Highlights
-
•
The genetic diversity of ASFVs was mainly caused by extensive genomic indels instead of point mutations.
-
•
Indels may be attributed much to the ectopic homologous recombination.
-
•
Extensive repeated elements in ASFV genomes facilitate the occurrence of homologous recombination.
Keywords: ASFV, Homologous recombination, Indel, Genetic diversity, Repeated element
Abstract
The African swine fever virus (ASFV) has severely influenced the swine industry of the world. Currently, there is no effective vaccine or drugs against the ASFV. How to effectively control the virus is challenging. In this study, we have analyzed all the publicly available ASFV genomes and demonstrated that there was a large genetic diversity of ASFV genomes. Interestingly, the genetic diversity was mainly caused by extensive genomic insertions and/or deletions (indels) instead of the point mutations. Further analyses showed that the indels may be attributed much to the homologous recombination, as supported by significant associations between the occurrence of extensive recombination events and the indels in the ASFV genomes. Besides, the homologous recombination also led to changes of gene content of ASFVs. Finally, repeated elements of dozens of nucleotides in length were observed to widely distribute and cluster in the adjacent positions of ASFV genomes, which may facilitate the occurrence of homologous recombination. This work highlighted the importance of homologous recombination in shaping the genetic diversity of the ASFVs, and could help understand the evolution of the virus.
1. Introduction
African swine fever virus (ASFV), the causative agent of African swine fever (ASF), is a complex, large, icosahedral multi-enveloped DNA virus. It is classified as the only member in the family Asfarviridae (Galindo and Alonso, 2017; Arias et al., 2018). The genome of the virus belongs to double-stranded DNA, with the size ranging from 170 kb to 190 kb (Dixon et al., 2013). ASFV mainly infect suids and soft ticks. The suids include domestic pigs and wild boars, and were reported as the natural hosts of the virus (Sanchez-Cordon et al., 2018; Costard et al., 2013). ASFV was firstly discovered in Kenya in 1921 (Arzt et al., 2010). It remained restricted in Africa till 1957, when it was reported in Spain and Portugal. Up to now, the virus has caused ASF outbreaks in more than fifty countries in Africa, Europe, Asia, and South America (Costard et al., 2013). The latest reports showed that the virus has caused outbreaks in nearly all provinces of China (Ge et al., 2018; World Animal Health Information and Analysis Department, 2018; Zhou et al., 2018; Food and Agriculture Organization of the United Nations, 2019). Because of the high lethality of ASFV in domestic pigs, the most commonly used strategies to control the virus were the massive culling campaigns and the restriction of pig movement (Sanchez-Cordon et al., 2018). Both strategies have resulted in a huge economic loss for pig industry and affected people’s livelihoods. How to effectively control the virus is still a great challenge for the globe.
The large genetic diversity of ASFVs, which was supposed to hinder the development of effective vaccines or drugs against the virus (Sanchez-Cordon et al., 2018; Escribano et al., 2013; Arabyan et al., 2018), has been investigated in many studies. The ASFV genome encodes over 150 proteins, including viral enzymes, viral transcription and replication-related proteins, structural proteins, other proteins involved in the virus assembly, the evading of host defence systems and the modulation of host cell function, etc (Dixon et al., 2013; Alejo et al., 2018; Kessler et al., 2018). For example, the transcription of the virus is independent on the host RNA polymerase because the virus contains relevant enzymes and factors (Dixon et al., 2013). The viral genome contains a conservative central region of about 12 kb and two variable ends, which results in the variable size of the genome (Dixon et al., 2013; Chapman et al., 2008; de Villiers et al., 2010). There are significant variations among the ASFV genomes due to the genomic insertion or deletion, such as the deletion of the multigene family (MGF) members (Dixon et al., 2013). Although much progress have been made on genetic diversity of the virus, the extent and mechanisms are still not clear. Besides, most of these studies either only investigated the genetic diversity of some common genes, such as p72 and p54 (Fraczyk et al., 2016; Michaud et al., 2013), or only used one to twelve isolate genomes (Dixon et al., 2013; Chapman et al., 2008; de Villiers et al., 2010). The number of discovered viral genomes has increased rapidly as the development of DNA sequencing technology. Therefore, a comprehensive study on the genetic diversity of ASFVs is necessary.
Homologous recombination, which has been reported to occur in several groups of viruses (Roossinck, 1997; Nagy and Bujarski, 1996; Wang et al., 2015), such as herpesvirus, retroviruses, and coronaviruses, has played an important role in viral evolution (Nagy and Bujarski, 1996). A few studies on several ASFV genes have suggested the occurrence of homologous recombination in the evolution of ASFVs (Dixon et al., 2013; Fraczyk et al., 2016). However, a comprehensive study on the homologous recombination in ASFV at the genomic scale is lacking, and the role of the recombination on the genetic diversity and the evolution of the virus is still unknown. In this study, we have systematically investigated the genomic diversity and the homologous recombination of ASFVs based on the analysis on all the publicly available ASFV genomes. The results demonstrated that the homologous recombination contributed much to the genetic diversity of ASFVs. This work would help to understand the evolution of the ASFV and thus facilitate the prevention and control of the virus.
2. Materials and methods
2.1. ASFV genome and alignment
All the ASFV genomic sequences with over 170, 000 bp were obtained from NCBI GenBank database on February 15, 2019 (Agarwala et al., 2016). After removing the genomic sequences derived from a patent, a total of 39 ASFV genomes were kept in the analysis (Table S1). The genomic sequences were aligned by MAFFT (version 7.127b) with the default parameters (Katoh and Standley, 2013). Unless otherwise specified, all the analyses in this study were conducted based on the alignment by MAFFT.
2.2. Gene prediction
Genes encoded in ASFV genomes were predicted with the help of GeneMarkS (version 4.28) (Besemer et al., 2001) with the default parameters, which is available at http://opal.biology.gatech.edu/GeneMark/. The protein sequences for these genes were also provided by GeneMarkS (Table S2).
2.3. Protein grouping
All the inferred proteins of ASFVs were grouped based on sequence homology using OrthoFinder (version 2.2.7) (Emms and Kelly, 2015) with the default parameters. Manual check was conducted to ensure that each protein group contains one type of protein. A total of 156 protein groups were obtained, including 146 groups with more than 2 proteins and 10 groups with only one protein (Table S3). To name each protein group, the proteins included in the group were blast against the ASFV proteins downloaded from NCBI protein database on February 20, 2019. The names of the blast best hits were used to infer the name of the protein group (Table S3). The functions of the protein groups were adapted from Dixon’s (Dixon et al., 2013) and Alejo’s work (Alejo et al., 2018) (Table S3).
2.4. Alignment of ASFV gene sets
To determine insertion and deletion events of genes between two ASFV genomes, an ASFV gene set was defined as all the genes encoded by the ASFV genome. The order of the gene, no matter which strand it was encoded, was determined by the coding region of the gene in the direction of 5′ end to the 3′ end on the plus strand. Each gene set was firstly sorted by the gene order. Then, each gene was named as the group name of the protein which the gene encoded. Finally, the global alignment of pairwise gene sets was conducted using the Needleman-Wunsch algorithm. To reduce the uncertainty of grouping MGF genes, the genes of the same MGF family were considered to be the same in the alignment.
2.5. Detection of homologous recombination events
RDP (version 4) (Martin and Rybicki, 2000) was used to detect the recombination events in the aligned ASFV genomes. A total of nine methods, i.e., RDP, GENECONV, Bootscan, Maxchi, Chimaera, SiSscan, PhylPro, LARD, 3Seq, were used to infer the recombination events with the default parameters. Only the recombination events with significant p-values (<0.05) were recorded for each method. For each recombination event, RDP outputted the recombination region, the recombinant virus, the potential major and minor parents, and the support by each method. For robustness, only the recombination events which were detected by at least two methods were used for further analysis (Table S4).
2.6. Searching for retrotransposon in ASFV genomes
All retrotransposons in the databases of RepBase (Version 23.10) (Genetic Information Research Institute, 2018) and TREP (Wicker et al., 2007) were downloaded on November 11, 2018. All ASFV genomes were searched against these retrotransposons using blastn (Altschul et al., 1997). No hits were obtained under the e-value cutoff of 0.001.
2.7. Phylogenetic tree inference and genotype determination
Maximum-likelihood phylogenetic trees were inferred using MEGA (version 5.0) (Tamura et al., 2013) with the default values of parameters. Bootstrap analysis was conducted with 100 replicates. The phylogenetic tree was visualized using Dendroscope (version 2.4) (Huson et al., 2007). To illustrate the recombination event, several maximum-likelihood phylogenetic trees were built based on genomic sequences with and without the recombination regions. To determine the genotype of ASFVs analyzed, the C-terminal sequences (478 bp) of B646 L gene of the ASFVs were used to build the maximum-likelihood phylogenetic tree. The genotype of each ASFV was assigned based on previous studies (Quembo et al., 2018; Bishop et al., 2015; Boshoff et al., 2007).
2.8. Statistics analysis
All the statistical analyses were conducted in R (version 3.2.5) (R Core Team, 2018). The t-test was used to test whether the ratios of the gaps in the recombination regions were similar to those in other regions, and whether the number of indels in the recombination regions was similar to that in other regions. The paired t-test was used to test whether the genomic differences caused by the insertions and deletions (indels) were similar to those caused by the point mutations, and whether the number of repeated elements in the windows (1000-10,000bp in length) including recombination was similar to those without recombination. The t-test and paired t-test was conducted by the function of t.test() in R.
3. Results
3.1. ASFV genomes
A total of 39 genome sequences of ASFVs were obtained from the NCBI GenBank database, which were listed in Table S1. They were mostly isolated from Africa and Europe during the years from 1950 to 2016. Besides, two isolates from China in 2018, i.e., 2018/AnhuiXCGQ and SY18, were also included. The size of the ASFV genomes ranged from 170,101 bp to 193,886 bp, averaged at 186,588 bp. The viral isolate Kenya_1950 had the largest size, while the isolate BA71 V had the smallest size. No increasing or decreasing trend in the genome size was observed from 1950 to 2018 (Fig. S1), suggesting the dynamic changes of the viral genomes.
3.2. Widespread indels in ASFV genomes
Pairwise comparisons between ASFV genomes were conducted after the multiple sequence alignment of 39 genomes. The pairwise genomic differences between ASFV genomes ranged from 3 to 52,251 bp (Fig. S2), with an average of 22,646 bp, which accounts for more than 10% of the genome alignment. Interestingly, the genomic differences caused by the insertions and deletions (indels) were much larger than those caused by the point mutations (p-value < 2.2e-16 in the paired t-test) (Fig. S3A). Specifically, the genomic differences caused by indels ranged from 3 to 37,837bp, with an average of 14,292bp; while that caused by point mutations ranged from 0 to 17,035bp, with an average of 8,354bp. Among these point mutations, a median ratio of 78.5% happened in the coding regions, and a median ratio of 43.3% belonged to non-silent substitutions (Fig. S4). For robustness of the results, we also conducted the analysis based on the genome alignment by ClustalW (Larkin et al., 2007), and found that the indels caused larger genomic differences than the point mutations did (p-value = 3.2e-9 in the paired t-test) (Fig. S3B).
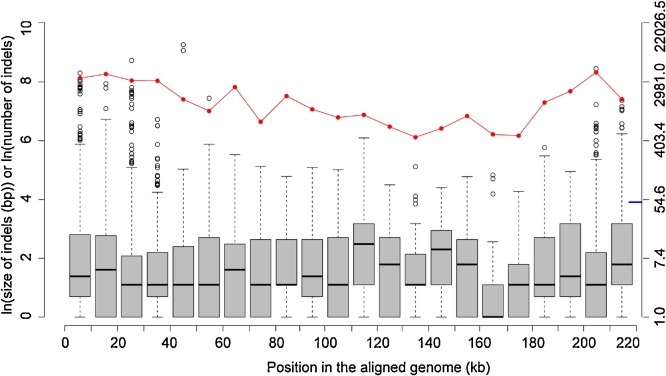
The size and the number of indels in ASFV genomes were also analyzed. 70% of indels were no longer than 10 bp, and about 9% of indels were more than 50 bp (Fig. S5). The number of indels in each genome ranged from 804 to 1062, with a median of 988. The occurrence of indels was much more frequent in both ends of the genome, especially in the 5′ end (indicated by the red line in Fig. 1 ). The distribution of the indel size was similar along the genome, but the large indels with over 50 bp (marked by a blue line in Fig. 1) were mostly observed in both ends.
Fig. 1.
The number of indels of varying sizes in intervals of 10 kb of the multiple sequence alignments of ASFV genomes (red curve), and the distribution of indel sizes in these intervals. For clarity, the natural logarithm of the indel size, or the number of indels, was used. The Y-axis on the right side indicates the actual numbers corresponding to those in the Y-axis on the left side. Position for an indel is defined as the middle position of the indel. The short blue line refers to the indel size of 50 bp. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
The impact of the indels on gene function for each genome was further analyzed. The number of indels which occurred in coding regions was counted for each genome. About 30% of indels were located in the coding regions (Fig. S6). Among them, about 70% of indels had length of 3 or multiple of 3 (colored in blue in Fig. S6), which were likely to cause amino acid indels; the remaining indels were likely to cause changes of reading frames (colored in red in Fig. S6).
3.3. Extensive homologous recombination among ASFV genomes
As numerous indels have been revealed in the ASFV genomes, then, we investigated the mechanism of generating indels. According to the results in previous studies, three factors may contribute to the extensive indels in ASFVs: replication slippage, retrotransposition and recombination. Replication slippage mainly produced duplications of short genetic sequences (Viguera et al., 2001) and may cause short indels, but it is unlikely to generate large indels observed in ASFVs. Retrotransposition can result in duplication of large genetic sequences or genes (Wicker et al., 2007), but no retrotransposons were observed in the analyzed ASFV genomes (as described in Materials and Methods).
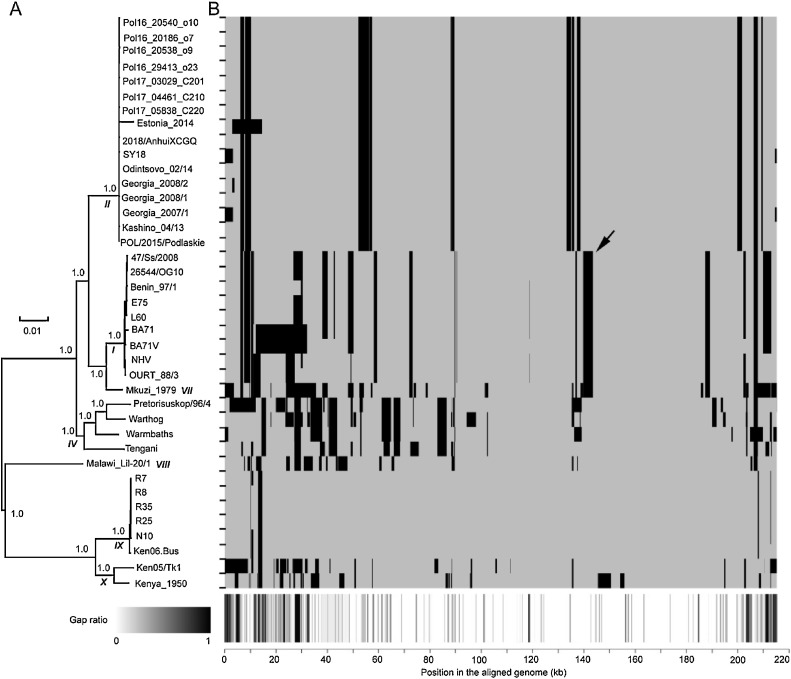
Finally, we investigated the role of recombination in the generation of indels in the ASFV genomes. The analyses on the recombination showed that there were a total of 171 unique recombination events, and each ASFV genome had 4–31 recombination events (Fig. 2 & Table S4). The virus isolate Mkuzi_1979 experienced the largest number of recombination events. On average, each virus experienced a median of 14 recombination events. The sizes of recombination region ranged from 50 to 20,010 bp. The ratio of recombination region in each genome, i.e., the proportion of genomic regions involved in the recombination events, ranged from 1% to 24%. In total, the regions in the ASFV genomes involved in all recombination events covered a total of 121,107 nucleotide sites, accounting for 56% of the aligned genome.
Fig. 2.
Recombination of ASFV genomes. (A) The maximum likelihood phylogenetic tree of ASFVs based on the genome sequences. The topology of the tree was the same as that of the standard p72 tree for genotype determination (Fig. S7). The genotypes of the ASFVs were indicated by the bold and italic numbers. The numbers beside the nodes indicated percent bootstrap support for each node over 100 replications. The scale bar represents the number of nucleotide substitutions per site. (B) Recombination regions (in black) in the genome of ASFVs. The bottom panel shows the ratio of gap in each position of the aligned genome. The panel uses the grayscale color bar at the bottom-left. The black arrow refers to the recombination event displayed in Fig. 3.
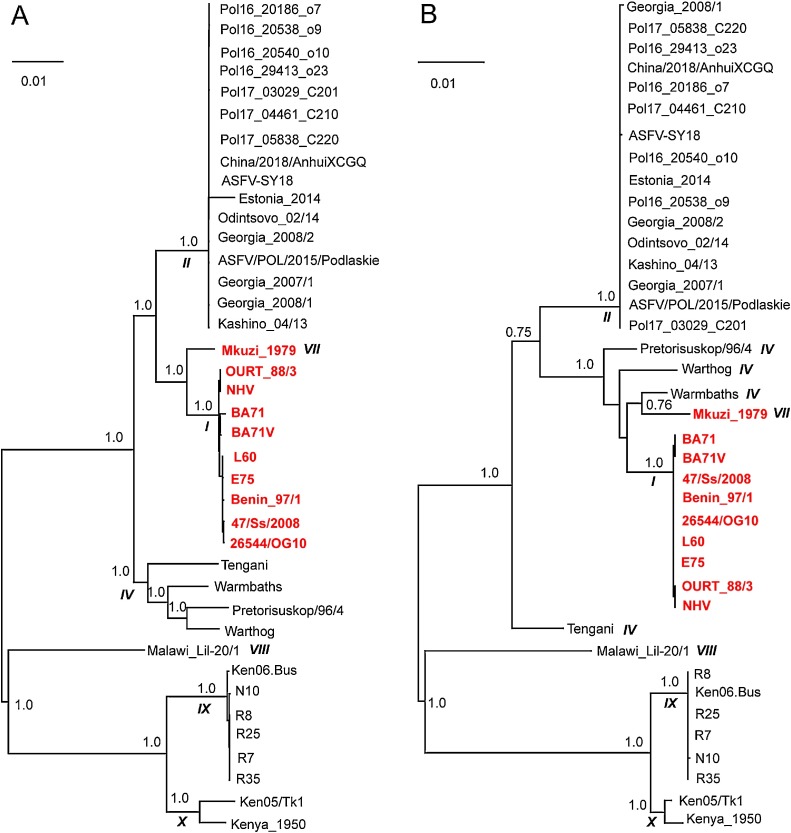
Most recombination events were genotype-specific. There were 7 genotypes among the ASFVs analyzed (Figs. 2A and S7). The genotype II constituted nearly half of ASFVs, including the isolates from East Europe and China in recent years. More than ten recombination events were genotype II-specific (Fig. 2B). The genotype IX, which included six viruses from Uganda and Kenya, had the least recombination events. Fig. 3 illustrates the recombination event in genotype I and VII (colored in red), including two viral isolates from Africa (Mkuzi_1979 and Benin_97/1) and eight viral isolates from Europe. These 10 viral isolates formed a separate lineage in the phylogenetic tree. The recombination region ranged from 139,742to 143,561 bp of the genome alignment, located in the central conservative region of the genome (shown by the black arrow in Fig. 2B). In the phylogenetic tree built with genomic sequences without the recombination regions, the recombinants are the neighbors of a clade containing viruses from Eastern Europe countries (Fig. 3A); while in the tree built with genomic sequences of the recombination regions, the recombinants are the descendants of viruses from Africa (Fig. 3B).
Fig. 3.
An example of a recombination event happened in 10 ASFVs (colored in red). Figure (A) refers to the maximum-likelihood phylogenetic tree built with genome sequences without the recombination region. Figure (B) refers to the phylogenetic tree built with genome sequences of the recombination region. The numbers beside the nodes indicated percent bootstrap support for each node over 100 replications. The scale bar represents the number of nucleotide substitutions per site. The numbers in bold and italic referred to the genotype of the viruses included in the node. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Most recombination events happened at both ends, especially at the 5′ end (Fig. 2B). Interestingly, the recombination event in the aligned genomes was observed to be consistent with the ratio of the gap in the genome (the bottom of Fig. 2). Almost all the recombination events happened in or close to the gap-rich regions where the indels were observed. The ratios of the gaps in the recombination regions were found to be significantly higher than those in other regions (p-value < 0.001 in the t-test) (Fig. S8). Further comparison of the number of indels in the recombination regions and other regions showed that for indels of varying length, such as those greater than 5, 10, or 50 bp, the number of indels in the recombination regions was much larger than those in other regions (p-values < 0.001 in the t-test) (Fig. S9).
3.4. Homologous recombinations lead to gene content variation
Then, we investigated the functional consequences of the recombination events in terms of gene content. In 150 of 171 recombination events, there was at least one gene included in the recombination region. The genes included in the recombination region of the recombinants were compared to those of the inferred major parent virus, which was supposed to provide the larger fraction of the recombinant sequence except the recombination region. In 41 of 171 recombination events, there were at least one gene difference in the recombination region between the recombinant and the major parent virus, either by gene insertion, deletion or replacement (Table 1 ). For example, in the ninth recombination event, a gene L83 L was inserted in the recombination region of the recombinant virus Ken05/Tk1, compared to that of the major parent virus Kenya_1950. In another recombination event, the major parent virus Kenya_1950 encoded the genes of DP148R and DP71 L in the recombination region, the latter of which was lost in the recombinant virus Pretorisuskop/96/4. Interesting to note, the genes of the MGF families were involved in the gene content variation of the recombination region in 24 of these 41 recombination events.
Table 1.
Gene content variation in the recombination region between the major parent and the recombinant virus in two recombination events. Genes highlighted in black bold referred to those different between the recombinant virus and the major parent virus in the recombination region.
| Recombination event 9 (1, 9042) | |||||||
|---|---|---|---|---|---|---|---|
| Major parent virus (Kenya_1950) |
Recombinant virus (Ken05/Tk1) |
||||||
| Gene | Strand | Regiona | Lenb | Gene | Strand | Region | Len |
| MGF_360 | – | 3498, 4657 | 1095 | MGF_360 | – | 3340, 4657 | 1071 |
| MGF_360 | – | 4896, 5966 | 1071 | MGF_360 | – | 4896, 5966 | 1071 |
| MGF_360 | – | 6062, 7176 | 1101 | MGF_360 | – | 6062, 7176 | 1098 |
| p22 | + | 7339, 7883 | 525 | p22 | + | 7339, 7883 | 531 |
| L83L | – | 8050, 8381 | 324 | ||||
| Recombination event 34 (206146, 207790) | |||||||
|---|---|---|---|---|---|---|---|
| Major parent virus (Kenya_1950) |
Recombinant virus (Pretorisuskop/96/4) |
||||||
| Gene | Strand | Region | Len | Gene | Strand | Region | Len |
| DP148R | + | 205778,206553 | 774 | DP148R | + | 205830,206547 | 714 |
| DP71L | – | 206530, 207359 | 558 | ||||
Region in the aligned genome.
Length of the gene (bp).
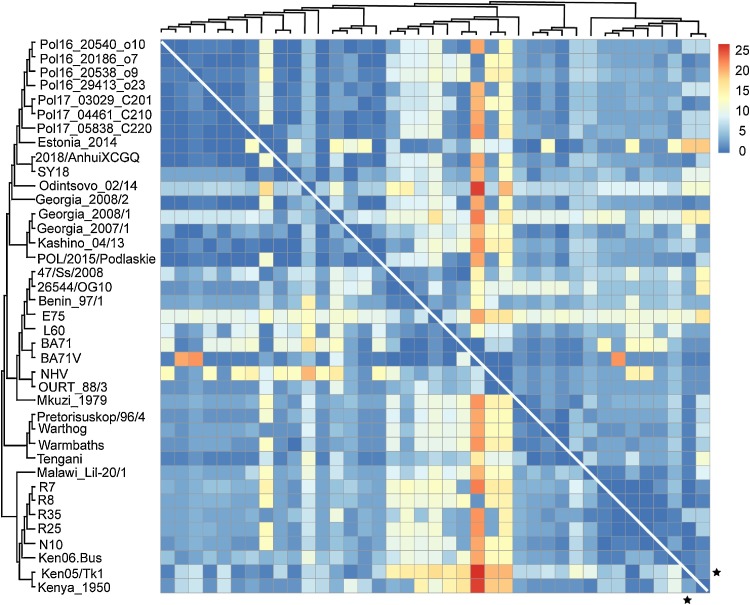
We further compared the gene content between the ASFV genomes. The ASFV genome encoded 128–154 genes, with an average of 145 genes (Table S2). After pairwise alignment of the gene set encoded by genomes, the number of different genes between genomes was calculated. On average, there were 11 different genes between the gene set of genomes, which was about 7% of the gene set. The number of gene deletions and insertions between genomes was further calculated and shown in Fig. 4 . The matrix referred to the number of gene deletions when comparing the gene set of ASFV genomes in the row to those in the column. It showed that in most cases there were both gene deletions and insertions when comparing pairwise gene sets of ASFVs. Even for the virus Ken05/Tk1, which encoded the largest number of genes among all ASFVs analyzed, there were still gene insertions when comparing to other genomes (marked by the black star). Among the 34 genes which were involved in gene insertions or deletions, the member of MGF families, especially the MGF-110 and MGF-360, accounted for nearly 70% of gene insertions or deletions (Fig. S10). Unfortunately, most of them had unknown functions. Besides, the genes of DP71 L, which was reported to have the function of neurovirulence, and p22, which was reported to be one of the antigen protein, were also widely involved in gene insertions or deletions.
Fig. 4.
Pairwise comparisons of the gene set encoded by ASFV genomes. Each element of the matrix, which was colored according to the legend on the top right, referred to the number of gene deletions when comparing the gene set of ASFVs listed in the phylogenetic tree in the left to those in the top of the matrix. Both the phylogenetic trees on the left and on the top were the same and were adapted from that shown in Fig. 2A. The black stars referred to the viral isolate Ken05/Tk1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.5. An abundance of repeated elements in ASFV genomes
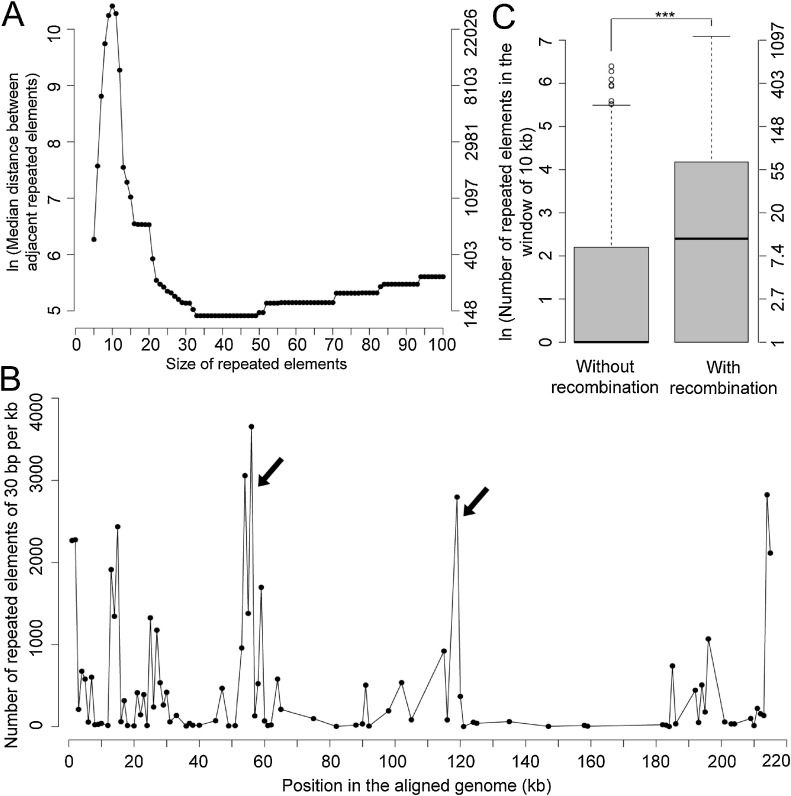
Repeated elements could facilitate the homologous recombination. In this study, lots of repeated elements ranging from 5 to 100 bp were identified, and then the distribution of the repeated elements in the ASFV genomes was analyzed. As shown in Fig. S11, the number of repeated elements in ASFV genomes decreased monotonously as the size of elements increased. Then, the distances between adjacent elements for a given repeated element was investigated (Fig. 5 A). As the size of the elements increased from 5 to 10, the average distance between the adjacent elements also increased because the number of repeated elements in the genome decreased and the repeated elements became more disperse in the genome. Interestingly, the average distance decreased as the size of the elements increased from 11 to 33; it reached to the minimum (136 bp) when the size was 33; then the distance kept unchanged as the size increased from 33 to 49; finally, it increased as the size of repeated element increased from 50 to 100. It should be noted that the average distance was still less than 300 bp even for the repeated elements of 100 bp. These phenomena suggested that the repeated elements of 11 bp or larger tended to cluster in the genome, especially for those of 33–49 bp.
Fig. 5.
Distribution of the repeated elements. (A)The median distance between adjacent repeated elements versus the size of repeated elements. (B) Number of the repeated elements with the size of 30 bp in each kb of the multiple genome sequence alignments. (C) Comparison of the number of repeated elements (30 bp in length) in the window of 10,000 bp with and without recombination in viral genomes. For clarity, the natural logarithm of the median distance between adjacent repeated elements (A) and the number of repeated elements (C) was used. The Y-axis in the right side (A & C) indicates the actual numbers corresponding to those in the Y-axis on the left side. “***”,p-value < 0.001.
For example, when the size of elements was 30 bp, each genome had a median of 427 types of elements which repeated at least two times in the genome. Some elements appeared for over ten times in the genome, such as the element “AGGCGTTAAACATTAAAATTATTACTACTG” in the viral strain BA71 V. The region covered by repeated elements accounted for 1%–3% of the genome in ASFVs. The median distance between repeated elements was 170 bp, suggesting they tended to cluster in adjacent regions. Fig. 5B shows the distribution of repeated elements in the aligned genome. Most repeated elements were located at both ends of the genome. Besides, there were two clusters of repeated elements in the positions of around 55 kb and 120 kb (marked by black arrows), respectively.
Finally, the contribution of repeated elements to the recombination was investigated. For elements of 10 or more nucleotides, the number of repeated elements in the windows (1000-10,000bp in length) including recombination was significantly larger than those without recombination (p-values < 0.001 in the paired t-test) (Table S5). Fig. 5C showed the comparison of the number of repeated elements (30 bp in length) in the windows of 10,000 bp with and without the recombination in viral genomes. The windows including the recombination had a mean of 72 repeated elements, which was four times of that in the windows without recombination.
4. Discussion
This work systematically analyzed the genetic diversity of ASFVs. Large diversity was observed among the genomes and the genes of ASFVs, which may lead to diverse phenotype, such as the diversity in antigen and virulence. Indels were found to have a larger contribution to the genetic diversity of ASFVs than the point mutations. This was similar to that observed in a previous study by Lin, during which the author found that insertion/deletion of simple sequence repeat (SSR) could cause large genetic variations in phages (Lin, 2016). Compared to point mutations, indels could introduce a larger variation to the genome, and cause a more severe damage to the genome structures, which may lead to the death of viruses (Wang et al., 2016; Singh et al., 2019; Sharma et al., 2018). Therefore, only few indels were observed in viruses with small genomes, such as influenza viruses (Taubenberger and Kash, 2010) and hepatitis C viruses (HCV) (Torres-Puente et al., 2007). However, it was more robust for the indels to occur inside the viruses with large genomes, such as ASFVs (Dixon et al., 2013), poxviruses (Elde et al., 2012) and phages (Lin, 2016), because the viruses with large genomes had lots of repeated elements (such as SSRs) and duplicated genes (such as MGFs). Moreover, indels may provide a more efficient way of survival than the point mutations under the natural selection pressure (Dixon et al., 2013; Lin, 2016; Elde et al., 2012), since the virus with indels could rapidly change its phenotype, such as antigen, virulence, or ability of replication and transcription. For example, the deletion of some MGF genes in ASFV could reduce the viral replication or virulence, which may help with the viral infection of soft ticks (Dixon et al., 2013; Burrage et al., 2004).
As Dixon et al. pointed out, gene families are commonly involved in the evolution of DNA viruses with large genomes (Dixon et al., 2013). For example, the poxviruses can rapidly acquired fitness via recurrent amplification of a key anti-host defence gene (Elde et al., 2012). The ASFV genomes have five MGF families, each of which has 3–22 MGF genes (Dixon et al., 2013). Previous studies have shown that most genome variation in ASFVs was as the result of gain or loss of MGF genes (Dixon et al., 2013). This study found that the member of MGF families were frequently involved in recombinations. They accounted for more than 2/3 of gene insertions or deletions when comparing the proteome of ASFVs. Although most of them had unknown functions until now, they are supposed to play important roles in rapidly changing the phenotypes of the virus.
Several factors could contribute to the indels, including replication slippage, retrotransposition and recombination (Zhang, 2003). The replication slippage may introduce short indels which were widely observed in ASFV genomes, but it is unlikely to cause large indels. This study suggested that the ectopic homologous recombination, during which the segments with unequal length were exchanged (Freitas-Junior et al., 2000), may contribute much to the extensive indels observed in ASFV genomes (Fig. 6 A). As a proof, significant associations were observed between the occurrence of recombination events and the indels. The clustered repeated elements observed in ASFV genomes may facilitate the homologous recombination (Fig. 5). Taken together, the homologous recombination should be the effective strategy of ASFVs to generate the genetic diversity, which further leads to the diverse phenotypes, including antigen, virulence, replication and transcription ability, and the “weapons” of escaping from the host immunity (Fig. 6B).
Fig. 6.
Homologous recombination leads to (A) the indels, and (B) the genetic diversity of ASFVs.
The widespread distribution of repeated elements in the ASFV genomes may have important implications for the viral evolution. The short repeated elements may facilitate the replication slippage, leading to short indels. For example, Dixon et al. have identified short tandem repeats within the ASFV genes (Dixon et al., 2013), such as E183 L and B602 L, which cause large variations of these genes. Besides the short repeated elements, there are also an abundance of long repeated elements, such as those longer than 30 bp. The clustered long repeated elements can facilitate the ectopic homologous recombination, which lead to large indels including the gene insertions/deletions.
This work provides some insights into the prevention and control of the ASFVs. Since the virus can rapidly change its phenotypes, such as the antigen, traditional methods of developing vaccines or drugs may be ineffective, as was demonstrated in previous studies (Escribano et al., 2013; Arabyan et al., 2018; Sanchez et al., 2019). Identification of the conservative antigenic epitopes or drug targets may help for development of effective vaccines and drugs. Besides, development of drugs targeting host proteins instead of viral proteins may be an alternative strategy (Han et al., 2017). Moreover, since the recombination play a large role in shaping the genetic diversity of the virus, coupling the drugs which inhibit the recombination process with the traditional drugs or vaccines, may help prevention and control of the viral infection.
There were some limitations to this study. Firstly, the number of ASFV genomes was limited, which hindered a comprehensive analysis on the evolution of ASFV genomes. Previous studies have identified over twenty genotypes of ASFVs (Boshoff et al., 2007; Bastos et al., 2003), among which only seven genotypes were included in this study. Fortunately, the isolates included in this study covered a long time period from 1950 to 2018, and also covered a large area including Africa, Europe and China, which were the major areas of the ASFV circulation. Besides, the genotype II, the most widely spread genotype in recent years (Ge et al., 2018; Zhou et al., 2018; Quembo et al., 2018; Garigliany et al., 2019), were also included and constituted nearly half of all ASFVs. Thus the results based on these isolates could reflect the genetic diversity of the ASFVs to a large extent. Secondly, the location and size of the indels observed in ASFV genomes may be affected by the sequence alignment algorithm. Two common methods, i.e., MAFFT and ClustalW, for the alignment of ASFV genomes were used in this study. In both methods, the indels were observed to contribute much more to the genetic diversity than the point mutations did (Fig. S3), suggesting the robustness of the results. Lastly, the extensively repeated elements in ASFV genomes could facilitate the frequent occurrence of recombination events. However, some of recombination events cannot be detected by the recombination detection method because of the exchange between the genomic segments with small indels. Such kinds of recombination events are difficult to detect. Increasing the sensitivity of the recombination detection method can help detect them, but may also bring false positives. Therefore, the sensitivity and specificity should be balanced in the recombination detection methods.Overall, this work provided a systematic view of the genetic diversity of ASFVs. Extensive homologous recombination detected in this study may contribute much to the widespread indels observed in ASFV genomes, which further lead to the large genetic diversity of ASFVs. The results on the causes of the diversity of ASFVs would help with the understanding of the evolution of the virus and thus facilitate the prevention and control of ASFVs.
Declaration of Competing Interest
The authors have declared that no competing interests exist.
Acknowledgements
This work was supported by the National Key Plan for Scientific Research and Development of China (2016YFD0500300 and 2017YFD0500104), the Hunan Provincial Natural Science Foundation of China (2018JJ3039), the National Natural Science Foundation of China (31671371) and the Chinese Academy of Medical Sciences (2016-I2M-1-005).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2019.08.003.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Agarwala R., Barrett T., Beck J. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejo Alí, Matamoros Tania, Guerra Milagros. A proteomic atlas of the African swine fever virus particle. J. Virol. 2018;92(23) doi: 10.1128/JVI.01293-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabyan E., Hakobyan A., Kotsinyan A. Genistein inhibits African swine fever virus replication in vitro by disrupting viral DNA synthesis. Antiviral Res. 2018;156:128–137. doi: 10.1016/j.antiviral.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias M., Jurado C., Gallardo C. Gaps in African swine fever: analysis and priorities. Transbound. Emerg. Dis. 2018;65:235–247. doi: 10.1111/tbed.12695. [DOI] [PubMed] [Google Scholar]
- Arzt J., White W.R., Thomsen B.V. Agricultural diseases on the move early in the third millennium. Vet. Pathol. 2010;47:15–27. doi: 10.1177/0300985809354350. [DOI] [PubMed] [Google Scholar]
- Bastos A.D., Penrith M.L., Cruciere C. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch. Virol. 2003;148:693–706. doi: 10.1007/s00705-002-0946-8. [DOI] [PubMed] [Google Scholar]
- Besemer J., Lomsadze A., Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop R.P., Fleischauer C., de Villiers E.P. Comparative analysis of the complete genome sequences of Kenyan African swine fever virus isolates within p72 genotypes IX and X. Virus Genes. 2015;50:303–309. doi: 10.1007/s11262-014-1156-7. [DOI] [PubMed] [Google Scholar]
- Boshoff C.I., Bastos A.D.S., Gerber L.J. Genetic characterisation of African swine fever viruses from outbreaks in southern Africa (1973-1999) Vet. Microbiol. 2007;121:45–55. doi: 10.1016/j.vetmic.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Burrage T.G., Lu Z., Neialan J.G. African swine fever virus multigene family 360 genes affect virus replication and generalization of infection in Ornithodoros porcinus ticks. J. Virol. 2004;78:9. doi: 10.1128/JVI.78.5.2445-2453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman D.A., Tcherepanov V., Upton C. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 2008;89:397–408. doi: 10.1099/vir.0.83343-0. [DOI] [PubMed] [Google Scholar]
- Costard S., Mur L., Lubroth J. Epidemiology of African swine fever virus. Virus Res. 2013;173:191–197. doi: 10.1016/j.virusres.2012.10.030. [DOI] [PubMed] [Google Scholar]
- de Villiers E.P., Gallardo C., Arias M. Phylogenomic analysis of 11 complete African swine fever virus genome sequences. Virology. 2010;400:128–136. doi: 10.1016/j.virol.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Dixon L.K., Chapman D.A.G., Netherton C.L. African swine fever virus replication and genomics. Virus Res. 2013;173:3–14. doi: 10.1016/j.virusres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Elde N.C., Child S.J., Eickbush M.T. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. 2012;150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano J.M., Galindo I., Alonso C. Antibody-mediated neutralization of African swine fever virus: myths and facts. Virus Res. 2013;173:101–109. doi: 10.1016/j.virusres.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations . 2019. ASF Situation in Asia Update. [Google Scholar]
- Fraczyk M., Wozniakowski G., Kowalczyk A. Evolution of African swine fever virus genes related to evasion of host immune response. Vet. Microbiol. 2016;193:133–144. doi: 10.1016/j.vetmic.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior L.H., Bottius E., Pirrit L.A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P-falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Galindo I., Alonso C. African swine fever virus: a review. Viruses. 2017;9 doi: 10.3390/v9050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigliany M., Desmecht D., Tignon M. Phylogeographic analysis of African swine fever virus, Western Europe, 2018. Emerg. Infect Dis. 2019;25:184–186. doi: 10.3201/eid2501.181535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S.Q., Li J.M., Fan X.X. Molecular characterization of African swine fever virus, China, 2018. Emerg. Infect Dis. 2018;24:2131–2133. doi: 10.3201/eid2411.181274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetic Information Research Institute . 2018. Repbase. [Google Scholar]
- Han L., Li K., Jin C.Z. Human enterovirus 71 protein interaction network prompts antiviral drug repositioning. Sci. Rep. 2017;7 doi: 10.1038/srep43143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Richter D.C., Rausch C. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C., Forth J.H., Keil G.M. The intracellular proteome of African swine fever virus. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-32985-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lin T.Y. Simple sequence repeat variations expedite phage divergence: mechanisms of indels and gene mutations. Mutat. Res.-Fund Mol. Mech. 2016;789:48–56. doi: 10.1016/j.mrfmmm.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Martin D., Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Michaud V., Randriamparany T., Albina E. Comprehensive phylogenetic reconstructions of African swine fever virus: proposal for a new classification and molecular dating of the virus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Bujarski J.J. Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J. Virol. 1996;70:415–426. doi: 10.1128/jvi.70.1.415-426.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quembo C.J., Jori F., Vosloo W. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transbound. Emerg. Dis. 2018;65:420–431. doi: 10.1111/tbed.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Roossinck M.J. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 1997;35:191–209. doi: 10.1146/annurev.phyto.35.1.191. [DOI] [PubMed] [Google Scholar]
- Sanchez E.G., Perez-Nunez D., Revilla Y. Development of vaccines against African swine fever virus. Virus Res. 2019;265:150–155. doi: 10.1016/j.virusres.2019.03.022. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cordon P.J., Montoya M., Reis A.L. African swine fever: a re-emerging viral disease threatening the global pig industry. Vet. J. 2018;233:41–48. doi: 10.1016/j.tvjl.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U., Gupta S., Venkatesh S. Comparative genetic variability in HIV-1 subtype C p24 Gene in early age groups of infants. Virus Genes. 2018;54:647–661. doi: 10.1007/s11262-018-1588-6. [DOI] [PubMed] [Google Scholar]
- Singh M., Kishore A., Maity D. A proline insertion-deletion in the spike glycoprotein fusion peptide of mouse hepatitis virus strongly alters neuropathology. J. Biol. Chem. 2019;294:8064–8087. doi: 10.1074/jbc.RA118.004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger J.K., Kash J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Puente M., Cuevas J.M., Jimenez-Hernandez N. Contribution of insertions and deletions to the variability of hepatitis C virus populations. J. Gen. Virol. 2007;88:2198–2203. doi: 10.1099/vir.0.82855-0. [DOI] [PubMed] [Google Scholar]
- Viguera E., Canceill D., Ehrlich S.D. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu D., Shi W. Origin and possible genetic recombination of the middle east respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. mBio. 2015;6:e01280–01215. doi: 10.1128/mBio.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Pan Q.H., Gendron P. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- Wicker T., Sabot F., Hua-Van A. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- World Animal Health Information and Analysis Department . 2018. African Swine Fever (ASF) Report N°4. October 5 - 18, 2018. [Google Scholar]
- Zhang J.Z. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. [Google Scholar]
- Zhou X.T., Li N., Luo Y.Z. Emergence of african swine fever in China, 2018. Transbound. Emerg. Dis. 2018;65:1482–1484. doi: 10.1111/tbed.12989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.