Abstract
Impaired insulin secretion from the pancreatic β-cells is central in the pathogenesis of type 2 diabetes (T2D), and microRNAs (miRNAs) are fundamental regulatory factors in this process. Differential expression of miRNAs contributes to β-cell adaptation to compensate for increased insulin resistance, but deregulation of miRNA expression can also directly cause β-cell impairment during the development of T2D. miRNAs are small noncoding RNAs that posttranscriptionally reduce gene expression through translational inhibition or mRNA destabilization. The nature of miRNA targeting implies the presence of complex and large miRNA–mRNA regulatory networks in every cell, including the insulin-secreting β-cell. Here we exemplify one such network using our own data on differential miRNA expression in the islets of T2D Goto-Kakizaki rat model. Several biological processes are influenced by multiple miRNAs in the β-cell, but so far most studies have focused on dissecting the mechanism of action of individual miRNAs. In this Perspective we present key islet miRNA families involved in T2D pathogenesis including miR-200, miR-7, miR-184, miR-212/miR-132, and miR-130a/b/miR-152. Finally, we highlight four challenges and opportunities within islet miRNA research, ending with a discussion on how miRNAs can be utilized as therapeutic targets contributing to personalized T2D treatment strategies.
Introduction
Currently, there are 2,654 mature microRNAs (miRNAs) in the human genome (mirbase.org, release 22.1) (1). Mature miRNAs are small (∼19–23 nucleotides) noncoding RNAs involved in posttranscriptional gene regulation, fundamentally required to specify cell identity and to adjust cell function. Therefore, perturbed miRNA expression is often associated with development of human diseases (2), including type 2 diabetes (T2D) and associated complications.
Canonical miRNAs may be encoded as individual genes (monocistronic), as gene clusters (polycistronic), or within introns of a host gene and are transcribed by RNA polymerase II to form primary miRNAs (see top of Fig. 1). These are processed by the microprocessor complex containing Drosha and DGCR8 to cleave the primary miRNAs into hairpin-structured pre-miRNAs (3). Once exported into the cytosol by Exportin 5, the RNase III-type enzyme Dicer produces double-stranded miRNAs that are handed over to Argonaute (AGO), which selects one strand to become the mature miRNA. AGO together with the mature miRNA forms the RNA-induced silencing complex (RISC). Generally, nucleotides at positions 2–7 of the miRNAs known as the seed region pair with target mRNA, most commonly in the 3′ untranslated region (UTR). This causes posttranscriptional repression through mRNA cleavage, translational repression, and/or mRNA destabilization. Although the majority of the miRNA binding sites occur in the 3′ UTR, recent studies using CLIP-based techniques show miRNA target sites also present in protein-coding sequences (CDSs) (4), which opens new possibilities for miRNA regulation.
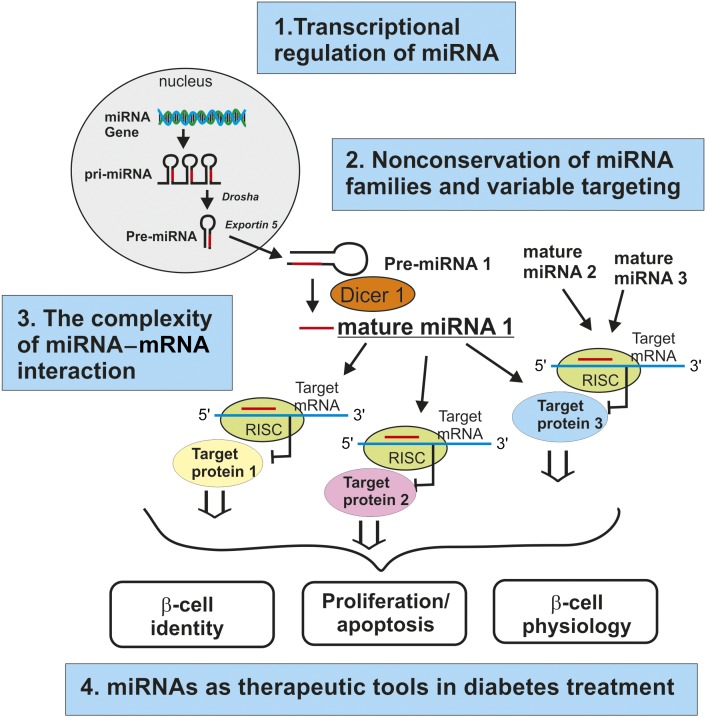
Figure 1.
Summary of future challenges and opportunities in islet miRNA research. The diagram illustrates miRNA biogenesis from gene to the mature form, the miRNA–mRNA interaction network, and the processes influenced by miRNAs in the β-cell (β-cell identity, β-cell proliferation/apoptosis, and β-cell physiology). For each field of these biological processes involving miRNAs, we have added the four areas of challenges and opportunities discussed in the text. pri-miRNA, primary miRNA.
The pairing of miRNAs to their target mRNA is central to the function of miRNAs. Due to the short seed region, one miRNA can have hundreds of different targets. On the other hand, one target can be influenced by several miRNAs. Although miRNAs themselves are highly conserved among species, their targets can vary. Considering this, it is important to discuss the concepts of miRNA cluster and miRNA family. A miRNA cluster refers to a group of miRNAs transcribed from the same gene cluster, whereas miRNAs belonging to the same family share the same seed sequence and hence also share the same targets but do not necessarily need to be transcribed from the same genomic location (3). Examples of a miRNA family abundant in the pancreatic islet cells are the miR-200 family (miR-200a, miR-200b, and miR-429 on chromosome 1 and miR-200c and miR-141 on chromosome 12) (5). Several computational target prediction tools are available primarily based on seed sequence complementarity and RNA folding energy minimization (6). However, a remaining challenge with genome-wide miRNA target searches is to minimize false-positive predictions.
The pancreas contains ∼1 million islets of Langerhans spread throughout the organ. Although the total islet mass only constitutes 1–2% of the whole pancreas, the islets are central for our survival, as they control blood glucose homeostasis. The main cell types within the pancreatic islets are the insulin-secreting β-cells and glucagon-secreting α-cells. A contributing factor in diabetes pathogenesis is when the balance between insulin secretion during hyperglycemia and glucagon secretion during hypoglycemia is disturbed. The role of the pancreatic β-cell in diabetes development has been questioned, but recent studies support the view that classical T2D is a combination of increased insulin resistance in target tissues and impaired β-cell compensation (7). This was reinforced by the new classification derived from analyses of a large Swedish cohort (n = 8,980; All New Diabetics In Scania [ANDIS] cohort) comprising newly diagnosed diabetes patients (8). A major finding is that the patients may be classified into five distinct subgroups, wherein among the nonautoimmune diabetes, 80% exhibit reduced insulin secretion capacity. Again, this supports the idea that impaired β-cell function not only contributes to the progression of the disease but is the primary driver of T2D.
It has been 15 years since the work on the first islet-abundant miRNA, miR-375, was published (9). Today we know that miR-375 has multiple functions in the islet β-cell including roles in development, proliferation, and secretion (9–12). In addition, many other miRNAs have been demonstrated to have central roles in the pancreatic β-cells (13,14). Thus, it is not surprising that several miRNAs are differentially expressed in islets from human T2D donors (15–17) and in diabetic animal models (18,19). Levels of miRNAs can be regulated by metabolic substrates that are increased in the blood prior to diabetes development, such as glucose and fatty acids (20). Moreover, miRNAs have the ability to act as rheostats, changing expression of the target genes to an optimal level (21). Hence, miRNAs are ideal to be part of β-cell adjustments to compensate for an increased demand to secrete more insulin. Indeed, many described differentially expressed islet miRNAs in diabetes have this role. Moreover, changes in the expression of other miRNAs before or during diabetes development are caused by genetic and/or regulatory defects of the miRNA expression leading to the inability of β-cells to secrete enough insulin.
In this Perspective we will first focus on the current views on how miRNAs are involved in the regulation of insulin secretion and development of T2D. Then we will try to summarize future challenges and opportunities in islet miRNA research.
Importance of miRNAs in the Pancreatic β-Cells
The significance of miRNAs in the development and function of pancreatic β-cells has been shown by studying β-cell–specific Dicer1 knockout mice. Several different models of β-cell–specific Dicer1 knockout mice have been generated, and they all provide different clues how miRNAs collectively operate in the β-cell (Table 1). Knockout of Dicer1 during early development reduces β-cell mass (22), whereas knockout in adulthood results in functional defects (23,24). When Dicer1 was deleted under the Ins1 promotor, the mice developed full-blown diabetes (25). The contribution of miRNAs in specifying cellular identity is clearly exemplified in the β-cell where miRNAs can repress the expression of “disallowed” genes. This was demonstrated in a study using tamoxifen-induced deletion of Dicer1 under the Pdx1 promotor (24). Together, these studies show the importance of miRNAs in multiple processes from development, cell fate, proliferation, and maintenance to insulin production and secretion.
Table 1.
Summary of different mouse models used to delete Dicer1 and their phenotypes
| Dicer knockout model | Phenotype | Reference |
|---|---|---|
| Pdx-Cre Dicer1flox/flox | Reduced β-cell mass | Lynn et al. (22) |
| RIP-Cre Dicer1flox/flox | Reduced β-cell mass | Kalis et al. (25) |
| Reduced insulin content | ||
| Reduced number of insulin granules | ||
| Reduced β-cell function | ||
| RIP‐CreER Dicer1LoxP/LoxP (tamoxifen injected in 1- to 5-month-old animals) | Reduced insulin content | Melkman-Zehavi et al. (23) |
| Reduced β-cell function | ||
| PdxCreER:Dicerlox/lox (tamoxifen injected in 7- to 8-week-old animals) | Reduced expression of “disallowed genes” | Martinez-Sanchez et al. (24) |
| Reduced β-cell function | ||
| Reduced insulin content | ||
| Reduced β-cell mass |
Global miRNA ablation upon deletion or knockdown of Dicer1 in β-cells unequivocally demonstrates that multiple pathways in β-cell biology are influenced by miRNAs. However, to obtain detailed information on specific miRNAs and their function in β-cells, the focus has been on single miRNAs or miRNA families and a few of their targets. The most abundant miRNA in the islet, miR-375, was also the first miRNA detected in pancreatic islet (9). In this pioneering study, overexpression of miR-375 resulted in reduced exocytosis and thereby reduced insulin secretion. Later miR-375 deletion resulted in a phenotype with reduced β-cell mass (12), whereas a mouse model with a mild overexpression had no obvious phenotype (11). Modulation of miR-375 in cell lines and primary rodent cells have suggested roles in proliferation, insulin biosynthesis, ion channel activity, and exocytosis (10,26,27). Also, human islet cells losing their identity could be recovered to a β-cell phenotype by addition of miR-375 (28). As previously hypothesized with regard to the fine-tuning function of miRNAs (21), miR-375 is a typical miRNA that needs to be expressed at optimal levels in the cell, i.e., too high or too low expression would be detrimental for cellular functions.
Other highly abundant miRNAs in islets are the members of the miR-29 family (miR-29a/b/c). These miRNAs are transcribed by two different miRNA clusters and contain a common seed sequence but can exhibit differential regulation and therefore do not always have the same function (29). The miR-29 family members have multiple functions in the β-cell, and downregulation increases expression of the disallowed monocarboxylate transporter gene Mct1 (30), increases exocytosis through targeting Onecut2 (31) and Stx1 (32), and increases apoptosis by binding to Mcl-1 mRNA (31).
The miR-7 and the miR-200 family are other examples of islet abundant miRNAs. miR-7 is highly conserved among species and is derived from three different precursors. β-cell–specific overexpression of miR-7a in mice results in reduced insulin secretion through targeting genes involved in vesicle fusion and SNARE activity such as Snca, Cspa, and Cplx1 (11). The miR-200 family consists of five members, and β-cell–specific deletion in mice of either miR-200a/miR-200b/miR-429 or miR-200c/miR-141 shows that these miRNAs strongly regulate β-cell survival (33). Overexpression of miR-200 induces apoptosis and development of T2D in mice and vice versa: deletion of miR-200 protects the β-cells from apoptosis and ameliorates T2D.
The stimulus-secretion coupling, which describes how glucose uptake results in increased insulin secretion through increased metabolism, electrical activity, intracellular Ca2+ concentration, and exocytosis, is central in β-cell function (34). Previous reviews from our group have highlighted the importance of miRNAs in the different subprocesses involved in insulin secretion (13), while another review emphasized miRNA involvement in β-cell survival and β-cell development (35). In summary, we have good knowledge today of how single miRNAs and specific miRNA families impact the expression of specific genes in the β-cell and how this influences insulin secretion. Next, we need to get a grip on the complexity in which miRNAs influence β-cell function, and for that we need to investigate how groups of miRNAs can affect single and/or multiple biological pathway(s).
miRNAs in Islets of Healthy Subjects and Subjects With Diabetes
Few studies have investigated global differential expression of miRNAs in human islets between donors without diabetes (ND) and donors with T2D (16,17). In the work by Kameswaran et al. (16), high-throughput sequencing of small RNAs was performed on islets from relatively few individuals (3 ND and 4 T2D), with a follow-up by quantitative PCR of hits in a larger cohort (islets from ∼15 donors in each group or sorted α- or β-cells from 3–4 donors). The expression of 16 miRNAs was identified to be downregulated in T2D islets (of these, 7 were transcribed from the DLK1-MEG3 locus at chromosome 14q32, a known region for genomic imprinting) and was shown to be regulated by epigenetic changes. The majority of these miRNAs had higher expression in β-cells compared with α-cells. Target validation using AGO2 immunoprecipitation identified targets within pathways important for cell survival (16). In another study using global Taqman miRNA arrays, Locke et al. (17) found miR-187 expression to be differentially upregulated in human T2D islets (11 T2D vs. 9 ND), followed by a validation cohort of 10 in each group using individual Taqman miRNA quantitative PCR assays. This miRNA was also observed to be upregulated in the work by Kameswaran et al. (16). Interestingly, miR-187 expression correlated negatively with insulin secretion in human islets, and overexpression of miR-187 in rat islets resulted in reduction of insulin secretion (17).
Due to the scarcity of human islets for research, valuable information regarding differential expression of miRNAs in T2D islets was instead derived from rodent models. Zhao et al. (19) compared miRNA expression in islets from diabetes-resistant (B6) and diabetes-susceptible (BTBR) ob/ob mice and found enrichment not only of miR-375 but also of miR-127, miR-153, and the miR-200 family in islets compared with liver and adipose tissue. Interestingly, in human tissue, miR-375 and miR-127 were also shown to be more enriched in islets than in liver and muscle (15). Data from the B6 and BTBR mice also suggest that miR-184 expression in islets is suppressed by increased obesity, whereas miR-132/miR-212, miR-133a, miR-185, miR-152, miR-126-5p, and miR-34a/b increased in expression (19). Expression of miR-184 was later shown to be reduced in human islets from T2D donors, and miR-184 was suggested to be part of β-cell compensation during development of the disease (36).
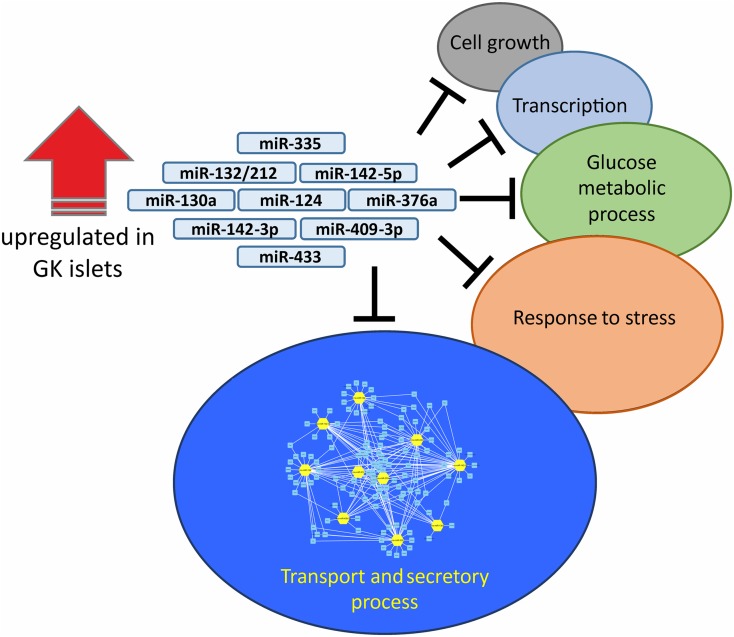
In rats, a global miRNA expression analyses in the pancreatic islets of the T2D model Goto-Kakizaki (GK) rat showed predominantly upregulation of miRNAs in the GK islets compared with Wistar controls (18). Among the top upregulated miRNAs validated were miR-132/212, miR-142-3p/5p, miR-130a, miR-124, miR-335, miR-376a, miR-433, and miR-409-3p. Collectively, although these miRNAs predominantly influence the expression of genes involved in transport and secretory processes, many potential target genes are also present in a wide array of biological networks (Fig. 2). Indeed, mechanistic studies on some of the upregulated GK miRNAs and their target genes revealed not only regulation of exocytotic genes, e.g., miR-335 targeting of Snap25, syntaxin-binding protein 1 (Stxbp1), and synaptotagmin 11 (Syt11) (37), but also regulation of genes involved in energy metabolism, e.g., regulation of pyruvate dehydrogenase E1 α (Pdha1) and glucokinase (Gck) by miR-130a/miR130b/miR-152 (38).
Figure 2.
The collective predicted mRNA targets of upregulated miRNAs in the GK islets were filtered to include only glucose-regulated genes and were subjected to GO enrichment analysis (18). The miRNA-mediated negative regulation of genes within each term is exemplified by the black “T” symbol. Among the genes within enriched GO terms, the majority belong to transport and secretory processes; the unique network (created in Cytoscape, version 3.7.2 [75]) in which the 10 miRNAs (yellow hubs) negatively regulate target genes (light blue nodes) is shown.
Several studies have investigated differential expression in T2D for specific miRNAs (17,36,39,40). In some cases, it has been a follow-up from global profiling in islets from different diabetic animal models. miR-184 is one of the miRNAs downregulated in human T2D islets (36). A reduced expression of miR-184 in rodent models had impact on the expression of Ago2 and miR-375, which improved β-cell proliferation and increased insulin secretion. Thus, miR-184 is a typical example of a miRNA that participates in β-cell compensation during diabetes development. Of note, compensatory miRNAs seem to primarily impact β-cell proliferation and/or apoptosis (20). Expectedly, some of these compensatory miRNAs were first discovered in experiments investigating expansion of β-cell mass during pregnancy. One such example is miR-338. This miRNA was shown by Jacovetti et al. (41) to be downregulated during pregnancy. Moreover, inhibition of miR-338 increases proliferation and protects against β-cell apoptosis. Finally, the authors could show that this miRNA is reduced in high-fat diet–fed and ob/ob mice, clearly demonstrating a compensatory role in diabetes development. Interestingly, DLK1 is upregulated during pregnancy (42). The genetic region coding for DLK1 encodes many miRNAs, and the region is highly methylated in T2D islets. Also, as mentioned above, expression of miRNAs from this region is downregulated in T2D islets (16). Interestingly, experiments performed in mouse insulinoma βTC6 cells in which DNA methyltransferases in this region were activated resulted in increased methylation, reduced miRNA expression, and increased sensitivity to cytokine-induced β-cell death (43). Altogether, the DLK1-MEG3 region and the miRNAs involved have the capacity to promote β-cell expansion and protect against β-cell death prior to diabetes development; failure leads to full-blown diabetes. To this group of compensatory miRNAs also belong miR-375 (12) and miR-132/miR-212 (44). Meanwhile, other differentially expressed miRNAs in diabetes contribute to the β-cell dysfunction (20), including miR-130a/b and miR-152 (38). This is also true for miR-7 (39), miR-204 (45), and miR-200c (33).
Recent focus in the field has been to identify islet cell type–specific or enriched miRNAs, e.g., with function in β-cells and subsequent validation in insulin-secreting cell lines or primary single β-cells. Only few studies have focused on α-cell–specific miRNAs. Two studies using sorted human α- and β-cells detected very few α-cell–specific miRNAs (16,46). In a more recent work, the α- and β-cell miRNA expression profiles between high-fat diet–fed obese hyperglycemic mice and low-fat diet–fed controls were compared (47), wherein miR-132 was identified to be highly differentially expressed in the α-cells between the two groups. Since glucagon secretion is also impaired in T2D and there is a potential for α-cell to β-cell transdifferentiation (48), understanding the role of miRNAs in α-cell biology is of great interest. One of the difficulties facing this area of research is the lack of a satisfactory α-cell line model.
Future Challenges and Opportunities in miRNA Research
Today we have good knowledge of a small group of miRNAs that show up in several studies to be either islet abundant (e.g., miR-375, miR-200 family, miR-127) or differentially expressed in islets in T2D (e.g., miR-7, miR-184, miR-187, miR-132/212, miR-130a/b, and miR-152). Still, current knowledge about islet miRNAs is limited, and there are enormous challenges and opportunities in miRNA research. For instance, one of the limitations of current findings is that most of the knowledge we have on miRNAs and their targets is from highly abundant pancreatic islet cell miRNAs (14). Meanwhile, even low/moderately expressed miRNAs are also perturbed in diabetes (19,38,44). However, investigating individual effects of such less expressed miRNAs is expected to generate marginal phenotypes with small effect size. To circumnavigate this, a combinatorial approach in manipulating the levels of multiple differentially expressed miRNAs could provide a key in truly understanding the impact of low/moderately expressed miRNAs in islet cell pathophysiology. Finally, there are great possibilities for clinical utilization of miRNAs in therapeutics and as biomarkers, but this also comes with a few challenges.
Below we exemplify four areas of interest for future islet miRNA research (also summarized in Fig. 1): 1) transcriptional regulation of miRNA expression, 2) nonconservation of miRNA families and variable targeting, 3) the complexity of miRNA–mRNA interaction network, and 4) use of miRNAs as therapeutic tools in diabetes treatment.
Transcriptional Regulation of miRNAs
Several islet studies have shown different environmental stimuli that can regulate miRNA expression such as glucose (18,45,49), fatty acids (44), and cytokines (50–52) via islet-specific transcription factors. Many intronic miRNAs are coregulated with their host coding genes. However, most miRNAs are under independent transcriptional control of specific transcription factors. Currently, a few detailed analyses of signaling pathways involved in miRNA transcriptional regulation in islets have been done. For miR-132/miR-212 cluster, we and others demonstrated that its transcriptional regulation is dependent on the presence of cAMP and is a PKA-dependent process involving the transcriptional coregulator CRTC1 together with CREB (53,54), as well as the novel transcription factor CAMTA1 (55). For other miRNAs, it has, e.g., been shown that miR-204 expression is increased through glucose-mediated regulation of thioredoxin-interacting protein (TXNIP) (45), while miR-184 expression is reduced through glucose-regulated changes in AMPK activity (49). Moreover, the transcription factor NeuroD1 cooperates with Pdx1 to regulate the transcription of miR-375 (56). Understanding detailed transcriptional regulation mechanisms of miRNAs is central for deeper understanding of their roles in diabetes development.
Another aspect is how miRNA expression can be regulated by changes in the epigenome and expression of other noncoding RNAs. The finding that several miRNA genes present in the DLK1-MEG3 cluster are highly methylated in T2D (16) suggests epigenetic regulation of miRNA expression. Moreover, differential DNA methylation between sexes is associated with differential expression of miR-660 and miR-532 (57). Other types of noncoding RNAs have also been suggested to regulate miRNA expression. There is initial evidence that long noncoding RNA H19 might repress let-7 (58) and that the circular RNAs ciRS7 and circHIPK3 can sequester miR-7 (59,60). The mathematical integration of posttranscriptional networks (containing gene expression data, expression of noncoding RNAs including miRNAs and epigenetic data) will be one of the future challenges, but also an opportunity for a better understanding of the complex regulation of the pancreatic islet cells. However, a comprehensive and accurate understanding of biological processes may only be attained by careful validation using traditional molecular biology tools.
Nonconservation of miRNA Families and Variable Targeting
The miRNA families, just like any genetic element, arose from evolutionary processes over long timescales. Remarkably, while strong purifying selection associated with their regulatory roles resulted in gain of new functional miRNAs, their loss in different genomes has also been shown to be frequent (61). This heterogeneity in rates of miRNA evolution across phyla resulted in species specificity of many miRNA families. Indeed, comparing the well-characterized genomes of humans and mice, there are more than 600 mature miRNAs that can only be found in humans (1). An interesting example of a human-specific miRNA is miR-941, which was shown to be highly expressed in pluripotent stem cells and is involved in brain development and function (62). Characterization of regulatory functions of the nonconserved human-specific miRNAs in mouse and rat diabetes models is a challenge, similar to hurdles posed by primate-specific long noncoding RNAs in islet research (63). Another challenge comes from the nature of miRNA–target interactions in which variable targeting can arise from differences in 3′ UTR sequences of the same gene target in different organisms (64). Although most mammalian mRNAs are conserved miRNA targets (65), it was estimated that ∼50% of the predicted miRNA target sites in humans are not conserved in other organisms (66). In one large-scale study, many experimentally determined noncanonical and nonconserved sites were identified for a number of miRNAs including let-7c, miR-103, miR-106b, miR-141, miR-15a, miR-16, miR-17-5p, miR-192, miR-20, miR-200a, and miR-215 (67). In human and chicken primary chondrocytes, a significant number of nonconserved targets of miR-140 were identified using mRNA expression profiles after manipulation of miR-140 levels (68). Even in closely related species such as rats and mice, the highly enriched β-cell miR-375 modulates voltage-gated sodium channels differently resulting in variable shifts in steady‐state inactivation properties of the channel (27).
Complexity of miRNA–mRNA Interaction Network
The development of mathematics moving into life science comes with many possibilities to fully dissect complex cellular networks. miRNAs have been suggested to act as rheostats in biological systems (21), making them ideal entities in computational modeling for better understanding of their roles. The fact that a single miRNA may have many different targets and that a single mRNA may be targeted by multiple miRNAs means a complex regulatory network of miRNA–mRNA interactions. Remarkably, miRNAs are known to target multiple genes belonging to the same biological pathway (69). In our own pathway analysis of collective targets of the 10 upregulated miRNAs in the islets of GK rats (18), we found that Gene Ontology (GO) terms clustered as “transport and secretory related genes” were enriched (Fig. 2). However, we also found other enriched biological processes, each of which has its own unique set of miRNA–mRNA interaction networks. Consequently, the complexity of miRNA-mediated regulation of insulin secretion starts to emerge.
In the future we need more of these networks in which we can also integrate genetic and epigenetic factors. This is a mathematical challenge encompassing a huge number of coregulatory processes, but with novel bioinformatics approaches there are great possibilities to solve this problem. However, one must always consider the quality of input data required in such endeavors, lest one only succumbs to the most common pitfall of “big” data analytics: “garbage in, garbage out.” For instance, the miRNA seed sequence is very short, giving rise to a high number of predicted false-positive targets, emphasizing why biological validation of targets is necessary for accurate network models.
miRNAs as Therapeutic Tools in Diabetes Treatment
Due to the fact that miRNAs can impact insulin secretion through multiple cellular pathways, they also hold promise to become excellent therapeutic targets. Currently several candidates are in phase 1 and phase 2 clinical trials, e.g., a locked nucleic acid (LNA)-based drug to inhibit miR-92 has potential in wound healing (70). In attempts to treat T2D, studies in animal models have used strategies that could be potentially translated to humans (71–73). Different approaches to silence the miRNAs using chemically modified antagomirs have been employed in these studies. LNA molecules with modified backbones are more stable in blood (74), which might be the most favorable for future studies. However, like any drug-development strategies, a huge challenge is tissue-specific delivery of these RNA-based therapeutics. In our work, systemic injection of antagomir-132 resulted in its delivery in the pancreatic islets resulting in reduced miR-132 levels and subsequent glycemic improvement in the mice (72). Although a promising “proof-of-concept” work in the field, specific delivery into the relevant cells/tissue of RNA-based drugs is highly desired. Another great opportunity utilizing miRNAs as drug targets is the simple base-pairing mechanism of miRNA-mediated regulation, which allows for designing sequences tailored for specific gene variants. With that said, there are still many challenges with RNA-based inhibitors of miRNAs. Current complications with specificity and off-target effects, sensitivity of inhibitors, innate immune responses, and the exact tissue/cell delivery require further development of antagomirs before use. However, the concept of using miRNA inhibition offers a novel approach in personalized diagnostic and treatment strategies in diabetes and associated complications.
Concluding Remarks
We hope this Perspective will help put renewed focus on miRNAs in islet research and the possibility of using miRNA antagomirs in treatment of T2D. We wanted to highlight some of the challenges and opportunities that miRNA research in islets, and in general, will be facing in the coming years. During the 15 years since the first discovery of an islet-enriched miRNA, an immense amount of data and knowledge have been gathered. Let us hope for a new prosperous miRNA period, keeping in mind that we are also moving into the era of personalized medicine.
Article Information
Acknowledgments. The authors thank colleagues and friends for fruitful discussions in this area.
Funding. The authors are supported by grants from the Swedish Foundation for Strategic Research (IRC-LUDC), the Swedish Research Council (project grant to L.E., SFO-EXODIAB), Region Skåne-ALF, the Swedish Diabetes Foundation, the Diabetes Wellness Network Sweden, Albert Påhlsson Foundation, Crafoord Foundation, Byggmästare Olle Engqvist Foundation, Syskonen Svenssons Fond, and the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement no. 667191 (T2DSystems).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Footnotes
This article is part of a special article collection available at https://diabetes.diabetesjournals.org/collection/small-noncoding-RNAs-in-diabetes.
References
- 1.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019;47(D1):D155–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul P, Chakraborty A, Sarkar D, et al. Interplay between miRNAs and human diseases. J Cell Physiol 2018;233:2007–2018 [DOI] [PubMed] [Google Scholar]
- 3.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 2019;20:5–20 [DOI] [PubMed] [Google Scholar]
- 4.Zhang K, Zhang X, Cai Z, et al. A novel class of microRNA-recognition elements that function only within open reading frames. Nat Struct Mol Biol 2018;25:1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet 2016;17:719–732 [DOI] [PubMed] [Google Scholar]
- 6.Lukasik A, Wójcikowski M, Zielenkiewicz P. Tools4miRs - one place to gather all the tools for miRNA analysis. Bioinformatics 2016;32:2722–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halban PA, Polonsky KS, Bowden DW, et al. β-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Diabetes Care 2014;37:1751–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahlqvist E, Storm P, Käräjämäki A, et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 2018;6:361–369 [DOI] [PubMed] [Google Scholar]
- 9.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004;432:226–230 [DOI] [PubMed] [Google Scholar]
- 10.Eliasson L. The small RNA miR-375 - a pancreatic islet abundant miRNA with multiple roles in endocrine beta cell function. Mol Cell Endocrinol 2017;456:95–101 [DOI] [PubMed] [Google Scholar]
- 11.Latreille M, Herrmanns K, Renwick N, et al. miR-375 gene dosage in pancreatic β-cells: implications for regulation of β-cell mass and biomarker development. J Mol Med (Berl) 2015;93:1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poy MN, Hausser J, Trajkovski M, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A 2009;106:5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esguerra JLS, Nagao M, Ofori JK, Wendt A, Eliasson L. MicroRNAs in islet hormone secretion. Diabetes Obes Metab 2018;20(Suppl. 2):11–19 [DOI] [PubMed] [Google Scholar]
- 14.LaPierre MP, Stoffel M. MicroRNAs as stress regulators in pancreatic beta cells and diabetes. Mol Metab 2017;6:1010–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolmeson C, Esguerra JL, Salehi A, Speidel D, Eliasson L, Cilio CM. Differences in islet-enriched miRNAs in healthy and glucose intolerant human subjects. Biochem Biophys Res Commun 2011;404:16–22 [DOI] [PubMed] [Google Scholar]
- 16.Kameswaran V, Bramswig NC, McKenna LB, et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab 2014;19:135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Locke JM, da Silva Xavier G, Dawe HR, Rutter GA, Harries LW. Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia 2014;57:122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esguerra JLS, Bolmeson C, Cilio CM, Eliasson L. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One 2011;6:e18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao E, Keller MP, Rabaglia ME, et al. Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mamm Genome 2009;20:476–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eliasson L, Regazzi R. Micro(RNA) management and mismanagement of the islet. J Mol Biol 2020;432:1419–1428 [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes 2007;56:2938–2945 [DOI] [PubMed] [Google Scholar]
- 23.Melkman-Zehavi T, Oren R, Kredo-Russo S, et al. miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. EMBO J 2011;30:835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Sanchez A, Nguyen-Tu MS, Rutter GA. DICER inactivation identifies pancreatic β-cell “disallowed” genes targeted by microRNAs. Mol Endocrinol 2015;29:1067–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalis M, Bolmeson C, Esguerra JL, et al. Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS One 2011;6:e29166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 2008;57:2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salunkhe VA, Esguerra JL, Ofori JK, et al. Modulation of microRNA-375 expression alters voltage-gated Na+ channel properties and exocytosis in insulin-secreting cells. Acta Physiol (Oxf) 2015;213:882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nathan G, Kredo-Russo S, Geiger T, et al. MiR-375 promotes redifferentiation of adult human β cells expanded in vitro. PLoS One 2015;10:e0122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics 2012;44:237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pullen TJ, da Silva Xavier G, Kelsey G, Rutter GA. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol Cell Biol 2011;31:3182–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roggli E, Gattesco S, Caille D, et al. Changes in microRNA expression contribute to pancreatic β-cell dysfunction in prediabetic NOD mice. Diabetes 2012;61:1742–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagge A, Dahmcke CM, Dalgaard LT. Syntaxin-1a is a direct target of miR-29a in insulin- producing β-cells. Horm Metab Res 2013;45:463–466 [DOI] [PubMed] [Google Scholar]
- 33.Belgardt BF, Ahmed K, Spranger M, et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med 2015;21:619–627 [DOI] [PubMed] [Google Scholar]
- 34.Rorsman P, Ashcroft FM. Pancreatic β-cell electrical activity and insulin secretion: of mice and men. Physiol Rev 2018;98:117–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaviani M, Azarpira N, Karimi MH, Al-Abdullah I. The role of microRNAs in islet β-cell development. Cell Biol Int 2016;40:1248–1255 [DOI] [PubMed] [Google Scholar]
- 36.Tattikota SG, Rathjen T, McAnulty SJ, et al. Argonaute2 mediates compensatory expansion of the pancreatic b cell. Cell Metab 2014;19:122–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salunkhe VA, Ofori JK, Gandasi NR, et al. MiR-335 overexpression impairs insulin secretion through defective priming of insulin vesicles. Physiol Rep 2017;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ofori JK, Salunkhe VA, Bagge A, et al. Elevated miR-130a/miR130b/miR-152 expression reduces intracellular ATP levels in the pancreatic beta cell. Sci Rep 2017;7:44986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latreille M, Hausser J, Stützer I, et al. MicroRNA-7a regulates pancreatic β cell function. J Clin Invest 2014;124:2722–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sebastiani G, Po A, Miele E, et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol 2015;52:523–530 [DOI] [PubMed] [Google Scholar]
- 41.Jacovetti C, Abderrahmani A, Parnaud G, et al. MicroRNAs contribute to compensatory β cell expansion during pregnancy and obesity. J Clin Invest 2012;122:3541–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winkel L, Bagge A, Larsen L, et al. Trefoil factor 3 in perinatal pancreas is increased by gestational low protein diet and associated with accelerated β-cell maturation. Islets 2018;10:e1472186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kameswaran V, Golson ML, Ramos-Rodríguez M, et al. The dysregulation of the DLK1-MEG3 locus in islets from patients with type 2 diabetes is mimicked by targeted epimutation of its promoter with TALE-DNMT constructs. Diabetes 2018;67:1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nesca V, Guay C, Jacovetti C, et al. Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia 2013;56:2203–2212 [DOI] [PubMed] [Google Scholar]
- 45.Xu G, Chen J, Jing G, Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat Med 2013;19:1141–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein D, Misawa R, Bravo-Egana V, et al. MicroRNA expression in alpha and beta cells of human pancreatic islets. PLoS One 2013;8:e55064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dusaulcy R, Handgraaf S, Visentin F, Vesin C, Philippe J, Gosmain Y. miR-132-3p is a positive regulator of alpha-cell mass and is downregulated in obese hyperglycemic mice. Mol Metab 2019;22:84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuoka TA, Kawashima S, Miyatsuka T, et al. Mafa enables Pdx1 to effectively convert pancreatic islet progenitors and committed islet α-cells into β-cells in vivo. Diabetes 2017;66:1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Sanchez A, Nguyen-Tu MS, Cebola I, et al. MiR-184 expression is regulated by AMPK in pancreatic islets. FASEB J 2018;32:2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grieco FA, Schiavo AA, Brozzi F, et al. The miRNAs miR-211-5p and miR-204-5p modulate ER stress in human beta cells. J Mol Endocrinol 2019;63:139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grieco FA, Sebastiani G, Juan-Mateu J, et al. MicroRNAs miR-23a-3p, miR-23b-3p, and miR-149-5p regulate the expression of proapoptotic BH3-only proteins DP5 and PUMA in human pancreatic β-cells. Diabetes 2017;66:100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roggli E, Britan A, Gattesco S, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes 2010;59:978–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malm HA, Mollet IG, Berggreen C, et al. Transcriptional regulation of the miR-212/miR-132 cluster in insulin-secreting β-cells by cAMP-regulated transcriptional co-activator 1 and salt-inducible kinases. Mol Cell Endocrinol 2016;424:23–33 [DOI] [PubMed] [Google Scholar]
- 54.Shang J, Li J, Keller MP, et al. Induction of miR-132 and miR-212 expression by glucagon-like peptide 1 (GLP-1) in rodent and human pancreatic β-cells. Mol Endocrinol 2015;29:1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mollet IG, Malm HA, Wendt A, Orho-Melander M, Eliasson L. CAMTA1-Calcium- Calmodulin Transcriptional Activator 1, a new player in the regulation of microRNAs and insulin secretion. Diabetologia 2015;58:S88–S88 [Google Scholar]
- 56.Keller DM, McWeeney S, Arsenlis A, et al. Characterization of pancreatic transcription factor Pdx-1 binding sites using promoter microarray and serial analysis of chromatin occupancy. J Biol Chem 2007;282:32084–32092 [DOI] [PubMed] [Google Scholar]
- 57.Hall E, Volkov P, Dayeh T, et al. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol 2014;15:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez-Parra C, Jacovetti C, Dumortier O, et al. Contribution of the long noncoding RNA H19 to β-cell mass expansion in neonatal and adult rodents. Diabetes 2018;67:2254–2267 [DOI] [PubMed] [Google Scholar]
- 59.Stoll L, Sobel J, Rodriguez-Trejo A, et al. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metab 2018;9:69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015;5:12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomson RC, Plachetzki DC, Mahler DL, Moore BR. A critical appraisal of the use of microRNA data in phylogenetics. Proc Natl Acad Sci U S A 2014;111:E3659–E3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu HY, He L, Fominykh K, et al. Evolution of the human-specific microRNA miR-941. Nat Commun 2012;3:1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esguerra JL, Eliasson L. Functional implications of long non-coding RNAs in the pancreatic islets of Langerhans. Front Genet 2014;5:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics 2008;24:325–332 [DOI] [PubMed] [Google Scholar]
- 67.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010;11:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nicolas FE, Pais H, Schwach F, et al. mRNA expression profiling reveals conserved and non-conserved miR-140 targets. RNA Biol 2011;8:607–615 [DOI] [PubMed] [Google Scholar]
- 69.Backes C, Kehl T, Stöckel D, et al. miRPathDB: a new dictionary on microRNAs and target pathways. Nucleic Acids Res 2017;45(D1):D90–D96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet 2019;10:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song Y, Jin D, Jiang X, Lv C, Zhu H. Overexpression of microRNA-26a protects against deficient β-cell function via targeting phosphatase with tensin homology in mouse models of type 2 diabetes. Biochem Biophys Res Commun 2018;495:1312–1316 [DOI] [PubMed] [Google Scholar]
- 72.Bijkerk R, Esguerra JLS, Ellenbroek JH, et al. In vivo silencing of microRNA-132 reduces blood glucose and improves insulin secretion. Nucleic Acid Ther; 2019;29:67–72 [DOI] [PubMed] [Google Scholar]
- 73.Mulder NL, Havinga R, Kluiver JL, Groen AK, Kruit JK. AAV8-mediated gene transfer of microRNA-132 improves beta-cell function in mice fed a high fat diet. J Endocrinol 2018;240:123–132 [DOI] [PubMed] [Google Scholar]
- 74.Elmén J, Thonberg H, Ljungberg K, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res 2005;33:439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]