L, Sciacca C, Beecher-Jones T, Böhm O, Pencek R, Jones D, Obeticholic Acid PBC Monotherapy Study Group. A randomized trial of obeticholic acid monotherapy in patients with primary biliary cholangitis. Hepatology 67: 1890–1902, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Krag E, Phillips SF. Active and passive bile acid absorption in man. Perfusion studies of the ileum and jejunum. J Clin Invest 53: 1686–1694, 1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kramer W, Burckhardt G, Wilson FA, Kurz G. Bile salt-binding polypeptides in brush-border membrane vesicles from rat small intestine revealed by photoaffinity labeling. J Biol Chem 258: 3623–3627, 1983. [PubMed] [Google Scholar]
- 211.Kramer W, Corsiero D, Friedrich M, Girbig F, Stengelin S, Weyland C. Intestinal absorption of bile acids: Paradoxical behaviour of the 14 kDa ileal lipid-binding protein in differential photoaffinity labelling. Biochem J 333 (Pt 2): 335–341, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kramer W, Girbig F, Glombik H, Corsiero D, Stengelin S, Weyland C. Identification of a ligand-binding site in the Na+/bile acid cotransporting protein from rabbit ileum. J Biol Chem 276: 36020–36027, 2001. [DOI] [PubMed] [Google Scholar]
- 213.Kramer W, Girbig F, Gutjahr U, Kowalewski S. Radiation-inactivation analysis of the Na+/bile acid co-transport system from rabbit ileum. Biochem J 306 (Pt 1): 241–246, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kramer W, Girbig F, Gutjahr U, Kowalewski S, Jouvenal K, Müller G, Tripier D, Wess G. Intestinal bile acid absorption. Na(+)-dependent bile acid transport activity in rabbit small intestine correlates with the coexpression of an integral 93-kDa and a peripheral 14-kDa bile acid-binding membrane protein along the duodenum-ileum axis. J Biol Chem 268: 18035–18046, 1993. [PubMed] [Google Scholar]
- 215.Kramer W, Nicol SB, Girbig F, Gutjahr U, Kowalewski S, Fasold H. Characterization and chemical modification of the Na(+)-dependent bile-acid transport system in brush-border membrane vesicles from rabbit ileum. Biochim Biophys Acta 1111: 93–102, 1992. [DOI] [PubMed] [Google Scholar]
- 216.Kubitz R, Dröge C, Kluge S, Stindt J, Häussinger D. Genetic variations of bile salt transporters. Drug Discov Today Technol 12: e55–e67, 2014. [DOI] [PubMed] [Google Scholar]
- 217.Kumar DP, Rajagopal S, Mahavadi S, Mirshahi F, Grider JR, Murthy KS, Sanyal AJ. Activation of transmembrane bile acid receptor TGR5 stimulates insulin secretion in pancreatic β cells. Biochem Biophys Res Commun 427: 600–605, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kwong E, Li Y, Hylemon PB, Zhou H. Bile acids and sphingosine-1-phosphate receptor 2 in hepatic lipid metabolism. Acta Pharm Sin B 5: 151–157, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lammert F, Paigen B, Carey MC. Localization of the ileal sodium-bile salt cotransporter gene (Slc10a2) to mouse chromosome 8. Mamm Genome 9: 173–174, 1998. [DOI] [PubMed] [Google Scholar]
- 220.Lan T, Haywood J, Dawson PA. Inhibition of ileal apical but not basolateral bile acid transport reduces atherosclerosis in apoE−/− mice. Atherosclerosis 229: 374–380, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lan T, Haywood J, Rao A, Dawson PA. Molecular mechanisms of altered bile acid homeostasis in organic solute transporter-alpha knockout mice. Dig Dis 29: 18–22, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lan T, Rao A, Haywood J, Kock ND, Dawson PA. Mouse organic solute transporter alpha deficiency alters FGF15 expression and bile acid metabolism. J Hepatol 57: 359–365, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Landrier J-F, Eloranta JJ, Vavricka SR, Kullak-Ublick GA. The nuclear receptor for bile acids, FXR, transactivates human organic solute transporter-alpha and -beta genes. Am J Physiol Gastrointest Liver Physiol 290: G476–G485, 2006. [DOI] [PubMed] [Google Scholar]
- 224.Landrier J-F, Grober J, Demydchuk J, Besnard P. FXRE can function as an LXRE in the promoter of human ileal bile acid-binding protein (I-BABP) gene. FEBS Lett 553: 299–303, 2003. [DOI] [PubMed] [Google Scholar]
- 225.Landrier JF, Thomas C, Grober J, Zaghini I, Petit V, Poirier H, Niot I, Besnard P. The gene encoding the human ileal bile acid-binding protein (I-BABP) is regulated by peroxisome proliferator-activated receptors. Biochim Biophys Acta 1735: 41–49, 2005. [DOI] [PubMed] [Google Scholar]
- 226.Lazaridis KN, Pham L, Tietz P, Marinelli RA, deGroen PC, Levine S, Dawson PA, LaRusso NF. Rat cholangiocytes absorb bile acids at their apical domain via the ileal sodium-dependent bile acid transporter. J Clinlnvest 100: 2714–2721, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lazaridis KN, Tietz P, Wu T, Kip S, Dawson PA, LaRusso NF. Alternative splicing of the rat sodium/bile acid transporter changes its cellular localization and transport properties. Proc Natl Acad Sci U S A 97: 11092–11097,2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Le T-A, Chen J, Changchien C, Peterson MR, Kono Y, Patton H, Cohen BL, Brenner D, Sirlin C, Loomba R, San Diego Integrated NAFLD Research Consortium (SINC). Effect of colesevelam on liver fat quantified by magnetic resonance in nonalcoholic steatohepatitis: A randomized controlled trial. Hepatology 56: 922–932, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lee H, Zhang Y, Lee FY, Nelson SF, Gonzalez FJ, Edwards PA. FXR regulates organic solute transporters alpha and beta in the adrenal gland, kidney, and intestine. J Lipid Res 47: 201–214, 2006. [DOI] [PubMed] [Google Scholar]
- 230.Lee Y-K, Schmidt DR, Cummins CL, Choi M, Peng L, Zhang Y, Goodwin B, Hammer RE, Mangelsdorf DJ, Kliewer SA. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol Endocrinol 22: 1345–1356, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102: 1016–1023, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lenicek M, Duricova D, Komarek V, Gabrysova B, Lukas M, Smerhovsky Z, Vítek L. Bile acid malabsorption in inflammatory bowel disease: Assessment by serum markers. Inflamm Bowel Dis 17: 1322–1327, 2011. [DOI] [PubMed] [Google Scholar]
- 233.Li H, Chen F, Shang Q, Pan L, Shneider BL, Chiang JYL, Forman BM, Ananthanarayanan M, Tint GS, Salen G, Xu G. FXR-activating ligands inhibit rabbit ASBT expression viaFXR-SHP-FTF cascade. Am J Physiol Gastrointest Liver Physiol 288: G60–G66, 2005. [DOI] [PubMed] [Google Scholar]
- 234.Li H, Liu Y, Zhang X, Xu Q, Zhang Y, Xue C, Guo C. Medium-chain fatty acids decrease serum cholesterol via reduction of intestinal bile acid reabsorption in C57BL/6J mice. Nutr Metab (Lond) 15: 37, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li H, Xu G, Shang Q, Pan L, Shefer S, Batta AK, Bollineni J, Tint GS, Keller BT, Salen G. Inhibition of ileal bile acid transport lowers plasma cholesterol levels by inactivating hepatic farnesoid X receptor and stimulating cholesterol 7 alpha-hydroxylase. Metab Clin Exp 53: 927–932, 2004. [DOI] [PubMed] [Google Scholar]
- 236.Li T, Apte U. Bile acid metabolism and signaling in cholestasis, inflammation, and cancer. Adv Pharmacol 74: 263–302, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Li T, Chen W, Chiang JYL. PXR induces CYP27A1 and regulates cholesterol metabolism in the intestine. J Lipid Res 48: 373–384, 2007. [DOI] [PubMed] [Google Scholar]
- 238.Li T, Chiang JYL. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am J Physiol Gastrointest Liver Physiol 288: G74–G84, 2005. [DOI] [PubMed] [Google Scholar]
- 239.Li T, Francl JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, Erickson SK, Chiang JYL. Glucose and insulin induction of bile acid synthesis: Mechanisms and implication in diabetes and obesity. J Biol Chem 287: 1861–1873,2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Li-Hawkins J, Gåfvels M, Olin M, Lund EG, Andersson U, Schuster G, Björkhem I, Russell DW, Eggertsen G. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest 110: 1191–1200, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lin MC, Gong YZ, Geoghegan KF, Wilson FA. Characterization of a novel 14 kDa bile acid-binding protein from rat ileal cytosol. Biochim Biophys Acta 1078: 329–335, 1991. [DOI] [PubMed] [Google Scholar]
- 242.Lin MC, Kramer W, Wilson FA. Identification of cytosolic and microsomal bile acid-binding proteins in rat ileal enterocytes. J Biol Chem 265: 14986–14995, 1990. [PubMed] [Google Scholar]
- 243.Lin MC, Mullady E, Wilson FA. Timed photoaffinity labeling and characterization of bile acid binding and transport proteins in rat ileum. Am J Physiol 265: G56–G62, 1993. [DOI] [PubMed] [Google Scholar]
- 244.Lionarons DA, Boyer JL, Cai S-Y. Evolution of substrate specificity for the bile salt transporter ASBT (SLC10A2). J Lipid Res 53: 1535–1542,2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lorenzo-Zúñiga V, Bartolí R, Planas R, Hofmann AF, Viñado B, Hagey LR, Hernández JM, Mañé J, Alvarez MA, Ausina V, Gassull MA. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology 37: 551–557,2003. [DOI] [PubMed] [Google Scholar]
- 246.Lozano E, Monte MJ, Briz O, Hernández-Hernández A, Banales JM, Marin JJG, Macias RIR. Enhanced antitumour drug delivery to cholangiocarcinoma through the apical sodium-dependent bile acid transporter (ASBT). J Control Release 216: 93–102, 2015. [DOI] [PubMed] [Google Scholar]
- 247.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell 6: 507–515, 2000. [DOI] [PubMed] [Google Scholar]
- 248.Lücke C, Zhang F, Rüterjans H, Hamilton JA, Sacchettini JC. Flexibility is a likely determinant of binding specificity in the case of ileal lipid binding protein. Structure 4: 785–800, 1996. [DOI] [PubMed] [Google Scholar]
- 249.Lundåsen T, Andersson E-M, Snaith M, Lindmark H, Lundberg J, Östlund-Lindqvist A-M, Angelin B, Rudling M. Inhibition of intestinal bile acid transporter Slc10a2 improves triglyceride metabolism and normalizes elevated plasma glucose levels in mice. PLoS One 7: e37787, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lundåsen T, Gälman C, Angelin B, Rudling M. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med 260: 530–536, 2006. [DOI] [PubMed] [Google Scholar]
- 251.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest 116: 1102–1109, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ma L, Jüttner M, Kullak-Ublick GA, Eloranta JJ. Regulation of the gene encoding the intestinal bile acid transporter ASBT by the caudal-type homeobox proteins CDX1 and CDX2. Am J Physiol Gastrointest Liver Physiol 302: G123–G133, 2012. [DOI] [PubMed] [Google Scholar]
- 253.Ma Y, Huang Y, Yan L, Gao M, Liu D. Synthetic FXR agonist GW4064 prevents diet-induced hepatic steatosis and insulin resistance. Pharm Res 30: 1447–1457, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science 296: 1313–1316, 2002. [DOI] [PubMed] [Google Scholar]
- 255.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science 284: 1362–1365, 1999. [DOI] [PubMed] [Google Scholar]
- 256.Martins RM, Silva CAD, Becker CM, Samios D, Christoff M, Bica CID. Anionic surfactant aggregation with (Hydroxypropyl)cellulose in the presence of added salt. J Braz Chem Soc 17: 944–953, 2006. [Google Scholar]
- 257.Martoni CJ, Labbé A, Ganopolsky JG, Prakash S, Jones ML. Changes in bile acids, FGF-19 and sterol absorption in response to bile salt hydrolase active L. reuteri NCIMB 30242. Gut Microbes 6: 57–65, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Nakamura T, Itadani H, Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun 298: 714–719, 2002. [DOI] [PubMed] [Google Scholar]
- 259.Maruyama T, Tanaka K, Suzuki J, Miyoshi H, Harada N, Nakamura T, Miyamoto Y, Kanatani A, Tamai Y. Targeted disruption of G proteincoupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol 191: 197–205, 2006. [DOI] [PubMed] [Google Scholar]
- 260.McGettigan BM, McMahan RH, Luo Y, Wang XX, Orlicky DJ, Porsche C, Levi M, Rosen HR. Sevelamer improves steatohepatitis, inhibits liver and intestinal farnesoid X receptor (FXR), and reverses innate immune dysregulation in a mouse model of non-alcoholic fatty liver disease. J Biol Chem 291: 23058–23067, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Meihoff WE, Kern F. Bile salt malabsorption in regional ileitis, ileal resection, and mannitol-induced diarrhea. J Clin Invest 47: 261–267, 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mekjian HS, Phillips SF, Hofmann AF. Colonic secretion of water and electrolytes induced by bile acids: Perfusion studies in man. J Clin Invest 50: 1569–1577, 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Metry M, Felton J, Cheng K, Xu S, Ai Y, Xue F, Raufman J-P, Polli JE. Attenuated accumulation of novel fluorine (19F)-labeled bile acid analogues in gallbladders of fibroblast growth factor-15 (FGF15)-deficient mice. Mol Pharm 15: 4827–4834, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Miethke AG, Zhang W, Simmons J, Taylor AE, Shi T, Shanmukhappa SK, Karns R, White S, Jegga AG, Lages CS, Nkinin S, Keller BT, Setchell KDR. Pharmacological inhibition of apical sodium-dependent bile acid transporter changes bile composition and blocks progression of sclerosing cholangitis in multidrug resistance 2 knockout mice. Hepatology 63: 512–523, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Miyata M, Matsuda Y, Nomoto M, Takamatsu Y, Sato N, Hamatsu M, Dawson PA, Gonzalez FJ, Yamazoe Y. Cholesterol feeding prevents hepatic accumulation of bile acids in cholic acid-fed farnesoid X receptor (FXR)-null mice: FXR-independent suppression of intestinal bile acid absorption. Drug Metab Dispos 37: 338–344, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Miyata M, Yamakawa H, Hamatsu M, Kuribayashi H, Takamatsu Y, Yamazoe Y. Enterobacteria modulate intestinal bile acid transport and homeostasis through apical sodium-dependent bile acid transporter (SLC10A2) expression. J Pharmacol Exp Ther 336: 188–196, 2011. [DOI] [PubMed] [Google Scholar]
- 267.Miyata M, Yamakawa H, Hayashi K, Kuribayashi H, Yamazoe Y, Yoshinari K. Ileal apical sodium-dependent bile acid transporter protein levels are down-regulated through ubiquitin-dependent protein degradation induced by bile acids. Eur J Pharmacol 714: 507–514, 2013. [DOI] [PubMed] [Google Scholar]
- 268.Montagnani M, Tsivian M,Neri F, Zvi IB, Mantovani I, Nanni P, Benevento M, Simoni P, Marangoni A, Pariali M, Fato R, Bergamini C, Leoni S, Azzaroli F, Mazzella G, Nardo B, Roda E, Aldini R. A new model for portal protein profile analysis in course of ileal intraluminal bile acid infusion using an in situ perfused rat intestine. Med Chem 7: 257–264, 2011. [DOI] [PubMed] [Google Scholar]
- 269.Monteiro I, David ES, Ferraris RP. Ontogenetic development of rat intestinal bile acid transport requires thyroxine but not corticosterone. Pediatr Res 55: 611–621, 2004. [DOI] [PubMed] [Google Scholar]
- 270.Moore RH, Chothe P, Swaan PW. Transmembrane domain V plays a stabilizing role in the function of human bile acid transporter SLC10A2. Biochemistry 52: 5117–5124, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mottino AD, Hoffman T, Dawson PA, Luquita MG, Monti JA, Sánchez Pozzi EJ, Catania VA, Cao J, Vore M. Increased expression of ileal apical sodium-dependent bile acid transporter in postpartum rats. Am J Physiol Gastrointest Liver Physiol 282: G41–G50, 2002. [DOI] [PubMed] [Google Scholar]
- 272.Mueller M, Thorell A, Claudel T, Jha P, Koefeler H, Lackner C, Hoesel B, Fauler G, Stojakovic T, Einarsson C, Marschall H-U, Trauner M. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J Hepatol 62: 1398–1404, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Mullins JG, Beechey RB, Gould GW, Campbell FC, Shirazi-Beechey SP. Characterization of the ileal Na+/bile salt co-transporter in brush border membrane vesicles and functional expression in Xenopus laevis oocytes. Biochem J 285 (Pt 3): 785–790, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Muthusamy S, Malhotra P, Hosameddin M, Dudeja AK, Borthakur S, Saksena S, Gill RK, Dudeja PK, Alrefai WA. N-glycosylation is essential for ileal ASBT function and protection against proteases. Am J Physiol Cell Physiol 308: C964–C971, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Nakahara M, Furuya N, Takagaki K, Sugaya T, Hirota K, Fukamizu A, Kanda T, Fujii H, Sato R. Ileal bile acid-binding protein, functionally associated with the farnesoid X receptor or the ileal bile acid transporter, regulates bile acid activity in the small intestine. J Biol Chem 280: 42283–42289, 2005. [DOI] [PubMed] [Google Scholar]
- 276.Nakajima A, Seki M, Taniguchi S, Ohta A, Gillberg P-G, Mattsson JP, Camilleri M. Safety and efficacy of elobixibat for chronic constipation: Results from a randomised, double-blind, placebo-controlled, phase 3 trial and an open-label, single-arm, phase 3 trial. Lancet Gastroenterol Hepatol 3: 537–547, 2018. [DOI] [PubMed] [Google Scholar]
- 277.Nakao N, Kaneda H, Tsushima N, Ohta Y, Tanaka M. Characterization of primary structure and tissue expression profile of the chicken apical sodium-dependent bile acid transporter mRNA. Poult Sci 94: 722–727, 2015. [DOI] [PubMed] [Google Scholar]
- 278.Neimark E, Chen F, Li X, Magid MS, Alasio TM, Frankenberg T, Sinha J, Dawson PA, Shneider BL. c-Fos is a critical mediator of inflammatory-mediated repression of the apical sodium-dependent bile acid transporter. Gastroenterology 131: 554–567, 2006. [DOI] [PubMed] [Google Scholar]
- 279.Neimark E, Chen F, Li X, Shneider BL. Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology 40: 149–156, 2004. [DOI] [PubMed] [Google Scholar]
- 280.Nevens F, Andreone P, Mazzella G, Strasser SI, Bowlus C, Invernizzi P, Drenth JPH, Pockros PJ, Regula J, Beuers U, Trauner M, Jones DE, Floreani A, Hohenester S, Luketic V, Shiffman M, van Erpecum KJ, Vargas V, Vincent C, Hirschfield GM, Shah H, Hansen B, Lindor KD, Marschall H-U, Kowdley KV, Hooshmand-Rad R, Marmon T, Sheeron S, Pencek R, MacConell L, Pruzanski M, Shapiro D, POISE Study Group. A placebo-controlled trial of obeticholic acid in primary biliary cholangitis. N Engl J Med 375: 631–643, 2016. [DOI] [PubMed] [Google Scholar]
- 281.Nizamutdinov D, DeMorrow S, McMillin M, Kain J, Mukherjee S, Zeitouni S, Frampton G, Bricker PCS, Hurst J, Shapiro LA. Hepatic alterations are accompanied by changes to bile acid transporterexpressing neurons in the hypothalamus after traumatic brain injury. Sci Rep 7:40112, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Noble CL, Abbas AR, Lees CW, Cornelius J, Toy K, Modrusan Z, Clark HF, Arnott ID, Penman ID, Satsangi J, Diehl L. Characterization of intestinal gene expression profiles in Crohn’s disease by genome-wide microarray analysis. Inflamm Bowel Dis 16: 1717–1728, 2010. [DOI] [PubMed] [Google Scholar]
- 283.Nowicki MJ, Shneider BL, Paul JM, Heubi JE. Glucocorticoids upregulate taurocholate transport by ileal brush-border membrane. Am J Physiol 273: G197–G203, 1997. [DOI] [PubMed] [Google Scholar]
- 284.Oelkers P, Dawson PA. Cloning and chromosomal localization of the human ileal lipid-binding protein. Biochim Biophys Acta 1257: 199–202, 1995. [DOI] [PubMed] [Google Scholar]
- 285.Oelkers P, Kirby LC, Heubi JE, Dawson PA. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2). J Clin Invest 99: 1880–1887, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Okuwaki M, Takada T, Iwayanagi Y, Koh S, Kariya Y, Fujii H, Suzuki H. LXR alpha transactivates mouse organic solute transporter alpha and beta via IR-1 elements shared with FXR. Pharm Res 24:390–398, 2007. [DOI] [PubMed] [Google Scholar]
- 287.Out C, Dikkers A, Laskewitz A, Boverhof R, van der Ley C, Kema IP, Wolters H, Havinga R, Verkade HJ, Kuipers F, Tietge UJF, Groen AK. Prednisolone increases enterohepatic cycling of bile acids by induction of Asbt and promotes reverse cholesterol transport. J Hepatol 61: 351–357, 2014. [DOI] [PubMed] [Google Scholar]
- 288.Out C, Patankar JV, Doktorova M, Boesjes M, Bos T, de Boer S, Havinga R, Wolters H, Boverhof R, van Dijk TH, Smoczek A, Bleich A, Sachdev V, Kratky D, Kuipers F, Verkade HJ, Groen AK. Gut microbiota inhibit Asbt-dependent intestinal bile acid reabsorption via Gata4. J Hepatol 63: 697–704, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Palmer M, Jennings L, Silberg DG, Bliss C, Martin P. A randomised, double-blind, placebo-controlled phase 1 study of the safety, tolerability and pharmacodynamics of volixibat in overweight and obese but otherwise healthy adults: Implications for treatment of non-alcoholic steatohepatitis. BMC Pharmacol Toxicol 19: 10, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Pan DH, Chen F, Neimark E,Li X, Shneider BL. FTF and LRH-1, two related but different transcription factors in human Caco-2 cells: Their different roles in the regulation of bile acid transport. Biochim Biophys Acta 1732: 31–37, 2005. [DOI] [PubMed] [Google Scholar]
- 291.Pan W, Song I-S, Shin H-J, Kim M-H, Choi Y-L, Lim S-J, Kim W-Y, Lee S-S, Shin J-G. Genetic polymorphisms in Na+-taurocholate co-transporting polypeptide (NTCP) and ileal apical sodium-dependent bile acid transporter (ASBT) and ethnic comparisons of functional variants of NTCP among Asian populations. Xenobiotica 41: 501–510, 2011. [DOI] [PubMed] [Google Scholar]
- 292.Pandak WM, Ren S, Marques D, Hall E, Redford K, Mallonee D, Bohdan P, Heuman D, Gil G, Hylemon P. Transport of cholesterol into mitochondria is rate-limiting for bile acid synthesis via the alternative pathway in primary rat hepatocytes. J Biol Chem 277: 48158–48164, 2002. [DOI] [PubMed] [Google Scholar]
- 293.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: Natural ligands for an orphan nuclear receptor. Science 284: 1365–1368, 1999. [DOI] [PubMed] [Google Scholar]
- 294.Patti M-E, Houten SM, Bianco AC, Bernier R, Larsen PR, Holst JJ, Badman MK, Maratos-Flier E, Mun EC, Pihlajamaki J, Auwerx J, Goldfine AB. Serum bile acids are higher in humans with prior gastric bypass: Potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring) 17: 1671–1677, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Pattni SS, Brydon WG, Dew T, Johnston IM, Nolan JD, Srinivas M, Basumani P, Bardhan KD, Walters JRF. Fibroblast growth factor 19 in patients with bile acid diarrhoea: A prospective comparison of FGF19 serum assay and SeHCAT retention. Aliment Pharmacol Ther 38: 967976, 2013. [DOI] [PubMed] [Google Scholar]
- 296.Pattni SS, Brydon WG, Dew T, Walters JRF. Fibroblast growth factor 19 and 7α-hydroxy-4-cholesten-3-one in the diagnosis of patients with possible bile acid diarrhea. Clin Transl Gastroenterol 3: e18, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Pavek P, Pregnane X. Receptor (PXR)-mediated gene repression and cross-talk of PXR with other nuclear receptors via coactivator interactions. Front Pharmacol 7: 456, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Pawlikowska L, Groen A, Eppens EF, Kunne C, Ottenhoff R, Looije N, Knisely AS, Killeen NP, Bull LN, Elferink RPJO, Freimer NB. A mouse genetic model for familial cholestasis caused by ATP8B1 mutations reveals perturbed bile salt homeostasis but no impairment in bile secretion. Hum Mol Genet 13: 881–892, 2004. [DOI] [PubMed] [Google Scholar]
- 299.Pellicciari R, Fiorucci S, Camaioni E, Clerici C, Costantino G, Maloney PR, Morelli A, Parks DJ, Willson TM. 6alpha-Ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem 45: 3569–3572, 2002. [DOI] [PubMed] [Google Scholar]
- 300.Perumpail BJ, Khan MA, Yoo ER, Cholankeril G, Kim D, Ahmed A. Clinical epidemiology and disease burden of nonalcoholic fatty liver disease. World J Gastroenterol 23: 8263–8276, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Pineda Torra I, Claudel T, Duval C, Kosykh V, Fruchart J-C, Staels B. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol Endocrinol 17: 259–272, 2003. [DOI] [PubMed] [Google Scholar]
- 302.Pizones Ruiz-Henestrosa VCM, Bellesi FA, Camino NA, Pilosof AMR. The impact of HPMC structure in the modulation of in vitro lipolysis: The role of bile salts. Food Hydrocoll 62: 251–261, 2017. [Google Scholar]
- 303.Popescu IR, Helleboid-Chapman A, Lucas A, Vandewalle B, Dumont J, Bouchaert E, Derudas B, Kerr-Conte J, Caron S, Pattou F, Staels B. The nuclear receptor FXR is expressed in pancreatic beta-cells and protects human islets from lipotoxicity. FEBS Lett 584: 2845–2851, 2010. [DOI] [PubMed] [Google Scholar]
- 304.Potter GD, Sellin JH, Burlingame SM. Bile acid stimulation of cyclic AMP and ion transport in developing rabbit colon. J Pediatr Gastroen-terol Nutr 13:335–341, 1991. [DOI] [PubMed] [Google Scholar]
- 305.Praslickova D, Torchia EC, Sugiyama MG, Magrane EJ, Zwicker BL, Kolodzieyski L, Agellon LB. The ileal lipid binding protein is required for efficient absorption and transport of bile acids in the distal portion of the murine small intestine. PLoS One 7: e50810, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Prawitt J, Abdelkarim M, Stroeve JHM, Popescu I, Duez H, Velagapudi VR, Dumont J, Bouchaert E, van Dijk TH, Lucas A, Dorchies E, Daoudi M, Lestavel S, Gonzalez FJ, Oresic M, Cariou B, Kuipers F, Caron S, Staels B. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes 60: 1861–1871, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Puleston J, Morgan H, Andreyev J. New treatment for bile salt malabsorption. Gut 54: 441–442, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Quinn CM, Jessup W, Wong J, Kritharides L, Brown AJ. Expression and regulation of sterol 27-hydroxylase (CYP27A1) in human macrophages: A role for RXR and PPARgamma ligands. Biochem J 385: 823–830, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rao A, Haywood J, Craddock AL, Belinsky MG, Kruh GD, Dawson PA. The organic solute transporter alpha-beta, OSTalpha-Ostbeta, is essential for intestinal bile acid transport and homeostasis. Proc Natl Acad Sci USA 105: 3891–3896, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Rao A, Kosters A, Mells JE, Zhang W, Setchell KDR, Amanso AM, Wynn GM, Xu T, Keller BT, Yin H, Banton S, Jones DP, Wu H, Dawson PA, Karpen SJ. Inhibition of ileal bile acid uptake protects against nonalcoholic fatty liver disease in high-fat diet-fed mice. Sci Transl Med 8: 357ra122, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Raufman J-P, Metry M, Felton J, Cheng K, Xu S, Polli J. A 19F magnetic resonance imaging-based diagnostic test for bile acid diarrhea. MAGMA 92: 163–171, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Reiss AB, Martin KO, Rojer DE, Iyer S, Grossi EA, Galloway AC, Javitt NB. Sterol 27-hydroxylase: Expression in human arterial endothelium. J Lipid Res 38: 1254–1260, 1997. [PubMed] [Google Scholar]
- 313.Renga B, Mencarelli A, Vavassori P, Brancaleone V, Fiorucci S. The bile acid sensor FXR regulates insulin transcription and secretion. Biochim Biophys Acta 1802: 363–372, 2010. [DOI] [PubMed] [Google Scholar]
- 314.Renner O, Harsch S, Schaeffeler E, Schwab M, Klass DM, Kratzer W, Stange EF. Mutation screening of apical sodium-dependent bile acid transporter (SLC10A2): Novel haplotype block including six newly identified variants linked to reduced expression. Hum Genet 125: 381–391,2009. [DOI] [PubMed] [Google Scholar]
- 315.Repa JJ, Lund EG, Horton JD, Leitersdorf E, Russell DW, Dietschy JM, Turley SD. Disruption of the sterol 27-hydroxylase gene in mice results in hepatomegaly and hypertriglyceridemia. Reversal by cholic acid feeding. J Biol Chem 275: 39685–39692, 2000. [DOI] [PubMed] [Google Scholar]
- 316.Reuben A. The biliary cycle of Moritz Schiff. Hematology 42: 500–505,2005. [DOI] [PubMed] [Google Scholar]
- 317.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol 30: 332–338, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Root C, Smith CD, Sundseth SS, Pink HM, Wilson JG, Lewis MC. Ileal bile acid transporter inhibition, CYP7A1 induction, and antilipemic action of 264W94. J Lipid Res 43: 1320–1330, 2002. [PubMed] [Google Scholar]
- 319.Rosen H, Gonzalez-Cabrera PJ, Sanna MG, Brown S. Sphingosine 1-phosphate receptor signaling. Annu Rev Biochem 78: 743–768, 2009. [DOI] [PubMed] [Google Scholar]
- 320.Rössel P, Sortsøe Jensen H, Qvist P, Arveschoug A. Prognosis of adult-onset idiopathic bile acid malabsorption. Scand J Gastroenterol 34: 587–590, 1999. [DOI] [PubMed] [Google Scholar]
- 321.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 72: 137–174, 2003. [DOI] [PubMed] [Google Scholar]
- 322.Sabit H, Mallajosyula SS, MacKerell AD, Swaan PW. Transmembrane domain II of the human bile acid transporter SLC10A2 coordinates sodium translocation. J Biol Chem 288: 32394–32404, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Sadik R, Abrahamsson H, Ung K-A, Stotzer P-O. Accelerated regional bowel transit and overweight shown in idiopathic bile acid malabsorption. Am J Gastroenterol 99: 711–718, 2004. [DOI] [PubMed] [Google Scholar]
- 324.Saeki T, Kuroda T, Matsumoto M, Kanamoto R, Iwami K. Effects of Cys mutation on taurocholic acid transport by mouse ileal and hepatic sodium-dependent bile acid transporters. Biosci Biotechnol Biochem 66: 467–470, 2002. [DOI] [PubMed] [Google Scholar]
- 325.Saeki T, Matoba K, Furukawa H, Kirifuji K, Kanamoto R, Iwami K. Characterization, cDNA cloning, and functional expression of mouse ileal sodium-dependent bile acid transporter. J Biochem 125: 846–851, 1999. [DOI] [PubMed] [Google Scholar]
- 326.Saeki T, Mizushima S, Ueda K, Iwami K, Kanamoto R. Mutational analysis of uncharged polar residues and proline in the distal one-third (Thr130-Pro142) of the highly conserved region of mouse Slc10a2. Biosci Biotechnol Biochem 73: 1535–1540, 2009. [DOI] [PubMed] [Google Scholar]
- 327.Saeki T, Munetaka Y, Ueda K, Iwami K, Kanamoto R. Effects of Ala substitution for conserved Cys residues in mouse ileal and hepatic Na+-dependent bile acid transporters. Biosci Biotechnol Biochem 71: 1865–1872, 2007. [DOI] [PubMed] [Google Scholar]
- 328.Saeki T, Sato K, Ito S, Ikeda K, Kanamoto R. Importance of uncharged polar residues and proline in the proximal two-thirds (Pro107-Ser128) of the highly conserved region of mouse ileal Na+-dependent bile acid transporter, Slc10a2, in transport activity and cellular expression. BMC Physiol 13: 4, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Sarwar Z, Annaba F, Dwivedi A, Saksena S, Gill RK, Alrefai WA. Modulation of ileal apical Na+-dependent bile acid transporter ASBT by protein kinase C. Am J Physiol Gastrointest Liver Physiol 297: G532–G538, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Sauter G. Serum concentrations of 7alpha-hydroxy-4-cholesten-3-one reflect bile acid synthesis in humans. Hepatology 24: 123–126, 1996. [DOI] [PubMed] [Google Scholar]
- 331.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, Angelin B, Hyötyläinen T, Oresic M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17: 225–235, 2013. [DOI] [PubMed] [Google Scholar]
- 332.Schaffner CA, Mwinyi J, Gai Z, Thasler WE, Eloranta JJ, Kullak-Ublick GA. The organic solute transporters alpha and beta are induced by hypoxia in human hepatocytes. Liver Int 35: 1152–1161, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Schiff ER, Small NC, Dietschy JM. Characterization of the kinetics of the passive and active transport mechanisms for bile acid absorption in the small intestine and colon of the rat. J Clin Invest 51: 1351–1362, 1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Schmidt DR, Holmstrom SR, Fon Tacer K, Bookout AL, Kliewer SA, Mangelsdorf DJ. Regulation of bile acid synthesis by fat-soluble vitamins A and D. J Biol Chem 285: 14486–14494, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Schölmerich J, Becher MS, Schmidt K, Schubert R, Kremer B, Feldhaus S, Gerok W. Influence of hydroxylation and conjugation of bile salts on their membrane-damaging properties—studies on isolated hepatocytes and lipid membrane vesicles. Hepatology 4: 661–666, 1984. [DOI] [PubMed] [Google Scholar]
- 336.Schote AB, Turner JD, Schiltz J, Muller CP. Nuclear receptors in human immune cells: Expression and correlations. Mol Immunol 44: 1436–1445, 2007. [DOI] [PubMed] [Google Scholar]
- 337.Schwarz M, Lund EG, Setchell KD, Kayden HJ, Zerwekh JE, Bjorkhem I, Herz J, Russell DW. Disruption of cholesterol 7alpha-hydroxylase gene in mice. II. Bile acid deficiency is overcome by induction of oxysterol 7alpha-hydroxylase. J Biol Chem 271: 18024–18031, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Schwarz M, Russell DW, Dietschy JM, Turley SD. Marked reduction in bile acid synthesis in cholesterol 7alpha-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. J Lipid Res 39: 1833–1843, 1998. [PubMed] [Google Scholar]
- 339.Seward DJ, Koh AS, Boyer JL, Ballatori N. Functional complementation between a novel mammalian polygenic transport complex and an evolutionarily ancient organic solute transporter, OSTalpha-OSTbeta. J Biol Chem 278: 27473–27482, 2003. [DOI] [PubMed] [Google Scholar]
- 340.Shah S, Sanford UR, Vargas JC, Xu H, Groen A, Paulusma CC, Grenert JP, Pawlikowska L, Sen S, Elferink RPJO, Bull LN. Strain background modifies phenotypes in the ATP8B1-deficient mouse. PLoS ONE 5: e8984, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Sharma V, Hiller M. Loss of enzymes in the bile acid synthesis pathway explains differences in bile composition among mammals. Genome Biol Evol 10: 3211–3217, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, Stoffel M. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet 27: 375–382, 2001. [DOI] [PubMed] [Google Scholar]
- 343.Shneider BL, Dawson PA, Christie DM, Hardikar W, Wong Mh, Suchy FJ. Cloning and molecular characterization of the ontogeny of a rat ileal sodium-dependent bile acid transporter. J Clin Invest 95: 745–754, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Shneider BL, Setchell KD, Crossman MW. Fetal and neonatal expression of the apical sodium-dependent bile acid transporter in the rat ileum and kidney. Pediatr Res 42: 189–194, 1997. [DOI] [PubMed] [Google Scholar]
- 345.Shneider BL, Spino C, Kamath BM, Magee JC, Bass LM, Setchell KD, Miethke A, Molleston JP, Mack CL, Squires RH, Murray KF, Loomes KM, Rosenthal P, Karpen SJ, Leung DH, Guthery SL, Thomas D, Sherker AH, Sokol RJ, Childhood Liver Disease Research Network. Placebo-controlled randomized trial of an intestinal bile salt transport inhibitor for pruritus in Alagille syndrome. Hepatol Commun 2: 1184–1198,2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Sievänen E. Exploitation of bile acid transport systems in prodrug design. Molecules 12: 1859–1889, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Simon FR, Sutherland J, Sutherland E. Identification of taurocholate binding sites in ileal plasma membrane. Am J Physiol 259: G394–G401, 1990. [DOI] [PubMed] [Google Scholar]
- 348.Simrén M, Bajor A, Gillberg P-G, Rudling M, Abrahamsson H. Randomised clinical trial: The ileal bile acid transporter inhibitor A3309 vs. placebo in patients with chronic idiopathic constipation—a doubleblind study. Aliment Pharmacol Ther 34: 41–50, 2011. [DOI] [PubMed] [Google Scholar]
- 349.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102: 731–744, 2000. [DOI] [PubMed] [Google Scholar]
- 350.Sinha J, Chen F, Miloh T, Burns RC, Yu Z, Shneider BL. beta-Klotho and FGF-15/19 inhibit the apical sodium-dependent bile acid transporter in enterocytes and cholangiocytes. Am J Physiol Gastrointest Liver Physiol 295: G996–G1003, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Slattery SA, Niaz O, Aziz Q, Ford AC, Farmer AD. Systematic review with meta-analysis: The prevalence of bile acid malabsorption in the irritable bowel syndrome with diarrhoea. Aliment Pharmacol Ther 42: 3–11, 2015. [DOI] [PubMed] [Google Scholar]
- 352.Soler DM, Ghosh A, Chen F, Shneider BL. A single element in the 3’UTR of the apical sodium-dependent bile acid transporter controls both stabilization and destabilization of mRNA. Biochem J 462: 547–553, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Srivastava A. Progressive familial intrahepatic cholestasis. J Clin Exp Hepatol 4: 25–36, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Staels B. A review of bile acid sequestrants: Potential mechanism(s) for glucose-lowering effects in type 2 diabetes mellitus. Postgrad Med 121: 25–30, 2009. [DOI] [PubMed] [Google Scholar]
- 355.Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol 101: 47–64, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A 98:3369–3374,2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Stelzner M, Hoagland V, Somasundaram S. Distribution of bile acid absorption and bile acid transporter gene message in the hamster ileum. Pflugers Arch 440: 157–162, 2000. [DOI] [PubMed] [Google Scholar]
- 358.Stengelin S, Apel S, Becker W, Maier M, Rosenberger J, Bewersdorf U, Girbig F, Weyland C, Wess G, Kramer W. The rabbit ileal lipid-binding protein. Gene cloning and functional expression of the recombinant protein. EurJ Biochem 239: 887–896, 1996. [DOI] [PubMed] [Google Scholar]
- 359.Stravitz RT, Sanyal AJ, Pandak WM, Vlahcevic ZR, Beets JW, Dawson PA. Induction of sodium-dependent bile acid transporter messenger RNA, protein, and activity in rat ileum by cholic acid. Gastroenterology 113: 1599–1608, 1997. [DOI] [PubMed] [Google Scholar]
- 360.Stroeve JHM, Brufau G, Stellaard F, Gonzalez FJ, Staels B, Kuipers F. Intestinal FXR-mediated FGF15 production contributes to diurnal control of hepatic bile acid synthesis in mice. Lab Invest 90:1457–1467, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, Pandak WM, Dent P, Spiegel S, Shi R, Xu W, Liu X, Bohdan P, Zhang L, Zhou H, Hylemon PB. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology 55: 267–276, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Styles NA, Falany JL, Barnes S, Falany CN. Quantification and regulation of the subcellular distribution of bile acid coenzyme A: Amino acid N-acyltransferase activity in rat liver. J Lipid Res 48: 1305–1315, 2007. [DOI] [PubMed] [Google Scholar]
- 363.Suchy FJ, Balistreri WF. Uptake of taurocholate by hepatocytes isolated from developing rats. Pediatr Res 16: 282–285, 1982. [DOI] [PubMed] [Google Scholar]
- 364.Suchy FJ, Balistreri WF, Heubi JE, Searcy JE, Levin RS. Physiologic cholestasis: Elevation of the primary serum bile acid concentrations in normal infants. Gastroenterology 80: 1037–1041, 1981. [PubMed] [Google Scholar]
- 365.Suchy FJ, Courchene SM, Balistreri WF. Ontogeny of hepatic bile acid conjugation in the rat. Pediatr Res 19: 97–101, 1985. [DOI] [PubMed] [Google Scholar]
- 366.Sultan M, Rao A, Elpeleg O, Vaz FM, Abu-Libdeh B, Karpen SJ, Dawson PA. Organic solute transporter-β (SLC51B) deficiency in two brothers with congenital diarrhea and features of cholestasis. Hepatology 68: 590–598, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Sun AQ, Ananthanarayanan M, Soroka CJ, Thevananther S, Shneider BL, Suchy FJ. Sorting of rat liver and ileal sodium-dependent bile acid transporters in polarized epithelial cells. Am J Physiol 275: G1045–G1055, 1998. [DOI] [PubMed] [Google Scholar]
- 368.Sun A-Q, Balasubramaniyan N, Chen H, Shahid M, Suchy FJ. Identification of functionally relevant residues of the rat ileal apical sodium-dependent bile acid cotransporter. J Biol Chem 281: 16410–16418, 2006. [DOI] [PubMed] [Google Scholar]
- 369.Sun A-Q, Balasubramaniyan N, Xu K, Liu CJ, Ponamgi VM, Liu H, Suchy FJ. Protein-protein interactions and membrane localization of the human organic solute transporter. Am J Physiol Gastrointest Liver Physiol 292: G1586–G1593, 2007. [DOI] [PubMed] [Google Scholar]
- 370.Sun A-Q, Salkar R, Sachchidanand, Xu S, Zeng L, Zhou M-M, Suchy FJ. A 14-amino acid sequence with a beta-turn structure is required for apical membrane sorting of the rat ileal bile acid transporter. J Biol Chem 278: 4000–4009, 2003. [DOI] [PubMed] [Google Scholar]
- 371.Sun A-Q, Zhu L, Luo Y, Xu S, Lin J, Suchy FJ. Human Organic Solute Transporter (hOST): Protein interaction and membrane sorting process. Int J Biochem Mol Biol 3: 290–301, 2012. [PMC free article] [PubMed] [Google Scholar]
- 372.Suzuki T, Oba K, Igari Y, Matsumura N, Watanabe K, Futami-Suda S, Yasuoka H, Ouchi M, Suzuki K, Kigawa Y, Nakano H. Colestimide lowers plasma glucose levels and increases plasma glucagon-like PEPTIDE-1 (7–36) levels in patients with type 2 diabetes mellitus complicated by hypercholesterolemia. J Nippon Med Sch 74: 338–343, 2007. [DOI] [PubMed] [Google Scholar]
- 373.Takahashi S, Fukami T, Masuo Y, Brocker CN, Xie C, Krausz KW, Wolf CR, Henderson CJ, Gonzalez FJ. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J Lipid Res 57: 2130–2137, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Tarling EJ, Clifford BL, Cheng J, Morand P, Cheng A, Lester E, Sallam T, Turner M, de Aguiar Vallim TQ. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J Clin Invest 127: 3741–3754, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Mac-chiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K. TGR5-mediatedbile acid sensing controls glucose homeostasis. Cell Metab 10: 167–177, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Thomas C, Landrier JF, Gaillard D, Grober J, Monnot M-C, Athias A, Besnard P. Cholesterol dependent downregulation of mouse and human apical sodium dependent bile acid transporter (ASBT) gene expression: Molecular mechanism and physiological consequences. Gut 55: 1321–1331, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ticho AL, Lee H, Gill RK, Dudeja PK, Saksena S, Lee D, Alrefai WA. A novel bioluminescence-based method to investigate uptake of bile acids in living cells. Am J Physiol Gastrointest Liver Physiol 315: G529–G537, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Ticho AL, Malhotra P, Dudeja PK, Gill RK, Alrefai WA. Bile acid receptors and gastrointestinal functions. Liver Res, 2019. DOI: 10.1016/j.livres.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Tiessen RG, Kennedy CA, Keller BT, Levin N, Acevedo L, Gedulin B, van Vliet AA, Dorenbaum A, Palmer M. Safety, tolerability and pharmacodynamics of apical sodium-dependent bile acid transporter inhibition with volixibat in healthy adults and patients with type 2 diabetes mellitus: A randomised placebo-controlled trial. BMC Gastroenterol 18:3,2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Tochtrop GP, Bruns JL, Tang C, Covey DF, Cistola DP. Steroid ring hydroxylation patterns govern cooperativity in human bile acid binding protein. Biochemistry 42: 11561–11567, 2003. [DOI] [PubMed] [Google Scholar]
- 381.Tochtrop GP, DeKoster GT, Covey DF, Cistola DP. A single hydroxyl group governs ligand site selectivity in human ileal bile acid binding protein. J Am Chem Soc 126: 11024–11029, 2004. [DOI] [PubMed] [Google Scholar]
- 382.Tochtrop GP, Richter K, Tang C, Toner JJ, Covey DF, Cistola DP. Energetics by NMR: Site-specific binding in a positively cooperative system. Proc Natl Acad Sci U S A 99: 1847–1852, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Toke O, Monsey JD, Cistola DP. Kinetic mechanism of ligand binding in human ileal bile acid binding protein as determined by stopped-flow fluorescence analysis. Biochemistry 46: 5427–5436, 2007. [DOI] [PubMed] [Google Scholar]
- 384.Toke O, Monsey JD, DeKoster GT, Tochtrop GP, Tang C, Cistola DP. Determinants of cooperativity and site selectivity in human ileal bile acid binding protein. Biochemistry 45: 727–737, 2006. [DOI] [PubMed] [Google Scholar]
- 385.Tolle-Sander S, Lentz KA, Maeda DY, Coop A, Polli JE. Increased acyclovir oral bioavailability via a bile acid conjugate. Mol Pharm 1: 40–48, 2004. [DOI] [PubMed] [Google Scholar]
- 386.Torcello-Gómez A, Fernández Fraguas C, Ridout MJ, Woodward NC, Wilde PJ, Foster TJ. Effect of substituent pattern and molecular weight of cellulose ethers on interactions with different bile salts. Food Funct 6: 730–739, 2015. [DOI] [PubMed] [Google Scholar]
- 387.Turpin ER, Fang H-J, Thomas NR, Hirst JD. Cooperativity and site selectivity in the ileal lipid binding protein. Biochemistry 52: 4723–4733, 2013. [DOI] [PubMed] [Google Scholar]
- 388.Vagne M, Mutt V. Entero-oxyntin: A stimulant of gastric acid secretion extracted from porcine intestine. Scand J Gastroenterol 15: 17–22, 1980. [DOI] [PubMed] [Google Scholar]
- 389.Van de Wiel S, Merkx M, Van de Graaf S. Real time monitoring of intracellular bile acid dynamics using a genetically encoded FRET-based bile acid sensor. J Vis Exp: e53659, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.van de Wiel SMW, de Waart DR, Oude Elferink RPJ, van de Graaf SFJ. Intestinal farnesoid X receptor activation by pharmacologic inhibition of the organic solute transporter α-β. Cell Mol Gastroenterol Hepatol 5: 223–237,2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.van der Mark VA, de Waart DR, Ho-Mok KS, Tabbers MM, Voogt HW, Oude Elferink RPJ, Knisely AS, Paulusma CC. The lipid flippase heterodimer ATP8B1-CDC50A is essential for surface expression of the apical sodium-dependent bile acid transporter (SLC10A2/ASBT) in intestinal Caco-2 cells. Biochim Biophys Acta 1842: 2378–2386, 2014. [DOI] [PubMed] [Google Scholar]
- 392.van der Velden LM, Golynskiy MV, Bijsmans ITGW, van Mil SWC, Klomp LWJ, Merkx M, van de Graaf SFJ. Monitoring bile acid transport in single living cells using a genetically encoded Förster resonance energy transfer sensor. Hepatology 57: 740–752, 2013. [DOI] [PubMed] [Google Scholar]
- 393.van Tilburg AJ, de Rooij FW, van den Berg JW, van Blankenstein M. Primary bile acid diarrhoea without an ileal carrier defect: Quantification of active bile acid transport across the ileal brush border membrane. Gut 32:500–503, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol 183: 6251–6261, 2009. [DOI] [PubMed] [Google Scholar]
- 395.Vijayvargiya P, Camilleri M, Carlson P, Lueke A, O’Neill J, Burton D, Busciglio I, Donato L. Performance characteristics of serum C4 and FGF19 measurements to exclude the diagnosis of bile acid diarrhoea in IBS-diarrhoea and functional diarrhoea. Aliment Pharmacol Ther 46: 581–588, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Vijayvargiya P, Camilleri M, Shin A, Saenger A. Methods for diagnosis of bile acid malabsorption in clinical practice. Clin Gastroenterol Hepatol 11: 1232–1239, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Vivian D, Cheng K, Khurana S, Xu S, Dawson PA, Raufman J-P, Polli JE. Design and evaluation of a novel trifluorinated imaging agent for assessment of bile acid transport using fluorine magnetic resonance imaging. J Pharm Sci 103: 3782–3792, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Vivian D, Cheng K, Khurana S, Xu S, Kriel EH, Dawson PA, Raufman J-P, Polli JE. In vivo performance of a novel fluorinated magnetic resonance imaging agent for functional analysis of bile acid transport. Mol Pharm 11: 1575–1582, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Vivian D, Cheng K, Khurana S, Xu S, Whiterock V, Witter D, Lentz KA, Santone KS, Raufman J-P, Polli JE. Design and characterization of a novel fluorinated magnetic resonance imaging agent for functional analysis of bile acid transporter activity. Pharm Res 30: 1240–1251,2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Vodenlich AD, Gong YZ, Geoghegan KF, Lin MC, Lanzetti AJ, Wilson FA. Identification of the 14 kDa bile acid transport protein of rat ileal cytosol as gastrotropin. Biochem Biophys Res Commun 177: 1147–1154, 1991. [DOI] [PubMed] [Google Scholar]
- 401.Wagner M, Halilbasic E, Marschall H-U, Zollner G, Fickert P, Langner C, Zatloukal K, Denk H, Trauner M. CAR and PXR agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology 42: 420–430, 2005. [DOI] [PubMed] [Google Scholar]
- 402.Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24: 41–50, 2016. [DOI] [PubMed] [Google Scholar]
- 403.Walters JRF, Johnston IM, Nolan JD, Vassie C, Pruzanski ME, Shapiro DA. The response of patients with bile acid diarrhoea to the farnesoid X receptor agonist obeticholic acid. Aliment Pharmacol Ther 41: 54–64, 2015. [DOI] [PubMed] [Google Scholar]
- 404.Walters JRF, Tasleem AM, Omer OS, Brydon WG, Dew T, le Roux CW. A new mechanism for bile acid diarrhea: Defective feedback inhibition of bile acid biosynthesis. Clin Gastroenterol Hepatol 7: 1189–1194, 2009. [DOI] [PubMed] [Google Scholar]
- 405.Walz DA, Wider MD, Snow JW, Dass C, Desiderio DM. The complete amino acid sequence of porcine gastrotropin, an ileal protein which stimulates gastric acid and pepsinogen secretion. J Biol Chem 263: 14189–14195, 1988. [PubMed] [Google Scholar]
- 406.Wang DQ-H, Tazuma S, Cohen DE, Carey MC. Feeding natural hydrophilic bile acids inhibits intestinal cholesterol absorption: Studies in the gallstone-susceptible mouse. Am J Physiol Gastrointest Liver Physiol 285: G494–G502, 2003. [DOI] [PubMed] [Google Scholar]
- 407.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell 3: 543–553, 1999. [DOI] [PubMed] [Google Scholar]
- 408.Wang L, Lee Y-K, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, Moore DD. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell 2: 721–731, 2002. [DOI] [PubMed] [Google Scholar]
- 409.Wang W, Seward DJ, Li L, Boyer JL, Ballatori N. Expression cloning of two genes that together mediate organic solute and steroid transport in the liver of a marine vertebrate. Proc Natl Acad Sci USA 98: 9431–9436, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Wang Y, Aoki H, Yang J, Peng K, Liu R, Li X, Qiang X, Sun L, Gurley EC, Lai G, Zhang L, Liang G, Nagahashi M, Takabe K, Pandak WM, Hylemon PB, Zhou H. The role of sphingosine 1-phosphate receptor 2 in bile-acid-induced cholangiocyte proliferation and cholestasis-induced liver injury in mice. Hepatology 65: 2005–2018, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439: 484–489, 2006. [DOI] [PubMed] [Google Scholar]
- 412.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest 113: 1408–1418,2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Wedlake L, A’Hern R, Russell D, Thomas K, Walters JRF, Andreyev HJN. Systematic review: The prevalence of idiopathic bile acid malabsorption as diagnosed by SeHCAT scanning in patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther 30: 707–717, 2009. [DOI] [PubMed] [Google Scholar]
- 414.Weerachayaphorn J, Mennone A, Soroka CJ, Harry K, Hagey LR, Kensler TW, Boyer JL. Nuclear factor-E2-related factor 2 is a major determinant of bile acid homeostasis in the liver and intestine. Am J Physiol Gastrointest Liver Physiol 302: G925–G936, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Weihrauch D, Kanchanapoo J, Ao M, Prasad R, Piyachaturawat P, Rao MC. Weanling, but not adult, rabbit colon absorbs bile acids: Flux is linked to expression of putative bile acid transporters. Am J Physiol Gastrointest Liver Physiol 290: G439–G450, 2006. [DOI] [PubMed] [Google Scholar]
- 416.Weinman SA. Electrogenicity of Na(+)-coupled bile acid transporters. Yale J Biol Med 70: 331–340, 1997. [PMC free article] [PubMed] [Google Scholar]
- 417.Weinman SA, Carruth MW, Dawson PA. Bile acid uptake via the human apical sodium-bile acid cotransporter is electrogenic. J Biol Chem 273: 34691–34695, 1998. [DOI] [PubMed] [Google Scholar]
- 418.West KL, Zern TL, Butteiger DN, Keller BT, Fernandez ML. SC-435, an ileal apical sodium co-dependent bile acid transporter (ASBT) inhibitor lowers plasma cholesterol and reduces atherosclerosis in guinea pigs. Atherosclerosis 171: 201–210, 2003. [DOI] [PubMed] [Google Scholar]
- 419.Wheeler SG, Hammond CL, Jornayvaz FR, Samuel VT, Shulman GI, Soroka CJ, Boyer JL, Hinkle PM, Ballatori N. OSTα−/− mice exhibit altered expression of intestinal lipid absorption genes, resistance to age-related weight gain, and modestly improved insulin sensitivity. Am J Physiol Gastrointest Liver Physiol 306: G425–G438, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Wojtal KA, Eloranta JJ, Hruz P, Gutmann H, Drewe J, Staumann A, Beglinger C, Fried M, Kullak-Ublick GA, Vavricka SR. Changes in mRNA expression levels of solute carrier transporters in inflammatory bowel disease patients. Drug Metab Dispos 37: 1871–1877, 2009. [DOI] [PubMed] [Google Scholar]
- 421.Wong BS, Camilleri M, Carlson P, McKinzie S, Busciglio I, Bondar O, Dyer RB, Lamsam J, Zinsmeister AR. Increased bile acid biosynthesis is associated with irritable bowel syndrome with diarrhea. Clin Gastroenterol Hepatol 10: 1009–15.e3, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Wong MH, Oelkers P, Craddock AL, Dawson PA. Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J Biol Chem 269: 1340–1347, 1994. [PubMed] [Google Scholar]
- 423.Wong MH, Oelkers P, Dawson PA. Identification of a mutation in the ileal sodium-dependent bile acid transporter gene that abolishes transport activity. J Biol Chem 270: 27228–27234, 1995. [DOI] [PubMed] [Google Scholar]
- 424.Wong MH, Rao PN, Pettenati MJ, Dawson PA. Localization of the ileal sodium-bile acid cotransporter gene (SLC10A2) to human chromosome 13q33. Genomics 33: 538–540, 1996. [DOI] [PubMed] [Google Scholar]
- 425.Wu T, Bound MJ, Standfield SD, Gedulin B, Jones KL, Horowitz M, Rayner CK. Effects of rectal administration of taurocholic acid on glucagon-like peptide-1 and peptide YY secretion in healthy humans. Diabetes Obes Metab 15: 474–477, 2013. [DOI] [PubMed] [Google Scholar]
- 426.Wu Y, Aquino CJ, Cowan DJ, Anderson DL, Ambroso JL, Bishop MJ, Boros EE, Chen L, Cunningham A, Dobbins RL, Feldman PL, Harston LT, Kaldor IW, Klein R, Liang X, McIntyre MS, Merrill CL, Patterson KM, Prescott JS, Ray JS, Roller SG, Yao X, Young A, Yuen J, Collins JL. Discovery of a highly potent, nonabsorbable apical sodium-dependent bile acid transporter inhibitor (GSK2330672) for treatment of type 2 diabetes. J Med Chem 56: 5094–5114, 2013. [DOI] [PubMed] [Google Scholar]
- 427.Xia X, Roundtree M, Merikhi A, Lu X, Shentu S, LeSage G. Degradation of the apical sodium-dependent bile acid transporter by the ubiquitin-proteasome pathway in cholangiocytes. J Biol Chem 279: 44931–44937, 2004. [DOI] [PubMed] [Google Scholar]
- 428.Xiao Y, Yan W, Zhou K, Cao Y, Cai W. Glucocorticoid treatment alters systemic bile acid homeostasis by regulating the biosynthesis and transport ofbile salts. Dig LiverDis 48: 771–779, 2016. [DOI] [PubMed] [Google Scholar]
- 429.Xie C, Jiang C, Shi J, Gao X, Sun D, Sun L, Wang T, Takahashi S, Anitha M, Krausz KW, Patterson AD, Gonzalez FJ. An intestinal farnesoid X receptor-ceramide signaling axis modulates hepatic gluconeogenesis in mice. Diabetes 66: 613–626, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A 98: 3375–3380, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Xu S, Soroka CJ, Sun A-Q, Backos DS, Mennone A, Suchy FJ, Boyer JL. A novel Di-Leucine motif at the N-terminus of human organic solute transporter beta is essential for protein association and membrane localization. PLoS One 11: e0158269, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: Trends, predictions, riskfactors and prevention. Nat Rev Gastroenterol Hepatol 15: 11–20, 2018. [DOI] [PubMed] [Google Scholar]
- 433.Zaghini I, Landrier J-F, Grober J, Krief S, Jones SA, Monnot M-C, Lefrere I, Watson MA, Collins JL, Fujii H, Besnard P. Sterol regulatory element-binding protein-1c is responsible for cholesterol regulation of ileal bile acid-binding protein gene in vivo. Possible involvement of liver-X-receptor. J Biol Chem 277: 1324–1331, 2002. [DOI] [PubMed] [Google Scholar]
- 434.Zanzoni S, Assfalg M, Giorgetti A, D’Onofrio M, Molinari H. Structural requirements for cooperativity in ileal bile acid-binding proteins. J Biol Chem 286: 39307–39317, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Zelcer N, van de Wetering K, de Waart R, Scheffer GL, Marschall H-U, Wielinga PR, Kuil A, Kunne C, Smith A, van der Valk M, Wijnholds J, Elferink RO, Borst P. Mice lacking Mrp3 (Abcc3) have normal bile salt transport, but altered hepatic transport of endogenous glucuronides. J Hepatol 44: 768–775, 2006. [DOI] [PubMed] [Google Scholar]
- 436.Zhang EY, Phelps MA, Banerjee A, Khantwal CM, Chang C, Helsper F, Swaan PW. Topology scanning and putative three-dimensional structure of the extracellular binding domains of the apical sodium-dependent bile acid transporter (SLC10A2). Biochemistry 43: 11380–11392, 2004. [DOI] [PubMed] [Google Scholar]
- 437.Zhang JH, Nolan JD, Kennie SL, Johnston IM, Dew T, Dixon PH, Williamson C, Walters JRF. Potent stimulation of fibroblast growth factor 19 expression in the human ileum by bile acids. Am J Physiol Gastrointest Liver Physiol 304: G940–G948, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Zhang Y, Ge X, Heemstra LA, Chen W-D, Xu J, Smith JL, Ma H, Kasim N, Edwards PA, Novak CM. Loss of FXR protects against diet-induced obesity and accelerates liver carcinogenesis in ob/ob mice. Mol Endocrinol 26: 272–280, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad SciU S A 103: 1006–1011,2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Zheng X, Pan Y, Acharya C, Swaan PW, Polli JE. Structural requirements of the ASBT by 3D-QSAR analysis using aminopyridine conjugates of chenodeoxycholic acid. Bioconjug Chem 21: 2038–2048,2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Zhou X, Levin EJ, Pan Y, McCoy JG, Sharma R, Kloss B, Brnni R, Quick M, Zhou M. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature 505: 569–573, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Zimmerman AW, van Moerkerk HT, Veerkamp JH. Ligand specificity and conformational stability of human fatty acid-binding proteins. Int J Biochem Cell Biol 33: 865–876, 2001. [DOI] [PubMed] [Google Scholar]
- 443.Zollner G, Wagner M, Moustafa T, Fickert P, Silbert D, Gumhold J, Fuchsbichler A, Halilbasic E, Denk H, Marschall H-U, Trauner M. Coordinated induction of bile acid detoxification and alternative elimination in mice: Role of FXR-regulated organic solute transporteralpha/beta in the adaptive response to bile acids. Am J Physiol Gastrointest Liver Physiol 290: G923–G932, 2006. [DOI] [PubMed] [Google Scholar]