Abstract
To better understand the evolution of hepadnaviruses, we sampled bats from Guizhou, Henan and Zhejiang provinces, China, and rodents from Zhejiang province. Genetically diverse hepadnaviruses were identified in a broad range of bat species, with an overall prevalence of 13.3%. In contrast, no rodent hepadnaviruses were identified. The newly discovered bat hepadnaviruses fell into two distinct phylogenetic groups. The viruses within the first group exhibited high diversity, with some closely related to viruses previously identified in Yunnan province. Strikingly, the newly discovered viruses sampled from Jiyuan city in the second phylogenetic group were most closely related to those found in bats from West Africa, suggestive of a long-term association between bats and hepadnaviruses. A co-phylogenetic analysis revealed frequent cross-species transmission among bats from different species, genera, and families. Overall, these data suggest that there are likely few barriers to the cross-species transmission of bat hepadnaviruses.
Keywords: Hepadnaviruses, Bats, Rodents, Phylogeny, Evolution, Cross-species transmission
Highlights
-
•
Diverse hepadnaviruses are identified in a broad range of bat species in China.
-
•
Some of them were closely related to those previously identified in China.
-
•
The viruses from Jiyuan were most closely related to Gabon bat hepadnaviruses.
-
•
Newly discovered viruses did not clustered by bat species or geographic location.
-
•
Frequent cross-species transmission among different bat species was observed.
1. Introduction
Hepadnaviruses (family Hepadnaviridae) are partially double-stranded DNA (dsDNA) viruses that replicate their DNA by reverse transcription of an RNA intermediate (Urban et al., 2010). At present, the family contains two genera -Avihepadnavirus, Orthohepadnavirus- as well as one floating virus, White sucker hepatitis B virus, sampled from a fish (https://talk.ictvonline.org/taxonomy). Recently, two additional hepadnaviruses were identified in fish and frogs and await classification (Dill et al., 2016). Hepadnaviruses possess very small (3.0–3.3 kb) partially dsDNA genomes that exhibit a circular conformation by base pairing in a cohesive overlap between the 5’ ends of the two DNA strands (Seeger et al., 2013). The orthohepadnavirus genome contains four overlapping open reading frames (ORFs) encoding the viral polymerase (P), core (C), surface (S), and X proteins, while the genus Avihepadnavirus lacks the ORF X. Hepadnaviruses are well known because the prototype virus of the family, human hepatitis B virus (HBV), causes around 250 million chronic infections worldwide and at least 887,000 deaths each year (http://www.who.int/mediacentre/factsheets/fs204/en).
Orthohepadnaviruses infect mammals including humans. Although hepadnaviruses were discovered in ground squirrels (genus Marmota) in the USA (Summers et al., 1978; Marion et al., 1980; Testut et al., 1996), they have been not observed in other rodents or in other geographic localities. Notably, however, orthohepadnaviruses have recently been identified in both Old and New World bats: specifically, long-fingered bat HBV (LBHBV) in Miniopterus fuliginosus from Myanmar (He et al., 2013), tent-making bat hepatitis B virus (TBHBV) in Uroderma bilobatum from Panama, roundleaf bat HBV (RBHBV) and horseshoe bat HBV (HBHBV) in Hipposideros cf. ruber and Rhinolophus alcyone from Gabon (Drexler et al., 2013), and HBHBV-like viruses in H. pomona, R. affinis and R. sinicus from Yunnan province, China (He et al., 2015; Wang et al., 2017a). These results suggest that bats may be important natural hosts for hepadnaviruses.
Hepadnaviruses are believed to be ancient pathogens, with an evolutionary history characterized by virus-host co-divergence across multiple vertebrate classes (Dill et al., 2016, Lauber et al., 2017) as well as frequent cross-species transmission (Geoghegan et al., 2017). In addition, ancient endogenous hepadnaviruses have been identified in a diverse range of vertebrates (Katzourakis and Gifford, 2010, Gilbert and Feschotte, 2010, Cui et al., 2014). Importantly, bat hepadnaviruses occupy diverse positions in the orthohepadnavirus phylogeny (Rasche et al., 2016), indicative of multiple cross-species transmission events. Indeed, TBHBVs harbored by tent-making bats from Panama form a sister-group to all known orthohepadnaviruses and can infect human hepatocytes (Drexler et al., 2013). However, because hepadnaviruses have only been identified in a narrow range of animal hosts, the origin and evolutionary history of these important mammalian viruses are unclear.
Bats and rodents comprise some 20% and 42% of living mammalian species, respectively (Wilson and Reeder, 2005). An increasing number of studies have demonstrated the importance of bats and rodents in the diversity and evolution of viruses, including arenaviruses (Grande-Perez et al., 2016), coronaviruses (Drexler et al., 2014), and hantaviruses (Holmes and Zhang, 2015). In addition, because of their often close proximity to humans, (re)emerging infectious diseases from bats and rodents present a continual threat to human health and agricultural production (Calisher et al., 2006, Meerburg et al., 2009). To better understand the diversity and evolution of orthohepadnaviruses, we screened bats and rodents sampled from three provinces in China.
2. Results
2.1. Collection of bats and rodents, and the detection of hepadnaviruses
During 2014–2016, 797 bats were captured from Anlong county of Guizhou province, Jiyuan city, Lushi and Neixiang counties of Henan province, and Longquan city of Zhejiang province, China ( Fig. 1). The numbers, species, and geographic distributions of these bats captured are described in Table 1. In addition, during 2015–2016, 462 rodents (23 Apodemus agrarius, 34 Rattuse dwardsi, 33 R. flavipectus, 3 R. losea, 154 R. norvegicus, 162 Niviventer confucianus, 36 N. fulvescens, and 17 Berylmys bowersi) and 148 Asian house shrews (Suncus murinus) were collected from Longquan city, and Lucheng district and Wencheng county of Wenzhou city, Zhejiang province. The liver tissues from these animals were then screened for hepadnaviruses.
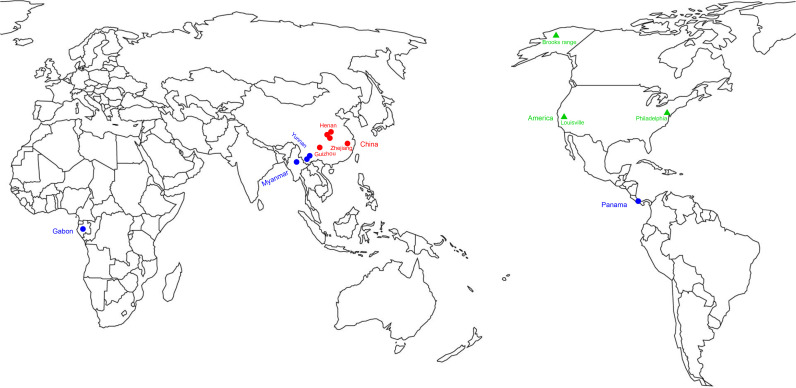
Fig. 1.
Map showing the location of trap sites (red circles) in which bats, rodents, shrews were captured in China, in this study and the location of known hepadnaviruses by their associated bat and rodent hosts (blue circles for bats and green triangles for rodents).
Table 1.
Prevalence of hepadnavirus in bats by species and location in China.
| Species | Guizhou |
Henan |
Zhejiang | Total (%) | ||
|---|---|---|---|---|---|---|
| Anlong | Jiyuan | Lushi | Neixiang | Longquan | ||
| Rhinolophus affinis | – | – | – | 0/3 | 0/4 | 0/7 (0) |
| R. ferrumequinum | – | 8/132 | 0/39 | 0/8 | – | 8/179 (5.0) |
| R. luctus | – | – | 0/2 | 0/5 | 1/1 | 1/8 (12.5) |
| R. macrotis | 0/10 | – | – | – | – | 0/10 (0) |
| R. monoceros | – | – | 0/2 | 1/1 | 27/69 | 28/72(38.9) |
| R. pearsonii | 0/20 | – | – | 3/7 | 3/21 | 6/48 (12.5) |
| R. pusillus | 0/9 | 9/26 | 2/60 | 14/43 | 1/1 | 26/139(18.7) |
| R. rex | 0/6 | – | – | – | – | 0/6 (0) |
| R. sinicus | 3/19 | – | – | – | 5/11 | 8/30 (26.7) |
| Hipposideros armiger | 0/45 | – | – | – | 19/47 | 19/92 (20.7) |
| H. Pomona | 0/3 | – | – | – | – | 0/3 (0) |
| Miniopterus schreibersii | 6/68 | – | – | 2/19 | 0/2 | 8/89 (9.0) |
| Myotis chinensis | 1/1 | 1/1 (100) | ||||
| M. davidii | 1/26 | – | – | 0/1 | – | 1/27 (3.7) |
| M. siligorensis | 0/46 | – | – | – | – | 0/46 (0) |
| Murina leucogaster | – | 0/2 | 0/5 | 0/21 | – | 0/28 (0) |
| M.sp | – | 0/3 | – | – | – | 0/3 (0) |
| Barbastella beijingensis | – | 0/2 | – | – | – | 0/2 (0) |
| Ia io | 0/2 | – | – | – | – | 0/2 (0) |
| Plecotus auritus | – | 0/2 | – | – | – | 0/2 (0) |
| Rousettus leschenaulti | 0/3 | – | – | – | – | 0/3 (0) |
| Total (%) | 11/258 (4.3) | 17/167 (10.2) | 2/108 (1.9) | 20/108 (18.5) | 56/156 (35.9) | 106/797 (13.3) |
Note: ‘‘-’’ indicates that no animals were captured.
PCR products of the expected size for the P gene were detected in 11 bats from Guizhou, 39 bats from Henan, and 56 bats from Zhejiang (Table 1 and S1). Genetic analysis of these sequences revealed that they were members of the genus Orthohepadnavirus. Hepadnavirus DNA was detected in 10 species of bats, with an overall detection rate of 13.3% (106/797). Notably, the detection rate was higher in R. monoceros (38.9%), H. armiger (20.7%), R. pusillus (18.7%), R. pearsonii (12.5), and Miniopterus schreibersii (9%). Due to variation in the dominant species, the prevalence of hepadnaviruses was highest in Longquan city (35.9%) in Zhejiang province, followed by Neixiang county (18.5%) and Jiyuan (10.2%) city in Henan province. Strikingly, viral DNA was not identified in any mice, rats or Asian house shrews tested here.
To better characterize the newly discovered bat hepadnaviruses, the complete genome was recovered from 40 viral DNA positive bat samples based on viral characteristics (see below), host species and sampling sites (Table S1).
2.2. Evolutionary relationships among newly identified and known hepadnaviruses
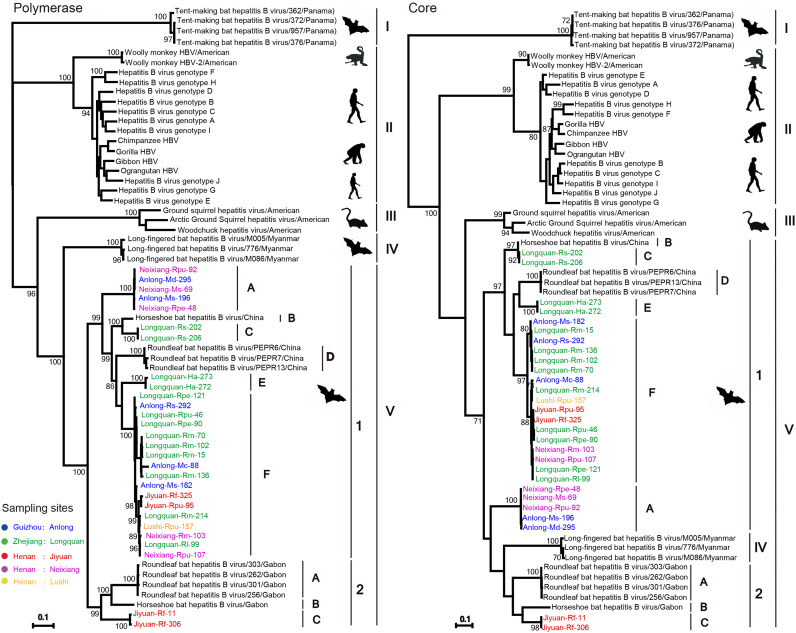
Hepadnavirus phylogenetic trees were estimated based on the nucleotide sequences of the P, C, S and X genes using an maximum likelihood (ML) approach. In the P gene tree ( Fig. 2), all known orthohepadnaviruses including the bat viruses newly identified here fell into five clades. Bat viruses comprised clades I, IV and V, while primate and rodent viruses formed the clades II and III, respectively. All the bat viruses newly identified here fell into clade V. This clade, comprising only hepadnaviruses from the Old World, was further divided into two phylogenetic groups. The first group could be further separated into six lineages (denoted A-F), highlighting the extensive diversity of bat hepadnaviruses. The viruses in these lineages, which were identified in M. schreibersii and M. davidii bats sampled from Anlong (Guizhou province) and M. schreibersii, R. pearsonii, and R. pusillus from Neixiang (Henan province), formed a distinct lineage (denoted A) and possessed 99.4–100% nucleotide identity. Both lineages C and E were sampled in Longquan in Zhejiang province, and found in R. sinicus and H. armiger bats, respectively. Strikingly, lineage F comprised viruses identified in eight bat species from three families (Miniopteridae, Rhinolophidae and Vespertilionidae) at all investigating sites, with 95.5–100% nucleotide identity. In addition, this group contained two other lineages (B and D) comprising viruses previously sampled from roundleaf bats (H. pomona) and horseshoe bats (R. affinis and R. sinicus) in Yunnan province, China (He et al., 2015; Wang et al., 2017a). The nucleotide differences between the six lineages ranged from 77.4% to 92.8%. Remarkably, within the second group, the bat viruses identified in R. ferrumequinum from Jiyuan in Henan province formed a sister group to viruses identified in horseshoe (R. alcyone) and roundleaf (H. cf. ruber) bats sampled from Gabon in West Africa (Drexler et al., 2013). The newly identified viruses exhibited 20.3–20.5% nucleotide difference from the Gabonese viruses ( Table 2). Clade V showed a sister relationship to clade IV, with the latter comprising only LBHBVs identified in M. fuliginosus sampled from Myanmar (He et al., 2013). Finally, the most divergent clade I only contained TBHBVs. In sum, these phylogenetic data reveal an evolutionary history characterized by extensive genetic diversity, frequent cross-species transmission, in situ evolution and a widespread geographic distribution (Fig. 1, Fig. 2).
Fig. 2.
Phylogenetic analysis of the nucleotide sequences of the P and C genes of orthohepadnaviruses. Host taxa and phylogenetic groups are indicated. Bootstrap values (> 70%) are shown at relevant nodes. The trees were mid-point rooted for clarity only. The scale bar depicts the number of nucleotide substitutions per site.
Table 2.
Nucleotide identities between the bat viruses newly identified here and known hepadnaviruses.
| Lineage (No. of viruses) | Genome nucleotides identity (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HBV/ayw | WMHBV | WHV | TBHBV | LBHBV | HBHBV-Gabon | RBHBV-Gabon | HBHBV-China | RBHBV-China | Within lineage | |
| 1A (7) | 64.2–64.3 | 65.6–65.8 | 64.5–64.7 | 62.5–62.7 | 72.2–72.4 | 74.9–75.0 | 73.9–74.1 | 79.3–79.6 | 77.4–77.6 | 99.4–100 |
| 1C (2) | 65.7 | 65.5–65.6 | 65.1–65.2 | 63.7–63.8 | 72.0 | 76.2 | 76.5 | 92.7–92.8 | 81.8 | 99.8 |
| 1E (2) | 64.6 | 65.4 | 64.9 | 62.3–62.4 | 71.6 | 74.8–74.9 | 74.2 | 83.4–83.5 | 82.4 | 99.7 |
| 1F (23) | 63.9–64.5 | 64.7–65.0 | 65.0–65.4 | 63.1–63.8 | 70.4–71.2 | 74.6–74.5 | 74.8–75.8 | 84.7–85.9 | 81.7–82.9 | 95.5–100 |
| 2C (6) | 66.0–66.2 | 66.0–66.1 | 66.1–66.2 | 63.8–63.9 | 72.0–72.3 | 79.6–79.7 | 79.5–79.7 | 76.1–76.3 | 74.5–74.7 | 99.2–99.9 |
Notes: HBV – human hepatitis B virus; WMHBV – Woolly monkey hepatitis virus; WHV – Woodchuck hepatitis virus; TBHBV – tent-making hepatitis B virus; LBHBV – long-fingered hepatitis B virus; HBHBV– horseshoe hepatitis B virus; RBHBV – roundleaf hepatitis B virus.
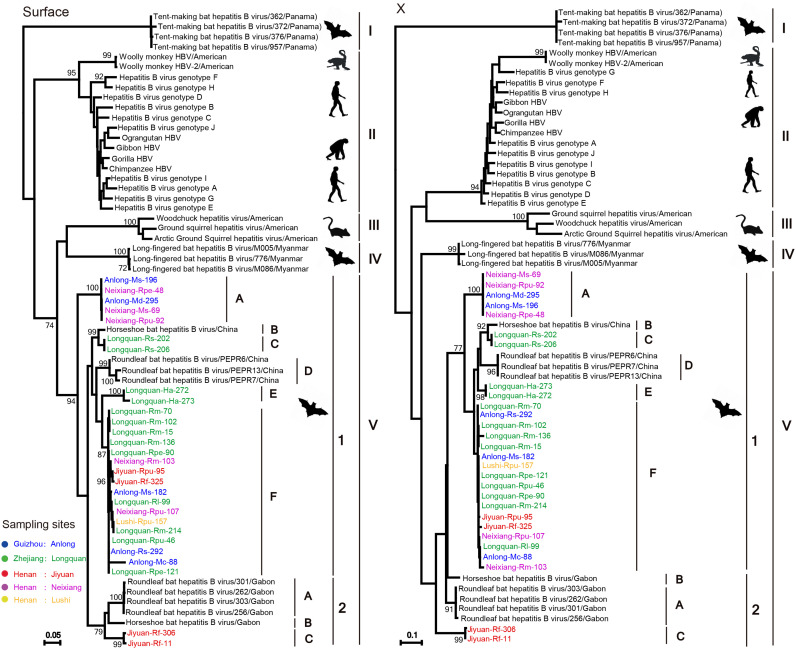
Although the topology of the C gene tree was similar to that of the P gene tree (Fig. 2), the clade IV viruses (LBHBVs) identified in the P gene now fell into clade V, compatible with recombination. In addition, lineage A fell into a different phylogenetic position in the C gene tree, and occupied a basal position in clade V in the S gene tree ( Fig. 3). Finally, the phylogenetic position of the second group of clade V also changed in the X gene tree, although this may reflect the short length of the sequences used. Indeed, despite the apparent phylogenetic incongruence in these trees, there was no statistically significant support for recombination from the RDP analysis, so that the occurrence and pattern of recombination in these data remains uncertain.
Fig. 3.
Phylogenetic analysis of the nucleotide sequences of the S and X genes of orthohepadnaviruses. Host taxa and phylogenetic groups are indicated. Bootstrap values (> 70%) are shown at relevant nodes. The trees were mid-point rooted for clarity only. The scale bar depicts the number of nucleotide substitutions per site.
2.3. Genome features of the newly identified and known hepadnaviruses
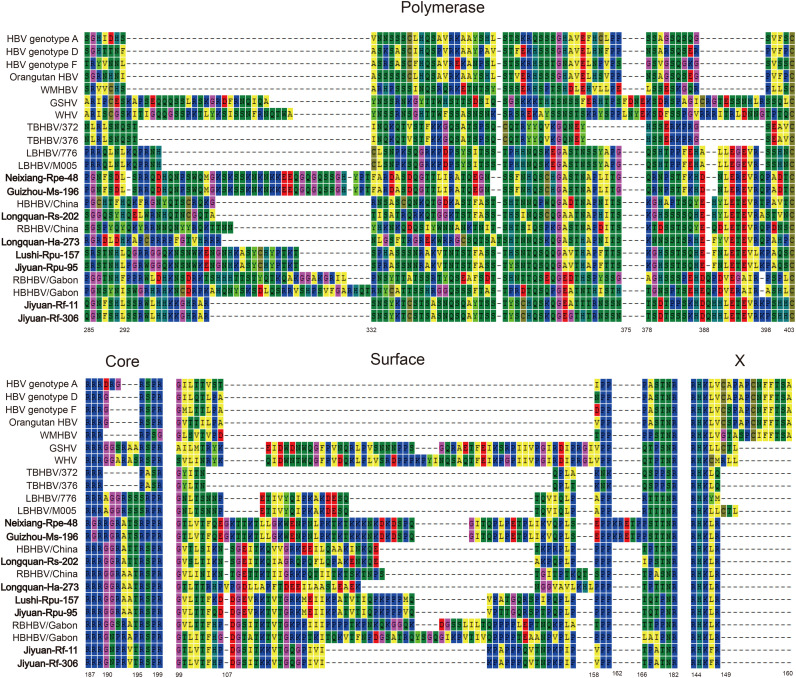
The genomic features of the newly identified bat viruses are described in Table 3. The genome organization of the newly identified bat viruses was similar to that of other orthohepadnaviruses, with P, C, S and X ORFs and a G+C content of 49.6–51.81%. The genome size (3272-3341nt) of the newly identified bat viruses was similar to that of known bat and rodent viruses, although larger than that of primate hepadnaviruses. The P gene exhibited substantial length variation: bat hepadnaviruses (2562-2700nt), rodent hepadnaviruses (2646-2655nt), primate hepadnaviruses (2499-2538nt), and TBHBV (2484nt). This length variation was mainly due to deletions at amino acids 292−332 and 388−398 ( Fig. 4). For the PreC/C gene the bat and rodent hepadnaviruses had the same gene length (654nt), with the exception of WHV (657nt) and TBHBV (567nt).
Table 3.
Key genome features of the bat hepadnaviruses newly identified here.
| Lineage | Genome size (nt) | G+C (%) | DR | ORF | Location (nt) | Length (nt) | Length (aa) |
|---|---|---|---|---|---|---|---|
| 1A | 3341 | 49.6–49.87 | TTCACCTGTGC | Pol | 2311–3341, 1–1639 | 2670 | 889 |
| PreS/S | 2867–3341, 1–836 | 1311 | 436 | ||||
| PreC/C | 1821–2474 | 654 | 217 | ||||
| X | 1381–1809 | 429 | 142 | ||||
| 1C | 3272 | 50.95 | TTCACCTGTGC | Pol | 2311–3272, 1–1639 | 2601 | 866 |
| PreS/S | 2867–3272, 1–836 | 1242 | 413 | ||||
| PreC/C | 1821–2474 | 654 | 217 | ||||
| X | 1381–1809 | 429 | 142 | ||||
| 1E | 3275 | 51.15–51.30 | TTCACCTGTGC | Pol | 2314–3276, 1–1642 | 2604 | 867 |
| PreS/S | 2870–3276,1–839 | 1245 | 414 | ||||
| PreC/C | 1824–2473 | 651 | 216 | ||||
| X | 1384–1812 | 429 | 142 | ||||
| 1F | 3320 | 50.95–51.81 | TTCACCTGTGC | Pol | 2314–3320, 1–1642 | 2649 | 882 |
| PreS/S | 2870–3320, 1–839 | 1290 | 429 | ||||
| PreC/C | 1824–2477 | 654 | 217 | ||||
| X | 1384–1812 | 429 | 142 | ||||
| 2C | 3293 | 51.32–51.43 | TTCACCTGTGC | Pol | 2314–3293, 1–1642 | 2622 | 873 |
| PreS/S | 2873–3293, 1–842 | 1263 | 420 | ||||
| PreC/C | 1824–2477 | 654 | 217 | ||||
| X | 1387–1812 | 426 | 141 | ||||
| RBHBV/China | 3278 | 47.83–48.02 | TTCACCTGTGC | Pol | 2314–3278, 1–1642 | 2607 | 868 |
| PreS/S | 2870–3278, 1–839 | 1248 | 415 | ||||
| PreC/C | 1824–2477 | 654 | 217 | ||||
| X | 1384–1812 | 429 | 140 | ||||
| RBHBV/Gabon | 3368 | 53.15–53.36 | TTCACCTGTGC | Pol | 2147–3368,1–1478 | 2700 | 899 |
| PreS/S | 2706–3368, 1–675 | 1338 | 445 | ||||
| PreC/C | 1657–2310 | 654 | 217 | ||||
| X | 1220–1645 | 426 | 141 | ||||
| HBV/ayw | 3182 | 48.49 | TTCACCTCTGC | Pol | 2309–3182, 1–1625 | 2499 | 832 |
| PreS/S | 2850–3182, 1–837 | 1170 | 389 | ||||
| PreC/C | 1816–2454 | 639 | 212 | ||||
| X | 1376–1840 | 465 | 154 |
Note: The numbering of the bat virus genomes newly identified here is based on RBHBV/China.
Fig. 4.
Genome features of orthohepadnaviruses. The main regions responsible for differences in genome size among the viruses analyzed are shown with alignment numbering. The viruses documented in this study are shown in bold.
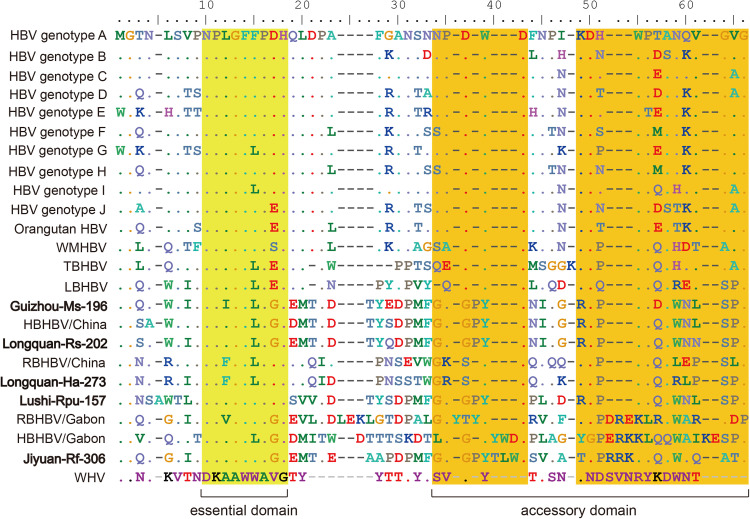
The variation in the preS/S gene length was also evident among bat, rodent and primate viruses. Compared with bat viruses, primate hepadnaviruses contained more amino acid deletions at positions 107−158. It is believed that the N-terminal amino acid residues of large surface protein (preS1 domain) are essential for HBV infectivity (Neurath et al., 1986). Comparison of amino sequences revealed that the newly identified bat viruses had high identity within a short amino acid motif (NPLGFFPDH) that is conserved among primate viruses ( Fig. 5). In particular, virus Lushi-Rpu-157 shared the same amino acid residues as the primate hepadnaviruses, and exhibited high levels of sequence identity to primate viruses within adjacent accessory domains (motifs NPDWD and NKDHWPEANKVGVG). As the lineage including Lushi-Rpu-157 have a broad range of bat hosts and geographic distribution, further studies are needed to evaluate their infectivity in human hepatocytes. Finally, unlike the other genes, the length of X gene was shorter in bat viruses (408-435nt) and rodent viruses (417-426nt), compared to primate viruses (465 nt).
Fig. 5.
Alignment of the essential and accessory domain in preS1. Essential (light yellow) and accessory (dark yellow) domains are compared among the hepadnaviruses from primates, bats and rodents. The viruses documented in this study are shown in bold.
2.4. Co-phylogenetic relationships of hepadnaviruses and their mammalian hosts
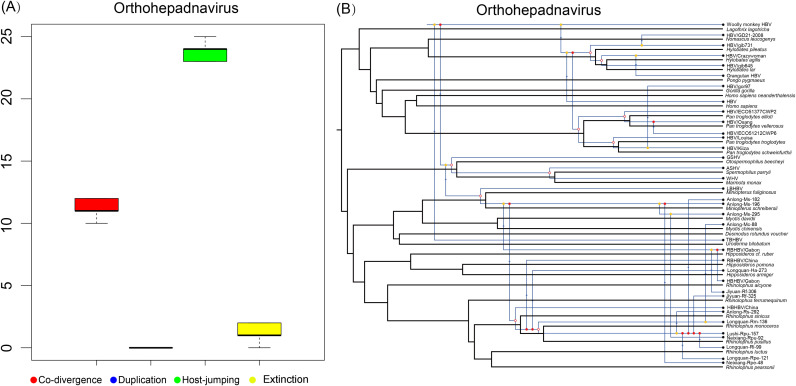
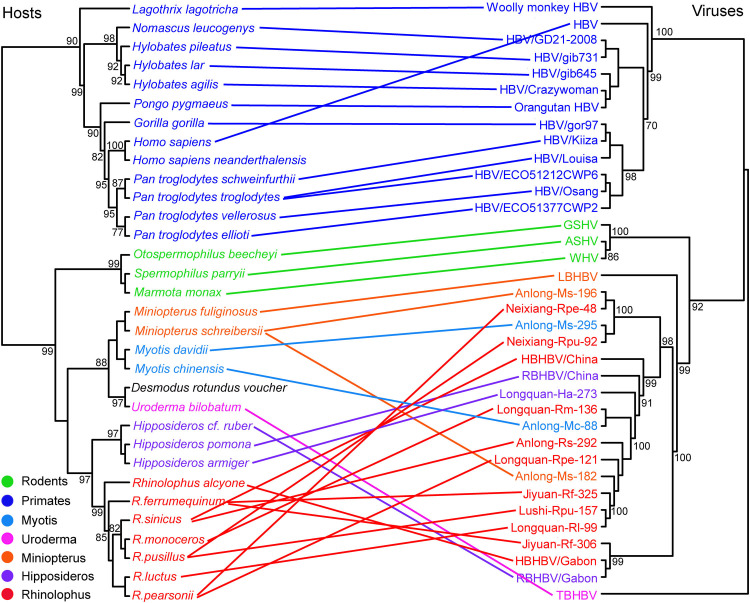
To better understand orthohepadnavirus evolution we performed a co-phylogenetic analysis of all these viruses and their mammalian hosts using a heuristic event-based method ( Fig. 6). The tree topologies of viruses and their hosts exhibited a strong co-divergence at the level of host order. However, there was a clear incongruence between bat hepadnaviruses and their bat hosts at the family, genus, species levels, especially for the newly identified viruses of lineage F. A similar incongruence between primate viruses and rodent viruses and their respective hosts was also evident. Combined, our co-phylogenetic analysis of orthohepadnaviruses and their hosts provided evidence for 10–12 co-divergence events, 23–25 host switches, 0 lineage duplications, 0–2 losses and 0 failure to diverge events (Fig. 6B). Overall, therefore, our co-phylogenetic analysis did not identify significant congruence between the phylogenetic trees of orthohepadnaviruses and their mammalian hosts (P < 0.05), although the presence of hepadnaviruses in diverse vertebrates makes it likely that these virus-host associations have been established for millions of years. A relatively high frequency of cross-species transmission on a background of co-divergence was also apparent in virus-host tanglegrams (Fig. S1).
Fig. 6.
Co-phylogenetic analyses of hepadnaviruses and their associated mammal hosts. (A) Box plots indicate the relative frequency of different co-phylogenetic events. (B) One reconciliation of the hepadnavirus tree is shown in blue while the corresponding host phylogeny is shown in black. The host tree was based on mitochondrial cytochrome b (mt-cyt b) gene sequences, and the hepadnavirus tree was based on the P gene. Filled circles at the nodes indicate co-divergence events, empty circles mark lineage duplication events, arrows indicate host-switching events, and dotted lines show loss events.
We also examined the frequency of cross-species transmission by examining the distribution of bat viruses in China. We identified hepadnaviruses from 10 species of bats sampled in five locations in three Chinese provinces (Fig. 1). Notably, within the first group of clade V the same virus was identified in several bat species from the same geographic locality. For example, lineage A was present in four bats species from three families sampled in both Guizhou and Henan provinces (Table S1). Similarly, lineage F of group 2 was identified in 11 bat species from three families. Notably, although Anlong (Guizhou province) is > 1102 km distant from Jiyuan, Lushi and Neixiang in Henan province in central China, and from Longquan (Zhejiang province) located in the eastern costal region (> 1420 km distant), the newly identified lineage F was found in all these regions, suggesting that these viruses face relatively limited host and geographic barriers. More notable was that the viruses identified in R. ferrumequinum bats sampled from Jiyuan city (Henan province) within group 2 were more closely related to the viruses sampled from Gabon in West Africa than to Chinese bat viruses, including others from Jiyuan. This phylogenetic pattern, and the relatively long branch lengths involved, again suggests that these hepadnaviruses have been associated with bats for a substantial period of time.
3. Discussion
Bats have received increasing attention because of their role in the evolution and transmission of a broad range of viruses (Epstein et al., 2010, Guo et al., 2013, Leroy et al., 2005, Li et al., 2005, Tong et al., 2012). However, the role bats have played in the evolution of hepadnaviruses is less clear (He et al., 2013, He et al., 2015; Drexler et al., 2013; Wang et al., 2017a). Herein, we describe diverse hepadnaviruses in 11 bat species representing four families sampled from five geographic locations in three Chinese provinces, with an overall prevalence of 13.3% (Fig. 1; Table 1). Hence, these data reveal both the high genetic diversity and wide geographic distribution of hepadnaviruses in diverse Chinese bats.
Hepadnaviruses have been found in seven bat species sampled from Myanmar, Gabon, Panama, and China (He et al., 2013, He et al., 2015; Drexler et al., 2013; Wang et al., 2017a). However, they were not identified in 17 bat species from Australia, Brazil, Germany and Papua New Guinea (Drexler et al., 2013). In the current study, 797bats representing 21 species from five families were captured, and hepadnaviruses were identified in all predominant species from all five locations (Table 1); this is a far bigger sample size than previous studies in China (He et al., 2013; Wang et al., 2017a). Additionally, viral prevalence was high in bats from almost all sampling locations, with the exception of Lushi county in Henan province. Hence, combined with previous studies, these data indicate that bats are an important natural reservoir of orthohepadnaviruses.
Although rodents comprise the largest order of mammals (Wilson and Reeder, 2005) and are a major source of zoonotic infectious diseases (Meerburg et al., 2009; Han et al., 2015), to date hepadnaviruses have only been discovered in three species of ground squirrels in the USA (Summers et al., 1978, Marion et al., 1980, Testut et al., 1996). We surveyed 462 rodents from 8 species and 148 Asian house shrews from Zhejiang province. Although our previous studies have demonstrated the extensive diversity of a broad range of viruses (arenaviruses, coronaviruses, hantaviruses, rotaviruses) in rodents and shrews from Longquan and Wenzhou in Zhejiang province (Li et al., 2016, Li et al., 2015; Wang et al., 2015, Wang et al., 2017b Lin et al., 2012), no hepadnavirus was identified in these small mammals, even though these viruses are presentin the bats sampled from the same location (i.e. Longquan).
Numerous studies have described the cross-species transmission of DNA viruses (Das et al., 2017, Tao et al., 2013, Kerr et al., 2015, Bandín and Dopazo, 2011, Franzo et al., 2017, Sarker et al., 2015, Scagliarini et al., 2013). Although a combination of co-divergence and host switching has been described in primate orthohepadnaviruses (Geoghegan et al., 2017, Rasche et al., 2016, Locarnini et al., 2013, Littlejohn et al., 2016), the exact evolutionary history of hepadnaviruses in their mammalian hosts is uncertain and clearly complex to resolve. Our co-phylogenetic analysis of hepadnaviruses and their hosts revealed that the cross-species transmission of orthohepadnaviruses has occurred relatively frequently in the evolutionary history of these viruses (Fig. 6). Most notably, the same bat hepadnavirus can be recovered in multiple bat species from the same geographic locality, such as seven bat species from two families sampled in Longquan. This suggests that there are few barriers to prevent the cross-species transmission of hepadnaviruses in bats.
Despite this frequent cross-species transmission, the co-divergence between hepadnaviruses and their hosts was evident at the level of mammalian orders, suggesting that virus host jumping has occurred on a background of an ancient virus-host association. This is further supported by the observation that the viruses harbored by R. ferrumequinumbats from Jiyuan in Henan province were most closely related to those identified in the R. alcyone and H. cf. ruber bats sampled from Gabon in West Africa (Drexler et al., 2013) (Figs. 2 and 3). The Rhinolophidae are estimated to have diverged from the ancestor of the Hipposideridae and Rhinonycteridae approximately 42 million years ago during the Eocene (Foley et al., 2015). Although an African origin of the Hipposideridae and Rhinolophidae was recently proposed (Foley et al., 2015), more researchers have argued for an Asian origin (Bogdanowicz and Owen, 1992, Teeling et al., 2005, Stoffberg et al., 2010, Ravel et al., 2014). In addition, the ancestor of the superfamily Noctilionoidea, to which U. bilobatum bats belong, is also likely to have originated in the Old World (Teeling et al., 2005). Thus, although the most divergent bat hepadnaviruses - the TBHBV clade - have to date only been sampled in the New World, it is possible that they are also present in the Old World. Indeed, our identification of divergent and widespread bat viruses in China indicates that the origin and evolution of orthohepadnaviruses should be interpreted with great caution.
4. Materials and methods
4.1. Trapping of small mammals and sample collection
During 2014–2016, bats were captured with mist nets in caves of natural roosts in Guizhou, Henan and Zhejiang provinces, China (Fig. 1). In addition, rodents and shrews were trapped with cages during 2015–2016 in Longquan city, Lucheng district and Wencheng county of Wenzhou city, Zhejiang province, China. Species were initially identified by morphological examination, and confirmed by analysis of the mt-cyt b gene (Guo et al., 2013). All animals were anesthetized before necropsy with every effort made to minimize suffering. Liver samples were collected from these mammals for screening hepadnaviruses and were stored at −80 °C before further processing.
The ethics committee of the National Institute for Communicable Disease Control and Prevention of the Chinese Center for Disease Control and Prevention (CDC) approved the study. All animals were treated in strict accordance to the Guidelines for Laboratory Animal Use and Care of the Chinese CDC and the Rules for the Implementation of Laboratory Animal Medicine (1998) from the Ministry of Health, China, based on the protocols reviewed by the National Institute for Communicable Disease Control and Prevention.
4.2. DNA extraction and hepadnavirus detection
Total DNA was extracted using the DNA Extraction kit (Omega bio-tek, USA) according to protocols suggested by the manufacturer. Viral DNA was detected by Pan-PCR targeting the conserved region of the P gene of hepadnaviruses. Complete genomes of hepadnaviruses were then amplified by nested PCR. All primers were designed based on conserved regions of known hepadnavirus genome sequences. Sequences were assembled and manually edited to produce the final viral genomes using the SeqMan program (DNASTAR, Madison, WI). The mt-cyt b gene was sequenced as described previously (Guo et al., 2013).
PCR amplicons (< 700 bp), which were purified using the QIAquick Gel Extraction kit (Qiagen, Valencia, USA) according to the manufacturer's recommendations, were subjected to direct sequencing. Purified DNA (> 700 bp) was cloned into pMD18-T vector (TaKaRa, Dalian, China), and subsequently transformed into JM109-143 competent cells. Three or more positive clones were chosen for sequencing.
4.3. Phylogenetic analysis
The MEGA program package (version 6) was used to obtain sequence alignments using the Clustal W program, with gaps and ambiguously aligned regions removed using Gblocks (v0.91b) (Talavera and Castresana, 2007). Phylogenetic trees of orthohepadnaviruses were estimated using the ML method implemented in PhyML version 3.0 (Guindon et al., 2010) with bootstrap support values calculated from 1000 replicate trees, employing the best-fit model of nucleotide substitution and a Subtree Pruning and Regrafting (SPR) branch-swapping algorithm. Phylogenetic analysis was performed using the four hepadnavirus ORFs: (i) Polymerase = 2165 nt; (ii) Surface = 746 nt; (iii) Core = 499 nt; (vi) X = 315 nt.
4.4. Recombination analysis
Putative recombination events in the orthohepadnaviruses were initially assessed based on obvious phylogenetic incongruence among the four gene trees. Subsequently, the full genome alignment of all bat virus sequences were scanned using the RDP, GENECONV, BootScan methods available within RDP, Version 4 (Martin et al., 2010). Only sequences with significant evidence (P < 0.05) of recombination by at least two methods and confirmed by phylogenetic analysis were taken to represent strong evidence for recombination.
4.5. Co-phylogenetic analysis of hepadnavirus and their hosts
To determine the relative frequency of co-divergence and cross-species transmission in the evolutionary histories of the orthohepadnaviruses we performed co-phylogenetic analyses of hepadnaviruses and their mammalian hosts using the heuristic event-based method available in the Jane software package (Conow et al., 2010). We reconstructed patterns of co-divergence (and consequently cross-species transmission) using arbitrary weights of 0 for co-divergence and a weight of 1 for duplication, host switching, lineage loss and failure to diverge as described previously (Geoghegan et al., 2017, Lin et al., 2017). We performed the Jane analysis with 100 generations and a population size of 100 as parameters for the genetic algorithm. To test the probability of observing the inferred number of co-divergence events by chance, we employed the random tip mapping method with the following parameters: generation number = 100, population size = 100, and sample size = 50. Finally, we used TreeMap (2.0b) to create tanglegrams of the hepadnavirus and host phylogenies as described previously (Jackson and Charleston, 2004).
Acknowledgements
This study was supported by the Special National Project on Research and Development of Key Biosafety Technologies (2016YFC1201900, 2016YFC1200101), and National Natural Science Foundation of China (Grants 81672057, 81290343, 81611130073). ECH is funded by an NHMRC Australia Fellowship (GNT1037231).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2017.11.005.
Appendix A. Supplementary material
Fig. S1.
Tanglegram illustrating the evolutionary associations between orthohepadnaviruses and their hosts. The host tree was estimated using mitochondrial cytochrome b gene sequences (left), while the virus tree was estimated using the polymerase gene (right).
Supplementary material
References
- Bandín I., Dopazo C.P. Host range, host specificity and hypothesized host shift events among viruses of lower vertebrates. Vet. Res. 2011;42:67. doi: 10.1186/1297-9716-42-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanowicz W., Owen R.D. Phylogenetic analyses of the bat family Rhinolophidae. J. Zool. Syst. Evol. 1992;30:142–160. [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conow C., Fielder D., Ovadia Y., Libeskind-Hadas R. Jane: a new tool for the cophylogeny reconstruction problem. Algorithms Mol. Biol. 2010;5:16. doi: 10.1186/1748-7188-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Zhao W., Huang Z., Jarvis E.D., Gilbert M.T., Walker P.J., Holmes E.C., Zhang G. Low frequency of paleoviral infiltration across the avian phylogeny. Genome Biol. 2014;15:539. doi: 10.1186/s13059-014-0539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Fearnside K., Sarker S., Forwood J.K., Raidal S.R. A novel pathogenic aviadenovirus from red-bellied parrots (Poicephalusrufiventris) unveils deep recombination events among avian host lineages. Virology. 2017;502:188–197. doi: 10.1016/j.virol.2016.12.031. [DOI] [PubMed] [Google Scholar]
- Dill J.A., Camus A.C., Leary J.H., Di Giallonardo F., Holmes E.C., Ng T.F. Distinct viral lineages from fish and amphibians reveal the complex evolutionary history of hepadnaviruses. J. Virol. 2016;90:7920–7933. doi: 10.1128/JVI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antivir. Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Geipel A., Konig A., Corman V.M., van Riel D., Leijten L.M., Bremer C.M., Rasche A., Cottontail V.M., Maganga G.D., Schlegel M., Muller M.A., Adam A., Klose S.M., Carneiro A.J., Stocker A., Franke C.R., Gloza-Rausch F., Geyer J., Annan A., Adu-Sarkodie Y., Oppong S., Binger T., Vallo P., Tschapka M., Ulrich R.G., Gerlich W.H., Leroy E., Kuiken T., Glebe D., Drosten C. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. USA. 2013;110:16151–16156. doi: 10.1073/pnas.1308049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J.H., Quan P.L., Briese T., Street C., Jabado O., Conlan S., Ali Khan S., Verdugo D., Hossain M.J., Hutchison S.K., Egholm M., Luby S.P., Daszak P., Lipkin W.I. Identification of GBV-D, a novel GB-like flavivirus from old world frugivorous bats (Pteropusgiganteus) in Bangladesh. PLoS Pathog. 2010;6:e1000972. doi: 10.1371/journal.ppat.1000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley N.M., Thong V.D., Soisook P., Goodman S.M., Armstrong K.N., Jacobs D.S., Puechmaille S.J., Teeling E.C. How and why overcome the impediments to resolution: lessons from rhinolophid and hipposiderid bats. Mol. Biol. Evol. 2015;32:313–333. doi: 10.1093/molbev/msu329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo G., Tucciarone C.M., Cecchinato M., Drigo M. Canine parvovirus type 2 (CPV-2) and Feline panleukopenia virus (FPV) codon bias analysis reveals a progressive adaptation to the new niche after the host jump. Mol. Phylogenet. Evol. 2017;114:82–92. doi: 10.1016/j.ympev.2017.05.019. [DOI] [PubMed] [Google Scholar]
- Geoghegan J.L., Duchene S., Holmes E.C. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 2017;13:e1006215. doi: 10.1371/journal.ppat.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Feschotte C. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 2010;8:e1000495. doi: 10.1371/journal.pbio.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande-Perez A., Martin V., Moreno H., de la Torre J.C. Arenavirus quasispecies and their biological implications. Curr. Top. Microbiol. Immunol. 2016;392:231–276. doi: 10.1007/82_2015_468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guo W.P., Lin X.D., Wang W., Tian J.H., Cong M.L., Zhang H.L., Wang M.R., Zhou R.H., Wang J.B., Li M.H., Xu J., Holmes E.C., Zhang Y.Z. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013;9:e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B.A., Schmidt J.P., Bowden S.E., Drake J.M. Rodent reservoirs of future zoonotic diseases. Proc. Natl. Acad. Sci. USA. 2015;112:7039–7044. doi: 10.1073/pnas.1501598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Fan Q., Yang F., Hu T., Qiu W., Feng Y., Li Z., Li Y., Zhang F., Guo H., Zou X., Tu C. Hepatitis virus in long-fingered bats Myanmar. Emerg. Infect. Dis. 2013;19:638–640. doi: 10.3201/eid1904.121655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhang F., Xia L., Hu T., Chen G., Qiu W., Fan Q., Feng Y., Guo H., Tu C. Identification of a novel Orthohepadnavirus in pomona roundleaf bats in China. Arch. Virol. 2015;160:335–337. doi: 10.1007/s00705-014-2222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C., Zhang Y.Z. The evolution and emergence of hantaviruses. Curr. Opin. Virol. 2015;10:27–33. doi: 10.1016/j.coviro.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Jackson A.P., Charleston M.A. A cophylogenetic perspective of RNA-virus evolution. Mol. Biol. Evol. 2004;21:45–57. doi: 10.1093/molbev/msg232. [DOI] [PubMed] [Google Scholar]
- Katzourakis A., Gifford R.J. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr P.J., Liu J., Cattadori I., Ghedin E., Read A.F., Holmes E.C. Myxoma virus and the Leporipoxviruses: an evolutionary paradigm. Viruses. 2015;7:1020–1061. doi: 10.3390/v7031020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Seitz S., Mattei S., Suh A., Beck J., Herstein J., Borold J., Salzburger W., Kaderali L., Briggs J.A.G., Bartenschlager R. Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe. 2017;22(387–399):e386. doi: 10.1016/j.chom.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Delicat A., Paweska J.T., Gonzalez J.P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Li K., Lin X.D., Huang K.Y., Zhang B., Shi M., Guo W.P., Wang M.R., Wang W., Xing J.G., Li M.H., Hong W.S., Holmes E.C., Zhang Y.Z. Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology. 2016;494:168–177. doi: 10.1016/j.virol.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Lin X.D., Wang W., Shi M., Guo W.P., Zhang X.H., Xing J.G., He J.R., Wang K., Li M.H., Cao J.H., Jiang M.L., Holmes E.C., Zhang Y.Z. Isolation and characterization of a novel arenavirus harbored by rodents and shrews in Zhejiang province, China. Virology. 2015;476:37–42. doi: 10.1016/j.virol.2014.11.026. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lin X.D., Guo W.P., Wang W., Zou Y., Hao Z.Y., Zhou D.J., Dong X., Qu Y.G., Li M.H., Tian H.F., Wen J.F., Plyusnin A., Xu J., Zhang Y.Z. Migration of Norway rats resulted in the worldwide distribution of Seoul hantavirus today. J. Virol. 2012;86:972–981. doi: 10.1128/JVI.00725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.D., Wang W., Hao Z.Y., Wang Z.X., Guo W.P., Guan X.Q., Wang M.R., Wang H.W., Zhou R.H., Li M.H., Tang G.P., Wu J., Holmes E.C., Zhang Y.Z. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn M., Locarnini S., Yuen L. Origins and evolution of hepatitis B virus and hepatitis D virus. Cold Spring Harb. Perspect. Med. 2016;6:a021360. doi: 10.1101/cshperspect.a021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locarnini S., Littlejohn M., Aziz M.N., Yuen L. Possible origins and evolution of the hepatitis B virus (HBV) Semin. Cancer Biol. 2013;23:561–575. doi: 10.1016/j.semcancer.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Marion P.L., Oshiro L.S., Regnery D.C., Scullard G.H., Robinson W.S. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc. Natl. Acad. Sci. USA. 1980;77:2941–2945. doi: 10.1073/pnas.77.5.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerburg B.G., Singleton G.R., Kijlstra A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009;35:221–270. doi: 10.1080/10408410902989837. [DOI] [PubMed] [Google Scholar]
- Neurath A.R., Kent S.B., Strick N., Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429–436. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- Rasche A., Souza B., Drexler J.F. Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr. Opin. Virol. 2016;16:86–94. doi: 10.1016/j.coviro.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Ravel A., Marivaux L., Qi T., Wang Y.Q., Beard K.C. New chiropterans from the middle Eocene of Shanghuang (Jiangsu Province, Coastal China): new insight into the dawn horseshoe bats (Rhinolophidae) in Asia. Zool. Scr. 2014;43:1–23. [Google Scholar]
- Sarker S., Moylan K.G., Ghorashi S.A., Forwood J.K., Peters A., Raidal S.R. Evidence of a deep viral host switch event with beak and feather disease virus infection in rainbow bee-eaters (Meropsornatus) Sci. Rep. 2015;5:14511. doi: 10.1038/srep14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scagliarini A., Gallina L., Battilani M., Turrini F., Savini F., Lavazza A., Chiari M., Coradduzza E., Peli A., Erdelyi K., Alberti A. Cervuselaphus papillomavirus (CePV1): new insights on viral evolution in deer. Vet. Microbiol. 2013;165:252–259. doi: 10.1016/j.vetmic.2013.03.012. [DOI] [PubMed] [Google Scholar]
- Seeger C., Zoulim F., Mason W.S. Hepadnaviruses. In: Knipe D.M., Cohen J.I., Griffin D.E., Lamb R.A., Martin M.A., Racaniello V.R., Roizman B., editors. Vol. 2. Lippincott Williams & Wilkins; Philadelphia, PA: 2013. pp. 2185–2221. (Fields Virology). [Google Scholar]
- Stoffberg S., Jacobs D.S., Mackie I.J., Matthee C.A. Molecular phylogenetics and historical biogeography of Rhinolophus bats. Mol. Phylogenet. Evol. 2010;54:1–9. doi: 10.1016/j.ympev.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Summers J., Smolec J.M., Snyder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc. Natl. Acad. Sci. USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tao Y., Shi M., Conrardy C., Kuzmin I.V., Recuenco S., Agwanda B., Alvarez D.A., Ellison J.A., Gilbert A.T., Moran D., Niezgoda M., Lindblade K.A., Holmes E.C., Breiman R.F., Rupprecht C.E., Tong S. Discovery of diverse polyomaviruses in bats and the evolutionary history of the polyomaviridae. J. Gen. Virol. 2013;94:738–748. doi: 10.1099/vir.0.047928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E.C., Springer M.S., Madsen O., Bates P., O'Brien S., J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Testut P., Renard C.A., Terradillos O., Vitvitski-Trepo L., Tekaia F., Degott C., Blake J., Boyer B., Buendia M.A. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J. Virol. 1996;70:4210–4219. doi: 10.1128/jvi.70.7.4210-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Li Y., Rivailler P., Conrardy C., Castillo D.A., Chen L.M., Recuenco S., Ellison J.A., Davis C.T., York I.A., Turmelle A.S., Moran D., Rogers S., Shi M., Tao Y., Weil M.R., Tang K., Rowe L.A., Sammons S., Xu X., Frace M., Lindblade K.A., Cox N.J., Anderson L.J., Rupprecht C.E., Donis R.O. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S., Schulze A., Dandri M., Petersen J. The replication cycle of hepatitis B virus. J. Hepatol. 2010;52:282–284. doi: 10.1016/j.jhep.2009.10.031. [DOI] [PubMed] [Google Scholar]
- Wang B., Yang X.L., Li W., Zhu Y., Ge X.Y., Zhang L.B., Zhang Y.Z., Bock C.T., Shi Z.L. Detection and genome characterization of four novel bat hepadnaviruses and a hepevirus in China. Virol. J. 2017;14:40. doi: 10.1186/s12985-017-0706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X.D., Guo W.P., Zhou R.H., Wang M.R., Wang C.Q., Ge S., Mei S.H., Li M.H., Shi M., Holmes E.C., Zhang Y.Z. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X.D., Liao Y., Guan X.Q., Guo W.P., Xing J.G., Holmes E.C., Zhang Y.Z. Discovery of a highly divergent coronavirus in the Asian house shrew from China illuminates the origin of the alphacoronaviruses. J. Virol. 2017;98 doi: 10.1128/JVI.00764-17. (764-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.E., Reeder D.M. 3rd ed. Johns Hopkins University Press; Baltimore, Maryland: 2005. Mammal Species of the World. A Taxonomic and Geographic Reference. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material