Abstract
Background
Recent studies suggest that microRNAs (miRNAs) can participate in depression pathogenesis by altering a host of genes that are critical in corticolimbic functioning. The present study focuses on examining whether alterations in the miRNA network in the amygdala are associated with susceptibility or resiliency to develop depression-like behavior in rats.
Methods
Amygdala-specific altered miRNA transcriptomics were determined in a rat depression model following next-generation sequencing method. Target prediction analyses (cis- and trans) and qPCR-based assays were performed to decipher the functional role of altered miRNAs. miRNA-specific target interaction was determined using in vitro transfection assay in neuroblastoma cell line. miRNA-specific findings from the rat in vivo model were further replicated in postmortem amygdala of major depressive disorder (MDD) subjects.
Results
Changes in miRNome identified 17 significantly upregulated and 8 significantly downregulated miRNAs in amygdala of learned helpless (LH) compared with nonlearned helpless rats. Prediction analysis showed that the majority of the upregulated miRNAs had target genes enriched for the Wnt signaling pathway. Among altered miRNAs, upregulated miR-128-3p was identified as a top hit based on statistical significance and magnitude of change in LH rats. Target validation showed significant downregulation of Wnt signaling genes in amygdala of LH rats. A discernable increase in expression of amygdalar miR-128-3p along with significant downregulation of key target genes from Wnt signaling (WNT5B, DVL, and LEF1) was noted in MDD subjects. Overexpression of miR-128-3p in a cellular model lead to a marked decrease in the expression of Dvl1 and Lef1 genes, confirming them as validated targets of miR-128-3p. Additional evidence suggested that the amygdala-specific diminished expression of transcriptional repressor Snai1 could be potentially linked to induced miR-128-2 expression in LH rats. Furthermore, an amygdala-specific posttranscriptional switching mechanism could be active between miR-128-3p and RNA binding protein Arpp21 to gain control over their target genes such as Lef1.
Conclusion
Our study suggests that in amygdala a specific set of miRNAs may play an important role in depression susceptibility, which could potentially be mediated through Wnt signaling.
Keywords: amygdala, miRNA, stress, depression, Wnt signaling, epigenetics
Significance Statement.
The amygdala is part of the limbic system involved in memory modulation, fear, and stress response as well as emotional regulation. Recent studies suggest that miRNAs are novel epigenetic modifiers that can participate in depression pathogenesis by altering a host of genes critical in neuronal functions. In this context, the present study focused on examining whether alterations in miRNA networks in amygdala are associated with susceptibility (learned helplessness) or resiliency (nonlearned helplessness) to develop depression in rodents. Our findings suggest large-scale changes in miRNA expression in amygdala of rats showing depression-like behavior. We also identified adaptive or maladaptive miRNAs that are critical for vulnerability or resiliency to develop depression. Our findings also suggest that the disruption in the Wnt signaling pathway, specifically the canonical Wnt/β-catenin pathway, may play a critical role in deciding vulnerability to develop depression.
Introduction
Major depressive disorder (MDD) is one of the most prevalent psychiatric disorders associated with significant disability (Lepine, 2001; Kessler et al., 2003), morbidity, and mortality (Soloff et al., 2000; Miret et al., 2013). About 40% of MDD patients do not respond to the currently available medications (Wong and Licinio, 2004; Nierenberg et al., 2006). Studies have shown that the duration of untreated depression might have a substantial impact on the clinical outcomes (Ghio et al., 2015). This is partially a result of poor understanding of the molecular pathophysiology underlying MDD. It is becoming increasingly evident that MDD may result from disruptions across whole cellular networks, leading to aberrant information processing in the circuits that regulate mood, cognition, and neurovegetative functions (Wong and Licinio, 2004; Drevets et al., 2008).
The role of the amygdala in stress and emotional processing is well documented (Drevets et al., 2008; Diego and Antonella, 2017). Studies have suggested that increased severity of depression may cause morphological changes in the amygdala due to abnormal stress response mechanisms (Vyas et al., 2002; Roozendaal et al., 2009). Direct efferent projections from amygdala to supraoptical and paraventricular nuclei further make amygdala a susceptible target associated with mood disorders. Recently, microRNAs (miRNAs), a prominent class of small noncoding RNAs, have emerged as a major regulator of neural plasticity and higher brain functioning (Dwivedi, 2011; Im and Kenny, 2012). By modulating translation and/or stability of a large number of mRNA targets in a coordinated and cohesive fashion, they are able to regulate entire genetic circuitries (Lages et al., 2012). These miRNAs are highly expressed in neurons, and, because they can regulate the expression of hundreds of target mRNAs, neuronal miRNA pathways can create an extremely powerful mechanism to dynamically adjust the protein content of neuronal compartments even without the need for new gene transcription (Schratt, 2009; Hussaini et al., 2014). In the past, several studies have identified MDD-specific miRNA signatures in prefrontal cortex (Belzeaux et al., 2012; Smalheiser et al., 2012; Song et al., 2015). Moreover, repatterning of interconnected miRNA networks has been identified in fronto-cortical areas resulting in alteration of many cellular pathways (Smalheiser et al., 2012; Roy et al., 2017b). Interestingly, no study, except one in a juvenile mouse model (Shen et al., 2019), has examined an amygdala-based discriminative role of persistent stress in altering the miRNA transcriptomics. Furthermore, no report to our knowledge has shown the relationship between transcriptome-wide miRNA changes and altered gene regulatory network(s) in amygdala to develop depression-like behavior. Individually, a few miRNAs (miR-34, miR-15a, and miR-135a) in amygdala, have been found to serve as key regulators in stress-related behavior, including fear memory, anxiety, and impulsivity (Pietrzykowski and Spijker, 2014; Andolina et al., 2016; Volk et al., 2016; Murphy et al., 2017; Mannironi et al., 2018).
Since the ability to cope with stress is critical at the human level, parallel studies of the effects of uncontrollable stress have been performed in animals, with results of proactive interference with the acquisition of escape–avoidance responding (Seligman and Maier, 1967). This phenomenon is termed learned helplessness (LH) and has been used extensively as an animal model of stress-induced behavioral depression (Petty and Sherman, 1979; Sherman et al., 1982; Henn et al., 1993). In the present study, the LH model was used to determine the potential role of miRNAs and the pathways that they regulate in susceptibility or resiliency to develop depression-like behavior in rodents. Our findings indicate that miRNA-mediated Wnt signaling system in amygdala may confer stress susceptibility to develop depression. In fact, a comprehensive review article by Maguschak and Ressler (Maguschak and Ressler, 2012) has provided substantial evidence from studies showing significant implication of Wnt/β-catenin signaling in mood regulation. In this context, identification of miR-128-3p, as one of the most significantly upregulated miRNAs in susceptible (LH) rats, appeared to be closely associated with Wnt signaling disruption by targeting select Wnt ligands (Wnt3/5), intracellular signal transducer (Dvl1), and pathway specific downstream transcription factor (Lef1). In addition, our study showed a mechanistic association between transcription repressor Snai1 and miR-128-3p, where reduced expression of Snai1 might cause induced expression of miR-128-3p in LH rats (Qian et al., 2012). Moreover, repression of Arpp21 gene, which acts as counteracting RNA binding protein and protects the repressive effect of miR-128-3p on its targets, might be nullified in LH rats. These results suggest a complex and dynamic intracellular switching mechanism on target genes at posttranscriptional level in amygdala that might be conferring susceptibility to depression in LH rats.
Materials and Methods
Detailed methods are provided in the Supplementary section.
Animals and Induction of LH Behavior
All animal-related experiments were performed in postnatal day-90 male Holtzman rats (Envigo, Indianapolis, IN) under the guidelines of Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. The induction of LH behavior (Figure 1A) has been used by us in previous studies and provides a model of long-term depression-like behavior. The method of LH induction is detailed in the Supplementary section. Briefly, rats were given random inescapable tail shocks on days 1 and 7 followed by escape tests on days 2, 8, and 14. Based on escape latency scores, rats were divided into 2 groups: learned helplessness (LH, escape latency ≥20 seconds; n = 10) and nonlearned helplessness (NLH, escape latency <20 seconds; n = 10). We also included another group, tested control (TC, n = 8), which was tested for escape latency; however, they were not subjected to inescapable shock. Rats were decapitated 24 hours after the last escape latency test. Amygdala was precisely dissected out from the whole-mount brain tissue sections under temperature-regulated cryostat (Leica Biosystems Inc., IL) and stored at −80oC until further analysis. The steps that we performed to collect the brain sections are as follows. Initially, a coronal cut was made approximately at bregma −1 and a second cut was made at approximately bregma −2.75) (Paxinos and Watson, 1998). Later, dissected brain samples were suspended in a 1:1 mix of optimal cutting temperature compound (Tissue-Tek; Sakura Finetek USA) and 30% sucrose solution (50 mL NaPO4 [1000 nm], 150 mL 3% saline, 300 mL 50% sucrose) and sliced into 300-μm sections on glass slides. From each mounted slide, amygdala was carved out bilaterally from both sides of the brain using a blunted needle. A similar procedure was followed to collect the rat hippocampal tissues.
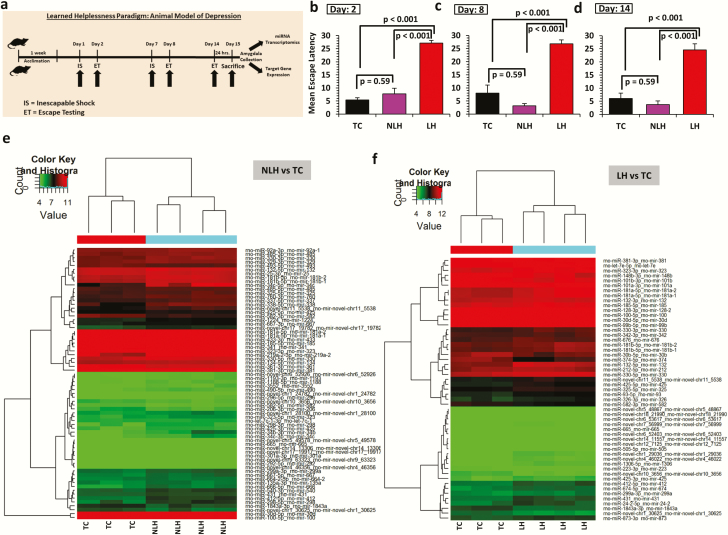
Figure 1.
(a) Schematic diagram showing induction of learned helplessness behavior (LH) in rats. Schematic diagram of the timeline followed as part of the stress paradigm to induce LH behavior in rats. (b-d) Mean escape latencies in nonlearned helpless (NLH) and LH rats compared with tested control (TC) rats. Bar diagrams represent escape latencies in TC, NLH, and LH rats measured on days 2, 8, and 14. Data are the mean ± SEM. Overall group comparisons are as follows: day 2: df = 2,25, F = 61.57; day 8: df = 2,25, F = 47.79; and day 14: df = 2,25, F = 38.99. Individual group comparison showed significantly higher escape latencies for LH rats compared with TC and NLH rats on days 2 (P < .001), 8 (P < .001), and 14 (P < .001). NLH rats did not show any significant difference in escape latency compared with the TC group at any time point (day 2: P = .37; day 8: P = .13; day 14: P = .27). (e) Expression-related changes in miRNA transcriptome based on NLH vs TC group comparison. Hierarchical clustering of miRNAs represents group-wise (NLH vs TC) expression changes plotted with a heat map. Individual samples are represented in each column, whereas each row demonstrates miRNAs. (f) Expression-related changes in miRNA transcriptome based on LH vs TC group comparison. Hierarchical clustering of miRNAs represents group-wise (LH vs TC) expression changes plotted with a heat map. Individual samples are represented in each column, whereas each row demonstrates miRNAs.
miRNA Sequencing in Rat Amygdala
The methods for RNA isolation, quality control, library preparation, and miRNA sequencing (Gopalakrishnan et al., 2015; Schmidt et al., 2018) are detailed in the supplementary section. Sequencing was carried out using the Illumina HiSeq 2000 according to the manufacturer’s instructions. The image analysis and base calling were performed using Solexa pipeline version 1.8 (Off-Line Base Caller software, version 1.8), and trimmed short reads (pass Solexa CHASTITY quality filter, trimmed 3′-adaptor bases) were aligned to rat pre-miRNA sequences in miRBase 21 using novoalign software. All sequences that fail to meet the quality cutoff, read length <16 nt and copy number <2 are discarded as unusable reads. For each microRNA sequence-based profile, the copy number of small RNA sequence reads were used to estimate expression level of each microRNA. The total number of valid sequence reads for each profile was used as the scaling factor for normalization [(total sequence reads, given)/(total sequence reads, reference)], as this number would given an indication of the total amount of RNA in the given sample. One profile was used as the reference, and the values of each of the other profiles are divided by the scaling factor. Finally, the miRNA expression levels were measured and normalized as transcripts per million of total aligned miRNA reads (TPM). After normalization the usable reads are firstly aligned to the latest sanger microRNA database to identify all the known microRNA species in sequencing samples and proceed for further expression analysis. Lastly, profile differences between two groups of samples were determined following a fold change calculation (i.e., the ratio of the group averages) and P values were computed. P values were adjusted for multiple testing using the Benjamini and Hochberg FDR correction method, and those with an adjusted P < .05 were considered significant. Fold change was calculated as a ratio between sample averages of transcripts per million for the groups compared.
In Silico Target Prediction
miRNAs whose expression level showed significant changes (P ≤ .05) were selected for further investigation. MiRWalk2.0 software was used to predict targets for the selected miRNAs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment function in miRWalk2.0 was used to determine significant pathways targeted by differentially expressed miRNAs.
Validation of miRNAs and Expression Analysis of Target Genes by qPCR
Using first-strand cDNA as template, gene expression assays were performed following the qPCR method (detailed in the supplementary information). First-strand cDNA synthesis was conducted based on oligo dT priming for mRNA and poly-A tailing method for miRNA expression study (Roy et al., 2017a). Fold change was calculated using Livak’s ΔΔCt calculation method (Livak and Schmittgen, 2001). Statistical significance (P ≤ .05) was determined using an independent sample t test.
In Vitro Cell Transfection
Double-stranded RNA oligos (Dharmacon) mimicking endogenous miR-128-3p (Mimic-128) and antisense miR-128-3p (Anti-128) were transfected into SH-SY5Y neuroblastoma cell lines (ATCC CRL2266) using lipofectamine RNAiMAX (Invitrogen), and results were compared with the control cell line group (Wang et al., 2018). Cells were harvested at 36 hours posttransfection for miRNA and target mRNA expression analysis as detailed in the supplementary information.
Human Postmortem Brain Studies
Amygdala obtained from 20 MDD and 22 nonpsychiatric control subjects were used. Psychological autopsy was performed as discussed in the supplementary information. Characteristics of these subjects are shown in supplementary Table 1. Toxicology and presence of antidepressants were examined by analysis of urine and blood samples. Brain pH was measured as described earlier (Harrison et al., 1995). RNA from amygdala was isolated and quality was checked as described in the supplementary section. Expression levels of miR-128-3p and several target genes were determined using qRT-PCR using human-specific primers as described in supplementary Table 8. Group differences were analyzed by independent sample t test, and effects of confounding variables were determined either by independent sample t test or by Pearson correlation coefficient method.
Results
Escape Latencies
As shown in Figure 1B–D, significant differences in escape latencies (P < .001) between TC, NLH, and LH groups were observed when tested on day 2 (df = 2,25, F = 61.57, day 8 [df = 2,25, F = 47.79]) and day 14 (df = 2,25, F = 38.99). Individual group comparisons showed significantly higher escape latencies (P < .001) for LH rats compared with TC and NLH rats on days 2, 8, and 14. NLH rats did not show any significant differences in escape latencies compared with the TC group at any time point (day 2: P = .37; day 8: P = .13; day 14: P = .27). Rats that had LH or NLH behavior on days 2 and 7 showed the same behavior when tested on day 14.
MiRNA Expression Changes Across NLH and LH Groups
Hierarchical clustering demonstrating differential miRNA expression profile between TC and NLH (Figure 1E) and TC and LH groups (Figure 1F) is shown as heat maps. Comparison of NLH vs TC group showed 60 significantly upregulated miRNAs (50 known and 10 novel miRNAs) and 10 significantly downregulated miRNAs (8 known and 2 novel miRNAs) (supplementary Table 2). An LH vs TC group comparison showed 37 significantly upregulated (29 annotated and 8 novel) and 16 significantly downregulated (12 annotated and 4 novel) miRNAs (supplementary Table 2). Comparison of LH and NLH rats showed an elevated expression of 17 miRNAs (15 annotated and 2 novel) and a contrasting set of 8 downregulated miRNAs (5 annotated and 3 novel) (supplementary Table 2). A differential expression profile of all miRNAs from LH vs NLH groups is presented as a heat map in Figure 2A. Fold changes and directionality of 20 significantly altered (both up- and downregulated) annotated miRNAs from LH vs NLH groups are presented as a separate composite heat map to show their corresponding expression changes across all 3 comparisons (NLH vs TC, LH vs TC, and LH vs NLH) (Figure 2B).
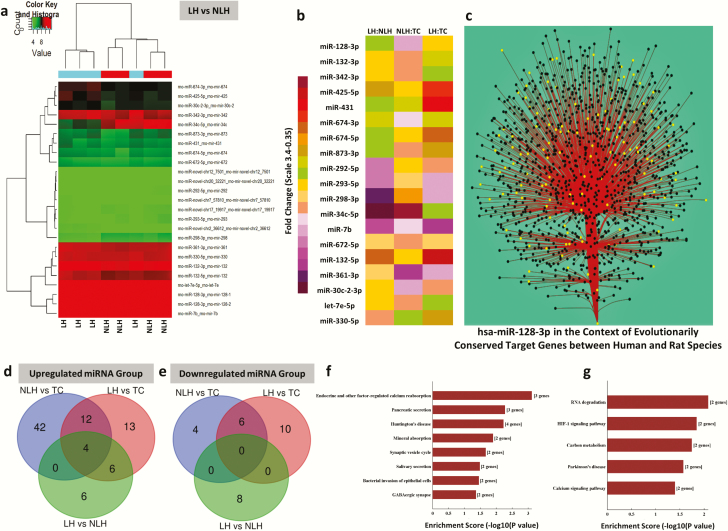
Figure 2.
(a) Expression-related changes in miRNA transcriptome associated with learned helplessness (LH) vs non-learned helplessness (NLH) group comparison. Hierarchical clustering of miRNAs representing group-wise (LH vs NLH) expression changes plotted with a heat map. Individual samples are represented in each column, whereas each row demonstrates miRNAs. (b) Heat map of significantly dysregulated top-ranking miRNAs in LH vs NLH group comparison. The list of significantly altered annotated miRNAs (including both up- and downregulated) from the LH vs the NLH group is presented with a separate composite heat map to show their corresponding expression changes across all 3 groups (NLH vs TC, LH vs TC, and LH vs NLH). (c) An interaction network between has-miR-128-3p and its predicted targets based on Cytoscape diffusion algorithm. Targets were determined using TargetScan’s prediction scheme with stringent selectivity to conserve site context++ scores only. The primary interaction is shown with main stem originating from miR-128-3p at the base and branching out to join the axial nodes (black circle); the edges (red solid line) are bundled to increase visual clarity. Use of diffusion algorithm produces a list of nodes ranked by the heat they accumulated (yellow circle). Theoretically, higher ranking is determined for the nodes, which have many stronger connections. (d) Venn diagram of the overlapping and nonoverlapping class of miRNAs (upregulated) derived from 3 group comparisons. Venn diagram showing significantly upregulated miRNAs either shared or exclusively represented by 2 distinct behavioral phenotypes related to either depression susceptibility or resiliency. (e) Venn diagram of the overlapping and nonoverlapping class of mRNAs (downregulated) derived from 3 group comparisons. A set of significantly downregulated miRNAs is represented on this Venn diagram, showing either distinct or overlapping relationship with depression or resiliency phenotype. (f) Cellular pathways affected by targets of upregulated miRNAs derived from LH vs NLH group comparison. Canonical biological pathways associated with genes that are predicted to be targets of significantly upregulated miRNAs in LH vs NLH rats. (g) Cellular pathways affected by targets of downregulated miRNAs derived from the LH vs the NLH group comparison. Canonical biological pathways associated with genes that are predicted to be targets of significantly downregulated miRNAs in LH vs NLH rats.
Based on both statistical significance and fold-change, miR-132-5p and miR-128-3p were identified as top-ranking miRNAs from the list of 17 upregulated miRNAs in the LH vs NLH group. However, based on a consensus from 3 different target prediction software tools (miRWalk, miRanda, RNA22, and TargetScan), miR-128-3p outnumbered predicted targets (2160) compared with miR-132-5p (432) by 5 times. A diffusion output–based miR-target network map following TargetScan prediction algorithm for miR-128-3p is shown in Figure 2C. This includes a set of 684 targets that are evolutionarily conserved between human and rat (supplementary Table 3).
Next, unique and overlapping sets of miRNAs associated with behavioral phenotypes were identified. Analysis showed that 13 upregulated (Figures 2D) and 10 downregulated miRNAs (Figures 2E) were specifically associated with vulnerability to depression (LH behavior). On the other hand, the 42 upregulated and 4 downregulated miRNAs were specifically associated with resiliency to depression (NLH group) (Figure 2D–E, respectively). However, when the lists of upregulated miRNAs were compared between NLH vs TC and LH vs NLH, 7 unique miRNAs (miR-298-5p, miR-666-5p, miR-338-5p, miR-219a-2-3p, miR-34c-5p, miR-34b-3p, and miR-34c-3p) were found to be upregulated in NLH rats compared with TC rats, whereas significantly upregulated miR-34c-5p from NLH vs TC was found to be downregulated in the LH vs NLH group. This indicates that these 7 miRNAs may potentially be associated with behavioral phenotype associated with depression susceptibility. On the other hand, 6 significantly altered miRNAs (let-7e-5p, miR-132-3p, miR-128-3p, miR-674-5p, miR-873-3p, and miR-342-3p) were identified that were upregulated in LH rats compared with either NLH or TC rats. These 6 miRNAs might be associated in augmenting depression susceptibility. We also determined a separate set of 4 miRNAs (miR-672-5p, miR-361-3p, miR-674-3p, and miR-30c-2-3p) that were uniquely upregulated in LH vs NLH rats but completely absent in the other 2 comparison groups (LH vs TC and NLH vs TC).
Pathway and gene ontology (GO) prediction in the LH vs NLH group based on targets of differentially altered miRNAs
Kyoto Encyclopedia of Genes and Genomes-based pathway analysis of genes predicted as targets of 15 significantly upregulated miRNAs in LH vs NLH rats revealed the involvement of 8 specific molecular pathways (Figure 2F), which included the ones associated with synaptic signaling and GABA. On the other hand, the 5 downregulated miRNAs and their targets from LH vs NLH comparison predicted 4 significantly altered pathways (Figure 2G), of which the identification of RNA degradation and calcium signaling are the most notable ones. Besides pathway analysis, the functional annotation of predicted target genes of differentially expressed miRNAs from the NLH vs LH group identified a rich set of neuronal functionalities when all 3 components (biological pathway, cellular component, and molecular function) were considered under the GO prediction scheme. The bar diagram presented in supplementary Figure 1A shows top 10-fold enrichment values collected from all predicted target genes regulated by 15 upregulated miRNAs. Supplementary Figure 1B represents the GO annotation associated with genes predicted as targets of 5 downregulated miRNAs in the LH compared with the NLH group. Although 3 different domains of functional clustering showed the potential involvement of several cellular and biochemical processes, the changes represented by cellular components in both up- and downregulated miRNA groups seem quite interesting stemming from their relation to synaptic functions and neuronal growth.
Prediction of Genes and Downstream Pathways Targeted by Top 10 Significantly Altered miRNAs Associated With LH Group
As part of the miRWalk prediction scheme, 3 different target prediction algorithms were used to identify the most putative targets of top 10 significantly upregulated miRNAs (rno-miR-132-3p, rno-miR-672-5p, rno-miR-132-5p, rno-miR-128-3p, rno-miR-674-5p, rno-miR-425-5p, rno-miR-361-3p, rno-miR-674-3p, rno-miR-30c-2-3p, and rno-miR-431) from the LH vs the NLH group. Further screening of the predicted targets for functional clustering with P value adjustment (supplementary Table 4) led us to identify Wnt signaling (highlighted with light green in supplementary Table 4) as the most conserved pathway significantly (FDR correction) targeted by 6 out of 10 miRNAs (rno-miR-132-3p, rno-miR-128-3p, rno-miR-361-3p, rno-miR-674-3p, rno-miR-30c-2-3p, and rno-miR-431). In addition, mapping of this pathway (supplementary Figure 1C) at the individual gene level led us to identification of 109 cross-sectional genes targeted by all 6 upregulated miRNAs from the LH vs the NLH group (supplementary Table 5). Besides Wnt signaling, an additional top 4 significantly affected pathways (axon guidance, MAP kinase, neurotrophin, and VEGF signaling) were also identified as targets of these altered miRNAs (supplementary Table 3). Further analysis using predicted targets from Wnt signaling identified 6 prominent genes: Wnt3, Wnt3a, Wnt5a, Wnt5b, Wnt9b, and Dvl1 (supplementary Figure 1D). Lastly, in a more conservative way, targets predicted from 3 different target prediction databases were used to identify pathways (besides Wnt signaling) based on their closely associated role in neuropsychiatric disorders, including MDD. The result showed the gene set enrichment for (1) chemokine and cytokine pathways, (2) cholecystokinin A receptor signaling pathway, (3) integrin signaling pathway, (4) heterotrimeric G-protein coupled signaling pathway, and (5) epidermal growth factor receptor signaling pathways (supplementary Figure 1E).
Expression of Important miRNA Target Genes From Wnt Signaling Pathway
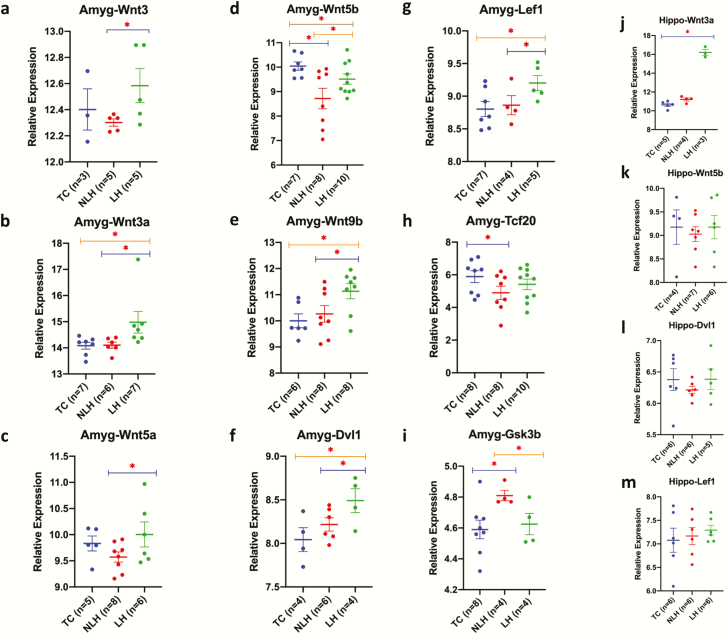
Next, in a follow-up experiment, expression levels of key target genes in the Wnt signaling pathway in amygdala of TC, NLH, and LH rats were determined. Gapdh was used as endogenous control. No significant differences were found in Gapdh expression between any groups (NLH vs TC: P = .29; LH vs TC: P = .12; LH vs NLH: P = .12). Significant expression downregulation was found for Wnt3 (P = .03), Wnt3a (P = .04), Wnt5a (P = .04), Wnt5b (P = .04), Wnt9b (P = .03), and Dvl1 (P = .04) genes in the NLH vs the LH group. Although Lef1 was not identified as a predicted target of altered miRNAs, its expression was tested due to its critical role in the Wnt signaling pathway as an endpoint transcription factor. Interestingly, significantly low expression of Lef1 (P = .05) was observed in the LH vs NLH comparison. Besides this, the expression of Tcf20 was downregulated in LH rats compared with NLH rats, although it was not statistically significant. As an antagonizing factor in the Wnt pathway, expression of Gsk3β was also tested. Gsk3β expression was significantly upregulated in LH vs NLH rats (P = .017). Comparison between LH and TC group of rats showed significant downregulation of Wnt3a (P = .03), Wnt9b (P = .008), Dvl1 (P = .02), and Lef1 (P = .01) genes. However, the other 2 genes, Wnt3 and Wnt5a, did not show statistically significant expression change but demonstrated a marked decrease in LH vs TC rats. In contrast, a nonsignificant upregulation was noted for Wnt5b and Tcf20 in LH vs TC rats. No significant change was noticed for Gsk3β expression in LH rats compared with TC rats. Lastly, the comparison between NLH and TC rats showed a significant increase in expression for Wnt5b (P = .007) and Tcf20 genes (P = .04). However, no significant changes were noted for Wnt3, Wnt3a, Wnt5a, and Wnt9b genes in NLH compared with TC rats. Unlike others, the expression of Gsk3β gene was significantly (P = .01) downregulated in NLH compared with TC rats. All comparisons associated with target gene expression in amygdala are presented in Figure 3A–I.
Figure 3.
Amygdala and hippocampus-based expression changes in miRNA targets across 3 comparison groups (nonlearned helplessness [NLH], learned helplessness [LH], and tested control [TC]). (a–i) All data are normalized with Gapdh and presented as the mean ± SEM. Significance level is indicated with an asterisk sign (*P ≤0.05). qPCR-based expression changes in miRNA targets Wnt3, Wnt3a, Wnt5a, Wnt5b, Wnt9b, Dvl1, Lef1, Tcf20, and Gsk3β in LH and NLH groups independently compared with TC. Expression-related significant differences in Wnt3 (P = .033), Wnt3a (P = .042), Wnt5a (P = .045), Wnt5b (P = .044), Wnt9b (P = .031), Dvl1 (P = .047), Lef1 (P = .053), and Gsk3b (P = .026) genes were noted in the LH vs the NLH group. Significant changes in expression were also noted for Wnt5b (P = .007), Tcf20 (P = .045), and Gsk3b (P = .017) genes when the NLH group was compared with the TC group. Group comparison between LH and TC rats identified significant changes in expression of Wnt3a (P = .03), Wnt5b (P = .043), Wnt9b (P = .008), Dvl1 (P = .029), and Lef1 (P = .019) genes. (j–m). All data are normalized with Gapdh and presented as the mean ± SEM. qPCR-based expression change of miRNA targets Wnt3a, Wnt5b, Dvl1, and Lef1 in hippocampus of LH and NLH groups independently compared with TC. Expression-related significant differences were only noted in Wnt3a (P < .005) gene when the LH group was compared with the TC group.
To examine if alterations in Wnt signaling pathway occurred selectively in amygdala of LH rats, expression of Wnt3a, Wnt5b, Dvl1, and Lef1 genes was also examined in the hippocampus. qPCR results did not show significant expression changes for Wnt5b, Dvl1, and Lef1 in either NLH and LH rats compared with TC rats, although a significant downregulation in Wnt3a expression was noticed only in LH vs TC rats (P < .005) but not between NLH vs LH rats (Figure 3J–M).
Wnt Signaling Pathway as a Major Target of miR-128-3p
Although the 2 most significantly upregulated miRNAs (miR-132-3p and miR-672-5p) from the top 10 miRNA list showed a sizeable number of predicted targets (44 and 34 genes, respectively) from the Wnt signaling pathway, miR-128-3p showed the maximum number of target genes (49) from this pathway (Figure 3B). In addition, our detailed bioinformatic analysis based on the TarPmiR (miRWalk) prediction algorithm identified miR-128-3p as the master regulator of the Wnt signaling pathway. In supplementary Figure 2A, a subset of genes (highlighted with circles) mapped on Wnt signaling pathway is shown as hot spot targets of miR-128-3p. Prediction algorithm-based miRNA target scanning (miRWalk; V.3) identified more than a thousand genes as putative targets of rat miR-128-3p (supplementary Table 6). However, following a selective inclusion strategy to find out conserved potential targets, 12 genes (Wnt1, Wnt3, Wnt3a, Wnt5a, Wnt5b, Wnt9a, Wnt9b, Lrp6, Gsk3b, Dvl [1 and 2], Axin, and Tcf) with key importance to Wnt signaling were identified (supplementary Figure 1B). In addition, Gsk3β interacting protein, Gskip (which acts as negative regulator of Gsk3β), was also identified as a target of miR-128-3p (supplementary Figure 2B).
Validation of miR-128-3p Predicted Target Genes Using in Vitro Cellular Model
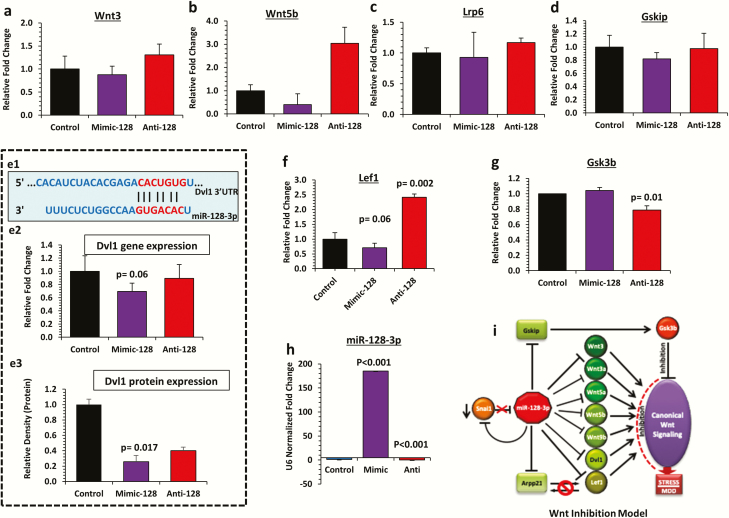
Inducing and knocking down the expression of miR-128-3p in SH-SY5Y neuroblastoma cell line was critical to validate its target gene expression. The putative targets of miR-128-3p (Wnt3, Wnt5b, Lrp6, and Gskip) showed marked expression downregulation in the miR-128-3p overexpression group (mimic-128), which were reversed when miR-128-3p was knocked down (anti-128) (Figure 4A–D). The most notable change was found in Wnt5b gene, with 60% downregulation in mimic-128, which was increased by 2-fold under miR-128-3p inhibition. Interestingly, ectopic overexpression of miR-128-3p (mimic) in SH-SY5Y cell line (Figure 4E1) decreased (30%) Dvl1 expression (P = .06), whereas antisense oligo transfection (anti-miR-128-3p) masked this effect (Figure 4E2). At the protein level, western-blot results from mimic-128 transfected cell line showed 75% reduction in Dvl1 expression (P = .017) compared with control. Sponging the inhibitory effect of endogenous miR-128-3p with anti-128 transfection was able to unmask repression on target gene (Figure 4E3). Expression-related reversal at the transcript level was also found for Lef1 gene (1.4-fold increase; P = .002) compared between the anti-128 and control groups (Figure 4F). Similar to in vivo results, a nearly significant (P = .06) downregulation (30%) was noted for Lef1 gene in a mimic vs control experiment (Figure 4F). Next, using an in vitro cell line system, we were able to mimic the expression-related changes for Gsk3β as was seen in LH vs TC rats. Under miR-128 overexpression, Gsk3β showed a slight increase in expression compared with the control cell line, although it did not reach statistical significance (Figure 4G). However, the endogenous expression of Gsk3β gene was significantly (P = .01) decreased by 22% in anti vs control cell line (Figure 4G). To determine the accuracy and efficacy of miR oligo transfection, we have presented the expression levels of miR-128-3p in both overexpressed (mimic-128) and knockdown (anti-128) SH-SY5Y cells (Figure 4H). Both the transfected groups showed significant changes in miR-128-3p expression that were inversely related to either mimic-128 (P < .001) or anti-128 (P < .001) oligo transfection compared with controls (Figure 4H). Based on in vivo and in vitro results, a miR-128-3p–mediated Wnt inhibition model is proposed, which could be relevant in its susceptibility to develop stress-induced depression (Figure 4I). This model has been described in detail in the Discussion section.
Figure 4.
Functional validation of miR-128-3p targets in SH-SY5Y cell line and proposed Wnt inhibition model. (a–g) Bar diagram represents the relative expression of Wnt3, Wnt5b, Lrp6, Gskip, Dvl1, Lef1, and Gsk3b transcripts in SH-SY5Y cell line transiently transfected with mimic miR-128-3p overexpression oligo (mimic-128) or anti-miR-128-3p overexpression oligo (anti-128) and compared with control cells. GAPDH was used as a normalizer for target gene expression. Values are represented as mean ± SEM. (e1) Diagram representing predicted target site interaction between rat miR-128-3p seed region and Dvl1 3’ untranslated region (UTR). (e2) Dvl1 gene expression showed significant (P = .06) change when the mimic-128 group was compared with control. (e3) Immunoblot analysis of Dvl1 protein expression in SH-SY5Y cell line miR-128-3p gain (mimic-128) and loss (anti-128) of function mutations. A significant (P = .017) decrease in Dvl1 protein level was noted in the mimic-128 group compared with control. Conversely, a marked increase in Dvl1 protein expression was identified in the anti-128 group compared with control. (f) A significantly (P = .002) higher expression of Lef1 gene was observed in anti-128 transfected cell line compared with the control group. (g) Expression of Gsk3b gene was significantly (P = .01) lower in anti-128 transfected cell line compared with control. (h) Expression level of miR-128-3p in SH-SY5Y cell line individually transfected with miR-128-3p oligo (mimic-128) and miR-128-3p anti-sense oligo (anti-128). Compared with the control group, the mimic-128 transfected cell line showed a significant (P < .001) increase in miR-128-3p expression, whereas significantly (P < .001) reduced miR-128-3p expression was observed in the anti-128 transfected group. (i) The cellular model of canonical Wnt pathway inhibition partially mediated via miR-128-3p overexpression under stress-induced maladaptive changes.
mRNA Expression Profile of Snai1, Arpp21, R3hdm1 Genes Associated With miR-128-3p
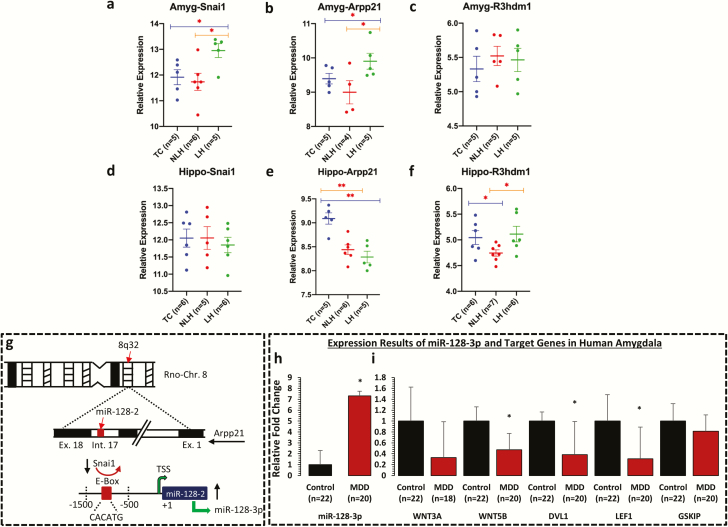
To examine the locus-specific transcriptional control of miR-128-3p, expression of Snai1 gene was tested as a transcriptional repressor of pri-miR-128-2 in amygdala of LH, NLH, and TC rats. Interestingly, approximately 50% reduction in expression of Snai1 was found in LH rats compared with NLH (P = .011) or TC rats (P = .015), which was elevated in NLH rats compared with TC rats (13%); however, this increase was not statistically significant (Figure 5A). Besides this, expression levels of Arpp21 and R3hdm1 genes were examined, which harbor miR-128-2 and miR-128-1 coding units in their intronic regions, respectively, and counter miR-128-3p effects by acting as RNA binding proteins. A significant decrease (30%) in expression for Arpp21 was noted in LH rats compared with TC rats (P = .05). However, this change was contrasted in NLH rats compared with TC rats (31% increase) but was not statistically significant (P = .14) (Figure 5B). On the contrary, a significant downregulation was noted for Arpp21 in NLH vs LH rats (P = .02). For R3hdm1, none of the groups (LH vs TC, NLH vs TC, and NLH vs LH) showed any significant change in its expression (Figure 5C).
Figure 5.
mRNA expression profile of Snai1, Arpp21, and R3hdm1 in amygdala and hippocampus of LH, NLH, and TC rats and expression profile of miR-128-3p and its target genes in amygdala of MDD subjects. qPCR-based expression changes in Snai1, Arpp21, and R3hdm1 genes in amygdala and hippocampus of LH and NLH groups independently compared with tested controls (TC). All data are Gapdh normalized and presented as the mean ± SEM. (a–c) mRNA-based expression profile in amygdala demonstrated significant changes in Snai1 gene when the LH group was independently compared with the TC (P = .015) or NLH (P = .011) groups. Group comparison between LH and TC rats identified significant changes in expression of Arpp21 (P = .05). Similarly, Arpp21 demonstrated significant change (P = .02) when LH rats were compared with NLH rats. (d–f) In hippocampus, group comparison between NLH and TC rats identified significant change in expression of Arpp21 (P = .001). Comparison between LH and TC rats also identified significant changes in expression of Arpp21 (P < .005). For R3hdm1 gene, group comparison between NLH and TC demonstrated significant difference in expression (P = .02). Significant change in expression was also noted for R3hdm1 (P = .01) when LH rats were compared with NLH rats. (g) Schematic diagram showing the underlying model of miR-128-3p transcriptional regulation being coordinated by transcriptional repressor Snai1 from rat chromosome 8. This zinc finger family transcriptional repressor binds with E-box (CACATG) motif of rat miR-128-2 coding upstream promoter and can impose a repressed state on miR-128-3p expression via Arpp21 intronic axis on chromosome 8. (h) Representative graph demonstrates the relative transcript abundance of miR-128-3p in amygdala of MDD subjects (n = 20) compared with nonpsychiatric control subjects (n = 22). Data are normalized using U6 gene expression values and represented as ±SEM. Level of significance was determined using independent-sample t test (df = 40, f = 13.47, t = 2.01, P = .04). (i) GAPDH normalized relative expression levels of WNT3A, WNT5B, DVL1, LEF1, and GSKIP analyzed in amygdala of MDD subjects. All data demonstrate the mean ± SEM (all genes were analyzed using 22 control subjects and 20 depressed subjects except WNT3A, control = 22 and MDD = 18). The level of significance was determined using independent-sample t test (WNT3A df = 38, f = 0.83, t = −1.75, P = .86; WNT5B df = 40, f = 0.45, t = −2.73, P = .01; DVL1 df = 40, f = 9.63, t = −2.28, P = .03; LEF1 df = 40, f = 0.57, t = −2.25, P = .03; and GSKIP df = 40, f = 0.10, t = −0.68, P = .49). Significance level is indicated with an asterisk sign (*P ≤0.05, **P ≤0.005).
To further investigate if the changes are specific to amygdala, expression of Arpp21 and R3hdm1 were examined in hippocampus. In hippocampus, none of the comparison groups showed any significant alterations in the expression of Snai1 gene (Figure 5D). In contrast to amygdala, the expression of Arpp21 in hippocampus was significantly increased in both NLH (P = .001) and LH (P < .005) rats compared with TC rats (Figure 5E). No significant changes in expression of Arpp21 were found between LH and NLH rats. Similarly, a significant increase (P = .02) was found in R3hdm1 gene in NLH vs TC rats (Figure 5F), which was lower in NLH vs LH rats (P = .01).
Expression of miR-128-3p and Its Target Genes in Amygdala of MDD Subjects
Expression of miR-128-3p and its target genes were examined in amygdala of MDD and nonpsychiatric control subjects. There were no significant differences in age (P = .19), postmortem interval (P = .48), brain pH (P = .92), and RNA integrity number (P = .97) between the 2 groups (supplementary Table 1). There were 13 males and 9 females in the nonpsychiatric control group and 10 males and 10 females in the MDD group. As shown in Figure 5H, a significant (P = .04) increase (approximately 6.5-fold) in the expression of miR-128-3p was observed in the MDD group. On the other hand, expressions of WNT5B (P = .01), DVL1 (P = .03), and LEF1 (P = .03) were significantly lower in the MDD group, except WNT3A (P = .86) and GSKIP (P = .49) genes, which were lower but did not reach significance (Figure 5I). There were no significant effects of age, brain pH, postmortem interval, and RNA integrity number on expression of miR-128-3p or its target genes, except significant positive correlations were noted between pH and the expression of WNT3A, WNT5B, and GSKIP genes (supplementary Table 7). Also, the expression of miR-128-3p and its target genes were not influenced by suicide, alcohol abuse, or antidepressant treatment (supplementary Figure 3).
Discussion
This is the first study to our knowledge examining the role of amygdalar miRNAs in susceptibility or resistance to depression. Our transcriptome-wide miRNA analysis in amygdala revealed differential regulation of 53 miRNAs (37 up- and 16 downregulated) in LH rats compared with TC rats, which could possibly be associated with molecular changes conferring maladaptive stress response, whereas comparison between NLH vs TC, which showed alterations in 70 miRNAs (60 up- and 10 downregulated), could be associated with stress resiliency. On the other hand, comparison of LH vs NLH rats demonstrated alterations in 25 miRNAs (17 up- and 8 downregulated) that could uniquely be associated with vulnerability to develop stress-induced depression and might act as priming factors to develop such vulnerability.
Our results suggest that some of the miRNAs (e.g., miR-425, 330, and 132) that were dysregulated in amygdala of LH rats had previously been reported to be altered in peripheral blood of MDD patients (Maffioletti et al., 2016; Fang et al., 2018). Alteration in miR-674-3p expression in LH rats is in line with a previous report showing similar changes in an animal model of posttraumatic stress disorder (Balakathiresan et al., 2014). Interestingly, upregulated miR-473-3p and miR-34c-5p in LH rats appear to be related to cognitive impairment in suicidal subjects (Lopez et al., 2014). On the other hand, miR-873-3p, which was upregulated in amygdala of LH rats, was reported earlier to target 2 important polyamine genes, SAT1 and SMOX, implicated in MDD and suicidal behavior (Lopez et al., 2014). The study, primarily focused on identifying abnormalities in metabolic functions of polyamine pathways, showed that expression of SAT1 and SMOX genes are reduced in depressed suicidal brain (Lopez et al., 2014). In the same study, a cross-platform miRNA prediction scheme identified a list of shared miRNAs that targeted SAT1 and SMOX genes. Further, in silico examination identified miR-873 as one of the targeting miRNAs to regulate both SAT1 and SMOX genes. Together, these results led the authors to propose a relationship between a reduced level of SAT1 and SMOX genes and a pathogenic role of miR-873 in depression/suicide. In the present study, we also identified upregulation of miR-873-3p in amygdala of LH rats. Together with the previous study, our study suggests a possible role of miR-873-3p and associated molecular pathways in depression pathology.
Our finding demonstrates significant involvement of miR-128-3p in LH behavior, which was found to be the most significantly upregulated miRNA in LH compared with NLH rats. The role of miR-128-3p is well conserved across higher vertebrates as demonstrated by our in-silico miR-target network construction based on human homologue of miR-128-3p, which showed a well-conserved set of 684 predicted target genes between rat and human species. Several reports from the past have shown an important role of miR-128-3p in various neurobiological processes, including neuronal differentiation, dendritic excitability, and increased dendritic spine formation (Lin et al., 2011; Tan et al., 2013). Interestingly, altered expression of miR-128-3p has been shown in schizoaffective disorder and MDD as well as in response to and the treatment response to selective serotonin reuptake inhibitor treatment to MDD patients (Perkins et al., 2007; Bocchio-Chiavetto et al., 2013).
GO analysis revealed several important cellular pathways affected by altered miRNAs in LH rats. Of them, Wnt signaling was most strongly associated with the top 10 significantly upregulated miRNAs, including miR-128-3p in the LH group compared with the NLH group. The Wnt signaling pathway primarily consists of a canonical Wnt/β-catenin axis (Mulligan and Cheyette, 2017). Multiple genes, including Wnt3, Wnt3a, Wnt5a, Wnt5b, Wnt9b, Dvl1, Tcf, and Gskip, were found to be the targets of these miRNAs. In this study, mRNA expression for Wnt3 was significantly downregulated in LH compared with NLH rats. Similar changes were noted for Wnt3a, Wnt5a, and Wnt9a ligands. Previous studies have shown that disruption of the Wnt pathway may be associated with depressive behavior (Zhang et al., 2013), and activation of this pathway may have antidepressant effects in a mouse model of depression (Kaidanovich-Beilin et al., 2004; Gould et al., 2007). A recent study also implicates Wnt2 and Wnt3 in depression (Zhou et al., 2016) where it was shown that upregulation of Wnt2 and Wnt3 can have antidepressant effect, whereas their downregulation can lead to depressive behavior. These findings complement our results and suggest that disruption in Wnt signaling can have a detrimental effect in developing a depression-like phenotype. In our in-vitro model system, a targeted decrease in Gskip expression via miR-128-3p was observed. Gskip acts as a negative regulator of Gsk3β (Tago et al., 2000; Chou et al., 2006). Therefore, miR-128-3p–mediated decrease in Gskip can be linked as an additional mechanism to dysregulate the Wnt pathway via Gsk3β. Additionally, alterations in expression of Dvl1 and Lef1 in LH rats provided evidence for their destabilizing effects on Wnt signaling. Activated Dvl1 disintegrates the β-catenin-destruction complex, thus activating canonical Wnt signaling. On the other hand, Lef1 forms an intranuclear heteromeric complex with Tcf and in combination acts as a transcription factor. Activation of canonical Wnt signaling results in translocation of β-catenin to induce transcription of early response genes via Tcf/Lef complex (Mulligan and Cheyette, 2017). Since Dvl1 and Lef1 expression are reduced in LH rats, which largely represent the inadequate functioning of these 2 key molecules in Wnt signaling pathway, a dysfunctional state for the same can be anticipated in LH behavior. Interestingly, the findings of altered Wnt signaling genes in LH rats were restricted to amygdala as no significant changes were noted in hippocampus. This suggests that miRNA-mediated regulation of Wnt signaling and their participation in inducing LH behavior could be brain region specific.
To understand the translational significance of the present finding, expression status of miR-128-3p was examined in the amygdala of MDD subjects. In addition, expression of 5 target genes of miR-128-3p was also examined. Not only was the expression of miR-128-3p upregulated, but significant reciprocal downregulation of target genes WNT5B, DVL1, and LEF1 was also observed in amygdala of MDD subjects compared with nonpsychiatric controls. The 2 other genes, (i.e., WNT3A and GSKIP) showed a sharp decline in their expression in MDD subjects but were not statistically significant. These findings directly implicate amygdala-based miR-128-3p and associated Wnt signaling in depressive behavior.
Although much work has been done to understand the regulatory mechanisms behind changes in miR-128-3p expression, none of them have been directly linked to psychiatric illnesses. Studies have shown that mature miR-128-3p can be transcribed from 2 different intronic loci. Chromosome 13 encodes for precursor-miR-128-1 and chromosome 8 encodes for precursor-miR-128-2 (Franzoni et al., 2015). The present study for the first time to our knowledge reports an expression-associated change in transcriptional repressor Snai1 selectively in amygdala of rats showing depressive behavior. Snai1 acts as a transcriptional repressor and can suppress miR-128-2 expression by binding to miR-128-2–specific promoter motif (Figure 5G) (Dong et al., 2014). In this regard, we noted a significant downregulation of Snai1 mRNA in amygdala of LH rats compared with NLH rats. This change was specific to amygdala as no alteration was observed in hippocampus. This suggests that the upregulation in miR-128-3p expression could possibly be associated with reduced expression of transcriptional repressor Snai1 in amygdala of LH rats. On the other hand, being a direct target of miR-128-3p (Qian et al., 2012), Snai1 can create a negative feedback loop to achieve a context-specific dynamic regulation. Therefore, upregulated expression of miR-128-3p in amygdala of LH rats may potentially transduce Snai1→miR-128-2 transcriptional axis. This mechanism has been represented as a schematic diagram in Figure 4I.
Interestingly, expression-associated changes were noted in Arpp21 gene in amygdala of LH rats. Arpp21 gene harbors miR-128-2 and miR-128-1 coding units in its intronic region and acts as an RNA-binding protein to counteract the repressive effect of miR-128 on its target genes. The expression of Arpp21 was lower in amygdala of LH rats compared with TC and NLH rats. On the contrary, significantly upregulated expression of Arpp21 was found in hippocampus of LH compared with TC or NLH rats. Decrease in Arpp21 expression suggests that Arpp21 may be less effective in protecting its target transcripts from miR-128-3p repression, thus creating a complex intracellular switching mechanism. Interestingly, a previous report has shown a direct interaction of Arpp21 and mRNA transcripts of Lef1 from Wnt signaling pathway (Rehfeld et al., 2018). Therefore, it is quite possible that due to higher expression of miR-128-3p in amygdala, the switching mechanism may possibly help miR-128-3p to mask the protective effect of Arpp21 on Lef1 and gain a pervasive control to induce a posttranscriptional repression (Figure 4I).
Altogether, the present study suggests large-scale changes in miRNA expression in amygdala of rats showing depression-like behavior. We also identified miRNAs that are critical for vulnerability or resiliency to develop depression. The findings also suggest that miRNA-mediated disruption in Wnt signaling pathway, specifically the canonical Wnt/β-catenin pathway, may play a critical role in deciding vulnerability to develop depression. This effect could be mediated through miR-128-3p regulation of Wnt3, Wnt3a, Wnt5b, Dvl1, and Lef1 genes. Mechanistically, these genes could be acting in a coordinated manner and suggest the role of miR-128-3p as a master regulator. Based on these results, miRNA influenced Wnt signaling inhibition model is proposed, which can selectively trigger depressive behavior. Further studies are needed to distinguish the role of specific molecular components in inducing or causing resiliency to develop depression.
Our study has 3 major implications that could be translationally important in stress-induced depression pathophysiology. First is the identification of miR-128-3p in amygdala as a valuable target for pharmacological intervention, especially in light of the recent advancement in antagomir-based therapeutics. Second is amygdala-specific epigenetic perturbation of Wnt signaling in depressed brain. For instance, Wnt signaling has been used for therapeutic interventions in various pathophysiological incidences. Therefore, identifying the Wnt pathway as downstream target of miRNA holds a promising clue for tailored pharmacogenomic treatment for depression. Lastly, highlighting amygdala as a salient brain area under miRNA-mediated epigenetic control in mood dysregulation and its implication in stress responsiveness can be considered a potential breakthrough in depression pathology. To our knowledge, no other report has shown the role of amygdalar miRNAs in Wnt-mediated stress susceptibility to depression.
Supplementary Material
Acknowledgments
This work was supported by grants from National Institute of Mental Health (R01MH082802; 1R01MH101890; R01MH100616; 1R01MH107183-01; R01MH118884-01A1) to Dr Dwivedi. The sponsoring agency had no role in study design, collection, analysis, interpretation of data, and in the writing of the manuscript.
Conflict of Interest
None.
References
- Andolina D, Di Segni M, Bisicchia E, D’Alessandro F, Cestari V, Ventura A, Concepcion C, Puglisi-Allegra S, Ventura R (2016) Effects of lack of microRNA-34 on the neural circuitry underlying the stress response and anxiety. Neuropharmacology 107:305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakathiresan NS, Chandran R, Bhomia M, Jia M, Li H, Maheshwari RK (2014) Serum and amygdala microRNA signatures of posttraumatic stress: fear correlation and biomarker potential. J Psychiatr Res 57:65–73. [DOI] [PubMed] [Google Scholar]
- Belzeaux R, Bergon A, Jeanjean V, Loriod B, Formisano-Tréziny C, Verrier L, Loundou A, Baumstarck-Barrau K, Boyer L, Gall V, Gabert J, Nguyen C, Azorin JM, Naudin J, Ibrahim EC (2012) Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl Psychiatry 2:e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, Giovannini C, Bignotti S, Tardito D, Corrada D, Milanesi L, Gennarelli M (2013) Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol 23:602–611. [DOI] [PubMed] [Google Scholar]
- Chou HY, Howng SL, Cheng TS, Hsiao YL, Lieu AS, Loh JK, Hwang SL, Lin CC, Hsu CM, Wang C, Lee CI, Lu PJ, Chou CK, Huang CY, Hong YR (2006) GSKIP is homologous to the Axin GSK3beta interaction domain and functions as a negative regulator of GSK3beta. Biochemistry 45:11379–11389. [DOI] [PubMed] [Google Scholar]
- Diego A, Antonella B (2017) The key role of the amygdala in stress. In: Amygdala and emotional modulation of multiple memory systems (Ferry B, ed), pp 187–213. London, UK: IntechOpen Limited. [Google Scholar]
- Dong Q, Cai N, Tao T, Zhang R, Yan W, Li R, Zhang J, Luo H, Shi Y, Luan W, Zhang Y, You Y, Wang Y, Liu N (2014) An axis involving SNAI1, microRNA-128 and SP1 modulates glioma progression. Plos One 9:e98651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Furey ML (2008) Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct Funct 213:93–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. (2011) Evidence demonstrating role of microRNAs in the etiopathology of major depression. J Chem Neuroanat 42:142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Qiu Q, Zhang S, Sun L, Li G, Xiao S, Li X (2018) Changes in miRNA-132 and miR-124 levels in non-treated and citalopram-treated patients with depression. J Affect Disord 227:745–751. [DOI] [PubMed] [Google Scholar]
- Franzoni E, Booker SA, Parthasarathy S, Rehfeld F, Grosser S, Srivatsa S, Fuchs HR, Tarabykin V, Vida I, Wulczyn FG (2015) miR-128 regulates neuronal migration, outgrowth and intrinsic excitability via the intellectual disability gene Phf6. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghio L, Gotelli S, Cervetti A, Respino M, Natta W, Marcenaro M, Serafini G, Vaggi M, Amore M, Belvederi Murri M (2015) Duration of untreated depression influences clinical outcomes and disability. J Affect Disord 175:224–228. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan K, Kumarasamy S, Mell B, Joe B (2015) Genome-wide identification of long noncoding RNAs in rat models of cardiovascular and renal disease. Hypertension 65:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TD, Einat H, O’Donnell KC, Picchini AM, Schloesser RJ, Manji HK (2007) Beta-catenin overexpression in the mouse brain phenocopies lithium-sensitive behaviors. Neuropsychopharmacology 32:2173–2183. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Heath PR, Eastwood SL, Burnet PW, McDonald B, Pearson RC (1995) The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vulnerability and comparison with their encoded proteins. Neurosci Lett 200:151–154. [DOI] [PubMed] [Google Scholar]
- Henn FA, Johnson J, Edwards E, Muneyyirci J (1993) Animal models of depression. Clin Neurosci 1:152–156. [Google Scholar]
- Hussaini SM, Choi CI, Cho CH, Kim HJ, Jun H, Jang MH (2014) Wnt signaling in neuropsychiatric disorders: ties with adult hippocampal neurogenesis and behavior. Neurosci Biobehav Rev 47:369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Kenny PJ (2012) MicroRNAs in neuronal function and dysfunction. Trends Neurosci 35:325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidanovich-Beilin O, Milman A, Weizman A, Pick CG, Eldar-Finkelman H (2004) Rapid antidepressive-like activity of specific glycogen synthase kinase-3 inhibitor and its effect on beta-catenin in mouse hippocampus. Biol Psychiatry 55:781–784. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, Rush AJ, Walters EE, Wang PS; National Comorbidity Survey Replication (2003) The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). JAMA 289:3095–3105. [DOI] [PubMed] [Google Scholar]
- Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel JP (2012) MicroRNAs: molecular features and role in cancer. Front Biosci (Landmark Ed) 17:2508–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepine JP. (2001) Epidemiology, burden, and disability in depression and anxiety. J Clin Psychiatry 62(Suppl 13):4–10; discussion 11–12. [PubMed] [Google Scholar]
- Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, Ratnu VS, Boskovic Z, Kobor MS, Sun YE, Bredy TW (2011) The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci 14:1115–1117. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Lopez JP, Fiori LM, Gross JA, Labonte B, Yerko V, Mechawar N, Turecki G (2014) Regulatory role of miRNAs in polyamine gene expression in the prefrontal cortex of depressed suicide completers. Int J Neuropsychopharmacol 17:23–32. [DOI] [PubMed] [Google Scholar]
- Maffioletti E, Cattaneo A, Rosso G, Maina G, Maj C, Gennarelli M, Tardito D, Bocchio-Chiavetto L (2016) Peripheral whole blood microRNA alterations in major depression and bipolar disorder. J Affect Disord 200:250–258. [DOI] [PubMed] [Google Scholar]
- Maguschak KA, Ressler KJ (2012) A role for WNT/β-catenin signaling in the neural mechanisms of behavior. J Neuroimmune Pharmacol 7:763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannironi C, Biundo A, Rajendran S, De Vito F, Saba L, Caioli S, Zona C, Ciotti T, Caristi S, Perlas E, Del Vecchio G, Bozzoni I, Rinaldi A, Mele A, Presutti C (2018) miR-135a regulates synaptic transmission and anxiety-like behavior in amygdala. Mol Neurobiol 55:3301–3315. [DOI] [PubMed] [Google Scholar]
- Miret M, Ayuso-Mateos JL, Sanchez-Moreno J, Vieta E (2013) Depressive disorders and suicide: epidemiology, risk factors, and burden. Neurosci Biobehav Rev 37:2372–2374. [DOI] [PubMed] [Google Scholar]
- Mulligan KA, Cheyette BN (2017) Neurodevelopmental perspectives on wnt signaling in psychiatry. Mol Neuropsychiatry 2:219–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CP, Li X, Maurer V, Oberhauser M, Gstir R, Wearick-Silva LE, Viola TW, Schafferer S, Grassi-Oliveira R, Whittle N, Hüttenhofer A, Bredy TW, Singewald N (2017) MicroRNA-mediated rescue of fear extinction memory by miR-144-3p in extinction-impaired mice. Biol Psychiatry 81:979–989. [DOI] [PubMed] [Google Scholar]
- Nierenberg AA, Fava M, Trivedi MH, Wisniewski SR, Thase ME, McGrath PJ, Alpert JE, Warden D, Luther JF, Niederehe G, Lebowitz B, Shores-Wilson K, Rush AJ (2006) A comparison of lithium and T(3) augmentation following two failed medication treatments for depression: a STAR*D report. Am J Psychiatry 163:1519–1530; quiz 1665. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C (1998) The rat brain in stereotaxic coordinates. 4th edition. Sydney, Australia: Academic Press. [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM (2007) microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol 8:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty F, Sherman AD (1979) Reversal of learned helplessness by imipramine. Commun Psychopharmacol 3:371–373. [PubMed] [Google Scholar]
- Pietrzykowski AZ, Spijker S (2014) Impulsivity and comorbid traits: a multi-step approach for finding putative responsible microRNAs in the amygdala. Front Neurosci 8:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P, Banerjee A, Wu ZS, Zhang X, Wang H, Pandey V, Zhang WJ, Lv XF, Tan S, Lobie PE, Zhu T (2012) Loss of SNAIL regulated miR-128-2 on chromosome 3p22.3 targets multiple stem cell factors to promote transformation of mammary epithelial cells. Cancer Res 72:6036–6050. [DOI] [PubMed] [Google Scholar]
- Rehfeld F, Maticzka D, Grosser S, Knauff P, Eravci M, Vida I, Backofen R, Wulczyn FG (2018) The RNA-binding protein ARPP21 controls dendritic branching by functionally opposing the miRNA it hosts. Nat Commun 9:1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, McEwen BS, Chattarji S (2009) Stress, memory and the amygdala. Nat Rev Neurosci 10:423–433. [DOI] [PubMed] [Google Scholar]
- Roy B, Dunbar M, Shelton RC, Dwivedi Y (2017a) Identification of MicroRNA-124-3p as a putative epigenetic signature of major depressive disorder. Neuropsychopharmacology 42:864–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Wang Q, Palkovits M, Faludi G, Dwivedi Y (2017b) Altered miRNA expression network in locus coeruleus of depressed suicide subjects. Sci Rep 7:4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. (2009) Fine-tuning neural gene expression with microRNAs. Curr Opin Neurobiol 19:213–219. [DOI] [PubMed] [Google Scholar]
- Schmidt JK, Block LN, Golos TG (2018) Defining the rhesus macaque placental miRNAome: Conservation of expression of placental miRNA clusters between the macaque and human. Placenta 65:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman ME, Maier SF (1967) Failure to escape traumatic shock. J Exp Psychol 74:1–9. [DOI] [PubMed] [Google Scholar]
- Shen M, Song Z, Wang JH (2019) microRNA and mRNA profiles in the amygdala are associated with stress-induced depression and resilience in juvenile mice. Psychopharmacology (Berl) 236:2119–2142. [DOI] [PubMed] [Google Scholar]
- Sherman AD, Sacquitne JL, Petty F (1982) Specificity of the learned helplessness model of depression. Pharmacol Biochem Behav 16:449–454. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Rizavi HS, Torvik VI, Turecki G, Dwivedi Y (2012) MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One 7:e33201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloff PH, Lynch KG, Kelly TM, Malone KM, Mann JJ (2000) Characteristics of suicide attempts of patients with major depressive episode and borderline personality disorder: a comparative study. Am J Psychiatry 157:601–608. [DOI] [PubMed] [Google Scholar]
- Song MF, Dong JZ, Wang YW, He J, Ju X, Zhang L, Zhang YH, Shi JF, Lv YY (2015) CSF miR-16 is decreased in major depression patients and its neutralization in rats induces depression-like behaviors via a serotonin transmitter system. J Affect Disord 178:25–31. [DOI] [PubMed] [Google Scholar]
- Tago K, Nakamura T, Nishita M, Hyodo J, Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H, Akiyama T (2000) Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev 14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- Tan CL, Plotkin JL, Venø MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O’Carroll D, Greengard P, Schaefer A (2013) MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science 342:1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk N, Pape JC, Engel M, Zannas AS, Cattane N, Cattaneo A, Binder EB, Chen A (2016) Amygdalar MicroRNA-15a is essential for coping with chronic stress. Cell Rep 17:1882–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas A, Mitra R, Shankaranarayana Rao BS, Chattarji S (2002) Chronic stress induces contrasting patterns of dendritic remodeling in hippocampal and amygdaloid neurons. J Neurosci 22:6810–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Roy B, Turecki G, Shelton RC, Dwivedi Y (2018) Role of complex epigenetic switching in tumor necrosis factor-α upregulation in the prefrontal cortex of suicide subjects. Am J Psychiatry 175:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ML, Licinio J (2004) From monoamines to genomic targets: a paradigm shift for drug discovery in depression. Nat Rev Drug Discov 3:136–151. [DOI] [PubMed] [Google Scholar]
- Zhang K, Song X, Xu Y, Li X, Liu P, Sun N, Zhao X, Liu Z, Xie Z, Peng J (2013) Continuous GSK-3β overexpression in the hippocampal dentate gyrus induces prodepressant-like effects and increases sensitivity to chronic mild stress in mice. J Affect Disord 146:45–52. [DOI] [PubMed] [Google Scholar]
- Zhou WJ, Xu N, Kong L, Sun SC, Xu XF, Jia MZ, Wang Y, Chen ZY (2016) The antidepressant roles of Wnt2 and Wnt3 in stress-induced depression-like behaviors. Transl Psychiatry 6:e892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.